Abstract
In silico simulation of high-throughput sequencing data is a technique used widely in the genomics field. However, there is currently a lack of effective tools for creating simulated data from nanopore sequencing devices, which measure DNA or RNA molecules in the form of time-series current signal data. Here, we introduce Squigulator, a fast and simple tool for simulation of realistic nanopore signal data. Squigulator takes a reference genome, a transcriptome, or read sequences, and generates corresponding raw nanopore signal data. This is compatible with basecalling software from Oxford Nanopore Technologies (ONT) and other third-party tools, thereby providing a useful substrate for development, testing, debugging, validation, and optimization at every stage of a nanopore analysis workflow. The user may generate data with preset parameters emulating specific ONT protocols or noise-free “ideal” data, or they may deterministically modify a range of experimental variables and/or noise parameters to shape the data to their needs. We present a brief example of Squigulator's use, creating simulated data to model the degree to which different parameters impact the accuracy of ONT basecalling and downstream variant detection. This analysis reveals new insights into the nature of ONT data and basecalling algorithms. We provide Squigulator as an open-source tool for the nanopore community.
Nanopore sequencing is an increasingly important genomic technology. Devices from Oxford Nanopore Technologies (ONT) have the ability to analyze both short and long native DNA and RNA molecules, with countless potential applications across the life sciences. An ONT device measures the displacement of ionic current as a DNA or RNA molecule passes through a nanoscale protein pore. The device records time-series current signal data (commonly referred to as “squiggle” data), which can be “basecalled” into sequence reads or analyzed directly in a variety of contexts (Wang et al. 2021).
Data simulation is an essential tool for data scientists and software developers in many scientific domains, including genomics (Escalona et al. 2016). The availability of reference data generated in silico with controlled parameters enables the user to test, debug, optimize, and validate new analysis methods in absence of confounding experimental and biological variables. Simulated data can also be used to develop and test new hypotheses or models, to inform experimental design, or to be a ground truth during benchmarking studies, among other applications (Escalona et al. 2016). There are a range of existing tools for high-throughput sequencing data simulation, including ART (Huang et al. 2012), GemSIM (McElroy et al. 2012), pIRS (Hu et al. 2012), and FASTQSim (Shcherbina 2014). These have helped catalyze important developments across genomics, transcriptomics, metagenomics, etc. (Escalona et al. 2016). A variety of tools now also support long-read sequencing data simulation, including Badread (Wick 2019), NanoSim (Yang et al. 2017), Trans-NanoSim (Hafezqorani et al. 2020), TKSM (Karaoğlanoğlu et al. 2024), and PBsim (Ono et al. 2021). However, these tools generate sequence reads only and cannot be used to create realistic nanopore sequencing data with accompanying raw signal. To our knowledge, the only existing tool for raw nanopore data simulation is DeepSimulator, which uses a neural network architecture to generate realistic signal data (Li et al. 2020).
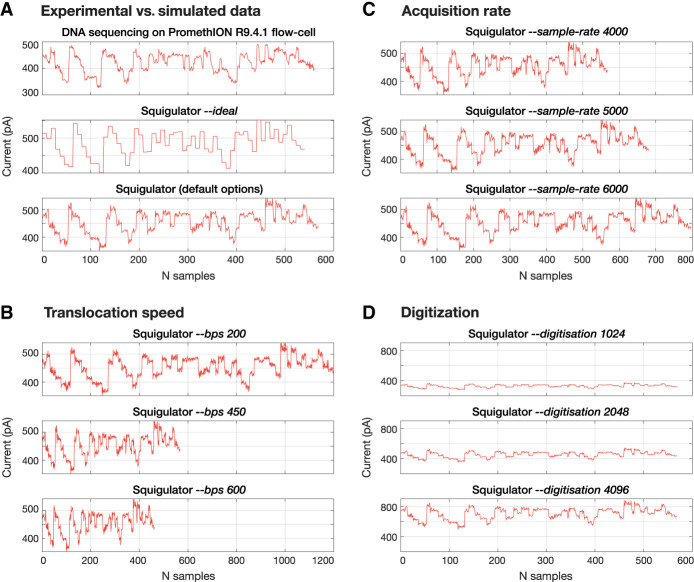
Here, we introduce Squigulator (squiggle simulator), a fast and simple tool for in silico generation of nanopore current signal data that emulates the properties of real data from a nanopore device. The simulated data are compatible with ONT basecalling software and open-source tools for signal data analysis, providing a useful substrate for developers and data scientists in the growing nanopore community. Squigulator uses existing ONT pore models, which model the expected current level as a given DNA/RNA subsequence occupies a nanopore, and applies empirically determined noise functions to generate realistic signal data from a reference sequence/s (Fig. 1A). To determine the properties of their simulated data set, users can flexibly adjust the noise parameters; technical variables like DNA translocation speed (Fig. 1B), data acquisition rate (Fig. 1C), and digitization (Fig. 1D); and pseudoexperimental variables like coverage depth and read-length distribution. They may opt to create “ideal” signal data lacking any noise or to select from preset configurations that recapitulate the parameters of specific ONT sequencing protocols. This capacity for deterministic parameter control is an important advantage of Squigulator, enabling parameter exploration during algorithm development.
