The publication of a draft sequence of 90% of the human genome1,2 heralds an exciting era in human genetics research. In the past 20 years, efforts have focused on mapping and cloning the genes for about 1000 human genetic disorders. This has led to the development of comprehensive services for prenatal diagnosis, carrier testing, and presymptomatic testing of mendelian disorders such as cystic fibrosis and the muscular dystrophies.3 Although this progress has been important to families affected by these diseases, it has had a limited effect on public health. All this could change if knowledge of 2.9 gigabases of the human genome sequence allows us to identify susceptibility genes for common diseases such as diabetes, asthma, and cancer. It may also lead to the identification of genetic variants that define a patient's response to a particular drug. If the promise of the genome sequence is even partially fulfilled, the next decade will see genetics spreading rapidly beyond the confines of specialist centres to impact on the diagnosis and management of common disorders in primary care.
Summary points
Genetic factors contribute substantially to the risk of developing many common diseases
Susceptibility genes for common disorders are being sought by genome scans and association studies in large patient cohorts
The publication of the sequence of the human genome and the discovery of millions of single nucleotide DNA polymorphisms have enhanced the prospects for identifying complex disease genes
Knowledge of such genes would permit identification of individuals at risk of particular diseases, improved preventive medicine, and tailoring of treatment to specific genetic profiles and disease subtypes
Single nucleotide polymorphism genotyping is likely to become part of the routine management of some common diseases within the next decade
Methods
This article is based primarily on reviews in Nature, Nature Genetics, Science, and New England Journal of Medicine (1999-2001) and on 20 years of personal experience in the mapping and cloning of genes for human disease and in the molecular diagnosis of human genetic disorders.
Are common diseases “genetic”?
Our risk of contracting common diseases is generally thought to be determined largely by environment and lifestyle. However, there is strong epidemiological evidence that genes contribute to overall risk. In multiple sclerosis, for example, the siblings of an affected person have a 25-fold increase in risk of developing the disease compared with the general population. Since relatives often share a common environment, the inheritability of a disease can also be assessed by comparing concordance rates for a disease in monozygotic and dizygotic twins. A higher concordance in monozygotic twins than dizygotic twins would suggest that the risk is determined mainly by genes. In multiple sclerosis, concordance is 30% for monozygotic twins and 3% for dizygotic twins, which shows that genes are important but that there is also a substantial environmental component.
Multiple sclerosis is an example of a complex disorder in which both genes and environment contribute to pathogenesis. There is also evidence that multiple genes contribute to disease susceptibility, each of which may confer a small increase in risk (perhaps up to fivefold). These conditions are also therefore referred to as polygenic disorders, and the susceptibility genes are called complex disease genes.
Why are complex disease genes important?
The modest increase in risk conferred by these genes, and the difficulties and costs involved in finding them, lead to the question of whether the investment is worth while. One important reason for finding them is that it might allow us to determine which people are at risk of a particular disorder.4 Such knowledge, although uncomfortable for people “at risk,” would be useful if it meant that they could avoid the environmental triggers that convert genetic susceptibility into disease. In addition, the identity of such genes should reveal much about the molecular pathways that lead to the disease state and thus identify new and relevant targets for drug treatment. The research would also lead to a molecular taxonomy of diseases that would permit drugs to be tailored to disease subtypes, greatly increasing their effectiveness.5
How can complex disease genes be found?
Genome scans
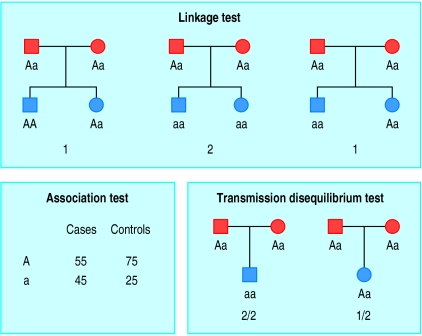
The search for complex disease genes begins by finding the chromosomal locations of the genes for disease susceptibility using linkage analysis.6,7 Figure 1 shows the principle of this approach. Families in which sibling pairs are affected with the disorder are typed with DNA polymorphisms (common variations of DNA sequence) to find a polymorphism that is coinherited with the disease in the pedigree. If excess sharing of the alleles of the polymorphism is found in large numbers of affected sibling pairs, the polymorphism is probably linked (close) to a gene that confers susceptibility to that disease. To find polymorphisms that are linked to the disease requires typing 200-300 families of affected sibling pairs with 300-400 polymorphisms that are evenly spaced along the human genome. The process is known as a genome scan.
Figure 1.
