In this study, Crain et al. describe the impact of histone H4 lysine 20 monomethylation (H4K20me1) on gene expression in Drosophila, showing that Set8, the histone lysine methyltransferase responsible for H4K20me1 deposition, and H4K20me1 reader l(3)mbt regulate gene expression and occupy chromatin through H4K20me1-independent mechanisms. These findings indicate that H4K20me1 does not have a significant role in regulating gene expression and may be dispensable for the function of Set8 and L(3)mbt in development and disease.
Keywords: chromatin, Drosophila melanogaster, epigenetics, gene expression, genomics, H4K20me1, L(3)mbt, Set8
Abstract
Monomethylation of lysine 20 of histone H4 (H4K20me1) is catalyzed by Set8 and thought to play important roles in many aspects of genome function that are mediated by H4K20me binding proteins. We interrogated this model in a developing animal by comparing in parallel the transcriptomes of Set8null, H4K20R/A, and l(3)mbt mutant Drosophila melanogaster. We found that the gene expression profiles of H4K20A and H4K20R larvae are markedly different than Set8null larvae despite similar reductions in H4K20me1. Set8null mutant cells have a severely disrupted transcriptome and fail to proliferate in vivo, but these phenotypes are not recapitulated by mutation of H4K20, indicating that the developmental defects of Set8null animals are largely due to H4K20me1-independent effects on gene expression. Furthermore, the H4K20me1 binding protein L(3)mbt is recruited to the transcription start sites of most genes independently of H4K20me even though genes bound by L(3)mbt have high levels of H4K20me1. Moreover, both Set8 and L(3)mbt bind to purified H4K20R nucleosomes in vitro. We conclude that gene expression changes in Set8null and H4K20 mutants cannot be explained by loss of H4K20me1 or L(3)mbt binding to chromatin and therefore that H4K20me1 does not play a large role in gene expression.
The prevailing model for how histone post-translational modifications (PTMs) regulate gene expression is that “writer” enzymes establish where and when histone PTMs are deposited in the genome, and “reader” proteins bind to these PTMs to activate or repress transcription by modulating the recruitment of transcription factors (Strahl and Allis 2000; Jenuwein and Allis 2001; Kouzarides 2007; Rothbart and Strahl 2014). Support for this model primarily comes from experiments that infer histone PTM function through genetic manipulation of individual writers or readers (Kouzarides 2007; Henikoff and Shilatifard 2011). However, this approach has a major caveat: Most writers have nonhistone substrates, and readers interact with many proteins other than modified histones (Clarke 2013; Zhang et al. 2015). Determining the roles of specific histone PTMs in genome function and development would benefit from a comparative analysis of mutations in a writer, its target histone residue, and a reader of the modified histone residue in a single animal model system. Such a combined approach is only feasible in Drosophila melanogaster, where the generation of histone mutant genotypes can be achieved (McKay et al. 2015). To better understand the role of H4K20 methylation in genome function, we used genomic approaches in Drosophila to examine in parallel gene expression phenotypes resulting from mutations of histone H4 lysine 20 (H4K20), the H4K20-specific monomethyltransferase Set8, and an H4K20me binding protein, L(3)mbt.
Methylation of histone H4 lysine 20 (H4K20me) has been implicated in the regulation of DNA replication (Jørgensen et al. 2007; Tardat et al. 2007; Huen et al. 2008; Takawa et al. 2012; Li et al. 2016; Brustel et al. 2017; Pellegrino et al. 2017; Shoaib et al. 2018; Hayashi-Takanaka et al. 2021), gene expression (Karachentsev et al. 2005; Barski et al. 2007; Kalakonda et al. 2008; Congdon et al. 2010; Beck et al. 2012; Yang et al. 2012; Veloso et al. 2014; Lv et al. 2016; Yu et al. 2019; Morgan and Shilatifard 2020), and DNA damage repair (Jørgensen et al. 2007; Sakaguchi and Steward 2007; Dulev et al. 2014) in both humans and flies, suggesting an evolutionarily conserved role for H4K20me in critical aspects of genome function and stability. Indeed, the human H4K20 monomethyltransferase Set8 (also known as KMT5A) can replace essentially all developmental functions of Drosophila Set8 (Crain et al. 2022). Different methylation states of H4K20 (i.e., K20me1, K20me2, and K20me3) are thought to mediate different genomic functions. Set8 catalyzes H4K20me1, the preferred substrate for subsequent dimethylation and trimethylation by Suv4-20 enzymes (Schotta et al. 2008; Yang et al. 2008; Southall et al. 2014; Weirich et al. 2016). Thus, loss of Set8 depletes all H4K20 methyl states and results in pleiotropic phenotypes and lethality in both flies and mice (Karachentsev et al. 2005; Oda et al. 2009; Crain et al. 2022). Therefore, current models in the field posit that Set8 regulates genome functions mainly through deposition of H4K20me1.
The genomic functions of H4K20me are thought to be mediated by proteins that recognize various methylation states of H4K20, including lethal (3) malignant brain tumor [L(3)mbt], Orc1, 53BP1, enhancer of zeste [E(z)], and the Msl complex (Kalakonda et al. 2008; Kim et al. 2010; Kuo et al. 2012; Sakaguchi et al. 2012; Tuzon et al. 2014; Weaver et al. 2019). Loss of L(3)mbt in Drosophila results in a somatic-to-germline shift in gene expression, leading to overproliferation of brain tissue (Bonasio et al. 2010; Janic et al. 2010). L(3)mbt contains three MBT domains that share homology with the “Royal” family of chromatin-interacting proteins and bind H4K20me1/2 in vitro (Maurer-Stroh et al. 2003; Min et al. 2007; Trojer et al. 2007; Kalakonda et al. 2008; Qi et al. 2010; Sakaguchi et al. 2012). L(3)mbt interacts in vivo with several partner proteins as a member of the LINT and MybMuvB/dREAM complexes, executing transcriptional repression seemingly independently of H4K20me (Meier et al. 2012; Blanchard et al. 2014; Coux et al. 2018; Yamamoto-Matsuda et al. 2022). Thus, the connection between L(3)mbt histone binding of H4K20me and its regulation of gene expression is incompletely understood.
Numerous previous studies suggest functional connections between H4K20me, Set8, and L(3)mbt. For instance, depletion of Set8 or L(3)mbt results in a loss of H4K20me1 and causes defects in DNA replication, chromatin organization, and transcription, implying that H4K20me1 plays a causal role in these processes (Beck et al. 2012; Sakaguchi et al. 2012). However, Set8 has nonhistone substrates (e.g., p53 [Shi et al. 2007] and PCNA [Takawa et al. 2012] and noncatalytic functions [e.g., in cell cycle entry) (Yin et al. 2008; Zouaz et al. 2018), and L(3)mbt colocalizes promiscuously with several different monomethylated and dimethylated histone residues (Blanchard et al. 2014). Furthermore, Drosophila mutants expressing unmodifiable H4K20A or H4K20R are phenotypically distinct from Set8null mutants (Crain et al. 2022), and loss of L(3)mbt function does not recapitulate many of the cell proliferation defects in Set8null animals despite a 60% reduction in H4K20me1 (Sakaguchi et al. 2012). Thus, whether and how H4K20me mediates the functions of Set8 and L(3)mbt remain unclear.
H4K20me1's role in gene expression in mammals and flies is also not easily reconciled into a simple model. In mammalian cells, H4K20me1 is found in the body of actively transcribed genes (Barski et al. 2007; Beck et al. 2012), suggesting a role for H4K20me1 in stimulating transcription (Kapoor-Vazirani and Vertino 2014; Veloso et al. 2014; Nikolaou et al. 2017; Shoaib et al. 2021) that was also suggested for Drosophila H4K20me1 (Lv et al. 2016; Yu et al. 2019; Huang et al. 2021). Conversely, H4K20me1 has been implicated in chromatin compaction (Lu et al. 2008) and gene repression (Kalakonda et al. 2008; Abbas et al. 2010; Congdon et al. 2010; Liu et al. 2010) via either its association with the transcriptional repressor L(3)mbt or its localization in inactive regions of the genome (Nishioka et al. 2002; Kalakonda et al. 2008; Sakaguchi et al. 2012). Here we determined whether the gene expression phenotypes arising upon removal of Set8 or L(3)mbt can be attributed to loss of H4K20me1. By leveraging null and hypomorphic alleles of Set8 and l(3)mbt with our ability to engineer fully histone mutant genotypes (e.g., H4K20A and H4K20R), we demonstrate that the major roles of Set8 and L(3)mbt in gene expression and cell proliferation do not require H4K20me. Our data suggest that phenotypes resulting from mutating H4K20 are due to effects on H4 binding proteins rather than loss of H4K20me.
Results
H4K20me1 is correlated with actively transcribed genes in Drosophila larva
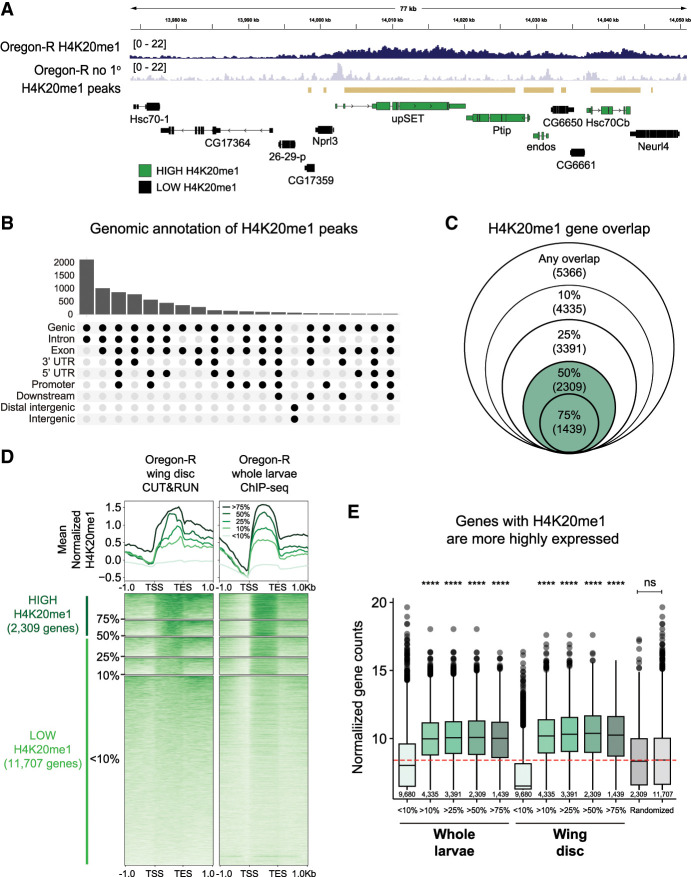
Although H4K20me1 ChIP-seq data sets exist for Drosophila (The modENCODE Consortium et al. 2010; Lv et al. 2016), the genome-wide relationship between H4K20me1 and gene expression has not been elucidated in flies. To determine this relationship, we first performed CUT&RUN genomic occupancy profiling in wild-type Oregon-R third instar wing imaginal discs using an H4K20me1-specific antibody. We measured enrichment of H4K20me1 signal over a no primary antibody control using a sliding window method and found the H4K20me1 signal in broad peaks primarily enriched in gene bodies, consistent with previous studies in mammalian cells (Fig. 1A,B; Barski et al. 2007; Beck et al. 2012). Using a metagene analysis, we found that the H4K20me1 signal overlapped by at least one base pair with 5366 protein-coding genes and that the amount of overlap varied among genes. For instance, 2309 protein-coding genes were covered at least 50% by H4K20me1, and 1439 had 75% coverage (the H4K20me1 “high” category) (Fig. 1C,D). The remaining 11,707 protein-coding genes did not have a H4K20me1 signal that was statistically enriched compared with control or were covered <50% by an H4K20me1 peak (the H4K20me1 “low” category) (Fig. 1C,D). We also found that the proportion of H4K20me1 coverage of each gene correlated positively with the amount of H4K20me1 signal at each gene (Fig. 1D).
