Abstract
Human immunodeficiency virus type 1 integrase (HIV-1 IN) is thought to have several putative roles at steps prior to integration, such as reverse transcription and nuclear transport of the preintegration complex (PIC). Here, we investigated new functional aspects of HIV-1 IN in the context of the viral replication cycle through point mutagenesis of Ser, Thr, Tyr, Lys, and Arg residues conserved in IN, some of which are located at possible phosphorylation sites. Our results showed that mutations of these Ser or Thr residues had no effect on reverse transcription and nuclear transport of PIC but had a slight effect on integration. Of note, mutations in the conserved KRK motif (amino acids 186 to 189), proposed previously as a putative nuclear localization signal (NLS) of HIV-1 IN, did not affect the karyophilic property of HIV-1 IN as shown by using a green fluorescent protein fusion protein expression system. Instead, these KRK mutations resulted in an almost complete lack of viral gene expression due to the failure to complete reverse transcription. This defect was complemented by supplying wild-type IN in trans, suggesting a trans-acting function of the KRK motif of IN in reverse transcription. Mutation at the conserved Tyr 143 (Y143G) resulted in partial impairment of completion of reverse transcription in monocyte-derived macrophages (MDM) but not in rhabdomyosarcoma cells. Similar effects were obtained by introducing a stop codon in the vpr gene (ΔVpr), and additive effects of both mutations (Y143G plus ΔVpr) were observed. In addition, these mutants did not produce two-long terminal repeat DNA, a surrogate marker for nuclear entry, in MDM. Thus, the possible impairment of Y143G might occur during the nuclear transport of the PIC. Taken together, our results identified new functional aspects of the conserved residues in HIV-1 IN: i) the KRK motif might have a role in efficient reverse transcription in both dividing and nondividing cells but not in the NLS function; ii) Y143 might be an important residue for maintaining efficient proviral DNA formation in nondividing cells.
Retroviruses establish a proviral state in which a double-stranded DNA copy of the viral genomic RNA is integrated into the host genome in a stable manner, through several steps following binding and entry into the target cell. These early events include uncoating, reverse transcription, nuclear transport of the viral genome, and integration. The viral enzyme integrase (IN) is encoded by the pol gene and the attachment (att) site located at the U3 and U5 termini of the viral DNA and is required for integration, which is the last event (6, 14, 43, 47, 53, 56, 57, 61, 66, 74). The detailed mechanism of retroviral integration has been elucidated from in vitro studies using recombinant IN protein and a synthetic DNA substrate mimicking the viral att sites. These studies, using in vitro assays, have contributed much information toward the currently accepted mechanism of retroviral integration (reviewed in references 34, 42, and 75). Mutational and structural studies of human immunodeficiency virus type 1 (HIV-1) IN have identified three functional domains: a central catalytic core domain, an N-terminal zinc binding domain, and a C-terminal nonspecific DNA binding domain. The core domain contains the highly conserved D,D35E motif, which is directly involved in the catalytic activities of IN (7, 23, 46, 48). The N-terminal domain contains a highly conserved HHCC motif, which binds to zinc. Through a tetrahedral attachment to the HHCC motif, zinc enhances both multimerization and enzymatic activities of HIV-1 IN in vitro (5, 8, 21, 79). The C terminus, consisting of a structure that closely resembles Src homology 3 domains, possesses sequence- and metal ion-independent DNA binding activity (20, 51). Each domain has been demonstrated to form a dimer or higher multimerization state of IN (8, 19, 20), which might be required for its full activity (13, 21, 22, 66, 70, 73).
Genetic analysis of HIV-1 IN has demonstrated multiple effects of mutations at steps distinct from integration. These steps include correct viral particle formation (24, 59), uncoating (54, 59), and reverse transcription (49, 54). During the early events of the infection cycle, prior to integration, around 50-100 protomers of IN exist as one of the major components of the preintegration complex (PIC). This is composed of the viral genome, the matrix protein (MA), viral protein R (Vpr), and other viral and host proteins (3, 4, 26, 27). A small number of protomers of IN are thought to be sufficient for the integration reaction. The multiple effects of IN mutations suggest that IN has roles during the early events prior to integration, such as reverse transcription and nuclear transport of the PIC. Several important aspects of these putative roles in the context of the viral replication cycle still remain to be determined. Cellular or viral cofactors may participate in the steps leading to integration in vivo. The host factors HMG-I(Y) (26, 36), Ini1 (integrase interactor 1) (40, 41), and BAF (barrier-to-autointegration factor) (12) are associated with IN and stimulate integration activity. Recently, it has been reported that DNA-dependent protein kinase is involved in the completion of the integration process (18). On the other hand, HIV-1 IN has been shown to interact with importin alpha-karyopherin, the cellular nuclear localization signal (NLS) receptor, and to facilitate nuclear transport of the PIC (31). Transport of the PIC from the cytoplasm into the nucleus is thought to be a key step in establishing proviral DNA formation of HIV-1 in nondividing cells such as macrophages (reviewed in reference 3). It has been reported that phosphorylation of a Tyr residue on MA governs HIV nuclear import (32). However, results pertaining to MA function in the nuclear import process are controversial (29, 30). The NLS presented in MA might be weak and insufficient to ensure effective import of the PIC (4, 58). In addition, Vpr is thought to act as a key regulator of nuclear import of HIV-1, enhancing the affinity of the putative NLS to the host nuclear pore complex (1, 2, 15, 28, 30, 35, 52, 64). These observations suggest that proteins other than MA, most probably IN, possess the NLS, enabling HIV-1 to infect nondividing cells with Vpr helper function.
Phosphorylation of viral proteins also plays an important role in the regulation of the viral life cycle. Identification of the virion-associated kinase and the mitogen-activated protein kinase (known as ERK or MAPK) suggested that an MAPK signal transduction pathway in the host cell might regulate an early step in HIV-1 infection (38). The phosphorylation status of IN after infection and its role(s) during early events in the retroviral infection cycle are poorly understood. In this study, we generated a series of HIV-1 IN mutant clones carrying single-amino acid substitutions at conserved Ser, Thr, or Tyr residues; some of them are located at a possible motif for phosphorylation by casein kinase II (CK-II) or protein kinase C (PKC). Furthermore, we introduced point or deletion mutations in the conserved Lys and Arg residues, reported to be the putative NLS of HIV-1 IN (31). Genetic analysis of HIV-1 IN mutants in the present study showed that in the context of the viral infection cycle, the KRK motif in HIV-1 IN has a trans-acting function which is important for the completion of reverse transcription in both dividing and nondividing cells. In addition, Y143 may be a key residue for efficient proviral formation in nondividing cells.
MATERIALS AND METHODS
Construction of mutants.
