Abstract
We have identified a nuclear structure that is induced after infection with the autonomous parvovirus H-1. Using fluorescence microscopy, we observed that the major nonstructural protein (NS1) of H-1 virus which is essential for viral DNA amplification colocalized with virus-specific DNA sequences and sites of ongoing viral DNA replication in distinct nuclear bodies which we designated H-1 parvovirus-associated replication bodies (H-1 PAR-bodies). In addition, two cellular proteins were shown to accumulate in H1 PAR-bodies: (i) the proliferating cell nuclear antigen (PCNA) which is essential for chromosomal and parvoviral replication and (ii) the NS1-interacting small glutamine-rich TPR-containing protein (SGT), suggesting a role for the latter in parvoviral replication and/or gene expression. Since many DNA viruses target preexisting nuclear structures, known as PML-bodies, for viral replication and gene expression, we have determined the localization of H-1 PAR- and PML-bodies by double-fluorescence labeling and confocal microscopy and found them to be spatially unrelated. Furthermore, H-1 PAR-bodies did not colocalize with other prominent nuclear structures such as nucleoli, coiled bodies, and speckled domains. Electron microscopy analysis revealed that NS1, as detected by indirect immunogold labeling, was localized in ring-shaped electron-dense nuclear structures corresponding in size and frequency to H-1 PAR-bodies. These structures were also clearly visible without immunogold labeling and could be detected only in infected cells. Our results suggest that H-1 virus does not target known nuclear bodies for DNA replication but rather induces the formation of a novel structure in the nucleus of infected cells.
Autonomous and adeno-associated parvoviruses have linear, single-stranded DNA genomes of approximately 5,000 nucleotides flanked with terminal palindromes. They target the nucleus for replication, gene transcription, and finally assembly of progeny virions (1, 4, 18, 48). To mount a lytic infection, the adeno-associated viruses (AAV) depend on helper virus functions provided by viruses of either the adenovirus or herpesvirus family (for a review see reference 5). This dependence is also reflected by the recruitment of AAV into adenovirus replication centers, where they are able to utilize cellular as well as helper virus proteins for their replication (65). In the absence of helper virus and genotoxic stress, AAV integrate into the host genome in a site-specific manner (6).
Infections with autonomous parvoviruses, to which the well-studied minute virus of mice (MVM) and the closely related H-1 virus belong, is strictly dependent on host cell functions expressed during the S-phase of the cell cycle but does not require helper virus factors (18, 62, 68). The major nonstructural protein NS1, a phosphoprotein with mainly nuclear localization, is essential for viral DNA replication (19, 50) and also transactivates transcription from the viral P38 capsid gene promoter (22, 34, 49). Infection with H-1 virus or MVM finally leads to cell death, and accumulation of NS1 was shown to be the major cause for this effect (10). The precise molecular mechanism of NS1-associated cytotoxicity is not yet understood; however, several virus- or NS1-induced cellular disturbances that could cause cytotoxicity, including cell cycle arrest (44, 45), altered expression and phosphorylation patterns of cellular proteins (2), and for the human lymphoblastoid cell line U937, induction of apoptosis (47), have been reported.
Our study aimed at identifying the subnuclear compartment utilized by autonomous parvovirus H-1 for replication. Since NS1 is known to be essential for the initiation of virus replication, we have used NS1 as an initial marker for sites of virus replication and analyzed the nuclear localization of NS1 in infected human NBE cells. NS1 was distributed nonhomogenously throughout the nucleus. A fraction of the protein was present diffusely in the nucleoplasm yet excluded from the nucleoli, whereas another fraction accumulated strongly in body-like structures. We further demonstrated that these bodies are the sites of H-1 DNA replication and coined the term H-1 parvovirus-associated replication bodies (H-1 PAR-bodies).
Many DNA viruses target PML-bodies for viral replication and thereby often induce changes in the protein composition of these bodies (for reviews, see references 23 and 36). In noninfected cells, PML-bodies are thought to be involved in transcriptional regulation (for a recent review, see reference 35). The question of why viruses target PML-bodies is still open. It was suggested that PML-bodies might function as a depository or reservoir for proteins needed for viral replication. Evidence for the involvement of PML-bodies in putative antiviral defense mechanisms came from the observation that PML-bodies increase in size and number when cells are treated with interferon (25, 26, 32, 38, 60). It was thus suggested that viruses modify PML-bodies in order to adapt these structures to their needs and/or to avoid these putative defense mechanisms (36). Through extensive confocal microscopic analysis of double-fluorescence labeling experiments, we have therefore determined the spatial relationship of H-1 PAR-bodies and the following nuclear structures: (i) PML-bodies, (ii) nucleoli, known to be involved in rRNA transcription, processing, and more (53); (iii) coiled bodies, thought to be involved in snRNP biogenesis (35); and (iv) speckled domains, which serve as storage sites for splicing factors such as SC-35 (39). Analysis of H-1 PAR-bodies at the ultrastructural level revealed a ring-shaped electron-dense structure found only in infected cells. Thus, our results show that H-1 PAR-bodies represent a distinct nuclear structure that is induced after parvovirus infection and serves as site of H-1 virus DNA replication.
MATERIALS AND METHODS
Viruses and mammalian cell lines.
Parvovirus H-1 was collected after infection of NBE cells (56) at a multiplicity of infection of 5 PFU per cell, and purified by cesium chloride gradient centrifugation, as previously described (11). Otherwise, all infections were performed with a multiplicity of 10 PFU per cell.
The cell line NBE was grown in modified Eagle's medium supplemented with 5% fetal calf serum (Life Technologies) at 37°C in a 10% CO2 atmosphere. Synchronization of NBE cells was achieved by an isoleucine-aphidicolin double-block protocol as previously described (13, 46). In brief, cells were seeded in medium lacking isoleucine. After 48 h, the medium was replaced by fresh complete medium containing 12 mg of aphidicolin (Sigma) per ml. After 10 h, the cells were infected or mock treated for 1 h and further incubated in the presence of aphidicolin. Another 9 h later, the cells were released from the aphidicolin block by addition of fresh aphidicolin-free medium.
Immunofluorescence.
Cells were grown on coverslips, fixed in 1% formaldehyde for 10 min at room temperature, dehydrated with methanol for 5 min and acetone for 2 min, and permeabilized with 1% saponin for 15 min at room temperature. After being washed with phosphate-buffered saline (PBS), the cells were preincubated with 1% goat serum in PBS, followed by two washing steps, one with 350 mM NaCl–0.2% Tween 20–0.2% NP-40 in PBS and the other with 2 mM MgCl2 in PBS. Primary and secondary antibodies were successively incubated for 1 h each, followed by three PBS washes. After staining with DAPI (4′,6′-diamino-2-phenylindole; 250 μg/ml in H2O) for 1 min, coverslips were mounted onto glass slides in Elvanol (polyvinyl alcohol; molecular weight, 77,000 to 79,000; ICN). For the detection of PCNA, cells were treated as described above except that incubation in formalin was omitted.