Testing for linkage: the two alleles of the polymorphism are indicated as A and a. Each parent is heterozygous for the polymorphism. If the two alleles were inherited randomly by the pairs of affected siblings in each family, we would expect them to share one of two (50%) parental alleles on average. The affected sibling pairs share four of six alleles (67%). Association test: the frequency (%) of the rarer allele of the single nucleotide polymorphism (allele a) is higher in cases than in controls. Transmission disequilibrium test: on average, we would expect each parent who was heterozygous (genotype Aa) to transmit alleles A and a to their affected offspring with equal frequency. In this example, allele a has been transmitted three out of a possible four times (75%), instead of the expected 50%
This approach has been used to “map” susceptibility genes for many complex diseases (table). However, the linkages reported by one group have often not been replicated by others. Failure to replicate linkages may be due to a lack of statistical power (that is, typing insufficient affected sibling pairs) or a false positive result in the original study. There might also be different sets of susceptibility genes operating in different populations. A degree of scepticism is appropriate until a linkage has been replicated in at least one large independent study.
Once linkage has been confirmed, the search for the critical gene within the linked region can begin. In common disorders, the region of linkage is generally large; thus, we may be looking for one gene in a region of 20-30 million base pairs, which is likely to contain 500-1000 genes. How can the relevant gene be identified in this genetic haystack?
Search for association
The next step is based on the premise that the susceptibility is caused by a mutation (variation in DNA sequence) in a gene that alters either its expression or the structure of the protein for which it codes. The mutation is likely to be common in the general population. This is in contrast with mutations in genes for mendelian single gene disorders, which are very rare. Also, it is hoped, but not yet proved, that one specific mutation will account for the susceptibility in most patients from a particular population.7 The mutation is likely to be a single nucleotide polymorphism (SNP), which is a common DNA sequence variant that alters only one base in a particular sequence of DNA. The second phase of the gene hunt is therefore to search the linked region for a single nucleotide polymorphism that shows association with the disease phenotype.5,7
The association can be tested in a conventional case-control study by looking for a difference in the frequency of one allele of a single nucleotide polymorphism between patients with the disease and suitably matched controls (fig 1). However, a common pitfall that can lead to false positive associations is a poor match of ethnic or geographical origins between cases and controls; frequencies of single nucleotide polymorphisms can vary widely between different populations, even within the British Isles. The other big problem is that sample numbers are often too small, which can lead to false positive (or false negative) results.
An alternative, family based approach, tests for preferential transmission of one allele of the single nucleotide polymorphism from heterozygous parents to an affected offspring. This is known as the transmission disequilibrium test. It avoids some of the problems involved in the selection of the control population (fig 1).
Both approaches are based on the assumption that the single nucleotide polymorphism being tested is the actual sequence variant that causes the genetic susceptibility or that it is in linkage disequilibrium with the true susceptibility allele. In linkage disequilibrium, one of the two alleles of the single nucleotide polymorphism is always (or almost always) associated with the true susceptibility allele and thus also shows association with the disease. The search for association may be random (testing single nucleotide polymorphisms at regular intervals across the critical region) or use a candidate gene approach, which tests single nucleotide polymorphisms within genes of particular interest. Candidate genes are selected on the basis of having a known or predicted function and expression profile that is consistent with the disease phenotype. Both approaches require testing of single nucleotide polymorphisms at frequent intervals because, in general, linkage disequilibrium is maintained over small genomic regions of about 5000-50 000 bp of DNA.
The importance of finding complex disease genes has led to initiatives to collect DNA samples from large numbers of patients with common disorders. In Britain, a prospective study of 500 000 adults from the general population has been established to examine the interaction of genes with environmental and lifestyle risk factors.
New technology
The search for the critical gene can be guided and prioritised by an electronic investigation of the region using bioinformatic analysis.8 The sequence of most of the linked region will be known but is of limited use unless it can be deciphered. Annotation is the process of converting raw DNA sequence data into biological knowledge. It predicts which bits of the sequence encode actual genes and what kinds of proteins are encoded by these genes. Gene prediction programs scan the sequence for general properties of protein coding sequences, while others search all available sequence databases for homology to known genes from other organisms. Much annotation has already been done, and gene maps for all chromosomal regions are available on public databases such as the National Centre for Biotechnology Information. The analysis may also allow the function of a new human gene to be deduced if its structure is homologous to a gene of known function in another organism. The databases also have information on the expression pattern of genes in tissues and organs, which provides further clues to possible function.
