Abstract
Activating signal co-integrator complex 1 (ASCC1) acts with ASCC-ALKBH3 complex in alkylation damage responses. ASCC1 uniquely combines two evolutionarily ancient domains: nucleotide-binding K-Homology (KH) (associated with regulating splicing, transcriptional, and translation) and two-histidine phosphodiesterase (PDE; associated with hydrolysis of cyclic nucleotide phosphate bonds). Germline mutations link loss of ASCC1 function to spinal muscular atrophy with congenital bone fractures 2 (SMABF2). Herein analysis of The Cancer Genome Atlas (TCGA) suggests ASCC1 RNA overexpression in certain tumors correlates with poor survival, Signatures 29 and 3 mutations, and genetic instability markers. We determined crystal structures of Alvinella pompejana (Ap) ASCC1 and Human (Hs) PDE domain revealing high-resolution details and features conserved over 500 million years of evolution. Extending our understanding of the KH domain Gly-X-X-Gly sequence motif, we define a novel structural Helix-Clasp-Helix (HCH) nucleotide binding motif and show ASCC1 sequence-specific binding to CGCG-containing RNA. The V-shaped PDE nucleotide binding channel has two His-Φ-Ser/Thr-Φ (HXT) motifs (Φ being hydrophobic) positioned to initiate cyclic phosphate bond hydrolysis. A conserved atypical active-site histidine torsion angle implies a novel PDE substrate. Flexible active site loop and arginine-rich domain linker appear regulatory. Small-angle X-ray scattering (SAXS) revealed aligned KH-PDE RNA binding sites with limited flexibility in solution. Quantitative evolutionary bioinformatic analyses of disease and cancer-associated mutations support implied functional roles for RNA binding, phosphodiesterase activity, and regulation. Collective results inform ASCC1's roles in transactivation and alkylation damage responses, its targeting by structure-based inhibitors, and how ASCC1 mutations may impact inherited disease and cancer.
Keywords: RNA, binding protein, phosphodiesterase: cancer, genomics, DNA repair, structural biology, crystallography, small angle X-ray scattering (SAXS), conformational change, inhibition mechanism
Chemotherapy with alkylating agents is one of the most common forms of treatment prescribed for cancer. Unfortunately, tumor resistance to alkylation therapy can emerge. This resistance mechanism is partly governed by alkylation damage response proteins, such as seen for the transcriptional co-activator Activating Signal Cointegrator 1 (ASC-1) complex subunit 1 (ASCC1) (1) which acts in ALKBH3-mediated alkylation damage repair in DNA and RNA. Importantly, members of the AlkB homolog (ALKBH) family of non-heme Fe (II) and 2-oxoglutarate-dependent dioxygenases perform a direct reversal of alkylation damage. ALKBH enzymes demethylate alkylated bases, for example, 1-methyladenine (1meA) and 3-methylcytosine (3meC) by oxidizing the N-linked methyl moiety, leading to the formation of an unstable methyl-iminium intermediate that hydrolyzes to form formaldehyde and a non-alkylated base (2). ALKBH3, with iron and 2-oxoglutarate, specifically can repair 1meA and 3meC alkylation lesions in single-stranded (ss) DNA and RNA (3, 4).
ASCC1 was shown to be constitutively present in nuclear foci and leaves the nuclear foci in response to an alkylation damage signal (1). Knockdown of ASCC1 has impaired alkylation response and improper recruitment of ASCC components in the damage site suggesting ASCC1 may be involved in regulation. Earlier work performed on unraveling ASCC1 functions implicate ASCC1 as an active member of the ASC-1-ALKBH3 complex for nucleic acid alkylation damage reversal in DNA damage responses (DDR). Yet, ASCC1 enzymatic activities in its transcriptional activation complex and RNA damage responses have been enigmatic.
Besides acting in alkylation damage, null/truncations mutations in ASCC1 are implicated in diseases such as Barrett’s esophagus and esophageal adenocarcinoma, spinal muscular atrophy with congenital bone fractures 2 (SMABF2), and rheumatoid arthritis (Table S1) (5, 6, 7, 8, 9, 10, 11, 12, 13, 14). Almost all the known pathogenic ASCC1 variants are predicted to entail either premature truncation or complete absence of the ASCC1 protein suggesting the full-length protein is necessary for its function(s).
ASCC1 has both an N-terminal K-Homology (KH) domain and a C-terminal phosphodiesterase (PDE) domain, an unstudied combination that is unknown in other proteins. The ∼70 amino acid KH domain, named by its identification in human heterogeneous nuclear ribonucleoprotein K (hnRNP K), binds 4 bases of single-stranded (ss) DNA or RNA with sequence specificity (15, 16, 17, 18). The KH GXXG sequence motif acts in interaction with the backbone of nucleotide bases. Found in archaea, bacteria, and eukaryotes, KH domains typically act in splicing, transcription regulation, and translational control. The ASCC1 PDE domain has sequence homology to 2H phosphodiesterases, which are widely present in all domains of life (19). They play essential roles in nucleic acid metabolism, including transesterification, hydrolysis of cyclic phosphates, and RNA ligation. Thus, PDE domains are also referred to as RNA ligase-like domains. These 2H PDEs have two conserved H-Φ-S/T-Φ motifs (where Φ represents a hydrophobic amino acid) separated by approximately 80 amino acids (20). These motifs are often referred to as HXT motifs. Like other members of the PDE superfamily, the ASCC1 PDE domain contains two HXT motifs (21).
Here, we sought to understand ASCC1 mechanisms and activities and how its uncommonly linked KH and PDE domains are functionally positioned. We reasoned that combining biological impacts of high and low ASCC1 expression in cancer, with structural and evolutionary sequence analysis would inform potential functional roles for its structurally identified features. We, therefore, examined The Cancer Genome Atlas (TCGA) data for the relationship of ASCC1 overexpression in tumors to patient survival. Combined high-resolution X-ray structures of Human (Hs) and Alvinella pompejana (Ap) ASCC1 and small angle X-ray scattering (SAXS) measurements in solution revealed the first protein structure with both KH and PDE domains and defined a stabilized conformational state of linked KH and PDE domains, with limited flexibility. Comparative computational structural analyses with other KH and PDE domains support and extend our understanding of their structure-function relationships. We identify a previously unrecognized structural Helix-Clasp-Helix four-nucleotide-binding motif in the KH domain and predict a novel PDE substrate implied by an atypical rotamer for the histidine in the second His-X-Thr motif defining the cyclic nucleotide-binding pocket. Quantitative evolutionary action analyses of cancer variants of unknown significance (VUS) and mapping these and a published ASCC1 germline-associated disease mutation (12) onto our X-ray crystal structure, implicate particular mutations in disrupting functional roles in structurally identified features for RNA binding, phosphodiesterase activity, and regulation. These collective results support a key role for ASCC1 in an RNA alkylation response important for human health and cancer biology.
Results
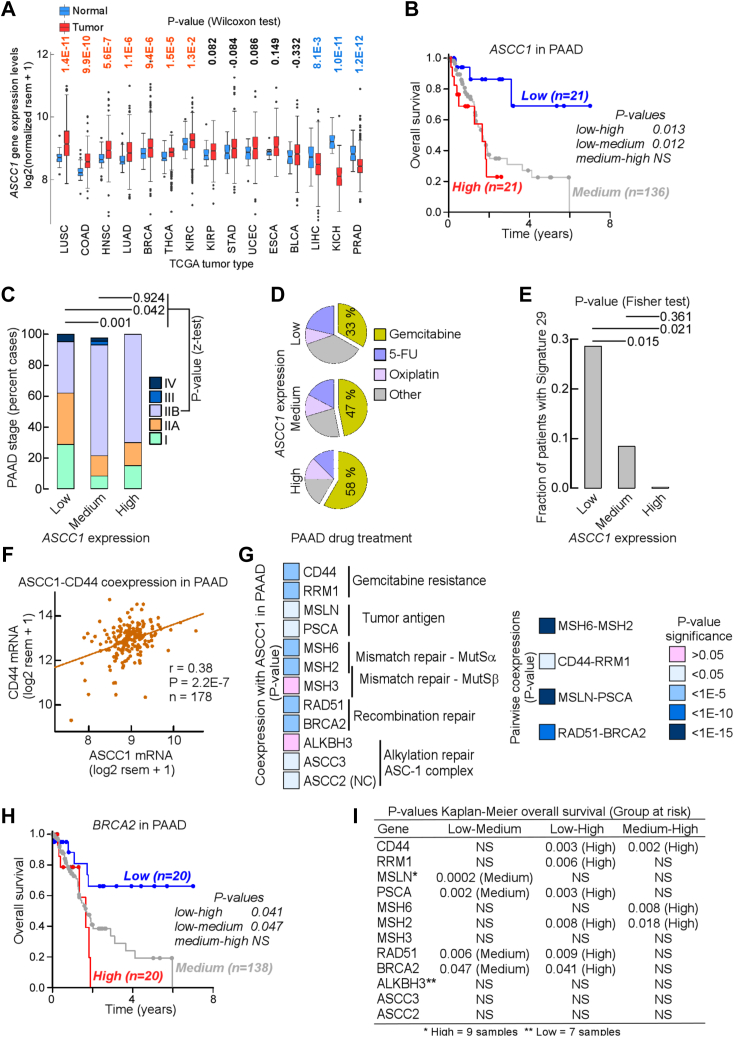
ASCC1 overexpression is a potential prognostic marker in pancreatic cancer
Given the role of ASCC1 in damage responses and RNA processing, we reasoned that alterations in its expression or inheritance of rare germline variants, such as missense mutation N290S in cancer precursor Barrett's esophagus, might promote cancer (12). We therefore performed bioinformatics analyses on global gene expression, patient survival, and gene co-expression patterns from the publicly available TCGA cohort (22). We first compared ASCC1 RNA levels in 15 tumor types and matched controls in a total of ∼7000 tumor samples. We limited our study to those types with 10 or more matched controls for statistical comparison. In 7 out of 15 tumor types, ASCC1 RNA levels were significantly higher in the tumor than in control tissues, including lung malignancies, in particular squamous cell carcinoma and adenocarcinoma (Fig. 1A). In 3 out of 15 tumor types, ASCC1 RNA levels were lower in the tumor than in controls; the difference was particularly striking in kidney chromophobe tumors, where the distribution of values was nearly non-overlapping between tumors and controls. In the remaining five tumor types, ASCC1 RNA levels between tumor and normal samples were indistinguishable. Differential expression between tumor types is common and may reflect differences in cancer etiology, tumor maintenance, and treatment. Together, these analyses suggest that ASCC1 RNA levels are higher in the tumor than in normal tissue in almost half of TCGA tumor types.
Figure 1.
ASCC1 overexpression is a potential prognostic marker in pancreatic cancer.A, box plots of ASCC1 mRNA levels in tumors and matched controls in TCGA. p-values from Wilcoxon tests. B, Kaplan–Meier overall survival curve for TCGA patients with pancreatic adenocarcinoma (PAAD) expressing low (g_low, blue), medium (g_medium, gray), or high (g_high, red) ASCC1 mRNA levels. p-values from pairwise comparisons with Holm-Sidak-corrected log-rank tests. C, bar graph of percent samples expressing Low, Medium, or High levels of ASCC1 mRNA with given AJCC PAAD tumor stage. Pairwise p-values from z-tests. D, pie charts of percent PAAD patients expressing Low, Medium, or High levels of ASCC1 mRNA treated with one or more of 3 main chemotherapy drugs. E, bar graph of fraction of PAAD samples expressing Low, Medium, or High levels of ASCC1 mRNA displaying a pattern of somatic single base substitutions (SBSs) conforming to Signature 29. p-values from Fisher exact tests. The fraction for the High group was 0.0 but is shown for reference. F, Cartesian plot and linear regression for coexpression of ASCC1 (x-axis) and CD44 (y-axis) mRNA levels in PAAD samples. p-value from Fisher-Snedecor distribution. G, left: check box of p-values for linear regression of coexpression between ASCC1 and given gene in PAAD. Genes are grouped by functional categories. Right: check box of p-values for linear regression of coexpression between pairs of functionally linked genes. p-value from Fisher-Snedecor distribution. Except for ASCC1, which displayed a negative correlation (NC), all other regression coefficients were positive. H, Kaplan–Meier overall survival curve for TCGA PAAD patients expressing Low (g_low, blue), Medium (g_medium, gray), or High (g_high, red) BRCA2 mRNA levels. p-values from pairwise comparisons with Holm-Sidak-corrected log-rank tests. I, p-values for Kaplan-Meier overall survival curves and group(s) at risk for PAAD patients expressing Low (g_low), Medium (g_medium) or High (g_high) levels of mRNA for genes from Panel G. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head-neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid cancer; UCEC, uterine corpus endometrial carcinoma.
To assess whether RNA expression levels would be associated with differences in survival time, we performed a Kaplan-Meier survival analysis. For each of the 33 types of tumors with RNA expression and survival data available in TCGA, we divided the patients into two groups: an “m_high” group in which ASCC1 RNA levels were higher than the mean and an “m_low” group where ASCC1 RNA levels were lower than the mean. Differences in survival were found in 5 types of tumors (Fig. S1, A–E), with four malignancies displaying worse survival in patients with high ASCC1 expression (Fig. S1, A–D). The difference in survival was particularly acute in pancreatic cancer (PAAD) (Fig. S1A), where the hazard ratio indicated a ∼3.7 fold-increased risk of poor prognosis in high ASCC1-expressing patients. Since survival analyses are sensitive to the choice of cutoff in the data, we performed a second analysis where patients were divided into three groups: a “g_high” group in which ASCC1 RNA levels were higher than one standard deviation above the mean; a “g_low” group where ASCC1 RNA levels were lower than the mean minus one standard deviation; and a “g_medium” group where ASCC1 RNA levels were within one standard deviation of the mean (Figs 1B and S2, A–F). In this analysis, differences in survival were seen in 7 types of tumors, with two malignancies displaying worse survival in patients with high (adrenocortical carcinoma, ACC) or medium-high (PAAD) ASCC1 expression, and one malignancy (kidney clear renal cell carcinoma, KIRC) exhibiting worse survival in patients with low ASCC1, similar to the first analysis (Figs. 1B, S1 and S2, and Table S2).
Given the consistency between the first and second analyses, we choose PAAD to gain additional insight into the relationships between gene expression and patient survival. We stratified patients with PAAD by tumor grade, which indicated that in the g_medium and g_high groups the percent patients with IIB tumor stage was substantially greater than in the g_low group, implying an overall more advanced malignancy (Fig. 1C). Next, to understand the impacts of cancer treatment, we investigated and found the chemotherapy regimen for patients with PAAD included gemcitabine, 5-fluorouracyl and oxiplatin; however, the percentage of patients receiving gemcitabine increased from the g_low to the g_medium to the g_high groups, although differences did not reach statistical significance (Fig. 1D).
