Abstract
Accumulated findings have indicated that hepatitis B virus (HBV) DNA integrates into the cellular DNA of HBV-infected chronic hepatitis tissues. The integrated sequence (IS) of HBV DNA at the virus-cell junction is conserved in a 25-bp region which is adjacent to direct repeat 1. A cellular protein which we purified from the nuclear extract of HepG2 cells binds to the IS and was designated IS binding protein 3 (ISBP3). The amino acid sequence of ISBP3 was determined and found to be identical to that of transcription initiation factor Yin and Yang 1 (YY1). An antibody against C-terminal amino acids of YY1 recognized ISBP3 in a Western blot analysis and an electrophoretic mobility shift assay. Furthermore, ISBP3 also interacted with Y3, which corresponds to the YY1 binding sequence, to enhance intramolecular recombination of polyomavirus DNA. Although YY1 is known as a transcription factor, the IS did not exhibit any effect on the transcription of precore and pregenome RNAs. The possible involvement of YY1 in the intramolecular recombination of linear replicative HBV DNA has been examined (Y. Hayashi et al., unpublished data). Data suggest that YY1 is involved in the joining reaction between HBV DNA and cellular DNA to form the virus-cell junction.
Accumulated findings have indicated that hepatitis B virus (HBV) DNA frequently integrates into the cellular DNA of HBV-infected chronic hepatitis tissues (3, 27, 33). For instance, random HBV DNA integration was detected in tissues from 15 of 16 chronic hepatitis cases (33). A detailed examination of integrated HBV DNAs indicates that the integrated sequence (IS) of HBV DNA at the virus-cell junction often resides in a 25-bp region which is adjacent to direct repeat 1 (DR1), while HBV DNA is integrated randomly into the cellular DNA. Almost all of the open reading frame of the X gene is preserved in a chromosome of the host cell. Integration is an essential step in the retrovirus life cycle, and the process is catalyzed by the viral enzyme integrase (4, 8, 16). On the other hand, integration is not essential in the life cycle (10) of HBV and HBV proteins do not catalyze the process as does integrase. Therefore, we focused on a cellular protein which plays a role in HBV DNA integration by interacting with the IS.
In this study, we detected a cellular protein which bound to the IS of HBV DNA at the virus-cell junction in the nuclear extract derived from HepG2 cells and was designated IS binding protein 3 (ISBP3). The amino acid sequence of ISBP3 was determined to be identical to that of the transcription factor Yin and Yang 1 (YY1). An antibody against the C-terminal amino acids of YY1 recognized ISBP3. ISBP3 was able to bind to the YY1 consensus sequence, as well as IS, in an electrophoretic mobility assay. YY1 is a highly conserved zinc finger protein which is able to initiate, activate, or repress the transcription of many viral, as well as cellular, genes (5, 13, 23, 26, 28, 29). However, the IS did not exhibit any specific effect on the transcription of precore and pregenome RNAs. Recent observations revealed that YY1 was able to bind Y3 of polyomavirus DNA to enhance intramolecular recombination of viral DNA in vivo (1, 11, 21, 22, 31). We therefore carried out a comparative electrophoretic mobility shift assay using the IS and Y3 as probes or competitors. Based on our findings, we concluded that ISBP3, as well as YY1, binds to the IS of HBV DNA at the virus-cell junction. In another study, we examined the possible involvement of YY1 in the intramolecular recombination of linear replicative HBV DNA (Y. Hayashi et al., unpublished data) and in the joining reaction between HBV DNA and cellular DNA.
MATERIALS AND METHODS
DNA fragments.
Fragments A, B, and D were synthesized by PCR using pHBVX-1 as described previously (18, 37). Other fragments were synthesized by using a DNA synthesizer (Applied Biosystems, Inc., Foster City, Calif.) and annealed. The sequences of those fragments are shown in the figures or figure legends.
Cell line and preparation of nuclear extract.
HepG2 is a hepatoblastoma cell line maintained in DM-160AU medium supplemented with 10% fetal calf serum and kanamycin (60 μg/ml) at 37°C in 5% carbon dioxide. The nuclear extract of HepG2 cells was prepared essentially as described by Dignam et al. (9), except that protein was solubilized from nuclei with 0.2 M (NH4)2SO4 instead of 0.42 M NaCl. Protein concentration was measured as described by Bradford (2).
Electrophoretic mobility shift assay.
