Abstract
Cannabis is widely used for medicinal and recreational purposes. As a result, there is increased interest in its chemical components and their physiological effects. However, current information on cannabis chemistry is often outdated or scattered across many books and journals. To address this issue, we used modern metabolomics techniques and modern bioinformatics techniques to compile a comprehensive list of >6000 chemical constituents in commercial cannabis. The metabolomics methods included a combination of high- and low-resolution liquid chromatography–mass spectrometry (MS), gas chromatography–MS, and inductively coupled plasma–MS. The bioinformatics methods included computer-aided text mining and computational genome-scale metabolic inference. This information, along with detailed compound descriptions, physicochemical data, known physiological effects, protein targets, and referential compound spectra, has been made available through a publicly accessible database called the Cannabis Compound Database (https://cannabisdatabase.ca). Such a centralized, open-access resource should prove to be quite useful for the cannabis community.
Keywords: cannabis, metabolomics, database, cannabinomics, mass spectrometry
Introduction
Cannabis, also known as hemp, is a widely cultivated flowering annual plant consisting of two major species, Cannabis sativa and Cannabis indica. The term “hemp” is often reserved for those cannabis cultivars that are grown for nondrug use, while the term “cannabis” is used for those cultivars specifically grown for drug use. Evidence of cannabis/hemp use in human societies dates back more than 26,000 years ago where it was used by early Paleolithic societies as a source of fiber to make baskets in the present-day Czech Republic.1 The medicinal use of cannabis dates to more than 5000 years ago where it was prescribed by Chinese physicians to treat fatigue, rheumatism, and malaria.2 The recreational use of cannabis was first mentioned by Herodotus nearly 2500 years ago, when he described how Scythians used hemp vapor in their steam baths.3 Since then, cannabis has become one of the most widely cultivated medicinal plants in the world, with its flowers being particularly rich in cannabinoids and other psychoactive phytochemicals. The global market (legal and illicit) for cannabis is estimated to be worth $344 billion USD/yr.4 There are believed to be more than 250 million cannabis users worldwide, with the largest number being in Asia, followed by North America and Europe. Nearly 60 countries have legalized the sale of cannabis for medicinal use, while another 53 have officially ignored, decriminalized, or legalized its use or possession for recreational purposes. Given the widespread use of cannabis, the growing level of legalization for medicinal/recreational purposes and a growing concern over its safety, health effects, benefits, and harm, there is increased interest in understanding the chemistry and chemical composition of cannabis.
Cannabis is known to have a very complex chemical profile. Various studies have reported more than 550 different compounds in cannabis. These include >140 cannabinoids, 120 terpenes, 50 hydrocarbons, 46 phenolic or polyphenolic compounds, 34 sugars, 25 ketones and aldehydes along with many kinds of organic acids, fatty acids, amino acids, esters, lactones, phytosterols, alkaloids, vitamins, and biogenic amines.5,6 These molecules, along with other yet-to-be-identified bioactive molecules, likely contribute to the widely varying physiological and psychoactive effects of different cannabis strains and different cannabis products on humans.
The main psychoactive chemical in cannabis is tetrahydrocannabinol (THC). Several other cannabinoids, such as cannabidiol (CBD), contribute to cannabis’ well-known psychoactive and physiological effects. These chemicals may be released, consumed, or inhaled through a variety of means such as smoking, vaping, or ingestion of cannabis edibles. Cannabis consumption or inhalation can lead to a variety of mental and physical effects, including euphoria, an altered sense of time, increased appetite, difficulty concentrating, impaired short-term memory, and clumsiness. Medically, cannabis has shown promise in effectively treating several conditions, including chronic pain, nausea, anxiety, and inflammation.7,8 The onset of effects is typically felt within a few minutes when smoked and within about 30–60 min when eaten. These effects last for 2–6 h depending on the amount used and the cultivar’s concentration of THC or CBD. Each cannabis compound, on its own, may have a well-defined physiological effect but certain combinations of chemicals (as found in plant extracts) can have a synergistic physiological effect or a so-called “entourage effect”.9,10 In addition to these hard-to-define entourage effects, the chemical profile and concentrations of each compound can vary dramatically among cannabis cultivars, and they can even vary depending on the age of the plant, harvest conditions, storage conditions, and growth conditions.11,12
While a large body of research exists that has focused on characterizing the chemical compounds in cannabis, there is no single resource that catalogues all known cannabis constituents. Instead, most of the information is scattered across several textbooks, monographs, and scientific journals.6,7 Furthermore, most of this information incompletely describes the precise structures, nomenclature, and other essential properties of these chemicals. It is also notable that remarkably little work has gone into accurately quantifying important cannabis compounds in different cannabis cultivars or developing reagents or techniques to enable their quantification.
Historically, most cannabis chemical composition studies have been performed using targeted chemical analyses aimed at characterizing specific classes of compounds (i.e., cannabinoids only, terpenes only, and volatiles only).13−17 While targeted analytic approaches are very accurate, they require considerable skill, are rather limited in their chemical scope, and often require a great deal of time and manual effort. With the development of quantitative, targeted metabolomics approaches, it has been possible to achieve far more comprehensive chemical coverage (with less analytical effort) of plants, plant products, extracts, biofluids, and other organisms.18,19 Thanks to significant advances in analytical techniques such as liquid chromatography (LC), gas chromatography (GC), tandem mass spectrometry (MS–MS), and nuclear magnetic resonance spectroscopy (NMR), metabolomics methods can routinely identify and quantify hundreds of compounds from a single sample.19−22 Indeed, metabolomics has already enabled the determination of extensive inventories of small-molecule metabolites for a wide range of organisms and biofluids.23,24 It has also been applied toward the characterization of cannabis chemicals in a number of studies.25−27
For instance, a recent review highlighted nearly 20 different metabolomic (or “cannabinomic”) studies that have been conducted on cannabis since 2008.25 These studies employed a variety of techniques including NMR, GC–MS, LC–MS, and GC coupled with flame ionization detection (GC-FID) to primarily perform chemotaxonomy, aiming at distinguishing different cannabis strains, cultivars, or chemovars. These investigations typically report 15–45 cannabis metabolites.25 In addition, other studies have made substantial efforts in understanding the chemical composition of cannabis at various levels, ranging from different parts of the plant itself,28,29 to cannabis extracts,30 and finally to the finished edible products.31 More recently, several very comprehensive cannabis metabolomic studies have been published that have substantially “raised the bar” regarding compound coverage. Specifically, Delgado-Povedano et al.26 used a combination of high-resolution LC–MS and GC–MS methods to identify 169 metabolites from more than a dozen classes of compounds in 17 cannabis cultivars. Antonelli et al.27 used high-resolution LC–MS-based lipidomics to putatively identify 189 different lipids, including 80 sulfolipids and 51 phospholipids from a single strain of Cannabis sativa. Mudge et al.32 used GC–MS-based metabolomics to positively identify 99 cannabis compounds, including 67 terpenoids from 33 different commercial cannabis chemovars, while Graves et al.33 employed comprehensive GC (GCxGC)–MS metabolomic methods to tentatively identify 536 compounds found in mainstream cannabis smoke. Unfortunately, only a small fraction of cannabis metabolomic studies published to date report actual concentrations or perform validated compound identification. Characterizing the thousands of compounds found in cannabis represents a difficult challenge for traditional analytical chemistry but one that is very well suited for the nascent field of metabolomics.34
To facilitate further research into cannabis chemistry, we believe it is critical to comprehensively and quantitatively characterize the chemical composition of cannabis (and different cultivars) using multiple metabolomic techniques. Such an undertaking would benefit cannabis cultivators/distributors, cannabis researchers, physicians, government regulators, and consumers. This is because it would create a centralized, comprehensive, and electronically accessible database of all measured or detectable metabolites/chemicals found in cannabis (and cannabis smoke). To create such a resource, we combined experimental metabolomic techniques with computer-aided text mining and computational genome-scale metabolic inference35−37 to compile essentially all the known chemicals (endogenous and exogenous) that can be detected in cannabis along with their respective concentrations, where possible. Experimentally, we used multiple quantitative, targeted metabolomics techniques including GC–MS,38 LC–MS/MS,18 and inductively coupled plasma–MS (ICP–MS)39 methods to identify and/or quantify 284 cannabis metabolites or metabolite species across a number of cannabis cultivars. In addition to using multiple metabolomics techniques, special care was taken in the selection of the extraction method(s) for these metabolites to ensure the highest level of recovery and the most comprehensive coverage of different classes of metabolites with different chemical or solubility properties. Most extractions were performed through liquid–liquid extraction, which separates compounds into two immiscible phases, aqueous and nonpolar organic. Depending on the metabolites of interest, we chose nonpolar solvents such as chloroform, methyl tert-butyl ether (MTBE), and hexane. Several solid–liquid extractions were also performed using solvents such as methanol and acetonitrile to help extract more polar metabolites from the powdered cannabis samples.18,40,41 There is no single solvent that can extract all the metabolites to cover the whole metabolome of the cannabis cultivars; therefore, a solvent system that combines both the polar and nonpolar solvents (e.g., mixture of hexane, methanol, and water in our method) is favored toward the extraction of various classes of metabolites.
To further enhance our experimental data, we conducted an extensive literature survey that was then combined with genome-scale metabolite inference. The entire data set is now housed in the Cannabis Compound Database (CCD) (https://cannabisdatabase.ca). This web-accessible source contains cannabis chemical concentration data, physicochemical data (including measured or predicted NMR and MS spectra), physiological/psychoactive effect data, cultivar/chemovar data, and extensive referential data for 6337 cannabis metabolites or metabolite species along with detailed data on 115 cannabis cultivars. We believe that such a centralized, open-access database will be particularly useful for the cannabis community, including researchers, producers, regulators, physicians, vendors, and users.
Materials and Methods
Reagents
Optima LC/MS-grade formic acid, Optima LC/MS-grade water, methanol, and acetonitrile, and Optima LC/MS-grade ammonium acetate were purchased from Fisher Scientific (Ottawa, CA). High-performance LC (HPLC)-grade pyridine and ethanol, phenyl isothiocyanate (PITC), 3-nitrophenylhydrazines (3-NPH), 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide (EDC), and butylated hydroxytoluene (BHT) were all purchased from Sigma-Aldrich (Oakville, CA). Octanol, tridecane, and hydrogen peroxide were purchased from Sigma-Aldrich (Burlington, MA). Indium standard and 15 mL metal-free tubes were purchased from PerkinElmer Inc. (Shelton, CT) and Thomas Scientific (Swedesboro, NJ), respectively. Chemical standards and isotope-labeled internal standards (ISTDs) for the common plant metabolomics assay were purchased from different vendors (details provided in Table S1). The vendors and/or concentrations of eight cannabinoid certified reference material standards (1 mL ampules), six standards used for peak identification, terpene mixtures 1 and 2 containing 21 and 16 compounds, respectively, 14 polyphenols, and 71 pesticides used for the cannabinoid, terpenoid, polyphenol, and pesticide assays are listed in Table S2. Nunc 96 DeepWell plates and MultiScreen “Solvinert” filter plates [hydrophobic, polytetrafluoroethylene (PTFE), 0.45 μm, clear, nonsterile] were bought from Sigma-Aldrich (Oakville, CA). The 0.22 μm Acrodisc syringe filters with a hydrophilic polypropylene (GHP) membrane were obtained from VWR (Radnor, PA).
