Abstract
A novel retrovirus, morphologically consistent with mammalian C-type retroviruses, was detected by electron microscopy in mitogen-stimulated peripheral blood mononuclear cell cultures from 163 koalas and in lymphoma tissue from 3 koalas. PCR amplified provirus from the blood and tissues of 17 wild and captive koalas, and reverse transcriptase-PCR demonstrated viral mRNA, viral genomic RNA, and reverse transcriptase activity in koala serum and cell culture supernatants. Comparison of viral sequences derived from genomic DNA and mRNA showed identity indicative of a single retroviral species—here designated koala retrovirus (KoRV). Southern blot analysis of koala tissue genomic DNA using labelled KoRV probes demonstrated banding consistent with an endogenous retrovirus. Complete and apparently truncated proviruses were detected in DNA of both clinically normal koalas and those with hematopoietic disease. KoRV-related viruses were not detected in other marsupials, and phylogenetic analysis showed that KoRV paradoxically clusters with gibbon ape leukemia virus (GALV). The strong similarity between GALV and KoRV suggests that these viruses are closely related and that recent cross-host transmission has occurred. The complete proviral DNA sequence of KoRV is reported.
Retroviruses comprise a large group of diploid RNA viruses that rely on reverse transcription as an essential part of their life cycle. Endogenous retroviruses (ERVs) and exogenous retroviruses have been demonstrated in a wide range of vertebrate species, and while some, particularly the exogenous viruses, may be pathogenic, many are not associated with disease (23, 37). ERVs, by definition, have become incorporated into the host genome by integration into germ line cells or early embryos and are transmitted as dominant Mendelian alleles. Exogenous (infectious) retroviruses are transmitted horizontally (3, 35).
The etiological role of retroviruses in a range of different diseases has been demonstrated in a number of vertebrate species (47). In some host species, interaction between ERVs and related exogenous retroviruses (helper viruses) is important in disease pathogenesis. For example, some feline leukemia virus (FeLV)-associated diseases of cats result from recombination between endogenous FeLV and exogenous FeLV strains (52). The presence of endogenous viruses may reduce the immune response to related pathogenic exogenous retroviruses, increasing the likelihood of disease, as demonstrated in chickens with avian leukosis or sarcoma virus infection (53). In contrast, expression of ERV genes may confer resistance to exogenous pathogenic viruses through receptor interference and other mechanisms (25, 45).
Lymphoma and leukemia have been long recognized as the most common form of neoplasia in both captive and free-living koalas. Mortality surveys of wild koalas indicate that these diseases account for around 3 to 5% of deaths in free-living koalas in the survey areas of New South Wales and southern Queensland (2, 7, 11, 24, 39, 55). However, anecdotal evidence from fauna parks in southeast Queensland suggests that up to 80% of mortalities in captive koalas may be attributable to lymphoma and a variety of leukemias (J. J. Hanger, unpublished data). The possible involvement of retroviruses in these diseases of koalas was suggested by a number of workers (7, 24). Type C retrovirus-like particles were seen by transmission electron microscopy (TEM) in tissues from a leukemic koala (8), in the blood of captive koalas abroad (61), and in mitogen-stimulated peripheral blood mononuclear cells (PBMCs) derived from 18 koalas of mixed clinical status (46). Retrovirus infection in koalas was confirmed with a report of the isolation and partial gene sequence of a retrovirus with homology to gibbon ape leukemia virus (GALV) and simian sarcoma virus (SSV) in both diseased and healthy koalas (41). Recently, sequence homologous to the pol region of murine leukemia virus (MLV)-related and GALV-related viruses was detected in the DNA of a wild-caught koala, but no clinical information was reported (37). We report the complete nucleotide sequence, phylogenetic analysis, and characterization of a novel type C ERV detected in koalas.
MATERIALS AND METHODS
PCR.
Genomic DNA was extracted from the blood and tissues of four wild and four captive koalas by methods previously described (50). Total RNA or polyadenylated RNA was extracted from tissues and blood by using either Trizol reagent (Life Technologies) or the Quick-Prep Micro mRNA Purification kit (Pharmacia Biotech), respectively. Koalas from which tissues were taken had either died or had been euthanized because of serious disease or injuries. Four koalas (one wild and three captive) from which tissues or blood were taken had leukemia or lymphoma. DNA for PCR and Southern hybridization controls was extracted from tissue samples from a mediastinal lymphoma in a dog, bone marrow from an eastern grey kangaroo (Macropus giganteus), and blood from a common wombat (Vombatus ursinus) and a southern hairy-nosed wombat (Lasiorhinus latifrons).
Initial PCR amplification of a 3-kb koala retrovirus (KoRV) sequence from genomic DNA was accomplished with an upstream primer, tRNAf (CAT TTG GGG CTC GTC CGG GAT), which annealed to the tRNApro primer binding site, and a downstream degenerate pro-pol primer, JPU2r [GC(ACG) GCC A(GAC)(CTG) A(AG)(GT) A(GTA)G TC(AG) TC] (bases in parentheses represent alternatives for degenerate bases), described previously (10). PCR was performed in 50-μl reaction mixtures with the Expand Long Template PCR system (Boehringer, Mannheim, Germany) in accordance with the manufacturer's recommendations. Cycling conditions were as follows: initial denaturation at 94°C for 3 min, 1 cycle; denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 68°C for 2 min, 10 cycles; followed by cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 68°C for 45 s plus 20 s for each consecutive cycle up to 2 min. A further 10 cycles with the same temperatures and 2-min extension times were performed followed by a final extension at 68°C for 7 min.
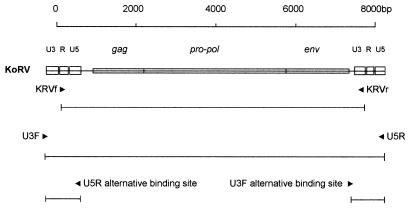
Touchdown PCR (46) was performed in 50-μl reaction mixtures using the Expand Long Template PCR system (Boehringer, Mannheim) in accordance with the manufacturer's recommendations. Two pairs of primers were used to amplify the putative koala provirus. One pair was designed to amplify the entire provirus, including both 5′ and 3′ long terminal repeats (LTRs): the forward primer U3F (AAT GAA GGA GGC AGA AAT CAT GAG GC) was designed to anneal to the first 26 bases of the U3 region of the LTR, and the reverse primer U5R (GGT AGT CCT CTG ACC TTG AGA) annealed to the last 21 bases of the U5 region (Fig. 1). The second pair of primers was designed to exclude repeated regions from the amplimer: the forward primer KRVf (GCG CCA GTC CCT CTA GAA GAC TGA) annealed to the first 24 bases of the 5′ repeated region (R) and the reverse primer KRVr (GCC GGG TGG ATT CTT TGG TCT CAT TT) annealed to the last 26 bases of the 3′ U3 region (Fig. 1). Cycling conditions for both primer pairs were as follows: denaturation at 94°C for 2 min (1 cycle); denaturation at 94°C for 30 s, annealing at 68°C for 1 min, and extension at 68°C for 4 min (2 cycles); followed by pairs of similar cycles, each with annealing at 66, 64, 62, and 60°C; and then 10 cycles with annealing at 58°C. This was followed by five similar cycles with annealing at 60°C with a 3-min extension at 68°C and then four cycles at the same temperatures with a 4-min extension time. The final extension at 68°C was for 7 min.
