Abstract
The replication of poliovirus, a positive-stranded RNA virus, requires translation of the infecting genome followed by virus-encoded VPg and 3D polymerase-primed synthesis of a negative-stranded template. RNA sequences involved in the latter process are poorly defined. Since many sequences involved in picornavirus replication form RNA structures, we searched the genome, other than the untranslated regions, for predicted local secondary structural elements and identified a 61-nucleotide (nt) stem-loop in the region encoding the 2C protein. Covariance analysis suggested the structure was well conserved in the Enterovirus genus of the Picornaviridae. Site-directed mutagenesis, disrupting the structure without affecting the 2C product, destroyed genome viability and suggested that the structure was required in the positive sense for function. Recovery of revertant viruses suggested that integrity of the structure was critical for function, and analysis of replication demonstrated that nonviable mutants did not synthesize negative strands. Our conclusion, that this RNA secondary structure constitutes a novel poliovirus cis-acting replication element (CRE), is supported by the demonstration that subgenomic replicons bearing lethal mutations in the native structure can be restored to replication competence by the addition of a second copy of the 61-nt wild-type sequence at another location within the genome. This poliovirus CRE functionally resembles an element identified in rhinovirus type 14 (K. L. McKnight and S. M. Lemon, RNA 4:1569–1584, 1998) and the cardioviruses (P. E. Lobert, N. Escriou, J. Ruelle, and T. Michiels, Proc. Natl. Acad. Sci. USA 96:11560–11565, 1999) but differs in sequence, structure, and location. The functional role and evolutionary significance of CREs in the replication of positive-sense RNA viruses is discussed.
Poliovirus, the archetypal picornavirus, is arguably one of the best characterized of all viruses. The availability of infectious molecular clones (33) has enabled the application of reverse genetics to understand the function of the nonstructural proteins and the noncoding regions (NCR) of the virus genome (reviewed in reference 41). The 7.4-kb single-stranded positive (messenger)-sense RNA genome encodes a single polyprotein and contains the necessary signals for virus replication in the cell cytoplasm. Posttranslational cleavage of the polyprotein by virus-encoded proteases (2Apro and 3Cpro) yields the four capsid proteins (VP1 to VP4), the RNA-dependent RNA polymerase (3Dpol), and the accessory proteins required for replication. The input RNA acts as a template for the synthesis of a negative strand which, in turn, is used as a template for synthesis of genome sense RNA. The virus-encoded protein VPg (3A) is implicated as a protein primer for both positive- and negative-sense strand initiation, as are RNA sequences occupying the 5′ NCR of the virus genome. A cloverleaf (CL) structure of 88 nucleotides (nt) at the 5′ end of the genome interacts with the virus 3CproDpol and either the VPg-containing precursor 3AB or the cellular poly-C binding protein type 2 (PCBP2) (2, 3, 11, 28). This 5′ NCR ribonucleoprotein complex is required for replication and may also be involved in the suppression of virus translation (10).
RNA sequences and structures within the 3′ NCR presumed to be involved in replication are much less well defined. Chimeric polioviruses in which the 3′ NCR is replaced by the analogous region of other picornaviruses generally retain viability (35), even though there is little sequence or structural homology between substituted 3′ NCR. These results are reflected in the cross-competition for cellular proteins seen between the poliovirus, coxsackievirus, and rhinovirus 3′ NCR in vitro (21). In contrast to the gross changes to 3′ NCR structure, defined mutations affecting formation of a pseudoknot (20, 25), or—in the rhinovirus type 14 3′ NCR—within a conserved sequence at the base of the structure or the terminal loop (22, 35), adversely affect replication. Characterization of revertants selected from such mutants have identified interacting RNA structures (25) or proteins (22). These results suggest that, although there is a degree of flexibility in the 3′ NCR sequences and structures that can be accommodated, this region of the genome plays an important function in the replication cycle of the virus. However, this conclusion is at odds with the observation that poliovirus or rhinovirus genomes from which the 3′ NCR is deleted retain the ability to replicate, albeit rather poorly (22, 39, 40). Therefore, the 3′ NCR cannot be an obligatory component for the initiation of negative-strand synthesis, which in vitro studies show can be primed by 3Dpol and VPg alone (29), but presumably influences the efficiency of the process.
The encapsidation of newly synthesized genomes completes the formation of infectious virions. The specificity of this process (5, 14, 32) can be readily monitored using subgenomic replicons, in which the capsid-encoding P1 region of the genome is replaced with a reporter gene such as that coding for chloramphenicol acetyl transferase (CAT) (30). We have used such replicons to demonstrate that RNA sequences involved in packaging are not located within the P1, 5′ NCR, 3′ NCR, or VPg-encoding regions (5, 30).
A characteristic of 5′ NCR sequences involved in translation and replication and of sequences in the 3′ NCR and an expected feature of encapsidation signals is that they all form secondary structures (1, 35–37). In an attempt to identify further functional stem-loop structures, we searched the polyprotein-coding region of the poliovirus genome for such elements. A candidate structure was identified computationally within the 2C-encoding region and disrupted by site-directed mutagenesis. Our results suggest that the 61-nt stem-loop is a location-independent cis-acting replication element (CRE) required in the positive sense for function. This is the first report of a CRE within the polyprotein-encoding region of poliovirus. The recent identification of an analogous element in human rhinovirus type 14 (HRV14) (18, 19), although distinctly different in sequence, structure, and location, and the presence of similar elements in other positive-sense RNA viruses may reflect a replication mechanism evolutionarily conserved by this widely divergent group of viruses.
MATERIALS AND METHODS
Computing.
RNA structural predictions used the Zuker energy minimization algorithm (42) as implemented in the MFOLD program of the Genetics Computer Group (GCG) suite (Wisconsin Package version 10.0; GCG Madison, Wis.). Complete genomes were searched for structural elements using overlapping windows generated by Perl batch scripts. Results were visualized using GCG PLOTFOLD, and localized structural RNA elements were identified subjectively by their consistently low P-Num value. Further analysis and structural predictions of the 2C loop used the RNAdraw (17) implementation of FoldRNA from the Vienna RNA package (12), which was used to generate Fig. 1B. Sequence alignments (Fig. 1C) were modified from data provided by Ann Palmenberg (personal communication).
FIG. 1.