Binding reactions were carried out with 10 μl of a buffer containing 12 mM HEPES/NaOH (pH 7.6), 60 mM KCl, 7.5 mM MgCl2, 12% glycerol, 0.1 mM EDTA, 0.3 mM dithiothreitol, 0.3 mM phenylmethylsulfonyl fluoride, 7 μg of poly(dA-dT) · poly(dA-dT), 3 × 104 cpm of a 32P-labeled probe, and 8 μg of nuclear extract and were kept at room temperature for 20 min before application to the gel. Electrophoresis was carried out with a 5% polyacrylamide slab gel at room temperature in a solution containing 22.5 mM Tris-HCl, 22.5 mM borate, and 1 mM EDTA at 150 V for 1.5 h with a prerun for 1 h. For the supershift assay, 300 ng of antibody was added to the reaction mixture, which was incubated on ice for 1 h prior to addition of the probe.
Purification of ISBP3.
Sequence-specific DNA affinity columns (5 ml) of wild-type and mutant DNAs were prepared by the method of Kadonaga and Tjian (15) using oligonucleotides made with positions 1676 to 1700 of HBV DNA. The sequence of the mutant IS DNA was derived by mutation from CACCATGCA to CTCGAAGGA. All purification procedures were carried out at 4°C. The nuclear extract (40 mg) supplemented with Tween 20 (final concentration of 0.1%) was applied to the mutant DNA column and washed with 25 ml of HEG buffer (20 mM HEPES/NaOH [pH 7.6], 0.2 mM EDTA, 20% glycerol, 0.1% Tween 20, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) containing 0.1 M NaCl. The flowthrough fraction was then diluted twofold with HEG buffer containing poly(dA-dT) · poly(dA-dT) at 13.5 μg/ml and loaded onto the DNA affinity column. The column was washed stepwise with 25 ml each of HEG buffer containing 0.05, 0.1, 0.15, 0.2, or 1 M NaCl. HEG buffer with 0.05 or 0.1 M NaCl was supplemented with poly(dA-dT) · poly(dA-dT) at a final concentration of 5 μg/ml. Binding activity was eluted with 0.15 M NaCl in HEG buffer. Throughout the purification step, DNA binding activity was monitored by electrophoretic mobility shift assay. The amount of protein was determined as described by Bradford (2) or silver staining on a polyacrylamide slab gel with a known concentration of bovine serum albumin as the standard. The N-terminal amino acid sequence was determined by automated Edman degradation and a Shimazu PPSQ-20 protein sequencer.
Western blot analysis.
Antibody against the 20 C-terminal amino acids of YY1 was obtained from Santa Cruz Biotechnology. Antibody against the N-terminal region of the YY1 protein was raised by injecting an oligopeptide corresponding to amino acids 19 to 35 of YY1 into a rabbit (Sawady Technology, Tokyo, Japan). The nuclear extract was separated by sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis and transferred electrophoretically onto an Immobilon membrane (Millipore Corporation, Bedford, Mass.). The membrane was then probed with antibodies and visualized by enhanced chemiluminescence immunodetection (Amersham Life Science, Buckinghamshire, England) in accordance with the manufacturer's instructions.
Plasmid DNAs, DNA transfection, and CAT activity assay.
To construct pEnh2/Cp-CAT and pEnh2/CpM1-CAT, HBV DNA fragments (nucleotides 1505 to 1700) were amplified by PCR using pHBVX-1 DNA (18) and primers with terminal HindIII sites. Products were then ligated into the HindIII site of pSV00CAT (Nippon Gene, Toyama, Japan). DNA transfection was carried out by calcium phosphate precipitation (32). The cells were plated at 6 × 106/10-cm-diameter dish, cultured for 16 h before transfection, incubated with DNA precipitates for 6 h at 37°C, and then subjected to a glycerol shock. After 2 days, cell extracts were prepared as described by Gorman et al. (12) and subjected to a chloramphenicol acetyltransferase (CAT) assay using [14C]chloramphenicol as the substrate (DuPont/NEN, Boston, Mass.). Chloramphenicol and its acetylated derivatives were separated by ascending thin-layer chromatography on a silica gel plate in acetic acid-methanol (95:5, vol/vol) and visualized by autoradiography.
RESULTS
Detection of binding proteins.