Figure 1.
Simulated nanopore signal data with tunable parameters. Examples show experimental and simulated data from a single DNA sequence, showcasing some of the different simulation parameters available in Squigulator. (A) Experimental nanopore raw signal data (top track) versus simulated data with no noise in the amplitude or time domains (‐‐ideal option; middle track) versus the -x dna-r9-prom preset configuration (bottom track), which is currently the default option in Squigulator. This configuration emulates standard sequencing of genomic DNA on an ONT PromethION R9.4.1 flow cell, matching the experimental data above. (B) Simulated data with varying DNA translocation speeds (‐‐bps option). (C) Simulated data with varying data acquisition rates (‐‐sample-rate option). (D) Simulated data with varying data digitization levels (‐‐digitisation option).
Results
Squigulator compatibility with nanopore analysis software
Squigulator is designed to produce simulated nanopore signal data that are compatible with any relevant software and can be used to recapitulate a complete nanopore analysis workflow. To test this capability, we created a data set that resembles a typical sequencing experiment run on the popular human reference individual NA12878 (see Supplemental Methods). Briefly, we used BCFtools consensus (Danecek et al. 2021) to incorporate high-confidence NA12878 variants (single-nucleotide variants [SNVs] and indels) annotated by the Genome in a Bottle Consortium (Zook et al. 2014) into the human reference genome sequence (GRCh38; FASTA format). This was performed in a haplotype-aware fashion, creating a diploid NA12878 reference in which heterozygous and homozygous variants are represented at an appropriate copy number. We then used Squigulator to generate simulated nanopore signal data from this custom reference, with default parameters. The read length, standard deviation, and read count were roughly matched to a real sequencing experiment performed on NA12878 genomic DNA (∼30× coverage) (see Supplemental Methods), which is used for comparison below.
We analyzed both the simulated and experimental NA12878 data sets via a typical analysis workflow. Signal data were basecalled with ONT's Guppy software (using the buttery-eel wrapper for SLOW5 data access) (Samarakoon et al. 2023a). Basecalled reads were then aligned to the reference genome using minimap2 (Li 2018), and SNVs were detected using each of two approaches: (1) with Nanopolish (Loman et al. 2015), which uses both the basecalled and raw-signal data to identify variants, and (2) with Clair3 (Zheng et al. 2022), which calls variants from the basecalled data alone. SNV detection performance was then evaluated with RTGtools vcfeval (see Supplemental Methods).
We observed broadly similar properties between the simulated and experimental NA12878 data sets at each workflow stage. The distribution of signal values was similar between the two raw data sets (Fig. 2A). After basecalling, they showed a similar read length, nucleotide composition, and quality score distributions (Fig. 2B,C). Minimap2 alignment statistics were similar (Supplemental Table S1), as were the patterns of mismatch and indel errors in reads aligned to GRCh38 (Fig. 2D,E). The most significant discrepancy between the simulated and experimental NA12878 data sets was a tail of basecalled reads with elevated mismatch errors in the experimental data, which was not recapitulated by Squigulator (Fig. 2F). However, modal error rates were similar between the two data sets (Fig. 2F), and variant detection tools were able to detect known NA12878 SNVs with accuracy >98% (F-score), with Clair3 showing superior performance to that of Nanopolish on both the simulated and experimental data sets (Supplemental Table S2). These results show the capacity of Squigulator to create simulated data that roughly emulate real experimental data and are compatible with relevant analysis software.
