Abstract
B cells can be activated by cognate antigen, anti-B-cell receptor antibody, complement receptors, or polyclonal stimulators like lipopolysaccharide; the overall result is a large shift in RNA processing to the secretory-specific form of immunoglobulin (Ig) heavy chain mRNA and an upregulation of Igh mRNA amounts. Associated with this shift is the large-scale induction of Ig protein synthesis and the unfolded protein response to accommodate the massive quantity of secretory Ig that results. Stimulation to secretion also produces major structural accommodations and stress, with extensive generation of endoplasmic reticulum and Golgi as part of the cellular architecture. Reactive oxygen species can lead to either activation or apoptosis based on context and the high or low oxygen tension surrounding the cells. Transcription elongation factor ELL2 plays an important role in the induction of Ig secretory mRNA production, the unfolded protein response, and gene expression during hypoxia. After antigen stimulation, activated B cells from either the marginal zones or follicles can produce short-lived antibody secreting cells; it is not clear whether cells from both locations can become long-lived plasma cells. Autophagy is necessary for plasma cell long-term survival through the elimination of some of the accumulated damage to the ER from producing so much protein. Survival signals from the bone marrow stromal cells also contribute to plasma cell longevity, with BCMA serving a potentially unique survival role. Integrating the various information pathways converging on the plasma cell is crucial to the development of their long-lived, productive immune response.
Keywords: immunoglobulin, antibody secreting cells, plasma cells, B-cell differentiation
I. INTRODUCTION
The clonal nature of antibody formation was clearly demonstrated in the early 1970s.1,2 Mature B lymphocytes develop in the bone marrow and migrate to lymph nodes or the white pulp of the spleen. These resting B cells express specific immunoglobulin receptors for antigen on their surfaces, i.e., the B-cell antigen receptor or BCR. It has been estimated that each B cell carries a low number, about 10, identical receptors (IgM and IgD) in association with the co-receptors CD79a and CD79b. Following stimulation with a number of different agents, B cells are activated to proliferate, differentiate into plasma cells, and secrete copious amounts of antibody molecules. These antibody secreting cells make immunoglobulins (Ig) that carry the specificity of the original responding B cell. A large amount of Ig in the gamma globulin fraction of serum is made by B and plasma cells, so much so that the amount approaches ~25% of the serum level of albumin, made by the ~three pound liver. There are both short-lived antibody secreting cells that only live for days and long-lived plasma cells, thought to be important contributors to the large amount of antibody that is made, reviewed in Ref. 3. Plasma cells are arrested in G1 of the cell cycle, held there by the cyclin dependent kinase inhibitor p18INK4c that acts on cdk4 and 6.4 Tumors of plasma cells like myeloma are able to bypass growth arrest and continue to secrete Ig while proliferating. The time course of mouse B-cell stimulation with naïve splenic B cells plus 10–50 ug/ml of LPS in vitro are summarized in the literature5–8 and here in Table 1. Approximately 25–50% of the naïve B cells are converted to B220loCD138+ cells in 72–86 h after LPS treatment.
TABLE 1:
Changes in naïve splenic B cells following LPS stimulation in vitro
| Igh mu |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hours > LPS | % S phase | 3H TdR incorp | J chain mRNA and protein | mRNA | % sec | Sec protein ng/ml* | Redox balance | ER resident proteins | Metabolism | Cyto and mito chaperones | CHOP mRNA | ELL2 mRNA |
| 0 | 1.2 | nd | 1× | 1 | <10 | – | 1 | 1 | 1 | 1 | 1× | 1× |
| 24 | 8.5 | 1× | 1× | 1× | <10 | 0.9 | 4× | 1.5× | 1.9X | 5× | 7× | 1× |
| 48 | 28.4 | 18× | 1× | 5× | 61 | – | 4× | 2.7× | 2.5× | 2.5× | 2× | 6× |
| 72 | 40 | 74× | 4× | 30× | >90 | 85 | 6× | 4× | 4× | 2.5× | 2× | 10× |
| 84–96 | 59 | 86× | 118× | 30× | >90 | 1100 | 11× | 6× | 3.7× | 2× | nd | nd |
| 120 | nd | 45× | 400× | 7300 | ||||||||
Supernatant from 4 × 106 cells/ml initial cell concentration. For references and further details see text.
Addition of other exogenous factors besides LPS can induce Ig secretion in vitro. These include CD40:CD40L engagement;9 CD40:CD40L+ IL10, which also induces isotype switching (10); CD27:CD70 interaction, which acts later than CD40 engagement, after proliferation, to promote IgE secretion;11,12 OX40 ligand cross-linking;13 addition of IL-5 (Igh mu production only) versus IL-5 plus IL-2, where Igh mu and J chain are both induced;14 addition of TNF-alpha to tonsular B cells within the first 24 h of culture;15 and IL-6 (BCDF) addition to human CESS or SKW B cells.16 IL-21 and Stat3 have emerged as potent inducers of terminal differentiation to antibody secretion in human cells.17 Exogenous inhibitors of Ig secretion include: LPS plus anti-Ig,18,19 LPS plus anti-Ig mu,20,21 and Interferon gamma.20
II. FACTORS LEADING TO ANTIBODY SECRETION
A. Early Drivers of Differentiation
Early on, opposing suites of transcription factors were shown to either maintain the B-cell program (e.g., Pax5, Bach2, Bcl6) or promote and facilitate differentiation to antibody secreting cells (e.g., IRF4, Blimp-1, XBP1).22 A number of excellent reviews on the action of IRF4, Blimp-1, and XBP1 have been published.17,22,23 Recently, several newly described pathways of inhibitors24,25 and activators26–28 of plasma cell differentiation have emerged. In Table 2, we list the genes described in the text that are involved in production of the various classes of B cells and long-lived plasma cells and briefly summarize their mode of action.
TABLE 2:
Factors important for antibody secretion and/or survival
| Expressed in or important for |
|||||
|---|---|---|---|---|---|
| Factor/gene | B cells | MZ cells | FO cells | LLPC | Comments |
| ABF1, GCN5 | + | Blocks further development if not shut off | |||
| ELL3 | + | On from stem cells stage, shut off after stimulation | |||
| EBF1 | + | + | + | Mediates choice of MZ vs FO fate | |
| TLRs, CD19/21 | + | Engagement actives cells | |||
| Bcl3 | + | Mediates choice of MZ or FO fate | |||
| CD40:CD40L | + | Interactions help activate cells | |||
| CHOP | + | + | + | Turns off Bcl2, turns on Atg5 | |
| Cytokines, BCR, NF-kB | + | + | + | Important for activation and survival | |
| IRF4, Blimp-1 | + | + | + | Help shut off Bcl6, pax5 | |
| ELL2 | + | + | + | Drives Igh secretory mRNA, UPR, BCMA, involved in hypoxia | |
| XBP1 | + | + | + | mRNA spliced by IRE1P to make transcription factor for UPR genes | |
| Zbtb20 | + | + | + | Promotes NF-kappaB activation | |
| CB2 stimulation | + | + | ? | Necessary for secretion, effects on survival not measured | |
| HDAC2 deletion | + | + | ? | Done with chicken DT40 cells, effects on survival not measured | |
| mTOR | + | + | ? | Necessary for secretion, effects on survival not measured | |
| Atg5 | + | Important for autophagy | |||
| BCMA | + | Induces MCL-1, an anti-apoptotic protein | |||
| CXCR4:CXCL12 | + | Important for retention in bone marrow | |||
| CD73 | + | + | Converts extracellular nucleosides to adenosine | ||
| hypoxia | ? | + | Promotes CD73 expression, modulates metabolism | ||
| Kruppel-like factor 2 | + | + | Control beta integrin expression | ||
MZ are marginal zone B cells, FO are follicular B cells, LLPC are long-lived plasma cells. See text for references and details.
Based on work with conditional knockout mice by a number of investigators, activation of IRF4 and Blimp appear very early in the differentiation of B to plasma cells. IRF4 is necessary for lymphocyte activation and immune responses in B and T cells;29 mice with a deletion in IRF4 in germinal center B cells are unable to differentiate into memory B cells and plasma cells.30 IRF4 is one of a few specific transcription factors necessary for myeloma cell survival, along with Blimp-1, ELL2, sub1, and CCNC/ cyclin C.31
High levels of IRF4 are required for the development of antibody secreting cells in a B-cell intrinsic fashion32 and to control the transcription of a number of plasma cell specific genes with an “interferon-like” signature.33 Low levels of IRF4 promote germinal center formation and class switch recombination.34 IRF4 can bind with high avidity to the 3′ enhancers of both kappa and lambda Ig light chains in conjunction with the ETS-family transcription factor PU.1 or the closely related factor Spi-B.35,36 IRF8 and IRF4 recognize the same DNA sequence and can both interact with PU.1. However, the outcome of these interactions is different; the complex of IRF8 and PU.1 (high in B cells) promotes Bcl6 and Pax5 gene expression while repressing Aicda and Blimp-1.24 Thus, the competition between IRF4 and IRF8 for PU.1 binding is in part responsible for controlling terminal B-cell differentiation.
Blimp-1 (aka PRDM1) is a negative transcription regulator that represses beta-interferon expression in other cells; in activated B cells, it is necessary to turn off Pax5, CIITA for MHC class II expression, c-myc,37 and Ets-1.38 Turning off c-myc stops cell proliferation; c-myc conditionally deficient B lymphocytes hypersecrete IgM and do not undergo Ig class switch recombination.39 Blimp-1 is thought to work by downregulating Pax5 expression, which then turns on XBP1, a transcription factor important in the upregulation of unfolded protein response (UPR) components.40 Ig secretion in a Blimp-1 knockout is low; this is thought to be indirectly through the downstream effects of Pax5,41 but may also involve the persistence of myc allowing cell cycle progression.