Additional educational resources
Collins FS. Shattuck lecture—medical and societal consequences of the human genome project. New Engl J Med 1999;341:28-37
Poste G, Bell J, Davies K, Goodfellow P, Hastie N, eds. Impact of genomics on healthcare. Br Med Bull 1999;55(2)
The human genome (special issue). Nature 2001;409:813-933
BMJ archive
Weatherall DJ. Single gene disorders or complex traits: lessons from the thalassaemias and other monogenic diseases. BMJ 2000;321:1117-20. http://bmj.com/cgi/content/full/321/7269/1117
Kwiatkowski D. Susceptibility to infection. BMJ 2000;321:1061-5. http://bmj.com/cgi/content/full/321/7268/1061
Josefen D. New gene implicated in type 2 diabetes. http://bmj.com/cgi/content/full/321/7265/854/a
Once a set of genes has been selected for study, they are screened for single nucleotide polymorphisms. The polymorphisms are then genotyped in large collections of patient DNA to determine whether they are associated with the disease. Most genes will contain several single nucleotide polymorphisms, some of which will already be known and entered on electronic databases. Additional single nucleotide polymorphisms can be found by scanning segments of the gene using one of the many methods now available for detecting mutations. An international consortium of pharmaceutical companies and the Wellcome Trust is searching for single nucleotide polymorphisms throughout the human genome and is making the data publicly available without restrictions. The consortium's December 2000 data release consists of 800 000 single nucleotide polymorphisms, and over 2.5 million have been submitted to the international National Center for Biotechnology Information database of single nucleotide polymorphisms.
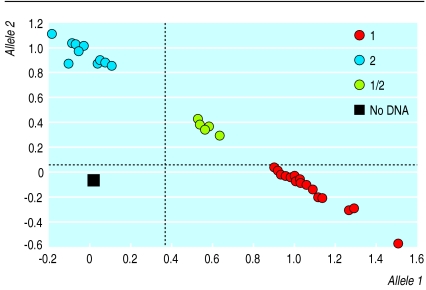
Analysis of genetic associations with common diseases requires technology that allows a high throughput. A typical study of one genetic locus might involve typing 1000 different single nucleotide polymorphisms in 2000 samples, generating 2 million genotypes. Various techniques are being developed for this purpose, including high density microarrays (or chips) and mass spectrometry.9,10 Figure 2 shows an example of one automated approach (TaqMan). This allows a plate of 96 DNA samples to be read in seconds, and the fluorescent signal in each well is instantly converted to a genotype. Existing techniques have the capacity to produce tens of thousands of genotypes from a single machine in one week. This means that once potentially useful genetic variants in a susceptibility gene have been identified, the technology is already in place to type them quickly and cheaply in large numbers of patients.
Useful websites
National Center for Biotechnology Information (www.ncbi.nlm.nih.gov)
National Coalition for Health Professional Education in Genetics (www.nchpeg.org)
Online mendelian inheritance in man (OMIM). A database and catalogue of human genes and genetic disorders compiled by Dr VA McKusick (www.ncbi.nlm.nih.gov/omim)
SNP Consortium Ltd (http://snp.cshl.org)
Figure 2.
TaqMan assay for genotyping single nucleotide polymorphisms: two short DNA probes are made, each of which is complementary to one of the two alleles of the polymorphism and labelled with a different fluorescent dye. A probe binds specifically to its target allele during the reaction, the dye is released, and the emitted fluorescence is measured. The ratio of the fluorescence from the two dyes in each sample tells us whether allele 1 (red), allele 2 (blue), or both alleles (green) are present
Progress and future perspectives
Progress in finding complex disease genes has been slow, but recent work gives grounds for optimism. In Alzheimer's disease, for example, a combination of linkage analysis and high plasma cholesterol concentrations in families with later onset Alzheimer's disease drew attention to the apolipoprotein E gene on chromosome 19. The e4 variant allele of apolipoprotein E was then shown to have a strong association with an increased risk of Alzheimer's disease.11 In white people, e4 heterozygotes have a twofold to threefold increased risk of Alzheimer's disease, and in e4 homozygotes the risk is 15-fold. Although this knowledge is not useful for predictive testing in unaffected individuals, since a cure for Alzheimer's disease is not yet available, it may help guide treatment. Patients with Alzheimer's disease who have the e4 subtype are less likely to benefit from treatment with the cholinomimetic drug tacrine than patients without this subtype.12
The past few months have seen considerable excitement in diabetes research. In 1996, a gene for type 2 diabetes was mapped by linkage analysis to human chromosome 2.13 Systematic analysis of single nucleotide polymorphisms in the linked region has now shown a strong association between a calpain protease gene, CAPN10 and type 2 diabetes.14 A strong association has also been reported between a single nucleotide polymorphism in the interleukin-12(p40) gene and type 1 diabetes.15 In these studies several single nucleotide polymorphisms in the candidate genes were strongly associated with the disease, so it is not yet clear which of them actually confers susceptibility.16 Final proof will depend on functional studies, such as showing a biological effect of a specific variant when it is transferred into cellular or animal models of the disease.