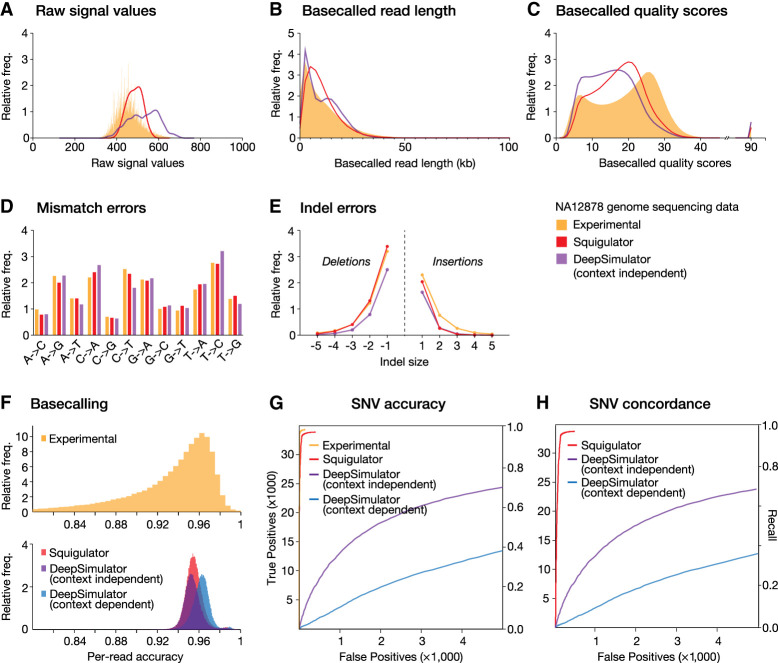
Figure 2.
Comparison of experimental and simulated NA12878 signal data sets. (A–C) Frequency histograms show distributions of raw signal values (A), basecalled read lengths (B), and Phred quality scores (C) in experimental data (orange) and simulated data sets from Squigulator (orange) or DeepSimulator (purple), based on the reference individual NA12878. A Guppy HAC basecalling model was used. (D,E) For the same data sets, bar charts show the relative frequencies of each possible base substitution (D), and line plots show the relative frequency of insertions and deletions of different sizes (E). Substitution and indels errors are determined relative to the GRCh38 reference genome after alignment with minimap2. (F) Guppy basecalling accuracy (HAC model), as measured by read:reference identity score distributions, for experimental (upper) and simulated (lower) data sets. Simulated data are from Squigulator (red) or DeepSimulator with context-independent (purple) or context-dependent (blue) settings. (G) ROC curves evaluate accuracy of SNV detection with Clair3 on the same data sets (colors as above). (H) ROC curves evaluate concordance of SNVs detected with real experimental NA12878 data set versus simulated data from Squigulator or DeepSimulator (colors as above). SUP basecalling was used to maximize accuracy of SNV detection. The left vertical axes in ROC curves show absolute numbers of detected SNVs, and right vertical axes show fraction of true positives detected (i.e., recall or sensitivity).
Comparison of Squigulator to DeepSimulator
There is currently only a single alternative tool for nanopore data simulation: DeepSimulator (Li et al. 2020). DeepSimulator generates simulated signal data via either of two approaches: (1) using a context-dependent Bi-LSTM trained model or (2) using a context-independent model that uses a k-mer model provided by ONT (see Supplemental Methods). Both approaches are designed to generate realistic nanopore signal data, although the context-independent model is considerably faster.