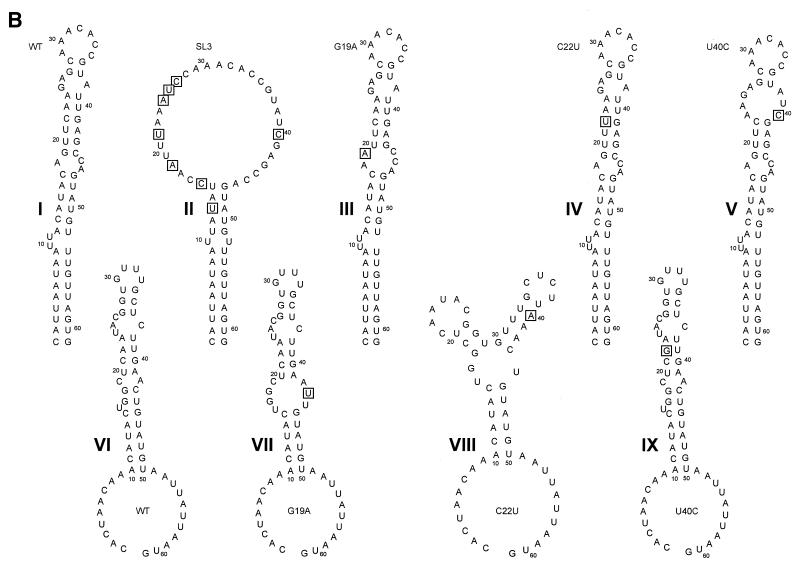
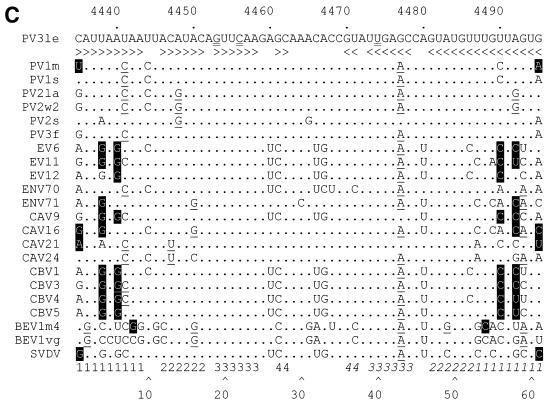
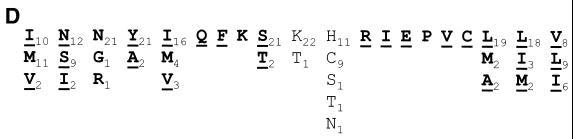
Sequence, structural, and covariance analysis of the poliovirus 2C loop. (A) P-Num output generated by GCG PLOTFOLD of MFOLD analysis of a 500-bp window (nt 4200 to 4700) of PV3 Leon. The region displaying a low P-Num value, corresponding to the 2C loop, is indicated. (B) Predicted RNA secondary structure of the positive (I to V)- and negative (VI to IX)-sense poliovirus 2C loop. I and VI, wild-type (WT) sequence; II, SL3 sequence; III and VII, G19A; IV and VIII, C22U; V and IX, U40C. In each structure sequences are numbered at intervals of 10 nucleotides from the first base shown (C4435). Mutations introduced into the sequence are boxed. (C) Covariance analysis of the 2C loop of enteroviruses. The RNA sequence of PV3 Leon (PV3le) is shown below the numbering of the region in the full-length genome and above a line indicating base-paired stems in the structure. Enterovirus sequence alignments (available from http://www.bocklabs.wisc.edu/seq.html) were provided by Ann Palmenberg (personal communication) and are indicated using abbreviations used in this work and the abbreviations EV (echovirus), ENV (enterovirus), BEV (bovine enterovirus), and SVDV (swine versicular disease virus). The sequences contributing to the 5′ and 3′ (italics) portions of stems 1 to 4 are indicated above a number scale for the 61-nt 2C loop. Covariant base pairs are indicated on black squares; other nucleotide substitutions that retain the predicted 2C loop structure are single underlined. The G19, C22, and U40 nucleotides within the third stem region of the 2C loop are double underlined in the PV3le sequence. (D) The amino acid sequence of the 2C loop product does not account for the observed sequence conservation. The polypeptide product (residues 108 to 127) of the PV3 2C loop is shown, together with the variation seen among the enteroviruses listed in panel C. Subscripted numbers indicate the frequency with which particular residues are present. Residues in boldface type are wholly or partially encoded by the predicted stem regions of the 2C loop. Underlined residues are those in which an alternate codon could be used, resulting in a nucleotide substitution within the base-paired stems of the 2C loop.
Cloning and mutagenesis. (i) Construction of mutations within the 2C loop.
The mutant pT7/SL3 was constructed by overlapping PCR mutagenesis. Primer pairs 43-0012+34-0067 and 43-0014+34-0069 (Table 1) were used to generate overlapping products using pT7/Rep3 (described in reference 5) as a template. Gel-purified products were annealed, extended using Klenow fragment, and used as a template for a second-round PCR using the primer pair 43-0012+34-0069 (Table 1). The resulting 2,167-nt product was digested with BstEII and NsiI and ligated with similarly prepared pT7/Rep3. The primers 43-0016, 43-0017, and 289 were used to confirm the sequence of the 1,246-bp BstEII-NsiI fragment in the resulting construct designated pT7/Rep3/SL3. The mutagenized 2C loop was rebuilt into the full-length infectious type 3 Leon cDNA (pT7/FLC) to generate pT7/SL3 on the unique 2.3-kb HindIII fragment (Fig. 2).
TABLE 1.
Sequences of primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| 289 | AGGGGTTGGAATGGGTGTCC |
| 2CLA | TTTAAAACGCGTTTAGTCGCAACTGATTTTCC |
| 2CLS | TTTAAAGCGCGCCCAGGAAATTTTGTTCAAC |
| 2Cmut | TAATAATTACATACA (G/A) TT (C/T) AAGAGCAAACACCGTAT (T/C) GAGCCAGTATGTTTG |
| 2CSA | TTTAAAGCGCGCACTAACAAACATACAGG |
| 2CSS | TTTAAAACGCGTCATTAATAATTACATACAG |
| 34-0067 | CAAACATACTGGCTCGATACGGTGTTTGGATTTAAATTGGATATAATTATTAATGGTG |
| 34-0069 | GGTCTCATCCACTGGATTTCACCGTTGCTCTGTGAGTGAAGTATGACC |
| 43-0012 | ACTATATTGAGTCACTCGGTGCTGCGTTCGG |
| 43-0014 | CACCATTAATAATTATATCCAATTTAAATCCAAACACCGTATCGAGCCAGTATGTTTG |
| 43-0016 | ACTATATTGAGTCACTCGGTG |
| 43-0017 | TGATGTTGACGATCCATCCT |
| 50.005 | ATATATGCTAGCCATATGGGTGATAGTTGGTTGAAAAAATTTAC |
| Else1 | GTTCAAGAGCAAACATCGTATTGAGCCAGTA |
| Else3 | TACATACAGTTCAAGTCCAAACACCGTATTGAG |
| Else5 | GTTGGAACACACCATCAATAATTACATACAG |
FIG. 2.
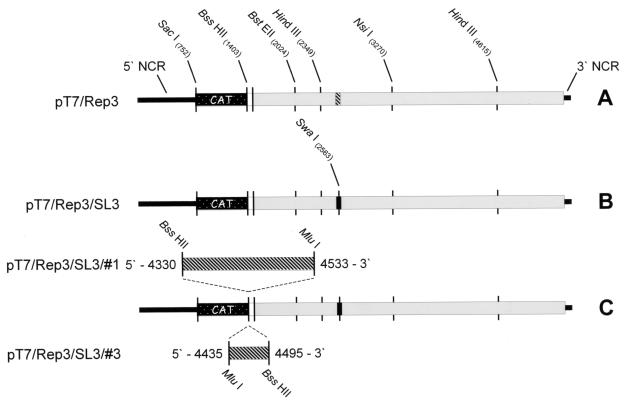
Poliovirus subgenomic replicons constructed during the analysis of the 2C loop. The open reading frame is indicated as a thick bar, with the CAT gene indicated as a stippled box, remaining P1 sequences shown in white, and the P2- and P3-encoding regions shown in grey. Wild-type 2C loop sequences are indicated with diagonal stripes, and the 2C loop containing the eight mutations is shown as a solid black box. Restriction sites of relevance to the work are indicated. (A) pT7/Rep3; (B) pT7/Rep3/SL3; (C) pT7/Rep3/SL3#1 and pT7/Rep3/SL3#3 showing the duplicated 2C loop insertions at 800% to better indicate the restriction sites used in the cloning.
