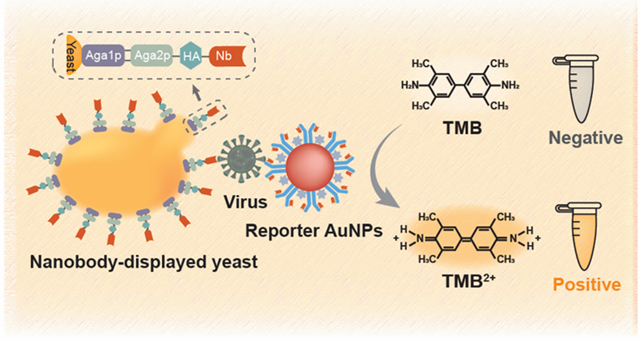
Abstract
The accurate and effective detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is essential to preventing the spread of infectious diseases and ensuring human health. Herein, a nanobody-displayed whole-cell biosensor was developed for colorimetric detection of SARS-CoV-2 spike proteins. Serving as bioreceptors, yeast surfaces were genetically engineered to display SARS-CoV-2 binding of llama-derived single-domain antibodies (nanobodies) with high capture efficiency, facilitating the concentration and purification of SARS-CoV-2. Gold nanoparticles (AuNPs) employed as signal transductions were functionalized with horseradish peroxidase (HRP) and anti-SARS monoclonal antibodies to enhance the detection sensitivity. In the presence of SARS-CoV-2 spike proteins, the sandwiched binding will be formed by linking engineered yeast, SARS-CoV-2 spike proteins, and reporter AuNPs. The colorimetric signal was generated by the enzymatic reaction of HRP and its corresponding colorimetric substrate/chromogen system. At the optimal conditions, the developed whole-cell biosensor enables the sensitive detection of SARS-CoV-2 spike proteins in a linear range from 0.01 to 1 μg/mL with a limit of detection (LOD) of 0.037 μg/mL (about 4 × 108 virion particles/mL). Furthermore, the whole-cell biosensor was demonstrated to detect the spike protein of different SARS-CoV-2 variants in human serum, providing new possibilities for the detection of future SARS-CoV-2 variants.
Keywords: nanobody, engineered yeast, gold nanoparticle, whole-cell biosensor, colorimetric detection, SARS-CoV-2
Graphical Abstract
1. INTRODUCTION
The current global pandemic associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused over 6.61 million deaths and drastically disrupted the daily life of many people worldwide.1,2 According to a recent study, symptomatic patients should be isolated within 24 h to control the spread of coronavirus disease.3 However, due to the similar symptoms between influenza and coronavirus, including muscle aches, chills, fever, headache, and diarrhea, it is difficult to distinguish between the two diseases based on clinical symptoms alone. Therefore, rapid detection of coronaviruses is crucial for promptly isolating infected cases, slowing the spread of the epidemic, and safeguarding public health.
Recently, many methods have been developed to detect SARS-CoV-2, such as reverse transcription-quantitative polymerase chain reaction (RT-qPCR),4 enzyme-linked immunosorbent assay (ELISA),5 and sequencing-based technologies.6 Although these methods have achieved high sensitivity for detecting viruses, the detection results may be significantly affected by the inefficient sample pretreatment method. Since the viral concentration can range widely from one complex sample to another (such as wastewater, human serum, and salvia), there is no gold standard method for concentrating SARS-CoV-2. The lack of practical methods for the rapid concentration and purification of SARS-CoV-2 viruses from complex samples remains a hindrance in developing an efficient detection assay.
Traditional immunoassays are based on the affinity reaction between antigens and antibodies to recognize and purify target analytes. By chemical cross-linking or physical adsorption, antibodies can be conjugated on solid-phase carriers to construct bioreceptor components for analyte capture.7 In the presence of external forces (such as magnetic fields or centrifugation), the bioreceptor-target conjugates can be separated from the complex matrix. Although such types of bioreceptors have been well described in the literature, there are still some limitations in their practical applications. First, the complicated production process and long manufacturing time make antibodies very pricey, limiting the widespread use of immunoassays in resource-limited regions.8 Second, it is difficult to control the conjugation orientations of antibodies for most chemical cross-linking and physical adsorption methods, such as EDC/NHS cross-linking method9 or electrostatic adsorption.10 Incorrectly oriented antibodies may result in a loss of binding efficiency between the antibodies and antigens. Therefore, there is an urgent need to develop low-cost, high-capture-efficiency, orientation controllable bioreceptor components.
With the development of synthetic biology, whole-cell biosensors provide a new solution to the above-mentioned problems.11 The whole-cell biosensor consists of a genetically encoded bioreceptor, which is usually based on mammalian cells,12 bacteria,13 or eukaryotic microorganisms,14 coupled to a signal transduction,. Notably, due to the high tolerance to the environment and known metabolic pathways, engineered yeasts have become one of the most attractive whole-cell biosensors.15,33 Relying on the yeast surface display, yeast-based whole-cell biosensors can address the drawbacks of traditionally manufactured bioreceptors. During the growth of engineered yeast, the nanobodies were correctly folded and the antigen binding sites were fully exposed on the cell surface, offering oriented and stable biomolecule recognition elements to develop an efficient detection platform.16,17 In addition, the cost of engineered yeasts is much lower than that of protein production, which makes it affordable for the large-scale manufacture of biosensing assays.
The choice of biomolecule recognition elements highly contributes to the sensitivity, functionality, and applications of whole-cell biosensors. Single domain antibodies, also known as nanobodies, offer several advantages over traditional antibodies.18 First, nanobodies are only 12–15 kDa, with a diameter of about 2.5 nm and a length of 4 nm, which is about one-tenth of the size of conventional antibodies. This feature facilitates expression and manipulation on the yeast surface. Second, nanobodies preserve complete antigen binding ability while providing higher thermal stability, broader pH tolerance, and higher sensitivity than conventional antibodies, enabling the application of biosensors in more extreme environments. Until now, despite numerous nanobody-related studies being developed for the treatment of SARS-CoV-2, the utilization of nanobodies on whole-cell biosensors for the detection of SARS-CoV-2 has not been explored.19,20
In this study, a whole-cell biosensor has been developed for the sensitive detection of SARS-CoV-2 spike proteins in human serum. The nanobody-displayed yeast (Yeast-Nb) and enzyme-functionalized gold nanoparticles (Reporter AuNPs) were employed as sensing elements. By the expression of nanobodies on the cell surface, the engineered yeasts (S. cerevisiae EBY 100) enable rapid purification and concentration of SARS-CoV-2 spike proteins from complex matrices. Upon forming sandwich bindings with reporter AuNPs, the enzymatic reaction was triggered to catalyze corresponding substrates, generating a colorimetric signal. Our whole-cell biosensor takes advantage of yeast cells and AuNPs, offering a low-cost, highly sensitive, and user-friendly detection of SARS-CoV-2.
