Abstract
We have previously shown that hepatitis B virus (HBV) replication is abolished in the liver of HBV transgenic mice by stimuli that induce alpha/beta interferon (IFN-α/β) in the liver. The present study was done to identify the step(s) in HBV replication that is affected by this cytokine in transgenic mice treated with the IFN-α/β inducer polyinosinic-polycytidylic acid [poly(I-C)]. Here we show that the pool of cytoplasmic HBV pregenomic RNA (pgRNA)-containing capsids is reduced 10-fold within 9 h after poly(I-C) administration, while there is no change in the abundance of HBV mRNA or in the translational status of cytoplasmic HBV transcripts. In addition, we show that the pool of HBV DNA-containing capsids is not reduced to the same degree until at least 15 h posttreatment, and we show that virus export is not accelerated and the half-life of virions in the serum is unchanged. These results indicate that IFN-α/β triggers intracellular events that either inhibit the assembly of pgRNA-containing capsids or accelerate their degradation, and that maturation and secretion of virus is responsible for clearance of HBV capsids and their cargo of replicative intermediates from the cytoplasm of the hepatocyte.
Hepatitis B virus (HBV) is a hepatotropic, noncytopathic DNA virus that causes acute and chronic necroinflammatory liver disease and hepatocellular carcinoma (7). We have previously shown that the intrahepatic induction of inflammatory cytokines inhibits HBV replication in transgenic mice (13) and that similar noncytopathic antiviral events occur in the liver of chimpanzees acutely infected with HBV (15). Recently, we showed that a single injection of the strong alpha/beta interferon (IFN-α/β) inducer polyinosinic-polycytidylic acid [poly(I-C)] (8, 28) clears HBV replicative intermediates from the hepatocyte cytoplasm of transgenic mice by an IFN-α/β-dependent pathway (22). Furthermore, we showed that infection of transgenic mice with unrelated hepatotropic viruses such as lymphocytic choriomeningitis virus, murine cytomegalovirus, and recombinant adenovirus also inhibits HBV replication via IFN-α/β-dependent mechanisms (3, 12, 22). These mechanisms must affect posttranscriptional steps in the viral life cycle, since the HBV capsids and their cargo of replicative intermediates rapidly disappear from the liver while the steady-state content of HBV RNA remains unchanged (3, 12, 22).
Several posttranscriptional steps (reviewed in references 11 and 25) could be affected by IFN-α/β. First, IFN-α/β may inhibit the translation of HBV transcripts into viral proteins, some of which, like the HBV core protein and the viral reverse transcriptase/DNA polymerase (RT/Pol), are essential for viral replication (33). Second, IFN-α/β may inhibit the encapsidation of viral pregenomic RNA (pgRNA) and RT/Pol into viral nucleocapsid particles. Third, IFN-α/β may inhibit either reverse transcription of encapsidated pgRNA into single-stranded DNA (ssDNA) or maturation of ssDNA into double-stranded DNA (dsDNA). Fourth, IFN-α/β may induce active degradation of pgRNA- and/or DNA-containing capsids. Finally, IFN-α/β may accelerate export of capsids out of the hepatocyte.
To determine which of these steps might be affected by IFN-α/β, we treated transgenic mice with the IFN-α/β inducer poly(I-C). In this report we demonstrate that IFN-α/β inhibits HBV replication at the level of pgRNA-containing capsids, either by preventing their assembly or by accelerating their degradation.
MATERIALS AND METHODS
HBV transgenic mice.
The HBV transgenic mice used in this study, lineages 1.3.32 and 1.3.46 (official designation, Tg[HBV 1.3 genome] Chi32 and Tg[HBV 1.3 genome] Chi46, respectively), have been previously described (14). These mice replicate HBV in the hepatocyte from an integrated greater-than-genome-length HBV transcriptional template. The level of HBV replication in the livers of these mice is comparable with that seen in the infected livers of patients with chronic hepatitis, and there is no evidence of cytopathology (14). Experiments were performed with mice that were matched for age (6 to 11 weeks), sex (male), and level of hepatitis B e antigen (HBeAg) in the serum (measured with a commercially available kit from Abbott Laboratories, Abbott Park, Ill.). HBV replication in both mouse lineages is equally sensitive to poly(I-C) treatment. The 1.3.46 lineage mice display a consistently higher level of HBV virions in the serum (14) and hence were used in this study for the experiments involving serum HBV DNA.
Poly(I-C) treatment.
Poly(I-C) was purchased from Sigma Chemical Co., St. Louis, Mo. (product no. P-0913). In all experiments, mice were injected once intravenously (i.v.) with 200 μg of poly(I-C) in a total volume of 200 μl of 0.9% NaCl solution (saline). Control animals were injected with saline only.
HBV DNA analysis.
Frozen liver tissue was processed as described previously (14), and 30 μg of total DNA was analyzed by Southern blotting after HindIII digestion (14). Encapsidated HBV DNA was extracted from 250 mg of frozen liver that was homogenized in 1 ml of homogenization buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris base [pH 8.0], 0.5% NP-40) by five to eight strokes in a motor-driven Potter-Elvehjem tissue grinder. The crude homogenate was cleared by centrifugation for 15 min at 12,000 rpm and 4°C in a SW60 rotor. The postmitochondrial supernatant was adjusted to 6 mM CaCl2, and 40 U of micrococcal nuclease (Amersham Pharmacia Biotech AB, Uppsala, Sweden) was used to digest nonencapsidated nucleic acids for 30 min at 37°C. The enzyme was inactivated by adding 20 mM EDTA, and a first 300-μl aliquot was removed for isolation of encapsidated RNA (see below). Encapsidated HBV DNA replicative intermediates were isolated from a second 300-μl aliquot of extract brought to 1% sodium dodecyl sulfate in 500 μl (total volume) and digested overnight at 37°C with 1 mg of proteinase K (Roche Molecular Biochemicals, Basel, Switzerland) per ml. Nucleic acids were extracted with equal volumes of phenol-chloroform and chloroform. Residual RNA was degraded by digestion with RNase A (0.01 mg/ml) for 30 min at 37°C. Nucleic acids were precipitated in the presence of 0.3 M sodium acetate with 1 volume of isopropanol. Nucleic acids were dissolved in 150 μl of Tris-EDTA (TE), and 10 μl was electrophoresed in a 1.3% agarose gel for Southern blot analysis as described elsewhere (14). All quantifications were done with a Cyclone storage phosphor system (Packard Instrument Company, Meriden, Conn.).