FIG. 1.
Graphic representation of KoRV provirus showing positions of primer pairs used to amplify proviral DNA. The primer pair U3F-U5R flanks the entire provirus, including both 5′ and 3′ LTRs, such that each primer binding site is duplicated in the provirus, whereas the primer pair KRVf-KRVr excludes amplification of repeated regions of the LTRs. The lines under each primer pair represent expected amplimers.
Cloning and sequencing.
PCR bands were resolved on a 0.9% agarose gel and then excised and extracted with the Qiaquick Gel Extraction kit (Qiagen). The fragments were cloned with the pGEM-T Easy Vector system (Promega), and automated sequencing was performed with M13 forward and reverse primers (Promega) or virus sequence-specific primers by using the BigDye Terminator system (Applied Biosystems, Perkin-Elmer). Gel separations were performed by the Australian Genome Research Facility.
Southern hybridization.
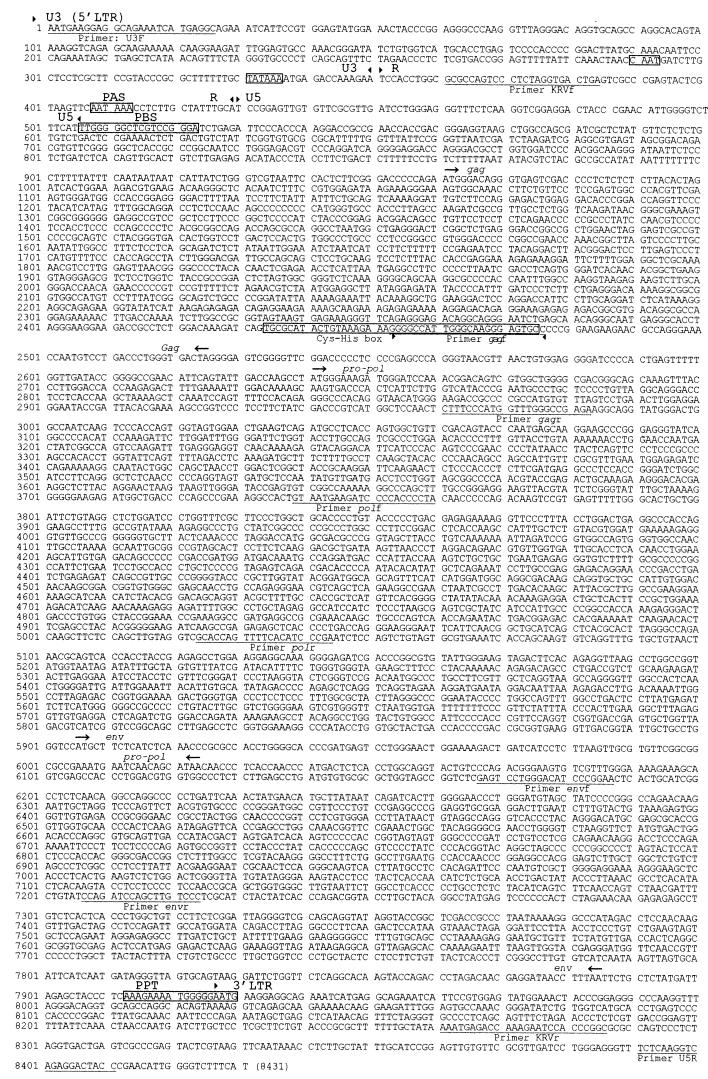
Southern hybridization was performed by digesting 10 μg of genomic DNA with PstI or HindIII and vacuum blotting the DNA after electrophoresis onto positively charged nylon membranes (Boehringer, Mannheim). Probe preparation and hybridization were performed with the Dig (digoxigenin) DNA Labeling and Detection kit (Boehringer, Mannheim). A 530-bp gag-pro-pol probe was prepared by radom hexameric primer extension with a PCR product derived from a plasmid containing the complete koala provirus. Positions of the primer pair (gagf-gagr) used to amplify this product are shown in Fig. 2. Hybridization was performed at 68°C in standard buffer in accordance with the manufacturer's recommendations. Hybridization of probes was detected by chemiluminescence with CDP-Star (Boehringer) after high-stringency washes (down to 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] 0.1% sodium dodecyl sulfate) at 68°C.
FIG. 2.
Complete nucleotide sequence of a KoRV provirus. The boundaries of the major sections of the genome are shown by arrowheads: 5′ LTR (U3-R-U5, boundaries based on GALV [GenBank accession no. M26927]), gag, pro-pol, env, and 3′ LTR. Sequences important in transcription and reverse transcription are shown in boxes: CAAT box, TATA box, Cys-His box, polyadenylation signal (PAS), polypurine tract (PPT) and tRNApro primer binding site (PBS). Primer sequences described in the text are underlined. Note that nucleotides 1 to 8411 were sequenced from one clone. Nucleotides 8412 to 8431 were inferred from the 5′ LTR.
Sequence analysis.
Initial sequence analysis was carried out on programs and databases made available by the Australian National Genomic Information Service (ANGIS). Original database searches were carried out with the BLASTn program. The GCG (Genetics Computer Group, University of Wisconsin, Madison) package programs GAP, HOMOLOGIES, and FRAMEALIGN were used for DNA and protein sequence comparisons. Predicted protein sequences were defined with the programs MAP and ETRANSLATE, while open reading frames (ORFs) and codon usage preference were assessed with FRAMES and CODONFREQUENCY. Protein similarities were calculated with the PAM 250 substitution matrix (51).
The maximum-likelihood phylogeny was estimated for sequences from KoRV and GALV and the murine leukemia-type viruses (Moloney MLV [MMLV; GenBank accession no. AF033811], Friend MLV [M93134], and FeLV [M18247]) and porcine ERV (PERV [U77599]). The likelihood analyses used an HKY+Γ model, which allows for uneven base composition and for different rates of transitions and transversions (19), with gamma-distributed rates across the sequence (62, 63). Base composition, transition/transversion ratio, and gamma shape parameter (γ) were estimated from the sequence data. A heuristic search in PAUP∗ [D. L. Swofford, PAUP∗: Phylogenetic Analysis Using Parsimony (∗ and other methods), version 4. Sinauer Assoc., Sunderland, Mass., 1999] was performed, with TBR branch swapping from an initial neighbor-joining tree. The analysis was repeated for pol, gag, and env genes, all aligned by eye. For the pol gene, the analysis was conducted for four separate alignments: (i) the complete gene (alignment length, 3959 bp), (ii) the conservative positions only (fast-changing sites excluded, 3,114 bp), (iii) the first and second codon positions only (2,076 bp), and (iv) the third codon positions only (1,038 bp). (Note that alignment length includes insertions.)
