Abstract
Adeno-associated virus (AAV) vectors can transduce cells by several mechanisms, including (i) gene addition by chromosomal integration or episomal transgene expression or (ii) gene targeting by modification of homologous chromosomal sequences. The latter process can be used to correct a variety of mutations in chromosomal genes with high fidelity and specificity. In this study, we used retroviral vectors to introduce mutant alkaline phosphatase reporter genes into normal human cells and subsequently corrected these mutations with AAV gene targeting vectors. We find that increasing the length of homology between the AAV vector and the target locus improves gene correction rates, as does positioning the mutation to be corrected in the center of the AAV vector genome. AAV-mediated gene targeting increases with time and multiplicity of infection, similar to AAV-mediated gene addition. However, in contrast to gene addition, genotoxic stress did not affect gene targeting rates, suggesting that different cellular factors are involved. In the course of these studies, we found that (i) vector genomes less than half of wild-type size could be packaged as monomers or dimers and (ii) packaged dimers consist of inverted repeats with covalently closed hairpins at either end. These studies should prove helpful in designing AAV gene targeting vectors for basic research or gene therapy.
Adeno-associated virus (AAV) is a dependent parvovirus with a 4.7-kb single-stranded linear DNA genome. Vectors based on AAV have been developed by replacing the viral genes with foreign DNA between the cis-acting terminal repeats. Typically, the foreign DNA encodes a transgene cassette with promoter and polyadenylation signals designed to express from an ectopic location. These vectors can transduce cells by a variety of mechanisms, including random chromosomal integration of the vector genome (29, 38) or episomal transgene expression (2, 7, 18). If transduction is performed in the presence of the AAV rep gene, vectors can also be designed to integrate at the site-specific integration locus of wild-type AAV located on human chromosome 19 (3, 24, 25, 33). In addition to these transduction mechanisms (which can be defined as gene addition strategies), we have shown that AAV vectors can be used to introduce specific genetic modifications at homologous chromosomal sequences in a gene targeting process (16, 27). The gene targeting rates produced by AAV vectors approach 1% at the single-copy hypoxanthine phosphoribosyl transferase (HPRT) locus in normal human cells, which is 3 to 4 logs higher than can typically be achieved in human cells with conventional gene targeting methods (27). Possible explanations for the high rates of AAV-mediated gene targeting include efficient nuclear delivery of vector genomes, homologous pairing promoted by the single-stranded vector genome, or effects of the vector inverted terminal repeats.
To simplify studies of AAV-mediated gene targeting, we recently developed an assay based on the correction of mutant neomycin phosphotransferase (neo) genes introduced by retroviral vectors at random chromosomal locations (16). This approach allowed us to control the structure of the targeted locus and compare correction rates of a variety of mutations in the neo gene. We found that AAV vectors could be used to correct substitution, insertion, and deletion mutations with high fidelity at multiple chromosomal positions. Here we have developed a similar approach in which mutant alkaline phosphatase (AP) genes introduced by retroviral vectors are corrected by AAV vectors, allowing us to measure gene targeting rates by histochemical staining in the absence of selection. We used this simplified assay to study vector design parameters and the effects of different transduction conditions on AAV-mediated gene targeting rates. We also observed effects on AAV packaging due to vector genome size.
MATERIALS AND METHODS
Vectors.
Plasmid pLAPSN (20) and its derivatives were used to generate retroviral vector stocks by transient transfection of PG13 cells (19), collection of conditioned medium 2 days later, and passage through 0.45-μm-pore-size filters. The various AP mutations shown in Fig. 1A were introduced into pLAPSN by standard techniques (31) and confirmed by DNA sequencing.
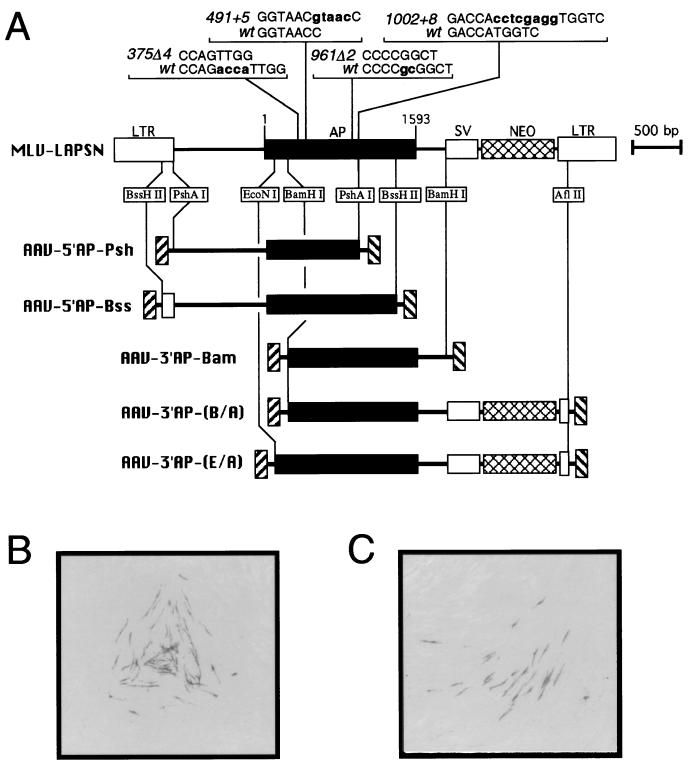
FIG. 1.