All chemicals were weighed on a Sartorius CPA225D electronic balance (Mississauga, CA) with a precision of 0.0001 g. Stock solutions of calibrants and ISTDs were prepared in water (or other organic solvents; Table S2). Calibration curve standards, covering the known concentration ranges or expected concentrations in cannabis and final working ISTD mixture solutions, were prepared by mixing and diluting corresponding stock solutions with water. All calibration curve standards and ISTD working solutions were aliquoted and stored in a Thermo Scientific −86 °C freezer (Waltham, MA.) until used (up to 2 months).
Commercial Cannabis Sample Acquisition
Six different commercially available cannabis cultivars were analyzed: Alien Dawg, Tangerine Dream, Sensi Star, Quadra, Gabriola (Frosty Monster), and Island Honey. These were purchased from a licensed cannabis distributor in Edmonton, Canada. These six cultivars were chosen because they were identified by the vendors as popular brands of commercial cannabis in Canada and presumed to be representative of the different types of cannabis consumed by Canadians. In Canada, recreational cannabis is approved for sale and is available at government-licensed retail distributors. Licensed cannabis sold in Western Canada is grown in licensed greenhouses. After purchase, each sample (3.5 g dry weight) was stored at room temperature (RT) until it was used for analysis. For all assays, one sample of each cultivar was collected, and three technical replicates were analyzed.
GC–MS Compound Identification and Quantification
Two GC–MS analyses were performed, a quantitative targeted analysis for terpenoids and a nontargeted qualitative analysis of other volatile or nonderivatizable compounds. The targeted terpenoid analysis was designed, calibrated, and tested to detect and fully quantify 28 and semiquantify 17 terpenoids over a 45 min GC–MS run. The sample preparation and analysis methods were adapted from a previously published method.38 In brief, 50 mg of dry cannabis plant material (for each of the six cultivars), previously ground via a pestle and mortar to a fine powder, was extracted using 1.0 mL of an ethyl acetate and ethanol mixture (1:1 v/v) containing 0.05% 1-octanol and 0.0125% tridecane as internal standards. The mixture was shaken at RT for 10 min at 300 rpm on a multitube vortexer (Fisher Scientific) in the dark and centrifuged at 11,000g for 10 min at 4 °C, and the supernatant was removed and used to quantify the terpenes.
The samples (2 μL injection volume) were delivered via a 7693 autosampler to a 7890B series GC coupled to a 5977A MSD mass detector (all from Agilent Technologies, Santa Clara, CA). An Agilent HP-5 MS capillary column (30 m × 250 μm inner diameter) was used to separate all compounds before MS analysis. Helium was used as the carrier gas, with a constant flow rate of 1 mL/min. The initial oven temperature was held at 60 °C for 3 min, increased to 65 °C at 3 °C/min (held for 7 min), increased to 90 °C at 1.8 °C/min, and then increased to 240 °C at 9 °C/min. Finally, the temperature was increased to 310 °C at 20 °C/min and held for 5 min. The injector temperature was 250 °C with a split ratio of 15:1. The mass spectrometer was set in full scan mode from 50–600 atomic mass units. The ionization energy was 70 eV. The ion source temperature was 230 °C, and the quadrupole temperature was 150 °C. The solvent delay was set to 5 min. The transfer line temperature was 280 °C. Data analysis was performed with Agilent MassHunter software. All terpenes were quantified using external six- to nine-point calibration curves (using authentic standards for each terpene) and linear regression. In instances in which authentic standards were not available, surrogate standards with near-identical chemical structures or properties were used. Calibration curves constructed using the surrogate standards were used to semiquantify the affected analytes. Retention indices (RIs) were calculated using a C8–C20 alkane mixture solution (Fluka, Sigma-Aldrich) as an external standard. For the untargeted GC–MS analysis of cannabis volatiles, the same extraction protocol, GC–MS separation, quantification, and data analysis as described above for terpenoid analysis were used.
Targeted LC–MS/MS Compound Identification and Quantification (Common Plant Metabolites)
A targeted, quantitative metabolite assay was used to detect and quantify a wide range of common plant metabolites using a combination of reversed-phase LC (RPLC) and direct flow injection (DFI) with MS/MS (RPLC–DFI–MS/MS). These MS-based analyses were performed on an AB SCIEX QTRAP 4000 mass spectrometer (Applied Biosystems/MDS Analytical Technologies, Foster City, CA). The assay is capable of detecting and quantifying 210 plant compounds, including amino acids, biogenic amines, glucose, organic acids, plant hormones, acylcarnitines, phosphatidylcholines (PCs), lysophosphatidylcholines (LysoPCs), sphingomyelins (SMs), and hydroxylated SM(OH)s. Details about the method, derivatization strategy, separation protocol, MS methods, calibration, and metabolite quantification process have been previously described.18 The method uses chemical derivatization (via 3-NPH for organic acids or PITC for amine-containing compounds), analyte extraction, and separation, combined with selective mass spectrometric detection using multiple reaction monitoring (MRM) pairs to identify and quantify metabolites. Isotope-labeled ISTDs along with other ISTDs are used for accurate metabolite quantification. For this study, cannabis metabolites were extracted from 25 mg of ground (mortar and pestle) cannabis cultivars using 1 mL of a chilled mixture of hexane and methanol (3:1, v/v) and then shaken and vortexed. A second aliquot of 25% aqueous methanol (0.5 mL) was added, and the mixture was vortexed and then centrifuged to produce 2 layers (a hexane and an aqueous layer) that were aliquoted into separate tubes. The hexane layer was used to quantify phospholipids, and the aqueous layer was used to quantify the remaining metabolites as described earlier.18
Targeted LC–MS/MS Compound Identification and Quantification (Cannabinoids)
A second targeted, quantitative LC–MS/MS assay was used to quantify cannabinoids in the different cultivars. The hexane layer (see the LC–MS method above, for extraction of common plant metabolites) was evaporated to dryness under a nitrogen stream and reconstituted with half its volume of acetonitrile. Then, 10 μL of the ISTD mixture (Table S2) and 100 μL of methanol were added to 50 μL of the sample and mixed completely before LC–MS/MS analysis. The cannabinoid assay was designed, calibrated, and tested to detect and quantify 16 cannabinoids over an 8 min LC–MS run. See the Supporting Information for more details about this LC–MS/MS assay.
Targeted LC–MS/MS Compound Identification and Quantification (Polyphenols)
A third targeted, quantitative metabolite assay using ultra-HPLC (UHPLC)–high-resolution MS (HRMS) was used to quantify the polyphenols in the six cannabis cultivars. Polyphenols were extracted using a modification of an organic solvent protocol.42 Samples were freeze-dried overnight and then ground to a fine powder (using a mortar and pestle). Next, 7.5 mg (dry weight) was weighed into a 1.5 mL microcentrifuge tube, and 10 μL of a mixture of nine isotopic-labeled ISTDs (Table S2) and 250 μL of 80% methanol in water (v/v) were added. Each tube was vortexed for 30 min and centrifuged for 5 min at 18,000g. The supernatant was filtered through 0.22 μm Acrodisc syringe filters with a GHP membrane before UHPLC–HRMS analyses (see the Supporting Information for details).
Targeted LC–MS/MS Compound Identification and Quantification (Pesticides)
A quantitative RPLC–atmospheric pressure ionization (API)–MS/MS method was developed to identify and quantify 71 pesticides at or above the limits of quantification (LOQs) specified for dried cannabis in Health Canada’s “Mandatory Cannabis Testing for Pesticide Active Ingredients—List and Limits”.43 Pesticides were isolated from dry cannabis using organic extraction as described elsewhere.44 The pesticides in 200 mg of plant material, ground using a Geno/Grinder (SPEX Sample Prep, Metuchen, NJ), were mixed with 1 mL of LC–MS-grade acetonitrile and sonicated for 15 min at RT in a water bath sonicator (Marshall Scientific, Hampton, NH). Sonicated samples were chilled at −20 °C for 2 h, then centrifuged at 1700g for 10 min at 4 °C. Supernatants were removed and filtered through a 0.2 μm PTFE filter. Then, 100 μL of filtrate was transferred to an LC vial fitted with a glass insert and diluted with 100 μL of LC–MS-grade methanol. The analysis was performed using an Agilent 1290 Infinity LC system (Agilent, Santa Clara, CA) connected to a Sciex Q-Trap 5500 mass detector (Sciex, Framingham, MA). The compounds were separated using a Zorbax Eclipse XDB-C18 solvent saver plus column (80 Å, 4.6 × 150 mm, 5 μm) (Agilent, Santa Clara, CA) before MS analysis.
Trace Elemental Analysis Using ICP–MS
ICP–MS is considered the gold standard for identifying and quantifying metal ions in biological samples and has been widely used in analyzing plant samples.45 Our ICP–MS analysis of cannabis plants was adapted from a previously described ICP–MS method.39 Briefly, 50 mg of dried cannabis was ground (via pestle and mortar) and accurately weighed into Posi-Lock, metal-free 1.5 mL microcentrifuge tubes (Thomas Scientific, Swedesboro, NJ). Next, 1 mL of digestion solvent (25% HNO3 and 5% H2O2 with 0.200 μg/L of indium as an internal standard) was added. The samples were heated in a 75 °C water bath for 1 h and then centrifuged at 21,000g for 10 min. Then, 150 μL of supernatant was pipetted into a 15 mL metal-free centrifuge tube, diluted 10 times with 1.35 mL of 5% H2O2 (made in ultrapure water), vortexed for 30 s, and then subjected to ICP–MS analyses.
All trace elemental analyses (41 metals) were performed on a PerkinElmer NexION 350× ICP–MS (Woodbridge, Canada), operating in a kinetic energy discrimination mode. Argon (ICP/MS grade, 99.99%) was used as a nebulizer (0.9 mL/min), an auxiliary (1 mL/min), and a plasma gas (15 mL/min). Helium (He) was used as a nonreactive collision gas (Cell gas A: 4.3) to eliminate/minimize chemical interference. The dwell time for each metal ion was set to 50 ms with a total integration time of 500 ms (10 sweeps per reading and three replicates). The uptake of samples/standards/QCs was done by a peristaltic pump using the following protocol: (1) sample flush for 50 s at 48 rpm, (2) read delay for 15 s at 20 rpm, (3) spectral analyses at 20 rpm, and (4) washing for 45 s at 24 rpm. An external calibration curve was used for the quantitation of all metal ions using a six- to nine-point calibration curve (for each metal) and linear regression. The performance of the ICP–MS was checked daily using a commercially prepared PerkinElmer NexION calibration solution (Millipore Sigma, Milwaukee, WI) to evaluate the sensitivity of the instrument. The NexION solution was also used to calibrate the mass spectrometer at low (Be), mid (In), and high (U) masses. Continuing calibration verification was run every 15 samples to monitor the validity of each calibration curve throughout the sequence.
