Abstract
Epigenetic transcriptional regulation frequently requires histone modifications. Some, but not all, of these modifications are able to template their own inheritance. Here I discuss the molecular mechanisms by which histone modifications can be inherited and relate these ideas to new results about epigenetic transcriptional memory, a phenomenon that poises recently-repressed genes for faster re-activation and has been observed in diverse organisms. Recently, we found that the histone H3 lysine 4 dimethylation that is associated with this phenomenon plays a critical role in sustaining memory and, when factors critical for the establishment of memory are inactivated, can be stably maintained through multiple mitoses. This chromatin-mediated inheritance mechanism may involve a physical interaction between an H3K4me2 reader, SET3C, and an H3K4me2 writer, Spp1− COMPASS. This is the first example of a chromatin-mediated inheritance of a mark that promotes transcription.
Keywords: Epigenetics, chromatin, transcription, memory
A genome encodes multitudes. The numerous phenotypes, morphologies and functions of cells in the tissues of a multicellular organism represent distinct expression states of the same genome 1. Likewise, unicellular organisms respond to changes in their environment by altering their gene expression. Thus, while the DNA sequence of the genome defines the phenotypic potential of an organism, that potential is realized through the regulation of gene expression, often through selective transcription. Changes in transcription can be rapid and transient but, under certain circumstances, cells undergo long-term changes in gene expression that persists in the absence of the initiating stimulus. For example, in multicellular organisms, transient developmental cues lead to the progressive establishment of stable and (generally) irreversible transcriptional programs that commit cells to differentiation and restrict their developmental potential 2,3. In single cell organisms, transcriptional responses can also result in stable, long-term changes in phenotype 4. For example, wild isolates of Saccharomyces cerevisiae5,6 as well as pathogenic yeasts such as Candida albicans 7,8 and Cryptococcus neoformans 9 exhibit switching between several alternative colony morphologies due to changes in transcription10,11. These meta-stable colony morphologies are stably inherited but their relative frequency can be modulated by environmental stimuli 12,13. Thus, the transcriptional responses to environmental stimuli can be either transient or longer-term; in some cases, they are inherited through mitosis and meiosis 14. Changes in transcription that are inherited through mitosis or meiosis are termed epigenetic transcriptional regulation.
Heritable transcriptional regulation can be mediated by transcription factor activity, DNA methylation, non-coding RNAs and histone modification. Such regulatory factors function either in trans or in cis; trans-acting factors such as transcription factors bind to genetically encoded DNA elements to promote or repress transcription, while cis-acting DNA methylation or histone modifications “mark” regulatory regions to impact transcription of nearby genes 15.
Chromatin as a regulator of transcription
How do cis-acting marks impact transcription? Transcription is a complex process involving the coordinated recruitment of numerous factors and complexes 16. Eukaryotic genomes are packaged into nucleosomes 17, which limit access to the DNA, inhibiting transcription 18,19 and many regulators of transcription impact DNA accessibility by regulating nucleosome occupancy 20. The first indication that this was the case was the discovery that enzymes that control acetylation or deacetylation of histones either promote or repress transcription 21,22 and that large ATP-dependent complexes that could slide or remove nucleosomes regulate transcription 23,24. Acetylation of lysine residues in histones promotes transcription 25,26, in part because it neutralizes their charge and reduces their affinity for DNA 27 and disrupts interactions between neighboring nucleosomes 28, making them easier to displace by transcription factors or RNA polymerase 29–32. Also, acetylated histones serve as binding sites to recruit coactivators – including acetyltransferases themselves and nucleosome remodeling complexes 33 - that possess bromodomains 34. Thus, histone deacetylation represses transcription both by stabilizing chromatin to limit access to the DNA and by preventing recruitment of bromodomain coactivators 35–37. Regulating nucleosome occupancy through nucleosome remodeling and histone modification plays a critical role in regulating transcription.
Myriad covalent modifications of histones have been identified 38, including methylation, phosphorylation and ubiquitylation on their unstructured terminal tails. Mutations in either the enzymes that catalyze these modifications or the amino acids that are modified perturb transcriptional regulation, highlighting their functional significance 38.
Whereas histone acetylation generally promotes transcription 26, histone methylation promotes both transcription and silencing, depending on which amino acid is methylated. The histone H3 lysine 9 methyltransferase from mammals, flies and yeast promotes transcriptional silencing 39,40. The histone H3 lysine 4 methyltransferase Trithorax and the histone H3 lysine 27 methyltransferase Polycomb play antagonistic roles during development 41; loss of Trithorax leads to decreased expression of critical transcription factors within the homeotic gene cluster, while loss of Polycomb has the opposite effect 41. Thus, these two marks are associated with alternative, stable transcriptional states that regulate differentiation; H3K4 methylation is associated with actively transcribed loci, while H3K27 methylation is associated with silent loci. Genomic analysis reveals that, in differentiated cells, H3K4 methylation (H3K4me) and H3K27 methylation (H3K27me) are generally mutually exclusive and that the boundary between these marks in the homeotic gene cluster in flies and mammals defines which genes are expressed and which are silenced 42. This is due to recruitment of co-activators by H3K4me or co-repressors by H3K27me, regulation of histone H3K27 acetylation by H3K27me and through changes in chromatin folding and compaction 43. In pluripotent stem cells, certain loci possess “bivalent” loci in with both H3K4me and H3K27me, reflecting their potential for transcription or silencing 42,44.
Regions near centromeres and telomeres are constitutively silent 45. This stable silencing requires histone deacetylation and, in most species, histone H3 lysine 9 methylation (H3K9me) 46,47. Reporter genes adjacent to telomeres and centromeres have been a critical tool for screens to identify molecular players essential for this type of stable silencing. For example, a chromosomal inversion in Drosophila that places the white gene beside the centromere of the X chromosome results in variegated silencing of white in a subset of cells in the eye 48,49. Likewise, reporter genes inserted near a centromere in S. pombe or near telomeres in S. cerevisiae show variegated silencing 50,51. ChIP sequencing reveals that H3K9me localizes at many constitutively repressed loci in the genomes of yeast, flies and mammals 52.
