Abstract
Entry into the host cell by enveloped viruses is mediated by fusion (F) or transmembrane glycoproteins. Many of these proteins share a fold comprising a trimer of antiparallel coiled-coil heterodimers, where the heterodimers are formed by two discontinuous heptad repeat motifs within the proteolytically processed chain. The F protein of human respiratory syncytial virus (RSV; the major cause of lower respiratory tract infections in infants) contains two corresponding regions that are predicted to form coiled coils (HR1 and HR2), together with a third predicted heptad repeat (HR3) located in a nonhomologous position. In order to probe the structures of these three domains and ascertain the nature of the interactions between them, we have studied the isolated HR1, HR2, and HR3 domains of RSV F by using a range of biophysical techniques, including circular dichroism, nuclear magnetic resonance spectroscopy, and sedimentation equilibrium. HR1 forms a symmetrical, trimeric coiled coil in solution (K3 ≈ 2.2 × 1011 M−2) which interacts with HR2 to form a 3:3 hexamer. The HR1-HR2 interaction domains have been mapped using limited proteolysis, reversed-phase high-performance liquid chromatography, and electrospray-mass spectrometry. HR2 in isolation exists as a largely unstructured monomer, although it exhibits a tendency to form aggregates with β-sheet-like characteristics. Only a small increase in α-helical content was observed upon the formation of the hexamer. This suggests that the RSV F glycoprotein contains a domain that closely resembles the core structure of the simian parainfluenza virus 5 fusion protein (K. A. Baker, R. E. Dutch, R. A. Lamb, and T. S. Jardetzky, Mol. Cell 3:309–319, 1999). Finally, HR3 forms weak α-helical homodimers that do not appear to interact with HR1, HR2, or the HR1-HR2 complex. The results of these studies support the idea that viral fusion proteins have a common core architecture.
Human respiratory syncytial virus (RSV) is an important cause of lower respiratory tract infections in infants, the elderly, and immunocompromised individuals. RSV is an enveloped virus with three transmembrane surface proteins, G, F, and SH. While all of these proteins are reported to be required for efficient fusion (26), the F (or fusion) glycoprotein is thought to be the major mediator of this event (33). Indeed, F is the only surface protein that is essential for viral replication in vitro and in vivo (5, 56, 60). F shares many features with homologous proteins in other paramyxoviruses (reviewed in reference 36). It is a type I glycoprotein that becomes active only after intracellular proteolytic cleavage, forming two disulfide-linked subunits, F1 and F2. The newly formed N terminus of F1 is believed to be the fusion peptide, and this sequence is separated from a transmembrane domain by ∼375 residues. Immediately adjacent to the fusion peptide and the transmembrane domains (see Fig. 1) are two heptad repeat (HR) regions, HR1 and HR2, respectively. All of these features are common in other fusion-mediating proteins of enveloped viruses.
FIG. 1.
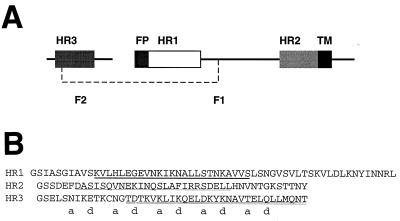
HR peptides from RSV fusion protein. (A) Schematic diagram of the RSV F protein showing F1 and F2 and the positions of HR1, HR2, HR3, the fusion peptide (FP), and the transmembrane region (TM). The dashed line represents a disulfide bond. (B) Sequences of the HR peptides used in this study. The N-terminal Gly-Ser sequence is derived from the pGEX-2T vector. The C-terminal Tyr residue in HR2 is nonviral and was designed to give the peptide an absorbance at 280 nm. Predictions of coiled-coil formation are derived from the program MultiCoil (65) and are represented as follows: strong dimer formation (double underline), moderate trimer formation (bold underline), and weak dimer or trimer formation (single underline).
A number of mutational studies have shown that the HR regions HR1 and HR2 are necessary for viral fusion (4, 12, 14, 49). Although their precise role in fusion is not yet known, structural studies on fragments from a range of viral fusion or transmembrane proteins have shown that these regions have a common, structurally important role. The structures of gp41 from simian immunodeficiency virus (SIV) and human immunodeficiency virus (HIV) (7, 10, 40, 55, 59), hemagglutinin (HA) from influenza A (6, 63), F from simian virus 5 (SV5 [1]), TM from Moloney murine leukemia virus (17), GP2 from Ebola virus (41, 58), gp21 from human T-cell leukemia virus type 1 (35), and HEF of influenza C virus (51) are all trimeric, centered around a trimeric coiled coil of the three HR1 regions. In some of these structures (e.g., gp41, HA, SV5 F, and Gp2), the HR2 region is resolved as a second layer of helices that packs in an antiparallel manner against the trimeric HR1 coiled coil.
Arguably, the best model for the mechanism of action of a fusion protein comes from studies of HA (for a recent review, see reference 13) for which structures of both a low pH (fusogenic [6]) and neutral pH (nonfusogenic [63]) form have been solved. These different forms have different conformations, suggesting that fusion is mediated by a conformational transition that occurs when a metastable neutral pH form of HA encounters the acidic environment in which fusion takes place. Although compelling analogies have been made on the basis of structural similarities between fusion proteins (see, for example, reference 10), it is not yet clear whether similar “spring-loaded” mechanisms are utilized by other viruses.
There is some evidence to suggest that HR regions from some paramyxovirus F proteins can associate with lipids, suggesting that these regions, while playing a structural role, might also assist in the fusion mechanism by binding to the membrane of target cells (2, 22, 44). Given that the HR regions form amphipathic helices in solution, and that these structures have a general tendency to bind lipids, such observations are perhaps not surprising; it remains to be confirmed whether this activity has biological significance in the case of the HR peptides.
Peptides corresponding to some of these HR regions have been found to specifically inhibit viral fusion and/or infection in vitro (27, 28, 37, 48, 61, 62, 67–69). Furthermore, it has been shown that a peptide (known as T-20 or DP-178) corresponding to the HR1 region of gp41 is safe and provides potent inhibition of HIV replication when injected into human patients (34). While these peptides could potentially bind to the lipid bilayer, host cell proteins, or various sites on the viral transmembrane proteins, a commonly invoked model involves direct binding to the complementary HR region in an intermediate form of the F glycoprotein (see, for example, reference 19). This model for inhibition is consistent with proposed mechanisms of fusion that involve changes in conformation of the F protein (see reference 1 and references therein). There is compelling evidence that the peptides and their synthetic analogues act in a dominant negative manner and inhibit fusion through interference with the intramolecular HR1 and HR2 interaction required for formation of the hexamer (see references 16, 18, 19, and 50 and references therein).
In RSV F, the positions of HR1 and HR2 are homologous to those found in all identified fusion proteins. HR1 (corresponding to the “core” peptide) lies immediately adjacent to the putative fusion peptide at the N terminus of F1. HR2 lies adjacent to the predicted transmembrane region of F1. A third heptad region, HR3, is predicted to form in the F2 subunit of F (37). Note that HR3 is not expected to correspond to putative leucine zipper regions that have been identified between the HR1 and HR2 regions in the F1 subunit of a number of other paramyxoviruses, including Sendai virus, rinderpest, measles, parainfluenza, and SV5 (20). The role of HR3 in RSV F has yet to be identified, and although HR2 has been shown to inhibit viral fusion (37), its mode of action and whether it interacts with HR1 and/or with HR3 have not yet been established.