To assess the support for the KoRV-GALV clade and the signal/noise ratio of the sequence data, a spectral analysis was performed (21, 22). A spectral plot represents the patterns in the sequence data that correspond to bipartitions in the data. These bipartitions represent possible clades, so the support for any given phylogenetic clade can be assessed from a spectral plot.
Cell culture.
Blood samples of 4 to 12 ml were collected into EDTA blood tubes from captive koalas kept in a number of wildlife parks in southeast Queensland. PBMCs were isolated from buffy coats with Ficoll-Hypaque (Pharmacia) by methods described previously (60). PBMCs were cultured in RPMI (Trace Scientific) containing 2 mM glutamine, 9% inactivated fetal calf serum, 1% inactivated koala serum (both inactivated at 56°C for 1 h), 100 IU of penicillin per ml, 100 μg of streptomycin per ml, 0.1 mM 2-mercaptoethanol, and 5 μg of concanavalin A per ml. PBMCs were cultured at a concentration of 5 × 105 cells/ml in 25-ml flasks and incubated in a humidified atmosphere of 5% CO2 in air at 37°C. After 2 days of incubation, the medium was replaced with fresh medium containing 100 IU of human recombinant interleukin-2 per ml, but without concanavalin A. This medium was replaced every 2 days until the cells and supernatant were harvested at between days 8 and 12 of culture.
RT-PCR.
Synthesis of cDNA from mRNA extracted from the blood or tissues was performed with the Universal Riboclone cDNA Synthesis kit (Promega). PCR of cDNA was performed with the primer pairs tRNAf (described above)-gagr, gagf-polr, and polf-KRVr (Fig. 2), and the cycling conditions were those described above for initial amplification of 3-kb koala provirus products. Viral RNA was extracted from cell-free culture supernatant and koala serum by using the Viral RNA Extraction kit (Qiagen) in accordance with the manufacturer's recommendations. Samples were treated with 1 U of RQ1 DNase (Promega) for 10 min at 37°C, which was then inactivated at 70°C for 15 min. First-strand cDNA synthesis and PCR were performed with the Access reverse transcriptase (RT)-PCR Kit (Promega) using the manufacturer's recommended procedure. RT reactions were amplified by PCR with gag, pol, and env primer pairs (Fig. 2). The cycling conditions were denaturation at 94°C 30 seconds; annealing 57°C, 1 minute; extension 68°C, 2 minutes for 40 cycles, followed by a final extension at 68°C of 5 min. Bands were resolved on a 2% agarose gel.
RT assay.
Cell culture supernatants from nine mitogen-stimulated PBMC cultures that displayed virions by electron microscopy were assayed for RT activity with raw MMLV RT (40 U; Promega) and supernatants derived from feline immunodeficiency virus (FIV)-infected cat lymphocyte cultures as positive controls. Culture medium incubated without cells was used as negative control. Supernatants were clarified by centrifugation at 1,000 × g for 10 min. A 30% concentration of polyethylene glycol 6000 to 8000 (0.5 volume) was added to 1.0-ml aliquots of the supernatant and incubated at 4°C overnight and then precipitated by centrifugation at 10,000 × g for 20 min. One hundred fifty microliters of virus lysis buffer (0.5% Triton-X 100, 20% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 800 mM NaCl, 50 mM Tris [pH 7.8]) was added to each precipitate, which was then incubated at room temperature for 30 min. Samples were vortexed briefly, and 40 μl was added to 90 μl of RT reaction buffer [final concentration: 0.1 M Tris (pH 8.3), 0.02 M dithiothreitol, 1 mM MnCl2, 0.1% Nonidet P-40, 0.1 M NaCl, 5 mM dNTP, 1 U of poly(rA) · oligo(dT)10, 1 uCi 3H-TTP]. Samples were incubated in quadruplicate in 96-well plates for 4 h at 37°C. Trichloroacetic acid (12.5 μl) (50%) was added, and samples were stored at 4°C for 30 min to precipitate the 3H-TTP. Samples were then harvested onto glass fiber discs, placed into 2 ml of scintillation fluid, and counted.
Alternatively, a more sensitive RT-PCR-based RT assay (38) was used by substituting the poly(rA) · oligo (dT) with 40 ng of kanamycin resistance gene mRNA (Promega), added as a template for reverse transcription, and 30 ng of oligo(dT)12–18 primer (Promega). Serum samples (25 μl) from 31 animals were tested by this method. Samples were incubated for 4 h at 37°C, and then 1-μl aliquots were used in a PCR with kanamycin template-specific primers and reagents in the Access RT-PCR kit (Promega) in accordance with the manufacturer's instructions. PCR bands were resolved on a 2% agarose gel, purified with the Qiaquick Gel Extraction kit (Qiagen), and sequenced as described above.
Electron microscopy.
Tissues for TEM were fixed in 3% glutaraldehyde in cacodylate buffer, postfixed in 1% osmium tetroxide, prepared by standard methods, and examined electron microscopically. PBMCs from the cell cultures were fixed in 3% glutaraldehyde in cacodylate buffer, embedded in 3% agarose, and then prepared as described above.
Nucleotide sequence accession number.
The complete provirus sequence reported here is contained in the GenBank nucleotide sequence database under accession no. AF151794.
RESULTS
Molecular characterization.
Initial PCR amplification of a 3-kb KoRV sequence from genomic DNA was accomplished with an upstream primer (tRNAf) which annealed to the tRNApro primer binding site and a downstream degenerate universal pro-pol primer (JPU2r) described previously by Colbatzky and Jacobs (10) (data not shown). Sequence analysis of this amplimer demonstrated close similarity to GALV sequences (13), which enabled the design of primers flanking other regions of the putative koala provirus. Definitive KoRV primers, which flank the entire provirus, were designed based upon sequences detected by these methods.
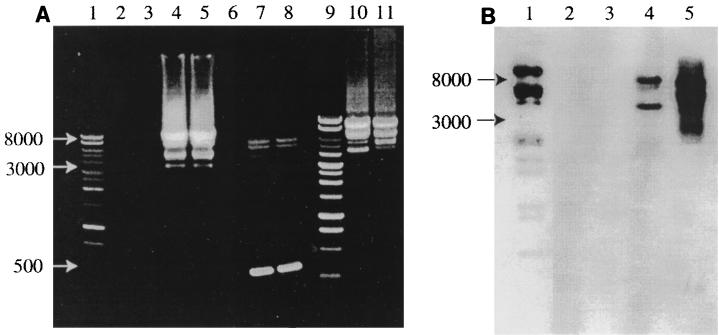
All 17 koalas amplified KoRV sequence from blood or tissue with a variety of KoRV-specific primers. PCR amplification of sequences with primers U3F and U5R resulted in expected bands of approximately 8.4 kb, representing putative full-length proviral amplimers, and 0.5 kb, representing amplification of the LTRs only. Primers KRVf and KRVr amplified products of approximately 8 kb, representing full-length provirus amplimers without repeated regions of the LTRs. However, a number of bands of intermediate size (4 to 7 kb) were consistently amplified with both primer pairs in PCRs. PCR analysis of multiple tissues from one koala (without hematopoietic disease) showed identical banding patterns between tissues. However, among koalas, all showed 8-kb (primers KRVf and KRVr) or 8.4-kb (primers U3F and U5R) bands, but the sizes and number of smaller bands varied. Southern hybridization of PCR blots with a KoRV pro-pol probe detected the 8.4- and 8-kb bands as well as some of the smaller bands, but not the 0.5-kb bands representing LTR-only amplimers (Fig. 3B). No PCR bands were obtained from tissues taken from a dog or a kangaroo (Fig. 3A). With primers tRNAf and JPU2r, a weak 3-kb band was amplified from DNA derived from the blood of a common wombat and a southern hairy-nosed wombat. Subsequent sequencing showed that these amplimers were not derived from retrovirus.