Genomes are therefore divided into regions having distinct histone marks that correlate with different transcriptional states. Histone modifications are generally directed by cis-acting genetic information. For example, histone acetyltransferases are recruited to enhancers or promoters by sequence-specific transcription factors 53. Likewise, at the silent mating type locus in budding yeast, which is silenced by histone deacetylation, the histone deacetylase Sir2/Sir4 is recruited through binding of the Origin Replication Complex and Sir1 to elements that flank the locus 54–56. In the case of histone methylation, the mechanism is related but less simple 46. While the enzymes responsible for H3K27 and H3K9 methylation can be recruited to chromosomal loci by sequence-specific transcription factors 57,58, other recruitment mechanisms also occur. H3K9 methylation near centromeres in S. pombe depends on low level transcription of pericentromeric repeats that results in double stranded RNAs that are processed by Dicer 59. These small RNAs are incorporated into an RNAi transcriptional silencing complex that recruits the H3K9 methyltransferase Clr4 to the transcribed locus 60,61. Likewise, H3K27me is stimulated by recruitment of PcG complexes by both sequence-specific DNA binding proteins and mechanisms such as RNA and other histone modifications 62,63.
H3K4 methylation has been proposed to reflect transcription; in yeast, the sole H3K4 methyltransferase Set1/COMPASS is recruited through interaction with the Paf1 complex (Paf1C) 64, which binds to elongating RNA polymerase II (RNAPII). Loss of Paf1C leads to global loss of H3K4 methylation 64. Furthermore, Paf1C physically interacts with the E3 ubiquitin-conjugation/ligase Rad6/Bre1, which mediates ubiquitination of H2B lysine 123 65. This mark is also required for all H3K4 methylation in budding yeast 66,67. These results suggest that active RNAPII recruits Set1/COMPASS, leading to H3K4 methylation.
Heritable chromatin states
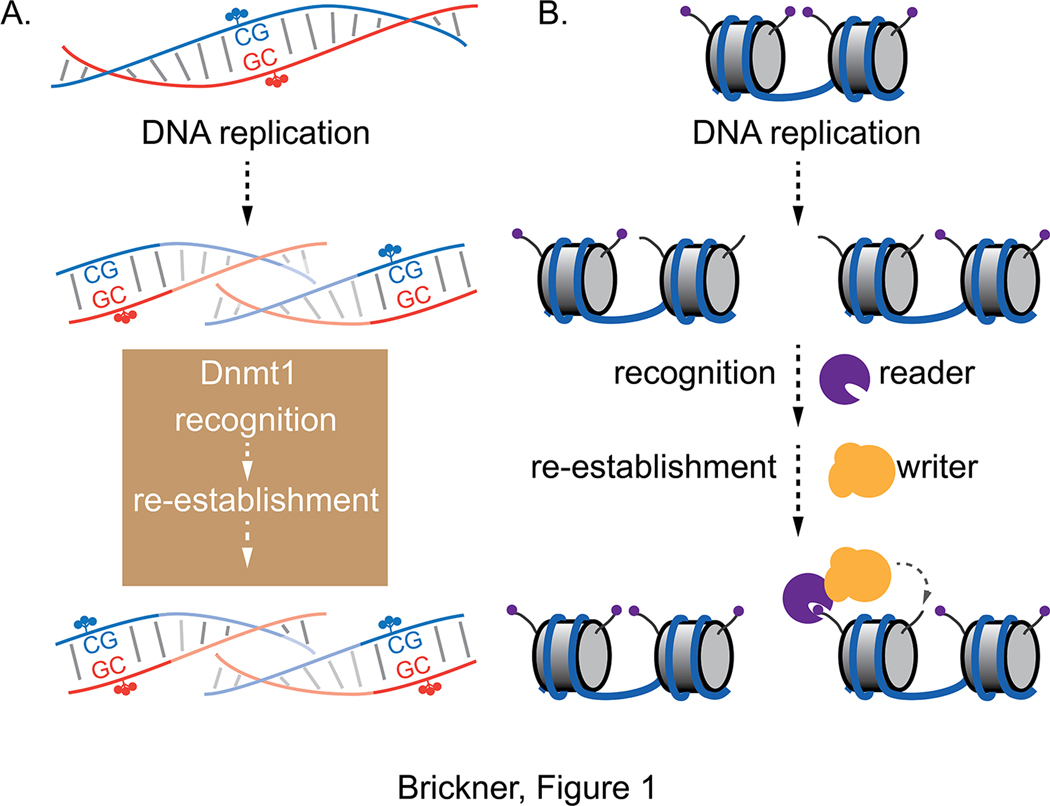
The heritability of DNA methylation and its requirement for the maintenance of certain epigenetic states suggests a general conceptual model for the inheritance of cis-acting information: following DNA replication, hemi-methylated CpG sites are recognized and re-methylated on the complementary cytosine (Figure 1A). Therefore, a mechanism that both recognizes a cis-acting mark and stimulates its re-establishment might allow that mark to be faithfully inherited at that location. Is there evidence that histone post-translational modifications are heritable? In other words, can cis-acting histone marks be perpetuated in the absence of trans-acting factors? Studies of silencing of telomeres, centromeres and special loci like the hidden mating type loci (HM) in flies and yeast suggested that histone modification-dependent transcriptional states are heritable in the absence of some of the trans-acting factors necessary for their establishment. For example, although Sir1 facilitates recruitment of the Sir2/Sir4 HDAC to the silent HM loci through interaction with sequence-specific DNA binding proteins 68, loss of Sir1 does not lead to immediate loss of silencing. Rather, in sir1 mutant strains, there are two stable populations, one that continues to silence the HM loci and the other that expresses the HM loci 69. This suggests that chromatin domains defined by deacetylated histones are epigenetically inherited.
