Abstract
Here we present the genomic sequence, with analysis, of a pathogenic fowlpox virus (FPV). The 288-kbp FPV genome consists of a central coding region bounded by identical 9.5-kbp inverted terminal repeats and contains 260 open reading frames, of which 101 exhibit similarity to genes of known function. Comparison of the FPV genome with those of other chordopoxviruses (ChPVs) revealed 65 conserved gene homologues, encoding proteins involved in transcription and mRNA biogenesis, nucleotide metabolism, DNA replication and repair, protein processing, and virion structure. Comparison of the FPV genome with those of other ChPVs revealed extensive genome colinearity which is interrupted in FPV by a translocation and a major inversion, the presence of multiple and in some cases large gene families, and novel cellular homologues. Large numbers of cellular homologues together with 10 multigene families largely account for the marked size difference between the FPV genome (260 to 309 kbp) and other known ChPV genomes (178 to 191 kbp). Predicted proteins with putative functions involving immune evasion included eight natural killer cell receptors, four CC chemokines, three G-protein-coupled receptors, two β nerve growth factors, transforming growth factor β, interleukin-18-binding protein, semaphorin, and five serine proteinase inhibitors (serpins). Other potential FPV host range proteins included homologues of those involved in apoptosis (e.g., Bcl-2 protein), cell growth (e.g., epidermal growth factor domain protein), tissue tropism (e.g., ankyrin repeat-containing gene family, N1R/p28 gene family, and a T10 homologue), and avian host range (e.g., a protein present in both fowl adenovirus and Marek's disease virus). The presence of homologues of genes encoding proteins involved in steroid biogenesis (e.g., hydroxysteroid dehydrogenase), antioxidant functions (e.g., glutathione peroxidase), vesicle trafficking (e.g., two α-type soluble NSF attachment proteins), and other, unknown conserved cellular processes (e.g., Hal3 domain protein and GSN1/SUR4) suggests that significant modification of host cell function occurs upon viral infection. The presence of a cyclobutane pyrimidine dimer photolyase homologue in FPV suggests the presence of a photoreactivation DNA repair pathway. This diverse complement of genes with likely host range functions in FPV suggests significant viral adaptation to the avian host.
Within the Chordopoxvirinae subfamily (poxviruses of vertebrates) of the family Poxviridae, only members of the Avipoxvirus genus infect nonmammalian hosts (118). Avipoxviruses are a large family of cytoplasmic DNA viruses which infect more than 60 species of wild birds representing 20 families (169). Variability in restriction enzyme profiles of viral DNA suggests significant genomic differences among family members (169). Cross-infection studies also suggest genetic differences among viruses, which are reflected as a wide range of pathogenic effects (absence of clinical disease, local pox lesions, local and generalized infection, and generalized infection with death) and a lack of cross protection, depending on the specific virus-host combination (46, 169).
Fowlpox virus (FPV), the prototypical member of the Avipoxvirus genus, infects chickens and turkeys. Poxvirus diseases of poultry and other domestic birds (canaries and pigeons) have significant economic impact worldwide, with losses resulting from a drop in egg production in layers, reduced growth rates in broilers, blindness, and in some cases death (46, 170). Two forms of disease are associated with different routes of infection. The most common, the cutaneous form, occurs following infection by biting arthropods that serve as mechanical vectors for viral transmission. The disease is characterized by an inflammatory process with hyperplasia of the epidermis and feather follicles, scab formation, and desquamation of the degenerated epithelium, and it predisposes the host to secondary bacterial infections. The second, or diphtheric, form involves droplet infection of the mucous membranes of the mouth, the pharynx, the larynx, and sometimes the trachea. The prognosis with this form of the disease is poor because lesions often cause death by asphyxiation (169–171).
Vaccination with live-attenuated viruses (FPV and canarypox virus [CaPV]) and nonattenuated viruses (pigeonpox virus) is used to control this disease (59, 77, 136, 182). Fowlpox and pigeonpox vaccines are applied by comb scarification, by the wing-web stick method, or by feather follicle immunization. Vaccination confers protective immunity 10 to 14 days after infection. Problems related to safety and efficacy of commercial FPV vaccines remain (9, 24, 29, 65).
Multivalent recombinant FPV vaccines as well as FPV vaccines which incorporate immune response modifiers have been constructed (28, 96). Recombinant FPV vaccines expressing foreign antigens have been used to immunize animals against other avian and mammalian diseases (26, 83, 112, 121, 124, 125, 187). Because FPV and CaPV undergo abortive replication in mammalian cells, their use as host range-restricted mammalian expression vectors has been suggested (164, 165).
The FPV genome, containing 260 to 309 kbp of double-stranded DNA, is larger than other described chordopoxvirus (ChPV) genomes (45, 115, 120). Past work on FPV genomics, much of which used highly tissue culture-passaged FPV strains, has provided genetic information on approximately one-third of the viral genome, including some viral genes with putative immune evasion and host range functions (16, 18, 20, 57, 93, 127, 150, 163, 166, 191). The rational design of safer and more effective FPV vaccines and FPV-based expression vectors will require complete information on viral genes associated with viral virulence and host range and a more complete understanding of how these genes function in viral pathogenesis, immune evasion, and avian host range. Here we report the genomic sequence and analysis of a highly pathogenic strain of FPV.
MATERIALS AND METHODS
FPV DNA isolation, cloning, and sequencing.
FPV genomic DNA was extracted from primary chicken embryo fibroblasts infected with a pathogenic FPV strain (fowlpox challenge virus; Animal Health Inspection Service Center for Veterinary Biologics, Ames, Iowa). Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509I endonuclease (New England Biolabs, Beverly, Mass.). DNA fragments of 1.5 to 2.5 kbp were isolated after separation on agarose gels, cloned into the dephosphorylated EcoRI site of pUC19 plasmids, and grown in Escherichia coli DH10B cells (Gibco BRL, Gaithersburg, Md.). Double-stranded pUC19 plasmids were purified by the alkaline lysis method in accordance with the manufacturer's instruction (5′→3′, Inc., Boulder, Colo.). DNA templates were sequenced from both ends with M13 forward and reverse primers, using dideoxy chain terminator sequencing chemistries (135) and an Applied Biosystems (ABI) PRISM 377 automated DNA sequencer (Perkin-Elmer, Foster City, Calif.). ABI sequence software (version 3.3) was used for lane tracking and trace extraction. Chromatogram traces were base called with Phred (64), which also produced a quality file containing a predicted probability of error at each base position. The sequences were assembled with Phrap (63), with the quality files and default settings being used to produce a consensus sequence, with some subsequent manual editing being performed by the Consed sequence editor (72). An identical sequence was assembled with the TIGR assembler, using quality files and clone length constraints (160). Gap closure was achieved by primer walking of gap-spanning clones and sequencing of PCR products. The final DNA consensus sequence represented on average sixfold redundancy at each base position.
DNA sequence analysis.
Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed as previously described (1). Open reading frames (ORFs) longer than 30 amino acids with a methionine start codon (155, 156) were evaluated for coding potential by the use of the Hexamer (ftp.sanger.ac.uk/pub/rd) and Glimmer (134) computer programs. Minor ORFs were excluded. Gene families were analyzed and annotated as previously described (1). Early-promoter sequences were predicted as follows. Fifteen-base DNA motifs with similarity to the vaccinia virus (VV) early-promoter consensus sequence (51, 118) were selected from regions located upstream of initiation codons of 30 FPV homologues of VV virus early genes. These motifs were used to generate a scoring matrix (PROFILEMAKE) (55), and this matrix was used to search 100 bases upstream of all FPV ORFs (MOTIFSEARCH) (55). Positive ORFs found by MOTIFSEARCH (P = 0.001) were further verified by visual inspection, and those that had substitutions at the most-conserved residues were excluded (14 genes).
Virus abbreviations.
Virus names are abbreviated in this article as follows: African swine fever virus, ASFV; Amsacta moorei entomopoxvirus, AmEPV; canarypox virus, CaPV; chordopoxvirus, ChPV; cowpox virus, CPV; ectromelia virus, ECT; entomopoxvirus, EPV; fowlpox virus, FPV; Heliothis armigera entomopoxvirus, HaEPV; lumpy skin disease virus, LSV; Lymantria dispar nuclear polyhedrosis virus, LdNPV; molluscum contagiosum virus, MCV; myxoma virus, MYX; orf virus, OV; Paramecium bursaria chlorella virus, PBCV; rabbit fibroma virus, RFV; rabbitpox virus, RPV; reticuloendotheliosis virus, REV; swinepox virus, SPV; tanapoxvirus, TPV; vaccinia virus, VV; and variola virus, VAR.
Nucleotide and protein sequence databases.
Accession numbers presented are from the GenBank, SwissProt, or PIR database unless otherwise noted.
Nucleotide sequence accession number.
The FPV genome sequence has been deposited in GenBank under accession no. AF198100.
RESULTS AND DISCUSSION
Organization of the FPV genome.
The FPV genome was assembled into a contiguous sequence of 288,539 bp, which is slightly smaller in size than previous estimates of 299 to 309 kbp for low-passage-number FPV field strains (45, 115). Because the hairpin loops were not sequenced, the left-most nucleotide of the assembled sequence was arbitrarily designated base 1. The nucleotide composition is 69% A+T and is uniformly distributed over the entire length of the FPV genome. Six small regions (102 to 315 bp in length) with higher C+G content (50%) are located in the terminal genomic regions (nucleotides 3219 to 5618 and 28222 to 285321). The total composition of all FPV ORFs reflects a bias for residues with A- and T-rich codons. Ile, Leu, Lys, Asn, Tyr, and Phe constitute 45% of all encoded amino acids.
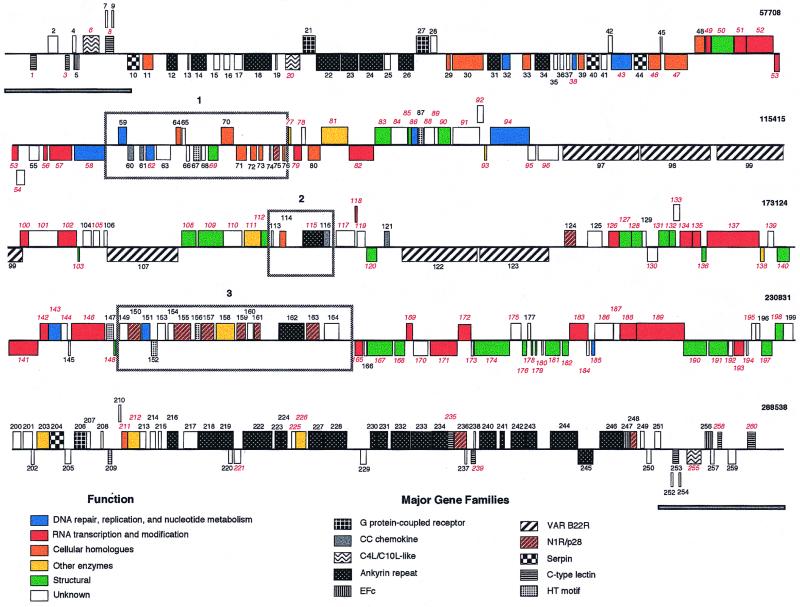
FPV encodes 260 putative genes of 60 to 1,949 amino acids in length (Fig. 1; Table 1). Predicted ORFs represent an 85% coding density, with an average ORF length of 943 nucleotides. One hundred and one FPV ORFs have been assigned similarity or putative function based on homologies with other viral or cellular genes. FPV has a genomic organization similar to that of other known ChPVs (71, 108, 141, 142). There is no evidence of introns, both strands are protein encoding, and ORFs frequently occur in head-to-tail tandem arrays (Fig. 1). Fifty-two ORFs partially overlap other ORFs. Within the terminal 50 kbp of the genome, most ORFs (74% of them in these regions) are transcriptionally oriented toward their respective termini. As seen in other poxviruses, the FPV genome contains a central coding region bounded by two identical inverted terminal repeat (ITR) regions of approximately 9.5 kbp each (Fig. 1). The 3′ 148 codons of ORFs FPV010 and FPV251 mark the boundary between the ITR and the central coding region (Fig. 1). The terminal 1,877 nucleotides are noncoding.
FIG. 1.
Linear map of the FPV genome. ORFs are numbered from left to right based on the position of the methionine initiation codon. ORFs transcribed to the right are located above horizontal lines; ORFs transcribed to the left are below. VV homologues are indicated with red italicized numbers. Genes with similar functions and members of gene families are colored according to the figure key. ITRs are represented as gray bars below the ORF map. Boxed regions 1 to 3 indicate novel coding regions at junction sites of major genome rearrangements, and they correspond to similarly numbered regions shown in Fig. 4.
TABLE 1.
