Abstract
Nervous systems of bilaterian animals generally consist of two cell types: neurons and glial cells. Despite accumulating data about the many important functions glial cells serve in bilaterian nervous systems, the evolutionary origin of this abundant cell type remains unclear. Current hypotheses regarding glial evolution are mostly based on data from model bilaterians. Non-bilaterian animals have been largely overlooked in glial studies and have been subjected only to morphological analysis. Here, we provide a comprehensive overview of conservation of the bilateral gliogenic genetic repertoire of non-bilaterian phyla (Cnidaria, Placozoa, Ctenophora, and Porifera). We overview molecular and functional features of bilaterian glial cell types and discuss their possible evolutionary history. We then examine which glial features are present in non-bilaterians. Of these, cnidarians show the highest degree of gliogenic program conservation and may therefore be crucial to answer questions about glial evolution.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13064-024-00184-4.
Keywords: Glia, Evolution, Glial cells missing, Non-bilaterians, Gliogenesis, Neurogenesis, Cnidaria
Background
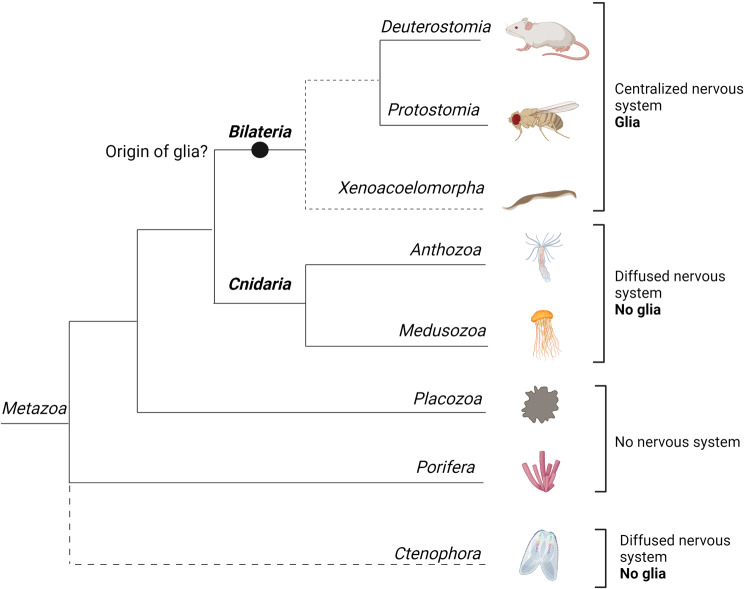
Metazoans are broadly subdivided into bilaterians (Protostomia and Deuterostomia) and non-bilaterians (Porifera (sponges), Placozoa, Ctenophora (comb jellies), and Cnidaria). Nervous systems of bilaterian animals are generally composed of two cell types: neurons and glial cells. Among non-bilaterians only ctenophorans and cnidarians possess neurons forming a diffuse nervous system, but these phyla are believed to lack glia [1, 2] (Fig. 1).
Fig. 1.
Phylogeny of Metazoa and distribution of glial genes. A metazoan phylogenetic tree of main animal groups includes the unresolved positions of Ctenophora and Xenacoelomorpha indicated by dotted branches [3]
Neurons are electrically active, which makes them the primary functional component of NSs. Although glia are considered supportive cells, they participate in almost every process in the nervous system of bilaterians, including neurotransmission, homeostasis, and also the important function of producing neurons [4, 5]. In the course of metazoan evolution, glial cells have acquired greater morphological and functional diversity [6]. This remarkable diversity is especially obvious in higher bilaterians such as vertebrates, as they include distinct types even within one glial cell class [7].
The emergence of glial cells is one of the key novelties in evolution of nervous systems. Important unanswered questions in glial biology are “Where did glial cells originate?”, and “What were their first functions?” Tracing origins of the first glia and identifying their original functions are therefore crucial to understand nervous system evolution.
Extensive studies of glia have mostly been confined to a few bilaterian model organisms, i.e., Caenorhabditis elegans (nematode) and Drosophila melanogaster (fruit fly) among invertebrates, Danio rerio (zebra fish), Mus musculus (mouse), Rattus norvegicus (rat), and Homo sapiens (human) among vertebrates. Some animals from other lineages, including non-bilaterians, have been screened for cells morphologically similar to glia. Glia-like cells have also been suggested in Xenacoelomorpha [2]. If this group of animals represents the earliest divergent bilaterian lineage, as several molecular phylogenetic analyses suggest, the emergence of glia-like cells may be traced back to the last common bilaterian ancestor. Overall glia are hypothesized to have evolved coincidentally with the appearance of the central nervous system (CNS), i.e., after the common bilaterian ancestor diverged from the Cnidaria [6, 8] (Fig. 1).
This view is being challenged by accumulating data collected from non-bilaterian animals. It was suggested that neurons and glia may have evolved at the same time, and that they have a common evolutionary origin [9]. This contrasts with the hypothesis that neuron-bearing, non-bilateral animals, Ctenophora and Cnidaria, have purely neuronal nervous systems [10]. Both frameworks assume that glial cells first emerged as metabolic support units for sensory neurons/organs, and that they then acquired an axonal support function, ensuring higher speed and more precise neuronal signaling by ensheathing axons. Finally, glial cells acquired other functions such as immune support.
Here we discuss genetic and cellular physiological features exhibited by bilaterian glial cells, and we also extend the investigation to common glial traits conserved in non-bilaterians that lack glial cells. In this review, we provide an overview of current knowledge about the extent to which the gliogenic genetic program in bilaterians is conserved and what function it serves in non-bilaterians. In the first part of the review, we provide a detailed examination of bilaterian glial cells, and their molecular signatures and functions. In the second part, we discuss conservation of the gliogenic program in non-bilaterians.
Main text
Morphology and physiology of bilaterian glial cells
Bilaterian glial cells are numerous and diverse, therefore defining them is challenging. Nevertheless, collectively, glia are considered non-neuronal cells of nervous systems. According to Shai Shaham [11], there are three primary characteristics of glial cells: morphology - glial cells are associated with neurons; physiology - glial cells do not conduct fast currents, do not possess neurotransmitter-laden vesicles and do not form presynaptic structures; origin (development) - glia, together with neurons, arise from neuroectoderm during embryogenesis.
Morphology is the most widely used means of identifying glia and is often the only method. Despite morphological variations observed in glial cells of different species (reviewed in [1]), close association with neurons is generally regarded as a common glia-specific feature. In addition, glial processes usually ensheath axons and creep through nerve bundles [12].
The astonishing morphological diversity of bilaterian glia seems related to diverse functions these cells perform: providing energy to neurons, maintaining the extracellular environment of neurons, immune response, serving as stem cells to generate glia and neurons in the adult brain, formation of the blood-brain barrier (BBB) [13–15]. On the other hand, the diversity of functions and glial cell types increases with the complexity of the nervous system. The increasing number of glia (from 10% of total brain volume in model invertebrates to over 50% in mammals), glia-to-neuron ratio, and glial cell complexity seems to support the idea that the earliest neurons did not need glial cells [6]. This led to a general definition of glia cells as “homeostatic cells of the nervous system” [6]. According to this definition, glial cells exist as housekeeping cells, whereas neurons serve as information processing units. At the same time, studies on C. elegans suggest that the first glia may have emerged at sensory receptive endings to control/support neuronal processing of incoming information about the environment [16]. Mammalian glial cells also assist in information processing by modulating synaptic activity and connectivity [17, 18]. Assigning a single function to glial cells in higher animals and using it as a universal diagnostic feature for all glial cells is therefore problematic.
Certain physiological features are helpful to consider when defining glial cells. Unlike neurons, glial cells are not known to generate action potentials. Nor has complete synaptic machinery been identified in glial cells. Other features tend to be glial cell type-specific, as it is possible to distinguish various glial cell types among model deuterostomes and protostomes.
As more molecular data are collected, classification of glia becomes more complex. In vertebrates, the term “glia” usually includes microglia and macroglia, i.e., radial glia, astrocytes, myelin-producing oligodendrocytes, and Schwann cells. There are several less numerous populations of glia, particularly in mammals, such as NG2 glia, known for their expression of a neurexin cell adhesion molecule NG2 and glial progenitor potential [19], pituicytes, tanycytes, and others residing in specific brain areas and resembling astrocytes transcriptionally [20–22]. In this review, we do not consider these glial subgroups separately.
In Drosophila seven types of glia have been identified [23], whereas C. elegans glia were divided into three groups (sheath glia, socket glia, and mesodermally derived glial cells) [16]. In other invertebrates, radial glia are often the only glial type reported [24]. On the other hand, more thorough examination of several non-model bilaterians reveals glial cells actively involved in neurotransmitter metabolism, which is an astrocytic feature [25]. Therefore, in order to trace glial origins, it is important to consider characteristics of different bilaterian glial types.
Bilaterian glia cell types
Radial glia
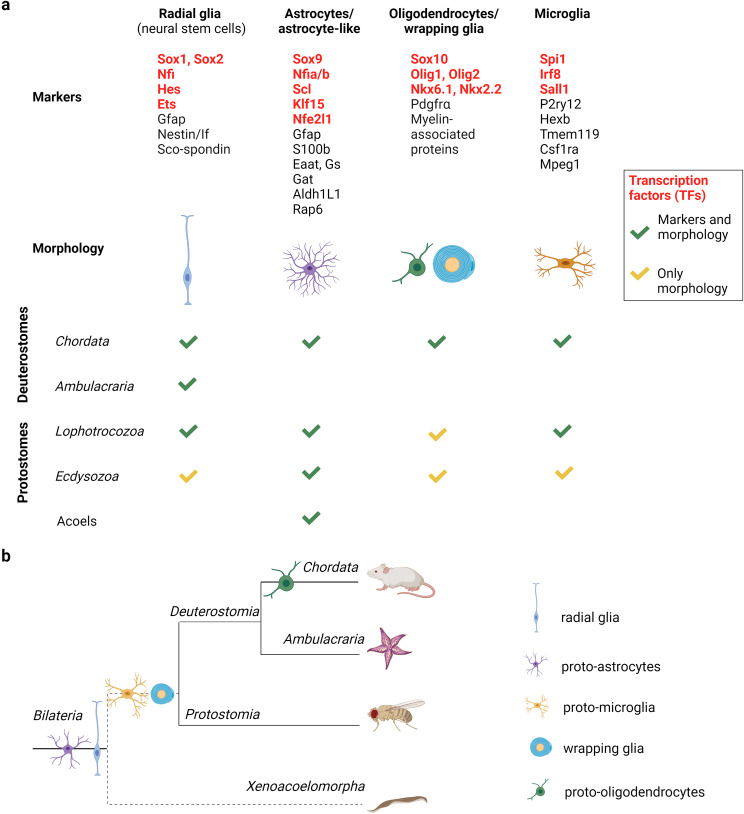
Radial glia are neural stem cells and as such, are sometimes not considered strictly glial. They express stem cell markers such as Sox [15]. These cells give rise to neurons and astrocytes in addition to forming scaffolds used by newly generated neurons to travel to their final destination. Radial glia secrete Reissner’s fiber components and several markers generally considered specific to astrocytes, such as glutamate transporters and intermediate filament proteins, including glial fibrillary acidic protein (Gfap) and Nestin [26]. Radial glia are elongated cells, and extend long processes through the neuropil. They are the earliest type of glia to develop in vertebrates, but are sparse in adult mammalian brains. Radial glia have been reported in both deuterostomes (Vertebrata, Echinodermata, Hemichordata) and protostomes (Annelida, Arthropoda) [27, 28]. They are the only type of glia identified so far in Echinodermata [24], and they perform a phagocytic function in addition to their neurogenic and scaffolding functions [29]. Given that radial glial cells are present throughout the Bilateria, that they can generate both neurons and glial cells, and are the first neural cell type to develop, they may have been the first glia to emerge in animals with nervous systems.