Cannabis Compounds in the Literature
We conducted an extensive literature review of known cannabis compounds (including smoke-derived compounds and combustion products) and their concentrations using a number of open-access literature search engines such as Google Scholar (https://scholar.google.ca/), PubMed (https://pubmed.ncbi.nlm.nih.gov/), and ScienceDirect (https://www.sciencedirect.com/). We also used several in-house text-mining software packages that were originally developed for the Human Metabolome Project (HMP) and the Human Metabolome Database (HMDB),46 namely, PolySearch47 and PolySearch2.48 These programs can take simple keywords (i.e., “cannabis”, “chemical”, and “metabolite”) as input and rapidly create hyperlinked lists of abstracts and papers containing information about cannabis metabolites and their corresponding concentration data from PubMed (and other data sources). The text mining was conducted based on various criteria such as compound categories, physiological effects and counter effects, and the detection status of the metabolite (quantified or detected only). The search terms included, but were not limited to, combinations of the keywords (cannabis, marijuana, “cannabis plant”) with (metabolite, cannabinoids, terpenes or terpenoids, polyphenols, “fatty acids”, “primary metabolites”, esters, alkanes, phytoestrogens, herbicides and fungicides, smoke, combustion) and (allergen, analgesic, antibacterial, anticancer, anxiolytic, irritant, psychoactive, sedative, stimulant) and (detected, quantified). This work led to the identification of ∼500 papers, abstracts, and textbooks with relevant chemical information on cannabis chemicals. From these documents, PolySearch2 compiled a ranked list of cannabis metabolites by measuring word co-occurrence frequency using terms such as “cannabis”, “marijuana”, or “hemp” in conjunction with words such as “concentration”, “identification”, “quantification”, “mM”, or “micromole”. PolySearch2 also extracted key sentences from the abstracts and then labeled and hyperlinked the metabolites mentioned in the text.
All literature-derived data regarding cannabis compounds, along with their concentrations and references, were manually compiled, compared, and their names “normalized” to match HMDB,46 Chemical Abstracts Service (CAS), and PubChem identifiers. The manually derived compound data was further annotated using an in-house program called DataWrangler,46 which automatically generates names, synonyms, partial descriptions, structures, chemical taxonomies, physical property data, and bioavailability data. The information generated by DataWrangler was manually checked by five different scientists (the annotation team) with postgraduate degrees in biochemistry, natural product chemistry, and/or chemistry. All compound structures, names, synonyms, and descriptions were extensively reviewed, cross-checked, and, if necessary, redone manually by the annotation team. After the manual checking phase was complete, the data were then entered into the CCD. Concentration data were cross-checked manually to identify any large discrepancies (>3×) among the entered values. Those that exceeded this threshold were reanalyzed to see if data entry errors had occurred. For highly discrepant values, a “majority wins” scheme was used to select the most consistent value(s). On the other hand, if our experimental data matched best with one of the discrepant values, then that value was selected over other literature-reported value(s).
Genome-Scale Inference of Expected Cannabis Compounds
A common set of essential or conserved “primary” plant metabolites was determined by carefully analyzing the published reaction network and metabolome of several diverse, but well-studied plants, including Arabidopsis thaliana, Solanum lycopersicum (tomato), and Oryza sativa (rice). This process involved comparing and consolidating the metabolite/pathway information found in AraCyc,49 PlantCyc,50 the Plant Metabolic Network,50 and various Kyoto Encyclopedia of Genes and Genomes plant pathway collections.51 This information, along with the recently published cannabis genome sequence,52 and other publicly available plant metabolite, protein, and pathway data from PathBank37 and UniProt53 (which provided additional details on plant lipids) were used to generate a genome-scale compilation of highly conserved or “expected” cannabis metabolites. These expected metabolites correspond to endogenously produced compounds that are essential to all known plants based on well-known or well-characterized biochemical pathways or reactions found in all higher plant cells. Many expected compounds correspond to common, high-abundance, so-called primary metabolites as well as lipids, transient intermediates, or low-abundance compounds that are not easily measurable or normally measured in metabolomics experiments.
Construction of the CCD
All data collected via the experimental assays (described above), literature surveys, and genome-scale metabolic inference were entered into the CCD. The CCD was implemented using a Ruby on Rails (version 4.2.0) web framework incorporating a MySQL relational database (version 15.1 Distribution 10.4.6-MariaDB) to manage all the metabolite data, including descriptions, synonyms, physicochemical properties, concentrations, spectra, and external references. The CCD was built using the framework used to develop the HMDB46 and is therefore similar in appearance and structure. The CCD uses the model–view–controller architecture in which internal data logic is separated from user input and data presentation. The raw information stored in the database is dynamically extracted and rendered into web pages. The CCD is hosted on a DigitalOcean server equipped with 4 CPUs, 80 GB of disk space, and 8 GB of RAM.
Results and Discussion
This study’s central aim was to identify, quantify, and comprehensively describe all the chemicals detectable in commercial cannabis (including combustion products) using a combination of comprehensive, quantitative experimental approaches, literature mining, genome-scale metabolic inference, and computer-aided and manual annotation. This entire data set is housed in the CCD, a compilation of all known molecules that could potentially be present in cannabis products, irrespective of whether they are endogenous or exogenous (i.e., pesticides). This section is divided into three subsections covering the following: (1) experimental metabolomics results; (2) literature review and genomic inference, and (3) a detailed description of the CCD.
Experimental Results
All data presented in Tables 1–6, and Tables S3–14 of the Supporting Information were obtained experimentally by our lab via one untargeted and six different targeted, fully quantitative analyses performed on six commercial cannabis samples (Figure 1). These include the following: (1) a targeted GC–MS assay for terpenoids (Table S3); (2) an untargeted GC–MS assay for terpenoids and other volatile compounds (Table S4); four targeted LC–MS/MS assays for (3) cannabinoids (Table S5); (4) polyphenols and phenolic acids (Table S6); (5) pesticides (Table S7); (6) primary plant metabolites (Table S8); and (7) a targeted ICP–MS assay for metal ions (Table S9). These data are listed in the CCD, with this paper as the reference. Details for some methods not described above, along with the validation data, are provided in the Supporting Information (Tables S10–S14). In addition to concentration data, details about each of the chemical classes identified by our assays, including detailed descriptions, biological functions, scent or aroma characteristics, health effects, nomenclature or nomenclature variations, physicochemical data and characteristics, and NMR and/or MS spectra, are provided in the CCD (https://cannabisdatabase.ca).
Table 1. Summary of the Types of Assays, Classes of Metabolites Assayed, Platforms, and Numbers of Metabolites Covered by the Different Methods, Analyzed, Detected, and Quantified in This Study.
| types of analytes detected | platform | type of assay | total # analytes covered | analytes
detected and/or quantified in cannabis
cultivars |
||
|---|---|---|---|---|---|---|
| total | min | max | ||||
| cannabinoids | LC–MS/MSa | targeted | 16 | 16 | 16 | 16 |
| terpenoids | GC–MS | targeted | 45 | 45 | 22 | 33 |
| volatiles and nonderivatizables | GC–MS | untargeted | NA | 104 | 104 | 104 |
| polyphenols and phenolic acids | UHPLC–HRMS | targeted | 19 | 17 | 14 | 16 |
| pesticides | RPLC–API–MS/MS | targeted | 71 | 14 | 3 | 6 |
| metal ions | ICP–MS | targeted | 16 | 16 | 14 | 16 |
| common plant metabolites: amino acids, biogenic amines, glucose, organic acids, plant hormones, acylcarnitines, PCs, LysoPCs, SMs, and SM(OH)s | RPLC–DFI–MS/MS | targeted | 210 | 72 | 72 | 72 |
| total | 377 | 284 | 141 | 159 | ||
Abbreviations: LysoPC—lysophosphatidylcholine, PC—phosphatidylcholine, SM—sphingomyelin, SM(OH)—hydroxylated sphingomyelin; GC–MS—gas chromatography–mass spectrometry; LC–MS/MS—liquid chromatography–MS/MS; UHPLC-HRMS—ultra-high-performance LC–high-resolution MS; ICP–MS—inductively coupled plasma–MS.
Table 6. Averages of Metals Plus Standard Deviations Detected from the Least to Most by ICP–MS in the Six Cannabis Cultivars.
| metal ion | Cannabis Compound Database # | average ± SD |
|---|---|---|
| cesium (Cs) | CDB006361 | 0.041 (1)ang/g |
| thallium (Tl) | CDB006363 | 5.4 ± 3.5 ng/g |
| titanium (Ti) | CDB006357 | 1.23 ± 0.21 μg/g |
| molybdenum (Mo) | CDB006360 | 3.35 ± 2.54 μg/g |
| barium (Ba) | CDB006362 | 3.612 ± 2.31 μg/g |
| copper (Cu) | CDB005281 | 7.93 ± 5.48 μg/g |
| rubidium (Rb) | CDB006358 | 9.98 ± 3.22 μg/g |
| boron (B) | CDB006355 | 31.8 ± 4.64 μg/g |
| zinc (Zn) | CDB005206 | 70.14 ± 38.6 μg/g |
| strontium (Sr) | CDB006359 | 95.6 ± 68.4 μg/g |
| manganese (Mn) | CDB005163 | 125.1 ± 34.3 μg/g |
| iron (Fe) | CDB005174 | 182 ± 78.6 μg/g |
| magnesium (Mg) | CDB005083 | 4.69 ± 1.11 mg/g |
| phosphorous (P) | CDB006356 | 9.66 ± 1.51 mg/g |
| calcium (Ca) | CDB005200 | 9.66 ± 4.9 mg/g |
| potassium (K) | CDB005090 | 40.77 ± 6.33 mg/g |
All metals were quantified in all cultivars except cesium (Cs) which was only quantified in one cultivar.
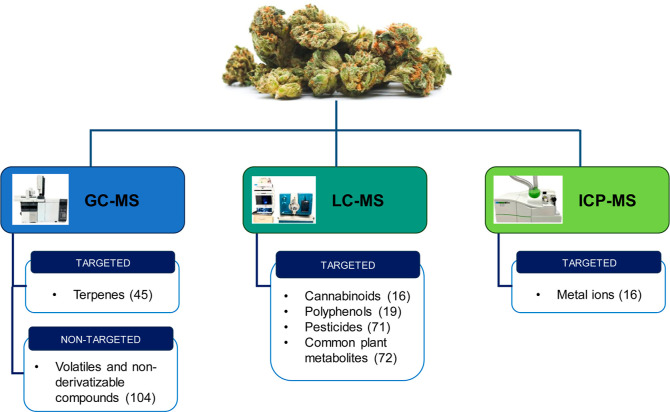
Figure 1.
Schematic representation of the platforms used in this study for the targeted and untargeted metabolomics assays.
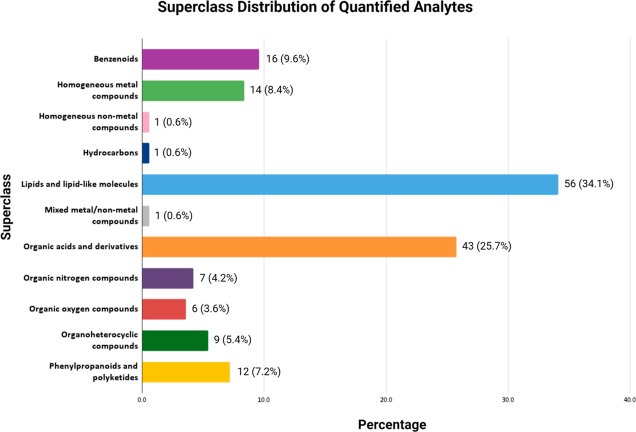
A total of 377 analytes were covered by the six targeted quantitative assays (Table 1). From the seven assays listed above, 284 metabolites were either identified (104) through our untargeted assay and/or detected and quantified (179) with our targeted assays in the six cannabis samples (Table 1). The quantified metabolites were grouped by their chemical superclass using the ClassyFire chemical taxonomy,54 and their distribution is shown in Figure 2. The most abundant class of metabolites belonged to “lipids and lipid-like molecules”, whereas the “homogeneous nonmetal compounds”, the “hydrocarbons”, and the “mixed metal/nonmetal compounds” were the least abundant class of compounds. Due to their extrinsic nature, the analytes from our pesticides assay were not used in this classification.