FPV ORFs
| ORF | Position (length, aa)a | Best match
|
Predicted structure and/or functionc | Promoter typed | FPV accession no.b | Corresponding ORF
|
Reference(s) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BlastP score | % Identity | Length, aaa | Accession no.b | Speciese | VV | MCV | ||||||
| FPV001 | 2491–1877 (205) | 121 | 29 | 134 | AF021350 | Rattus norvegicus | C-type lectin family; TM | A40R | 36 | |||
| FPV002 | 3367–4032 (222) | A06621 | 36 | |||||||||
| FPV003 | 4871–4503 (123) | 89 | 30 | 122 | Q07108 | Homo sapiens | C-type lectin family | E | C31685 | A40R | 36 | |
| FPV004 | 5125–5427 (101) | A06621 | 36 | |||||||||
| FPV005 | 5589–5224 (122) | EFc family | E | E31685 | 36 | |||||||
| FPV006 | 5931–7184 (418) | 250 | 27 | 298 | P03296 | VV | C4L/C10L-like family | P14361 | C10L | 36 | ||
| FPV007 | 7599–7814 (72) | D00295 | ||||||||||
| FPV008 | 7681–8181 (167) | 197 | 29 | 127 | AB015628 | Gallus gallus | C-type lectin family; SP | P14370 | A40R | |||
| FPV009 | 8048–8245 (66) | D00295 | ||||||||||
| FPV010 | 10190–9126 (355) | 354 | 30 | 357 | AB006423 | H. sapiens | Serpin family | P14369 | ||||
| FPV011 | 11112–10279 (278) | 596 | 43 | 275 | S32367 | Bos taurus | α-SNAP | E | ||||
| FPV012 | 13033–12041 (331) | 212 | 37 | 127 | Q01485 | H. sapiens | Ankyrin repeat family | |||||
| FPV013 | 13890–13711 (60) | |||||||||||
| FPV014 | 15222–13912 (437) | 293 | 28 | 334 | U13616 | H. sapiens | Ankyrin repeat family | |||||
| FPV015 | 16176–15646 (177) | TM | ||||||||||
| FPV016 | 17068–16355 (238) | |||||||||||
| FPV017 | 17913–17179 (245) | V-type Ig domain | ||||||||||
| FPV018 | 20091–17992 (700) | 288 | 24 | 509 | X69063 | Mus musculus | Ankyrin repeat family; TM | L | ||||
| FPV019 | 20487–20176 (104) | |||||||||||
| FPV020 | 22302–21025 (426) | 210 | 24 | 283 | L22579 | VAR | C4L/C10L-like family | C10L | ||||
| FPV021 | 22393–23352 (320) | 560 | 36 | 288 | AF100204 | Macaca mulatta | G-protein-coupled receptor family; TM | E | ||||
| FPV022 | 25111–23378 (578) | 289 | 27 | 395 | L35601 | Drosophila melanogaster | Ankyrin repeat family | |||||
| FPV023 | 26496–25195 (434) | 303 | 36 | 206 | AB002377 | H. sapiens | Ankyrin repeat family | |||||
| FPV024 | 28352–26565 (596) | 394 | 26 | 456 | L35601 | D. melanogaster | Ankyrin repeat family | |||||
| FPV025 | 29066–28458 (203) | |||||||||||
| FPV026 | 30754–29447 (436) | 248 | 33 | 214 | U21734 | Caenorhabditis elegans | Ankyrin repeat family | |||||
| FPV027 | 30796–31803 (336) | 541 | 36 | 296 | AF100204 | Macaca mulatta | G-protein-coupled receptor family; TM | E | ||||
| FPV028 | 31857–32396 (180) | |||||||||||
| FPV029 | 33396–33025 (124) | 201 | 36 | 112 | AF151905 | H. sapiens | Conserved hypothetical protein | E | ||||
| FPV030 | 35950–33500 (817) | 1,334 | 40 | 716 | P22413 | H. sapiens | Alkaline phosphodiesterase; TM | AJ006408 | 93 | |||
| FPV031 | 37100–36078 (341) | 218 | 31 | 185 | L35601 | D. melanogaster | Ankyrin repeat family | AJ006408 | 93 | |||
| FPV032 | 37858–37163 (232) | 374 | 40 | 211 | Z46266 | C. elegans | DNase II | AJ006408 | 93 | |||
| FPV033 | 39535–38675 (287) | 600 | 43 | 284 | U39412 | H. sapiens | α-SNAP | AJ006408 | 93 | |||
| FPV034 | 40823–39579 (415) | 305 | 30 | 271 | AF102552 | R. norvegicus | Ankyrin repeat family | E | AJ006408 | 93 | ||
| FPV035 | 41414–41010 (135) | |||||||||||
| FPV036 | 41913–41455 (153) | |||||||||||
| FPV037 | 42406–41921 (162) | TM | ||||||||||
| FPV038 | 42840–42406 (145) | 439 | 61 | 136 | M89913 | H. sapiens | dUTP pyrophosphatase | L | F2L | |||
| FPV039 | 43418–42894 (175) | 133 | 27 | 143 | AF115380 | R. norvegicus | Bcl-2; TM | E | ||||
| FPV040 | 44476–43466 (337) | 150 | 19 | 290 | L40377 | H. sapiens | Serpin family; TM | E | ||||
| FPV041 | 45178–44561 (206) | TM | ||||||||||
| FPV042 | 45222–45476 (85) | |||||||||||
| FPV043 | 46957–45266 (564) | 1,553 | 52 | 570 | X84740 | H. sapiens | DNA ligase | Z29716 | A50R | 150 | ||
| FPV044 | 48067–46994 (358) | 259 | 26 | 374 | L28101 | H. sapiens | Serpin family | Z29716 | 150 | |||
| FPV045 | 49173–49391 (73) | 150 | ||||||||||
| FPV046 | 49228–48119 (370) | 717 | 42 | 360 | AB000199 | R. norvegicus | Hydroxysteroid dehydrogenase | E | Z29716 | A44L | MC152R | 150 |
| FPV047 | 51118–49283 (612) | 784 | 35 | 542 | AF071542 | H. sapiens | Semaphorin; TM, SP | A39R | 150 | |||
| FPV048 | 51568–52350 (261) | 441 | 44 | 196 | AL034374 | H. sapiens | GNS1/SUR4; TM | 18 | ||||
| FPV049 | 52429–52890 (154) | 390 | 50 | 148 | P33814 | VAR | Late transcription factor VLTF-2 | I | S42254 | A1L | MC103L | 18 |
| FPV050 | 52914–54569 (552) | 1,667 | 57 | 550 | Q08517 | SPV | Rifampicin resistance protein | S42253 | D13L | MC102L | 18 | |
| FPV051 | 54604–55470 (289) | 937 | 55 | 285 | U60315 | MCV | mRNA capping enzyme, small subunit | S42252 | D12L | MC101L | 18 | |
| FPV052 | 55548–57458 (637) | 2,093 | 61 | 633 | U60315 | MCV | NPH-I, transcription termination factor | L | S42251 | D11L | MC100R | 18 |
| FPV053 | 58142–57468 (225) | 400 | 39 | 212 | U60315 | MCV | mutT motif; gene expression regulator | P32817 | D10R | MC099R | 18 | |
| FPV054 | 58821–58129 (231) | 472 | 47 | 207 | P04311 | VV | mutT motif | E | D9R | MC098R | 18 | |
| FPV055 | 59898–59074 (275) | 97 | 24 | 183 | P13596 | R. norvegicus | V-type Ig domain | P21975 | 18, 163 | |||
| FPV056 | 60547–60065 (161) | 515 | 56 | 161 | U60315 | MCV | RNA polymerase subunit RPO18 | L | P21967 | D7R | MC097R | 18, 163 |
| FPV057 | 62435–60537 (633) | 2,626 | 78 | 631 | U60315 | MCV | Early transcription factor VETFS | L | P21966 | D6R | MC095R | 18, 163 |
| BlastP score | % Identity | Length, aaa | Accession no.b | Speciese | VV | MCV | ||||||
| FPV058 | 64791–62419 (791) | 2,364 | 57 | 785 | S47250 | RFV | NTPase; DNA replication | L | P21969 | D5R | MC094R | 163 |
| FPV059 | 65732–66388 (219) | 314 | 33 | 239 | P27707 | H. sapiens | Deoxycytidine kinase | P21974 | 163 | |||
| FPV060 | 67002–66439 (188) | 82 | 24 | 124 | U48722 | H. sapiens | CC chemokine family; TM, SP | P21973 | 163 | |||
| FPV061 | 67765–67379 (129) | 65 | 32 | 43 | U74585 | Herpesvirus | CC chemokine family; TM | E | P21972 | 163 | ||
| FPV062 | 68517–67864 (218) | 705 | 58 | 216 | P32941 | RFV | Uracil DNA glycosylase; TM | E | P21968 | D4R | MC093R | 163 |
| FPV063 | 69767–68568 (400) | E | P21971 | 163 | ||||||||
| FPV064 | 69939–70538 (200) | 365 | 44 | 163 | X71973 | H. sapiens | Glutathione peroxidase | MC066L | ||||
| FPV065 | 70545–70877 (111) | L | ||||||||||
| FPV066 | 71223–70858 (122) | |||||||||||
| FPV067 | 71611–71342 (90) | HT motif family | ||||||||||
| FPV068 | 72382–71984 (133) | 119 | 38 | 86 | U17055 | PBCV-1 | E | |||||
| FPV069 | 73268–72459 (270) | 172 | 29 | 251 | P25952 | RFV | Virion protein | D3R | MC092R | |||
| FPV070 | 73394–74212 (273) | 621 | 43 | 276 | X74504 | Mus musculus | T10 gene product | |||||
| FPV071 | 75489–74623 (289) | 504 | 38 | 291 | AF132150 | D. melanogaster | Conserved hypothetical protein | |||||
| FPV072 | 76236–75679 (186) | 290 | 41 | 146 | P19093 | Cavia porcellus | β-NGF | |||||
| FPV073 | 76782–76261 (174) | 71 | 26 | 110 | AF110798 | H. sapiens | IL-18 binding protein; SP | |||||
| FPV074 | 77351–77040 (104) | TM | ||||||||||
| FPV075 | 77954–77358 (199) | 129 | 29 | 131 | AF017791 | HaEPV | N1R/p28 family; TM | |||||
| FPV076 | 78457–78026 (144) | 133 | 27 | 118 | P34128 | Bungarus multicinctus | β-NGF | |||||
| FPV077 | 78569–78943 (125) | 290 | 42 | 120 | U60315 | MCV | Glutaredoxin | L | G4L | MC059L | ||
| FPV078 | 79590–79898 (103) | 119 | 26 | 109 | P21024 | VV | TM | L | G3L | MC057L | ||
| FPV079 | 79596–78922 (225) | 339 | 31 | 240 | U60315 | MCV | Putative elongation factor | G2R | MC058R | |||
| FPV080 | 81019–79931 (363) | 178 | 34 | 134 | AJ007836 | Oncorhynchus mykiss | TGF-β; TM | |||||
| FPV081 | 81091–82968 (626) | 1,393 | 43 | 623 | U60315 | MCV | Metalloprotease | L | H48563 | G1L | MC056L | 16 |
| FPV082 | 85003–82958 (682) | 1,544 | 46 | 682 | U60315 | MCV | RNA helicase/NPH-II | G48563 | I8R | MC050R | 16 | |
| FPV083 | 85036–86298 (421) | 1,713 | 74 | 421 | D86731 | CaPV | Virion core protein | L | F48563 | I7L | MC049L | 16 |
| FPV084 | 86304–87473 (390) | 1,529 | 69 | 391 | D86731 | CaPV | E48563 | I6L | MC048L | 16 | ||
| FPV085 | 87477–87719 (81) | 315 | 72 | 81 | D86731 | CaPV | TM, SP | L | P18521 | I5L | MC047L | 16 |
| FPV086 | 87732–88280 (183) | 612 | 65 | 180 | D78347 | CaPV | Thymidine kinase | E | P10052 | J2R | 16 | |
| FPV087 | 88360–88632 (91) | 239 | 55 | 79 | D86731 | CaPV | HT motif family | B48563 | 16 | |||
| FPV088 | 88668–89537 (290) | 817 | 55 | 287 | D86731 | CaPV | DNA-binding phosphoprotein | EI | AJ223385 | I3L | MC046L | 16, 127 |
| FPV089 | 89541–89735 (65) | 204 | 63 | 68 | D86731 | CaPV | TM | L | AJ223385 | I2L | MC045L | 127 |
| FPV090 | 89745–90677 (311) | 1,077 | 65 | 301 | U60315 | MCV | Virion protein | L | AJ223385 | I1L | MC044L | 127 |
| FPV091 | 90845–92812 (656) | 386 | 23 | 453 | U60315 | MCV | TM | AJ223385 | O1L | MC042L | 127 | |
| FPV092 | 92757–93149 (131) | 215 | 29 | 131 | U60315 | MCV | L | AJ223385 | E11L | MC041L | 127 | |
| FPV093 | 93433–93152 (94) | 283 | 49 | 95 | U60315 | MCV | Potential redox protein ERV1 | AJ223385 | E10R | MC040R | 127 | |
| FPV094 | 93460–96423 (988) | 2,674 | 51 | 1,004 | U60315 | MCV | DNA polymerase | E | P21402 | E9L | MC039L | 127, 19 |
| FPV095 | 97236–96421 (272) | 673 | 50 | 264 | U94848 | VV | TM | E | E8R | MC038R | ||
| FPV096 | 98944–97232 (571) | 1,594 | 50 | 566 | U60315 | MCV | E6R | MC037R | ||||
| FPV097 | 104808–99073 (1912) | 2,544 | 35 | 1,896 | L22579 | VAR | VAR B22R family; TM | E | MC035R | |||
| FPV098 | 110282–104877 (1802) | 2,556 | 36 | 1,806 | L22579 | VAR | VAR B22R family; TM | E | MC035R | |||
| FPV099 | 116372–110526 (1949) | 4,316 | 48 | 1,916 | Y15035 | CPV | VAR B22R family | MC035R | ||||
| FPV100 | 116439–116984 (182) | 639 | 63 | 180 | U60315 | MCV | RNA polymerase subunit RPO30 | E4L | MC034L | |||
| FPV101 | 117040–119190 (717) | 696 | 29 | 704 | AF035773 | VAR | TM | E2L | MC032L | |||
| FPV102 | 119180–120595 (472) | 1,222 | 49 | 465 | U60315 | MCV | Poly(A) polymerase PAPL | E1L | MC031L | |||
| FPV103 | 120936–120595 (114) | 210 | 46 | 97 | P07396 | VV | DNA-binding virion core phosphoprotein | L | F17R | MC030R | ||
| FPV104 | 121013–121642 (210) | 97 | 31 | 130 | U80056 | AmEPV | L | |||||
| FPV105 | 121770–122213 (148) | 464 | 53 | 147 | U60315 | MCV | F15L | MC025L | ||||
| FPV106 | 122506–122718 (71) | |||||||||||
| FPV107 | 128102–122772 (1777) | 2,483 | 34 | 1,817 | U18339 | VAR | VAR B22R family; TM | E | MC035R | |||
| FPV108 | 128284–129414 (377) | 873 | 44 | 373 | P25392 | MCV | Virion envelope protein; membrane | P36316 | F13L | MC021L | 35 | |
| FPV109 | 129455–131344 (630) | 433 | 28 | 604 | P21053 | VV | Virion release | P36317 | F12L | MC019L | 35 | |
| FPV110 | 131387–132739 (451) | 158 | 23 | 234 | U60315 | MCV | E | P36700 | F11L | MC018L | 35 | |
| FPV111 | 132820–134151 (444) | 1,299 | 55 | 421 | U52849 | MCV | Ser/Thr protein kinase; virus assembly | L | F10L | MC017L | ||
| FPV112 | 134129–134767 (213) | 405 | 39 | 213 | P33869 | VAR | Putative membrane protein; TM | F9L | MC016L | |||
| FPV113 | 134861–135058 (66) | E | ||||||||||
| FPV114 | 135430–135978 (183) | 518 | 58 | 174 | U80192 | Arabidopsis thaliana | HAL3 domain | |||||
| BlastP score | % Identity | Length, aaa | Accession no.b | Speciese | VV | MCV | ||||||
| FPV115 | 137148–138773 (542) | 340 | 28 | 412 | L40632 | Mus musculus | Ankyrin repeat family | E | M1L | |||
| FPV116 | 138798–139157 (120) | 95 | 31 | 57 | P14844 | R. norvegicus | CC chemokine family; SP | |||||
| FPV117 | 139706–141025 (440) | 588 | 31 | 438 | U60315 | MCV | TM | G5R | MC060R | |||
| FPV118 | 141030–141218 (63) | 235 | 69 | 63 | U60315 | MCV | RNA polymerase subunit RP07 | G5.5R | MC061R | |||
| FPV119 | 141221–141784 (188) | 379 | 39 | 176 | U60315 | MCV | G6R | MC062R | ||||
| FPV120 | 142783–141755 (343) | 604 | 37 | 395 | U60315 | MCV | Virion core protein | L | G7L | MC065L | ||
| FPV121 | 143187–143549 (121) | 69 | 25 | 82 | P10147 | H. sapiens | CC chemokine family; SP | |||||
| FPV122 | 150066–144457 (1870) | 2,506 | 34 | 1,862 | L22579 | VAR | VAR B22R family; TM | E | MC035R | |||
| FPV123 | 155397–150100 (1766) | 2,329 | 34 | 1,834 | AF012825 | ECT | VAR B22R family; TM | E | U17141 | MC035R | ||
| FPV124 | 156366–157232 (289) | 173 | 39 | 129 | AF017791 | HaEPV | N1R/p28 family | U17141 | ||||
| FPV125 | 158143–159177 (345) | 96 | 33 | 115 | P31836 | Bos taurus | V-type Ig domain; SP | E | ||||
| FPV126 | 159602–160381 (260) | 1,098 | 78 | 260 | U60315 | MCV | VLTF-1; TM | E, I, L | P15908 | G8R | MC067R | 20 |
| FPV127 | 160397–161404 (336) | 572 | 38 | 340 | P32998 | VAR | Myristylated protein; TM | L | P15909 | G9R | MC068R | 20 |