We reasoned that chemotherapy-induced DNA replication arrest may prompt resistance mechanisms involving DNA repair, such that the landscape of single base substitutions genome-wide might differ among the g_low, g_medium and g_high groups. Mutations have been categorized into “individual mutational signatures” that reflect the type of DNA damage and associated repair processes (23, 24). Of the thirty distinct mutational signatures originally identified (25), Signature 29 displayed the largest differences among the three groups, with nearly 30% PAAD samples with low ASCC1 expression (the g_low) carrying the mutational signature, compared to ∼10% in the g_medium group and none in the g_high group (Fig. 1E). Signature 29 is characterized by a bias in C>A substitutions, and it has been observed only in gingivo-buccal oral squamous cell carcinoma (https://cancer.sanger.ac.uk/signatures/signatures_v2/). Thus, it is possible that, in PAAD, Signature 29 mutations might arise from error-prone repair following chemotherapy.
The absence of Signature 29 mutations in the g_high group suggested activation of error-free repair of chemotherapy-related DNA lesions, which might have led to chemoresistance and poor survival. To explore this hypothesis, we conducted an mRNA coexpression analysis between ASCC1 and other genes. Coexpression was highly significant with CD44, a marker of resistance to gemcitabine (26) (Fig. 1F) and RRM1 (Fig. 1G), which encodes the large subunit of ribonucleotide reductase, a target of gemcitabine activity. ASCC1 was also coexpressed with the two tumor-coding antigens, mesothelin (MSLN) and prostate stem cell antigen (PSCA) (Fig. 1G), the mismatch repair MutSα complex, which acts upon postreplicative single-base mismatches and dinucleotide insertion/deletion loops, and the BRCA2-RAD51 subcomplex, active in error-free recombination repair of double-strand breaks at stalled replication forks. Moreover, coexpression was highest when pairs of related genes were compared, such as MSH6 versus MSH2, MSLN versus PSCA and RAD51 versus BRCA2 (Fig. 1G). High co-expression of homologous recombination (HR) and mismatch repair proteins with ASCC1 is consistent with our hypothesis that these error-free pathway proteins may contribute in limiting Signature 29 mutations in cancers with high ASCC1 expression.
Finally, we examined the pattern of overall survival in PAAD patients expressing low (g_low), medium (g_medium), or high (g_high) levels of mRNA for each of the coexpression analysis genes, with the expectation that these would be similar to that observed for ASCC1. For BRCA2 (Fig. 1H), PSCA and RAD51 both the g_medium and g_high groups were at higher risk than g_low, whereas for CD44, MSH6, MSH2 the g_high group was at risk (Fig. 1I). Interestingly, there were no differences among groups for MSH3, ALKBH3, ASCC2 and ASCC3 (Fig. 1I), in accordance with the results from the coexpression analysis (Fig. 1G). In summary, given that reactivation of tumor markers such as CD44, MSLN, and PSCA are known to occur in response to chemoresistance, our data support the idea that levels of gene expression of ASCC1 act as prognostic markers in PAAD patients, by informing on the tumor response to chemotherapy, most likely to gemcitabine in our case. Interestingly, the absence of connection with ALKBH3, ASCC2, and ASCC3 suggests an independent function for ASCC1, at least in PAAD.
ASCC1 is physiologically co-expressed with PGAM4
Having seen that ASCC1 expression is often dysregulated in cancer and that in pancreatic cancer it is co-expressed with markers of chemoresistance, we wondered whether there was also a physiological pattern of co-expression between ASCC1 and other genes that would be common to most, if not all, tumor types and normal tissues. To this end, we performed a global study on gene co-expression between RNA levels of ASCC1 and RNA levels of all genes for which RNA-Seq data were available in TCGA, which included ∼20,000 genes in 33 tumor types, for a total of ∼660,000 correlations. We ranked by p-value both positive and negative correlations together, which showed that strengths of correlation extended up to a -log10 value of 102 (Fig. S3A), with similar numbers of positive and negative correlations. To assess the most common occurrence of positive correlations, we ranked the number of tumor types for which a significant positive correlation between a gene and ASCC1 was found. Occurring in 23/33 types of tumors, the top 22 correlating genes were unsurprisingly located on chromosome 10, as is the ASCC1 gene (10q22.1) (Fig. S3B). A common mechanism for ASCC1 overexpression in cancer may thus be through amplification of a part, or all, of chromosome 10, given a reported linkage of chromosome 10 with cancer. Chromosome 10 is often lost in several types of brain tumors (27, 28, 29), neuroblastoma (30), and melanoma (31).
Intriguingly, the top correlation not on chromosome 10 was with PGAM4, a gene on chromosome Xq21.1, and its neighboring gene, PGK1 (Fig. S3, B and C), two genes coding for interacting members of the phosphoglycerate family (Fig. S3D). These correlations were also revealed in matched controls (Fig. S3E). Investigation of a PGAM4 function for any phenotype was inconclusive. Silencing PGAM4 with shRNAs inhibits glycolysis and tumor growth in mice xenografts derived from glioma cells (32), while we found that, in the TCGA cohort, patients with low PGAM4 expression are at high risk. A hint as to an ASCC1 role in RNA processing, PGAM4 is uncommonly formed by retro-positioning, where mRNA is reverse-transcribed and inserted back into the DNA (33). In summary, our analysis indicates that ASCC1 is physiologically co-expressed with genes coding for enzymes interacting with other glycolytic enzymes at the triose stage as well as the pentose phosphate pathway; however, the nature of such correlations remains to be elucidated.
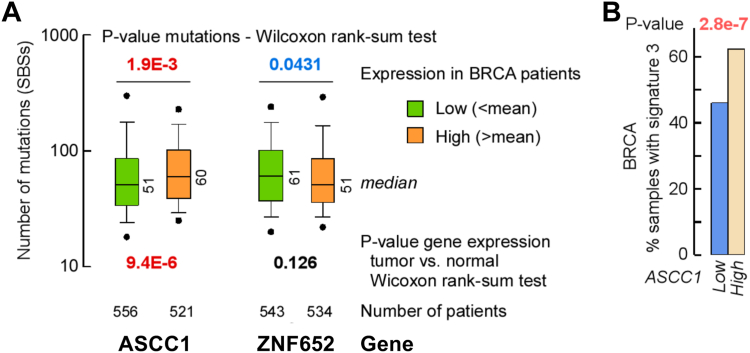
High ASCC1 RNA expression is associated with high mutational load and mutations arising from deficiency in homologous recombination repair
To further assess the involvement of ASCC1 in tumors, we tested whether patients with cancer and a higher expression of ASCC1 (the m_high group) would also incur a greater load of single base substitutions (SBSs) than those with lower expression (the m_low group). We, therefore, queried the RNA-Seq data from TCGA and the base variants called from exome-wide sequencing and curated by COSMIC (34). We found a match in patient codes between these two databases for 24/36 types of tumor (Table S3). The results indicated that the m_high group incurred a higher load of SBSs in bladder, breast, and lung carcinomas than the m_low group, whereas the opposite was observed in sarcoma and endometrial carcinomas. A similar analysis whereby patients were categorized into three groups, that is, g_low, g_medium and g_high, stressed the significantly higher mutational load for the g_group in breast carcinoma (Table S4). Notably, ASCC1 is overexpressed in breast carcinoma (Fig. 1A). By contrast, no increase in mutations was observed in the m_high group for our negative control ZNF652, whose expression between the m_high and m_low groups were indistinguishable in bladder, breast, and lung carcinomas (Fig. 2A and Table S3).
Figure 2.
High ASCC1 expression increases mutational load and the fraction of mutations conforming to homologous recombination deficiency.A, box plot of the number of SBSs in BRCA patients for the low (m_low, green) and high (m_high, orange) groups for ASCC1, which is overexpressed in the tumor compared to matched control, and ZNF652, whose expression does not differ between tumor and matched control. p-values from Wilcoxon tests. B, percentage of patients expressing high (m_high, above mean, tan) or low (m_low, below mean, blue) levels of ASCC1 in which single base substitutions conforming to Signature 3 from exome-wide sequencing were observed in breast invasive carcinoma (BRCA). p-value from Fisher’s exact test.
Next, we compared the mutational signatures of TCGA patients who displayed high (m_high or g_high) ASCC1 RNA levels, with those in which ASCC1 RNA levels were medium (g_medium) or low (m_low or g_low) (Tables S5 and S6). The most striking result was observed for breast invasive carcinoma (BRCA), in which samples with high levels of ASCC1 RNA were 35% more likely to carry Signature 3 mutations than those with low levels of ASCC1 RNA (Fig. 2B). Signature 3 comprises a complex set of SBSs dominated by C>N changes (23) and in breast cancer has been linked to mutations arising from DNA lesions repaired by mechanisms that operate when homologous recombination repair is compromised (35). The large difference between 46% and 62% BRCA samples displaying Signature 3 mutations (Fig. 2B) was unexpected, as one would predict BRCA to have high Signature 3, independent of ASCC1. To further elucidate relationships between ASCC1 gene expression and Signature 3 mutations, we extended the comparison between high/low gene expression to all genes (∼20,000) for which RNA-Seq data were available in BRCA patients, with the expectation that DNA repair-related genes would dominate the outcome. A total of 119 genes were found to be significantly associated with Signature 3 mutations when highly expressed. A gene set enrichment analysis (GSEA) on these 119 hits revealed predominant enrichment in mitochondria-associated genes (3.49-fold) and genes coding for centromeric structures (2.57-fold enrichment; Fig. S3F). Based on this linkage with mitochondria and cell division genes, we hypothesize that ASCC1-related Signature 3 may originate in part from induction of DNA repair mechanisms for oxidative damage that disfavor homologous recombination and for which ASCC1 is part.
The ASCC1 crystal structure reveals the side-by-side orientation of KH and PDE domains
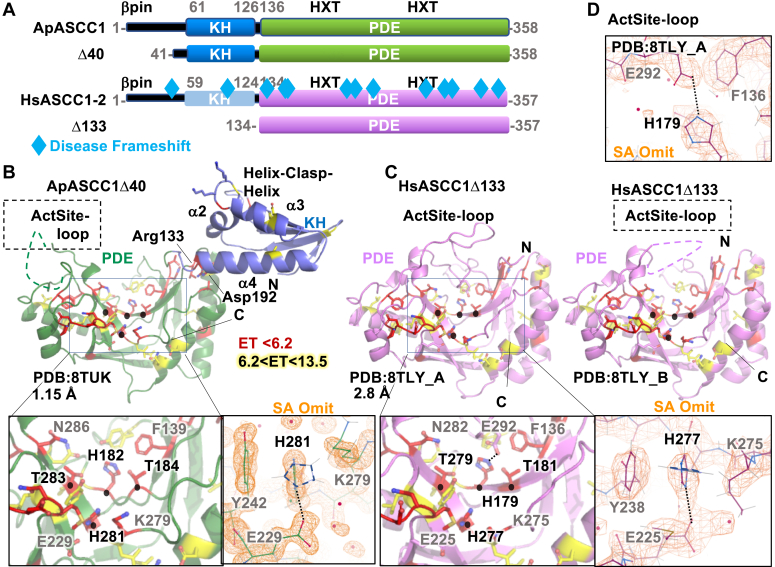
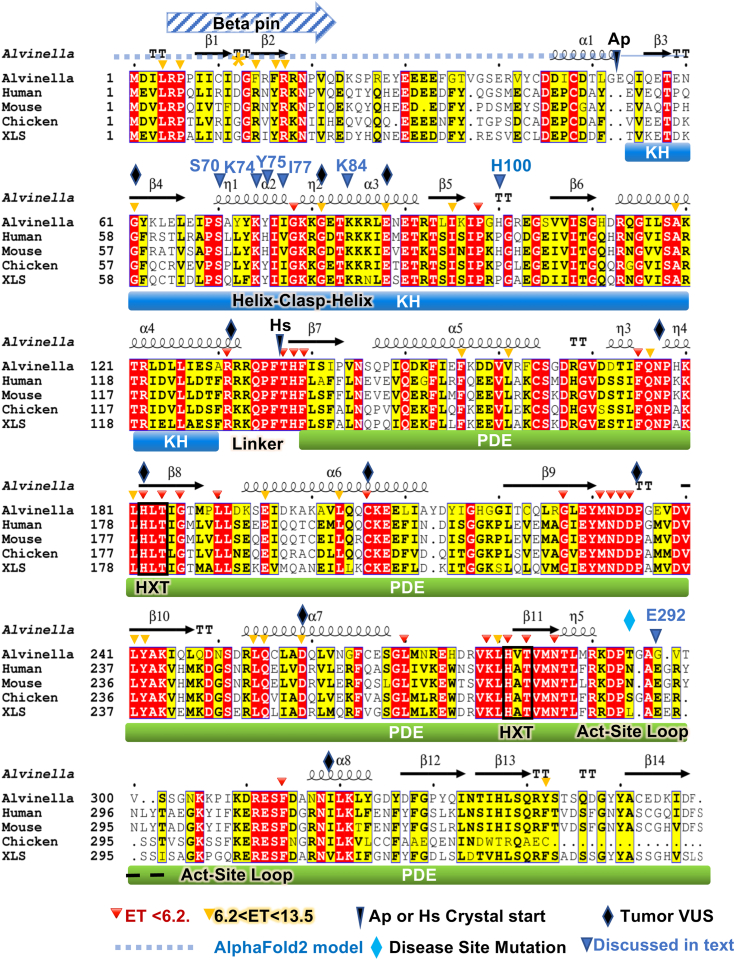
To probe the domain structural architecture and functional features of ASCC1, we initiated crystallographic analysis. The overall ASCC1 sequence and apparent domain architecture are conserved in higher order eukaryotes, with an N-terminal region predicted to be disordered, a KH domain, and an RNA ligase-like/PDE domain (Figs. 3A and 4) (1). We analyzed HsASCC1 isoform 2 and thermostable ApASCC1, separated by 500 million years in evolution and thereby allowing identification of conserved structural features. Proteins from A. pompejana, deep-sea extremophile worms that live on hydrothermal vents, often behave well for structural studies and have high sequence similarity to human proteins (36, 37); ApASCC1 has 46% identify to HsASCC1 (Fig. 4). We determined crystal structures for two constructs, ApASCC1Δ40 with both KH and PDE domains and HsASCC1 PDE domain (HsASCC1Δ133) to 1.15 Å and 2.8 Å resolution respectively (Figs. 3, B–D, 4, and S4, and Tables S7 and S8). The Wilson B-factors were 16 and 44, respectively for ApASCC1 and HsASCC1 structures, consistent with the hypothesis that proteins from hyperthermophilic organisms are better for crystallization.