Figure 1. Read-write mechanisms for replicating chromatin marks.
A. Following DNA replication, hemi-methylated CpG sites are recognized by Dnmt1, which re-establishes symmetric methylation. This protein possesses the ability to both recognize the appropriate substrate and to catalyze the reaction. B. Because histone H3/H4 nucleosome cores are reincorporated nearby but randomly distributed between the daughter products of DNA replication, approximately half of the nucleosomes contain parental, local H3/H4 tetramers and half contain new H3/H4 tetramers. The parental nucleosomes can be recognized by reader proteins that bind to specific histone marks. Recognition by readers leads to recruitment of writers, which re-establish these marks on neighboring nucleosomes after DNA replication. C. Inheritance of H3K9 methylation. Both the writer Clr4/Suv29h and the reader Swi6/HP1 can bind H3K9me3. Because Swi6 physically interacts with Clr4, this provides both a direct and an indirect mechanism for Clr4 recruitment to H3K9me3-marked loci. Binding of H3K9me3 to Clr4 also stimulates catalytic activity of the enzyme. D. Inheritance of H3K27 methylation. The PRC2 Polycomb complex contains both a reader (EED; light green) and a writer (EZH2; dark green). Binding of EED to H3K27me3 both recruits PRC2 to chromosomal sites with this mark and stimulates EZH2 catalytic activity to promote methylation of lysine 27 on neighboring nucleosomes.
Additional compelling evidence for epigenetically heritable chromatin modifications comes from studies of H3K9 methylation in S. pombe. Tethering of a the H3K9 methyltransferase Clr4 to an ectopic locus is sufficient to stimulate nearby H3K9 methylation and transcriptional silencing 70,71. In cells lacking a putative H3K9 demethylase Epe1, this mark can be maintained for many generations upon removing the tethered Clr4 71. Inheritance requires the endogenous Clr4 71 and is dependent on the density of H3K9me3 72, but is independent of the RNAi machinery 71. Thus, while wild type cells (having Epe1) require constant re-establishment of H3K9 methylation, this mark can mediate its own inheritance.
For histone modifications to be inherited, two conditions must be met. First, nucleosomes must be reincorporated into chromatin near their original location following DNA replication. Classic pulse-chase experiments 73 as well as more recent global experiments74 reveal that nucleosomes are partitioned equally between the two daughter chromosomes after DNA replication. Furthermore, the location of nucleosomes is quite stable over many cell divisions, particularly in silent regions 75. This suggests that nucleosomes are preferentially reincorporated near their original location. Indeed, several histone chaperones associate with PCNA and the DNA replication machinery and have been proposed to facilitate efficient reincorporation 76,77. These results suggest that, following DNA replication, approximately half of nucleosomes will be marked as they were before S-phase and the other half will be unmarked.
Second, to facilitate re-establishment of histone marks after DNA replication, the histone modifications on parental nucleosomes must be recognized (by a “reader”) and that must be coupled to recruitment of the enzyme that modifies new nucleosomes (“writer”). This is referred to as a read-write mechanism (Figure 1B). Read-write mechanisms have been defined for heritable histone deacetylation, H3K9 methylation and H3K27 methylation. Deacetylated nucleosomes of the HM loci or telomeres are recognized by the Sir3 protein, which both binds to unacetylated histone H4 lysine 16 78 and recruits the Sir2 histone deacetylase. Likewise, H3K9 methylation is recognized by both the HP1 protein and a chromodomain within the Clr4 H3K9 methyltransferase. Because HP1 interacts with Clr4, this protein functions as a reader, while Clr4 is both a reader and writer. The recognition of H3K9 methylation stimulates the methyltransferase activity 79,80, promotes spreading of the mark 81 and is essential for inheritance of H3K9 methylation 71 (Figure 1C). Finally, H3K27methylation can also stimulate its own inheritance through a read-write mechanism involving two subunits of the PRC2 complex. The EED subunit of PRC2 binds to H3K27me3. This leads to both recruitment of PCR2 to nucleosomes that possess H3K27me3 as well as stimulation of the catalytic activity of the methyltransferase EZH2 82,83 (Figure 1D). Thus, read-write mechanisms are a common strategy to faithful replication of histone modifications.
Epigenetic transcriptional memory
To-date, the best examples of heritable chromatin states are those associated with stable silencing. Histone modifications associated with active transcription are, for the most part, not heritable; histone acetylation is unstable and nucleosomes in actively transcribed regions are not reincorporated faithfully at the same location through multiple cell divisions 75. However, modifications that reflect previous transcription can be inherited. Here, I highlight a phenomenon whereby a heritable histone modification over an inactive gene both reflects previous expression and promotes future transcription. This mark (H3K4me2) is associated with both active and poised loci, although the molecular requirements and heritability in these two circumstances are different.