| FPV128 | 161408–162136 (243) | 919 | 69 | 243 | U60315 | MCV | Myristylated membrane protein; TM | L | P15910 | L1R | MC069R | 20 |
| FPV129 | 162174–162461 (96) | TM | L | P15911 | MC070R | 20 | ||||||
| FPV130 | 163359–162457 (301) | 739 | 50 | 281 | U60315 | MCV | L | P15912 | L3L | MC072L | 20 | |
| FPV131 | 163385–164143 (253) | 494 | 40 | 253 | U60315 | MCV | DNA-binding virion core protein VP8; TM | L | P15913 | L4R | MC073R | 20 |
| FPV132 | 164147–164533 (129) | 191 | 41 | 113 | P07615 | VV | Putative membrane protein; TM | L | P15914 | L5R | 20 | |
| FPV133 | 164487–164930 (148) | 290 | 37 | 145 | U60315 | MCV | L | P15915 | J1R | MC075R | 20 | |
| FPV134 | 164966–165889 (308) | 865 | 55 | 294 | U60315 | MCV | Poly(A) polymerase PAPS | M17418 | J3R | MC076R | 57 | |
| FPV135 | 165889–166446 (186) | 560 | 55 | 181 | P07391 | VV | RNA polymerase subunit RPO22 | M17418 | J4R | MC077R | 57 | |
| FPV136 | 166852–166442 (137) | 356 | 48 | 132 | U60315 | MCV | Membrane protein | M17418 | J5L | MC078L | 57 | |
| FPV137 | 166893–170753 (1287) | 5,074 | 72 | 1,289 | U60315 | MCV | RNA polymerase subunit RPO147 | J6R | MC079R | |||
| FPV138 | 171265–170768 (166) | 463 | 51 | 169 | U60315 | MCV | Protein-tyrosine phosphatase | L | H1L | MC082L | ||
| FPV139 | 171281–171850 (190) | 541 | 49 | 191 | U60315 | MCV | TM | H2R | MC083R | |||
| FPV140 | 173017–172037 (327) | 356 | 31 | 316 | AF124516 | LSV | Virion envelope protein p35; TM | L | H3L | MC084L | ||
| FPV141 | 175414–173021 (798) | 2,331 | 56 | 798 | U60315 | MCV | RNA polymerase-associated protein RAP94 | L | L46396 | H4L | MC085L | 191 |
| FPV142 | 175558–176079 (174) | 80 | 26 | 149 | S62819 | ORF virus | VLTF-4 | E, I | L46396 | H5R | MC086R | 191 |
| FPV143 | 176083–177030 (316) | 936 | 57 | 303 | L22579 | VAR | DNA topoisomerase | L | L46396 | H6R | MC087R | 191 |
| FPV144 | 177038–177493 (152) | 223 | 40 | 145 | U94848 | VV | Putative 17-kDa protein | E, L | H7R | MC088R | ||
| FPV145 | 177770–177462 (103) | 89 | 28 | 114 | U60315 | MCV | SP | L | MC089L | |||
| FPV146 | 177778–180330 (851) | 2,425 | 54 | 814 | U60315 | MCV | mRNA capping enzyme, large subunit | E, L | D1R | MC090R | ||
| FPV147 | 180392–180703 (104) | HT motif family | ||||||||||
| FPV148 | 181122–180706 (139) | 141 | 31 | 133 | U60315 | MCV | Virion protein | D2L | MC091L | |||
| FPV149 | 181390–181947 (186) | |||||||||||
| FPV150 | 182017–182844 (276) | 302 | 37 | 159 | U41315 | H. sapiens | N1R/p28 family | |||||
| FPV151 | 182887–183591 (235) | 396 | 38 | 239 | X77731 | Mus musculus | dCK | |||||
| FPV152 | 183987–183607 (127) | HT motif family | ||||||||||
| FPV153 | 184086–184709 (208) | |||||||||||
| FPV154 | 184937–185386 (150) | |||||||||||
| FPV155 | 185418–186641 (408) | N1R/p28 family | ||||||||||
| FPV156 | 186900–187295 (132) | HT motif family | ||||||||||
| FPV157 | 187344–188276 (311) | 383 | 41 | 167 | U41315 | H. sapiens | N1R/p28 family | E | ||||
| FPV158 | 188369–189760 (464) | 1,432 | 56 | 462 | D31902 | Monodelphis domestica | Photolyase | |||||
| FPV159 | 189902–190624 (241) | 160 | 34 | 130 | AF017791 | HaEPV | N1R/p28 family | |||||
| FPV160 | 190708–191175 (156) | |||||||||||
| FPV161 | 191226–191696 (157) | 94 | 24 | 127 | AF017791 | HaEPV | N1R/p28 family; TM | |||||
| FPV162 | 193077–194885 (603) | 499 | 31 | 464 | X69063 | Mus musculus | Ankyrin repeat family | E | ||||
| FPV163 | 195121–195909 (263) | 171 | 32 | 137 | AF017791 | HaEPV | N1R/p28 family | E | ||||
| FPV164 | 196404–197552 (383) | |||||||||||
| FPV165 | 199299–198625 (225) | 956 | 77 | 225 | U60315 | MCV | Late transcription factor VLTF-3 | I | A2L | MC104L | ||
| FPV166 | 199514–199299 (72) | 154 | 42 | 69 | U60315 | MCV | L | MC105L | ||||
| FPV167 | 201502–199532 (657) | 2,074 | 61 | 654 | U60315 | MCV | Virion core protein P4b | L | P17355 | A3L | MC106L | 17 |
| FPV168 | 202431–201568 (288) | Immunodominant virion protein | L | AJ005164 | A4L | 18 | ||||||
| FPV169 | 202470–202970 (167) | 472 | 57 | 166 | U60315 | MCV | RNA polymerase subunit RPO19 | E, L | A08272 | A5R | MC108R | |
| BlastP score | % Identity | Length, aaa | Accession no.b | Speciese | VV | MCV | ||||||
| FPV170 | 204092–202971 (374) | 781 | 38 | 373 | P20985 | VV | A08272 | A6L | MC109L | |||
| FPV171 | 206225–204099 (709) | 2,526 | 65 | 709 | U60315 | MCV | Early transcription factor VETFL | L | A7L | MC110L | ||
| FPV172 | 206291–207193 (301) | 721 | 45 | 298 | U60315 | MCV | Intermediate transcription factor VITF-3 | A8R | MC111R | |||
| FPV173 | 207388–207161 (76) | 189 | 47 | 69 | U60315 | MCV | TM | L | A9L | MC112L | ||
| FPV174 | 210064–207392 (891) | 1,948 | 43 | 891 | U60315 | MCV | Virion core protein P4a | L | A20158 | A10L | MC113L | |
| FPV175 | 210082–210903 (274) | 654 | 47 | 304 | U60315 | MCV | L | A20158 | A11R | MC114R | ||
| FPV176 | 211422–210910 (171) | 243 | 40 | 169 | P33837 | VAR | Virion protein | L | A12L | MC115L | ||
| FPV177 | 211437–211640 (68) | SP | ||||||||||
| FPV178 | 211848–211636 (71) | 100 | 35 | 59 | U60315 | MCV | Virion protein; SP | L | A13L | MC117L | ||
| FPV179 | 212190–211918 (91) | 167 | 43 | 87 | U60315 | MCV | Virion envelope protein; TM | L | A14L | MC118L | ||
| FPV180 | 212676–212386 (97) | 79 | 40 | 35 | P33840 | VAR | L | A15L | MC120L | |||
| FPV181 | 213769–212663 (369) | 844 | 44 | 369 | U60315 | MCV | Putative myristylated membrane protein; TM | L | A16L | MC121L | ||
| FPV182 | 214381–213788 (198) | 294 | 36 | 179 | P16711 | VV | Phosphorylated virion membrane protein; TM | L | A17L | MC122L | ||
| FPV183 | 214396–215781 (462) | 1,269 | 53 | 458 | U60315 | MCV | DNA helicase; transcriptional elongation | A18R | MC123R | |||
| FPV184 | 216018–215755 (88) | 159 | 52 | 73 | P33842 | VAR | L | A19L | MC124L | |||
| FPV185 | 216366–217664 (433) | 489 | 27 | 429 | U60315 | MCV | Processivity factor | E | A20R | MC126R | ||
| FPV186 | 216367–216029 (113) | 255 | 45 | 113 | U60315 | MCV | TM | A21L | MC125L | |||
| FPV187 | 217667–218134 (156) | 318 | 43 | 143 | P33845 | VAR | A22R | MC127R | ||||
| FPV188 | 218148–219296 (383) | 1,034 | 53 | 381 | P20998 | VV | Intermediate transcription factor VITF-3 | A23R | MC128R | |||
| FPV189 | 219326–222808 (1161) | 4,855 | 76 | 1,156 | U60315 | MCV | RNA polymerase subunit RPO132 | E | A24R | MC129R | ||
| FPV190 | 224610–222751 (620) | 281 | 25 | 348 | U60315 | MCV | A-type inclusion protein | L | A25L | MC130L | ||
| FPV191 | 226070–224649 (474) | 471 | 29 | 475 | U60315 | MCV | A-type inclusion protein | L | A26L | MC133L | ||
| FPV192 | 226496–226074 (141) | 322 | 43 | 141 | U60315 | MCV | TM | A28L | MC134L | |||
| FPV193 | 227419–226514 (302) | 693 | 45 | 300 | P33812 | VAR | RNA polymerase subunit RPO35 | A29L | MC135L | |||
| FPV194 | 227618–227397 (74) | 118 | 37 | 58 | P21088 | VV | A30L | MC136L | ||||
| FPV195 | 227797–228135 (113) | 160 | 30 | 107 | U60315 | MCV | A31R | MC138R | ||||
| FPV196 | 228139–228498 (120) | L | ||||||||||
| FPV197 | 229395–228493 (301) | 725 | 56 | 246 | U60315 | MCV | Virion assembly protein | L | A32L | MC140L | ||
| FPV198 | 229584–230102 (173) | 121 | 25 | 173 | S61094 | VV | C-type lectin like; TM | A34R | MC143R | |||
| FPV199 | 230160–230816 (219) | 113 | 25 | 108 | U18697 | Ginglymostoma cirratum | V-type Ig domain; TM | |||||
| FPV200 | 230794–231588 (265) | 92 | 28 | 128 | U18697 | G. cirratum | V-type Ig domain | L | ||||
| FPV201 | 231612–232460 (283) | 169 | 22 | 286 | U60315 | MCV | E | AF006064 | MC144R | 76 | ||
| FPV202 | 232649–232350 (100) | |||||||||||
| FPV203 | 232786–233640 (285) | 245 | 26 | 247 | JQ1743 | RFV | Tyrosine PK; TM | E | AF006064 | 76 | ||
| FPV204 | 233679–234704 (342) | 243 | 22 | 347 | AB006423 | H. sapiens | Serpin family | AF006064 | 76 | |||
| FPV205 | 235366–234713 (218) | TM | ||||||||||
| FPV206 | 235481–236404 (308) | 513 | 35 | 299 | P32249 | H. sapiens | G-protein-coupled receptor family | E | ||||
| FPV207 | 236417–236716 (100) | |||||||||||
| FPV208 | 237605–237805 (67) | |||||||||||
| FPV209 | 238466–238077 (130) | HT motif family | E | |||||||||
| FPV210 | 238970–239167 (66) | |||||||||||
| FPV211 | 239109–239483 (125) | 125 | 36 | 61 | D30783 | H. sapiens | EGF-like protein; TM | C11R | ||||
| FPV212 | 239489–240397 (303) | 731 | 45 | 302 | AB000450 | H. sapiens | Ser/Thr PK | B1R | ||||
| FPV213 | 240451–240936 (162) | TM | ||||||||||
| FPV214 | 241278–241649 (124) | 91 | 29 | 124 | U94848 | VV | Putative 13.7-kDa protein | |||||
| FPV215 | 241765–241986 (74) | TM | ||||||||||
| FPV216 | 242384–243271 (296) | 124 | 24 | 275 | L35601 | D. melanogaster | Ankyrin repeat family | |||||
| FPV217 | 243703–244686 (328) | 234 | 27 | 230 | AF081810 | LdNPV | ||||||
| FPV218 | 244729–246111 (461) | 298 | 26 | 349 | L35601 | D. melanogaster | Ankyrin repeat family | E | ||||
| FPV219 | 246144–247445 (434) | 295 | 31 | 275 | L35601 | D. melanogaster | Ankyrin repeat family | E | ||||
| FPV220 | 247453–247103 (117) | TM | ||||||||||
| FPV221 | 248001–247453 (183) | 79 | 30 | 180 | P21067 | VV | L | A47L | ||||
| FPV222 | 248089–250329 (747) | 433 | 25 | 641 | X69063 | Mus musculus | Ankyrin repeat family | E | ||||
| FPV223 | 250544–250966 (141) | 181 | 33 | 141 | AF153912 | TPV | Ankyrin repeat family | |||||
| FPV224 | 250974–251411 (146) | 131 | 31 | 143 | X62907 | A. thaliana | Ankyrin repeat family | |||||
| FPV225 | 251791–252102 (104) | 118 | 32 | 101 | Y11842 | CPV | B20R | |||||
| FPV226 | 252108–252983 (292) | 477 | 34 | 291 | P16913 | VV | Ser/Thr PK | B1R | ||||
| FPV227 | 253039–254121 (361) | 189 | 32 | 153 | U42580 | PBCV-1 | Ankyrin repeat family | |||||
| FPV228 | 254214–255788 (525) | 316 | 29 | 395 | X16609 | H. sapiens | Ankyrin repeat family; TM | E | ||||
| FPV229 | 257507–256968 (180) | |||||||||||
| FPV230 | 257639–258202 (188) | 200 | 33 | 157 | U50071 | C. elegans | Ankyrin repeat family | E | 166 | |||
| BlastP score | % Identity | Length, aaa | Accession no.b | Species | VV | MCV | ||||||
| FPV231 | 258169–258936 (256) | 209 | 31 | 193 | U21734 | C. elegans | Ankyrin repeat family | |||||
| FPV232 | 259206–260651 (482) | 274 | 29 | 294 | L35601 | D. melanogaster | Ankyrin repeat family | |||||
| FPV233 | 260697–262232 (512) | 243 | 26 | 318 | L35601 | D. melanogaster | Ankyrin repeat family | E | ||||
| FPV234 | 262266–263549 (428) | 259 | 29 | 330 | L35601 | D. melanogaster | Ankyrin repeat family | P14367 | ||||
| FPV235 | 263571–263999 (143) | 108 | 26 | 56 | X54868 | H. sapiens | C-type lectin family; TM | P14372 | A40R | 166 | ||
| FPV236 | 264005–264844 (280) | 170 | 29 | 159 | L26342 | RFV | N1R/p28 family | P14365 | 166 | |||
| FPV237 | 265059–264859 (67) | P14366 | 166 | |||||||||
| FPV238 | 265612–265794 (61) | D00295 | 166 | |||||||||
| FPV239 | 265666–265178 (163) | 262 | 31 | 152 | M88072 | G. gallus | C-type lectin family; SP | E | P14371 | A40R | 166 | |
| FPV240 | 265785–267014 (410) | 256 | 29 | 234 | Q01485 | H. sapiens | Ankyrin repeat family | E | P14360 | 166 | ||
| FPV241 | 267416–267733 (106) | 106 | 33 | 72 | X69065 | Mus musculus | Ankyrin repeat family | |||||
| FPV242 | 268121–269194 (358) | 229 | 26 | 335 | Q01485 | H. sapiens | Ankyrin repeat family; TM | |||||
| FPV243 | 269266–270051 (262) | 115 | 24 | 238 | Y15035 | CPV | Ankyrin repeat family | |||||
| FPV244 | 271027–273030 (668) | 368 | 32 | 348 | U43965 | H. sapiens | Ankyrin repeat family | |||||
| FPV245 | 274352–273045 (436) | 371 | 30 | 340 | L35601 | D. melanogaster | Ankyrin repeat family | |||||
| FPV246 | 274734–276509 (592) | 330 | 33 | 274 | X16609 | H. sapiens | Ankyrin repeat family | E | ||||
| FPV247 | 276565–276936 (124) | EFc familye | E | |||||||||
| FPV248 | 277051–277503 (151) | 130 | 30 | 139 | AF017791 | HaEPV | N1R/p28 family | E | P14364 | 166 | ||
| FPV249 | 277887–278201 (105) | P14363 | 166 | |||||||||
| FPV250 | 278855–278436 (140) | 210 | 42 | 95 | L22174 | MDV | US ORF2 | P14362 | 166 | |||
| FPV251 | 278971–279414 (148) | 187 | 35 | 150 | U60474 | MYX | Serpin family | P14369 | 166 | |||
| FPV252 | 280492–280295 (66) | D00295 | 166 | |||||||||
| FPV253 | 280859–280359 (167) | 179 | 24 | 166 | M88072 | G. gallus | C-type lectin family; SP | P14370 | 166 | |||
| FPV254 | 280941–280726 (72) | D00295 | 166 | |||||||||
| FPV255 | 282609–281356 (418) | 252 | 27 | 296 | U18337 | VAR | C4L/C10L-like family | P14361 | C10L | 166 | ||
| FPV256 | 282951–283316 (122) | EFc family | E | E31685 | ||||||||
| FPV257 | 283415–283113 (101) | A06621 | ||||||||||
| FPV258 | 283669–284037 (123) | 89 | 30 | 122 | Q07108 | H. sapiens | C-type lectin family | E | C31685 | A40R | ||
| FPV259 | 285173–284508 (222) | A06621 | ||||||||||
| FPV260 | 286049–286663 (205) | 121 | 29 | 134 | AF021350 | R. norvegicus | C-type lectin family; TM | A40R | ||||
aa, amino acids.
