Abstract
Background
Recently developed blood markers for Alzheimer's disease (AD) detection have high accuracy but usually require ultra-sensitive analytic tools not commonly available in clinical laboratories, and their performance in clinical practice is unknown.
Methods
We analyzed plasma samples from 290 consecutive participants that underwent lumbar puncture in routine clinical practice in a specialized memory clinic (66 cognitively unimpaired, 130 participants with mild cognitive impairment, and 94 with dementia). Participants were classified as amyloid positive (A +) or negative (A-) according to CSF Aβ1–42/Aβ1–40 ratio. Plasma pTau217, pTau181, Aβ1–42 and Aβ1–40 were measured in the fully-automated LUMIPULSE platform. We used linear regression to compare plasma biomarkers concentrations between A + and A- groups, evaluated Spearman’s correlation between plasma and CSF and performed ROC analyses to assess their diagnostic accuracy to detect brain amyloidosis as determined by CSF Aβ1–42/Aβ1–40 ratio. We analyzed the concordance of pTau217 with CSF amyloidosis.
Results
Plasma pTau217 and pTau181 concentration were higher in A + than A- while the plasma Aβ1–42/Aβ1–40 ratio was lower in A + compared to A-. pTau181 and the Aβ1–42/Aβ1–40 ratio showed moderate correlation between plasma and CSF (Rho = 0.66 and 0.69, respectively). The areas under the ROC curve to discriminate A + from A- participants were 0.94 (95% CI 0.92–0.97) for pTau217, and 0.88 (95% CI 0.84–0.92) for both pTau181 and Aβ1–42/Aβ1–40. Chronic kidney disease (CKD) was related to increased plasma biomarker concentrations, but ratios were less affected. Plasma pTau217 had the highest fold change (× 3.2) and showed high predictive capability in discriminating A + from A-, having 4–7% misclassification rate. The global accuracy of plasma pTau217 using a two-threshold approach was robust in symptomatic groups, exceeding 90%.
Conclusion
The evaluation of blood biomarkers on an automated platform exhibited high diagnostic accuracy for AD pathophysiology, and pTau217 showed excellent diagnostic accuracy to identify participants with AD in a consecutive sample representing the routine clinical practice in a specialized memory unit.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13195-024-01513-9.
Keywords: Plasma, Biomarkers, Alzheimer, Blood, Amyloid, Tau
What is already known on this topic
Blood biomarkers have shown high accuracy to detect AD pathophysiology. The feasibility of those biomarkers in different platforms and the influence of comorbidities in their concentrations needs to be further studied.
What this study adds
We analyzed the feasibility and diagnostic performance of blood AD biomarkers using a fully-automated platform in the setting of a memory clinic and assessed the impact of comorbidities on their diagnostic performance.
How this study might affect research, practice, or policy: The measurement of plasma AD biomarkers in an automated platform yields high accuracy to detect AD pathophysiology and would be easy to implement. Plasma pTau217 was the single most accurate marker for clinical implementation.
Introduction
Early and accurate diagnosis is becoming an increasing priority with the recent developments of disease-modifying therapies for Alzheimer’s Disease (AD). Pathophysiological biomarkers in cerebrospinal fluid (CSF) and positron emission tomography (PET) imaging with amyloid and tau tracers have extensively proven to be useful to detect the disease pathophysiology but are either expensive and/or invasive [1], which can delay the diagnosis and access to treatment.
Measurement of AD biomarkers in blood through reliable high-throughput platforms would simplify the diagnostic process. This is now technically possible thanks to the development of sensitive technologies that can consistently quantify brain-derived molecules that are present in blood in very low concentrations [2–4]. Amyloid-β (Aβ) peptides and different isoforms of phosphorylated tau (pTau) in blood have shown high accuracy for detecting AD pathophysiology in previous research studies [5–12]. How all these plasma markers are affected by different comorbidities is also starting to be understood thanks to large well-characterized cohorts [13–15]. Thus, blood-based markers have the potential to be of great use in screening, early diagnosis, tracking progression, and ultimately, monitoring the efficacy of treatment [16–20]. Of all the plasma biomarkers evaluated, pTau217 has demonstrated a promising profile in identifying amyloidosis, showing the largest fold changes in symptomatic AD patients and the most predictive ability to identify cognitive decline [21–29]. In previous immunoassay studies CSF pTau217 showed better correlation with amyloid-PET and tau-PET than pTau181 [30]. Subsequent research has revealed comparable efficacy of pTau217 in both plasma and CSF for the identification of AD neuropathology and for distinguishing pathopshysiological AD from other neurodegenerative diseases [31–33]. However, most of the existing studies have assessed each of these plasma markers separately, measuring them on different platforms or through techniques not widely available in clinical laboratories, which limits their potential to be widely applied in the clinical routine. The implementation of blood AD markers in a fully-automated platform would facilitate their reproducibility and accessibility in clinical laboratories [34].
The fully-automated platform LUMIPULSE G, extensively used to measure CSF AD biomarkers in clinical laboratories world-wide, has recently launched developed assays to measure pTau217, pTau181, Aβ1-42 and Aβ1-40 in plasma. In this study, our aim was to assess the feasibility and diagnostic performance of pTau217, pTau181, Aβ1-42 and Aβ1-40 in plasma in the LUMIPULSE fully-automated platform in a cohort of well characterized consecutive individuals assessed in a memory clinic.
Methods
Study participants and clinical classification
We included all consecutive individuals who underwent lumbar puncture for the analysis of AD CSF biomarkers assessed at the Sant Pau Memory Unit (SPIN cohort, Barcelona, Spain) as part of their diagnostic work-up [35] between January 2021 and December 2021 (shown in flowchart in Supplementary Material (Fig. 1). The study was approved by the Sant Pau Ethics Committee (Protocol code: EC/22/202/6880) following the standards for medical research in humans recommended by the Declaration of Helsinki. All participants or their legally authorized representative gave written informed consent to participate in biomarkers research studies.
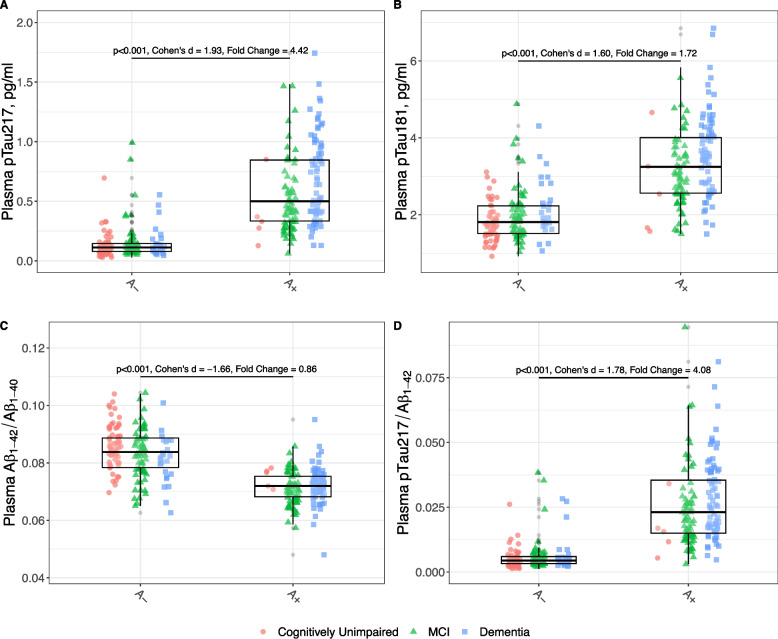
Fig. 1.
Levels of plasma biomarkers and their ratios according to the A status in CSF. All p-values are derived from multivariate linear model, adjusted for the effects of age, sex, APOE ε4 status, chronic kidney disease stage, vascular risk factors and clinical stage. For better visualization, two outliers exceeding 2 pg/mL were excluded from the pTau217 boxplot for the A + group in the dementia stages. Not adjusted by other variables effect sizes are shown (Cohen’s d). Plasma biomarkers Cohen’s d and Fold Change calculated in log-transformed data. pTau217: phosphorylated tau 217, pTau181: phosphorylated tau 181. Aβ1–42: Amyloid β1–42. Aβ1–40: Amyloid β1–40. MCI: Mild cognitive impairment
At the time of CSF and plasma acquisition, participants had a diagnosis of dementia, mild cognitive impairment (MCI), or were cognitively unimpaired (CU). The clinical diagnosis was established after a thorough neurological and neuropsychological evaluation [35]. A more extensive description about how cognitively unimpaired participants were recruited and classified is provided in Supplementary Material (Text 1 & Fig. 1). To assess the impact of vascular risk factors and comorbidities, we collected information about the presence of high blood pressure, dyslipidemia, diabetes, obstructive sleep apnea and history of stroke. Participants were also classified according to the estimated glomerular filtrate rate (eGFR) in different stages (1–5) of chronic kidney disease (CKD) using CKD-EPI formula.