For immunolocalization, the following antibodies were used: for NS1, the polyclonal antiserum SP8 (8) and the monoclonal antibody 3D9 (a generous gift from N. Salomé and D. Pintel); for PML-bodies, the commercially available monoclonal PML antibody (Santa Cruz Biotechnology), a monoclonal SP100 antibody (a generous gift from G. Maul), and a polyclonal SP100 antiserum (61); for SGT, the affinity-purified polyclonal antiserum AC1.2 (21); for PCNA, a commercially available monoclonal antibody (Upstate Biotechnology, Lake Placid, N.Y.); for No38, a polyclonal antiserum from guinea pig (a generous gift from M. Schmidt-Zachmann) (55); for speckled domains, the commercially available monoclonal antibody SC-35 (Sigma, Munich, Germany) (59); for coiled bodies, a polyclonal p80/coilin antiserum (a generous gift from A. Lamond); for NS2, the SP6 antiserum (8); and for capsid proteins, a polyclonal serum (29).
Visualization of DNA replication and in situ localization of viral nucleic acids.
To detect ongoing DNA replication, cells were labeled with bromodeoxyuridine (BrdU) at a final concentration of 10 μM in the medium for 20 min, fixed with 3.7% formaldehyde for 10 min, and then washed with PBS and H2O. To denature double-stranded DNA, the preparations were incubated with 7.4% HCl for 10 min and then washed with H2O and PBS. Replicating DNA was visualized by immunofluorescence after incubation with a BrdU-specific antibody (Becton Dickinson) and a secondary antibody coupled to tetramethyl rhodamine isocyanate (TRITC) as described above.
To localize viral nucleic acids, fluorescence in situ hybridization (FISH) was performed essentially as described previously (31). Briefly, after immunolocalization of NS1, the preparations were denatured with 70% formamide (pH 7) in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 73°C for 3 min, followed by incubation in 50% formamide (pH 7) at 73°C for equilibration. The infectious H-1 virus DNA clone pSR19 (24) was biotinylated via nick translation as previously described (33). Labeled DNA (200 ng) was denatured with 50% formamide (pH 7) in 2× SSC at 75°C for 5 min and applied onto the specimen followed by renaturation overnight at 37°C. Hybridization signals resisting stringent washing conditions (65°C in 0.1× SSC) were visualized using streptavidin-fluorescein isothiocyanate (FITC) (Dianova).
Fluorescence microscopy.
After immunolabeling and mounting, specimens were analyzed by conventional epifluorescence microscopy (Leica; 40× and 63× objectives with immersion oil), as well as confocal laser scanning microscopy (Zeiss LSM310 or LSM510 UV; 63× Plan-Apochromat objective). For the colocalization studies, optical sections were acquired simultaneously in order to avoid voxel shift. For the detection of FITC and TRITC signals, the 488- and 543-nm excitation lines of a argon-krypton laser were applied. For data acquisition and imaging, the LSM510 UV software version 2.3 and Adobe Photoshop 4.4 were used.
Electron microscopy.
Synchronized and infected NBE cells were fixed for 15 min with 2% formaldehyde (freshly prepared from paraformaldehyde), rinsed several times with PBS containing 50 mM NH4Cl, permeabilized with 0.5% Triton X-100 in PBS for 5 min, briefly washed with PBS, and incubated for 1 h with the anti-NS1 polyclonal serum SP8 at a dilution of 1:1,000 or the monoclonal antibody 3D9 at a dilution of 1:5. After three washes with PBS, the cells were incubated for 2 h with nanogold-coupled secondary antibodies. Silver enhancement, fixation, and embedding in Epon were carried out as described previously (52). Data were acquired with an electron microscope (EM910; LEO Optics, Oberkochen, Germany).
RESULTS
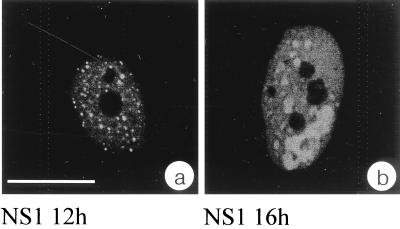
To identify the subnuclear compartment in which parvovirus H-1 DNA replication proceeds, we have analyzed the distribution of the virus-induced protein NS1, which is known to be essential for replication of the viral genome. For this, NS1 was detected by indirect immunofluorescence in time course experiments using synchronized NBE cells infected with H-1 virus. Cells were infected while being blocked at the G1/S border through isoleucine depletion followed by aphidicolin treatment and subsequently released into the mitotic cycle. The earliest time point at which NS1 could be detected by immunofluorescence using the monoclonal antibody 3D9 was 8 h after removal of aphidicolin. In the course of infection, increasing amounts of NS1 accumulated in the nucleoplasm of infected cells. Using epifluorescence or confocal microscopy, we noticed that NS1 strongly accumulated in distinct nuclear bodies. In addition, NS1 was also distributed diffusely throughout the nucleoplasm at lower concentrations but absent from nucleoli (Fig. 1a). This particular nuclear distribution of NS1 was observed in cells starting at 12 h after release from aphidicolin block (Fig. 1a) up to the latest time point tested (18 h postrelease [data not shown]). NS1 accumulation continued during the course of infection (Fig. 1b). Since we did not monitor the fate of individual sites of NS1 accumulation, it is not clear whether neighboring bodies merged over time or whether single bodies increased more in size than others in a given time interval.
FIG. 1.
NS1 localizes in the nucleoplasm, accumulates in nuclear bodies, and is absent from nucleoli. Immunofluorescence experiments were performed with synchronized and H-1-infected NBE cells at 12 h (a) and 16 h (b) after release of the aphidicolin block. Panels represent confocal sections showing NS1 immunolocalized with the monoclonal antibody 3D9 and detected via an FITC-conjugated secondary antibody. Bar, 20 μm in both panels.
H-1 viral DNA sequences and ongoing replication are confined to the same nuclear structure in which NS1 accumulates.