Astrocytes/astrocyte-like
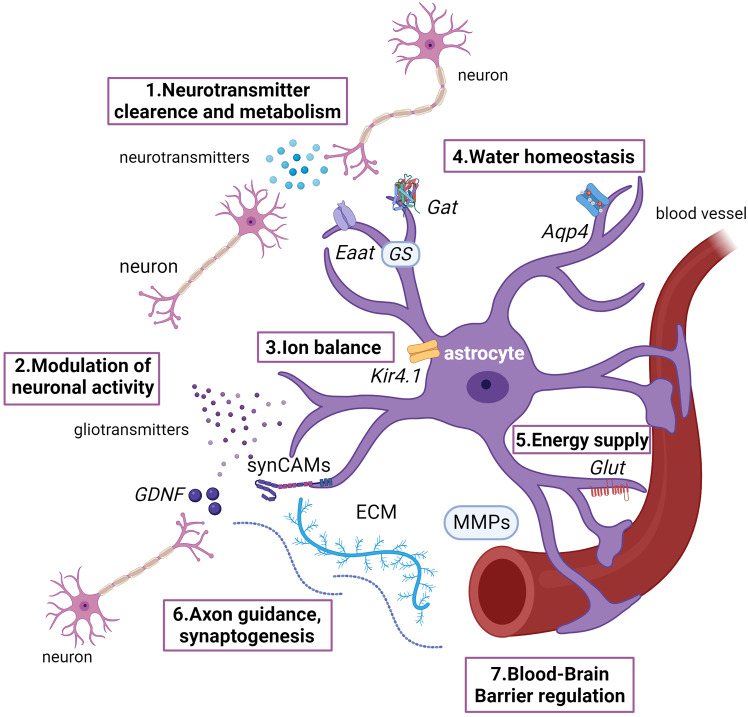
Astrocytes are glial cells in the most classical sense. They are closely associated with neurons, and are the primary cells maintaining homeostasis of vertebrate CNS. Astroglia fulfill many functions in the nervous system such as regulating ionic and neurotransmitter composition of the neuronal environment [30, 31]; maintaining water homeostasis [32]; energetic support of neurons [33]; maintaining the blood-brain barrier [14, 34]; synaptogenesis [35], axon guidance [36]; and phagocytosis [37, 38] (Fig. 2). Each of these functions is evidenced by expression of genes such as ion channels, including potassium channels (Kir4.1) [39, 40], GABA and glutamate transporters, glutamine synthetase (Gat, Eaat, Gs) [41, 42]; aquaporin channels (Aqp4) [43]; glucose transporters (Glut) [44]; BBB-regulating factors such as matrix metalloproteinases (MMPs) and glial-derived neurotrophic factor (GDNF) [14]; ECM proteins that are synaptogenic, e.g., thrombospondins, and axon guiding, e.g., SynCAMs factors [45, 46]; cell death abnormality (Ced) pathway components involved in engulfment and phagocytosis [47] (Fig. 2). A metabolic enzyme, Aldh1l1 (aldehyde dehydrogenase 1 family member L1), and intracellular signaling Rab6 are pan-astrocytic markers in rodents [47, 48]. Some of these markers, especially those involved in glutamate metabolism, have been successfully used to identify glial presence in invertebrate bilaterians, e.g., planarians [25]. Several transcription factors (TFs) including Nfe2l1(bZIP TF) regulated by Sox9 [49], Klf15 (Kruppel-like family), and Scl (bHLH TF) [50] are involved in astrocyte development. Astrocytes demonstrate “classical” glial morphology: multiple processes extending into the neuropil, surrounding synapses, and neuronal processes. Based on these criteria in addition to expression of Gfap, astrocyte-like glia were identified in early-branching bilaterians - acoels [51, 52]. Glial cells that morphologically and functionally resemble astrocytes are also found in zebrafish (deuterostomes), C .elegans, and Drosophila (protostomes). Because astrocyte-like glial cells are present throughout bilaterian lineages, astroglia may have been the first true glia type to evolve.
Fig. 2.
Astrocyte functional diversity. Each function is fulfilled by astrocyte-expressed, secreted proteins
Oligodendrocytes, Schwann cells, wrapping glia
Myelin production is the major function of oligodendrocytes in the CNS and Schwann cells in the PNS of vertebrates. The appearance of myelin correlates with jaw development, which suggests that it is a recent invention, from an evolutionary point of view [2]. Myelin-associated proteins are used as oligodendrocyte and Schwann cell markers [53, 54]. At the same time, oligodendrocyte lineage markers are present in species that do not possess myelin. These include TFs that drive oligodendrogenesis such as Sox10 and Olig2, as well as a well-known marker of oligodendrocyte progenitors - platelet-derived growth factor receptor (PDGFRα) [55] (Fig. 3). This may indicate a common gliogenic program dating back to invertebrates. Even among vertebrates, not all oligodendrocytes produce myelin. The common feature that oligodendrocytes do share, however, is axonal ensheathment. Glial cells covering axons with their membranes are found in various animals, including protostomes [2]. Ensheathment of axons is hypothesized to have evolved to allow increased conduction speed and precision of neuronal signaling by blocking electrical crosstalk between axons [9, 56]. Simultaneously, these cells may have provided nutrients for neurons. Oligodendrocytes share the function of providing metabolic support to neurons with astrocytes and express the same glutamate and GABA transporters [57]. Oligodendrocytes and astrocytes form networks via gap junctions, contributing to ionic buffering during neuronal activity [58]. Therefore, distinguishing between oligodendrocytes and astrocytes may be trivial only in vertebrates, as functions of both these cell types are performed by the same cells in animals with simpler nervous systems.
Fig. 3.
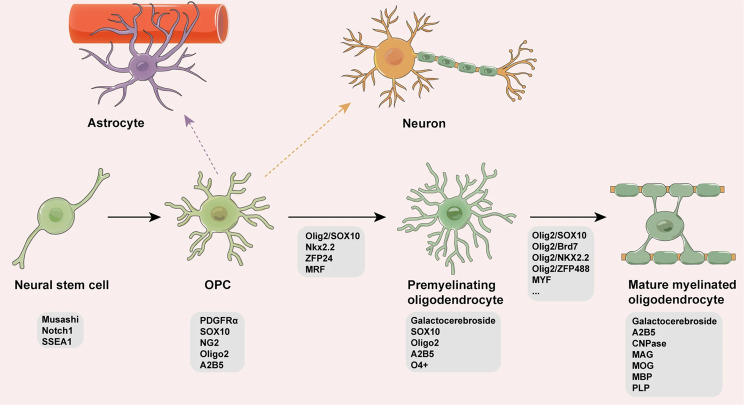
Developmental program of mammalian oligodendrocytes. Neural stem cells give rise to oligodendrocyte progenitor cells (OPCs), which begin to express oligodendrocyte markers (Sox10, PDGFRa). OPCs potentially differentiate into astrocytes, neurons, and oligodendrocytes, although the former two signaling pathways are under debate. Olig2 and Sox10 genes, among others, drive oligodendrocyte maturation. Myelin constituents, such as MBP (myelin basic protein) and PLP (proteolipid protein), are expressed by mature myelinated oligodendrocytes. Image taken from [59]
When considering evolutionary roots of glia, it is helpful to consider the gliogenic vs. neurogenic program. It is generally believed that oligodendrocytes arise from the same progenitors as neurons [60]. If this is the case, only the astrogenic program should be searched for in organisms with simple nervous systems, in order to reveal glial evolution. However, some progenitors produce both oligodendrocytes and astrocytes [61]. Radial glia produce all three cell types, depending on the developmental stage [62]. Oligodendrocyte progenitor cells (OPCs) may also generate all three cell types (Fig. 3). Therefore, it is rather difficult to identify a strictly gliogenic program.
Microglia
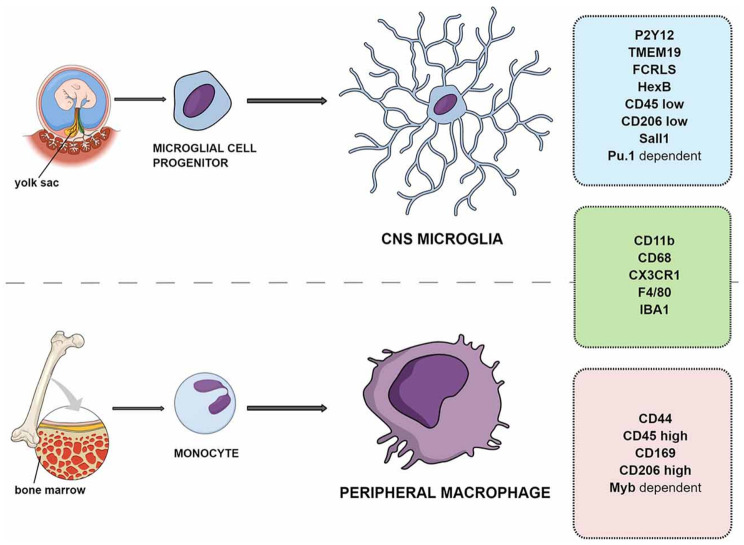
Unlike macroglia, which originate within the ectoderm, vertebrate microglia have a mesodermal origin. Similarly, C. elegans possesses 6 mesodermally derived glial-like cells in the nerve ring (GLRs) serving as connections between neurons and muscles [16]. The function of GLRs is not clear, but they engulf dead cells [63]. In mammals, microglia serve as resident macrophages of the CNS [64]. Because one of the main functions of microglia is debris clearance and degradation, markers of phagocytic pathways such as P2ry12 and lysosomal enzymes such as Hesb are abundantly expressed by these cells [47, 65]. To this end, microglia and astrocytes share the function of synapse pruning and engulfment [66]. Similarly, both cell types react to injury and inflammation. Even more so, astrocytes assume the role of phagocytes in case of microglial dysfunction [67]. Like other neural cells, vertebrate microglia express various ion channels, neurotransmitter receptors, and transporters [68]. In general, however, the transcriptome of microglia differs significantly from that of macroglia in that it is enriched with markers related to immune system processes and macrophages, e.g. CD45, CD68, but also microglia-specific markers, e.g. TMEM119 [65, 69] (Fig. 4). The transcriptional program driving microglial identity is drastically different from that of macroglia [70]. Spi1 (or Pu.1 - Ets-domain TF) and Irf8 (interferon regulatory factor family) are the main TFs driving microgliogenesis in various vertebrate species [71]. Morphologically, resting microglia resemble astroglia in that cells extend processes from the central soma. Upon activation in response to injury microglia change their morphology and aggregate at the lesion site. The complexity of the ramified structure of microglia varies with the overall trend of increasing in evolution. However, unlike astrocytes, human microglia do not display the most complex morphology compared to other vertebrates [71]. Apart from vertebrates, insects (Arthropoda), leech (Annelida), and molluscs are reported to have microglial cells [2]. Among these, leech microglia have been studied most thoroughly, albeit using few molecular markers [72]. Surprisingly, no other glial cell types have been identified in leeches. It is not clear to what extent the vertebrate microglial molecular program and functions are conserved in invertebrates, due to the paucity of studies.
Fig. 4.
Mammalian microglia and peripheral macrophage ontogeny and markers. The two cell types share a majority of markers, although their origins differ. Image taken from [73]
Figure 5 summarizes glial cells and their marker genes found in major model animals. With the emergence of molecular identifiers for each glial cell type, it has become possible to identify various glial cells in bilaterian lineages other than vertebrates and insects (Fig. 5a). Nevertheless, few glial markers have been searched for in non-model organisms. Based on data obtained mostly from histological studies, it is likely that radial glia were the first glial type to evolve. These cells combine features of both neurons and glia and give rise to both. The first true glia to emerge may have been astrocyte/oligodendrocyte-like cells that assumed several key functions, including metabolic support for and electrical insulation of neurons. Glial cells reminiscent of microglia can be found in basal bilaterians, but were likely to have emerged later in evolution (Fig. 5b).
Fig. 5.
Bilaterian glial cell types: molecular markers, morphology, and evolutionary processes. (a) Glial cell type conservation in bilaterian phyla. Molecular markers and morphological features of major glial cell types are shown. (b) Evolutionary tree of glial cell types based on available bilaterian studies
Phylogenetic distribution of glia-related genes
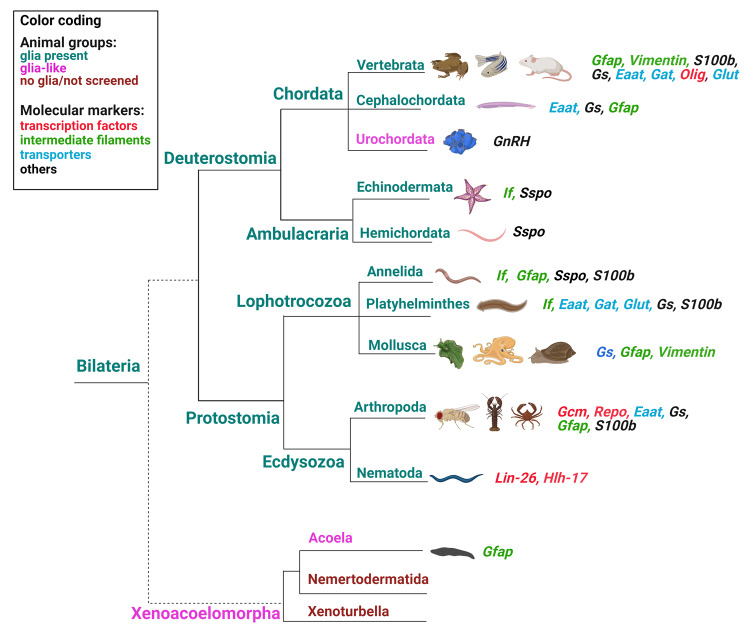
Molecular markers have long been used to identify cell types and to identify their functional characteristics. These have become even more prevalent with the emergence of omics techniques. The most widely used glial marker is an intermediate filament (If) protein, Gfap, which was first identified in vertebrates. Gfap immunostaining has been used to identify potential glia in various invertebrates, including an early-branching bilaterian, Symsagittifera roscoffensis (Xenacoela) [51] (Fig. 6). Immunostaining with GFAP antibodies documented the distribution of putative glial cells; however, Gfap labels only a subset of astroglia in vertebrates [47], and is also expressed in non-glial cells [74]. In addition, Gfap is not expressed in other glial types, like oligodendrocytes [47]. Intermediate filaments including Gfap are prominent features of glial cells in several bilaterians [75] (Fig. 6). Apart from Gfap, another intermediate filament protein, vimentin, is expressed in glial cells in the snail Megalobulimus abbreviatus [76]. In mammalians, vimentin is expressed in radial glia and astrocytes during early differentiation stages, and is later replaced by Gfap [77]. Overall, it is less glia-specific than Gfap, even in invertebrates [78]. Interestingly, intermediate filament-1 protein (If-1) is expressed in planarian glia [25].
Fig. 6.