2. MATERIALS AND METHODS
2.1. Materials.
S. cerevisiae EBY100 strain was purchased from ATCC (MYA-4941, Manassas, VA). All DNA primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Restriction enzymes, polymerase chain reaction (PCR) kits, and golden gate assembly kits were purchased from New England Biolabs (Ipswich, MA). A panel of SARS-CoV-2 spike proteins was received from BEI Resources: Alpha variant (NR-55,277, B.1.1.7), Beta variant (NR-55,278, B.1.1351), Delta variant (NR-55,614, B.1.617.2), Omicron variant (NR-58,646, BA.4), D614G (NR-55,418), and Y114del (NR-55,415). The wild-type SARS-CoV-2 (Wuhan-1) spike protein, the mouse anti-SARS-CoV-2 spike protein S1 monoclonal antibodies, goat antimouse IgG antibodies, and other reagents used in this study were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). All chemical reagents were analytical grade, and RNase-free water was used throughout this study.
All colorimetric experiments were conducted on the Synergy HTX multimode reader (BioTek Instruments, Winooski, VT). The cell density was measured by a Nanodrop 2000c Spectrophotometer (Thermo Scientific, Waltham, MA).
2.2. Nanobody Source.
Nanobodies to SARS-CoV-2 used in the present work were developed by Pavan et al.21 The Nb selected for the development of the present diagnosis assay were obtained after panning an immune library with the full-length, locked, prefused spike protein of the Wuhan-Hu-1 strain SARS-CoV-2. The three selected clones (Nb-33, Nb-45, and Nb-46) recognized the spike protein but no receptor binding domain (RBD) by enzyme-linked immunoassay (ELISA). Nb-33 gives a positive band in Western Blot (WB) with spike protein under native conditions and by peptide array analysis seems to be directed to an epitope located at the S2 domain and does not possess neutralizing activity, Nb-45 also recognized the spike protein in a native gel, possesses low neutralizing activity, and according to biliverdin blocking assays can be directed to the NTD region. Finally, Nb-46 is not neutralizing, directed to a conformational epitope in Spike (negative by WB).21
2.3. Construction of Nanobody-Display Vectors.
The nanobody-display vectors were constructed using golden gate assembly,22 which consisted of three parts: the backbone, nanobody sequence, and Aga2-HA tag. All of these fragments were amplified by PCR with primers consisting of assembly junctions and BsaI restriction sites. The backbone was amplified from the pWL34 plasmid (a golden gate derivative of pAG304GPD, yeast TRP1 integrating vector, containing pGPD promoter and tCYC1 terminator), which was kindly provided by Dr. Clay Wright at Virginia Tech.23 The nucleic acid sequences of SARS-CoV-2 nanobodies were codon optimized for yeast and ordered from Twist Bioscience Company (San Francisco, CA). The sequence of the Aga2-HA tag was cloned from the pCTcon2 plasmid, which was a gift from Dane Wittrup (Addgene plasmid #41843).24 The PCR products were digested by the BsaI enzyme, cleaned, and assembled into the nanobody-display vectors using a golden gate assembly, prior to transformation into E. coli TOP10 chemically competent cell. The sequence of all assembly junctions and inserts in final vectors was confirmed by Sanger sequencing.
2.4. Transformation of the Nanobody Expression Cassette.
The nanobody-display vector was digested with PmeI, to separate the E. coli plasmid elements from the yeast integrating fragment, which contained the “Aga2-HA-nanobody” gene expression cassette and homologous arms of the TRP1 locus on both sides. The nanobody expression cassette was transformed into yeast competent cells (S. cerevisiae EBY100) through the LiAc/SS carrier DNA/PEG method,25 and then integrated into the yeast genome through homologous recombination at the TRP1 locus. Briefly, the digest reaction containing the nanobody expression cassette was mixed with lithium acetate (36 μL,1 M), PEG MW 3350 (240 μL, 50%), and single-stranded carrier DNA (50 μL, 2 mg/mL), and the mixture was then added into the competent yeast solution. After vortexing, the pellet was incubated at 42 °C for 40 min under agitation of 200 rpm. Afterward, the yeast transformation mixture was centrifuged (13 000 × g, 30 s) and washed with sterilized water. Finally, the yeast suspensions were plated on SDCAA-TRP selective plate and incubated at 30 °C for 3 days. The engineered yeasts were screened using yeast colony PCR, to confirm correct homologous recombination had occurred.
2.5. Preparation of Nanobody-Displayed Yeast.
One single colony of nanobody-displayed yeasts was inoculated into YPAD media (3 mL) and incubated at 30 °C. After overnight growth, the culture was diluted 100-fold in YPAD (50 mL) media and cultured at 30 °C until the OD600 reached 3.0. The yeasts were washed three times with phosphate buffered saline (PBS, 10 mM, pH 7.4) and stored at 4 °C for further experiments.
2.6. Preparation of Reporter AuNPs.
AuNPs were synthesized as described in our previous study.26 Trisodium citrate (25 mL, 38.8 mM) was added to a chloroauric acid solution (250 mL, 1 mM) and heated for 30 min. During this time, the mixture changed from light yellow to dark red. After that, the solution was slowly cooled to generate citrate modified AuNPs (21.5 nM).
Reporter AuNPs were prepared according to the previous reports.27 Briefly, potassium carbonate (0.2 M) was used to adjust the pH of the AuNPs solution to 8.3. Then anti-SARS-CoV-2 antibodies (10 μL, 1 mg/mL) and HRP enzymes (10 μL, 1 mg/mL) were simultaneously added to the AuNPs (1 mL). The mixture was gently shaken at 4 °C overnight. After that, bovine serum albumin (BSA, 1%) was added and incubated at room temperature for 1 h to block the unreacted active sites. The final product was collected and washed by centrifugation (10 100 × g, 30 min) and resuspended in PBS (10 mM, pH 7.4, with 1% BSA). The reporter AuNPs were stored at 4 °C for further experiments.
2.7. Detection of SARS-CoV-2 Using the Colorimetric Whole-Cell Biosensors.
SARS-CoV-2 spike protein solution at various concentrations (10 μL) was incubated with nanobody-displayed yeast solution (490 μL, 107 cells/mL) at 37 °C for 1 h. After centrifugation (3 000 × g, 1 min) and washing with PBS containing 0.05% Tween-20 (PBST, pH 7.4, 10 mM), the yeast-spike protein composites were resuspended in PBST (480 μL). Subsequently, the reporter AuNPs labeled with anti-SARS antibody and HRP (20 μL) were added to the mixture and incubated at 37 °C for 1 h to form sandwiched bindings. After centrifugation and washing, the obtained pellets were resuspended into 3,3′,5,5′-tetramethylbenzidine/hydrogen peroxide substrate (TMB-H2O2, 100 μL) substrate and incubated at 37 °C for 15 min. The enzymatic reaction was stopped using H2SO4 (100 μL, 0.18 M). The absorbance intensities at 450 nm were measured by using a plate reader.
