Abstract
Objective:
Suicide is a major public health problem and it has a prominent genetic component. We performed a genome-wide association study (GWAS) of suicidal behaviour severity.
Methods:
Suicide behaviour severity was assessed within the Schedules for Clinical Assessment in Neuropsychiatry in our mood disorder sample (n=3506) for the GWAS. We also performed polygenic risk score analyses to explore genetic sharing between suicidal behaviour severity and a number of phenotypes, including bipolar disorder, major depressive disorder, alcoholism, post-traumatic stress disorder, impulsivity, insomnia, educational attainment, loneliness, maltreatment, and amygdala volume.
Results:
We did not detect genome-wide significant findings at the single-marker or gene level. We report a number of suggestive single-marker and gene-based findings. Our polygenic risk score analyses did not yield significant findings with these phenotypes.
Conclusions:
Larger sample sizes are required to detect moderate effects.
Keywords: Suicidality, genome-wide association study, bipolar disorder, major depressive disorder, suicidal behaviour severity
Introduction
Suicide claims over 800,000 lives worldwide each year, and 3–5% of adults attempt suicide at least once in their lifetime, making it a serious public health issue (Kessler et al. 1999; Weissman et al. 1999; Szadoczky et al. 2000; WHO 2014). Whilst the mortality rates of cancers, HIV, diabetes, and cardiovascular diseases have been declining steadily, rates of suicide have declined much more slowly over the years (Statcan 2017). Lifetime prevalence of suicide attempt is over 30% in both bipolar disorder and major depressive disorder; thus it is important to study risk factors of suicide in mood disorders (Dong, Lu, et al. 2019; Dong, Zeng, et al. 2019). Suicidal thinking (ideation) and behaviour/attempt (SIB) are very complex and are likely affected by many variables. Family history appears to be an important risk factor for suicide (Turecki and Brent 2016).
Family, twin, and adoption studies support a prominent genetic component for SIB (Tidemalm et al. 2011; Zai, de Luca, et al. 2012). Heritability of suicide attempt/completion is estimated to be at 43% (95% confidence interval: 27–60%) (McGuffin et al. 2010), with the genetic component for suicidality at least partly independent of that for the underlying psychiatric disorders (Zai, de Luca, et al. 2012). A number of promising genetic markers have arisen from genetic association studies of SIB (Zai, de Luca, et al. 2012; Sokolowski et al. 2014). Our group reported in a meta-analysis that the Brain-derived Neurotrophic Factor (BDNF) rs6265 Met66 allele was associated with suicide attempts (Zai, Manchia, et al. 2012). Evidence for its receptor Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2) has also been growing (Pulay and Rethelyi 2016). Meta-analyses have supported the roles of the serotonin transporter gene (SLC6A4) promoter polymorphism (HTTLPR) (Li and He 2007), FK506 binding protein 5 (FKBP5) rs3800373 and rs1360780, Corticotropin-releasing Hormone Receptor 1 (CRHR1) rs2664008, rs1724425, and rs878886 (De la Cruz-Cano 2017), Catechol-O-methyltransferase (COMT) rs4680 (Sadeghiyeh et al. 2017), tryptophan hydroxylase 1 (TPH1) rs1800532 and rs1799913. In addition, the Acid Phosphatase (ACP1) rs300774 identified in a suicide attempt genome-wide association studies (GWAS) (Willour et al. 2012) was replicated recently in two samples (Pawlak et al. 2016; Li et al. 2017). Among GWAS of SIB conducted (Bani-Fatemi et al. 2016; Coon et al. 2018; Kimbrel et al. 2018; Brick et al. 2019), more recent GWASs of suicide attempt have identified additional significant markers. These include rs4809706 in the iPSYCH Danish population birth cohort study (Erlangsen et al. 2020), centrosomal protein 162 rs12524136 from US military cohorts (Stein et al. 2017), olfactory receptor family 7 subfamily G member 2 rs12972618 for high suicide attempt risk in a patient sample from the Vanderbilt University Medical Centre (Ruderfer et al. 2020), Cortexin Domain Containing 1 rs72740082 from the Yale-Penn study on the genetics of drug and alcohol dependence (Levey et al. 2019), and interacts with SUPT6H, CTD assembly factor 1 rs138689899 from the Psychiatric Genomics Consortium (Mullins et al. 2019). Aside from the replication of ACP1 rs300774, other GWAS findings have not been successfully replicated. Though earlier candidate genes findings in BDNF and NTRK2 have not been implicated in suicidality GWAS, neurodevelopment in the form of gene-based analysis or polygenic risk scores calculated from neurodevelopmental genes has been implicated in SIB (Pulay and Rethelyi 2016; Sokolowski et al. 2016).
While most GWASs focussed on the binary phenotype of suicide attempts, few have looked into quantitative measures of suicidality in GWASs. Examining the continuous variable of the severity of SIB may offer more statistical power to detect an effect size than suicide attempt (Bhandari et al. 2002; Altman and Royston 2006). Strawbridge et al. (2019) performed a GWAS on suicidality that included suicidal ideation and suicide attempt in the UK Biobank sample. They found zinc finger CCHC-type containing 7 rs62535711 to be genome-wide significant (Strawbridge et al. 2019). We previously published a GWAS of suicidal behaviour severity in bipolar disorder (BD) patients (Zai et al. 2015) and suicidality scores in major depressive disorder patients (Schosser et al. 2011). Here, we provide an updated GWAS of suicide severity and SCAN suicidality scores across bipolar disorder and major depressive disorder (MDD) patients where suicidal behaviour severity was assessed using the same instrument.