DNA fragments for mutagenesis of HIV-1 IN were derived from the HIV-1 pNL43lucΔenv vector (54, 62) in which the env gene is defective, allowing the formation of pseudotypes, and the nef gene is replaced with the firefly luciferase (Luc) gene. For mutagenesis of Thr and Tyr residues (T66A, T93A, T125A, and Y143G), a 1.6-kb fragment of the pNL43lucΔenv vector, spanning the KpnI and SalI sites (nucleotides [nt] 4154 to 5785), was subcloned into pBluescript SK(+) (Stratagene, Calif.) (pSKnlK/S). To introduce mutations at each residue, four mutagenic primers were designed to span the NsiI and AflII sites (nt 4377 to 4743). Products amplified by PCR with each mutagenic primer pair were digested with NsiI and AflII. Then, the mutant fragments were ligated to NsiI-AflII-digested pSKnlK/S. After confirmation of mutation by DNA sequence analysis, KpnI-SalI fragments (nt 4154 to 5785) containing each mutation were ligated to the fragment spanning nt 1507 to 4154 to generate the 4.2-kb SpeI-SalI fragment (nt 1507 to 5785). The SpeI/SalI fragments, containing each mutation, were inserted back into the pNL43lucΔenv vector. For mutagenesis of the Ser residues (S195A and S283A) or Lys and Arg residues (K186Q, ΔKRK, and K211N), mutagenic primers were designed to span the AflII and NdeI sites (nt 4743 to 5172) of the pNL43lucΔenv vector. The products amplified by PCR using each mutagenic primer pair, were subcloned into an AflII-NdeI-digested pGEM5-5Zf(+) vector (Promega, Madison, Wis.), containing pNL43lucΔenv (nt 1507 to 5122 [SpeI-NdeI fragment]). The 3.6-kb fragment of pNL43lucΔenv spanning the SpeI and NdeI sites (nt 1505 to 5122), which contained each mutation, were inserted back into the pNL43lucΔenv vector. DNA sequence analysis showed that the K211N mutant fragment contained an additional mutation of G to A at nt 4874, resulting in a Gly-to-Arg substitution at amino acid 189. We therefore termed this mutant K211N G/R. For preparation of the Vpr-defective clone (ΔVpr), a stop codon (TAA) was introduced in the vpr open reading frame at nt 5624 to 5626 using mutagenic primers 5′-TAAGCTAACTTAAGAGTGAA-3′ (nt 5624 to 5645). This mutation site was 7 nt downstream to the end of the vif open reading frame. To generate the double mutant, Y143G&ΔVpr, the NdeI-SalI fragment (nt 5124 to 5785) which contained the ΔVpr mutation was replaced with the corresponding region of the SpeI-SalI pBluescript SK(+) vector containing the Y143G mutation. The SpeI-SalI fragment containing both mutations was inserted back into the pNL43lucΔenv vector. To prepare the eukaryotic expression vector for HIV-1 IN fused to green fluorescent protein (GFP), the entire coding region of HIV-1 IN was amplified by PCR and inserted into the pCMX-SAH/Y145F vector (kindly provided by Kazuhiko Umesono and Hidesato Ogawa, Kyoto University, Kyoto, Japan) at the SalI and BamHI sites. Primers used for amplification of HIV-1 IN were as follows: GFPIN-sense (5′-ACGCGTCGACGTCGGCCATAGCGGCCTTTTTAGATGGAATAGAT-3′) and GFPIN-antisense (5′-CGCGGATCCGCGTTAATCCTCATCCTGTCTACT-3′). Similarly, IN-coding regions carrying each mutation were amplified by PCR using each mutant plasmid (pNL43lucΔenv) as a template. The amplified region and cloning junctions were confirmed by DNA sequencing.
Cell culture and isolation of human MDMs and PBLs.
COS, human rhabdomyosarcoma (RD), and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Human monocyte-derived macrophages (MDMs) and peripheral blood lymphocytes (PBLs) were obtained from peripheral blood. Briefly, peripheral blood was obtained from the buffy coats of healthy, HIV-seronegative blood donors, and peripheral blood mononuclear cells were separated over a Ficoll-Hypaque gradient (Ficoll-Paque PLUS; Amersham, Pharmacia Biotech, Tokyo, Japan). Peripheral blood mononuclear cell suspensions were incubated for adhesion onto the plastic of a tissue culture dish in RPMI 1640 (GIBCO BRL, Long Island, N.Y.) containing 10% human AB serum (Sigma, St. Louis, Mo.) for 2 h. Nonadherent cells (PBLs) were then stimulated with 1% phytohemagglutinin (Difco Laboratories, Detroit, Mich.). These cells were grown in RPMI 1640 medium containing 2 ng of recombinant interleukin 2 (Shionogi, Osaka, Japan) per ml and 10% fetal bovine serum. Adherent cells were detached with Cell Dissociation Solution (Sigma) and cultured in RPMI 1640 medium, supplemented with 10% human AB serum. The cells were >98% macrophages as judged by fluorescence-activated cell sorter analysis using an anti CD14 monoclonal antibody (DAKO, Kyoto, Japan) and nonspecific esterase activity.
Virus preparation and infection.
Pseudotype viruses were generated by cotransfection of COS cells with the pNL43lucΔenv vector, containing each IN mutation, and an amphotropic Moloney murine leukemia virus (MuLV) envelope expression vector (pJD-1) or a macrophage-tropic HIV envelope expression vector (pJR-FL), using Lipofectamine (GIBCO BRL). The culture supernatants (5 ml) of the transfected COS cells were harvested 48 h posttransfection, filtered through 0.45-μm-pore-size filters, and used as virus preparations. Each virus preparation was treated with DNase I (40 μg/ml; Worthington) in the presence of 10 mM MgCl2 at 37°C for 1 h to avoid DNA contamination. An aliquot of each virus preparation was incubated at 65°C for 1 h and used as a heat-inactivated control. To monitor the amount of virus in each preparation, HIV-1 p24 antigen levels were determined by an enzyme immunoassay system (EIA-II; Abbott Diagnostika, Wiesbaden-Delkengeim, Germany). To monitor viral gene expression from each plasmid vector, luciferase activity in transfected COS cells was also measured. At 48 h posttransfection, COS cells were lysed with 1 ml of 1× luciferase lysis buffer (Promega). One microliter of each cell lysate was subjected to the luciferase assay. RD cells (5 × 104), MDMs (5 × 105) or PBLs (1 × 106) were infected with an aliquot (1 ml) of DNase-treated virus. The infection proceeded in the presence of Polybrene (10 μg/ml) at 37°C. After incubation for 3 h, the viruses were removed and the cells were overlaid with fresh media and incubated at 37°C.
Luciferase activity.
For luciferase analysis, infected cells were harvested at 4 days postinfection. The total cell pellets from each well were washed twice with phosphate-buffered saline (PBS) and lysed with 200 μl (RD), 150 μl (MDM) or 100 μl (PBL) of 1× luciferase lysis buffer (Promega). Ten microliters of each cell lysate was subjected to the luciferase assay.
Analysis of HIV-1 DNA synthesis and formation of two-LTR circles.
Total cells were harvested from each well at 2 and 6 days postinfection for RD cells and harvested from each well at 3 days post-infection for MDMs. After washing with PBS, nucleic acids were extracted as described elsewhere (78). Briefly, cells were disrupted in urea lysis buffer (4.7 M urea, 1.3%, sodium dodecyl sulfate [SDS], 0.23 M NaCl, 0.67 mM EDTA [pH 8.0], 6.7 mM Tris-HCl [pH 8.0]) and subjected to phenol-chloroform extraction and ethanol precipitation. The resulting DNA pellet was resuspended in 50 μl of water. An aliquot (5 μl) of each sample was subjected to PCR using primer pairs specific for the R-U5 region of HIV-1 (M667/AA55) or the R-gag region (M667/M661) as described elsewhere (78). Detection of HIV-1 DNA sequences by each primer pair was performed using 35 cycles of 95°C for 1 min, 65°C for 2 min, and 72°C for 2 min. For HIV-1 DNA standards, 10 to 25,000 copies of linearized HIV-1 JR-CSF DNA were amplified in parallel. Amplified products were resolved on a 2% agarose gel and stained with Syber-green (FMC Bioproduct, Rockland, Maine). To normalize the amount of cellular DNA in the samples, a primer pair complementary to the first exon of the human β-globin gene was used. For detection of human β-globin DNA, 20 cycles of amplification were performed under the same conditions as those for the HIV-1 DNA amplification. A standard curve for human β-globin DNA was obtained by amplifying known amounts of cellular DNA from RD or MDM cells in parallel. Quantitative analysis of amplified products was performed using the Epi-Light UV FA1100 system with a Luminous Imager (Aisin Cosmos R&D Co.). To further examine the rate of viral DNA synthesis, real-time quantitative PCR (TaqMan PCR detection; Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) was used. Fluorogenic probes were designed to anneal to the target between the sense primer and the antisense primer in the R-U5 and the R-gag region: 5′-TAGTGTGTGCCCGTCTGTTGTGTGAC-3′ and 5′-CCGAACAGGGACTTGAAAGCGAAA-3′, respectively. To analyze the two-long terminal repeat (two-LTR) circular DNA in nuclei, PCR was performed using specific primers as previously described (24). The amplified products of the two-LTR junction sequence were further subjected to nested PCR with the following internal primers: 5′-AATCTCTAGCAGT-3′ and 5′-GTCAGTGGATATCTGATCCCTG-3′. The PCR products were detected as described above.