FIG. 3.
(A) PCRs using primer pairs KRVf-KRVr and U3F-U5R resolved on a 1% agarose gel stained with ethidium bromide. Lanes: 1 and 9, 1-kb-ladder molecular weight markers; 2, no-DNA PCR control; 3, kangaroo DNA PCR control; 4, lymphoma DNA amplified with primer pair KRVf-KVRr; 5, blood DNA from the same koala amplified with the same primers; 7 and 8, the same samples amplified with primer pair U3F-U5R, showing amplimers representing full-length and truncated proviruses, as well as LTRs only (0.5-kb bands); 10 and 11, KRVf-KRVr amplimers from blood DNA from two different koalas. (B) Southern blot of a similar PCR gel probed with a KoRV pol probe. Lanes: 1 digoxigenin-labelled molecular weight marker; 2, kangaroo DNA PCR control; 3, dog lymphoma DNA PCR control; 4, koala DNA U3F-U5R PCR products; 5, koala DNA KRVf-KRVr PCR products.
Sequence analysis of clones of the 8.4- and 8-kb fragments revealed a complete provirus of 8,431 bp with the structure of a simple type C mammalian retrovirus. Common features of retroviral genomes were identified, including 5′ and 3′ LTRs, a tRNAPRO primer binding site; a CAAT box; a TATA box; gag, pro-pol, and env coding sequences; a Cys-His box (protein translation, CAYCKERGHWAREC), a polypurine tract, and a polyadenylation signal (Fig. 2). There are three ORFs on two forward-reading strands corresponding to gag, pol, and env ORFs with the typical structure of gag and pro-pol in the same frame, while the 5′ end of env overlaps the 3′ end of pol in an adjacent frame. A splice donor site was located at bases 564 to 570, and a presumptive splice acceptor was located at bases 5701 to 5709 (based upon sequencing of a spliced env transcript). There are several small ORFs, with predicted protein lengths greater than 30 amino acids, whose significance, if any, has yet to be determined. Partial sequence analysis of some of the clones of smaller PCR bands showed them to have homology to the full-length provirus clones; however, complete sequencing of these clones was not performed, and the deleted regions were not defined.
KoRV sequence comparison (GCG) demonstrated a 78% nucleotide similarity across the complete genome between KoRV (8,431 bp) and GALV (8,513 bp; M26927) and strong similarity across the three major coding regions with GALV (Table 1). The predicted proteins from the gag, pol, and env ORFs all showed the greatest similarity to GALV or a novel strain of GALV (GALVX).
TABLE 1.
Comparison of the size, nucleic acid identity, and amino acid residue numbers of GALV strain SEATO/SF (M269297), GALV 5′ LTR strain SF (J02196), and KoRV (AF151794)
| Sequence compared | Length (nucleotides) | Nucleotide % identity | Amino acid residue no. |
|---|---|---|---|
| Provirus | |||
| KoRV | 8,431 | 78 | NAa |
| GALV | 8,513 | ||
| LTR | |||
| KoRV | 505 | 69 | NA |
| GALV | 494 | ||
| gag | |||
| KoRV | 1,566 | 82 | 522 |
| GALV | 1,560 | 520 | |
| pol | |||
| KoRV | 3,384 | 82 | 1128 |
| GALV | 3,384 | 1128 | |
| env | |||
| KoRV | 1,980 | 74 | 660 |
| GALV | 1,995 | 665 |
NA, not applicable.
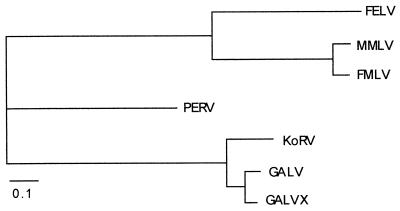
All phylogenetic analyses grouped the KoRV sequence with GALV sequences, to the exclusion of PERV and the MLV group. Figure 4 shows the maximum-likelihood tree for the pol gene. The maximum likelihood trees for the env and gag sequences were the same as that shown in Fig. 4. Separate analysis of an alignment of only the 1st and 2nd codon positions of pol also gave the tree in Fig. 4, as did an alignment of only 3rd codon positions. Spectral analysis indicated strong support for a KoRV-GALV clade, with little conflicting signal (Fig. 5).
FIG. 4.
Maximum likelihood phylogeny for the pol gene of KoRV, GALV or GALVX, MLV-type viruses (MMLV, FMLV, and FeLV), and PERV. The transition/transversion ratio (3.1) and gamma shape parameter (0.44) were estimated from the data [D. L. Swofford, PAUP∗: Phylogenetic Analysis Using Parsimony (∗ and other methods), version 4. Sinauer Assoc., Sunderland, Mass., 1999]. The results are the same for the gag and env genes and for all alternative alignments of the pol gene (see Materials and Methods).
FIG. 5.
Spectral plot showing the relative support for branches in the pol phylogeny (Fig. 4). The frequency of patterns in the sequence data that support a partition (clade) is given above the x axis, and the frequencies of patterns that support a conflicting bipartition are given below. The KoRV-GALV clade has strong support and little conflicting signal. Spectral analysis was performed with the Spectrum package (9).
Multiple sequence alignment of two full and two partial (spanning 3 kb of gag-pro-pol) provirus sequences from four different koalas showed greater than 98.5% identity between all sequences (data not shown). Three of the koalas were captive koalas from southeast Queensland, but were unrelated, and one wild koala was a resident in a geographically isolated region of New South Wales. RT-PCR of mRNA extracted from the blood of a clinically normal captive koala, with three primer pairs spanning the retroviral genome from the tRNA primer binding site to the 3′ LTR, resulted in bands of the expected length with 98.5% sequence identity to proviral sequences (data not shown). RT-PCR of the same sample with primer pair KRVf-KRVr (Fig. 1) amplified a band of approximately 2.6 kb, which sequencing showed to represent spliced env transcripts.
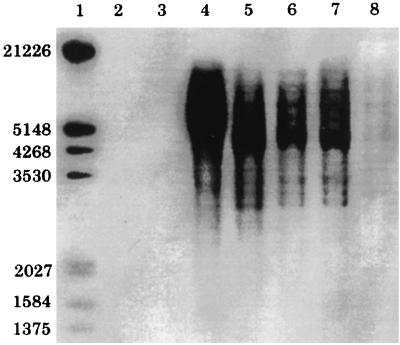
Southern hybridization of genomic DNA digested with PstI and HindIII from a range of tissues taken from two koalas and hybridized with a gag-pro-pol-specific probe demonstrated a banding pattern, as well as smearing in the 3- to 12-kb range (Fig. 6). One koala was affected by lymphoid neoplasia, which involved multiple tissues; the other was not affected by neoplastic or hematopoietic disease. Probes did not bind to DNA from lymphoma tissue from a dog or bone marrow from an Eastern Grey kangaroo.