Figure 3.
ASCC1 has tandem KH and PDE domains with uncommon Histidine rotamer and an active site loop.A, domain schematic of Alvinella (Ap) and Human (Hs, Isoform 2) ASCC1 and constructs used in this study. ASCC1 germline mutations that cause frameshifts in ASCC1 (Table S1) are mapped onto the human schematic. B, the crystal structure of ApASCC1Δ40 shows the relative position of the KH and PDE domains and their functional motifs. The two HXT motifs (black spheres) cluster in adjacent anti-parallel beta strands at the bottom of the V-shaped groove in the PDE domain on the same side as the phosphate-interacting GKKG motif in the KH domain. Conserved residues ranked by ET analysis localize to the PDE 2HXT groove, the hinge, and the KH domain. Left zoom inset shows conserved residues in the PDE including the HXT motifs. Right panel with simulated Annealing (SA) omit maps (orange, contoured at 1.5s) shows an uncommon rotamer position of the Histidine in the second HXT motif due to steric sandwiching between a Tyr and Lys and hydrogen bonded to a glutamate. The overlaid His side chain outline (dashed) shows the closest preferred rotamer. C, crystal structures of HsASCC1Δ133 reveal functional motifs in the PDE domain, depicted as in B. There were two molecules in the asymmetric unit (chain A and B). The comparison revealed an active site loop coordinated to the first HXT motif and that is only present in the A chain. D, ActSite-Loop coordination to His179 in the first HXT motif, overlaid with SA omit map (orange, contoured at 1.5s).
Figure 4.
ASCC1 Sequence, secondary structure, and residue function for multi-sequence alignment. ApASCC1Δ40 crystal structure is mapped onto full-length sequence alignment. Fig. S1 maps secondary structure onto crystal structure and positions of tumor VUS with EA score >80 and E292 disease mutant.
As the first protein structure with both KH and PDE domains, the ApASCC1 crystal structure indicated a side-by-side architecture, where the KH domain is fixed to one edge of the PDE V-shaped domain (Fig. 3B) and not a beads-on-a-string relationship as is often observed for multi-domain proteins. Although we cannot exclude a contribution of crystal packing to the observed domain–domain conformation, the interface between domains was well ordered. Both domains individually follow the expected architecture for their respective structural family. Conserved in KH domain family members and shown to interface with ssRNA or ssDNA, the GXXG sequence motif (15, 16) is accessible on one side of the KH domain. The ApASCC1 PDE domain structurally resembles a valley, or V-shape, formed by three beta sheets. Three helices lie on the outer edges of the V. Marking the active site, the two PDE-conserved His-x-Thr (HXT) motifs are located centrally, at the bottom of the V.
To identify functional residues, we carried out Evolutionary Tracing (ET) analysis and mapped them onto the ASCC1 structures, as we previously did for other DNA repair structures (38, 39, 40). The two top-ranked tiers include residues in the known motifs (HXT and GXXG) but also mapped to the possible RNA-binding path in the KH domain, the PDE active site, and unexpectedly the interface connecting the KH and PDE domains. Arg133-Asp192 pair connects KH α4 helix and PDE α6 helix. Arg131 on the end of a KH domain helix makes a salt bridge to a Glu195, hydrogen bonds to two main chain contacts, and packs against Pro189 on the PDE domain. In turn, Glu195 hydrogen bonds Tyr72 and packs against a Pro69 on the KH domain. Finally, the aliphatic chain of a KH domain Arg132 packs against that of Asn272 on the PDE domain. These three contiguous basic residues in the hinge are highly conserved and may also serve to C-cap the α4 helix.
In the HsASCC1 crystal, there were two molecules in the asymmetric unit (Fig. 3C). The HsASCC1 PDE domain crystal structure overlaid well onto the ApASCC1 PDE domain with a Root Mean Square Deviation (RMSD) of 0.89 Å over all residues, supporting the applicability of the ApASCC1 as a model for the human protein (Fig. S7, panel in top left corner). In the A chain, there was traceable density for a loop that lay over the PDE active site (Fig. 3D). Coordination from Glu292 to His179 in the first HXT motif suggests that it would block catalytic residues and would have a potential inhibitory effect, which we have called ActSite-loop. Indicating flexibility in the ActSite-loop, we did not observe density for the ActSite-loop in the B chain of the HsASCC1 crystal nor in the ApASCC1.
Notably, the histidine in the second HXT was modeled in an atypical, albeit allowed rotamer (41). In a His-Glu pair and packed between a tyrosine and aliphatic chain of lysine, the histidine is orthogonal to a more preferred orientation (Fig. 3, B and C). This atypical rotamer is found in both Ap and HsASCC1, indicating this is an evolutionarily conserved structural feature. Highlighting that uncommon stereochemistry is a weakness in machine learning prediction programs, AlphaFold2 predicted this histidine in the more canonical rotamer and accommodated the preferred rotamer by erroneously shifting the neighboring active site side chains and main chain.
KH structural comparative analysis
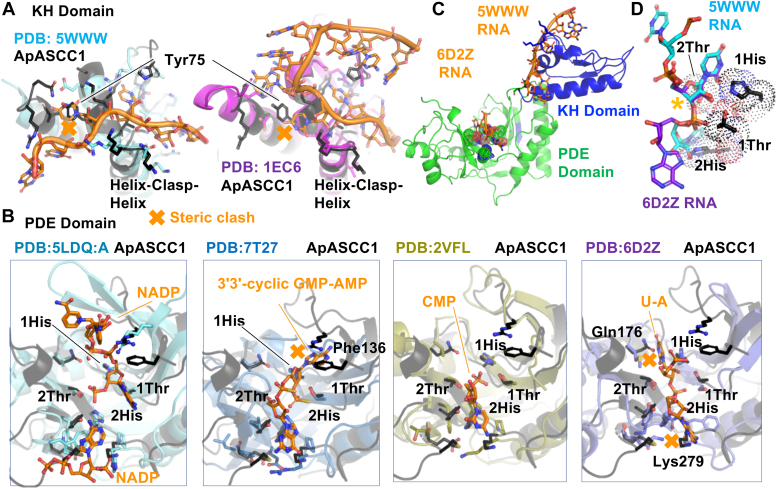
To learn from related structures, we ran the ApASCC1 KH and PDE domains separately through DALI (42) and Foldseek (43). Inputting the full-length protein only gave results for the individual domains. In Foldseek, the N-terminal KH domain pulled up 333 hits, while the C-terminal PDE domain pulled up 66 hits. In the top 20 hits, the sequence identity ranged from 18 to 30% and 16 to 20%, respectively. We aligned ASCC1 with representative orthologs that were close in similarity (Fig. 5, A and B and S5, S7, and S8) and looked for clues to ASCC1 function.
Figure 5.
ASCC1 overlaid with structural orthologs highlights conserved and unique ASCC1 structural elements.A, ApASCC1 KH domain overlaid onto structural orthologs complexed to RNA, suggests which critical motifs are conserved and which are inconsistent with current ASCC1 structure. KH domains were superimposed over whole ASCC1 KH domain. Additional overlays are in Supporting information. B, ApASCC1 PDE domain overlaid onto structural orthologs complexed to RNA or ligand suggests which critical motifs are conserved and which are inconsistent with current ASCC1 structure. PDE domains were superimposed based on the two HXT motifs (labeled as 1 and 2 in figure). Additional overlays are in Supporting information. C, ApASCC1 KH and PDE domain with superimposed substrate based upon superimposition of protein domains as in A and B. D, Superimposition of base 1 (∗, base numbering based on Fig. 6) from KH-bound RNA onto PDE-bound RNA, relative to ApASCC1 HXT motifs. Sugar puckers were both C2′ endo.
The KH domain is a small, ∼70 residue domain associated with binding to RNA and ssDNA (Figs. 5 and S5). The ASCC1 KH domain topology is well conserved, with a beta sheet on one side and three helices on the other (15, 16). The RMSD was as low as 0.5 Å. KH domains have a GXXG sequence motif that has been shown to directly interact with the phosphodiester backbone of the oligonucleotides. The XX is typically a basic residue (lys or arg), but can also include residues that can H-bond to the phosphodiester backbone. To probe how ASCC1 KH domain might bind RNA, we examined three KH domain structures complexed to RNA. Based on the overlay, ApASCC1 could bind similarly as Nova-1 (PDB:2ANN) and Nova-2 (PDB:1EC6), but not MEX-3D (PDB:5WWW) (44, 45, 46). Notably, ASCC1-conserved Tyr75 suggests an ASCC1-specific recognition of the first base, potentially selecting amide-containing adenine or cytosine bases (Fig. 5A).
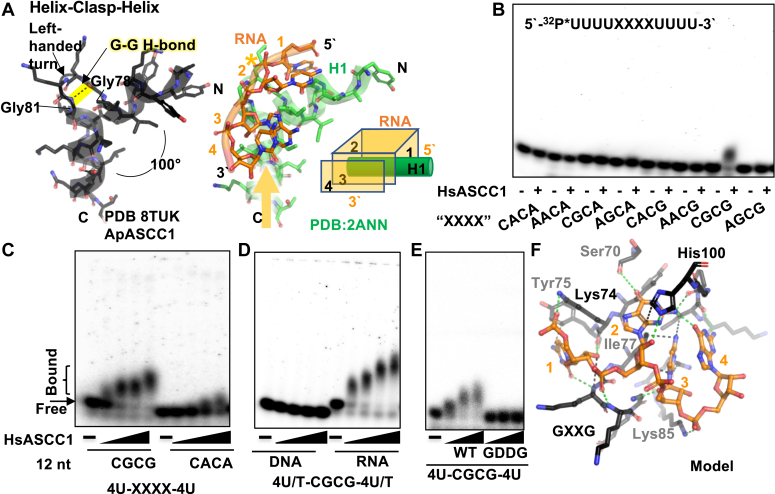
In examining the KH domains in complex with RNA or DNA, we observed that the oligonucleotide path can vary from straight to hairpins, with four of the bases positioned similarly. Even the phosphodiester can be shifted by as much as 3 Å, as observed in MEX-3C (PDB:5WWW) (Fig. 5A and S5). Notably, we found a striking vignette centered at the GXXG motif where three of the bases, like three sides of a box, wrap the helix N-terminal to the GXXG motif (Fig. 6A). The 5′ base #1 may or may not stack, the central base #2 is never stacked, and base #3 is often stacked with base #4. Suggesting an interface role, the positive dipole of the helix C-terminal to the GXXG is pointed towards the phosphate of the middle base, indicated by an ∗ in Figure 6A These two helices, on either side of the GXXG motif, form a 100° helical corner, with the glycines making a main chain H-bond that resembled a clasp. Hence, we expand the GXXG sequence motif to include the two helices, as a functional Helix-Clasp-Helix (HCH) structural motif conserved in KH domains. It is notably distinct from the two well-known nucleic acid two-helix motifs, Helix-Turn-Helix (HTH) and Helix-hairpin-Helix (HhH) (Fig. S6) (47, 48). For either motif, the two helices do not overlay with the two helices of the KH domain HCH. Not forming a helical corner, the HTH helices criss-cross each other to interface with the minor groove of duplex DNA, while the helices in the HhH motif are at a 30° angle (49). We note that the C-terminal helix of the HhH is forming a similar basic dipole interaction with phosphodiester, although neither the oligonucleotide path nor both helices overlay. Based on this analysis, the HCH is a newly defined functional motif for interaction with the phosphodiester. In addition to the electrostatic GXXG interaction with the phosphodiester, the N-terminal helix acts as a steric wedge to splay out three bases, while the C-terminal helix provides a basic dipole to interface with the phosphodiester.
Figure 6.
KH Domain Helix-Clasp-Helix structural motif for nucleotide binding is distinct from other two helix nucleic acid interaction motifs.A, structural motif, Helix-Clasp-Helix, found in KH domain of ASCC1 or in representative Nova-1 KH domain bound to RNA (PDB:2ANN). The GXXG motif is positioned by the two helices. In Nova-1, the amino-terminal dipole (yellow arrow) of the second helix is directed towards the RNA phosphate backbone (∗). B, biochemical analysis showing HsASCC1 bound specifically to CGCG-containing RNA. EMSA of 116 nM HsASCC1 incubated with 10 nM radiolabeled 12 nt UUUUXXXXUUUU RNA with XXXX representing specific sequence. C, HsASCC1 sequence specificity for CGCG. Dose-dependent EMSA with varying ASCC1 concentration (60, 120, 240, 360 nM) and 10 nM radiolabeled RNA containing CGCG or CACA. Quantitation in Fig. S4B. D, HsASCC1 sequence specificity for RNA. Dose-dependent EMSA with varying ASCC1 concentration (60, 120, 240, 360 nM) and 10 nM radiolabeled RNA or DNA containing CGCG. Quantitation in Fig. S4C. E, HsASCC1 RNA-binding requires KH domain GXXG motif. Dose-dependent EMSA with varying WT or mutant (GDDG) ASCC1 concentration (60, 120, 240, 360 nM), 10 nM radiolabeled RNA containing CGCG. Quantitation in Fig. S4D. F, Homology-based and energy minimized computational model of ApASCC1 bound to CGCG RNA. Major interactions are colored green (H-bonds, polar) or black (van der Waals). Orientation similar to Figure 5A. Sidechains interacting with RNA are labeled.
To decipher sequence specificity, we examined the overlay of the ASCC1 KH domain with KH domain-bound RNA (PDB:2ANN). For base 1, we predicted that ASCC1-specific Tyr75 would mandate an amide from cytosine or adenine. Based on how well-packed Tyr75 is in ASCC1, it is unlikely to shift out of the way. We predicted that His100 would stack on base 2, but this would require a purine, either adenine or guanine. For base 3, ASCC1-specific Lys84 would be selected for a pyrimidine with a carbonyl. A main chain carbonyl (Ile97) would select cytosine over uracil. Stacking with base, base 4 would likely require a purine. Other potential interacting side chains are not well-conserved. Thus, our structural analysis suggested that ASCC1 would have specificity for RNA containing C/A-A/G-C-A/G.