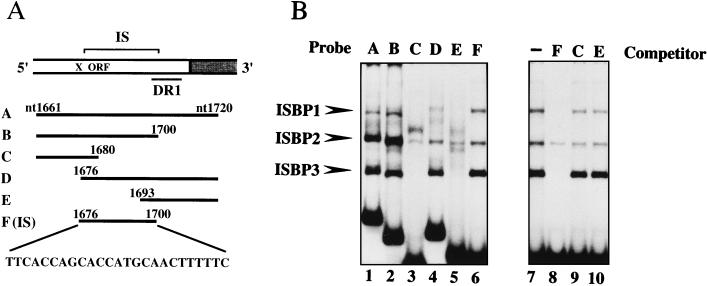
As a 25-bp region which is adjacent to DR1 of HBV DNA is frequently observed as an IS at the virus-cell junction (32) (Fig. 1A), we performed an electrophoretic mobility shift assay using end-labeled 60-bp DNA fragment A containing the IS and DR1 as a probe (Fig. 1A). The nuclear extract derived from HepG2 cells was found to contain several proteins which were able to bind to fragment A (Fig. 1B, lane 1). To define the binding sites of these proteins, we prepared a series of subfragments as probes for the electrophoretic mobility shift assay, as shown in Fig. 1A. All shifted bands were related to DNA fragment F, which corresponds exactly to the IS (Fig. 1B, lane 6). We tentatively named these binding proteins ISBP1, -2, and -3. Of these proteins, ISBP1 and -3 exhibited the same sequence-specific binding activity, since the formation of complexes was completely prevented by the addition of an excess amount of the unlabeled IS but not by DNA fragments C and E (Fig. 1B, lanes 7 to 10). A strong shifted band of ISBP3 was obtained compared to that of ISBP1, while ISBP2 was a nonspecific DNA binding protein. We therefore focused on ISBP3 for further analysis.
FIG. 1.
Detection of IS binding proteins. (A) Schematic illustration of the IS-containing region of HBV DNA adjacent to DR1. DR1 is one of two 11-bp direct repeats. The probes used in the electrophoretic mobility shift assay are shown. The numbers indicate nucleotide (nt) positions on the HBV DNA of the adr subtype (18). Probe F (IS) corresponds to the IS (nucleotides 1676 to 1700). (B) Electrophoretic mobility shift assay (lanes 1 to 6) with radiolabeled probes (60 fmol) and the nuclear extract derived from HepG2 cells (8 μg). Shifted bands are indicated with arrowheads. Competition assay without a competitor (lane 7) and with a 300-fold excess of the IS (lane 8), fragment C (lane 9), and fragment E (lane 10).
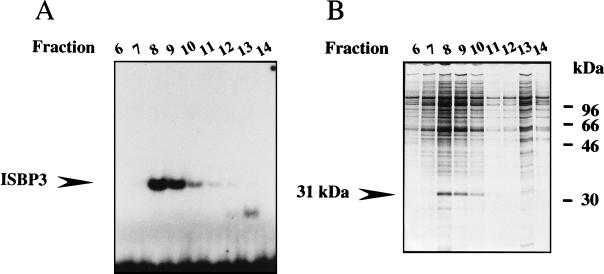
Affinity purification of ISBP3.
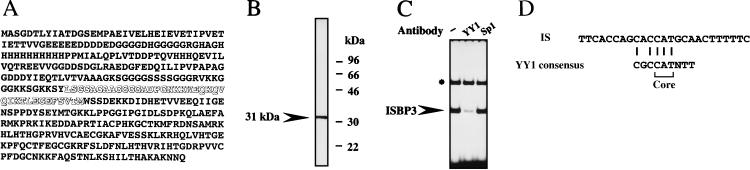
To characterize ISBP3 further, we attempted to purify it from the nuclear extract of HepG2 cells by monitoring the DNA binding activity using the electrophoretic mobility shift assay. After the nuclear extract was loaded onto the mutant DNA column, the flowthrough fraction was applied to the sequence-specific DNA affinity column. IS binding activity was eluted with loading buffer containing 150 mM NaCl. We identified a protein comigrating with the binding activity in the DNA affinity column (Fig. 2). The molecular mass of this protein was 31 kDa as determined by SDS-polyacrylamide gel electrophoresis. The sequence of the 38 N-terminal amino acids of this 31-kDa protein from the gel was determined. The sequence was completely identical to the middle portion of YY1 (amino acids 186 to 223) (Fig. 3A). A polyclonal antibody against the 20 C-terminal amino acids of YY1 recognized the 31-kDa protein in an active fraction of the DNA affinity column in a Western blot analysis (Fig. 3B). In addition, this antibody prevented formation of the complex in the electrophoretic mobility shift assay (Fig. 3C). Figure 3D shows that the binding sequence of ISBP3 has some homology with the YY1 consensus sequence including the 5′-CAT-3′ core, which is conserved in all of the known YY1 binding sites (14). YY1 is known as a multifunctional transcription factor with an apparent molecular mass of 68 kDa which plays a role in transcriptional initiation, as well as in transcriptional regulation (29). The results of the present study indicate that 31-kDa ISBP3 corresponds to the C-terminal half of the YY1 molecule, which retains the DNA binding activity.