Methods
Samples
The characteristics of the samples included in this study have been described previously (Scott et al. 2009; Schosser et al. 2011; Zai et al. 2015) and are shown in Table 1. BD patients (Sample CA2, n = 428) at CAMH, and an independent sample of 483 BD cases (IOP) from the Institute of Psychiatry, Psychology & Neuroscience in London, UK. The participants were of self-reported European ancestry and were at least 18 years old when they were enrolled through advertisements in hospitals, family physicians’ offices, clinics, or patient support groups. Their BD diagnoses according to DSM-IV and ICD-10 criteria were confirmed using the Schedules for Clinical Assessment in Neuropsychiatry (SCAN). Exclusion criteria included a diagnosis of intravenous drug dependency, reported intravenous drug use, the presence of mood incongruent psychotic symptoms, manic episodes only in conjunction with or as a result of alcohol, substance abuse, substance dependence, medical illnesses, or medication. Their suicidality was assessed using the SCAN Suicide Severity item (6.011) as follows: 0 for non-suicidal, 1 for deliberately considering suicide or self-harm, 2 for injuring self or making an attempt without serious consequences; 3 for serious self-harm or attempt; 4 for an attempt at suicide designed to be lethal. We also derived SCAN Suicidality (SSU) scores as described previously (Schosser et al. 2011). SSU a combination of the SCAN Suicide Severity item (6.011) and the SCAN Loathing of Life item (6.012), where individuals who scored 1 or above in the Suicide Severity item would have the SSU score the same as the Suicide Severity item score plus three, and those who scored 0 would have the SSU score the same as the Loathing of Life score. These two measures are highly correlated within each of our three samples, with R values of more than 0.90. The distributions of the Suicide Severity item and SSU scores are shown in Supplementary Figure S1. This work has been approved by the institutional review boards and adheres to the ethical standards of the national and institutional committees on human experimentation as well as the Declaration of Helsinki in 1975 (revised in 2008).
Table 1.
Demographic information for the 3 sample sets of European ancestry included in the current study.
| Sample | CA2 (Canada) | IOP (UK) | RAD (RADIANT) |
|---|---|---|---|
|
| |||
| Diagnosis | Bipolar disorder | Bipolar disorder | Major depressive disorder |
| Total n | 313 | 406 | 2787 |
| % Female (n) | 0.60 (188) | 0.67 (270) | 0.71 (1973) |
| Age (mean ±SD) | 43.67 ±12.41 | 47.32 ±11.04 | 44.89±12.11 |
| % Ever >5 alcoholic drinks almost every day (n) | 0.36 (112) | 0.32 (128) | N/A |
| SCAN Suicide Severity (6.011) (n (%)) | |||
| 0 | 112 (36%) | 119 (29%) | 1188 (43%) |
| 1 | 108 (34%) | 162 (40%) | 1134 (41%) |
| 2 | 52 (17%) | 59 (15%) | 249 (9%) |
| 3 | 13 (4%) | 16 (4%) | 64 (2%) |
| 4 | 28 (9%) | 50 (12%) | 152 (5%) |
| SCAN Suicidality (SSU) Score (N(%)) | |||
| 0 | 36 (12%) | 52 (13%) | 373 (13%) |
| 1 | 22 (7%) | 11 (3%) | 133 (5%) |
| 2 | 21 (7%) | 23 (5%) | 347 (13%) |
| 3 | 33 (10%) | 33 (8%) | 335 (12%) |
| 4 | 108 (34%) | 162 (40%) | 1134 (41%) |
| 5 | 52 (17%) | 59 (15%) | 249 (9%) |
| 6 | 13 (4%) | 16 (4%) | 64 (2%) |
| 7 | 28 (9%) | 50 (12%) | 152 (5%) |
MDD patients came from the RADIANT (RAD) studies DeCC and DeNt, with suicidal behaviour severity was assessed during a depressive episode. The DeCC (Depression Case Control) sample consists of 1346 cases (69.3% women) of recurrent depression fulfilling DSM-IV and/or ICD-10 criteria of at least moderate severity ascertained from three UK clinical sites (London, Cardiff, and Birmingham) (Cohen-Woods et al. 2009), with a mean age of onset of 22.9 years (SD 10.8 years). Subjects were identified from psychiatric clinics, hospitals, general medical practices, and from volunteers responding to media advertisements. The DeNt (Depression Network) affected sibling pair linkage study (Farmer et al. 2004; McGuffin et al. 2005) consists of recurrent depression cases with moderate or more severe symptoms. Participants were enrolled from the same three UK sites as DeCC, plus four additional sites in Europe (Aarhus, Bonn, Dublin, and Lausanne) and one in the USA (St. Louis). Only the 898 probands from the families with available GWAS data were included in the present study. A further sample of 163 recurrent depression cases was included in the present study; these participants were enrolled in Bonn and Lausanne, using the same protocol as the DeNt study. Overall, all RADIANT participants fulfilled DSM-IV and/or ICD-10 criteria for moderate or more severe depression (Schosser et al. 2011). The RADIANT study coordinators were trained by AEF, thus ensuring consistency in the clinical data being collected. Exclusion criteria included a history or family history of schizophrenia or bipolar disorder, mood-incongruent psychosis, or mood symptoms that were related to alcohol or substance use. Each participant in the RADIANT study had his/her most severe episode of depression collected retrospectively using SCAN, which included the same two questions on a loathing of life (6.012) and suicide severity (6.011) mentioned above (Figure S1).
Most of the genotyping on the Samples CA2 and IOP were performed with the IlluminaSentrix Human Hap550 Beadchip (Illumina Inc., San Diego, CA, USA) at Illumina Inc. (San Diego, CA, USA), with 290 CA2 participants genotyped at the Genome Quebec facility (Montreal, Quebec, Canada). Genotyping of the RAD sample was performed with the Illumina HumanHap610-Quad BeadChip at the Centre National de Génotypage, France.