Western blot analysis.
Viruses were concentrated in an ultracentrifuge (1 h at 315,000 × g using a Beckman TLX-100 centrifuge with a TLA-100.4 rotor), and the pellets were resuspended in PBS. Viral proteins containing approximately 10 ng of p24 were subjected to SDS–12% polyacrylamide gel electrophoresis. Following blotting of proteins onto a nitrocellulose membrane (ATTO, Tokyo, Japan), the membrane was first incubated with antiserum from AIDS patients (provided by Y. Koyanagi, Tohoku University, Sendai, Japan) followed by horseradish peroxidase-conjugated anti-human immunoglobulin. HIV-1 proteins were visualized using an enhanced chemiluminescence detection system (Amersham, Pharmacia Biotechnology).
Fluorescence microscopy.
HeLa cells (4 × 104) were seeded onto poly-d-lysine-coated eight-well Culture Slides (Becton Dickinson Labware, Bedford, Mass.) and transfected with the indicated plasmids using Effectene Transfection Reagent (Qiagen, Hilden, Germany). At 24 h posttransfection, cells were washed once with PBS and fixed with acetone for 5 min. After washing with PBS, cells were mounted in 90% glycerol–50 mM NaHCO3 and covered with a coverslip. Confocal microscopy was performed with an OLIMPAS BX50 fluorescence microscope. One representative medial section was mounted using Adobe Photoshop software.
RESULTS
Construction of HIV-1 IN mutants.
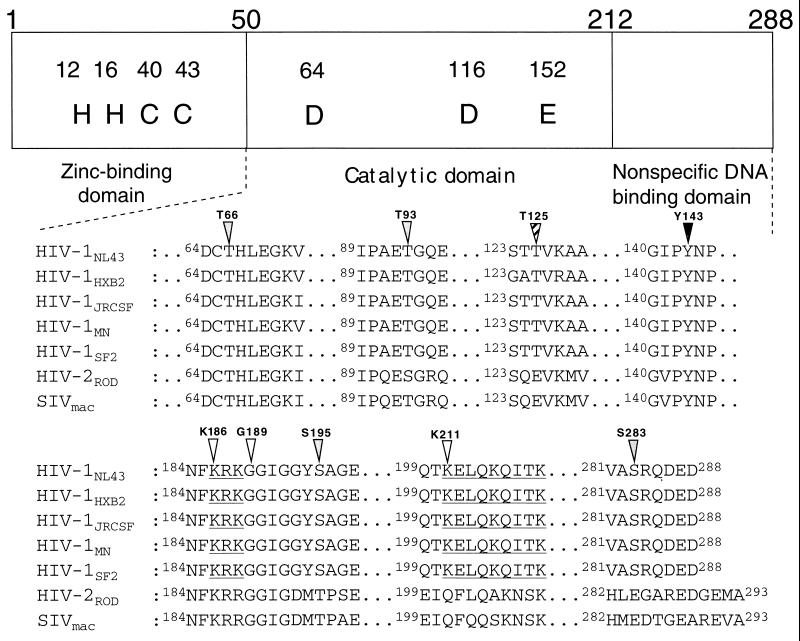
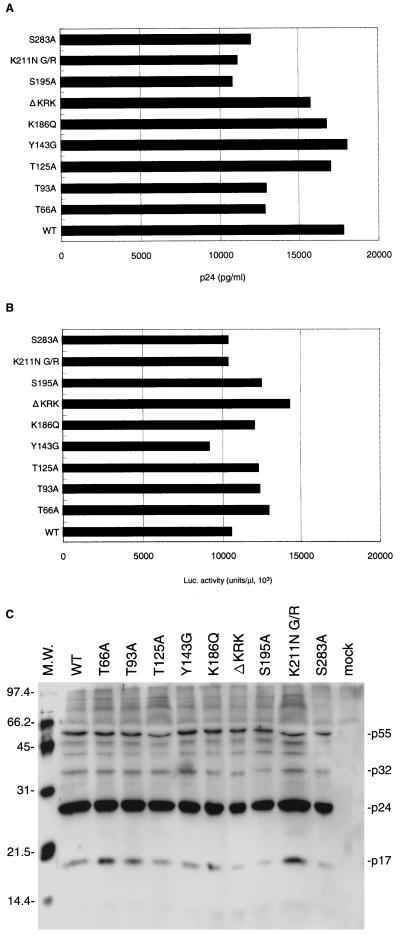
Genetic analysis of HIV-1 IN showed that mutations had multiple effects on the viral life cycle, suggesting that IN might have multiple roles in the early steps preceding integration. These include uncoating, reverse transcription, and nuclear transport of the PIC. Recent study has shown that phosphorylation of viral proteins might be important in regulating viral infectivity (38). To examine the possible involvement of HIV-1 IN phosphorylation in the putative functions of IN, we generated HIV-1 mutants carrying single-amino-acid substitutions at Ser, Thr, or Tyr residues which are conserved among HIV-1 strains and HIV-related lentiviruses (Fig. 1). These included Ser-to-Ala substitutions at position 195 or 283 (S195A or S283A), Thr-to-Ala substitutions at position 66, 93, or 125 (T66A, T93A, or T125A), and a Tyr-to-Gly substitution at position 143 (Y143G) (Fig. 1 and Table 1). Some of these conserved Ser and Thr residues were located in the possible phosphorylation motifs, (S/T)XX(D/E) for CK-II (Fig. 1 [shaded triangle]) and (S/T)X(R/K) for PKC (Fig. 1 [striped triangle]). These motifs are well conserved among HIV-1 strains, although all are not completely conserved in other HIV-1-related lentiviruses. In addition, we generated mutations in the conserved Lys- and Arg-rich motifs spanning position 186 to 188 and position 211 to 219 of HIV-1 IN (Fig. 1 [open triangles]), since these motifs are located in the putative NLS of IN (31) (Fig. 1 [underlined]). The mutations included a Lys-to-Glu substitution at position 186 (K186Q), a Lys-to-Asn substitution at position 211 with an additional substitution of Gly to Arg at position 189 (K211N G/R), and deletion of the KRK motif at position 186 to 188 (ΔKRK). A series of point and deletion mutations were introduced into the IN gene through site-directed mutagenesis of the SpeI-SalI subgenomic fragments and reconstruction of the mutations into the HIV-1 pNL43lucΔenv vector (54, 62). This vector contained a defective env gene, allowing formation of HIV-1 (amphotropic) pseudotypes, and the nef gene was replaced by the firefly luciferase gene, allowing efficient monitoring of HIV-1 expression. We prepared a pseudotype virus of each IN mutant by cotransfection of COS cells with the pNL43lucΔenv vector carrying each IN mutation and the amphotropic Moloney MuLV envelope expression vector (pJD-1). All mutants had comparable levels of p24 in culture supernatants harvested from transfected COS cells (Fig. 2A) and comparable levels of luciferase (Luc) activity in cell lysates of transfected COS cells (Fig. 2B). Thus, mutations had no significant effect on transfected proviral gene expression or virus release. In order to verify that the gag-pol polyprotein processing had been completed in mutant virus particles, we performed a Western blot analysis of viral proteins contained in virus particles using antiserum from human AIDS patients. No apparent difference between parental (wild-type [WT]) and mutant viruses was observed in the profiles of viral proteins (Fig. 2C). The profile of viral proteins of each mutant was also verified by using specific monoclonal antibodies to p24 and reverse transcriptase (RT) (data not shown). These results showed that none of the mutations significantly affected the proviral gene expression, virus particle release, or gag-pol polyprotein processing.