To compare the performance of Squigulator and DeepSimulator, we repeated the experiment above but this time generating simulated NA12878 data sets using DeepSimulator (see Supplemental Methods). We assessed the read length, nucleotide composition, quality score distributions, Guppy basecalling accuracy, patterns of mismatch and indel errors, and Clair3 SNV detection, as above, with the real experimental NA12878 data set as a point of comparison (Fig. 2A–H). Although there were modest differences in each comparison, the Squigulator and DeepSimulator data sets showed broadly similar accuracy to real experimental data with equivalent basecalling models (Fig. 2F). However, inspecting alignments to the reference genome, we observed that basecalled data from DeepSimulator showed many reproducible errors, whereas the errors in Squigulator data and experimental data were distributed more randomly. As a result, Clair3 called an abundance of erroneous SNVs on the DeepSimulator data sets and missed many true SNVs (Supplemental Fig. S1A). We also observed many examples of false-positive SNVs called in both the real experimental data and Squigulator data but missing from DeepSimulator data (Supplemental Fig. S1B). As a result, the NA12878 SNV accuracy with DeepSimulator data (context-dependent, 0.625; context-independent, 0.769 F-scores; Guppy SUP) was poor by comparison to that of the Squigulator (0.987) or real experimental data (0.996) (Fig. 2G), and SNVs called from the real experimental NA12878 data set showed a higher concordance to those called with the Squigulator data (0.985) than the DeepSimulator data (0.624, 0.769) (Fig. 2H). Therefore, although the ONT basecalling process itself uses context-dependent neural network methods comparable to those of DeepSimulator, the k-mer-based method used by Squigulator appears to generate more realistic simulated data.
In addition to data quality, we compared the time and resources used by the two simulation tools. Running with 16 CPUs, Squigulator took just 156 sec to generate ∼30× sequencing data on Chr 22 (with peak RAM usage of 0.5 GB) or 68 min for an entire human genome (3.4 GB RAM) (Supplemental Table S3). To generate an equivalent Chr 22 data set on the same system, DeepSimulator took about 20 times longer (50 min) in a context-independent mode and about 3000 times longer (131 h) in a context-dependent mode (Supplemental Table S3). DeepSimulator's peak RAM usage was also about ninefold and about 206-fold higher than that of Squigulator, in a context-independent and a context-dependent mode, respectively. Although these analyses show that DeepSimulator is able to generate simulated nanopore signal data, Squigulator requires a fraction of the time and memory and generates data that more closely resemble real experimental data.
We also performed similar comparisons between Badread (Wick 2019), a popular simulator for generating long, noisy sequence reads, and Squigulator data basecalled with Guppy (see Supplemental Methods). Although, Badread sequence data were preferable to those of DeepSimulator, Squigulator data showed the highest concordance to real experimental data (Supplemental Fig. S2A,B).
Using Squigulator for parameter exploration
Although they showed similar patterns, we observed higher overall error rates during basecalling and SNV detection with the simulated versus experimental data sets above (Supplemental Table S1). Anticipating that these processes may be affected by noise/warping in the amplitude and time domains, we next used Squigulator to model the impact of each noise parameter. To do so, we generated alternative simulated data sets in which the degree of noise in one domain was systematically modified, while keeping the other static, and then repeated the above analysis workflow with each data set (see Supplemental Methods).
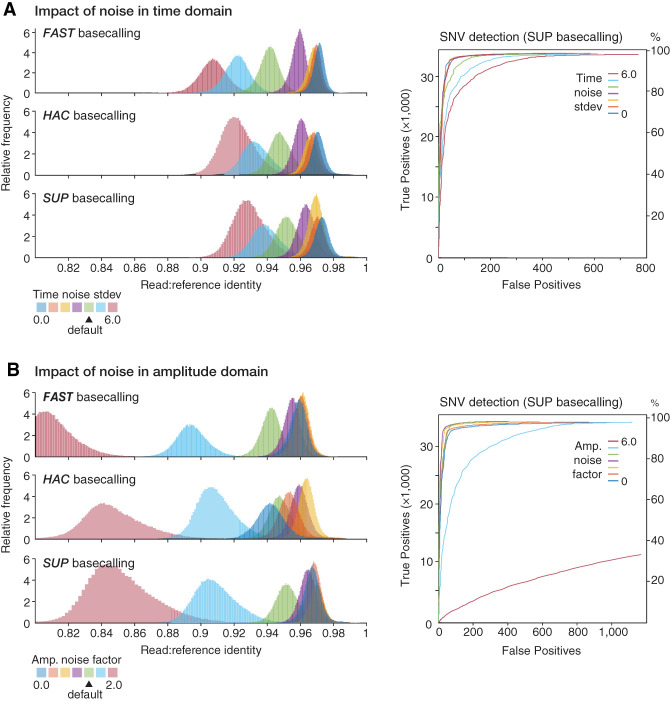