Figure 2.
Distribution of the 179 detected and quantified metabolites across chemical super classes.
Terpenoids are particularly important phytochemicals as they often generate the characteristic scent or aroma of many flowers, plants, or trees, which are used to either attract or repel insects or other plant predators. For cannabis, the characteristic aroma or flavor associated with particular cultivars is often defined by the terpenoid composition. Our targeted GC–MS assay was designed to detect 45 terpenoids, which included 22 monoterpenoids and 23 sesquiterpenoids. The assay had an intra- and inter-day precision of ≤15% coefficient of variation (CV) and a recovery ranging between 82 and 119%. As can be seen from Table S3, an average of 27 terpenoids in each cultivar were detected and quantified with the least (22/45 compounds) in the “Island Honey” cultivar and the most (33/45 compounds) in the Sensi Star cultivar. The most abundant terpenoid, (E)-β-farnesene, was found in the Gabriola cultivar and in 4/6 cultivars (Table 2). The other most abundant terpenoids in Gabriola were eudesma-3,7(11)-diene, trans-caryophyllene, and R-(+)-limonene. The other most abundant terpenoids included (−)-α-bisabolol, linalool, β-myrcene, trans-nerolidol, and terpinolene. Our terpenoid analysis demonstrated that the six cultivars have very different terpenoid compositions, which could explain the varied aromas and flavors associated with each cultivar (Table 2). The values found with these cannabis terpenoids agree with those published elsewhere.55,56
Table 2. Most Abundant Terpenoids Found in the Six Cannabis Cultivars and Their Flavor and Aroma Profiles.
| terpenoid | Cannabis Compound Database # | aroma/flavor | cultivar (mg/g dry weight) |
|||||
|---|---|---|---|---|---|---|---|---|
| Alien Dawg | Tangerine Dream | Sensi Star | Quadra | Gabriola | Island Honey | |||
| (–)-α-bisabolol | CDB000119 | balsamica | 0.26 | 0.5 | 0.4 | 0.46 | 0.45 | 0.39 |
| trans-caryophyllene | CDB000712 | sweet, woody, spicy, clove, skunky smell of smoke | 1.86 | 0.85 | 0.78 | 1.02 | 2.37 | 1.32 |
| eudesma-3,7(11)-diene | CDB000219 | found in essential oil of hops | 1.32 | NDb | 0.39 | 0.77 | 1.58 | 0.59 |
| (E)-β-farnesene C | CDB000352 | woody, citrus, herbal, sweet | 1.54 | 2.21 | ND | 1.01 | 3.1 | ND |
| R-(+)-limonene | CDB000069 | citrus, orange, fresh, sweet | 0.86 | 0.26 | 0.18 | 1.38 | 2.48 | 0.12 |
| linalool | CDB000089 | citrus, floral, sweet, bois de rose, woody, green blueberry | 0.11 | 0.29 | ND | 0.89 | 0.7 | 0.19 |
| β-myrcene | CDB000573 | peppery, terpenic, spicy, balsamic, plastic, skunky smell of smoke | 1.17 | 1.71 | 0.75 | 0.38 | 0.42 | 0.44 |
| trans-nerolidol | CDB000351 | floral, green, waxy, citrus, woody | ND | 1.09 | ND | ND | ND | 1.1 |
| terpinolene | CDB000209 | herbal, fresh woody sweet, pine, citrus | ND | ND | 1.89 | ND | ND | ND |
Sources for scents: for most compounds, information about scents can be found in the CCB as well as the good scent company (http://www.thegoodscentscompany.com/) and the ContaminantDB (https://contaminantdb.ca/).
ND—not detected.
For the untargeted GC–MS terpenoid assay, the analytes were putatively identified at a confidence level of two. The identification criteria were based on a combination of the RIs and matching the experimental electron impact (EI)–MS fragmentation patterns of the corresponding compounds with the EI–MS spectra in the NIST20 GC–MS library. For those not found in NIST20 library, further matching was based on the scale published by the Metabolomics Standard Initiative and further refined by Schymanski et al., 2014.57 This approach allowed us to detect 104 volatile terpenoids, which include 1 hemiterpenoid, 34 monoterpenoids, 46 sesquiterpenoids, 1 diterpenoid, 1 alkanes, 10 esters, and 10 unclassified volatile organic compounds (Table S4). On average, 59 molecules were detected in the analyzed samples, with Tangerine Dream having the lowest number, while Sensi Star had the highest number. Sensi Star also had the highest number of detected and quantified terpenoids in our targeted assay. As this was a qualitative assay in which volatile terpenoids were not quantified, we cannot comment precisely on how the detected compounds contribute to the flavor and aroma of the cultivars. While the compound coverage of our GC–MS terpenoid assay was quite broad, the use of a longer column (and a longer runtime) along with a higher-resolution mass spectrometer could have increased compound coverage by another 20–30%.
Cannabinoids play a pivotal role in cannabis bioactivity and in human health. Our LC–MS/MS targeted cannabinoids assay detected and quantified 16 cannabinoids (Table S5). Among them, tetrahydrocannabinolic acid (THCA) (not THC) was the most abundant compound found in all cultivars, with concentrations ranging from 133 mg/g in the Tangerine Dream and Gabriola cultivars to 162 mg/g in the Sensi Star and Alien Dawg cultivars. The nonpsychoactive THCA is expected to be in higher levels in dry cannabis than that in THC since THCA is decarboxylated to THC with heating (by cooking or smoking).58,59 Cannabidivarin (CBDV) was the least abundant compound found in low concentrations ranging from 0.312 μg/g in the Alien Dawg cultivar to 1.58 μg/g in the Quadra cultivar. The averages with standard deviation (±SD) of cannabinoids (from the least to the most) quantified in the six cultivars are shown in Table 3. The calibration curves range from 0.0625 to 12.5 μg/mL for all the analytes. The recovery rates from spiking plant samples with low (0.125 μg/mL), medium (1.25 μg/mL), and high (6.25 μg/mL) concentrations ranged from 80 to 120% with precision values of <20%. The assay had an intra- and inter-day precision of 20% CV. The cannabinoids’ concentrations found in these cultivars fall within those reported elsewhere.60
Table 3. Averages of Cannabinoids Quantified (from the Least to Most) in the Six Cultivars Using a Targeted LC–MS/MS Metabolomics Assay.
| cannabinoid | average concentration ± SD (mg/g) |
|---|---|
| cannabidivarin (CBDV) | 0.002 ± 0.001 |
| cannabidivarinic acid (CBDVA) | 0.063 ± 0.028 |
| 11-nor-9-carboxy-tetrahydrocannabinol (THC–COOH) | 0.087 ± 0.059 |
| cannabicyclolic acid (CBLA) +cannabichromenic acid (CBCA) | 0.099 ± 0.017 |
| cannabinolic acid (CBNA) | 0.176 ± 0.027 |
| cannabidiolic acid (CBDA) | 0.227 ± 0.039 |
| tetrahydrocannabivarin (THCV) | 0.227 ± 0.080 |
| cannabigerolic acid; (CBGA) | 0.317 ± 0.112 |
| cannabidiol (CBD) | 0.385 ± 0.246 |
| cannabinol (CBN) | 0.412 ± 0.226 |
| cannabichromene (CBC) | 0.605 ± 0.402 |
| cannabigerol (CBG) | 0.738 ± 0.456 |
| cannabicyclol (CBL) | 0.778 ± 0.581 |
| Δ9-tetrahydrocannabinol (THC) | 4.792 ± 1.908 |
| tetrahydrocannabinolic acid (THCA) | 148.833 ± 19.364 |
Polyphenols and phenolic acids are important phytochemicals as they often play a role in plant coloration as well as providing defenses against ultraviolet radiation, drought, other abiotic stresses, or attacks by pathogens. Our LC–MS phenolic assay was designed to detect and quantify 19 free polyphenols and phenolic acids (aglycones only). As can be seen from Table S6, our phenolic assay measured a minimum of 15 compounds for two cultivars (Alien Dawg and Gabriola) and a maximum of 16 compounds for the remaining four cultivars (Quadra, Tangerine Dream, Sensi Star, and Island Honey). Two polyphenols, gallocatechin and pungenol, were not detected or quantified in any cultivar. The averages (±SD) from the least to most polyphenols quantified in the six cultivars are shown in Table 4. The least abundant polyphenol was isorhamnetin in the Island Honey cultivar, while the least averaged polyphenol was naringenin. The highest averaged polyphenol in all cultivars and the most abundant polyphenol was catechin in the Quadra cultivar. While several studies report total phenolic content in cannabis using the traditional antioxidant Folin–Ciocalteu method,61−63 there is a scarcity of articles reporting the concentration of specific polyphenols.64 Hence, our study is quite unique for reporting an exceptionally broad coverage of cannabis polyphenols. However, the performance of our assay was somewhat limited due to the lack of availability of authentic isotopically labeled phenolic standards. The use of chemo-selective labeling methods65 could potentially increase the analytical coverage by a factor of two or more.
Table 4. Averages of Phenolic Compounds from the Least to Most in Cannabis Cultivars Detected by LC–MS/MS.
| polyphenol | Cannabis Compound Database # | avg ± SD (μg/g) |
|---|---|---|
| gallocatechin | CDB006404 | NDa |
| pungenol | CDB006403 | ND |
| naringenin | CDB004939 | 0.73 ± 0.16 (2)b |
| isorhamnetin | CDB004938 | 0.94 ± 0.49 (6) |
| apigenin | CDB000370 | 0.94 ± 0.88 (6) |
| gallic acid | CDB004972 | 0.96 ± 0.20 (6) |
| caffeic acid | CDB006367 | 1.59 ± 0.75 (6) |
| protocatecuic aldehyde | CDB006366 | 2.43 ± 1.2 (6) |
| kaempferol | CDB000373 | 6.40 ± 3.9 (6) |
| piceol | CDB005786 | 6.41 ± 0.38 (3) |
| vanillic acid | CDB006148 | 9.33 ± 5.4 (5) |
| quercetin | CDB000372 | 9.96 ± 8.9 (6) |
| ferulic acid | CDB000044 | 13.33 ± 4.13 (6) |
| p-coumaric acid | CDB000177 | 15.03 ± 12.2 (6) |
| taxifolin | CDB006368 | 16.8 ± 0.43 (6) |
| myricetin | CDB004941 | 19.72 ± 1.2 (6) |
| vanillin | CDB005772 | 27.86 ± 1.5 (6) |
| 3,4-dihydroxybenzoic acid | CDB006364 | 31.7 ± 10.4 (6) |
| catechin | CDB006365 | 145 ± 15 (6) |
ND—not detected.
(#)—number of cultivars where compound was detected.
Because cannabis is an intensely cultivated crop, it is susceptible to a wide range of bacterial, fungal, parasitic, or insect attacks. Therefore, cannabis crops are often treated with a wide range of herbicides or pesticides. Most legal cannabis sold in Canada is grown indoors in specialized and highly regulated greenhouses, which limits the need for herbicides. In other countries, cannabis (legal and illegal) is grown both outdoors and indoors, requiring a greater use of herbicides, pesticides, and other biocides. Our targeted LC–MS assay was designed to detect and quantify 71 pesticides in cannabis cultivars, aiming to identify as many approved pesticides as possible, irrespective of the region or country of cultivation (Table S7). The calibration curves ranged from 0.1 to 500 ng/mL, and the 20 min chromatographic separation achieved a CV ≤ 10% and accuracy between 80 and 120%. Only 14/71 compounds could be quantified, with the lowest number (3) detected in the Gabriola cultivar and the highest (6) detected for the Alien Dawg cultivar. Most pesticides fell below the LOQ of our in-house assay (Table S7). Among those detected, the least abundant pesticides were ethoprophos, malathion, mevinphos, and MGK-264 found in three cultivars, and the most abundant pesticide was methoprene in the Alien Dawg cultivar (Table S7). While some pesticides were detected in all six cultivars, only benzovindiflupyr, metalaxyl, and methoprene in the Alien Dawg cultivar were at concentrations > LOQ set by Health Canada43 (Table S7). The measurement of pesticides in cannabis is crucial for ensuring the safety and quality of commercial cannabis products in both North America and Europe. Other jurisdictions have different rules regarding the use of different pesticides or herbicides, as well as their tolerable limits. Cannabis products grown or prepared illegally are more likely to have higher (and potentially unsafe or toxic) concentrations of these pesticides.