FIG. 1.
Diagram of the domain structure and mutations of HIV-1 IN. The amino acid sequences of HIV-1 strains (NL43, HXB2, JR-CSF, MN, and SF2), HIV-2ROD and SIVmac239 were obtained from GenBank. The accession numbers of HIV-1 NL43, HXB2, JR-CSF, MN, SF2, HIV-2ROD, and SIVmac239 are M19921, K03455, M38429, M17449, K02007, M15390, and M33262, respectively. Portions of the central catalytic domain and the C-terminal domain are shown. Symbols above the sequence are indicated positions of mutation (shaded triangle, possible site of phosphorylation by CK-II; striped triangle, possible site of phosphorylation by PKC; closed triangle, Tyr residue; open triangle, residues conserved in putative NLS (30) indicated with underlines.
TABLE 1.
Mutation of the HIV-1 integrase
| Mutant | Nucleotide change | Amino acid change |
|---|---|---|
| T66A | 4425aca→Gcaa | 66Thr→Alab |
| T93A | 4506aca→Gca | 93Thr→Ala |
| T125A | 4602aca→Gca | 125Thr→Ala |
| Y143G | 4656tac→GGG | 143Tyr→Gly |
| K186Q | 4785aaa→Caa | 186Lys→Gln |
| G189R/K211N | 4794ggg→Agg/4860aaa→aaC | 189Gly→Arg/211Lys→Asn |
| ΔKRK | 4785aaaagaaaa→deletion | 186LysArgLys→deletion |
| S195A | 4812agt→GCt | 195Ser→Ala |
| S283A | 5076agt→GCt | 283Ser→Ala |
Altered nucleotides are indicated by capital letters. Positions of nucleotide are numbered according to the NL43 sequences.
Numbers refer to amino acid residues of NL43 IN.
FIG. 2.
Gene expression of each mutant proviral DNA after transfection of COS cells and viral protein profiles. Pseudotype viruses were generated by cotransfection of COS cells with pNL43lucΔenv vector containing either of mutations in IN and an amphotropic Moloney MuLV envelope expression vector (pJD-1), using Lipofectamine (GIBCO BRL). Culture supernatants (5 ml) of the transfected COS cells were harvested at 48 h posttransfection. (A) p24 levels in culture supernatants were determined by an enzyme immunoassay system (EIA-II; Abbott Diagnostika). (B) Luciferase activity in transfected COS cells were measured at 2 days posttransfection. Cells were washed with PBS and lysed with 1 ml of cell lysis buffer (Promega). One microliter of each cell lysate was subjected to the luciferase assay. (C) Virus particles in culture supernatants (5 ml) of COS cells were precipitated at 48 h posttransfection by ultracentrifuge (1 h at 315,000 × g using a Beckman TLX-100 centrifuge). Viral proteins were separated by SDS–12% PAGE. Culture supernatants of mock-transfected COS cells were precipitated and subjected to SDS-PAGE in parallel as a negative control. After blotting of proteins to nitrocellulose membrane (ATTO), the membrane was subjected to a reaction with serum from a patient and then incubated with horseradish peroxidase-conjugated anti-human immunoglobulin. Viral proteins were visualized by using the enhanced chemiluminescence detection system (Amersham). Positions of the major viral proteins are indicated by their sizes (in kilodaltons) relative to those of molecular weight (M.W.) markers.
Proviral formation of HIV-1 IN mutants in dividing cells.
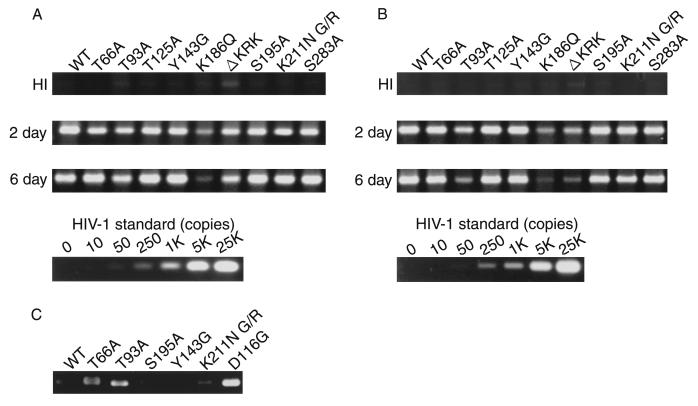
We previously showed that the single-round infection system with HIV-1 (amphotropic) pseudotypes was useful for estimating the integration efficiency in vivo by monitoring levels of de novo synthesized viral DNA and luciferase activity, expressed in infected cells (53, 54). We first assessed whether HIV-1 IN mutants were able to synthesize viral DNA following infection of susceptible RD cells. At 2 and 6 days postinfection, total DNA was harvested from infected RD cells, and an aliquot of each DNA sample was subjected to quantitative PCR analysis (Fig. 3A and B). We monitored the formation of various species of viral DNA using primers M667/AA55 for the early viral DNA species (R/U5) or M667/M661 for the formation of complete or nearly complete viral DNA (R/gag) (78). An aliquot of the same DNA sample was also subjected to real-time quantitative PCR analysis (TaqMan PCR detection; Perkin-Elmer Applied Biosystems Division) using the R/U5 or R/gag specific internal probe. Relative to the WT, the T66A, T93A, T125A, Y143G, S195A, K211N G/R, and S283A mutants showed comparable levels of early (Fig. 3A) and complete or nearly complete viral DNA (Fig. 3B) at 2 days postinfection. These results indicated that none of these mutations affected reverse transcription. The stability of de novo synthesized viral DNA was monitored by measuring the level at 6 days postinfection. This level varied among mutants, probably due to their different integration activities. On the other hand, K186Q and ΔKRK mutants produced very low levels of R/gag (∼10% of the WT level), indicating severe impairment of reverse transcription prominently at the late step in these mutants.
FIG. 3.
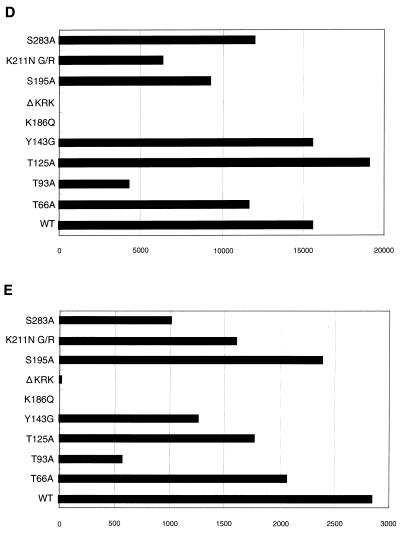
Analysis of IN mutants in dividing cells. Each virus was prepared by cotransfection of COS cells with pNL43lucΔenv vector and pJD-1. The DNase-treated supernatants containing ∼10 ng of p24 were inoculated into RD cells or PBLs. At 2 or 6 days postinfection, as indicated on the left, the entire cell culture was harvested. Total DNA was extracted from infected RD cells and subjected to PCR analysis with the primer pairs for R/U5 (A) and R/gag (B) and the two-LTR circle (C). For HIV-1 DNA standards, 50 to 100,000 copies of linearized HIV-1 JR-CSF DNA were amplified in parallel. Amplified products were resolved on 2% agar gel and visualized by Syber-Green staining (FMC Bioproduct). Virus treated at 65°C for 30 min prior to inoculation was used as a heat-inactivated control (HI). After 4 days of infection, the entire cells were harvested and washed with PBS. The cell pellets were resuspended with 200 μl (for RD cells) (D) and 100 μl (for PBLs) (E) of luciferase lysis buffer (Promega Corp.). Ten microliters of each cell lysate was subjected to luciferase assay as described in Materials and Methods. Luciferase activities are shown in units per microliter.