A yeast-based enzyme-linked immunosorbent assay (yeast-based ELISA) was utilized for comparison to AuNPs-based detection. Instead of reporter AuNPs, the mouse anti-SARS-CoV-2 antibodies (primary antibodies, 200 μL, 1 μg/mL) were used to label the yeast-spike protein composites and incubated at room temperature for 1 h. The conjugates were washed with PBST twice and resuspended in HRP-conjugated goat antimouse IgG antibody solution (secondary antibodies, 200 μL, 1 μg/mL), incubating at room temperature for 1 h. After centrifugation and washing, the obtained pellets were resuspended into the TMB-H2O2 (100 μL) substrate and incubated at 37 °C for 15 min. The enzymatic reaction was stopped using H2SO4 (100 μL, 0.18 M). The absorbance intensities at 450 nm (A450 nm) were measured using a plate reader.
To eliminate the influence of the background signal, a normalized signal-to-noise ratio (S/N) was used as a parameter to indicate the signal intensity in further experiments. The normalized S/N is obtained by dividing the A450 nm of the experimental group (with spike protein) by the A450 nm of the control group (without spike protein).
2.8. Detection of SARS-CoV-2 in Spiked Serum Samples.
To demonstrate the performance in complex biological samples, normal human serum (Thermo Fisher Scientific Inc., Waltham, MA) spiked with various concentrations of spike protein was analyzed with the developed whole-cell biosensor. The specificity of the biosensor was investigated using spike proteins from different SARS-CoV-2 variants, including Alpha, Beta, Delta, and Omicron and two amino acid mutations (D614G and Y114del). To reduce the matrices effect, the analyzed samples were 10-fold diluted with PBS before detection.
2.9. Statistical Analysis.
Student t tests were used to evaluate the statistical significance between the mean OD450 nm from the positive versus negative samples. One asterisk (*) and two asterisks (**) indicated the experimental group of data had a difference (0.01 < p-value <0.05) and significant difference (p-value <0.01) from the control group, respectively.
3. RESULTS AND DISCUSSION
3.1. Principle of the Nanobody-Displayed Whole-Cell Biosensor to Detect SARS-CoV-2.
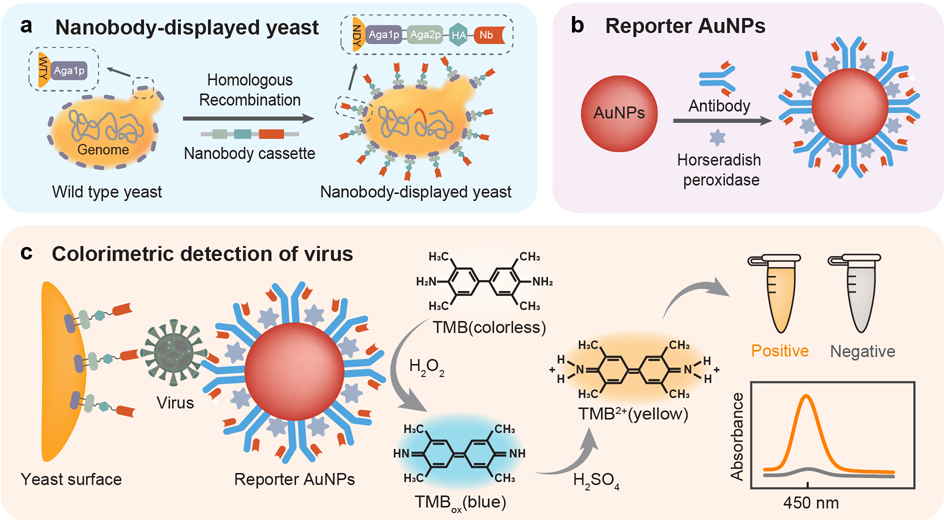
The detection principle of the nanobody-displayed whole-cell biosensor is illustrated in Figure 1. Briefly, the developed whole-cell biosensor is constituted of two components: a bioreceptor component to recognize and concentrate viruses, and a signal transduction component to generate a colorimetric signal for readouts. Besides, the SARS-CoV-2 spike proteins are regarded as detection target analytes, which can be specifically recognized by the nanobodies on the yeast surface.
Figure 1.
Schematic illustration of the nanobody-displayed whole-cell biosensor for the colorimetric detection of SARS-CoV-2. (a) Design of nanobody-displayed engineered yeast. (b) Functionalization of reporter AuNPs. (c) Colorimetric detection of SARS-CoV-2.
The nanobody-displayed yeasts (yeasts-Nb, also known as engineered yeasts) were designed as the bioreceptor components of the biosensor. As shown in Figure 1a, the yeast genome was engineered through homologous recombination with a nanobody expression cassette containing the genes for the Aga2 protein (Aga2p), hemagglutinin (HA, a surface glycoprotein) tag, and SARS-CoV-2 binding nanobody. Naturally, the mating agglutinin protein Aga2p is harnessed to mediate cell–cell contact on the yeast surface.28,29 In this study, the Aga2p serves as an anchor protein to link the Aga1 protein (Aga1p) via disulfide bonds, tethering the nanobodies on the yeast surface and ensuring the directional arrangement of the nanobodies. Additionally, the HA tag is fused to the C-terminus of Aga2p, serving as the epitope tag to characterize the expression of nanobodies on the yeast surface.
Due to the ease-of-synthesis and good biocompatibility, gold nanoparticles (AuNPs) are ideal solid phase carriers for biosensor signal transductions.30,31 To construct the signal transduction component for the whole-cell biosensor, the AuNPs were functionalized with anti-SARS-CoV-2 monoclonal antibodies and enzymes (horseradish peroxidases, HRP) via physical absorption. The functionalization process of the AuNPs is shown in Figure 1b. After physical absorption, the functionalized AuNPs have dual functions of recognizing target viruses and providing signal outputs, so they are defined as “reporter AuNPs”.
The principle of the whole-cell biosensor for the colorimetric detection of SARS-CoV-2 is shown in Figure 1c. In the presence of SARS-CoV-2 spike proteins, engineered yeasts could easily recognize and concentrate target analytes with simple centrifugation. After labeling with reporter AuNPs, the sandwich immunocomplex was formed between engineered yeast and reporter AuNPs. Upon addition of the enzyme substrate, H2O2 and 3,3, 5,5-tetramethylbenzidine (TMB) as chromogen, HRP on the reporter AuNPs catalyzed the formation of colorimetric products, and the solution color changed from colorless to blue. After the reaction was stopped using H2SO4, the absorbance intensities at 450 nm were recorded. By measuring the increased absorbance after the enzymatic reaction, the developed whole-cell biosensor can quantitatively determine the concentration of SARS-CoV-2 spike protein.