After a full evaluation that included analysis of AD CSF biomarkers, participants were classified according to their etiologic diagnosis, as Alzheimer disease (AD), other neurodegenerative diseases (OtherDem), not neurodegenerative diseases (OtherNotDeg) or CU. A proportion of participants' diagnosis was classified as “uncertain” as they had an unclear etiological diagnosis after a full initial evaluation and required clinical follow-up.
Cognitively unimpaired participants were patients with no evidence of cognitive impairment after a thorough neuropsychological evaluation and healthy volunteers interested in research.
Sample collection and analysis
Blood samples were collected in EDTA-K2 tubes and subsequently centrifuged (2000 rpm × 10 min, 4ºC) within 2 h after extraction. Plasma was aliquoted and stored at -80ºC until analysis. CSF samples were obtained through lumbar puncture, and were also centrifuged, aliquoted and stored at -80ºC until analysis. Blood and CSF samples were collected simultaneously. Full protocol for CSF and blood sample collection in our center has been previously reported [35, 36].
All plasma samples were measured in the Lumipulse fully-automated platform G600II using commercially available kits (Fujirebio Europe, Ghent, Belgium) for pTau181, Aβ1-42 and Aβ1-40 between July and August 2022 with the same lot of reagents. Plasma pTau217 was analyzed between August and September 2023 in another aliquot of the same samples using a novel assay recently developed by Fujirebio. On the day of the analysis, plasma samples were brought to room temperature, mixed thoroughly, centrifuged for 5 min at 2000 g, and subsequently transferred to specific cuvettes for analysis in the Lumipulse platform.
CSF markers Aβ1–42, Aβ1–40, pTau181 and tTau were used in the diagnostic assessment of patients and measured in routine runs scheduled twice a month throughout 2021 following previously reported methods [37]. According to CSF markers, all participants were classified as amyloid positive (A + , CSF Aβ1–42/Aβ1–40 < 0.062) or negative (A-), and as tau positive (T + , pTau181 > 63 pg/mL) or negative (T-). Validation of these cutoff values has been described elsewhere [37]. To validate that our main findings would not be overly influenced by analytical error in CSF measurements, we performed additional sensitivity analyses excluding participants ± 10% of the CSF cutoff points (Supplementary Material, Figs. 1–3).
DNA was extracted from full blood using standard procedures, and APOE was genotyped following previously reported methods [35]. Briefly, direct DNA sequencing of exon 4 was performed routinely for all participants in the SPIN cohort, followed by visual analysis of the resulting electropherogram to identify the two coding polymorphisms that encode the three possible APOE isoforms.
Statistical analysis
Data normality was assessed with the Shapiro–Wilk test. Non-normally distributed variables were log-transformed when necessary. Kruskal–Wallis and Wilcoxon rank sum test were used in continuous variables that were not normal distributed. Linear regression models and ANCOVA adjusted by age and sex were performed for group comparison. We used Chi Square test to assess differences in categorical variables and Fisher’s exact test in group comparisons with small number of observations, and Spearman test to assess the correlation between plasma and CSF markers.
Diagnostic accuracy of plasma biomarkers was assessed through receiver operating characteristic (ROC) analysis. We calculated the areas under the curve (AUC) of individual markers and that of logistic regression models that combined them with each other and with clinical variables. A basic model that included Age, Sex and APOEε4 status was used as a reference to assess the added diagnostic value of plasma markers. We compared the accuracy of individual markers and regression models using DeLong's test adjusted by multiple comparisons using Bonferroni method. We evaluated the sensitivity, specificity, and Youden’s J index of a range of cutoffs to discriminate A + from A- participants. We analyzed the concordance of pTau217 with CSF amyloidosis. We followed a previously reported approach [38] to stratify our cohort in low, medium, and high risk of having CSF amyloidosis. Using predictive models, according to the risk of the participants of being A + , we selected conservative (97.5% sensitivity/specificity) and more lenient (95% sens/spec) cutoffs. We bootstrapped to assess the cutoff robustness. All tests were performed in R statistical software version 4.2.1. Alpha threshold was set at 0.05 for all analysis.
Results
Study participants and clinical classification
We included 290 participants who were syndromically classified as cognitively unimpaired (CU, n = 66), having mild cognitive impairment (MCI, n = 130) or a clinical diagnosis of dementia (n = 94). Table 1 shows the etiologic diagnoses in each category, main demographic characteristics, and biomarker measures in each group. CU participants were younger than those with MCI (p < 0.001) and those with dementia (p < 0.001). There were more female participants (62%). The proportion of A + , A + T + and APOEε4 positive increased according to the clinical stage. More extensive demographics details, including stratification by clinical diagnosis or A status, can be found in Supplementary Material (Tables 1 & 2).
Table 1.
Demographics and plasma biomarker concentrations
| Cognitively Unimpaired, N = 66 | MCI, N = 130 | Dementia, N = 94 | P value | |
|---|---|---|---|---|
| Age | 57 (12) | 73 (11) | 75 (9) | < 0.001 |
| Sex (Female) | 46 (70%) | 77 (59%) | 57 (61%) | 0.3 |
| MMSE | 29.0 (1.0) | 26.0 (4.0) | 22.0 (5.8) | < 0.001 |
| APOEε4 | 15 (23%) | 32 (25%) | 36 (38%) | 0.042 |
| Plasma pTau217 (pg/mL) | 0.11 (0.08) | 0.23 (0.35) | 0.46 (0.63) | < 0.001 |
| Plasma pTau181 (pg/mL) | 1.71 (0.55) | 2.32 (1.28) | 3.35 (1.86) | < 0.001 |
| Plasma Aβ1-42 (pg/mL) | 25.0 (4.6) | 23.0 (5.0) | 24.6 (6.2) | 0.028 |
| Plasma Aβ1-40 (pg/mL) | 290 (65) | 301 (66) | 319 (72) | < 0.001 |
| Plasma Aβ1-42/Aβ1-40 | 0.085 (0.010) | 0.076 (0.014) | 0.074 (0.008) | < 0.001 |
| A | < 0.001 | |||
| A + | 5 (7.6%) | 66 (51%) | 69 (73%) | |
| AT | < 0.001 | |||
| A-T- | 61 (92%) | 64 (49%) | 25 (27%) | |
| A + T- | 2 (3.0%) | 17 (13%) | 12 (13%) | |
| A + T + | 3 (4.5%) | 49 (38%) | 57 (61%) | |
| Etiology | < 0.001 | |||
| CU | 56 (85%) | 0 (0%) | 0 (0%) | |
| AD | 4 (6.1%) | 43 (33%) | 50 (53%) | |
| OtherNotDeg | 1 (1.5%) | 37 (28%) | 11 (12%) | |
| OtherDem | 3 (4.5%) | 18 (14%) | 20 (21%) | |
| Uncertain | 2 (3.0%) | 32 (25%) | 13 (14%) | |
| VRF | 39 (60%) | 102 (82%) | 75 (85%) | < 0.001 |
| HBP | 24 (37%) | 71 (57%) | 49 (56%) | 0.021 |
| DM | 11 (17%) | 28 (23%) | 20 (23%) | 0.6 |
| DLP | 21 (32%) | 64 (52%) | 55 (63%) | 0.001 |
| eGFR(mL/min/1.73m3) | < 0.001 | |||
| > 90 | 36 (64%) | 29 (23%) | 13 (14%) | |
| 60–90 | 18 (32%) | 91 (71%) | 68 (72%) | |
| < 60 | 2 (3.6%) | 8 (6.3%) | 13 (14%) |
Median (IQR); n (%)
Kruskal–Wallis rank sum test was used to compare continuous variables that were not normally distributed; Pearson’s Chi-squared test was used to compare categorical variables; Fisher’s exact test was used to compare categorical variables with small number of observations
Unless otherwise specified, values are presented as median (IQR)
CU Cognitively Unimpaired, MCI Mild cognitive impairment, AD Alzheimer disease, OtherNotDeg Other not degenerative, OtherDem Other diseases, VRF one or more vascular risk factors, HBP High blood pressure, DM Diabetes mellitus, DLP Dyslipidemia, eGFR Estimated glomerular filtration rate
Measures of pTau217, pTau181, Aβ1–42 and Aβ1–40 in plasma
All plasma measures for pTau217, pTau181, Aβ1–42 and Aβ1–40 were above their lower limit of quantification. The plasma concentration ranges in the study were 0.03 to 2.84 for pTau217, 0.92 to 9.63 pg/mL for pTau181, 14.06 to 50.84 pg/mL for Aβ1–42 and 180.41 to 569.89 pg/mL for Aβ1–40. Inter-assay coefficients of variation are shown in Supplementary Material (Table 3).