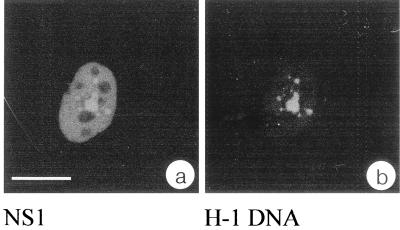
Since NS1 is essentially required for viral genome amplification and viral gene transcription, we tested whether viral nucleic acid sequences were localized within the nuclear structures in which NS1 accumulated. For this, synchronized, H-1 virus-infected NBE cells were subjected to a combination of FISH and immunolocalization. Examination of the specimens using confocal laser scanning microscopy revealed a strict and exclusive localization of viral nucleic acid sequences within the NS1-containing nuclear bodies for all cells analyzed (Fig. 2). It is likely that most of the signal represents hybridization of the probe to amplified viral DNA, but contribution of viral transcripts cannot be excluded. In mock-infected cells, no signals corresponding to NS1 or H-1 viral sequences were detected.
FIG. 2.
H-1 sequences colocalize with NS1 in nuclear bodies. Synchronized and infected NBE cells were simultaneously subjected to immunolocalization using the anti-NS1 polyclonal serum SP8 and FISH with the biotinylated probe pSR19. NS1 was detected via an FITC-conjugated secondary antibody (a), and H-1 genomes were visualized using streptavidin-Cy3 (b). The equivalence in size and shape of the signals present in both channels of one typical confocal section indicate a complete colocalization of NS1 and H-1 genomes. Bar, 20 μm in all panels.
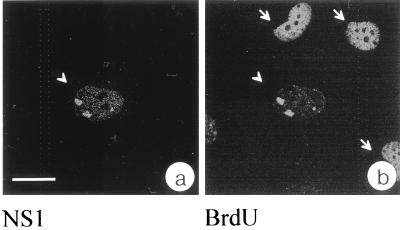
To ascertain the presence of viral DNA in the identified nuclear structure and to test whether viral DNA is replicated there, we performed BrdU labeling experiments using synchronized and infected NBE cells that were released for 17 h from the aphidicolin block and pulse-labeled with BrdU for 20 min prior to fixation. The specimens were incubated with the anti-NS1 serum SP8 and the corresponding secondary antibody for immunolocalization of NS1. After acid treatment for denaturation of DNA, incorporated BrdU was detected by indirect immunofluorescence using a primary BrdU-specific antibody. In all infected cells, however, a strict colocalization of DNA replication with NS1-containing bodies was observed (Fig. 3). Strict colocalization of NS1 and BrdU incorporation was also observed when no DNA denaturation step was performed (data not shown). The protocol used here to visualize BrdU incorporation also permitted the detection of chromosomal DNA replication (Fig. 3b). In infected cells, however, DNA replication was observed only in the nuclear bodies which accumulated NS1; no incorporation of BrdU into chromosomal DNA was detected. These results indicated that in cells where parvovirus DNA replication took place, chromosomal DNA replication had been either completed or blocked. Biochemical evidence for the downregulation of cellular DNA replication during parvoviral infection has been reported previously (18). Our results demonstrate this effect for the first time on a single-cell level. On the basis of these results, we conclude that the bodies in which NS1 accumulated are also the nuclear structure in which H-1 virus DNA replication proceeds and thus refer to this structure as H-1 PAR-bodies.
FIG. 3.
Parvoviral DNA replication colocalizes with NS1 in nuclear bodies. NS1 was localized via the polyclonal antiserum SP8 and an FITC-conjugated secondary antibody (a). Replication was monitored by incorporation of BrdU and indirect immunofluorescence using a TRITC-conjugated secondary antibody (b). The cell in the center of the confocal section (arrowhead) exhibits a colocalization of NS1 and viral DNA replication. Arrows point to cells in which chromosomal replication is still in process. Bar, 20 μm.
We also analyzed the subcellular localization of the smaller nonstructural protein NS2 using the SP6 antibody; we found NS2 to be mainly present in the cytoplasm but also homogenously distributed in the nucleus of infected cells. We could not detect a specific accumulation of NS2 in H-1 PAR-bodies (U. Bodendorf and C. Cziepluch, unpublished results). This pattern of NS2 distribution is in agreement with results reported by others (17) and ourselves (7) for the NS2 protein expressed in cells infected by the closely related virus MVM. When we analyzed the localization of capsid proteins using a polyclonal serum raised against recombinant capsid protein, we observed a predominantly diffuse nuclear staining in immunofluorescence experiments. It is, however, known that progeny single-stranded DNA accumulation is linked to capsid formation (51, 63). We therefore expect that capsid assembly also takes place in close proximity to H-1 PAR-bodies. We are currently attempting to raise an antibody directed against assembled capsids, which might allow the detection of assembled capsids close to H-1 PAR-bodies.
PCNA accumulates in H-1 PAR-bodies.
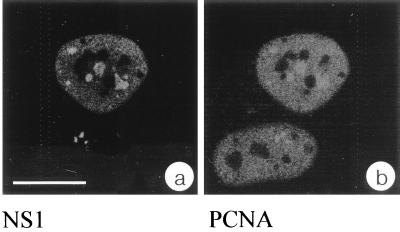
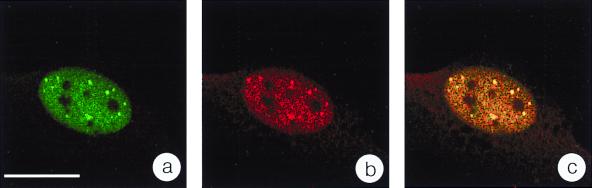
PCNA is required for polymerase δ-driven chromosomal (27, 28) and most likely also parvoviral (12) DNA replication and can therefore be used as a representative cellular marker for DNA replication. This prompted us to test whether PCNA was present in H-1 PAR-bodies. Confocal analysis of indirect double-immunofluorescence experiments showed that PCNA was distributed throughout the nucleoplasm of infected cells excluding nucleoli. In addition PCNA was concentrated in specific nuclear structures, showing a clear colocalization with NS1 and therefore H-1 PAR-bodies (Fig. 4). Since PCNA is an essential component of the cellular replication machinery, this finding confirms the results obtained from the BrdU incorporation experiments (Fig. 3) and strongly supports the idea that H-1 PAR-bodies represent the sites of autonomous parvovirus DNA replication. It is not known whether PCNA accumulates in H-1 PAR-bodies through direct interaction with the viral genome or with other cellular constituents such as replication factor C, which is known to load PCNA onto DNA (9).
FIG. 4.
The essential replication factor PCNA accumulates with NS1 in H-1 PAR-bodies. Double immunofluorescence was performed with synchronized and H-1-infected NBE cells 14 h after release from aphidicolin block. NS1 was immunolocalized with the anti-NS1 monoclonal antibody 3D9 and an FITC-conjugated secondary antibody (a), and PCNA was localized with a polyclonal anti-PCNA antibody and a TRITC-conjugated secondary antibody (b). Confocal images of both channels from the same confocal plane clearly indicate complete colocalization of the signals. Bar, 20 μm.