Serum HBV DNA analysis.
Serum HBV DNA was quantified by Southern blot analysis. Aliquots (50 μl) of serum were digested with proteinase K (1 mg/ml) in a total volume of 500 μl containing 50 mM Tris base (pH 8.0) and 1% sodium dodecyl sulfate at 37°C overnight. Nucleic acids were extracted as described for encapsidated DNA except for the addition of 10 μg of Escherichia coli tRNA during precipitation. Nucleic acids were dissolved in 30 μl of TE; 10 μl was loaded onto a 1.3% agarose gel (1× Tris-acetate-EDTA) and electrophoresed for 15 to 30 min at 5 V/cm. The gel was then blotted in 1.5 M NaCl–0.5 M NaOH by vacuum blotting (VacuGene; Amersham Pharmacia Biotech AB) for 1 h onto a nylon membrane (Magnagraph, Osmonics Laboratory Products, Minnetonka, Minn.). Subsequent Southern blotting was performed as described elsewhere (14).
HBV RNA analysis.
Total liver RNA was isolated from frozen liver tissue, and 20-μg aliquots were analyzed by Northern blotting as described previously (14). Encapsidated RNA was extracted from the first 300-μl aliquot of extract (see “Serum HBV DNA analysis” above) by addition of 200 μl of prewarmed (65°C) 1.67× guanidinium thiocyanate (GTC) solution (7 M GTC, 0.042 M sodium citrate [pH 7.3], 0.84% sarcosyl). Subsequent extraction steps were performed as described elsewhere (14), using 1/10 of the volumes. Before precipitation, 0.1 volume of 10× DNase buffer (500 mM Tris-HCl [pH 7.6], 100 mM MgCl2) was added, and DNA was removed by digestion with 1 U of RNase-free DNase I (Promega, Madison, Wis.) at 37°C for 20 min. After phenol-chloroform extraction, 0.1 volume of 3 M sodium acetate (pH 7.0) was added, and the RNA was precipitated with 1 volume of isopropanol. The RNA was dissolved in 150 μl of H2O, and 70 μl was used for Northern blot analysis as described elsewhere (14).
Polyribosome analysis.
Polyribosomal extracts were prepared essentially as described previously (1). One gram of fresh liver tissue was homogenized in 4 ml of buffer A (250 mM KCl, 10 mM MgCl2, 20 mM HEPES [pH 7.5], 10% sucrose, 2 mM dithiothreitol, 150 μg of cycloheximide per ml, 200 U of RNasin per ml), using a motor driven Potter-Elvehjem tissue grinder. A postmitochondrial supernatant was prepared by centrifugation of the lysate for 15 min at 4°C and 12,000 rpm. The supernatant (1.2 ml) was loaded on a 50 to 10% sucrose gradient prepared in RNasin-free buffer A to which heparin-sodium salt (0.5 mg/ml) (Sigma product no. H-3149) was added. Gradient centrifugation was carried out for 4 h at 4°C and 27,000 rpm. Fractions of ∼500 μl were collected by dripping from the punched bottom of the centrifugation tube. Individual fractions were brought to 2 ml at a final concentration of 1.5 M CsCl–10 mM Tris base (pH 7.0)–25 mM EDTA and were layered onto a 3 M CsCl cushion. Free RNA was pelleted by centrifugation for 12 h at 4°C and 35,000 rpm, while HBV capsids remained in the supernatant. The pellets were dissolved in GTC, and RNA was extracted as described previously (14). The RNA of each fraction was dissolved in 60 μl of H2O, and 10 μl was used for Northern blot analysis as described elsewhere (14).
BrdU labeling in vivo.
5′-Bromo-2′-deoxyuridine (BrdU; Sigma product no. B-9285) was dissolved in H2O to yield a 100 mM stock solution. For in vivo labeling, an osmotic pump (model 1003D; Alza Scientific Products, Palo Alto, Calif.) was filled with the 100 mM BrdU solution and surgically implanted into the peritoneal cavity. Osmotic pumps hold 100 μl and, according to the manufacturer's instructions, deliver 1 μl/h for 3 days. To rapidly achieve high serum BrdU concentrations, 3 mg of BrdU in saline was injected intraperitoneally at the time of surgery (9). For detection of BrdU-labeled HBV DNA, DNA from 100 μl of serum was extracted as described above and dissolved in 30 μl of TE. Then 10 μl of DNA was supplemented with 2.5 μl (2.5 μg/μl) of sheared salmon sperm DNA, denatured with 1.3 μl of 1 N NaOH for 1 min, and neutralized with 2.6 μl of 1 M Tris base (pH 7). The DNA samples were then incubated for 1 h at room temperature with 5 μl of anti-BrdU antibody (0.1 μg/μl; Roche Molecular Biochemicals), 6 μl of 5× Tris-buffered saline, and 1.6 μl of H2O. To increase the gel retardation of the BrdU-labeled DNA maximally, the molecular weight of the DNA-antibody complexes was increased by incubation with 1 μl of a secondary anti-mouse antibody (1 μg/μl; product no. 04-6100, Zymed, South San Francisco, Calif.) for another hour. Five microliters of loading buffer (50% glycerol, 1× Tris-borate-EDTA, 0.025% each xylene cyanole, and bromophenol blue) was added, and the samples were loaded onto a 0.8% agarose gel and run at 1.3 V/cm overnight in 1× Tris-borate-EDTA. The nucleic acids were then blotted as described for serum HBV DNA analysis and subjected to Southern blot hybridization as described elsewhere (14).
RESULTS
Poly(I-C) inhibits HBV replication but not HBV gene expression in livers of transgenic mice.