3.2. Characterization of Engineered Yeasts and Reporter AuNPs.
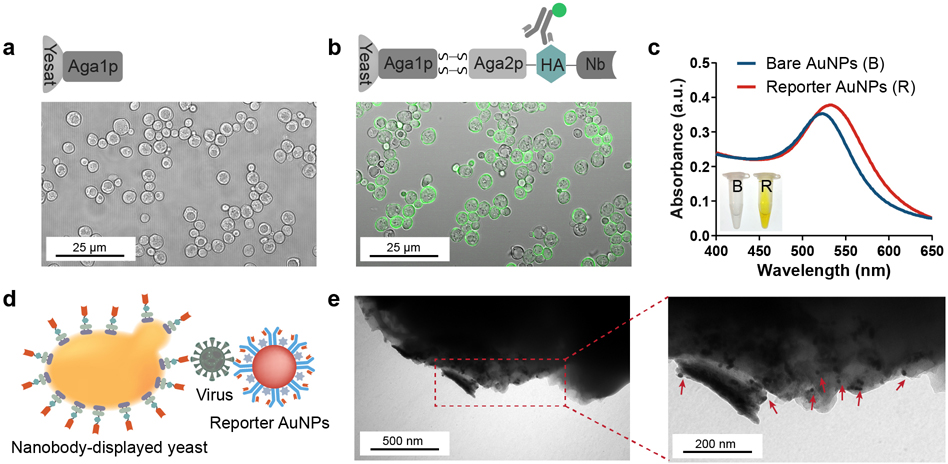
To determine whether nanobodies were expressed on the cell surface, the HA tag was inserted between the Aga2p and the nanobody. Confocal microscopy was used to characterize the expression of the nanobodies on the yeast surface. As shown in Figure 2a, b, while the wild-type yeast can only express Aga1p on the surface, the engineered yeast can secret Aga2p-HA-nanobody fusions from the cells and tether the fusions with a disulfide bond between Aga1p and Aga2p. Fluorescently labeled anti-HA antibodies were added to the solutions of wild-type and engineered yeasts to determine the nanobody expression. After incubation for 1 h and washing three times, the yeasts were collected and placed under a confocal microscope. The results showed that the engineered yeasts have green fluorescence but none of the wild-type yeasts were stained green, indicating that the nanobodies were successfully expressed on the surface of engineered yeasts. According to the previous report, the expression level of nanobodies on the yeast surfaces is around 104–105 copies per cell.24,32 Besides, the flow cytometer results from other literature showed that the yeast display system has a high expression level of nanobodies.33
Figure 2.
Characterization of the nanobody-displayed colorimetric whole-cell biosensor. Confocal image of (a) wild-type yeast and (b) engineered yeast. (c) UV–vis absorption spectra of bare AuNPs and reporter AuNPs. (Inset: photograph of bare AuNPs and reporter AuNPs after enzymatic substrate catalysis. Left side: bare AuNPs. Right side: reporter AuNPs) (d) Schematic illustration of “yeast-virus-AuNPs” theoretical immunocomplex. (e) TEM images of the engineered yeast surface after capturing spike proteins and labeling with reporter AuNPs.
As the signal transduction component, the size of the AuNPs was determined by transmission electron microscopy (TEM). As shown in Figure S1, the diameter of AuNPs is around 17 nm. In addition, the successful modification of reporter AuNPs was characterized by UV-absorption spectra. Compared to bare AuNPs, the spectra of the reporter AuNPs showed a slight red shift along with broadening (Figure 2c),34 indicating that the AuNPs were effectively functionalized with antibodies and HRP enzymes. To verify the catalytic effect of reporter AuNPs, TMB/H2O2 substrates were added to the AuNPs solutions and incubated for 10 min before the reaction was terminated with H2SO4. From the results (inset photograph in Figure 2c), the color of the bare AuNP solution remains colorless, while the reporter AuNPs successfully catalyzed the colorimetric reaction.
Once the SARS-CoV-2 spike proteins were captured by engineered yeasts, the reporter AuNPs were introduced to form a “yeast-virus-AuNP” sandwich immunocomplex (Figure 2d). To verify the immunocomplex more visually, the engineered yeasts and reporter AuNPs were successively added to the spike protein solution, and the complexes were characterized using TEM (Figure 2e). The engineered yeasts without spike protein and the reporter AuNPs were regarded as the control (Figure S2). It was found that the reporter AuNPs were indeed attached to the yeast surface in the presence of spike protein, proving the reliability of the theoretical immunocomplex.
3.3. Determination of Assay Conditions.
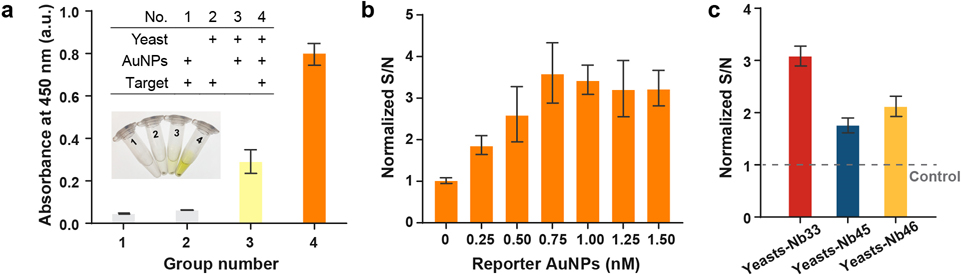
To verify the detection principle of the nanobody-displayed whole-cell biosensor, control tests were performed in the absence of sensing components or target analytes. As shown in Figure 3a, in the absence of engineered yeasts (No.1) or reporter AuNPs (No.2), the absorbance intensities at 450 nm (A450 nm) were lower than 0.1, indicating that the sandwich binding would not be formed without any of the sensing components. In the presence of target analytes (No.4), a visible color change was observed from colorless to yellow (the inset photograph in Figure 3a), and A450 nm was around 0.8, which is 2.5 times higher than the group without the target analytes (No.3). Despite there being a significant difference between negative group (No.3) and positive group (No.4), there was a slight response signal in the negative group, which might be due to nonspecific adsorption between the engineered yeasts and the reporter AuNPs. To eliminate the influence of background signal, normalized signal-to-noise ratio (S/N) was employed as a parameter to indicate the signal intensity in further experiments. The normalized S/N is obtained by dividing the A450 nm of the experimental group (with spike protein) by that of the control group (without spike protein).