Quality control procedures were applied to individual SNP data in the RAD sample as described previously (Lewis et al. 2010; Schosser et al. 2011). Individuals were excluded if they had more than 2% missing genotypes, the mismatch between genotype-assigned and reported sex, abnormal heterozygosity, relatedness (up to 2nd-degree) with any other participants, or non-European ancestry as determined using principal components analysis with reference data from HapMap CEU, JPT, CHB, YRI, and GIH populations with EIGENSTRAT (Price et al. 2006). SNPs were excluded if they had less than 1% minor allele frequency, genotype frequencies deviating significantly from Hardy–Weinberg equilibrium (p < 1E – 6). Principal component analysis was performed again after QC procedures, and three ancestry-informative principal components, as determined by the Scree plot, were used as covariates in the analysis. The data set of 4465 participants (2905 being depression cases) genotyped on 464,759 SNPs went through the whole-genome imputation described below.
We applied comparable quality control measures for the CA2 and IOP samples separately using PLINK (Purcell et al. 2007) and R (R Development Core Team 2008). Briefly, participants were excluded if they had more than 5% missing genotypes, the mismatch between genotype-assigned sex and reported sex, abnormal heterozygosity, relatedness to any other participants. SNPs of which genotypes deviated significantly from Hardy-Weinberg Equilibrium (p < 1E – 6) were excluded from subsequent analyses, as were those with less than 1% minor allele frequency or more than 5% missing genotypes. We ran a principal component analysis of the genotypes to retain the discrete cluster that corresponded to European ancestry. Principal component analysis was performed again after QC procedures, and two ancestry-informative principal components, as determined by the Scree plot, were used as covariates in the analysis. After quality control, 508,883 SNPs for 313 CA2 bipolar disorder cases, and 515,024 SNPs for 406 IOP bipolar disorder cases went through whole-genome imputation.
Whole-genome imputation was performed using Minimac4 implemented on the Michigan Imputation Server with haplotype phasing using Eagle v2.4, 1000 Genomes Phase 3 data (for CA2 and IOP) or Haplotype Reference Consortium data (for RAD) as the reference panel, and minimum R-squared threshold of 0.3 (Das et al. 2016), with additional filtering for SNPs with R-squared value of at least 0.6. Whole-genome imputation and quality control yielded 8,701,619 SNPs for the 313 CA2 cases, 8,724,065 SNPs for the 406 IOP cases, and 7,472,311 SNPs for the 2787 RAD cases.
Our total sample size of 3506 has enough power to detect 3% of the variance of suicide severity that is accounted for by the single-nucleotide polymorphisms (SNPs; alpha = 5E – 8, minor allele frequency 1%, additive model) (Gauderman and Morrison 2006).
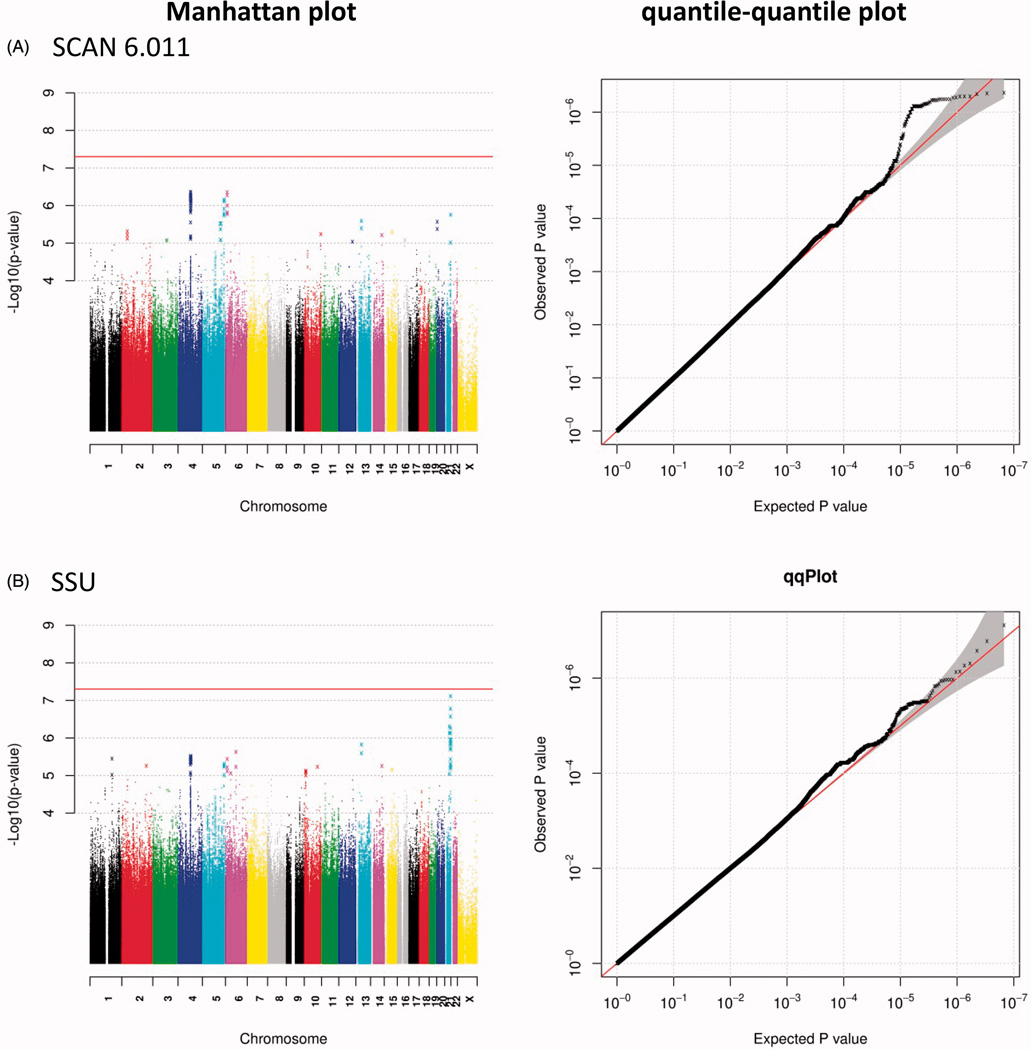
For 6,737,359 SNPs that are included in all three samples, we conducted analyses of suicide severity scores and suicidality scores separately for the three sample sets using PLINK, with covariates including sex, age, and ancestry-informative principal components for all three sample sets, as well as the sub-studies within the RAD sample set. Then we performed a meta-analysis using sample size, the direction of effect, and p-value to derive an overall p-value for each marker (METAL software (Willer et al. 2010)). The quantile-quantile plots and the Manhattan plots were graphed in R, with the significance threshold for single-SNP analysis set at 5E – 8.