FIG. 2.
Purification of ISBP3. (A) DNA binding activity of fractions eluted from sequence-specific DNA affinity column chromatography in an electrophoretic mobility shift assay using the IS as a probe. The position of ISBP3 is indicated. (B) SDS-polyacrylamide gel electrophoresis of eluted proteins. Proteins were subjected to SDS–12.6% polyacrylamide gel electrophoresis and stained with silver.
FIG. 3.
Recognition of ISBP3 by anti-YY1 antibody. (A) Amino acid sequence of YY1. The sequence of the 38 N-terminal amino acids of ISBP3 is indicated by open letters. This sequence is identical to the middle portion of YY1 (amino acids 186 to 223). (B) Western blot analysis using the active fraction eluted by DNA affinity column chromatography and a polyclonal antibody against the 20 C-terminal amino acids of YY1. (C) Electrophoretic mobility shift assay without an antibody (lane 1) and with 300 ng of an anti-YY1 antibody (lane 2) or an anti-Sp1 antibody (lane 3). The asterisk indicates ISBP2, a nonspecific DNA binding protein. (D) Comparison of the nucleotide sequence of the IS and the YY1 consensus sequence. The core is the sequence conserved in all of the known YY1 binding sites. Identical nucleotides are shown by vertical lines.
Detection of YY1.
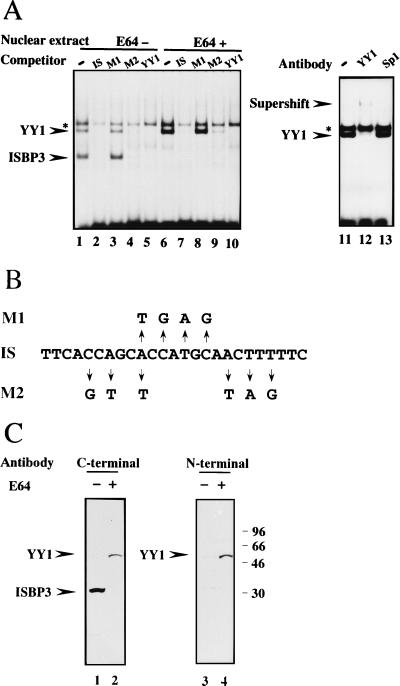
To test the possibility that ISBP3 is a degradation product of YY1, we added various protease inhibitors to buffers during preparation of the nuclear extract and performed the electrophoretic mobility shift assay. As a result, another shifted band with mobility lower than that of ISBP3 was predominantly detected in the nuclear extract prepared using buffers supplied with cysteine protease inhibitor E-64 while the shifted band due to ISBP3 disappeared (Fig. 4A, compare lanes 1 and 6). Complex formation was prevented by addition of the unlabeled IS, M2, and YY1 consensus oligonucleotide but not by addition of M1 (Fig. 4A, lanes 7 to 10). M1 and M2 are point mutant sequences derived from the IS which are shown in Fig. 4B. This sequence specificity of DNA binding activity is exactly the same with that of ISBP3 (Fig. 4A). An antibody against the 20 C-terminal amino acids of YY1 supershifted this complex, whereas an antibody against Sp1 had no effect on it (Fig. 4A, lanes 11 to 13). From these results, we concluded that ISBP3 is a degradation product of YY1 produced during preparation of the nuclear extract. Our finding is consistent with the previous report that two complexes due to YY1 and a degradation product were detected in the nuclear extract derived from HeLa cells (17).
FIG. 4.
Detection of YY1. (A) Electrophoretic mobility shift assay using the nuclear extract prepared with buffers supplied without E64 (lanes 1 to 6) or with E64 (lanes 6 to 13). Lanes: 1 and 6, without a competitor; 2 to 6 and 7 to 10, with a 300-fold excess of the IS, M1, M2, and YY1 consensus sequences, respectively; 11, without an antibody; 12, with anti-YY1 antibody; 13, with anti-Sp1 antibody. A supershifted band and ISBP2 (asterisk) are indicated. (B) Nucleotide sequences of IS, M1, and M2. (C) Western blot analysis using nuclear extract without E64 (lanes 1 and 3) or with E64 (lanes 2 and 4) and an antibody against the C-terminal 20 amino acids of YY1 (lanes 1 and 2) or against the N-terminal region of YY1, amino acids 19 to 36 (lanes 3 and 4).