Correlation between plasma and CSF biomarkers
As per inclusion criteria, all participants had CSF biomarkers measures, and we explored the correlation between both matrices. The correlation between plasma and CSF was moderate for pTau181 (Rho = 0.66, p < 0.001) and low for Aβ1–42 (Rho = 0.26, p = 0.007), and Aβ1–40 (Rho 0.11, p = 0.06). When using ratios, the correlation was moderate for Aβ1–42/Aβ1–40 (Rho = 0.69, p < 0.001). The correlation between plasma pTau217 and CSF pTau181 was high (Rho = 0.75, p < 0.001). Both plasma pTau217 and pTau181 correlated with age (Rho = 0.42, p < 0.001 and Rho = 0.45, p < 0.001 respectively). Detailed correlations within clinical subgroups and partial correlations are shown in Supplementary Material (Figs. 4 and 5).
Association between plasma biomarkers and amyloid status in CSF
We assessed the differences in plasma biomarkers between CSF amyloid positive and amyloid negative individuals considering other variables in a multivariate model. We studied the effect of age, sex, APOE status (APOE ε4 +), renal function measured by the estimated glomerular filtration rate (eGFR), vascular risk factors (presence of at least one of the following: high blood pressure, diabetes mellitus, dyslipidemia, history of stroke, obstructive sleep apnea with CPAP) and clinical status (CU, MCI and Dementia). As shown in Fig. 1, the log transformed multivariate model confirmed that the A + group had higher plasma concentrations of pTau217 (fold-change 4.42, p < 0.001) and pTau181 (fold-change 1.72, p < 0.001) compared to the A- group. Similar results were seen using the ratios pTau217/Aβ1–42 and Aβ1–42/Aβ1–40. The plasma pTau217/Aβ1–42 ratio was higher in A + compared to A- (fold-change 4.08, p < 0.001). The plasma Aβ1–42/Aβ1–40 ratio was lower in A + compared to A- (fold-change 0.86, p < 0.001). We found lower plasma concentrations of Aβ1–42 in A + compared to the A- group (fold-change 0.92, p < 0.001) and no differences in Aβ1–40 concentrations between A status (not shown). To assess the influence of the values close to CSF Aβ1–42/Aβ1–40 cutoff, we conducted a sensitivity analysis by excluding participants with values close to the cutoff point (± 10%) (Supplementary Material, Figs. 1– 3). We found no significant influence of these observations in our analysis.
Effect of other variables on plasma biomarkers
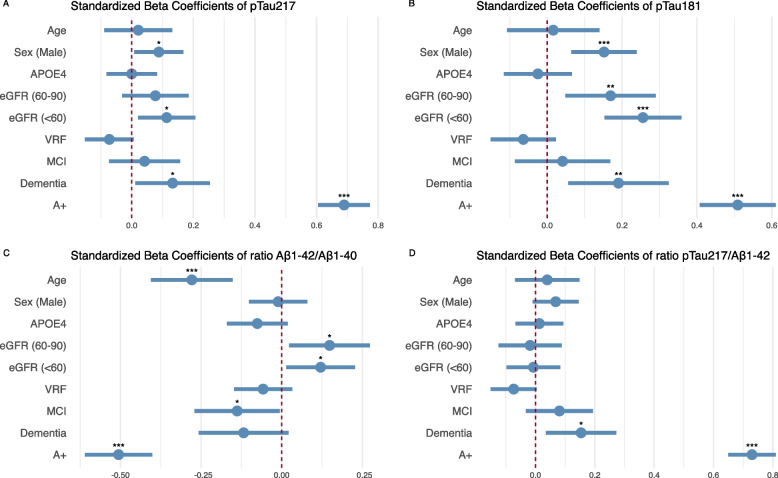
We assessed whether log transformed plasma pTau217, pTau181, Aβ1-42 and Aβ1-40 were affected by other variables in the multivariate model. As seen in Fig. 2, amyloid positivity was the variable with the largest effect on all plasma markers. We also observed that decreased renal function was associated with higher concentrations of pTau217 (p = 0.019) and pTau181 (p < 0.001) and higher Aβ1–42/Aβ1–40 ratio (p < 0.001). Male sex was associated with higher pTau217 (p = 0.033) and pTau181 (p = 0.0009), and age was associated with lower Aβ1–42/Aβ1–40 ratio. Our model had and adjusted R2 value of 0.62 for pTau217, 0.52 for pTau181 and 0.48 for Aβ1–42/Aβ1–40. All the presented coefficients were obtained from the model in which all the listed variables were included. Complete forest plot shown in Supplementary Material (Fig. 6).
Fig. 2.
Effect of different variables on plasma pTau217, pTau181, Aβ1–42/Aβ1–40 and pTau217/ Aβ1–42. Dots and bars represent the standardized beta coefficients of each variable in a multivariate regression model. Lines represent the 95% confidence interval for each standardized beta coefficient. Red vertical dashed lines indicate a null effect. We can see the effect size of A positivity adjusted by other variables. pTau217: phosphorylated tau 217. pTau181: phosphorylated tau 181. Aβ1–42: Amyloid β1–42. Aβ1–40: Amyloid β1–40. MCI: mild cognitive impairment. VRF: vascular risk factors. eGFR: estimated glomerular filtration rate
To further investigate the association of pTau217 and pTau181 with renal function, we performed a subanalysis stratifying by estimated glomerular filtration rate. We found that pTau181 concentration in plasma was higher as renal function decreased (< 60 vs 60-90 mL/min/1.73m2 and < 60 vs. > 90 mL/min/1.73m2, p < 0.001). Aβ1–42 and Aβ1–40 concentrations in plasma were also higher as renal function decreased (p < 0.001). We also observed marginally significant differences in pTau217 concentrations in plasma samples from patients with low renal function (< 60 mL/min/1.73m2) compared to those with normal renal function (> 90 mL/min/1.73m2, p = 0.047). However, those differences were lost when using the Aβ1–42/Aβ1–40 or the pTau217/Aβ1–42 ratios.
Diagnostic accuracy of plasma biomarkers and their combinations for the discrimination of A + from A-
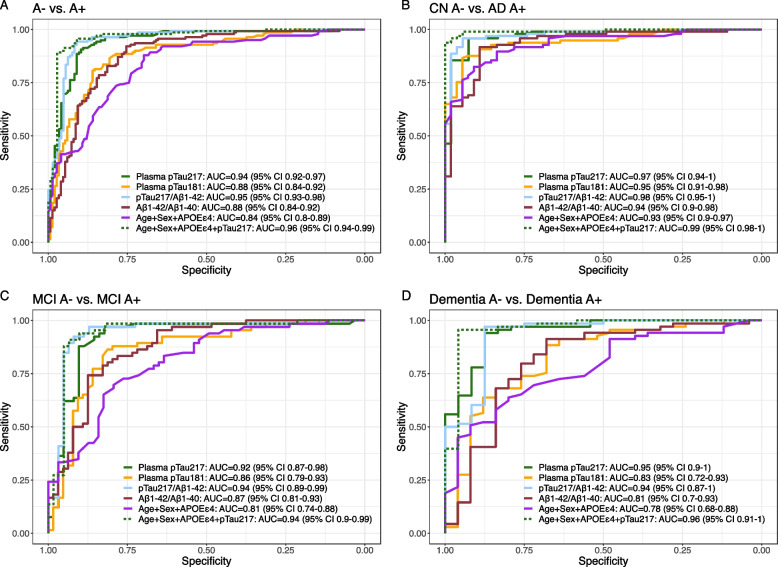
In the whole sample, the AUC to discriminate A + from A- participants were 0.94 (95% Cl 0.92–0.97) for pTau217, and 0.88 (95% CI 0.84–0.92) for both pTau181 and Aβ1–42/Aβ1–40 (Fig. 3). The diagnostic accuracy of pTau217 to detect amyloid positivity was not outperformed by any other individual plasma biomarker, their ratios or their combinations. Aβ1–42 and Aβ1–40 individually had poor diagnostic accuracy, yielding AUCs below 0.70. Detailed two-by-two comparisons can be found as Supplementary Table 4. Sensitivity, specificity and Youden indices yielded by individual plasma markers are shown in Supplementary Fig. 7.
Fig. 3.