The cellular NS1-interacting protein SGT colocalizes with NS1 in H-1 PAR-bodies.
Rat and human SGT proteins were previously identified as cellular interacting partners for NS1 of H-1 virus. Neither the function of SGT nor the function of the NS1-SGT complex is understood. It has been speculated that SGT might be involved in housekeeping processes, since sgt transcripts are present in all rat and human tissues tested. In noninfected cells, SGT is distributed diffusely in the cytoplasm as well as the nucleus but is essentially absent from nucleoli (21, 30). We determined the nuclear localization of SGT in infected cells by immunofluorescence microscopy. SGT was present throughout the nucleoplasm and excluded from nucleoli, and most strikingly accumulated with NS1 in H-1 PAR-bodies (Fig. 5). Since NS1 and SGT are able to interact directly in vitro, it is tempting to speculate that SGT is also recruited to H-1 PAR-bodies through direct interaction with NS1 in vivo. When we investigated the subcellular distribution of SGT in adenovirus-infected cells with or without AAV coinfection, we found that the distribution of SGT was not altered (data not shown). It therefore appears that SGT is specifically recruited only to the DNA replication sites of autonomous parvovirus H-1. The accumulation of SGT in H-1 PAR-bodies point to a specific role for SGT in replication and/or gene expression of autonomous parvoviruses.
FIG. 5.
NS1 and the cellular NS1-interacting protein SGT colocalize in H-1 PAR-bodies. Double immunofluorescence was performed with synchronized and H-1-infected NBE cells 14 h after release from aphidicolin block. NS1 detected with FITC results in a green signal (a). SGT was immunolocalized with the polyclonal anti-SGT serum AC1.2 and was stained using a TRITC-conjugated secondary antibody (b). (c) Merged confocal images of panels a and b, showing the colocalization of fractions of SGT and NS1. Bar, 20 μm.
H-1 PAR-bodies represent a distinct nuclear structure.
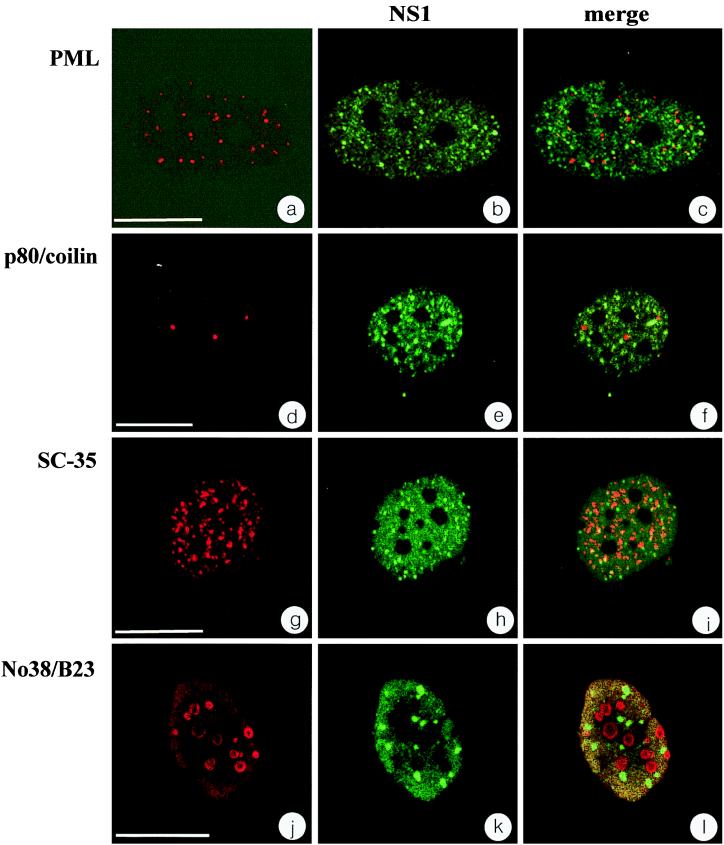
Viruses with double-stranded DNA genomes such as adenovirus, simian virus 40, or herpes simplex virus type 1 were shown to replicate in association with PML-bodies, and it was therefore of interest to determine the relationship between H-1 PAR- and PML-bodies. To do so, confocal analysis of double-immunofluorescence experiments was performed, using synchronized, H-1 virus-infected NBE cells. At 14 h after release from aphidicolin block, the PML-bodies were clearly visible in both uninfected and infected cells as detected with a monoclonal PML antibody (Fig. 6a and c). H-1 PAR-bodies were detected with the SP8 serum directed against NS1 (Fig. 6b). The nuclear localization of PML- and H-1 PAR-bodies indicated that the two structures were essentially distinct from each other and did not colocalize (Fig. 6c). The same results were obtained with an independent monoclonal antibody directed against SP100 (data not shown). However, it cannot be excluded that other protein components besides PML and SP100 are common to both structures.
FIG. 6.
H-1 PAR-bodies form a distinct nuclear compartment. A set of different nuclear bodies were colocalized with NS1 in infected NBE cells 14 h after release from aphidicolin block. Shown are PML bodies recognized by an anti-PML antibody (a and c), coiled bodies recognized by an anti-p80/coilin antibody (d and f), speckled domains recognized by an anti-SC-35 antibody (g and i), and nucleoli recognized by an anti-No38/B23 antibody (j and l). (b, e, h, and k) Immunolocalization of NS1 in the same confocal plain as shown in the left column, detected with FITC. Overlays of the channels from the left and center columns (c, f, i, and l) exhibit differential staining of red and green signals, providing evidence for spatial separation of H-1 PAR-bodies from all nuclear bodies investigated in this study. Bars: a, 15 μm; d, g, and j, 20 μm.
Up to 18 h after release from aphidicolin block, we did not observe a significant change in the number of PML-bodies in virus-infected cells compared to noninfected cells (data not shown). We did, however, observe that the number of PML-bodies was decreased in asynchronous NBE cells at 48 h postinfection or in cells that expressed high amounts of NS1 after transient transfection when DNA condensation and other signs of induced cell death were visible (data not shown). It is therefore possible that NS1, though not localized in PML-bodies, disturbs PML and SP100 protein accumulation and/or PML-body stability through its known cytotoxic effects (10, 64).
Furthermore, we investigated the spatial distribution of H-1 PAR-bodies with respect to a set of different nuclear structures such as coiled bodies, speckled domains, and nucleoli (Fig. 6d to l). Antibodies directed against p80/coilin, SC-35, and No38/B23 were used for the detection of coiled bodies (Fig. 6d and f), speckled domains (Fig. 6g and i), and nucleoli (Fig. 6j and l), respectively. As illustrated in Fig. 6f, i, and l, H-1 PAR-bodies did not show any colocalization with either of the nuclear structures tested. Taken together, our results show that H-1 PAR-bodies represent a distinct nuclear structure that can be distinguished spatially from those previously described, including PML-bodies, coiled bodies, speckled domains, and nucleoli.