Phylogenetic distribution of major glial markers. Presence of glia is inferred from reports on cells morphologically and genetically similar to well-developed glia of vertebrates. Glial markers identified in each phylum are specified and color-coded. Representative species screened for glial markers are enumerated in Supplementary Table S1 and accompanied by literature references
Other vertebrate glial markers searched for and used to identify glia in invertebrates include glutamine synthetase (Gs) in the lobster (Panulirus argus) [79] and Aplysia [80], S100 calcium-binding protein B (S100b) in the giant prawn (Macrobrachium rosenbergi) [81] and flatworm (Christensenella minuta) [82], transporters for glutamate (Eaat), GABA (Gat), and glucose (Glut) in planaria (Schmidtea mediterranea) [25] (Fig. 6). Expression of Eaat, Gs, Gfap/vimentin/If genes was explored in the lancelet, documenting glia in the Cephalochordata [83]. Sco-spondin (Sspo), an extracellular matrix (ECM) glycoprotein, the main component of the Reissner’s fiber [84], is another glial marker. It is secreted by radial glia and was used to identify glia in both deuterostomes and protostomes [27, 85]. Some glial markers seem phylum/class-specific and have been used to identify glia in certain animals. For example, gonadotropin-releasing hormone (GnRH), is expressed in glia of a urochordate, Ciona intestinalis [86], while Aplysia glia secrete a protein called Ag [87] (Fig. 6).
Gliogenic program in bilaterians
Expression of glial molecular markers, i.e., functional genes, is driven by TFs and extracellular cues. Therefore, by unraveling the genetic developmental program driving gliogenesis, it may be possible not only to define glia more precisely, but also to trace back their evolutionary origins. In Drosophila, it has been possible to identify TFs that drive expression of glial-specific markers, the so-called ‘binary switches’, such as Gcm and Repo [88, 89], demonstrating that invertebrates have a more robust intrinsic system for neuro/gliogenesis than had been thought previously. The vertebrate situation is more complex, particularly among mammals. No specific TFs responsible for glial cell fate acquisition have been identified per se, even though some, such as bHLHs, Sox group E, Ets and NfI family, are obviously involved [49, 90, 91]. Therefore, gliogenic programs in protostomes (insects) and deuterostomes (vertebrates) do not appear to be homologous.
Glial cells missing (Gcm)
Gcm is a master regulator of gliogenesis in Drosophila [92–94]. Gcm is indispensable for gliogenesis: a mutation in this gene turns presumptive glia into neurons [92]. All lateral glia of Drosophila rely on Gcm for their development, but specific subtypes of glial cells are established via different mechanisms [23]. Drosophila has two Gcm genes with partially redundant functions [95, 96]. Gcm2 seems less important for glial differentiation, but is involved in macrophage development [95, 96]. Both Gcm genes promote postembryonic neurogenesis, in addition to glial development in Drosophila CNS [97]. The neuro/gliogenic functions of Gcm homologs have also been shown in other taxonomic classes. Gcm neural expression and function has been demonstrated in crayfish (crustaceans) [98]. It is not clear whether crustacean Gcm-expressing cells are glial. Nevertheless, as in vinegar flies, crustacean Gcm seems to be co-expressed with Repo, which is an important gliogenic TF under Gcm regulation in Drosophila. Sea urchins (Echinodermata) also possess Gcm, although it is not expressed in neural tissue [99]. It functions in pigment cell specification instead [100]. Importantly, however, in both Drosophila and Strongylocentrotus purpuratus, Gcm expression is driven by Notch [99, 101]. In bilaterians, Notch-Delta signaling promotes gliogenesis [102, 103] by activating glial genes and sustaining neuronal precursors in both vertebrates and invertebrates [104–106]. Hes/hairy are primary downstream targets of Notch, inhibiting neurogenic bHLH TFs such as Atonal and Achaete–Scute complex [102, 106, 107]. In Drosophila, Notch drives Gcm expression to give rise to subperineurial glia (SPG) [101]. However, Notch has an opposite effect on Gcm in the sensory organ precursor lineage of the peripheral nervous system (PNS) [108]. Likewise, there is no unified effect of Notch on glial differentiation in vertebrates, where Notch promotes certain glial types such as astrocytes and radial glia, but not oligodendrocytes (reviewed in [109]). Functions of Gcm homologs in vertebrates are still debated. Gcma (or Gcm1) and Gcmb (or Gcm2) have been isolated in mammals [110]. These are expressed predominantly in tissues other than neural tissue: Ggma is expressed in placenta [111, 112], Gcmb in parathyroid glands [113]. However, both genes induce generation of neural stem cells [114]. The neurogenic role of Gcma is conserved in chickens [114, 115]. Moreover, rodent Gcma induces gliogenesis and drives astrocyte differentiation [116, 117]. In zebrafish, one Gcm gene was isolated and named Gcmb, due to its similarity to mammalian Gcmb. It is expressed in macrophages and contributes to pharyngeal cartilage formation [118]. In C. elegans, all glial cells express the Zn-finger transcription factor, Lin-26 [119]. Epithelial cells also express Lin-26 and are transformed into neurons in Lin-26 mutants, highlighting the conserved linear relationships between glia, neurons, and epithelia. The function of Lin-26 is reminiscent of the function of Gcm. However, Lin-26 and Gcm proteins are not clearly homologous. A definite role for Lin-26-related genes in gliogenesis is currently unknown except in C. elegans, suggesting that this mechanism may have been acquired independently in that lineage. In bilaterians that have only a single Gcm gene, its neural and gliogenic functions are not yet clear. In lineages that have two Gcm genes, homologs of Drosophila Gcm1 show gliogenic functions. In both protostomes and deuterostomes, Gcm is tightly regulated by Notch [101, 114, 120]. Thus, the Notch-regulated Gcm program seems conserved in bilaterians.
Reversed polarity (Repo)
Repo is a homeobox gene downstream from Gcm in Drosophila, which ensures terminal differentiation of glial cells [121]. Repo, in turn, drives expression of glial-specific markers, including Pointed, which is also regulated by Hairy [88, 122]. In addition to Pointed, Tramtrack69, Loco, and M84 are effectors of Repo, which participate in glial differentiation and morphogenesis in Drosophila [88]. Repo mutants demonstrate reduced cell number and poorly differentiated glia [123]. Repo is not known to have a gliogenic role in animals other than in insects. Vertebrates lack the gene for Repo altogether.
Oligodendrocyte transcription factor (Olig)
Olig belongs to group A bHLH genes. Olig1 and Olig2 promote oligodendrocyte differentiation in mammals [124, 125]. Olig genes are among several TFs that couple neuronal and oligodendrocyte specification [60, 126]. In addition, a subpopulation of astrocytes expresses Olig2 [127]. These findings indicate that Olig TFs are not specific oligodendrocyte genes. The primary function of oligodendrocytes is to myelinate neurons to ensure fast action potential propagation. Myelin is a new invention, from an evolutionary point of view, as it is associated only with vertebrates [2, 128]. On the other hand, axonal ensheathment by glial membranes is observed in invertebrates as well. This suggests that the first functions of glia may have been metabolic support for neurons. Interestingly, C. elegans possesses a bHLH gene, Hlh-17, and regulation of its expression is similar to that of mammalian Olig2 [129]. C. elegans single-cell data confirm its mostly glial expression, although it is also expressed in neurons [130]. Therefore, Olig-like genes may have emerged as TFs with glio/neurogenic functions in the common ancestor of deuterostomes and protostomes.
Hairy and Enhancer of split (Hes)
Hes are involved in many developmental processes, including suppression of proneural bHLH genes, and promotion of gliogenesis [131, 132]. Mammalian Hes genes are homologs of Drosophila Hairy and Enhancer of Split. Vertebrate Hes1, Hes3, and Hes5 particularly promote gliogenesis at a later stage of the developing brain, and control production of neural stem cells at an earlier neurogenic stage [133]. Interestingly, Hes5 is specifically expressed in mammalian Muller glial cells [134]. Its expression is regulated by Gcm genes at an early stage to induce neural stem cell generation, and it is later replaced by activation by Notch [114]. Hes genes are known effectors of Notch in mammals, as well as Drosophila [133, 135, 136]. Gcm, Hes, and Notch could therefore be important for gliogenesis in various lineages, even though Gcm-Notch synergy driving glia generation in Drosophila seems independent of Hes [137].
Nuclear factor I (NfI)
NfI genes are CCAAT box element-binding TFs [138]. In vertebrates, the NfI family is composed of four genes: NfIa, NfIb, NfIc and NfIx. These are important for development of various tissues, including the nervous system [138, 139]. NfIa, NfIb, and NfIc promote differentiation of radial glial cells into both glia and neurons [140]. NfIa directly induces expression of glial-specific genes [141], and initiates gliogenesis under the control of Sox9 [142]. Notch induces NfIa to drive gliogenesis via Hes genes [141]. A single NfI gene is present in Amphioxus, C. elegans, and Drosophila [138, 143, 144]], but in these animals, NfI does not appear to have a gliogenic function.
SRY-box transcription factor (Sox)
SoxE is a group of genes belonging to a high mobility group (HMG)-box Sox family, which serves various functions, including nervous system development. In mammals, SoxE genes including Sox8, Sox9, and Sox10 are essential for glia generation [145], among which, Sox9 is a major neuron-glia switch, as it directly regulates expression of NfIa, is indispensable for astrogenesis, and prevents neurogenesis [90, 91, 142]. Sox8 and Sox10 contribute to oligodendrogenesis [145]. Sox10 is induced by Olig2 and interacts with Olig1, driving expression of myelin genes and suppressing expression of astrocyte-related genes [91]. SoxE genes also have gliogenic functions in jawless vertebrates (lampreys) [55]. Even though these animals lack oligodendrocytes, they possess the genetic regulatory network required for oligodendrogenesis, including SoxE and Olig orthologs. Gliogenic functions of SoxE genes have not been reported in protostomes. In crustaceans, SoxE orthologs regulate gonad and embryo development [146, 147]. In Drosophila an ortholog of vertebrate Sox8,9,10 is expressed in the gut and gonads, and is required for intestinal epithelium function [148, 149]. The gliogenic function of class E Sox genes seems specific only in chordates.
Erythroblast transformation specific (Ets) family
Ets proteins belonging to the group A bHLH TF family, regulate various developmental processes, including glial cell differentiation. In Drosophila, an Ets TF, Pointed (Pnt), is activated by Gcm via Repo to induce expression of glial markers in several glial cell types [88, 150, 151]. Vertebrate homologs of Pnt, Ets-1 and Ets-2, drive radial glia formation in Xenopus [152]. In mammals, two Ets family TFs (Ets1 and Fli1) are expressed mainly in astrocytes and oligodendrocytes [153]. Other Ets family members drive gliogenesis in rodent peripheral nervous systems and promote oligodendrocyte proliferation [154, 155]. Therefore, the gliogenic nature of Ets is common to both protostomes and deuterostomes, necessitating further investigation regarding its conserved function in non-bilaterians.
Extracellular signaling pathways regulating gliogenesis: In addition to intrinsic factors, extracellular cues influence cell fate acquisition. Various signaling pathways have been implied in bilaterian gliogenesis [156]. As already mentioned, Notch-Delta signaling is a major, versatile pathway in bilaterian gliogenesis [102, 103]. The Notch function of maintaining a pool of stem cells in the nervous system in the form of either glio-neuro-precursors or mature glial cells seems conserved among bilaterians. Other signaling pathways driving gliogenesis in vertebrates include JAK-STAT [157–159], which talks with Notch-Delta to drive glial differentiation [160], BMP signaling [161, 162], and Hedgehog [25, 156]. Control of glial cell differentiation by these signal pathways is well known in vertebrates. Considering the presence of these signal genes in early-branching animals, the function of these signals in pre-bilateral animals should be an interesting subject of study.
Repertoires of “gliogenic” genes in non-bilaterians
Extant lineages of non-bilaterian animals include Placozoa, Porifera (sponges), Ctenophora (comb jellies), and Cnidaria, of which Ctenophora and Cnidaria possess nervous systems (Fig. 1). While Cnidaria is clearly the closest sister group to Bilateria, phylogenetic relationships among other non-bilaterians are still debated [3, 163, 164]. Recently, these phyla have been actively studied, particularly in the context of nervous system evolution. Thanks to advancements in sequencing and molecular techniques, it has been possible to address questions regarding nervous systems of ancestral metazoans. Prior to investigating gliogenic program conservation in non-bilaterians, it is necessary to characterize nervous systems of these animals, as discussed below.