In addition to single-marker analysis, we conducted gene-based analysis for each sample separately and then meta-analysed the results across the samples using MAGMA (de Leeuw et al. 2015), with the significance threshold for 18,601 genes set at 2.7E – 6. We further obtained standardised polygenic risk scores for each sample without p-value thresholding (that is, p-value threshold of 1) using PRSice (Euesden et al. 2015), and ran linear regression analyses in R with age, sex, ancestry-informative principal components, and sub-studies as covariates. Then we ran meta-analyses using the effect estimates and standard errors from the three samples using the R metafor package. Previous studies reported associations between suicidality and a number of phenotypes, including impulsivity (Anestis et al. 2014), alcoholism (Darvishi et al. 2015), educational attainment (Dong, Lu, et al. 2019), insomnia (Dong, Zeng, et al. 2019), loneliness (Dong et al. 2018), post-traumatic stress disorder (Krysinska and Lester 2010), and amygdala volume (Wang et al. 2019). We used the summary statistics for 10 such phenotypes to calculate the polygenic risk scores: alcohol dependence (for Europeans (Walters et al. 2018)), bipolar disorder (Stahl et al. 2019), impulsivity [F1.50.BIS_NonP.csv – top 10 K SNPs for BIS from 23&Me], insomnia (from UKB (Jansen et al. 2019)), post-traumatic stress disorder (PTSD, Freeze 1 (Duncan et al. 2018)), major depressive disorder (PGC-MDD2 excluding 23&Me data (Wray et al. 2018)), educational attainment (Lee et al. 2018), childhood maltreatment (Freeze 2 with UKB data (Dalvie et al. 2020)), loneliness (Gao et al. 2017), and amygdala volume (ENIGMA2 (Hibar et al. 2015)).
Results
We found no genome-wide significant results in our single-marker analysis (Figure 1). The suggestive findings (p < 1E – 5) for Suicide Severity are shown in Table 2, Supplementary Table S1, and Supplementary Figure S2, and those for SCAN Suicidality are shown in Table 2, Supplementary Table S2, and Supplementary Figure S3. We also included genes for which they are shown to be expression quantitative trait loci according to the Genotype Tissue Expression (GTEx; (Battle et al. 2017)) portal in Supplementary Tables S1 and S2. Among the top findings for SCAN Suicide Severity (6.011), a number of correlated SNPs are located in the sperm tail PG-rich repeat-containing 2 (STPG2) gene at chromosomal location 4q23 and their genotypes are associated with STPG2 gene expression according to the GTEx portal. Another set of correlated SNPs are located in the mitochondrial calcium uniporter regulator 1 (MCUR1) gene at 6p23 and their genotypes are associated with MCUR1 gene expression (GTEx). The third set of SNPs is located in the Rho GTPase activating protein 26 (ARHGAP26) gene at 5q31.3. SNPs in STPG2 and MCUR1 are also among the top findings for SSU scores. There are also correlated SNPs in the USP6 N-terminal like (USP6NL) gene at 10p14 that are also associated with USP6NL gene expression (GTEx) and others upstream of the Thrombospondin Type Laminin G Domain And EAR Repeats (TSPEAR) gene at 21q22.3 that are also associated with TSPEAR gene expression (GTEx).
Figure 1.
Manhattan and quantile–quantile plots for the analysis of (A) the SCAN Suicide Severity (6.011) and (B) SCAN Suicidality SSU scores.
Table 2.
Summary of results from our genome-wide association study of SCAN Suicide Severity (6.011) and SCAN Suicidality (SSU) scores.
| Phenotype | SNP | Gene | eQTLa | CHR | BP | Z-score | p | Direction |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 6.011 | rs9654143 | STPG2 | STPG2/RAP1GDS1 | 4 | 98,689,152 | −5.054 | 4.33E − 07 | ––– |
| 6.011 | rs9647614 | MCUR1 | 6 | 13,843,271 | 4.892 | 9.98E − 07 | +++ | |
| 6.011 | rs61246028 | LINC00400 | 13 | 43,756,955 | 4.703 | 2.57E − 06 | +++ | |
| 6.011 | rs79099900 | RASSF2 | 20 | 4,804,781 | −4.692 | 2.71E − 06 | ––– | |
| 6.011 | rs74961332 | POFUT2 | POFUT2 | 21 | 46,702,822 | 4.779 | 1.76E − 06 | +–+ |
| SSU | rs9654143 | STPG2 | STPG2/RAP1GDS1 | 4 | 98,689,152 | −5.054 | 4.33E − 07 | +++ |
| SSU | rs9647614 | 6 | 13,843,271 | 4.892 | 9.98E – 07 | ––– | ||
| SSU | rs61246028 | LINC00400 | 13 | 43,756,955 | 4.703 | 2.57E − 06 | +++ | |
| SSU | rs77916371 | PTPN21 | 14 | 88,969,691 | −4.522 | 6.13E − 06 | ––– | |
| SSU | rs11071625 | LOC107984782 | 15 | 61,967,903 | −4.562 | 5.07E − 06 | ––– | |
|
|
||||||||
| Gene Name | Gene ID | Number of SNPs | CHR | N | ZSTAT | p | NPERM | |
|
|
||||||||
| 6.011 | ACE | 1636 | 61 | 17 | 3506 | 3.7384 | 9.26E − 05 | 1000 |
| 6.011 | GFRA1 | 2674 | 666 | 10 | 3506 | 4.1658 | 1.55E − 05 | 1000 |
| 6.011 | STPG2 | 285555 | 1887 | 4 | 3506 | 3.9027 | 4.76E − 05 | 1000 |
| 6.011 | IQCA1 | 79781 | 373 | 2 | 3506 | 4.1165 | 1.92E − 05 | 1000 |
| SSU | GFRA1 | 2674 | 666 | 10 | 3506 | 3.9624 | 3.71E − 05 | 1000 |
| SSU | STPG2 | 285555 | 1887 | 4 | 3506 | 3.8058 | 7.07E − 05 | 1000 |
| SSU | DNASE2B | 58511 | 92 | 1 | 3506 | 3.7905 | 7.52E − 05 | 1000 |
The upper panel shows the top five SNPs for each phenotype, and the bottom panel shows the top suggestive gene-based findings for these phenotypes.