Ultrastructure of H-1 PAR-bodies.
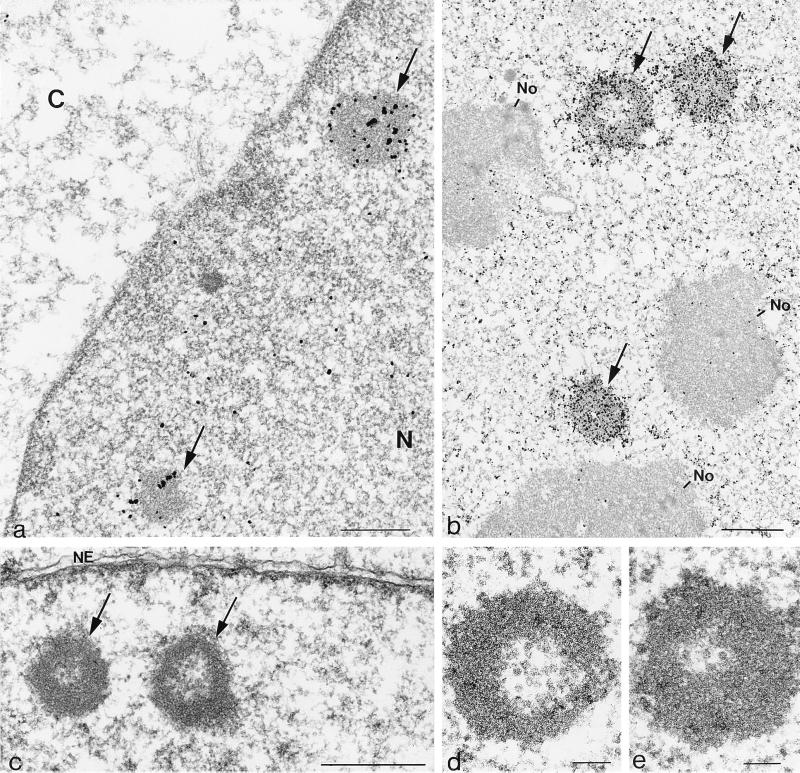
To investigate whether the H-1 PAR-bodies detected by light microscopy correspond to structures visible by electron microscopy in ultrathin sections, we performed immunogold localization of NS1. NBE cells blocked at the G1/S transition were infected with H-1 virus or mock treated, released for 17 h into the mitotic cycle, fixed in paraformaldehyde, and processed for sectioning. The preparations were then incubated with an NS1-specific antibody, either the antibody 3D9 (Fig. 7a) or the antiserum SP8 (Fig. 7b), and the corresponding gold-conjugated secondary antibodies. The results obtained with both NS1 antibodies were very similar and in agreement with the data obtained by confocal laser scanning analysis of immunofluorescence experiments. NS1 was found both diffusely in the nucleoplasm and also strongly accumulated in electron-dense nuclear structures, with diameters of approximately 1 to 5 μm (Fig. 7a and b). At 17 h after release from aphidicolin block, the nucleoli appeared to be morphologically intact and essentially free of gold particles (Fig. 7b). Using the 3D9 antibody to detect NS1, the overall staining with gold was less intense and the electron-dense structure of the H-1 PAR-bodies could readily be seen (Fig. 7a).
FIG. 7.
Electron micrographs of ultrathin sections through infected NBE cells showing the ultrastructure of H-1 PAR-bodies. (a and b) Immunogold localization of NS1 with the monoclonal antibody 3D9 (a) and the polyclonal antibody SP8 (b) to electron dense structures identified as H-1 PAR-bodies; (c and d) ultrastructure of H-1 PAR-bodies. Bars: 0.5 μm (a and c), 1 μm (b), and 0.1 μm (d and e).
When infected cells were directly analyzed by electron microscopy without indirect immunogold labeling, we detected ring-shaped structures composed of a less electron-dense core surrounded by material of higher electron density which corresponded in size and number to H-1 PAR-bodies (Fig. 7c). Since the ring-shaped structure was apparent in the preparations that were not treated with antibodies, this most probably reflects the native structure of H-1 PAR-bodies more closely. Our analysis did not yield evidence that H-1 PAR-bodies were generally localized in close proximity to other nuclear structures that could be identified by electron microscopy. In preparations of uninfected cells, no structures resembling H-1 PAR-bodies were detectable, indicating that H-1 PAR-bodies are induced by virus infection.
At higher magnifications, we detected in the internal space of H-1 PAR-bodies structures which were very similar in size and shape to empty viral particles (Fig. 7d and e). The magnifications achieved here, however, do not allow an unequivocal identification of single viral capsids. Further investigations with antibodies specifically directed against assembled capsids are therefore required to acertain whether progeny viruses concentrate in H-1 PAR-bodies.
H-1 PAR-bodies appears to be very similar, if not identical, in structure to electron-dense entities previously observed in H-1 virus-infected NBE cells and termed “nucleolar inclusions” (1) and “hollow spheres” (58). In contrast with their original interpretation as functionally irrelevant structures, the present work clearly points to these structures as the sites at which parvovirus DNA replication takes place. Furthermore, our data do not support the original assignment of these structures as nucleolar derivatives.
DISCUSSION
Virus-encoded and cellular proteins form H-1 PAR-bodies.
This study led to the identification of a nuclear structure that is distinct from known nuclear structures such as coiled bodies, speckled domains, nucleoli, and PML-bodies by both its spatial distribution and at least some of its protein constituents. This new type of nuclear body was identified by analyzing the localization of NS1, which is known to be essential for parvovirus replication (16, 19). A fraction of NS1 accumulated in specific nuclear bodies which we identified as the sites of viral DNA replication since H-1 virus nucleic acid sequences and ongoing DNA synthesis were colocalized there. These structures were termed H-1 PAR-bodies. Our findings are reminiscent of recent observations concerning Aleutian mink disease virus-infected Crandell feline kidney cells which show that NS1 localizes to discrete sites of viral DNA replication. While NS1 from Aleutian mink disease virus appeared to be exclusively localized at these sites (42, 43), we have reproducibly detected a fraction NS1 from H-1 virus outside of H-1 PAR-bodies by both immunofluorescence and immunogold labeling using two different NS1 antibodies. The function of this NS1 pool is unknown, as is the mechanism which regulates the nuclear distribution of NS1. It has previously been shown that biochemical activities of NS1 are regulated by phosphorylation in vitro (40, 41) and in vivo (15). It is thus possible that the nuclear distribution of NS1 is also regulated through differential phosphorylation.