To test if ASCC1 can bind to our predicted RNA sequence, we designed eight RNAs, UUUUXXXXUUUU (12 nucleotides in length), where XXXX represents the 8 possible combinations of C/A-A/G-C-A/G (Table S9). By electrophoretic mobility shift analysis (EMSA) under stringent 150 mM KCl conditions, we surprisingly observed specific binding of wildtype HsASCC1 to only one sequence containing CGCG (Fig. 6B). Supporting this finding, we observed dose-dependent binding (Fig. 6C for gel and Fig. S6B for quantification) as low as 60 nM protein. Suggesting that the protein was coming on and off the RNA and ultimately complicating quantitative analysis, the mobility of the RNA shifted with protein concentration, and the band was spread out. Many KH domains also bind DNA, so we tested if ASCC1 binds to DNA containing CGCG. We did not observe any significant shift in DNA mobility, in contrast to the respective RNA substrate (Fig. 6D for gel and Fig. S6C for quantification). As both KH or PDE domains could interact with RNA, we next tested if mutation of the KH domain GXXG motif to GDDG is sufficient to prevent RNA binding. Indicating that RNA binding was indeed dependent on the ASCC1 KH domain, we did not observe any significant shift in UUUUCGCGUUUU when we added the GDDG mutant, in contrast to wild-type (WT) control (Fig. 6E for gel and Fig. S6D for quantification). Thus, our EMSA analysis supports ASCC1 binding is RNA- and sequence-specific through its KH domain, with the caveat that the KH domain may interact with other sequences or methylated RNA.
To understand this specificity, we homology modeled the ApASCC1 KH domain bound to CGCG and relaxed the model using energy minimization protocols in Rosetta (Fig. 6F). Consistent with sequence specificity, there were multiple side and main chain interactions (H-bonding, polar, and van der Waals) to the bases. Interacting side chains were highly conserved, except His100. His100 is positioned to clamp down the RNA, and a lysine/arginine nearby in other organisms could serve this role, as we observed for Arg177 in APE1 (50). We observed interactions with the 2`-OH of the RNA sugar for bases 1, 2, and maybe 3. Supporting sequence specificity for the guanines, the N2 of guanine base 2 interacted with the O4 of guanine base 4. Explaining the requirement for the smallest base, cytosine base 3 was sterically fit into a tight pocket. This computational model of ApASCC1 bound to RNA showed how RNA and sequence specificity could be carried out.
PDE structural comparative analysis
Next, we overlaid ASCC1 PDE with structurally close orthologs, based on DALI and Foldseek (Figs. 5B, S7 and S8). In contrast to the KH domain structural conservation, the V-shaped PDE domains varied, such that global superimposition did not always overlay HXT motifs. The RMSD range was much higher, with the lowest RMSD of 0.8 Å. Thus, we superimposed based only on the two HXT motifs. We found that the first HXT motif overlaid well, but the atypical rotamer conformation of the ASCC1 histidine of the second HXT motif was not observed in any other PDE domain. Neighboring residues that stabilize the rotamer position are also not conserved, suggesting their preferred orientation is not a crystallographic artifact.
To find clues to ASCC1 PDE function, we looked for structural compatibility with the four PDE structures in complex with ligands, AKAP18, EcLigT, Phage PDE, and HsUsb1 (Fig. 5B). Although AKAP18 (PDB:2VFL) (51) is structurally most similar to ASCC1 and CMP would fit with no clash in ASCC1, the second histidine is 3 Å away from that in ASCC1. Based on overlay with EcLigT (PDB:5LDQ:A) (52), NADP also does not clash, but ASCC1 lacks some of the NADP-interacting residues. For 3′3′-cyclic GMP-AMP bound by Phage PDE (PDB: 7T27) (53) and for dinucleotide bound by HsUsb1 (PDB:6D2Z) (54), there was both steric clash and lack of similar side chains for interaction. ApASCC1 Phe136 and PHE139 would clash with the adenine base of the 3′3′-cyclic GMP-AMP, while ApASCC1 Gln 176 would clash with the first uridine base in the dinucleotide. The diversity of known PDE binders, their lack of structural compatibility with the ASCC1 PDE domain, and the rotamer uniqueness of the histidine in the second HXT suggest either that the rotamer and active site are positioned to negatively regulate the catalytic mechanism or the ASCC1 substrate is structurally distinct from those of other PDE’s.
Although our ApASCC1 structure appeared sterically incompatible with substrates bound to other PDE domains, we cannot exclude the possibility that the substrate induces a conformational change that allows binding. We tested for 2′-5′ phosphodiesterase activity. Unfortunately, we found neither ASCC1 of human nor Alvinella origin caused detectable cleavage of 2′-5′ linked oligoadenylates 2-5A (2′-5′p3A3) (Fig. S9) in HPLC-based assays. However, under the same experimental conditions, positive control protein RVA VP3-CTD cleaved >95% 2′-5′p3A3, as expected (55). We further tested ASCC1 for ability to cleave possible alternative substrates 2′,3′-cGAMP, 2′,3′-cAMP, and 3′,5′-cAMP. However, we found no detectable cleavage of these potential substrates by ASCC1, consistent with our working hypothesis that the ASCC1 substrate will be distinct from other 2H phosphodiesterases.
Connected KH and PDE domains in ASCC1 suggest functional interaction, such as that the KH domain holds RNA for the PDE domain. Consistent with this hypothesis, the RNA binding side of the KH domain is on the same side of ApASCC1 as the PDE active site, marked by the two HXT motifs (Fig. 5C). KH domains can bind the nucleic acid either straight across the HCH motif, or as a hairpin that curls around. These structures are consistent with two models for KH-PDE interaction. Model 1 is that the KH domain rigidly holds a nucleic acid substrate and feeds the end to the PDE active site; even AlphaFold2 correctly predicted the exact orientation of the KH and PDE domains to each other (56). Requiring flexibility between the KH and PDE domains, model 2 is that the nucleic acid-bound KH domain flips into the PDE domain, like a waffle maker with RNA in the middle. The three conserved arginines in the linker may move toward KH-bound RNA, disrupt the helix that they were C-capping, and the linker region now can serve as a phosphate-induced hinge, as seen in kinases (57). Supporting the second model, the sugar puckers of the KH domain-bound RNA and the PDE-bound RNA are C2′ endo, typical for DNA and not for RNA, and can overlay on each other (Fig. 5D). However, protein-protein clashes in the simple RNA overlay imply that the KH domain would need to release the RNA before the PDE domain could bind RNA.
SAXS analysis indicates stabilized KH-PDE domains with potential for flexibility and validates AlphaFold2-predicted N-terminal beta pin
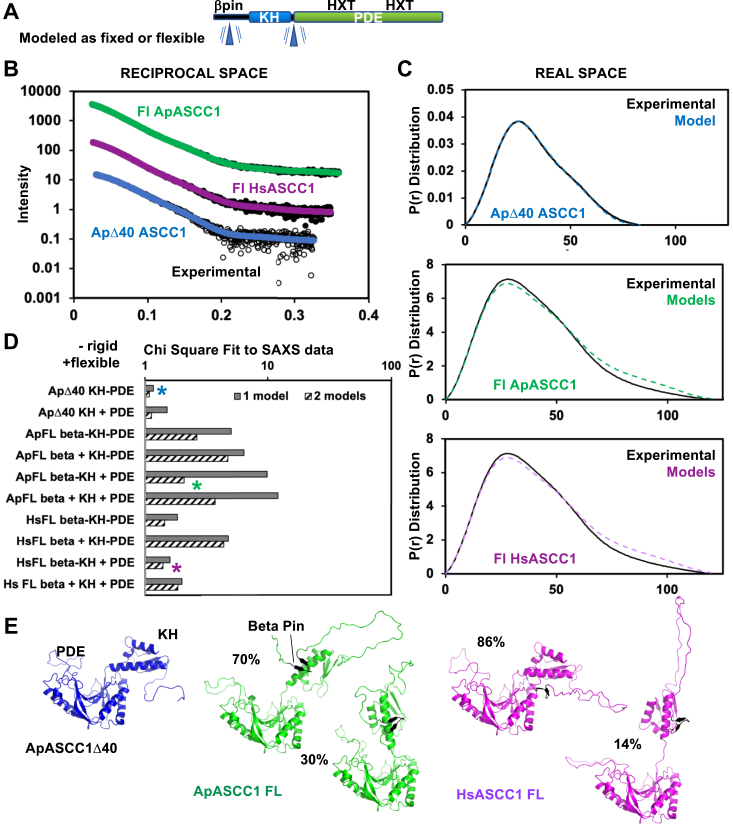
We next measured ASCC1 structural dynamics with SAXS analysis in solution. While the above KH-PDE feeding Model 1 could be consistent with flexibly or rigidly linked KH-PDE domains, waffle maker Model 2 requires flexibility. We also wanted to test the side-by-side orientation of KH-PDE domains, as the crystal structure may have been due to crystal packing. We conducted SAXS analysis on three constructs: ApASCC1Δ40 (corresponding to the crystal structure), full-length ApASCC1, and full-length HsASCC1. SAXS data were collected at SIBYLS Beamline 12.3.1 at the Advanced Light Source synchrotron, under size exclusion chromatography (SEC)-Multi-Angle Light Scattering (MALS)–SAXS mode (Fig. 7 and Table S10). The main peak of ApASCC1Δ40 and full length HsASCC1 eluted as a single peak, while there was a small shoulder right before for full-length ApASCC1, for which we used evolving factor analysis to separate out the signal for the main peak. Guinier plots were linear (Fig. S10), and the SAXS molecular weight (MW) were consistent with the theoretical MW of a monomer. Interestingly, the quantitative flexibility metric, the Porod Exponent (PX) was 3 and ∼4 for HsASCC1 and ApASCC1 respectively (Table S10) (58, 59). As indicated by a PX of 4, the greater rigidity indicated in ApASCC1 may be related to the hyperthermic environment in which A. pompejana live.
Figure 7.
SAXS analysis indicates stabilized KH-PDE domains with the potential for flexibility and supports the betapin model.A, ASCC1 domain schematic indicating with a blue triangle which regions were allowed to be flexible in the SAXS modeling. B, experimental reciprocal space SAXS data (black) for ApASCC1Δ40, ApASCC1 full-length (FL), and HsASCC1 FL SAXS curves predicted from best-fitting models (colored, see C) are overlaid. Initial models were built based on crystal structures and AlphaFold2 models and fit to the experimental SAXS data. C, experimental SAXS data and models, as described in (B), in real space. D, lowest chi square metrics of SAXS-fit atomic models to experimental SAXS data. During modeling, beta pin, KH, and PDE domains were internally kept rigid, but the domains relative to each other were either kept rigid (−) or allowed to move (+), as indicated by blue triangle in (A). Models (∗) with the lowest chi square were selected for further analysis. E, best SAXS-fit atomic models were consistent with ASCC1 dominant KH-PDE conformation being similar to the crystal orientation, potential for limited flexibility between KH and PDE domains, and a beta pin predicted by AlphaFold2 to reside next to the KH domain.
To test whether KH and PDE domains were rigid relative to each other or were flexibly linked, we generated atomic models using AlphaFold2 predictions (60) and replaced regions with the relevant crystal structure when possible (Fig. 7A). Predicted models had an almost identical orientation of the two domains, but the PDE domain was incorrectly predicted around His281 (ApASCC1)/His277 (HsASCC1) with the atypical rotamer, as mentioned earlier. Unexpectedly for the full-length models, AlphaFold2 predicted an N-terminal two-stranded beta hairpin, attached to the KH domain, which we term beta pin. Supporting its presence, the sequence is highly conserved and occurs in both HsASCC1 and ApASCC1 structure predictions. Thus, we included the beta pin in our analysis. Maintaining the beta pin, KH domain, and PDE domain as rigid bodies, we kept these domains in the same orientation or allowed these domains to move relative to each other in four separate categories (Fig. 7, A and D). Calculated theoretical SAXS from atomistic models were compared to the experimental SAXS data as single or ensembles of 2, 3, or 4 models. For all analyses, there was little significant improvement for ensembles of 3 or 4 models, so we focused on fits for single or two-state models. For ApASCC1Δ40, the best fit was to a single model where KH and PDE domains were kept fixed (Fig. 7E). For full-length models, the best fit was to two-state models. Ensembles that allowed the beta pin to move away from the KH domain led to worse fits, validating the beta pins existence and its positioning. The beta pin is not close to the RNA binding interface in the KH domain. With the beta pin fixed relative to the KH domain, there was a slight preference for allowing the KH and PDE domains to move relative to each other. Although not definitively better, this improvement was observed in both ApASCC1 and HsASCC1. Inspection of the two models showed a predominant model similar to a rigidly fixed KH-PDE consistent with the ApASCC1 crystal conformation, with a 20 to 30% population of a model where the two domains are rotated. First, these results support the existence of a beta pin attachment to the KH domain, as predicted by AlphaFold2, with implications for ASCC1 N-terminal interaction with ASCC3. Second, they are consistent with ASCC1 having segmental flexibility, where certain conformations are stabilized but have potential for flexibility, which we have observed previously for FEN1-PCNA and ubiquitinated PCNA (61, 62, 63). Small differences between experimental and predicted SAXS curves calculated from the models, suggest that the conformational landscape is not fully modeled.
Tumor-associated and genetic site mutations target conserved structural elements on ASCC1
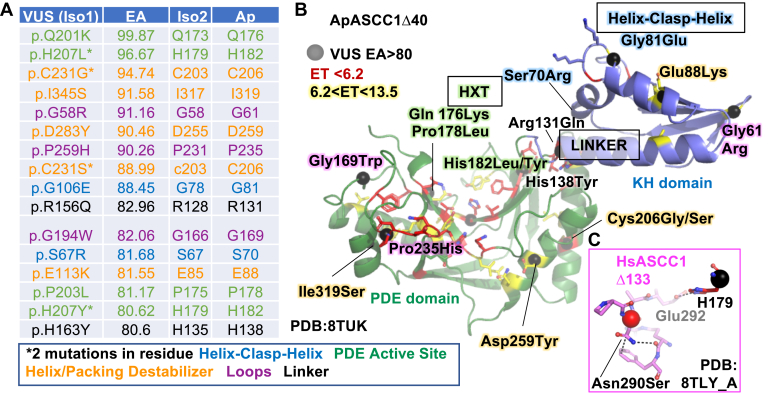
To provide a structural rationale for disease and tumor-associated single-site mutations, we mapped them onto the ApASCC1 structure (Fig. 8). We collected the tumor-associated mutations, also known as VUS from the Catalogue of Somatic Mutations In Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic, CosmicMutantExport.tsv file, v. 92) (34). Applying Evolutionary Action analysis, which combines evolutionary conservation and impact of changing one residue to another (64), we examined the 16 VUS predicted to be the most severe, with an EA score >80 (Table S11). These VUS mapped to both KH and PDE domains, and many to sites of high functional significance. Using the ApASCC1 numbering for comparison to the structure with Human Isoform 1 (HsI) in parentheses, Helix-Clasp-Helix residues Gly81Glu (HsI Gly106) in the glycine-glycine clasp and Ser70Arg (HsI 67), identified in our RNA model, would likely disrupt RNA binding by the KH domain. Likely inactivating the PDE catalytic activity, His182Leu or His182Tyr (HsI His207) would replace the histidine in the first HXT motif, required for abstracting a hydrogen from the substrate to initiate the catalytic reaction. Gln176Lys (HsI 201) and Pro178Leu (HsI 203) would destabilize the structure supporting His182 (HsI His207). Arg131Gln (HsI Arg156), and His138Tyr (HsI 163) map to the linker, potentially disrupting the domain-domain orientation and its regulation. Five mutations mapping within helices will likely disrupt protein stability. Three mutations replace loop-promoting residues, Gly and Pro. Particularly interesting, Pro235 is part of a highly evolutionarily conserved loop, of unknown functional significance.