All mutants showed comparable levels of Luc activity following transfection of COS cells and produced comparable levels of viral DNA after infection, with the exception of the K186Q and ΔKRK mutants. Thus, we could estimate the relative integration efficiency directly by measuring Luc activity following infection of RD cells. We repeated this experiment more than five times with independently prepared viruses, and a representative experiment is shown in Fig. 3D. The level of Luc activity in T125A was always high, ranging from 100 to 120% of the WT level. On the other hand, T66A, T93A, S195A, S283A, Y143G, and K211N G/R mutants showed varied Luc activities of 60 to 80%, 25 to 55%, 60 to 80%, 40 to 80%, 85 to 100%, and 40 to 60% of the WT level, respectively. Furthermore, we performed PCR analysis of de novo synthesized viral DNA using primer pairs which amplify sequences unique to the two-LTR-containing circular DNA. The two-LTR-containing circular DNA is thought to be produced by host nuclear enzymes as an alternative to correct integration by IN and is formed in the absence of functional IN. The PCR amplification of this structure is therefore confirmation of a specific abrogation of integration. The HIV-1 IN catalytic-site mutant D116G (54) was used as a control for the integration-defective mutant. We clearly detected an amplified fragment corresponding to the two-LTR circle junction in DNA samples from RD cells infected with T66A, T93A, K211N G/R, and D116G mutants. However, it was only weakly detected in the WT, S195A, and Y143G samples (Fig. 3C). Thus, the lower levels of Luc activity associated with T66A, T93A, and K211N G/R most probably represent lowered integration efficiencies induced by each mutation. On the other hand, as expected from the low level of viral DNA synthesis, K186Q and ΔKRK did not exhibit detectable levels of Luc activity. We also examined these mutants using human PBLs as primary dividing target cells for virus infection (Fig. 3E). In most mutants, the Luc activity relative to WT levels was similar to that obtained in RD cells, although there were slight differences in the magnitude of the effect of each mutation between RD cells and PBLs. However, the magnitude also varied among PBLs isolated from different blood donors (data not shown). For example, the level of Y143G, which showed 85 to 100% of WT level in RD cells, was always reduced to around 50% of the WT level. Thus, there are slight differences in the magnitude of the effect of each mutation on proviral formation between in vitro-adapted (RD) and primary (PBL) dividing cells.
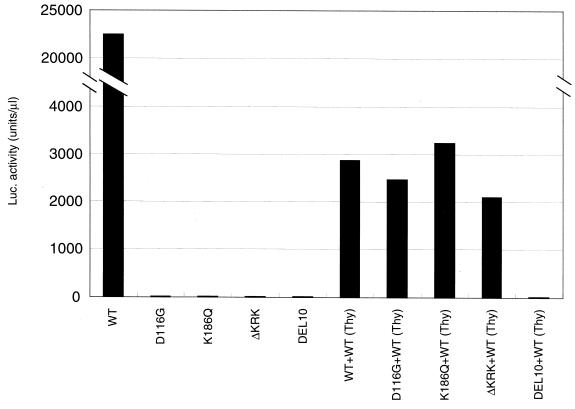
Of note, an almost complete lack of proviral formation by K186Q and ΔKRK in RD cells was found in primary cells, PBLs and MDMs (data not shown). K186 to 188 overlapped with the central polypurine tract (cPPT) which is thought to function as a second priming site for viral plus-strand DNA (10). To examine whether abrogation of viral DNA synthesis was due to the destruction of the cPPT function by K186Q or ΔKRK mutations, we performed the trans complementation test in vivo as described previously (54). Briefly, we prepared pseudotype viruses by cotransfection of COS cells with the pJD-1 vector, the pNL43lucΔenv vector (WT, D116G, K186Q, or ΔKRK), and the pNL43thyΔenv vector, which is identical to the pNL43lucΔenv vector except that it carries the mouse thy-1.2 gene instead of the luc gene (62). As shown in Fig. 4, defects in the trans-acting function, such as a catalytic site mutant of IN, D116G was efficiently complemented. In contrast, defects in the cis-acting function were not complemented as seen in DEL10 in which both the U3att and U5att regions were deleted (53). Luc activity after infection of RD cells with K186Q or ΔKRK was restored by this complementation (Fig. 4) to levels as efficient as those of D116G or the WT. Thus, viral RNA containing the K186Q or ΔKRK mutations could be efficiently reverse transcribed, integrated, and efficiently expressed if WT IN were provided. These results suggest that low levels of K186Q and ΔKRK reverse transcription might not be simply due to a perturbation of the structure or function of the cPPT. Alternatively, we suggest that the KRK motif is critical for HIV-1 IN to function as an antiterminator of the central plus strand termination, as has been previously proposed (11).
FIG. 4.
trans-complementation of K186Q and ΔKRK. Pseudotype viruses were obtained by cotransfection of COS cells with pJD-1 and WT or mutant pNL43lucΔenv vector. In rescue experiments, pseudotype viruses were prepared by cotransfection with the pJD-1 vector and pairs of each of two different mutant pNL43lucΔenv vectors, indicated below the columns. For pairs of mutant and WT vectors, pNL43thyΔenv vector (61), which contains the WT IN and replaces the mouse thy1.2 gene with the luc gene, was used. Infection was performed as described for Fig. 3. A 1-ml aliquot of each virus was inoculated into 5 × 104 RD cells. At 3 days postinfection, the entire culture was harvested and subjected to the luciferase assay as described in Fig. 3. Luciferase activity was determined after subtraction of background level.
Proviral formation of HIV-1 IN mutants in nondividing cells.
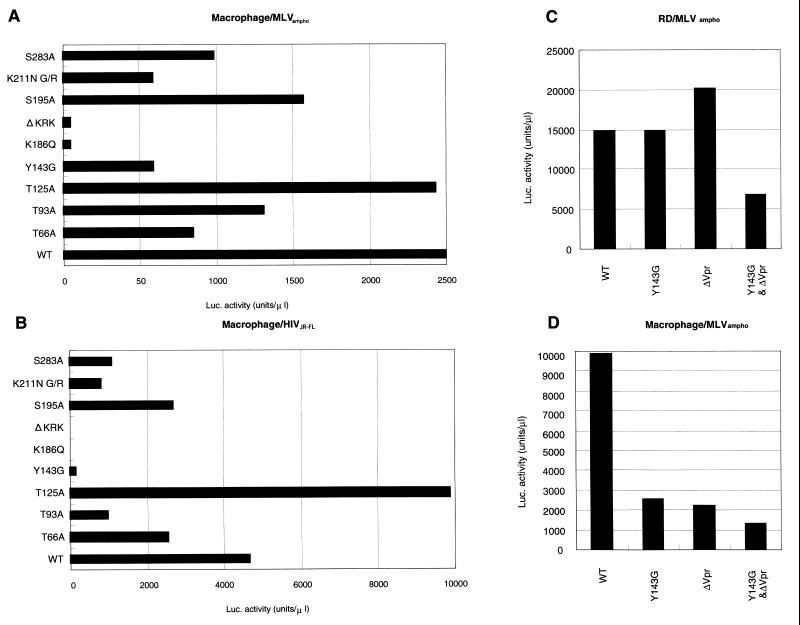
A feature of HIV-1 distinct among retroviruses is their highly efficient proviral formation and replication in nondividing cells. To examine the effects of each HIV-1 IN mutation on this, we examined the efficiency of proviral formation of HIV-1 IN mutants using human primary MDMs as nondividing cells for virus infection. To estimate the relative integration efficiency of each mutant, Luc activity was measured in the infected MDMs at 4 days postinfection (Fig. 5A). Luc activity of each mutant relative to that in WT MDMs was similar to that obtained in RD cells or PBLs (Fig. 3D and E). Of note, relative Luc activity of Y143G mutant in MDMs (20 to 30% of WT level) was significantly lower than that obtained in RD cells (85 to 100% of WT level). In addition, neither K186Q nor ΔKRK produced significant levels of Luc activity in MDMs as was observed in RD cells and PBLs. We reproduced similar results using independent MDMs isolated from more than 10 different healthy blood donors. Furthermore, we examined each mutation by pseudotyping with the HIV-1 macrophage-tropic envelope (pJR-FL). In all but mutant Y143G, pseudotyping with the pJR-FL envelope (Fig. 5B) did not significantly alter their relative Luc activities (compare Fig. 5A and B). However, in the case of Y143G, relative Luc activity was significantly reduced by pseudotyping with the pJR-FL envelope, most probably due to a lowered multiplicity of infection (MOI). Thus, we found that the effect of Y143G was unique in that the magnitude of the effect was significantly enhanced in nondividing cells but only weakly detected in dividing cells.