Diagnostic accuracy of plasma biomarkers for the discrimination of A + from A- categories. pTau217: phosphorylated tau 217, pTau181: phosphorylated tau 181. Aβ1–42: Amyloid β1–42. Aβ1–40: Amyloid β1–40. CN, cognitively unimpaired. MCI, mild cognitive impairment
The diagnostic performance of pTau217 was also high across different clinical categories. In the MCI group, the accuracy of pTau217 (AUC = 0.92, 95% CI 0.87–0.98) was not significantly different than that pTau181 (AUC = 0.86, 95% CI 0.79–0.93) or that of the basic model with Age, Sex and APOE ε4 (AUC = 0.81, 95% CI 0.74–0.88). However, the addition of pTau217 to Age, Sex and APOE ε4 increased the accuracy of the model significantly from 0.81 (95% CI 0.74–0.88) to 0.94 (95% CI 0.9–0.99). pTau217 also showed very high accuracies (AUC = 0.97; 95% CI 0.94–1.00) to discriminate A + patients with a diagnosis of AD from different A- clinical groups (CN, other dementias and other not degenerative, Supplementary Fig. 8). The performance of plasma biomarkers to discriminate A + T + from A-T- participants yielded similar results (Supplementary Fig. 9).
Cutoffs application
Table 2 shows the accuracy of different thresholds for pTau217, pTau181 and Aβ1–42/Aβ1–40 to detect amyloid positivity with a sensitivity and specificity of 97.5%, 95% and 90%. As pTau217 was the individual biomarker with highest diagnostic accuracy, we assessed the performance of selected cutoffs for this marker in different clinical groups, stratifying by decade. We found that the cutoff that had a global sensitivity of 95% (0.186 pg/mL) yielded accuracies above 84% across all decades and clinical groups when the amyloidosis prevalence was above 20%. In groups with lower amyloidosis prevalence (i.e. CN) the cutoff with specificity of 95% (0.388 pg/mL) showed higher accuracies. We observed that the cutoff that had a specificity of 95% showed a progressive decrease in accuracy with age in the MCI group (accuracy 84% in 60–69, 75% in 70–79 and 62% over 80 years). The negative predictive value of this cutoff was low over 70 years (58%). Detailed information on the accuracy of cutoffs in distinct clinical groups and decades can be found in Supplementary Material (tables 5, 6, 7 & 8).
Table 2.
Thresholds for plasma pTau217, pTau181 and Aβ1–42/Aβ1–40 to detect A + participants
| Threshold | Sensitivity | Specificity | NPV | PPV | Youden | Accuracy |
|---|---|---|---|---|---|---|
| Accuracy of plasma pTau217 | ||||||
| 0.130 | 97.8% | 65.5% | 96.9% | 73.1% | 0.634 | 81.3% |
| 0.186 | 95.0% | 82.1% | 94.4% | 83.5% | 0.770 | 88.4% |
| 0.247 | 90.6% | 89.7% | 90.9% | 89.4% | 0.803 | 90.1% |
| 0.249 | 89.9% | 89.7% | 90.3% | 89.3% | 0.796 | 89.8% |
| 0.388 | 69.8% | 95.2% | 76.7% | 93.3% | 0.650 | 82.7% |
| 0.552 | 45.3% | 97.2% | 65.0% | 94.0% | 0.426 | 71.8% |
| Accuracy of plasma pTau181 | ||||||
| 1.573 | 97.9% | 32.0% | 94.1% | 57.3% | 0.299 | 63.8% |
| 1.740 | 95.0% | 44.7% | 90.5% | 61.6% | 0.397 | 69.0% |
| 2.125 | 90.0% | 71.3% | 88.4% | 74.6% | 0.613 | 80.3% |
| 2.815 | 64.3% | 90.0% | 73.0% | 85.7% | 0.543 | 77.6% |
| 3.345 | 47.9% | 94.7% | 66.0% | 89.3% | 0.425 | 72.1% |
| 3.840 | 31.4% | 97.3% | 60.3% | 91.7% | 0.288 | 65.5% |
| Accuracy of Aβ1–42/Aβ1–40 | ||||||
| 0.086 | 97.9% | 41.3% | 95.4% | 60.9% | 0.392 | 68.6% |
| 0.080 | 95.0% | 65.3% | 93.3% | 71.9% | 0.603 | 79.7% |
| 0.078 | 90.0% | 75.3% | 89.0% | 77.3% | 0.653 | 82.4% |
| 0.073 | 65.0% | 90.0% | 73.4% | 85.8% | 0.550 | 77.9% |
| 0.070 | 37.9% | 94.7% | 62.0% | 86.9% | 0.325 | 67.2% |
| 0.067 | 20.7% | 97.3% | 56.8% | 87.9% | 0.180 | 60.3% |
pTau217: phosphorylated tau 217, pTau181: phosphorylated tau 181. Aβ1–42: Amyloid β1–42. Aβ1–40: Amyloid β1–40. NPV, negative predictive value. PPV, positive predictive value. Threshold units for pTau are in pg/mL
We conducted a supervised decision tree analysis to determine the potential of various biomarkers and demographic factors in correctly identifying individuals with amyloidosis. This analysis incorporated plasma pTau217, pTau181, ratio Aβ1–42/ Aβ1–40, Age, Sex, APOE ε4 allele presence, and clinical diagnosis group (cognitively unimpaired, mild cognitive impairment, and dementia). The most effective discriminators for amyloidosis were plasma pTau217 followed by the Aβ1–42/ Aβ1–40 ratio when pTau217 was high (Supplementary Fig. 10). Other variables were deemed less critical for amyloidosis detection and thus excluded from the decision tree. This algorithm exhibited a misclassification rate of 7.4%, with a sensitivity of 91%, specificity of 94%, overall accuracy of 93%, positive predictive value (PPV) of 94%, and negative predictive value (NPV) of 91%, accompanied by a false-negative rate (FNR) of 9.4% and a false-positive rate (FPR) of 5.6%.
Potential of plasma pTau217 to predict Alzheimer disease pathophysiology
We assessed the predictive capability of pTau217 to properly classify participants with amyloidosis in our dataset, defined as CSF Aβ1–42/Aβ1–40 < 0.062. As the model that combined pTau217 with Age, Sex and APOEε4 did not perform better than pTau217 alone in any comparison, we chose the simplest predictive model with pTau217. We bootstrapped to obtain robust predictive cuttoffs. We found no differences between the initial prediction and the mean of the 1000 predictive iterations, with a perfect correlation between them (Rho = 1, p < 0.001) thus reinforcing the robustness of our initial predictions.
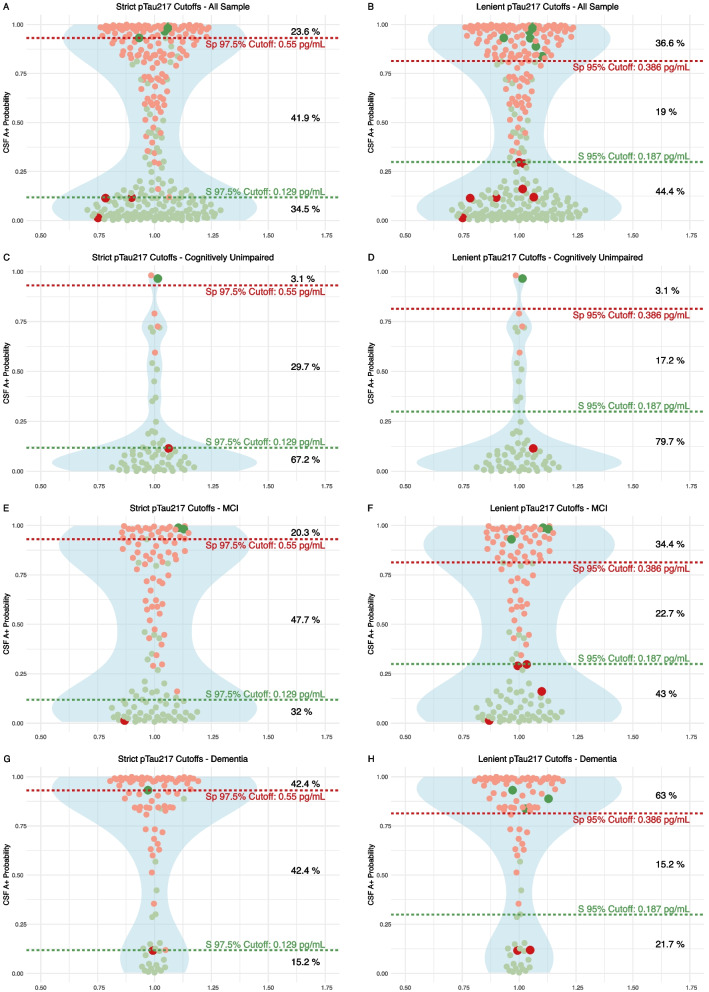
To facilitate the clinical implementation of plasma biomarkers while ensuring accuracy, we applied a two-threshold approach to classify participants into three groups, those with high, medium, and low likelihood of being CSF amyloid positive. Following this approach, those with a medium risk would benefit from a confirmation with gold standard tests like CSF or Amyloid PET. Figure 4 shows the thresholds in two different scenarios based on two levels of restrictiveness in sensitivity and specificity (97.5% and 95%). Using a highly accurate combination of cutoffs (one for 97.5% Sens and another for 97.5% Spec), only 41.9% of patients in the whole sample would require an additional test, with a global misclassification rate of 4.2%. Using less restrictive cutoffs (95% Sens/Spec), the proportion could be reduced to 19% with a global misclassification rate of 6%. Similar results were found in the subgroup of patients with CU, MCI and Dementia.