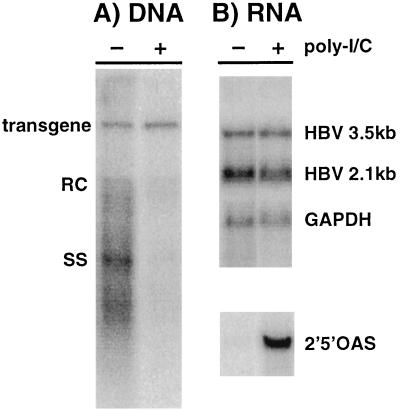
We recently showed that a single i.v. injection of 200 μg of poly(I-C) noncytopathically eliminates HBV DNA replicative intermediates and nucleocapsid particles from the hepatocyte cytoplasm by an IFN-α/β-dependent mechanism (22). As shown in Fig. 1, the inhibitory effect of poly(I-C) on HBV replication was associated with the induction of 2′,5′-oligoadenylate synthetase (2′5′OAS) RNA (a known marker of IFN-α/β induction), and it was not associated with a reduction in the steady-state level of HBV RNA in the liver. This indicates that transcription and RNA turnover are not affected under these conditions.
FIG. 1.
Poly(I-C) inhibits HBV replication but not gene expression in livers of transgenic mice. Two groups (three mice per group) of age (8 to 10 weeks)-, sex (male)-, and serum HBeAg-matched mice from lineage 1.3.32 were injected either with saline (−) or with a single i.v. dose (200 μg) of poly(I-C) (+). Twenty hours later, the mice were sacrificed; following extraction, total hepatic RNA and DNA were analyzed for HBV gene expression and replication by Northern and Southern blot analysis; a representative sample is shown. (A) Southern blot analysis was performed with 30 μg of total hepatic DNA. All DNA samples were RNase treated before gel electrophoresis. Bands corresponding to the integrated transgene, relaxed circular (RC), and single-stranded (SS) HBV DNA replicative forms are indicated. The integrated transgene can be used to normalize the amount of DNA bound to the membrane. The filter was hybridized with a 32P-labeled HBV-specific probe. (B) Northern blot analysis was performed with 20 μg of total hepatic RNA. The membrane was hybridized with 32P-labeled HBV-, 2′5′OAS-, and GAPDH-specific DNA probes. Bands corresponding to the 3.5- and 2.1-kb viral mRNAs are indicated.
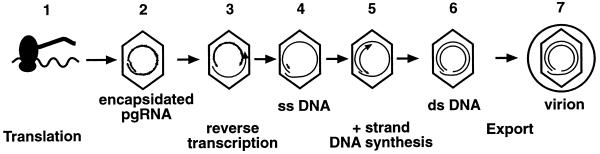
Based on these results, we designed experiments to determine which posttranscriptional step(s) in the viral life cycle is interrupted by poly(I-C). As shown in Fig. 2, the HBV transcripts are first translated into the viral gene products (step 1). Viral pgRNA is encapsidated along with the viral RT/Pol (step 2). Pregenomic RNA is reverse transcribed by the RT/Pol and digested by the RNase H activity of the enzyme, leaving a short RNA segment at the 5′ end of the newly formed ssDNA (steps 3 and 4). Further capsid maturation involves transfer of the primer to the 3′ end of the ssDNA followed by plus-strand DNA synthesis (step 5), yielding mature capsids containing dsDNA (step 6). Mature capsids are targeted for envelopment and are subsequently exported out of the cell as virions (step 7).
FIG. 2.
Schematic representation of the posttranscriptional, cytoplasmic steps in the viral life cycle in livers of transgenic mice. Step 1, the HBV transcripts are translated into the viral gene products; step 2, viral pgRNA is encapsidated along with the viral RT/Pol; step 3, pgRNA is reverse transcribed by the RT/Pol and digested, leaving a short 5′ fragment and ssDNA of minus-strand polarity (step 4). Further capsid maturation involves transfer of the RNA fragment to the 3′ end of the ssDNA, where it serves as the primer for subsequent plus-strand DNA synthesis (step 5), which produces mature capsids containing dsDNA (step 6). Mature capsids are targeted for envelopment and are subsequently exported out of the cell as virions (step 7).
Poly(I-C) does not alter the translational status of HBV transcripts.
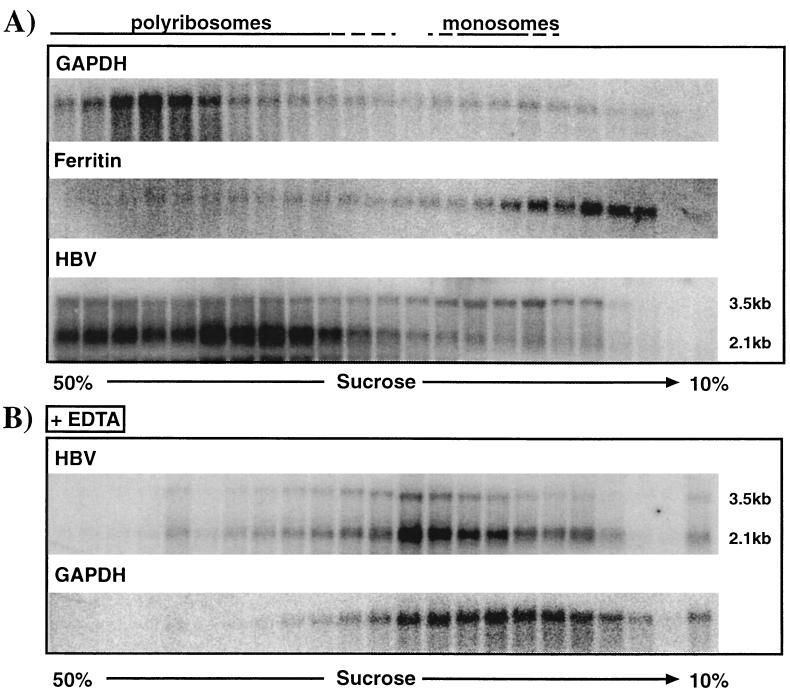
To determine whether poly(I-C) treatment inhibits HBV RNA translation, we compared the polyribosomal distribution of HBV transcripts in the livers of transgenic mice that were treated either with saline or with poly(I-C). First, we had to confirm that polyribosome analysis of mouse liver mRNA appropriately tests the translational status of the HBV transcripts. As shown in Fig. 3A for representative samples of saline-injected transgenic mice, the polyribosomal distribution of the highly expressed glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was mostly found in high-density fractions, representing polyribosomes. In contrast, the ferritin transcript, which is known to be translationally inactive in the absence of iron treatment (1), was predominantly found in very low density fractions, representing monosomes or ribosome-free RNA. These results indicate that analysis of the polyribosome distribution of mouse liver mRNAs allows discrimination between translationally active and inactive transcripts.