The micro-RNA, miR-155, is required for the initiation of plasma cell development from B cells because it normally inhibits PU.1 expression through a sequence in the 3’UTR of PU.1 mRNA; this then leads to Pax5 downregulation with the subsequent downstream events as in Blimp-1 mediated suppression of Pax5.28 Removing the target in PU.1 mRNA obviated the repression by miR-155. This study was important in that it also established that there is a network of cellular adhesion genes modulated by PU.1 controlling B- and T-cell interactions.
The AP-1 transcription factor Fra1 appears to inhibit Blimp-1 expression by binding to its promoter. When Fra1 is deleted in B cells, there are more plasma cells and an exacerbated antibody response; so Fra1 acts as a negative regulator of plasma cell differentiation.25
In mature plasma cells, the endoplasmic reticular (ER) response is unique from that seen in other cells.42 The unfolded protein response (UPR) in many cells typically has three arms, i.e. the IRE-1/ XBP1 pathway, an ATF6 pathway, and the PERK pathway.43 But PERK knockout mice secrete normal amounts of Ig, while PERK protein expression is not changed significantly between B and plasma cells.44,45 In addition, ATF6 (activating transcription factor 6) is not necessary for the development of antibody secreting cells; thus, when B cells are stimulated to secrete antibody, the primary pathway for ER remodeling appears to reside in the IRE1 to XBP1 pathway.46 Aggregation and then auto phosphorylation of IRE1 causes it to acquire the ability to specifically cleave and then splice XBP1 mRNA; the newly spliced XBP1 RNA species encodes a novel XBP1 protein with transcriptional activity on its own promoter and other UPR promoters containing the element UPRE.44 In an XBP1 conditional deletion, the mice show defects in plasma cell development47 and low levels of secretory Ig.48 But it has been argued that the consequences of XBP1 deletion alone are relatively mild.49 Antibody secreting cells are present in normal frequencies in resting and immunized animals and Ig secretion is reduced but not eliminated in conditional XBP1 knockouts. Thus, the gene regulatory program controlling plasma cell differentiation may proceed relatively normally in the absence of XBP1.49
On further analysis, the low levels of Igh mRNA in XBP1−/− mice result from the eightfold increased levels of IRE1-P over control; the highly abundant IRE1-P cleaves the Igh mu secretory mRNA.50 This is a process similar to the previously described pathway51 in which IRE1-P can act to cleave its own mRNA, as well as other RNAs in a process called RIDD.52 Only XBP1 mRNA is spliced, not cleaved, by IRE1-P to form a new functional RNA.53 A double deletion of XBP1 and IRE1 restores IgM secretion by inhibiting Ig mRNA degradation.50 Mutations in the IRE1 nuclease function cause only a twofold reduction in Ig secretion.54 Taken together, this leads to a conclusion that some Ig secretion can occur without the unusual cleavage and splicing of XBP1 and there may be other proteins that allow for the upregulation of the UPR besides the spliced mRNA encoded XBP1. As we discuss below, ELL2, a transcription elongation factor, has a role in enhancing the transcription of other UPR proteins through the UPR element26 thereby linking production of the Igh secretory mRNA and the buildup of the UPR. Activation of the mTOR pathway can also bypass XBP1 for Ig secretion.27
B. Production of Igh Secretory mRNA and Transcription Elongation
Antibody secreting cells differentiate from B cells, in which the Ig molecules are predominantly on the surface of the cell as antigen receptor, into antibody secreting factories. The shift in protein expression is reflected in the level of the production of the two Ig heavy chain mRNA isoforms, encoding either the secretory-specific or the membrane-specific forms; these are derived by alternative processing of the primary transcript of the immunoglobulin heavy chain (Igh) gene. The regulation of alternative Igh RNA processing has served as a model system for revealing the competition between cis and trans-acting RNA factors influencing splicing and polyadenylation, reviewed in Refs. 55 and 56.
ELL3, a shorter transcription elongation factor and homolog of ELL2, is expressed at a higher level in B cells and is reduced when B cells begin to secrete Ig. After IRF4 and Blimp-1 induction, there is a rise in the level of the transcription elongation factor ELL2 (eleven-nineteen lysine rich leukemia gene 2). ELL2 binds to the RNAP-II on the Igh gene, impels high levels of Igh mRNA production, and drives alternative processing at the promoter proximal, secretory-specific poly(A) site {sec poly (A)}.57 It does this by enhancing modifications to RNA polymerase II, activating histone modifications, and subsequent downstream events.58 The Igh sec poly(A) site, essentially hidden in B cells, is found by a complex of elongation factors including ELL2 and polyadenylation factors in plasma cells. Thus, elongation enhances the production of the shorter Igh secretory mRNA because the poly(A) site comes first in the transcript. This leads to a massive increase in efficient mRNA processing from only a twofold increase in the number of polymerase passes over the gene.59 Both the 5’ splice site in the last sec exon and the sec poly(A) sites in Igh are weak and in competition;60 but splicing is decreased in plasma cells.61 A shift in the relative distribution of serine/arginine-rich proteins is seen from the activating type (ASF/SF2) in B cells to the repression type (SRp20) in plasma cells.61 The factors hnRNP F and snRNP U1A protein, both of which block the secretory-specific poly(A) site from functioning, are reduced in plasma cells,62–65 thereby allowing the Igh secretory poly(A) site to function better. Numerous genes contain multiple poly(A) sites;66 many are subject to regulation;67,68 Blimp1 is one such gene69 although the functional significance of the different isoforms is not clear. It has been estimated that up to 20% of the human genes may be arranged with a competition of splicing and polyadenylation sites as seen in the Igh genes.70,71 The advancing developmental stage is generally correlated with use of promoter distal poly(A) sites in those multi-poly(A) site genes.72,73 Yet, the opposite situation applies as the B cell terminally differentiates into plasma cells; thus, the changes seen in the expression of elongation factors may hold the key to understanding this poly(A) site choice paradox. The amounts of the polyadenylation factor CstF-64 also increase following B-cell stimulation74 as cells go from Go to an actively growing state75 that can stimulate overall polyadenylation.
Transfection of B cells with either Blimp-1 or IRF4 cDNA caused elevated expression of the mRNA for transcription elongation factor ELL2.31,34,41 But in luciferase constructs driven by the ELL2 promoter, IRF4 was most efficient at driving its transcription, along with NF-kB p65.26 Studies of the ELL2 promoter (–1142 to +154) show the presence of several putative NF-kB binding sites and cyclic AMP response elements; the promoter responds to these and to an HTLV-1 viral oncoprotein, Tax, in T cells.76 The effect of Blimp-1 on ELL2 transcription may be more indirect.
The addition to B cells of exogenous cDNA for ELL2 stimulates alternative RNA processing resulting in the use of the Igh secretory-specific poly(A) site and the skipping of weak alternative exons in Igh and test substrates.57 ELL2 drives the association of positive transcription elongation factor b (pTEFb) to RNAP-II, thereby increasing phosphorylation of ser-2 on the carboxyl-termini of the elongating polymerase near the start of the Igh locus.57–59 The binding of ELL2 with the highly phosphorylated carboxyl-terminal domain of RNA polymerase II, polyadenylation factors, Dot1L (the histone H3K79 methylase), and the histone H3 K79me3 modifications are seminal to the changes in RNA processing seen at the Igh locus.
Using deep mRNA sequencing, the knockdown of ELL2 by siRNA in a plasma cell line was shown to influence several other genes besides Igh secretory specific mRNA processing, namely, several splicing factors, cyclin B2 (Ccnb2), and the B-cell maturation antigen (Tnfrsf17) aka BCMA.77 Long-term survival of plasma cells is impaired by the lack of BCMA in a knockout mouse.78 But loss of BCMA alone in −/− mice does not alter humoral responses (T-independent or T-dependent) nor the formation of short-lived plasma cells,79 yet loss of ELL2 in mice does.26
The B-cell-specific ELL2 conditional knockout mice (ell2loxp/loxp CD19cre/+) exhibit curtailed humoral responses both in NP-ficol and NP-KLH immunized animals; recall responses were also diminished.26 The number of immature and recirculating B cells in the bone marrow was increased in the conditional knockouts while plasma cells in spleen and bone marrow are reduced relative to control animals. Production of LPS ex vivo stimulated B220loCD138+ cells from ELL2 deficient mouse spleens is fourfold less than from control splenic B cells. The resulting cells have a paucity of secreted Igh, and distended, abnormal appearing ER. IRE1-alpha is efficiently phosphorylated but the amounts of Ig kappa, ATF6, BiP, Cyclin B2, OcaB (BOB1, Pou2af1), and XBP1 mRNAs, unspliced and spliced, are severely reduced in ELL2 deficient cells. Transcription from the cyclin B2 and the canonical UPR promoter elements in luciferase reporters are upregulated by ELL2 cDNA. Thus, ELL2 has direct effects on those genes. BCMA expression is curtailed in the knockouts by as yet undefined mechanisms.
C. Other Genetic Factors for Secretion
Bright, for B-cell regulator of Ig heavy chain transcription, binds to the A:T rich sequences in the Igh enhancer region and the 5’ flanks of some Vh promoters; Bright is induced after B-cell stimulation80 to enhance Igh transcription. Constitutive Bright expression skews B cells toward marginal zone cells at the expense of follicular cells81 while deletion of Bright causes embryonic lethality and loss of B lineage development.82
Recently ABF-1, aka activated B-cell factor or musculin, was found to suppress plasma cell differentiation.83 ABF-1 was previously shown to bind to the E-box element in DNA, form a homo- or heterodimer with E12 or E47, and repress E47 transcriptional activity.84 While plasma cell differentiation was suppressed by overexpression of ABF-1, memory and germinal center cell formation was enhanced; Blimp-1 downregulates ABF-1 expression.83 Thus, the E-box in the Igh enhancer and modulation of Igh transcription may play a role in the germinal center, memory versus plasma cell fate decision. Interestingly, Myc has also been hypothesized to act through binding to E-box elements,85 which represent a crucial point of regulation.