H-1 PAR-bodies contain not only viral proteins but also cellular factors. PCNA is an essential factor of the cell DNA replication machinery thought to be required for autonomous parvoviral DNA replication (12). In agreement with this finding, we have observed an accumulation of this factor in H-1 PAR-bodies. It may be expected, but remains to be shown, that other cellular factors which are equally important for parvovirus DNA replication such as the parvovirus initiation factor, replication protein A (12), and the high-mobility-group proteins (20) also accumulate in H-1 PAR-bodies. The present study further demonstrated that SGT, a cellular protein previously identified for its ability to interact with NS1 in vitro and in the yeast two-hybrid system (21), accumulates in H-1 PAR-bodies. This finding suggests a functional role for SGT in autonomous parvoviral DNA replication and/or gene expression, and we are therefore investigating whether the presence of SGT in H-1 PAR-bodies is essential for virus replication. Since SGT and NS1 can interact directly in vitro, it is possible that recruitment of SGT to H-1 PAR-bodies is mediated by NS1. It was previously observed that a fraction of SGT is modified in the presence of NS1, most likely by phosphorylation (21), and we are therefore testing whether recruitment of SGT into H-1 PAR-bodies is regulated by phosphorylation.
H-1 PAR-bodies have a distinct ultrastructure.
H-1 PAR-bodies were identifiable in ultrathin sections through immunogold labeling of NS1 and displayed a characteristic ultrastructure with a less electron-dense core surrounded by material of higher electron density. These structures are not artifacts due to immunogold labeling since they could also be detected in unstained preparations. The ultrastructure of H-1 PAR-bodies fits nicely with a recently suggested unified model for cellular transcription and DNA replication factories. In these factories, the enzymatic functions are proposed to be concentrated in the center, while the nucleic acids, i.e., genes and transcripts, are found surrounding the core (14). Accordingly the outer ring of H-1 PAR-bodies, in which NS1 accumulates, might contain newly synthesized viral DNA and transcripts. In this respect, H-1 PAR-bodies could serve as a model for cellular transcription and replication factories, allowing the investigation of parameters which regulate the assembly and stability of such centers.
In previous studies, ultrastructural changes induced by parvovirus H-1 in NBE cells have been thoroughly analyzed (1, 58). The most pronounced ultrastructural alterations observed were reported to be (i) margination of the nuclear chromatin, (ii) modifications and eventual breakdown of nucleoli, and (iii) appearance of doughnut-shaped bodies of assumed nucleolar origin (1). Subsequent studies performed with H-1 virus-infected NBE cells synchronized by a double methotrexate block revealed that the earliest morphological changes occurred in nucleoli at 12 h postinfection, when fragmentation and loss of nucleolar fibrous matter were detected. Between 18 and 36 h postinfection, electron-dense hollow spheres thought to be composed of fibrous and/or granular nucleolar elements and to contain incomplete H-1 virus particles were observed. These structures were interpreted as being by-products of nucleolar breakdown and were considered irrelevant for H-1 virus propagation (58). The ultrastructure of H-1 PAR-bodies resembles the doughnut-shaped nucleolar inclusions observed by al-Lami and coworkers (1) and the hollow spheres described by Singer and Toolan (58) so closely that we consider them to be identical. Additional evidence for the identity of H-1 PAR-bodies and these previously observed structures comes from experiments in which the nuclear sites of ongoing viral DNA replication were localized. Our results show that BrdU incorporation is confined to H-1 PAR-bodies (Fig. 4). When cells were pulse-labeled with [3H]thymidine, Singer and Rhode also observed incorporated label only in the electron-dense structures and not within intact nucleoli (57). The present work questions the conclusions drawn from these previous studies by showing that the nuclear bodies induced by parvovirus infection are unlikely to be of nucleolar origin. Indeed, we could detect H-1 PAR-bodies when nucleoli appeared to be still intact, and we found no evidence for a relationship between H-1 PAR-bodies and nucleoli (Fig. 6 and 7). Our data, however, do not exclude the possibility that some proteins present in nucleoli are also recruited to H-1 PAR-bodies. A good candidate protein is nucleolin, which has indeed been shown to form a specific complex with the minus strand of MVM DNA (3). Finally, our finding that cellular (PCNA) and viral (NS1) factors known to be essential for parvovirus growth accumulate with replicating DNA in H-1 PAR-bodies argues against the previously accepted view that parvovirus-induced nuclear structures are not relevant to the virus life cycle.
Why are H-1 PAR-bodies induced after H-1 infection?
Viruses with double-stranded DNA genomes target PML-bodies for the transcription of their early gene products and also initiate DNA replication close to these sites (36). Data presented in this study demonstrate that H-1 PAR-bodies are the sites for H-1 virus DNA replication and that they represent a novel nuclear structure that is distinct from PML-bodies and other known nuclear structures. These results were unexpected and raise the question of why this virus induces specific nuclear structures instead of utilizing preexisting ones.
One possible reason for the fact that the growth of H-1 virus does not require their targeting to PML-bodies may be traced back to the proposed role of these bodies in cellular antiviral defense mechanisms. Viruses with double-stranded DNA genomes express so-called early proteins shortly after infection. These viral products often alter host cell physiology and in particular drive quiescent cells into a replicative phase in order to support viral DNA synthesis. It may be speculated that an infection with PML-associated viruses is rapidly sensed by target cells, which then have the opportunity to mount an antiviral response. This defense mechanism needs to be circumvented by the virus for it to replicate, which could be achieved through the targeting and modification of PML-bodies. Indeed, the immediate-early gene products encoded by some but not all DNA viruses induce the dispersion of proteins present in PML-bodies (36). Furthermore, mutant herpes simplex 1 viruses that are not able to disrupt PML-bodies were shown to be impaired in expression of viral genes and also in the reactivation of latent viral genomes (37). It remains to be determined whether the dispersion of PML-body proteins is required to adapt these sites to specific viral needs or to modify putative cellular antiviral responses (36). In contrast to double-stranded DNA viruses, autonomous parvoviruses cause opportunistic infections because their genes are unable to be expressed immediately after infection due to the single strandedness of their genome and have to wait until host cells reach S-phase to undergo genomic conversion to duplex replicative forms that can be expressed and amplified (19). This dependence may delay the time at which infected cells are induced to mount an antiviral response, saving parvoviruses the need for early targeting and modification of PML-bodies. In this respect it is worth pointing out that parvoviruses were indeed reported to be poor inducers of antiviral effectors such as interferon (54, 66, 67). Nevertheless, it should be stated that at late stages of infection, H-1 virus had an effect on PML-bodies, which were found to decrease in number. It is tempting to relate this late alteration to the intracellular accumulation of the NS1 protein, which is known to have a cytotoxic function (10). The precise molecular mechanism of NS1-associated cytotoxicity is poorly understood, although several parvovirus- or NS1-induced cellular disturbances, including cell cycle arrest, DNA cleavage (44, 45), dysregulation of cellular promoters (64), and altered expression and phosphorylation of specific cellular proteins (2), have been reported. It is therefore possible that late in infection, PML and SP100 proteins are also targets for these pleiotropic cytopathic effects of NS1.