In addition to measuring secondary metabolites (terpenoids and polyphenols), we also measured many primary metabolites, i.e., those compounds involved in the growth, development, and reproduction. Our LC–MS/MS plant metabolite assay detected and quantified 72 compounds in the six cultivars (Table S8). The detectable compounds and their average concentrations (±SD) from the lowest to highest are listed in Table 5. Compounds in several subclasses were detected, including 31 amino acids and analogues, 13 organic acids, 7 biogenic amines, 3 plant hormones, 1 fatty acids and conjugates, 5 glycerophosphocholines, 6 phenolic compounds, 2 carbohydrate and conjugates, and 4 other subclasses. The least abundant compound was phenylethylamine (0.048 μg/g) in the Sensi Star cultivar. The most abundant compound was asparagine (36.3 mg/g) in the Alien Dawg cultivar. The most abundant primary metabolites (>0.1–20 mg/g) were amino acids, glucose, glycerate, choline, and four organic acids (Table 5).
Table 5. Average of Primary Metabolites Arranged by Sub-Classes and from the Least to Most in Cannabis Cultivars Detected by LC–MS/MS.
| Cannabis Compound Database # | metabolites | chemical subclass | amount (mg/g) avg ± SD | Cannabis Compound Database # | metabolites | chemical sublass | amount (mg/g) avg ± SD |
|---|---|---|---|---|---|---|---|
| CDB000060 | methionine | amino acids, peptides, and analogues | 0.011 ± 0.003 | CDB000235 | aspartic acid | amino acids, peptides, and analogues | 1.038 ± 0.479 |
| CDB000083 | tyrosine | amino acids, peptides, and analogues | 0.07 ± 0.021 | CDB004853 | glutamine | amino acids, peptides, and analogues | 3.26 ± 1.855 |
| CDB006139 | glycine | amino acids, peptides, and analogues | 0.108 ± 0.043 | CDB000117 | arginine | amino acids, peptides, and analogues | 5.25 ± 1.55 |
| CDB000134 | phenylalanine | amino acids, peptides, and analogues | 0.2 ± 0.108 | CDB004813 | asparagine | amino acids, peptides, and analogues | 20.56 ± 9.22 |
| CDB000135 | threonine | amino acids, peptides, and analogues | 0.211 ± 0.054 | CDB006327 | kynurenine | amino acids, peptides, and analogues | 0.0008 ± 0.0006 |
| CDB000120 | lysine | amino acids, peptides, and analogues | 0.237 ± 0.111 | CDB006328 | creatine | amino acids, peptides, and analogues | 0.0028 ± 0.001 |
| CDB000132 | isoleucine | amino acids, peptides, and analogues | 0.258 ± 0.156 | CDB006325 | asymmetric dimethylarginine | amino acids, peptides, and analogues | 0.0032 ± 0.002 |
| CDB000136 | valine | amino acids, peptides, and analogues | 0.350 ± 0.143 | CDB006322 | trans-hydroxyproline | amino acids, peptides, and analogues | 0.003 ± 0.0008 |
| CDB000133 | leucine | amino acids, peptides, and analogues | 0.179 ± 0.079 | CDB006330 | methylhistidine | amino acids, peptides, and analogues | 0.004 ± 0.002 |
| CDB004871 | tryptophan | amino acids, peptides, and analogues | 0.377 ± 0.249 | CDB | total dimethylarginine | amino acids, peptides, and analogues | 0.007 ± 0.002 |
| CDB000128 | alanine | amino acids, peptides, and analogues | 0.380 ± 0.077 | CDB004788 | betaine | amino acids, peptides, and analogues | 0.012 ± 0.004 |
| CDB000130 | glutamic acid | amino acids, peptides, and analogues | 0.382 ± 0.162 | CDB006324 | acetyl-ornithine | amino acids, peptides, and analogues | 0.017 ± 0.005 |
| CDB000131 | serine | amino acids, peptides, and analogues | 0.483 ± 0.113 | CDB006323 | methionine-sulfoxide | amino acids, peptides, and analogues | 0.022 ± 0.018 |
| CDB000059 | histidine | amino acids, peptides, and analogues | 0.629 ± 0.281 | CBD005105 | ornithine | amino acids, peptides, and analogues | 0.033 ± 0.032 |
| CDB000105 | proline | amino acids, peptides, and analogues | 1.025 ± 0.899 | CDB004869 | citrulline | amino acids, peptides, and analogues | 0.051 ± 0.02 |
| CDB000126 | phenylethyl-amine | phenethylamines | 0.0002 ± 0.0002 | CBD005134 | α-aminoadipic acid | amino acids, peptides, and analogues | 0.071 ± 0.035 |
| CDB005121 | histamine | amines | 0.0005 ± 0.0003 | CDB006321 | taurine | organosulfonic acids and derivatives | 0.003 ± 0.002 |
| CDB004900 | spermine | amines | 0.0008 ± 0.0005 | CDB006164 | glyceric acid | carbohydrate and conjugates | 0.204 ± 0.087 |
| CDB004914 | putrescine | amines | 0.001 ± 0.0003 | CDB005078 | glucose | carbohydrate and conjugates | 8.273 ± 8.605 |
| CDB004901 | spermidine | amines | 0.0031 ± 0.0027 | CDB006137 | carnitine | quaternary ammonium salts | 0.0045 ± 0.002 |
| CDB006329 | trimethyl-amine N-oxide | aminoxides | 0.0031 ± 0.0003 | CDB006345 | choline | quaternary ammonium salts | 1.069 ± 0.197 |
| CDB006168 | tyramine | phenethylamines | 0.0134 ± 0.014 | CDB005162 | shikimic acid | alcohols and polyols | 0.002 ± 0.0004 |
| CDB004839 | uric acid | purine and purine derivatives | 0.0202 ± 0.007 | CDB005178 | abscisic acid | sesquiterpenoids | 0.002 ± 0.001 |
| CDB005315 | jasmonic acid | lineolic acids and derivatives | 0.014 ± 0.007 | CDB006354 | methylmalonic acid | dicarboxylic acids and derivatives | 0.0003 ± 0.00006 |
| CDB004817 | lactic acid | α-hydroxy acids and derivatives | 0.0246 ± 0.014 | CDB004809 | fumaric acid | dicarboxylic acids and derivatives | 0.025 ± 0.025 |
| CDB006332 | β-hydroxy butyric acid | β-hydroxy acids and derivatives | 0.0072 ± 0.002 | CDB006138 | succinic acid | dicarboxylic acids and derivatives | 0.038 ± 0.012 |
| CDB006173 | malic acid | β-hydroxy acids and derivatives | 0.533 ± 0.263 | CDB005136 | glutaric acid | dicarboxylic acids and derivatives | 0.043 ± 0.024 |
| CDB004829 | pyruvic acid | α-keto acids and derivatives | 0.003 ± 0.0005 | CDB006339 | oxalic acid | dicarboxylic acids and derivatives | 0.287 ± 0.565 |
| CDB004819 | α-ketoglutaric acid | ɣ-keto acids and derivatives | 0.0019 ± 0.0011 | CDB004795 | aconitic acid | tricarboxylic acids and derivatives | 0.539 ± 0.407 |
| CDB006335 | propionic acid | carboxylic acids | 0.0153 ± 0.008 | CDB006159 | citric acid | tricarboxylic acids and derivatives | 5.81 ± 3.22 |
| CDB006405 | PC aa C36:0a | glycerophospho-cholines | 0.0004 ± 0.0001 | CDB006333 | HPHPA | phenylpropanoic acids | 0.0001 ± 0.000004 |
| CDB006406 | PC aa C36:6 | glycerophospho-cholines | 0.0015 ± 0.0003 | CDB006334 | homovanillic acid | methoxyphenols | 0.0004 ± 0.0005 |
| CDB006342 | LysoPC a C18:1 | glycerophospho-cholines | 0.0017 ± 0.0005 | CDB006336 | p-hydroxy-phenylacetic acid | 1-hydroxy-2-unsubstituted benzenoids | 0.002 ± 0.0006 |
| CDB004990 | LysoPC a C16:0 | glycerophospho-cholines | 0.0061 ± 0.002 | CDB006338 | hippuric acid | benzoic acids and derivatives | 0.0013 ± 0.0009 |
| CDB006341 | LysoPC a C18:2 | glycerophospho-cholines | 0.008 ± 0.004 | CDB004930 | salicylic acid | benzoic acids and derivatives | 0.016 ± 0.007 |
| CDB006337 | butyric acid | fatty acids and conjugates | 0.0195 ± 0.011 | CDB004927 | benzoic acid | benzoic acids and derivatives | 0.061 ± 0.028 |
Abbreviations: HPHPA—3-(3-hydroxyphenyl)-3-hydroxypropanoic acid; LysoPC—lysophosphatidylcholine; PC—phosphatidylcholine.
Metal ions and trace elements play a crucial role in the growth and development of plants as they serve as essential components of various enzymes and proteins. Our ICP–MS assay detected 16 metal ions in our cannabis cultivars (Table S9). Cesium was the most challenging to detect, found only in the Island Honey cultivar. The metal concentrations in all cultivars were within reported ranges,66 and the most abundant were K (avg. 40 mg/g) > Ca > P > Mg (4.7 mg/g). The least abundant metals found in all cultivars were thallium—Tl (5 ng/g) < Mo < Ti < Cu (7.9 μg/g) (Table 6). Although found in low levels, metal toxicity from smoking cannabis could be a concern. Accumulation of certain metals, like arsenic (As), Tl, or lead (Pb), can be toxic when inhaled through smoking. Our ICP–MS assay is able to detect 41 elements, including As, Tl, and Pb. No As or Pb and only minute amounts of Tl (ng/g) were detected and quantified in our cultivars, suggesting that they are all safe for human consumption. Smoking cannabis grown in different soil and environmental conditions may lead to higher metal content, potentially posing health risks.