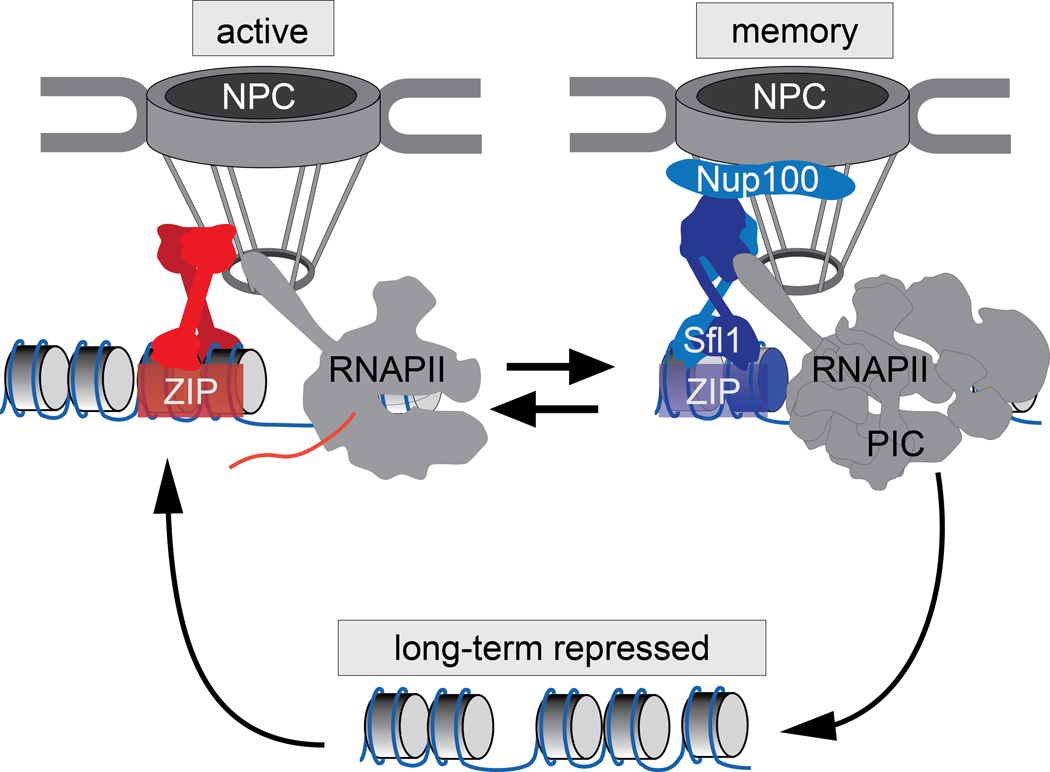
Some inducible genes are more rapidly or strongly induced in cells that have previously expressed them 14. This phenomenon, called epigenetic transcriptional memory, was discovered in budding yeast 84, but has since been observed in flies 85 and human cells 86 and related phenomena occur in worms 87 and plants 88,89. The general features of this type of memory are the requirement for interaction with nuclear pore proteins, H3K4me2 and recruitment of poised RNAPII. This type of memory persists through mitosis for between 4 and 15 cell divisions, depending on the system. One of the best-understood models for memory is the yeast INO1 gene 90,91, encoding the inositol 3-phosphate synthase enzyme, which is only expressed in the absence of exogenous inositol. This gene has also been a model to understand the role of the nuclear pore complex (NPC) in regulating transcription and chromatin structure 92.
When cells are starved for inositol, the transcriptional activation of the INO1 gene is coupled with repositioning to the nuclear periphery and a physical interaction with the NPC 92,93 (Figure 2, left). The interaction with the NPC requires two transcription factors (Cbf1 and Put3) that bind to cis-acting DNA zip codes upstream of the promoter, as well as several nuclear pore proteins 93–95 (Figure 2, left). Disrupting the interaction with the NPC leads to a defect in INO1 transcription 93,94.
Figure 2. INO1 transcriptional states.
Long-term repressed INO1 localizes in the nucleoplasm and is neither associated with RNAPII nor H3K4 methylation. Upon activation, INO1 is recruited to the nuclear pore complex (NPC) through the action of the Put3 and Cbf1 TFs (red proteins), which bind to cis-acting DNA zip codes upstream of INO1. Upon transcriptional repression, INO1 remains associated with the NPC. During memory, interaction with the NPC requires different factors: the Sfl1 TF and the nuclear pore protein Nup100. This interaction leads to changes in chromatin modifications (H3K4me2 and H2A.Z incorporation; indicated in blue) and recruitment of a pre-initiation form of RNAPII.
If inositol is added back, INO1 is immediately repressed 84,96. However, the gene remains associated with the NPC for approximately four generations (8h), both in the cells that had been expressing INO1 and in their daughters, granddaughters, and great granddaughters 84 (Figure 2, right). Ultimately, this interaction is lost and INO1 repositions to the nucleoplasm. Thus, the interaction of recently-repressed INO1 with the NPC is an epigenetically heritable state. Importantly, this interaction is mediated by a different transcription factor called Sfl1 97. This TF binds upon repression and is necessary and sufficient to mediate interaction with the NPC97. Also, the nuclear pore protein Nup100 is required for localization to the periphery only during memory and has no role in targeting active INO1 to the periphery 96. Finally, the systems controlling positioning of active and recently repressed INO1 are independent of each other. Thus, genes can be targeted to the NPC by at least two mechanisms and the INO1 gene utilizes both under different circumstances.
Transcriptional memory maintains INO1 in a poised state (Figure 2). The RNAPII preinitiation complex associates with the INO1 promoter during memory, leading to faster induction and faster adaptation if the cells are challenged with inositol starvation 96–98. Also, memory is associated with distinct histone modifications over the promoter compared with those associated with either the long-term repressed or active promoter. Whereas the long-term repressed INO1 promoter shows low histone acetylation and H3K4 methylation and the active INO1 promoter shows high acetylation and H3K4me3, during memory, the promoter and 5’ end of the INO1 gene show low levels of acetylation and high levels of H3K4me2, but not H3K4me3 86,97,98. During memory, an alternative form of the Set1/COMPASS histone methyltransferase lacking the Spp1 subunit, is recruited 97. Spp1− COMPASS catalyzes dimethylation, but not trimethylation, of H3K4 99. Furthermore, the histone variant H2A.Z is incorporated upstream of the promoter during memory 84,96. Poised RNAPII, H3K4me2 and H2A.Z are also associated with the promoters of other yeast genes that exhibit memory as well as genes in human cells that exhibit memory 86,97,98,100.
The interaction with the NPC is required for these chromatin changes. Loss of Nup100 or Sfl1 leads to loss of H3K4me2 and H2A.Z incorporation during memory 86,96,97. In contrast, loss of these factors has no effect on the H2A.Z at the +1 nucleosome of the repressed INO1 promoter or H3K4 methylation over the active INO1 promoter96,97.