Fig. 4.
Plasma pTau217 predictive models. Strict and lenient cutoffs in the whole sample and in the distinct clinical groups. We illustrate the implementation of various cutoff thresholds within our sample, denoted by dashed lines in red and green at distinct Y-axis levels, each line representing the associated sensitivity and specificity values. Displayed in red dots are individuals with amyloid positivity in cerebrospinal fluid (CSF) and in green those amyloid negatives. Those participants with pTau217 concentrations above dashed red lines are classified as high risk of amyloid CSF positivity. The medium risk category is between dashed red and green lines. Below dashed green line are the participants classified as low risk. Observations that have been incorrectly categorized into high or low risk groups are represented by distinct sizes and colors. To the right, the corresponding percentages of the sample assigned to each risk category are presented. pTau217: phosphorylated tau 217, pTau181: phosphorylated tau 181. Aβ1–42: Amyloid β1–42. Aβ1–40: Amyloid β1–40. Model 1 included only plasma pTau217 for predictions. S, sensitivity. Sp, specificity
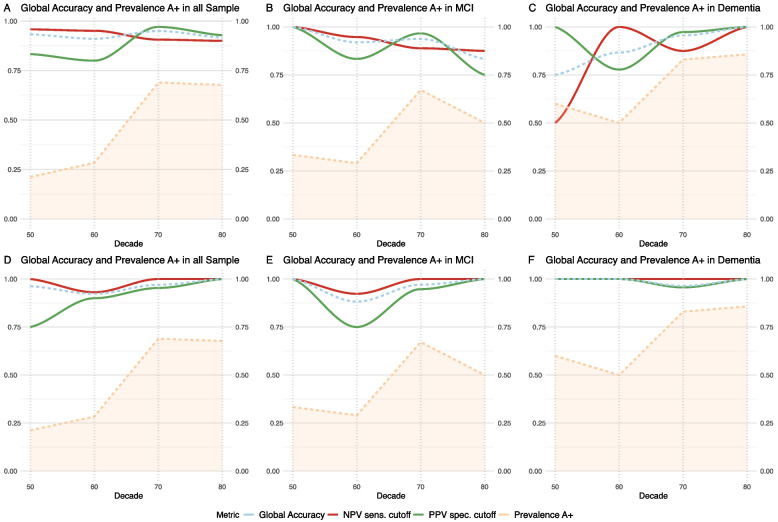
We finally assessed the robustness of those cutoffs in the whole sample, considering the increase of prevalence of amyloidosis with age in our population. In Fig. 5, we show the NPV, PPV, and global accuracy of pTau217 to detect A positivity by decades using cutoff combinations with 95% and 97.5% sensitivity and specificity in different clinical groups. We found high global accuracy (75%-100%) of the two-threshold application in all the scenarios, with variations of PPV and NPV according to CSF amyloid positive prevalence in our sample.
Fig. 5.
Negative predictive value, positive predictive value and global accuracy of pTau217 for the combination of cutoffs with 95% (A, B, C) and 97.5% (D, E, F) sensitivity and specificity and their relationship with age and prevalence of CSF Amyloidosis. The figure illustrates the association between the prevalence of cerebrospinal fluid (CSF) Amyloidosis (depicted by a dashed orange line), Positive Predictive Value (PPV, red line), Negative Predictive Value (NPV, green line), and global accuracy (dashed blue line) across different decades of life (X-axis) within our study cohort. In panels A-C, the PPV was determined using a cutoff for 95% specificity set at 0.386 pg/mL, while the NPV was ascertained using a 95% sensitivity cutoff at 0.187 pg/mL. In panels D-F, the PPV was determined using a cutoff for 97.5% specificity set at 0.55 pg/mL, while the NPV was ascertained using a 97.5% sensitivity cutoff at 0.129 pg/mL. The term 'global accuracy' in this context refers to the proportion of participants accurately classified as either positive or negative, based on the application of these two cutoffs. It should be noted that this calculation of global accuracy excludes participants categorized as 'indeterminate' (falling within the grey zone), for whom a confirmatory test is recommended. pTau217: phosphorylated tau 217, Sens, sensitivity. Spec, specificity. NPV, negative predictive value. PPV, positive predictive value. MCI, mild cognitive impairment
Discussion
In this study, we found that the concentration of plasma pTau217, plasma pTau181, and the ratio Aβ1–42/Aβ1–40 measured in a fully automated platform, yielded excellent accuracy to detect the AD pathophysiology in the setting of the routine clinical practice of a memory clinic. Of all plasma markers, the recently developed assay that measures pTau217 was the most accurate followed by the Aβ1–42/Aβ1–40 ratio and pTau181. We also found that different comorbidities had a mild significant effect on plasma markers of AD, but the amyloid status was the single variable with the largest effect on their concentration. In patients with advanced chronic kidney disease, the use of ratios could reduce the impact of having higher plasma concentrations associated to low renal function. Furthermore, we applied predictive models to obtain stratification profiles that showed the potential to reduce the need of more costly or invasive procedures by approximately 60 to 80%.
The performance of plasma markers to detect the AD pathophysiology has been assessed in previous studies using different analytical platforms, with AUCs ranging from 0.70 to 0.96 for pTau181 [7, 23, 39–43], and from 0.64 to 0.86 for Aβ1–42/Aβ1–40 [5, 8, 10]. Plasma pTau231 and pTau217 have shown to better capture the earliest cerebral Aβ changes in CU, before overt Aβ plaque pathology is present [26] For plasma pTau217, accuracies have varied depending on the platform, yet the performance has consistently been high in discriminating amyloidosis in MCI and predicting progression, typically outperforming that of other plasma pTau isoforms [7, 23, 33, 44]. Most research studies reported better accuracies with the use of composite measures that combined two or more markers and/or clinical or genetic information [45–47]. In our study, plasma pTau217, the Aβ1–42/Aβ1–40 ratio and pTau181 measured with a fully automated platform showed high diagnostic performance to detect amyloid positivity. Of these, pTau217 was the marker that showed higher accuracy and, importantly, it was not outperformed by composite measures indicating that it is a good candidate for its implementation as a single biomarker.
Automated platforms have revolutionized CSF analysis by resolving critical analytical factors and, similarly, they hold promise for transforming plasma testing, enhancing both precision and efficiency in biomarker quantification. Recent studies have assessed the diagnostic performance of pTau181 and Aβ1–42/Aβ1–40 in the Lumipulse platform. Janelidze et al. reported an AUC of 0.7 for pTau181 for the identification of CSF amyloidosis in MCI [23]and of 0.74 to detect progression to dementia. However, Wilson et al. reported a higher accuracy of 0.96 [40] for the discrimination between Aβ- CU and Aβ + AD patients. Another recent study using the Lumipulse platform analyzed the accuracy to detect AD of plasma pTau181 and Aβ1–42/Aβ1–40 ratio in cognitively unimpaired participants and found that Aβ1–42/Aβ1–40 ratio was the most cost-effective (AUC 0.9 for A + and 0.89 for A + T +), followed by pTau181 that showed an AUC of 0.76 for A + and 0.86 for A + T + [48]. Our results are in line with previous studies, and show a global accuracy of 0.88 for plasma pTau181 and for the plasma Aβ1–42/Aβ1–40 ratio. A variety of reasons could explain the minor discrepancies between studies, including differences in preanalytical conditions [49], in the kits that were used, characteristics of the sample and cohorts, and the design of the studies. Our design including consecutive patients that underwent a lumbar puncture for routine diagnostic work-up in a memory clinic setting, provides information about the potential implementation of plasma markers in this context.