In this paper, we have attempted to address these deficiencies. We have produced recombinant peptides corresponding to HR1, HR2, and HR3 and have studied their conformations and their ability to form stable complexes with themselves and with each other. We show that HR1 does form a helical trimer and that this trimer interacts with monomeric HR2 to form a hexameric complex that we believe to be the core of a fusion active trimeric F protein. HR3 does not appear to interact with HR1, HR2, or the HR1-HR2 hexamer, and instead, it is anticipated to have an independent structural role.
MATERIALS AND METHODS
Production of HR1-HR2 peptides.
Synthetic genes were constructed for the production of recombinant HR1, HR2, and HR3. Oligonucleotides (∼70 bases; Gibco Life Sciences) with overlaps of 20 bp were designed such that they corresponded to the amino acid sequences of HR1, HR2, and HR3. HR2 contains an additional nonviral C-terminal tyrosine residue to enable detection of the peptide at 280 nm. Codon usage reflected that of highly expressed Escherichia coli genes.
The synthetic genes were generated in a single step using PCR and were further amplified using PCR. These inserts were then cloned, via BamHI and EcoRI restriction sites, into the E. coli expression vector pGEX-2T (Pharmacia), creating C-terminal fusions with glutathione-S-transferase (GST). The GST fusion proteins were expressed in the host strain BL21(DE3) grown in Luria broth. Cells were grown at 25 or 37°C, and expression of the recombinant proteins was induced at an A600 of between 0.6 and 0.8 by the addition of isopropyl-β-d-thiogalactoside (0.1 to 0.4 mM). After a further 3 to 4 h, cells were harvested by centrifugation and stored at −80°C. In the case of 15N-labelled HR1, cells were grown in minimal media containing 15N-labelled NH4Cl as the sole nitrogen source, according to the method of Cai et al. (8).
Cell pellets were resuspended in lysis buffer containing Tris-HCl (50 mM, pH 8), EDTA (1 mM), NaCl (150 mM), phenylmethylsulfonyl fluoride (0.5 μM), and β-mercaptoethanol (2 mM). The suspension was incubated on ice for 30 min, treated with Triton X-100 to 1%, and further incubated on ice for 30 min. Cells were lysed by sonication and pelleted by centrifugation. The supernatant was loaded onto an S-hexylglutathione column (Pharmacia) preequilibrated in phosphate-buffered saline (PBS) (20 mM NaH2PO4, 150 mM NaCl, pH 7.4) containing 1 mM EDTA and 0.5 μM phenylmethylsulfonyl fluoride. The column was washed, and the recombinant peptides were cleaved from the column with thrombin (20 U per 1-liter culture) at 37°C for 1 h. The eluted peptides were purified to homogeneity by reversed-phase high-performance liquid chromatography (HPLC), and their identity was confirmed using electrospray-mass spectrometry (ES-MS). Molar extinction coefficients were estimated using the method described by Gill and von Hippel (23).
Preparation of HR3-IAM.
HR3 peptide was dissolved in a solution containing 0.5 M Tris-HCl (pH 8.0), 1 mM EDTA, 6 M GdnHCl, and 5 mM dithiothreitol, and the solution was incubated at room temperature for 2 h in order to fully reduce any disulfide bonds. Reduced HR3 was then treated with 20 mM iodoacetamide for 40 min at room temperature in the dark, and the alkylation reaction was quenched by the addition of excess β-mercaptoethanol. The modified peptide was desalted and purified by reversed-phase HPLC using a Waters radial compression semipreparative column and a linear acetonitrile-trifluoroacetic acid gradient. The mass of the modified peptide corresponded to the addition of a single acetamide group.
CD spectropolarimetry.
Circular dichroism (CD) spectra were recorded on a Jasco J-720 spectropolarimeter over the wavelength range of 184 to 260 nm with a resolution of 0.5 nm and a bandwidth of 1 nm. The temperature was maintained at 25°C using a water-jacketed cell holder. Final spectra were the sums of three to five scans accumulated at a speed of 20 nm min−1 with a response time of 1 s and were baseline corrected. Path lengths were 0.1, 1, or 10 mm for >50 μM, 20 to 50 μM, or <20 μM solutions, respectively.
Peptide samples (10 to 20 μM) were prepared by dissolving each peptide in water and then adding 20 mM NaH2PO4 (pH 6.0 or 7.4) either with or without 150 mM NaCl. The proportions of different secondary structural elements were predicted using the programs Prosec (Aviv; with reference to the standard spectra of Yang et al. [66]) and CDsstr (30).
Analytical ultracentrifugation.
Sedimentation equilibrium experiments were carried out at 25°C using a Beckman Optima XL-A ultracentrifuge equipped with an Anti-60 rotor. Samples were either dialyzed against PBS (20 mM NaH2PO4, pH 7.4, 150 mM NaCl) or were passed through an analytical gel filtration column (Superdex Peptide PC 3.2/30; Pharmacia) using PBS as a buffer and at a flow rate of 50 to 100 μl/min. Samples were prepared at three different protein concentrations (by serial dilution) so that they had absorbances at 280 or 230 nm of ∼1.0, 0.3, and 0.1 (in 1.2-cm-path-length cells). Data were recorded at three different speeds in double-sector cells as absorbance-versus-radius scans (0.001-cm increments, 10 scans). Scans were collected at intervals of 3 h and compared to ensure that the sample reached equilibrium. Analysis of the data was carried out using the NONLIN (29) and OMMENU (47) software, and the final parameters were determined by a nonlinear least-squares fit to models incorporating either a single, nonassociating species or a reversible oligomerization reaction. The goodness-of-fit was determined by examination of the χ2 values and the residuals derived from the fit. The partial specific volume of each peptide was calculated from the amino acid sequence using the program SEDNTERP (25), and the solvent density was calculated from tabulated values for each major solution component.
1H NMR spectroscopy.
Peptides were dissolved in a solution (0.55 ml) containing a 95:5 ratio of H2O to D2O and 20 μM d4-(trimethylsilyl)propionic acid (d4-TSP) and adjusted to pH 5.5 using 0.1 M NaOH. Nuclear magnetic resonance (NMR) spectra were recorded at 25°C on a Bruker DRX600 equipped with a triple resonance (HCN) probe and three-axis pulsed-field gradients. One-dimensional 1H data were acquired with 16K complex data points and 64 to 128 scans and over a spectral width of 7,200 Hz. The solvent signal was suppressed using a WATERGATE sequence (45) immediately prior to data acquisition. 15N-HSQC (43) and HNHA (57) experiments were recorded using published sequences. All spectra were acquired with spectral widths of 1,800 and 7,200 Hz in the 15N and 1H dimensions, respectively. Acquisition times of 142 ms (1H, F3), 8.9 ms (1H, F2), and 26 ms (15N, F1) were used in the HNHA experiment.