Chromatin-dependent transcriptional memory
These chromatin changes play an essential role in promoting INO1 memory. Mutations that inactivate the Set1/COMPASS histone methyltransferase or the SWR1 complex - which exchanges H2A.Z for H2A - lead to loss of memory 86,96,97. Likewise, deletion of the HTZ1 gene or mutation of the lysine 4 in histone H3 also disrupt memory 84,97. Finally, while H2A.Z incorporation requires H3K4me2, H3K4 dimethylation is independent of H2A.Z, suggesting that H3K4me2 is a critical upstream step 98. In fact, the SET3C complex, which binds preferentially to H3K4me2 101, is essential for both maintaining H3K4me2 and for binding of RNAPII 86,97. Importantly, this role is specific to memory; loss of Set3 has no effect on H3K4me2/3 or RNAPII binding over the active INO1 promoter 86,97. Finally, conditional inactivation of Set1/COMPASS during memory leads to loss of H3K4 methylation and RNAPII association 97. Thus, H3K4me2 is continuously required for RNAPII poising.
To understand the molecular function of H3K4me2, we employed conditional genetics. The recruitment of preinitiation RNAPII to the INO1 promoter during memory requires the Mediator kinase Cdk8 97,98. Either conditional depletion of Cdk8 97 or inhibiting an analog sensitive mutant Cdk898 leads to rapid loss of RNAPII from the poised INO1 promoter. Inhibition by the analog is rapidly reversible, allowing us to test if continuous RNAPII binding is critical for maintaining the INO1 promoter in a poised state. If so, then RNAPII should fail to be recruited upon removing the inhibitor. However, RNAPII was recruited back to the recently repressed INO1 promoter following removal of the Cdk8 inhibitor, suggesting that the conditions necessary for RNAPII poising are maintained in the absence of RNAPII 98.
Given that RNAPII binding is not required to maintain the INO1 promoter in a poised state, we next tested if H3K4me2 is required. To do this, we combined temporary inhibition of Cdk8, which leads to loss of RNAPII binding, with inactivation of SET3C, which leads to rapid loss of H3K4me2 97. Thus, we assessed the ability of RNAPII to be recruited back to the INO1 promoter after removal of the Cdk8 inhibitor in the presence and absence of H3K4me2. In the absence of H3K4me2, RNAPII failed to associate with the INO1 promoter 98, suggesting that recruitment of poised RNAPII during memory requires H3K4me2.
How does H3K4me2 promote RNAPII recruitment? Our data suggest that H3K4me2 functions in a positive feedback loop during memory. While loss of Sfl1 or Nup100 prevents H3K4me2 from being deposited during memory, conditional removal of H3K4me2 disrupts Sfl1 binding 98. Therefore, Sfl1-mediated interaction with the NPC is required for H3K4 dimethylation and H3K4me2 is required for continued Sfl1 binding. This suggests that H3K4me2 permits Sfl1 binding and Sfl1 interacts with Cdk8+ Mediator 102 to promote RNAPII recruitment.
Chromatin as a source of heritable transcriptional memory
Could H3K4me2 be the source of epigenetically heritable information? In other words, is H3K4me2 inherently stable and capable of being re-established after DNA replication? We first assessed the stability of H3K4me2 associated with active transcription by examining this mark over the INO1 promoter in a mutant that lacks memory. In such a mutant, H3K4me2 is rapidly lost upon repressing transcription 98. This suggests that H3K4me2 per se, is neither stable nor heritable.
If H3K4me2 associated with memory is epigenetically heritable, it should persist in the absence of trans-acting initiating factors 103. To assess the heritability of H3K4me2 during memory, Sfl1 was depleted by auxin-induced degradation 104 after establishing INO1 memory. Under these conditions, interaction with the NPC, RNAPII binding and H2A.Z were lost, confirming that Sfl1 function was disrupted 98. However, H3K4me2 was maintained without dilution through four cell divisions 98. Thus, while interaction with the NPC is essential for establishment of H3K4me2 during memory, this mark can be maintained and re-established through cell divisions in the absence of this interaction. This is reminiscent of the perpetuation of silent chromatin upon loss of Sir1 on HM silencing or removal of tethered Clr4 and suggests that H3K4me2 may stimulate its own inheritance.
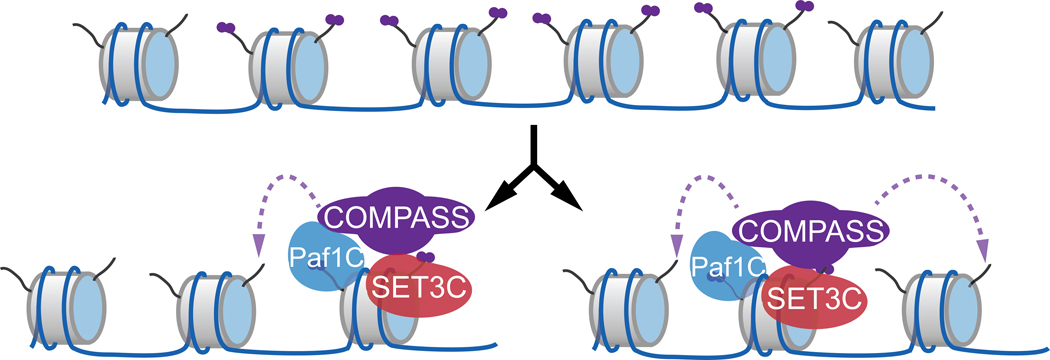
How might H3K4me2 be inherited during memory? Could this involve a read-write mechanism? During memory, the writer of H3K4me2 is Spp1− COMPASS and the putative reader is SET3C 86,97, a histone deacetylase complex with a PHD finger that recognizes H3K4me2 101. Simultaneous inactivation of Sfl1 and SET3C led to loss of H3K4me2 during memory 98, consistent with this complex facilitating inheritance. Furthermore, co-immunoprecipitation revealed that SET3C physically interacts with Spp1− COMPASS 98. This interaction suggests a read-write model in which SET3C recognizes H3K4me2-marked nucleosomes after DNA replication and recruits Spp1− COMPASS to re-establish this mark, facilitating both its spreading and inheritance (Figure 3).