Figure 1.
H4K20me1 is enriched in gene bodies in Drosophila melanogaster. (A) Genome browser shot of a representative locus depicting the H4K20me1 CUT&RUN signal. Gold bars indicate peaks of H4K20me1 signal in wild-type Oregon-R relative to no primary antibody control as defined by merged 150 bp sliding windows. (B) Upset plot depicting the frequency of H4K20me1 peaks overlapping specific genomic features. The histogram at the top indicates the number of peaks that overlap each of the genomic features shown below each bar. (C) Venn diagram depicting the number of genes covered by H4K20me1 peaks. The outer circle indicates >1 bp overlap of a gene covered by an H4K20me1 peak, followed by 10%, 25%, 50%, and 75% coverage. Green indicates our H4K20me1 “high” category. (D) Heat map (bottom) and summary metaplot (top) of H4K20me1 coverage in gene bodies scaled to 1 kb as well as ±1 kb unscaled sequence. The plots are organized by gene sets defined in C for Oregon-R wing disc CUT&RUN in this study (left) and for Oregon-R whole-larvae ChIP-seq from the modEncode database (right). Genes with >50% overlap with H4K20me1 peaks are considered high H4K20me1 (dark green; left). Genes with <50% overlap with H4K20me1 are considered low H4K20me1 (light green; left). Each gene row is ordered by mean H4K20me1 in the wing disc data set. Summary plot depicts the mean signal at each position in the metaplot (50 bp bins) for each set of genes. (E) Box plot of average normalized gene counts of each of the gene sets defined in C in whole-larvae RNA-seq (this study), wing disc RNA-seq (Armstrong et al. 2020), or a random set of genes of the indicated size. The red dotted line indicates the median RNA abundance in whole larvae. Significance was determined by Wilcoxon sum rank test with Benjamin–Hochberg multiple testing correction. (****) P < 0.0001 for each indicated set compared with both genes covered by <10% H4K20me1 peak and the randomized gene set.
We then performed RNA sequencing of Oregon-R whole third instar larvae to assess whether H4K20me1 correlates with RNA abundance. Whole-animal RNA-seq was required due to the difficulty of obtaining enough material from individual wing discs of some mutant genotypes (see below). Our H4K20me1 CUT&RUN data in wing disc tissue correlate well with a previous modENCODE H4K20me1 ChIP-seq data set in whole larvae, suggesting that the distribution of H4K20me1 within genes is consistent across tissues (Fig. 1D). We found that genes in the H4K20me1 high category were more highly expressed compared with genes in the H4K20me1 low category in both whole-larval RNA-seq (this study) and wing disc RNA-seq data (Fig. 1E; Armstrong et al. 2020). High H4K20me1 genes and low H4K20me1 genes were also significantly different than random gene sets of equal size (Fig. 1E). Together, these data suggest that H4K20me1 deposition in gene bodies is positively correlated with active transcription in developing Drosophila larvae.
Modification of H4K20 is not required for transcription
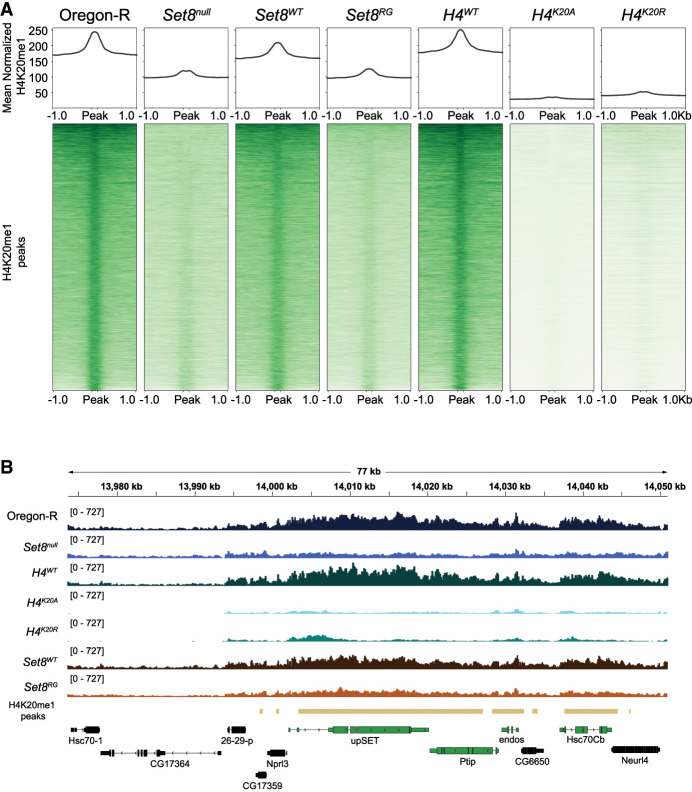
Since H4K20me1 is positively associated with gene expression, we asked whether loss of H4K20me1 would result in downregulation of genes with H4K20me1 by performing H4K20me1-specific CUT&RUN in Set8null third instar wing imaginal discs. Consistent with previous studies, Set8null mutants have a reduction in H4K20me1 levels across gene bodies throughout the genome (Fig. 2A,B; Nishioka et al. 2002; Karachentsev et al. 2005; Beck et al. 2012). Surprisingly, Set8null wing discs from wandering third instar larvae have residual H4K20me1 as assessed by CUT&RUN (Fig. 2A) despite Set8null larval brains lacking H4K20me1 signal by Western blot (Crain et al. 2022). CUT&RUN is a more sensitive assay, and others have reported H4K20me1 in Set8null third instar wandering salivary glands (Karachentsev et al. 2005). We conclude that previous studies using Western blots lacked the sensitivity to detect low H4K20me1 levels in Set8null mutants.
Figure 2.
Loss of H4K20me in Set8 and H4K20 mutants. (A) Heat (bottom) and summary (top) metaplots of spike-in-normalized H4K20me1 signal at H4K20me1 peaks in the indicated genotypes. Each peak is scaled to 200 bp and flanked by 1 kb of unscaled sequence. The summary plot depicts mean signal at each position in the metaplot (50 kb bins) for all peaks. (B) Representative locus depicting H4K20me1 CUT&RUN coverage in wing discs. Gold bars indicate peaks of H4K20me1 signal in Oregon-R relative to no primary antibody control as defined by merged 150 bp sliding windows. Green and black genes indicate H4K20me1 “high” and “low” categories, respectively.
To ask whether the decreased H4K20me1 levels in Set8null mutants were associated with a change in gene expression, we performed total RNA-seq in Set8null larvae and compared these data with the Oregon-R whole-larvae RNA-seq data set described in Figure 1. We chose to assess gene expression in whole mutant larvae even though we measured H4K20me1 in wing discs because Set8null animals have small, morphologically perturbed wing discs containing few cells (Karachentsev et al. 2005), making it challenging to obtain enough high-quality material for RNA-seq even though we obtained enough for CUT&RUN. Moreover, there is no evidence for tissue-specific patterns of H4K20me1 deposition in chromatin (see Fig. 1). We therefore considered the loss of H4K20me1 that we observed in wing discs to be representative of other tissues.
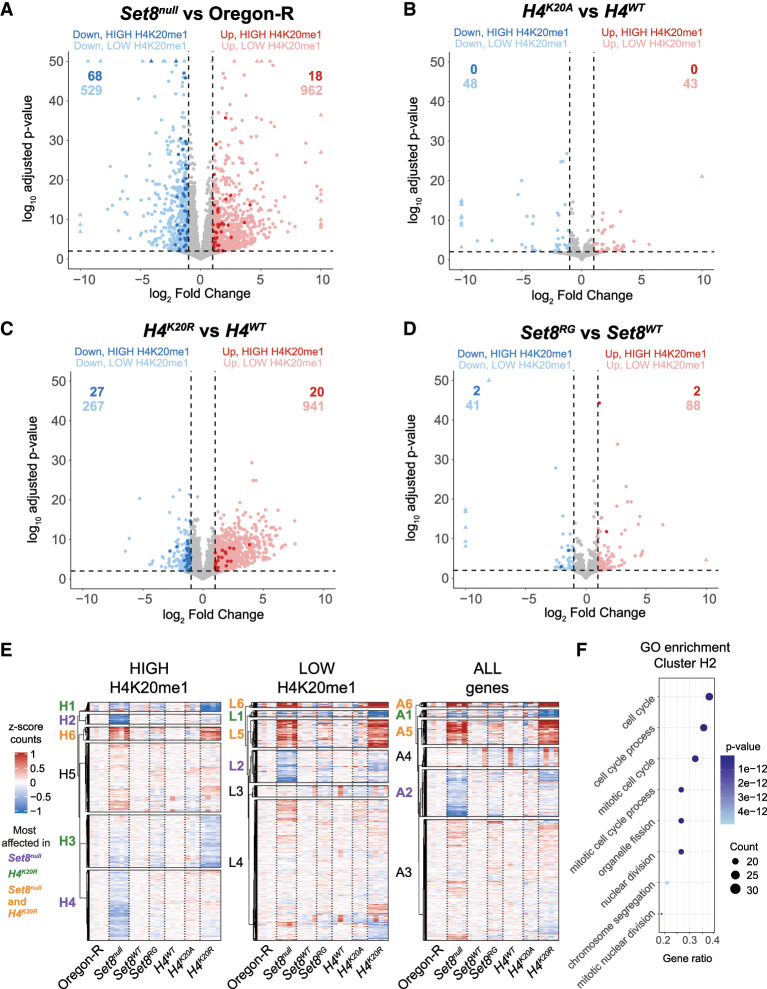
Differential expression analysis of Set8null relative to Oregon-R wild-type control revealed that Set8null animals had 1491 differentially expressed protein-coding genes (DEGs; 962 upregulated and 529 downregulated), indicating that loss of Set8 significantly disrupts the Drosophila transcriptional program (Fig. 3A; Supplemental Table S1). These results correlate well with another recently published RNA-seq data set from Set8null wandering third instar Drosophila larva (Supplemental Fig. S1A; Bamgbose et al. 2024). The majority of DEGs (962; 65% of total DEGs) are upregulated, and only 86 Set8null DEGs (18 upregulated and 68 downregulated) are in the high H4K20me1 category (Fig. 3A, dark dots). These 86 represent 5.8% of all Set8null DEGs and only 3.7% of genes in the high H4K20me1 category, indicating that loss of H4K20me1 is not predictive of either an increased or decreased change in gene expression (Fig. 3A; Supplemental Table S2). We also included in our analyses an RNA-seq data set that we generated from larvae expressing catalytic-deficient Set8 (Set8R634G; referred to here as Set8RG) (Fig. 3D; Crain et al. 2022). Only two genes in the H4K20me1 high category are downregulated in Set8RG despite a reduction in H4K20me1 CUT&RUN levels like that in Set8null (Fig. 2A,B). These data indicate that H4K20me1 does not play a causal, global role in transcriptional control.