Figure 8.
Cancer and Germline mutations have implied impacts on functional structural elements.A, table of VUS, with EA >80 calculated based on HsASCC1 Isoform 1 (Iso1) with corresponding residues in Iso2 and ApASCC1 and colored according to functional impact. B, VUS positions mapped onto ApASCC1Δ40 structure (black sphere). C, Barrett’s oesophagus/osephageal adenocarcinoma germline site mutation (Asn290Ser), mapped onto ActSite-loop on HsASCC1Δ133 structure, likely disrupts ActSite-loop coordination to His179.
We mapped a disease site mutant, Asn290Ser, identified in a study of Barrett esophagus and esophageal adenocarcinoma (12), to the ActSite-loop in the HsASCC1 structure (Figs. 4 and 8C), Asn290 sidechain makes a main chain H-bond and π-stacks with Phe340, which a serine would not be able to do. Thus, the Asn290Ser likely disrupts the ActSite-loop structure, particularly for nearby Glu292, which makes a H-bond to active site His179 (Fig. 3C). Although this study did not mention whether this variant is recessive or dominant, we predict a dysregulated ASCC1 mutant would be dominant.
Discussion
With no previously reported experimental ASCC1 structures and limited knowledge of its activities and regulation, ASCC1 has been, until now, perhaps the most enigmatic of the ASC-1 complex proteins associated with multiple RNA-driven processes: ASCC1, ASCC2, ASCC3, and TRIP4 with ALKBH3. ASCC1, along with ASCC2, ASCC3, and TRIP4, contribute to resolving internally stalled ribosomes due to alkylation damage or otherwise difficult-to-translate mRNAs (65, 66, 67). ASCC2 binds to K63-linked ubiquitin chains to signal alkylation damage or stalled ribosomes and recruit the other ASCC components (68). ASCC3, which directly interacts with ASCC1, acts as a DNA/RNA helicase (1, 69). Like the base repair enzyme NEIL1, the direct alkylation reversal enzyme ALKBH3 suppresses transcription-associated DNA damage at highly expressed genes (70, 71). Thyroid Hormone Receptor Interactor 4 (TRIP4) is part of the Ribosome Quality-control Trigger (RQT) complex to clear stalled ribosomes on nascent mRNA, and it has been reported to transactivate NF-Kappa-B, SRF, and AP-1 (66, 72, 73). However, TRIP4 competes with ALKBH3 for binding to ASCC3. It does not appear to be critical for alkylation damage responses, suggesting that the ASC-1 complex is dynamic and is utilized for multiple functions in the cell (74). Using AlphaFold-multimer (75), we modeled an assembly of ASCC1, ASCC2, ASCC3, and ALKBH3 (Fig. S11). The ALKBH3 RNA path, based on a recent complex structure (76), is predicted to be distant from where the helicases are likely to bind RNA. The ASCC1 N-terminal beta pin is predicted to tether ASCC1 to the second helicase domain of ASCC3. In the model, ALKBH3 packs between the two helicase domains with two N-terminal interfaces serving as a tether on the opposite face of the SEC63 domain of the second helicase to the ASCC1 beta pin. Suggesting functional significance of this feature, SEC63 in Brr2p can regulate the second helicase activity (77). As previously shown (78), ASCC2 interacts with the ASCC3 N-terminus, which is loosely tethered to the two-helicase core. Based on an overlay with the yeast structure of ASCC2, ASCC3, and collided ribosomes, all three proteins are not in contact, but are nearby to predicted RNA locations. It will be interesting to test this model and see the molecular mechanisms as to how all three proteins and RNA can be simultaneously bound to ASCC3, work modularly, and be coordinated, in part, through ASCC3.
The significance of ASCC1 is found in the diseases associated with ASCC1 and/or ASC-1. ASCC1 and TRIP4 mutations are associated with a severe form of spinal muscular atrophy (SMA) with congenital Bone Fractures 2 (SMABF2) (6, 7, 8, 9, 10, 11, 13, 79, 80, 81, 82). Via the ASC-1 complex, SMA is linked to the fatal neurodegenerative disease Amyotrophic Lateral Sclerosis (ALS). Multiple proteins, whose mutations can cause ALS, act in the functional association of the ASC-1 complex with the RNA Polymerase II and U1 small nuclear ribonucleoprotein particle (RNAP II/U1 snRNP) machinery (73, 83). As SMA-causative mutation in ASCC1 or ALS-causative mutation in FUS can disrupt the association between the ASC-1 complex and the RNAP II/U1 snRNP machinery, this disruption may be a central nexus for SMA and ALS. This notion provocatively implies that multiple complex activities of this machinery in RNA capping, transcriptional regulation, splicing, DNA repair, and stress responses may underlie the pathogenesis of these diseases. Furthermore, many genes whose defects cause SMA and ALS encode RNA/DNA binding proteins with low complexity regions likely to promote aggregation or phase condensation, as do ALS mutations in reactive oxygen defense enzyme superoxide dismutase (84). In fact, ASCC1 can assemble into nuclear speckles (1) and has a disordered N-terminal region with low sequence complexity, which is predicted to include a beta pin (Fig. 7E), suggesting the possibility of its forming multi-valent interactions and super-stoichiometric condensates acting in higher-level response mechanisms. These studies, which reveal interwoven protein and nucleic acid interactions from nano to mesoscale, highlight the importance of foundational structural information on ASCC1 suitable to select mutations to test and dissect its potential biochemical and cellular activities in multiple contexts.
One biological commonality of ALS and SMA pathogenesis, to test, may be an overwhelmed DDR due to accumulation of RNA/DNA damage, coupled to loss of functional DDR machinery interactions, as seen for cancer and degenerative disease-causing interactions in the nucleotide excision repair machinery (85). Notably, neurons, such as those affected in SMA, are extremely metabolically active and post-mitotic, so lack replication-associated repair and the homologous recombination repair pathway to deal with high levels of DNA and RNA damage-causing reactive oxygen species. This idea of overwhelmed and defective DDR is supported by a similar GSEA enrichment in mitochondria-associated genes and genes coding for centromeric structures for genes that, when highly expressed, are associated with Signature 3 mutations, such as we found for ASCC1 in breast cancer. These data suggest that the main drivers of Signature 3 mutations may be overproduction of reactive oxygen species and other oxidants by mitochondria, together with high rates of cell division: two processes likely associated with replication stress and tolerance to DNA damage in cancer cells (86, 87). In turn, replication stress may overwhelm the DNA repair capacity of the tumor cell, particularly homologous recombination repair, with DNA damage being shunted to alternative pathways for repair that are not dependent on extensive homology, such as non-homologous recombination and alternative end-joining pathways. Therefore, as a co-transcriptional regulator, ASCC1 is likely to be among the ∼0.5% candidate genes genome-wide that may be at the core of genetic instability hallmark in cancer, and that may include an RNA role in DDR alternatives to homologous recombination repair. In pancreatic cancer, ASCC1 is strongly coexpressed with markers of resistance to chemotherapy, independently of its role in the ASC-1 complex, and it will be of interest to assess its putative role in DNA damage repair by replication-blocking agents such as gemcitabine.
Our ApASCC1 and HsASCC1 structures reveal the two domains suitable to recognize RNA and process associated cyclic nucleotides: a KH four-base nucleotide-binding domain joined to a 2H PDE domain. ASCC1 furthermore plays a central role in the nuclear alkylation response, which could include damage to RNA. Often overlooked, RNA is damaged 30-fold more often than DNA by MMS, and 10-fold more alkylated RNA is repaired than alkylated DNA (88). Notably, all RNAs can be modified by alkylation damage as a result of endogenous or exogenous agents, and unregulated alkylated RNA is potentially genotoxic and can affect transcription, RNA processing, mRNA turnover, and translation (89). So, ASCC1 structural features and their potential roles in RNA and DNA alkylation responses merit experimental examination for their biological and medical significance.
One striking insight from our comparative structural analysis of the ApASCC1 KH domain was the Helix-Clasp-Helix (HCH) motif in the KH domain for sculpting nucleotide positioning (Fig. 6). Although the GXXG motif is well described (15, 16, 90), the role of the helices brought together by the glycine-glycine clasp was not. The N-terminal α-helix provides a central support for three bases to wrap around it like three sides of a box, while the C-terminal α-helix donates a positive dipole for phosphate backbone recognition (Fig. 6A). Importantly, the HCH recognition motif is distinct from established two-helix motifs that have proven informative in understanding and predicting protein-nucleic acid interactions: the HTH motif for binding to the DNA major groove with sequence recognition (91, 92) or the DNA minor groove in repair proteins (93, 94), and the HhH motif for DNA phosphate backbone binding, as discovered in exonuclease III (48) and seen with two hairpin turns in the FEN1 replication-repair nuclease and the XPG nucleotide excision repair nuclease (95, 96). In contrast to HTH and HhH motifs, the HCH motif identified here breaks normal base stacking – base 2 is always unstacked. Combining our structural and binding analysis, we propose that the ASCC1 KH domain has RNA binding properties with novel specificity for CGCG sequences in comparison to other KH domains (97). However, our limited study cannot exclude ASCC1 binding to other sequences. ASCC-1 specific residue Tyr75 may also be important. Alkylation of bases can dramatically transform its negative nature into one positively charged that could interact with pi stacking of Tyr75. Suggestive as to the significance of CGCG-binding, it is identified as part of the most activated regulatory element, TCTCGCGAGA, and occurs in 1% of promoters (98, 99).
The three PDE X-ray crystal structures reveal a conserved V-shaped catalytic channel open on two ends, similar to other PDE structures. It is ∼14 Å wide at the V bottom between Tyr242 and Thr187, but ∼30 Å wide at the V top between Arg133 and Arg311 with the two His-X-Thr pairs along the floor and the KH domain perched on the rim along one edge. Entry of RNA bound to the KH domain into the PDE active site channel would require displacing the flexible ActSite-loop, which appears suitable to enforce specificity for substrate binding. The active site channel is ∼25 Å deep from His282 to Arg133 and ∼23 Å in length from Lys279 to Lys180. These dimensions are consistent with cyclic dinucleotide binding based upon superpositions with PDE superfamily member structures with bound nucleotides. Yet, the open-ended channel and our observation that some interacting residues are missing or not positioned properly suggest a different substrate from any known PDE structures. The ASCC1-specific rotamer for the histidine in the second HXT motif supports this hypothesis. In other PDE proteins, it has been proposed that the first histidine initiates the PDE reaction by abstracting a hydrogen and the second histidine donates a hydrogen to finish the reaction. The second histidine’s position could easily be substrate-specific. As an aside, our experimental structures highlight a challenge for protein structure prediction algorithms. Not only did AlphaFold2 mispredict this atypical histidine rotamer, its attempt to accommodate a more canonical rotamer caused ripple effects across the active site and produced a prediction that topologically looked fine, but could not be used for design of small molecule inhibitors.
A feature with regulatory implications, the Arg-Arg-Gln-Pro-Phe-Thr-His linker C-caps the α4 helix, which ends the KH domain, and links to the first PDE domain β-stand. We hypothesize that RNA substrate bound by the KH domain could adjust Arg-Thr-His positions resulting in potential destabilization of the α4 C-terminal end and remodeling of the current domain conformations, as seen for protein kinase A and superfamily ATPase FlaI (57, 100). The SAXS measurements validate the potential for the linker to be primarily stabilized but also form a hinge between KH and PDE domains.
Currently, the fixed position of the KH and PDE domains in solution, as conserved from HsASCC1 and ApASCC1, places the binding sites along a single face of ASCC1, supporting the notion that they can provide bipartite combined specificity for binding to both sites. In support of a regulatory ActSite-loop function, analogous flexible loops forming interactions that block ss-nt binding have seen for regulated inhibition of the XPD translocase and EXO5 nuclease (101, 102) as distinct to the inhibition of other DNA repair proteins by ssDNA mimicry (103, 104). We therefore identify at least four structural features likely to promote ASCC1 substrate specificity: potential dual binding to KH and PDE, potential for the linker to switch conformations upon interaction with an RNA phosphate backbone, the need for displacement of the observed regulatory loop interaction with the critical active site His, and distinct positions for conserved side chains and the unexpected active site histidine torsion angle. Together, these results point to a likely enzymatic activity for this domain of ASCC1, which to date remains elusive.
Here, combined structural and quantitative evolutionary action analyses of cancer-associated VUS corroborate key functional roles for the HCH, Arg linker, PDE domain ActSite-loop, and PDE domain two HXT active sites. For ASCC1 the location of disease-associated frameshift mutations throughout both domains and to the end of the PDE domain supports the biological importance of its intact structural architecture characterized here. Furthermore, the AlphaFold2 model of the ASCC Alkylation Response Complex for ASCC1 and ALKBH3 combined with ASCC2 and ASCC3 tethers enzymes ASCC1 and ALKBH3 on opposite sides of ASCC3 helicase SEC63 domain, suggestive of their co-regulation for recognition and repair of alkylated RNA. This merits further investigations.
Overall, we expect that these ASCC1 crystal structures and X-ray scattering results combined with bioinformatic and computational structural analyses offer specific insights and ideas for experimentally testing ASCC1 activities and biological functions. An intriguing idea is that ASCC1 could act on endogenous RNA fragments with 2′,3′-cyclic phosphate ends derived from partial RNase A digestion of 45S ribosomal RNA during RNA stress to prevent undesired RIG-I activation of the innate immune system (105). In any case, the combined KH and PDE binding site structure and architecture will inform experiments to elucidate ASCC1 roles in transactivation and alkylation damage responses as well as how ASCC1 mutations may promote inherited disease and cancer and its up-regulation in some cancers may be targeted by structure-based inhibitors.