AP gene targeting vectors. (A) Diagrams of retroviral and AAV vectors used for AP gene targeting. The murine leukemia virus (MLV) retroviral vector pLAPSN is shown, with the positions of the LTRs, AP gene, SV40 promoter (SV), and neo gene indicated. The AP mutations used in this study are shown above the AP gene along with the corresponding wild-type (wt) sequences, with numbering starting at nucleotide position 1 of the AP reading frame. Differences between the wild-type and mutant sequences are shown in bold lowercase letters. Maps of five AAV vectors containing wild-type portions of the AP gene are shown below the pLAPSN map, with regions of homology and the restriction enzymes used to generate these AP fragments indicated. (B and C) Examples of AP+ foci produced in fibroblasts containing the 375Δ4 mutation by gene targeting with the AAV-5′AP-Bss vector in our standard assay (see Materials and Methods). Photomicrographs obtained with a dissecting microscope of clonal foci containing several AP+ cells are shown.
AAV vector stocks were made by transient transfection of TtetA2 packaging cells or 293T cells with AAV vector plasmids as described elsewhere (17). All stocks were treated with nuclease and purified on CsCl gradients. Any residual adenovirus was heat inactivated at 56°C for 1 h. Unless otherwise stated, AAV vector stock particle numbers were based on the amount of full-length, single-stranded vector DNA genomes detected by alkaline Southern blot analysis (17). AAV vector plasmids pA2-5′AP-Psh, pA2-5′AP-Bss, pA2-3′AP-Bam, pA2-3′AP-(B/A), and pA2-3′AP-(E/A) were made by inserting the restriction fragments from pLAPSN shown in Fig. 1A into the BglII site of pTR (30) and used to generate the corresponding vectors AAV-5′AP-Psh, AAV-5′AP-Bss, AAV-3′AP-Bam, AAV-3′AP-(B/A), and AAV-3′AP-(E/A), respectively. When necessary, these fragments were first given compatible cohesive ends by attaching BclI linkers (31). The AAV gene addition vector AAV-LAPSN (28) used in the experiments in Fig. 3c contains the murine leukemia virus long terminal repeat (LTR) driving expression of a functional AP gene, as well as a simian virus 40 (SV40) promoter driving neo, and thus is analogous to the retroviral vector pLAPSN.
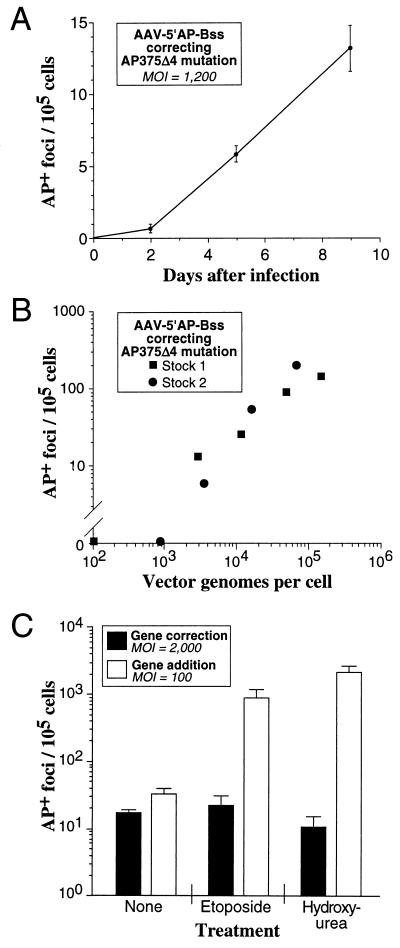
FIG. 3.
Effects of MOI, time, and genotoxic stress on gene targeting. AP gene correction assays were performed on MHF2 fibroblasts containing the AP375Δ4 target mutation with the vector AAV-5′AP-Bss. Experimental conditions were the same as our standard assay except for the modifications noted below. (A) The length of time before histochemical staining for AP expression was varied from 2 to 9 days after exposure to the AAV vector. Each point represents the mean of three independent measurements with standard error bars shown. (B) The MOI was varied as indicated, and the experiment was performed with two different AAV-5′AP-Bss stocks. (C) The cells were treated for 20 h with medium containing etoposide (3 μM), hydroxyurea (40 mM), or no drug immediately prior to exposure to the AAV vector. Parallel gene addition transduction experiments were performed with the AAV-LAPSN vector (28) under the same conditions except at an MOI of 100 instead of 2,000 vector genomes per cell (to generate similar numbers of AP+ foci in untreated samples). Values shown are the mean of three independent measurements with standard error bars shown.
For Fig. 4, fractions were collected directly from CsCl gradients, their densities were determined by refractometry, and the vector DNA within each fraction was released, separated on alkaline agarose gels, and quantitated by PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) analysis of Southern blots as described elsewhere (17). For Fig. 5, AAV vector DNA was purified from dialyzed CsCl fractions as described elsewhere (32). The light DNA samples were purified from the monomer peak fractions of AAV-3′AP-Bam and AAV-5′AP-Psh shown in Fig. 4 (densities of 1.367 and 1.366 g/ml, respectively). The heavy DNA samples were purified from fractions with higher densities than the dimer peaks shown in Fig. 4 to minimize copurifying monomer vector DNA (densities of 1.401 g/ml for AAV-3′AP-Bam and 1.404 g/ml for AAV-5′AP-Psh).