In contrast to ABF-1, ectopic Zbtb20 expression in primary B cells facilitates terminal B-cell differentiation to antibody secreting cells. In plasma cell lines, Zbtb20 induces cell survival but blocks cell cycle progression. Zbtb20 induction depends directly on IRF4 binding to the Zbtb20 promoter; but not on Blimp-1 expression.86 Depending on the adjuvant used for immunization with an antigen, Zbtb20 deficient cells responded differently. Adjuvants that activate TLR2 and TRL4 restored antibody production through induction of other survival programs,87 leading to the conclusion that plasma cell lifetimes are driven by the primary activation conditions. Zbtb20 represses IkB alpha after TLR activation, suppressing a suppressor to promote NF-kB activation.88
In chicken DT40 B cells, a knockout of histone deacetylase 2 (HDAC2), but not histone deactylase −1, caused the B cells to begin to secrete Ig mu, changing transcription and mRNA levels.89 Histone deacetylase-2 does this by controlling the expression of the Igh and light chain genes through EBF1, Pax5, and Aiolos downregulation and by E2A upregulation of their expression.90 In chicken DT40 B cells, deficiency of GCN5, a ubiquitous histone acetyl transferase that normally promotes transcriptional activation, caused a decrease in IgM surface and secreted protein expression; light chain was unaffected. By chromatin immuno-precipitation, GCN5 was bound to the Igh mu gene but not the light chain gene.91 Thus, the acetylation state of the histones also regulates Igh expression.
There are two cannabinoid receptors, i.e., CB1 and CB2; while CB1 is expressed in the nervous system, CB2 is expressed predominantly in B and T lymphocytes and in monocytes with B cells having the highest levels of the receptor. Cannabinoids directly induce B-cell class switching from IgM to IgE through a mechanism involving CB2 receptors, thereby biasing toward Th2-type immunity.92 Initially, both delta9-tetrahydrocannabinol (THC) and anandamide (an endogenous cannabinoid) were shown to be immunosuppressive in primary and secondary antibody formation in vitro via their dose-dependent action on the CB2 receptor.93 But the opposite conclusion was reached using a human B-cell line that required IL-6 to induce Ig secretion; antagonism of the CB2 pathway suppressed IgM secretion implying that activation of CB2 stimulates secretion.94 With the recent interest in the medical uses of marijuana in myeloma therapy, this topic should be explored further.
D. Expansion of the Endoplasmic Reticulum and Chaperones
It has been assumed that expansion of the endoplasmic reticulum and chaperones in the unfolded protein response are independent of the shift to Igh sec mRNA production but recent data indicate there is a tighter linkage in their control than was previously thought. When a B-cell line (L29mu+) was stimulated to convert to the plasma cell phenotype, it did so in a multistep process; proteomic analyses showed that the metabolic capacity and secretory machinery were put into place prior to the mass production of Ig that normally follows (Table 1 and Ref. 8). When B cells deficient in secretory-specific mu protein (AID−/− mus−/− or mus−/−) were stimulated with mitogens, they showed reduced ability to differentiate into B220loCD138+ antibody secreting cells and reduced survival.95 But the absence of secretory Ig in the knockouts did not prevent XBP1 accumulation, or XBP1 splicing, which was then assumed to precede upregulation of secreted Ig.44,95 However, in the ELL2 conditional knockouts, overall XBP1 production and its cytoplasmic splicing by IRE1-P, as well as ATF6, BiP, and cyclin B2, are reduced along with Igh. ELL2 may influence a potential coordination of regulation in Ig secretory protein production and the secretory apparatus itself.
The mechanistic protein target of rapamycin (aka mTOR) acts as a serine-threonine kinase on several proteins including: ribosomal protein S6, Akt, and 4EBP1, a binding protein for eukaryotic translation initiation factor eIF4E; dissociating eIF4E from the binding protein on phosphorylation activates mRNA translation. When mTOR was either knocked out in B cells or expressed at reduced levels by a hypomorphic knockin mutation, the mice showed decreased germinal center formation, lack of both somatic hypermutation and heavy chain class switching, and reduced ability to develop high-affinity antibodies for specific antigens.96 Germinal center functions are therefore subject to regulation via mTOR.
The kinase mTOR is negatively regulated by the tuberous sclerosis complex (TSC) and deletion of it makes mTOR hyperactive. When mice were conditionally deleted for XBP1 and the tuberous sclerosis complex there was enhanced Ig synthesis and differentiation into plasma cells by the double knockouts.27
The typical abnormalities in the ER seen in XBP1 and ELL2 knockout CD138+ plasma cells were gone in the XBP1 and tuberous sclerosis complex double knockouts. Thus, hyperactive mTOR may compensate for loss of XBP1.27
The balance between Igh and light chain protein synthesis must be maintained as well. The treatment of plasma cells with an siRNA to lambda light chain caused an imbalance of H and L amounts, reduction in Ig secretion, and induction of effector caspases leading to cell death.97 Thus, if the unfolded protein response is not fully balanced, or perhaps too vigorously induced, antibody secreting cells may die.
III. FACTORS IN LONGEVITY OF ANTIBODY SECRETING CELLS
The intrinsic survival capacity of short-lived antibody secreting cells from the spleen versus those found in the bone marrow was found to be similar in vitro in a supportive stromal environment; this suggests that extrinsic factors in the environment of the spleen versus bone marrow may have the greatest contribution to their differential survival.98 But long-lived plasma cells have intrinsic CXCR4, which allows them to interact with CXCL12 in bone marrow stroma, retaining them there. However, manipulating CXCL12 by addition or neutralization had minimal effects on the survival of the antibody secreting cells in vitro;98 thus, other factors also must play a role. Kruppel-like factor 2 controls homeostasis of B cells and homing of plasma cells to the bone marrow presumably through its action on beta-integrin expression.99 Soluble survival factors like BAFF and APRIL (tumor-necrosis factor ligands) in the bone marrow help antibody secreting cells so they do not undergo apoptosis and seem most effective in the bone marrow.100 Most striking is that most of the long-lived plasma cells appear to come from the follicular B cells and undergo a series of steps to arrive in the bone marrow where the environment is hypoxic. This leads to the question—are Ig secretion, location, and hypoxia linked in long-term plasma cell survival?
A. The B-Cell Location and History Influence Longevity
1. Marginal Zone Cell Responses
B cells mature in the bone marrow and travel to specific regions of the lymph node or white pulp of the spleen. Specific surface markers are associated with the B cells located in different regions of the spleen or lymph node, which may alter their functioning,23 for example, the marginal zone (MZ), B cells are CDd1+, CD9+, and CD21+, versus those in the lymph node or splenic follicles FO. CD1 is involved in presentation of lipids to T cells. CD9 can modulate cell adhesion, while CD21, in association with CD19, is the C3d complement receptor, which can co-activate B cells when engaged along with antigen: Ig complexes. In the marginal zone, there are also dendritic cells and macrophages to produce cytokines and cell surface ligands. B cells entering the MZ tend to react to T-independent, bacterial cell wall antigens, produce primarily IgM, and display a lower activation threshold than their follicular B-cell counterparts, most likely through the co-receptor engagements. This leads to a heightened propensity for cell differentiation that may contribute to an accelerated primary antibody response.101,102 If activated plasmablasts arise and remain in the marginal zone of the lymph node, there is a high probability that they will produce short-lived plasma cells, which will undergo apoptosis in a brief period of time (days) following stimulation. This may be because they either self-destruct through failure to remove an overload of internally derived reactive oxygen species (ROS) or because they lack receptors for external survival signals, or both. The apoptosis of most short-lived plasma cells is effector caspase-3 and −9 independent. Cell death occurs in a poorly understood mechanism that is triggered by excessive protein production; meanwhile, caspase-12 links ER stress and apoptosis of plasma cells.103
2. Follicular B-Cell Responses
Follicular B cells (FOs) express lower levels of CD21 (complement receptor 2) than do MZ B cells and can be divided into two types, i.e., type I and type II.104 Follicular type I B cells express high IgD and low IgM. The type II follicular B cells express high levels of surface IgM, IgD, and CD23a; their unique responses to calcium require the Kruppel-like factor 2 gene to distinguish them from marginal zone B cells.99 CD23a is the low-affinity receptor for the Fc portion of IgE; it is not clear how this plays into the reactivity of these cells but it may facilitate antigen presentation to B cells by CD11c+ cells. Follicular B cells require CD40-CD40L dependent T-cell help to promote effective primary immune responses, to undergo antibody isotype switching, and to establish high-affinity B-cell memory105 as they pass through the germinal center. Once activated, FOs undergo class switching and acquire homing signals like CXCR4; they may then travel to specific niches in the bone marrow where CXCL12 expressing stromal cells and soluble factors provide for their survival (months to years) as long-lived plasma cells or memory cells.106
The relationship of B-cell memory and long-lived plasma cells (LLPC in Table 2) is still a matter of active investigation. It is clear the initial interactions at the cell surface and in the germinal center are key.107 Expression of CD73, which catalyzes the conversion of extracellular nucleosides to adenosine, is high in mature germinal center cells and influences the establishment of the long-lived plasma cell compartment.108 Memory B cells are long lived and retain the ability to self-renew and can be divided into at least two varieties. CD80+ PD-L2+ memory cells differentiate into antibody secreting cells rapidly without new germinal center formation while CD80-PD-L2 memory cells robustly seed the germinal centers and produce antibody secreting cells more slowly.109 Both CD80 and PD-L2 are members of the B7 family of surface molecules and can be co-inhibitory or co-stimulatory. Viral particles can also drive memory B cells to differentiate into secondary plasma cells,110 presumably through interactions with surface receptors.