Experimental procedures
Materials
2′-5′ linked oligoadenylates 2-5A (2′-5′p3A3) was synthesized from ATP as described in (55). Substrates 2′,3′-cAMP, and 2′,3′-cGAMP were obtained from Sigma Aldrich, and 3′,5′-cAMP was obtained from Cayman Chemicals.
Bioinformatic analyses
Gene expression data in cancer were obtained from RNA-seq experiments conducted during the TCGA project, as previously done (106). Files containing normalized rsem data (https://doi.org/10.5281/zenodo.7885655) were generated using the TCGA Assembler v. 2.0 (https://github.com/compgenome365/TCGA-Assembler-2) and processed using in-house scripts (https://doi.org/10.5281/zenodo.7874703 and https://doi.org/10.5281/zenodo.7874772). Box plots were generated in R using libraries “ggplot2”, “ggpibr”, “grid”, and “extrafont”, which compute and display the interquartile range (IQR) from Q1 to Q3 (25–75% percentiles), median (center line), whiskers extending to the minimum (Q1–1.5∗IQR) and maximum (Q3 + 1.5∗IQR), outliers (dots), and p-values from Wilcoxon rank-sum tests. Normality tests on gene expression data using https://www.statskingdom.com/ indicated that their patterns did not follow normal distributions.
For survival analyses, patient data for each type of tumor were separated two different ways. In the first analysis, we separated into two groups, “m_high”, which contained samples with mRNA expression for a gene above the mean, and “m_low”, which contained samples with mRNA expression for a gene below the mean. Kaplan-Meier survival curves were generated using R libraries “dplyr”, “survival”, and “survminer” according to the Cox proportional-hazards regression model, with p-values assessed from nonparametric log-rank tests. In the second analysis, we accounted for average levels of ASCC1. We divided patients into three groups: a “g_high” group in which ASCC1 RNA levels were higher than one standard deviation above the mean; a “g_low” group where ASCC1 RNA levels were lower than one standard deviation below the mean; and a “g_medium” group where ASCC1 RNA levels were within one standard deviation of the mean. Otherwise, we did the same statistical analysis as the first analysis.
Data for mutational signatures were retrieved from the Catalogue Of Somatic Mutations In Cancer (COSMIC, https://cancer.sanger.ac.uk/signatures/) (34) and processed using custom scripts. A master file (signature_profile_sample_t.txt.zip) was used in which each patient sample contained the percent of single base substitutions (SBSs) mapping to each of the 30 mutational signatures. The set of signatures were from 2015, version 2, (https://cancer.sanger.ac.uk/signatures/signatures_v2/). The master file was queried to compute the number of samples containing a given signature for the gene of interest, both for the g_high and the g_low samples. The difference in the number of g_high and g_low samples was assessed by Fisher's exact test. For gene coexpression analyses between ASCC1 and all other genes, we applied a Bonferroni correction to statistical p-values and deemed significant regression coefficients with p-values <2.5E-06. Statistical p-values were obtained from the F distribution using the c++ Boost library custom c++ utilities (https://github.com/abacolla/linearRegression).
To generate the set of genes for gene set enrichment analysis (GSEA), we selected those genes in which the difference in the number of patients with Signature 3 mutations between the g_low and g_high groups exceeded a p-value of 1.0E-6 (Fisher’s exact test). Since Signature 13, which has been attributed to the activity of AID/APOBEC family of cytosine deaminases, is often associated with Signature 3, genes with positive Signature 3 were further filtered against co-occurrence with Signature 13 mutations (p-value < 0.05; Fisher’s exact test), which for ASCC1 were not significantly represented. For pathway analyses we used the Database for Annotation, Visualization and Integrated Discovery (DAVID, at https://david.ncifcrf.gov).
Molecular cloning
DNA regions encoding HsASCC1 and ApASCC1 were PCR-amplified from synthetic genes (g-blocks, Integrated DNA Technologies) and codon optimized for expression in Escherichia coli (E. coli) cells. The A. pompejana genomic assembly was previously published (71). DNA regions of protein of interest were cloned into a modified pET expression vector 1B (Addgene), designed for ligation-independent cloning under the control of T7 promoter, which directed the synthesis of fusion proteins with TEV-cleavable N-terminal 6xHis-tags. For ApASCC1 crystallization, we truncated the N-terminal region (residues 1–40) predicted to be disordered (http://www.pondr.com/) (107). All constructs were confirmed by sequencing.
HsASCC1 PDE domain (residues 134–357) was cloned into the NdeI and XhoI sites of vector pET28a. The construct was engineered to have a 6xHis-tag with a cleavable thrombin site.
Individual KH or PDE domains of HsASCC1 or ApASCC1 were cloned by PCR amplifying the corresponding DNA regions and cloned into 1B ligation-independent vector with TEV cleavable 6xHis-tag.
Site-directed mutants of HsASCC1 and ApASCC1 were created through PCR amplification of the wildtype (WT) plasmid using primers designed to introduce specific amino acid mutations. The PCR products were then treated with DPN1 enzyme to cleave the methylated parental template strand. The resulting mutated DNA fragments were transformed into E. coli (E. coli) for propagation, and the mutants were confirmed by sequencing analysis.
Protein purification
ApASCC1 Δ40 purification
ApASCC1Δ40 for crystallization was expressed and purified from E. coli Rosetta (DE3) pLysS Cell lines (Novagen). Protein expression was induced at an O.D 0.4 to 0.6 at 600 nm with 0.1 mM IPTG, grown overnight at 20 °C. Cells were lysed in Lysis Buffer containing 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM βME, 20 mM Imidazole and 5% [v/v] glycerol, supplemented with protease inhibitor tablets (Themo Fisher), DNaseI 10 μg/ml (Roche) and RNaseA 10 μg/ml (Themo Fisher). Cells were lysed using sonicator on ice. Proteins were purified with Ni2+-NTA beads (Qiagen). The 6xHis-tag was cleaved off by TEV protease during overnight dialysis into buffer containing 50 mM Tris-HCl pH 8.0, 100 mM NaCl 4 °C. Protease-treated samples were reloaded on a fresh Ni2+-NTA gravity flow column to remove the 6xHis-tag, uncleaved protein and the 6xHis-tagged protease. The flow-through was further purified on a Superdex 200 10/600 Gl column (GE Healthcare) in 20 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 2 mM 2-mercaptoethanol. Protein was concentrated to 30 mg/ml. All the TEV cleaved ASCC1 proteins have SerAsnAla residues at the N-Terminus. ASCC1 protein sequences in this study are provided in Table S7.
Human ASCC1Δ133 purification
HsASCC1Δ133 was expressed in the E. coli (E. coli) Rosetta (DE3) cell line. Protein expression was induced at an O.D. of 0.4 to 0.6 at 600 nm with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and cells were grown overnight at 20 °C. The cell pellet was resuspended, sonicated not above 10 °C in Lysis Buffer A (50 mM Tris-HCl pH 8, 500 mM NaCl, 5% [v/v] glycerol, 0.05% Tween-20, 1 mM DTT, 0.2 mM PMSF, 0.2 μg/ml aprotinin, 0.2 μg/ml leupeptin, 0.2 μg/ml pepstatin, 1 μg/ml DNase, 100 μg/ml lysozyme), and centrifuged. Soluble HsASCC1Δ133 was purified on a HisTrap column (GE Healthcare), a Superdex 200 HiLoad 16/300 Gl (GE Healthcare) column and a Q sepharose column. For crystallization trials, HsASCC1Δ133 retained the 6xHis tag and was dialyzed against 15 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 1 mM DTT, 1 mM EDTA and 5% glycerol and concentrated to 13 mg/ml.
Hs ASCC1 full length from Sf9 cells
The full-length HsASCC1 protein was expressed and purified from Sf9 insect cells. Sf9 cells were infected with ASCC1 baculovirus for 52 h, and lysates were prepared using a Dounce homogenizer with lysis buffer containing 50 mM HEPES pH 7.5, 500 mM NaCl, 2% glycerol, 2 mM BME, 20 mM Imidazole, and EDTA-free protease inhibitor tablets (Thermo Fisher). After centrifugation, the soluble ASCC1 was purified with a HisTrap column (Cytiva). The protein was further purified using a Superdex 200 increase 10/600 Gl SEC column in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, and 2 mM BME. Fractions containing at least 95% pure ASCC1 were pooled and concentrated for SAXS experiments.
HS ASCC1 and ApASCC1 for biochemical assays
HsASCC1 full-length and ApASCC1 full-length proteins used biochemical assays were expressed and purified from E. coli (E. coli) Rosetta (DE3) cell line. Protein expression was induced at an O.D. of 0.4 to 0.6 at 600 nm with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and cells were grown overnight at 20 °C. Cells were lysed in a lysis buffer containing 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM βME, 20 mM Imidazole, supplemented with protease inhibitor tablets (Thermo Fisher # A 32963). Cells were lysed on ice by either a sonicator or high-pressure cell disruptor (Constant System Limited). Proteins were purified with Ni2+-NTA beads (Qiagen). Proteins were further purified on a Superdex 200 10/600 Gl column (GE Healthcare) in 20 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 2 mM 2-mercaptoethanol.
Rotavirus A VP3 C-terminal domain (RVA VP3-CTD) protein was expressed and purified as previously described (55).
Macromolecular crystallography
ASCC1 formed crystals by hanging drops within 1 week. The HsASCC1 PDE domain Δ133 (13.4 mg/ml) was crystallized in 12.5% MPEG 2K, 200 mM imidazole/malate (I/M) pH 5.0, 5% saturated KCl at room temperature. For cryoprotection, 25% ethylene glycol was added to the mother liquor. The ApASCC1Δ40 crystal form grew from protein (14 mg/ml), 15% MPEG 2K, 200 mM I/M pH 5.0. 2.5% KCl (saturated), 0.6% 2-mercaptoethanol at 15 °C. For cryo protection, crystals were soaked for about 2 s in 15% MPEG 2K, 200 mM I/M pH 5.0, 2.5% KCl (saturated) with 60% ethylene glycol mixed 1:2 (cryo: reservoir).
Diffraction data were collected at SSRL 9-2 from protein crystals at 100K with X-ray wavelength at 0.97946 Å and processed with HKL2000 (HKL Research). The structure of the HsASCC1 PDE domain was determined by molecular replacement with human AKAP18 from PDB code 2VFK (51) as a search model using the program Phaser (108). ApASCC1 crystals were subsequently phased using HsASCC1 PDE domain. Manual fitting was carried out in Coot (109) according to the 2Fo-Fc and Fo-Fc electron density maps, and further refinement was carried out with PHENIX (110). Structure figures were primarily created in Pymol (Schrodinger). Simulated annealing omit map figures were created in Coot (109).
Electrophoretic mobility shift assays
In a standard reaction, 200 fmol of the 32P-labeled RNA oligos (IDT, Table S9) were incubated at room temperature for 10 min in a 20 μl reaction mixture containing 20 mM Hepes-KOH, pH 7.5, 150 mM KCl and 5% glycerol plus 100 μg/ml BSA and Rnasin (0.3 unit) with various amounts of purified full length human ASCC1 as described in the figure legends. After the incubation, the samples were loaded on an 8% non-denaturing polyacrylamide gel (19:1 acrylamide:bis-acrylamide) and electrophoresis was performed at 150 V for 2 to 2.5 h at room temperature with EMSA running buffer (50 mM Tris, 380 mM glycine, 2 mM EDTA). The gels were exposed on Phosphor Imager screen for 12 to 16 h and scanned using Typhoon FLA 7000 Phosphor Imager and analyzed. No significant nuclease activity was detected.
Modeling RNA to ASCC1 KH domain (Rosetta relax)
In the absence of an available crystal structure with RNA, we positioned the RNA by aligning the KH1 domain of the neuronal splicing factor Nova-1 (PDB ID: 2ANN (45)) onto the KH domain of our structure. Considering the sequence-specific interactions observed between RNA and the Helix-Clasp-Helix region of the KH domain in our experimental findings, we introduced mutations to the RNA, altering four consecutive nucleic acid residues to CGCG using the swapna module in Chimera. Subsequently, the modeled structure was subjected to relaxation in Rosetta (111, 112, 113, 114), involving energy minimization through necessary adjustments to the backbone and sidechain, employing gradient-based minimization of torsional degrees of freedom. A total of five relaxation cycles were conducted, each consisting of four rounds of repacking followed by minimization in torsion space. The repulsive contribution to the total energy was scaled to 2%, 25%, 55%, and 100% in the first, second, third, and last rounds, respectively. The best scoring pose from the final cycle was chosen, resulting in the final computational model with bound RNA.
Activity assays
Activity assays, sample processing, and HPLC analyses were performed as described previously (55) with a few modifications. Briefly, 10 μM of the substrates was incubated with 1 μM enzyme in a reaction mixture containing 20 mM HEPES buffer (pH 7.4), and 1 mM DTT by incubation at 30 °C for 30 min. Substrates incubated under similar conditions in the absence of enzyme served as controls. Reactions were stopped by heating at 95 °C for 5 min. Samples were centrifuged at 18,000g for 15 min at 4 °C. Supernatants were collected and analyzed by HPLC. The intact peak of the substrate was analyzed on a 1260 Infinity II Agilent Technologies HPLC instrument equipped with an Infinitylab Poroshell 120 C18 analytical column (4.6 by 150 mm, 4 μm; Agilent Technologies). Five microliters of processed samples were injected onto the C18 column, at a flow rate of 1 ml/min, and eluted with a linear gradient (0–40%) of eluent B over 20 min and then 3 min of 40% eluent B, followed by equilibration to initial conditions (100% eluent A). Eluent A was 50 mM ammonium phosphate buffer (pH 6.8), and eluent B was 50% methanol in water. HPLC was maintained at 40 °C and spectra were recorded at 256 nm using the method described previously (55). Open Lab CDS software was used to analyze and calculate the areas under the peaks in HPLC spectra.