The data were processed by zero-filling once, linear predicting (for indirectly acquired dimensions), and applying either Lorentzian-Gaussian or squared shifted sine bell window functions prior to Fourier transformation. A polynomial baseline correction was applied in the directly detected dimension. The 1H frequency scale of all spectra was directly referenced to d4-TSP at 0.00 ppm, while the 15N frequency scale was indirectly referenced to liquid NH3 by using the method described by Live et al. (39). 3JHNHα coupling constants were extracted from the HNHA spectrum as described by Vuister and Bax (57).
Limited proteolysis.
Peptides were dissolved in PBS (pH 7.4) to a final concentration of 1 mg ml−1 and were treated with thermolysin (thermolysin-to-peptide ratio, 1:10 [wt/wt]). The reaction was monitored using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Digested peptides from time points at 60 and 120 min were separated and purified using reversed-phase HPLC on a Vydac C18 column (4.1 mm by 250 mm) with a linear acetonitrile-trifluoroacetic acid gradient. After lyophilization, the masses of peptides in the main peaks were determined by ES-MS on a Finnegan LCQ mass spectrometer, using an aqueous solution of 50% MeOH–1% acetic acid and a direct infusion rate of 3 μl min−1.
RESULTS
Production of HR peptides.
The sequence of the F protein (SWISS-PROT accession number P13843) of RSV (subgroup B, strain 18537) was submitted to the program MultiCoil (65) in order to determine whether any regions of the protein were likely to form dimeric and trimeric coiled coils. Three such regions were identified, all of which corresponded closely to previously identified sequences of HR1, HR2, and HR3 regions (RSV A, strain Long [37]). However, the details of the predictions for each of these regions differed. HR1 was predicted to be trimeric, HR2 was weakly predicted to be either dimeric or trimeric, and HR3 was strongly predicted to be dimeric. The score obtained for the predicted HR3 coiled coil was far in excess of those obtained for HR1 and HR2, or any predicted non-HR1-HR2 coiled-coil regions from all other paramyxovirus F proteins that were subjected to this analysis (data not shown). The HR1, HR2, and HR3 regions of RSV F and the sequences of the recombinant peptides used in this study are shown in Fig. 1. To increase the likelihood of the recombinant peptides forming native-like structure, their lengths were chosen such that all residues in each region that had a nonzero propensity to form coiled coils were included, i.e., the peptides were designed to extend beyond the core (underlined) sequences. It is notable that the region of HR2 that was predicted to have the highest likelihood of forming a coiled coil (Fig. 1B, thin underline) corresponded exactly with the sequence that is reported to most strongly inhibit syncytium formation (37). Synthetic genes corresponding to the protein sequences of HR1, HR2, and HR3 were assembled and subcloned into the pGEX-2T vector. The peptides were expressed as GST fusion proteins, purified using glutathione affinity chromatography, cleaved with thrombin, and purified to homogeneity using reversed-phase HPLC. Note that the HR3 sequence contains a single cysteine residue. To avoid the potential problem of disulfide formation, a cysteine-modified form of this peptide was generated by either iodoacetamide treatment (to form HR3-IAM) or reduction with the reducing agent Tris(2-carboxyethyl)phosphine (TCEP). The masses of the purified peptides were confirmed using electrospray mass spectrometry.
CD analysis of HR peptides.
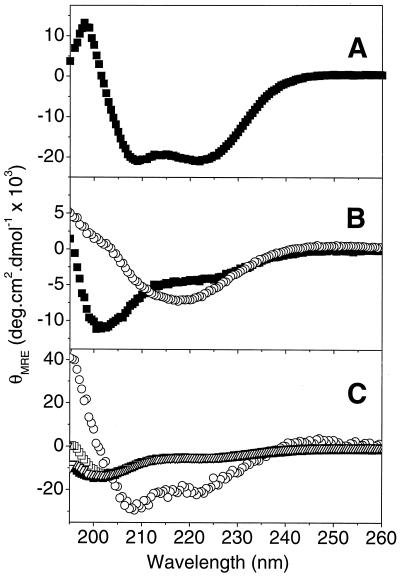
Far-UV CD spectropolarimetry is a good indicator of the type of secondary structure contained in proteins and peptides. The far-UV CD spectrum of HR1 (Fig. 2A) exhibited minima at 208 and 222 nm, indicating that it is predominantly α-helical (38 to 55% helical content depending on the program used to predict secondary structure content). The appearance of the spectrum was not affected by changes in protein concentration in the range of 2 to 120 μM, indicating that there were no major concentration-dependent folding transitions taking place across this range. In contrast to HR1, HR2 appeared to be largely unfolded, with a characteristic minimum at ∼200 nm (Fig. 2B). A shoulder was observed in the HR2 spectrum at 222 nm, suggesting the presence of a small amount of secondary structure. The HR2 secondary structure was predicted to be a mixture of α-helix and β-sheet by the programs CDsstr and Prosec. Quantitation of the HR2 secondary structure elements was not attempted due to sample-to-sample variations and differences in output between the two programs. However, after extended periods at room temperature or at high protein concentrations, marked changes were observed in the HR2 spectrum (Fig. 2B). A broad minimum appeared at ∼218 nm together with a maximum at 190 nm; these features are characteristic of β-sheet conformations, and the secondary structure prediction software estimated a β-sheet content of ∼70%.
FIG. 2.
Far-UV CD spectra of HR1, HR2, and HR3. Shown are 20 μM HR1 (A); 20 μM HR2 normal (■) and aged (○) (B); and HR3-IAM at concentrations of 1.8 μM (■), 20 μM (▿), 33 μM (□), and 110 μM (○) (C). Peptides were buffered in a solution containing 150 mM sodium chloride and 20 mM sodium phosphate (pH 7.4). Spectra are the averages of three to five scans and are baseline corrected.
While HR3 also appeared to be largely unfolded by far-UV CD at low concentrations (8 to 9% helix at 2 μM), the CD spectrum of this peptide was strongly concentration dependent. The α-helical content of HR3-IAM was found to increase significantly at concentrations above 20 μM, with estimates of 49 to 57% α-helix at 110 μM (Fig. 2C). Essentially identical results were observed for HR3 in the presence of TCEP (HR3-TCEP; data not shown). For all peptides, there were no apparent differences between spectra recorded at pHs 6.0 and 7.4 or between spectra recorded in the presence or absence of 150 mM NaCl (data not shown).
Oligomeric state of HR peptides.