FIG. 3.
Polyribosome isolation from livers of HBV transgenic mice. Polyribosomes were harvested as described in Materials and Methods. (A) Serial fractions from sucrose gradients of polyribosomal liver extracts were analyzed by Northern blotting for the distribution of GAPDH, ferritin, and HBV 3.5- and 2.1-kb transcripts as described in Materials and Methods. (B) Polyribosomal liver extracts were treated with EDTA prior to sucrose density centrifugation, and fractions were analyzed by Northern blotting for HBV 3.5- and 2.1-kb transcripts and GAPDH transcripts as described above.
Figure 3A also shows that the 3.5-kb HBV pregenomic RNA was present in both the high-density and low-density fractions, suggesting that not all of the 3.5-kb messages are translationally active. It is important to note that the 3.5-kb RNA represents not only pgRNA but also slightly longer transcripts that are translated to produce HBeAg (36). Since it is theoretically possible that these two RNA species are differentially represented in the sucrose fractions, we performed an RNase protection assay of these samples to distinguish between pgRNA- and HBeAg-encoding transcripts. Such analysis did not reveal any differential distribution of these transcripts (data not shown).
To further confirm that the HBV and GAPDH transcripts were indeed associated with ribosomes, we treated polyribosomal extracts with 15 mM EDTA, a procedure known to release transcripts from ribosomes (21). As shown in Fig. 3B, the HBV and GAPDH transcripts moved into the lower-density fractions after EDTA treatment, confirming their polyribosomal association in Fig. 3A.
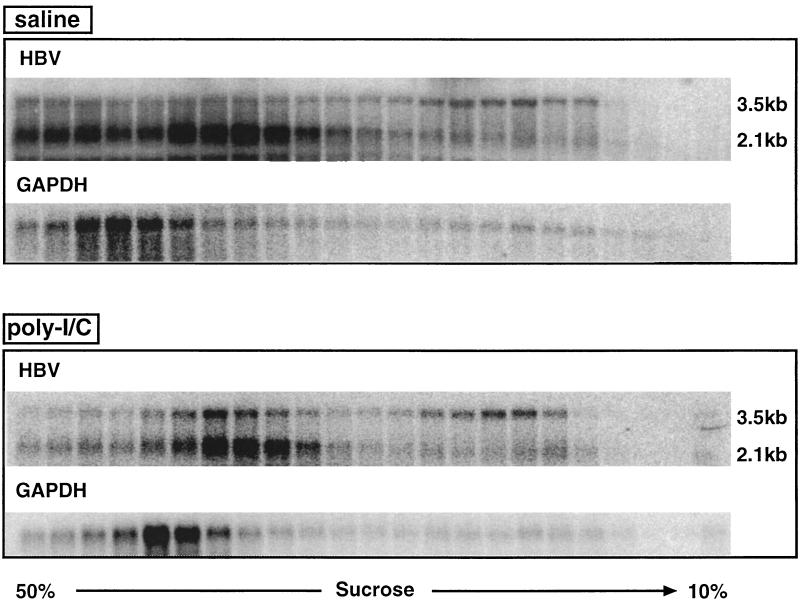
Next, we analyzed the polyribosomal distribution of HBV transcripts in the liver of age (8 to 10 weeks)-, sex (male)-, and HBeAg-matched transgenic mice from lineage 1.3.32 that were injected either with saline or with 200 μg poly(I-C). Mice were sacrificed 20 h after injection, when the content of HBV replicative intermediates was profoundly reduced (Fig. 2A). As shown in Fig. 4, the relative distribution of the HBV 3.5- and 2.1-kb transcripts in the sucrose gradients compared to the GAPDH distribution was the same in poly(I-C)-treated mice as in saline-injected controls. In particular, there is no accumulation of HBV transcripts following poly(I-C) treatment in low-density fractions where translationally silent transcripts would be expected as shown for ferritin in Fig. 3A. These results indicate that the translational status of the HBV transcripts is not changed by poly(I-C) treatment, suggesting that inhibition of translation (Fig. 2, step 1) does not account for the elimination of HBV capsids and replicative intermediates from the hepatocyte cytoplasm.
FIG. 4.
Poly(I-C) treatment does not alter the translational status of HBV transcripts. The polyribosomal distributions of HBV 3.5- and 2.1-kb and GAPDH transcripts in livers of HBV transgenic mice were analyzed 20 h after a single i.v. injection of saline or 200 μg of poly(I-C) as described in the legend to Fig. 3.
Poly(I-C) eliminates pgRNA-containing capsids from the hepatocyte cytoplasm.
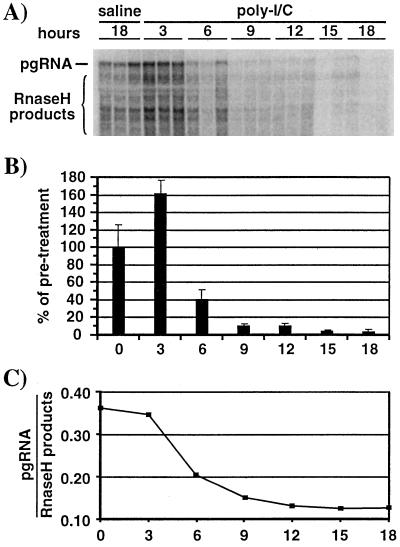
To identify the posttranslational step(s) in capsid maturation that may be inhibited by poly(I-C), we first examined the effect of poly(I-C) on the abundance of immature, RNA-containing capsids in the hepatocyte cytoplasm (Fig. 2, step 2). Age (7 to 11 weeks)-, sex (male)-, and serum HBeAg-matched mice from lineage 1.3.32 were injected either with saline or with poly(I-C). Groups of three mice were sacrificed at different time points after injection, and their livers were analyzed for encapsidated RNA (Fig. 5A) and total liver RNA (not shown) by Northern blot analysis. The results of the Northern blot analysis of encapsidated RNA (Fig. 5A) were quantified by phosphorimaging analysis and displayed as graphs in Fig. 5B and C. The level of encapsidated pgRNA decreased by 60% within 6 h and about 10-fold within 9 h after poly(I-C) injection (Fig. 5B), indicating that HBV RNA-containing capsids are eliminated from the hepatocyte cytoplasm after IFN-α/β induction. As expected, the steady-state level of total cellular HBV RNA did not significantly change throughout the course of the experiment (data not shown). In two out of three liver specimens at 3 h after poly(I-C) injection, we observed an increased level of encapsidated pgRNA (Fig. 5A and B) which also resulted in a higher level of replicative DNA intermediates isolated from the same mouse livers (Fig. 6A and B). In follow-up experiments, however, we did not observe increased levels of encapsidated RNA or replicative DNA intermediates at this time after poly(I-C) treatment (data not shown). Therefore, the increased level of HBV replication in these samples is most likely due to mouse to mouse variability in HBV gene expression.