FIG. 6.
Southern blot of koala genomic DNA digests. Lanes: 1, digoxigenin-labelled molecular weight marker; 2, dog genomic DNA digest (control); 3, kangaroo genomic DNA digest (control); 4 to 7, DNA digests from a koala with lymphoma; 4, 10 μg of bone marrow DNA digested with PstI; 5, 10 μg of bone marrow DNA digested with HindIII; 6, 10 μg of lymphoma DNA digested with HindIII; 7, 10 μg of lung DNA digested with HindIII; 8, 2 μg of bone marrow DNA digested with HindIII from a koala without lymphoma.
Viral RNA.
Viral RNA extracted from either serum or cell-free PBMC culture supernatant was reverse transcribed and analyzed by PCR with the primer pairs gagf-gagr, polf-polr, and envf-envr (Fig. 2). Of the 10 koalas examined, all three primer pairs amplified bands of the expected lengths in 2 koalas (in both serum and PBMC culture supernatant), but in 3 koalas, only amplimers of gag and env or only env was detected. Viral RNA was not detected by RT-PCR in serum or supernatant in one koala; however, in another koala, pro-pol transcripts were detected in bone marrow, but not in blood or serum.
RT assay.
RT assays with supernatants from PBMCs cultured in vitro from nine koalas, with and without lymphoid neoplasia, showed optimal RT activity in the presence of manganese. When the conventional 3H-TTP incorporation RT assay was used, weak activity levels were demonstrated; culture supernatants derived from nine koalas showed a mean activity of 5,663 dpm (standard deviation [SD], 2,749; range, 1,965 to 10,432) compared to negative controls (924 dpm [n = 4; SD, 539]), positive control FIV supernatant (42,198 dpm [n = 4; SD, 11,821]) and positive control raw MMLV RT 24,630 dpm [n = 4; SD, 5,674). When the more sensitive qualitative RT-PCR-based RT assay was applied to koala serum, 9 of 31 (29%) reactions amplified the expected size fragment of kanamycin cDNA, indicating in vivo RT activity in the serum of these animals.
Electron microscopy.
TEM demonstrated characteristic type C retrovirus-like particles associated with lymphoma in three koalas or with mitogen-stimulated cultured PBMCs derived from 163 of 166 koalas. Particles were 80 to 110 nm in diameter, enveloped, and contained an electron-dense spherical core. Budding forms were observed, with characteristic C-shaped capsid assembly at the cytoplasmic membrane. For three koalas, PBMCs grew poorly in cell culture and virus particles were unable to be demonstrated by TEM.
DISCUSSION
From the data presented, it is clear that a provirus (KoRV) related to the simian type C retroviruses is present in the DNA of koalas. The observation that retrovirus-like particles were produced by both tumor cells in vivo and cultured PBMCs in vitro suggests that proviral transcription and virus assembly occur. This is supported by the detection of retroviral transcripts homologous to proviral sequences in the blood of koalas and the detection of viral RNA and RT activity in serum and in PBMC culture supernatants.
Provirus was detected in all koalas and in all tissues by PCR, and virus-like particles were demonstrated by TEM in 98% of mitogen-stimulated PBMC cultures. Southern hybridization of genomic DNA demonstrated similar intensity and banding patterns across a range of tissues in koalas with and without lymphoid neoplasia. These data suggest that KoRV is an ERV, although exogenous forms may also exist. Full-length provirus sequences were detected and contained all of the basic genetic elements common to type C retroviruses (44, 58), namely, flanking LTRs; gag, pro-pol, and env genes; a tRNA primer binding site; a polypurine tract; a CAAT box; a TATA box; a Cys-His box; and a polyadenylation signal. The sequences showed the most similarity to the GALV-related simian retroviruses and also showed ORFs and spliced env transcripts consistent with the type C mammalian group of retroviruses.
While full-length provirus is present in koalas, there is also strong evidence for the presence of truncated proviruses. Undersized PCR products derived from primers that span the complete proviral genome were detected in all animals, although the molecular size ranges of these products differed between animals. These amplimers hybridized to labelled KoRV pro-pol, and partial sequencing confirmed their KoRV derivation, suggesting that they represent truncated proviruses. Tissue-, animal-, or population-specific truncated KoRV variants may have arisen in the koala genome either via recombination excision events or novel superinfection or reinsertion events. Considering the marked variation between koalas in the lengths of apparently truncated proviruses detected by PCR, it appears that either KoRV is not stable, in contrast to most other endogenous viruses (3), or PCR detection is confounded by the presence of related exogenous virus.
Detection of some viral transcripts containing gag and/or env sequences and failure to detect viral transcripts containing pro-pol sequences by RT-PCR of serum and cell-free culture supernatants from some koalas may be due to transcription and expression of truncated proviruses and assembly of virions containing defective genomic RNA. In humans, there are numerous examples of expression of defective or truncated endogenous proviruses in both normal and transformed tissues (1, 18, 34, 35, 57, 59). Furthermore, the expression of the retroviral Gag protein alone may be sufficient to cause assembly and budding of virus-like particles from the cell (58). Clearly, in the koala genome, there are multiple copies of proviruses, including both full-length and truncated proviruses, at least some of which are expressed both in vivo and in vitro.
The inability to detect strong RT activity in the serum or PBMC culture supernatants of some koalas may be explained by a number of possibilities. First, inhibition of RT activity by tissue-derived specific inhibitors has been demonstrated with some human ERVs (59). A second possibility is that virions may not contain detectable RT because of a defective or absent pro-pol gene. This is consistent with the observed expression of defective or truncated proviruses and has been previously demonstrated for HERV-K, which encodes a functional enzyme with weak activity (57). A third possibility is that assay conditions may not have been optimal for the detection of KoRV RT activity.
The DNA sequence data from gag, pol, and env unambiguously support a grouping of KoRV and GALV, to the exclusion of PERV and the MLV group. A similar grouping was also obtained from an independent phylogenetic study that compared pol fragments derived from a wide range of host taxa (37). This is an intriguing grouping, because gibbons (Hylobates spp.) and koalas are taxonomically distant, making virus acquisition from a common ancestor unlikely and because gibbons and koalas are from remote biogeographic regions, making direct natural transmission improbable. A preliminary investigation of small numbers of animals failed to detect KoRV-like retroviruses in other marsupials, including an eastern grey kangaroo (Macropus giganteus) and common wombats (Vombatus ursinus) and southern hairy-nosed wombats (Lasiorhinus latifrons) (data not shown). Wombats are the closest living relatives of koalas, although still relatively distantly related. Other studies have also failed to demonstrate KoRV-like viruses in marsupials other than koalas (23, 37, 40). This suggests that endogenization of KoRV in koalas postdates the divergence of koalas from other marsupials.