Literature Review and Genomic Inference
A computer-aided literature survey was conducted using PolySearch47 and PolySearch2.48 Chemical composition data was extracted from more than 480 scientific articles. This “‘bibliomic’” effort yielded data for another 2107 cannabis metabolites or metabolite species. The experimentally acquired metabolite data was then combined with genome-scale metabolite inference—a technique commonly used to fill in the metabolic “holes” for other metabolomes, such as the human metabolome,46 the yeast metabolome,35 and the Escherichia coli metabolome.36 This method uses known, organism-specific metabolic pathways37 and known, organism-specific gene/protein reactions to provide data on metabolites that are known to exist but are not normally measured via NMR or MS techniques. This led to the addition of another 3969 metabolites or metabolite species. All the acquired compound data was then annotated by a team of curators, through careful review of the literature, to include known or reported physiological and psychoactive effect data, known protein (human) target data, and metabolic/signaling pathways. These data were further supplemented with computationally calculated physicochemical and spectral data using methods described previously.46
Cannabis Compound Database
In addition to our experimental data, all identified, detected, quantified, mined from the literature, and genomically inferred compounds in cannabis have been deposited into a freely accessible database called the CCD, available at https://cannabisdatabase.ca. The CCD contains a complete list of cannabis chemicals including their common and International Union of Pure and Applied Chemistry (IUPAC) names, their structure in multiple formats, basic descriptions, chemical ontology, physicochemical properties (predicted or measured), their reference spectra (NMR, GC–MS, and LC–MS), their Kovats RIs, collisional cross-section (CCS), pathway information (as derived from PathBank),37 and references to scientific literature (to the best of our knowledge) of all cannabis compounds that have ever been identified, quantified, or reported either in this paper or in existing scientific literature. Currently, the CCD contains information on 891 unique, experimentally detected compounds with well-defined structures and names. The CCD also contains 6331 compounds that have been computationally inferred from detailed genomic analysis, biochemical pathway analysis, and a comparison to other plant metabolomes.
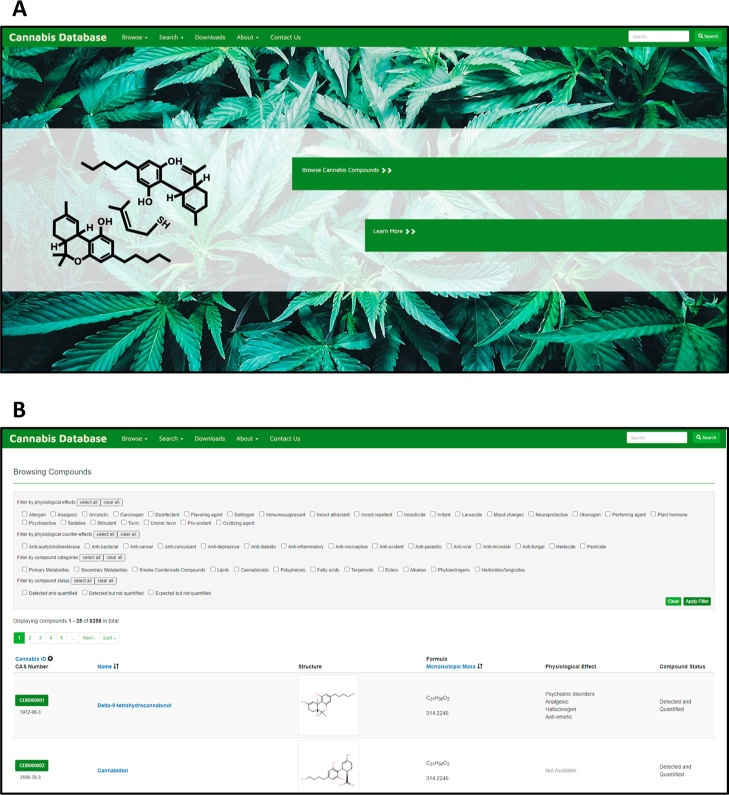
The CCD has been structured and designed after other popular, user-friendly databases developed by our group such as the HMDB,46 Yeast metabolome Database (YMDB),35 or Microbial Metabolome Database (MiMeDB).67 A screenshot of the CCD homepage is shown in Figure 3A. The navigation panel consists of five menu options: Browse, Search, Downloads, About, and Contact Us. Below the navigation panel are two colored hyperlinked bars, (i) Browse Cannabis Compounds and (ii) Learn More. Clicking on them takes the user to the compound browser or the summary of the CCD, respectively.
Figure 3.
Screenshot of the CCD showing the (A) CCD Homepage layout and (B) “Browsing Compounds” viewing page.
Under the Browse tab, there are options to browse by (i) Compounds, (ii) Protein Targets, (iii) Cannabis Cultivars, and (iv) Pathways. The “Compounds” option displays a table with compound details that can be sorted or filtered (Figure 3B). Clicking on a compound name or Cannabis ID leads to a detailed MetaboCard (Figure S1A). Each MetaboCard contains 11 data fields, including: (i) CCDB Record Information, (ii) Cannabis Compound Identification (including a detailed description, the structure, and other chemical features of the compound), (iii) Chemical Taxonomy, (iv) Ontology, (v) Physical Properties, (vi) Spectra, (vii) Pathways, (viii) Protein Targets, (ix) Concentrations Data, (x) External Links, and xi) References.
Also, under the Browse menu, users may select “Protein Targets”, which lists known human protein receptor targets of various cannabis compounds, “Cannabis Cultivars”, which brings users to detailed descriptions for 115 known cannabis cultivars, and “Pathways”, which comprises both human and plant pathways. The Search tab offers options to search by compound structure, molecular weight, general text, sequence, and spectra. Results from ChemQuery and a GC–MS search are shown in Figure S1B,C, respectively. Users can download CCD data files, available in comma-separated value (CSV), structure data file (SDF), or extensible markup language (XML) formats. The About tab provides an overview of the CCD, database statistics, and information regarding its Findable, Accessible, Interoperable and Resuable (FAIR) compliance.
The CCD was assembled using a combination of computer-aided literature mining, data mining of open-access databases, manual annotations, and automated genome annotation. The same quality control procedures implemented for all our group’s databases were applied.46,67,68 This included curator training, data provenance tracking, continuous data quality and completeness review, and secondary checks by independent reviewers. The CCD is continuously updated with minor changes made regularly without any formal announcement. The current version of this database is CCD v. 1.0. Subsequent large-scale updates will be assigned new version numbers (2.0, 3.0, etc.) along with their corresponding update dates.
The CCD is FAIR compliant.69 Details concerning its FAIRness are provided in the About tab under the “FAIR Compliance” submenu. To ensure findability, all entries in the CCD have a unique identifier consisting of a six-digit CCD identifier. To ensure accessibility, CCD provides not only a well-supported web-based user interface with extensive search functions but also an API located under the About menu tab, to support programmatic access to the data. To ensure interoperability, all the CCD compounds are interlinked to major external sites such as HMDB, FooDB, ChemSpider,70 KNApSAcK,71 and CHEBI. All the molecular or physiological data in the CCD have clear references to other established references, meta-data, or data resources. To ensure reusability, all data in CCD are extensively sourced with clear provenance. The data CCD are released under the Creative Commons 4.0 License Suite according to the Attribution BY and noncommercial (NC) licensing conditions.
Limitations and Future Plans
The total number of chemical compounds present in the CCD is not a static value, and there are certainly far more compounds in cannabis than that currently listed in the CCD. As advancements in technology and instrument sensitivity continue to occur, it is expected that the number of compounds in the CCD will inevitably increase. This is because lower abundance metabolites that were previously undetectable will now be able to be identified. As these are identified, we will endeavor to include them in future iterations of the CCD. Likewise, as more data is gathered, measured, and published regarding the physiological or physicochemical properties of compounds already in the CCD, we intend to add this information to the corresponding CCD entries as quickly as possible. While the CCD does not currently have a mechanism for data deposition, interested parties can contact us to request to have their data deposited into the CCD, and we will handle it on their behalf. Their contributions, as seen in other databases managed by our group, can significantly enhance this resource. We expect that, over the coming decade, the CCD will become progressively more comprehensive and inclusive, providing researchers with even more information about the diverse range of compounds that can be found in cannabis—along with more information about their molecular targets and physiological effects.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Institutes for Health Research (CIHR), the Canada Foundation for Innovation (CFI), and Genome Canada.
Glossary
Abbreviations
- API
application programming interface
- BHT
butylated hydroxytoluene
- CBC
cannabichromene
- CC
Creative Commons
- CBD
cannabidiol
- CBDA
cannabidiolic acid
- CBDV
cannabidivarin
- CBDVA
cannabidivarinic acid
- CBG
cannabigerol
- CBGA
cannabigerolic acid
- CBL
cannabicyclol
- CBN
cannabinol
- CDB
Cannabis Compound Database
- CV
coefficient of variation
- CCV
continuing calibration verification
- CSV
comma-separated value
- DFI
direct flow injection
- EDC
carbodiimide
- GC–MS
gas chromatography–mass spectrometry
- GC-FID
gas chromatography with flame ionization detection
- HMDB
Human Metabolome Database
- HMP
Human Metabolome Project
- ICP-MS
inductively coupled plasma-mass spectrometry
- In
indium
- ISTD
internal standard
- IUPAC
International Union of Pure and Applied Chemistry
- KED
kinetic energy discrimination
- LC–MS
liquid chromatography–mass spectrometry
- MiMeDB
Microbial Metabolome Database
- MRM
multiple reaction monitoring
- NMR
nuclear magnetic resonance spectroscopy
- 3-NPH
3-nitrophenylhydrazines
- PCs
phosphatidylcholines
- PITC
phenylisothiocyanate
- QC
quality control
- RIs
retention indices
- RPLC
reversed-phase liquid chromatography
- SDF
structure data file
- SM
sphingomyelin
- SM (OH)
hydroxylated sphingomyelin
- Δ8-THC
Δ8-tetrahydrocannabinol
- THC
Δ9-tetrahydrocannabinol
- THCA
tetrahydrocannabinolic acid
- THCV
tetrahydrocannabivarin
- UHPLC-HRMS
ultra-high-performance liquid chromatography–high-resolution mass spectrometry
- XML
extensible markup language
- YMDB
Yeast Metabolome Database
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.3c06616.
Chemical standards and isotope-labeled internal standards used to identify and quantify common plant metabolites, targeted GC–MS analysis of terpenoids in cannabis, untargeted GC–MS analysis of terpenoids and volatile compounds, cannabinoids quantified using a targeted LC–MS/MS metabolomics assay, targeted LC–MS/MS analysis of phenolic compounds found in cannabis, quantitative RPLC–API–MS/MS method to determine the presence of pesticides in cannabis, common plant metabolites quantified by LC–MS/MS, trace elemental analysis using ICP–MS and validation data, and montage of the CCD(PDF)
The authors declare no competing financial interest.
This paper was published ASAP on January 5, 2024, with an incorrect spelling to the author name Shirin Zahraei. The corrected version was reposted February 2, 2024.
Supplementary Material
References
- Sommano S. R.; Chittasupho C.; Ruksiriwanich W.; Jantrawut P. The cannabis terpenes. Molecules 2020, 25 (24), 5792. 10.3390/molecules25245792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand E. J.; Zhao Z. Cannabis in Chinese medicine: Are some traditional indications referenced in ancient literature related to cannabinoids?. Front. Pharmacol 2017, 8, 108. 10.3389/fphar.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner T. F. Marijuana in ancient Greece and Rome? The literary evidence. Bull. Hist. Med. 1973, 47 (4), 344–355. [PubMed] [Google Scholar]
- Naville S.$340 Billion: The Global Cannabis Market. https://www.gbnews.ch/340-billion-the-global-cannabis-market/ (accessed 2023-04-04).