Figure 3. Proposed mechanism of replication of H3K4me2 during transcriptional memory.
Following DNA replication, SET3C recognizes the H3K4me2 mark near the promoter of genes that exhibit memory. Through interaction with Spp1− COMPASS, potentially involving the Paf1C complex subunit Leo1, SET3 recruits COMPASS to this site and re-establishes H3K4me2.
These observations suggest that the same mark (H3K4me2) at the same location in the genome can be either unstable or heritable, depending on the pathway by which it is deposited. While all H3K4 methylation in budding yeast requires COMPASS, at least two distinct mechanisms can lead to COMPASS-mediated H3K4me2; H3K4me2 during active transcription and H3K4me2 during memory. H3K4 dimethylation during active transcription requires RNAPII, but H3K4 dimethylation during memory does not; inactivation of Cdk8 or Sfl1 led to loss of RNAPII but did not affect H3K4me2 during memory 98. H3K4me2 during memory requires Nup100 and Set3C, but these factors are not required for H3K4me2 over transcribed genes 86,97. Likewise, a subunit of the Paf1C, Leo1, is specifically required for H3K4 methylation during memory 98. Therefore, the heritability and functional impact of H3K4 methylation depends on additional, context-specific factors.
Future work will determine the molecular basis of the switch from RNAPII-dependent, unstable H3K4me2 to RNAPII-independent, heritable H3K4me2 during memory. We hypothesize that this switch is regulated by transcription factors that mediate interaction with nuclear pore proteins. But it is unclear what regulates these transcription factors and how the interaction with nuclear pore proteins impacts histone methylation. Nup98, the Nup100 homolog in flies and mammals, physically interacts with the Trithorax and Set1/COMPASS complexes and impacts H3K4 methylation in those organisms 105,106. This suggests that nuclear pore proteins may play critical and conserved roles in regulating epigenetically heritable histone methylation. Also, it will be important to define the molecular mechanism by which SET3C and Spp1− COMPASS physically interact to mediate inheritance of H3K4me2. Integrating how transcription factors, nuclear pore proteins and chromatin modifications stimulate RNAPII poising to lead to changes in future gene expression will have a broad impact on basic cell biology, developmental biology and genetics.
Acknowledgements
The author acknowledges support from the National Institute of General Medical Science (R35GM136419) and past and present members of the Brickner laboratory who have contributed to the discoveries described.
Footnotes
Competing interests
The author declares no competing interests.
REFERENCES
- 1.LASKEY RA & GURDON JB. 1970. Genetic Content of Adult Somatic Cells tested by Nuclear Transplantation from Cultured Cells. Nature 228: 1332–1334. [DOI] [PubMed] [Google Scholar]
- 2.Skora AD & Spradling AC. 2010. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proc National Acad Sci 107: 7389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gifford CA, Ziller MJ, Gu H, et al. 2013. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell 153: 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanney DL 1958. EPIGENETIC CONTROL SYSTEMS. Proc National Acad Sci 44: 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuthan M, Devaux F, Janderová B, et al. 2003. Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol Microbiol 47: 745–754. [DOI] [PubMed] [Google Scholar]
- 6.Voordeckers K, Maeyer DD, Zande E, et al. 2012. Genetic network for colony morphology in yeast. Mol Microbiol 86: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slutsky B, Buffo J. & Soll DR. 1985. High-frequency switching of colony morphology in Candida albicans. Science 230: 666–9. [DOI] [PubMed] [Google Scholar]
- 8.Slutsky B, Staebell M, Anderson J, et al. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169: 189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman DL, Fries BC, Franzot SP, et al. 1998. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci U S A 95: 14967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nantel A, Dignard D, Bachewich C, et al. 2002. Transcription Profiling of Candida albicans Cells Undergoing the Yeast-to-Hyphal Transition. Mol Biol Cell 13: 3452–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Šťovíček V, Váchová L, Begany M, et al. 2014. Global changes in gene expression associated with phenotypic switching of wild yeast. Bmc Genomics 15: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramírez-Zavala B, Reuß O, Park Y-N, et al. 2008. Environmental Induction of White–Opaque Switching in Candida albicans. Plos Pathog 4: e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granek JA & Magwene PM. 2010. Environmental and Genetic Determinants of Colony Morphology in Yeast. Plos Genet 6: e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Urso A. & Brickner JH. 2017. Epigenetic transcriptional memory. Curr Genet 63: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinberg D. & Vales LD. 2018. Chromatin domains rich in inheritance. Science 361: 33–34. [DOI] [PubMed] [Google Scholar]
- 16.Nikolov DB & Burley SK. 1997. RNA polymerase II transcription initiation: A structural view. Proc National Acad Sci 94: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornberg RD & Thomas JO. 1974. Chromatin Structure: Oligomers of the Histones. Science 184: 865–868. [DOI] [PubMed] [Google Scholar]
- 18.Cedar H. & Felsenfeld G. 1973. Transcription of chromatin in vitro. J Mol Biol 77: 237–254. [DOI] [PubMed] [Google Scholar]
- 19.Lorch Y, LaPointe JW & Kornberg RD. 1987. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49: 203–210. [DOI] [PubMed] [Google Scholar]
- 20.Pazin MJ, Kamakaka RT & Kadonaga JT. 1994. ATP-Dependent Nucleosome Reconfiguration and Transcriptional Activation from Preassembled Chromatin Templates. Science 266: 2007–2011. [DOI] [PubMed] [Google Scholar]
- 21.Brownell JE, Zhou J, Ranalli T, et al. 1996. Tetrahymena Histone Acetyltransferase A: A Homolog to Yeast Gcn5p Linking Histone Acetylation to Gene Activation. Cell 84: 843–851. [DOI] [PubMed] [Google Scholar]
- 22.Taunton J, Hassig CA & Schreiber SL. 1996. A Mammalian Histone Deacetylase Related to the Yeast Transcriptional Regulator Rpd3p. Science 272: 408–411. [DOI] [PubMed] [Google Scholar]
- 23.Peterson CL & Herskowitz I. 1992. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68: 573–583. [DOI] [PubMed] [Google Scholar]