There are two possible explanations for the close relationship between KoRV and GALV. Either KoRV and GALV share a more recent common ancestor than all other ERVs sequenced to date, and therefore the grouping reflects the true evolutionary history of these sequences, or the grouping is an artifact of sequence analysis. The potential causes of spurious phylogenetic signal include recombination between strains, base composition bias, sequence saturation, and molecular convergence. Each of these artifacts can be discounted on the basis of detailed DNA sequence analysis, suggesting that the KoRV-GALV grouping represents a true biological relationship.
Recombination between virus lineages can cause unusual phylogenetic groupings, because different regions of the genome have different phylogenetic histories. However, there is no evidence that recombination is causing the KoRV-GALV grouping, because all viral genes gave the same phylogenetic signal. There is little evidence of saturation (true signal obscured by multiple hits) in the DNA sequences (Fig. 5). Nor is there any evidence for base composition bias pulling KoRV and GALV together, because the base composition of these sequences does not differ noticeably from the base compositions of the Friend MLV, MMLV, and PERV sequences. The KoRV-GALV grouping is unlikely to be caused by molecular convergence, which would require independent acquisition of many identical substitutions in both KoRV and GALV lineages and would expect to result in a strong conflicting signal in the data set. However, spectral analysis shows little conflicting signal (Fig. 5). Strong support for the KoRV-GALV grouping from both the 1st and 2nd codon positions, which predominantly cause nonsynonymous changes, and 3rd codon positions, which are mostly “silent,” provides further evidence against molecular convergence, since a convergent signal would be expected only in the nonsynonymous changes.
Some ERV phylogenies closely match the phylogeny of their hosts, indicating that the viruses are evolving as an endogenous element of the host genome, rarely infecting other hosts. Examples include the MLV-related retroviruses in reptiles and amphibians (23, 37) and some ERVs of great apes (27). Because the phylogeny does not match the order of branching of the mammalian tree (37) (Fig. 4), KoRV and GALV could only be considered to be strictly host tracking if the KoRV-GALV ancestor represents a novel insertion into the ancestral mammalian genome independently of the other mammalian ERVs sequenced to date. The similarity of KoRV and GALV to FeLV, the MLVs, and PERV would then be due to the similarity of the free-living viruses that inserted into the mammalian genome multiple times at least 130 million years ago, at the time of metatherian and eutherian mammal divergence (26). This could be confirmed by identifying the host genome insertion points for these viruses: if insertion is random, then independent acquisitions of ERVs will occur at different loci (27). However, the depth of molecular divergence in the KoRV-GALV clade is less than half that observed in the MLV clade, so the host-tracking hypothesis would require that the rate of molecular evolution in the KoRV-GALV lineage was dramatically slower than that of the MLV genomes. Given that GALV is exogenous, it seems highly unlikely that its rate of molecular evolution would have decreased to such an extent. Indeed, it would be expected that the rate of molecular evolution in an actively infectious virus would be greater than that of an endogenous virus (49). A host-tracking explanation for the KoRV-GALV grouping is therefore inconsistent with the sequence data.
There seems to be no reason to cast doubt on the phylogenetic relationship between KoRV and GALV to the exclusion of other retroviruses sequenced to date. There are two possible explanations for the relationship: inheritance from the genome of the last common ancestor of gibbons and koalas or cross-species transmission. The former explanation is unlikely, given the DNA sequence data and the preliminary assessment that KoRV-like viruses are absent in other marsupials. It seems most likely that the KoRV-GALV grouping is the result of a relatively recent cross-species transmission of a KoRV-like virus into gibbon and/or koala populations. Because gibbons and koalas are from distinct biogeographical regions with very different faunas, direct cross-infection in the wild is improbable. However, natural transfer via an intermediate host, for example, a mobile species such as rodents, bats, or birds or an arthropod vector, cannot currently be ruled out. A scenario suggested by Martin and coworkers (37) is that transmission was dependent upon an intermediate host, possibly a species of Asian rodent in which ERVs related to the primate type C viruses have been detected (6, 30).
KoRV has been isolated from both captive and wild koalas, but to our knowledge, GALV has been detected only in captive gibbons. This observation suggests that iatrogenic infection of captive gibbons with a KoRV-like virus derived either from live captive koalas, from tissue or fomites, is also a possible mechanism of recent cross-species transfer. If this observation holds true, it would imply that GALV infection in gibbons is a recent artifact of captivity. Alternatively species mixing and opportunities for either direct or indirect cross-infection between koalas and gibbons may have been promoted by indigenous trade between southeast Asia and mainland Australia in precolonial times. This mechanism has been proposed as a transmission method of an Australian marsupial arthropod parasite, Heterodoxus spiniger, to Asian dogs (16).
Whether iatrogenic or natural transmission has occurred, pinpointing the date of such a host jump through “molecular clock” dating may help to resolve the path of cross-species infection. However, dating virus divergence times from molecular data is made difficult by lack of a suitable calibration rate. There are no currently known divergence dates that can be used to calibrate the rate of change in KoRV or GALV. Given that substitution rates can vary enormously for different virus lineages (49) and between genes within a lineage (32), it is prudent to be wary of assuming a molecular clock to date virus divergences. However, the relatively low degree of divergence between the env genes of KoRV and the various GALV strains supports a recent divergence. In view of the apparently widespread endogenous infection of koalas and the suggestion that the exogenous GALV viruses are relatively recently acquired by simians (33), the possibility remains that the GALV group originated in koalas and spread to gibbons by a yet to be discovered mechanism. The parallel finding that macropodid herpesviruses paradoxically cluster with the simian herpesviruses (36) also supports the possibility of an occult epidemiological link between Australian marsupials and old-world primates.
Lymphoid malignancy is a relatively common disease of both free-living and captive koalas (11, 55), and its clinical similarity to retrovirus-associated disease in other mammals has been noted (7, 24). The ability of GALV and SSV to cause neoplasia in their host species has been demonstrated in both natural (28, 54, 56) and experimental infections (12, 29). In view of the relatively high prevalence of hematopoietic neoplasia and related diseases in koalas (4, 11, 61) and anecdotal evidence for lymphoid neoplasia epizootics in captive koala colonies, it is reasonable to suspect a pathogenic role for KoRV. However, KoRV was detected in all diseased and healthy koalas tested and appears to be endogenous. Although this lack of clinical correlation does not rule out a pathogenic role for the virus, it implies that other factors are necessary for disease to occur. Potential cofactors include superinfection with KoRV exogenous pathogenic variants, recombination with other retroviruses, interactions with host haplotypes, interactions with somatic mutations, or coinfection with other viruses, such as herpesviruses.
Serious consideration should be given to the zoonotic potential of the GALV-KoRV group. Early experimental work on GALV and more recent investigation of GALV as a vector for gene therapy have demonstrated multiple host and tissue tropisms (17). The broad in vitro host range of GALV, its apparently well-conserved receptor (42), and the evidence presented for taxonomic cross-transmission in vivo suggest that the members of the GALV-KoRV group have the potential for future host jumping. Furthermore, simian type C-related virus sequences and cross-reactive proteins have been detected in humans (14, 17, 20, 31), and a novel strain of GALV (GALVX) has been isolated from a human cell line (5, 43). Continuing studies are aimed at further defining the epizootiology of the KoRV-GALV group and at clarifying the pathogenic and zoonotic potential of this virus.