Figure 3.
The Set8 and H4K20 mutant transcriptomes differ. (A–D) Volcano plots depicting the relationship between the log2 fold change (x-axis) and log10 adjusted P-value (y-axis) of gene expression in the indicated comparisons. Blue dots indicate significantly downregulated genes (log2FC < −1 and FDR < 0.01), and red dots indicate significantly upregulated genes (log2FC > 1 and FDR < 0.01). Darker-shaded dots indicate genes in the high H4K20me1 category. (E) Clustered heat maps of average centered normalized counts. Cluster identifiers are colored based on whether the expression of genes within that cluster is most affected in Set8null (purple), H4K20R (green), or both Set8null and H4K20R (orange). Black cluster identifiers indicate no enrichment of gene expression changes in any genotype. (F) GO enrichment summary for cluster H2. The top eight GO terms are shown on the y-axis. The x-axis indicates the ratio of genes from cluster H2 that intersect with each GO term over all genes in cluster H2. The size of the dot indicates the number of intersecting genes, and the color indicates the P-value of each term. Full lists of GO terms for clusters in G are in Supplemental Tables S3–S5.
To investigate the role of H4K20me1 in gene expression more directly, we performed H4K20me1-specific CUT&RUN in wing imaginal discs and RNA sequencing of third instar whole larvae in H4K20 mutant (H4K20A and H4K20R) and H4WT control genotypes. Both H4K20A and H4K20R have a strong reduction of H4K20me1 genome-wide despite the presence of replication-independent His4r in these genotypes (Fig. 2A). His4r is a single-copy gene located outside of the replication-dependent histone gene array, and its expression is not replication-coupled but encodes a protein with an amino acid sequence identical to that of replication-dependent H4. We and others have shown previously that His4r can partially compensate for the absence of replication-dependent H4 (Armstrong et al. 2018; Copur et al. 2018; Faragó et al. 2021; Crain et al. 2022). Nevertheless, our H4K20me1 CUT&RUN data indicate that His4r is a minor contributor to H4K20me1 amounts in the genome.
Despite levels of H4K20me1 lower than in Set8null (Fig. 2A), H4K20A animals have only a small number of DEGs (91 total; 43 upregulated and 48 downregulated) relative to control (Fig. 3B; Supplemental Table S1). Remarkably, no H4K20A DEGs are in the high H4K20me1 category (Fig. 3B; Supplemental Table S2). H4K20R animals have a larger number of DEGs (1208 total; 941 upregulated and 267 downregulated) compared with H4K20A (Fig. 3C; Supplemental Table S1). Still, only 3.9% (47) of the H4K20R DEGs are in the high H4K20me1 category, and only 57.4% (27) of those DEGs are downregulated in H4K20R (Supplemental Table S2). Given that H4K20me1 levels are severely depleted in H4K20A and H4K20R animals, these data suggest that transcript levels of most protein-coding genes are not sensitive to reduction of H4K20me1 but are instead sensitive to the residue identity at position 20 on the H4 tail. Mutation of H4K20 results in loss of all lysine modifications, not just H4K20me1. H4K20me3 is enriched in constitutive heterochromatin, which contains transposons and piRNAs (Agredo and Kasinski 2023). Therefore, we evaluated the expression of transposons and piRNAs in H4K20 and Set8null mutants. Whereas loss of Set8 results in derepression of transposons and piRNAs to an extent similar to what we observed previously in H3K9R mutants (Supplemental Fig. S1B,C; Penke et al. 2016), there was no substantial effect in the H4K20 mutants (Supplemental Fig. S1D,E). Together, these data further emphasize that the level of H4K20me1 is not causal for gene expression in either euchromatin or heterochromatin despite H4K20me1 enrichment at transcriptionally active protein-coding genes (Fig. 1). They also emphasize that the differences in both the gene expression and developmental phenotypes of H4K20A and H4K20R mutants (Crain et al. 2022) arise from something unrelated to H4K20 methylation.
H4K20A/R and Set8null gene expression profiles are distinct
Although H4K20me1 is not required for expression of most genes, a subset of genes enriched for H4K20me1 might depend on H4K20me1 and be drivers of gene expression cascades. We addressed this question by comparing normalized gene counts in Set8null, H4K20A, and H4K20R mutants through k-means clustering. By applying this method first to genes in the high H4K20me1 category, we found distinct clusters of downregulated genes—two that were specific to Set8null (clusters H2 and H4) (Fig. 3E) and two containing genes that were most affected in H4K20R (clusters H1 and H3) (Fig. 3E). We also found that most upregulated genes in the high H4K20me1 category were shared between Set8null and H4K20R (cluster H6) (Fig. 3E).
As with the high H4K20me1 category, we found that the low H4K20me1 category of genes contained clusters of downregulated genes that were most affected in Set8null (cluster L2) or H4K20R (cluster L1) (Fig. 3E). We also found clusters that were upregulated in both Set8null and H4K20R but not in H4K20A or Set8RG (clusters L5 and L6) (Fig. 3E), suggesting that changes in the expression of genes in these clusters do not arise from loss of H4K20me1. Moreover, even when we considered all genes independently of H4K20me status, the resulting cluster patterns were remarkably consistent with when we considered only the high or low H4K20me1 categories. Finally, the small number of H4K20A or Set8RG upregulated or downregulated genes did not show a clustering pattern like either Set8null or H4K20R for either the H4K20me1 high or low category. Thus, when comparing transcriptomes using either DEGs or k-means clustering of normalized counts, each mutant displays a distinct gene expression profile that cannot easily be explained by loss of H4K20me.
Since we observed distinct gene expression patterns in Set8null and H4K20R mutants, we were curious whether genes in each of the identified clusters were enriched in specific biological processes. Therefore, we performed gene ontology (GO) enrichment analysis on each DEG cluster and grouped significant GO terms by semantic similarity (Supplemental Figs. S2–S4; Supplemental Tables S3–S5). We were especially interested in clusters H2 and H4 that contained a subset of downregulated genes in the high H4K20me1 category that were specific to Set8null. Cluster H2 is enriched for GO terms related to the cell cycle, cell proliferation, cell differentiation, and development (Fig. 3F; Supplemental Fig. S2; Supplemental Table S3). Cluster H4 is enriched for GO terms related to gene expression, RNA processing/splicing, and histone modification (Supplemental Fig. S2; Supplemental Table S3). Clusters H2 and H4 were also represented in clusters of genes with low H4K20me1 and all genes (L2 and A2), suggesting that these changes in gene expression are H4K20me1-independent (Fig. 3E; Supplemental Figs. S3, S4; Supplemental Tables S4, S5). Clusters H1 and H3, containing genes downregulated primarily in H4K20R, are enriched for GO terms related to ubiquitin-dependent catabolism and development (H1) as well as metabolism, cell differentiation, cell signaling, growth, and development (H3) (Supplemental Fig. S2; Supplemental Table S3). Although both Set8null-specific (H2 and H4) and H4K20R-specific (H1 and H3) clusters are enriched for genes involved in development and cell differentiation, the misregulated genes themselves are distinct. Together, we conclude that the Set8null and H4K20 mutant gene expression profiles are overlapping yet contain unique features and thus are not primarily driven by loss of H4K20me1.
Cell proliferation defects upon removal of Set8 are largely independent of loss of H4K20me
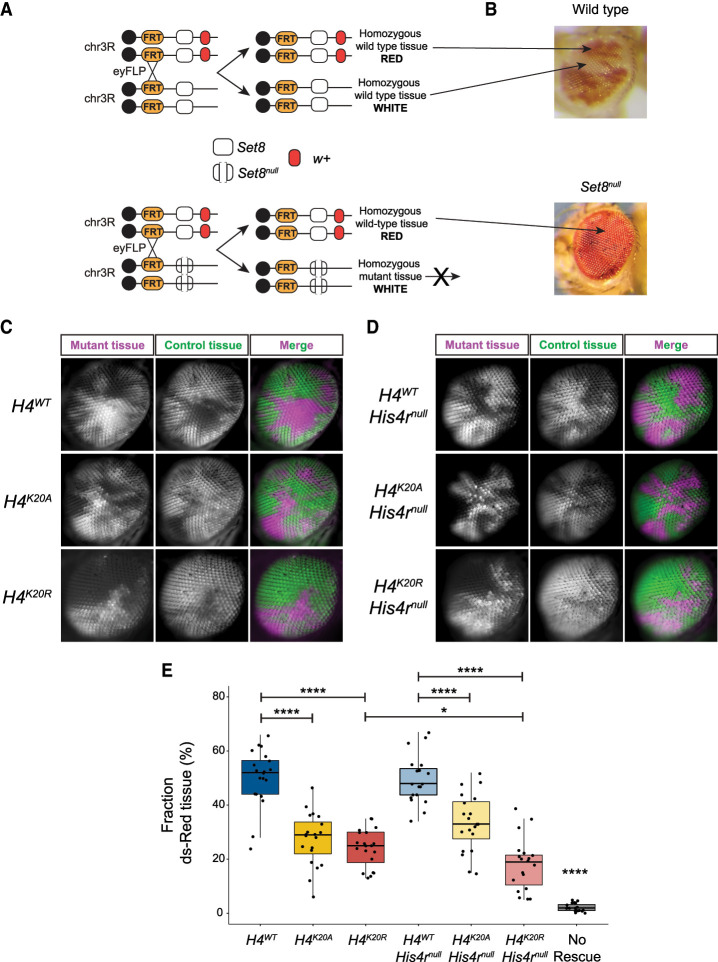
One of the unique features of the Set8null transcriptome is the downregulation of genes involved in the cell cycle (clusters H2, L2, and A2) (Fig. 3E,F). We also found GO terms related to growth in H4K20R (cluster H3) (Fig. 3E). We therefore assessed the cell proliferation capacity of Set8null versus H4K20 mutant cells. We generated mosaic tissue in the Drosophila eye by inducing mitotic recombination with the FLP/FRT system using eyeless-FLP, which expresses FLP recombinase throughout eye development. In this assay, populations of homozygous mutant cells are generated next to populations of homozygous wild-type cells very early in development. During growth of the eye imaginal disc (the precursor of the adult eye), these clones of cells compete to populate the tissue, resulting in patches of fully mutant (white) and fully wild-type (red) tissue in the adult eye (Fig. 4A). In this assay, Set8null mutant cells are never present in the adult eye, indicating a strong proliferation defect upon removal of Set8 (Fig. 4B).