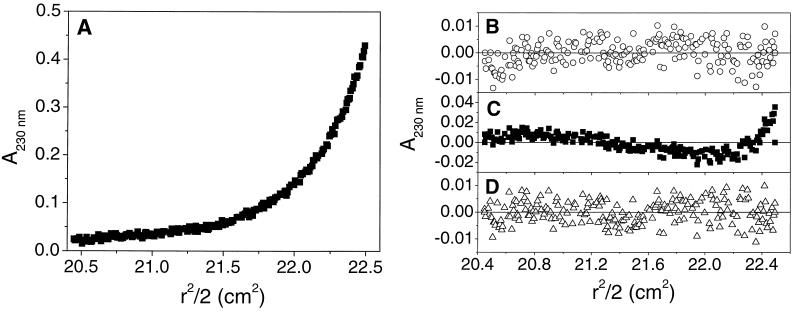
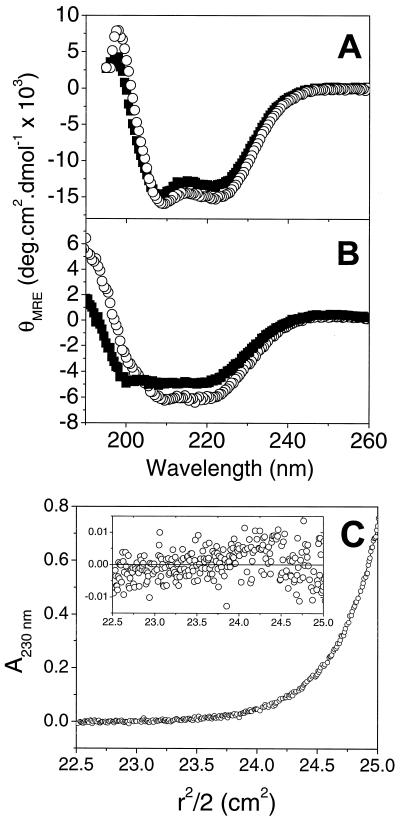
Sedimentation equilibrium experiments were carried out to determine the oligomeric states of the HR peptides. The HR1 peptide was studied over two different concentration ranges (with three different loading concentrations in each case) and at three different rotor speeds. Data from the first concentration range fitted well to a model incorporating a single, ideal species, yielding a solution molecular mass of 18,000 Da (with 95% confidence limits of 16,900 and 19,000 Da). This value corresponds well to that expected for a trimer of HR1, based on the amino acid sequence (18,273 Da). The high quality of the single-species fit suggested that HR1 was exclusively trimeric over the entire concentration range of the experiment. Thus, in order to determine an estimate for the trimerization constant, it was necessary to use lower peptide loading concentrations (Fig. 3A). Although a single-species model yielded a good fit to the nine data sets used (Fig. 3B, χ2 = 0.023), the weight-average molecular weight obtained was lower than expected for a trimer (15,400 Da). A monomer↔dimer model yielded fits to the data that were significantly worse, as indicated by the nonrandom residuals (Fig. 3C) and increased value of χ2 (0.046). A monomer↔trimer model yielded the best fit to the data, exhibiting random residuals (Fig. 3D), a lower value of χ2 (0.021), and an association constant of 2.2 × 1011 M−2. Models corresponding to tetramers or higher oligomeric states yielded poor quality fits (data not shown). An association constant of this magnitude implies that ∼90% of the peptide would be present as trimer in a 20 μM solution (the concentration of most of the CD experiments). Furthermore, >99% of the peptide would be present as trimer at a peptide concentration of 1 mM (the concentration of the NMR experiments; see below).
FIG. 3.
HR1 is trimeric in solution. (A) A representative data set from the sedimentation equilibrium analysis of HR1, shown as a plot of A280 against radial position (150 mM NaCl, 20 mM NaH2PO4, pH 7.4). A total of nine data sets were recorded and fitted globally to a range of models. A fit of the data to a monomer↔trimer model is shown. Residuals from the fit of this data set to single-species (B), monomer↔dimer (C), and monomer↔trimer (D) models are also shown, demonstrating that the monomer↔trimer model fits best to the data.
HR2 sedimentation equilibrium data recorded at loading concentrations of 20 μM were well-fitted by a single-species model with a solution molecular mass of 4,710 Da (95% confidence limits of 4,620 and 4,810 Da). This corresponds well to the theoretical molecular mass of an HR2 monomer (4,787 Da; data not shown). At higher concentrations, this peptide appeared to form a large molecular weight aggregate that rapidly sedimented at low rotor speeds. This observation is consistent with results from one-dimensional 1H NMR experiments, in which increases in protein concentration caused a substantial broadening of all resonances (data not shown).
Data from the HR3 peptides HR3-IAM and HR3-TCEP fitted best to a monomer-dimer model with a dimerization constant of ∼1,300 M−1 (data not shown).
NMR studies of HR1.
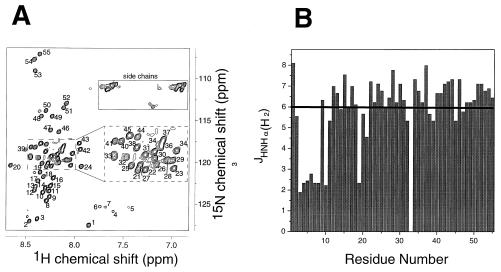
A 15N-HSQC spectrum of HR1 (Fig. 4A) revealed approximately 55 major cross-peaks arising from backbone amide protons (in addition, cross-peak 33 probably comprises two overlapped signals). This is consistent with the amino acid sequence from which a maximum of 57 signals would be expected (the N-terminal residue will not appear). The relatively narrow spread of chemical shifts in the 1H dimension is consistent with a mixture of α-helical secondary structure and random coil, as predicted from the CD data. In order to accurately estimate the secondary structure content of the HR1 domain, a three-dimensional HNHA spectrum (57) was recorded. From this experiment, 3JHNHα scalar coupling constants may be extracted for each nonoverlapped residue. The values of these coupling constants depend on the φ angle for that residue. Thus, coupling constants of less than ∼6 Hz are indicative of α-helical secondary structure, while values greater than 8 Hz indicate an extended β-conformation. The values of 3JHNHα are plotted against peak number in Fig. 4B. Twenty of the 54 residues measured (peak 33 was not used because of overlap) displayed a 3JHNHα value of <6 Hz, suggesting that ∼35% of the residues are in a helical conformation under these conditions. This corresponds well to the lower estimates of α-helical content from CD data. In addition, given that the HR1 peptide is trimeric in solution, the presence of only 55 signals in the HSQC spectrum implies that the trimer is completely symmetrical. In other words, all of the three chains comprising the trimer adopt identical conformations and are in the same orientation with respect to each other.
FIG. 4.
NMR data on the HR1 domain of RSV. (A) 15N-HSQC spectrum of HR1 (1 mM, pH 5.5). Arbitrary peak numbers are indicated on the spectrum. The side chains of Asn and Gln residues, together with aliased signals from Arg and Lys residues, are boxed. The inset panel shows an expansion of the most overlapped region of the spectrum. (B) Graph of 3JHNHα against peak number, measured from the HNHA spectrum of HR1. The solid line indicates the limiting value of 3JHNHα below which a residue is very likely to be in a helical conformation.
HR1 and HR2 form a 3:3 hexamer.
A far-UV CD spectrum of an equimolar mixture of HR1 and HR2 was recorded. This spectrum was compared with a theoretical spectrum obtained by summing experimental spectra of equivalent amounts of HR1 and HR2 alone (Fig. 5A). Small but significant differences between the two spectra were observed, suggesting that secondary structure is induced by an interaction between these two peptides. Specifically, the experimental spectrum shows an increased positive signal at 195 nm (indicative of a more folded structure), an increase in the magnitude of the minimum at 222 nm, and a shift of the minimum at 204 nm towards 208 nm; all these are characteristics of α-helical structure. Estimates of secondary structure content indicated that these changes correspond to an increase in α-helical content of 4 to 7%. Similarly, differences were observed between a spectrum of a mixture of HR1 and “aged” HR2 and a summed data set created from spectra of the individual components (Fig. 5B). In this case, an increased maximum at 190 nm and the appearance of a double minimum at 208 and 222 nm also suggest an increase in α-helical structure, demonstrating that HR1 is able to “rescue” the aged HR2.