Both noise parameters had a significant impact on basecalling accuracy. Accuracy was negatively correlated with noise in the time domain (Fig. 3A) and surpassed real experimental data when noise was minimized (Supplemental Fig. S3A). In fact, basecalling was much more sensitive to dwell-time noise (i.e., standard deviation) than to fixed changes in the dwell-time mean (Supplemental Fig. S3B,C). This indicates that consistency in DNA translocation speed is more important to Guppy than the actual speed itself (within a sensible range). Noise in the amplitude domain was also detrimental to basecalling accuracy; however, reducing amplitude noise to zero did not lead to optimum results (Fig. 3B). Instead, Guppy performed best when a small amount of noise was applied, presumably reflecting properties of the neural network models involved, which are trained on real experimental data. These trends were not consistent between Guppy's FAST, HAC, or SUP models, with HAC being more sensitive to changes in the degree of amplitude noise than SUP or FAST (Fig. 3B). As expected, differences in basecalling accuracy at the read level resulting from noise in the time and amplitude domains manifested in corresponding differences in the accuracy of SNV detection Clair3 (Fig. 3B,C). Overall, this analysis provides a simple demonstration of how simulated data with tunable parameters may be used to inform algorithm development and optimization in the nanopore field.
Figure 3.
Modeling the impact of time and amplitude noise on sequencing accuracy. (A, left) Guppy basecalling accuracy, as measured by read:reference identity score distributions, for repeated experiments in which warping in the time domain (i.e., variation in the DNA translocation speed) was varied, whereas other parameters were held at default. Higher values indicate increasing noise and default value is ‐‐dwell-std = 4 (green distribution). Experiment was repeated with Guppy's FAST (upper), HAC (middle), and SUP (lower) basecalling models. (Right) ROC curves assess accuracy of SNV detection by Clair3 on the same data sets (colors are matched). (B) Same analysis as above but with noise in the amplitude domain (‐‐amp-noise) varied, whereas other parameters are static.
Discussion
The genomics field has benefitted from the availability of many different tools for in silico data simulation, which have been developed in parallel to new sequencing technologies and applications (Escalona et al. 2016). Squigulator addresses the need for a simple and effective tool for creating realistic simulated data from nanopore sequencing experiments.
The most popular tool currently available for nanopore data simulation, NanoSim (Yang et al. 2017) generates basecalled sequencing data (FASTQ format) but does not generate current signal data, which is the primary output from a nanopore device. Therefore, NanoSim cannot be used to recapitulate a complete nanopore analysis workflow and has no utility for development and benchmarking of signal-level analyses. In contrast, we have shown above how Squigulator can be used to generate realistic signal data, suitable to evaluate every stage of a nanopore bioinformatics workflow.
Another alternative, DeepSimulator (Li et al. 2020), uses a neural network architecture to generate realistic nanopore signal data. This approach is computationally expensive but is designed to best emulate the subtleties of experimental data on which it was modeled. In contrast, Squigulator's simple, deterministic framework is designed to be fast and lightweight and to grant the user maximum control over all variables and noise parameters. Our comparison experiments show that simulated data from Squigulator is more similar to matched experimental data than can be achieved with DeepSimulator's either context-dependent or context-independent modes. We note that the test data set selected here was highly similar to DeepSimulator's internal training data, that is, NA12878 genome sequencing data with ONT R9.4.1 chemistry.
Generating realistic data is not the limit of Squigulator's capabilities. Rather than choosing a preset parameter configuration modeled on a given ONT sequencing protocol, the user may instead tune each experimental variable and/or noise parameter to assess the impact on their analysis workflow. The pore-model input framework also allows the user to provide their own custom pore model. This means Squigulator is, in theory, suitable for simulating data from any alternative nanopore system in which the user has access to a k-mer-based pore model (e.g., a hypothetical solid-state nanopore device with voltage sensing). This need not be limited to DNA or RNA; nanopore protein sequencing is similarly amenable to k-mer-based biophysical modeling (Motone et al. 2023) and may, therefore, be simulated with Squigulator.