T:B interactions are important not only for activating plasma cells, but also for modulating their activities. CD28, B7.1, and B7.2 deficient mice each produce higher levels of antibody than do wild-type mice. They do this by increased production of both long- and short-lived plasma cells, more antibodies per cell, higher levels of survival factors, and lower levels of the apoptosis inducer caspase-12.111
The widely studied lipopolysaccharide (LPS) activation of splenic B cells ex vivo through TLR4 may most closely resemble the MZ plasma-blast reactions while B-cell receptor engagement by anti-IgM may mimic follicular cell responses, based on surface markers engaged. It is interesting that in murine lupus models, the anti-DNA antibodies arise from short-lived plasma blasts (TLR activation) while the anti-RNA and anti-cardiolipin antibodies arise from long-lived plasma cells,112,113 so in this case the triggering antigens and the longevity of the antibody secreting cells differ based on their anatomical location and the nature of receptors engaged.
3. Deciding Where to Go and What to Do
A few transcription factors have recently been implicated in directing the MZ versus FO fate. Overexpression of the Bright transcription factor for Igh drives cells to the marginal zones versus the follicles,81 so the level of surface Igh may play a role in where to go. Bcl-3 functions as a nuclear transcription cofactor and may promote or repress expression of some genes. Loss of Bcl-3 in B cells increased the marginal zone pool while decreasing follicular cells; both types were more responsive to LPS but more prone to apoptosis following B-cell receptor stimulation.114 Early B-cell factor 1 (EBF1) binds DNA and functions as a tissue-specific regulator of chromatin structure at B-cell-specific genes.115 While EBF1 is important in early B-cell development, it is also responsible for enhancing the production and maintenance of marginal zone, B-1 B, follicular, and germinal center B cells.116 EBF1 deficiency blocks signaling via the BAFF receptor and B-cell receptor dependent Akt pathways; the deficiency can be rescued by c-myb or Bcl-xL overexpression.117 It is also a previously unrecognized target of somatic hyper-mutation in lymphomas.118
B. The Role of Reactive Oxygen Species (ROS)
As mature B cells transition to activated antibody secreting cells, they are signaled through a variety of ligands and corresponding receptors that may differ based on their expression in various locations, MZ versus FO, for example. But all antibody secreting cells undergo rapid and extensive enlargement of their endoplasmic reticulum in order to accommodate increased immunoglobulin production and secretion, the hallmarks of the antibody secreting cell. The increased production of immunoglobulin results in the accumulation of reactive oxygen species such as H2O2. Several redundant antioxidant pathways are activated in response to the H2O2; these pathways may be necessary to the process of plasma cell differentiation, immunoglobulin secretion, and long-term survival.119 But the cellular sources of the ROS and the functional role of antioxidants (internal and external) in the response are not clear. It is also not well understood how ROS-mediated induction of endoplasmic reticular stress contributes to longevity since there are both positive and negative effects of reactive oxygen species on NF-kB expression.120
Mitochondrial metabolism and misfolded protein formation in cells secreting large amounts of protein can induce stress and block cell survival. Mitochondria make superoxide, aka hyperoxide or O2–, as a result of the activation of the electron transport chain making ATP; superoxide is converted to H2O2 and further converted to H2O and O by superoxide dismutases, catalases, or peroxidases. Small molecules such as vitamin C, uric acid, glutathione, and polyphenol antioxidants scavenge free radicals to prevent damage from these reactive oxygen species. While their levels are key to survival, they have not been well studied in plasma cell differentiation. Damage to mitochondria by H2O2 can cause apoptosis. Bcl-2 proteins are layered on the surface of mitochondria and activate Bax, which punches holes in mitochondrial membranes causing cytochrome C to leak out; Bcl2 binds to Apaf-1 to form apoptosomes. Meanwhile, Bcl6 suppresses Bcl2 via Miz1 and this can lead to apopotosis in lymphoma cells.121
In cell membranes, NADPH conversion to NADP+ also generates H2O2; NADPH is also found in mitochondria and the endoplasmic reticulum. Lipoxygenase action also stimulates H2O2 production.122 In large quantities, reactive oxygen species like H2O2, superoxide, NADPH oxidase, and disulfide related oxidative species (collectively ROS) can cause organelle and DNA damage leading to apoptosis. In smaller amounts. they are utilized by the cell for signaling and survival.123 It is interesting to note that autophagy, most likely in response to the need to remove cells damaged by ROS, is necessary for sustainable immunoglobulin production in plasma cells.124
Many disulfide bonds in Igs are formed when massive amounts of protein are made. But B-cell differentiation per se does not cause massive thioldisulfide stress to the cells. The steady state levels of protein-glutathione mixed substrates are maintained at very low levels, even in fully differentiated cells, and the overall protein redox state is not affected until late in differentiation, when large-scale IgM production has started and the ER stress response has been activated.125
The B-cell receptor (BCR), CD40 and CXCR4 signaling pathways in B cells require reactive oxygen species to activate their downstream targets like JNK, p38, and Akt. High levels of ROS might contribute to excessive B-cell activation or to malignancy123 versus cell death, which one might have expected. Thus, the role of ROS in differentiating short-versus long-term antibody secreting cells is still enigmatic.
C. Signaling through the BCR
At first glance, signal transduction through the surface Ig serving as antigen receptor should be different for MZ IgM+ plasmablasts and long-lived plasma cells since the bone marrow antibody secreting cells have typically undergone isotype switching and have down-modulated the expression of the alpha (CD79a) co-receptor.126 The cytoplasmic tail of IgM is much shorter than that of IgG and is less able to interact with signaling molecules without the benefit of the CD79a and CD79b co-receptors. Restoring CD79a and b expression to plasma cells increases surface expression of the BCR by an order of magnitude while still allowing them to secrete copious quantities of Ig.127 In addition, FO derived bone marrow plasma cells have higher affinity BCRs than MZ cells, having passed through the germinal centers and experienced affinity maturation, so the balance between affinity of the surface receptors versus amounts is different. Thus, long-lived plasma cells may not be stimulated until recall antigen rises to a high level or the epitopes are very specific, leading to the cell’s inherent quiescence and thus survival.
D. Survival Factors
B-cell activating factor (BAFF, TNFRSF13B) is produced by monocytes, dendritic cells, and bone marrow stromal cells. BAFF is first expressed as a membrane-bound protein, which is cleaved to generate a soluble factor. It is the natural ligand of three tumor necrosis factor receptors named BAFF-R (BR3), TACI (transmembrane activator and calcium modulator and cyclophilin ligand interactor), and BCMA (B-cell maturation antigen, TNFRSF17), all of which have differing binding affinities for BAFF.78 These receptors are expressed mainly on mature B lymphocytes, increase as cells mature, with BCMA expression on plasma cells most important for long-term survival.78 TACI has higher affinity for a proliferation-inducing ligand (APRIL) than it does for BAFF.100 BCMA displays an intermediate binding phenotype and will bind either BAFF or APRI, but signaling through BAFF-R and BCMA stimulates B lymphocytes to undergo proliferation and to counter apoptosis.128
Mice with deficiencies in BAFF, APRIL, BAFFR, and TACI all have multiple deficiencies in B-cell maturation and development while the BCMA−/− mice show impaired survival only of long-lived bone marrow plasma cells,128 indicating its unique function for that compartment. BCMA induces high levels of Mcl-1 in bone marrow but not spleen. Mcl-1 is an anti-apoptotic protein; in mice where it is deficient, the plasma cell compartment is depleted.129 ELL2 expression links secretory Igh mRNA production and BCMA expression in CD138+B220 low cells.26,77
E. Autophagy in Maintaining Survival
The B-cell conditional knockout mouse of Atg5, autophagy related gene 5, had plasma cells with more endoplasmic reticulum, higher expression of Blimp-1, and more Ig secretion; this was associated with less intracellular ATP and fewer antigen specific long-lived bone marrow plasma cells.124 The defects occur post germinal center; therefore, autophagy must play a role in maintaining plasma cell homeostasis. There is a balance between Ig synthesis with secretion and long-lived plasma cell viability. Secrete a lot and die, or control the secretion and reduce stress and live. The ER is the specific target of auto-phagosomal degradation (reticulophagy) by autophagy related gene 5 while mitochondria and ribosomes are spared. Autophagy related gene 5 transgenic mice are more tolerant to oxidative damage and cell death induced by oxidative stress, and this tolerance was reversible by treatment with an autophagy inhibitor.130 In contrast to the experiments with culturing of different populations of antibody secreting cells with stromal cells, the autophagy results argue for cell intrinsic regulation of viability.
F. CHOP
CHOP (C/EBP homologous protein aka DNA damage inducible gene 3) is downstream of PERK in the ER stress pathway; as shown in Table 1, it is induced early after B-cell activation in vitro and then declines.42 PERK−/− mice can secrete Ig. CHOP, BiP, and XBP1 share an ER stress response element in their promoters.43 CHOP causes Bcl2 inhibition (thus inhibiting growth), activation of beclin1 (involved in autophagy), and upregulation of autophagy related gene 5.131 It is not clear if all of these functions of CHOP are knocked out in the PERK−/− mice. CHOP deficient animals have plasma cells that secrete Ig at a lower rate than wild type, but antibody secreting cell differentiation and lifespan appear normal in the CHOP−/− knockout mice. This appears paradoxical since CHOP seems required for expression of genes like autophagy related gene 5 for plasma cell viability.132 As a counter to CHOP, Bcl-xL protects the plasma cells from CHOP-dependent apoptosis, connecting the UPR and cell death.133 In summary, the interactions of the balance of the ER stress pathways and autophagy mediated by CHOP to control ROS damage to the ER are emerging as noteworthy but still unresolved pathways for understanding the survival of long-lived plasma cells.