A broadly similar strategy was used to introduce the Else 1.2, 2.1, and 3.2 mutations into pT7/Rep3 using the primers Else1, Else3, and Else5, respectively, in combination with primer 34-0069 (Table 1), with the product from this reaction being used together with primer 43-0012 in the second-round PCR. Likewise, the remaining mutations were generated using the degenerate primer 2Cmut in combination with primer 34-0069. In each case mutations were identified and confirmed by DNA sequencing of the entire rebuilt BstEII-NsiI fragment. Where applicable, mutations were exchanged into pT7/FLC on the unique 2.3-kb HindIII fragment.
(ii) Construction of replicons with 2C loop duplications.
pT7/FLC was used as a template for the PCR primer pair 2CSS+2CSA or 2CLS+2CLA to generate DNA fragments encompassing nt 4435 to 4495 and nt 4330 to 4533, respectively. Each primer included either BssHII or compatible MluI restriction sites, and the double-digested product was inserted into the unique BssHII site in pT7/Rep3 to generate an in-frame fusion between the CAT reporter gene and the sequences encoding the carboxyl terminus of VP1 (see reference 5). Recombinants were screened by restriction digest for insert orientation and sequenced to confirm they were in frame and that no errors had been introduced during the amplification and cloning.
RNA transfection, analysis of genome replication, and recovery of revertant viruses. (i) T7 transfections.
T7 transfections of Ohio-HeLa cells were essentially conducted as described previously (9), with the exception of transfections for RNase protection assays which used a double-pulse electroporation schedule. Where necessary RNA was labeled with [14C]rATP, incorporation was quantified by scintillation counting, and RNA amounts were normalized prior to transfection of 2 to 4 μg per dish. Transfected cells were grown and maintained as described previously (23).
(ii) In vitro translations.
RNA was translated in vitro using a rabbit reticulocyte lysate (Promega) used essentially according to the manufacturer's instructions. Each 20-μl reaction mixture consisted of 70% (by volume) rabbit reticulocyte extract, programmed with approximately 0.5 μg of RNA and supplemented with HeLa S10 extracts (prepared and used according to reference 7), 20 μM amino acids, [35S]methionine (1,200 Ci/ml; Amersham), and RNasin (40 U/μl; Promega). Products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
(iii) CAT assays.
CAT assays of poliovirus replicon-derived constructs were performed as described previously (5). Where necessary, guanidine hydrochloride (GuHCl) was added to growth media at a final concentration of 4 mM. CAT assays were quantified by phosphorimager. The construction and characterization of the control pT7/Δpol/Rep3 replicon bearing a deletion within the 3D polymerase encoding regions has been described previously (35).
(iv) Assays of positive- and negative-sense strand synthesis.
Synthesis of positive- and negative-sense strands from pT7/Rep3-derived replicons was achieved using a two-step RNase protection assay (26) as modified by McKnight and Lemon (19). Briefly, 6 × 107 transfected cells were lysed in 100 μl of direct lysate buffer (4 M guanidine thiocyanate, 25 mM sodium citrate, and 0.5% Sarkosyl), and chromosomal DNA was removed by centrifugation (15,000 × g; 15 min). For negative-sense strand analysis, 50 μl of this lysate was self-annealed at 37°C for 16 h, RNase and subsequently proteinase K digested, phenol-chloroform extracted, precipitated (see below), and resuspended in 45 μl of direct lysis buffer. Aliquots (10 and 45 μl, respectively) of the positive- or negative-sense strand RNA preparations, made up to a final volume of 50 μl with direct lysate buffer, were incubated for 16 h at room temperature with 3 × 105 cpm of a 32P-labeled strand-specific probe (see below). RNase digestion was performed in a total volume of 500 μl, containing 10 mM Tris (pH 7.5), 5 mM EDTA, 300 mM NaCl, and 5 μl of Ambion RNase cocktail for 30 min at 37°C. Proteinase K digestion was performed at 37°C for 30 min by the addition of 20 μl of 10% sodium dodecyl sulfate and 10 μl of proteinase K (20 mg/ml; Ambion). Protected RNA was precipitated by the addition of 2 μl of glycogen (20 mg/ml; Roche), 500 μl of isopropanol, and centrifugation for 15 min at 15,000 × g. The resultant pellet was resuspended in RNA loading buffer and electrophoresed on a 7% acrylamide gel containing urea which was fixed and dried prior to exposure on a Bio-Rad phosphorimager screen.
Positive- and negative-sense strand-specific probes were generated from pBSCAT, a pBSII KS-based vector containing nt 1 to 144 of the CAT gene. pBSCAT was linearized with NotI or SalI and used as a template to generate [32P]CTP-labeled transcripts with T7 or T3 polymerase as described previously (22). Probes were gel purified on a 7% acrylamide-urea gel prior to use. The positive- and negative-sense probes were 195 nt and 168 nt, respectively, in length, and both protected a CAT-specific fragment of 113 nt.
(v) Selection of revertant viruses.
Revertants were selected using two strategies. In vitro-synthesized T7 runoff RNA from the nonviable genomes containing two mutations within the 2C loop were serially diluted, transfected into Ohio-HeLa cells in six-well plates, overlaid with agarose, and incubated for 2 to 3 days at 37°C. Individual plaques were picked and subjected to two further rounds of plaque purification before further analysis. T7-generated RNA from 2Cmut6 (2C loop treble mutant) and pT7/SL3 were transfected into Ohio-HeLa cells in a T25 flask and overlaid with 5 ml of Dulbecco's modified Eagle medium containing 5% fetal calf serum. Five hundred microliters of tissue culture supernatant was repeatedly passaged onto fresh cells every 3 days until a cytopathic effect became evident (2Cmut6) or for seven passages (pT7/SL3). Virus was subjected to two rounds of plaque purification before further characterization.
(vi) Sequence analysis of revertant viruses.
Virus was amplified in Ohio-HeLa cells, and virus RNA was isolated from 200 μl of cell culture supernatant by Trizol extraction (GibcoBRL). One quarter of the RNA was reverse transcribed using avian myeloblastosis virus reverse transcriptase (Promega) and primer 34-0069 (Table 1), and half of this reaction mixture was used as a template for a PCR with primers 50.005 and 2CLA. The resulting product was sequenced by the automated DNA sequencing facility of the Institute of Virology, University of Glasgow.
RESULTS
Identification and covariance analysis of an RNA structural element within the poliovirus genome.