SAXS
SAXS coupled with multi-angle light scattering (MALS) in line with size-exclusion chromatography (SEC-MALS-SAXS) experiments were collected at the Advanced Light Source beamline 12.3.1 in Lawrence Berkley National Laboratory (115, 116, 117, 118). X-ray wavelength was set at λ = 1.127 Å, and the sample-to-detector distance was 2100 mm, resulting in scattering vectors spanning from 0.01 < q (Å−1) < 0.45. Briefly, a SAXS flow cell was directly coupled with an online Agilent 1260 Infinity HPLC system using a Shodex KW803 column. The column was equilibrated with a running buffer (25 mM HEPES pH 7.5, 200 mM NaCl, 2 mM 2-mercaptoethanol), as indicated above, with a 0.65 ml/min flow rate. 80 μl of each sample was loaded onto the SEC, the elution was passed through a Wyatt MALS instrument, and then 3-s X-ray exposures were collected continuously during a 25-min elution. Based on the MALS chromatogram, the SAXS frames recorded before or after the protein elution peak were used to subtract all other frames. Subsequently, SEC-SAXS frames were deconvoluted in single SAXS profiles using SCÅTTER (119) or RAW (120). We built HsASCC1 and ApASCC1 atomic models using AlphaFold2 predictions (60) and replaced regions with the relevant crystal structure when possible. Ensembles of models were generated using BILBOMD (121) and MultiFOXS (122) with input generated by CHARMM-GUI (123). Structures were drawn with Pymol (Schrodinger).
Data availability
A. pompejanaASCC1 complete coding sequence is available at GenBank with record number OQ857929 (https://www.ncbi.nlm.nih.gov/nuccore/OQ857929). The TCGA RNA-Seq normalized rsem data, TCGA clinical data, and mutational signature files used for the analyses are available at https://doi.org/10.5281/zenodo.7885656 (‘tumortypernaSeq.tar’, ‘cliDat_tcga_18.tar’ and ‘signatureProfileSample.txt.zip’ files); source codes for TCGA analyses (tcgaAnalyses and sumbitMPI) are available at https://doi.org/10.5281/zenodo.7874703 and https://doi.org/10.5281/zenodo.7874772, respectively. Crystal structures have been deposited to the PDB under coordinates 8TUK and 8TLY for ApASCC1Δ40 and HsASCC1Δ133, respectively. SAXS data has been deposited in Simple Scatter Database (https://simplescattering.com/) (73) under XSWQ5ILA, XSA5YQYE, and XSVL1FQL for full-length HsASCC1, full-length ApASCC1, and ApASCC1Δ40, respectively.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Nima Mosammaparst is an editorial board member.
Acknowledgments
We thank advice from Roland Dunbrack on alternative histidine rotamers and support from Arnold Groehler and Orlando D. Schärer when testing PDE functions. The cancer bioinformatic results here are based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author contributions
S. E. T., A. B., N. M., and J. A. T. conceptualization; S. E. T., D. J. R., N. C., R. T., A. B., A. S. A., A. H. S., and M. H. data curation; S. E. T., A.S., A. A., N. C., A. B., A. S. A., A. H. S. formal analysis; S. E. T., R. H. S., N. M., I. I., O. L., and J. A. T. funding acquisition; S. E. T., D. J. R., A. A., N. C., M-S. T., R. T., A. B., P. K., R. H. S., A. S. A., A. H. S., and M. H. investigation; S. E. T., N. C., R. T., and J. A. T. methodology; S. E. T. and J. A. T.project administration; S. E. T., N. C., M-S. T., J. M. S., A. H. S., and J. A. T. resources; S. E. T., N. C., A. B., P. K., T. P., I. I., A. S. A., O. L. software; S. E. T., A.S., R. H. S., N. M., O. L., and J. A. T., and M. H. supervision; S. E. T., A. A., R. H. S., A. H. S., and J. A. T. validation; S. E. T., A. A., N. C., A. B., R. H. S., A. H. S., and J. A. T. visualization; S. E. T., A. A., N. C., R. T., A. B., R. H. S., T. P., I. I., A. H. S., and J. A. T. writing–original draft; S. E. T., N. C., R. T., A. B., R. H. S., N. M., T. P., I. I., A. H. S., and J. A. T. writing–review & editing.
Funding and additional information
This work was supported the following grants: NCI P01 CA092584 (J. A. T., N. M., S. E. T., I. I.) and R35 CA220430 (J. A. T.), NIAID R01 AI104887 (R. H. S.), and NCI R01 CA193318 and R01 CA227001 (N. M.), NIGMS R35 GM139382 (I. I), NSF MCB-2027902 (I. I.), Robert A. Welch Chemistry Chair (G-0010, J. A. T.) and the Cancer Prevention and Research Institute of Texas RP180813 (J. A. T.). N. M. acknowledges support from the Siteman Cancer Center, the Barnard Foundation, Centene Corporation, and an American Cancer Society Research Scholar Award (RSG-18-156-01-DMC). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory and the SSRL Structural Molecular Biology Program, is supported by the U.S. DOE, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515, DOE Office of Biological and Environmental Research, and by the NIGMS (P41GM103393). This research used resources of the Advanced Light Source, which are DOE Office of Science User Facilities under contract no. DE-AC02-05CH11231. The SIBYLS beamline 12.3. is supported by the DOE-BER IDAT program and the NIH supported ALS-ENABLE (P30 GM124169). This work used HPC resources at the Texas Advanced Computing Center (TACC) at The University of Texas at Austin (URL: http://www.tacc.utexas.edu).
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Contributor Information
Susan E. Tsutakawa, Email: setsutakawa@lbl.gov.
John A. Tainer, Email: JTainer@mdanderson.org.
Supporting information
References
- 1.Soll J.M., Brickner J.R., Mudge M.C., Mosammaparast N. RNA ligase-like domain in activating signal cointegrator 1 complex subunit 1 (ASCC1) regulates ASCC complex function during alkylation damage. J. Biol. Chem. 2018;293:13524–13533. doi: 10.1074/jbc.RA117.000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 3.Falnes P.O., Bjoras M., Aas P.A., Sundheim O., Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundheim O., Vågbø C.B., Bjørås M., Sousa M.M., Talstad V., Aas P.A., et al. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohm J., Malfatti E., Oates E., Jones K., Brochier G., Boland A., et al. Novel ASCC1 mutations causing prenatal-onset muscle weakness with arthrogryposis and congenital bone fractures. J. Med. Genet. 2019;56:617–621. doi: 10.1136/jmedgenet-2018-105390. [DOI] [PubMed] [Google Scholar]
- 6.Bradinova I., Andonova S., Vazharova R., Tomova S., Balabanski L., Savov A. Spinal muscular atrophy with congenital bone fractures 2 caused by a rare loss-of-function ASCC1 gene mutation in two Bulgarian Roma patients. Clin. Genet. 2022;102:78–79. doi: 10.1111/cge.14130. [DOI] [PubMed] [Google Scholar]
- 7.Giuffrida M.G., Mastromoro G., Guida V., Truglio M., Fabbretti M., Torres B., et al. A new case of SMABF2 diagnosed in stillbirth expands the prenatal presentation and mutational spectrum of ASCC1. Am. J. Med. Genet. A. 2020;182:508–512. doi: 10.1002/ajmg.a.61431. [DOI] [PubMed] [Google Scholar]
- 8.Knierim E., Hirata H., Wolf N.I., Morales-Gonzalez S., Schottmann G., Tanaka Y., et al. Mutations in subunits of the activating signal cointegrator 1 complex are associated with prenatal spinal muscular atrophy and congenital bone fractures. Am. J. Hum. Genet. 2016;98:473–489. doi: 10.1016/j.ajhg.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu W., Liang M., Su J., Wang J., Li L., Zhang S., et al. Novel compound heterozygous pathogenic variants in ASCC1 in a Chinese patient with spinal muscular atrophy with congenital bone fractures 2: evidence supporting a "Definitive" gene-disease relationship. Mol. Genet. Genomic Med. 2020;8 doi: 10.1002/mgg3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marais A., Bertoli-Avella A.M., Beetz C., Altunoglu U., Alhashem A., Mohamed S., et al. Further clinical and genetic evidence of ASC-1 complex dysfunction in congenital neuromuscular disease. Eur. J. Med. Genet. 2022;65 doi: 10.1016/j.ejmg.2022.104537. [DOI] [PubMed] [Google Scholar]
- 11.Oliveira J., Martins M., Pinto Leite R., Sousa M., Santos R. The new neuromuscular disease related with defects in the ASC-1 complex: report of a second case confirms ASCC1 involvement. Clin. Genet. 2017;92:434–439. doi: 10.1111/cge.12997. [DOI] [PubMed] [Google Scholar]
- 12.Orloff M., Peterson C., He X., Ganapathi S., Heald B., Yang Y.R., et al. Germline mutations in MSR1, ASCC1, and CTHRC1 in patients with Barrett esophagus and esophageal adenocarcinoma. JAMA. 2011;306:410–419. doi: 10.1001/jama.2011.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosano K.K., Wegner D.J., Shinawi M., Baldridge D., Bucelli R.C., Dahiya S., et al. Biallelic ASCC1 variants including a novel intronic variant result in expanded phenotypic spectrum of spinal muscular atrophy with congenital bone fractures 2 (SMABF2) Am. J. Med. Genet. A. 2021;185:2190–2197. doi: 10.1002/ajmg.a.62219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shamseldin H.E., Kurdi W., Almusafri F., Alnemer M., Alkaff A., Babay Z., et al. Molecular autopsy in maternal-fetal medicine. Genet. Med. 2018;20:420–427. doi: 10.1038/gim.2017.111. [DOI] [PubMed] [Google Scholar]
- 15.Valverde R., Edwards L., Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 16.Grishin N.V. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siomi H., Choi M., Siomi M.C., Nussbaum R.L., Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 18.Siomi H., Matunis M.J., Michael W.M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remus B.S., Jacewicz A., Shuman S. Structure and mechanism of E. coli RNA 2',3'-cyclic phosphodiesterase. RNA. 2014;20:1697–1705. doi: 10.1261/rna.046797.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasr F., Filipowicz W. Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1",2"-cyclic phosphate. Nucleic Acids Res. 2000;28:1676–1683. doi: 10.1093/nar/28.8.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacolla A., Tainer J.A. Robust computational approaches to defining insights on the interface of DNA repair with replication and transcription in cancer. Methods Mol. Biol. 2022;2444:1–13. doi: 10.1007/978-1-0716-2063-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Zhao S., Zhao X., Cao L., Karnad A., Kumar A.P., et al. Gemcitabine resistance of pancreatic cancer cells is mediated by IGF1R dependent upregulation of CD44 expression and isoform switching. Cell Death Dis. 2022;13:682. doi: 10.1038/s41419-022-05103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berzero G., Di Stefano A.L., Ronchi S., Bielle F., Villa C., Guillerm E., et al. IDH-wildtype lower-grade diffuse gliomas: the importance of histological grade and molecular assessment for prognostic stratification. Neuro Oncol. 2021;23:955–966. doi: 10.1093/neuonc/noaa258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Gines C., Cerda-Nicolas M., Gil-Benso R., Pellin A., Lopez-Guerrero J.A., Callaghan R., et al. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme. Clin. Neuropathol. 2005;24:209–218. [PubMed] [Google Scholar]
- 29.Stichel D., Ebrahimi A., Reuss D., Schrimpf D., Ono T., Shirahata M., et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018;136:793–803. doi: 10.1007/s00401-018-1905-0. [DOI] [PubMed] [Google Scholar]
- 30.Jiang C.Y., Xu X., Jian B.L., Zhang X., Yue Z.X., Guo W., et al. Chromosome 10 abnormality predicts prognosis of neuroblastoma patients with bone marrow metastasis. Ital. J. Pediatr. 2021;47:134. doi: 10.1186/s13052-021-01085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong L.N., Chin L. Chromosome 10, frequently lost in human melanoma, encodes multiple tumor-suppressive functions. Cancer Res. 2014;74:1814–1821. doi: 10.1158/0008-5472.CAN-13-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu B., Nie X.H., Yin R., Ding P., Su Z.Z., Qiu S., et al. PGAM4 silencing inhibited glycolysis and chemoresistance to temozolomide in glioma cells. Cell Biol. Int. 2023;47:776–786. doi: 10.1002/cbin.11983. [DOI] [PubMed] [Google Scholar]
- 33.Wang W., Zheng H., Fan C., Li J., Shi J., Cai Z., et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polak P., Kim J., Braunstein L.Z., Karlic R., Haradhavala N.J., Tiao G., et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin D.S., Didonato M., Barondeau D.P., Hura G.L., Hitomi C., Berglund J.A., et al. Superoxide dismutase from the eukaryotic thermophile Alvinella pompejana: structures, stability, mechanism, and insights into amyotrophic lateral sclerosis. J. Mol. Biol. 2009;385:1534–1555. doi: 10.1016/j.jmb.2008.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longo M.A., Roy S., Chen Y., Tomaszowski K.H., Arvai A.S., Pepper J.T., et al. RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles. Nat. Commun. 2023;14:4445. doi: 10.1038/s41467-023-40096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosey C.A., Houl J.H., Katsonis P., Balapiti-Modarage L.P.F., Bommagani S., Arvai A., et al. Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors. Prog. Biophys. Mol. Biol. 2021;163:171–186. doi: 10.1016/j.pbiomolbio.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lees-Miller J.P., Cobban A., Katsonis P., Bacolla A., Tsutakawa S.E., Hammel M., et al. Uncovering DNA-PKcs ancient phylogeny, unique sequence motifs and insights for human disease. Prog. Biophys. Mol. Biol. 2021;163:87–108. doi: 10.1016/j.pbiomolbio.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsutakawa S.E., Bacolla A., Katsonis P., Bralic A., Hamdan S.M., Lichtarge O., et al. Decoding cancer variants of unknown significance for helicase-nuclease-RPA complexes Orchestrating DNA repair during transcription and replication. Front Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.791792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapovalov M.V., Dunbrack R.L., Jr. Statistical and conformational analysis of the electron density of protein side chains. Proteins. 2007;66:279–303. doi: 10.1002/prot.21150. [DOI] [PubMed] [Google Scholar]