In conclusion, we have identified through immunofluorescence experiments H-1 PAR-bodies as the site of H-1 virus DNA replication and have characterized these nuclear bodies also at the ultrastructural level. More recent results obtained in our laboratory show that similar bodies are found in MVM-infected mouse cells (T. Bashir and C. Cziepluch, unpublished results). This finding might indicate that H-1 PAR bodies represent a prototype for structures also induced in the nucleus of cells infected by other autonomous parvoviruses.
ACKNOWLEDGMENTS
We acknowledge the generous gift of antibodies from A. Lamond (University of Dundee), G. Maul (Wistar University, Philadelphia, Pa.), D. Pintel (University of Missouri, Columbia), and H. Will (HPI, Hamburg, Germany) and from N. Salomé (University of Missouri, Columbia), M. Schmidt-Zachmann (DKFZ, Heidelberg, Germany), and T. Sternsdorf (HPI). The expert support of H. Spring (DKFZ) with acquisition of data by confocal microscopy is gratefully acknowledged. We thank T. Bashir for help with FACS analyses in the initial phase of the project. We are very grateful to W. W. Franke (DKFZ) for critical discussion of the electron microscopic data and comments on the manuscript. We also thank T. Bashir, U. Bodendorf, and N. Salomé (DKFZ) for comments on the manuscript. Finally, we thank H. zur Hausen for continuous encouragement and support.
REFERENCES
- 1.al-Lami F, Ledinko N, Toolan H W. Electron microscope study of human NB and SMH cells infected with the parvovirus H-1: involvement of the nucleolus. J Gen Virol. 1969;5:485–492. [Google Scholar]
- 2.Anouja F, Wattiez R, Mousset S, Caillet Fauquet P. The cytotoxicity of the parvovirus minute virus of mice nonstructural protein NS1 is related to changes in the synthesis and phosphorylation of cell proteins. J Virol. 1997;71:4671–4678. doi: 10.1128/jvi.71.6.4671-4678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrijal S, Perros M, Gu Z, Avalosse B L, Belenguer P, Amalric F, Rommelaere J. Nucleolin forms a specific complex with a fragment of the viral (minus) strand of minute virus of mice DNA. Nucleic Acids Res. 1992;20:5053–5060. doi: 10.1093/nar/20.19.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns K I, Bohenzky R A. Adeno-associated viruses: an update. Adv Virus Res. 1987;32:243–306. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 6.Berns K I, Linden R M. The cryptic life style of adeno-associated virus. Bioessays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 7.Bodendorf U, Cziepluch C, Jauniaux J C, Rommelaere J, Salome N. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J Virol. 1999;73:7769–7779. doi: 10.1128/jvi.73.9.7769-7779.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockhaus K, Plaza S, Pintel D J, Rommelaere J, Salome N. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J Virol. 1996;70:7527–7534. doi: 10.1128/jvi.70.11.7527-7534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee C G, Phillips B, Finkelstein J, Yao N, O'Donnell M, Hurwitz J. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc Natl Acad Sci USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caillet Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y Q, de Foresta F, Hertoghs J, Avalosse B L, Cornelis J J, Rommelaere J. Selective killing of simian virus 40-transformed human fibroblasts by parvovirus H-1. Cancer Res. 1986;46:3574–3579. [PubMed] [Google Scholar]
- 12.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens K E, Pintel D J. The two transcription units of the autonomous parvovirus minute virus of mice are transcribed in a temporal order. J Virol. 1988;62:1448–1451. doi: 10.1128/jvi.62.4.1448-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook P R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 15.Corbau R, Salom N, Rommelaere J, Nuesch J P. Phosphorylation of the viral nonstructural protein NS1 during MVMp infection of A9 cells. Virology. 1999;259:402–415. doi: 10.1006/viro.1999.9786. [DOI] [PubMed] [Google Scholar]
- 16.Cotmore S F, Christensen J, Nuesch J P, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotmore S F, Tattersall P. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology. 1990;177:477–487. doi: 10.1016/0042-6822(90)90512-p. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 19.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 20.Cotmore S F, Tattersall P. High-mobility-group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J Virol. 1998;72:8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cziepluch C, Kordes E, Poirey R, Grewenig A, Rommelaere J, Jauniaux J C. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J Virol. 1998;72:4149–4156. doi: 10.1128/jvi.72.5.4149-4156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doerig C, Hirt B, Beard P, Antonietti J P. Minute virus of mice non-structural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J Gen Virol. 1988;69:2563–2573. doi: 10.1099/0022-1317-69-10-2563. [DOI] [PubMed] [Google Scholar]
- 23.Doucas V, Evans R M. The PML nuclear compartment and cancer. Biochim Biophys Acta. 1996;1288:M25–M29. doi: 10.1016/s0304-419x(96)00028-5. [DOI] [PubMed] [Google Scholar]
- 24.Faisst S, Faisst S R, Dupressoir T, Plaza S, Pujol A, Jauniaux J C, Rhode S L, Rommelaere J. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J Virol. 1995;69:4538–4543. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grotzinger T, Jensen K, Guldner H H, Sternsdorf T, Szostecki C, Schwab M, Savelyeva L, Reich B, Will H. A highly amplified mouse gene is homologous to the human interferon-responsive Sp100 gene encoding an autoantigen associated with nuclear dots. Mol Cell Biol. 1996;16:1150–1156. doi: 10.1128/mcb.16.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guldner H H, Szostecki C, Grotzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- 27.Jonsson Z O, Hubscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 28.Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 29.Kestler J, Neeb B, Struyf S, Van Damme J, Cotmore S F, D'Abramo A, Tattersall P, Rommelaere J, Dinsart C, Cornelis J J. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum Gene Ther. 1999;10:1619–1632. doi: 10.1089/10430349950017626. [DOI] [PubMed] [Google Scholar]
- 30.Kordes E, Savelyeva L, Schwab M, Rommelaere J, Jauniaux J C, Cziepluch C. Isolation and characterization of human SGT and identification of homologues in saccharomyces cerevisiae and caenorhabditis elegans. Genomics. 1998;52:90–94. doi: 10.1006/geno.1998.5385. [DOI] [PubMed] [Google Scholar]