ACKNOWLEDGMENTS
We thank Steven Barker for his intellectual input, Kevin Bradley and Anne Williams (Dreamworld, Gold Coast, Australia), Ray and Jennifer Chafer (Wild World, Cairns, Australia), Vere Nicholson (Currumbin Sanctuary, Gold Coast, Australia), Paul O'Callagan and Galit Tzipori (Lone Pine Sanctuary, Brisbane, Australia), and Rosie Booth for assistance with collection of samples. We also thank Majeed Godhussi for assistance with TEM and Rodney Williams and Erica Maddock for preparation of photographs.
This project was supported by grants from the Australian Koala Foundation and Dreamworld, Australia.
REFERENCES
- 1.Andersson A, Svensson A, Rolny C, Andersson G, Larsson E. Expression of human endogenous retrovirus ERV-3 (HERV-R) mRNA in normal and neoplastic tissues. Int J Oncol. 1998;12:309–313. doi: 10.3892/ijo.12.2.309. [DOI] [PubMed] [Google Scholar]
- 2.Backhouse T C, Bolliger A. Morbidity and mortality in the koala (Phascolarctos cinereus) Aust J Zool. 1961;9:24–37. [Google Scholar]
- 3.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, et al., editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–436. [PubMed] [Google Scholar]
- 4.Booth R J, Blanshard W. Diseases of koalas. In: Fowler M E, Miller R E, editors. Zoo and wild animal medicine, current therapy 4. W. B. Philadelphia, Pa: Saunders; 1999. pp. 321–333. [Google Scholar]
- 5.Burtonboy G, Delferriere N, Mousset B, Heusterspreute M. Isolation of a C-type retrovirus from an HIV infected cell line. Arch Virol. 1993;130:289–300. doi: 10.1007/BF01309661. [DOI] [PubMed] [Google Scholar]
- 6.Callahan R, Meade C, Todaro G J. Isolation of an endogenous type C virus related to the infectious primate type C viruses from the Asian rodent Vandeleuria oleracea. J Virol. 1979;30:124–131. doi: 10.1128/jvi.30.1.124-131.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canfield P J, Brown A S, Kelly W R, Sutton R H. Spontaneous lymphoid neoplasia in the koala (Phascolarctos cinereus) J Comp Pathol. 1987;97:171–178. doi: 10.1016/0021-9975(87)90037-5. [DOI] [PubMed] [Google Scholar]
- 8.Canfield P J, Sabine J M, Love D N. Virus particles associated with leukemia in a koala. Aust Vet J. 1988;65:327–328. doi: 10.1111/j.1751-0813.1988.tb14518.x. [DOI] [PubMed] [Google Scholar]
- 9.Charleston M. Spectrum: spectral analysis phylogenetic data. Bioinformatics. 1998;14:98–99. doi: 10.1093/bioinformatics/14.1.98. [DOI] [PubMed] [Google Scholar]
- 10.Colbatzky F, Jacobs R M. Seventy-Third Conference of Research Workers in Animal Diseases. 1992. Detection of retroviral-like elements in genomic DNA of dogs with and without lymphoma; p. 51. [Google Scholar]
- 11.Connolly J H, Canfield P J, Hemsley S, Spencer A J. Lymphoid neoplasia in the koala. Aust Vet J. 1998;76:819–825. doi: 10.1111/j.1751-0813.1998.tb12337.x. [DOI] [PubMed] [Google Scholar]
- 12.Deinhardt F, Wolfe L, Northrop R, Marczynska B, Ogden J, McDonald R, Falk L, Shramek G, Smith R, Deinhardt J. Simian sarcoma virus: oncogenicity, focus assay, presence of associated virus and comparison with avian and feline sarcoma virus-induced neoplasia in marmoset monkeys. Bibl Haematol. 1973;39:258–262. doi: 10.1159/000427850. [DOI] [PubMed] [Google Scholar]
- 13.Delassus S, Sonigo P, Wain-Hobson S. Genetic organisation of gibbon ape leukemia virus. Virology. 1989;173:205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- 14.Derks J P A, Hofmans L, Bruning H W, Rood J J V. Synthesis of a viral protein with molecular weight of 30 000 (p30) by leukemic cells and antibodies cross-reacting with simian sarcoma virus p30 in serum of a chronic myeloid leukemia patient. Cancer Res. 1982;42:681–686. [PubMed] [Google Scholar]
- 15.Eglitis M A, Schneiderman R D, Rice P M, Eiden M V. Evaluation of retroviral vectors based on the gibbon ape leukemia virus. Gene Ther. 1995;2:486–492. [PubMed] [Google Scholar]
- 16.Flannery T F. The future eaters. Sydney, Australia: Reed New Holland Books; 1994. p. 170. [Google Scholar]
- 17.Gallo R C, Wong-Staal F. Molecular biology of primate retroviruses. In: Klein G, editor. Viral oncology. New York, N.Y: Raven Press; 1980. pp. 399–431. [Google Scholar]
- 18.Gotzinger N, Sauter M, Roemer K, Mueller-Lantzsch N. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J Gen Virol. 1996;77:2983–2990. doi: 10.1099/0022-1317-77-12-2983. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 20.Hehlmann R, Schetters H, Erfle V, Leib-Mosch C. Detection and biochemical characterization of antigens in human leukemic sera that cross-react with primate C-type viral proteins (Mr 30 000) Cancer Res. 1983;43:392–399. [PubMed] [Google Scholar]
- 21.Hendy M D, Penny D. Spectral analysis of phylogenetic data. J Classif. 1993;10:5–23. [Google Scholar]
- 22.Hendy M D, Penny D, Steel M A. A discrete Fourier analysis for evolutionary trees. Proc Natl Acad Sci USA. 1994;91:3339–3343. doi: 10.1073/pnas.91.8.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. Retroviral diversity and distribution in vertebrates. J Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuschele W P, Hayes J R. Acute leukemia in a New South Wales koala. Cancer Res. 1961;21:1394–1395. [PubMed] [Google Scholar]
- 25.Ikeda H, Sugimura H. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 1989;63:5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janke A, Xiufeng X, Arnason U. The complete mitochondrial genome of the wallaroo Macropus robustus and the phylogenetic relationship among monotremata, marsupialia and eutheria. Proc Natl Acad Sci USA. 1997;94:1276–1281. doi: 10.1073/pnas.94.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson W E, Coffin J M. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawakami T G, Huff S D, Buckley P M, Dungworth D L, Snyder S P. C-type virus associated with gibbon lymphosarcoma. Nat New Biol. 1972;246:170–171. doi: 10.1038/newbio235170a0. [DOI] [PubMed] [Google Scholar]
- 29.Kawakami T G, McDowell T S. Factors regulating the onset of chronic myelogenous leukemia in gibbons. Cold Spring Harbor Conf Cell Proliferation. 1980;7:719–727. [Google Scholar]
- 30.Leiber M M, Sherr C J, Todaro G J, Benveniste R E, Callahan R, Coon H G. Isolation from the Asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc Natl Acad Sci USA. 1975;81:2315–2317. doi: 10.1073/pnas.72.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leib-Mosch C, Brack R, Werner T, Erfle V, Hehlmann R. Isolation of SSAV-related endogenous sequences from human DNA. Virology. 1986;155:666–677. doi: 10.1016/0042-6822(86)90226-6. [DOI] [PubMed] [Google Scholar]