FIG. 5.
Viral gene expression after infection of MDMs. Each virus was prepared by cotransfection to COS cells with pNL43lucΔenv vector and pJD-1 (A, C, and D) or HIV-1 macrophage-tropic envelope expression vector (pJR-FL) (B). The supernatants harvested and treated with DNase at 48 h posttransfection were used to inoculate MDMs (A, B, and D) or RD cells (C). At 4 days postinfection, the entire cells were harvested and washed with PBS. Cell pellets were lysed with 150 μl of luciferase lysis buffer (Promega). Ten microliters of each cell lysate was subjected to the luciferase assay as described in Materials and Methods.
Analysis of Y143G mutant with or without additional vpr mutation.
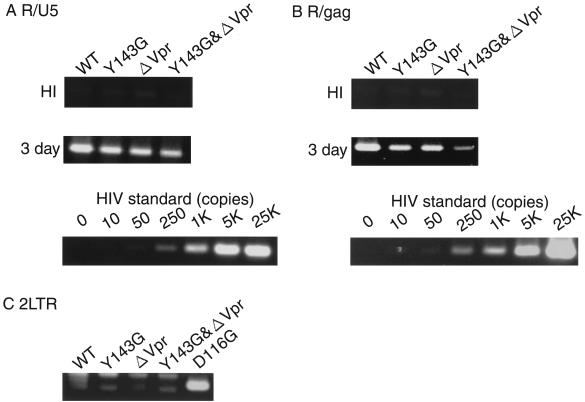
Transport of the PIC from the cytoplasm into the nucleus is thought to be a key step in establishing HIV-1 proviral DNA formation in nondividing cells. Recent evidence suggests that Vpr plays a key regulatory role in this process by binding to karyopherin alpha, a cellular receptor for NLS, thereby increasing its affinity for NLS (63, 64). The genetic analysis of HIV-1 IN mutants described above suggests a partial abrogation of nuclear transport function associated with the Y143G mutation. Firstly, to analyze Vpr-defective HIV-1 under our experimental conditions, we generated a Vpr-defective HIV-1 mutant (ΔVpr) by introducing a stop codon (TAA) in the vpr open reading frame at nt 5624 to 5626 to avoid alteration of the vif gene. Mutants carrying either the single mutation Y143G or ΔVpr produced levels of Luc activity equivalent to WT levels after infection of RD cells (Fig. 5C). However, less activity (20 to 30% of WT levels) was observed after infection of MDMs (Fig. 5D). This result suggests that the effect of Y143G mutation on proviral DNA formation in MDMs might be equivalent to that of ΔVpr mutation. To confirm this further, we examined levels of de novo synthesized viral DNA after infection of MDMs with Y143G or ΔVpr (Fig. 6). The level of the early product (R/U5) produced by Y143G or ΔVpr was ∼50% of the WT level at 3 days postinfection (Fig. 6A). Both mutants produced significantly lower levels of the late product (R/gag), less than 30% of the WT level at 3 days postinfection (Fig. 6B), reaching 50 to 80% of the WT level at 4 to 6 days postinfection (data not shown). These results suggest that Y143G or ΔVpr might initiate reverse transcription with almost the same efficiency as the WT. However the completion of reverse transcription was delayed by Y143G or ΔVpr mutations. Finally, we determined the level of the two-LTR circle form of viral DNA in DNA samples harvested at 4 days after infection of MDMs. No significant band corresponding to the amplified two-LTR junction was detected in any sample, except in the DNA sample infected in parallel with D116G (Fig. 6C). Since the level of R/gag products produced by Y143G or ΔVpr reached around 50 to 80% of the WT level at 4 or 6 days postinfection (data not shown), the lack of detection of the two-LTR junction probably mainly reflects retardation in the nuclear transport of de novo synthesized DNA. Thus, under our experimental conditions, the effect of the Y143G mutation on formation of proviral DNA was equivalent to that of ΔVpr in dividing and nondividing cells.
FIG. 6.
Quantitative analysis of de novo synthesized viral DNA and formation of two-LTR circles after infection of MDMs. Infection of MDMs with virus was performed as described in Fig. 3. At 3 (A and B) and 4 (C) days postinfection, as indicated on the left, the entire cell culture was harvested and PCR analysis was performed as described in Materials and Methods.
Finally, to examine the additive effect of Y143G and ΔVpr mutations on proviral DNA formation, we generated a double-mutant HIV-1, carrying both mutations (Y143G&ΔVpr). As shown in Fig. 5D, Luc activity produced by Y143G&ΔVpr was reduced to less than 10% of the WT level in MDMs, concomitant with the lower rate of synthesis of R/gag products (Fig. 6B). In addition, Y143G&ΔVpr showed a significant reduction in Luc activity (40% of the WT level) in RD cells, while the single mutants, Y143G or ΔVpr, produced levels of Luc activity equivalent to the WT level (Fig. 5C). Thus, we demonstrated that the effects of the Y143G mutation were similar to ΔVpr mutation and had synergistic effects on proviral formation at steps prior to integration. These results indicate that Y143 might be a critical residue for efficient proviral formation in nondividing cells, most probably by facilitating nuclear transport of the PIC in combination with Vpr.
Nuclear localization of HIV-1 IN fused to GFP.
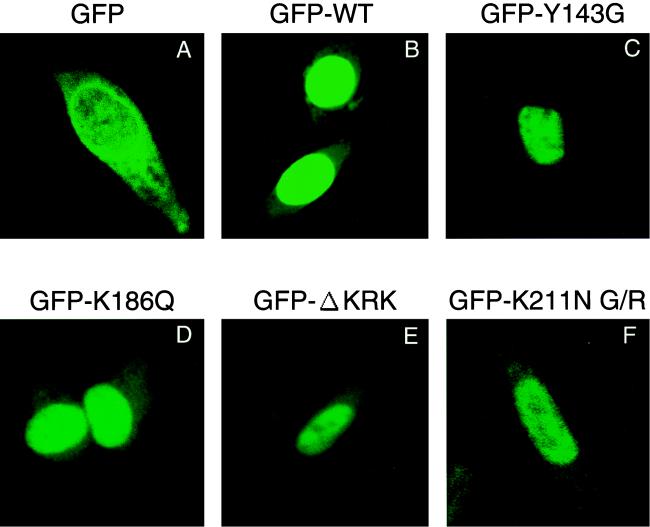
To more directly address the karyophilic properties of HIV-1 IN and the effect of each mutation on these properties, we generated constructs expressing full-length HIV-1 IN fused to GFP protein (pGFP-IN). To examine the subcellular localization of HIV-1 IN, HeLa cells were transfected with the pGFP-IN vector and subjected to analysis by confocal microscopy. At 24 h posttransfection, GFP-IN accumulated almost exclusively in the nucleus (Fig. 7B), while GFP control protein without IN was uniformly scattered throughout both the cytoplasm and the nucleus (Fig. 7A). Thus, we confirmed that HIV-1 IN possesses strong karyophilic properties. Finally, we examined the effect of IN mutations on the karyophilic properties of HIV-1 IN under our experimental conditions. We found that all IN mutants tested here (Y143G, K186Q, ΔKRK, and K211N G/R) were almost exclusively localized to the nucleus, as seen in the WT (Fig. 7C to F). Thus, none of these mutations significantly affected the karyophilic properties of HIV-1 IN under this experimental condition. An earlier result suggests that the NLS of HIV-1 IN is bipartite and consists of the KRK and residues 212 to 219 (31). However, we found that the IN, which lacks the C-terminal region, residues 181 to 288, retained the karyophilic properties (data not shown). Thus, lack of defect in the karyophilic property of HIV-1 IN by a single mutation at KRK or the K212N mutation might not reflect the bipartite function of the NLS. These results indicate that perhaps none of these amino acids is directly involved in the NLS function of HIV-1 IN. Together with our genetic analysis, this leads us to suggest that Y143 is involved in the regulation of karyophilic properties or the putative NLS in the PIC, effects of which apparently only occur in the context of the natural course of viral infection.