- 31.Kurz A, Lampel S, Nickolenko J E, Bradl J, Benner A, Zirbel R M, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 33.Lichter P, Ledbetter S A, Ledbetter D H, Ward D C. Fluorescence in situ hybridization with Alu and L1 polymerase chain reaction probes for rapid characterization of human chromosomes in hybrid cell lines. Proc Natl Acad Sci USA. 1990;87:6634–6638. doi: 10.1073/pnas.87.17.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorson C, Burger L R, Mouw M, Pintel D J. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J Virol. 1996;70:834–842. doi: 10.1128/jvi.70.2.834-842.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matera A G. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 36.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 37.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 38.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 39.Misteli T, Spector D L. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 40.Nuesch J P, Corbau R, Tattersall P, Rommelaere J. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J Virol. 1998;72:8002–8012. doi: 10.1128/jvi.72.10.8002-8012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nuesch J P, Dettwiler S, Corbau R, Rommelaere J. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J Virol. 1998;72:9966–9977. doi: 10.1128/jvi.72.12.9966-9977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oleksiewicz M B, Costello F, Huhtanen M, Wolfinbarger J B, Alexandersen S, Bloom M E. Subcellular localization of Aleutian mink disease parvovirus proteins and DNA during permissive infection of Crandell feline kidney cells. J Virol. 1996;70:3242–3247. doi: 10.1128/jvi.70.5.3242-3247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oleksiewicz M B, Wolfinbarger J B, Bloom M E. A comparison between permissive and restricted infections with Aleutian mink disease parvovirus (ADV): characterization of the viral protein composition at nuclear sites of virus replication. Virus Res. 1998;56:41–51. doi: 10.1016/s0168-1702(98)00053-7. [DOI] [PubMed] [Google Scholar]
- 44.Op De Beeck A, Anouja F, Mousset S, Rommelaere J, Caillet-Fauquet P. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 1995;6:781–787. [PubMed] [Google Scholar]
- 45.Op De Beeck A, Caillet Fauquet P. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J Virol. 1997;71:5323–5329. doi: 10.1128/jvi.71.7.5323-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedrali-Noy G, Spadari S, Miller-Faures A, Miller A O, Kruppa J, Koch G. Synchronization of HeLa cell cultures by inhibition of DNA polymerase alpha with aphidicolin. Nucleic Acids Res. 1980;8:377–387. doi: 10.1093/nar/8.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rayet B, Lopez-Guerrero J A, Rommelaere J, Dinsart C. Induction of programmed cell death by parvovirus H-1 in U937 cells: connection with the tumor necrosis factor alpha signalling pathway. J Virol. 1998;72:8893–8903. doi: 10.1128/jvi.72.11.8893-8903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redemann B E, Mendelson E, Carter B J. Adeno-associated virus Rep protein synthesis during productive infection. J Virol. 1989;63:873–882. doi: 10.1128/jvi.63.2.873-882.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhode S L. trans-activation of parvovirus P38 promoter by the 76K noncapsid protein. J Virol. 1985;55:886–889. doi: 10.1128/jvi.55.3.886-889.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhode S L, Paradiso P R. Parvovirus replication in normal and transformed human cells correlates with the nuclear translocation of the early protein NS1. J Virol. 1989;63:349–355. doi: 10.1128/jvi.63.1.349-355.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards R G, Armentrout R W. Early events in parvovirus replication: lack of integration by minute virus of mice into host cell DNA. J Virol. 1979;30:397–399. doi: 10.1128/jvi.30.1.397-399.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose O, Grund C, Reinhardt S, Starzinski-Powitz A, Franke W W. Contactus adherens, a special type of plaque-bearing adhering junction containing M-cadherin, in the granule cell layer of the cerebellar glomerulus. Proc Natl Acad Sci USA. 1995;92:6022–6026. doi: 10.1073/pnas.92.13.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 54.Schlehofer J R, Rentrop M, Mannel D N. Parvoviruses are inefficient in inducing interferon-beta, tumor necrosis factor-alpha, or interleukin-6 in mammalian cells. Med Microbiol Immunol (Berlin) 1992;181:153–164. doi: 10.1007/BF00202055. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt-Zachmann M S, Hugle-Dorr B, Franke W W. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shein H M, Enders J F. Multiplication and cytopathogenicity of simian vacuolating virus 40 in cultures of human tissue. Proc Soc Exp Biol Med. 1962;109:495–503. doi: 10.3181/00379727-109-27246. [DOI] [PubMed] [Google Scholar]
- 57.Singer I I, Rhode S L. Ultrastructural studies of H-1 parvovirus replication. VI. Simultaneous autoradiographic and immunochemical intranuclear localization of viral DNA synthesis and protein accumulation. J Virol. 1978;25:349–360. doi: 10.1128/jvi.25.1.349-360.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singer I I, Toolan H W. Ultrastructural studies of H-1 parvovirus replication. I. Cytopathology produced in human NB epithelial cells and hamster embryo fibroblasts. Virology. 1975;65:40–54. doi: 10.1016/0042-6822(75)90005-7. [DOI] [PubMed] [Google Scholar]
- 59.Spector D L, Fu X D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stadler M, Chelbi-Alix M K, Koken M H, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin M C, Schindler C, et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- 61.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 62.Tattersall P. Replication of the parvovirus MVM. I. Dependence of virus multiplication and plaque formation on cell growth. J Virol. 1972;10:586–590. doi: 10.1128/jvi.10.4.586-590.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tullis G E, Burger L R, Pintel D J. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J Virol. 1993;67:131–141. doi: 10.1128/jvi.67.1.131-141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanacker J M, Rommelaere J. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin Virol. 1995;6:291–297. [Google Scholar]
- 65.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wiedbrauk D L, Bloom M E, Lodmell D L. Mink parvoviruses and interferons: in vitro studies. J Virol. 1986;60:1179–1182. doi: 10.1128/jvi.60.3.1179-1182.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiedbrauk D L, Hadlow W J, Ewalt L C, Lodmell D L. Interferon response in normal and Aleutian disease virus-infected mink. J Virol. 1986;59:514–517. doi: 10.1128/jvi.59.2.514-517.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolter S, Richards R, Armentrout R W. Cell cycle-dependent replication of the DNA of minute virus of mice, a parvovirus. Biochim Biophys Acta. 1980;607:420–431. doi: 10.1016/0005-2787(80)90152-5. [DOI] [PubMed] [Google Scholar]