FIG. 5.
The interaction of HR1 and HR2 induces the formation of α-helix. Far-UV CD spectra of HR1 with fresh HR2 (A) and with aged HR2 (B). In both cases, experimental data (○) and simulated data (■) of noninteracting HR1 and HR2 are shown. (C) A representative data set from the sedimentation equilibrium analysis of the HR1-HR2 mixture, shown as a plot of A280 against radial position (20 mM NaH2PO4, 150 mM NaCl, pH 7.4). A total of 11 data sets were fitted globally to a range of models. The best fit was obtained using a single-species model, and this fit is shown for the plotted data set. The residuals for the fit are shown in the inset panel. Note that the scales of the main and inset panels are different.
Solutions of HR1 and HR2 either were separately dialyzed against PBS and mixed in a 1:1 molar ratio or were coinjected and collected as a single peak from an analytical gel filtration column. These solutions were subjected to sedimentation equilibrium analysis, and the resulting data were well fitted by a single-species model (Fig. 5C) with a solution molecular mass of 31,800 Da (with 95% confidence limits of 29,900 and 33,800 Da). This corresponds closely to a hexamer containing three molecules of HR1 and three molecules of HR2 (32,634 Da).
HR3 does not interact further with HR1, HR2, or the HR1-HR2 complex.
When HR3 peptides were mixed with HR1, HR2, or a solution containing HR1 and HR2, far-UV CD spectra showed no change in secondary structure content (data not shown). This suggests that there is unlikely to be an interaction between HR3 and the other HR peptides. Similarly, sedimentation equilibrium experiments conducted on solutions of HR3-IAM mixed with HR1, HR2, or HR1-HR2 gave no indication of any additional interaction. The molecular weights obtained from fits to single-species models corresponded to the expected weight-average molecular weights of mixtures of monomeric HR3-IAM with trimeric HR1, monomeric HR3-IAM with monomeric HR2, and monomeric HR3-IAM with the hexameric HR1-HR2 complex, respectively (data not shown).
Identification of the HR1-HR2 interface.
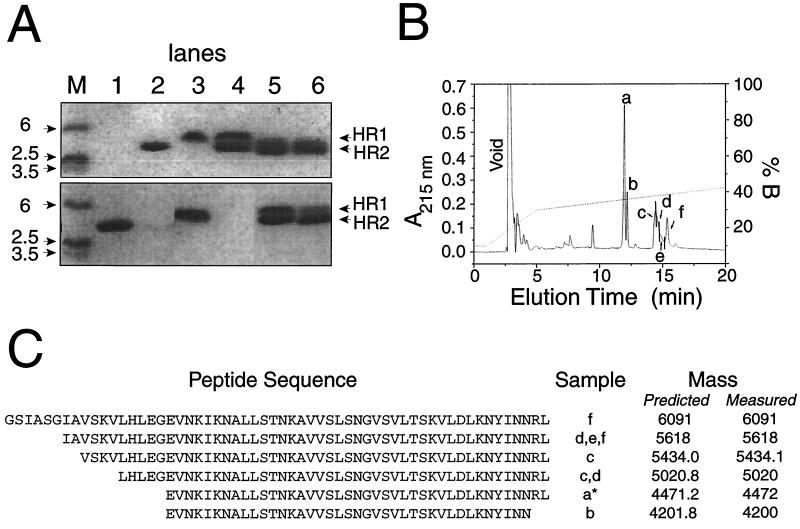
In order to determine which regions of HR1 and HR2 interact, the complex of these two peptides was subjected to limited proteolysis using thermolysin. Regions protected either by a high level of secondary structure and/or by the formation of the HR1-HR2 hexamer were expected to be comparatively resistant to proteolytic cleavage. Analysis by SDS-PAGE (Fig. 6A) showed that over the course of a 1- to 2-h incubation, the band corresponding to HR1 disappeared, the band corresponding to HR2 remained unchanged, and no significant bands of smaller size appeared (the limit of detection by this system is approximately 2.5 kDa). Aliquots corresponding to 60- and 120-min incubation times were separated by reversed-phase HPLC (Fig. 6B), and the major peaks were analyzed by ES-MS. Peptides corresponding to full-length HR1, full-length HR2, and shorter forms of HR1 (but not HR2) could be identified (Fig. 6C). The cleavage sites in HR1 that gave rise to the shorter forms of the peptide were all located in the N-terminal region, suggesting that all of HR2 and the C-terminal residues of HR1 are involved in the HR1-HR2 interaction.
FIG. 6.
Identification of the interaction domains of HR1 and HR2 by partial thermolysin proteolysis. (A) SDS-PAGE gel showing the following. (Top) Lane M, molecular weight markers; lane 1, thermolysin; lane 2, HR2; lane 3, HR1; lane 4, HR1-HR2; lane 5, HR1-HR2 after 60 min of thermolysin digestion; lane 6, HR1-HR2 after 60 min of thermolysin digestion. (Bottom) Lane M, molecular weight markers; lane 1, HR2 at time zero; lane 2, HR2 at 10 min; lane 3, HR1 at time zero; lane 4, HR1 at 10 min; lane 5, HR1-HR2 at 0 min; lane 6, HR1-HR2 at 10 min. (B) Reversed-phase HPLC chromatogram of a 60-min thermolytic digest of HR1-HR2. Peptides were identified by ES-MS from samples taken at times indicated by a to f. (C) Sequences of peptides identified by ES-MS. ∗, The major component of the peak was uncleaved HR2. No cleaved forms of HR2 were identified.
In contrast to the results obtained with HR1-HR2 mixtures, thermolysin treatment of solutions of HR1 and HR2 in isolation resulted in comparatively rapid degradation of both species (Fig. 6A). These data support the conclusion that protection of HR1 and HR2 in the mixed solution is due to the formation of HR1-HR2 hetero-oligomers and not the formation of independent HR1 trimers and/or aggregates of HR2.
Short form of HR1.
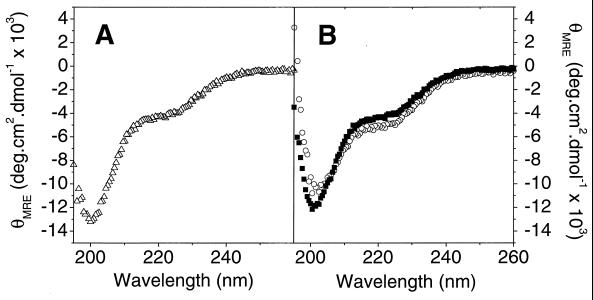
A recombinant peptide corresponding to the putative HR2-interacting region of HR1 (HR118–58) was also produced. Far-UV CD analysis revealed that HR118–58 was significantly less helical than HR1 (Fig. 7A). Data acquired from sedimentation equilibrium experiments of HR118–58 could not be adequately described by either a single-species model or a monomer↔trimer model, but appeared to be aggregating in a nonspecific manner (data not shown).