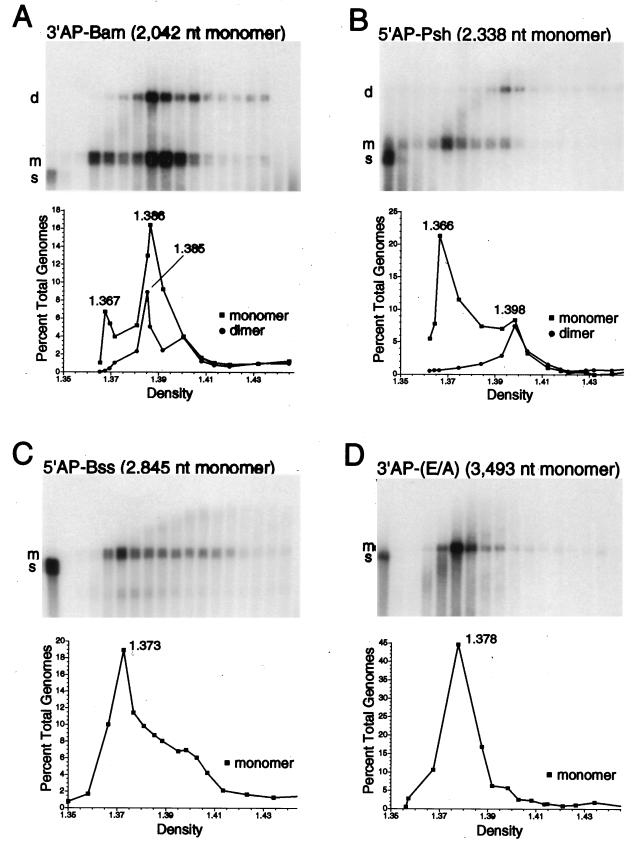
FIG. 4.
Monomer and dimer vector genome forms are packaged in particles with different densities. Vector stocks of AAV-3′AP-Bam, AAV-5′AP-Psh, AAV-5′AP-Bss, and AAV-3′AP-(E/A) were purified on CsCl gradients, fractions were collected, and the encapsidated DNA present in each fraction was isolated, separated as single-stranded molecules on alkaline agarose gels, transferred to nylon membranes, and probed for vector sequences as described elsewhere (17). The sizes in nucleotides (nt) of full-length, monomer vector genomes of each vector are shown above the blots. The density of each fraction was determined by refractometry. The results of this alkaline Southern blot analysis are shown for each vector stock, with fractions increasing in density from left to right. The first two lanes of blots A and B represent 1.0 ng and 100 pg of standards consisting of the same DNA fragment present in each vector genome but lacking the inverted terminal repeats. The first lanes of blots C and D represent analogous 1.0-ng standards. These standards were mixed with lightest gradient fraction before loading on the gel to adjust for salt concentrations. The positions of vector genome dimers (d), monomers (m), and standards (s) are indicated at the left of each blot. In the graphs below, the amounts of monomer forms (squares) and dimer forms (circles) present in each fraction were determined and plotted as a percentage of the total genome content of the gradient (dimers and monomers) versus the density (grams per milliliter of each fraction). Density values of the fractions containing peak values of monomer and dimer forms are indicated.
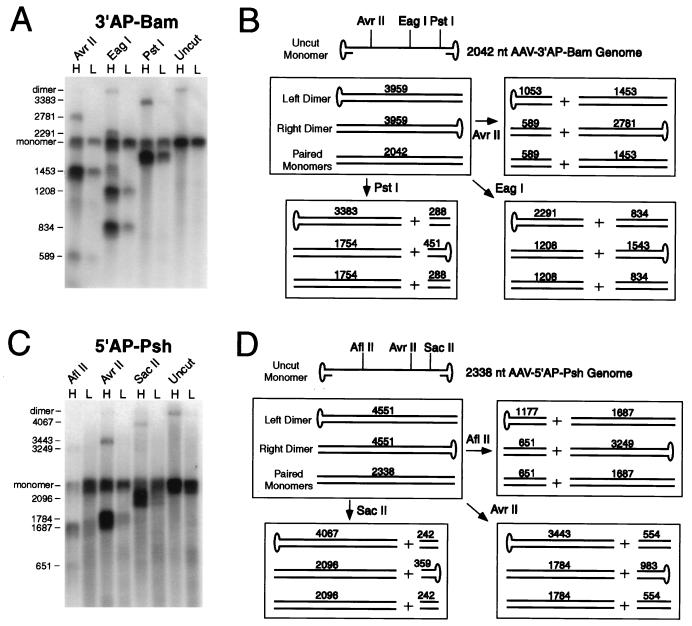
FIG. 5.
Restriction analysis of vector dimer forms. Encapsidated vector DNA molecules from light (L) and heavy (H) CsCl gradient fractions of AAV-3′AP-Bam and AAV-5′AP-Psh vector stocks were purified, digested with the indicated restriction endonucleases, and analyzed by alkaline gel electrophoresis of denatured molecules. (A and C) Southern blot analysis of these digests for AAV-3′AP-Bam and AAV-5′AP-Psh, respectively. Undigested samples were also run on each gel. The sizes of the major single-stranded digestion products and of full-length monomer and dimer genome forms are shown in nucleotides at the left of each blot. Denatured plasmid restriction fragments of known size were run on each gel as size standards (not shown). (B and D) Each box shows the predicted structure and size (in total nucleotides [nt] of denatured molecules) of the predicted left dimer, right dimer, and paired monomer forms present in undigested DNA samples and samples digested with the indicated restriction endonucleases for AAV-3′AP-Bam and AAV-5′AP-Psh, respectively. Left and right dimer forms differ in the location of covalently closed hairpin ends.