Figure 4.
Set8 but not H4K20 mutant cells fail to proliferate. (A,B) Diagram of w+/w− mosaic eye generation using FLP-FRT-mediated mitotic recombination. (A) The Set8 wild-type (white box) control experiment results in an adult eye with an equal mixture of red and white tissue. (B) The Set8null (open white box) experiment results in only red tissue because homozygous Set8null cells fail to proliferate. (C) Single optical sections of genetically mosaic eyes composed of clonal populations of cells that are homozygous for Act5C-GFP and HisC+ (green) or homozygous for ΔHisCcadillac and lacking Act5C-GFP (magenta) rescued by the indicated control or H4K20 mutant transgenes. (D) Same as C except in a His4rnull mutant background. The right column shows merged images of the pseudocolored histone mutant clones (magenta) and sister histone wild-type clones (green). (E) Quantification of mosaic eye clones in C and D. Box plot showing the fraction of mutant tissue (magenta) for 19–20 eyes of each genotype (10 males and nine to 10 females each). Black dots indicate measurements from individual eyes. Significance was determined by Student's t-test. (****) P < 0.0001, (*) P < 0.05.
Assessing the proliferation capacity of histone mutant cells using white-marked versus red-marked eye clones was not possible using our previously described histone gene replacement system (McKay et al. 2015), primarily because the ΔHisC deletion allele of the endogenous replication-dependent histone genes contains a copy of the white gene. To remedy this problem, we engineered a new histone gene deletion (ΔHisCcadillac) that replaces the white gene with dsRed driven by the ubiquitously expressed Act5C promoter, as well as a wild-type HisC+ chromosome marked with an Act5C-GFP transgene (Crain et al. 2024). Thus, we can assess proliferation of histone mutant cells (magenta) next to wild-type cells (green) during eye development using fluorescence microscopy. We found that ΔHisCcadillac homozygous mutant cells rescued by one copy of a control H4WT transgene proliferated normally, resulting in ∼50% of magenta tissue in the adult eye (Fig. 4C,E).
In contrast to Set8null cells, H4K20A cells were able to proliferate and populate the adult eye but not as well as H4WT control cells (Fig. 4C,E). This result is consistent with our previous observation that H4K20A mutant animals can complete development (Crain et al. 2022). Strikingly, H4K20R cells proliferate similarly to H4K20A cells despite the dramatically different gene expression profiles of these two mutants (Figs. 3B,C, 4C,E). Moreover, a defect in cell proliferation likely does not explain the failure of H4K20R mutant animals to complete development. Furthermore, H4K20A or H4K20R mutant cells lacking the His4r gene, which are therefore incapable of generating any H4K20me, also generated clones of magenta cells, indicating that the proliferation defect of Set8null cells is independent of loss of H4K20me (Fig. 4D,E). Together with our observation that cell proliferation genes are uniquely downregulated in Set8null animals, we conclude that Set8 functions in cell proliferation independently of H4K20me1 in Drosophila, likely via a target other than H4K20.
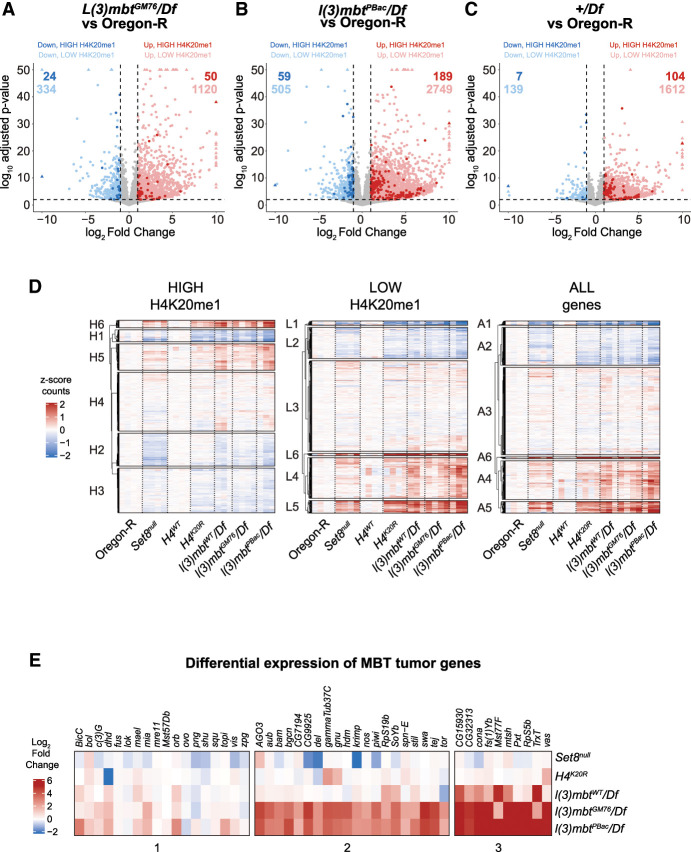
Gene expression changes in l(3)mbt mutants are not explained by H4K20me1 but correlate with gene expression changes in Set8null and H4K20R mutants
Because Set8null and H4K20R have disparate developmental phenotypes and gene expression changes that cannot be explained by loss of H4K20me1, we considered other interpretations of our data. Since Set8 and L(3)mbt bind each other directly (Kalakonda et al. 2008) and interact with the H4 tail, mutation of H4K20 might affect recruitment of these proteins to chromatin irrespective of loss of H4K20me. Thus, we asked whether mutation of l(3)mbt results in expression changes similar to Set8null or H4K20R mutants. We performed RNA sequencing of +/DfED10966 (Df) hemizygous control, l(3)mbtGM76/Df [referred to here as l(3)mbtGM76], and l(3)mbtPBac{Scarless-dsRed}/Df [referred to here as l(3)mbtPBac] mutant whole third instar larvae at 25°C and 29°C, where the l(3)mbt mutant lethality and brain overgrowth phenotypes arise (Bonasio et al. 2010; Janic et al. 2010). l(3)mbtGM76 is a null allele that contains a nonsense mutation in the second of the three L(3)mbt MBT domains (Yohn et al. 2003; Blanchard et al. 2014). l(3)mbtPBac is a CRISPR-engineered null allele that we generated containing a 1.8 kb insertion immediately upstream of the l(3)mbt start codon.
Loss of L(3)mbt function [l(3)mbtGM76 or l(3)mbtPBac] significantly disrupts the Drosophila transcriptome (1519 upregulated and 358 downregulated or 2938 upregulated and 564 downregulated, respectively) (Fig. 5A,B; Supplemental Table S6). The majority of DEGs are upregulated, consistent with the previously reported transcription repressor function of L(3)mbt (Trojer et al. 2007; Kalakonda et al. 2008; West et al. 2010; Coux et al. 2018). As with Set8null and H4K20R, neither the upregulated nor the downregulated DEGs in the l(3)mbt mutants are enriched for high H4K20me1 genes [3.9% in l(3)mbtGM76 and 7.1% in l(3)mbtPBac] (Fig. 5A,B, dark dots; Supplemental Table S7). Unexpectedly, we found that removing only one copy of L(3)mbt in +/Df results in many gene expression changes (1716 upregulated and 146 downregulated) relative to true wild-type Oregon-R, but with smaller effect sizes than l(3)mbtGM76 or l(3)mbtPBac (Fig. 5C). The Df is relatively small, deleting only 28 kb, including seven genes in addition to l(3)mbt: woc, mrt, TfIIA-L, CG5934, CG5938, CG14260, and CG14262. We found minimal differences in the transcriptomes of l(3)mbtGM76 versus a +/Df, suggesting that the effect that we observed in +/Df is due to haploinsufficiency of l(3)mbt. We only found a small number of gene expression changes in animals raised at 29°C relative to animals raised at 25°C despite a decrease in viability and brain tissue overgrowth at 29°C (Meier et al. 2012), suggesting that the phenotypes associated with heat stress are not due to large changes in gene expression.
Figure 5.
l(3)mbt, Set8, and H4K20R transcriptomes are similar. (A–C) Volcano plots depicting the relationship between log2FC (x-axis) and log10 adjusted P-value (y-axis) of gene expression in the indicated comparisons. Blue dots indicate significantly downregulated genes (log2FC < −1 and FDR < 0.01), and red dots indicate significantly upregulated genes (log2FC > 1 and FDR < 0.01). Darker-shaded dots indicate genes in the high H4K20me1 category. (D) Clustered heat maps of average centered normalized counts in the indicated genotypes. (E) Heat map of log2FC values of the indicated genotypes for genes involved in MBT formation (Janic et al. 2010).
To ask whether loss of L(3)mbt could explain gene expression changes in Set8null and H4K20R mutants, we compared normalized gene counts in l(3)mbt mutants with Set8null and H4K20R mutants using k-means clustering. We found several clusters of genes that had similar expression in l(3)mbt, Set8null, and H4K20R regardless of H4K20me1 status (Fig. 5D), suggesting that a subset of gene expression changes in Set8null or H4K20R results from altered L(3)mbt function. We observed several clusters that contained genes that were upregulated in each of the Set8null, H4K20R, and l(3)mbt mutants, suggesting that expression of these genes shares a common mechanism (Fig. 5D). Janic et al. (2010) identified a group of 48 genes that were upregulated in l(3)mbt loss-of-function mutants, resulting in tumors with a characteristic soma-to-germline transition phenotype. We investigated whether this group of genes was among the upregulated genes in our l(3)mbt mutants and whether Set8null and H4K20R shared these gene expression changes. We found that the majority of l(3)mbt tumor genes are upregulated in our l(3)mbt mutant data sets (even +/Df) but are not significantly changed in Set8null or H4K20R mutants (Fig. 5E). Therefore, disruption of L(3)mbt function from loss of H4K20me1 likely does not contribute substantially to the overgrowth phenotypes associated with MBTs.