FIG. 5.
Poly(I-C) treatment eliminates pgRNA-containing capsids from the hepatocyte cytoplasm. Age (7 to 11 weeks)-, sex (male)-, and serum HBeAg-matched mice from lineage 1.3.32 were injected either with saline or with a single i.v. injection of poly(I-C). Groups of three mice were sacrificed at the indicated times after injection, and the liver tissue was harvested. (A) Northern blot analysis was performed to detect encapsidated viral RNA as described in Materials and Methods. RNA isolated from identical amounts of liver tissue (35 mg) was loaded. (B) Graphic representation of encapsidated pgRNA. Signals for the saline samples were set to 100%. (C) Ratio of encapsidated pgRNA versus RNase H products plotted for each time point after poly(I-C) injection. Signals were quantified from the indicated band (pgRNA) and region (RNase H products) shown in the Northern blot above. Saline samples were considered to be preinjection (time zero) samples. The mean values for each group of mice are shown.
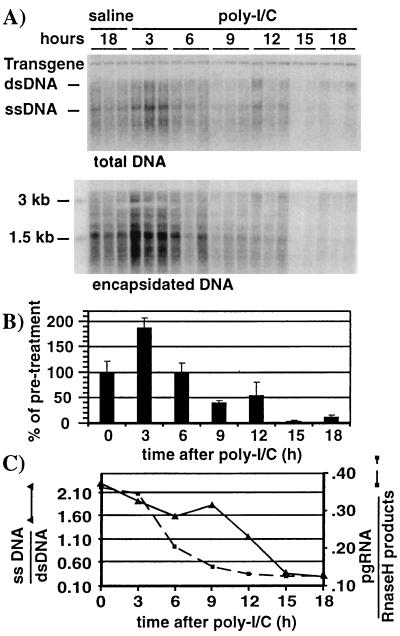
FIG. 6.
ssDNA-containing capsids are cleared after RNA-containing capsids but before mature capsids. Liver tissue samples from the mice represented in Fig. 5 were processed as follows. (A) Southern blot analysis was performed with 30 μg of total liver DNA as described in the legend to Fig. 2 (top), and encapsidated DNA from the equivalent of 5 mg of liver tissue was extracted and analyzed by Southern blotting as described in the Materials and Methods (bottom). Bands corresponding to the integrated transgene, dsDNA, and ssDNA HBV DNA replicative forms and size markers are indicated. (B) Graphic representation of HBV DNA at the indicated time points after poly(I-C) injection. Bars represent the combined signals for dsDNA and ssDNA. Mean values for each group of mice are shown. The value for the saline sample was set to 100%. (C) Ratios of ssDNA versus dsDNA were plotted at each time point after poly(I-C) injection (solid line). Ratios of encapsidated pgRNA versus RNase H products from Fig. 5C were also included (dotted line). HBV DNA signals were quantified by phosphorimaging analysis of the Southern blot shown in the top part of panel A. Signals were corrected for variations in the transgene.
As indicated in Fig. 2 (steps 2 and 3), newly formed cytoplasmic capsids contain full-length pgRNA. More mature capsids contain shorter RNA fragments which are generated by digestion of pgRNA by the RNase H function of HBV RT/Pol (29, 33). Thus, encapsidated full-length pgRNA is a marker of immature capsids (Fig. 2, step 2), while encapsidated RNase H products are markers of more mature capsids that are involved in reverse transcription (Fig. 1, step 3). Accordingly, by Northern blot analysis we detected a discrete band representing intact pgRNA and a smear of smaller transcripts representing RNase H digestion products. By plotting the ratio of encapsidated pgRNA versus RNase H products after poly(I-C) injection, we show that pgRNA-containing capsids are preferentially lost from the hepatocyte cytoplasm between 3 and 9 h after poly(I-C) injection (Fig. 5C). Thus, poly(I-C) treatment leads initially to the elimination of pgRNA-containing capsids (Fig. 2, step 2), and this is followed by the elimination of more mature capsids involved in reverse transcription (Fig. 2, step 3).
ssDNA-containing capsids are cleared after RNA-containing capsids but before mature capsids.
Reverse transcription converts the pgRNA in cytoplasmic capsids into ssDNA, yielding ssDNA-containing capsids as depicted in Fig. 2, step 4. The ssDNA-containing capsids subsequently mature into partially dsDNA-containing capsids (Fig. 2, step 6) through plus-strand DNA synthesis by the HBV RT/Pol (Fig. 2, step 5) (33). To monitor the kinetics by which ssDNA- and dsDNA-containing capsids are cleared from the hepatocyte cytoplasm after poly(I-C) injection, total DNA was extracted from the liver samples used for the RNA analysis (Fig. 5) and subjected to Southern blot analysis (Fig. 6A, top panel) As expected, Southern blot analysis of encapsidated DNA (Fig. 6A, bottom panel) yielded results equivalent to those for the total liver DNA, since all of the HBV replicative DNA intermediates in the liver are encapsidated. Therefore, the Southern blot of total DNA was used to compare the intensity of the ssDNA-specific band and the bands corresponding to the more mature double-stranded linear and relaxed circular DNA species (dsDNA) (Fig. 6A). The levels of HBV DNA replicative intermediates were corrected for loading according to the transgene signal, and they were plotted as a function of time after poly(I-C) injection. The combined level of dsDNA and ssDNA did not change from baseline during the first 6 h after poly(I-C) injection, then fell by 50% at 9 to 12 h, and finally decreased about 10-fold 15 h after poly(I-C) injection (Fig. 6B), indicating that DNA-containing capsids are eliminated from the hepatocyte cytoplasm in this time frame.