G. Hypoxia
1. The Bone Marrow Environment
Following stimulation and germinal center passage, long-lived plasma cells reside in bone marrow niches, which are believed to be hypoxic.134,135 Hypoxia may be a key contributor to long-lived plasma cell survival through degradation of c-myc, a key cell cycle regulator,134 to balancing ROS production and autophagy,135 and manipulating the bio-energetic profile of the cells. In many tissues HIF-1, hypoxia inducible factor, suppresses mitochondrial function in tumor cells, induces glucose transporters and hexokinase, and may modulate glycolytic flux.136 The switch between glycolysis and oxidative phosphorylation is controlled by the relative activities of two enzymes, i.e., pyruvate dehydrogenase and lactate dehydrogenase. More ATP is produced through oxidative phosphorylation in mitochondria than through anaerobic glycolysis in the Krebs cycle, which dominates when oxygen is limited.
In addition, CD73 has an important and previously unrecognized role in modulating the establishment of the long-lived plasma cell compartment through securing more adenosine for ATP.108 Although multiple cell types express CD73 and adenosine receptors, CD73 is markedly upregulated on both germinal center B and T cells. Hypoxic environments and increased extracellular ATP define the germinal center compartment as well. HIF-1alpha expression is seen in germinal center B cells and promotes CD73 expression.
2. Lessons from Multiple Myeloma
Multiple myeloma is a tumor of plasma cells that is responsible for almost 10% of hematological malignancies and is characterized by bone lesions and excess Ig secretion. Concentrations of Ig in the serum in patients can approach that of serum albumin.137 Most myelomas have a translocation of the Igh enhancer into genes like myc, ras, maf, cyclin D, etc., to activate expression of those as oncogenes138 while still secreting Ig. Thus, a cessation of growth is not a requirement for Ig secretion. Extrinsic cytokines made by the bone marrow stroma that aid in myeloma survival are IL-6, IL-10, IL-11, ciliary neurotropic factor, and oncostatin M leukemia inhibitory factor.139 IL-6 has been shown to be important for long-term survival of tonsular plasma cells expressing CD27 and CD38 in organ culture even from non-myeloma patients.140 Too much IL-6 made by the myeloma cells can activate osteoclasts, erode the bone, and cause the characteristic lesions seen on X-ray; in a mouse model, this is driven by c-myc or Bcl-XL.141
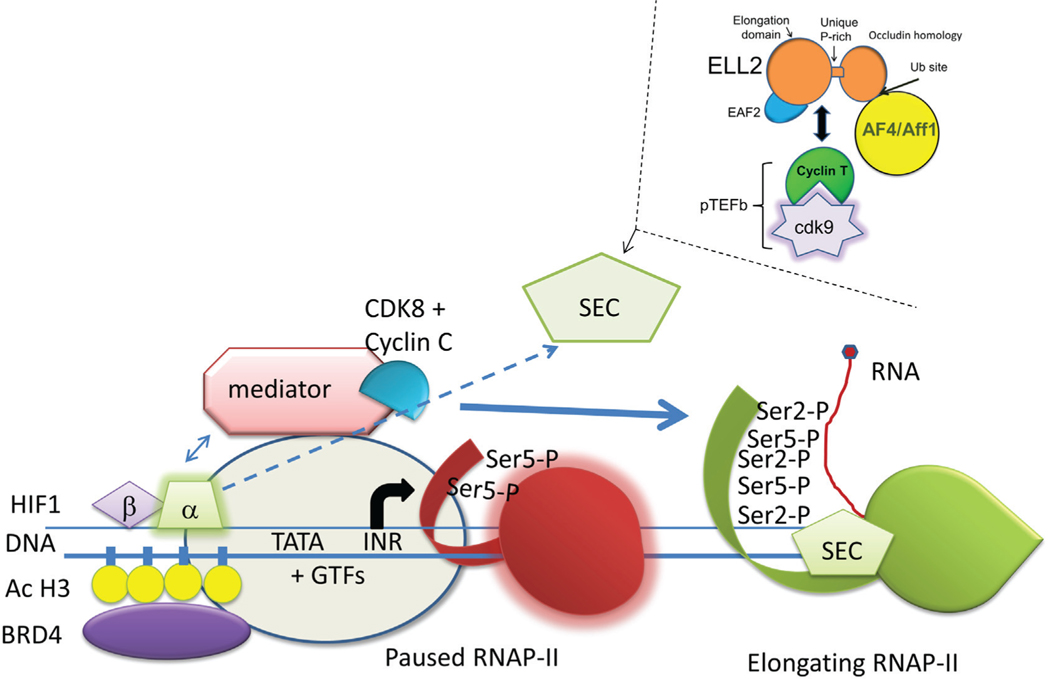
Intrinsic survival factors for myeloma cells include IRF4 and a limited set of transcription factors, namely, sub1, ELL2, CCNC, and PRDM1 (aka Blimp-1).31 BCMA and caspase-3 expression in myeloma cells are also strong survival factors. Many genes active in multiple myeloma undergo alternative splicing or polyadenylation patterns.31 As part of their survival strategy, these plasma cell tumors reside in hypoxic niches, which may present a unique target for anti-myeloma therapy.142 CCNC or cyclin C, in both myeloma and long-lived plasma cells, interacts with CDK8 in the mediator complex to phosphorylate the carboxyl terminus of RNAP-II. CCNC is important not only for myeloma survival, but also for the expression of Hif1 alpha and beta regulated genes. In hypoxia, Hif1 and CCNC act, along with the super elongation complex, composed of pTEFb and ELL2, to accelerate transcription of a subset of hypoxia inducible genes in many cell types.143 This is illustrated in Fig. 1. Suppression of Hif1 blocks tumor growth and stops bone destruction by excess IL-6 production.144
FIG. 1:
Hypoxia regulation with CCNC, cdk8, super elongation complex
IV. CONCLUSIONS
Ig secretion and longevity are influenced by a variety of parameters. Terminal Ig secreting cells are arrested in G1 in a normal immune response; tumor cells bypass this cell-cycle block and still secrete Ig. The evidence points to the fact that short-lived plasma cells “overcommit” to secretion and thereby cause damage to the ER with reactive oxygen species unless they receive signals from their environment to balance out secretion and stress. Use of antioxidants supplied exogenously may help to mitigate this damage. A balance between heavy and light chain protein synthesis is also crucial indicating tight coordinate control. Damage to the ER brought about by the stress imposed by secreting large amounts of Ig can be countered by autophagy, which helps to remove damaged organelles. The role of the cannabinoid receptor CB2 in influencing Ig secretion is also of interest based on the increasing legality of marijuana use.
Cells that have experienced T-cell help during their activation in the germinal centers are more likely to migrate to bone marrow niches by virtue of CXC4:CXCL12 interactions. Once in the bone marrow, expression of the surface receptor BCMA, in the Tnf family, stimulates survival from signals made by the stromal cells. Exogenous application of BAFF and APRIL survival signals in some critical circumstances may increase long-lived plasma cell longevity. Experiments point to the hypoxic environment of the bone as contributing to the long-term survival of plasma cells that reside there. The details of the relationship between hypoxia, autophagy, Ig secretion, and longevity will require further exploration. The transcription elongation factor ELL2 links secretion, the unfolded protein response, and hypoxia by virtue of its action on the RNA polymerase and interactions with mediator complex factors. ELL2 is important for normal plasma cell development and crucial for survival of myeloma cells. It may represent a promising target for therapy in myeloma, an as yet incurable disease.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation Grant No. MCB-0842725, National Institutes of Health shared resources Grant No. P30CA047904 to the University of Pittsburgh Cancer Institute, and internal funding from the School of Medicine and the Department of Immunology, University of Pittsburgh. We thank Dr. Florian Weisel and Dr. Kevin Nickerson for critically reading the manuscript.