L(3)mbt binds the genome independently of H4K20me
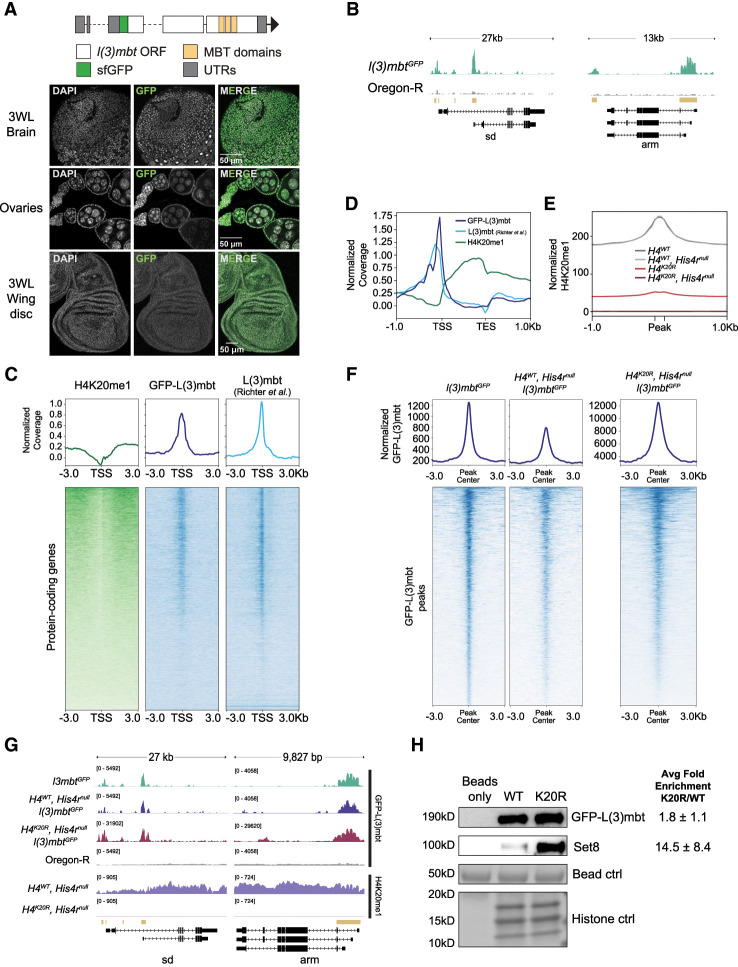
Shared gene expression changes in Set8null, H4K20R, and l(3)mbt mutants might result from loss of chromatin binding proteins, such as L(3)mbt. The L(3)mbt MBT domains preferentially bind to H4K20me1/2 peptides in vitro (Li et al. 2007; Min et al. 2007; Trojer et al. 2007), although recent in vivo studies found that L(3)mbt ChIP-seq peaks colocalize with other methylated histone lysines more than H4K20me1 (Blanchard et al. 2014). We asked whether L(3)mbt recruitment to the genome requires H4K20me using our H4K20R, His4rnull genotype lacking all modifiable H4K20. Since no L(3)mbt antibodies were available to us, we engineered N-terminal GFP- and FLAG-tagged alleles at the endogenous l(3)mbt locus to assess genome-wide binding of L(3)mbt (Fig. 6A). Animals expressing only GFP- or FLAG-tagged L(3)mbt are viable and display no obvious phenotypic abnormalities. Using anti-GFP-FLAG or anti-FLAG antibodies, we found that our epitope-tagged L(3)mbt proteins are expressed in third instar larval brains, adult ovaries, and wing imaginal discs in patterns and at levels consistent with previous studies (Fig. 6A; Richter et al. 2011; Meier et al. 2012; Coux et al. 2018; Yamamoto-Matsuda et al. 2022).
Figure 6.
L(3)mbt binds the genome independently of H4K20me. (A, top) Diagram of a GFP-tagged l(3)mbt allele generated using CRISPR. (Bottom) Confocal images of GFP-L(3)mbt accumulation in the nuclei of third instar larval brains, adult female ovaries, and third instar larval wing discs. (B) A representative locus depicting normalized GFP-L(3)mbt CUT&RUN coverage. Gold bars indicate peaks of GFP signal in l(3)mbtGFP (top) relative to Oregon-R control (bottom) as defined by merged 150 bp sliding windows. (C) Heat (bottom) and summary (top) metaplots of z-normalized wing disc H4K20me1 CUT&RUN, wing disc GFP-L(3)mbt CUT&RUN, and L(3)mbt ChIP-seq (Richter et al. 2011) signal at protein-coding genes. Plots are centered at the TSSs and flanked by 3 kb of unscaled sequence. Each row represents a single gene, and genes are ordered by mean H4K20me1. The summary plot depicts mean signal for H4K20me1 (green), GFP-L(3)mbt (blue), or L(3)mbt (light blue) at each position in the metaplot (50 bp bins). (D) Summary metaplot of z-normalized H4K20me1 (green), GFP-L(3)mbt CUT&RUN (blue), and L(3)mbt ChIP-seq (light blue) at high H4K20me1 genes. (E) Summary metaplot of spike-in-normalized H4K20me1 CUT&RUN coverage at H4K20me1 peaks in the indicated genotypes. Each peak is scaled to 200 bp and flanked by 1 kb of unscaled sequence. The summary plot depicts mean signal at each position in the metaplot (50 bp bins) for all peaks. (F) Summary (top) and heat (bottom) metaplots of spike-in-normalized GFP-L(3)mbt CUT&RUN signal. Plots are centered at the GFP-L(3)mbt peak, and each peak is flanked by 3 kb of unscaled sequence. Each row represents a single peak, and peaks are ordered by mean GFP-L(3)mbt in l(3)mbtGFP. (G) Representative locus depicting spike-in-normalized GFP-L(3)mbt and H4K20me1 CUT&RUN signal. Gold bars indicate peaks of GFP signal in l(3)mbtGFP relative to Oregon-R control as defined by merged 150 bp sliding windows. (H) Western blot using anti-GFP (top) or anti-Set8 (middle) antibodies following recombinant nucleosome binding assay with nuclear lysate from third instar larvae and either wild-type (WT) or H4K20R (K20R) recombinant nucleosomes. (Bottom) Histone proteins from Swift protein stain were used as loading control. Average binding of Set8 and GFP-L(3)mbt on K20R versus WT nucleosomes ±standard deviation from three biological replicates is shown at the right.
We performed CUT&RUN in third instar wing imaginal discs that express only GFP-L(3)mbt from the endogenous locus. We identified regions of the genome bound by L(3)mbt by measuring enrichment of GFP-L(3)mbt signal over an Oregon-R control (which lacks GFP). We found that L(3)mbt accumulates at 4052 well-defined peaks across the genome that are preferentially located at transcription start sites (TSSs) of genes enriched with H4K20me1 (Fig. 6B–D). Despite the correlation with H4K20me1-enriched genes, L(3)mbt is not enriched in gene bodies, unlike H4K20me1 (Fig. 6D). To corroborate this observation, we also analyzed the location of peaks in a previously published L(3)mbt ChIP-seq data set from third instar larval brains and found that L(3)mbt is enriched at transcription start sites of high H4K20me1 genes in both data sets (Fig. 6C,D).
We next performed anti-GFP-L(3)mbt CUT&RUN in a genotype that lacked all modifiable H4K20 (H4K20R, His4rnull). We found that H4K20R, His4rnull third instar wing discs have a genome-wide ablation of H4K20me1, while removal of His4r alone (H4WT, His4rnull) does not detectably affect total H4K20me1 levels (Fig. 6E). Since H4K20me increases the affinity of L(3)mbt for H4 tail peptides in vitro (Li et al. 2007; Min et al. 2007; Trojer et al. 2007), we hypothesized that loss of modifiable H4K20 would result in a reduction of GFP-L(3)mbt signal at GFP-L(3)mbt peaks. We observed that GFP-L(3)mbt is recruited to the genome at most of its binding sites when all H4K20 is mutated to Arg (Fig. 6F,G). Unexpectedly, we also observed a >10-fold increase of GFP-L(3)mbt binding across all peaks in H4K20R, His4rnull compared with H4WT, His4rnull (Fig. 6F,G). These data indicate that mutation of H4K20 to Arg results in altered, rather than the prevention of, accumulation of GFP-L(3)mbt on chromatin.
To determine how GFP-L(3)mbt directly associates with H4K20R nucleosomes, we performed in vitro binding assays using H4K20R recombinant nucleosomes (Skrajna et al. 2020). We found that GFP-L(3)mbt exhibited a modest increase (1.8-fold ± 1.1-fold) in binding to H4K20R nucleosomes compared with WT nucleosomes (Fig. 6H). Interestingly, we also observed a 14.5-fold (±8.4-fold) increase in the binding of Set8 to H4K20R nucleosomes relative to WT nucleosomes (Fig. 6H). These data suggest that the Lys-to-Arg substitution results in neomorphic effects likely due to increased H4 tail binding of L(3)mbt, Set8, and possibly other proteins, a phenomenon also found in oncohistone mutations such as H3K27M (Sahu and Lu 2022). Together, these data indicate that H4K20me is not necessary for L(3)mbt to bind chromatin in vivo.
Discussion
By combining genetic and genomic approaches in Drosophila melanogaster, we provide evidence that H4K20me1 is dispensable for key functions of Set8, including regulation of gene expression and cell proliferation, that have been previously attributed to H4K20me1. We also demonstrate that L(3)mbt functions in gene expression independently of binding H4K20me.
Set8 and L(3)mbt function in gene expression independent of H4K20me1
Our data do not support a model in which the primary functions of Set8 and L(3)mbt are mediated through the deposition and recognition of H4K20me1, respectively. We found that Set8- and l(3)mbt-null mutations have a greater effect on the Drosophila transcriptome than Set8RG, H4K20A, or H4K20R mutants. Remarkably, Set8RG and H4K20A mutants have minimal gene expression changes (none of which are in genes with the highest coverage of H4K20me1) despite a strong genome-wide reduction in H4K20me1. More genes change in expression in H4K20R mutants than in H4K20A mutants, but many of these changes do not correlate with gene expression changes in Set8null. Moreover, we demonstrate that cells lacking all modifiable H4K20 (H4K20A, His4rnull or H4K20R, His4rnull) can proliferate (in stark contrast to Set8null cells, which cannot), indicating that Set8 but not its catalytic activity on H4K20 is required for cell proliferation. Thus, the Set8 requirement for cell proliferation may reflect a noncatalytic role, which is increasingly being recognized as a feature of many histone-modifying enzymes (Morgan and Shilatifard 2023).