The electrophoretic mobility shift assay using mutated oligonucleotides as competitors revealed that the binding protein has affinity for M2 but not for M1 (Fig. 4A). These data indicate that the middle portion of the IS is essential for binding, consistent with the fact that the middle portion is similar to the YY1 consensus sequence (Fig. 3D).
To identify a protein with lower mobility, we performed the Western blot analysis using both nuclear extracts. The molecular mass of a protein predominantly detected in the nuclear extract prepared using E64-supplied buffers is 54 kDa, as estimated by SDS-polyacrylamide gel electrophoresis (Fig. 4C), and which is slightly lower than the reported molecular mass of YY1 purified from the nuclear extract of HeLa cells. However, this 54-kDa protein was recognized by both antibodies against the 20 C-terminal amino acids and against the N-terminal region (amino acids 19 to 35) of YY1 in the Western blot analysis (Fig. 4C). Moreover, the Northern blot analysis using a part of the YY1 cDNA to probe the HepG2 mRNA revealed a single 2.6-kb band (data not shown) corresponding to the size reported before (28). In addition, the molecular weight of YY1 calculated from its amino acids is 45,000, which is closer to 54 kDa than 68 kDa. From these results, we concluded that the 54-kDa protein is full-length YY1. The reason for this discrepancy between the molecular weights is not known. It may be the result of various modifications depending on the cell types. Hereafter, we call the 54-kDa protein YY1.
Effects of the IS on transcription of the HBV genome.
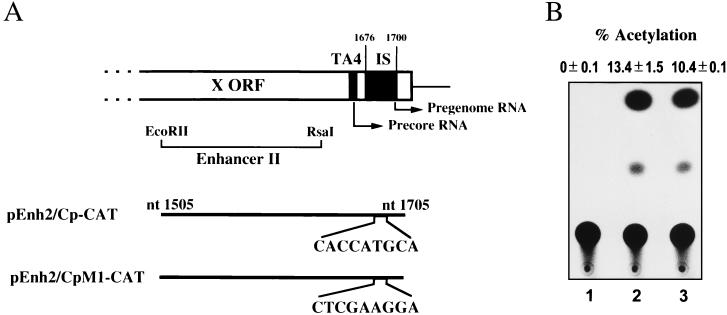
YY1 is known as a zinc finger protein which is able to initiate, activate, or repress the transcription of various genes. In some promoters, YY1 binding sites can positively modulate transcription if they are located near the transcriptional initiation site (13, 28). In the HBV genome, the IS is present near the transcriptional initiation site of two HBV RNAs, i.e., the precore and pregenome RNAs. To examine whether the IS has an effect on the transcriptional activity of the HBV genome, we constructed plasmids with the CAT-encoding gene under the control of TA4 and enhancer II. TA4 (nucleotide 1662 to 1667) is an incomplete TATA box in HBV DNA interacting with the TATA box binding protein and is sufficient to direct precise initiation of the precore and pregenome RNAs (6). The activity of TA4 is activated by enhancer II, which is adjacent to TA4 (7, 35, 39) (Fig. 5A). Transfection of the wild-type plasmid (pEnh2/Cp-CAT) into HepG2 cells resulted in high CAT activity. Even when a point mutant (pEnh2/CpM1-CAT) which failed to interact with YY1 in vitro was transfected, the CAT activity was not modulated (Fig. 5B). This result indicates that the IS-containing region has no effect on the transcription of the precore and pregenome RNAs in this context. This finding is consistent with the result showing that deletion of the IS-containing region exhibited no effect at all on the transcriptional activity in transfected HuH-7 cells (6).
FIG. 5.
Effects of the IS on transcription of precore and pregenome RNAs. (A) Schematic representation of plasmids containing the CAT-encoding gene controlled by enhancer II and TA4 of HBV DNA (pEnh2/Cp-CAT) or its mutant form (pEnh2/CpM1-CAT). The positions of the initiation sites of the precore and pregenome RNAs are indicated by arrows. nt, nucleotide. (B) CAT assay of lysates prepared from HepG2 cells transfected with pSV00CAT (lane 1), pEnh2/Cp-CAT (lane 2), or pEnh2/CpM1-CAT (lane 3).
Comparison of protein binding to IS and Y3.