ABBREVIATIONS:
- BCMA
B-cell maturation antigen, aka tnfrsf17, CD269
- Blimp-1
B lymphocyte maturation protein 1, aka PRDM1
- cKO
conditional knockout; for ELL2 this is CD19cre/+ ell2loxp/loxp in exon 1 or exon 3
- c-myc
cellular myelocytomatosis proto-oncogene
- ELL
eleven-nineteen lysine-rich leukemia gene in the ELL family of three genes (1, 2, 3), part of the biochemically defined super elongation complex
- ER
endoplasmic reticulum
- FO
follicular
- Igh
immunoglobulin heavy chain
- IRF4
interferon regulatory protein 4
- LLPC
long-lived plasma cell
- LPS
lipopolysaccharide, a pan B-cell activator
- mTOR
mammalian target of rapamycin
- MZ
marginal zone
- sec (lowercase)
secretory-specific form of Igh mRNA or protein
- SEC (uppercase)
super elongation complex
- pTEFb
positive transcription factor b composed of cdk9 (a kinase) and cyclin T1 or 2, the enzymatic part of SEC
- RNAP-II
eukaryotic RNA polymerase II
- ROS
reactive oxygen species
- UPR
unfolded protein response
- XBP1
X-box protein 1
REFERENCES
- 1.Bosma M, Weiler E. The clonal nature of antibody formation. I. clones of antibody-forming cells of poly-D-alanine specificity. J Immunol. 1970;104:203–14. [PubMed] [Google Scholar]
- 2.Lefkovits I Induction of antibody-forming cell clones in microcultures. Eur J Immunol. 1972;2:360–6. [DOI] [PubMed] [Google Scholar]
- 3.Manz RA, Radbruch A. Plasma cells for a lifetime? Eur J Immunol. 2002;32(4):923–7. [DOI] [PubMed] [Google Scholar]
- 4.Chen-Kiang S Cell-cycle control of plasma cell differentiation and tumorigenesis. Immunol Rev. 2003;194:39–47. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard C, Fridman WH, Sautes C. Effect of TGF-beta1 on cell cycle regulatory proteins in LPS-stimulated normal mouse B lymphocytes. J Immunol. 1997;159:4155–64. [PubMed] [Google Scholar]
- 6.Lamson GL, Koshland ME. Changes in J chain and mu chain mRNA expression as a function of B cell differentiation. J Exp Med. 1984;160:877–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amigorena S, Bonnerot C, Choquet D, Fridman WH, Teillaud JL. Fc gamma RII expression in resting and activated B lymphocytes. Eur J Immunol. 1989;19(8):1379–85. [DOI] [PubMed] [Google Scholar]
- 8.van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, Heck AJ. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18(2):243–53. [DOI] [PubMed] [Google Scholar]
- 9.Noelle R, Ledbetter A, Aruffo A. CD40 and its ligand, an essential ligand receptor pair for thymus-dependent B-cell activation. Immunol Today. 1992;13:431. [DOI] [PubMed] [Google Scholar]
- 10.Malisan F, Briere F, Bridon J-M, Harindranath N, Mills C, Max E#, Banchereau J, Martinez-Valdez H. Interleukin-10 induces immunoglobulin G isotype switch recombination in human CD40-activated naive B lymphocytes. J Exp Med. 1996;183:937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacquot S, Kobata T, Iwata S, Morimoto C, Schlossman S. CD154/CD40 and CD70/CD27 interactions have different and sequential functions in T cell-dependent B cell responses. J Immunol. 1997;159:2652–7. [PubMed] [Google Scholar]
- 12.Nagumo H, Agematsu K, Shinozaki K, Hokibara S, Ito S, Takamoto M, Nikaido T, Yasui K, Uehara Y, Yachie A, Komiyama A. CD27/CD 70 interaction augments IgE secretion by promoting the differentiation of memory B cells into plasma cells. J Immunol. 1998;161(12): 6496–502. [PubMed] [Google Scholar]
- 13.Stuber E, Neurath M, Calderhead D, Fell H, Strober W. Cross-linking of 0X40 ligand, a member of the TNF/NGF cytokine family induces proliferation and differentiation in murine splenic cells. Immunity. 1995;2:507–21. [DOI] [PubMed] [Google Scholar]
- 14.Matsui K, Nakanishi K, Cohen DI, Hada T, Furuyama J, Hamaoka T, Higashino K. B-cell response pathways regulated by IL-5 and IL-2. Secretory muH chain-mRNA and J chain mRNA expression are separately controlled events. J Immunol. 1989;142:2918–23. [PubMed] [Google Scholar]
- 15.Rodríguez C, Roldán E, Navas G, Brieva JA. Essential role of tumor necrosis factor-α in the differentiation of human tonsil in vivo induced B cells capable of spontaneous and high-rate immunoglobulin secretion. Eur J Immunol. 1993;23(5):1160–4. [DOI] [PubMed] [Google Scholar]
- 16.Kikutani H, Taga T, Akira S, Kishi H, Miki Y, Saiki O, Yamamura Y, Kishimoto T. Effect of B cell differentiation factor (BCDF) on biosynthesis and secretion of Ig molecules in human B cell lines. J Immunol. 1985;134:990–5. [PubMed] [Google Scholar]
- 17.Moens L, Tangye SG. Cytokine-mediated regulation of plasma cell generation: IL-21 takes center stage. Frontiers in Immunology. 2014. Feb 18;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan D Molecular basis for the inhibition of LPS induced differentiation by anti-immunoglobulin. J Mol Cell Immunol. 1987;3(3):133–44. [PubMed] [Google Scholar]
- 19.Phillips C, Schimpl A, Dietrich-Goetz W, Clements J, Virtanen A. Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur J Immunol. 1996;26(12):3144–52. [DOI] [PubMed] [Google Scholar]
- 20.Chen U Analysis of cell proliferation and mu-RNA processing during activation of mouse B-cells by anti-mu and T lymphocytes. Mol Immunol. 1990;27(12):1249–57. [DOI] [PubMed] [Google Scholar]
- 21.Chen U, Scheuermann R, Wirth T, Gerster T, Roeder R, Harshman K, Berger C. Anti-IgM antibodies down modulate mu-enhancer activity and OTF2 levels in LPS-stimulated mouse splenic B-cells. Nucleic Acids Res. 1991;19:5981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin Immunol. 2011;23(5):341–9. [DOI] [PubMed] [Google Scholar]
- 23.Fairfax KA, Kallies A, Nutt SL, Tarlinton DM. Plasma cell development: from B-cell subsets to long-term survival niches. Semin Immunol. 2008;20(1):49–58. [DOI] [PubMed] [Google Scholar]
- 24.Carotta S, Willis SN, Hasbold J, Inouye M, Pang SH, Emslie D, Light A, Chopin M, Shi W, Wang H, Morse HC 3rd, Tarlinton DM, Corcoran LM, Hodgkin PD, Nutt SL. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014. Oct 20;211(11):2169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grötsch B, Brachs S, Lang C, Luther J, Derer A, Schlötzer-Schrehardt U, Bozec A, Fillatreau S, Berberich I, Hobeika E, Reth M, Wagner EF, Schett G, Mielenz D, David JP. The AP-1 transcription factor Fra1 inhibits follicular B cell differentiation into plasma cells. J Exp Med. 2014. Oct 20;211(11):2199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park KS, Bayles I, Szlachta-McGinn A, Paul J, Boiko J, Santos P, Liu J, Wang Z, Borghesi L, Milcarek C. Transcription enlongation factor ELL2 drives immunoglobulin secretory specific mRNA production and the unfolded protein response. J Immunol. 2014;193:4663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benhamron S, Pattanayak SP, Berger M, Tirosh B. mTOR activation promotes plasma cell differentiation and bypasses XBP-1 for immunoglobulin secretion. Mol Cell Biol. 2014. Oct 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu D, Nakagawa R, Lazzaro S, Staudacher P, Abreu-Goodger C, Henley T, Boiani S, Leyland R, Galloway A, Andrews S, Butcher G, Nutt SL, Turner M, Vigorito E. The miR-155—PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. The Journal of Experimental Medicine. 2014. Oct 20;211(11):2183–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittrucker HW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–3. [DOI] [PubMed] [Google Scholar]
- 30.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–82. [DOI] [PubMed] [Google Scholar]
- 31.Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, Zeng Y, Chen B, Epstein J, Staudt LM. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, Smyth GK, Tarlinton DM, Belz GT, corcoran LM, Kallies A, Nutt SL. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol. 2014. Apr 1;192(7):3200–6. [DOI] [PubMed] [Google Scholar]
- 33.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, Sciammas R. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38(5):918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sciammas R Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–36. [DOI] [PubMed] [Google Scholar]
- 35.Pongubala JM, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3’ enhancer activity. Mol Cell Biol. 1992. Jan 1;12(1):368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su GH, Ip HS, Cobb BS, Lu MM, Chen HM, Simon MC. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J Exp Med. 1996. Jul 1;184(1):203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SC, Bottaro A, Insel RA. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol Immunol. 2003;39(15):923–32. [DOI] [PubMed] [Google Scholar]
- 38.John SA, Clements JL, Russell LM, Garrett-Sinha LA. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor blimp-1. J Biol Chem. 2008. Jan 11;283(2):951–62. [DOI] [PubMed] [Google Scholar]
- 39.Fernández D, Ortiz M, Rodríguez L, García A, Martinez D, Moreno de Alborán I. The proto-oncogene c-myc regulates antibody secretion and Ig class switch recombination. J Immunol. 2013. Jun 15;190(12): 6135–44. [DOI] [PubMed] [Google Scholar]
- 40.Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM, Nitt SL. Initiation of plasma-cell differentiation is independent of the transcription factor blimp-1. Immunity. 2007;26:555–66. [DOI] [PubMed] [Google Scholar]
- 41.SShaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. [DOI] [PubMed] [Google Scholar]
- 42.Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones. 2010;15(3):281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takayanagi S, Fukuda R, Takeuchi Y, Tsukada S, Yoshida K. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones. 2013;18(1):11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002. Dec 13;277(50):49047–54. [DOI] [PubMed] [Google Scholar]
- 45.Gass JN, Jiang H-Y, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2008;45(4):1035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aragon IV, Barrington RA, Jackowski S, Mori K, Brewer JW. The specialized unfolded protein response of B lymphocytes: ATF6a-independent development of antibody-secreting B cells. Mol Immunol. 2012;51(3–4):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reimold A, Iwakoshi N, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–7. [DOI] [PubMed] [Google Scholar]