Similarly, we found that H4K20me is dispensable for L(3)mbt recruitment to the genome, including promoter-associated peaks. Methylated lysine residues other than H4K20me could help recruit L(3)mbt to specific promoters, consistent with recent work reporting L(3)mbt coenrichment with several different methylated histone residues at promoters (Blanchard et al. 2014). A similar situation occurs with other MBT domain-containing proteins such as Sfmbt (Klymenko et al. 2006). Instead, H4K20me could be required for positioning of L(3)mbt on chromatin or chromatin compaction after being recruited via other mechanisms (Li et al. 2007; Min et al. 2007; Trojer et al. 2007; Blanchard et al. 2014). We conclude that many, if not most, of the critical cellular and developmental functions of Set8 and L(3)mbt in Drosophila are likely mediated through targets other than H4K20.
The analysis of histone mutations in Drosophila has often supported a role for histone PTMs in gene regulation, most notably in the case of H3K27 mutations that disrupt Polycomb-mediated repression because of the failure to deposit H3K27me3 (Pengelly et al. 2013; McKay et al. 2015). However, in other cases, histone mutation (i.e., of H2A to block ubiquitylation) does not recapitulate the expected effect on Polycomb-mediated gene repression (Pengelly et al. 2015). Nevertheless, we were surprised to find that H4K20me1 had little impact on gene expression given that H4K20me1 is enriched in the bodies of expressed genes in both flies (this study; Lv et al. 2016) and humans (Barski et al. 2007; Congdon et al. 2010; Beck et al. 2012). Indeed, H4K20me1 is among the histone PTMs most highly correlated with active transcription (Wang et al. 2008). Set8 interacts with elongating RNA polymerase II (Li et al. 2011), so H4K20me1 accumulation in gene bodies may be a consequence of the Set8/RNA Pol II interaction without playing a regulatory role in transcription. Although we did not observe gene expression changes associated with mutation of H4K20 in our whole-larvae data sets, our analysis could lack the power to detect H4K20me1-dependent tissue-specific gene expression changes. For instance, H4K20me1 covers genes that are uniquely expressed in specific cell lines (Beck et al. 2012). H4K20me1 might promote a permissive environment in which tissue-specific transcription factors function more efficiently or help counteract other repressive chromatin domains. For instance, Lv et al. (2016) observed that Pc+, H3K27me3+, and H4K20me1+ genes were expressed, whereas Pc+, H3K27me3+, and H4K20me1− genes were repressed. Another proposed role of H4K20me1 is recruitment of the MSL complex to the TSSs to release paused polymerase into productive elongation (Kapoor-Vazirani and Vertino 2014; Nikolaou et al. 2017). Since all our data were collected from female larvae, we were not able to address this mechanism. Nevertheless, our data demonstrate that H4K20me has no uniform genome-wide regulatory role, either positively or negatively, in gene expression in Drosophila.
Mechanisms of H4K20 methylation
We detected low levels of H4K20me1 in Set8null animals by CUT&RUN, which was unexpected, as Set8 is currently the only known H4K20 monomethyltransferase, and previously we did not detect H4K20me1 signal by Western blot in Set8null mutants (Crain et al. 2022). We detected no H4K20me1 CUT&RUN signal in H4K20R, His4rnull animals, which contain no modifiable H4K20, indicating that the H4K20me1 antibody does not recognize other histone PTMs. Although our data cannot exclude that the H4K20me1 antibody binds also to unmodified H4K20, they raise the possibility of another H4K20 monomethyltransferase in flies. Karachentsev et al. (2005) also reported residual H4K20me1 in Set8null salivary glands by immunofluorescence, which they hypothesized was due to another H4K20 monomethyltransferase or stabilization of the monomethyl mark over multiple cell divisions. One possibility is that the H4K20 dimethyltransferase and trimethyltransferase Suv4-20 can generate H4K20me1 in the absence of Set8. Three studies reported that Suv4-20 preferentially uses H4K20me1 as a substrate in vitro but can also produce H4K20me1 (Yang et al. 2008; Southall et al. 2014; Weirich et al. 2016). Another possibility is the production of H4K20me1 by demethlyation of long-lived pools of H4K20me2,3 that are derived from maternally deposited Set8 in Set8null mutant animals. Although not yet identified in flies, H4K20 demethylase enzymes are present in other metazoans, including PHF8 in zebrafish and mammals and DPY-21 in Caenorhabditis elegans (Qi et al. 2010; Gu et al. 2016; Brejc et al. 2017). Regardless of the source of this small amount of H4K20me1 in Set8null mutants, our results clearly indicate that Set8 is responsible for the bulk of H4K20me1 in Drosophila.
The H4K20A and H4K20R mutant phenotypes differ
H4K20me1 has been implicated in many essential nuclear processes, and thus our previous observation that animals expressing only unmodifiable H4K20A histones can complete development was surprising (Crain et al. 2022). This result is consistent with the small number of gene expression changes in H4K20A mutants that we report here. However, only 20% of H4K20A animals and no H4K20R animals, reach adulthood, indicating that H4K20 is important for development (Crain et al. 2022). The more extensive gene expression changes in H4K20R animals could explain why they die during development. These changes do not overlap substantially with gene expression changes in Set8null mutants. In contrast, we observed similar gene expression profiles in H4K20R and l(3)mbt mutants. These changes are not driven by loss of H4K20me but rather by mutating histone H4K20 to Arg. Thus, these data indicate that H4K20me is not essential for Drosophila development but do not explain the phenotypic differences between H4K20A and H4K20R mutants or which processes are affected by mutating H4K20.
We suspect that the Lys-to-Ala and Lys-to-Arg amino acid changes in the H4 tail disrupt or change the interaction of nucleosome binding proteins irrespective of the absence of H4K20me. These proteins include Set8 and L(3)mbt, as we observed binding of L(3)mbt to both the H4K20R mutant genome and purified H4K20R nucleosomes, as well as increased binding of Set8 to purified H4K20R nucleosomes. Unfortunately, we were unable to produce H4K20A protein for reasons that are not clear, precluding our ability to test protein binding to purified H4K20A mutant nucleosomes. H4K20 mutations may also affect the H4 tail conformation and thus the availability in vivo of the H4 tail to chromatin binding proteins. Regardless, alteration of Set8 or L(3)mbt H4 tail binding alone is insufficient to explain the phenotypes of Set8null and l(3)mbt mutant animals because they are so different from the H4K20 mutant phenotypes (Crain et al. 2022). Future determination of the H4 tail and/or broader nucleosome interactome in H4K20A and H4K20R mutants should be informative in this regard.
Our study illustrates the power of using genomic analyses in a genetically tractable organism like Drosophila melanogaster to deconvolve the complex relationship between a writer (Set8) and a reader [L(3)mbt] of a particular histone PTM (H4K20me). Notably, as the chromatin field continues to identify pleiotropic functions of writer and reader proteins, revisiting other histone PTM/writer/reader paradigms using a similar type of analysis may prove fruitful in untangling the functions of histone PTMs in various biological processes.
Materials and methods
GFP-L(3)mbt immunofluorescence
Cuticles from l(3)mbtGFP larvae were inverted and fixed in 3.7% paraformaldehyde for 25 min, washed, and then blocked in 500 μL of 5% NGS in PBS for 30 min prior to staining with α-GFP Rb primary antibody (1:1000; Abcam ab6556) overnight at 4°C followed by rabbit Alexa 488 secondary antibody (1:1000) for 2 h at room temperature. Tissues were dissected off cuticles, mounted on a glass slide with a glass coverslip in 11 μL of Prolong, and left in the dark overnight before imaging on a Leica SP8 confocal microscope. Ovaries were dissected from 3 day old adult females, fixed, and stained as above.
Larvae collection and RNA extraction
Four replicates of eight third instar wandering larvae of each genotype were homogenized in TRIzol (Invitrogen) and flash-frozen in liquid nitrogen. RNA was isolated using the Direct-zol RNA minipreparation kit (Zymo).
RNA-seq library preparation and sequencing
RNA-seq libraries were prepared using the Universal Plus total RNA-seq with NuQuant (Tecan). The number of cycles required for library amplification was determined empirically using qPCR. DNA concentrations, fragment size distributions, and quality were determined using Qubit and Agilent TapeStation 4150. Libraries were pooled and sequenced PE100 on a NovaSeq.
Analysis of RNA-seq data
Paired-end FASTQ files from three to four replicates of each genotype were trimmed using bbduk and then passed to the quant function within Salmon (Patro et al. 2017) in mapping-based mode with the parameters validateMappings, seqBias, eVBOpt, and numBootstraps 30 and protein-coding transcript indexes and decoys that were generated from the dm6 Drosophila genome build. Quant files were imported into R using Txiimport (Soneson et al. 2015). Genotypes for all replicates were confirmed using sequencing data. Replicates with reads that did not match the intended genotype were discarded and not used for further analyses. For Figure 1E, counts for Oregon-R whole larvae (this study) and yw wing disc (Armstrong et al. 2020) were normalized using DESeq2 with a variance stabilizing transformation. Normalized counts from all replicates of a given genotype were averaged, and genes were binned based on percentage of H4K20me1 coverage and then plotted using ggboxplot from the ggpubr R package. Differential expression analysis was performed using DESeq2 (Love et al. 2014). Log2FC values were shrunk using the ashr method (Stephens 2017). Volcano plots and box plots were generated using ggplot (Wickham 2016).
CUT&RUN
Twenty third instar larval wing discs per replicate were dissected and processed as previously described (Uyehara et al. 2022) with rabbit α-GFP (1:100; Rockland 600-401-215) or mouse α-H4K20me1 (1:100; Thermo Fisher MA5-18067) and pAG-MNase (1:100; University of North Carolina Core Facility) (Salzler et al. 2023).
Library preparation and sequencing
DNA libraries of supernatant fractions were prepared using a ThruPLEX DNA-seq kit (Takara) and DNA Unique dual-index kit with associated protocols. DNA concentration, fragment distribution, and quality were determined by Qubit and Agilent TapeStation 4150. Libraries were pooled and sequenced PE75 on a NextSeq2000.
CUT&RUN sequencing data analysis
Data processing
Replicates from each genotype were processed using a Snakemake pipeline (https://github.com/snystrom/cutNrun-pipeline.git). Adapters were trimmed with bbduk, reads were aligned to dm6 with bowtie2 (Langmead and Salzberg 2012) and converted to Bam format, reads with a quality score of <30 for GFP CUT&RUN and a quality score of 5 for H4K20me1 CUT&RUN were removed via samtools (Li et al. 2009), and duplicate reads were kept for GFP CUT&RUN and removed for H4K20me1 CUT&RUN. Bam files were generated, sorted, and converted to bed format with bedtools (Quinlan and Hall 2010)
Peak calling
The following was performed using the csaw package (Lun and Smyth 2016) unless stated otherwise. Reads from Oregon-R H4K20me1 and Oregon-R no primary antibody were binned into 150 bp windows with a 50 bp slide. Background was calculated by binning reads into large 10 kb windows, and 150 bp windows were retained only if they were log2(2) higher than background. Window counts for each replicate were normalized for compositional bias, and differential binding was assessed using edgeR (Robinson et al. 2010). Windows that were significantly enriched (log2FC > 2 and FDR < 0.05) in Oregon-R H4K20me1 over Oregon-R no primary antibody were merged if they were within 1 kb of each other and FDR was <0.05. Subsequent merged windows were saved to bed format and used for downstream analyses. Peaks in GFP-L(3)mbt over Oregon-R negative control were determined as in H4K20me1 with the following exceptions: (1) They did not meet the log2FC of >3 significance cutoff and (2) peaks within 250 bp were merged.