Nervous system features of non-bilaterians
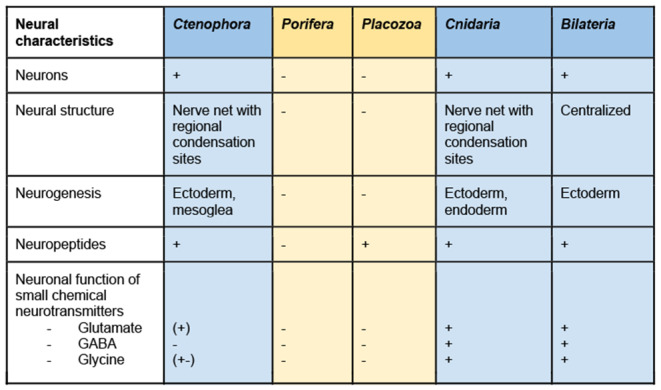
Placozoans and poriferans do not possess neurons, but their genomes contain pro-neural TFs and encode proteins required for synapse formation and neurotransmitter synthesis [165, 166]. Therefore, studies focused on unraveling functions of neuro-associated genes in these animals are expected to shed light on evolution of the nervous system. In addition, nerveless Placozoa and Porifera display epithelial contractile responses to neurotransmitters, such as short peptides, glutamate, GABA, and glycine [167–170]. The first nervous system probably heavily relied on peptidergic signaling, as evidenced by an extensive repertoire of neuropeptides in non-bilaterians with nervous systems, cnidarians and ctenophores [171, 172]. Cnidaria and Ctenophora both possess neurons organized into nerve nets with regional compartmentalization (Table 1). Phylogenomic analyses place Cnidaria as a sister group to all Bilateria, whereas Ctenophora could be one of the earliest-branching lineages of Metazoa. Recent molecular and structural studies on Ctenophora have revealed that they have, at least in part, unique neural characteristics, acquired independently from other metazoans [173, 174]. On the other hand, conserved neurogenic TFs (SoxB, bHLH) and vesicle secretion exist in Bilateria, Cnidaria and Ctenophora. Although it is still unclear whether chemical neurotransmitters are recruited in ctenophoran nervous systems. Current evidence suggests that glutamate and glycine, but not GABA, are involved in muscle contraction [175, 176]. Unlike ctenophores, cnidarians share all key features of bilaterian nervous systems, including a diverse repertoire of gene orthologs involved in neurogenesis and neural functions [177–179]. Cnidarian nervous systems generally consist of a nerve net and regional condensations in the oral (“nerve ring”) and aboral regions. Although cnidarian nervous systems are rich in neuropeptides, classical chemical neurotransmitters such as nitric oxide (NO), glutamate, GABA, glycine are also involved in neural functions (Table 1). Small transmitters, including glutamate, can perform both non-neuronal and neuronal functions, as may be the case in Cnidaria. Regardless, glutamate and glycine may have been recruited by neurons as neurotransmitters at some point in evolution [175]. More functional studies are required to understand whether this occurred with emergence of the first neurons, however. It is also debated whether acetylcholine and monoamines function as neurotransmitters in cnidarians, since a complete gene set of canonical pathways for synthesis of these molecules is absent [175, 180].
Table 1.
Neuronal features in non-bilaterians. Nerveless phyla are highlighted in yellow. Phyla possessing neurons are highlighted in blue
In summary, neuronal genes and modules can be found even in non-bilaterians without nervous systems, as described above. This suggests that many “neural” genes already existed before emergence of the nervous system, and that they acquired neural functions with emergence of the nervous system. Similarly, glial genes and modules may be present in lineages without glia. An interesting matter is to what extent the bilaterian gliogenic program is conserved in non-bilaterians. Glial and neuronal developmental programs are tightly intertwined, which complicates identification of strictly glial genes. Nevertheless, as discussed, several specific glial TFs and effector genes are present throughout Bilateria.
In this part of the review, we used comparative sequence analysis and the literature to survey orthologs of bilaterian glial TFs in non-bilaterians. It is common not to find specific orthologs for bilaterian genes in early-branching lineages. Orthologous family members should be analyzed instead. Functions of these non-bilaterian TFs can be assumed from their sequence similarities to bilaterian orthologs, but expression patterns and effector genes must be considered for accurate functional assessment. Therefore, in addition to sequence similarities, we explored expression patterns of some of these glial TF orthologs.
Glial transcription factors and signaling pathways in non-bilaterians
NfI
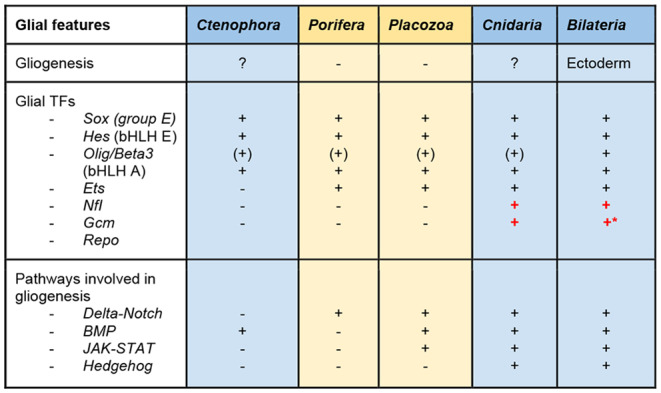
TFs belonging to the NfI family have a gliogenic function in vertebrates. As discussed before, a single member of the NfI family is present in invertebrates and does not seem to contribute to gliogenesis. Similarly, in the cnidarian, Nematostella vectensis, a single NfIx-like gene was identified (Table 2). It is expressed in the central region at blastula stage and is thought to be involved in endomesoderm specification [181]. It is unclear if the NfIx-like gene in Nematostella is expressed beyond this developmental stage and whether it has other functions. A single NfI-like gene was also identified in Porifera and Placozoa (Table 2). To date, no expression pattern of this gene has been reported.
Table 2.
Glial genes in metazoans. Nerveless phyla are highlighted in yellow. Phyla possessing neurons are highlighted in blue. Transcription factors that seem to be related to neural sophistication are shown in red. Asterisk: The Repo gene is present in some insects, but not in other bilaterians belonging to Lophotrochozoa and Deuterostomia
SoxE
Certain members of the SoxE group drive gliogenesis in bilaterians. SoxE genes can be found in genomes of non-bilaterians [182–184] (Table 2). In sponges, SoxE is expressed in choanocytes - flagellum-containing cells filtering particles out of the water [185]. Endodermal expression of two SoxE genes occurs in both Ctenophora [182–184] and Cnidaria [186–188]. The function of SoxE genes in these early-branching metazoans still remains to be explored, their broad expression pattern in endoderm does not suggest their involvement in development of nervous systems.
Group A and E bHLH genes
Well-known bilaterian gliogenic TFs, Olig and Hes, belong to Groups A and E of the bHLH family, respectively. Several members of Group A (including Atonal) and a single member of group E (Hes/Hey) were identified in the Porifera [189, 190] (Table 2). Poriferan Atonal is expressed in putative sensory cells and had strong proneural activity when it was over-expressed in Xenopus [191]. Oligo/Beta3-like and Hes ortholog were also identified in the Placozoa [192]. Three Hes genes are present in Ctenophora genomes (Table 2). Therefore, it is assumed that bHLH Groups A and E emerged during the very early phase of metazoan evolution. However, a significant expansion of these genes occurred in the Cnidaria. Thirty Group A genes and eleven Hes copies are present in the genome of a sea anemone, Nematostella. Nematostella has two Olig-like genes and one of them is expressed in the oral region of endoderm [193, 194]. Expression of Hes genes varies [186, 195]. Interestingly, one Hes-like gene (Nvhl3) is strongly expressed in a subset of cells, which is reminiscent of the Gcm expression pattern (see below).
Ets family
Both vertebrate (Ets1, Ets2) and invertebrate (Pointed) genes belonging to the Pointed Ets group, which contain an N-terminal Pointed domain, have a gliogenic function [152, 196]. In Nematostella, expression and a gene regulatory network (GRN) of a Pointed-containing Ets gene (NvERG) has been reported [197, 198]. NvErg GRN includes, but is not limited to nervous system components, such as SoxB, neuropeptides, and other members of the Ets family. Among members of an apical pole GRN of NvErg are orthologs of bilaterian glial TFs, Hes, SoxE genes, and Gcm. 12 Ets genes were identified in Nematostella, but expression and functions of most of them are still unknown.
Gcm and Repo
Gcm is a master regulator of gliogenesis in Drosophila, which also shows gliogenic potential in vertebrates. Our phylogenetic analysis of Gcm revealed that among non-bilaterians, the Gcm domain is highly conserved in the Cnidaria (Fig. 7, Table S2). Repo, another important TF driving gliogenesis under regulation of Gcm in Drosophila, is also conserved in the Cnidaria. In Nematostella, Gcm is expressed in a subset of cells in the ectoderm at early gastrula stage, and then expands to both endoderm and ectoderm [193, 194]. Repo is expressed in the oral region of ectoderm and may participate in specification of the oral nerve ring [193, 194]. Therefore, the two TFs seem to be expressed in different cells, which is unlike Drosophila Gcm.
Fig. 7.
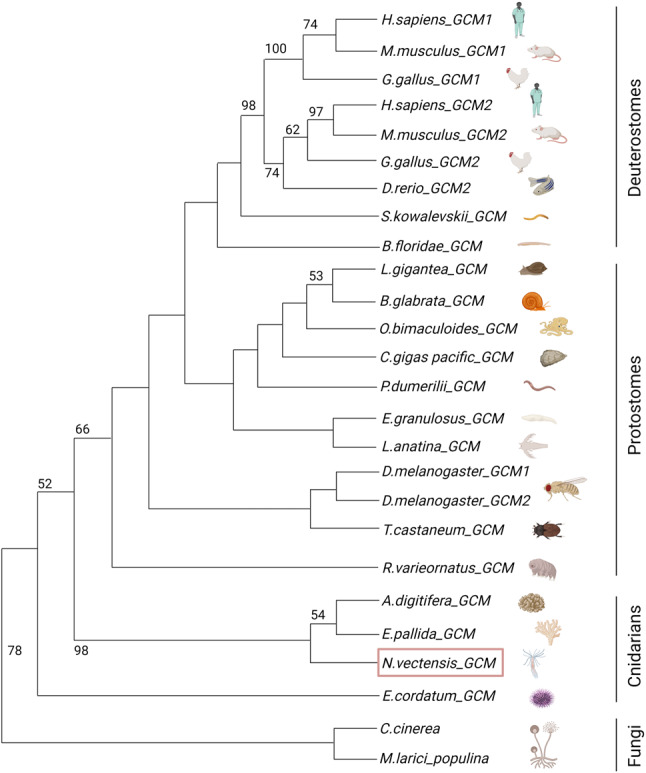
Phylogeny of Gcm in metazoans. Among non-bilaterians, Gcm is conserved only in the Cnidaria. Support values > 50 are shown at basal nodes. Nematostella Gcm is framed in red. Species screened for Gcm are listed in Supplementary Table 2. Bidirectional BLAST [199, 200] searches using the Drosophila melanogaster Gcm1 protein sequence were performed against databases of metazoan organisms. Fungal protein sequences with the highest similarity to animal Gcm were used as an outgroup. Sequences were aligned using MUSCLE (Mega7) and trimmed by eye, to include only the domain. The tree was constructed with the maximum likelihood (ML) method using PhyML (SeaView). Bootstrap support is based on 2000 replicates
In summary, orthologs of most families containing gliogenic TFs in bilaterians are also present in non-bilaterians (Table 2). However, among non-bilaterians a full complement of bilaterian gliogenic TFs are present only in the Cnidaria. Future research focusing on these TF gene regulatory networks and characterizing cells expressing orthologs of bilaterian glial markers in cnidarians should shed light on glial evolutionary roots.
Notch-Delta
This pathway is composed of the Notch receptor and its ligands (Delta/Jagged). Most Notch-Delta components are present in Porifera, Placozoa, and Cnidaria (Table 2). This pathway seems functional in all non-bilaterians except the Ctenophora, since the latter does not have Delta/Jagged [201]. In sponges, this pathway seems to be involved in sensory cell differentiation [191]. In Nematostella, Notch signaling regulates neural progenitors and restricts neurogenesis [195, 202, 203]. The function of cnidarian Notch is thus reminiscent of bilaterian Notch, which acts by repressing neuronal genes. In both Nematostella and Hydra, Notch-Delta is known to regulate development of nematocytes, Cnidaria-specific neurosensory cells.
Hedgehog
True Hedgehog (hh) genes containing both ‘hedge’ and ‘hog’ domains are absent in non-bilaterians, except the Cnidaria [204] (Table 2). Hog-domain proteins have been identified throughout the Metazoa. In Nematostella, Hedgehog gene expression analysis shows that true hh genes participate in gut formation, and hh-related genes are involved in neuronal development [205]. A more recent study demonstrated that germ cell development is dependent on Hedgehog in Nematostella [206].
Jak-Stat
The Jak-Stat pathway is composed of several proteins comprising specific domains, which together assemble into a functional system driving transcriptional responses to specific extracellular signals [207]. Non-bilaterian metazoans possess most Jak-Stat components (Table 2). The Ctenophora has the fewest conserved proteins, while the Cnidaria and Placozoa only lack one functional unit exclusively present in bilaterians [208]. There are no functional studies of Jak-Stat pathway in non-bilaterians.
Bmp family
Bmps belong to the Tgf-b superfamily and are involved in several aspects of neural development, including glial cell differentiation [209, 210]. All early-branching metazoans, except for the Porifera, have Bmp-like genes [211] (Table 2). In Cnidaria, Bmp signaling is involved in oral nervous system formation [194]. Given that Bmp genes are expressed in a neuron-rich aboral region of ctenophores, they may contribute to nervous system development in these animals as well [211].