According to GTEx portal.
The gene-based analysis yielded a number of suggestive findings (p < 1E – 4); Supplementary Figures S2 and S3; genes with p < 1E – 4 are shown in Supplementary Tables S3 and S4), including the GDNF family receptor alpha 1 (GFRA1, at 10q25.3), STPG2 genes for both suicide severity and suicidality score, as well as the angiotensin I converting enzyme (ACE, at 17q23.3) and IQ motif containing with AAA domain 1 (IQCA1, at 2q37.3) genes for suicide severity alone, and the Deoxyribonuclease 2 Beta (DNASE2B at 1p31.1) gene for suicidality score only. Polygenic risk score analyses did not yield significant findings for either suicide severity or suicidality score (Supplementary Table S5). Furthermore, we examined the top genome-wide significant findings from recent GWASs of a suicide attempt. For the previous genome-wide significant SNPs that were covered by our GWAS data, we did not find them to be nominally significant in our GWAS (p > 0.05) (Supplementary Table S6). We re-ran the analysis of the suggestive SNPs from the current study and previously reported top SNPs using ordinal regression (STATA v15); the results from ordinal regression were comparable to those from linear regression in plink in that for p-values of less than 0.7 the direction of effect were the same between the two regression methods, and for p-values of >0.005, the p-values were within an order of magnitude between the two regression methods (Supplementary Tables S7 and S8) for Suicide Severity and SCAN Suicidality, respectively).
Discussion
We report here a GWAS of suicidal behaviour severity in approximately 3500 mood disorder patients using the same instrument for assessing suicidality. The lack of statistically significant findings is likely the result of an insufficient sample size to detect small effect sizes for a highly complex phenotype. The mechanisms of the suggestive associations are unclear. The single-SNP and gene-based findings both point to the role of the STPG2 gene. The STPG2 gene has been implicated in autistic spectrum disorder (Connolly et al. 2017) and neurodevelopmental disorders (Asadollahi et al. 2017). This finding supports the previously reported associations of neurodevelopment genes in suicidality (Zai, Manchia, et al. 2012; Pulay and Rethelyi 2016; Sokolowski et al. 2016). In addition, a rare variant in STPG2 (rs1727284 G allele) was associated with alcohol consumption in a recent meta-analysis (Brazel et al. 2019), and another variant (rs11933823 C allele) was associated with adventurousness (Karlsson Linnér et al. 2019). The MCUR1 gene has been associated with intelligence (Savage et al. 2018). Abnormal splicing of the ARHGAP26 gene has been reported in schizophrenia (Glatt et al. 2011). The USP6NL gene has been associated with Alzheimer’s disease (Jun et al. 2017). The finding with GFRA1 was previously reported for GWAS of suicidality in the MDD sub-sample of our current study (Schosser et al. 2011). Increased IQCA1 gene expression has been reported in Alzheimer’s disease patients compared to healthy controls (Brooks and Mias 2019). As for the suggestive gene-level finding with ACE, a number of early genetic studies yielded mixed findings (Hong et al. 2002; Hishimoto et al. 2006; Fudalej et al. 2009; Sparks et al. 2009; Zai, de Luca, et al. 2012). Since there is growing evidence of angiotensin-receptor blockers as a possible risk factor for suicide (OR = 3.52) (Callreus et al. 2007; Mamdani et al. 2019), future suicide genetic studies that control for medications such as ACE blockers may help clarify some of the mixed findings. Future work may include an examination of pharmacogenetics of suicide in the context of ACE inhibitor/blocker medications.
Our findings need to be considered with caution due to a number of limitations. We proposed that severity score phenotype for suicidality might have larger effect sizes. However, we were unable to support this hypothesis with our data, probably because our sample size was insufficient to detect effects at single-marker, gene, or polygenic risk score levels. For polygenic risk score analysis of impulsivity, we only have the top 10,000 SNPs available, so we may not be getting the full picture of the genetic sharing between impulsivity and suicidality from this analysis. Our phenotypes of suicide severity and suicidality score pertain to the most severe depressive episode for each patient and might have missed some lifetime suicidal ideation or suicide attempts that did not occur in the most severe depressive episodes. Thus, our preliminary findings encourage additional studies on the role of neurodevelopment and medication use in suicidality.
Supplementary Material
Acknowledgements
The authors would like to thank the funding sources and participating sites for the study samples. The authors also gratefully acknowledge the contributing studies and the participants in those studies without whom this effort would not be possible.