Accession numbers are from the GenBank or SwissProt database.
Function was deduced either from the degree of amino acid similarity to known genes or by the presence of Prosite signatures. TM, a Z score of >1.96 was used for the prediction of transmembrane (TM) domains with the Memsat computer programs; SP, N-terminal signal peptide (Z score of >2.5 using Sigcleave).
Putative promoter. E, early; I, intermediate; L, late.
Similar to EFc ORF described in reference 36.
The remnant of an integrated avian reticuloendotheliosis virus (REV) genome in the FPV genome is represented by 253 nucleotides (232464 to 232717) that are similar (98% identity) to the long terminal repeat of a chicken B lymphoma-derived REV (accession no. M22223). However, REV env, gag, and pol genes were not found as has been reported for some FPV strains (76). The same long terminal repeat (98% identity over 200 nucleotides) is also found in several strains of Marek's disease virus (MDV), a herpesvirus of chickens. The fragmented remains of a ubiquitin gene are present in the FPV genome from nucleotides 74550 to 74220. Interestingly, the best match to this gene is chicken ubiquitin (accession no. M1110), which exhibits 54% identity over 76 amino acids with one frameshift, two in-frame stops, and two gaps.
ITRs.
The FPV genome contains identical ITRs of 9,520 nucleotides at both termini (Fig. 1). Within each ITR, a 1.7-kbp region contains 42 copies of a 31- to 32-bp tandem repeat (70 to 95% identical) between nucleotides 198 and 1835 as well as between nucleotides 286703 to 288340. Sizing of seven cloned fragments spanning this tandem-repeat region produced specific size classes of 1.7, 2.4, 3.3, 5.1, and 5.8 kbp in length, indicative of length polymorphism. Therefore, individual FPV genomes could be at least 8 kbp longer than the genomic sequence assembled here. Each ITR also contains 10 ORFs. These ITR sequence data are consistent with previous descriptions of FPV ITR regions (36, 166).
Gene expression regulatory elements.
FPV ORFs contain typical poxvirus promoter sequences upstream of their translation initiation codons. Sequences with similarity to the VV early-promoter consensus sequence (AAAAATGAAAAAAAA) have previously been noted in the 5′ untranslated regions of known and predicted FPV early genes (90, 91, 191). Fifty-six FPV ORFs contain putative early promoters (Table 1). Of these, 22 contain a poxvirus early transcriptional stop sequence (TTTTTXT, where X is any nucleotide) near the translational stop codon (50 bases upstream to 100 bases downstream) and lack the early stop sequence elsewhere in the ORF (189). As seen in other poxviruses, many genes with potential early promoters are members of gene families and/or putative host range genes (Table 1). Three of five homologues of VV intermediate genes (FPV088, FPV126, and FPV165) contain the VV intermediate-promoter sequence (AAAXAAX11–13TAAA) (10, 11, 118), and one (FPV049) contains a single-base substitution (AAAXAG). A total of 55 putative late FPV ORFs, including many of the conserved virion-associated poxvirus genes (Table 1), contain the VV late-promoter sequence (TAAATG) at the ATG codon (131). The TAAATG late promoter has been previously described to be located upstream of FPV late genes (17, 91, 163, 191), and it is known that early-late and late promoters can be exchanged between FPV and VV with no loss of temporal specificity (27).
Transcription and mRNA biogenesis.
FPV contains 26 genes involved in poxvirus transcriptional processes (Table 1). These include RNA polymerase subunits; mRNA transcription initiation, elongation, and termination factors; and the enzymes that direct posttranscriptional processing of viral mRNA (118). FPV RNA polymerase subunits include homologues of VV RPO147 (FPV137), RPO132 (FPV189), RAP94 (FPV141), RPO35 (FPV193), RPO30 (FPV100), RPO22 (FPV135), RPO19 (FPV169), RPO18 (FPV056), and RPO7 (FPV118). Homologues of all previously described early (E), intermediate (I), and late (L) poxvirus transcription factors (TFs) are found in FPV, including the following: VETFS (FPV057), VETFL (FPV171), VITF-3 (FPV172 and FPV188), VLTF-1 (FPV126), VLTF-2 (FPV049), VLTF-3 (FPV165), and VLTF-4 (FPV142) (87, 191). FPV079 and FPV183 encode elongation factors for late transcription (VV G2R and A18R) (22, 44, 186). Both transcriptional terminator NPH-1 (FPV052) and the RNA helicase NPH-II (FPV082) are present. FPV146 and FPV051 encode both subunits of the mRNA capping enzyme, and FPV102 and FPV134 encode both subunits of the poly(A) polymerase. FPV053 and FPV054 contain MutT-like motifs and are similar to VV D10R and D9R (85). D10R has recently been shown to be a negative regulator of viral transcription (149).
Nucleotide metabolism.
FPV contains homologues of thymidine kinase (FPV086), dUTP pyrophosphatase (FPV038), glutaredoxin (FPV077), two deoxycytidine kinases (dCKs; FPV059 and FPV151), and a putative DNase II (FPV032) (Table 1). Genes encoding dCK and DNase II are unique to FPV and have been previously described (86, 93). Interestingly, sequencing of the complete genome has revealed a second dCK gene (FPV151). These two FPV dCK genes are 42% identical to each other and exhibit 32% amino acid identity to cellular dCK (Table 1). The DNase II homologue, FPV032, is truncated compared to the previously described FPV gene, FPCEL-1 (93). FPV032 represents the largest subunit (α2) of cellular DNase II and includes the conserved histidine at the potential active site (99, 174). The function of this gene in the viral replication cycle is unknown; however, FPCEL-1 is not essential for viral growth in vitro (93). Cellular DNase II is thought to function in DNA catabolism during apoptosis (89, 168). FPV lacks other known poxvirus genes thought to be involved in nucleotide metabolism, including thymidylate kinase, thymidylate synthase, ribonucleotide reductase, guanylate kinase, and thioredoxin. This specific complement of nucleotide metabolism genes in FPV suggests that they have significance for cell and/or tissue tropism.
DNA replication and repair.
FPV contains homologues of ChPV genes involved in DNA replication and repair (118) (Table 1). These include a DNA ligase (FPV043), ATP-GTP binding protein (FPV058), uracil DNA glycosylase (FPV062), DNA polymerase (FPV094) (19), DNA topoisomerase (FPV143), processivity factor (FPV185), and replication-essential protein kinase (FPV212).
Interestingly, FPV158 is a homologue of class II cyclobutane pyrimidine dimer (CPD) photolyases. Although the gene has been previously described in the Entomopoxvirinae (1), this is the first description of a photolyase in a ChPV. FPV158 is most similar to marsupial photolyase (56% identity over 462 amino acids) (188) and is slightly less similar to Melanoplus sanguinipes entomopoxvirus (EPV) photolyase (54% identity over 448 amino acids) (1). Both class II photolyase Prosite signatures (PS01083 and PS01084) are present with a single conservative substitution at residue 302. Although the function of this FPV gene is unknown, CPD photolyase is a photoreactive enzyme that efficiently repairs UV-induced CPD lesions in DNA, using visible light as an energy source (75). Since EPVs have insect hosts and FPV is mechanically vectored by insects (48), the presence of a photolyase gene in both viral genomes is suggestive of a relationship between a viral phase in insects and/or the environment and the need for this type of virus-encoded DNA repair.
Protein modification.
FPV contains at least six genes with putative protein modification functions (Table 1). The homologues encoded include three serine/threonine protein kinases (PKs) (FPV111, FPV212, and FPV226), one tyrosine PK (FPV203), a tyrosine/serine protein phosphatase (FPV138), and a metalloprotease (FPV081). FPV212 and FPV226 are similar to the serine/threonine PKs B1R and B12R of VV. FPV111 is similar to VV F10L, a serine/threonine PK essential for phosphorylation of virus proteins during virion assembly (14, 54). FPV203 shows similarity to the product of a tyrosine PK-like ORF found in rabbit fibroma virus (RFV) (109); however, neither of these poxvirus proteins contains the critical Asp residue at the predicted active site (Prosite PS00109). FPV138 is a homologue of the VV H1L tyrosine/serine protein phosphatase, which is involved in VV assembly (54). FPV081 is a homologue of the VV protease G1L. This protein contains the characteristic amino-terminal His-XX-Glu-His inverted metalloprotease motif, and it may function in viral protein processing and virion morphogenesis (178).
Structural proteins.
FPV encodes homologues of at least 31 known VV structural proteins, and the majority of them are associated with the intracellular mature virus particle (IMV) (Table 1). FPV homologues of VV core proteins include FPV069 (D3R), FPV083 (I7L), FPV090 (I1L), FPV103 (F17R), FPV120 (G7L), FPV131 (L4R), FPV148 (D2L), FPV167 (A3L), FPV168 (A4L), FPV174 (A10L), and FPV176 (A12L) (15, 25). FPV homologues of VV IMV membrane-associated proteins include FPV050 (D13L), FPV085 (I5L), FPV128 (L1R), FPV140 (H3L), FPV178 (A13L), FPV179 (A14L), and FPV182 (A17L). FPV lacks homologues of VV IMV membrane proteins A27L, which is required for extracellular enveloped virion (EEV) envelopment and egress and for heparan sulfate binding (41, 130), and D8L, a cell surface binding protein (103). FPV structural proteins FPV120, FPV131, FPV167, FPV174, FPV176, and FPV182, like their VV homologues, contain the conserved AG proteolytic cleavage sites, which suggests that aspects of structural protein processing are conserved in FPV (173). FPV197 is the homologue of VV ATP-GTP binding protein A32L, which likely functions in virion assembly and DNA packaging (38).
FPV contains three genes that encode proteins potentially associated with EEVs (118, 123). FPV108, FPV109, and FPV198 are similar to VV F13L, F12L, and A34R, respectively (35). Missing from FPV are obvious homologues of VV EEV genes B5R, A33R, A36R, and A56R. EEV membrane proteins are involved with EEV formation, release, and infectivity (23, 111, 181). Since these functions may be associated with aspects of host range, the lack of well-conserved homologues of these genes in FPV is not surprising.
Homologues of five genes representing two conserved poxvirus gene families with putative structural functions are present in FPV. The genes encoding FPV112 and FPV128, homologues of VV F9L and L1R, respectively, comprise one gene family (142). The genes encoding FPV127, FPV136, and FPV181, homologues of VV G9R, J5L, and A16L, respectively, comprise a second, small gene family. G9R and A16L proteins are myristylated and potentially soluble (105), and J5L is thought to be an essential gene (190). Invariant cysteine residues and putative transmembrane domains unique to each family are conserved in these FPV ORFs (142).
FPV190 and FPV191 are homologues of poxvirus A-type inclusion (ATI) proteins (Table 1), insoluble proteins that constitute the protein matrix of ATIs. Cytoplasmic ATIs are thought to protect mature virions from environmental insults, and they may be of significance for FPV transmission in nature (40, 82, 128, 133).
Host-related functions.
FPV contains a significant number of putative host range genes that exhibit similarity to cellular genes and to other known poxvirus genes. This diverse complement of host range genes, some of which are novel, is suggestive of significant adaptation to the avian host. These genes may function in host immune evasion, immune modulation, and aspects of cell and/or tissue tropism or perform other cellular functions. Most of these genes are found in terminal regions of the FPV genome, although several groupings of them are more centrally located.
Immune evasion functions.
FPV080 is a homologue of the eukaryotic transforming growth factor β (TGF-β) (Table 1; Fig. 2A). To our knowledge, this is the first TGF-β gene found in a virus genome. Similarities to eukaryotic TGF-β include the 112-amino-acid peptide region of the active protein, Prosite signature PS00250 (with one mismatch), and cysteines necessary for intra- and interchain disulfide bond formation. TGF-β is a multifunctional peptide that both stimulates connective tissue cell growth and differentiation, particularly during neovascularization and wound healing, and suppresses proliferation of most other cell types (58). TGF-β also exhibits a range of immunomodulatory effects, including suppression of cellular and humoral immune mechanisms, specifically generation and/or activity of cytotoxic T lymphocytes, natural killer (NK) cells, and lymphokine-activated killer cells, generation and/or activity of lymphokines (interleukin-1 [IL-1], IL-6, tumor necrosis factor, and IL-2); and production of polyclonal antibodies (58). Chemoattractant and proinflammatory properties have also been associated with TGF-β (58). A role for FPV080 in suppression of the host immune response and/or cell growth and differentiation is likely.
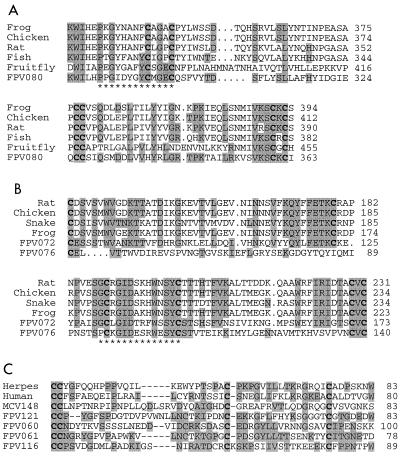
FIG. 2.
Multiple amino acid sequence alignments of proteins encoded by putative FPV immune evasion genes. Boldfaced letters represent conserved cysteine residues, asterisks mark Prosite signatures, and shaded residues indicate identity to amino acids of FPV proteins. Amino acid positions are indicated on the right. (A) Alignment of FPV080 with TGF-β genes. The Prosite signature is PS00250. Frog, Xenopus laevis, accession no. P17247; chicken, Gallus gallus, accession no. P30371; rat, Rattus norvegicus, accession no. P17246; fish, Oncorhynchus mykiss, accession no. AJ007836; fruit fly, Drosophila melanogaster, accession no. M77012. (B) Alignment of FPV072 and FPV076 sequences with those of NGFs. The Prosite signature is PS00248. Rat, Rattus norvegicus, accession no. P25247; chicken, Gallus gallus, accession no. P05200; snake, Bungarus multicinctus, accession no. P34128; frog, Xenopus laevis, accession no. P211617. (C) Alignment of FPV060, FPV061, FPV116, and FPV121 sequences with those of viral and cellular CC chemokines. Herpesvirus, Kaposi's sarcoma-associated herpes-like virus, accession no. U74585; human, Homo sapiens, accession no. P22362; MCV148, molluscum contagiosum virus, accession no. U60315.
FPV072 and FPV076 are similar to cellular β nerve growth factor (β-NGF) (Table 1; Fig. 2B). This is the first example of a virus encoding β-NGF-like genes. Both FPV proteins contain the six cysteine residues involved in intrachain disulfide bonding and the Prosite β-NGF family signature (PS00248) (Fig. 2B). β-NGF, a member of the neurotrophin protein family, stimulates neuronal survival, division, and differentiation and promotes survival of memory B lymphocytes and mast cells (30, 97, 167). Recently, β-NGF has been shown to be an autocrine survival factor for human immunodeficiency virus type 1-infected macrophages (68). An FPV-encoded β-NGF may be involved in promoting infected-cell survival. In addition, β-NGF has proinflammatory and immunomodulatory effects (5). β-NGF, which is produced by fibroblasts and keratinocytes in response to injury, induces differentiation, activation, and degranulation of mast cells and modifies expression of mast cell-derived immunoregulatory mediators and cytokines (34, 104, 138, 176, 177, 183). Conceivably, a virus-encoded β-NGF antagonist could have a role in inhibiting antiviral immune responses in FPV-infected skin and respiratory tract. Given that mast cells are initiators and amplifiers of innate immune responses, the presence of β-NGF homologues in FPV suggests that interference with early innate immune responses may be important for viral infection.