We searched the polyprotein-encoding region of poliovirus for candidate secondary structures that—by analogy with sequences in the 5′ and 3′ NCR or the known properties of RNA packaging signals—may have a role in virus replication. Searches were conducted using the GCG MFOLD program driven by a batch script generating overlapping 300- to 600-nt windows selected to minimize the influence of long-range interactions. Results were visualized using the GCG PLOTFOLD program and analyzed subjectively for defined regions displaying low P-Num values, indicative of local interactions (Fig. 1A). Comparative analysis of picornavirus genomes, including poliovirus type 3 (PV3), HRV2, HRV14, coxsackie B virus 4 (CBV4), enterovirus 70 (ENV70), and echovirus type 7 (EV7), identified a region within P2 which was predicted to form a broadly conserved structure in enteroviruses but was absent from rhinoviruses. Although the level of sequence conservation within this region (designated the 2C loop on account of the location) of the enteroviruses was not consistent with an RNA structure involved in determining the specificity of encapsidation, sequence alignments (see below) suggested that the structural conservation reflected a functionally important region of the genome and that it warranted further study.
Structural prediction and covariance analysis of the 2C loop.
The 2C loop in PV3 is predicted to consist of four distinct base-paired regions separated by short unpaired sequences or bulges (Figure 1B [structure I]). The element has a predicted structural energy of −61.8 kJ at 37°C. Alignment of this region of a representative group of enteroviruses (Fig. 1C) shows that the level of variation within the paired regions is not consistent throughout the stem-loop. The basal stem is relatively poorly conserved, with the level of conservation within the stem increasing towards the distal end of the structure. Notably, predicted base pairing in stem 3 is completely conserved. However, when complementary base pairing and covariance are taken into account it is clear that there is significant conservation of structure of stems 1 to 4 (Fig. 1C). In comparison, the loop (nt 29 to 35, corresponding to nt 4463-4469, inclusive) exhibits more extensive variation. The amino acids encoded by the nucleotides that contribute to stems 2, 3, and 4 do not dictate the high level of sequence conservation, as the residues are not encoded by triplets underrepresented in the codon table (Fig. 1D). These observations strongly suggest that the RNA sequence and structural conservation of the 2C loop per se are of significance for an aspect of the poliovirus life cycle.
Functional analysis of the 2C loop. (i) Disruption of the 2C loop causes a defect in poliovirus replication.
Taking advantage of the degeneracy of the triplet code, a total of eight substitutions were introduced to the 2C loop, the majority of them being in the third-base “wobble” position of codons, leaving the encoded product unaltered. The substitutions—C13U, A16C, G19A, C22U, G25A, A26U, G27C, and U40C—disrupt the structure of the 2C loop in the positive sense (Fig. 1B [structure II]) and were also predicted to prevent the correct folding of a weaker structure in the negative-sense strand (Fig. 1b [structure VI and data not shown]). (The numbering in these substitutions is relative to the first nucleotide of the predicted 2C loop; add 4,434 to identify the location in the genome. Mutations are indicated in terms of the original base, the position, and the substituted base.) They also introduced a unique SwaI site into this region of the poliovirus genome. The resulting subclone was sequenced to confirm that no further mutations had been introduced, and was rebuilt into an infectious PV3 cDNA (pT7/FLC) and the CAT-encoding replicon pT7/Rep3 (5) (Fig. 2A) to generate pT7/SL3 and pT7/Rep3/SL3, respectively.
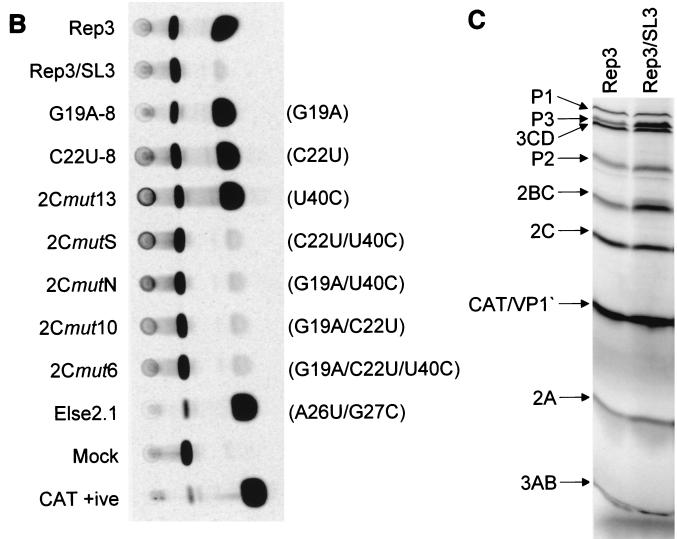
RNA generated in vitro from pT7/SL3 was transfected into subconfluent Ohio-HeLa monolayers but failed to show any signs of cytopathic effect within 7 days (data not shown). In vitro-generated T7 runoff RNA from pT7/Rep3/SL3 was similarly transfected and harvested at 8 h posttransfection, and the residual CAT activity in 50 μg of cell extract was assayed. Only trace levels of CAT activity could be detected (Fig. 3A) in the presence or absence of 4 mM GuHCl. The translation-only control, a pT7/FLC/Rep3-derived replicon bearing a deletion within the 3D polymerase-encoding region, and therefore incapable of replicating (35), generated a higher CAT signal than pT7/Rep3/SL3 (Fig. 3A) in the presence or absence of GuHCl.
FIG. 3.
(A) CAT assays of pT7/Rep/SL3. Fifty micrograms of total cell protein from cells transfected with normalized amounts of replicon RNA was assayed for CAT activity using monoacetylated chloramphenicol. Transfections were conducted in the presence (+) or absence (−) of 4 mM GuHCl and included a replication-competent positive control (Rep3) and a replicon bearing a deletion within the polymerase-encoding region (Δpol/Rep3) which is capable of translation only. (B) CAT assays of subgenomic replicons with mutations within the 2C loop. Fifty micrograms of total cell protein from cells transfected with normalized amounts of replicon RNA was assayed for CAT activity using monoacetylated chloramphenicol. The weak positive signals generated from Rep3/SL3, 2CmutS, 2CmutN, 2Cmut10, and 2Cmut6 are due to translation alone (compare with Rep3/SL3 and Δpol/Rep3 in panel A). (C) In vitro transcription and translation, in HeLa S10-supplemented rabbit reticulocyte lysates, of RNA generated from pT7/Rep3 and pT7/Rep3/SL3, showing the expected protein profiles.
To further confirm that pT7/Rep3/SL3 did not contain additional defects, possibly introduced during mutagenesis and cloning, T7-synthesized RNA was used to prime an in vitro translation reaction using a HeLa S10-supplemented rabbit reticulocyte lysate. The resulting protein profile was indistinguishable from pT7/Rep3, the parental replicon, with the expected P2 and P3 products, and the CAT-VP1′ fusion product clearly visible (Fig. 3C). This suggested that the open reading frame of pT7/Rep3/SL3 was intact, and supported the preliminary conclusions that disruption of the 2C loop introduced a defect in poliovirus replication.
(ii) Stem 3 contains critical sequences and structures for virus replication.
The eight mutations introduced into the 2C loop were designed to totally disrupt the structure from forming. Although the phenotype observed affirmed the importance of this region of the genome for replication, it obscured the identification of specific sequences critical for function. To better identify these, a range of mutations were introduced individually or in combination (Table 2). Mutations were selected based upon the sequence and structural conservation within this region of enterovirus genomes and taking into account the covariance analysis (Fig. 1C). None of the introduced substitutions altered the encoded 2C protein.