By plotting the ratio of ssDNA versus dsDNA as a function of time after poly(I-C) injection (Fig. 6C), we observed that the majority of ssDNA-containing capsids are eliminated from the hepatocyte cytoplasm between 9 and 15 h after poly(I-C) injection. This suggests that most of the ssDNA-containing capsids are cleared from the hepatocyte cytoplasm before the mature dsDNA-containing capsids.
Together, these and the foregoing observations with RNA-containing capsids (Fig. 5C) suggest that poly(I-C) eliminates immature RNA-containing capsids from the hepatocyte cytoplasm before the clearance of ssDNA- and dsDNA-containing capsids (Fig. 6C). These results suggest that poly(I-C) inhibits HBV replication by either preventing pgRNA encapsidation or reducing the stability of pgRNA-containing capsids. If this is correct, the preformed DNA-containing capsids should be cleared from the hepatocyte cytoplasm by export into the serum, and this would be followed by the elimination of virions from this compartment.
Determination of the export rate of HBV DNA in untreated transgenic mice.
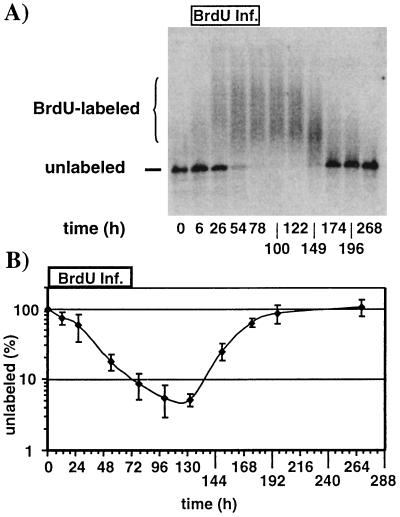
To determine whether poly(I-C) treatment affects virus export out of the hepatocyte, one has to first determine the production rate and half-life of virus in the serum of untreated transgenic mice. Thus, we monitored BrdU incorporation in virions circulating in the serum of untreated transgenic mice in which BrdU was continuously infused by an osmotic pump for 72 h. Inside the hepatocyte, the BrdU is converted to 5-bromo-2′-deoxyuridine-5′-triphosphate, which then can be incorporated into HBV minus-strand DNA during reverse transcription (Fig. 2, step 3) as well as in plus-strand DNA (Fig. 2, step 5). Subsequently, virions containing BrdU-labeled mature dsDNA are exported into the serum (Fig. 2, step 7). As shown in Fig. 7A for a representative BrdU-labeling experiment, BrdU-containing serum HBV DNA was detected by monitoring the ability of a BrdU-specific monoclonal antibody to retard the electrophoretic mobility of HBV DNA extracted from circulating virus particles. Antibody binding to the BrdU incorporated into HBV DNA occurs only on ssDNA; hence dsDNA isolated from HBV virions is denatured prior to the gel retardation experiment as described in Materials and Methods. BrdU free minus- and plus-strand virion DNA migrates as a single band in the gel retardation electrophoresis (Fig. 7A, unlabeled). BrdU-containing minus- and plus-strand DNA is retarded by the bound antibodies and is detected as a smear of high-molecular-weight DNA (Fig. 7A, BrdU-labeled). Control experiments were done with an irrelevant antibody, confirming the specificity of this assay (data not shown).
FIG. 7.
Determination of the viral half-life in sera of HBV transgenic mice. A group of four age (7 to 11 weeks)-, sex (male)-, and HBeAg-matched mice from lineage 1.3.46 were infused with BrdU as described in Materials and Methods. Serum samples were collected at the indicated time points. (A) Representative HBV-specific Southern blot analysis of a gel retardation experiment as described in Materials and Methods. Positions of unlabeled and BrdU-labeled DNA are indicated. (B) Levels of unlabeled serum HBV DNA were plotted on a log scale for each time point during BrdU infusion (0 to 72 h) and thereafter. The mean values of four mice are shown. Serum HBV DNA levels prior to BrdU infusion were set to 100%.
During the time of BrdU infusion, newly synthesized BrdU-containing HBV DNA was exported from hepatocytes and accumulated in the serum, where it was detectable as a high-molecular-weight DNA smear in the Southern blot shown in Fig. 7A. At the same time, preexisting virions were eliminated from the serum of untreated transgenic mice shown by the disappearance of the unlabeled, rapidly migrating HBV DNA forms (Fig. 7A). As the concentration of free BrdU eventually decreased over time and its incorporation into HBV DNA decreased, labeled DNA eventually disappeared from the serum as it was replaced by unlabeled DNA (Fig. 7A). The disappearance of unlabeled DNA from the serum was plotted as a function of time during BrdU labeling (Fig. 7B). From the linear part of the graph between 24 and 72 h, we determined that the viral half-life of HBV virions in the serum of transgenic mice under baseline conditions is about 16 to 18 h. This is also a measure of its export rate, since the steady-state level of HBV DNA in the serum of these mice was stable during the course of these experiments.
Poly(I-C) does not affect virus export or half-life.