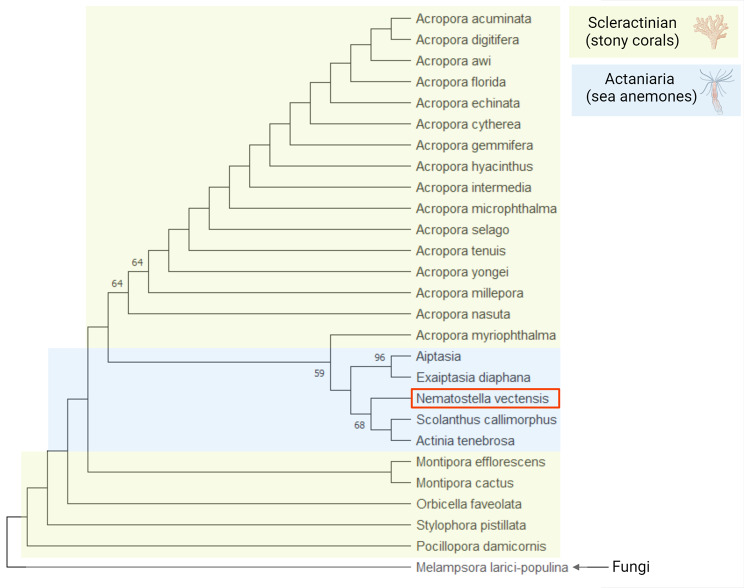
In summary, in addition to gliogenic TF conservation, the Cnidaria is the only non-bilaterian phylum characterized by conservation of all signaling pathways required for glial development in bilaterians (Table 2). Moreover, many of these show a conserved function of driving development of the nervous system. Most components of these pathways are present in the other three non-bilaterian phyla, but their functional description is still limited. The Cnidaria also possesses all functional glial genes, including GABA and glutamate transporters and enzymes required for their synthesis, glucose transporters, TRPM ion channels, aquaporins, etc [178, 212]. Thus, a complete set of neuronal and glial genes in the Cnidaria substantiates the possibility of simultaneous evolution of both cell types, as argued by Rey et al., 2020 [9]. Cnidarians are believed to lack glial cells based on morphological assessment of their representatives performed in the 1960s [213]. Horridge and Mackay performed electron microscopic analysis of ectodermal tissue of two cnidarians (jellyfish) and did not observe any cells ensheathing axons or associated with neurons otherwise. In contrast, our phylum-wide genome-wide analysis of bilaterian glial TF conservation in non-bilaterians confirmed that cnidarians have a complete genetic toolkit to drive gliogenesis (Table 2). Therefore, it is imperative to explore this matter in more detail. It is conceivable that if a cnidarian glia population exists, it may consist of just a few cells associated with a particular group of neurons. In addition, the Cnidaria consists of two clades, Anthozoa (sea anemone, coral) and Medusozoa (jellyfish) (Fig. 1), which differ dramatically, not only in their body shape and life cycle, but also genetic composition [214]. Incidentally, Cnidaria-specific phylogenetic analysis of Gcm revealed that it is highly conserved only in anthozoans (Fig. 8, Table S3). We could not identify Gcm orthologs in Aurelia aurita (scyphozoan), Hydra viridissima (hydrozoan), or Morbakka virulenta (cubozoan). Apparently, Gcm was lost in the Medusozoa, which is consistent with data showing that Hydra has lost more transcription factor families than Nematostella [215]. The starlet sea anemone, Nematostella vectensis, has become an intensely used cnidarian model. This animal is easy to culture in the lab. Its genome, transcriptome, and single-cell transcriptome are available. Numerous gene function manipulation techniques have been developed and transgenic lines of Nematostella have been established [178, 212, 216–218]. Accumulating data on neurogenesis, nervous system development and functioning, as well as neuronal type diversity in Nematostella make it possible to investigate gliogenic program conservation in this animal. Although no distinct glial cell clusters, or glial transcriptome signatures, have been reported in Nematostella [178], further analysis is required to verify the presence of glial function in pre-bilaterian animals. In Nematostella, a Gcm ortholog is expressed in a subset of cells during development [193]. Our recent studies in Nematostella demonstrated that Gcm controls expression of Eaat1, an astrocyte marker in bilaterians, which is expressed in supposedly neural cells with non-typical morphology [219]. Future experiments including electrophysiology, loss of function, and reporter lines should clarify genetic and physiological features of these cells.
Fig. 8.
Phylogeny of Gcm in Anthozoa (Cnidaria). This domain is highly conserved in stony corals and sea anemones. Support values > 50 are shown at basal nodes. Nematostella Gcm is framed in red. Cnidarian species screened for Gcm are listed in Supplementary Table 3. Bidirectional BLAST [199, 200] searches using Drosophila melanogaster Gcm1 protein sequence were performed using databases of metazoan organisms. Fungal protein sequences with the highest similarity to animal Gcm were used as an outgroup. Sequences were aligned using MUSCLE (Mega7) and trimmed by eye to include only the domain. The tree was constructed with the maximum likelihood (ML) method using PhyML (SeaView). Bootstrap support is based on 2000 replicates
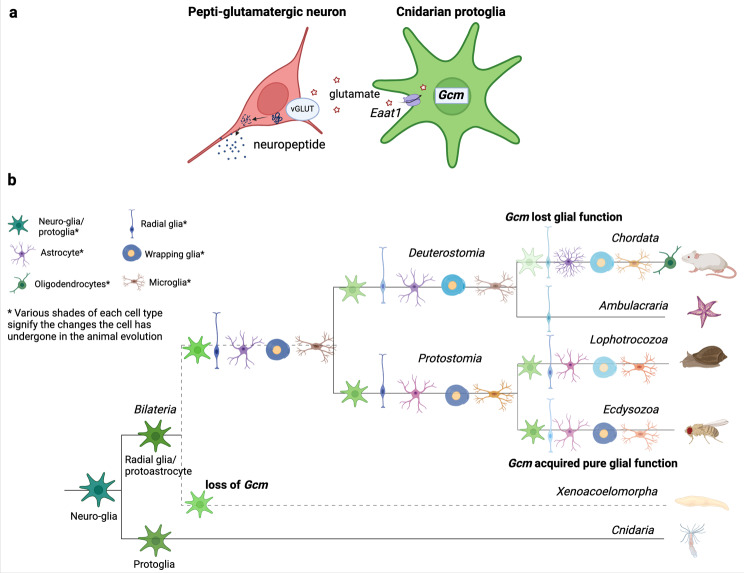
The Nematostella Gcm/Eaat1-expressing cells could represent “protoglia”, which has an important function of glutamate quenching for pepti-glutamatergic neurons (Fig. 9a). These cells could have complementarily combined the features of both glia and neurons and diversified into the distinct cell types later in evolution. The protoglial ancestral cell type had a specific molecular signature that can be traced in Nematostella and is conserved to a various degree in different bilaterian lineages. With functional segregation the Gcm-controlled program either maintained its glial regulation, which is the case in Drosophila, or was significantly modified to have only a potential to induce glial markers as is the case in vertebrates (Fig. 9b). The program kept being modified so that it is almost impossible to recognize it in the most advanced species. This explains the absence of obvious homologous glial cell types in Nematostella. Instead Gcm/Eaat1-expressing cells are a subset of neurons possessing glial features. Metabolic support of neurons, i.e. glutamate recycling, could have been the primary glial function that gave rise to a distinct cell type and separated glia from neurons.
Fig. 9.
Protoglia function and evolutionary development. (a) Schematic representation of cnidarian porotoglia and its interaction with neurons that use both glutamate and neuropeptides as neurotransmitters. (b) Evolutionary tree of glia based on the analysis of the gliogenic program conservation described in this review. The diversification of protoglia into various glial cell types with specialized functions is shown
Conclusions
In recent years, non-bilaterian animals have been extensively studied particularly in the context of neuronal development thanks to rapid development of ‘omics’ tools. However, glial program conservation has not been investigated in these animals. In this review, a comparative analysis shows that all constituents of the bilaterian gliogenic program are conserved in the Anthozoa (Cnidaria). A representative of this group, Nematostella vectensis, could be a useful model system to investigate functions of genes driving gliogenesis in bilaterians and to answer important questions about primordial glia. Until now, no cell clusters with a glial transcriptome signature have been identified in Nematostella. This is not surprising, given that no universal glial markers or unified glial genetic signatures are present among bilaterians. Instead, an organism-wide search of glial orthologs and clarification of their functions might reveal novel cell types in the sea anemone. Future studies should clarify functions of conserved bilaterian glial TFs and functional genes not only in Cnidaria, but also other non-bilaterians. This is paramount to reconstruct a more accurate picture of glial evolution.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Steven D. Aird for editing and proof-reading the manuscript.
Abbreviations
- Aldh1l1
Aldehyde dehydrogenase 1 family member L1
- Aqp4
Aquaporin channel 4
- BBB
Blood-brain barrier
- bHLH
Basic helix-loop-helix
- BMP
Bone morphogenetic protein
- Ced
Cell death abnormality
- CNS
Central nervous system
- Eaat
Excitatory amino acid transporter
- ECM
Extracellular matrix
- Ef1a
Elongation factor 1-alpha
- Est
Erythroblast transformation specific
- GABA
Gamma-Aminobutyric acid
- Gapdh
Glyceraldehyde 3-phosphate dehydrogenase
- Gat
GABA transporter
- Gcm
Glial cells missing
- GDNF
Glial-derived neurotrophic factor
- Gfap
Glial fibrillary acidic protein
- Glut
Glucose transporter
- GPCR
G protein-coupled receptor
- GS
Glutamine synthetase
- Hes
Hairy and enhancer of split
- Jak-Stat
Janus kinase-signal transducer and activator of transcription
- MMP
Matrix metalloproteinase
- NfI
Nuclear factor I
- NO
Nitric oxide
- NS
Nervous system
- Nv
Nematostella vectensis
- OPC
Oligodendrocyte precursor cell
- Olig
oligodendrocyte transcription factor
- PDGFR
platelet-derived growth factor receptor alpha
- PNS
peripheral nervous system
- Repo
reversed polarity
- RGC
radial glial cell
- TF
transcription factor
Author contributions
LS was a major contributor in writing the manuscript. HW contributed to conceptualization and structuring of the manuscript. Both authors read and approved the final manuscript.
Funding
This study was supported by Okinawa Institute of Science and Technology Graduate University.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hartline DK. The evolutionary origins of glia. Glia. 2011;59:1215–36. doi: 10.1002/glia.21149. [DOI] [PubMed] [Google Scholar]
- 2.Verkhratsky A, Ho MS, Parpura V. Evolution of Neuroglia. In: Verkhratsky A, Ho MS, Zorec R, Parpura V, editors. Neuroglia in neurodegenerative diseases. Singapore: Springer Singapore; 2019. pp. 15–44. [Google Scholar]
- 3.Kapli P, Telford MJ. Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci Adv. 2020;6. 10.1126/sciadv.abc5162. [DOI] [PMC free article] [PubMed]
- 4.Araque A, Navarrete M. Glial cells in neuronal network function. Philos Trans R Soc Lond B Biol Sci. 2010;365:2375–81. doi: 10.1098/rstb.2009.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimou L, Götz M. Glial cells as progenitors and stem cells: new roles in the healthy and diseased brain. Physiol Rev. 2014;94:709–37. doi: 10.1152/physrev.00036.2013. [DOI] [PubMed] [Google Scholar]
- 6.Verkhratsky A, Butt A, Neuroglia. Definition, classification, evolution, numbers, Development. Glial physiology and pathophysiology. John Wiley & Sons, Ltd, UK; 2013. pp. 73–104.