FIG. 7.
Confocal microscopic analysis of GFP-IN fusion proteins. HeLa cells were transfected with plasmid expressing GFP only (A), GFP fused to full-length WT HIV-1 IN (B), or GFP fused to the IN carrying the mutation Y143G (C), K186Q (D), ΔKRK (E), or K211N G/R (F) by using Effectene Transfection Reagent (Qiagen). At 24 h posttransfection, cells were fixed and examined with a confocal fluorescent microscope.
DISCUSSION
Genetic analyses of HIV-1 IN (9, 24, 49, 54, 59, 69, 77) suggest putative roles for IN at steps prior to integration, such as uncoating and reverse transcription. In addition, recent evidence indicating the karyophilic properties of HIV-1 IN suggests important roles of IN for nuclear transport of the PIC. In this study, to identify critical amino acids in HIV-1 IN required for putative functions at steps distinct from integration, we generated HIV-1 mutants carrying point or deletion mutations at conserved Ser, Thr, Tyr, Lys, or Arg residues in HIV-1 IN. None of these mutants affected levels of proviral gene expression, virus release, or viral protein processing in the virion particle (Fig. 2), indicating no apparent effect of these mutations in the late stages of the viral life cycle.
The phenotype of each mutant was evaluated using the human RD cell line or PBLs as target cells for in vitro-adapted and primary dividing cells, respectively. Since nuclear transport of HIV-1 PIC is a key step in establishing proviral DNA formation in nondividing cells such as macrophages, we also tested each mutant using human MDMs as primary nondividing cells. Phenotypes of mutants are divided into three groups according to the steps affected by each mutation: integration, reverse transcription, and nuclear transport of the PIC.
Residues affecting integration.
Motif analyses of HIV-1 IN revealed the existence of several possible phosphorylation sites on the conserved Ser or Thr residues, T66, T93, S195, and S283 for phosphorylation by CK-II and T125 for phosphorylation by PKC. Although it has been reported that avian retroviral IN can undergo phosphorylation at the Ser residue in the carboxyl terminus (37), phosphorylation of HIV-1 IN in the virus particle has not been reported. In addition, the phosphorylation status of IN after infection and its role(s) during the early events of retroviral infection cycle are poorly understood. Our genetic analyses showed that each of these mutations might affect the integration step to different degrees; T66A, T93A, T125A, S195A, and S283A mutants in RD cells varied from 60 to 80%, 25 to 55%, 100 to 120%, 60 to 80%, or 40 to 80% of the WT level, respectively. Some of the mutants have been examined previously using in vitro cell assays or in the context of virus replication. T66A has been shown to be defective in 3′ processing in vitro (22% of WT activity) and exhibits partially defective integration (42% of WT activity) (23). However, it has been reported that T66A replicates with kinetics similar to those of the WT (9). Consistent with our results, T125A has also been reported to replicate with kinetics similar to those of the WT (69). It has been reported that the K211 residue is involved in the putative NLS function of the IN (31). However, we could not detect any apparent defect in the karyophilic properties of HIV-1 IN in our experimental system (Fig. 7). K211N G/R showed a slight reduction of integration activity to 40 to 60% in dividing and nondividing cells. Our results are consistent with a previous report showing that K211N reduced integration activity to 32% of the WT level (77). Among the mutants, T93A showed the most prominent effect on integration activity (25 to 55% of the WT level). The loss of integration activity upon mutation of the T66, T93, T125, S195, S283, and K211 residues suggests that they form part of the active site. However, the magnitude of the effect of these mutations was not as dramatic as that of the catalytic site mutation, D116G, which showed less than 0.5% of the WT activity (54). In conclusion, phosphorylation at these sites, if it occurs, might have little effect on reverse transcription and nuclear transport of PIC and a slight but not essential role in integration.
Residues affecting reverse transcription.
The KRK motif, spanning residues 186 to 188 of HIV-1 IN, has been shown to be critical for interaction with cellular importin alpha, suggesting its putative role as the NLS of the PIC (31). However, we could not detect any apparent effects of K186Q or ΔKRK on the karyophilic properties of HIV-1 IN in our experimental system, using the GFP fusion protein expression system (Fig. 7). Instead, we found an almost complete lack of viral gene expression due to failure to complete viral DNA synthesis in RD cells, PBLs, and MDMs. We have previously reported that mutations within the HHCC motif of IN abolished viral gene expression completely, due to severe defects at or prior to the initiation of reverse transcription (54). Since the HHCC mutants showed WT levels of endogenous RT activity (54) and binding and entry to target cells were not affected (59), we hypothesized that the defects caused by the HHCC mutation occur at the uncoating step. Interestingly, defects in reverse transcription associated with K186Q and ΔKRK were found mainly at a late stage, most probably during plus strand synthesis (Fig. 3). Thus, the effects of K186Q and ΔKRK mutations on reverse transcription are different from those associated with HHCC mutations. The KRK motif overlaps with the cPPT which is thought to function as a second priming site for viral plus-strand DNA synthesis (10). However, trans-complementation of the KRK mutations by WT IN suggests that this defect is not simply due to a perturbation of the structure or function of the cPPT. Alternatively, we favor the idea that the KRK residues might represent a motif critical for the function of HIV-1 IN as an antiterminator of the central plus strand termination as has been previously proposed (11). Interestingly, a similar termination of plus strand synthesis proximal to the cPPT has recently been reported in the equine infectious anemia virus, suggesting this is a common feature among lentiviruses (68). Thus, it would be interesting to examine the exact mechanisms underlying the roles of IN in the complex mode of HIV-1 reverse transcription and their biological significance.
Residues affecting nuclear transport of PIC.