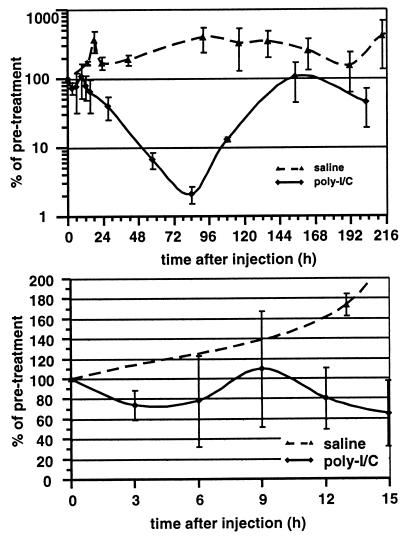
Next, we monitored serum HBV DNA levels after poly(I-C) or saline injection in transgenic mice. The serum HBV DNA levels were plotted against time after injection (Fig. 8). As expected, serum HBV DNA remained present at all times after saline injection, while virus was eventually cleared from the serum after poly(I-C) injection (Fig. 8, top panel). Figure 8 also shows that HBV replication was only transiently inhibited after a single poly(I-C) injection, since serum HBV levels returned to pretreatment levels at 154 h postinjection. Importantly, serum HBV DNA levels did not increase within 15 h after poly(I-C) injection (Fig. 8, lower panel), while HBV capsids were cleared from the cytoplasm of hepatocytes during this time interval (Fig. 6B). This suggests that the viral export rate is not increased after poly(I-C) injection, and hence increased export does not account for the depletion of capsids from the hepatocyte cytoplasm. Rather, the rate of clearance of virus from the serum after poly(I-C) injection followed the rate of clearance of unlabeled DNA during BrdU labeling in untreated transgenic mice, indicating that poly(I-C) did not change the half-life of virions in the sera of HBV transgenic mice.
FIG. 8.
Poly(I-C) does not affect virus export or half-life. Two groups of age (6 to 8 weeks)-, sex (male)-, and serum HBeAg-matched mice from the 1.3.46 lineage were injected either with saline (two mice) or with a single i.v. injection of poly(I-C) (three mice), and serum samples were analyzed at different time points after injection. Serum HBV DNA levels were plotted on a log scale for all time points measured after poly(I-C) injection (top). The serum HBV DNA levels up to 15 h after poly(I-C) injection were replotted on a linear scale (bottom). Mean values for relative serum HBV DNA levels in each group were plotted. Pretreatment serum HBV DNA levels were set to 100%.
DISCUSSION
In this study we showed that poly(I-C) suppresses the level of viral RNA-containing capsids in the liver without inhibiting transcription or translation of the viral RNA and without accelerating the export of preformed DNA-containing capsids out of the liver. Based on these observations, we conclude that poly(I-C) either inhibits the assembly or accelerates the degradation of HBV RNA-containing capsids in the cytoplasm of the hepatocyte. Since these effects coincided with the induction of IFN-α/β in the liver, and because we have recently reported that the antiviral effects of poly(I-C) are mediated by IFN-α/β (22), we conclude that the effects described herein were mediated by this cytokine. This is compatible with a recent report from our laboratory that treatment of duck HBV-infected primary duck hepatocyte cultures with duck IFN-α selectively eliminates pgRNA-containing capsids from the cells (30).
Contributions to viral clearance by other IFN-α/β-mediated effects such as inhibition of reverse transcription and/or viral DNA synthesis (i.e., capsid maturation) cannot be ruled out since they could not be experimentally measured in the transgenic mouse model. We observed, however, continuous virus export for at least up to 12 h after poly(I-C) treatment, indicating that virus production is not significantly reduced during clearance of capsids from the hepatocyte cytoplasm. This suggests that IFN-α/β does not inhibit capsid maturation as a mechanism to inhibit HBV replication in the transgenic mice.
Similarly, depletion of HBV capsids from the hepatocyte cytoplasm by increased viral export is very unlikely for two reasons. First, if the export rate was increased, one would expect mature DNA-containing capsids to be depleted from the hepatocytes before immature RNA-containing capsids, and this did not occur. Second, if the export rate was increased, one would expect to detect an increase in serum HBV DNA levels, and this did not occur (Fig. 8). This should have been easily detectable, had it occurred, because there are only about 107 to 108 virions/ml in the serum (14), while we calculate that there are about 109 capsids in the entire liver. The latter calculation is based on estimates from the Southern blot in Fig. 6A that there are at least 50 DNA-containing capsids per transgenic mouse hepatocyte and from our earlier results indicating that about 30% of these hepatocytes replicate HBV (14). Masking of increased virus export by reducing the half-life of virions in the serum is ruled out by our observation that the viral half-life did not significantly change after poly(I-C) treatment.
In summary, the results presented here suggest that IFN-α/β inhibits the formation of pgRNA-containing capsids or accelerates their degradation, while continued maturation of preexisting capsids and virus secretion accounts for the intracellular depletion of HBV capsids and replication intermediates. It is noteworthy that in parallel experiments (not shown), we found that elimination of pgRNA-containing capsids also occurred in mice that received repetitive injection of IL-12, a stimulus known to inhibit HBV replication by an IFN-γ-dependent mechanism (4). Although IFN-α/β and IFN-γ have both been shown to independently inhibit HBV replication in transgenic mice (22), these results indicate that similar intracellular steps in the HBV life cycle are targeted by these cytokines, suggesting that their antiviral activities may converge inside the cell.
The cytokine-induced signal transduction pathways and cellular protein(s) responsible for these effects are not known. IFN-α/β and IFN-γ interfere with various steps in the replication cycle of different viruses (reviewed in reference 35). For example, IFN-γ has been shown to directly inhibit replication of the poxviruses ectromelia virus and vaccinia virus as well as herpes simplex virus (HSV) by inducing nitric oxide synthase (18). The mechanisms that inhibit ectromelia and HSV replication are not well defined. Inhibition of vaccinia virus replication by inducible nitric oxide synthase occurs during late protein synthesis, DNA replication, and virus particle formation (17). IFN-α/β has been demonstrated to induce at least three different antiviral mechanisms (reviewed in reference 32). First, IFN-α/β can induce the dsRNA-dependent protein kinase (PKR) which phosphorylates the alpha subunit of eukaryotic initiation factor 2 (23) and inhibits translation initiation, thereby suppressing replication of RNA viruses such as encephalomyocarditis virus (24) or late gene expression of the DNA virus simian virus 40 (37). Second, IFN-α/β can induce the dsRNA-activated 2′5′OAS (2, 10) which leads to RNase L activation that can selectively reduce viral RNA during encephalomyocarditis virus infection (20) and seems to be the primary pathway to inhibit picornavirus replication (6). Third, IFN-α/β can induce the Mx proteins, GTPases in the dynamin superfamily (16), that interfere with transcription of negative-stranded RNA viruses such as vesicular stomatitis virus (31) or influenza virus (26, 27) but do not inhibit the replication of DNA viruses such as HSV (19, 27). Furthermore, IFN-α/β-responsive genes inhibit proper assembly of vaccinia virus by some unknown mechanism(s) (34) and prevent morphogenesis of HSV (5).