- 7.Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol. 2010;20:588–94. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Freeman MR, Rowitch DH. Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron. 2013;80:613–23. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rey S, Zalc B, Klämbt C. Evolution of glial wrapping: a new hypothesis. Dev Neurobiol. 2020 doi: 10.1002/dneu.22739. [DOI] [PubMed] [Google Scholar]
- 10.Verkhratsky A, Arranz AM, Ciuba K, Pękowska A. Evolution of neuroglia. Ann N Y Acad Sci. 2022;1518:120–30. doi: 10.1111/nyas.14917. [DOI] [PubMed] [Google Scholar]
- 11.Shaham S. Glia–neuron interactions in nervous system function and development. Curr Top Dev Biol; 2005. [DOI] [PubMed]
- 12.Radojcic T, Pentreath VW. Invertebrate glia. Prog Neurobiol. 1979;12:115–79. doi: 10.1016/0301-0082(79)90002-9. [DOI] [PubMed] [Google Scholar]
- 13.Sancho L, Contreras M, Allen NJ. Glia as sculptors of synaptic plasticity. Neurosci Res. 2020 doi: 10.1016/j.neures.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michinaga S, Koyama Y. Dual roles of astrocyte-derived factors in regulation of blood-brain barrier function after brain damage. Int J Mol Sci. 2019;20. 10.3390/ijms20030571. [DOI] [PMC free article] [PubMed]
- 15.Gebara E, Bonaguidi MA, Beckervordersandforth R, Sultan S, Udry F, Gijs P-J, et al. Heterogeneity of Radial Glia-Like cells in the adult Hippocampus. Stem Cells. 2016;34:997–1010. doi: 10.1002/stem.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oikonomou G, Shaham S. The glia of Caenorhabditis elegans. Glia. 2011;59:1253–63. doi: 10.1002/glia.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkhratsky A, Nedergaard M. Astroglial cradle in the life of the synapse. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130595. doi: 10.1098/rstb.2013.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perea G, Sur M, Araque A. Neuron-glia networks: integral gear of brain function. Front Cell Neurosci. 2014;8:378. doi: 10.3389/fncel.2014.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trotter J, Karram K, Nishiyama A. NG2 cells: Properties, progeny and origin. Brain Res Rev. 2010;63:72–82. doi: 10.1016/j.brainresrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suess U, Pliska V. Identification of the pituicytes as astroglial cells by indirect immunofluorescence-staining for the glial fibrillary acidic protein. Brain Res. 1981;221:27–33. doi: 10.1016/0006-8993(81)91061-1. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez E, Guerra M, Peruzzo B, Blázquez JL. Tanycytes: a rich morphological history to underpin future molecular and physiological investigations. J Neuroendocrinol. 2019;31:e12690. doi: 10.1111/jne.12690. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Leshkowitz D, Blechman J, Levkowitz G. Single-cell Molecular and Cellular Architecture of the mouse Neurohypophysis. eNeuro. 2020;7. 10.1523/ENEURO.0345-19.2019. [DOI] [PMC free article] [PubMed]
- 23.Yildirim K, Petri J, Kottmeier R, Klämbt C. Drosophila glia: few cell types and many conserved functions. Glia. 2019;67:5–26. doi: 10.1002/glia.23459. [DOI] [PubMed] [Google Scholar]
- 24.Mashanov V, Zueva O. Radial glia in echinoderms. Dev Neurobiol. 2019;79:396–405. doi: 10.1002/dneu.22659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang IE, Lapan SW, Scimone ML, Clandinin TR, Reddien PW. Hedgehog signaling regulates gene expression in planarian glia. Elife 2016;5. 10.7554/eLife.16996. [DOI] [PMC free article] [PubMed]
- 26.Campbell K, Götz M. Radial glia: multi-purpose cells for vertebrate brain development. Trends Neurosci. 2002;25:235–8. doi: 10.1016/S0166-2236(02)02156-2. [DOI] [PubMed] [Google Scholar]
- 27.Helm Conrad K, Anett B, Patrick K-S, Sabrina U, Elke K, Ioannis et al. Early evolution of radial glial cells in Bilateria. Proceedings of the Royal Society B: Biological Sciences. 2017;284:20170743. [DOI] [PMC free article] [PubMed]
- 28.Jurisch-Yaksi N, Yaksi E, Kizil C. Radial glia in the zebrafish brain: functional, structural, and physiological comparison with the mammalian glia. Glia. 2020;68:2451–70. doi: 10.1002/glia.23849. [DOI] [PubMed] [Google Scholar]
- 29.Mashanov VS, Zueva OR, Heinzeller T. Regeneration of the radial nerve cord in a holothurian: a promising new model system for studying post-traumatic recovery in the adult nervous system. Tissue Cell. 2008;40:351–72. doi: 10.1016/j.tice.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Boddum K, Jensen TP, Magloire V, Kristiansen U, Rusakov DA, Pavlov I, et al. Astrocytic GABA transporter activity modulates excitatory neurotransmission. Nat Commun. 2016;7:13572. doi: 10.1038/ncomms13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimelberg HK, Nedergaard M. Functions of astrocytes and their potential as therapeutic targets. Neurotherapeutics. 2010;7:338–53. doi: 10.1016/j.nurt.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin B-J, Zhang H, Binder DK, Verkman AS. Aquaporin-4-dependent K(+) and water transport modeled in brain extracellular space following neuroexcitation. J Gen Physiol. 2013;141:119–32. doi: 10.1085/jgp.201210883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown AM, Ransom BR. Astrocyte glycogen and brain energy metabolism. Glia. 2007;55:1263–71. doi: 10.1002/glia.20557. [DOI] [PubMed] [Google Scholar]
- 34.Limmer S, Weiler A, Volkenhoff A, Babatz F, Klämbt C. The Drosophila blood-brain barrier: development and function of a glial endothelium. Front Neurosci. 2014;8:365. doi: 10.3389/fnins.2014.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin KT, Eroglu C. Molecular mechanisms of astrocyte-induced synaptogenesis. Curr Opin Neurobiol. 2017;45:113–20. doi: 10.1016/j.conb.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joosten EA, Gribnau AA. Astrocytes and guidance of outgrowing corticospinal tract axons in the rat. An immunocytochemical study using anti-vimentin and anti-glial fibrillary acidic protein. Neuroscience. 1989;31:439–52. doi: 10.1016/0306-4522(89)90386-2. [DOI] [PubMed] [Google Scholar]
- 37.Morizawa YM, Hirayama Y, Ohno N, Shibata S, Shigetomi E, Sui Y, et al. Reactive astrocytes function as phagocytes after brain ischemia via ABCA1-mediated pathway. Nat Commun. 2017;8:28. doi: 10.1038/s41467-017-00037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.al-Ali SY, al-Hussain SM. An ultrastructural study of the phagocytic activity of astrocytes in adult rat brain. J Anat. 1996;188(Pt 2):257–62. [PMC free article] [PubMed] [Google Scholar]
- 39.Shigetomi E, Tong X, Kwan KY, Corey DP, Khakh BS. TRPA1 channels regulate astrocyte resting calcium and inhibitory synapse efficacy through GAT-3. Nat Neurosci. 2011;15:70–80. doi: 10.1038/nn.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen ML, Khakh BS, Skatchkov SN, Zhou M, Lee CJ, Rouach N. New insights on Astrocyte Ion channels: critical for Homeostasis and Neuron-Glia Signaling. J Neurosci. 2015;35:13827–35. doi: 10.1523/JNEUROSCI.2603-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang J, Takeuchi H, Doi Y, Kawanokuchi J, Sonobe Y, Jin S, et al. Excitatory amino acid transporter expression by astrocytes is neuroprotective against microglial excitotoxicity. Brain Res. 2008;1210:11–9. doi: 10.1016/j.brainres.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Ghirardini E, Wadle SL, Augustin V, Becker J, Brill S, Hammerich J, et al. Expression of functional inhibitory neurotransmitter transporters GlyT1, GAT-1, and GAT-3 by astrocytes of inferior colliculus and hippocampus. Mol Brain. 2018;11:4. doi: 10.1186/s13041-018-0346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubbard JA, Szu JI, Binder DK. The role of aquaporin-4 in synaptic plasticity, memory and disease. Brain Res Bull. 2018;136:118–29. doi: 10.1016/j.brainresbull.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Morgello S, Uson RR, Schwartz EJ, Haber RS. The human blood-brain barrier glucose transporter (GLUT1) is a glucose transporter of gray matter astrocytes. Glia. 1995;14:43–54. doi: 10.1002/glia.440140107. [DOI] [PubMed] [Google Scholar]
- 45.Frei JA, Stoeckli ET. SynCAMs extend their functions beyond the synapse. Eur J Neurosci. 2014;39:1752–60. doi: 10.1111/ejn.12544. [DOI] [PubMed] [Google Scholar]
- 46.Christopherson KS, Ullian EM, Stokes CCA, Mullowney CE, Hell JW, Agah A, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–33. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melzer L, Freiman TM, Derouiche A. Rab6A as a pan-astrocytic marker in mouse and human brain, and comparison with other glial markers (GFAP, GS, Aldh1L1, SOX9). Cells 2021;10. 10.3390/cells10010072. [DOI] [PMC free article] [PubMed]
- 49.Molofsky AV, Glasgow SM, Chaboub LS, Tsai H-H, Murnen AT, Kelley KW, et al. Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia. 2013;61:1518–32. doi: 10.1002/glia.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, et al. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci. 2009;29:11399–408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailly X, Laguerre L, Correc G, Dupont S, Kurth T, Pfannkuchen A, et al. The chimerical and multifaceted marine acoel Symsagittifera roscoffensis: from photosymbiosis to brain regeneration. Front Microbiol. 2014;5:498. doi: 10.3389/fmicb.2014.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bery A, Cardona A, Martinez P, Hartenstein V. Structure of the central nervous system of a juvenile acoel, Symsagittifera roscoffensis. Dev Genes Evol. 2010;220:61–76. doi: 10.1007/s00427-010-0328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scolding NJ, Frith S, Linington C, Morgan BP, Campbell AK, Compston DA. Myelin-oligodendrocyte glycoprotein (MOG) is a surface marker of oligodendrocyte maturation. J Neuroimmunol. 1989;22:169–76. doi: 10.1016/0165-5728(89)90014-3. [DOI] [PubMed] [Google Scholar]
- 54.Hai M, Muja N, DeVries GH, Quarles RH, Patel PI. Comparative analysis of Schwann cell lines as model systems for myelin gene transcription studies. J Neurosci Res. 2002;69:497–508. doi: 10.1002/jnr.10327. [DOI] [PubMed] [Google Scholar]
- 55.Yuan T, York JR, McCauley DW. Gliogenesis in lampreys shares gene regulatory interactions with oligodendrocyte development in jawed vertebrates. Dev Biol. 2018;441:176–90. doi: 10.1016/j.ydbio.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Nave K-A, Werner HB. Ensheathment and myelination of axons: evolution of glial functions. Annu Rev Neurosci. 2021;44:197–219. doi: 10.1146/annurev-neuro-100120-122621. [DOI] [PubMed] [Google Scholar]
- 57.Seifert G, Steinhäuser C. Heterogeneity and function of hippocampal macroglia. Cell Tissue Res. 2018;373:653–70. doi: 10.1007/s00441-017-2746-1. [DOI] [PubMed] [Google Scholar]
- 58.Wasseff SK, Scherer SS. Cx32 and Cx47 mediate oligodendrocyte:astrocyte and oligodendrocyte:oligodendrocyte gap junction coupling. Neurobiol Dis. 2011;42:506–13. doi: 10.1016/j.nbd.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou B, Zhu Z, Ransom BR, Tong X. Oligodendrocyte lineage cells and depression. Mol Psychiatry. 2021;26:103–17. doi: 10.1038/s41380-020-00930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, et al. Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/S0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 61.Chari D. Glial Cell Development, 2nd Edition. Neuropathol Appl Neurobiol. 2002;28:85–6.
- 62.Bergles DE, Richardson WD. Oligodendrocyte Development and Plasticity. Cold Spring Harb Perspect Biol. 2015;8:a020453. doi: 10.1101/cshperspect.a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Randy F, Stout AVAVP., Jr Caenorhabditis elegans glia modulate neuronal activity and behavior. Front Cell Neurosci. 2014 doi: 10.3389/fncel.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ginhoux F, Prinz M. Origin of microglia: current concepts and past controversies. Cold Spring Harb Perspect Biol. 2015;7:a020537. doi: 10.1101/cshperspect.a020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazzolini J, Le Clerc S, Morisse G, Coulonges C, Kuil LE, van Ham TJ, et al. Gene expression profiling reveals a conserved microglia signature in larval zebrafish. Glia. 2020;68:298–315. doi: 10.1002/glia.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vainchtein ID, Molofsky AV. Astrocytes and microglia: in sickness and in Health. Trends Neurosci. 2020;43:144–54. doi: 10.1016/j.tins.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Konishi H, Okamoto T, Hara Y, Komine O, Tamada H, Maeda M, et al. Astrocytic phagocytosis is a compensatory mechanism for microglial dysfunction. EMBO J. 2020;39:e104464. doi: 10.15252/embj.2020104464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 69.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016;113:E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holtman IR, Skola D, Glass CK. Transcriptional control of microglia phenotypes in health and disease. J Clin Invest. 2017;127:3220–9. doi: 10.1172/JCI90604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, et al. Cross-species single-cell analysis reveals divergence of the Primate Microglia Program. Cell. 2019;179:1609–e2216. doi: 10.1016/j.cell.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 72.Sharma K, Bisht K, Eyo UB. A comparative biology of microglia across species. Front Cell Dev Biol. 2021;9:652748. doi: 10.3389/fcell.2021.652748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jurga AM, Paleczna M, Kuter KZ. Overview of General and discriminating markers of Differential Microglia phenotypes. Front Cell Neurosci. 2020;14:198. doi: 10.3389/fncel.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hainfellner JA, Voigtländer T, Ströbel T, Mazal PR, Maddalena AS, Aguzzi A, et al. Fibroblasts can express glial fibrillary acidic protein (GFAP) in vivo. J Neuropathol Exp Neurol. 2001;60:449–61. doi: 10.1093/jnen/60.5.449. [DOI] [PubMed] [Google Scholar]
- 75.Beckers P, Helm C, Bartolomaeus T. The anatomy and development of the nervous system in Magelonidae (Annelida) – insights into the evolution of the annelid brain. BMC Evol Biol 2019;19. [DOI] [PMC free article] [PubMed]
- 76.Dos Santos PC, Gehlen G, Faccioni-Heuser MC, Achaval M. Detection of glial fibrillary acidic protein (GFAP) and vimentin (vim) by immunoelectron microscopy of the glial cells in the central nervous system of the snail Megalobulimus Abbreviatus: GFAP and vim in glial cells of Megalobulimus. Acta Zool. 2005;86:135–44. doi: 10.1111/j.1463-6395.2005.00195.x. [DOI] [Google Scholar]
- 77.Bramanti V, Tomassoni D, Avitabile M, Amenta F, Avola R. Biomarkers of glial cell proliferation and differentiation in culture. Front Biosci. 2010;2:558–70. doi: 10.2741/s85. [DOI] [PubMed] [Google Scholar]
- 78.Cardone B, Roots BI. Comparative immunohistochemical study of glial filament proteins (glial fibrillary acidic protein and vimentin) in goldfish, octopus, and snail. Glia. 1990;3:180–92. doi: 10.1002/glia.440030305. [DOI] [PubMed] [Google Scholar]
- 79.Linser PJ, Trapido-Rosenthal HG, Orona E. Glutamine synthetase is a glial-specific marker in the olfactory regions of the lobster (Panulirus argus) nervous system. Glia. 1997;20:275–83. doi: 10.1002/(SICI)1098-1136(199708)20:4<275::AID-GLIA1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 80.Levenson J, Endo S, Kategaya LS, Fernandez RI, Brabham DG, Chin J, et al. Long-term regulation of neuronal high-affinity glutamate and glutamine uptake in Aplysia. Proc Natl Acad Sci U S A. 2000;97:12858–63. doi: 10.1073/pnas.220256497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Allodi S, Bressan CM, Carvalho SL, Cavalcante LA. Regionally specific distribution of the binding of anti-glutamine synthetase and anti‐S100 antibodies and of Datura stramonium lectin in glial domains of the optic lobe of the giant prawn. Glia. 2006;53:612–20. doi: 10.1002/glia.20317. [DOI] [PubMed] [Google Scholar]
- 82.Biserova NM, Gordeev II, Korneva JV, Salnikova MM. Structure of the glial cells in the nervous system of parasitic and free-living flatworms. Biology Bull. 2010;37:277–87. doi: 10.1134/S106235901003009X. [DOI] [PubMed] [Google Scholar]
- 83.Bozzo M, Lacalli TC, Obino V, Caicci F, Marcenaro E, Bachetti T, et al. Amphioxus neuroglia: molecular characterization and evidence for early compartmentalization of the developing nerve cord. Glia. 2021 doi: 10.1002/glia.23982. [DOI] [PubMed] [Google Scholar]
- 84.Driever W. Developmental Biology: Reissner’s Fiber and straightening of the body Axis. Curr Biol. 2018;28:R833–5. doi: 10.1016/j.cub.2018.05.080. [DOI] [PubMed] [Google Scholar]
- 85.Viehweg J, Naumann WW, Olsson R. Secretory radial glia in the ectoneural system of the sea StarAsterias rubens(echinodermata) Acta Zool. 1998;79:119–31. doi: 10.1111/j.1463-6395.1998.tb01151.x. [DOI] [Google Scholar]