FPV060, FPV061, FPV116, and FPV121 exhibit similarity to the CC class of small soluble chemokines found in vertebrates (Table 1). The FPV genes contain the conserved pattern of four cysteines which are necessary for disulfide bond formation (Prosite PS00472), as well as other conserved residues (Fig. 2C). The FPV genes are similar in size (120 to 181 amino acids) to other known CC chemokines. Three of the products contain potential signal sequences at the N terminus, indicating that they may be secreted proteins. In general, CC chemokines attract T lymphocytes and NK cells to sites of infection (113). Other ChPVs modulate CC chemokine activity by secreting novel proteins that specifically bind CC chemokines and inhibit their effects in vitro and in vivo. These inhibitors are widespread among mammalian poxviruses, including VV, variola virus (VAR), cowpox virus (CPV), RFV, myxoma virus (MYX), and rabbitpox virus but is notably absent from FPV (4, 73, 94, 151). In molluscum contagiosum virus (MCV), a CC chemokine-like protein, MC148R, functions as a broad-spectrum CC and CXC chemokine antagonist (49). FPV's large repertoire of CC chemokine homologues functioning as antagonists could result in broad-range inhibition of normal CC chemokine function during host antiviral immune responses. Alternatively, as is the case for the viral macrophage-inhibitory protein 1 chemokine encoded by human herpesvirus 8, FPV chemokine homologues may function as agonists to modify normal host immune responses (47, 61).
FPV contains three genes encoding proteins with homology to G-protein-coupled receptors (Table 1). FPV021 and FPV027 are most similar to a monkey chemokine receptor protein (GPR1), while FPV206 is most closely related to the human Epstein-Barr virus-induced G-protein-coupled receptor (21). The highest level of amino acid similarity to cellular genes occurs at the seven transmembrane domains, the first cytoplasmic domain, and the second extracellular domain. The conserved acidic amino acid-Arg-aromatic amino acid triplets in the amino-terminal portion of the second aromatic loop, which have been implicated in the interaction with G proteins, are conserved in FPV021 and conservatively substituted in FPV027 and FPV206 (8). As with other G-protein-coupled receptors, the FPV proteins contain potential glycosylation sites at their carboxyl termini. G-protein-coupled receptors are integral membrane proteins that transduce extracellular signals to the intracellular environment through activation of the phosphatidylinositol-calcium second-messenger system (139). These receptors have been identified in the capripoxviruses and in swinepox virus, where their function is not known (37, 107). However, G-protein-coupled receptors encoded by several herpesvirus genomes are able to bind chemokines and invoke signal transduction responses that affect viral replication and pathogenesis in the host (2, 7, 13, 67).
FPV073 exhibits similarity to mammalian and ChPV IL-18-binding protein and contains potential N-glycosylation sites and a signal peptide (Table 1) (142, 184). Cellular and MCV IL-18bp homologues have been found to inhibit IL-18-dependent gamma interferon production (3, 185). IL-18 is a multifunctional proinflammatory cytokine of the IL-1 family that induces gamma interferon production, Th-1 responses, and NK cell activity, and it is important for effective host responses to VV infection in mice (50, 56, 79, 114, 161, 162). An anti-inflammatory function for FPV073 is likely.
FPV047 most closely resembles mammalian K/L-type semaphorins and the alcelaphine herpesvirus semaphorin homologue (accession no. U18243) (33% identity over 597 amino acids) (Table 1). Like the K/L-type semaphorin and alcelaphine herpesvirus semaphorin, FPV064 contains a potential amino-terminal signal sequence, a large semaphorin K/L domain, an immunoglobulin (Ig) domain, and a hydrophobic carboxyl terminus (62, 95). VV also encodes a K/L-like semaphorin homologue (A39R); however, the semaphorin domain is truncated and the Ig domain is absent (84). Semaphorins are a large family of secreted and membrane-associated proteins that act as axon guidance molecules during embryonic development and may affect organogenesis, vascularization, and angiogenesis (154). In addition, the CD100 semaphorin protein found on the surface of T lymphocytes functions in cell activation (52). The secreted VV A39R protein binds a plexin-like receptor found on lymphocytes and induces cytokine production and ICAM up-regulation in monocytes (43). The FPV semaphorin homologue may have a similar immunomodulatory function.
FPV contains eight ORFs (FPV001, FPV003, FPV008, FPV235, FPV239, FPV253, FPV258, and FPV260) with homology to C-type lectins NKG2 and CD94 proteins present on NK cells and CD69 protein present on lymphocytes (Table 1). Similar proteins have been described in poxviruses (VV and CPV) and African swine fever virus (ASFV) (122, 145, 179). Although the functions of these viral proteins are unknown, the VV C-type lectin protein, A40R, localizes to infected cell plasma membranes (179). C-type lectin cellular NK cell receptors bind class I major histocompatibility complex antigens and promote or inhibit immune activity through intracellular signaling pathways (66, 132, 175). It is conceivable that the expression of these proteins in FPV-infected cells interferes with normal immune surveillance or host responses.
FPV encodes five homologues of serine proteinase inhibitors (serpins) (FPV010, FPV040, FPV044, FPV204, and FPV251) (Table 1). All contain the serpin Prosite signature (PS00284) and exhibit 21 to 29% amino acid identity to each other. Serpin genes have been found in most ChPVs (rabbitpox virus, RFV, VV, VAR, CPV, MYX, and ectromelia virus [ECT]), where they perform host range functions involving anti-inflammatory activity and/or regulation of cellular apoptosis in specific cells through inhibition of IL-1β-converting enzyme, the cytotoxic-T-lymphocyte-derived protease granzyme B, and other caspases within the apoptosis-regulatory cascade (172).
Other host range functions.
The gene encoding FPV039 is the first reported poxvirus member of the Bcl-2 gene family. FPV039 resembles MCL1, a protein induced during monocyte/macrophage differentiation in myeloid leukemia cell lines, and BFL1 (29% identity over 134 amino acids), an antiapoptosis protein expressed specifically in the bone marrow, spleen, and thymus (88, 100) (Table 1; Fig. 3A). FPV039 contains one BH1 domain and one modified BH2 domain (Prosites PS01080 and PS01258) but lacks additional BH3 and BH4 domains. As with other viral Bcl-2 homologues, FPV039 may prevent a cellular apoptotic response to viral infection (12).
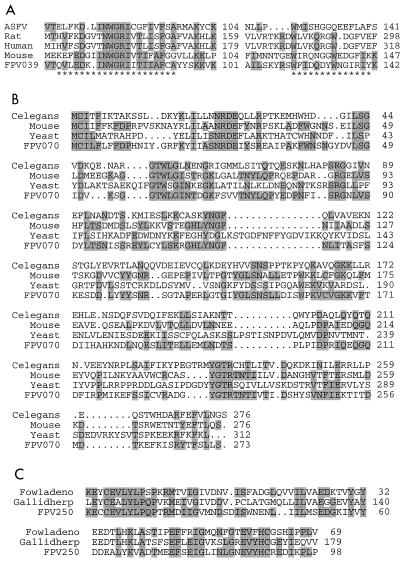
FIG. 3.
Multiple amino acid sequence alignments of proteins encoded by putative FPV host range genes. Asterisks mark Prosite signatures; shaded residues exhibit identity to amino acids to FPV proteins. Amino acid positions are indicated. (A) Alignment of FPV039 sequence with those viral and cellular Bcl-2 homologues. ASFV, accession no. Q07819; rat, Rattus norvegicus, accession no. AF115380; human, Homo sapiens, accession no. Q07820; mouse, Mus musculus, accession no. Q07440. (B) Alignment of FPV070 sequence with homologues of the mouse T10 protein. Celegans, Caenorhabditis elegans, accession no. Z78016; mouse, Mus musculus, accession no. X74504; yeast, S. cerevisiae, accession no. P53275. (C) Alignment of FPV250 sequence with those of proteins from avian viruses. Fowladeno, fowl adenovirus, accession no. AF007578; Gallidherp, Marek's disease virus, accession no. L22174.
FPV070 is a homologue of the mouse T10 gene and a yeast protein of unknown function (Table 1; Fig. 3B). This gene has not been previously found in a viral genome. T10 encodes a protein which is specifically expressed at high levels in epithelial cells of the trachea, esophagus, lung, and velopharyngeal region during early embryogenesis (74). The diphtheric form of FPV infection in chickens involves viral infection of the mucous membranes of the mouth, pharynx, and larynx and sometimes the trachea (169–171). An FPV T10 homologue may perform a host range function in epithelial cells of the respiratory tract.
FPV217 and FPV250 have similarities to genes of unknown function present in other viruses (Table 1; Fig. 3C). FPV217 is similar to a gene in Lymantria dispar nuclear polyhedrosis virus. FP250 is similar to putative proteins encoded by MDV and fowl adenovirus (44% identical over 99 amino acids) (32, 146). The presence of this homologue in three different avian DNA viruses suggests a significant avian host range function.
FPV211 contains an epidermal growth factor (EGF)-like domain which includes the six cysteine residues involved in disulfide bond formation, a potential signal peptide, and a transmembrane domain (Table 1). The similarity of FPV211 to secreted poxvirus EGF-like growth factors is based solely on the presence of the EGF domain. Poxvirus EGF-like growth factors are not essential for virus replication in vitro, influence virulence in vivo, and stimulate cell proliferation at sites of viral replication (110). FPV211 may contribute to the hyperplasia observed in FPV-infected tissue (169).
FPV contains 31 ORFs with ankyrin repeat motifs (Table 1). This large gene family is clustered at both ends of the genome, with two additional ORFs (FPV115 and FPV162) being found in more central locations (Fig. 1). Proteins encoded by FPV ankyrin family genes contain 1 to 12 copies of the ankyrin repeat motif (102), range in size from 104 to 747 amino acids, and are from 20 to 45% identical to each other depending on ORF size and alignment length. This level of amino acid identity is higher than that to ankyrin repeats of proteins found in a wide phylogenetic range of organisms. The ankyrin gene copy number may differ in FPV strains. Sequence from a genomic region in the right end of a highly passaged FPV strain contains nine fewer ankyrin genes than the number found here (166). Poxvirus ankyrin repeat genes have been associated with host range functions in MYX, CPV, and VV, and they may inhibit virus-induced apoptosis (70, 80, 119, 126, 153, 159). Ankyrin repeat motifs are clearly involved in mediating protein-protein interactions (101, 140). In CPV, which has a relatively broad host range, at least 16 ankyrin repeat genes have been identified (145). Loss or disruption of many of these genes in other orthopoxviruses that have a more restricted host range has suggested that loss of ankyrin genes may be associated with the narrowing of host range (6, 145).
FPV contains 10 ORFs (FPV075, FPV124, FPV150, FPV155, FPV157, FPV159, FPV161, FPV163, FPV236, and FPV248) with homology to N1R of RFV, p28 of ECT, and other ChPV and EPV genes (Table 1). Amino acid identity among FPV NR1/p28 family members is 20 to 38% and includes a conserved amino-terminal region with an invariant tryptophan residue. This domain is necessary for localization of RFV N1R to viral factories (31). FPV150 and FPV157, together with RFV N1R, ECT p28, and CPV and VAR homologues, contain a carboxyl-terminal C3HC4 RING finger. RING fingers are cysteine-rich zinc-binding motifs that are present in functionally diverse proteins, mediate protein-protein interactions, and help direct protein ubiquitination (81, 137). ECT p28 is a host range factor required for viral replication in mouse macrophages and for viral virulence in mice (143, 144); thus, a role in viral virulence and/or host range is likely for some members of this FPV family.
FPV064 encodes a homologue of cellular and MCV (MC066L) glutathione peroxidase (Table 1). FPV064 contains the glutathione peroxidase signature sequence (Prosites PS00460 and PS00763) including the active site for selenocysteine encoded by the opal codon (UGA). Cellular glutathione peroxidases reduce hydroxyperoxides with glutathione and are believed to provide protection from oxidative stress caused by ingested or endogenously formed hydroxyperoxides (157). MC066L protects human keratinocytes against cytotoxic effects of UV radiation and hydrogen peroxide and may permit efficient viral replication under conditions of environmental stress (148). A similar function for FPV064 is likely.
Cellular functions.
FPV114 shares a 180-amino-acid conserved domain with proteins found in plants (accession no. U80192), yeast (accession no. P36024 and X88900), roundworms (accession no. Z81069), and bacteria (accession no. P24285, P30197, Q04810, and D90910). FPV114 is most closely related to the yeast Hal3 and SIS2 genes and a putative Hal3 homologue from the plant Arabidopsis thaliana (Table 1). These proteins function as inhibitory subunits of cellular protein phosphatases, and they promote salt tolerance and affect growth (53). FPV114, roundworm, and bacterial homologues lack the amino- and carboxyl-terminal domains found in the yeast protein. Bacterial homologues function in DNA/pantothenate and lantibiotic metabolism (39, 92). The wide phylogenetic distribution of FPV114 homologues suggests that their function is highly conserved.
FPV048 encodes a 261-amino-acid protein that is similar to the members of the GNS1/SUR4 family of integral membrane proteins (Table 1). Similarities to the GNS1/SUR4 gene family include a defined motif (BLOCKs database signature BL01188) and a conserved protein structure consisting of an N-terminal region with two transmembrane domains, a central hydrophilic loop, a C-terminal region with one to three transmembrane domains, and the Prosite family signature (PS01188). The yeast GNS1 and SUR4 genes function in glucose metabolism, and they are suspected to have pleiotrophic functions in the cellular response to nutrient availability (60, 69, 129).
FPV011 and FPV033 are similar to the eukaryotic α-type soluble NSF attachment protein (α-SNAP) (Table 1). FPV033 has been previously described, but this is the first report of a second FPV α-SNAP homologue (93). FPV011 and FPV033 are similar in size (278 and 267 amino acids long, respectively) and exhibit 34% amino acid identity to each other over 249 amino acids. α-SNAPs are involved in vesicular trafficking, mediating intracellular membrane fusion by recruiting soluble NSF to membrane receptors (158). α-SNAPs and their yeast homologues (Sec17) are required for vesicular transport through the Golgi complex and for exocytosis (42, 117). The fact that FPV033 is not essential for growth in vitro suggests that it has a host range function (93).
FPV093 is the homologue of VV E10R, a protein that is conserved in many cytoplasmic DNA viruses and eukaryotes and contains a pattern of cysteine residues typical of glutaredoxin and thioredoxin redox-active centers (78). The homologue of this protein in ASFV, 9GL, has recently been shown to be involved in virion maturation and viral growth in swine macrophages (98).
FPV030 exhibits homology to human PC-1, which has alkaline phosphodiesterase and nucleotide pyrophosphatase activities and has been previously found in FPV (33, 93). The function of this conserved but nonessential FPV gene is unknown; however, it has been suggested that it may provide an external source of nucleotides or regulate signal transduction (93).
FPV046 encodes a homologue of 3-β-hydroxysteroid dehydrogenase (3βHSDH), previously described in FPV and other poxviruses (VV and MCV) (Table 1) (116, 142, 150). In VV, 3βHSDH has steroidogenic activity in vitro and is involved in viral virulence in vivo (116). Cellular 3βHSDH catalyzes the oxidative conversion of both δ(5)-ene-3-β-hydroxysteroid and ketosteroids, performing a crucial role in the biosynthesis of all classes of steroid hormones.
Two unrelated FPV ORFs, FPV029 and FPV071, have striking similarity to genes present in a diverse phylogenetic range of organisms (Table 1). FPV029 is similar to proteins of unknown function from yeast (accession no. P34222), bacteria (accession no. U67463 and AE000927), a roundworm (accession no. AF067936), a plant (accession no. AL031804), the fruit fly (accession no. AE0015722), and humans (accession no. AF151905). All of these proteins show several conserved domains, and the bacterial genes and the FPV037 ORF have similar lengths. FPV071 is similar to genes of unknown function from yeast (accession no. P40506), humans (accession no. AI391502), tomato (accession no. AI771876), and fruit fly (accession no. AF132150).
Gene families of unknown function.