Cell culture and transduction.
PG13 cells (19) and MHF2 normal human fibroblasts (repository no. GM05387; Coriell Institute for Medical Research, Camden, N.J.) (27) were propagated in Dulbecco's modified Eagle's medium supplemented with heat-inactivated 10% fetal bovine serum at 37°C. Proviral target sites were introduced into MHF2 cells by transduction with pLAPSN-derived retroviral vector stocks, followed by selection in G418 (0.6 mg of active compound per ml). The polyclonal, transduced cell populations were all derived from >104 independent transduction events, as determined by plating dilutions of the transduced cells for selection 1 day after exposure to retroviral vector stocks and counting the number of G418-resistant colonies. Multiplicities of infection (MOIs) of ≤0.1 retroviral transducing units per cell were used to ensure that most cells contained a single retroviral vector provirus.
To measure AP gene correction rates in our standard assay, MHF2-derived target cells were seeded on day 1 at 105 cells/well in 48-well plates, infected with AAV vectors on day 2 at an MOI of 2,000 vector particles/cell (assuming no increase in cell number), treated with trypsin, and plated in two 6-cm-diameter dishes on day 3 (0.25 or 99.75% of cells). The 0.25% plating was stained on day 4 and used to calculate the total number of viable cells per well (values ranged from 5 × 104 to 1.5 × 105 viable cells/original well, and exposure to AAV vector stocks had no effect on plating efficiencies). The 99.75% plating was cultured until day 9 and then stained histochemically for AP expression as described previously (9). Gene correction rates were calculated as the number of AP+ foci/105 viable cells. In some experiments, the day of AP staining was varied (Fig. 3A), the MOI was varied (Fig. 3B and Table 1), or the cells were treated for 20 h immediately prior to the addition of vector with etoposide or hydroxyurea (Fig. 3C). Experiments included a control for each target cell population that did not receive vector and always failed to produce AP+ foci. In addition, each AAV vector used for gene targeting failed to produce AP+ foci when used to infect cells without target loci. These controls confirmed that all AP+ foci represented gene targeting events rather than reversion of target loci or targeting vector AP mutations.
TABLE 1.
Gene correction rates at different monomer and dimer genome MOIs
| AP target mutationa | AAV vector | Gradient fraction | MOI (vector genomes/cell)
|
AP+ foci/105 cellsb | |
|---|---|---|---|---|---|
| Monomer | Dimer | ||||
| 961Δ2 | None | 0 | |||
| 961Δ2 | None | 0 | |||
| 961Δ2 | None | 0 | |||
| 961Δ2 | 3′AP-Bam | Heavy | 2,000 | 1,500 | 20 |
| 961Δ2 | 3′AP-Bam | Middle | 2,000 | 800 | 28 |
| 961Δ2 | 3′AP-Bam | Middle | 2,000 | 800 | 16 |
| 961Δ2 | 3′AP-Bam | Middle | 2,000 | 800 | 26 |
| 961Δ2 | 3′AP-Bam | Light | 2,000 | 100 | 63 |
| 375Δ4 | None | 0 | |||
| 375Δ4 | None | 0 | |||
| 375Δ4 | None | 0 | |||
| 375Δ4 | 5′AP-Psh | Heavy | 2,000 | 800 | 27 |
| 375Δ4 | 5′AP-Psh | Heavy | 2,000 | 800 | 24 |
| 375Δ4 | 5′AP-Psh | Heavy | 2,000 | 800 | 31 |
| 375Δ4 | 5′AP-Psh | Heavy | 2,000 | 800 | 38 |
| 375Δ4 | 5′AP-Psh | Light | 3,000 | 180 | 34 |
| 375Δ4 | 5′AP-Psh | Light | 3,000 | 180 | 18 |
All targets were in MHF2 normal human fibroblasts.
Each value is from a single experiment using our standard AP correction assay (see Materials and Methods).
RESULTS
AP gene correction strategy.
We used bicistronic retroviral vectors to introduce mutant AP genes into normal human fibroblasts and then corrected these mutations by gene targeting with AAV vectors (Fig. 1A). The retroviral vectors were based on pLAPSN (20), which expresses (i) AP from the murine leukemia virus LTR promoter and (ii) neomycin phosphotransferase from an internal SV40 promoter that served as a selectable marker. Four different mutations (2- and 4-bp deletions, as well as 5- and 8-bp insertions) were placed into the AP gene of pLAPSN, and polyclonal G418-resistant fibroblast populations containing >104 independent proviral insertions were generated for each of these mutations. The high complexity of these polyclonal populations allowed us to avoid position effects that might otherwise influence gene targeting rates if individual clones were compared.
Five different AAV vectors were constructed, each of which contained an incomplete portion of the wild-type AP gene (Fig. 1A). These vectors were all homologous to the retroviral target loci except for the mutation to be corrected, and they varied in length of homology to the target site as well as positioning of the mutations to be corrected. All of the AAV targeting vectors and retroviral target loci except AAV-3′AP-(E/A) contained nonfunctional AP genes, and so correction of the retroviral target mutations by AAV targeting vectors could be scored histochemically as foci of AP+ purple cells (Fig. 1B and C). The AAV-3′AP-(E/A) vector contained a partially functional AP fragment, and so this vector was used only in vector packaging experiments, not in gene targeting studies. We used this simple and convenient AP targeting assay to study several parameters relevant to gene targeting.