- 86.Okawa N, Shimai K, Ohnishi K, Ohkura M, Nakai J, Horie T, et al. Cellular identity and Ca2 + signaling activity of the non-reproductive GnRH system in the Ciona intestinalis type A (Ciona robusta) larva. Sci Rep. 2020;10:18590. doi: 10.1038/s41598-020-75344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lockhart ST, Levitan IB, Pikielny CW. Ag, a novel protein secreted from Aplysia glia. J Neurobiol. 1996;29:35–48. doi: 10.1002/(SICI)1097-4695(199601)29:1<35::AID-NEU3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 88.Yuasa Y, Okabe M, Yoshikawa S, Tabuchi K, Xiong W-C, Hiromi Y, et al. Drosophila homeodomain protein REPO controls glial differentiation by cooperating with ETS and BTB transcription factors. Development. 2003;130:2419–28. doi: 10.1242/dev.00468. [DOI] [PubMed] [Google Scholar]
- 89.Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev. 2005;122:849–62. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 90.Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–89. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Glasgow SM, Zhu W, Stolt CC, Huang T-W, Chen F, LoTurco JJ, et al. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 2014;17:1322–9. doi: 10.1038/nn.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hosoya T, Takizawa K, Nitta K, Hotta Y. Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–36. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- 93.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–80. doi: 10.1016/S0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 94.Soustelle L, Giangrande A. Glial differentiation and the gcm pathway. Neuron Glia Biol. 2007;3:5–16. doi: 10.1017/S1740925X07000464. [DOI] [PubMed] [Google Scholar]
- 95.Kammerer M, Giangrande A. Glide2, a second glial promoting factor in Drosophila melanogaster. EMBO J. 2001;20:4664–73. doi: 10.1093/emboj/20.17.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alfonso TB, Jones BW. Gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol. 2002;248:369–83. doi: 10.1006/dbio.2002.0740. [DOI] [PubMed] [Google Scholar]
- 97.Chotard C, Leung W, Salecker I. Glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–51. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 98.Junkunlo K, Söderhäll K, Söderhäll I. A transcription factor glial cell missing (gcm) in the freshwater crayfish Pacifastacus leniusculus. Dev Comp Immunol. 2020;113:103782. doi: 10.1016/j.dci.2020.103782. [DOI] [PubMed] [Google Scholar]
- 99.Ransick A, Davidson EH. cis-regulatory processing of notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 100.Wessel GM, Kiyomoto M, Shen T-L, Yajima M. Genetic manipulation of the pigment pathway in a sea urchin reveals distinct lineage commitment prior to metamorphosis in the bilateral to radial body plan transition. Sci Rep. 2020;10:1973. doi: 10.1038/s41598-020-58584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Udolph G, Rath P, Chia W. A requirement for Notch in the genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development. 2001;128:1457–66. doi: 10.1242/dev.128.8.1457. [DOI] [PubMed] [Google Scholar]
- 102.Fischer A, Gessler M. Delta-Notch–and then? Protein interactions and proposed modes of repression by Hes and hey bHLH factors. Nucleic Acids Res. 2007;35:4583–96. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou Z-D, Kumari U, Xiao Z-C, Tan E-K. Notch as a molecular switch in neural stem cells. IUBMB Life. 2010;62:618–23. doi: 10.1002/iub.362. [DOI] [PubMed] [Google Scholar]
- 104.Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–90. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- 105.Ge W, Martinowich K, Wu X, He F, Miyamoto A, Fan G, et al. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J Neurosci Res. 2002;69:848–60. doi: 10.1002/jnr.10364. [DOI] [PubMed] [Google Scholar]
- 106.Wheeler SR, Stagg SB, Crews ST. Multiple notch signaling events control Drosophila CNS midline neurogenesis, gliogenesis and neuronal identity. Development. 2008;135:3071–9. doi: 10.1242/dev.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Monastirioti M, Giagtzoglou N, Koumbanakis KA, Zacharioudaki E, Deligiannaki M, Wech I, et al. Drosophila Hey is a target of notch in asymmetric divisions during embryonic and larval neurogenesis. Development. 2010;137:191–201. doi: 10.1242/dev.043604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van De Bor V, Giangrande A. Notch signaling represses the glial fate in fly PNS. Development. 2001;128:1381–90. doi: 10.1242/dev.128.8.1381. [DOI] [PubMed] [Google Scholar]
- 109.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 110.Kim J, Jones BW, Zock C, Chen Z, Wang H, Goodman CS, et al. Isolation and characterization of mammalian homologs of the Drosophila gene glial cells missing. Proc Natl Acad Sci U S A. 1998;95:12364–9. doi: 10.1073/pnas.95.21.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schreiber J, Riethmacher-Sonnenberg E, Riethmacher D, Tuerk EE, Enderich J, Bösl MR, et al. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol Cell Biol. 2000;20:2466–74. doi: 10.1128/MCB.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamada K, Ogawa H, Honda S-I, Harada N, Okazaki T. A GCM motif protein is involved in placenta-specific expression of human aromatase Gene*. J Biol Chem. 1999;274:32279–86. doi: 10.1074/jbc.274.45.32279. [DOI] [PubMed] [Google Scholar]
- 113.Liu Z, Yu S, Manley NR. Gcm2 is required for the differentiation and survival of parathyroid precursor cells in the parathyroid/thymus primordia. Dev Biol. 2007;305:333–46. doi: 10.1016/j.ydbio.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hitoshi S, Ishino Y, Kumar A, Jasmine S, Tanaka KF, Kondo T, et al. Mammalian gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat Neurosci. 2011;14:957–64. doi: 10.1038/nn.2875. [DOI] [PubMed] [Google Scholar]
- 115.Soustelle L, Trousse F, Jacques C, Ceron J, Cochard P, Soula C, et al. Neurogenic role of gcm transcription factors is conserved in chicken spinal cord. Development. 2007;134:625–34. doi: 10.1242/dev.02750. [DOI] [PubMed] [Google Scholar]
- 116.Iwasaki Y, Hosoya T, Takebayashi H, Ogawa Y, Hotta Y, Ikenaka K. The potential to induce glial differentiation is conserved between Drosophila and mammalian glial cells missing genes. Development. 2003;130:6027–35. doi: 10.1242/dev.00822. [DOI] [PubMed] [Google Scholar]
- 117.Saito A, Kanemoto S, Kawasaki N, Asada R, Iwamoto H, Oki M, et al. Unfolded protein response, activated by OASIS family transcription factors, promotes astrocyte differentiation. Nat Commun. 2012;3:967. doi: 10.1038/ncomms1971. [DOI] [PubMed] [Google Scholar]
- 118.Hanaoka R, Ohmori Y, Uyemura K, Hosoya T, Hotta Y, Shirao T, et al. Zebrafish gcmb is required for pharyngeal cartilage formation. Mech Dev. 2004;121:1235–47. doi: 10.1016/j.mod.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 119.Labouesse M, Hartwieg E, Horvitz HR. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. 1996;122:2579–88. doi: 10.1242/dev.122.9.2579. [DOI] [PubMed] [Google Scholar]
- 120.Umesono Y, Hiromi Y, Hotta Y. Context-dependent utilization of notch activity in Drosophila glial determination. Development. 2002;129:2391–9. doi: 10.1242/dev.129.10.2391. [DOI] [PubMed] [Google Scholar]
- 121.Trébuchet G, Cattenoz PB, Zsámboki J, Mazaud D, Siekhaus DE, Fanto M, et al. The Repo Homeodomain transcription factor suppresses hematopoiesis in Drosophila and preserves the glial fate. J Neurosci. 2019;39:238–55. doi: 10.1523/JNEUROSCI.1059-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, et al. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–32. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- 124.Dai J, Bercury KK, Ahrendsen JT, Macklin WB. Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J Neurosci. 2015;35:4386–402. doi: 10.1523/JNEUROSCI.4962-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–93. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 126.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/S0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 127.Ohayon D, Aguirrebengoa M, Escalas N, Soula C. Transcriptome profiling of the Olig2-expressing astrocyte subtype reveals their unique molecular signature. bioRxiv 2020:2020.10.15.340505. 10.1101/2020.10.15.340505. [DOI] [PMC free article] [PubMed]
- 128.Salzer JL. Schwann cell myelination. Cold Spring Harb Perspect Biol. 2015;7:a020529. doi: 10.1101/cshperspect.a020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S. mls-2 and vab-3 control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. Elegans. Development. 2008;135:2263–75. doi: 10.1242/dev.019547. [DOI] [PubMed] [Google Scholar]
- 130.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–7. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kageyama R, Ohtsuka T, Kobayashi T. The hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–51. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 132.Gazave E, Guillou A, Balavoine G. History of a prolific family: the Hes/Hey-related genes of the annelid Platynereis. Evodevo. 2014;5:29. doi: 10.1186/2041-9139-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(Suppl 1):S97–103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 134.Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–22. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- 135.de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, et al. Conservation of the notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–48. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 136.Kageyama R, Ohtsuka T. The notch-hes pathway in mammalian neural development. Cell Res. 1999;9:179–88. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- 137.Magadi SS, Voutyraki C, Anagnostopoulos G, Zacharioudaki E, Poutakidou IK, Efraimoglou C, et al. Dissecting hes-centred transcriptional networks in neural stem cell maintenance and tumorigenesis in Drosophila. Development. 2020;147. 10.1242/dev.191544. [DOI] [PubMed]
- 138.Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/S0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 139.Campbell CE, Piper M, Plachez C, Yeh Y-T, Baizer JS, Osinski JM, et al. The transcription factor nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harris L, Genovesi LA, Gronostajski RM, Wainwright BJ, Piper M. Nuclear factor one transcription factors: divergent functions in developmental versus adult stem cell populations. Dev Dyn. 2015;244:227–38. doi: 10.1002/dvdy.24182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–68. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 142.Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fletcher CF, Jenkins NA, Copeland NG, Chaudhry AZ, Gronostajski RM. Exon structure of the nuclear factor I DNA-binding domain from C. Elegans to mammals. Mamm Genome. 1999;10:390–6. doi: 10.1007/s003359901008. [DOI] [PubMed] [Google Scholar]
- 144.Lazakovitch E, Kalb JM, Matsumoto R, Hirono K, Kohara Y, Gronostajski RM. nfi-I affects behavior and life-span in C. Elegans but is not essential for DNA replication or survival. BMC Dev Biol. 2005;5:24. doi: 10.1186/1471-213X-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Reiprich S, Wegner M. From CNS stem cells to neurons and glia: Sox for everyone. Cell Tissue Res. 2015;359:111–24. doi: 10.1007/s00441-014-1909-6. [DOI] [PubMed] [Google Scholar]
- 146.Wan H, Liao J, Zhang Z, Zeng X, Liang K, Wang Y. Molecular cloning, characterization, and expression analysis of a sex-biased transcriptional factor sox9 gene of mud crab Scylla paramamosain. Gene. 2021;774:145423. doi: 10.1016/j.gene.2021.145423. [DOI] [PubMed] [Google Scholar]
- 147.Hu Y, Jin S, Fu H, Qiao H, Zhang W, Jiang S, et al. Functional analysis of a SoxE gene in the oriental freshwater prawn, Macrobrachium nipponense by molecular cloning, expression pattern analysis, and in situ hybridization (De Haan, 1849) 3 Biotech. 2020;10:10. doi: 10.1007/s13205-019-1996-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jin Z, Chen J, Huang H, Wang J, Lv J, Yu M, et al. The Drosophila Ortholog of mammalian transcription factor Sox9 regulates intestinal homeostasis and regeneration at an appropriate level. Cell Rep. 2020;31:107683. doi: 10.1016/j.celrep.2020.107683. [DOI] [PubMed] [Google Scholar]
- 149.Phochanukul N, Russell S. No backbone but lots of sox: Invertebrate Sox genes. Int J Biochem Cell Biol. 2010;42:453–64. doi: 10.1016/j.biocel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 150.Klämbt C. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development. 1993;117:163–76. doi: 10.1242/dev.117.1.163. [DOI] [PubMed] [Google Scholar]
- 151.Peco E, Davla S, Camp D, Stacey SM, Landgraf M, van Meyel DJ. Drosophila astrocytes cover specific territories of the CNS neuropil and are instructed to differentiate by Prospero, a key effector of Notch. Development. 2016;143:1170–81. doi: 10.1242/dev.133165. [DOI] [PubMed] [Google Scholar]
- 152.Kiyota T, Kato A, Kato Y. Ets-1 regulates radial glia formation during vertebrate embryogenesis. Organogenesis. 2007;3:93–101. doi: 10.4161/org.3.2.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tabula Muris Consortium. Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018;562:367–72. [DOI] [PMC free article] [PubMed]
- 154.Hagedorn L, Paratore C, Brugnoli G, Baert JL, Mercader N, Suter U, et al. The ets domain transcription factor Erm distinguishes rat satellite glia from Schwann cells and is regulated in satellite cells by neuregulin signaling. Dev Biol. 2000;219:44–58. doi: 10.1006/dbio.1999.9595. [DOI] [PubMed] [Google Scholar]
- 155.Ahmad ST, Rogers AD, Chen MJ, Dixit R, Adnani L, Frankiw LS, et al. Capicua regulates neural stem cell proliferation and lineage specification through control of ets factors. Nat Commun. 2019;10:2000. doi: 10.1038/s41467-019-09949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sauvageot CM, Stiles CD. Molecular mechanisms controlling cortical gliogenesis. Curr Opin Neurobiol. 2002;12:244–9. doi: 10.1016/S0959-4388(02)00322-7. [DOI] [PubMed] [Google Scholar]
- 157.Lee HS, Han J, Lee S-H, Park JA, Kim K-W. Meteorin promotes the formation of GFAP-positive glia via activation of the Jak-STAT3 pathway. J Cell Sci. 2010;123:1959–68. doi: 10.1242/jcs.063784. [DOI] [PubMed] [Google Scholar]
- 158.He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, et al. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci. 2005;8:616–25. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–83. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 160.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between notch and JAK–STAT signalling. Nat Cell Biol. 2004;6:547–54. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 161.Gross RE, Mehler MF, Mabie PC, Zang Z, Santschi L, Kessler JA. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron. 1996;17:595–606. doi: 10.1016/S0896-6273(00)80193-2. [DOI] [PubMed] [Google Scholar]
- 162.Gomes WA, Mehler MF, Kessler JA. Transgenic overexpression of BMP4 increases astroglial and decreases oligodendroglial lineage commitment. Dev Biol. 2003;255:164–77. doi: 10.1016/S0012-1606(02)00037-4. [DOI] [PubMed] [Google Scholar]
- 163.Shen X-X, Hittinger CT, Rokas A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 2017;1:s41559–017. doi: 10.1038/s41559-017-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li Y, Shen X-X, Evans B, Dunn CW, Rokas A. Rooting the animal tree of life. Mol Biol Evol. 2021 doi: 10.1093/molbev/msab170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–60. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 166.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–6. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Romanova DY, Heyland A, Sohn D, Kohn AB, Fasshauer D, Varoqueaux F, et al. Glycine as a signaling molecule and chemoattractant in Trichoplax (Placozoa): insights into the early evolution of neurotransmitters. NeuroReport. 2020;31:490–7. doi: 10.1097/WNR.0000000000001436. [DOI] [PubMed] [Google Scholar]
- 168.Ryan JF, Chiodin M. Where is my mind? How sponges and placozoans may have lost neural cell types. Philos Trans R Soc Lond B Biol Sci. 2015;370. 10.1098/rstb.2015.0059. [DOI] [PMC free article] [PubMed]
- 169.Senatore A, Reese TS, Smith CL. Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J Exp Biol. 2017;220:3381–90. doi: 10.1242/jeb.162396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Varoqueaux F, Williams EA, Grandemange S, Truscello L, Kamm K, Schierwater B, et al. High cell diversity and Complex Peptidergic Signaling Underlie Placozoan Behavior. Curr Biol. 2018;28:3495–e5012. doi: 10.1016/j.cub.2018.08.067. [DOI] [PubMed] [Google Scholar]
- 171.Sachkova MY, Nordmann E-L, Soto-Àngel JJ, Meeda Y, Górski B, Naumann B et al. The unique neuronal structure and neuropeptide repertoire in the ctenophore Mnemiopsis leidyi shed light on the evolution of animal nervous systems. bioRxiv 2021:2021.03.31.437758. 10.1101/2021.03.31.437758.