- 42.Holm L. Dali server: structural unification of protein families. Nucleic Acids Res. 2022;50:W210–W215. doi: 10.1093/nar/gkac387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Kempen M., Kim S.S., Tumescheit C., Mirdita M., Lee J., Gilchrist C.L.M., et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2023;42:243–246. doi: 10.1038/s41587-023-01773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis H.A., Musunuru K., Jensen K.B., Edo C., Chen H., Darnell R.B., et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 45.Teplova M., Malinina L., Darnell J.C., Song J., Lu M., Abagyan R., et al. Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure. 2011;19:930–944. doi: 10.1016/j.str.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L., Wang C., Li F., Zhang J., Nayab A., Wu J., et al. The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity. J. Biol. Chem. 2017;292:16221–16234. doi: 10.1074/jbc.M117.797746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tubbs J.L., Pegg A.E., Tainer J.A. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thayer M.M., Ahern H., Xing D., Cunningham R.P., Tainer J.A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Batra V.K., Oertell K., Beard W.A., Kashemirov B.A., McKenna C.E., Goodman M.F., et al. Mapping functional substrate-enzyme interactions in the pol beta active site through chemical biology: structural responses to acidity modification of incoming dNTPs. Biochemistry. 2018;57:3934–3944. doi: 10.1021/acs.biochem.8b00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mol C.D., Hosfield D.J., Tainer J.A. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means. Mutat. Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 51.Gold M.G., Smith F.D., Scott J.D., Barford D. AKAP18 contains a phosphoesterase domain that binds AMP. J. Mol. Biol. 2008;375:1329–1343. doi: 10.1016/j.jmb.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myllykoski M., Kursula P. Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hobbs S.J., Wein T., Lu A., Morehouse B.R., Schnabel J., Leavitt A., et al. Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity. Nature. 2022;605:522–526. doi: 10.1038/s41586-022-04716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomura Y., Roston D., Montemayor E.J., Cui Q., Butcher S.E. Structural and mechanistic basis for preferential deadenylation of U6 snRNA by Usb1. Nucleic Acids Res. 2018;46:11488–11501. doi: 10.1093/nar/gky812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asthana A., Gaughan C., Dong B., Weiss S.R., Silverman R.H. Specificity and mechanism of coronavirus, rotavirus, and mammalian two-histidine phosphoesterases that antagonize antiviral innate immunity. mBio. 2021;12 doi: 10.1128/mBio.01781-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander L.T., Lepore R., Kryshtafovych A., Adamopoulos A., Alahuhta M., Arvin A.M., et al. Target highlights in CASP14: analysis of models by structure providers. Proteins. 2021;89:1647–1672. doi: 10.1002/prot.26247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oruganty K., Talathi N.S., Wood Z.A., Kannan N. Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:924–929. doi: 10.1073/pnas.1207104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rambo R.P., Tainer J.A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brosey C.A., Tainer J.A. Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr. Opin. Struct. Biol. 2019;58:197–213. doi: 10.1016/j.sbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsutakawa S.E., Yan C., Xu X., Weinacht C.P., Freudenthal B.D., Yang K., et al. Structurally distinct ubiquitin- and sumo-modified PCNA: implications for their distinct roles in the DNA damage response. Structure. 2015;23:724–733. doi: 10.1016/j.str.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsutakawa S.E., Van Wynsberghe A.W., Freudenthal B.D., Weinacht C.P., Gakhar L., Washington M.T., et al. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17672–17677. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Querol-Audi J., Yan C., Xu X., Tsutakawa S.E., Tsai M.S., Tainer J.A., et al. Repair complexes of FEN1 endonuclease, DNA, and Rad9-Hus1-Rad1 are distinguished from their PCNA counterparts by functionally important stability. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8528–8533. doi: 10.1073/pnas.1121116109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katsonis P., Lichtarge O. A formal perturbation equation between genotype and phenotype determines the evolutionary action of protein-coding variations on fitness. Genome Res. 2014;24:2050–2058. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stoneley M., Harvey R.F., Mulroney T.E., Mordue R., Jukes-Jones R., Cain K., et al. Unresolved stalled ribosome complexes restrict cell-cycle progression after genotoxic stress. Mol. Cell. 2022;82:1557–1572.e7. doi: 10.1016/j.molcel.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hashimoto S., Sugiyama T., Yamazaki R., Nobuta R., Inada T. Identification of a novel trigger complex that facilitates ribosome-associated quality control in mammalian cells. Sci. Rep. 2020;10:3422. doi: 10.1038/s41598-020-60241-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Best K., Ikeuchi K., Kater L., Best D., Musial J., Matsuo Y., et al. Structural basis for clearing of ribosome collisions by the RQT complex. Nat. Commun. 2023;14:921. doi: 10.1038/s41467-023-36230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brickner J.R., Soll J.M., Lombardi P.M., Vagbo C.B., Mudge M.C., Oyeniran C., et al. A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature. 2017;551:389–393. doi: 10.1038/nature24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dango S., Mosammaparast N., Sowa M.E., Xiong L.J., Wu F., Park K., et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liefke R., Windhof-Jaidhauser I.M., Gaedcke J., Salinas-Riester G., Wu F., Ghadimi M., et al. The oxidative demethylase ALKBH3 marks hyperactive gene promoters in human cancer cells. Genome Med. 2015;7:66. doi: 10.1186/s13073-015-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bacolla A., Sengupta S., Ye Z., Yang C., Mitra J., De-Paula R.B., et al. Heritable pattern of oxidized DNA base repair coincides with pre-targeting of repair complexes to open chromatin. Nucleic Acids Res. 2021;49:221–243. doi: 10.1093/nar/gkaa1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Narita M., Denk T., Matsuo Y., Sugiyama T., Kikuguchi C., Ito S., et al. A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation. Nat. Commun. 2022;13:6411. doi: 10.1038/s41467-022-34097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung D.J., Sung H.S., Goo Y.W., Lee H.M., Park O.K., Jung S.Y., et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol. Cell Biol. 2002;22:5203–5211. doi: 10.1128/MCB.22.14.5203-5211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia J., Hilal T., Bohnsack K.E., Chernev A., Tsao N., Bethmann J., et al. Extended DNA threading through a dual-engine motor module of the activating signal co-integrator 1 complex. Nat. Commun. 2023;14:1886. doi: 10.1038/s41467-023-37528-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2022 doi: 10.1101/2021.10.04.463034. [preprint] [DOI] [Google Scholar]
- 76.Zhang L., Duan H.C., Paduch M., Hu J., Zhang C., Mu Y., et al. The molecular basis of human ALKBH3 mediated RNA N(1) -methyladenosine (m(1) A) demethylation. Angew. Chem. Int. Ed. Engl. 2024;63 doi: 10.1002/anie.202313900. [DOI] [PubMed] [Google Scholar]
- 77.Cordin O., Hahn D., Alexander R., Gautam A., Saveanu C., Barrass J.D., et al. Brr2p carboxy-terminal Sec63 domain modulates Prp16 splicing RNA helicase. Nucleic Acids Res. 2014;42:13897–13910. doi: 10.1093/nar/gku1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jia J., Absmeier E., Holton N., Pietrzyk-Brzezinska A.J., Hackert P., Bohnsack K.E., et al. The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations. Nat. Commun. 2020;11:5535. doi: 10.1038/s41467-020-19221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voraberger B., Mayr J.A., Fratzl-Zelman N., Blouin S., Uday S., Kopajtich R., et al. Investigating the role of ASCC1 in the causation of bone fragility. Front Endocrinol. (Lausanne) 2023;14 doi: 10.3389/fendo.2023.1137573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharova M., Guseva D., Kurenkov A., Novoselova O., Murtazina A., Skoblov M. Congenital myopathy as a new phenotype caused by two undescribed variants in ASCC1 gene. Am. J. Med. Genet. A. 2022;188:3100–3105. doi: 10.1002/ajmg.a.62898. [DOI] [PubMed] [Google Scholar]
- 81.Nair D., Li D., Erdogan H., Yoon A., Harr M.H., Bergant G., et al. Discovery of a neuromuscular syndrome caused by biallelic variants in ASCC3. HGG Adv. 2021;2 doi: 10.1016/j.xhgg.2021.100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meunier J., Villar-Quiles R.N., Duband-Goulet I., Ferreiro A. Inherited defects of the ASC-1 complex in congenital neuromuscular diseases. Int. J. Mol. Sci. 2021;22:6039. doi: 10.3390/ijms22116039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chi B., O'Connell J.D., Iocolano A.D., Coady J.A., Yu Y., Gangopadhyay J., et al. The neurodegenerative diseases ALS and SMA are linked at the molecular level via the ASC-1 complex. Nucleic Acids Res. 2018;46:11939–11951. doi: 10.1093/nar/gky1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pratt A.J., Shin D.S., Merz G.E., Rambo R.P., Lancaster W.A., Dyer K.N., et al. Aggregation propensities of superoxide dismutase G93 hotspot mutants mirror ALS clinical phenotypes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4568–E4576. doi: 10.1073/pnas.1308531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu J., Yan C., Dodd T., Tsai C.L., Tainer J.A., Tsutakawa S.E., et al. Dynamic conformational switching underlies TFIIH function in transcription and DNA repair and impacts genetic diseases. Nat. Commun. 2023;14:2758. doi: 10.1038/s41467-023-38416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaillard H., Garcia-Muse T., Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 87.Bacolla A., Ye Z., Ahmed Z., Tainer J.A. Cancer mutational burden is shaped by G4 DNA, replication stress and mitochondrial dysfunction. Prog. Biophys. Mol. Biol. 2019;147:47–61. doi: 10.1016/j.pbiomolbio.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vagbo C.B., Svaasand E.K., Aas P.A., Krokan H.E. Methylation damage to RNA induced in vivo in Escherichia coli is repaired by endogenous AlkB as part of the adaptive response. DNA Repair (Amst) 2013;12:188–195. doi: 10.1016/j.dnarep.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 89.Thapar R., Bacolla A., Oyeniran C., Brickner J.R., Chinnam N.B., Mosammaparast N., et al. RNA modifications: reversal mechanisms and cancer. Biochemistry. 2019;58:312–329. doi: 10.1021/acs.biochem.8b00949. [DOI] [PubMed] [Google Scholar]
- 90.Auweter S.D., Oberstrass F.C., Allain F.H. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson W.F., Ohlendorf D.H., Takeda Y., Matthews B.W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981;290:754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- 92.Harrison S.C., Aggarwal A.K. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 93.Tubbs J.L., Latypov V., Kanugula S., Butt A., Melikishvili M., Kraehenbuehl R., et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daniels D.S., Woo T.T., Luu K.X., Noll D.M., Clarke N.D., Pegg A.E., et al. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 95.Tsutakawa S.E., Sarker A.H., Ng C., Arvai A.S., Shin D.S., Shih B., et al. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proc. Natl. Acad. Sci. U. S. A. 2020;117:14127–14138. doi: 10.1073/pnas.1921311117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G., et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yadav M., Singh R.S., Hogan D., Vidhyasagar V., Yang S., Chung I.Y.W., et al. The KH domain facilitates the substrate specificity and unwinding processivity of DDX43 helicase. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.015824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ernst J., Melnikov A., Zhang X., Wang L., Rogov P., Mikkelsen T.S., et al. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat. Biotechnol. 2016;34:1180–1190. doi: 10.1038/nbt.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reindl S., Ghosh A., Williams G.J., Lassak K., Neiner T., Henche A.L., et al. Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol. Cell. 2013;49:1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hambarde S., Tsai C.L., Pandita R.K., Bacolla A., Maitra A., Charaka V., et al. EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart. Mol. Cell. 2021;81:2989–3006.e9. doi: 10.1016/j.molcel.2021.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yan C., Dodd T., He Y., Tainer J.A., Tsutakawa S.E., Ivanov I. Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nat. Struct. Mol. Biol. 2019;26:397–406. doi: 10.1038/s41594-019-0220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Putnam C.D., Shroyer M.J., Lundquist A.J., Mol C.D., Arvai A.S., Mosbaugh D.W., et al. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 104.Zhao W., Vaithiyalingam S., San Filippo J., Maranon D.G., Jimenez-Sainz J., Fontenay G.V., et al. Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA mimicry. Mol. Cell. 2015;59:176–187. doi: 10.1016/j.molcel.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jung S., von Thulen T., Yang I., Laukemper V., Rupf B., Janga H., et al. A ribosomal RNA fragment with 2',3'-cyclic phosphate and GTP-binding activity acts as RIG-I ligand. Nucleic Acids Res. 2020;48:10397–10412. doi: 10.1093/nar/gkaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Syed A., Filandr F., Patterson-Fortin J., Bacolla A., Ravindranathan R., Zhou J., et al. Novobiocin blocks nucleic acid binding to Poltheta and inhibits stimulation of its ATPase activity. Nucleic Acids Res. 2023;51:9920–9937. doi: 10.1093/nar/gkad727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conway P., Tyka M.D., DiMaio F., Konerding D.E., Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khatib F., Cooper S., Tyka M.D., Xu K., Makedon I., Popovic Z., et al. Algorithm discovery by protein folding game players. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18949–18953. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nivon L.G., Moretti R., Baker D. A Pareto-optimal refinement method for protein design scaffolds. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tyka M.D., Keedy D.A., Andre I., Dimaio F., Song Y., Richardson D.C., et al. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 2011;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rosenberg D.J., Hura G.L., Hammel M. Size exclusion chromatography coupled small angle X-ray scattering with tandem multiangle light scattering at the SIBYLS beamline. Methods Enzymol. 2022;677:191–219. doi: 10.1016/bs.mie.2022.08.031. [DOI] [PubMed] [Google Scholar]
- 116.Dyer K.N., Hammel M., Rambo R.P., Tsutakawa S.E., Rodic I., Classen S., et al. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol. Biol. 2014;1091:245–258. doi: 10.1007/978-1-62703-691-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Classen S., Hura G.L., Holton J.M., Rambo R.P., Rodic I., McGuire P.J., et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. J. Appl. Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Classen S., Rodic I., Holton J., Hura G.L., Hammel M., Tainer J.A. Software for the high-throughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. J. Synchrotron Radiat. 2010;17:774–781. doi: 10.1107/S0909049510028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tully M.D., Tarbouriech N., Rambo R.P., Hutin S. Analysis of SEC-SAXS data via EFA deconvolution and Scatter. J. Vis. Exp. 2021 doi: 10.3791/61578. [DOI] [PubMed] [Google Scholar]
- 120.Hopkins J.B., Gillilan R.E., Skou S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 2017;50:1545–1553. doi: 10.1107/S1600576717011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pelikan M., Hura G.L., Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A. pompejanaASCC1 complete coding sequence is available at GenBank with record number OQ857929 (https://www.ncbi.nlm.nih.gov/nuccore/OQ857929). The TCGA RNA-Seq normalized rsem data, TCGA clinical data, and mutational signature files used for the analyses are available at https://doi.org/10.5281/zenodo.7885656 (‘tumortypernaSeq.tar’, ‘cliDat_tcga_18.tar’ and ‘signatureProfileSample.txt.zip’ files); source codes for TCGA analyses (tcgaAnalyses and sumbitMPI) are available at https://doi.org/10.5281/zenodo.7874703 and https://doi.org/10.5281/zenodo.7874772, respectively. Crystal structures have been deposited to the PDB under coordinates 8TUK and 8TLY for ApASCC1Δ40 and HsASCC1Δ133, respectively. SAXS data has been deposited in Simple Scatter Database (https://simplescattering.com/) (73) under XSWQ5ILA, XSA5YQYE, and XSVL1FQL for full-length HsASCC1, full-length ApASCC1, and ApASCC1Δ40, respectively.