- 32.Leitner T, Albert J. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc Natl Acad Sci USA. 1999;96:10752–10757. doi: 10.1073/pnas.96.19.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowenstine L J, Lerche N W. Retrovirus infections of non-human primates: a review. J Zoo Anim Med. 1988;19:168–187. [Google Scholar]
- 34.Lower R, Boller K, Hasenmaier B, Korbmacher C, Mueller-Lantzsch N, Lower H, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahony T J, Smith G A, Thomson D M. Macropodid herpesviruses 1 and 2 occupy unexpected molecular phylogenic positions within the Alphaherpesvirinae. J Gen Virol. 1999;80:433–436. doi: 10.1099/0022-1317-80-2-433. [DOI] [PubMed] [Google Scholar]
- 37.Martin J, Herniou E, Cook J, Waugh O'Neill R, Tristem M. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J Virol. 1999;73:2442–2449. doi: 10.1128/jvi.73.3.2442-2449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maudru T, Peden K. Elimination of background signals in a modified polymerase chain reaction-based reverse transcriptase assay. J Virol Methods. 1997;66:247–261. doi: 10.1016/s0166-0934(97)00067-0. [DOI] [PubMed] [Google Scholar]
- 39.McKenzie R A. Observations on diseases of free-living and captive koalas (Phascolarctos cinereus) Aust Vet J. 1981;57:243–246. doi: 10.1111/j.1751-0813.1981.tb02670.x. [DOI] [PubMed] [Google Scholar]
- 40.Montali R J, Bush M, Cromie R, Holland S M, Maslow J N, Worley M, Witebsky F G, Phillips T M. Primary Mycobacterium avium complex infections correlate with lowered cellular immune reactivity in Matschie's tree-kangaroos (Dendrolagus matschiei) Infect Dis. 1998;178:1719–1725. doi: 10.1086/314517. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien T M, Hanger J J, McKee J J, Robinson W F. Proceedings of a Conference on the Status of the Koala in 1997. Brisbane, Australia: Australian Koala Foundation; 1997. The isolation, characterisation and partial gene sequence of a retrovirus from koalas; pp. 106–113. [Google Scholar]
- 42.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 43.Parent I, Qin Y, Vandenbroucke A T, Walen C, Delferriere N, Godfroid E, Burtonboy G. Characterisation of a C-type retrovirus isolated from an HIV infected cell line: complete nucleotide sequence. Arch Virol. 1998;143:1077–1092. doi: 10.1007/s007050050357. [DOI] [PubMed] [Google Scholar]
- 44.Petropoulos C. Retroviral taxonomy, protein structures, sequences, and genetic maps. In: Coffin J M, et al., editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 757–805. [Google Scholar]
- 45.Rasmussen H B. Interactions between exogenous and endogenous retroviruses. J Biomed Sci. 1997;4:1–8. doi: 10.1007/BF02255587. [DOI] [PubMed] [Google Scholar]
- 46.Robinson W, O'Brien T, Hanger J, McKee J. Proceedings of a Conference on the Status of the Koala in 1996. Brisbane, Australia: Australian Koala Foundation; 1996. Leukaemia in koalas (Phascolarctos cinereus): the presence of viral particles in cultured lymphocytes; pp. 97–99. [Google Scholar]
- 47.Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Coffin J M, et al., editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 475–586. [PubMed] [Google Scholar]
- 48.Roux K H. Optimization and troubleshooting in PCR. In: Dieffenbach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 53–57. [Google Scholar]
- 49.Salemi M, Lewis M, Egan J F, Hall W W, Desmyter J, Vandamme A. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc Natl Acad Sci USA. 1999;96:13253–13258. doi: 10.1073/pnas.96.23.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Schwartz R M, Dayhoff M O. Matrices for detecting distance relationships. In: Dayhoff M O, editor. Atlas of protein sequence and structure. 5, Suppl. 3. Washington, D.C.: National Biomedical Research Foundation; 1978. pp. 353–358. [Google Scholar]
- 52.Sheets R L, Pandey R, Jen W-C, Roy-Burman P. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol. 1993;67:3118–3125. doi: 10.1128/jvi.67.6.3118-3125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith E J. Endogenous avian leukemia viruses. In: de Boer G F, editor. Avian leukosis. Boston, Mass: Martinus Nijhoff Publishing; 1987. pp. 101–120. [Google Scholar]
- 54.Synder S P, Dungworth D L, Kawakami T G, Callaway E, Lau D T L. Two cases of lymphosarcoma in gibbons (Hylobates lar) with associated C-type virus. J Natl Cancer Inst. 1973;51:89–94. doi: 10.1093/jnci/51.1.89. [DOI] [PubMed] [Google Scholar]
- 55.Spencer A J, Canfield P J. Lymphoid neoplasia in the koala (Phascolarctos cinereus): a review and classification of 31 cases. J Zoo Wildl Med. 1996;27:303–314. [Google Scholar]
- 56.Theilen G H, Gould D, Fowler M, Dungworth D L. C-type virus in tumor tissue of a woolly monkey (Lagothrix sp.) with fibrosarcomas. J Natl Cancer Inst. 1971;47:881–889. [PubMed] [Google Scholar]
- 57.Tonjes R R, Lower R, Boller K, Denner J, Hasenmaier B, Kirsch H, Konig H, Korbmacher C, Limbach C, Lugert R, Phelps R C, Scherer J, Thelen K, Lower J, Kurth R. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:s261–s267. doi: 10.1097/00042560-199600001-00039. [DOI] [PubMed] [Google Scholar]
- 58.Vogt V M. Retroviral virions and genomes. In: Coffin J M, et al., editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 59.Wilkinson D A, Mager D L, Leong J A C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 60.Wilkinson R, Kotlarski I, Barton M. Koala lymphoid cell: analysis of antigen-specific responses. Vet Immunol Immunopathol. 1992;33:237–247. doi: 10.1016/0165-2427(92)90184-r. [DOI] [PubMed] [Google Scholar]
- 61.Worley M, Rideout B, Shima A, Janssen D. Proceedings of the American Association of Zoo Veterinarians. 1993. Opportunistic infections, cancer and hematologic disorders associated with retrovirus infection in the koala (Phascolarctos cinereus) pp. 181–182. Saint Louis, Mo. [Google Scholar]
- 62.Yang Z. Maximum likelihood phylogenetic estimate from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 63.Yang Z, Goldman N, Friday A. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol. 1994;11:316–324. doi: 10.1093/oxfordjournals.molbev.a040112. [DOI] [PubMed] [Google Scholar]