- 24.Cairns BR 1998. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci 23: 20–5. [DOI] [PubMed] [Google Scholar]
- 25.Hebbes TR, Thorne AW & Crane-Robinson C. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. Embo J 7: 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dion MF, Altschuler SJ, Wu LF, et al. 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A 102: 5501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong L, Schroth GP, Matthews HR, et al. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biological Chem 268: 305–14. [PubMed] [Google Scholar]
- 28.Luger K, Mäder AW, Richmond RK, et al. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- 29.Allfrey VG, Faulkner R. & Mirsky AE. 1964. ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS*. Proc National Acad Sci 51: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vettese-Dadey M, Grant PA, Hebbes TR, et al. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. Embo J 15: 2508–18. [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson JD, Lowary PT & Widom J. 2001. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 307: 977–85. [DOI] [PubMed] [Google Scholar]
- 32.Tse C, Sera T, Wolffe AP, et al. 1998. Disruption of Higher-Order Folding by Core Histone Acetylation Dramatically Enhances Transcription of Nucleosomal Arrays by RNA Polymerase III. Mol Cell Biol 18: 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan AH, Prochasson P, Neely KE, et al. 2002. Function and Selectivity of Bromodomains in Anchoring Chromatin-Modifying Complexes to Promoter Nucleosomes. Cell 111: 369–379. [DOI] [PubMed] [Google Scholar]
- 34.Dhalluin C, Carlson JE, Zeng L, et al. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496. [DOI] [PubMed] [Google Scholar]
- 35.Kimura A, Umehara T. & Horikoshi M. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet 32: 370–377. [DOI] [PubMed] [Google Scholar]
- 36.Suka N, Luo K. & Grunstein M. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–83. [DOI] [PubMed] [Google Scholar]
- 37.Braunstein M, Rose AB, Holmes SG, et al. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev 7: 592–604. [DOI] [PubMed] [Google Scholar]
- 38.Bannister AJ & Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rea S, Eisenhaber F, O’Carroll D, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. [DOI] [PubMed] [Google Scholar]
- 40.Lachner M, O’Carroll D, Rea S, et al. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–20. [DOI] [PubMed] [Google Scholar]
- 41.Schuettengruber B, Chourrout D, Vervoort M, et al. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–45. [DOI] [PubMed] [Google Scholar]
- 42.Schuettengruber B, Bourbon H-M, Croce LD, et al. 2017. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171: 34–57. [DOI] [PubMed] [Google Scholar]
- 43.Eskeland R, Leeb M, Grimes GR, et al. 2010. Ring1B Compacts Chromatin Structure and Represses Gene Expression Independent of Histone Ubiquitination. Mol Cell 38: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein BE, Mikkelsen TS, Xie X, et al. 2006. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- 45.Grewal S. 2013. Epigenetic genome control by heterochromatin and RNAi machinery. Epigenet Chromatin 6: O32–O32. [Google Scholar]
- 46.Grewal SI & Moazed D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802. [DOI] [PubMed] [Google Scholar]
- 47.Yamada T, Fischle W, Sugiyama T, et al. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–85. [DOI] [PubMed] [Google Scholar]
- 48.Muller HJ 1930. Types of visible variations induced by X-rays inDrosophila. J Genet 22: 299–334. [Google Scholar]
- 49.Sinclair DAR, Mottus RC & Grigliatti TA. 1983. Genes which suppress position-effect variegation in Drosophila melanogaster are clustered. Molec. Gen. Genet. 191: 326–333. [Google Scholar]
- 50.Gottschling DE, Aparicio OM, Billington BL, et al. 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- 51.Allshire RC, Javerzat J-P, Redhead NJ, et al. 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157–169. [DOI] [PubMed] [Google Scholar]
- 52.Richards EJ & Elgin SC. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500. [DOI] [PubMed] [Google Scholar]
- 53.Kadosh D. & Struhl K. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–71. [DOI] [PubMed] [Google Scholar]
- 54.Moazed D, Kistler A, Axelrod A, et al. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci U S A 94: 2186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rusche LN, Kirchmaier AL & Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516. [DOI] [PubMed] [Google Scholar]
- 56.Laurenson P. & Rine J. 1992. Silencers, silencing, and heritable transcriptional states. Microbiol Rev 56: 543–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laprell F, Finkl K. & Müller J. 2017. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356: 85–88. [DOI] [PubMed] [Google Scholar]
- 58.Wang X. & Moazed D. 2017. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 356: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Volpe TA, Kidner C, Hall IM, et al. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–7. [DOI] [PubMed] [Google Scholar]
- 60.Noma K, Sugiyama T, Cam H, et al. 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–80. [DOI] [PubMed] [Google Scholar]
- 61.Verdel A, Jia S, Gerber S, et al. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kassis JA & Brown JL. 2013. Chapter Three Polycomb Group Response Elements in Drosophila and Vertebrates. Adv Genet 81: 83–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laugesen A, Højfeldt JW & Helin K. 2019. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol Cell 74: 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogan NJ, Dover J, Wood A, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–9. [DOI] [PubMed] [Google Scholar]
- 65.Kim J. & Roeder RG. 2009. Direct Bre1-Paf1 Complex Interactions and RING Finger-independent Bre1-Rad6 Interactions Mediate Histone H2B Ubiquitylation in Yeast*. J Biol Chem 284: 20582–20592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Z-W & Allis CD. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. [DOI] [PubMed] [Google Scholar]
- 67.Dover J, Schneider J, Tawiah-Boateng MA, et al. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–71. [DOI] [PubMed] [Google Scholar]
- 68.Chien C, Buck S, Sternglanz R, et al. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75: 531–541. [DOI] [PubMed] [Google Scholar]
- 69.Pillus L. & Rine J. 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59: 637–647. [DOI] [PubMed] [Google Scholar]