An effective tool for in silico data generation should reduce friction in bioinformatic software development. Speed and simplicity are essential to achieve this aim. Therefore, Squigulator is designed to be easy to install and use. It has no external dependencies apart from the ubiquitous zlib software library and can be compiled on Linux, MacOS, and Windows or simply run using precompiled binaries provided for common systems. Squigulator is also exceptionally fast in comparison to other similar tools. We have found that generating 1 Gb of simulated data takes ∼2 min when running on 16 CPU threads compared with ∼45 min with DeepSimulator's low-accuracy context-independent mode and ∼90 h with its context-dependent mode (a comparison with NanoSim was not possible because we were unable to complete the installation). This combination of speed, simplicity, and realistic, tunable data (see above) make Squigulator the ideal tool for nanopore data simulation.
Squigulator outputs nanopore signal data in binary SLOW5 (BLOW5) format, an open-source community-centric alternative to ONT's native FAST5 format (Gamaarachchi et al. 2022). This feature is essential to the tool's high performance, because the writing speed for BLOW5 is considerably faster than what is attainable with FAST5. The BLOW5 format is now compatible with the latest ONT basecalling models and approaches, via buttery-eel (a SLOW5 wrapper for ONT's production basecaller, Guppy) (Samarakoon et al. 2023a) or SLOW5-enabled forks for open-source basecallers Dorado (https://github.com/hiruna72/slow5-dorado) or Bonito (https://github.com/Psy-Fer/bonito). BLOW5 format is also compatible with a growing variety of open-source software (Simpson et al. 2017; Gamaarachchi et al. 2020; Bao et al. 2021; Zhang et al. 2021; Senanayake et al. 2023; Shih et al. 2023). Users may alternatively decide to convert their BLOW5 files back to ONT's native FAST5 format using slow5tools (Samarakoon et al. 2023b), before proceeding to their analysis workflow.
Methods
Squigulator methodology
Squigulator generates simulated nanopore signal data based on an input reference genome or transcriptome sequence (FASTA format) or directly from a set of basecalled reads (FASTQ or FASTA format). To do so, it uses an idealized “pore model” specifying the expected current signal reading associated with every possible DNA or RNA k-mer, as appropriate to the specific nanopore protocol being emulated. Pore models are available for ONT protocols (https://github.com/nanoporetech/kmer_models), covering a range of different flow-cell versions (e.g., R9, R10), kits (e.g., DNA vs. RNA sequencing) and ONT devices (e.g., PromethION vs. MinION). The user can also provide their own custom pore model, enabling simulation of data from existing or future non-ONT nanopore systems. We also provide a “how to” guide on how to model realistic noise parameters from scratch, for a new sequencing protocol: https://hasindu2008.github.io/squigulator/docs/profile.html#determining-parameters-for-a-profile.
Squigulator generates sequential signal values corresponding to sequential k-mers in the provided reference sequence. The number of signal values per k-mer is dictated by the translocation speed and data acquisition rate for a specified ONT sequencing protocol (e.g., 450 nt/sec and 400 kHz for DNA sequencing on R9.4.1 flow cells) or provided manually by the user (Fig. 1). Other technical variables relevant to nanopore sequencing, such as digitization, range, offset, etc., can be similarly modified. This process generates perfect, noise-free signal reads when the ‐‐ideal option is selected (Fig. 1A). By default, however, Squigulator applies noise to the data to produce realistic, rather than ideal, signal reads (Fig. 1A). The noise functions applied are empirically determined statistical distributions modeled on stochasticity in the amplitude domain and warping in the time domain observed during ONT experiments. The user can select from a menu of preset parameter configurations modeled on specific ONT protocols. Additional preset configurations will be added for further protocols from ONT as these are released. Alternatively, they can use the ‐‐ideal-time or ‐‐ideal-amp options to limit noise to just a single domain, or they can manually adjust each variable independently. The user can also modify several pseudoexperimental variables, including coverage depth, read-length mean, variation, etc., to determine the dimensions of their simulated data set. Detailed descriptions of the software usage and options are provided at GitHub (https://hasindu2008.github.io/squigulator/docs/man.html).