Together with the commercially available pTau181 and Aβ1–42/Aβ1–40, we evaluated the performance of plasma pTau217, recently developed for the same automated platform, and found that it outperformed pTau181 and Aβ1–42/Aβ1–40 in the detection of amyloid positivity. In previous research studies, plasma pTau217 has consistently shown exceptionally high accuracy across different platforms and has demonstrated strong correlations with other markers of AD (CSF biomarkers, amyloid PET and Tau PET) and with neuropathology [30]. The fact that it has shown greatest fold changes and effect sizes compared to other pTau isoforms, makes it a perfect candidate for its implementation in clinical settings, as small analytical variations (5–10%) would not substantially affect its diagnostic performance. Its implementation on a fully automated platform would not only simplify the process but also enhance accessibility for clinical laboratories. In our study using a fully automated platform, we found that A + patients had 4.42 times higher concentrations of plasma pTau217. This magnitude of effect, combined with the advantages of automation, makes this assay particularly promising for integration into standard clinical practices.
To facilitate a more rational utilization of plasma biomarkers, we followed the methodology delineated by Brum et al. stratifying the risk of having AD pathopshysiology [38]. By implementing this strategy, it is feasible to define threshold values that are highly sensitive and specific to either detect or rule out CSF amyloidosis minimizing the probability of misclassification. This would also allow for selecting those patients that fall within an intermediate likelihood category or ‘grey zone’ and that would benefit of further etiological investigations (CSF biomarkers, amyloid or Tau PET). This stratification framework represents a pragmatic and flexible approach for the incorporation of plasma biomarkers in clinical settings. An open point of discussion is the optimal context of use —primary care, general neurology, or specialized memory clinics— and the management strategies for participants identified as high or low risk. Our study showed that following this approach, pTau217 had an excellent performance in the context of a specialized memory clinic, but these classifications will need to be contextualized within other clinical settings [4, 34]. The effect of comorbidities as CKD on plasma biomarker concentrations points in the same direction as recently published studies [15, 50], in which the use of ratios could attenuate the effect of CKD. Moreover, we found that the effect of renal dysfunction on plasma pTau217 concentrations was significantly less than that of the amyloid positivity status, suggesting that the actual impact of CKD on the diagnostic performance of this marker would be minimal. When we assessed the impact of renal dysfunction on different plasma pTau biomarker concentrations, we found that the effect size of eGFR < 60 mL/min/1.73m3 in pTau181 was double that observed in pTau217, with standardized beta coefficients of 0.25 and 0.11, respectively (Fig. 2), both statistically significant. When we compared the AUC of plasma pTau217 and pTau181 in the subset of patients with eGFR < 60 mL/min/1.73m3, although higher in pTau217 (AUC 0.83, CI 95% 0.64–1) than in pTau181 (AUC 0.75, CI 95% 0.53–0.97), accuracy did not differ between both biomarkers, probably because of low statistical power to capture differences in this subset in our cohort, as only 11 patients were A- and 12 patients A + .
One of the strengths of our study is that we included all consecutive participants from routine clinical practice that underwent lumbar puncture throughout one year in our memory clinic including a variety of diagnoses. This approach reduces the risk of selection biases and ensures a reliable representation of the population assessed in the setting of a specialized memory clinic, also providing relevant information on their potential implementation in the routine diagnostic work-up in this context. Other strengths in our study are the fact that all markers were measured using the same batch of reagents and that the clinical information available allowed us to analyze the potential impact of comorbidities and perform sub analyses within distinct clinical stages.
Limitations
Our study also has some limitations. First, as the inclusion criteria required that participants had received a lumbar puncture for CSF biomarkers, the extrapolation to other contexts of use different than specialized memory units, such as primary care or population screening programs, should be made cautiously. Second, we could not compare plasma pTau217 with its counterpart in CSF as the assay was specifically developed for plasma. Another limitation is the lack of Amyloid/Tau PET or neuropathological confirmation in our participants, and although the CSF biomarker cutoffs in our center were validated against amyloid PET [37], we cannot be certain about how using a different gold-standard might affect our results. Finally, this is a single-center study, and even though this increases uniformity and we performed a bootstrapped cross-validation to ensure the predictive robustness of our models, the accuracy and true predictive power of the cutoffs derived from our results need to be verified in diverse datasets with comparable characteristics.
Conclusions
Our study provides evidence that plasma markers can reliably be measured in an automated platform, and highlights plasma pTau217 as the most promising plasma biomarker, showing great potential for the detection of AD pathophysiology in the context of a memory clinic. With the arrival of disease-modifying treatments into clinical practice, it is urgent to have easily accessible and efficient diagnostic methods to identify patients that could benefit from these therapies. The implementation of plasma biomarkers in readily accessible fully automated platforms will streamline the diagnosis and enhance the accessibility of disease modifying therapies.
Supplementary Information
Acknowledgements
We are grateful to all participants in the study and their families. We also thank all the clinical team members that were involved in the selection and assessment of participants in the SPIN cohort, and the laboratory teams for sample handling, biomarker analyses, and structural support. We also thank Fujirebio for providing technical support and the reagents necessary to complete the study.
Authors’ contribution
DA and AL designed the study. JA, NZ, SR-G, IR-B, RF, MC-I, IB, II-G, MS-S, AL, JF, MT, DA acquired data relevant for the study. MC-I, II-G, JF, AL, MT, DA contributed vital reagents/tools/patents. MC-I, II-G, MS-S, JF, AL, DA obtained funding for the study. DA and JA performed statistical analysis. DA, JA contributed in analysis and interpretation of data. DA, AL, JF participated in study supervision or coordination. JA and DA drafted the first version of the manuscript.
Funding
This study was supported by the Fondo de Investigaciones Sanitario (FIS), Instituto de Salud Carlos III (PI21/00791 to IIG, PI14/01126, PI17/01019 and PI20/01473 to JF, PI13/01532 and PI16/01825 to RB, PI18/00335 to MCI, PI19/00882 to MS-S, PI18/00435, PI22/00611 and INT19/00016 to DA, PI17/01896 and AC19/00103to AL) and the CIBERNED program (Program 1, Alzheimer Disease to AL), jointly funded by Fondo Europeo de Desarrollo Regional, Unión Europea, “Una manera de hacer Europa”.
This work was also supported by the National Institutes of Health (NIA grants 1R01AG056850-01A1; R21AG056974; and R01AG061566 to JF), by Generalitat de Catalunya (2017-SGR-547, SLT006/17/125 to DA, SLT006/17/119 to JF, SLT002/16/408 to AL) and “Marató TV3” foundation grants 20141210 to JF, 044412 to RB and 20142610 to AL. This work was also supported by a grant from the Fundació Bancaria La Caixa to RB (DABNI project). Fundació Catalana Síndrome de Down and Fundació Víctor Grífols i Lucas partly supported this work. Horizon 21 Consortium is partly funded by Jérôme Lejeune Foundation (Clinical and trial outcome measures for dementia in individuals with Down syndrome).
The reagents necessary to complete the study were funded by Fujirebio-Europe.
The sponsors of the study did not take part in the design and conduct of the study; collection, management, analysis, and interpretation of the data; writing and review of the report; or the decision to submit the article for publication.
Availability of data and materials
Raw anonymized data and code for statistical analysis are available upon reasonable request. All requests should be sent to the corresponding author detailing the study hypothesis and statistical analysis plan. The steering committee of this study will decide whether data/code sharing is appropriate based on the novelty and scientific rigor of the proposal. All applicants will be asked to sign a data access agreement.
Declarations
Ethics approval and consent to participate
The ethics committee of Hospital Sant Pau approved all procedures included in this study following the standards for medical research in humans recommended by the Declaration of Helsinki. All participants or their legally authorised representative gave written informed consent before enrolment in the study.
Consent for publication
All authors revised the manuscript for content and provided critical feedback.
Competing interests
Daniel Alcolea is employed by Hospital de la Santa Creu i Sant Pau and received research grants from Pla Estratègic de Recerca i Innovació en Salut (PERIS SLT006/17/125), and from Instituto de Salud Carlos III (PI18/00435 and INT19/00016). He participated in advisory boards from Fujirebio-Europe and Roche Diagnostics and received speaker honoraria from Fujirebio-Europe, Roche Diagnostics, Nutricia, Zambon S.A.U., Esteve, and from Krka Farmacéutica S.L.
Javier Arranz is employed by Biomedical Research Institute Sant Pau. He is funded by a “Rio Hortega” research grant from the Institute of Health Carlos III. Declarations of interest: none.
Nuole Zhu is employed by Biomedical Research Institute Sant Pau. He is funded by a “Rio Hortega” research grant from the Institute of Health Carlos III. Declarations of interest: none.
Sara Rubio-Guerra is employed by Hospital de la Santa Creu i Sant Pau. Declarations of interest: none.