- 48.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J Exp Med. 2005. Aug 15;202(4):505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taubenheim N, Tarlinton DM, Crawford S, Corcoran LM, Hodgkin PD, Nutt SL. High rate of antibody secretion is not integral to plasma cell differentiation as revealed by XBP-1 deficiency. J Immunol. 2012. Oct 1;189(7):3328–38. [DOI] [PubMed] [Google Scholar]
- 50.Benhamron S, Hadar R, Iwawaky T, So J-S, Lee A-H, Tirosh B. Regulated IRE-1 dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur J Immunol. 2014;44:867–76. [DOI] [PubMed] [Google Scholar]
- 51.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186(3):323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000. Nov 1;14(21):2725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002. Feb 15;16(4):452–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwawaki T, Akai R, Kohno K. IRE1a disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLOS One. 2010;5(9):e13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson ML. Mechanisms controlling production of membrane and secreted immunoglobulin during B cell development. Immunol Res. 2007;37(1):33–48. [DOI] [PubMed] [Google Scholar]
- 56.Borghesi L, Milcarek C. From B cell to plasma cell: regulation of V(D)J recombination and antibody secretion. Immunol Res. 2006;36(1–3):27–32. [DOI] [PubMed] [Google Scholar]
- 57.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10(10):1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milcarek C, Albring M, Langer C, Park KS. The eleven-nineteen lysine-rich leukemia gene (ELL2) influences the histone H3 modifications accompanying the shift to secretory Immunoglobulin heavy chain mRNA production. J Biol Chem. 2011. Aug 9;286: 33795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shell SA, Martincic K, Tran J, Milcarek C. Increased phosphorylation of the carboxyl terminal domain of RNA polymerase II and loading of polyadenylation and co-transcriptional factors contribute to regulation of the Ig heavy chain mRNA in plasma cells. J Immunol. 2007;179:7663–73. [DOI] [PubMed] [Google Scholar]
- 60.Peterson ML, Perry RP. The regulated production of mu-m and mu-s mRNA is dependent on the relative efficiencies of mu-s poly(A) site usage and the Cmu4 to M1 splice. Mol Cell Biol. 1989;9:726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruce SR, Dingle RWC, Peterson ML. B-cell and plasma-cell splicing differences: A potential role in regulated immunoglobulin RNA processing. RNA. 2003. Oct 1;9(10):1264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster L, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B-cells. Mol Cell Biol. 2001;21:1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Milcarek C, Martincic K, Chung-Ganster L-H, Lutz CS. The snRNP-associated U1A levels change following IL-6 stimulation of human B-cells. Mol Immunol. 2003;39:809–14. [DOI] [PubMed] [Google Scholar]
- 64.Alkan SA, Martincic K, Milcarek C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem J. 2006;393:361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma J, Gunderson SI, Phillips C. Non-snRNP U1A levels decrease during mammalian B-cell differentiation and release the IgM secretory poly(A) site from repression. RNA. 2006. Jan 1;12(1):122–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucl Acids Res. 2005. Jan 12;33(1):201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edwalds-Gilbert G, Veraldi K, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25: 2547–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wires RNA. 2011;2:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tunyaplin C, Shapiro MA, Calame KL. Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter. Nucleic Acids Res. 2000. Dec 15;28(24):4846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3’-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008. Jan 15;28(2):849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007. Feb 1;17(2):156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji Z, Tian B. Reprogramming of 3’UTR untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009. Dec 23;4(12):e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3’ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009. April 28;106(17):7028–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takagaki Y, Manley J. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–71. [DOI] [PubMed] [Google Scholar]
- 75.Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze M, Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the Go to S phase transition. Proc Natl Acad Sci U S A. 1998;95(19):11095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mann MC, Strobel S, Fleckenstein B, Kress AK. The transcription elongation factor ELL2 is specifically upregulated in HTLV-1-infected T-cells and is dependent on the viral oncoprotein tax. Virology. 2014;464–465(0):98–110. [DOI] [PubMed] [Google Scholar]
- 77.Benson MJ, Äijö T, Chang X, Gagnon J, Pape UJ, Anantharaman V, Aravind L, Pursiheimo JP, Oberdoerffer S, Liu XS, Lahesmaa R, Lähdesmäki H, Rao A. Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc Nat Acad Sci U S A. 2012. Oct 2;109(40):16252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin LL, Mantchev GT, Bram RJ, Noelle RJ. BCMA Is Essential for the Survival of Long-lived Bone Marrow Plasma Cells. J Exp Med. 2004. Jan 5;199(1):91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu S, Lam KP. B-cell maturation protein, which binds the tumor necrosis factor famliy members BAFF and APRIL, is dispensible for humoral immune responses. Mol Cell Biol. 2001;21:4067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nixon JC, Rajaiya J, Webb CF. Mutations in the DNA-binding domain of the transcription factor, bright, act as dominant negative proteins and interfere with immunoglobulin transactivation. J Biol Chem. 2004. Sep 27;279:52465–72. [DOI] [PubMed] [Google Scholar]
- 81.Oldham AL, Miner CA, Wang HC, Webb CF. The transcription factor Bright plays a role in marginal zone B lymphocyte development and autoantibody production. Mol Immunol. 2011;49:367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Webb CF, Bryant J, Popowski M, Allred L, Kim D, Harriss J, Schmidt C, Miner CA, Rose K, Cheng HL, Griffin C, Tucker PW The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Mol Cell Biol. 2011. Mar 1;31(5):1041–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiu YK, Lin IY, Su ST, Wang KH, Yang SY, Tsai DY, Hsieh YT, Lin KI. Transcription factor ABF-1 suppresses plasma cell differentiation but facilitates memory B cell formation. J Immunol. 2014. Jul 28. [DOI] [PubMed] [Google Scholar]
- 84.Massari ME, Rivera RR, Voland JR, Quong MW, Breit TM, van Dongen JJ, de Smit O, Murre C. Characterization ofABF-1, a novel basic helix-loop-helix transcription factor expressed in activated B lymphocytes. Mol Cell Biol. 1998. June 1;18(6):3130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grandori C, Eisenman RN. Myc target genes. Trends Biochem Sci. 1997;22(5):177–81. [DOI] [PubMed] [Google Scholar]
- 86.Chevrier S, Emslie D, Shi W, Kratina T, Wellard C, Karnowski A, Erikci E, Smyth GK, Chowdhury K, Tarlinton D, Corcoran LM. The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J Exp Med. 2014. Apr 7;211:827–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Bhattacharya D. Adjuvant-specific regulation of long-term antibody responses by ZBTB20. J Exp Med. 2014. May 5;211(5):841–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Zhang P, Bao Y, Han Y, Wang Y, Zhang Q, Zhan Z, Meng J, Li Y, Li N, Zhang WJ, Cao X. Zinc finger protein ZBTB20 promotes toll-like receptor-triggered innate immune responses by repressing IκBα gene transcription. Proc Nat Acad Sci U S A. 2013. June 17;110:11097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takami Y, Kikuchi H, Nakayama T. Chicken histone deacetylase-2 controls the amount of the IgM H-chain at the steps of both transcription of its gene and alternative processing of its pre-mRNA in the DT40 cell line. J Biol Chem. 1999;34:23977–90. [DOI] [PubMed] [Google Scholar]
- 90.Nakayama M, Suzuki H, Yamamoto-Nagamatsu N, Barman HK, Kikuchi H, Takami Y, Toyonaga K, Yamashita K, Nakayama T. HDAC2 controls IgM H- and L-chain gene expressions via EBF1, Pax5, Ikaros, Aiolos and E2A gene expressions. Genes Cells. 2007;12(3):359–73. [DOI] [PubMed] [Google Scholar]
- 91.Kikuchi H, Nakayama M, Kuribayashi F, Imajoh-Ohmi S, Nishitoh H, Takami Y, Nakayama T. GCN5 is involved in regulation of immunoglobulin heavy chain gene expression in immature B cells. Gene. 2014;544(1):19–24. [DOI] [PubMed] [Google Scholar]
- 92.Agudelo M, Newton C, Widen R, Sherwood T, Nong L, Friedman H, Klein TW. Cannabinoid receptor 2 (CB2) mediates immunoglobulin class switching from IgM to IgE in cultures of murine-purified B lymphocytes. J Neuroimmune Pharm. 2008;3(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eisenstein TK, Meissler JJ, Wilson Q, Gaughan JP, Adler MW. Anandamide and A9-tetrahydrocannabinol directly inhibit cells of the immune system via CB2 receptors. J Neuroimmunol. 2007;189(1–2):17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feng R, Milcarek CA, Xie X-Q. Antagonism of cannabinoid receptor 2 pathway suppresses IL-6 induced immunoglobulin IgM secretion. BMC Pharm Tox. 2014;15:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumazaki K, Tirosh B, Maehr R, Boes M, Honjo T, Ploegh HL. AID−/−μs−/− mice are agammaglobulinemic and fail to maintain B220-CD138+ plasma cells. J Immunol. 2007. Feb 15;178(4):2192–203. [DOI] [PubMed] [Google Scholar]
- 96.Zhang S, Pruitt M, Tran D, Du Bois W, Zhang K, Patel R, Hoover S, Simpson RM, Simmons J, Gary J, Snapper CM, Casellas R, Mock BA. B cell-specific deficiencies in mTOR limit humoral immune responses. J Immunol. 2013. Aug 15;191(4):1692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou P, Ma X, Iyer L, Chaulagain C, Comenzo RL. One siRNA pool targeting the lambda constant region stops lambda light-chain production and causes terminal endoplasmaic reticulum stress. Blood. 2014;123:3440–51. [DOI] [PubMed] [Google Scholar]