Funding
This work was supported by a joint grant from the Medical Research Council, UK and GlaxoSmithKline [G0701420]; by financial support from the NIHR Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, King’s College London. The GENDEP study was funded by a European Commission Framework 6 grant, EC Contract Ref. [LSHB-CT-2003–503428]. GlaxoSmithKline funded the collection of the DeNt cohort of depression cases, the genotyping of all RADIANT cases (with the MRC), and both the collection and genotyping of the GSK-Munich cohort of depression cases. GMH was supported by Barts Charity [OGA007973] and International Bipolar Foundation. This project was supported by funding sources including: Canadian Institutes for Health Research [JLK (MOP-115097), JBV (MOP-84391), VdL (MOP-115689, MOP-119332)], Brain and Behaviour Research Foundation (NARSAD Young Investigator Awards) [AKT, CCZ], and CAMH Foundation [JLK, AKT, CCZ]. CF was supported by Fondazione Umberto Veronesi. The collection of the IOP and CA2 samples was supported by funding from GlaxoSmithKline. This work was also supported by funding sources and participating studies that contributed to the summary statistics data for the polygenic risk score analyses. In addition, the Substance Use Disorders Working Group of the Psychiatric Genomics Consortium (PGC-SUD) is supported by funds from NIDA and NIMH [MH109532] and, previously, with analyst support from NIAAA [U10AA008401 (COGA)]. All genotype and loneliness phenotype data were collected as part of the Health and Retirement Study (http://hrsonline.isr.umich.edu/). The Health and Retirement Study genetic data is sponsored by the National Institute on Aging [U01AG009740, RC2AG036495, and RC4AG039029] and was conducted by the University of Michigan.
Disclosure statement
JLK: has received honoraria from Novartis, Roche and Eli Lilly corporations, and is an unpaid member of the scientific advisory board of AssureRx Corp. JLK & CCZ: authors in patents on suicide markers. PM was employee of GlaxoSmithKline when that research was performed. CF, AKT, EK, GMH, RSZ, VdL, JS, NK, CML, GB, AF, PM, IJ, LJ, and JBV reported no conflict of interest related to this manuscript. The funding sources had no direct involvement in the study design, data collection, analysis, or interpretation in the present study.
Footnotes
Supplemental data for this article can be accessed at http://dx.doi.org/10.1080/15622975.2021.1907711
References
- Altman DG, Royston P. 2006. The cost of dichotomising continuous variables. BMJ. 332(7549):1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anestis MD, Soberay KA, Gutierrez PM, Hernandez TD, Joiner TE. 2014. Reconsidering the link between impulsivity and suicidal behavior. Pers Soc Psychol Rev. 18(4):366–386. [DOI] [PubMed] [Google Scholar]
- Asadollahi R, Zweier M, Gogoll L, Schiffmann R, Sticht H, Steindl K, Rauch A. 2017. Genotype-phenotype evaluation of MED13L defects in the light of a novel truncating and a recurrent missense mutation. Eur J Med Genet. 60(9): 451–464. [DOI] [PubMed] [Google Scholar]
- Bani-Fatemi A, Graff A, Zai C, Strauss J, De Luca V. 2016. GWAS analysis of suicide attempt in schizophrenia: main genetic effect and interaction with early life trauma. Neurosci Lett. 622:102–106. [DOI] [PubMed] [Google Scholar]
- Battle A, Brown CD, Engelhardt BE, Montgomery SB, 2017. Genetic effects on gene expression across human tissues. Nature. 550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari M, Lochner H, Tornetta P. 2002. Effect of continuous versus dichotomous outcome variables on study power when sample sizes of orthopaedic randomized trials are small. Arch Orthop Trauma Surg. 122(2):96–98. [DOI] [PubMed] [Google Scholar]
- Brazel DM, Jiang Y, Hughey JM, Turcot V, Zhan X, Gong J, Batini C, Weissenkampen JD, Liu M, Barnes DR, et al. 2019. Exome chip meta-analysis fine maps causal variants and elucidates the genetic architecture of rare coding variants in smoking and alcohol use. Biol Psychiatry. 85(11): 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick LA, Marraccini ME, Micalizzi L, Benca-Bachman CE, Knopik VS, Palmer RHC. 2019. Overlapping genetic effects between suicidal ideation and neurocognitive functioning. J Affect Disord. 249:104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks LRK, Mias GI. 2019. Data-driven analysis of age, sex, and tissue effects on gene expression variability in Alzheimer’s disease. Front Neurosci. 13:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callreus T, Agerskov Andersen U, Hallas J, Andersen M. 2007. Cardiovascular drugs and the risk of suicide: a nested case-control study. Eur J Clin Pharmacol. 63(6):591–596. [DOI] [PubMed] [Google Scholar]
- Cohen-Woods S, Gaysina D, Craddock N, Farmer A, Gray J, Gunasinghe C, Hoda F, Jones L, Knight J, Korszun A, et al. 2009. Depression Case Control (DeCC) Study fails to support involvement of the muscarinic acetylcholine receptor M2 (CHRM2) gene in recurrent major depressive disorder. Hum Mol Genet. 18(8):1504–1509. [DOI] [PubMed] [Google Scholar]
- Connolly S, Anney R, Gallagher L, Heron EA. 2017. A genome-wide investigation into parent-of-origin effects in autism spectrum disorder identifies previously associated genes including SHANK3. Eur J Hum Genet. 25(2):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon H, Darlington TM, DiBlasi E, Callor WB, Ferris E, Fraser A, Das SC, Crowell SE, Chen D, Anderson JS, et al. 2018. Genome-wide significant regions in 43 Utah high-risk families implicate multiple genes involved in risk for completed suicide. Mol Psychiatry. 25:3077–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvie S, Maihofer AX, Coleman JRI, Bradley B, Breen G, Brick LA, Chen C-Y, Choi KW, Duncan LE, Guffanti G, et al. 2020. Genomic influences on self-reported childhood maltreatment. Transl Psychiatry. 10(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvishi N, Farhadi M, Haghtalab T, Poorolajal J. 2015. Alcohol-related risk of suicidal ideation, suicide attempt, and completed suicide: a meta-analysis. PLoS One. 10(5): e0126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. 2016. Next-generation genotype imputation service and methods. Nat Genet. 48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cruz-Cano E 2017. Association between FKBP5 and CRHR1 genes with suicidal behavior: a systematic review. Behav Brain Res. 317:46–61. [DOI] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D. 2015. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Wang SB, Li Y, Xu DD, Ungvari GS, Ng CH, Chow IHI, Xiang YT. 2018. Prevalence of suicidal behaviors in patients with major depressive disorder in China: a comprehensive meta-analysis. J Affect Disord. 225:32–39. [DOI] [PubMed] [Google Scholar]