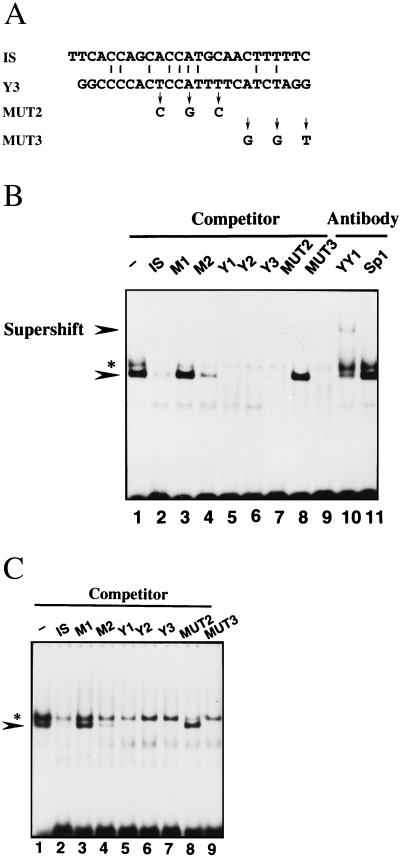
Bourgaux et al. (1) recently constructed an assay system for intramolecular recombination using modified polyomavirus DNA (RmI) and 3T6 cells (1, 11, 21, 22, 31). RmI is a circular viral DNA into which a 1,628-bp segment of mouse DNA was inserted and is converted into unit length viral DNA in the system. Polyomavirus DNA carries three YY1 binding sites, Y1, Y2, and Y3. Y3 is immediately adjacent to the recombination hot spot and has been shown to enhance intramolecular recombination of the viral DNA (11). Point mutations which abolished binding of YY1 to Y3 in vitro strongly depressed recombination, indicating that enhancement of Y3 was mediated by YY1 (11). They described YY1 as a protein which interacts with Y1, Y2, Y3, and MUT2 but not with MUT3. MUT2 and MUT3 are mutant derivatives of Y3; their sequences are shown in Fig. 6A. They also reported that the complex of YY1 and the Y3 probe was supershifted by a human monoclonal antibody against YY1 in an electrophoretic mobility shift assay. To confirm that the DNA binding proteins of the IS and Y3 exhibit the same binding specificity, we performed an electrophoretic mobility shift assay using an oligonucleotide corresponding to Y3 and the IS as probes. As previously reported, we observed complex formation, which was prevented by the addition of unlabeled Y1, Y2, Y3, and MUT2 but not by addition of MUT3 (Fig. 6B). Moreover, the anti-YY1 antibody supershifted the complex, while the anti-Sp1 antibody failed to affect it (Fig. 6B). When IS was used as a probe in the electrophoretic mobility shift assay, exactly the same binding specificity was observed (Fig. 6C). These results clearly indicate that YY1 interacts with IS in HBV DNA, as well as the Y3 site of polyomavirus DNA.
FIG. 6.
Comparison of binding proteins for the IS and Y3. (A) Nucleotide sequences of IS, Y3, MUT2, and MUT3. Y1, -2, and -3 are three YY1 binding sites in polyomavirus DNA. MUT2 and MUT3 are mutant derivatives from Y3. The sequence of Y1 is 6′-GCTTCAGAAGATGGCGGAGGGCCT-3′, and that of Y2 is 6′-TGCGGCTCCCATTTTGAAAATTCA-3′. (B) Electrophoretic mobility shift assay using the nuclear extract prepared with E64-containing buffers and radiolabeled Y3 as a probe. Lanes: 1, without a competitor; 2 to 9, with a 300-fold excess of the IS, M1, M2, Y1, Y2, Y3, MUT2, or MUT3, respectively; 10, with anti-YY1 antibody; 11, with anti-Sp1 antibody. A supershifted band is indicated. The asterisk indicates ISBP2, a nonspecific DNA binding protein. (C) Electrophoretic mobility shift assay using the radiolabeled IS as a probe. Lanes: 1, without a competitor; 2 to 9, with a 300-fold excess of the IS, M1, M2, Y1, Y2, Y3, MUT2, and MUT3, respectively.
DISCUSSION
In this study, we focused on the 54-kDa protein binding to integrated HBV DNA at the virus-cell junction. This protein has been detected as ISBP3 in the nuclear extract prepared from HepG2 cells using E64-containing buffer, although its molecular weight is slightly lower than that of YY1 purified from HeLa cells (28). The 54-kDa protein binds to the YY1 consensus sequence, as well as the IS, in an electrophoretic mobility shift assay. The complex of the protein and the IS was supershifted by addition of the anti-YY1 antibody. This protein was recognized by antibodies against both the 20 C-terminal amino acids and the N-terminal region (amino acids 19 to 35) of YY1 in a Western blot analysis. In addition, Northern blot analysis revealed a single 2.6-kb band (data not shown), suggesting that no other mRNA species was produced by alternative splicing. Taking these observations together, we concluded that the 54-kDa protein is YY1.