Iñigo Rodríguez-Baz is employed by Biomedical Research Institute Sant Pau. He is funded by a “Rio Hortega” research grant from the Institute of Health Carlos III. Declarations of interest: none.
Rosa Ferrer is employed by Hospital de la Santa Creu i Sant Pau. Declarations of interest: none.
María Carmona-Iragui is employed by Hospital de la Santa Creu i Sant Pau. Declarations of interest: none.
Isabel Barroeta is employed by Hospital de la Santa Creu i Sant Pau. Declarations of interest: none.
Dr. Illán-Gala is a senior Atlantic Fellows for Equity in Brain Health at the Global Brain Health Institute (GBHI), and is supported with funding from GBHI, Alzheimer’s Association, and Alzheimer’s Society (GBHI ALZ UK-21–720973 and AACSF-21–850193). Dr Illán-Gala was also supported by the Juan Rodés Contract (JR20/0018) and Fondo de Investigaciones Sanitario, (PI21/00791) from Instituto de Salud Carlos III.
Ignacio Illán-Gala reported receiving personal fees from Nutricia, Esteve, UCB, and Neuraxpharm Spain outside the submitted work.
Miguel Santos-Santos is employed by Hospital de la Santa Creu i Sant Pau. His research is supported by funding from the Spanish Institute of Health Carlos III (Juan Rodés contract JR18-00018; Fondo de investigación sanitaria grant PI19/00882), the Alzheimer’s Association clinician scientist fellowship (AACSF-22–972945), and the National Institutes of Health (R01AG080470).
Juan Fortea is employed by Hospital de la Santa Creu i Sant Pau and received research grants from Institute of Health Carlos III, National Institutes of Health, Fundació La Marató de TV3, and Pla Estratègic de Recerca i Innovació en Salut (PERIS). Dr. Fortea has served as a consultant for Novartis and Lundbeck, has received honoraria for lectures from Roche, NovoNordisk, Esteve and Biogen and served at advisory boards for AC Immune, Zambon and Lundbeck.
Alberto Lleó is employed by Hospital de la Santa Creu i Sant Pau and received research grants from CIBERNED, Institute of Health Carlos III, Generalitat de Catalunya (PERIS and AGAUR) and Fundación Tatiana and BBVA. He participated in advisory boards from Biogen, Eisai, Fujirebio-Europe, Novartis, NovoNordisk, Nutricia, Otsuka Pharmaceutical, and Zambón, and received speaker honoraria from Lilly, Biogen, KRKA and Zambon.
Mireia Tondo is employed by Hospital de la Santa Creu i Sant Pau and has received research grants from Instituto de Salud Carlos III (PI18/00164; PI21/00140) and has served as consultant in Araclon.
Footnotes
The original version of this article was revised: the authors corrected the Funding section to mention “project PI22/00611”.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/27/2024
A Correction to this paper has been published: 10.1186/s13195-024-01538-0
Contributor Information
Mireia Tondo, Email: mtondo@santpau.cat.
Daniel Alcolea, Email: dalcolea@santpau.cat.
References
- 1.Fargo KN, Carrillo MC, Weiner MW, et al. The crisis in recruitment for clinical trials in Alzheimer’s and dementia: An action plan for solutions. Alzheimer’s and Dementia. 2016;12:1113–5. 10.1016/j.jalz.2016.10.001. 10.1016/j.jalz.2016.10.001 [DOI] [PubMed] [Google Scholar]
- 2.Teunissen CE, Verberk IMW, Thijssen EH, et al. Blood-based biomarkers for Alzheimer’s disease: towards clinical implementation. Lancet Neurol. 2022;21:66–77. 10.1016/S1474-4422(21)00361-6. 10.1016/S1474-4422(21)00361-6 [DOI] [PubMed] [Google Scholar]
- 3.Alcolea D, Delaby C, Muñoz L, et al. Use of plasma biomarkers for AT(N) classification of neurodegenerative dementias. J Neurol Neurosurg Psychiatry. 2021;92:1206–14. 10.1136/jnnp-2021-326603 [DOI] [PubMed] [Google Scholar]
- 4.Alcolea D, Beeri MS, Rojas JC, et al. Blood Biomarkers in Neurodegenerative Diseases: Implications for the Clinical Neurologist. Neurology. 2023. 10.1212/WNL.0000000000207193. 10.1212/WNL.0000000000207193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554:249–54. 10.1038/nature25456 [DOI] [PubMed] [Google Scholar]
- 6.Janelidze S, Stomrud E, Palmqvist S, et al. Plasma β-amyloid in Alzheimer’s disease and vascular disease. Sci Rep. 2016;6. 10.1038/srep26801. [DOI] [PMC free article] [PubMed]
- 7.Ashton NJ, Puig-Pijoan A, Milà-Alomà M, et al. Plasma and CSF biomarkers in a memory clinic: Head-to-head comparison of phosphorylated tau immunoassays. Alzheimer’s Dement Published Online First. 2022. 10.1002/alz.12841. 10.1002/alz.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schindler SE, Bollinger JG, Ovod V, et al. High-precision plasma β-amyloid 42/40 predicts current and future brain amyloidosis. Neurology. 2019;93:E1647–59. 10.1212/WNL.0000000000008081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montoliu-Gaya L, Strydom A, Blennow K, et al. Blood biomarkers for alzheimer’s disease in down syndrome. J Clin Med. 2021;10. 10.3390/jcm10163639. [DOI] [PMC free article] [PubMed]
- 10.Janelidze S, Teunissen CE, Zetterberg H, et al. Head-to-Head Comparison of 8 Plasma Amyloid-β 42/40 Assays in Alzheimer Disease. JAMA Neurol. 2021;78:1375–82. 10.1001/jamaneurol.2021.3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayoumy S, Verberk IMW, den Dulk B, et al. Clinical and analytical comparison of six Simoa assays for plasma P-tau isoforms P-tau181, P-tau217, and P-tau231. Alzheimers Res Ther. 2021;13. 10.1186/s13195-021-00939-9. [DOI] [PMC free article] [PubMed]
- 12.Illán-Gala I, Lleo A, Karydas A, et al. Plasma Tau and Neurofilament Light in Frontotemporal Lobar Degeneration and Alzheimer Disease. Neurology. 2021;96:e671–83. 10.1212/WNL.0000000000011226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mielke MM, Dage JL, Frank RD, et al. Performance of plasma phosphorylated tau 181 and 217 in the community. Nat Med. 2022;28:1398–405. 10.1038/s41591-022-01822-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichet Binette A, Janelidze S, Cullen N, et al. Confounding factors of Alzheimer’s disease plasma biomarkers and their impact on clinical performance. Alzheimer’s and Dementia Published Online First. 2022. 10.1002/alz.12787. 10.1002/alz.12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syrjanen JA, Campbell MR, Algeciras-Schimnich A, et al. Associations of amyloid and neurodegeneration plasma biomarkers with comorbidities. Alzheimer’s Dement. 2022;18:1128–40. 10.1002/alz.12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashton NJ, Janelidze S, Mattsson-Carlgren N, et al. Differential roles of Aβ42/40, p-tau231 and p-tau217 for Alzheimer’s trial selection and disease monitoring. Nat Med. 2022;28:2555–62. 10.1038/s41591-022-02074-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verberk IMW, Slot RE, Verfaillie SCJ, et al. Plasma Amyloid as Prescreener for the Earliest Alzheimer Pathological Changes. Ann Neurol. 2018;84:648–58. 10.1002/ana.25334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pichet Binette A, Palmqvist S, Bali D, et al. Combining plasma phospho-tau and accessible measures to evaluate progression to Alzheimer’s dementia in mild cognitive impairment patients. Alzheimers Res Ther. 2022;14. 10.1186/s13195-022-00990-0. [DOI] [PMC free article] [PubMed]
- 19.Palmqvist S, Tideman P, Cullen N, et al. Prediction of future Alzheimer’s disease dementia using plasma phospho-tau combined with other accessible measures. Nat Med Published Online First. 2021. 10.1038/s41591-021-01348-z. 10.1038/s41591-021-01348-z [DOI] [PubMed] [Google Scholar]
- 20.Mattsson-Carlgren N, Salvadó G, Ashton NJ, et al. Prediction of Longitudinal Cognitive Decline in Preclinical Alzheimer Disease Using Plasma Biomarkers. JAMA Neurol. 2023;80:360–9. 10.1001/jamaneurol.2022.5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teunissen CE, Thijssen EH. Plasma p-tau217: From ‘new kid’ to most promising candidate for Alzheimer’s disease blood test. Brain. 2020;143:3170–80. 10.1093/BRAIN/AWAA329. 10.1093/BRAIN/AWAA329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvadó G, Ossenkoppele R, Ashton NJ, et al. Specific associations between plasma biomarkers and postmortem amyloid plaque and tau tangle loads. EMBO Mol Med. 2023;15. 10.15252/emmm.202217123. [DOI] [PMC free article] [PubMed]
- 23.Janelidze S, Bali D, Ashton NJ, et al. Head-to-head comparison of 10 plasma phospho-tau assays in prodromal Alzheimer’s disease. Brain. Published Online First. 2022. 10.1093/brain/awac333. [DOI] [PMC free article] [PubMed]
- 24.Ashton NJ, Brum WS, Di Molfetta G, et al. Diagnostic accuracy of the plasma ALZpath pTau217 immunoassay to identify Alzheimer’s disease pathology. medRxiv. Published Online First: 12 July 2023. 10.1101/2023.07.11.23292493.