TABLE 2.
Replication of mutants used in this study
| Mutanta | RNA substitution(s) | Replicationb
|
|
|---|---|---|---|
| CAT replicon | Virus | ||
| SL3 | C13U, A16C, G19A, C22U, G25A, A26U, G27C, U40C | − | − |
| Else 1.2 | C34U | + | ND |
| Else 2.1 | A26U, G27C | + | ND |
| Else 3.2 | U4C | + | ND |
| G19A-8 | G19A | + | + |
| C22U-8 | C22U | + | + |
| C22U-11 | C22U | + | + |
| 2Cmut13 | U40C | + | + |
| 2CmutS | C22U, U40C | − | − |
| 2CmutN | G19A, U40C | − | − |
| 2CmutF | G19A, U40C | − | − |
| 2Cmut10 | G19A, C22U | − | − |
| 2Cmut6 | G19A, C22U, U40C | − | − |
The mutants listed were tested for replication competence in a full-length cDNA backbone or as subgenomic replicons and are referred to in the text using the prefix pT7/FLC or pT7/Rep3, respectively.
+, positive; −, negative; ND, not determined.
Stem 3, the base pairing of which is totally conserved in the enteroviruses, was the focus of these mutagenesis studies. Individual substitution of G19A, C22U, or U40C in both replicons and full-length genomes exhibited phenotypes indistinguishable from the parental unmodified pT7/Rep3 or pT7/FLC (G19A-8, C22U-8/11, and 2Cmut13) (Table 2 and Fig. 3B). In contrast, the substitution of any two of these nucleotides, or all three together, destroyed replication of both replicons and full-length genomes (2CmutS, 2CmutN/F, 2Cmut10, and 2Cmut6) (Table 2 and Fig. 3B). Substitutions elsewhere in the 2C loop, including the basal stem (U4C), the terminal bulge and stem 4 (A26U + G27C; Else 2.1), or the terminal loop (C34U), did not have a measurable effect on the replication of a CAT-containing replicon (Fig. 3B; Table 2) or, where tested, on virus viability (Table 2). These results suggested that stem 3 within the 2C loop can accommodate a limited amount of variation but that, in agreement with the sequence analysis, this region contains sequences critical for poliovirus replication.
(iii) The 2C loop is required in the positive sense for function.
Although originally identified in the genome sense, the 2C loop can also fold to form a structure in the negative-sense strand, albeit with reduced stability (Fig. 1B [structure VI]). The disruption of either or both structures could be responsible for the deleterious effect on virus replication. The predicted structures in the positive- and negative-sense strands of all possible combinations of G19A, C22U, and U40C were modeled (Fig. 1B) to identify any structure in which there was significant modification in the absence of a distinct alteration of phenotype.
In the sense strand structures, the substitution of C22U had no effect upon the predicted structure, as C22 is base paired to G41, and remains paired in a predicted fold of the mutant (Fig. 1B [structure IV]). G19A and U40C both introduced minor alterations to the predicted structure of the sense strand; G19A opened the central bulge loop, and U40C increased the size of the terminal bulge loop and in doing so reduced the length of the third stem (Fig. 1B [structures III and V]). However, the overall shape and structure of the stem-loop was predicted to remain largely unaltered. In the negative-sense strand structures, the U40C substitution had no effect on the predicted structure as the mutation restored base pairing to this section of the stem (Fig. 1B [structure IX]). The G19A mutation had a more significant effect on the structure, opening a bulge loop and so reducing the length of the basal stem (Fig. 1B [structure VII]). However, the C22U mutation, which had no effect upon the sense strand structure, was predicted to introduce a major change into the antisense structure, converting the single stem-loop into a bifurcated stem (Fig. 1B [structure VIII]). Since both virus and replicon bearing a C22U mutation replicated with a wild-type phenotype, this suggested that the 2C loop was required in the positive strand for function.
Selection of revertants from 2C loop mutations.
Although the stem 3 double mutants (2CmutS, 2CmutN, and 2Cmut10) were nonviable (Table 2), it was possible to select revertants at a low frequency, suggesting the acquisition of a compensating mutation. In comparison to pT7/FLC, which routinely generates 106 PFU of transfected RNA per μg, 2Cmut10 generated plaques at a rate approximately 2 to 3 log10 units lower than control pT7/FLC. In contrast, plaques were produced from 2CmutS and 2CmutN/2CmutF at a rate approximately 5 to 6 log10 units lower than the control pT7/FLC (data not shown). In all cases the plaques generated were indistinguishable in phenotype from those produced by pT7/FLC (data not shown). A total of four revertant viruses were recovered and subjected to two rounds of plaque purification, and the sequence of a 200-nt window spanning the 2C loop was determined. Each sequenced virus contained a single reversion of one of the introduced mutations (Table 3). In the case of the 2Cmut10 mutation (G19A and C22U) two plaques were characterised, one contained a reversion of U22 to C and the other A19 to G. No other mutations were observed within the region of the genome sequenced.
TABLE 3.
Reversion of the mutants used in this study
| Clone | Substitution(s) | Reversion(s) |
|---|---|---|
| 2CmutS | C22U, U40C | C40U |
| 2CmutN | G19A, U40C | C40U |
| 2Cmut10#1 | G19A, C22U | U22C |
| 2Cmut10#2 | G19A, C22U | A19G |
| 2Cmut6 | G19A, C22U, U40C | U22C, C40U |
The 2C loop stem 3 treble mutant (2Cmut6; G19A, C22U, and U40C) could not be recovered by direct plaque assay of the T7 transfection. However, three repeated blind passages of transfected Ohio-HeLa cells resulted in a cytopathic effect, and four viruses from two initial transfections were plaque purified and the 2C loop was sequenced. All the recovered viruses carried two reversions, of U22 to C and C40 to U, resulting in the retention of only the original G19A mutation, which alone had already been shown not to influence replication (Fig. 3A; Table 2). Repeated attempts to recover revertants from pT7/SL3, including up to seven blind passages of tissue culture supernatant, were unsuccessful (data not shown).
The 2C loop functions when provided in cis.
If the 2C loop contributes a critical function to virus replication it may be possible to restore functionality to a genome bearing a lethal mutation within this region by supplying an additional, unmodified copy of the sequence in cis.
Two versions of the unmodified 2C loop were amplified from a PV3 Leon cDNA (pT7/FLC) and cloned into the unique BssHII site in pT7/SL3, located at the junction of the CAT reporter gene and the remaining P1 sequences (Fig. 2). The amplified sequences corresponded to nt 4435 to 4495, defined as the 2C loop, and nt 4330 to 4533, a longer region spanning the entire 2C loop, generating pT7/SL3#3 and pT7/SL3#1, respectively. The reading frame of the CAT-P1 junction was maintained in both constructs, and the orientation and sequence of the introduced PCR product were verified prior to further studies.