Figure 3.
Determination of assay conditions. (a) Comparison of control tests (inset: corresponding photographs). (b) The effect of reporter AuNP concentration on the normalized detection signal. (c) Determination of three nanobody-displayed engineered yeasts. Error bars were calculated from three independent experiments. Note: yeast-Nb33 were used in Figure 3a, b.
Reporter AuNPs are one of the key factors that determines the detection performance. To achieve higher sensitivity, the amounts of reporter AuNPs were investigated for the detection of SARS-CoV-2. After spike proteins were captured by engineered yeasts (107 cells/mL), various amounts of reporter AuNPs (from 0.25 nM to 1.5 nM) were added to generate colorimetric signals. Figure 3b shows the normalized S/N ratio at different concentrations of reporter AuNPs. As the reporter AuNPs concentration increased, the response signal gradually increased. When the concentration of reporter AuNPs exceeds 0.75 nM, the response signal reaches a plateau. Therefore, the optimized reporter AuNPs concentration for the developed biosensor was 0.75 nM.
Three nanobodies were screened as the candidates for the whole-cell biosensors using the full-length prefused and locked SARS-CoV-2 spike protein (Table S1).33 To construct the yeast-based bioreceptor components, three nanobody (Nb-33, Nb-45, and Nb-46) expression cassettes were assembled into the yeast genome through homologous recombination, respectively. These nanobody-displayed yeasts were named yeast-Nb33, yeast-Nb45, and yeast-Nb46. The signal responses of these yeast-based bioreceptor components were determined by detecting SARS-CoV-2 spike protein at the concentration of 0.800 μg/mL. As shown in Figure 3c, the yeast-Nb33 achieved the highest signal response among these bioreceptors. According to the ELISA results from our collaborator, the Nb-33 achieved the lowest half-maximal effective concentration (EC50) toward spike protein among the three nanobodies.21 This result indicated that Nb-33 has a higher capture efficiency and could be regarded as an ideal candidate for SARS-CoV-2 detection. During culturing, yeast-Nb33 grew at the slowest rate, which might indicate there are more nanobodies expressed on the yeast surface.
3.4. Analytical Performance of the Whole-Cell Biosensor.
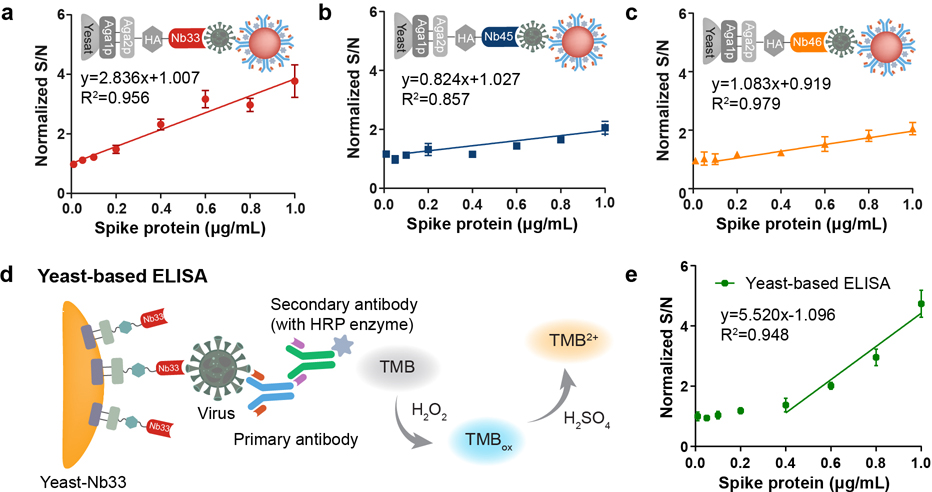
For whole-cell biosensing detection, the nanobody-displayed yeasts were employed as the bioreceptor component to capture spike proteins, and the reporter AuNPs served as the signal transduction components to convert the biological recognition to colorimetric signals. Herein, the sensitivity and quantitative range of the three nanobody-displayed whole-cell biosensors were studied using different spike protein concentrations. The linear detection range (LDR) and the limit of detection (LOD) were investigated (Figure 4a–c). Significantly, the normalized S/N value increased with an increasing concentration of spike proteins. The yeast-Nb33 offers a wider LDR from 0.010 μg/mL to 1.000 μg/mL, while the yeast-Nb45 and yeast-Nb46 have an LDR between 0.100 μg/mL to 1.000 μg/mL. In addition, the LOD was estimated as the control signal plus three times the standard deviation, which was calculated to be 0.037 μg/mL (corresponding to approximately 4 × 108 virion particles/mL) for yeasts-Nb33, 0.393 μg/mL (≈ 4.3 × 109 virion particles/mL) for yeasts-Nb45, and 0.327 μg/mL (≈ 3.6 × 109 virion particles/mL) for yeasts-Nb46. Taking advantage of the wider LDR and lower LOD, yeast-Nb33 was used as the bioreceptor for future experiments.
Figure 4.
Sensitivity analysis of the developed whole-cell biosensor. Calibration plots of nanobody-displayed whole-cell biosensors for detecting SARS-CoV-2 spike proteins using (a) yeast-Nb33, (b) yeast-Nb45, and (c) yeast-Nb46. (d) Schematic illustration of a yeast-based ELISA. (e) Calibration plots of yeast-based ELISA methods used to detect spike proteins.
For comparison, the yeast-based enzyme-linked immunosorbent assay (yeast-based ELISA) was also investigated to detect spike proteins. Since yeast-Nb33 offered a better performance in the previous experiments, it was chosen to capture and concentrate spike proteins in the yeast-based ELISA method. As shown in Figure 4d, the mouse anti-SARS-CoV-2 primary antibody and HRP-conjugated goat antimouse IgG secondary antibody were used as the signal transduction component. After being captured by yeast-Nb33, the yeast-virus complex was recognized with the primary antibodies followed by labeling with the secondary antibodies to generate colorimetric signals. The LDR and LOD of the yeast-based ELISA method for the detection of SARS-CoV-2 spike proteins were 0.4–1.000 μg/mL and 0.402 μg/mL (≈ 4.3 × 109 virion particles/mL) (Figure 4e), respectively. Compared with the reporter AuNPs, despite the primary antibody-secondary antibody structure having a higher normalized S/N value due to the lower steric hindrance, the narrow LDR makes it difficult to detect spike proteins at low concentrations. In contrast, benefiting from the integration of the recognition element and the signal reporting molecule in the same solid phase platform, the use of the reporter AuNPs eliminates the time required to label secondary antibodies and improves the LOD by at least 10-fold, which is favorable for further real sample detection.