- 25.Cullen NC, Janelidze S, Mattsson-Carlgren N, et al. Test-retest variability of plasma biomarkers in Alzheimer’s disease and its effects on clinical prediction models. Alzheimer’s Dement. 2023;19:797–806. 10.1002/alz.12706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milà-Alomà M, Ashton NJ, Shekari M, et al. Plasma p-tau231 and p-tau217 as state markers of amyloid-β pathology in preclinical Alzheimer’s disease. Nat Med. 2022;28:1797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu L, Boyle PA, Janelidze S, et al. Plasma p-tau181 and p-tau217 in discriminating PART, AD and other key neuropathologies in older adults. Acta Neuropathol. 2023;146:1–11. 10.1007/s00401-023-02570-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pais MV, Forlenza OV, Diniz BS. Plasma Biomarkers of Alzheimer’s Disease: A Review of Available Assays, Recent Developments, and Implications for Clinical Practice. J Alzheimers Dis Rep. 2023;7:355–80. 10.3233/ADR-230029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack CR, Wiste HJ, Algeciras-Schimnich A, et al. Predicting amyloid PET and tau PET stages with plasma biomarkers. Brain. 2023;146:2029–44. 10.1093/brain/awad042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Therriault J, Vermeiren M, Servaes S, et al. Association of Phosphorylated Tau Biomarkers with Amyloid Positron Emission Tomography vs Tau Positron Emission Tomography. JAMA Neurol. 2023;80:188–99. 10.1001/jamaneurol.2022.4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Therriault J, Servaes S, Tissot C, et al. Equivalence of plasma p-tau217 with cerebrospinal fluid in the diagnosis of Alzheimer’s disease. Alzheimer’s Dement Published Online First. 2023. 10.1002/alz.13026. 10.1002/alz.13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groot C, Cicognola C, Bali D, et al. Diagnostic and prognostic performance to detect Alzheimer’s disease and clinical progression of a novel assay for plasma p-tau217. Alzheimers Res Ther. 2022;14. 10.1186/s13195-022-01005-8. [DOI] [PMC free article] [PubMed]
- 33.Palmqvist S, Janelidze S, Quiroz YT, et al. Discriminative Accuracy of Plasma Phospho-tau217 for Alzheimer Disease vs Other Neurodegenerative Disorders. J Am M Assoc. 2020;324:772–81. 10.1001/jama.2020.12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansson O, Edelmayer RM, Boxer AL, et al. The Alzheimer’s Association appropriate use recommendations for blood biomarkers in Alzheimer’s disease. Alzheimer’s Dement. 2022;18:2669–86. 10.1002/alz.12756. 10.1002/alz.12756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alcolea D, Clarimón J, Carmona-Iragui M, et al. The Sant Pau Initiative on Neurodegeneration (SPIN) cohort: a data set for biomarker discovery and validation in neurodegenerative disorders. Alzheimer’s Dement. 2019;5:597–609. 10.1016/j.trci.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansilla A, Canyelles M, Ferrer R, et al. Effects of storage conditions on the stability of blood-based markers for the diagnosis of Alzheimer’s disease. Clin Chem Lab Med. 2023;61:1580–9. 10.1515/cclm-2023-0245 [DOI] [PubMed] [Google Scholar]
- 37.Alcolea D, Pegueroles J, Muñoz L, et al. Agreement of amyloid PET and CSF biomarkers for Alzheimer’s disease on Lumipulse. Ann Clin Transl Neurol. 2019;6:1815–24. 10.1002/acn3.50873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brum WS, Cullen NC, Janelidze S, et al. A two-step workflow based on plasma p-tau217 to screen for amyloid β positivity with further confirmatory testing only in uncertain cases. Nat Aging Published Online First. 2023. 10.1038/s43587-023-00471-5. 10.1038/s43587-023-00471-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baiardi S, Quadalti C, Mammana A, et al. Diagnostic value of plasma p-tau181, NfL, and GFAP in a clinical setting cohort of prevalent neurodegenerative dementias. Alzheimers Res Ther. 2022;14. 10.1186/s13195-022-01093-6. [DOI] [PMC free article] [PubMed]
- 40.Wilson EN, Young CB, Ramos Benitez J, et al. Performance of a fully-automated Lumipulse plasma phospho-tau181 assay for Alzheimer’s disease. Alzheimers Res Ther. 2022;14. 10.1186/s13195-022-01116-2. [DOI] [PMC free article] [PubMed]
- 41.Thijssen EH, La Joie R, Strom A, et al. Plasma phosphorylated tau 217 and phosphorylated tau 181 as biomarkers in Alzheimer’s disease and frontotemporal lobar degeneration: a retrospective diagnostic performance study. Lancet Neurol. 2021;20:739–52. 10.1016/S1474-4422(21)00214-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karikari TK, Pascoal TA, Ashton NJ, et al. Blood phosphorylated tau 181 as a biomarker for Alzheimer’s disease: a diagnostic performance and prediction modelling study using data from four prospective cohorts. Lancet Neurol. 2020;19:422–33. 10.1016/S1474-4422(20)30071-5 [DOI] [PubMed] [Google Scholar]
- 43.Sarto J, Ruiz-García R, Guillén N, et al. Diagnostic Performance and Clinical Applicability of Blood-Based Biomarkers in a Prospective Memory Clinic Cohort. Neurology. 2022. 10.1212/WNL.0000000000201597. 10.1212/WNL.0000000000201597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brickman AM, Manly JJ, Honig LS, et al. Plasma p-tau181, p-tau217, and other blood-based Alzheimer’s disease biomarkers in a multi-ethnic, community study. Alzheimer’s and Dementia. 2021;17:1353–64. 10.1002/alz.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cullen NC, Leuzy A, Palmqvist S, et al. Individualized prognosis of cognitive decline and dementia in mild cognitive impairment based on plasma biomarker combinations. Nat Aging. 2021;1:114–23. 10.1038/s43587-020-00003-5 [DOI] [PubMed] [Google Scholar]
- 46.Janelidze S, Palmqvist S, Leuzy A, et al. Detecting amyloid positivity in early Alzheimer’s disease using combinations of plasma Aβ42/Aβ40 and p-tau. Alzheimer’s Dement. 2022;18:283–93. 10.1002/alz.12395 [DOI] [PubMed] [Google Scholar]
- 47.Palmqvist S, Stomrud E, Cullen N, et al. An accurate fully automated panel of plasma biomarkers for Alzheimer’s disease. Alzheimer’s Dement. 2023;19:1204–15. 10.1002/alz.12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez-Dubarbie F, Guerra-Ruiz A, López-García S, et al. Accuracy of plasma Aβ40, Aβ42, and p-tau181 to detect CSF Alzheimer’s pathological changes in cognitively unimpaired subjects using the Lumipulse automated platform. Alzheimers Res Ther. 2023;15:163. 10.1186/s13195-023-01319-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musso G, Cosma C, Zaninotto M, et al. Pre-analytical variability of the Lumipulse immunoassay for plasma biomarkers of Alzheimer’s disease. Clin Chem Lab Med. 2023;61:E53–6. 10.1515/cclm-2022-0770. 10.1515/cclm-2022-0770 [DOI] [PubMed] [Google Scholar]
- 50.Janelidze S, Barthélemy NR, He Y, et al. Mitigating the Associations of Kidney Dysfunction With Blood Biomarkers of Alzheimer Disease by Using Phosphorylated Tau to Total Tau Ratios. JAMA Neurol. Published Online First. 2023. 10.1001/jamaneurol.2023.0199. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw anonymized data and code for statistical analysis are available upon reasonable request. All requests should be sent to the corresponding author detailing the study hypothesis and statistical analysis plan. The steering committee of this study will decide whether data/code sharing is appropriate based on the novelty and scientific rigor of the proposal. All applicants will be asked to sign a data access agreement.