Effects of homology length and mutation type on gene correction rates.
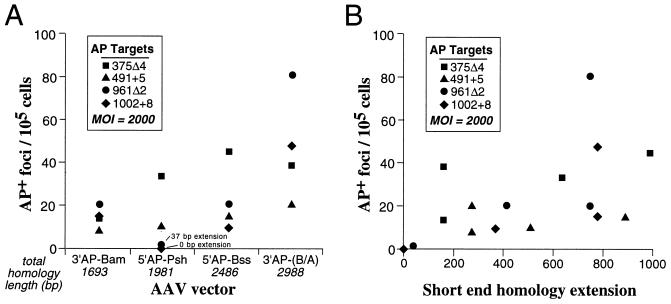
Fibroblast cell populations containing each of the four AP mutations shown in Fig. 1A were infected with four different AAV targeting vectors, for a total of 16 possible combinations. We used our standard AP correction assay, which consisted of infection with an AAV targeting vector at an MOI of 2,000 particles/cell and then staining for AP expression 7 days later (see Materials and Methods). Figure 2A shows the gene correction rates from these experiments plotted for each AAV targeting vector. The total homology between the targeting vectors and the target loci varied from 1,693 to 2,988 bp, and increasing homology length appeared to increase targeting rates. However, even the shortest homology vector, AAV-3′AP-Bam, was able to correct all four mutations, with targeting rates only two- to fivefold lower than that of the longest homology vector, AAV-3′AP-(B/A).
FIG. 2.
Correction of AP mutations with different AAV targeting vectors. Standard AP gene correction assays were performed on polyclonal fibroblast populations containing each of the indicated AP target mutations with AAV-3′AP-Bam, AAV-5′AP-Psh, AAV-5′AP-Bss, and AAV-3′AP-(B/A). Each value represents the average from at least two independent experiments. (A) Correction rates were plotted for each AAV targeting vector, with the total length of homology each AAV vector shared with the target locus indicated. The data are arranged with increasing total homology length from left to right. (B) The data in panel A are plotted relative to the short end homology extension for each correction assay. The short end homology extension is the distance of the corrected mutation from the end of the short side flanking homology.
A more dramatic effect of homology on gene targeting occurred when the mutation being corrected was positioned near one end of the AAV vector genome. Figure 2B plots the same gene correction values as in Fig. 2A versus the distance of the corrected mutation from the end of the short side flanking homology (called the short end homology extension). In the case of the AAV-5′AP-Psh vector, two of the mutations had small short end homology extensions of 0 or 37 bp, which resulted in significantly lower targeting rates. All other data points had short end homology extensions of >150 bp, and although there appeared to be some increase in gene correction rates with longer extensions, these effects were not consistent.
All four mutations could be corrected by AAV-mediated gene targeting. If one excludes the two data points with short end homology extensions below 150 bp, then the average gene correction rates were 13.8, 24.3, 33.4, and 41.1 (AP+ foci/105 cells) for mutations 491+5, 1002+8, 375Δ4, and 961Δ2, respectively. Although these correction rates are all within threefold of each other, it is worth noting that both insertion mutations were corrected at lower rates. A similar result was obtained when AAV vectors were used to correct mutations in the neo gene, and a 1-bp insertion was corrected less efficiently than a 1-bp deletion or several substitution mutations (16). More controlled experiments with defined insertions and deletions placed at the same position in target loci will be required to determine if a true bias exists against correcting insertion mutations.
Effects of different transduction conditions on AAV-mediated gene targeting.
We studied the effects of time, MOI, and genotoxic stress on gene targeting rates. As shown in Fig. 3A and B, correction of the AP375Δ4 mutation by AAV-mediated gene targeting increased with longer times after vector exposure and higher MOIs. In Fig. 3C, etoposide and hydroxyurea were used to induce genotoxic stress prior to vector exposure. Etoposide is an inhibitor of topoisomerase II which can enhance enzyme-mediated DNA cleavage, and hydroxyurea prevents DNA synthesis by inhibiting ribonucleotide reductase and depleting deoxynucleotide pools. A transient exposure to these agents prior to the addition of AAV vectors induces cellular functions involved in DNA repair and/or synthesis that result in higher gene addition rates by AAV vectors (26). Since some of these same cellular functions might be involved in AAV-mediated gene targeting, we tested their effects on correction of the AP375Δ4 mutation. No effect on gene targeting was observed with either drug, although parallel experiments demonstrated a significant increase in gene addition rates with the same treatments. In Fig. 3C, we adjusted MOIs to produce similar numbers of AP+ foci in untreated gene addition and gene targeting samples. At an MOI of 2,000 particles/cell, gene addition still increased with genotoxic stress (14.8-fold increase for etoposide treatment and 59-fold increase for hydroxyurea treatment).
Small vector genomes can be packaged as dimers.
We routinely analyzed the genome content of AAV vector stocks by alkaline gel electrophoresis of each CsCl gradient fraction and observed that in some cases a dimer-sized vector genome band was present (see Materials and Methods). Because these stocks were digested extensively with micrococcal nuclease before being loaded on the gradients, these bands represent encapsidated genomes. Packaged dimers appeared only when the vector genome size was less than half the size of wild-type AAV (<2.5 kb). The dimer-sized genomes are clearly present in the gradients from stocks of AAV-3′AP-Bam and AAV-5′AP-Psh, with genome sizes of 2,042 and 2,338 nucleotides, respectively, but were missing from stocks of AAV-5′AP-Bss and AAV-3′AP(E/A), with genome sizes of 2,845 and 3,493 nucleotides, respectively (Fig. 4).