Squigulator implementation
Squigulator is developed in C programming language. The only external dependency of Squigulator is the ubiquitous library zlib. The Slow5lib library (Gamaarachchi et al. 2022) for reading and writing nanopore signal data is included inside the source code of Squigulator itself. The minimum compiler requirement is a C99 or higher C compiler that supports POSIX extensions (e.g., gcc, clang, or icc). Therefore, the software can be easily compiled on Linux, MacOS and Windows (through Windows Subsystem for Linux).
The core random number generator used by Squigulator is a simple uniform random number generator based on the simple Multiplicative Linear Congruential Generator (LCG). This provides the basis for generating normal distributions (using Box-Muller transform) and gamma distributions, where appropriate. Sampling the genome at random positions for reads is modeled as a uniform random distribution. Normal distributions are used for modeling noise along amplitude domains of the signal, where the mean and standard deviation of each k-mer in the pore-model form a random number generator. Normal distributions are also used for generating noise along the time domain (dwell), and other signal metadata such as offset and median_before. Read length variability is modeled using a gamma distribution. In each case, the most appropriate distribution type was selected based on empirical exploration of real experimental ONT data.
Benchmarking data sets
The experimental data set used in benchmarking experiments was generated by sequencing genomic DNA from the human NA12878 reference sample on an ONT PromethION device. Unsheared DNA libraries were prepared using the ONT LSK109 ligation library prep, and two R9.4.1 flow cells were used to generate ∼30× genome coverage. The data are available at the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA744329. All other benchmarking data sets can be created using Squigulator (see Supplemental Methods).
Software availability
With the exception of ONT's commercially available Guppy basecaller, all software used in this project is free and open source, including Squigulator (https://github.com/hasindu2008/squigulator). Squigulator experiments were performed using the following GitHub commit: 7422d7384be428ac334caa61c019473f31f1e633. Squigulator is also available as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Derrick Lin and Tim Ho for providing HPC support. We acknowledge the following funding support: Australian Medical Research futures fund grants MRF1173594, MRF2016008, and MRF2023126 (to I.W.D.) and Australian Research Council Discovery Early Career Researcher Award (DECRA) fellowship DE230100178 (to H.G.).
Author contributions: H.G. developed Squigulator with contributions from all other authors. H.S., K.L., and J.M.F. conducted rigorous user testing and feedback. I.W.D., H.G., and J.M.F. performed benchmarking experiments. H.G. and I.W.D. generated the figures and wrote the manuscript, with input from other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278730.123.
Freely available online through the Genome Research Open Access option.
Competing interest statement
I.W.D. manages a fee-for-service sequencing facility at the Garvan Institute of Medical Research that is a customer of Oxford Nanopore Technologies (ONT), but has no further financial relationship. H.G., J.M.F., and I.W.D. have previously received travel and accommodation expenses to speak at ONT conferences. H.G. and I.W.D. have paid consultant roles with Sequin PTY. The authors declare no other competing financial or nonfinancial interests.