YY1 is known as a multifunctional transcription initiation factor of various viral and cellular genes (5, 13, 23, 26, 28, 29), and YY1 binding sites are often found near the sites of transcriptional initiation (13, 28). The IS to which YY1 binds is close to the initiation sites of the precore and pregenome RNAs, suggesting the possibility that YY1 regulates the promoter activity by interacting with the IS. However, the IS had no effect on the transcription of the RNAs in transfected HepG2 cells. In addition, Chen et al. demonstrated that the region carrying the IS had no effect on transcriptional activity using deletion mutants and HuH-7 cells (6). Although the mechanism by which YY1 activates, represses, or initiates transcription has not been fully elucidated, the context of the regulatory region and/or cell type is probably the primary factor. As the IS exhibited no effect on the transcription of genes in either HepG2 or HuH-7 cells, it is not a specific event for HepG2 cells. YY1 binding to the IS seems to be implicated in another function in this context.
Recently, the Y3 site, the third YY1 binding site in polyomavirus DNA, has been found to enhance intramolecular recombination of the viral DNA. Furthermore, it has been indicated that YY1 mediates this enhancement through binding to Y3 (11). Interestingly, we demonstrated that the same binding protein, YY1, interacts with both the IS and Y3. Comparing their surrounding DNA sequences, the situations are similar in the following points. (i) Both sequences are present immediately next to or exactly in the integration (recombination) hot spot in the viral DNA. (ii) Both sequences are close to two direct repeats. (iii) Both sequences are operative not in a chromosome of the host cell but in the viral DNA. Some of these common situations are likely essential for YY1 binding sequences to be involved in integration (recombination) because only a limited number of the YY1 binding sequences in the viral DNA are involved in integration (recombination). Actually, in the polyomavirus recombination system, point mutations which abolish binding of YY1 to Y3 in vitro strongly depressed recombination at proximal sites while having no effect on recombination at distal sites (11), indicating that it is necessary for YY1 binding sequences to be located near the recombination hot spot in order to enhance recombination. Thus, in the case of HBV DNA, YY1 may be essential for integration, consisting of intermolecular recombination between the viral and cellular DNAs.
Why is integration of HBV DNA prone to occur at the IS? The HBV genome consists of an incomplete double strand, and the minus strand has a breakage at the 5′ termini located near the IS (36, 24, 25, 34, 38) (Fig. 1A). This situation appears to facilitate joining of the 5′ terminus into a break in the double strand of the cellular DNA produced accidentally. It is possible that the break at the 5′ terminus in the viral genome is one of the reasons underlying the predisposition for integration to occur in the IS.
How would YY1 be involved in integration? Since YY1 has been shown to bend DNA (20), YY1 presumably creates a tertiary structure of HBV DNA suitable for integration by bending it. Alternatively, as YY1 has been found to associate with other cellular transcription factors, for example, Sp1, c-Myc, and E1A (19, 28, 30), YY1 bound to the IS might associate with other transcription factors bound to the cellular DNA. In this way, YY1 could bring HBV DNA near to the cellular DNA, which would allow the DNA strand of HBV to transfer into that of the host cell. If YY1 associates with a repair protein which binds to a double-strand break in the cellular DNA, the efficiency of the end-joining reaction would increase. The recombination functions of the IS and YY1, which binds to the IS, remain obscure. In the replication cycle of duck hepatitis B virus DNA, the double-stranded linear form of the viral DNA is generated as a minor replicative intermediate which is efficiently converted to covalently closed circular DNA by intramolecular recombination (38). Construction of an assay system for recombination of linear replicative HBV DNA and identification of the proteins that associate with YY1 would help us to better understand their roles in recombination and integration as well. Such an experiment has been carried out in another study, the results of which suggest that the YY1 binding site is required for intramolecular recombination between terminally repeated sequences of linear replicative HBV DNA (Hayashi et al., unpublished data).
ACKNOWLEDGMENTS
This work was supported partly by a grant-in-aid from the Ministry of Education, Science, Sports and Culture and a grant-in-aid from the Ministry of Health and Welfare, Japan, to K.K.