3.5. Detection on Serum Samples Spiked with the Spike Protein.
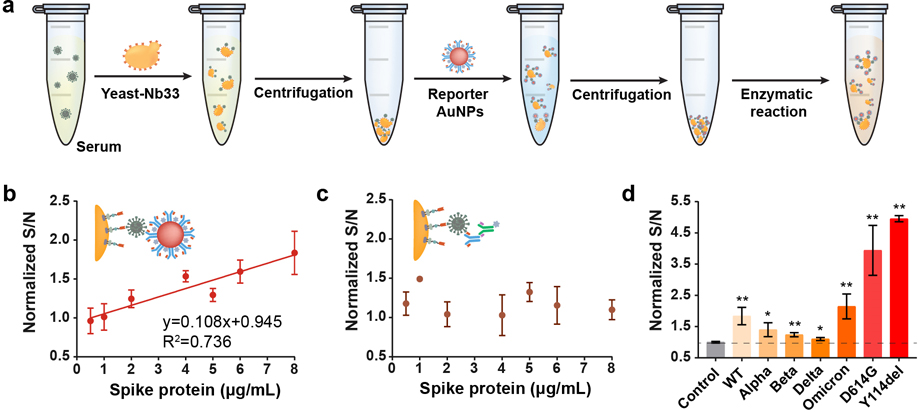
To validate the performance in a complex biological sample, a total of 7 human serum samples were spiked with various concentrations of spike proteins (from wild-type strain) and analyzed with the developed whole-cell biosensor. The workflow of detecting SARS-CoV-2 in human serum is shown in Figure 5a. First, the yeast-Nb33 was added to the spiked human serum to capture spike proteins. After gentle centrifugation, the yeast-virus complexes can be purified and enriched from the serum matrix and then reconstituted in PBS. Subsequently, the reporter AuNPs were conjugated with yeast-virus complexes to form the sandwich bindings. After the unbound AuNPs were washed away, the enzymatic reaction was activated in the presence of the colorimetric substrates, and the absorbance intensities were recorded as signal outputs.
Figure 5.
The developed whole-cell biosensor for detecting SARS-CoV-2 in human serum. (a) Workflow of the SARS-CoV-2 detection. (b) Calibration plots of the whole-cell biosensor for the detection of spike proteins (wild-type) in human serum. (c) Yeast-based ELISA method for the detection of spike protein (wild-type) in human serum. (d) The signal response of the whole-cell biosensor toward the spike proteins (8 μg/mL) from different SARS-CoV-2 variants in human serum.
As depicted in Figure 5b, the response signals exhibited a gradual increase from 0.5 to 8 μg/mL, and the LOD in human serum was 2.5 μg/mL (≈ 2.7 × 1010 virion particles/mL or 5.56 nM), which is very similar to the viral concentration found in patients severely infected with the disease,35,36 indicating that the proposed biosensor has potential to be used as an alternative diagnosis technique in clinical application. It should be underlined that the complex matrix (such as globulin proteins, electrolytes, and antibodies) of human serum makes detection quite challenging. Through engineered yeasts have a superior ability for interference resistance, but the primary antibody-secondary antibody structure of the yeast-based ELISA method was affected by the complex matrix, leading to invalid results (Figure 5c). These data demonstrate the reliability of nanobody-displayed yeast and the stability of gold nanoparticles in the practical application. Taking these advantages, the proposed whole-cell biosensor achieves a higher sensitivity compared to some other reported bio-sensors.37–39
Since the nanobodies were screened using the spike epitopes outside the RBD of wild-type SARS-CoV-2, they should be relatively conserved and might be able to target different variants. Next, we investigated whether the whole-cell biosensor could detect SARS-CoV-2 variants. Human serum spiked with different variants, including Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), and Omicron (BA.4), was analyzed with the whole-cell biosensor. The final concentration of the spike proteins was 8 μg/mL. As shown in Figure 5d, the developed biosensors have response signals for all these variants, with differences (0.01 < p-value <0.05) between the experimental and control groups. In addition, some of the amino acid mutations (D614G and Y114del) carried by many variants were also selected for specificity testing. Notably, the whole-cell biosensor can still detect mutated spike proteins (p-value <0.01), demonstrating that the target region of the nanobody is relatively conserved, which is beneficial for dealing with the emergence of mutated virulent strains in the future.
CONCLUSION
In conclusion, we developed a nanobody-displayed whole-cell biosensor with high sensitivity and specificity to detect the SARS-CoV-2 spike protein in human serum. Benefiting from synthetic biology, it is convenient to construct nanobody-displayed yeasts as programmed bioreceptors to capture and concentrate target analytes from complex matrices. Additionally, the developed biosensor adequately utilizes the high specific surface area of the AuNPs and robustness of the HRP enzymatic system, effectively improving the sensitivity and interference resistance. Using our developed whole-cell biosensor, we were able to detect SARS-CoV-2 spike proteins at a concentration of 0.037 μg/mL (about 4 × 108 virion particles/mL) within 2.5 h. The highlights of this work mainly focus on the following issues: (1) engineered yeasts are utilized as the bioreceptors of whole-cell biosensors, ensuring that the nanobodies are optimally oriented for affinity capture; (2) the use of the reporter AuNPs with highly efficient catalytic activity is conducive to improving the sensitivity and interference resistance of the assay; and (3) the developed whole-cell biosensor can detect different variants with the advantage of low-cost and user-friendly, demonstrating the potential application in high-throughput screening of SARS-CoV-2 in the future.
To further improve the detection sensitivity and shorten the detection time, several methods can be used in the future. First, the orientations of antibodies on the AuNPs surface can be controlled through protein A mediated immobilization or biotin/avidin mediated interaction.40 Second, simultaneous expression of multiple nanobodies on the yeast surface can improve the capture efficiency of target analytes and hence provide a better detection sensitivity. Moreover, the nanobody-displayed yeasts and reporter AuNPs can be introduced to recognize the target at the same time, forming a sandwich structure by the one-pot method to shorten the reaction time. Future works will focus on applying yeast-based biosensors for the detection of viruses.
Supplementary Material
ACKNOWLEDGMENTS
This work is financially supported by the NIH NIGMS (R35GM147069), USDA NIFA (2023–67017-40044), USDA NIFA Hatch (VA-160143), and a start-up grant from Virginia Tech. The following SARS-CoV-2 spike proteins were obtained through BEI Resources, NIAID, NIH: B.1.1.7 (NR-55,277, Alpha variant), B.1.1351 (NR-55,278, Beta variant), B.1.617.2 (NR-55,614, Delta variant), BA.4 (NR-58,646, Omicron variant), D614G (NR-55,418), and Y114del (NR-55,415).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.3c05900.