FPV097, FPV098, FPV099, FPV107, FPV122, and FPV123 are homologues of VAR B22R (Table 1). B22R homologues are also present in CPV, ECT, and MCV but are absent from VV (6, 71). FPV gene family members exhibit 34 to 52% amino acid identity to each other and 32 to 36% identity to the other poxvirus homologues, with the highest level of similarity being in the carboxyl-terminal regions. Several features make these FPV B22R homologues notable. They represent the largest genes in FPV (1,766 to 1,949 amino acids), and they comprise 12% of the viral genome. FPV contains multiple B22R homologues, while other poxviruses either contain a single copy of the gene or lack it (71, 108, 142). FPV B22R homologues are present in a central genomic region, while orthopoxvirus homologues are located in the terminal regions of their respective genomes (Fig. 1) (71, 108). Although no function has yet been assigned to any of these ORFs, it has been suggested that they are type II membrane proteins (108, 145).
FPV017, FPV055, FPV125, FPV199, and FPV200 have similarity to V-type Ig domains of diverse proteins (Table 1). All five proteins contain conserved Ig domain cysteines and surrounding residues. FPV017 and FPV199 are notably similar to each other (25% amino acid identity), as are FPV055 and FPV125 (37% identity over 270 amino acids). Cellular members of the Ig superfamily include secreted and membrane-bound receptors and cell adhesion proteins (180). Ig domain-containing proteins, including hemagglutinin, cytokine receptor, and HLA antigen homologues, are present in other poxviruses (141, 142, 147, 152).
FPV067, FPV087, FPV147, FPV152, FPV156, and FPV209 comprise the His-X-X-Thr motif gene family (HT motif). These genes exhibit 18 to 34% identity over 69 to 102 amino acids and contain the HT motif at residues 19 to 33 or 51 to 65. FPV147, FPV152, and FPV209 also have HX4–5T motifs upstream of the primary HT motif. The HT family ORFs have no significant similarity to other sequences in the database.
FPV006 and FPV255 are 35% identical to FPV020 over 283 amino acids. All three FPV ORFs are similar to VV C10L and C4L and homologues present in CPV and VAR (Table 1). Like their orthopoxvirus homologues, these FPV ORFs are located in the terminal genomic regions. Although their function has not been determined, C10L and C4L are dispensable for virus growth in cell culture (126).
Relationship of FPV to other ChPVs.
FPV resembles other ChPVs in overall genome structure and composition (the presence of a central conserved core of genes, ITRs, and large numbers of homologues). However, compared to those of other ChPVs, the genome of FPV exhibits large-scale genomic rearrangement, more extensive gene families, and the presence of novel host range genes. Genomic comparisons of FPV and VV have shown major rearrangement of blocks of genes (115). Analysis of the complete FPV genomic sequence reveals that FPV contains at least two major genomic rearrangements in the conserved colinear core of genes present in VV, VAR, and MCV (71, 108, 142) (Fig. 4). A 12-kbp FPV genomic region containing ORFs FPV049 to FPV058 (comparable to VV A1L to D5R) is inverted and translocated toward the left end of the genome relative to VV. A 56-kbp FPV genomic region containing ORFs FPV077 to FPV112 (comparable to VV G4L to F9L) is inverted relative to VV. At the junction sites of these major rearrangements there are novel coding regions of 5 to 17.5 kbp (see boxed areas 1 to 3 in Fig. 1 and regions 1 to 3 in Fig. 4). Genes within these junction regions are predominantly homologues of cellular genes and/or are members of gene families. This clustering of cellular homologues and gene families in central genomic locations has not been previously observed in the subfamily Chordopoxvirinae, in which these types of genes are generally found in terminal variable regions of the genome (71, 106, 141). This observation suggests that blocks of genes may have translocated from terminal variable regions of the genome to central regions during large-scale rearrangements of FPV. FPV genome colinearity with genomes of other ChPVs is also interrupted at multiple sites by insertions or deletions of individual genes and multiple copies of B22R gene family members (Fig. 1 and 4 and Table 1).
FIG. 4.
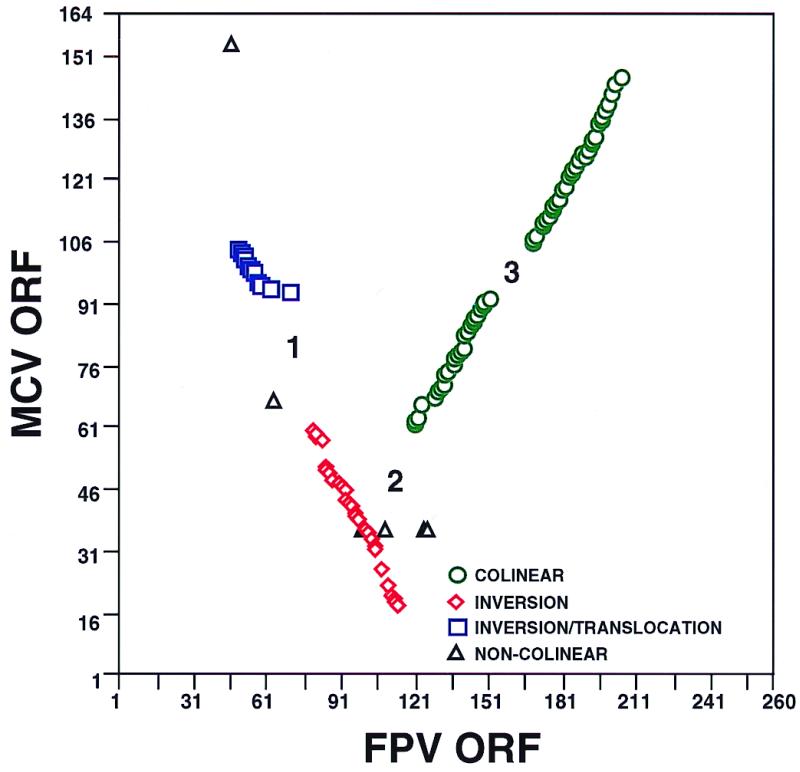
Comparison of gene orders of FPV and MCV homologues. Symbols represent homologous genes. Green circles, colinear genes; red diamonds, inverted genes; blue squares, inverted and translocated genes; black triangles, noncolinear genes. Noncolinear genes include FPV046 (hydroxysteroid dehydrogenase) and FPV064 (glutathione peroxidase), as well as FPV097, FPV098, FPV099, FPV107, FPV122, and FPV123 (VAR B22R homologues). Areas numbered 1 to 3 indicate novel coding regions at junction sites of major genome rearrangement and correspond to similarly numbered boxes in Fig. 1.
The FPV genome (260 to 309 kbp) is larger than other completely sequenced ChPV genomes (178 to 191 kbp). This size difference is due largely to the presence of multiple and, in some cases, large gene families. In other ChPVs, gene families contain fewer members (e.g., genes encoding ankyrin and serpins) or are represented as a single gene (e.g., genes encoding N1R/p28, B22R, CC chemokine, and NKG2-like proteins). Notably, the FPV ankyrin repeat family (31 genes), N1R/p28 family (10 genes), and B22R family (6 genes) comprise 32% of the total genome. In addition, cellular homologues novel to FPV are often found in multiple copies (e.g., β-NGF, α-SNAP, and dCK). It has been suggested that the size of the ankyrin repeat multigene family may affect poxvirus host range (145). The large number of FPV multigene family members, together with the wide avian host range of FPV, provides support for the role of gene families in host range (169).
Conclusions.
FPV genome analysis provides basic knowledge of viral functions, including mRNA biogenesis, DNA replication and repair, nucleotide metabolism, protein processing, manipulation of cellular responses, viral virulence, and host range, which underlie FPV interactions with its avian host and the environment. An improved understanding of these interactions will permit the engineering of novel vaccine viruses and expression vectors with enhanced efficacy and greater versatility. Additionally, the identification and characterization of FPV virulence and host range genes will contribute novel concepts to our overall understanding of pathogen-host interactions, information that is likely to have a broad impact on future strategies for controlling avian infectious diseases in general.
ACKNOWLEDGMENTS
We thank Scott Taylor for providing the NVSL challenge strain of FPV; A. Ciupryk and G. Smoliga for excellent technical assistance; and W. H. Martinez, F. P. Horn, and R. G. Breeze for interest and encouragement.
REFERENCES
- 1.Afonso C L, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahuja S K, Murphy P M. Molecular piracy of mammalian interleukin-8 receptor type B by herpesvirus saimiri. J Biol Chem. 1993;268:20691–20694. [PubMed] [Google Scholar]
- 3.Aizawa Y, Akita K, Taniai M, Torigoe K, Mori T, Nishida Y, Ushio S, Nukada Y, Tanimoto T, Ikegami H, Ikeda M, Kurimoto M. Cloning and expression of interleukin-18 binding protein. FEBS Lett. 1999;445:338–342. doi: 10.1016/s0014-5793(99)00148-9. [DOI] [PubMed] [Google Scholar]
- 4.Alcami A, Symons J A, Collins P D, Williams T J, Smith G L. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J Immunol. 1998;160:624–633. [PubMed] [Google Scholar]
- 5.Aloe L, Simone M D, Properzi F. Nerve growth factor: a neurotrophin with activity on cells of the immune system. Microsc Res Tech. 1999;45:285–291. doi: 10.1002/(SICI)1097-0029(19990515/01)45:4/5<285::AID-JEMT12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Antoine G, Scheiflinger F, Dorner F, Falkner F G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 7.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 8.Attwood T K, Eliopoulos E E, Findlay J B C. Multiple sequence alignment of protein families showing low sequence homology: a methodological approach using database pattern-matching discriminators for G-protein-linked receptors. Gene. 1991;98:153–159. doi: 10.1016/0378-1119(91)90168-b. [DOI] [PubMed] [Google Scholar]
- 9.Back A, Soncini R A, Ruthes O, Madureira S, Jr, Flores R. An atypical fowl pox outbreak in broilers in southern Brazil. Avian Dis. 1995;39:902–906. [PubMed] [Google Scholar]
- 10.Baldick C J, Jr, Keck J G, Moss B. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate-class promoters. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldick C J, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry M, McFadden G. Apoptosis regulators from DNA viruses. Curr Opin Immunol. 1998;10:422–430. doi: 10.1016/s0952-7915(98)80116-7. [DOI] [PubMed] [Google Scholar]
- 13.Beisser P S, Grauls G, Bruggeman C A, Vink C. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J Virol. 1999;73:7218–7230. doi: 10.1128/jvi.73.9.7218-7230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betakova T, Wolffe E J, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J Virol. 1999;73:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binns M, Mason C, Boursnell M. A 39,000 Mr immunodominant protein of fowlpox virus contains multiple copies of a 12 amino acid repeat sequence. J Gen Virol. 1990;71:2883–2888. doi: 10.1099/0022-1317-71-12-2883. [DOI] [PubMed] [Google Scholar]
- 16.Binns M M, Boursnell M E G, Skinner M A. Gene translocations in poxviruses: the fowlpox virus thymidine kinase gene is flanked by 15 bp direct repeats and occupies the locus which in vaccinia virus is occupied by the ribonucleotide reductase large subunit gene. Virus Res. 1992;24:161–172. doi: 10.1016/0168-1702(92)90004-s. [DOI] [PubMed] [Google Scholar]
- 17.Binns M M, Boursnell M E G, Tomley F M, Campbell J. Analysis of the fowlpoxvirus gene encoding the 4b core polypeptide and demonstration that it possesses efficient promoter sequences. Virology. 1989;170:288–291. doi: 10.1016/0042-6822(89)90380-2. [DOI] [PubMed] [Google Scholar]
- 18.Binns M M, Britton B S, Mason C, Boursnell M E. Analysis of the fowlpox virus genome region corresponding to the vaccinia virus D6 to A1 region: location of, and variation in, non-essential genes in poxviruses. J Gen Virol. 1990;71:2873–2881. doi: 10.1099/0022-1317-71-12-2873. [DOI] [PubMed] [Google Scholar]
- 19.Binns M M, Stenzler L, Tomley F M, Campbell J, Boursnell M E. Identification by a random sequencing strategy of the fowlpoxvirus DNA polymerase gene, its nucleotide sequence and comparison with other viral DNA polymerases. Nucleic Acids Res. 1987;15:6563–6573. doi: 10.1093/nar/15.16.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binns M M, Tomley F M, Campbell J, Boursnell M E G. Comparison of a conserved region in fowlpox virus and vaccinia virus genomes and the translocation of the fowlpox virus thymidine kinase gene. J Gen Virol. 1988;69:1275–1283. doi: 10.1099/0022-1317-69-6-1275. [DOI] [PubMed] [Google Scholar]
- 21.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black E P, Moussatche N, Condit R C. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- 23.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaxland J D. Atypical fowl pox in chickens. Vet Rec. 1976;99:222. doi: 10.1136/vr.99.11.222-b. [DOI] [PubMed] [Google Scholar]
- 25.Boulanger D, Green P, Smith T, Czerny C-P, Skinner M A. The 131-amino-acid repeat region of the essential 39-kilodalton core protein of fowlpox virus FP9, equivalent to vaccinia virus A4L protein, is nonessential and highly immunogenic. J Virol. 1998;72:170–179. doi: 10.1128/jvi.72.1.170-179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle D B. Disease and fertility control in wildlife and feral animal populations: options for vaccine delivery using vectors. Reprod Fertil Dev. 1994;6:393–400. doi: 10.1071/rd9940393. [DOI] [PubMed] [Google Scholar]
- 27.Boyle D B. Quantitative assessment of poxvirus promoters in fowlpox and vaccinia virus recombinants. Virus Genes. 1992;6:281–290. doi: 10.1007/BF01702566. [DOI] [PubMed] [Google Scholar]
- 28.Boyle D B, Heine H G. Recombinant fowlpox virus vaccines for poultry. Immunol Cell Biol. 1993;71:391–397. doi: 10.1038/icb.1993.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle D B, Pye A D, Coupar B E H. Comparison of field and vaccine strains of Australian fowlpox viruses. Arch Virol. 1997;142:737–748. doi: 10.1007/s007050050115. [DOI] [PubMed] [Google Scholar]
- 30.Bradshaw R A, Blundell T L, Lapatto R, McDonald N Q, Murray-Rust J. Nerve growth factor revisited. Trends Biochem Sci. 1993;18:48–52. doi: 10.1016/0968-0004(93)90052-o. [DOI] [PubMed] [Google Scholar]
- 31.Brick D J, Burke R D, Schiff L, Upton C. Shope fibroma virus RING finger protein N1R binds DNA and inhibits apoptosis. Virology. 1998;249:42–51. doi: 10.1006/viro.1998.9304. [DOI] [PubMed] [Google Scholar]
- 32.Brunovskis P, Velicer L F. The Marek's disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 33.Buckley M F, Loveland K A, McKinstry W J, Garson O M, Goding J W. Plasma cell membrane glycoprotein PC-1. J Biol Chem. 1990;265:17506–17511. [PubMed] [Google Scholar]
- 34.Bullock E D, Johnson E M., Jr Nerve growth factor induces the expression of certain cytokine genes and bcl-2 in mast cells. Potential role in survival promotion. J Biol Chem. 1996;271:27500–27508. doi: 10.1074/jbc.271.44.27500. [DOI] [PubMed] [Google Scholar]
- 35.Calvert J G, Ogawa R, Yanagida N, Nazerian K. Identification and functional analysis of the fowlpox virus homolog of the vaccinia virus p37K major envelope antigen gene. Virology. 1992;191:783–792. doi: 10.1016/0042-6822(92)90254-m. [DOI] [PubMed] [Google Scholar]
- 36.Campbell J I A, Binns M W, Tomley F M, Boursnell M E G. Tandem repeated sequences within the terminal region of the fowlpox virus genome. J Gen Virol. 1989;70:145–154. doi: 10.1099/0022-1317-70-1-145. [DOI] [PubMed] [Google Scholar]
- 37.Cao J X, Gershon P D, Black D N. Sequence analysis of HindIII Q2 fragment of capripoxvirus reveals a putative gene encoding a G-protein-coupled chemokine receptor homologue. Virology. 1995;209:207–212. doi: 10.1006/viro.1995.1244. [DOI] [PubMed] [Google Scholar]
- 38.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen N-Y, Jiang S-Q, Klein D A, Paulus H. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993;268:9448–9465. [PubMed] [Google Scholar]
- 40.Cheville N F. Cytopathologic changes in fowlpox (turkey origin) inclusion body formation. Am J Pathol. 1966;49:723–737. [PMC free article] [PubMed] [Google Scholar]