Average signal plots
For Figure 1D, coverage files for three biological replicates of H4K20me1 Oregon-R wing disc and no primary antibody Oregon-R CUT&RUN and two biological replicates of H4K20me1 Oregon-R whole-larvae ChIP-seq and input controls (GSE47254) (The modENCODE Consortium et al. 2010) were normalized by reads per genome content (RPGC) and averaged using deeptools BigWigAverage (Ramírez et al. 2016). The ratio of each averaged H4K20me1 file over control was calculated using deeptools BigWigCompare (Ramírez et al. 2016) and then z-normalized. For Figure 6, C and D, coverage files for three biological replicates of Oregon-R H4K20me1 and Oregon-R no primary control wing disc CUT&RUN, two biological replicates of l(3)mbtGFP and Oregon-R control wing disc CUT&RUN, and two biological replicates of l(3)mbt and input wild-type brain ChIP-seq (GSE29206) (Richter et al. 2011) were processed as above.
For Figures 2A and 6E, individual genome coverage files (three replicates; two each for H4WT, H4K20A, and H4K20R, His4rnull) (see below) for each genotype were normalized using yeast spike-in DNA by the spike-in-normalized reads per million mapped reads in the negative control (SRPMC) method (DeBerardine et al. 2023). The ratio of fly to yeast reads was calculated (reads per spike-in [RPS]), RPS values between each sample and its control were calculated (relative signal), and the relative signal for each sample was RPGC-scaled. The SRPMC scaling factor was used in bedtools genomeCoverage (Quinlan and Hall 2010) to produce scaled bedgraph files, which were subsequently converted to BigWig coverage files using ucsctools wigToBigWig. One replicate of H4WT was removed due to an exceptionally high yeast spike-in count, one replicate of H4K20A was removed because it contained wild-type reads at H4K20, and one replicate of H4K20R, His4rnull was removed due to an exceptionally low read count. The remaining replicates of each genotype were averaged using deeptools bigWigAverage (Ramírez et al. 2016) using 1 bp bins. Metaplots were generated using the computeMatrix and plotHeatmap or plotProfile functions in deeptools (Ramírez et al. 2016). Two biological replicates of each l(3)mbt genotype for α-GFP CUT&RUN were processed as above.
Mitotic eye clone generation and quantification
eyFLP; Actin5C-GFP/CyO; + or eyFLP; Actin5C-GFP/CyO; His4rnull females were crossed to yw; ΔHisCcadillac/CyO; 12xHistoneTransgene (HTG) or yw; ΔHisCcadillac/CyO; 12xHTG, His4rnull males. Nineteen to 20 adults (nine to 10 males and females each) were aged 1–2 days after eclosion, placed in a 96 well dish containing molten 1% agarose, and cooled to solidify. Images were obtained on a Leica M205 FCA fluorescent microscope using GFP and RFP bandpass filters. Quantification of mutant clone size was determined as described by Crain et al. (2024).
Recombinant nucleosome binding assays
Nuclei from 100–120 l(3)mbtGFP third instar wandering larvae were collected as previously described (Leatham-Jensen et al. 2019). Nucleus pellets were resuspended in 500 µL of BB420 buffer (20 mM HEPES at pH 7.5, 420 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 10% glycerol, 0.1% NP-40), homogenized 10 times with a Dounce homogenizer, and then centrifuged at 17,000g for 30 min. The supernatant was moved to a new 1.5 mL tube and brought to a final salt concentration of 120 mM (BB120). The H4K20R mutant was cloned using site-directed mutagenesis. Nucleosomes containing FLAG-tagged H2A and either WT H4 or H4K20R were assembled, and binding assays were performed as previously described (Skrajna et al. 2020). Briefly, 20 µL of anti-FLAG-conjugated magnetic beads (Millipore Sigma) was incubated with 20 µg of FLAG-tagged WT or H4K20R nucleosomes or an equivalent volume of BB120 (beads-only control) for 2 h at 4°C. Beads were washed three times with BB120. The nuclear lysate was split equally between the three tubes, and the volume was increased to 500 μL with BB120 prior to incubation for 2 h at 4°C. Beads were washed twice with BB120, followed by a 30 min wash in BB120 at 4°C. Following the last wash, bound proteins were eluted with 25 µL of gel loading buffer, and samples were boiled for 5 min. Samples were run on a 4%–20% gradient SDS-PAGE gel (Bio-Rad) and transferred to a nitrocellulose membrane (Bio-Rad Quick transfer). Total protein staining was performed using Swift protein stain (G-Biosciences). Membranes were blocked in 5% milk in TBS-Tween for 1 h prior to incubation with α-GFP (1:1000; Rockland) or α-Set8 (1:1000; Novus Biologicals) primary antibody overnight at 4°C, followed by rabbit HRP secondary antibody (1:10,000) for 1 h at room temperature. Blots were incubated in SuperSignal West Pico chemiluminescent substrate (Fisher) for 5 min and then imaged on an Amersham imager (GE). The entire blot was autoscaled for brightness/contrast prior to quantification by densitometry in Fiji.
Data availability
High-throughput sequencing data sets have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE268819, GSE268820, and GSE268821. The code for peak calling and differential expression analysis is available at https://zenodo.org/doi/10.5281/zenodo.11550660.
Supplementary Material
Acknowledgments
We thank Markus Nevil and Harmony Salzler for critical reading of the manuscript; Gabrielle Budziszewski for production of the H4K20R histone plasmid; Alexsandra Skrajna for providing reagents, protocols, and troubleshooting advice for nucleosome binding assays; and the University of North Carolina (UNC) High-Throughput Sequencing Facility sequencing core for RNA-seq. This work was supported by National Institutes of Health grants T32GM007092 to A.T.C., R35GM133498 to R.K.M., and R35GM145258 to R.J.D., and in part by a UNC Creativity Hub Award to Corbin Jones.
Author contributions: A.T.C. and R.J.D. conceptualized the study. A.T.C. and M.B.B. performed the experiments and data analysis. C.A.H. contributed to CUT&RUN and RNA-seq experiments. A.T.C. and M.B.B. performed bioinformatic analyses. R.K.M. provided expertise, troubleshooting advice, and reagents for nucleosome binding assays. M.H. assembled recombinant nucleosomes and performed extensive troubleshooting. A.T.C. and R.J.D. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351698.124.
Competing interest statement
The authors declare no competing interests.
References
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. 2010. CRL4Cdt2 regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21. 10.1016/j.molcel.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agredo A, Kasinski AL. 2023. Histone 4 lysine 20 tri-methylation: a key epigenetic regulator in chromatin structure and disease. Front Genet 14: 1243395. 10.3389/fgene.2023.1243395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RL, Penke TJR, Strahl BD, Matera AG, McKay DJ, MacAlpine DM, Duronio RJ. 2018. Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila. Genome Res 28: 1688–1700. 10.1101/gr.239913.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RL, Das S, Hill CA, Duronio RJ, Nordman JT. 2020. Rif1 functions in a tissue-specific manner to control replication timing through its PP1-binding motif. Genetics 215: 75–87. 10.1534/genetics.120.303155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamgbose G, Bordet G, Lodhi N, Tulin A. 2024. Mono-methylated histones control PARP-1 in chromatin and transcription. Elife 13: RP91482. 10.7554/eLife.91482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D. 2012. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 26: 325–337. 10.1101/gad.177444.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard DP, Georlette D, Antoszewski L, Botchan MR. 2014. Chromatin reader L(3)mbt requires the Myb-MuvB/DREAM transcriptional regulatory complex for chromosomal recruitment. Proc Natl Acad Sci 111: E4234–E4243. 10.1073/pnas.1416321111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Reinberg D. 2010. MBT domain proteins in development and disease. Semin Cell Dev Biol 21: 221–230. 10.1016/j.semcdb.2009.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, Bian Q, Uzawa S, Wheeler BS, Anderson EC, King DS, Kranzusch PJ, Preston CG, Meyer BJ. 2017. Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell 171: 85–102.e23. 10.1016/j.cell.2017.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustel J, Kirstein N, Izard F, Grimaud C, Prorok P, Cayrou C, Schotta G, Abdelsamie AF, Déjardin J, Méchali M, et al. 2017. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J 36: 2726–2741. 10.15252/embj.201796541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SG. 2013. Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem Sci 38: 243–252. 10.1016/j.tibs.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC. 2010. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem 110: 609–619. 10.1002/jcb.22570 [DOI] [PubMed] [Google Scholar]
- Copur Ö, Gorchakov A, Finkl K, Kuroda MI, Müller J. 2018. Sex-specific phenotypes of histone H4 point mutants establish dosage compensation as the critical function of H4K16 acetylation in Drosophila. Proc Natl Acad Sci 115: 13336–13341. 10.1073/pnas.1817274115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux R-X, Teixeira FK, Lehmann R. 2018. L(3)mbt and the LINT complex safeguard cellular identity in the Drosophila ovary. Development 145: dev160721. 10.1242/dev.160721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain AT, Klusza S, Armstrong RL, Santa Rosa P, Temple BRS, Strahl BD, McKay DJ, Matera AG, Duronio RJ. 2022. Distinct developmental phenotypes result from mutation of Set8/KMT5A and histone H4 lysine 20 in Drosophila melanogaster. Genetics 221: iyac054. 10.1093/genetics/iyac054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain AT, Nevil M, Leatham-Jensen MP, Reeves KB, Matera AG, McKay DJ, Duronio RJ. 2024. Redesigning the Drosophila histone gene cluster: an improved genetic platform for spatiotemporal manipulation of histone function. Genetics (in press). [Google Scholar]
- DeBerardine M, Booth GT, Versluis PP, Lis JT. 2023. The NELF pausing checkpoint mediates the functional divergence of Cdk9. Nat Commun 14: 2762. 10.1038/s41467-023-38359-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulev S, Tkach J, Lin S, Batada NN. 2014. SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Rep 15: 1163–1174. 10.15252/embr.201439434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faragó A, Ürmösi A, Farkas A, Bodai L. 2021. The histone replacement gene His4r is involved in heat stress induced chromatin rearrangement. Sci Rep 11: 4878. 10.1038/s41598-021-84413-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Hitzel J, Moll F, Kruse C, Malik RA, Preussner J, Looso M, Leisegang MS, Steinhilber D, Brandes RP, et al. 2016. The histone demethylase PHF8 is essential for endothelial cell migration. PLoS One 11: e0146645. 10.1371/journal.pone.0146645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y, Hayashi Y, Hirano Y, Miyawaki-Kuwakado A, Ohkawa Y, Obuse C, Kimura H, Haraguchi T, Hiraoka Y. 2021. Chromatin loading of MCM hexamers is associated with di-/tri-methylation of histone H4K20 toward S phase entry. Nucleic Acids Res 49: 12152–12166. 10.1093/nar/gkab1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. 2011. Histone modification: cause or cog? Trends Genet 27: 389–396. 10.1016/j.tig.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Huang J, Gujar MR, Deng Q, Y Chia S, Li S, Tan P, Sung W-K, Wang H. 2021. Histone lysine methyltransferase Pr-set7/SETD8 promotes neural stem cell reactivation. EMBO Rep 22: e50994. 10.15252/embr.202050994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MSY, Sy SM-H, van Deursen JM, Chen J. 2008. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem 283: 11073–11077. 10.1074/jbc.C700242200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. 2010. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 330: 1824–1827. 10.1126/science.1195481 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Jørgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sørensen CS. 2007. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345. 10.1083/jcb.200706150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, et al. 2008. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27: 4293–4304. 10.1038/onc.2008.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Vertino PM. 2014. A dual role for the histone methyltransferase PR-SET7/SETD8 and histone H4 lysine 20 monomethylation in the local regulation of RNA polymerase II pausing. J Biol Chem 289: 7425–7437. 10.1074/jbc.M113.520783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 19: 431–435. 10.1101/gad.1263005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S. 2010. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol 17: 1027–1029. 10.1038/nsmb.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20: 1110–1122. 10.1101/gad.377406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 484: 115–119. 10.1038/nature10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatham-Jensen M, Uyehara CM, Strahl BD, Matera AG, Duronio RJ, McKay DJ. 2019. Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLoS Genet 15: e1007932. 10.1371/journal.pgen.1007932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. 2007. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell 28: 677–691. 10.1016/j.molcel.2007.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, Gui B, Shang Y. 2011. The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem 286: 13925–13936. 10.1074/jbc.M111.223198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. 2016. Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila. Nucleic Acids Res 44: 7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512. 10.1038/nature09272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. 2008. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol 15: 1122–1124. 10.1038/nsmb.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Smyth GK. 2016. Csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res 44: e45. 10.1093/nar/gkv1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Han Z, Chen H, Yang B, Yang X, Xia Y, Pan C, Fu L, Zhang S, Han H, et al. 2016. A positive role for polycomb in transcriptional regulation via H4K20me1. Cell Res 26: 529–542. 10.1038/cr.2016.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. 2003. The Tudor domain “Royal family”: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci 28: 69–74. 10.1016/S0968-0004(03)00004-5 [DOI] [PubMed] [Google Scholar]
- McKay DJ, Klusza S, Penke TJR, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD, et al. 2015. Interrogating the function of metazoan histones using engineered gene clusters. Dev Cell 32: 373–386. 10.1016/j.devcel.2014.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier K, Mathieu E-L, Finkernagel F, Reuter LM, Scharfe M, Doehlemann G, Jarek M, Brehm A. 2012. LINT, a novel dL(3)mbt-containing complex, represses malignant brain tumour signature genes. PLoS Genet 8: e1002676. 10.1371/journal.pgen.1002676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. 2007. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol 14: 1229–1230. 10.1038/nsmb1340 [DOI] [PubMed] [Google Scholar]
- The modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MAJ, Shilatifard A. 2020. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat Genet 52: 1271–1281. 10.1038/s41588-020-00736-4 [DOI] [PubMed] [Google Scholar]
- Morgan MAJ, Shilatifard A. 2023. Epigenetic moonlighting: catalytic-independent functions of histone modifiers in regulating transcription. Sci Adv 9: eadg6593. 10.1126/sciadv.adg6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou KC, Moulos P, Harokopos V, Chalepakis G, Talianidis I. 2017. Kmt5a controls hepatic metabolic pathways by facilitating RNA Pol II release from promoter-proximal regions. Cell Rep 20: 909–922. 10.1016/j.celrep.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201–1213. 10.1016/S1097-2765(02)00548-8 [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295. 10.1128/MCB.01768-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino S, Michelena J, Teloni F, Imhof R, Altmeyer M. 2017. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep 19: 1819–1831. 10.1016/j.celrep.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699. 10.1126/science.1231382 [DOI] [PubMed] [Google Scholar]
- Pengelly AR, Kalb R, Finkl K, Müller J. 2015. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev 29: 1487–1492. 10.1101/gad.265439.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke TJ, McKay DJ, Strahl BD, Matera AG, Duronio RJ. 2016. Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev 30: 1866–1880. 10.1101/gad.286278.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu G-Q, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507. 10.1038/nature09261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Oktaba K, Steinmann J, Müller J, Knoblich JA. 2011. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat Cell Biol 13: 1029–1039. 10.1038/ncb2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Strahl BD. 2014. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 1839: 627–643. 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu V, Lu C. 2022. Oncohistones: hijacking the histone code. Annu Rev Cancer Biol 6: 293–312. 10.1146/annurev-cancerbio-070120-102521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A, Steward R. 2007. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol 176: 155–162. 10.1083/jcb.200607178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A, Joyce E, Aoki T, Schedl P, Steward R. 2012. The histone H4 lysine 20 monomethyl mark, set by PR-Set7 and stabilized by L(3)mbt, is necessary for proper interphase chromatin organization. PLoS One 7: e45321. 10.1371/journal.pone.0045321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzler HR, Vandadi V, McMichael BD, Brown JC, Boerma SA, Leatham-Jensen MP, Adams KM, Meers MP, Simon JM, Duronio RJ, et al. 2023. Distinct roles for canonical and variant histone H3 lysine-36 in polycomb silencing. Sci Adv 9: eadf2451. 10.1126/sciadv.adf2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callén E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22: 2048–2061. 10.1101/gad.476008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646. 10.1016/j.molcel.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaib M, Walter D, Gillespie PJ, Izard F, Fahrenkrog B, Lleres D, Lerdrup M, Johansen JV, Hansen K, Julien E, et al. 2018. Histone H4K20 methylation mediated chromatin compaction threshold ensures genome integrity by limiting DNA replication licensing. Nat Commun 9: 3704. 10.1038/s41467-018-06066-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaib M, Chen Q, Shi X, Nair N, Prasanna C, Yang R, Walter D, Frederiksen KS, Einarsson H, Svensson JP, et al. 2021. Histone H4 lysine 20 mono-methylation directly facilitates chromatin openness and promotes transcription of housekeeping genes. Nat Commun 12: 4800. 10.1038/s41467-021-25051-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrajna A, Goldfarb D, Kedziora KM, Cousins EM, Grant GD, Spangler CJ, Barbour EH, Yan X, Hathaway NA, Brown NG, et al. 2020. Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res 48: 9415–9432. 10.1093/nar/gkaa544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4: 1521. 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Cronin NB, Wilson JR. 2014. A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases. Nucleic Acids Res 42: 661–671. 10.1093/nar/gkt776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. 2017. False discovery rates: a new deal. Biostatistics 18: 275–294. 10.1093/biostatistics/kxw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403: 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Takawa M, Cho H-S, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, et al. 2012. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res 72: 3217–3227. 10.1158/0008-5472.CAN-11-3701 [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. 2007. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426. 10.1083/jcb.200706179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129: 915–928. 10.1016/j.cell.2007.03.048 [DOI] [PubMed] [Google Scholar]
- Tuzon CT, Spektor T, Kong X, Congdon LM, Wu S, Schotta G, Yokomori K, Rice JC. 2014. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep 8: 430–438. 10.1016/j.celrep.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyehara CM, Leatham-Jensen M, McKay DJ. 2022. Opportunistic binding of EcR to open chromatin drives tissue-specific developmental responses. Proc Natl Acad Sci 119: e2208935119. 10.1073/pnas.2208935119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. 2014. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24: 896–905. 10.1101/gr.171405.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903. 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver TM, Liu J, Connelly KE, Coble C, Varzavand K, Dykhuizen EC, Musselman CA. 2019. The EZH2 SANT1 domain is a histone reader providing sensitivity to the modification state of the H4 tail. Sci Rep 9: 987. 10.1038/s41598-018-37699-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich S, Kudithipudi S, Jeltsch A. 2016. Specificity of the SUV4-20H1 and SUV4-20H2 protein lysine methyltransferases and methylation of novel substrates. J Mol Biol 428: 2344–2358. 10.1016/j.jmb.2016.04.015 [DOI] [PubMed] [Google Scholar]
- West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. 2010. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem 285: 37725–37732. 10.1074/jbc.M110.139527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. Ggplot2: elegant graphics for data analysis (use R!), 2nd ed. Springer, Cham, Switzerland. [Google Scholar]
- Yamamoto-Matsuda H, Miyoshi K, Moritoh M, Yoshitane H, Fukada Y, Saito K, Yamanaka S, Siomi MC. 2022. Lint-O cooperates with L(3)mbt in target gene suppression to maintain homeostasis in fly ovary and brain. EMBO Rep 23: e53813. 10.15252/embr.202153813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, Mizzen CA. 2008. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem 283: 12085–12092. 10.1074/jbc.M707974200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. 2012. SET8 promotes epithelial–mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J 31: 110–123. 10.1038/emboj.2011.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yu VC, Zhu G, Chang DC. 2008. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 7: 1423–1432. 10.4161/cc.7.10.5867 [DOI] [PubMed] [Google Scholar]
- Yohn CB, Pusateri L, Barbosa V, Lehmann R. 2003. L(3)malignant brain tumor and three novel genes are required for Drosophila germ-cell formation. Genetics 165: 1889–1900. 10.1093/genetics/165.4.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu L, Li X, Hu X, Song H. 2019. The histone H4K20 methyltransferase PR-Set7 fine-tunes the transcriptional activation of Wingless signaling in Drosophila. J Genet Genomics 46: 57–59. 10.1016/j.jgg.2018.06.009 [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang Y, Shi X. 2015. Emerging roles of lysine methylation on non-histone proteins. Cell Mol Life Sci 72: 4257–4272. 10.1007/s00018-015-2001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouaz A, Fernando C, Perez Y, Sardet C, Julien E, Grimaud C. 2018. Cell-cycle regulation of non-enzymatic functions of the Drosophila methyltransferase PR-Set7. Nucleic Acids Res 46: 2834–2849. 10.1093/nar/gky034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data sets have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE268819, GSE268820, and GSE268821. The code for peak calling and differential expression analysis is available at https://zenodo.org/doi/10.5281/zenodo.11550660.