FIG. 7.
Far-UV CD spectra of HR118–58. Far-UV CD spectra of HR118–58 (▵) (A) and an equimolar mixture of experimental data (○) (B). Also shown in panel B is a simulated spectrum of noninteracting HR118–58 and HR2 (■). Total protein concentrations were 20 μM in each case. Peptides were buffered in 20 mM NaH2PO4, 150 mM NaCl (pH 7.4). Spectra are the average of three to five scans and are baseline corrected.
When a solution containing equimolar amounts of HR2 and HR118–58 was analyzed by far-UV CD, a small increase in α-helical content (4 to 15%) was observed (Fig. 7B). In addition, sedimentation equilibrium analysis of a mixture of HR2 and HR118–58 revealed an increase of the weight-average molecular weight of the mixture over the weight-average molecular weight of HR118–58 alone (data not shown). Taken together, these observations suggest that HR118–58 and HR2 interact, although the nature of the interaction could not be clearly defined due to the self-association of HR118–58.
DISCUSSION
The striking similarities in gross structural features of viral fusion proteins are suggestive of similar mechanisms of action and have led to speculation of common ancestry (1, 9, 51, 58). One of the most conserved of these features is an HR that forms a trimeric coiled-coil core (termed here the HR1 region) arranged along the central axis of the fusion protein and perpendicular to the viral membrane. The data presented here demonstrate that the corresponding region of the RSV fusion protein (HR1) forms a symmetrical trimer that contains a significant amount of α-helix.
The α-helical content of HR1 is estimated to be ∼35% from NMR scalar coupling measurements and using the CD secondary structure prediction program Prosec. This equates to ∼20 residues existing in an α-helical conformation and is comparable to the level of 47% helical content reported for a slightly larger peptide corresponding to the HR1 region of a related paramyxovirus, SV5 (31). Both estimates of helical content are low compared with NMR studies in which the Newcastle disease virus (NDV) HR1 peptide was shown to be monomeric and highly helical (69). However, it should be noted that a much shorter peptide (23 amino acids) was used in the latter studies and that the experiments were carried out in the presence of SDS micelles, which are known to promote α-helical content in peptides that correspond to amphipathic helices in proteins.
Although 20 amino acids constitute a relatively small α-helix in comparison to published HR1 structures, it is reasonable to expect a proportion of random coil in these peptides given that they are isolated peptides and thus likely to have frayed ends. The trimming of HR1 by proteolysis of the HR1-HR2 complex residues (Fig. 6) also suggests that not all of the HR1 region in the peptide studied is involved in coiled-coil formation. In light of these results, we expected that the proteolytically clipped form of HR1 (HR118–58) would be correspondingly more helical, although this was not the case. Instead, HR118–58 was significantly less helical, showed a tendency to self-associate nonspecifically, and displayed only limited evidence of interaction with HR2. These data are consistent with the results of Lamb and colleagues (31), who have reported that the trimming of eight residues from the N terminus of the HR1 peptide of a related paramyxovirus, SV5, virtually halved the α-helical character of that peptide. The untrimmed SV5 peptide used in their subsequent studies was slightly larger than our full-length HR1 peptide and included two hydrophobic residues that have been subsequently shown to form part of a nonpredicted, stuttered 3-4-4-4-3 HR at the C terminus of the SV5 coiled-coil (1). Pneumovirus F sequence alignments (not shown) also show conservation of a hydrophobic amino acid equivalent to the penultimate residue in our HR1 sequence (corresponding to I180 in SV5 and V207 in human RSV), supporting the proposal that the RSV coiled coil adopts a comparable structure (1). If so, the absence of the complete C-terminal turn in our HR1 peptide may also contribute to the slightly lower-than-expected α-helical content in the untrimmed peptide.
We also examined a C-terminally truncated form of HR1 lacking seven residues that were removed by thrombin cleavage as part of the purification process (data not shown). This peptide, although still maintaining helical character, was significantly less helical than HR1 (data not shown). Such decreases in helical character may result from the elimination of key residues that specifically direct trimer formation. In general, the HR1 regions are irregular, containing a high proportion of hydrophobic residues apart from the actual hydrophobic repeat. These additional hydrophobic residues are involved in interactions with HR2 regions. It has been shown that the buried core residues of the HR1 region in gp41 specifically direct trimer formation (53). Thus, it is possible that the loss of a few key residues could reduce or eliminate such specificity, and the high proportion of hydrophobic residues remaining in HR118–58 could cause nonspecific aggregation, preventing helical formation. Using sedimentation equilibrium experiments, the association constant of trimerization was estimated to be 2.2 × 1011 M−2. This is essentially identical to estimates of 1.5 × 1011 M−2 for SIV e-gp41 (7) and 4.5 × 1011 M−2 for HIV e-gp41 (64), constructs which contain both the HR1 and the HR2 regions as well as the intervening residues of gp41. Given that our estimate of the HR1 association constant does not account for any contribution by HR2, we would expect the association constant for an equivalent construct of the RSV F protein to be significantly larger.
The formation of a trimer by HR1 is consistent with cross-linking studies of RSV F (52). However, sedimentation equilibrium studies of the HR1 peptide from gp41 suggested that this peptide underwent a monomer↔dimer↔tetramer association (38). This observation is consistent with earlier reports for isolated gp41 HR1 peptide and peptide-fusion protein constructs (3, 46, 54) as well as whole-protein studies (see reference 42 and references therein), but is in conflict with later observations of the trimer core (7, 55, 59, 64). These discrepancies may be partially attributable to the susceptibility of differing techniques to artifact (e.g., see reference 3), and in addition, it is notable that the HR1 peptide sequences used by various groups were not identical. The propensity to form specific oligomers could be dependent on a few key residues which differ from study to study or can be modified by other parts of the protein (42). It is well-known that the oligomerization specificity of coiled coils can be switched by the mutation of even single residues (see, for example, reference 24).
The largely unstructured conformation of HR2 is a common feature of several other HR2 peptides from other viral fusion or transmembrane proteins (21, 37, 62). The notable exception to this is the HR2 region from NDV, which forms helical structure with a strip of leucine residues aligned along one face (68). The tendency for the HR2 regions to self-associate under some conditions has also been noted for the SV HR2 (SV-465 [21]). This property is not surprising, given that the role of this region under native conditions is to form part of the hydrophobic core of the F protein. It is usually the case that both the HR1 and HR2 peptides exhibit antiviral activity that is specific to the virus of origin, but that the HR2 peptides are significantly more potent. However, it does not appear to be the unstructured nature of the HR2 peptides that confers this potency. For example, the NDV HR2 peptide is both helical and has antiviral activity (68). In addition, a short form of the gp41 analogue DP-178 has been forced into an α-helical conformation through the use of one or two lactam bridges; these helical peptides are significantly more potent than the unstructured, unmodified forms of the same peptide (32). Because the mechanism of peptide inhibition is not yet known, it is difficult to account for the increased inhibitory activity of HR2 peptides. It may simply be that only monomeric HR peptides are active, in which case the effective concentrations of free monomer are much higher for HR2 than for HR1, which undergoes a monomer↔trimer equilibrium (7). Alternatively, if inhibition occurs after the assembly of an F trimer, increased potency may arise from the lower energetic cost of displacing HR2 from a position on the outside of the HR1 trimer, compared to the cost of complete disassembly of the trimer or hexamer (which would be required to displace HR1).