References
- Bao Y, Wadden J, Erb-Downward JR, Ranjan P, Zhou W, McDonald TL, Mills RE, Boyle AP, Dickson RP, Blaauw D, et al. 2021. SquiggleNet: real-time, direct classification of nanopore signals. Genome Biol 22: 298. 10.1186/s13059-021-02511-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience 10: giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalona M, Rocha S, Posada D. 2016. A comparison of tools for the simulation of genomic next-generation sequencing data. Nat Rev Genet 17: 459–469. 10.1038/nrg.2016.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamaarachchi H, Lam CW, Jayatilaka G, Samarakoon H, Simpson JT, Smith MA, Parameswaran S. 2020. GPU accelerated adaptive banded event alignment for rapid comparative nanopore signal analysis. BMC Bioinformatics 21: 343. 10.1186/s12859-020-03697-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamaarachchi H, Samarakoon H, Jenner SP, Ferguson JM, Amos TG, Hammond JM, Saadat H, Smith MA, Parameswaran S, Deveson IW. 2022. Fast nanopore sequencing data analysis with SLOW5. Nat Biotechnol 40: 1026–1029. 10.1038/s41587-021-01147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezqorani S, Yang C, Lo T, Nip KM, Warren RL, Birol I. 2020. Trans-NanoSim characterizes and simulates nanopore RNA-sequencing data. GigaScience 9: giaa061. 10.1093/gigascience/giaa061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Yuan J, Shi Y, Lu J, Liu B, Li Z, Chen Y, Mu D, Zhang H, Li N, et al. 2012. pIRS: profile-based Illumina pair-end reads simulator. Bioinformatics 28: 1533–1535. 10.1093/bioinformatics/bts187 [DOI] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, Marth GT. 2012. ART: a next-generation sequencing read simulator. Bioinformatics 28: 593–594. 10.1093/bioinformatics/btr708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoğlanoğlu F, Orabi B, Flannigan R, Chauve C, Hach F. 2024. TKSM: highly modular, user-customizable, and scalable transcriptomic sequencing long-read simulator. Bioinformatics 40: btae051. 10.1093/bioinformatics/btae051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang S, Bi C, Qiu Z, Li M, Gao X. 2020. DeepSimulator1.5: a more powerful, quicker and lighter simulator for nanopore sequencing. Bioinformatics 36: 2578–2580. 10.1093/bioinformatics/btz963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735. 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- McElroy KE, Luciani F, Thomas T. 2012. GemSIM: general, error-model based simulator of next-generation sequencing data. BMC Genomics 13: 74. 10.1186/1471-2164-13-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motone K, Kontogiorgos-Heintz D, Wee J, Kurihara K, Yang S, Roote G, Fang Y, Cardozo N, Nivala J. 2023. Multi-pass, single-molecule nanopore reading of long protein strands with singe-amino acid sensitivity. bioRxiv 10.1101/2023.10.19.563182 [DOI]
- Ono Y, Asai K, Hamada M. 2021. PBSIM2: a simulator for long-read sequencers with a novel generative model of quality scores. Bioinformatics 37: 589–595. 10.1093/bioinformatics/btaa835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon H, Ferguson JM, Gamaarachchi H, Deveson IW. 2023a. Accelerated nanopore basecalling with SLOW5 data format. Bioinformatics 39: btad352. 10.1093/bioinformatics/btad352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon H, Ferguson JM, Jenner SP, Amos TG, Parameswaran S, Gamaarachchi H, Deveson IW. 2023b. Flexible and efficient handling of nanopore sequencing signal data with slow5tools. Genome Biol 24: 69. 10.1186/s13059-023-02910-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake A, Gamaarachchi H, Herath D, Ragel R. 2023. DeepSelectNet: deep neural network based selective sequencing for oxford nanopore sequencing. BMC Bioinformatics 24: 31. 10.1186/s12859-023-05151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbina A. 2014. FASTQSim: platform-independent data characterization and in silico read generation for NGS datasets. BMC Res Notes 7: 533. 10.1186/1756-0500-7-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih PJ, Saadat H, Parameswaran S, Gamaarachchi H. 2023. Efficient real-time selective genome sequencing on resource-constrained devices. GigaScience 12: giad046. 10.1093/gigascience/giad046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. 2017. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14: 407–410. 10.1038/nmeth.4184 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. 2021. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol 39: 1348–1365. 10.1038/s41587-021-01108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR. 2019. Badread: simulation of error-prone long reads. J Open Source Software 4: 1316. 10.21105/joss.01316 [DOI] [Google Scholar]
- Yang C, Chu J, Warren RL, Birol I. 2017. NanoSim: nanopore sequence read simulator based on statistical characterization. GigaScience 6: 1–6. 10.1093/gigascience/gix010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li H, Jain C, Cheng H, Au KF, Li H, Aluru S. 2021. Real-time mapping of nanopore raw signals. Bioinformatics 37: i477–i483. 10.1093/bioinformatics/btab264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Li S, Su J, Leung AW-S, Lam T-W, Luo R. 2022. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Nat Comput Sci 2: 797–803. 10.1038/s43588-022-00387-x [DOI] [PubMed] [Google Scholar]
- Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, Salit M. 2014. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol 32: 246–251. 10.1038/nbt.2835 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.