- Aizpurua-Olaizola O.; Soydaner U.; Öztürk E.; Schibano D.; Simsir Y.; Navarro P.; Etxebarria N.; Usobiaga A. Evolution of the cannabinoid and terpene content during the growth of Cannabis sativa plants from different chemotypes. J. Nat. Prod. 2016, 79 (2), 324–331. 10.1021/acs.jnatprod.5b00949. [DOI] [PubMed] [Google Scholar]
- ElSohly M. A.; Slade D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78 (5), 539–548. 10.1016/j.lfs.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Berman P.; Futoran K.; Lewitus G. M.; Mukha D.; Benami M.; Shlomi T.; Meiri D. A new ESI-LC/MS approach for comprehensive metabolic profiling of phytocannabinoids in cannabis. Sci. Rep. 2018, 8 (1), 14280. 10.1038/s41598-018-32651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting P. F.; Wolff R. F.; Deshpande S.; Di Nisio M.; Duffy S.; Hernandez A. V.; Keurentjes J. C.; Lang S.; Misso K.; Ryder S.; Schmidlkofer S.; Westwood M.; Kleijnen J. Cannabinoids for medical use: A systematic review and meta-analysis. JAMA, J. Am. Med. Assoc. 2015, 313 (24), 2456–2473. 10.1001/jama.2015.6358. [DOI] [PubMed] [Google Scholar]
- Anand U.; Pacchetti B.; Anand P.; Sodergren M. H. Cannabis-based medicines and pain: A review of potential synergistic and entourage effects. Pain Manage. 2021, 11 (4), 395–403. 10.2217/pmt-2020-0110. [DOI] [PubMed] [Google Scholar]
- Russo E. B. The case for the entourage effect and conventional breeding of clinical cannabis: No “Strain,” no gain. Front. Plant Sci. 2019, 9, 1969. 10.3389/fpls.2018.01969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo E. B. Taming THC: potential cannabis synergy and phytocannabinoid-terpenoid entourage effects. Br. J. Pharmacol. 2011, 163, 1344–1364. 10.1111/j.1476-5381.2011.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zager J. J.; Lange I.; Srividya N.; Smith A.; Lange B. M. Gene networks underlying cannabinoid and terpenoid accumulation in cannabis. Plant Physiol. 2019, 180 (4), 1877–1897. 10.1104/pp.18.01506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eržen M.; Košir I. J.; Ocvirk M.; Kreft S.; Čerenak A. Metabolomic analysis of cannabinoid and essential oil profiles in different hemp (Cannabis sativa l.) phenotypes. Plants 2021, 10 (5), 966. 10.3390/plants10050966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happyana N.; Kayser O. Monitoring metabolite profiles of Cannabis sativa L.Trichomes during flowering period using 1H NMR-based metabolomics and real-time PCR. Planta Med. 2016, 82 (13), 1217–1223. 10.1055/s-0042-108058. [DOI] [PubMed] [Google Scholar]
- Citti C.; Battisti U. M.; Braghiroli D.; Ciccarella G.; Schmid M.; Vandelli M. A.; Cannazza G. A metabolomic approach applied to a liquid chromatography coupled to high-resolution tandem mass spectrometry method (HPLC-ESI-HRMS/MS): Towards the comprehensive evaluation of the chemical composition of cannabis medicinal extracts. Phytochem. Anal. 2018, 29 (2), 144–155. 10.1002/pca.2722. [DOI] [PubMed] [Google Scholar]
- Kriese U.; Schumann E.; Weber W. E.; Beyer M.; Brühl L.; Matthäus B. Oil content, tocopherol composition and fatty acid patterns of the seeds of 51 Cannabis sativa L. genotypes. Euphytica 2004, 137, 339–351. 10.1023/B:EUPH.0000040473.23941.76. [DOI] [Google Scholar]
- Muntendam R.; Happyana N.; Erkelens T.; Bruining F.; Kayser O. Time dependent metabolomics and transcriptional analysis of cannabinoid biosynthesis in Cannabis sativa var. Bedrobinol and Bediol grown under standardized condition and with genetic homogeneity. Online Int. J. Med. Plant. Res. 2012, 1, 31–40. 10.13140/RG.2.1.2263.5360. [DOI] [Google Scholar]
- Zheng J.; Johnson M.; Mandal R.; Wishart D. S. A comprehensive targeted metabolomics assay for crop plant sample analysis. Metabolites 2021, 11, 303. 10.3390/metabo11050303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- Dias D. A.; Jones O. A. H.; Beale D. J.; Boughton B. A.; Benheim D.; Kouremenos K. A.; Wolfender J. L.; Wishart D. S. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites 2016, 6 (4), 46. 10.3390/metabo6040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Cheng L. L.; Copié V.; Edison A. S.; Eghbalnia H. R.; Hoch J. C.; Gouveia G. J.; Pathmasiri W.; Powers R.; Schock T. B.; Sumner L. W.; Uchimiya M. NMR and metabolomics—A roadmap for the future. Metabolites 2022, 12 (8), 678. 10.3390/metabo12080678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.; Zhang Q.; Zhao X.; Yang Z.; Yang F.; Yang Y.; Tang J.; Laghi L. Metabolomic analysis of multiple biological specimens (feces, serum, and urine) by 1H-NMR spectroscopy from dairy cows with clinical mastitis. Animals 2023, 13 (4), 741. 10.3390/ani13040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti H.; Westler W. M.; Tonelli M.; Wedell J. R.; Markley J. L.; Eghbalnia H. R. Spin system modeling of nuclear magnetic resonance spectra for applications in metabolomics and small molecule screening. Anal. Chem. 2017, 89, 12201–12208. 10.1021/acs.analchem.7b02884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglada-Girotto M.; Handschin G.; Ortmayr K.; Campos A. I.; Gillet L.; Manfredi P.; Mulholland C. V.; Berney M.; Jenal U.; Picotti P.; Zampieri M. Combining CRISPRi and metabolomics for functional annotation of compound libraries. Nat. Chem. Biol. 2022, 18 (5), 482–491. 10.1038/s41589-022-00970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliferis K. A.; Bernard-Perron D. Cannabinomics: Application of metabolomics in cannabis (Cannabis sativa L.) research and development. Front. Plant Sci. 2020, 11, 554. 10.3389/fpls.2020.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Povedano M. M.; Sánchez-Carnerero Callado C.; Priego-Capote F.; Ferreiro-Vera C. Untargeted characterization of extracts from Cannabis sativa L. cultivars by gas and liquid chromatography coupled to mass spectrometry in high resolution mode. Talanta 2020, 208, 120384. 10.1016/j.talanta.2019.120384. [DOI] [PubMed] [Google Scholar]
- Antonelli M.; Benedetti B.; Cannazza G.; Cerrato A.; Citti C.; Montone C. M.; Piovesana S.; Laganà A. New insights in hemp chemical composition: A comprehensive polar lipidome characterization by combining solid phase enrichment, high-resolution mass spectrometry, and cheminformatics. Anal. Bioanal. Chem. 2020, 412 (2), 413–423. 10.1007/s00216-019-02247-6. [DOI] [PubMed] [Google Scholar]
- Toonen M. A. J.; Maliepaard C.; Reijmers T. H.; Van Der Voet H.; Dick Mastebroek H.; Van Den Broeck H. C.; Ebskamp M. J. M.; Kessler W.; Kessler R. W.. Predicting the chemical composition of fibre and core fraction of hemp (Cannabis sativa L.). Euphytica; Kluwer Academic Publishers, 2004; Vol. 140, pp 39–45. [Google Scholar]
- Ingallina C.; Sobolev A. P.; Circi S.; Spano M.; Fraschetti C.; Filippi A.; Di Sotto A.; Di Giacomo S.; Mazzoccanti G.; Gasparrini F.; Quaglio D.; Campiglia E.; Carradori S.; Locatelli M.; Vinci G.; Rapa M.; Ciano S.; Giusti A. M.; Botta B.; Ghirga F.; Capitani D.; Mannina L. Cannabis sativa L. inflorescences from monoecious cultivars grown in central Italy: An untargeted chemical characterization from early flowering to ripening. Molecules 2020, 25 (8), 1908. 10.3390/molecules25081908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Vyawahare R.; Lewis-Bakker M.; Clarke H. A.; Wong A. H. C.; Kotra L. P. Bioactive chemical composition of cannabis extracts and cannabinoid receptors. Molecules 2020, 25 (15), 3466. 10.3390/molecules25153466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H.; Shahidi F. Cannabis and cannabis edibles: A review. J. Agric. Food Chem. 2021, 69 (6), 1751–1774. 10.1021/acs.jafc.0c07472. [DOI] [PubMed] [Google Scholar]
- Mudge E. M.; Brown P. N.; Murch S. J. The terroir of cannabis: Terpene metabolomics as a tool to understand Cannabis sativa selections. Planta Med. 2019, 85 (09/10), 781–796. 10.1055/a-0915-2550. [DOI] [PubMed] [Google Scholar]
- Graves B. M.; Johnson T. J.; Nishida R. T.; Dias R. P.; Savareear B.; Harynuk J. J.; Kazemimanesh M.; Olfert J. S.; Boies A. M. Comprehensive characterization of mainstream marijuana and tobacco smoke. Sci. Rep. 2020, 10 (1), 7160. 10.1038/s41598-020-63120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert-Giesbrecht M.; Torres-Calzada C.; Wishart D. S.. Metabolomics of the cannabis plant. Cannabis Use, Neurobiology, Psychology, and Treatment; Elsevier, 2023; pp 3–19. 10.1016/B978-0-323-89862-1.00002-7. [DOI] [Google Scholar]
- Ramirez-Gaona M.; Marcu A.; Pon A.; Guo A. C.; Sajed T.; Wishart N. A.; Karu N.; Djoumbou Feunang Y.; Arndt D.; Wishart D. S. YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 2017, 45 (D1), D440–D445. 10.1093/nar/gkw1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajed T.; Marcu A.; Ramirez M.; Pon A.; Guo A. C.; Knox C.; Wilson M.; Grant J. R.; Djoumbou Y.; Wishart D. S. ECMDB 2.0: A richer resource for understanding the biochemistry of E. coli. Nucleic Acids Res. 2016, 44 (D1), D495–D501. 10.1093/nar/gkv1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Li C.; Marcu A.; Badran H.; Pon A.; Budinski Z.; Patron J.; Lipton D.; Cao X.; Oler E.; Li K.; Paccoud M.; Hong C.; Guo A. C.; Chan C.; Wei W.; Ramirez-Gaona M. PathBank: A comprehensive pathway database for model organisms. Nucleic Acids Res. 2020, 48 (D1), D470–D478. 10.1093/nar/gkz861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim E. A.; Wang M.; Radwan M. M.; Wanas A. S.; Majumdar C. G.; Avula B.; Wang Y. H.; Khan I. A.; Chandra S.; Lata H.; Hadad G. M.; Abdel Salam R. A.; Ibrahim A. K.; Ahmed S. A.; Elsohly M. A. Analysis of terpenes in Cannabis sativa L. using GC/MS: Method development, validation, and application. Planta Med. 2019, 85 (05), 431–438. 10.1055/a-0828-8387. [DOI] [PubMed] [Google Scholar]
- Foroutan A.; Guo A. C.; Vazquez-Fresno R.; Lipfert M.; Zhang L.; Zheng J.; Badran H.; Budinski Z.; Mandal R.; Ametaj B. N.; Wishart D. S. Chemical composition of commercial cow’s milk. J. Agric. Food Chem. 2019, 67 (17), 4897–4914. 10.1021/acs.jafc.9b00204. [DOI] [PubMed] [Google Scholar]
- Saini R. K.; Prasad P.; Shang X.; Keum Y. S. Advances in lipid extraction methods—A review. Int. J. Mol. Sci. 2021, 22 (24), 13643. 10.3390/ijms222413643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Ubeed H. M. S.; Bhuyan D. J.; Alsherbiny M. A.; Basu A.; Vuong Q. V. A comprehensive review on the techniques for extraction of bioactive compounds from medicinal cannabis. Molecules 2022, 27 (3), 604. 10.3390/molecules27030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Zhang L.; Wang L.; Zhao Y. Total phenols, flavonoids, and procyanidins levels and total antioxidant activity of different Korean pine (Pinus koraiensis) varieties. J. For. Res. 2019, 30 (5), 1743–1754. 10.1007/s11676-018-0744-0. [DOI] [Google Scholar]
- Health Canada . Mandatory Cannabis Testing for Pesticide Active Ingredients—Lists and Limits. 2019. https://www.canada.ca/content/dam/phac-aspc/documents/services/publications/drugs-health-products/cannabis-testing-pesticide-list-limits/Mandatory-Cannabis-Testing-List-and-limits.pdf#:~:text=Mandatory%20cannabis%20testing%20for%20pesticide%20active%20ingredients%E2%80%94List%20and,Phenothrin%200.025%200.05%20%2A%20Phosmet%200.01%200.02%20%2A (accessed 2023-06-01).