- Dong M, Lu L, Zhang L, Zhang Q, Ungvari GS, Ng CH, Yuan Z, Xiang Y, Wang G, Xiang YT. 2019. Prevalence of suicide attempts in bipolar disorder: a systematic review and meta-analysis of observational studies. Epidemiol Psychiatr Sci. 29:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Zeng LN, Lu L, Li XH, Ungvari GS, Ng CH, Chow IHI, Zhang L, Zhou Y, Xiang YT. 2019. Prevalence of suicide attempt in individuals with major depressive disorder: a meta-analysis of observational surveys. Psychol Med. 49(10):1691–1704. [DOI] [PubMed] [Google Scholar]
- Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Bierut LJ, Bisson J, et al. 2018. Largest GWAS of PTSD (N 20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 23(3): 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlangsen A, Appadurai V, Wang Y, Turecki G, Mors O, Werge T, Mortensen PB, Starnawska A, Borglum AD, Schork A, et al. 2020. Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Mol Psychiatry. 25(10): 2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. 2015. PRSice: polygenic Risk Score software. Bioinformatics. 31(9):1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer A, Breen G, Brewster S, Craddock N, Gill M, Korszun A, Maier W, Middleton L, Mors O, Owen M, et al. 2004. The Depression Network (DeNT) Study: methodology and sociodemographic characteristics of the first 470 affected sibling pairs from a large multi-site linkage genetic study. BMC Psychiatry. 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudalej S, Fudalej M, Kostrzewa G, Kuźniar P, Franaszczyk M, Wojnar M, Krajewski P, Płoski R. 2009. Angiotensin-converting enzyme polymorphism and completed suicide: an association in Caucasians and evidence for a link with a method of self-injury. Neuropsychobiology. 59(3):151–158. [DOI] [PubMed] [Google Scholar]
- Gao J, Davis LK, Hart AB, Sanchez-Roige S, Han L, Cacioppo JT, Palmer AA. 2017. Genome-Wide Association Study of loneliness demonstrates a role for common variation. Neuropsychopharmacology. 42(4):811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Morrison JM. 2006. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. http://hydra.usc.edu/gxe. [Google Scholar]
- Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. 2011. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 156B(4):382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, et al. 2015. Common genetic variants influence human subcortical brain structures. Nature. 520(7546):224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishimoto A, Shirakawa O, Nishiguchi N, Hashimoto T, Yanagi M, Nushida H, Ueno Y, Maeda K. 2006. Association between a functional polymorphism in the renin-angiotensin system and completed suicide. J Neural Transm. 113(12):1915–1920. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Wang YC, Tsai SJ. 2002. Association study of angiotensin I-converting enzyme polymorphism and symptomatology and antidepressant response in major depressive disorders. J Neural Transm. 109(9):1209–1214. [DOI] [PubMed] [Google Scholar]
- Jansen PR, Watanabe K, Stringer S, Skene N, Bryois J, Hammerschlag AR, de Leeuw CA, Benjamins JS, Muñoz-Manchado AB, Nagel M, et al. 2019. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 51(3):394–403. [DOI] [PubMed] [Google Scholar]
- Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, Buxbaum JD, Byrd GS, Carrasquillo MM, Crane PK, et al. 2017. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 13(7): 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, Lebreton M, Tino SP, Abdellaoui A, Hammerschlag AR, et al. 2019. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 51(2):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Borges G, Walters EE. 1999. Prevalence of and risk factors for lifetime suicide attempts in the National Comorbidity Survey. Arch Gen Psychiatry. 56(7):617–626. [DOI] [PubMed] [Google Scholar]
- Kimbrel NA, Garrett ME, Dennis MF, Hauser MA, Ashley-Koch AE, Beckham JC, 2018. A genome-wide association study of suicide attempts and suicidal ideation in U.S. military veterans. Psychiatry Res. 269:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysinska K, Lester D. 2010. Post-traumatic stress disorder and suicide risk: a systematic review. Arch Suicide Res. 14(1):1–23. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linnér R, et al. 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 50(8): 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey DF, Polimanti R, Cheng Z, Zhou H, Nunez YZ, Jain S, He F, Sun X, Ursano RJ, Kessler RC, Smoller JW, et al. 2019. Genetic associations with suicide attempt severity and genetic overlap with major depression. Transl Psychiatry. 9(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, Weale ME, Schosser A, Paredes UM, Rivera M, et al. 2010. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 167(8): 949–957. [DOI] [PubMed] [Google Scholar]
- Li D, He L. 2007. Meta-analysis supports association between serotonin transporter (5-HTT) and suicidal behavior. Mol Psychiatry. 12(1):47–54. [DOI] [PubMed] [Google Scholar]
- Li J, Yoshikawa A, Meltzer HY. 2017. Replication of rs300774, a genetic biomarker near ACP1, associated with suicide attempts in patients with schizophrenia: relation to brain cholesterol biosynthesis. J Psychiatr Res. 94:54–61. [DOI] [PubMed] [Google Scholar]
- Mamdani M, Gomes T, Greaves S, Manji S, Juurlink DN, Tadrous M, Kennedy SH, Antoniou T. 2019. Association between angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, and suicide. JAMA Netw Open. 2(10):e1913304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Perroud N, Uher R, Butler A, Aitchison KJ, Craig I, Lewis C, Farmer A. 2010. The genetics of affective disorder and suicide. Eur Psychiatry. 25(5):275–277. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, Gill M, Korszun A, Maier W, Middleton L, et al. 2005. Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet. 14(22):3337–3345. [DOI] [PubMed] [Google Scholar]