- 172.Hayakawa E, Watanabe H, Menschaert G, Holstein TW, Baggerman G, Schoofs L. A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella Vectensis. PLoS ONE. 2019;14:e0215185. doi: 10.1371/journal.pone.0215185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Norekian TP, Moroz LL. Comparative neuroanatomy of ctenophores: neural and muscular systems in Euplokamis dunlapae and related species. J Comp Neurol. 2020;528:481–501. doi: 10.1002/cne.24770. [DOI] [PubMed] [Google Scholar]
- 174.Burkhardt P, Colgren J, Medhus A, Digel L, Naumann B, Soto-Angel JJ, et al. Syncytial nerve net in a ctenophore adds insights on the evolution of nervous systems. Science. 2023;380:293–7. doi: 10.1126/science.ade5645. [DOI] [PubMed] [Google Scholar]
- 175.Moroz LL, Romanova DY, Kohn AB. Neural versus alternative integrative systems: molecular insights into origins of neurotransmitters. Philos Trans R Soc Lond B Biol Sci. 2021;376:20190762. doi: 10.1098/rstb.2019.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–14. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Watanabe H. Back through time: how cnidarians and basal metazoans shed light on ancient nervous systems. Brain evolution by design. Tokyo: Springer; 2017. pp. 45–75. [Google Scholar]
- 178.Sebé-Pedrós A, Saudemont B, Chomsky E, Plessier F, Mailhé M-P, Renno J, et al. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell. 2018;173:1520–e3420. doi: 10.1016/j.cell.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 179.Oren M, Brickner I, Brikner I, Appelbaum L, Levy O. Fast neurotransmission related genes are expressed in non nervous endoderm in the sea anemone Nematostella Vectensis. PLoS ONE. 2014;9:e93832. doi: 10.1371/journal.pone.0093832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Goulty M, Botton-Amiot G, Rosato E, Sprecher SG, Feuda R. The monoaminergic system is a bilaterian innovation. Nat Commun. 2023;14:3284. doi: 10.1038/s41467-023-39030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Röttinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for endomesoderm specification: the inputs of ß-catenin/TCF signaling. PLoS Genet. 2012;8:e1003164. doi: 10.1371/journal.pgen.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miller DJ, Ball EE. Animal evolution: Trichoplax, trees, and taxonomic turmoil. Curr Biol. 2008;18:R1003–5. doi: 10.1016/j.cub.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 183.Jager M, Quéinnec E, Houliston E, Manuel M. Expansion of the SOX gene family predated the emergence of the Bilateria. Mol Phylogenet Evol. 2006;39:468–77. doi: 10.1016/j.ympev.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 184.Schnitzler CE, Simmons DK, Pang K, Martindale MQ, Baxevanis AD. Expression of multiple sox genes through embryonic development in the ctenophore Mnemiopsis leidyi is spatially restricted to zones of cell proliferation. Evodevo. 2014;5:15. doi: 10.1186/2041-9139-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fortunato S, Adamski M, Bergum B, Guder C, Jordal S, Leininger S, et al. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: multiple genes with unique expression patterns. Evodevo. 2012;3:14. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Magie CR, Pang K, Martindale MQ. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev Genes Evol. 2005;215:618–30. doi: 10.1007/s00427-005-0022-y. [DOI] [PubMed] [Google Scholar]
- 187.DuBuc TQ, Traylor-Knowles N, Martindale MQ. Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biol. 2014;12:24. doi: 10.1186/1741-7007-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shinzato C, Iguchi A, Hayward DC, Technau U, Ball EE, Miller DJ. Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol Biol. 2008;8:311. doi: 10.1186/1471-2148-8-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhou M, Yan J, Ma Z, Zhou Y, Abbood NN, Liu J, et al. Comparative and evolutionary analysis of the HES/HEY gene family reveal exon/intron loss and teleost specific duplication events. PLoS ONE. 2012;7:e40649. doi: 10.1371/journal.pone.0040649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fortunato SAV, Vervoort M, Adamski M, Adamska M. Conservation and divergence of bHLH genes in the calcisponge Sycon ciliatum. Evodevo. 2016;7:23. doi: 10.1186/s13227-016-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–61. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 192.Gyoja F. A genome-wide survey of bHLH transcription factors in the Placozoan Trichoplax adhaerens reveals the ancient repertoire of this gene family in metazoan. Gene. 2014;542:29–37. doi: 10.1016/j.gene.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 193.Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol. 2009;69:235–54. doi: 10.1002/dneu.20698. [DOI] [PubMed] [Google Scholar]
- 194.Watanabe H, Kuhn A, Fushiki M, Agata K, Özbek S, Fujisawa T, et al. Sequential actions of β-catenin and bmp pattern the oral nerve net in Nematostella vectensis. Nat Commun. 2014;5:5536. doi: 10.1038/ncomms6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marlow H, Roettinger E, Boekhout M, Martindale MQ. Functional roles of notch signaling in the cnidarian Nematostella vectensis. Dev Biol. 2012;362:295–308. doi: 10.1016/j.ydbio.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Albagli O, Klaes A, Ferreira E, Leprince D, Klämbt C. Function of ets genes is conserved between vertebrates and Drosophila. Mech Dev. 1996;59:29–40. doi: 10.1016/0925-4773(96)00568-0. [DOI] [PubMed] [Google Scholar]
- 197.Larroux C, Fahey B, Liubicich D, Hinman VF, Gauthier M, Gongora M, et al. Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evol Dev. 2006;8:150–73. doi: 10.1111/j.1525-142X.2006.00086.x. [DOI] [PubMed] [Google Scholar]
- 198.Amiel AR, Johnston H, Chock T, Dahlin P, Iglesias M, Layden M, et al. A bipolar role of the transcription factor ERG for cnidarian germ layer formation and apical domain patterning. Dev Biol. 2017;430:346–61. doi: 10.1016/j.ydbio.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 200.States DJ, Gish W. Combined use of sequence similarity and codon bias for coding region identification. J Comput Biol. 1994;1:39–50. doi: 10.1089/cmb.1994.1.39. [DOI] [PubMed] [Google Scholar]
- 201.He X, Wu F, Zhang L, Li L, Zhang G. Comparative and evolutionary analyses reveal conservation and divergence of the notch pathway in lophotrochozoa. Sci Rep. 2021;11:11378. doi: 10.1038/s41598-021-90800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Richards GS, Rentzsch F. Regulation of Nematostella neural progenitors by SoxB, notch and bHLH genes. Development. 2015;142:3332–42. doi: 10.1242/dev.123745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Layden MJ, Martindale MQ. Non-canonical notch signaling represents an ancestral mechanism to regulate neural differentiation. Evodevo. 2014;5:30. doi: 10.1186/2041-9139-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 205.Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. The hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan hedgehog pathway evolution. Dev Biol. 2008;313:501–18. doi: 10.1016/j.ydbio.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen C-Y, McKinney SA, Ellington LR, Gibson MC. Hedgehog signaling is required for endomesodermal patterning and germ cell development in the sea anemone Nematostella Vectensis. Elife 2020;9. 10.7554/eLife.54573. [DOI] [PMC free article] [PubMed]
- 207.Liongue C, Ward AC. Evolution of the JAK-STAT pathway. JAKSTAT. 2013;2:e22756. doi: 10.4161/jkst.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liongue C, Taznin T, Ward AC. Signaling via the CytoR/JAK/STAT/SOCS pathway: emergence during evolution. Mol Immunol. 2016;71:166–75. doi: 10.1016/j.molimm.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 209.Li H, Grumet M. BMP and LIF signaling coordinately regulate lineage restriction of radial glia in the developing forebrain. Glia. 2007;55:24–35. doi: 10.1002/glia.20434. [DOI] [PubMed] [Google Scholar]
- 210.See J, Mamontov P, Ahn K, Wine-Lee L, Crenshaw EB, 3rd, Grinspan JB. BMP signaling mutant mice exhibit glial cell maturation defects. Mol Cell Neurosci. 2007;35:171–82. doi: 10.1016/j.mcn.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pang K, Ryan JF, Baxevanis AD, Martindale MQ. Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS ONE. 2011;6:e24152. doi: 10.1371/journal.pone.0024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 213.Horridge GA, Mackay B. Naked axons and symmetrical synapses in Coelenterates. J Cell Sci. 1962;s3–103:531–41. doi: 10.1242/jcs.s3-103.64.531. [DOI] [PubMed] [Google Scholar]
- 214.Khalturin K, Shinzato C, Khalturina M, Hamada M, Fujie M, Koyanagi R, et al. Medusozoan genomes inform the evolution of the jellyfish body plan. Nat Ecol Evol. 2019;3:811–22. doi: 10.1038/s41559-019-0853-y. [DOI] [PubMed] [Google Scholar]
- 215.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, et al. The dynamic genome of Hydra. Nature. 2010;464:592–6. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Layden MJ, Rentzsch F, Röttinger E. The rise of the starlet sea anemone Nematostella Vectensis as a model system to investigate development and regeneration. Wiley Interdiscip Rev Dev Biol. 2016;5:408–28. doi: 10.1002/wdev.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Karabulut A, He S, Chen C-Y, McKinney SA, Gibson MC. Electroporation of short hairpin RNAs for rapid and efficient gene knockdown in the starlet sea anemone, Nematostella vectensis. Dev Biol. 2019 doi: 10.1016/j.ydbio.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 218.Renfer E, Amon-Hassenzahl A, Steinmetz PRH, Technau U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci U S A. 2010;107:104–8. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sheloukhova L, Watanabe H. Analysis of cnidarian gcm suggests a neuronal origin of glial EAAT1 function. Sci Rep. 2023;13:14790. doi: 10.1038/s41598-023-42046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.