- 41.Chung C-S, Hsiao J-C, Chang Y-S, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clary D O, Griff I C, Rothman J E. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- 43.Comeau M R, Johnson R, DuBose R F, Petersen M, Gearing P, VandenBos T, Park L, Farrah T, Buller R M, Cohen J I, Strockbine L D, Rauch C, Spriggs M K. A poxvirus-encoded semaphorin induces cytokine production from monocytes and binds to a novel cellular semaphorin receptor, VESPR. Immunity. 1998;8:473–482. doi: 10.1016/s1074-7613(00)80552-x. [DOI] [PubMed] [Google Scholar]
- 44.Condit R C, Xiang Y, Lewis J I. Mutation of vaccinia virus gene G2R causes suppression of gene A18R ts mutants: implications for control of transcription. Virology. 1996;220:10–19. doi: 10.1006/viro.1996.0280. [DOI] [PubMed] [Google Scholar]
- 45.Coupar B E H, Teo T, Boyle D B. Restriction endonuclease mapping of the fowlpox virus genome. Virology. 1990;179:159–167. doi: 10.1016/0042-6822(90)90285-y. [DOI] [PubMed] [Google Scholar]
- 46.Cunningham C H. Avian pox. In: Hofstad M S, Barnes H J, Calnek B W, Reid W M, Yoder H W Jr, editors. Diseases of poultry. Ames: Iowa State University Press; 1984. pp. 597–609. [Google Scholar]
- 47.Dairaghi D J, Fan R A, McMaster B E, Hanley M R, Schall T J. HHV8-encoded vMIP-I selectively engages chemokine receptor CCR8. J Biol Chem. 1999;274:21569–21574. doi: 10.1074/jbc.274.31.21569. [DOI] [PubMed] [Google Scholar]
- 48.DaMassa A J. The role of Culex tarsalis in the transmission of fowl pox virus. Avian Dis. 1965;10:57–66. [Google Scholar]
- 49.Damon I, Murphy P M, Moss B. Broad spectrum chemokine antagonistic activity of a human poxvirus chemokine homolog. Proc Natl Acad Sci USA. 1998;95:6403–6407. doi: 10.1073/pnas.95.11.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dao T, Ohashi K, Kayano T, Kurimoto M, Okamura H. Interferon-gamma-inducing factor, a novel cytokine, enhances Fas ligand-mediated cytotoxicity of murine T helper 1 cells. Cell Immunol. 1996;173:230–235. doi: 10.1006/cimm.1996.0272. [DOI] [PubMed] [Google Scholar]
- 51.Davison A J, Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 52.Delaire S, Elhabazi A, Bensussan A, Boumsell L. CD100 is a leukocyte semaphorin. Cell Mol Life Sci. 1998;54:1265–1276. doi: 10.1007/s000180050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Nadal E, Clotet J, Posas F, Serrano R, Gomez N, Arino J. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc Natl Acad Sci USA. 1998;95:7357–7362. doi: 10.1073/pnas.95.13.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derrien M, Punjabi A, Khanna M, Grubisha O, Traktman P. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinarello C A. IL-18: a TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J Allergy Clin Immunol. 1999;103:11–24. doi: 10.1016/s0091-6749(99)70518-x. [DOI] [PubMed] [Google Scholar]
- 57.Drillien R, Spehner D, Villeval D, Lecocq J-P. Similar genetic organization between a region of fowlpox virus DNA and the vaccinia virus HindIII J fragment despite divergent location of the thymidine kinase gene. Virology. 1987;160:203–209. doi: 10.1016/0042-6822(87)90061-4. [DOI] [PubMed] [Google Scholar]
- 58.Durum S K, Oppenheim J J. Proinflammatory cytokines and immunity. In: Paul W E, editor. Fundamental immunology. 3rd ed. New York, N.Y: Raven Press; 1993. pp. 801–835. [Google Scholar]
- 59.Eidson C S, Villegas P, Kleven S H. Efficacy of turkey herpesvirus vaccine when administered simultaneously with fowl pox vaccine. Poult Sci. 1975;54:1975–1981. doi: 10.3382/ps.0541975. [DOI] [PubMed] [Google Scholar]
- 60.El-Sherbeini M, Clemas J A. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-β-glucan in vitro. J Bacteriol. 1995;177:3227–3234. doi: 10.1128/jb.177.11.3227-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Endres M J, Garlisi C G, Xiao H, Shan L, Hedrick J A. The Kaposi's sarcoma-related herpesvirus (KSHV)-encoded chemokine vMIP-I is a specific agonist for the CC chemokine receptor (CCR)8. J Exp Med. 1999;189:1993–1998. doi: 10.1084/jem.189.12.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ensser A, Fleckenstein B. Alcelaphine herpesvirus type 1 has a semaphorin-like gene. J Gen Virol. 1995;76:1063–1067. doi: 10.1099/0022-1317-76-4-1063. [DOI] [PubMed] [Google Scholar]
- 63.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 64.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 65.Fatunmbi O O, Reed W M. Evaluation of a commercial quail pox vaccine (Bio-Pox Q) for the control of “variant” fowl poxvirus infections. Avian Dis. 1996;40:792–797. [PubMed] [Google Scholar]
- 66.Fukuda M, Hiraoka N, Yeh J-C. C-type lectins and sialyl Lewis X oligosaccharides: versatile roles in cell-cell interaction. J Cell Biol. 1999;147:467–470. doi: 10.1083/jcb.147.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J-L, Murphy P M. Human cytomegalovirus open reading frame US28 encodes a functional β chemokine receptor. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 68.Garaci E, Caroleo M C, Aloe L, Aquaro S, Piacentini M, Costa N, Amendola A, Micera A, Calio R, Perno C F, Levi-Montalcini R. Nerve growth factor is an autocrine factor essential for the survival of macrophages infected with HIV. Proc Natl Acad Sci USA. 1999;96:14013–14018. doi: 10.1073/pnas.96.24.14013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Arranz M, Maldonado A M, Mazon M J, Portillo F. Transcriptional control of yeast plasma membrane H+-ATPase by glucose. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- 70.Gillard S, Spehner D, Drillien R, Kirn A. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc Natl Acad Sci USA. 1986;83:5573–5577. doi: 10.1073/pnas.83.15.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 72.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:192–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 73.Graham K A, Lalani A S, Macen J L, Ness T L, Barry M, Liu L-Y, Lucas A, Clark-Lewis I, Moyer R W, McFadden G. The T1/35kDa family of poxvirus-secreted proteins bind chemokines and modulate leukocyte influx into virus-infected tissues. Virology. 1997;229:12–24. doi: 10.1006/viro.1996.8423. [DOI] [PubMed] [Google Scholar]
- 74.Halford S, Wilson D I, Daw S C, Roberts C, Wadey R, Kamath S, Wickremasinghe A, Burn J, Goodship J, Mattei M G. Isolation of a gene expressed during early embryogenesis from the region of 22q11 commonly deleted in DiGeorge syndrome. Hum Mol Genet. 1993;10:1577–1582. doi: 10.1093/hmg/2.10.1577. [DOI] [PubMed] [Google Scholar]
- 75.Hearst J E. The structure of photolyase: using photon energy for DNA repair. Science. 1995;268:1858–1859. doi: 10.1126/science.7604259. [DOI] [PubMed] [Google Scholar]
- 76.Hertig C, Coupar B E H, Gould A R, Boyle D B. Field and vaccine strains of fowlpox virus carry integrated sequences from the avian retrovirus, reticuloendotheliosis virus. Virology. 1997;235:367–376. doi: 10.1006/viro.1997.8691. [DOI] [PubMed] [Google Scholar]
- 77.Hitchner S B. Canary pox vaccination with live embryo-attenuated virus. Avian Dis. 1981;25:874–881. [PubMed] [Google Scholar]
- 78.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 79.Hyodo Y, Matsui K, Nayashi N, Tsutsui H, Kashiwamura S I, Yamauchi H, Hiroishi K, Takeda K, Tagawa Y-I, Iwakura Y, Kayagaki N, Kurimoto M, Okamura H, Hada T, Yagita H, Akira S, Nakanishi K, Higashino K. IL-18 up-regulates perforin-mediated NK activity without increasing perforin messenger RNA expression by binding to constitutively expressed IL-18 receptor. J Immunol. 1999;162:1662–1668. [PubMed] [Google Scholar]
- 80.Ink B S, Gilbert C S, Evan G I. Delay of vaccinia virus-induced apoptosis in nonpermissive Chinese hamster ovary cells by the cowpox virus CHOhr and adenovirus E1B 19K genes. J Virol. 1995;69:661–668. doi: 10.1128/jvi.69.2.661-668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joazeiro C A P, Wing S S, Huang H-K, Leverson J D, Hunter T, Liu Y-C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 82.Kato S, Cutting W. A study of the inclusion bodies of rabbit myxoma and fibroma virus and a consideration of the relationship between all pox virus inclusion bodies. Stanford Med Bull. 1959;17:34–45. [PubMed] [Google Scholar]
- 83.Kent S J, Zhao A, Best S J, Chandler J D, Boyle D B, Ramshaw I A. Enhanced T-cell immunogenicity and protective efficacy of a human immunodeficiency virus type 1 vaccine regimen consisting of consecutive priming with DNA and boosting with recombinant fowlpox virus. J Virol. 1998;72:10180–10188. doi: 10.1128/jvi.72.12.10180-10188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolodkin A L, Matthes D J, Goodman C S. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- 85.Koonin E V. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koonin E V, Senkevich T G. Fowlpox virus encodes a protein related to human deoxycytidine kinase: further evidence for independent acquisition of genes for enzymes of nucleotide metabolism by different viruses. Virus Genes. 1993;7:289–295. doi: 10.1007/BF01702589. [DOI] [PubMed] [Google Scholar]
- 87.Kovacs G R, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kozopas K M, Yang T, Buchan H L, Zhou P, Craig R W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krieser R J, Eastman A. The cloning and expression of human deoxyribonuclease II. J Biol Chem. 1998;273:30909–30914. doi: 10.1074/jbc.273.47.30909. [DOI] [PubMed] [Google Scholar]
- 90.Kumar S, Boyle D B. Mapping of a major early/late gene of fowlpox virus. Virus Res. 1990;15:175–185. doi: 10.1016/0168-1702(90)90007-x. [DOI] [PubMed] [Google Scholar]
- 91.Kumar S, Boyle D B. A poxvirus bidirectional promoter element with early/late and late functions. Virology. 1990;179:151–158. doi: 10.1016/0042-6822(90)90284-x. [DOI] [PubMed] [Google Scholar]
- 92.Kupke T, Stevanovic S, Sahl H-G, Gotz F. Purification and characterization of EpiD, a flavoprotein involved in the biosynthesis of the lantibiotic epidermin. J Bacteriol. 1992;174:5354–5361. doi: 10.1128/jb.174.16.5354-5361.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laidlaw S M, Anwar M A, Thomas W, Green P, Shaw K, Skinner M A. Fowlpox virus encodes nonessential homologs of cellular alpha-SNAP, PC-1, and an orphan human homolog of a secreted nematode protein. J Virol. 1998;72:6742–6751. doi: 10.1128/jvi.72.8.6742-6751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lalani A S, Ness T L, Singh R, Harrison J K, Seet B T, Kelvin D J, McFadden G, Moyer R W. Functional comparisons among members of the poxvirus T1/35kDa family of soluble CC-chemokine inhibitor glycoproteins. Virology. 1998;250:173–184. doi: 10.1006/viro.1998.9340. [DOI] [PubMed] [Google Scholar]
- 95.Lange C, Liehr T, Goen M, Gebhart E, Fleckenstein B, Ensser A. New eukaryotic semaphorins with close homology to semaphorins of DNA viruses. Genomics. 1998;51:340–350. doi: 10.1006/geno.1998.5256. [DOI] [PubMed] [Google Scholar]
- 96.Leong K H, Ramsay A J, Boyle D B, Ramshaw I A. Selective induction of immune responses by cytokines coexpressed in recombinant fowlpox virus. J Virol. 1994;68:8125–8130. doi: 10.1128/jvi.68.12.8125-8130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 98.Lewis T, Zsak L, Burrage T G, Lu Z, Kutish G F, Neilan J G, Rock D L. An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine. J Virol. 2000;74:1275–1285. doi: 10.1128/jvi.74.3.1275-1285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao T-H. The subunit structure and active site sequence of porcine spleen deoxyribonuclease. J Biol Chem. 1985;260:10708–10713. [PubMed] [Google Scholar]
- 100.Lin E Y, Orlofsky A, Berger M S, Prystowsky M B. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J Immunol. 1993;151:1979–1988. [PubMed] [Google Scholar]
- 101.Lin J-H, Makris A, McMahon C, Bear S E, Patriotis C, Prasad V R, Brent R, Golemis E A, Tsichlis P N. The ankyrin repeat-containing adaptor protein Tvl-1 is a novel substrate and regulator of Raf-1. J Biol Chem. 1999;274:14706–14715. doi: 10.1074/jbc.274.21.14706. [DOI] [PubMed] [Google Scholar]
- 102.Lux S E, John K M, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;34:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- 103.Maa J-S, Rodriguez J F, Esteban M. Structural and functional characterization of a cell surface binding protein of vaccinia virus. J Biol Chem. 1990;265:1569–1577. [PubMed] [Google Scholar]
- 104.Marshall J S, Gomi K, Blennerhassett M G, Bienenstock J. Nerve growth factor modifies the expression of inflammatory cytokines by mast cells via a prostanoid-dependent mechanism. J Immunol. 1999;162:4271–4276. [PubMed] [Google Scholar]
- 105.Martin K H, Grosenbach D W, Franke C A, Hruby D E. Identification and analysis of three myristylated vaccinia virus late proteins. J Virol. 1997;71:5218–5226. doi: 10.1128/jvi.71.7.5218-5226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Massung R F, Esposito J J, Liu L I, Qi J, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, Selivanov N A, Cavallaro K F, Kerlavage A R, Mahy B W J, Venter J C. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature. 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 107.Massung R F, Jayarama V, Moyer R W. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 108.Massung R F, Liu L-I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 109.Massung R F, McFadden G, Moyer R W. Nucleotide sequence analysis of a unique near-terminal region of the tumorigenic poxvirus, Shope fibroma virus. J Gen Virol. 1992;73:2903–2911. doi: 10.1099/0022-1317-73-11-2903. [DOI] [PubMed] [Google Scholar]
- 110.McFadden G, Graham K, Barry M. New strategies of immune modulation by DNA viruses. Transplant Proc. 1996;28:2085–2088. [PubMed] [Google Scholar]
- 111.McIntosh A A, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McMillen J K, Cochran M D, Junker D E, Reddy D N, Valencia D M. The safe and effective use of fowlpox virus as a vector for poultry vaccines. Dev Biol Stand. 1994;82:137–145. [PubMed] [Google Scholar]
- 113.Melchers F, Rolink A G, Schaniel C. The role of chemokines in regulating cell migration during humoral immune responses. Cell. 1999;99:351–354. doi: 10.1016/s0092-8674(00)81521-4. [DOI] [PubMed] [Google Scholar]
- 114.Micallef M J, Tanimoto T, Torigoe K, Nishida Y, Kohno K, Ikegami H, Kurimoto M. Simultaneous exposure to interleukin-18 and interleukin-10 in vitro synergistically augments murine spleen natural killer cell activity. Cancer Immunol Immunother. 1999;48:109–117. doi: 10.1007/s002620050554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mockett B, Binns M M, Boursnell M E G, Skinner M A. Comparison of the locations of homologous fowlpox and vaccinia virus genes reveals major genome reorganization. J Gen Virol. 1992;73:2661–2668. doi: 10.1099/0022-1317-73-10-2661. [DOI] [PubMed] [Google Scholar]