The stocks containing dimer-sized genomes also contained monomer-sized genomes, which were present across a broader range of densities. At low densities, only monomers were present, while at high densities a second peak containing both monomer- and dimer-sized genomes occurred. These results demonstrate that the amount of encapsidated DNA contributes to the density of vector virions, with a greater DNA content producing a higher density particle. Similar results were noted previously in wild-type AAV stocks containing variant genomes of different sizes (4). Presumably the denser peak contained virions with either one dimer-sized genome or two monomer-sized genomes, which would produce very similar total DNA contents. A faint smear could typically be seen extending from the lighter monomer peak to the denser dimer peak in these gradients (Fig. 4A and B). This signal corresponds to vector DNA molecules with sizes between one and two complete genome lengths, with the sizes approaching that of a complete dimer as the density increased. A similar phenomenon can be seen in Fig. 4C, where a smear extends above the monomer peak and increases in size at higher densities. No true dimer band is observed here, since full-length dimers of AAV-5′AP-Bss would be larger than the wild-type viral genome and exceed the packaging capacity of AAV.
Packaged vector dimers are inverted repeats with hairpins at either end.
The most likely structure of the dimer-sized genomes present in vector stocks would be head-to-head or tail-to-tail inverted dimers formed by DNA replication extending from the 3′-terminal repeat (13, 35). These molecules should base pair along most of their length and contain a covalently closed hairpin at the terminal repeat where DNA synthesis began. Analogous molecules may also be present in wild-type AAV stocks that contain small, “snap-back” variant genomes (4). Because the dimers form double-stranded molecules, they can be efficiently digested with restriction endonucleases, allowing a more detailed analysis.
We isolated encapsidated DNA from heavy and light CsCl gradient fractions of AAV-3′AP-Bam and AAV-5′AP-Psh vector stocks. The heavy fractions contained dimer-sized genomes as well as monomer-sized genomes, and the light fractions contained only monomer-sized genomes (Fig. 4A and B). The DNA molecules present in these fractions were digested under neutral conditions with restriction endonucleases that have sites located asymmetrically in the vector genome. The resulting restriction fragments were separated as single strands on alkaline agarose gels, transferred to nylon membranes, and probed for internal vector sequences. In each case the presence of head-to-head or tail-to-tail dimers should produce characteristic restriction fragments, both for dimers containing a closed hairpin at the left terminal repeat (left dimer) and for dimers containing a closed hairpin at the right terminal repeat (right dimer). Complementary monomer molecules could also hybridize to some extent under these conditions and produce specific restriction fragments.
The results of this analysis, including the structures of the left and right dimers and their predicted restriction digestion products, are shown in Fig. 5. The heavy gradient fractions of AAV-3′AP-Bam and AAV-5′AP-Psh produced the expected restriction fragments of left and/or right dimers for each set of enzymes, as well as the fragments expected from paired monomers. The dimer digestion products were absent from the light gradient fractions, while the paired monomer restriction fragments were still present. Because the restriction sites were located asymmetrically in the vector genome, each enzyme produced a large fragment for only one dimer orientation (left or right) and a small fragment for the other orientation. In most cases, only the larger fragment could be detected due to preferential hybridization with the probe. However, the use of different enzymes allowed us to demonstrate the presence of both left and right dimer forms in each vector stock. For example, in the case of AAV-3′AP-Bam, the 2,781-nucleotide fragment produced by AvrII was due to the right dimer, while the 3,383-nucleotide fragment produced by PstI was from the left dimer. Digestion of the AAV-3′AP-Bam heavy gradient fraction with EagI produced fragments due to both left (2,291-nucleotide) and right (1,543-nucleotide) dimer forms in the same reaction. It is also possible that some dimers have closed hairpins at both ends, since these molecules have been shown to arise during AAV replication (23, 37).
Full-length monomer genomes were present in both the light and heavy gradient fractions, and only a portion of these could be digested with restriction enzymes. The digested monomers presumably had hybridized with complementary monomers during the restriction digestion, allowing efficient cleavage of double-stranded molecules. In contrast, most of the full-length dimer forms present in the untreated samples were completely digested by restriction enzymes. This argues against the presence of packaged head-to-tail dimers (direct repeats) in these vector stocks, since these molecules are not self-complementary, and based on the results of monomer digestion, only a portion of single-stranded dimer genomes would be expected to hybridize with complementary dimers during restriction digestion. Single-stranded, head-to-tail dimer forms would therefore be expected to be largely resistant to restriction digestion and remain as full length dimers.
Gene targeting rates correlate with monomer vector genome levels.
We compared gene correction rates using different CsCl gradient fractions of AAV-3′AP-Bam and AAV-5′AP-Psh to determine if dimer or monomer genomes were preferentially used for gene targeting (Table 1). Although the analysis was complicated by the fact that all fractions contained monomer genomes, no significant contribution to gene targeting could be attributed to the presence of dimer genomes. This is most clearly demonstrated by comparing the gene correction rates of AAV-3′AP-Bam light and heavy fractions. An increase in the dimer MOI from 100 genomes/cell (light fraction) to 1,500 genomes/cell (heavy fraction) in the presence of a constant amount of monomer genomes did not increase gene correction rates. The conclusion that monomers are the preferred substrates for gene targeting is also supported by the data in Fig. 2, since the larger AAV-5′AP-Bss and AAV-3′AP-(B/A) vectors tended to produce higher gene correction rates, despite the absence of dimer forms in these stocks (the MOIs in Fig. 2 were all based on monomer forms).