Complex formation.
Sedimentation equilibrium data indicate that the complex formed by HR1 and HR2 is hexameric, with a solution molecular weight corresponding to three molecules of each peptide. This is consistent with studies of the HR peptides from gp41 (7, 64) and the paramyxovirus F protein from SV5 (31), although those complexes were shown to be highly helical. In this study, the complex formed by HR1 and HR2 is apparently less helical, despite the high affinity of the interaction (demonstrated by the ability of HR1 to rescue aggregated HR2) (Fig. 5B). CD data suggest that four to seven additional residues take up a helical conformation as a result of the interaction between HR1 and HR2. This implies that for the RSV F protein, either a core region consisting of the trimeric coiled-coil HR1 surrounded by helical HR2 is significantly shorter than has been observed for equivalent proteins in other viruses or additional constraints provided by other parts of the protein are required for full helix formation. Interestingly, the structure of the core region of the SV5 F protein shows that the HR2 domain contains both an α-helix and an extended conformation that stretches out along the HR2 region. Our studies demonstrate that RSV HR2 has a tendency to adopt a β-sheet conformation and is protected from proteolysis when in complex with HR1. Thus, it is probable that the RSV F protein core resembles more closely that of the SV5 F protein (1) than that of gp41 (7, 10), which has a highly helical HR2 region. When the residues corresponding to the interacting regions of HR1 and HR2 are superimposed onto the SV5 F structure, using the best sequence alignment of RSV HR1 and HR2 with SV5 HR1 and HR2 (Fig. 8), they are found to correspond almost exactly with interacting regions of the SV5 F structure. In addition, given that there is also some variation between the reported properties of the HR1 and HR2 peptides among the paramyxoviruses (1, 68), it may be that relatively more of the HR2 region is in an extended conformation in the RSV F protein. While we have not yet been able to experimentally determine the orientation of the HR2 peptides with respect to the HR1 parallel trimer, we believe they are likely to assume the antiparallel orientation found in most F and TM proteins (see, for example, references 1, 6, 7, 17, and 41).
FIG. 8.
Comparison of RSV HR1 and HR2 with SV5 F. (A) Ribbon diagram of the SV5 F structure (protein data bank identification code, 1svf [1]). HR1 regions are shown in gray, and HR2 regions are shown in black. Cα atoms corresponding to the interacting regions of HR1 and HR2 are shown in CPK format. (B) Sequence alignment of HR1 with the HR1 peptide used in the SV5 structure. (C) Sequence alignment of HR2 with the HR2 peptide used in the SV5 structure. Identical residues are represented by dots. Residues derived from the linker region of the GST fusion protein are in bold. Residues corresponding to the interacting regions of HR1 and HR2 are in italics.
Proteolysis data show that essentially all of the HR2 peptide is protected by complex formation, while only the C-terminal ∼70% of HR1 is protected, suggesting that our original HR1 construct extends beyond the complexed region. This may represent a flexible region immediately adjacent to the fusion peptide, as is found in HA, Ebola virus GP2, and human T-cell leukemia virus type 1 (11, 35, 41, 58). Alternatively, an extended form of HR1 that included the fusion peptide might result in an extension of the trimeric coiled-coil core in the manner of the HIV-1 and SV5 fusion proteins (1, 7, 10, 55, 59).
RSV HR3 does not appear to be further involved in complex formation.
RSV is strongly predicted to contain a third coiled-coil region (HR3) in the F2 subunit (Fig. 2) (37). We have shown that RSV HR3 is able to form dimeric helices in a concentration-dependent manner, although the affinity constant for this interaction was rather low. However, we found no evidence to suggest that HR3 can interact with HR1, HR2, or the HR1-HR2 complex. Although HR3 does not appear to contribute to the core region of the RSV F protein, it may have a structural role. For example, HR3-HR3 dimerization may be involved in inter- or intramolecular associations between or within F oligomers, respectively. Alternatively, it may be involved in interactions with other non-HR regions. Indeed, a cysteine residue located towards the N terminus of HR3 (just outside the strongly predicted coiled-coil region) is likely to connect the F2 and F1 subunits. The flanking HR region may serve to stabilize an interaction between these two subunits. To ensure that this cysteine residue was not itself a barrier to HR3-core interactions, a 30-residue synthetic peptide version of HR3, encompassing only the strongly predicted HR region, was also investigated. This peptide similarly failed to exhibit any signs of an interaction with the core HR peptides.
Additional HR or potential leucine zipper regions have been identified in a number of different paramyxovirus F proteins, most notably those located on the F1 subunit between the HR1 and HR2 regions (15, 48). However, there are some discrepancies regarding their role. In the case of Sendai virus, peptides corresponding to both this leucine zipper and a proposed internal fusion peptide were shown to associate in an as yet undefined manner with biologically active forms of Sendai F HR1 and HR2 (20–22, 44). However, although similar associations were shown between the leucine zipper region and HR1 and HR2 regions of SV5, it was concluded that the leucine zipper region did not form part of the HR1-HR2 core because no increase in stability of the complex was observed (15). More detailed biophysical analysis and/or structure determination may resolve these discrepancies.
In conclusion, this work supports a structural theme that is emerging for viral fusion proteins: a homotrimeric coiled-coil core that interacts with three complementary sequences to form a hexamer of heterodimers. Our data demonstrate that HR1 forms a strong symmetrical coiled-coil trimer that is able to form a hexamer with HR2. A third HR region in F2 does not appear to be further involved in the formation of the F protein core. It is likely that the structure of the RSV F protein will be similar in appearance to the closely related SV5 F protein, with a section of the HR2 region forming an extended conformation, although in the RSV F protein this extended conformation may comprise a larger proportion of the HR2 region.
ACKNOWLEDGMENTS
J.M.M. is the recipient of a U2000 Fellowship. J.P.M. is the recipient of an ARC Australian Research Fellowship.
We thank A. J. Mason and G. F. King for helpful discussions.