Information of nanobodies and TEM image of AuNPs and engineered yeasts (PDF)
Contributor Information
Yawen He, Department of Biological Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States.
Zhiyuan Xu, Department of Biological Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States.
Tom Kasputis, Department of Biological Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States.
Xue Zhao, Department of Biological Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States.
Itati Ibañez, Instituto de Química Física de los Materiales, Medio Ambiente y Energía (INQUIMAE), CONICET Universidad de Buenos Aires, Buenos Aires 1428, Argentina.
Florencia Pavan, Instituto de Química Física de los Materiales, Medio Ambiente y Energía (INQUIMAE), CONICET Universidad de Buenos Aires, Buenos Aires 1428, Argentina.
Marina Bok, Incuinta, Instituto Nacional de Tecnología Agropecuaria (INTA), Instituto de Virología e Innovaciones Tecnológicas, INTA-Consejo Nacional de Investigaciones Científicas y Técnicas (IVIT, INTA-CONICET), Buenos Aires 1712, Argentina.
Juan Pablo Malito, Incuinta, Instituto Nacional de Tecnología Agropecuaria (INTA), Instituto de Virología e Innovaciones Tecnológicas, INTA-Consejo Nacional de Investigaciones Científicas y Técnicas (IVIT, INTA-CONICET), Buenos Aires 1712, Argentina.
Viviana Parreno, Incuinta, Instituto Nacional de Tecnología Agropecuaria (INTA), Instituto de Virología e Innovaciones Tecnológicas, INTA-Consejo Nacional de Investigaciones Científicas y Técnicas (IVIT, INTA-CONICET), Buenos Aires 1712, Argentina; Department of Biomedical Sciences and Pathobiology, Virginia-Maryland College of Veterinary Medicine, Virginia Tech, Blacksburg, Virginia 24061, United States.
Lijuan Yuan, Department of Biomedical Sciences and Pathobiology, Virginia-Maryland College of Veterinary Medicine, Virginia Tech, Blacksburg, Virginia 24061, United States.
R. Clay Wright, Department of Biological Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States
Juhong Chen, Department of Biological Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States.
REFERENCES
- (1).Hu B; Guo H; Zhou P; Shi Z-L Characteristics of Sars-Cov-2 and Covid-19. Nature Reviews Microbiology 2021, 19 (3), 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Tao K; Tzou PL; Nouhin J; Gupta RK; de Oliveira T; Kosakovsky Pond SL; Fera D; Shafer RW The Biological and Clinical Significance of Emerging Sars-Cov-2 Variants. Nat. Rev. Genet. 2021, 22 (12), 757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chowell G; Dhillon R; Srikrishna D Getting to Zero Quickly in the 2019-Ncov Epidemic with Vaccines or Rapid Testing. medRxiv 2020, 2020.02. 03.20020271. [Google Scholar]
- (4).Vogels CB; Brito AF; Wyllie AL; Fauver JR; Ott IM; Kalinich CC; Petrone ME; Casanovas-Massana A; Catherine Muenker M; Moore AJ; et al. Analytical Sensitivity and Efficiency Comparisons of Sars-Cov-2 Rt–Qpcr Primer–Probe Sets. Nature microbiology 2020, 5 (10), 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Beavis KG; Matushek SM; Abeleda APF; Bethel C; Hunt C; Gillen S; Moran A; Tesic V Evaluation of the Euroimmun Anti-Sars-Cov-2 Elisa Assay for Detection of Iga and Igg Antibodies. Journal of Clinical Virology 2020, 129, No. 104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).La Rosa G; Brandtner D; Mancini P; Veneri C; Bonanno Ferraro G; Bonadonna L; Lucentini L; Suffredini E Key Sars-Cov-2 Mutations of Alpha, Gamma, and Eta Variants Detected in Urban Wastewaters in Italy by Long-Read Amplicon Sequencing Based on Nanopore Technology. Water 2021, 13 (18), 2503. [Google Scholar]
- (7).Gloag L; Mehdipour M; Chen D; Tilley RD; Gooding JJ Advances in the Application of Magnetic Nanoparticles for Sensing. Adv. Mater. 2019, 31 (48), No. 1904385. [DOI] [PubMed] [Google Scholar]
- (8).Ramos Peña AM; González-Valdez J; Aguilar O Protein a Chromatography: Challenges and Progress in the Purification of Monoclonal Antibodies. J. Sep. Sci. 2019, 42 (9), 1816–1827. [DOI] [PubMed] [Google Scholar]
- (9).Lei H-X; Niu C-C; Li T; Wan Y-F; Liang W-B; Yuan R; Liao P A Novel Electrochemiluminescent Immunoassay Based on Target Transformation Assisted with Catalyzed Hairpin Assembly Amplification for the Ultrasensitive Bioassay. ACS Appl. Mater. Interfaces 2019, 11 (34), 31427–31433. [DOI] [PubMed] [Google Scholar]
- (10).Eivazzadeh-Keihan R; Bahreinizad H; Amiri Z; Aliabadi HAM; Salimi-Bani M; Nakisa A; Davoodi F; Tahmasebi B; Ahmadpour F; Radinekiyan F; et al. Functionalized Magnetic Nanoparticles for the Separation and Purification of Proteins and Peptides. TrAC Trends in Analytical Chemistry 2021, 141, No. 116291. [Google Scholar]
- (11).Moraskie M; Roshid MHO; O’Connor G; Dikici E; Zingg J-M; Deo S; Daunert S Microbial Whole-Cell Biosensors: Current Applications, Challenges, and Future Perspectives. Biosens. Bioelectron. 2021, 191, No. 113359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Guerreiro MR; Freitas DF; Alves PM; Coroadinha AS Detection and Quantification of Label-Free Infectious Adenovirus Using a Switch-on Cell-Based Fluorescent Biosensor. Acs Sensors 2019, 4 (6), 1654–1661. [DOI] [PubMed] [Google Scholar]
- (13).Wu Y; Wang C-W; Wang D; Wei N A Whole-Cell Biosensor for Point-of-Care Detection of Waterborne Bacterial Pathogens. ACS synthetic biology 2021, 10 (2), 333–344. [DOI] [PubMed] [Google Scholar]
- (14).Miller RA; Lee S; Fridmanski EJ; Barron E; Pence J; Lieberman M; Goodson HV Scentsor”: A Whole-Cell Yeast Biosensor with an Olfactory Reporter for Low-Cost and Equipment-Free Detection of Pharmaceuticals. ACS sensors 2020, 5 (10), 3025–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Miettinen K; Leelahakorn N; Almeida A; Zhao Y; Hansen LR; Nikolajsen IE; Andersen JB; Givskov M; Staerk D; Bak Sør.; Kampranis SC A Gpcr-Based Yeast Biosensor for Biomedical, Biotechnological, and Point-of-Use Cannabinoid Determination. Nat. Commun. 2022, 13 (1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Venkatesh A; Brickner H; Looney D; Hall D; Aronoff-Spencer E Clinical Detection of Hepatitis C Viral Infection by Yeast-Secreted Hcv-Core: Gold-Binding-Peptide. Biosens. Bioelectron. 2018, 119, 230–236. [DOI] [PubMed] [Google Scholar]