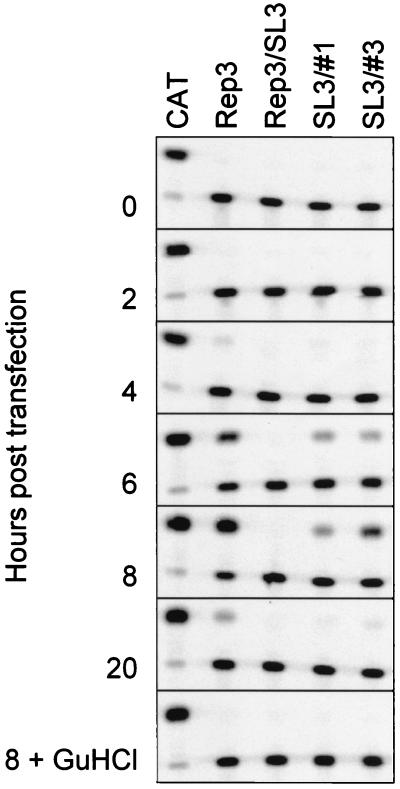
Equivalent amounts of T7-generated RNA from pT7/SL3#3 and pT7/SL3#1 were transfected into Ohio-HeLa cells, and replication was monitored by quantifying the CAT activity in 5 μg of total cell protein over a 20-h period. Normalized levels of RNA from pT7/Rep3 and pT7/SL3 were transfected in parallel as positive and negative controls, respectively (Fig. 4). As expected, pT7/Rep3 replicated well, resulting in maximal levels of CAT conversion at 8 h posttransfection. In contrast, pT7/SL3 generated no detectable CAT activity, consistent with the inability of this defective subgenomic replicon to replicate. The two replicons containing duplicated 2C loop sequences both produced low but significant levels of CAT activity, which peaked at 10 to 25% of pT7/Rep3 activity at 8 h posttransfection. To confirm that the signal observed was due to de novo replication of the input RNA, 4 mM GuHCl was included as a further control (Fig. 4), which totally inhibited the signals produced by pT7/Rep3, pT7/SL3#3, and pT7/SL3#1 when assayed at 8 h posttransfection. As a final control, pT7/Rep3/SL3#1 and pT7/Rep3/SL3#3 DNA were digested with SwaI and used to prime a T7 reaction. RNA produced from the latter failed to yield a CAT signal upon transfection of Ohio-HeLa cells (data not shown), thereby confirming that the CAT signal obtained previously derived from a replicon bearing the original (SL3) mutation within the 2C loop.
FIG. 4.
Replication of pT7/Rep3/SL3#1 and pT7/Rep3/SL3#3. Standardized levels of in vitro-generated RNA were transfected into Ohio-HeLa cells, samples were taken at the times indicated, and CAT activity in 5 μg of total cell protein was assayed. No replication at 8 h posttransfection was observed in the presence of 4 mM GuHCl (bottom panel).
2C loop mutants are defective in negative-sense-strand synthesis.
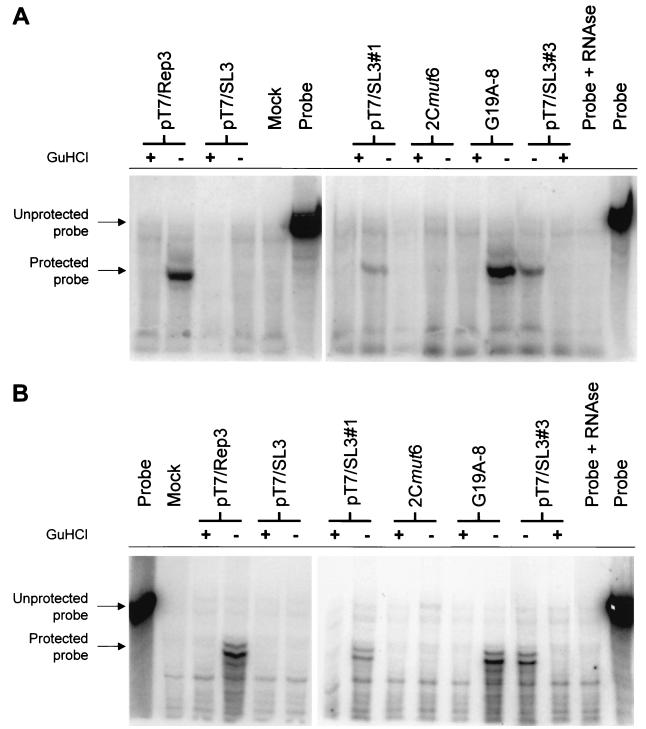
To better identify the replication defect in nonviable 2C loop mutants we analyzed the production of positive- and negative-strand RNA in cells transfected with replicon-derived RNA. An efficient double-pulse electroporation procedure was used to achieve maximal levels of transfection and was verified to not adversely affect the phenotype of the 2C mutants analyzed (data not shown). The results of positive- and negative-sense-strand synthesis from pT7/Rep3, pT7/Rep3/SL3, pT7/Rep3/SL3#1, pT7/Rep3/SL3#3, pT7/Rep3/G19A-8, and pT7/Rep3/2Cmut6 are shown in Fig. 5.
FIG. 5.
Positive (A)- and negative (B)-sense strand synthesis from poliovirus subgenomic replicons with modifications of the 2C loop. In each panel the unprotected and protected (113-nt) probes are indicated, as is the presence (+) or absence (−) of 4 mM GuHCl in the growth medium.
A positive signal was clearly detectable from pT7/Rep3 but was not produced in the presence of 4 mM GuHCl, so demonstrating that it was the product of de novo RNA synthesis. Neither of the replicons bearing mutations within the 2C loop, pT7/Rep3/SL3 and pT7/Rep3/2Cmut6, produced a positive-sense strand signal. In contrast, the replicons previously shown to generate CAT signals (Fig. 3B), pT7/Rep3/G19A-8, pT7/Rep3/SL3#1, and pT7/Rep3/SL3#3, all generated positive-sense strand hybridization signals (Fig. 5A).
Using the modified two-step RNase protection assay of Novak and Kirkegaard (26), negative-sense strand RNA synthesis was clearly demonstrable from pT7/Rep3 and pT7/Rep3/G19A-8 (Fig. 5B), both of which had already been shown to generate strong CAT signals (Fig. 3B). Both of the replicons bearing a duplication of the 2C loop, pT7/Rep3/SL3#1 and pT7/Rep3/SL3#3, also generated signals indicative of negative-sense strand synthesis. Again, 4 mM GuHCl prevented replication, resulting in the failure to detect any negative strands. Replicons that did not previously generate a CAT signal (Fig. 3B), pT7/Rep3/SL3 and pT7/Rep3/2Cmut6, did not generate detectable levels of negative-sense strands.
DISCUSSION
In our search for structural RNA elements involved in the replication and encapsidation of poliovirus RNA, we searched the polyprotein-encoding region for sequences capable of forming localized secondary structures. Several such structures were predicted, including a 61-nt stem-loop element (PV3 Leon nt 4435 to 4495) encoding residues 108 to 127 of the 2C protein (Fig. 1A and B [structure I]). Covariance analysis provided good evidence that the predicted structure was conserved within the enteroviruses (Fig. 1C), although the overall level of sequence conservation was higher than would be expected to account for the specificity of encapsidation previously observed (5). The significant level of nucleotide sequence conservation within stems 2 to 4 in aligned enterovirus genomes (Fig. 1C) was not a consequence of the available codons for the encoded protein product (Fig. 1D). This strongly implied that the sequence and structural conservation within the 2C loop were important in the enterovirus life cycle.