- 70.Audergon PNCB, Catania S, Kagansky A, et al. 2015. Restricted epigenetic inheritance of H3K9 methylation. Science 348: 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ragunathan K, Jih G. & Moazed D. 2015. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348: 1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DiPiazza ARC, Taneja N, Dhakshnamoorthy J, et al. 2021. Spreading and epigenetic inheritance of heterochromatin require a critical density of histone H3 lysine 9 tri-methylation. P Natl Acad Sci Usa 118: e2100699118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jackson V, Granner DK & Chalkley R. 1975. Deposition of histones onto replicating chromosomes. Proc National Acad Sci 72: 4440–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu C, Gan H, Serra-Cardona A, et al. 2018. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361: 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Escobar TM, Oksuz O, Saldaña-Meyer R, et al. 2019. Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication. Cell 179: 953–963.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gan H, Serra-Cardona A, Hua X, et al. 2018. The Mcm2-Ctf4-Polα Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol Cell 72: 140–151.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ransom M, Dennehey BK & Tyler JK. 2010. Chaperoning Histones during DNA Replication and Repair. Cell 140: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oppikofer M, Kueng S, Martino F, et al. 2011. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. Embo J 30: 2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Müller MM, Fierz B, Bittova L, et al. 2016. A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat Chem Biol 12: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Al-Sady B, Madhani HD & Narlikar GJ. 2013. Division of Labor between the Chromodomains of HP1 and Suv39 Methylase Enables Coordination of Heterochromatin Spread. Mol Cell 51: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang K, Mosch K, Fischle W, et al. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–8. [DOI] [PubMed] [Google Scholar]
- 82.Margueron R, Justin N, Ohno K, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiao L. & Liu X. 2015. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350: aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, et al. 2007. H2A.Z-Mediated Localization of Genes at the Nuclear Periphery Confers Epigenetic Memory of Previous Transcriptional State. Plos Biol 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pascual-Garcia P, Debo B, Aleman JR, et al. 2017. Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol Cell 66: 63–76 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Light WH, Freaney J, Sood V, et al. 2013. A Conserved Role for Human Nup98 in Altering Chromatin Structure and Promoting Epigenetic Transcriptional Memory. Plos Biol 11: e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maxwell CS, Kruesi WS, Core LJ, et al. 2014. Pol II Docking and Pausing at Growth and Stress Genes in C. elegans. Cell Reports 6: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lämke J, Brzezinka K, Altmann S, et al. 2015. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. Embo J 35: 162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lämke J. & Bäurle I. 2017. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol 18: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Culbertson MR, Donahue TF & Henry SA. 1976. Control of inositol biosynthesis in Saccharomyces cerevisiae; inositol-phosphate synthetase mutants. J Bacteriol 126: 243–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Culbertson MR & Henry SA. 1975. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics 80: 23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brickner JH & Walter P. 2004. Gene Recruitment of the Activated INO1 Locus to the Nuclear Membrane. Plos Biol 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahmed S, Brickner DG, Light WH, et al. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brickner DG, Ahmed S, Meldi L, et al. 2012. Transcription Factor Binding to a DNA Zip Code Controls Interchromosomal Clustering at the Nuclear Periphery. Dev Cell 22: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Randise-Hinchliff C, Coukos R, Sood V, et al. 2016. Strategies to regulate transcription factor–mediated gene positioning and interchromosomal clustering at the nuclear periphery. J Cell Biology 212: 633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Light WH, Brickner DG, Brand VR, et al. 2010. Interaction of a DNA Zip Code with the Nuclear Pore Complex Promotes H2A.Z Incorporation and INO1 Transcriptional Memory. Mol Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.D’Urso A, Takahashi Y, Xiong B, et al. 2016. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife 5: e16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sump B, Brickner DG, D’Urso A, et al. 2022. Mitotically heritable, RNA polymerase II-independent H3K4 dimethylation stimulates INO1 transcriptional memory. Elife 11: e77646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takahashi YH, Lee JS, Swanson SK, et al. 2009. Regulation of H3K4 Trimethylation via Cps40 (Spp1) of COMPASS Is Monoubiquitination Independent: Implication for a Phe/Tyr Switch by the Catalytic Domain of Set1. Mol Cell Biol 29: 3478–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sood V, Cajigas I, D’Urso A, et al. 2017. Epigenetic Transcriptional Memory of GAL Genes Depends on Growth in Glucose and the Tup1 Transcription Factor in Saccharomyces cerevisiae. Genetics 206: 1895–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim T. & Buratowski S. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5’ transcribed regions. Cell 137: 259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song W. & Carlson M. 1998. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. Embo J 17: 5757–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Berger SL, Kouzarides T, Shiekhattar R, et al. 2009. An operational definition of epigenetics. Gene Dev 23: 781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nishimura K, Fukagawa T, Takisawa H, et al. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. [DOI] [PubMed] [Google Scholar]
- 105.Franks TM, McCloskey A, Shokirev MN, et al. 2017. Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Gene Dev 31: 2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pascual-Garcia P, Jeong J. & Capelson M. 2014. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Reports 9: 433–42. [DOI] [PubMed] [Google Scholar]