DISCUSSION
In this report we have described an AP gene correction assay to study several parameters that influence AAV-mediated gene targeting. The assay is simple to perform, allows for sequence manipulations of both the target locus and targeting vector, and in principle can be applied to any cell line that can be transduced by retroviral vectors. By assaying polyclonal cell populations with target loci at multiple chromosomal locations, we were able to avoid position effects that might influence gene targeting and compare correction rates of different AP mutations. A major advantage of the AP system over a similar approach that we recently described based on correction of neo gene mutations (16) is that there is no selective pressure for corrected loci, and gene targeting is assayed instead by histochemical staining. A disadvantage of this system is that clones containing corrected loci cannot easily be separated from nontargeted cells and expanded for DNA analysis. However, our previous HPRT and neo gene targeting experiments have shown that >90% of targeted loci contain the expected genetic change with no secondary mutations (16, 27), and so we assume that most AP+ foci represent accurate gene correction events.
A comparison of AP gene correction rates obtained with different AAV vectors suggests that central positioning of the modification being introduced and increasing total homology with the target locus will lead to optimal gene targeting rates. Although our data set does not establish the ideal length of homology extension beyond the introduced modification, there were dramatic decreases in AP gene correction rates when this distance was 37 bp or less (Fig. 2B). The effects of total homology length are best illustrated by comparing the AP correction rates obtained with the smallest (AAV-3′AP-Bam; 1,693-bp homology) and largest (AAV-3′AP-[B/A]; 2,988-bp homology) AAV vectors. In these cases a 1.8-fold increase in total homology increased gene targeting rates two- to fivefold (Fig. 2A). A dependence on homology length was also observed with conventional gene targeting approaches based on electroporation or transfection of plasmid constructs (5, 36). However, the absolute targeting frequencies obtained by conventional methods were approximately 10−6 with 2- to 4-kb constructs (5), compared to 10−3 for AAV-mediated gene targeting (16, 27) (Fig. 3B).
To date, we have used AAV vectors to introduce several different types of genetic modifications, including substitutions (1 and 2 bp), insertions (1, 2, and 4 bp), and deletions (1, 5, 8, and 14 bp) at three different target loci (neo, HPRT, and AP [references 16 and 27 and this study]). The only modifications that were introduced at lower frequencies were a 1-bp deletion in the neo gene at about 10-fold-lower rates (16) and 5- or 8-bp deletions in the AP gene at 2- to 3-fold lower rates (Fig. 2). Thus, small insertion mutations (which are corrected by introducing deletions) may be more difficult to correct than other types of mutations. Although the basis for these differences is not understood, our results suggest that many of the mutations responsible for genetic diseases will be amenable to gene correction by AAV vectors. We have not determined if larger modifications can be efficiently introduced; however, the AAV packaging capacity of <5 kb will ultimately limit the size of insertion modifications, especially if substantial homology to the target locus must be included in the vector genome.
Transduction conditions are known to have significant effects on gene addition by AAV vectors, including time after vector exposure, MOI, and genotoxic stress (1, 8, 11, 12, 14, 15, 26, 28, 34). Similarly, we found that both longer times and higher MOIs also increased gene targeting by AAV vectors. A clear dependence on MOI was previously noted for HPRT and neo gene targeting as well (16, 27). The increase over time is presumably related to the persistence of AAV vector genomes as single-stranded, episomal molecules in the nuclei of normal human fibroblasts (28) and suggests that these genomes continue to participate in gene targeting reactions long after the initial infection with vector particles.
In contrast to its effects on gene addition, genotoxic stress induced by etoposide or hydroxyurea had no effect on gene targeting by AAV vectors. These treatments are thought to increase gene addition by promoting integration and/or second-strand synthesis of the vector genome (1, 8, 10, 26). Etoposide introduces chromosomal breaks that could serve as vector integration sites, and the removal of hydroxyurea allows nucleotide precursor pools to reaccumulate, which may promote second-strand synthesis. Apparently neither of these processes nor other cellular DNA repair and/or synthesis functions induced by genotoxic stress are involved in gene targeting. This is consistent with a model in which single-stranded, episomal vector genomes, rather than integrated or double-stranded forms, participate in the gene targeting reaction. The monomer and dimer data in Table 1 also support this model, since the double-stranded vector dimers did not appear to contribute to gene targeting. Given that different mechanisms are used, it may be possible to identify conditions that favor gene targeting without increasing gene addition and thereby increase the ratio of targeted to nontargeted transduction events. A dependence on single-stranded vector genomes also suggests that annealing of complementary input vector genomes may limit gene targeting in some situations.
Our observation that small vector genomes can be packaged as dimers is consistent with prior studies suggesting that the AAV packaging capacity is approximately 5 kb of DNA (21). When vector genomes are greater than one-half wild-type size (>2.5 kb), single-stranded, linear monomers are packaged, as with wild-type AAV. Smaller vector genomes can be packaged in relatively light capsids as single-stranded linear monomers or in heavier capsids as dimers or pairs of linear monomers (based on the equivalent densities of these types of particles). The encapsidated dimers are head-to-head or tail-to-tail repeats with covalently closed hairpins present at a terminus. These dimers are expected to form double-stranded molecules throughout their length, except for the small loop regions of the terminal repeats. However, this does not demonstrate that single-stranded DNA is not required for packaging, since these dimers may be encapsidated as unwound, single-stranded molecules. A previous study also concluded that AAV vector particles may contain two copies of small genomes, but no encapsidated dimer forms were described (6).