- Lorenzo R. D.; Tran D.; Butt C.; Winkler P.; Krepich S.; Borton C.. Analysis of the Canadian Cannabis Pesticides List Using Both ESI and APCI Techniques. https://www.cannabissciencetech.com/view/analysis-of-the-canadian-cannabis-pesticides-list-using-both-esi-and-apci-techniques (accessed 2023-08-15).
- Wilschefski S. C.; Baxter M. R. Inductively coupled plasma mass spectrometry: Introduction to analytical aspects. Clin. Biochem. Rev. 2019, 40 (3), 115–133. 10.33176/AACB-19-00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Guo A. C.; Oler E.; Wang F.; Anjum A.; Peters H.; Dizon R.; Sayeeda Z.; Tian S.; Lee B. L.; Berjanskii M.; Mah R.; Yamamoto M.; Jovel J.; Torres-Calzada C.; Hiebert-Giesbrecht M.; Lui V. W.; Varshavi D.; Varshavi D.; Allen D.; Arndt D.; Khetarpal N.; Sivakumaran A.; Harford K.; Sanford S.; Yee K.; Cao X.; Budinski Z.; Liigand J.; Zhang L.; Zheng J.; Mandal R.; Karu N.; Dambrova M.; Schiöth H.; Greiner R.; Gautam V. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50 (D1), D622–D631. 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D.; Knox C.; Young N.; Stothard P.; Damaraju S.; Wishart D. S. PolySearch: A web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 2008, 36, W399–W405. 10.1093/nar/gkn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Liang Y.; Wishart D. PolySearch2: A significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 2015, 43 (W1), W535–W542. 10.1093/nar/gkv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L. A.; Zhang P.; Rhee S. Y. AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 2003, 132 (2), 453–460. 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C.; Ginzburg D.; Zhao K.; Dwyer W.; Xue B.; Xu A.; Rice S.; Cole B.; Paley S.; Karp P.; Rhee S. Y. Plant Metabolic Network 15: A resource of genome-wide metabolism databases for 126 plants and algae. J. Integr. Plant Biol. 2021, 63 (11), 1888–1905. 10.1111/jipb.13163. [DOI] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45 (D1), D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.; Wang B.; Xie S.; Xu X.; Zhang J.; Pei L.; Yu Y.; Yang W.; Zhang Y. A high-quality reference genome of wild Cannabis sativa. Hortic. Res. 2020, 7 (1), 73. 10.1038/s41438-020-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A.; Martin M. J.; Orchard S.; Magrane M.; Agivetova R.; Ahmad S.; Alpi E.; Bowler-Barnett E. H.; Britto R.; Bursteinas B.; Bye-A-Jee H.; Coetzee R.; Cukura A.; da Silva A.; Denny P.; Dogan T.; Ebenezer T. G.; Fan J.; Castro L. G.; Garmiri P.; Georghiou G.; Gonzales L.; Hatton-Ellis E.; Hussein A.; Ignatchenko A.; Insana G.; Ishtiaq R.; Jokinen P.; Joshi V.; Jyothi D.; Lock A.; Lopez R.; Luciani A.; Luo J.; Lussi Y.; MacDougall A.; Madeira F.; Mahmoudy M.; Menchi M.; Mishra A.; Moulang K.; Nightingale A.; Oliveira C. S.; Pundir S.; Qi G.; Raj S.; Rice D.; Lopez M. R.; Saidi R.; Sampson J.; Sawford T.; Speretta E.; Turner E.; Tyagi N.; Vasudev P.; Volynkin V.; Warner K.; Watkins X.; Zaru R.; Zellner H.; Bridge A.; Poux S.; Redaschi N.; Aimo L.; Argoud-Puy G.; Auchincloss A.; Axelsen K.; Bansal P.; Baratin D.; Blatter M. C.; Bolleman J.; Boutet E.; Breuza L.; Casals-Casas C.; de Castro E.; Echioukh K. C.; Coudert E.; Cuche B.; Doche M.; Dornevil D.; Estreicher A.; Famiglietti M. L.; Feuermann M.; Gasteiger E.; Gehant S.; Gerritsen V.; Gos A.; Gruaz-Gumowski N.; Hinz U.; Hulo C.; Hyka-Nouspikel N.; Jungo F.; Keller G.; Kerhornou A.; Lara V.; Le Mercier P.; Lieberherr D.; Lombardot T.; Martin X.; Masson P.; Morgat A.; Neto T. B.; Paesano S.; Pedruzzi I.; Pilbout S.; Pourcel L.; Pozzato M.; Pruess M.; Rivoire C.; Sigrist C.; Sonesson K.; Stutz A.; Sundaram S.; Tognolli M.; Verbregue L.; Wu C. H.; Arighi C. N.; Arminski L.; Chen C.; Chen Y.; Garavelli J. S.; Huang H.; Laiho K.; McGarvey P.; Natale D. A.; Ross K.; Vinayaka C. R.; Wang Q.; Wang Y.; Yeh L. S.; Zhang J.; Ruch P.; Teodoro D. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49 (D1), D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou Feunang Y.; Eisner R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminf. 2016, 8 (1), 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig K. W. A chemotaxonomic analysis of terpenoid variation in Cannabis. Biochem. Syst. Ecol. 2004, 32 (10), 875–891. 10.1016/j.bse.2004.04.004. [DOI] [Google Scholar]
- Johnson A.; Stewart A.; El-Hakim I.; Hamilton T. J. Effects of super-class cannabis terpenes beta-caryophyllene and alpha-pinene on zebrafish behavioural biomarkers. Sci. Rep. 2022, 12 (1), 17250. 10.1038/s41598-022-21552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E. L.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H. P.; Hollender J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48 (4), 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Yamauchi T.; Shoyama Y.; Aramaki H.; Azuma T.; Nishioka I. Tetrahydrocannabinolic acid, a genuine substance of tetrahydrocannabinol. Chem. Pharm. Bull. 1967, 15 (7), 1075–1076. 10.1248/cpb.15.1075. [DOI] [PubMed] [Google Scholar]
- Iffland A. K.; Carus M.; Grotenhermen F.. Decarboxylation of Tetrahydrocannabinolic Acid (THCA) to Active THC; European Industrial Hemp Association, 2016; pp 1–3. [Google Scholar]
- Aizpurua-Olaizola O.; Omar J.; Navarro P.; Olivares M.; Etxebarria N.; Usobiaga A. Identification and quantification of cannabinoids in Cannabis sativa L. plants by high performance liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2014, 406 (29), 7549–7560. 10.1007/s00216-014-8177-x. [DOI] [PubMed] [Google Scholar]
- Ahmed M.; Ji M.; Qin P.; Gu Z.; Liu Y.; Sikandar A.; Iqbal M. F.; Javeed A. Phytochemical screening, total phenolic and flavonoids contents and antioxidant activities of citrullus colocynthis L. and Cannabis Sativa L. Appl. Ecol. Environ. Res. 2019, 17 (3), 6961–6979. 10.15666/aeer/1703_69616979. [DOI] [Google Scholar]
- Galasso I.; Russo R.; Mapelli S.; Ponzoni E.; Brambilla I. M.; Battelli G.; Reggiani R. Variability in seed traits in a collection of cannabis sativa L. genotypes. Front. Plant Sci. 2016, 7, 688. 10.3389/fpls.2016.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzoni D.; Skřivanová E.; Rebucci R.; Crotti A.; Baldi A.; Marchetti L.; Giromini C. Total phenolic content and antioxidant activity of in vitro digested hemp-based products. Foods 2023, 12 (3), 601. 10.3390/foods12030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo L.; Castaldo L.; Narváez A.; Graziani G.; Gaspari A.; Rodríguez-Carrasco Y.; Ritieni A. Analysis of phenolic compounds in commercial Cannabis sativa L. Inflorescences using UHPLC-Q-Orbitrap HRMs. Molecules 2020, 25 (3), 631. 10.3390/molecules25030631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Dong L.; Zhao C.; Zhang F.; Zhao S.; Zhan J.; Li J.; Li L. Development of a high-coverage quantitative metabolome analysis method using four-channel chemical isotope labeling LC-MS for analyzing high-salt fermented food. J. Agric. Food Chem. 2022, 70 (28), 8827–8837. 10.1021/acs.jafc.2c03481. [DOI] [PubMed] [Google Scholar]
- Mihoc M.; Pop G.; Alexa E.; Radulov I. Nutritive quality of romanian hemp varieties (Cannabis sativa L.) with special focus on oil and metal contents of seeds. Chem. Cent. J. 2012, 6, 122. 10.1186/1752-153X-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Oler E.; Peters H.; Guo A. C.; Girod S.; Han S.; Saha S.; Lui V. W.; LeVatte M.; Gautam V.; Kaddurah-Daouk R.; Karu N. MiMeDB: the Human Microbial Metabolome Database. Nucleic Acids Res. 2023, 51 (D1), D611–D620. 10.1093/nar/gkac868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; MacIejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le Di.; Pon A.; Knox C.; Wilson M. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46 (D1), D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. D.; Dumontier M.; Aalbersberg Ij. J.; Appleton G.; Axton M.; Baak A.; Blomberg N.; Boiten J. W.; da Silva Santos L. B.; Bourne P. E.; Bouwman J.; Brookes A. J.; Clark T.; Crosas M.; Dillo I.; Dumon O.; Edmunds S.; Evelo C. T.; Finkers R.; Gonzalez-Beltran A.; Gray A. J. G.; Groth P.; Goble C.; Grethe J. S.; Heringa J.; t Hoen P. A. C.; Hooft R.; Kuhn T.; Kok R.; Kok J.; Lusher S. J.; Martone M. E.; Mons A.; Packer A. L.; Persson B.; Rocca-Serra P.; Roos M.; van Schaik R.; Sansone S. A.; Schultes E.; Sengstag T.; Slater T.; Strawn G.; Swertz M. A.; Thompson M.; Van Der Lei J.; Van Mulligen E.; Velterop J.; Waagmeester A.; Wittenburg P.; Wolstencroft K.; Zhao J.; Mons B. Comment: The FAIR guiding principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence H. E.; Williams A. ChemSpider: An online chemical information resource. Chem. Educ. Today 2010, 87 (11), 1123–1124. 10.1021/ed100697w. [DOI] [Google Scholar]
- Afendi F. M.; Okada T.; Yamazaki M.; Hirai-Morita A.; Nakamura Y.; Nakamura K.; Ikeda S.; Takahashi H.; Altaf-Ul-Amin M..; Darusman L. K.; Saito K.; Kanaya S.; et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53, e1 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.