- 98.Minges Wols HA, Ippolito JA, Yu Z, Palmer JL, White FA, Le PT, Witte PL. The effects of microenvironment and internal programming on plasma cell survival. Int Immunol. 2007. Jul 1;19(7):837–46. [DOI] [PubMed] [Google Scholar]
- 99.Winkelmann R, Sandrock L, Porstner M, Roth E, Mathews M, Hobeika E, Reth M, Kahn ML, Schuh W, Jäck HM. B cell homeostasis and plasma cell homing controlled by Krüppel-like factor 2. ProcNat Acad Sci, USA. 2011. Jan 11;108(2):710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: A Tutorial on B Cell Survival. Ann Rev Imm. 2003;21(1):231–64. [DOI] [PubMed] [Google Scholar]
- 101.Lopes-Carvalho T, Foote J, Kearney JF. Marginal zone B cells in lymphocyte activation and regulation. Curr Opin Immunol. 2005;17(3):244–50. [DOI] [PubMed] [Google Scholar]
- 102.Lopes-Carvalho T, Kearney JF. Development and selection of marginal zone B cells. Immunol Rev. 2004;197(1):192–205. [DOI] [PubMed] [Google Scholar]
- 103.Auner HW, Beham-Schmid C, Dillon N, Sabbattini P. The life span of short-lived plasma cells is partly determined by a block on activation of apoptotic caspases acting in combination with endoplasmic reticulum stress. Blood. 2010;116:3445–55. [DOI] [PubMed] [Google Scholar]
- 104.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9(11):767–77. [DOI] [PubMed] [Google Scholar]
- 105.McHeyzer-Williams LJ, McHeyzer-Williams MG. n. Annu Rev Immunol. 2005;25:487–513. [DOI] [PubMed] [Google Scholar]
- 106.Radbruch A, Muehlinghaus G, Luger EO, Inamine A, Smith KGC, Dörner T, Hiepe Fl. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6(10):741–50. [DOI] [PubMed] [Google Scholar]
- 107.Good-Jacobson KL, Shlomchik MJ. Plasticity and heterogeneity in the generation of memory B cells and long-lived plasma cells: the influence of germinal center interactions and dynamics. J Immunol. 2010. Sep 15;185(6):3117–25. [DOI] [PubMed] [Google Scholar]
- 108.Conter LJ, Song E, Shlomchik MJ, Tomayko MM. CD73 Expression is dynamically regulated in the germinal center and bone marrow plasma cells are diminished in its absence. PLOS One. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15(7):631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zabel F, Mohanan D, Bessa J, Link A, Fettelschoss A, Saudan P, Kündig TM Bachmann MF. Viral particles drive rapid differentiation of memory B cells into secondary plasma cells producing increased levels of antibodies. J Immunol. 2014. May 12;192:5499–508. [DOI] [PubMed] [Google Scholar]
- 111.Njau MN, Kim JH, Chappell CP, Ravindran R, Thomas L, Pulendran B, Jacob J. CD28-B7 Interaction modulates short- and long-lived plasma cell function. J Immunol. 2012. Sep 15;189(6):2758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoyer BF, Moser K, Hauser AJ, Peddinghaus A, Voigt C, Eilat D, Radbruch A, Hiepe F, Manz RA. Short-lived plasmablasts and long-lived plasma cells contribute to chronic humoral autoimmunity in NZB/W mice. J Exp Med. 2004;199:1577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Z, Zou Y-R, Davidson A. Plasma cells in systemic lupus erythematosus: the long and short of it all. Eur J Immunol. 2011;41(3):588–91. [DOI] [PubMed] [Google Scholar]
- 114.Zhang X, Paun A, Claudio E, Wang H, Siebenlist U. The tumor promoter and NF-κB modulator Bcl-3 regulates splenic B cell development. J Immunol. 2013;191:5984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hagman J, Ramírez J, Lukin K. B Lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1. In: Murre C, editor. Epigenetic regulation of lymphocyte development. Berlin: Springer; 2012. p. 17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vilagos B, Hoffmann M, Souabni A, Sun Q, Werner B, Medvedovic J, Bilic I, Minnich M, Axelsson E, Jaritz M, Busslinger M. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med. 2012. Apr 9;209(4):775–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Györy I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E, Grosschedl R. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 2012. Apr 1;26(7):668–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jiang Y, Soong TD, Wang L, Melnick AM, Elemento O. Genome-wide detection of genes targeted by non-Ig somatic hypermutation in lymphoma. PLoS ONE. 2012;7(7):e40332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bertolotti M, Sitia R, Rubartelli A. On the redox control of B lymphocyte differentiation and function. Antiox Redox Signal. 2012;16:1139–49. [DOI] [PubMed] [Google Scholar]
- 120.Nakajima S, Kitamura M. Bidirectional regulation of NF-κB by reactive oxygen species: a role of unfolded protein response. Free Radical Biol Med. 2013;65(0):162–74. [DOI] [PubMed] [Google Scholar]
- 121.Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, Crespo M, Shen Q, Bhagat G, Califamo A, Chadburn A, Pasqualucci L, Dalla-Favera R. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2009. Jul 7;106(27):11294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Novo E, Parola M. Redox mechanisms in hepatic chronic would healing and fibrogenesis. Fibrogenesis Tissue Repair. 2008;1:5–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee RL, Westendorf J, Gold MR. Differential role of reactive oxygen species in the activation of mitogen-activated protein kinases and Akt by key receptors on B-lymphocytes: CD40, the B cell antigen receptor, and CXCR4. J Cell Commun Signal. 2007;1:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, Orfanelli U, Ponzoni M, Sitia R, Casola S, Cenci S. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. [DOI] [PubMed] [Google Scholar]
- 125.Hansen WR, Barcistress N, Taylor L, Curthoys NP. The 3’-nontranslated region of rat renal glutaminase mRNA contains a pH-responsive stability element. Am J Physiol Renal Physiol. 1996;40(1):F126–F31. [DOI] [PubMed] [Google Scholar]
- 126.Hombach J, Lottspeich F, Reth M. Identification of the genes encoding the IgM-α and Ig-β components of the IgM antigen receptor complex by amino-terminal sequencing. Eur J Immunol. 1990;20(12):2795–9. [DOI] [PubMed] [Google Scholar]
- 127.Price PW, McKinney EC, Wang Y, Sasser LE, Kandasamy MK, Matsuuchi L, Milcarek C, Deal RB, Culver DG, Meagher RB. Engineered cell surface expression of membrane immunoglobulin as a means to identify monoclonal antibody-secreting hybridomas. J Immunol Methods. 2009;343:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9(7):491–502. [DOI] [PubMed] [Google Scholar]
- 129.Peperzak V, Vikstrom I, Walker J, Glaser SP, LePage M, Coquery CM, Erickson LD, Fairfax K, Mackay F, Strasser A, Nutt SL, Tarlinton DM. Mcl-1 is essential for the survival ofplasma cells. Nat Immunol. 2013;14(3):290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Deegan S, Saveljeva S, Gorman A, Samali A. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci. 2013;70(14):2425–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Masciarelli S, Fra AM, Pengo N, Bertolotti M, Cenci S, Fagioli C, Ron D, Hendershot LM, Sitia R. CHOP-independent apoptosis and pathway-selective induction of the UPR in developing plasma cells. Mol Immunol. 2010;47(6):1356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gaudette BT, Iwakoshi NN, Boise LH. Bcl-xL protein protects from C/EBP homologous protein (CHOP)-dependent apoptosis during plasma cell differentiation. J Biol Chem. 2014. Aug 22;289(34):23629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wong WJ, Qiu B, Nakazawa MS, Qing G, Simon MC. MYC Degradation under low O2 tension promotes survival by evading hypoxia-induced cell death. Mol Cell Biol. 2013. Sep 1;33(17):3494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Levine B Cell biology: autophagy and cancer. Nature. 2007;446(7137):745–7. [DOI] [PubMed] [Google Scholar]
- 136.Marin-Hernandez A, Gallardo-Perez JC, Ralph SJ, Rodriguez-Enriquez S, Moreno-Sanchez R. Hif-1alpha modulates eneregy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev Med Chem. 2009;9:1084–101. [DOI] [PubMed] [Google Scholar]
- 137.Keren DF. Laboratory evaluation for monoclonal gammopthies: MGUS to myeloma. Warde Med Lab. 2010;21(2). [Google Scholar]
- 138.Chesi M, Bergsagel PL. Many multiple myelomas: making more of the molecular mayhem. ASH Educ Program Book. 2011. Dec 10;2011(1):344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gu ZJ, Costes V, Lu ZY, Zhang XG, Pitard V, Moreau JF, Bataille R, Wijdenes J, Rossi JF, Klein B. Interleukin-10 is a growth factor for human myeloma cells by induction of an oncostatin M autocrine loop. Blood. 1996;88(10):3972–86. [PubMed] [Google Scholar]
- 140.van Laar JM, Melchers M, Tang YK, van der Zouwen B, Mohammadi R, Fischer R, Margolis L, Fitzgerald W, Grivel JC, Breedveld FC, Lipsky PE, Grammer AC. Sustained secretion of immunoglobulin by long-lived human tonsil plasma cells. Am J Path. 2007;171:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boylan KLM, Gosse MA, Staggs SE, Janz S, Grindle S, Kansas GS, Van Ness BG. A transgenic mouse model of plasma cell malignancy shows phenotypic, cytogenetic, and gene expression heterogeneity similar to human multiple myeloma. Cancer Res. 2007 May 1, 2007;67(9):4069–78. [DOI] [PubMed] [Google Scholar]
- 142.Hu J, Van Valckenborgh E, Menu E, De Bruyne E, Vanderkerken K. Understanding the hypoxic niche of multiple myeloma: therapeutic implications and contributions of mouse models. Dis Models Mech. 2012 Nov 1, 2012;5(6):763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Galbraith MD, Allen MA, Bensard CL, Wang XD, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153(6):1327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Storti P, Bolzoni M, Donofrio G, Airoldi I, Guasco D, Toscani D, Martella E, Lazzaretti M, Mancini C, Agnelli L, Patrene K, Maiga S, Franceschi V, Colla S, Anderson J, Neri A, Amiot M, Aversa F, David Roodman G, Giulianai N. Hypoxia-inducible factor (HIF)-1[alpha] suppression in myeloma cells blocks tumoral growth in vivo inhibiting angiogenesis and bone destruction. Leukemia. 2013;27(8):1697–706. [DOI] [PubMed] [Google Scholar]