The encapsidated genome dimers are expected to be produced as replication intermediates in the presence of the AAV Rep proteins and adenovirus helper functions (13, 35). Packaging of these dimers may be associated with replication, since AAV particle assembly is linked to viral DNA synthesis (22, 39). In one model for packaging, preassembled AAV capsids incorporate vector genomes that sometimes exceed the packaging capacity, which results in portions of the vector genome remaining outside the capsid, and these exposed genome portions are subsequently removed by nucleases (6). This may result in the smear seen on some alkaline gels of intermediate-sized genomes packaged in particles of increasing density (Fig. 4).
ACKNOWLEDGMENTS
We thank Rong Dong, John Weller, and Richard Newton for technical assistance.
This work was supported by grants from the March of Dimes Birth Defects Foundation, Cystic Fibrosis Foundation, and National Institutes of Health.
REFERENCES
- 1.Alexander I E, Russell D W, Miller A D. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J Virol. 1994;68:8282–8287. doi: 10.1128/jvi.68.12.8282-8287.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertran J, Miller J L, Yang Y, Fenimore Justman A, Rueda F, Vanin E F, Nienhuis A W. Recombinant adeno-associated virus-mediated high-efficiency, transient expression of the murine cationic amino acid transporter (ecotropic retroviral receptor) permits stable transduction of human HeLa cells by ecotropic retroviral vectors. J Virol. 1996;70:6759–6766. doi: 10.1128/jvi.70.10.6759-6766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertran J, Yang Y, Hargrove P, Vanin E F, Nienhuis A W. Targeted integration of a recombinant globin gene adeno-associated viral vector into human chromosome 19. Ann N Y Acad Sci. 1998;850:163–177. doi: 10.1111/j.1749-6632.1998.tb10473.x. [DOI] [PubMed] [Google Scholar]
- 4.de la Maza L M, Carter B J. Molecular structure of adeno-associated virus variant DNA. J Biol Chem. 1980;255:3194–3203. [PubMed] [Google Scholar]
- 5.Deng C, Capecchi M R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong J Y, Fan P D, Frizzell R A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 7.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields Berry S C, Halliday A L, Cepko C L. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher K J, Gao G P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 12.Flannery J G, Zolotukhin S, Vaquero M I, LaVail M M, Muzyczka N, Hauswirth W W. Efficient photoreceptor-targeted gene expression in vivo by recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:6916–6921. doi: 10.1073/pnas.94.13.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauswirth W W, Berns K I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977;78:488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- 14.Hermonat P L, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herzog R W, Hagstrom J N, Kung S H, Tai S J, Wilson J M, Fisher K J, High K A. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue N, Hirata R K, Russell D W. High-fidelity correction of mutations at multiple chromosomal positions by adeno-associated virus vectors. J Virol. 1999;73:7376–7380. doi: 10.1128/jvi.73.9.7376-7380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue N, Russell D W. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J Virol. 1998;72:7024–7031. doi: 10.1128/jvi.72.9.7024-7031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik P, McQuiston S A, Yu X J, Pepper K A, Krall W J, Podsakoff G M, Kurtzman G J, Kohn D B. Recombinant adeno-associated virus mediates a high level of gene transfer but less efficient integration in the K562 human hematopoietic cell line. J Virol. 1997;71:1776–1783. doi: 10.1128/jvi.71.3.1776-1783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 22.Myers M W, Carter B J. Assembly of adeno-associated virus. Virology. 1980;102:71–82. doi: 10.1016/0042-6822(80)90071-9. [DOI] [PubMed] [Google Scholar]
- 23.Ni T H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palombo F, Monciotti A, Recchia A, Cortese R, Ciliberto G, La Monica N. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J Virol. 1998;72:5025–5034. doi: 10.1128/jvi.72.6.5025-5034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recchia A, Parks R J, Lamartina S, Toniatti C, Pieroni L, Palombo F, Ciliberto G, Graham F L, Cortese R, La Monica N, Colloca S. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc Natl Acad Sci USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell D W, Alexander I E, Miller A D. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell D W, Hirata R K. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell D W, Miller A D, Alexander I E. Adeno-associated virus vectors preferentially transduce cells in S phase. Proc Natl Acad Sci USA. 1994;91:8915–8919. doi: 10.1073/pnas.91.19.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutledge E A, Russell D W. Adeno-associated virus vector integration junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 34.Snyder R O, Miao C H, Patijn G A, Spratt S K, Danos O, Nagy D, Gown A M, Winther B, Meuse L, Cohen L K, Thompson A R, Kay M A. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- 35.Straus S E, Sebring E D, Rose J A. Concatemers of alternating plus and minus strands are intermediates in adeno-associated virus DNA synthesis. Proc Natl Acad Sci USA. 1976;73:742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 37.Ward P, Berns K I. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- 38.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Muzyczka N. In vitro packaging of adeno-associated virus DNA. J Virol. 1998;72:3241–3241. doi: 10.1128/jvi.72.4.3241-3247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]