REFERENCES
- 1.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Efraim I, Kliger Y, Hermesh C, Shai Y. Membrane-induced step in the activation of Sendai virus fusion protein. J Mol Biol. 1999;285:609–625. doi: 10.1006/jmbi.1998.2370. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein H B, Tucker S P, Kar S R, McPherson S A, McPherson D T, Dubay J W, Lebowitz J, Compans R W, Hunter E. Oligomerization of the hydrophobic heptad repeat of gp41. J Virol. 1995;69:2745–2750. doi: 10.1128/jvi.69.5.2745-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 5.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai M, Huang Y, Sakaguchi K, Clore G, Gronenborn A, Craigie R. An efficient and cost-effective isotope labelling protocol for labelling protein expressed in Escherichia coli. J Biomol NMR. 1998;11:97–102. doi: 10.1023/a:1008222131470. [DOI] [PubMed] [Google Scholar]
- 9.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 10.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Skehel J J, Wiley D C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S S, Lee C N, Lee W R, McIntosh K, Lee T H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chernomordik L V, Leikina E, Kozlov M M, Frolov V A, Zimmerberg J. Structural intermediates in influenza haemagglutinin-mediated fusion. Mol Membr Biol. 1999;16:33–42. doi: 10.1080/096876899294733. [DOI] [PubMed] [Google Scholar]
- 14.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutch R E, Leser G P, Lamb R A. Paramyxovirus fusion protein: characterization of the core trimer, a rod-shaped complex with helices in anti-parallel orientation. Virology. 1999;254:147–159. doi: 10.1006/viro.1998.9532. [DOI] [PubMed] [Google Scholar]
- 16.Eckert D M, Malashkevich V N, Hong L H, Carr P A, Kim P S. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 17.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 18.Ferrer M, Kapoor T M, Strassmaier T, Weissenhorn W, Skehel J J, Oprian D, Schreiber S L, Wiley D C, Harrison S C. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat Struct Biol. 1999;6:953–960. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 19.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh J K, Ovadia M, Shai Y. A leucine zipper motif in the ectodomain of Sendai virus fusion protein assembles in solution and in membranes and specifically binds biologically-active peptides and the virus. Biochemistry. 1997;36:15451–15462. doi: 10.1021/bi971152i. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh J K, Peisajovich S G, Ovadia M, Shai Y. Structure-function study of a heptad repeat positioned near the transmembrane domain of Sendai virus fusion protein which blocks virus-cell fusion. J Biol Chem. 1998;273:27182–27190. doi: 10.1074/jbc.273.42.27182. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh J K, Shai Y. Direct evidence that the N-terminal heptad repeat of Sendai virus fusion protein participates in membrane fusion. J Mol Biol. 1999;292:531–546. doi: 10.1006/jmbi.1999.3097. [DOI] [PubMed] [Google Scholar]
- 23.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez L, Jr, Woolfson D N, Alber T. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 25.Hayes D B, Laue T, Philo J. SEDNTERP. Durham: University of New Hampshire; 1995. [Google Scholar]
- 26.Heminway B R, Yu Y, Tanaka Y, Perrine K G, Gustafson E, Bernstein J M, Galinski M S. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology. 1994;200:801–805. doi: 10.1006/viro.1994.1245. [DOI] [PubMed] [Google Scholar]
- 27.Jiang S, Lin K, Strick N, Neurath A R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 28.Jiang S, Lin K, Strick N, Neurath A R. Inhibition of HIV-1 infection by a fusion domain binding peptide from the HIV-1 envelope glycoprotein GP41. Biochem Biophys Res Commun. 1993;195:533–538. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- 29.Johnson M L, Correia J J, Yphantis D A, Halvorson H R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson W C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 31.Joshi S B, Dutch R E, Lamb R A. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 32.Judice J K, Tom J Y, Huang W, Wrin T, Vennari J, Petropoulos C J, McDowell R S. Inhibition of HIV type 1 infectivity by constrained alpha-helical peptides: implications for the viral fusion mechanism. Proc Natl Acad Sci USA. 1997;94:13426–13430. doi: 10.1073/pnas.94.25.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahn J S, Schnell M J, Buonocore L, Rose J K. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology. 1999;254:81–91. doi: 10.1006/viro.1998.9535. [DOI] [PubMed] [Google Scholar]
- 34.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 35.Kobe B, Center R J, Kemp B E, Poumbourios P. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc Natl Acad Sci USA. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamb R A. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 37.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R., Jr Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawless M K, Barney S, Guthrie K I, Bucy T B, Petteway S R, Jr, Merutka G. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–13708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 39.Live D H, Davis D G, Agosta W C, Cowburn D. Long range hydrogen bonding mediated effects in peptides: 15N NMR study of gramicidin S in water and organic solvents. J Am Chem Soc. 1984;106:1939–1941. [Google Scholar]
- 40.Malashkevich V N, Chan D C, Chutkowski C T, Kim P S. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malashkevich V N, Schneider B J, McNally M L, Milhollen M A, Pang J X, Kim P S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McInerney T L, El Ahmar W, Kemp B E, Poumbourios P. Mutation-directed chemical cross-linking of human immunodeficiency virus type 1 gp41 oligomers. J Virol. 1998;72:1523–1533. doi: 10.1128/jvi.72.2.1523-1533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norwood T J, Boyd J, Heritage J E, Soffe N, Campbell I D. Comparison of techniques for 1H-detected heteronuclear 1H-15N spectroscopy. J Magn Reson. 1990;87:488–501. [Google Scholar]
- 44.Peisajovich S G, Samuel O, Shai Y. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J Mol Biol. 2000;296:1353–1365. doi: 10.1006/jmbi.2000.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 46.Rabenstein M, Shin Y K. A peptide from the heptad repeat of human immunodeficiency virus gp41 shows both membrane binding and coiled-coil formation. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 47.Ralston G B. The concentration dependence of the activity coefficient of the human spectrin heterodimer. A quantitative test of the Adams-Fujita approximation. Biophys Chem. 1994;52:51–61. doi: 10.1016/0301-4622(94)00065-4. [DOI] [PubMed] [Google Scholar]
- 48.Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reitter J N, Sergel T, Morrison T G. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rimsky L T, Shugars D C, Matthews T J. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal P B, Zhang X, Formanowski F, Fitz W, Wong C H, Meier-Ewert H, Skehel J J, Wiley D C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell R, Paterson R G, Lamb R A. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology. 1994;199:160–168. doi: 10.1006/viro.1994.1108. [DOI] [PubMed] [Google Scholar]
- 53.Shu W, Ji H, Lu M. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry. 1999;38:5378–5385. doi: 10.1021/bi990199w. [DOI] [PubMed] [Google Scholar]
- 54.Shugars D C, Wild C T, Greenwell T K, Matthews T J. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1996;70:2982–2991. doi: 10.1128/jvi.70.5.2982-2991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng M N, Collins P L. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vuister G, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J Amer Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 58.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 59.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 60.Whitehead S S, Bukreyev A, Teng M N, Firestone C Y, St. Claire M, Elkins W R, Collins P L, Murphy B R. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson I, Skehel J, Wiley D. Structure of the heamagglutinin membrane glycoprotein of influenza at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 64.Wingfield P T, Stahl S J, Kaufman J, Zlotnick A, Hyde C C, Gronenborn A M, Clore G M. The extracellular domain of immunodeficiency virus gp41 protein: expression in Escherichia coli, purification, and crystallization. Protein Sci. 1997;6:1653–1660. doi: 10.1002/pro.5560060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Wu C-S, Martinez H. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 67.Yao Q, Compans R W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 68.Young J K, Hicks R P, Wright G E, Morrison T G. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology. 1997;238:291–304. doi: 10.1006/viro.1997.8834. [DOI] [PubMed] [Google Scholar]
- 69.Young J K, Li D, Abramowitz M C, Morrison T G. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J Virol. 1999;73:5945–5956. doi: 10.1128/jvi.73.7.5945-5956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]