- 116.Moore J B, Smith G L. Steroid hormone synthesis by a vaccinia enzyme: a new type of virus virulence factor. EMBO J. 1992;11:1973–1980. doi: 10.1002/j.1460-2075.1992.tb05251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morgan A, Burgoyne R D. A role for soluble NSF attachment proteins (SNAPs) in regulated exocytosis in adrenal chromaffin cells. EMBO J. 1995;14:232–239. doi: 10.1002/j.1460-2075.1995.tb06996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 119.Mossman K, Lee S F, Barry M, Boshkov L, McFadden G. Disruption of M-T5, a novel myxoma virus gene member of poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J Virol. 1996;70:4394–4410. doi: 10.1128/jvi.70.7.4394-4410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muller H K, Wittek R, Schaffner W, Schumperli D, Menna A, Wyler R. Comparison of five poxvirus genomes by analysis with restriction endonucleases HindIII, BamI and EcoRI. J Gen Virol. 1978;38:135–147. doi: 10.1099/0022-1317-38-1-135. [DOI] [PubMed] [Google Scholar]
- 121.Nazerian K, Witter R L, Lee L F, Yanagida N. Protection and synergism by recombinant fowl pox vaccines expressing genes from Marek's disease virus. Avian Dis. 1996;40:368–376. [PubMed] [Google Scholar]
- 122.Neilan J G, Borca M V, Lu Z, Kutish G F, Kleiboeker S B, Carrillo C, Zsak L, Rock D L. An African swine fever virus ORF with similarity to C-type lectins is non-essential for growth in swine macrophages in vitro and for virus virulence in domestic swine. J Gen Virol. 1999;80:2693–2697. doi: 10.1099/0022-1317-80-10-2693. [DOI] [PubMed] [Google Scholar]
- 123.Ogawa R, Calvert J G, Yanagida N, Nazerian K. Insertional inactivation of a fowlpox virus homologue of the vaccinia virus F12L gene inhibits the release of enveloped virions. J Gen Virol. 1993;74:55–64. doi: 10.1099/0022-1317-74-1-55. [DOI] [PubMed] [Google Scholar]
- 124.Omar A R, Schat K A, Lee L F, Hunt H D. Cytotoxic T lymphocyte response in chickens immunized with a recombinant fowlpox virus expressing Marek's disease herpesvirus glycoprotein B. Vet Immunol Immunopathol. 1998;62:73–82. doi: 10.1016/s0165-2427(97)00159-1. [DOI] [PubMed] [Google Scholar]
- 125.Paoletti E. Applications of pox virus vectors to vaccination: an update. Proc Natl Acad Sci USA. 1996;93:11349–11353. doi: 10.1073/pnas.93.21.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Perkus M E, Goebel S J, Davis S W, Johnson G P, Limbach K, Norton E K, Paoletti E. Vaccinia virus host range genes. Virology. 1990;179:276–286. doi: 10.1016/0042-6822(90)90296-4. [DOI] [PubMed] [Google Scholar]
- 127.Pollitt E, Skinner M A, Heaphy S. Nucleotide sequence of the 4.3kbp BamHI-N fragment of fowlpox virus FP9. Virus Genes. 1998;17:5–9. doi: 10.1023/a:1008045914991. [DOI] [PubMed] [Google Scholar]
- 128.Purcell D A, Clarke J K, McFerran J B, Hughes D A. The morphogenesis of pigeonpox virus. J Gen Virol. 1972;15:79–83. doi: 10.1099/0022-1317-15-1-79. [DOI] [PubMed] [Google Scholar]
- 129.Revardel E, Bonneau M, Durrens P, Aigle M. Characterization of a new gene family developing pleiotropic phenotypes upon mutation in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1263:261–265. doi: 10.1016/0167-4781(95)00124-y. [DOI] [PubMed] [Google Scholar]
- 130.Rodriguez J F, Smith G L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ryan J C, Seaman W E. Divergent functions of lectin-like receptors on NK cells. Immunol Rev. 1997;155:79–89. doi: 10.1111/j.1600-065x.1997.tb00941.x. [DOI] [PubMed] [Google Scholar]
- 133.Sadasiv E C, Chang P W, Gulka G. Morphogenesis of canary poxvirus and its entrance into inclusion bodies. Am J Vet Res. 1985;46:529–535. [PubMed] [Google Scholar]
- 134.Salzberg S L, Delcher A L, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sarma D K, Sharma S N. Comparative immunity of fowlpox virus vaccines. J Vet Med Biol. 1988;35:19–23. doi: 10.1111/j.1439-0450.1988.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 137.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 138.Scholzen T, Armstrong C A, Bunnett N W, Luger T A, Olerud J E, Ansel J C. Neuropeptides in the skin: interactions between the neuroendocrine and skin immune systems. Exp Dermatol. 1998;7:81–96. doi: 10.1111/j.1600-0625.1998.tb00307.x. [DOI] [PubMed] [Google Scholar]
- 139.Schoneberg T, Schultz G, Gudermann T. Structural basis of G protein-coupled receptor function. Mol Cell Endocrinol. 1999;151:181–193. doi: 10.1016/s0303-7207(99)00017-9. [DOI] [PubMed] [Google Scholar]
- 140.Sedgwick S G, Smerdon S J. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 141.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 142.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of Molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 143.Senkevich T G, Koonin E V, Buller R M L. A poxvirus protein with a RING zinc finger motif is of crucial importance for virulence. Virology. 1994;198:118–128. doi: 10.1006/viro.1994.1014. [DOI] [PubMed] [Google Scholar]
- 144.Senkevich T G, Wolffe E J, Buller R M L. Ectromelia virus RING finger protein is localized in virus factories and is required for virus replication in macrophages. J Virol. 1995;69:4103–4111. doi: 10.1128/jvi.69.7.4103-4111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shchelkunov S N, Safronov P F, Totmenin A V, Petrov N A, Ryazankina O I, Gutorov V V, Kotwal G J. The genome sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology. 1998;243:432–460. doi: 10.1006/viro.1998.9039. [DOI] [PubMed] [Google Scholar]
- 146.Sheppard M, Werner W, Tsatas E, McCoy R, Prowse S, Johnson M. Fowl adenovirus recombinant expressing VP2 of infectious bursal disease virus induces protective immunity against bursal disease. Arch Virol. 1998;143:915–930. doi: 10.1007/s007050050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 148.Shisler J L, Senkevich T G, Berry M L, Moss B. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science. 1998;279:102–105. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 149.Shors T, Keck J G, Moss B. Down regulation of gene expression by the vaccinia virus D10 protein. J Virol. 1999;73:791–796. doi: 10.1128/jvi.73.1.791-796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Skinner M A, Moore J B, Binns M M, Smith G L, Boursnell M E G. Deletion of fowlpox virus homologues of vaccinia virus genes between the 3 β-hydroxysteroid dehydrogenase (A44L) and DNA ligase (A50R) genes. J Gen Virol. 1994;75:2495–2498. doi: 10.1099/0022-1317-75-9-2495. [DOI] [PubMed] [Google Scholar]
- 151.Smith C A, Davis Smith T, Smolak P J, Friend D, Hagen H, Gerhart M, Park L, Pickup D J, Torrance D, Mohler K, Schooley K, Goodwin R G. Poxvirus genomes encode a secreted, soluble protein that preferentially inhibits β chemokine activity yet lacks sequence homology to known chemokine receptors. Virology. 1997;236:316–327. doi: 10.1006/viro.1997.8730. [DOI] [PubMed] [Google Scholar]
- 152.Smith G L, Chan Y S. Two vaccinia virus proteins structurally related to the interleukin-1 receptor and the immunoglobulin superfamily. J Gen Virol. 1991;72:511–518. doi: 10.1099/0022-1317-72-3-511. [DOI] [PubMed] [Google Scholar]
- 153.Spehner D, Gillard S, Drillien R, Kirn A. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J Virol. 1988;62:1297–1304. doi: 10.1128/jvi.62.4.1297-1304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Spriggs M K. Shared resources between the neural and immune systems: semaphorins join the ranks. Curr Opin Immunol. 1999;11:387–391. doi: 10.1016/S0952-7915(99)80065-X. [DOI] [PubMed] [Google Scholar]
- 155.Staden R, McLachlan A D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stadtman T C. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 158.Stenbeck G. Molecules in focus soluble NSF-attachment proteins. Int J Biochem Cell Biol. 1998;30:573–577. doi: 10.1016/s1357-2725(97)00064-2. [DOI] [PubMed] [Google Scholar]
- 159.Sutter G, Ramsey-Ewing A, Rosales R, Moss B. Stable expression of the vaccinia virus K1L gene in rabbit cells complements the host range defect of a vaccinia virus mutant. J Virol. 1994;68:4109–4116. doi: 10.1128/jvi.68.7.4109-4116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sutton G G, White O, Adams M D, Kerlavage A R. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Tech. 1995;1:9–19. [Google Scholar]
- 161.Takeda K, Tsutsui H, Yoshimoto T, Adachi O, Yoshida N, Kishimoto T, Okamura H, Nakanishi K, Akira S. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 162.Tanaka-Kataoka M, Kunikata T, Takayama S, Iwaki K, Ohashi K, Ikeda M, Kurimoto M. In vivo antiviral effect of interleukin 18 in a mouse model of vaccinia virus infection. Cytokine. 1999;11:593–599. doi: 10.1006/cyto.1998.0453. [DOI] [PubMed] [Google Scholar]
- 163.Tartaglia J, Winslow J, Goebel S, Johnson G P, Taylor J, Paoletti E. Nucleotide sequence analysis of a 10.5 kbp HindIII fragment of fowlpox virus: relatedness to the central portion of the vaccinia virus HindIII D region. J Gen Virol. 1990;71:1517–1524. doi: 10.1099/0022-1317-71-7-1517. [DOI] [PubMed] [Google Scholar]
- 164.Taylor J, Paoletti E. Fowlpox virus as a vector in non-avian species. Vaccine. 1988;6:466–468. doi: 10.1016/0264-410x(88)90091-6. [DOI] [PubMed] [Google Scholar]
- 165.Taylor J, Weinberg R, Tartaglia J, Richardson C, Alkhatib G, Briedis D, Appel M, Norton E, Paoletti E. Nonreplicating viral vectors as potential vaccines: recombinant canarypox virus expressing measles virus fusion (F) and hemagglutinin (HA) glycoproteins. Virology. 1992;187:321–328. doi: 10.1016/0042-6822(92)90321-f. [DOI] [PubMed] [Google Scholar]
- 166.Tomley F, Binns M, Campbell J, Boursnell M. Sequence analysis of an 11.2 kilobase, near-terminal, BamHI fragment of fowlpox virus. J Gen Virol. 1988;69:1025–1040. doi: 10.1099/0022-1317-69-5-1025. [DOI] [PubMed] [Google Scholar]
- 167.Torcia M, Bracci-Laudiero L, Lucibello M, Nencioni L, Labardi D, Rubartelli A, Cozzolino F, Aloe L, Garaci E. Nerve growth factor is an autocrine survival factor for memory B lymphocytes. Cell. 1996;85:345–356. doi: 10.1016/s0092-8674(00)81113-7. [DOI] [PubMed] [Google Scholar]
- 168.Torriglia A, Perani P, Brossas J Y, Chaudun E, Treton J, Courtois Y, Counis M F. L-DNase II, a molecule that links proteases and endonucleases in apoptosis, derives from the ubiquitous serpin leukocyte elastase inhibitor. Mol Cell Biol. 1998;18:3612–3619. doi: 10.1128/mcb.18.6.3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tripathy D N. Pox. In: Calnek B W, Barnes H J, Beard C W, Reid W M, Yoder H W Jr, editors. Diseases of poultry. 9th ed. Ames: Iowa State University Press; 1991. pp. 583–596. [Google Scholar]
- 170.Tripathy D N, Cunningham C H. Avian pox. In: Hofstad M S, Barnes H J, Reid W M, Yoder H W J, editors. Diseases of poultry. 8th ed. Ames: Iowa State University Press; 1984. pp. 524–534. [Google Scholar]
- 171.Tripathy D N, Hanson L E. Pathogenesis of fowlpox in laying hens. Avian Dis. 1978;22:259–265. [PubMed] [Google Scholar]
- 172.Turner P C, Moyer R W. Control of apoptosis by poxviruses. Semin Virol. 1998;8:453–469. [Google Scholar]
- 173.VanSlyke J K, Whitehead S S, Wilson E M, Hruby D E. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology. 1991;183:467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- 174.Wang C-C, Lu S-C, Chen H-L, Liao T-H. Porcine spleen deoxyribonuclease II. J Biol Chem. 1998;273:17192–17198. doi: 10.1074/jbc.273.27.17192. [DOI] [PubMed] [Google Scholar]
- 175.Weis W I, Taylor M E, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 176.Welker P, Grabbe J, Grutzkau A, Henz B M. Effects of nerve growth factor (NGF) and other fibroblast-derived growth factors on immature human mast cells (HMC-1) Immunology. 1998;94:310–317. [PMC free article] [PubMed] [Google Scholar]
- 177.Weskamp G, Otten U. An enzyme-linked immunoassay for nerve growth factor (NGF): a tool for studying regulatory mechanisms involved in NGF production in brain and in peripheral tissues. J Neurochem. 1987;48:1779–1786. doi: 10.1111/j.1471-4159.1987.tb05736.x. [DOI] [PubMed] [Google Scholar]
- 178.Whitehead S S, Hruby D E. A transcriptionally controlled trans-processing assay: putative identification of a vaccinia virus-encoded proteinase which cleaves precursor protein P25K. J Virol. 1994;68:7603–7608. doi: 10.1128/jvi.68.11.7603-7608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wilcock D, Duncan S A, Traktman P, Zhang W-H, Smith G L. The vaccinia virus A40R gene product is a nonstructural, type II membrane glycoprotein that is expressed at the cell surface. J Gen Virol. 1999;80:2137–2148. doi: 10.1099/0022-1317-80-8-2137. [DOI] [PubMed] [Google Scholar]
- 180.Williams A F, Barclay A N. The immunoglobulin superfamily—domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 181.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Woodward H, Tudor D C. The immunizing effect of commercial pigeon pox vaccines on pigeons. Poult Sci. 1973;52:1463–1468. doi: 10.3382/ps.0521463. [DOI] [PubMed] [Google Scholar]
- 183.Woolf C J, Ma Q P, Allchorne A, Poole S. Peripheral cell types contributing to the hyperalgesic action of nerve growth factor in inflammation. J Neurosci. 1996;16:2716–2723. doi: 10.1523/JNEUROSCI.16-08-02716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xiang Y, Moss B. Identification of human and mouse homologs of the MC51L-53L-54L family of secreted glycoproteins encoded by the Molluscum contagiosum poxvirus. Virology. 1999;257:297–302. doi: 10.1006/viro.1999.9676. [DOI] [PubMed] [Google Scholar]
- 185.Xiang Y, Moss B. IL-18 binding and inhibition of interferon γ induction by human poxvirus encoded proteins. Proc Natl Acad Sci USA. 1999;96:11537–11542. doi: 10.1073/pnas.96.20.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xiang Y, Simpson D A, Spiegel J, Zhou A, Silverman R H, Condit R C. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J Virol. 1998;72:7012–7023. doi: 10.1128/jvi.72.9.7012-7023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yanagida N, Ogawa R, Li Y, Lee L F, Nazerian K. Recombinant fowlpox viruses expressing the glycoprotein B homolog and the pp38 gene of Marek's disease virus. J Virol. 1992;66:1402–1408. doi: 10.1128/jvi.66.3.1402-1408.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yasui A, Eker A P M, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994;13:6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zajac P, Spehner D, Drillien R. The vaccinia virus J5L open reading frame encodes a polypeptide expressed late during infection and required for viral multiplication. Virus Res. 1995;37:163–173. doi: 10.1016/0168-1702(95)00025-l. [DOI] [PubMed] [Google Scholar]
- 191.Zantinge J L, Krell P J, Derbyshire J B, Nagy E. Partial transcriptional mapping of the fowlpox virus genome and analysis of the EcoRI L fragment. J Gen Virol. 1996;77:603–614. doi: 10.1099/0022-1317-77-4-603. [DOI] [PubMed] [Google Scholar]