A feature of HIV-1 distinctive among retroviruses is their highly efficient proviral formation and replication in nondividing cells, such as macrophages. Nuclear transport of HIV-1 PIC is a key step in establishing proviral DNA formation in nondividing cells. One major object of the present study was to identify the critical residues of HIV-1 IN involved in its nuclear transport function. Since the nuclear import function is not strictly required for infection of dividing cells, we made the assumption that effects on proviral formation might be evident in MDMs but slight in RD cells or activated PBLs. Among the mutants tested here, we found that Y143G showed a significant reduction in viral gene expression (20 to 30% of the WT level) in MDMs while retaining high gene expression levels (80 to 100% of the WT level) in RD cells. Our results are in part consistent with previous reports in which point mutations at Y143 resulted in competent replication with slightly delayed kinetics in T-cell lines (67, 77). Y143 is well conserved among mammalian retroviral INs, although nonconservative amino acid substitutions occur naturally at an analogous position in some INs (23, 39, 44). A nonconservative amino acid substitution at this position in HIV-2 IN (Y143L) had no effect on oligonucleotide cleavage, strand joining, or disintegration reaction (72). Recently, involvement of the Y143 residue of HIV-1 IN in specific interactions with the att sequences has been reported using an in vitro photo-cross-linking assay (25). Of note, the effect of the Y143G mutation on proviral formation was evident in MDMs but slight in RD cells. We also demonstrated that this phenotype was quite similar to ΔVpr. Vpr is thought to be one of the key regulatory viral proteins possessing a nuclear transport function (1, 2, 15, 28, 30, 35, 52, 64). Thus, we suggest that the Y143G mutation might affect the nuclear transport function of the PIC, but not integration. Interestingly, in RD cells, one of these (Vpr and IN) functions might be sufficient for efficient proviral formation. However, both functions might be necessary, especially in MDMs and to a lesser degree in primary isolated PBLs. In the GFP-IN expression system, we did not see any effect of the Y143G mutation on the karyophilic properties of HIV-1 IN. It is still possible that the functional role of the Y143 residue involves regulation of the karyophilic properties or putative NLS in the PIC, the effect of which is apparent only in the context of the natural course of viral infection. Alternatively, it is possible that the effect is evident in nondividing cells. Due to poor transfection efficiency of the GFP-IN vector into MDMs, we are currently addressing this point using the lentivirus vector expression system. In addition, the exact region of the putative NLS in HIV-1 IN remains to be determined, since none of the mutations tested here showed any apparent effects on the karyophilic properties of HIV-1 IN in our experimental system. On the other hand, the lowered rate of completion of reverse transcription in these mutants suggests that reverse transcription of HIV-1 might be complete after nuclear entry. A similar correlation between nuclear entry and completion of reverse transcription has been reported in an avian retrovirus (50) and HIV-1 (55). Thus, it would be of interest to identify the nuclear cofactor(s) required for completion of HIV-1 reverse transcription.
In summary, the present study has identified residues that may be critical for the putative functional roles(s) of HIV-1 IN in the steps prior to integration in dividing and/or nondividing cells. Nondividing cells of the monocyte/macrophage lineage are regarded as the first target of HIV-1 infection and act as one of the reservoirs of HIV-1 in persistent infection in vivo (16, 17, 33, 45, 60, 65, 71, 76, 80). Thus, identification of the critical motif in HIV-1 IN or other components in the PIC might be important in revealing new aspects of the HIV-1 life cycle. Hopefully, it may also contribute to the development of new strategies for AIDS treatment and anti-HIV-1 drugs.
ACKNOWLEDGMENTS
We thank K. Umesono and H. Ogawa for providing the pCMX-SAH/Y145F vector; D. P. Grandgenett for supplying antiserum against HIV-1 IN and for good discussions; C. Depienne and K. Morizono for their helpful technical advice; and Y. Koyanagi for providing pJR-FL and serum from AIDS patients.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture; a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Culture; the Japan Health Sciences Foundation; and the Ministry of Science and Technology Agency of Japan.
REFERENCES
- 1.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 2.Bukrinskaya A G, Ghorpade A, Heinzinger N K, Smithgall T E, Lewis R E, Stevenson M. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc Natl Acad Sci USA. 1996;93:367–371. doi: 10.1073/pnas.93.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinsky M I, Haffar O K. HIV-1 nuclear import: in search of a leader. Front Biosci. 1997;2:578–587. doi: 10.2741/a213. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 6.Bushman F D, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 9.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charneau P, Clavel F. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J Virol. 1991;65:2415–2421. doi: 10.1128/jvi.65.5.2415-2421.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. HIV-1 reverse transcription. A termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow S A, Brown P O. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 15.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 16.Connor R I, Ho D D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelissen M, Mulder-Kampinga G, Veenstra J, Zorgdrager F, Kuiken C, Hartman S, Dekker J, van der Hoek L, Sol C, Coutinho R, et al. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J Virol. 1995;69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel R, Katz R A, Skalka A M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 19.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 20.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 21.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 22.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 27.Farnet C M, Wang B, Lipford J R, Bushman F D. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fletcher T M R, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 29.Fouchier R A, Meyer B E, Simon J H, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed E O, Englund G, Maldarelli F, Martin M A. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell. 1997;88:171–174. doi: 10.1016/s0092-8674(00)81836-x. [DOI] [PubMed] [Google Scholar]
- 31.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 33.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 34.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 35.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horton R, Mumm S, Grandgenett D P. Avian retrovirus pp32 DNA endonuclease is phosphorylated on Ser in the carboxyl-terminal region. J Virol. 1988;62:2067–2075. doi: 10.1128/jvi.62.6.2067-2075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacque J M, Mann A, Enslen H, Sharova N, Brichacek B, Davis R J, Stevenson M. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 1998;17:2607–2618. doi: 10.1093/emboj/17.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson M S, McClure M A, Feng D F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 41.Kalpana G V, Reicin A, Cheng G S, Sorin M, Paik S, Goff S P. Isolation and characterization of an oligomerization-negative mutant of HIV-1 integrase. Virology. 1999;259:274–285. doi: 10.1006/viro.1999.9767. [DOI] [PubMed] [Google Scholar]
- 42.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 43.Katzman M, Sudol M. Influence of subterminal viral DNA nucleotides on differential susceptibility to cleavage by human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1996;70:9069–9073. doi: 10.1128/jvi.70.12.9069-9073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koenig S, Gendelman H E, Orenstein J M, Dal Canto M C, Pezeshkpour G H, Yungbluth M, Janotta F, Aksamit A, Martin M A, Fauci A S. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 46.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaFemina R L, Callahan P L, Cordingley M G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y M, Coffin J M. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 52.Lu Y L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuda T, Planelles V, Krogstad P, Chen I S. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller M D, Wang B, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J Virol. 1995;69:3938–3944. doi: 10.1128/jvi.69.6.3938-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy J E, Goff S P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J Virol. 1989;63:319–327. doi: 10.1128/jvi.63.1.319-327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nadler S G, Tritschler D, Haffar O, Blake J, Bruce A G, Cleaveland J S. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura T, Masuda T, Goto T, Sano K, Nakai M, Harada S. Lack of infectivity of HIV-1 integrase zinc finger-like domain mutant with morphologically normal maturation. Biochem Biophys Res Commun. 1997;239:715–722. doi: 10.1006/bbrc.1997.7541. [DOI] [PubMed] [Google Scholar]
- 60.O'Brien W A, Pomerantz R J. HIV infection and associated diseases. In: Nathanson N, Ahmed R, Gonzalez-Scarano F, Griffin D E, Holmes K V, Murphy F A, Robinson H, Lippincott-Raven L, editors. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 815–836. [Google Scholar]
- 61.Panganiban A T, Temin H M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983;306:155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- 62.Planelles V, Haislip A, Withers-Ward E S, Stewart S A, Xie Y, Shah N P, Chen I S. A new reporter system for detection of retroviral infection. Gene Ther. 1995;2:369–376. [PubMed] [Google Scholar]
- 63.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J Biol Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 64.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin C G, Taddeo B, Haseltine W A, Farnet C M. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stetor S R, Rausch J W, Guo M J, Burnham J P, Boone L R, Waring M J, Le Grice S F. Characterization of (+) strand initiation and termination sequences located at the center of the equine infectious anemia virus genome. Biochemistry. 1999;38:3656–3667. doi: 10.1021/bi982764l. [DOI] [PubMed] [Google Scholar]
- 69.Taddeo B, Carlini F, Verani P, Engelman A. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J Virol. 1996;70:8277–8284. doi: 10.1128/jvi.70.12.8277-8284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Gent D C, Vink C, Groeneger A A, Plasterk R H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van't Wout A B, Koostra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varmus H E, Brown P O. Retroviruses. In: Howe M, Berg D, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 73.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vink C, van Gent D C, Elgersma Y, Plasterk R H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 76.Wiley C A, Schrier R D, Nelson J A, Lampert P W, Oldstone M B. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wiskerchen M, Muesing M A. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 79.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu K, Cordeiro M L, Atienza J, Robinson W E, Jr, Chow S A. Irreversible inhibition of human immunodeficiency virus type 1 integrase by dicaffeoylquinic acids. J Virol. 1999;73:3309–3316. doi: 10.1128/jvi.73.4.3309-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]