In view of these known antiviral effects, our demonstration that IFN-α/β selectively eliminates immature (i.e., nascent) viral nucleocapsids from the cytoplasm of cells that replicate HBV appears to expand the antiviral repertoire of this cytokine and define its anti-HBV activity very explicitly. Accordingly, efforts to identify cytokine-inducible cellular genes that inhibit HBV replication are currently under way.
ACKNOWLEDGMENTS
This work was supported by grants CA40489 (F.V.C.) and AI40696 (L.G.G.) from the National Institutes of Health. S.F.W. was partially supported by fellowships from the Ciba Geigy Jubiläumsstiftung and the Roche Research Foundation.
We are grateful to Jesse Summers for stimulating discussions and advice. We thank Carl Colburn for excellent technical assistance and Jennifer Newmann for assistance with manuscript preparation.
Footnotes
Manuscript no. 12744-MEM from the Scripps Research Institute.
REFERENCES
- 1.Aziz N, Munro H N. Both subunits of rat liver ferritin are regulated at a translational level by iron induction. Nucleic Acids Res. 1986;14:915–927. doi: 10.1093/nar/14.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll S S, Chen E, Viscount T, Geib J, Sardana M K, Gehman J, Kuo L C. Cleavage of oligoribonucleotides by the 2′,5′-oligoadenylate-dependent ribonuclease L. J Biol Chem. 1996;271:4988–4992. doi: 10.1074/jbc.271.9.4988. [DOI] [PubMed] [Google Scholar]
- 3.Cavanaugh V J, Guidotti L G, Chisari F V. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanaugh V J, Guidotti L G, Chisari F V. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee S, Hunter E, Whitley R. Effect of cloned human interferons on protein synthesis and morphogenesis of herpes simplex virus. J Virol. 1985;56:419–425. doi: 10.1128/jvi.56.2.419-425.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 7.Chisari F V, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 8.De Clercq E. Interferon induction by polynucleotides, modified polynucleotides, and polycarboxylates. Methods Enzymol. 1981;78:227–236. doi: 10.1016/0076-6879(81)78122-9. [DOI] [PubMed] [Google Scholar]
- 9.deFazio A, Leary J A, Hedley D W, Tattersall M H. Immunohistochemical detection of proliferating cells in vivo. J Histochem Cytochem. 1987;35:571–577. doi: 10.1177/35.5.3549891. [DOI] [PubMed] [Google Scholar]
- 10.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 11.Ganem D. Hepadnaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, Roberts M D, Chanock M D, editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 12.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B, Chisari F V. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti L G, Chisari F V. Cytokine-induced viral purging—role in viral pathogenesis. Curr Opin Microbiol. 1999;2:388–391. doi: 10.1016/s1369-5274(99)80068-x. [DOI] [PubMed] [Google Scholar]
- 14.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F V. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 16.Haller O, Frese M, Kochs G. Mx proteins: mediators of innate resistance to RNA viruses. Rev Sci Technol. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 17.Harris N, Buller R M, Karupiah G. Gamma interferon-induced, nitric oxide-mediated inhibition of vaccinia virus replication. J Virol. 1995;69:910–915. doi: 10.1128/jvi.69.2.910-915.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karupiah G, Xie Q W, Buller R M, Nathan C, Duarte C, MacMicking J D. Inhibition of viral replication by interferon-gamma-induced nitric oxide synthase. Science. 1993;261:1445–1448. doi: 10.1126/science.7690156. [DOI] [PubMed] [Google Scholar]
- 19.Landis H, Simon-Jodicke A, Kloti A, Di Paolo C, Schnorr J J, Schneider-Schaulies S, Hefti H P, Pavlovic J. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J Virol. 1998;72:1516–1522. doi: 10.1128/jvi.72.2.1516-1522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X L, Blackford J A, Hassel B A. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald C C, Williams D L. Proteins associated with the messenger ribonucleoprotein particle for the estrogen-regulated apolipoprotein II mRNA. Biochemistry. 1992;31:1742–1748. doi: 10.1021/bi00121a023. [DOI] [PubMed] [Google Scholar]
- 22.McClary H, Koch R, Chisari F V, Guidotti L G. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J Virol. 2000;74:2255–2264. doi: 10.1128/jvi.74.5.2255-2264.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meurs E, Chong K, Galabru J, Thomas N S, Kerr I M, Williams B R, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 24.Meurs E F, Watanabe Y, Kadereit S, Barber G N, Katze M G, Chong K, Williams B R, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5804–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 26.Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavlovic J, Zürcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitha P M. Interferon induction with insolubilized polynucleotides and their preparation. Methods Enzymol. 1981;78:236–242. doi: 10.1016/0076-6879(81)78123-0. [DOI] [PubMed] [Google Scholar]
- 29.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz U, Summers J, Staeheli P, Chisari F V. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J Virol. 1999;73:5459–5465. doi: 10.1128/jvi.73.7.5459-5465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwemmle M, Weining K C, Richter M F, Schumacher B, Staeheli P. Vesicular stomatitis virus transcription inhibited by purified MxA protein. Virology. 1995;206:545–554. doi: 10.1016/s0042-6822(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 32.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 33.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 34.Thornton A M, Buller R M, DeVico A L, Wang I M, Ozato K. Inhibition of human immunodeficiency virus type 1 and vaccinia virus infection by a dominant negative factor of the interferon regulatory factor family expressed in monocytic cells. Proc Natl Acad Sci USA. 1996;93:383–387. doi: 10.1073/pnas.93.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 36.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakobson E, Prives C, Hartman J R, Winocour E, Revel M. Inhibition of viral protein synthesis in monkey cells treated with interferon late in simian virus 40 lytic cycle. Cell. 1977;12:73–81. doi: 10.1016/0092-8674(77)90186-6. [DOI] [PubMed] [Google Scholar]