The pace of discovery of single nucleotide polymorphisms and the developments in high throughput genotyping should lead to many susceptibility genes for complex disorders being identified in the next 5-10 years. This knowledge might be applied in various clinical settings. To take a hypothetical example, a 45 year old woman who had hypertension diagnosed would have a buccal scrape sample sent for genetic testing to allow molecular classification of the disease as type I, II, or III. She would then be prescribed a specific drug and dietary regimen known to be effective in the relevant disease subtype. In a preventive setting, a 5 year old boy whose older sibling had developed asthma might be tested (with parental consent and appropriate counselling) for asthma susceptibility genes. If the results were positive, he might be prescribed prophylactic drugs to prevent asthma. Genotyping may therefore become part of the routine management of an expanding range of human diseases within the next 10 years.
Glossary
Association—The occurrence of a particular allele of a polymorphism in a group of patients more frequently than would be expected by chance
Complex disease—Both genetic and environmental factors contribute to pathogenesis. No clear mendelian pattern of inheritance is discernible
DNA polymorphism—A variation in DNA sequence that occurs in at least 1% of the general population. Most polymorphisms contribute to normal human diversity rather than disease, but a small proportion will contribute to susceptibility to common disorders
Linkage—Coinheritance of genetic markers (such as DNA polymorphisms) with the disease phenotype in families with multiple affected members. Consistent coinheritance of the marker with the disease in many families indicates that it is in close proximity to the actual disease gene, and is said to be “linked”
Polygenic disorder—Susceptibility to the disease is a consequence of the combined action of several different genes, each of which confers a moderate degree of risk
Single nucleotide polymorphism (SNP)—A DNA polymorphism that involves a change in a single base of a DNA sequence (for example, ACGT→AGGT). The human genome is likely to contain about 10 million single nucleotide polymorphisms
Table.
Chromosomal locations of some complex disease genes
| Disease | Chromosomal location |
|---|---|
| Alzheimer's disease | 12q, 19q13 |
| Asthma | 5q, 6p21, 12q |
| Type 1 diabetes | 5q, 6p21, 11p |
| Type 2 diabetes | 1q, 2q37 |
| Inflammatory bowel disease | 6p21, 12, 16q |
Acknowledgments
I thank P Braude, M Capra, R Robinson, and R Trembath for helpful comments on the manuscript, and Muddassar Mirza for providing the data in figure 2. Work in my laboratory is supported by the Wellcome Trust.
Footnotes
Competing interests: None declared.
References
- 1.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myerss EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Mathew CG. DNA diagnostics: goals and challenges. Br Med Bull. 1999;55:325–339. doi: 10.1258/0007142991902402. [DOI] [PubMed] [Google Scholar]
- 4.Collins FS. Shattuck lecture—medical and societal consequences of the human genome project. N Engl J Med. 1999;341:28–37. doi: 10.1056/NEJM199907013410106. [DOI] [PubMed] [Google Scholar]
- 5.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 6.Haines JL, Pericak-Vance MA. Approaches to gene mapping in complex human diseases. New York, Chichester: Wiley-Liss; 1998. [Google Scholar]
- 7.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 8.Searls DB. Bioinformatics tools for whole genomes. Annu Rev Genomics Hum Genet. 2000;1:251–279. doi: 10.1146/annurev.genom.1.1.251. [DOI] [PubMed] [Google Scholar]
- 9.Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21(suppl):42–47. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 10.Jackson PE, Scholl PF, Groopman JD. Mass spectrometry for genotyping: an emerging tool for molecular medicine. Mol Med Today. 2000;6:271–276. doi: 10.1016/s1357-4310(00)01698-1. [DOI] [PubMed] [Google Scholar]
- 11.Roses AD. Apolipoprotein E affects the rate of Alzheimer disease expression: beta-amyloid burden is a secondary consequence dependent on APOE genotype and duration of disease. J Neuropathol Exp Neurol. 1994;53:429–437. doi: 10.1097/00005072-199409000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Poirier J, Delisle MC, Quirion R, Aubert I, Farlow M, Lahiri D, et al. Apolipoprotein E4 allele as a predictor of cholinergic deficits and treatment outcome in Alzheimer disease. Proc Natl Acad Sci USA. 1995;92:12260–12264. doi: 10.1073/pnas.92.26.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanis CL, Boerwinkle E, Chakraborty R, Ellsworth DL, Concannon P, Stirling B, et al. A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 14.Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 15.Morahan G, Huang D, Ymer SI, Cancilla MR, Stephen K, Dabadghao P. Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat Genet. 2001;27:218–221. doi: 10.1038/84872. [DOI] [PubMed] [Google Scholar]
- 16.Altshuler D, Daly M, Kruglyak L. Guilt by association. Nat Genet. 2000;26:135–137. doi: 10.1038/79839. [DOI] [PubMed] [Google Scholar]