- Mullins N, Bigdeli TB, Børglum AD, Coleman JRI, Demontis D, Mehta D, Power RA, Ripke S, Stahl EA, Starnawska A, et al. 2019. GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am J Psychiatry. 176(8):651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak J, Dmitrzak-Weglarz M, Wilkosc M, Szczepankiewicz A, Leszczynska-Rodziewicz A, Zaremba D, Kapelski P, Rajewska-Rager A, Hauser J. 2016. Suicide behavior as a quantitative trait and its genetic background. J Affect Disord. 206:241–250. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- Pulay AJ, Rethelyi JM. 2016. Multimarker analysis suggests the involvement of BDNF signaling and microRNA biosynthesis in suicidal behavior. Am J Med Genet. 171(6): 763–776. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2008. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. ISBN 3–900051-07–0. http://www.R-project.org [Google Scholar]
- Ruderfer DM, Walsh CG, Aguirre MW, Tanigawa Y, Ribeiro JD, Franklin JC, Rivas MA. 2020. Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol Psychiatry. 25(10):2422–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghiyeh T, Hosseini Biouki F, Mazaheri M, Zare-Shehneh M, Neamatzadeh H, Poursharif Z. 2017. Association between catechol-O-methyltransferase Val158Met (158G/ A) polymorphism and suicide susceptibility: a meta-analysis. J Res Health Sci. 17(2):e00383. [PubMed] [Google Scholar]
- Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, Nagel M, Awasthi S, Barr PB, Coleman JRI, et al. 2018. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 50(7):912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosser A, Butler AW, Ising M, Perroud N, Uher R, Ng MY, Cohen-Woods S, Craddock N, Owen MJ, Korszun A, et al. 2011. Genomewide association scan of suicidal thoughts and behaviour in major depression. PLoS One. 6(7): e20690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, et al. 2009. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 106(18):7501–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M, Wasserman J, Wasserman D. 2014. Genome-wide association studies of suicidal behaviors: a review. Eur Neuropsychopharmacol. 24(10):1567–1577. [DOI] [PubMed] [Google Scholar]
- Sokolowski M, Wasserman J, Wasserman D. 2016. Polygenic associations of neurodevelopmental genes in suicide attempt. Mol Psychiatry. 21(10):1381–1390. [DOI] [PubMed] [Google Scholar]
- Sparks DL, Hunsaker JC, Amouyel P, Malafosse A, Bellivier F, Leboyer M, Courtet P, Helbecque N. 2009. Angiotensin I-converting enzyme I/D polymorphism and suicidal behaviors. Am J Med Genet B Neuropsychiatr Genet. 150B(2): 290–294. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar HA, et al. 2019. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 51(5): 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statcan. 2017. Age-standardized mortality rates are calculated using the 1991 population of Canada as standard population. Ottawa (Canada): Statistics Canada; CANSIM, table 102–0552. [Google Scholar]
- Stein MB, Ware EB, Mitchell C, Chen C-Y, Borja S, Cai T, Dempsey CL, Fullerton CS, Gelernter J, Heeringa SG, et al. 2017. Genomewide association studies of suicide attempts in US soldiers. Am J Med Genet B Neuropsychiatr Genet. 174(8):786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawbridge RJ, Ward J, Ferguson A, Graham N, Shaw RJ, Cullen B, Pearsall R, Lyall LM, Johnston KJA, Niedzwiedz CL, et al. 2019. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine. 41:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadoczky E, Vitrai J, Rihmer Z, Furedi J. 2000. Suicide attempts in the Hungarian adult population. Their relation with DIS/DSM-III-R affective and anxiety disorders. Eur Psychiatry. 15(6):343–347. [DOI] [PubMed] [Google Scholar]
- Tidemalm D, Runeson B, Waern M, Frisell T, Carlstrom E, Lichtenstein P, Langstrom N. 2011. Familial clustering of suicide risk: a total population study of 11.4 million individuals. Psychol Med. 41(12):2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Brent DA. 2016. Suicide and suicidal behaviour. Lancet. 387(10024):1227–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, et al. 2018. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 21(12):1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao Y, Edmiston EK, Womer FY, Zhang R, Zhao P, Jiang X, Wu F, Kong L, Zhou Y, et al. 2019. Structural and functional abnormities of amygdala and prefrontal cortex in major depressive disorder with suicide attempts. Front Psychiatry. 10:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman MM, Bland RC, Canino GJ, Greenwald S, Hwu HG, Joyce PR, Karam EG, Lee CK, Lellouch J, Lepine JP, Newman SC, et al. 1999. Prevalence of suicide ideation and suicide attempts in nine countries. Psychol Med. 29(1):9–17. [DOI] [PubMed] [Google Scholar]
- WHO. 2014. Preventing suicide, a global imperative. Geneva (Switzerland): World Health Organization Press. [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Seifuddin F, Mahon PB, Jancic D, Pirooznia M, Steele J, Schweizer B, Goes FS, Mondimore FM, MacKinnon DF, et al. 2012. A genome-wide association study of attempted suicide. Mol Psychiatry. 17(4): 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. 2018. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 50(5):668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai CC, de Luca V, Strauss J, Tong RP, Sakinofsky I, Kennedy JL. 2012. Genetic factors and suicidal behavior. In: Dwivedi Y, editor. The neurobiological basis of suicide. Boca Raton (FL): CRC Press. [PubMed] [Google Scholar]
- Zai CC, Manchia M, De Luca V, Tiwari AK, Chowdhury NI, Zai GC, Tong RP, Yilmaz Z, Shaikh SA, Strauss J, et al. 2012. The brain-derived neurotrophic factor gene in suicidal behaviour: a meta-analysis. Int J Neuropsychopharmacol. 15(8):1037–1042. [DOI] [PubMed] [Google Scholar]
- Zai CC, Goncalves VF, Tiwari AK, Gagliano SA, Hosang G, de Luca V, Shaikh SA, King N, Chen Q, Xu W, Strauss J, Breen G, et al. 2015. A genome-wide association study of suicide severity scores in bipolar disorder. J Psychiatr Res. 65:23–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.