REFERENCES
- 1.Bourgaux-Ramoisy D, Gendron D, Bourgaux P. A hotspot for promoter-dependent recombination in polyomavirus DNA. J Mol Biol. 1995;248:220–224. doi: 10.1016/s0022-2836(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Bréchot C, Hadchonuel M, Scotto J, Degos F, Charnay P, Trepo C, Tiollais P. Detection of hepatitis B virus DNA in liver and serum: a direct appraisal of the chronic carrier state. Lancet. 1981;ii:765–768. doi: 10.1016/s0140-6736(81)90182-3. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushmeyer S, Park K, Atchison M L. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 6.Chen I-H, Hung C-J, Ting L-P. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69:3647–3657. doi: 10.1128/jvi.69.6.3647-3657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S-T, Porte P L, Yee J-K. Mutational analysis of hepatitis B virus enhancer 2. Virology. 1993;196:652–659. doi: 10.1006/viro.1993.1521. [DOI] [PubMed] [Google Scholar]
- 8.Craigie R, Fujiwara T, Busman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R D. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem D, Varmus H E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 11.Gendron D, Delbecchi L, Bourgaux-Ramoisy D, Bourgaux P. An enhancer of recombination in polyomavirus DNA. J Virol. 1996;70:4748–4760. doi: 10.1128/jvi.70.7.4748-4760.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hariharan N, Kelley D E, Perry R P. δ, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyde-DeRuyscher R P, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadonaga J T, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz R A, Merkel G, KulKosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 17.Klug J, Beato M. Binding of YY1 to a site overlapping a weak TATA box is essential for transcription from the uteroglobin promoter in endometrial cells. Mol Cell Biol. 1996;16:6398–6407. doi: 10.1128/mcb.16.11.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi M, Koike K. Complete nucleotide sequence of hepatitis B virus DNA of subtype adr and its conserved gene organization. Gene. 1984;30:227–232. doi: 10.1016/0378-1119(84)90124-0. [DOI] [PubMed] [Google Scholar]
- 19.Lee J-S, Galvin K M, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci USA. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natesan S, Gilman M Z. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 21.Nault C, Fricker A, Delbecchi L, Bourgaux-Ramoisy D, Bourgaux P. Intramolecular recombination in polyomavirus DNA is a nonconservative process directed from the viral intergenic region. J Virol. 1994;68:5439–5447. doi: 10.1128/jvi.68.9.5439-5447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nault C, Veilleux S, Delbecchi L, Bourgaux-Ramoisy D, Bourgaux P. Intramolecular recombination in polyomavirus DNA is controlled by promoter elements. Nucleic Acids Res. 1994;22:485–491. doi: 10.1093/nar/22.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park K, Atchison M L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain μE1 site. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeger C, Maragos J. Identification and characterization of the woodchuck hepatitis virus origin of DNA replication. J Virol. 1990;64:16–23. doi: 10.1128/jvi.64.1.16-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seeger C, Maragos J. Identification of a signal necessary for initiation of reverse transcription of the hepadnavirus genome. J Virol. 1991;65:5190–5196. doi: 10.1128/jvi.65.10.5190-5195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–246. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 27.Shafritz D A, Shouval D, Shermann H I, Hadziyannis S J, Kew M C. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma. N Engl J Med. 1981;305:1067–1073. doi: 10.1056/NEJM198110293051807. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Seto E, Chang L-D, Shenk T. Transcriptional repression by YY1, a human GLI-kruppel-related protein and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 29.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcription regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shrivastava S, Saleque S, Kalpana G V, Artandi S, Goff S P, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 31.Sylla B S, Huberdeau D, Bourgaux-Ramoisy D, Bourgaux P. Site-specific excision of integrated polyoma DNA. Cell. 1984;37:661–667. doi: 10.1016/0092-8674(84)90398-2. [DOI] [PubMed] [Google Scholar]
- 32.Takada S, Koike K. Trans-activation function of a 3′ truncated X gene-cell fusion product from integrated hepatitis B virus DNA in chronic hepatitis tissues. Proc Natl Acad Sci USA. 1990;87:5628–5632. doi: 10.1073/pnas.87.15.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takada S, Gotoh Y, Hayashi S, Yoshida M, Koike K. Structural rearrangement of integrated hepatitis B virus DNA as well as cellular flanking DNA is present in chronically infected hepatic tissues. J Virol. 1990;64:822–828. doi: 10.1128/jvi.64.2.822-828.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavis J E, Ganem D. RNA sequences controlling the initiation and transfer of duck hepatitis B virus minus-strand DNA. J Virol. 1995;69:4283–4291. doi: 10.1128/jvi.69.7.4283-4291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Chen P, Wu X, Sun A-L, Wang H, Zhu Y-A, Li Z-P. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol. 1990;64:3977–3981. doi: 10.1128/jvi.64.8.3977-3981.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Will H, Reiser W T, Weimer T, Pfaff E, Buscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Summers J. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yee J-K. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science. 1989;246:658–661. doi: 10.1126/science.2554495. [DOI] [PubMed] [Google Scholar]