Extensive disruption of the predicted 2C loop structure, achieved by the introduction of multiple non-coding changes, abolished genome replication (Fig. 3A). The inability of the CAT-encoding subgenomic replicon pT7/Rep3/SL3 (containing eight substitutions within the 2C loop) to replicate was not due to the production of an aberrant translation product (Fig. 3C), implying that the 2C loop has a role in genome replication. Coinfection of pT7/Rep3/SL3-transfected cells with PV3 did not increase the CAT signal from the defective replicon, suggesting that the 2C loop could not function in trans (data not shown). Less drastic modification of the 2C loop highlighted the critical importance of the conserved stem 3, although this region could still accommodate single substitutions without a significant alteration of replicon phenotype (Fig. 3B; Table 2). Analysis of the predicted negative-sense stem-loops formed by these mutants, in particular, the substitution of C22U (C22U-8 [Fig. 1B and 3B]), strongly suggested that the element was required in the positive sense. Although we have not conducted extensive mutagenesis of other parts of the 2C loop, our preliminary studies suggest that neither stem 1 nor the terminal loop is critically important for function. Considering the variation present in the predicted subterminal bulge loop and stem 4 (Fig. 1C) of enteroviruses, it is possible that this region is of less importance for function, a conclusion supported by the phenotype of pT7/Rep3/Else 2.1 (A26U and G27A), which had no apparent deleterious effect upon replication (Fig. 3B; Table 2). Further studies of the importance of this region and the central bulge loop are currently under way. Taken together, the mutagenesis studies support a role for the 2C loop as a CRE required in the genome sense for poliovirus replication.
Further evidence supporting this conclusion was obtained by experiments involving the insertion of a duplicate 2C loop into a genome bearing the SL3 mutation. The addition of an unmodified sequence corresponding to PV3 nt 4435 to 4495 to pT7/Rep3/SL3 to generate pT7/Rep3/SL3#3 restored the ability to replicate. Although the overall level of replication, as indicated by CAT production, was reduced, the kinetics of replication were broadly similar, with the CAT signal being maximal at 8 h posttransfection (Fig. 4). Since the introduced copy of the 2C loop in pT7/Rep3/SL3#3 was >1.1 kb away from the original location (at nt 1403, compared to the native location at nt 2542 [Fig. 2]), this clearly demonstrates that the 2C loop CRE is not totally position dependent for function. It will be interesting to investigate whether the 2C loop CRE can be situated at other locations within the genome, such as the 5′ or 3′ NCR, and to determine if the level of replication is related to the distance from the native position, which may provide an insight into the function of this stem-loop.
Quantitative comparison of the levels of CAT activity and positive- and negative-strand synthesis by pT7/Rep3/SL3#1 and pT7/Rep3/SL3#3 demonstrated they were broadly in agreement. pT7/Rep3/SL3#1 and pT7/Rep3/SL3#3 generated 10 and 25%, respectively, of pT7/Rep3 CAT activity, with positive-sense strand RNA levels of 15 and 44% and negative-sense strand RNA levels of 27 and 59% (Fig. 4 and 5). This suggests that the presentation of the 2C loop in the larger insert (pT7/Rep3/SL3#1) may be suboptimal. This approach, using duplications in a pT7/Rep3/SL3 backbone, will provide a useful method to characterize the sequence and structural requirements for 2C loop function. In particular, we are interested in determining if stem 1, which is less well conserved (Fig. 1C), is required for function and whether the 2C loop can be functionally exchanged between enteroviruses.
Precedents already exist for cis-acting replication signals located in positive-sense RNA virus genomes other than at the extreme 5′ and 3′ regions. Most notable to the present study is the demonstration that HRV14 contains such an element, located within the region encoding VP1 (18). In addition to the location, the HRV14 CRE differs in sequence and structure and is slightly longer (69 to 96 nt) than the poliovirus CRE defined here (19). However, there are similarities between the apparent roles of the two CREs; both are required in the genome sense and are not position-dependent for function, and disruption of both leads to the failure to synthesize negative-sense strand RNA (Fig. 3A and 5B) (19). These similarities suggest that they may fulfill the same role in virus replication and, considering the conservation of this group of viruses, imply that other picornaviruses also contain such CREs. This suggestion is supported by the recent identification of a CRE within the capsid-encoding region of the Cardiovirus genus of the picornaviruses (16), which, in addition to exhibiting no homology with the rhinovirus CRE or the poliovirus sequence reported here, could not be functionally exchanged with the analogous region of HRV14.
Our preliminary analysis and published complete genome folding studies (27) suggest that localized RNA structural elements are not uncommon in picornavirus genomes; PV3 has an additional stem-loop of approximately 70 nt located in the region encoding 3Dpol, and the minor group rhinoviruses (e.g., HRV2) have an extensive region of predicted secondary structure located between nt 5080 and 5200 (D. J. Evans, unpublished results). Although functional data supporting a role in replication for these or other picornavirus structures has yet to be obtained, several other positive-sense RNA viruses contain CREs for which a replication function has been defined (reviewed in reference 8). Of these, the RNA3 intergenic region of the tripartite brome mosaic virus (BMV) genome and the S and M sites within the genome of bacteriophage Qβ, are the best characterized (6, 13, 24, 31, 38). In both cases it has been proposed that the internal stem-loop structure(s) acts as a nucleation site for the assembly of the replicase complex (and the Hfq host factor for Qβ) prior to the initiation of negative-sense strand synthesis. In the case of BMV RNA3, this process has been further dissected to demonstrate a BMV 1a helicase-mediated stabilization of the RNA template, presumably involved in the switch from translation to replication of the segment (38), a function recently ascribed in poliovirus to the interaction of 3CD with the 5′ NCR CL (10). Although the initiation site for poliovirus negative-sense strand synthesis is presumably the 3′ NCR, deletion of the NCR does not prevent virus replication (22, 39, 40) and the biochemical model for negative-sense strand initiation does not implicate sequences other than the poly(A) tail (29). In this context it is interesting that the Escherichia coli host factor (Hfq) bound at or near the S and M sites of bacteriophage Qβ recruits the 3′ end in a sequence-independent manner, requiring only a free 3′ end (6, 24). The CRE structures in other positive-stranded RNA viruses are less well characterized; RNA2 of the bipartite genome of flock house virus, a nodavirus, has been reported to contain a 58-nt CRE located centrally in the segment (4) and the JHM strain of mouse hepatitis virus, a coronavirus, also has a 58-nt structure required in the genome sense for replication (15, 34). Defective interfering MHV-JHM genomes bearing a modification of this sequence were temperature dependent for positive-sense genome replication (15).
Although the integrity of stem 3 is clearly important for 2C loop function, other domains of the structure may also contribute. McKnight and Lemon showed that extensive mutagenesis of the terminal loop of HRV14 CRE disrupted function (19), whereas in the present study a C34U substitution (Else 1.2) (Table 2) had no effect upon replication. It is notable that there is a level of conservation of the terminal loop sequence within certain enterovirus species, with both echoviruses and coxsackie B viruses having a G at position 33 (Fig. 1C). This may be an adaptation reflecting an interaction mediated by this region of the CRE with a virus or cellular protein. Our initial studies, involving the selection of revertants from nonviable stem 3 mutants, have not found any evidence for an interaction with poliovirus sequences or proteins, as no second-site revertants were identified. Although this could indicate an interaction with a cellular protein, it is more probably due to the base-paired nature of the nucleotides originally selected and a requirement to maintain stem 3 within the CRE. In preliminary experiments we have not been able to identify a specific interaction of the 2C loop with proteins from uninfected Ohio-HeLa cell extracts using electrophoretic mobility shift assays (data not shown), suggesting that if such binding occurs, it may also require virus proteins to form a ribonucleoprotein complex. Considering the nature of the mutations observed, the frequency with which revertants of 2CmutS and 2CmutN were recovered—3 log10 units lower than the other single-base reversion of 2Cmut10—was surprising. We currently attribute this lower recovery to the influence of the RNA secondary structure; certain mutations may destabilize the structure to a greater extent. It is also worth noting that, of the limited number of revertants analyzed, all the double and treble mutations involving a U40C substitution reverted to a U at this position, suggesting a critical role for this region of the 2C loop CRE. We are currently enzymatically determining the structure of the 2C loop, including the 2CmutS and 2CmutN variants, to test this hypothesis.
We have not been able to demonstrate encapsidation of the 2C loop-duplicated replicons pT7/Rep3/SL3#1 or pT7/Rep3/SL3#3 using our standard packaging assay (5) with a PV3 helper (data not shown). Although this may reflect a position-dependent role for the 2C loop in poliovirus packaging, we consider this more likely to be due to the relatively poor level of replication, at most 25% of wild type, exhibited by these replicons (Fig. 4). In support of this conclusion, we have observed that another poorly replicating subgenomic replicon (a PV3 replicon with a modified HRV14 3′ NCR in which the loop sequence was modified to GCUAU and which consistently produces ∼20% of wild-type CAT activity [see reference 35]) is also defective for packaging (data not shown).
In conclusion, we present evidence for a CRE consisting of a maximal 61 nt of the 2C-encoding region of poliovirus. The function of the CRE is not position dependent and probably involves the initiation or synthesis of negative-sense strand template RNA during poliovirus replication. Further studies are under way to better define the function of the 2C loop, to understand the biochemical mechanism involved, and to investigate the possible conservation of this replication strategy in picornaviruses and other positive-stranded RNA viruses.
ACKNOWLEDGMENTS
This work was supported by a Medical Research Council (MRC) Programme Grant (G9006199) and a BBSRC project grant (17/G11681). Andrew Richardson was the recipient of an MRC postgraduate studentship.
We appreciate the assistance of Alan Cann (University of Leicester) for providing access to the GCG suite.
REFERENCES
- 1.Andino R, Böddeker N, Silvera D, Gamarnik A V. Intracellular determinants of picornavirus replication. Trends Microbiol. 1999;7:76–82. doi: 10.1016/s0966-842x(98)01446-2. [DOI] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Ball L A, Li Y. cis-acting requirements for the replication of flock house virus-RNA 2. J Virol. 1993;67:3544–3551. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barclay W, Li Q, Hutchinson G, Moon D, Richardson A, Percy N, Almond J, Evans D. Encapsidation studies of poliovirus subgenomic replicons. J Gen Virol. 1998;79:1725–1734. doi: 10.1099/0022-1317-79-7-1725. [DOI] [PubMed] [Google Scholar]
- 6.Barrera I, Schuppli D, Sogo J M, Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 7.Barton D J, Flanegan J B. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional three-dimensional polymerase and infectious virus. J Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck K W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans D J, Almond J W. Design, construction and characterization of poliovirus antigen chimeras. Methods Enzymol. 1991;203:387–400. doi: 10.1016/0076-6879(91)03022-9. [DOI] [PubMed] [Google Scholar]
- 10.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 12.Hofacker I L, Fontana W, Stadler P F, Bonhoeffer L S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- 13.Janda M, Ahlquist P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc Natl Acad Sci USA. 1998;95:2227–2232. doi: 10.1073/pnas.95.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia X Y, Van Eden M, Busch M G, Ehrenfeld E, Summers D F. trans-encapsidation of a poliovirus replicon by different picornavirus capsid proteins. J Virol. 1998;72:7972–7977. doi: 10.1128/jvi.72.10.7972-7977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y N, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobert P E, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci USA. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matzura O, Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput Appl Biosci. 1996;12:247–249. doi: 10.1093/bioinformatics/12.3.247. [DOI] [PubMed] [Google Scholar]
- 18.McKnight K L, Lemon S M. Capsid coding sequence is required for efficient replication of human rhinovirus-14 RNA. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight K L, Lemon S M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melchers W J G, Hoenderop J G J, Slot H J B, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the 2 predominant hairpin loops in the coxsackie-B virus 3′-untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellits K H, Meredith J M, Rohll J B, Evans D J, Almond J W. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J Gen Virol. 1998;79:1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- 22.Meredith J M, Rohll J B, Almond J W, Evans D J. Similar interactions of the poliovirus and rhinovirus 3D polymerase with the 3′ untranslated region of rhinovirus 14. J Virol. 1999;73:9952–9958. doi: 10.1128/jvi.73.12.9952-9958.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minor P D. Growth, assay and purification of picornaviruses. In: Mahy B W J, editor. Virology: a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 25–41. [Google Scholar]
- 24.Miranda G, Schuppli D, Barrera I, Hausherr C, Sogo J M, Weber H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J Mol Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 25.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmenberg A C, Sgro J. Topological organisation of picornaviral genomes: statistical prediction of RNA structural signals. Semin Virol. 1997;8:231–241. [Google Scholar]
- 28.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 29.Paul A V, vanBoom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 30.Percy N, Barclay W S, Sullivan M, Almond J W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pogue G P, Marsh L E, Connell J P, Hall T C. Requirement for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology. 1992;188:742–753. doi: 10.1016/0042-6822(92)90529-x. [DOI] [PubMed] [Google Scholar]
- 32.Porter D C, Ansardi D C, Wang J, McPherson S, Moldoveanu Z, Morrow C D. Demonstration of the specificity of poliovirus encapsidation using a novel replicon which encodes enzymatically active firefly luciferase. Virology. 1998;243:1–11. doi: 10.1006/viro.1998.9046. [DOI] [PubMed] [Google Scholar]
- 33.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 34.Repass J F, Makino S. Importance of the positive-strand RNA secondary structure of a murine coronavirus defective interfering RNA internal replication signal in positive-strand RNA synthesis. J Virol. 1998;72:7926–7933. doi: 10.1128/jvi.72.10.7926-7933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′-untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlesinger S, Makino S, Linial M L. Cis-acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin Virol. 1994;5:39–49. doi: 10.1006/smvy.1994.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skinner M A, Racaniello V R, Dunn G, Cooper J, Minor P D, Almond J W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989;207:379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd S, Semler B L. Structure-infectivity analysis of the human rhinovirus genomic RNA 3′-noncoding region. Nucleic Acids Res. 1996;24:2133–2142. doi: 10.1093/nar/24.11.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wimmer E, Hellen C U T, Cao X M. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 42.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]