- (17).Venkatesh AG; Sun A; Brickner H; Looney D; Hall DA; Aronoff-Spencer E Yeast Dual-Affinity Biobricks: Progress Towards Renewable Whole-Cell Biosensors. Biosens. Bioelectron. 2015, 70, 462–468. [DOI] [PubMed] [Google Scholar]
- (18).Zare H; Aghamollaei H; Hosseindokht M; Heiat M; Razei A; Bakherad H Nanobodies, the Potent Agents to Detect and Treat the Coronavirus Infections: A Systematic Review. Molecular and Cellular Probes 2021, 55, No. 101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Pymm P; Adair A; Chan L-J; Cooney JP; Mordant FL; Allison CC; Lopez E; Haycroft ER; O’Neill MT; Tan LL; et al. Nanobody Cocktails Potently Neutralize Sars-Cov-2 D614g N501y Variant and Protect Mice. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (19), No. e2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schoof M; Faust B; Saunders RA; Sangwan S; Rezelj V; Hoppe N; Boone M; Billesbølle CB; Puchades C; Azumaya CM; et al. An Ultrapotent Synthetic Nanobody Neutralizes Sars-Cov-2 by Stabilizing Inactive Spike. Science 2020, 370 (6523), 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pavan MF; Bok M; Juan RBS; Malito JP; Marcoppido GA; Franco DR; Militello DA; Schammas JM; Bari S; Stone WB; López K; Porier DL; Muller J; Auguste AJ; Yuan L; Wigdorovitz A; Parreño V; Ibañez LI Nanobodies against Sars-Cov-2 Reduced Virus Load in the Brain of Challenged Mice and Neutralized Wuhan, Delta and Omicron Variants. bioRxiv, 2023,2023.03.14.532528. [Google Scholar]
- (22).Engler C; Kandzia R; Marillonnet S A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PloS one 2008, 3 (11), No. e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wright RC; Zahler ML; Gerben SR; Nemhauser JL Insights into the Evolution and Function of Auxin Signaling F-Box Proteins in Arabidopsis Thaliana through Synthetic Analysis of Natural Variants. Genetics 2017, 207 (2), 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chao G; Lau WL; Hackel BJ; Sazinsky SL; Lippow SM; Wittrup KD Isolating and Engineering Human Antibodies Using Yeast Surface Display. Nature protocols 2006, 1 (2), 755–768. [DOI] [PubMed] [Google Scholar]
- (25).Gietz RD; Schiestl RH High-Efficiency Yeast Transformation Using the Liac/Ss Carrier DNA/Peg Method. Nature protocols 2007, 2 (1), 31–34. [DOI] [PubMed] [Google Scholar]
- (26).Kasputis T; Hilaire SS; Xia K; Chen J Colorimetric Detection of Antimicrobial Resistance from Food Processing Facilities Using a Crispr System. ACS Food Science & Technology 2023, 3, 17. [Google Scholar]
- (27).Liao T; Yuan F; Yu H; Li Z An Ultrasensitive Elisa Method for the Detection of Procalcitonin Based on Magnetic Beads and Enzyme-Antibody Labeled Gold Nanoparticles. Analytical Methods 2016, 8 (7), 1577–1585. [Google Scholar]
- (28).Shusta EV; Pepper LR; Cho YK; Boder ET A Decade of Yeast Surface Display Technology: Where Are We Now? Combinatorial chemistry & high throughput screening 2008, 11 (2), 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lim S; Glasgow JE; Filsinger Interrante M; Storm EM; Cochran JR Dual Display of Proteins on the Yeast Cell Surface Simplifies Quantification of Binding Interactions and Enzymatic Bioconjugation Reactions. Biotechnology journal 2017, 12 (5), No. 1600696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hua Z; Yu T; Liu D; Xianyu Y Recent Advances in Gold Nanoparticles-Based Biosensors for Food Safety Detection. Biosens. Bioelectron. 2021, 179, No. 113076. [DOI] [PubMed] [Google Scholar]
- (31).Zhang L; Mazouzi Y; Salmain M; Liedberg B; Boujday S Antibody-Gold Nanoparticle Bioconjugates for Biosensors: Synthesis, Characterization and Selected Applications. Biosens. Bioelectron. 2020, 165, No. 112370. [DOI] [PubMed] [Google Scholar]
- (32).Helma J; Cardoso MC; Muyldermans S; Leonhardt H Nanobodies and Recombinant Binders in Cell Biology. J. Cell Biol. 2015, 209 (5), 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhao X; Rahman M; Xu Z; Kasputis T; He Y; Yuan L; Wright RC; Chen J Engineered Yeast Displaying Specific Norovirus-Binding Nanobodies for the Concentration and Detection of Human Norovirus in Food Matrix. J. Agric. Food Chem. 2023, 71, 8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Liu Y; Zhang L; Wei W; Zhao H; Zhou Z; Zhang Y; Liu S Colorimetric Detection of Influenza a Virus Using Antibody-Functionalized Gold Nanoparticles. Analyst 2015, 140 (12), 3989–3995. [DOI] [PubMed] [Google Scholar]
- (35).Paria D; Kwok KS; Raj P; Zheng P; Gracias DH; Barman I Label-Free Spectroscopic Sars-Cov-2 Detection on Versatile Nanoimprinted Substrates. Nano Lett. 2022, 22 (9), 3620–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pan Y; Zhang D; Yang P; Poon LL; Wang Q Viral Load of Sars-Cov-2 in Clinical Samples. Lancet infectious diseases 2020, 20 (4), 411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cennamo N; Pasquardini L; Arcadio F; Lunelli L; Vanzetti L; Carafa V; Altucci L; Zeni L Sars-Cov-2 Spike Protein Detection through a Plasmonic D-Shaped Plastic Optical Fiber Aptasensor. Talanta 2021, 233, No. 122532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mojsoska B; Larsen S; Olsen DA; Madsen JS; Brandslund I; Alatraktchi F. A. a. Rapid Sars-Cov-2 Detection Using Electrochemical Immunosensor. Sensors 2021, 21 (2), 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Pinals RL; Ledesma F; Yang D; Navarro N; Jeong S; Pak JE; Kuo L; Chuang Y-C; Cheng Y-W; Sun H-Y; Landry MP Rapid Sars-Cov-2 Spike Protein Detection by Carbon Nanotube-Based near-Infrared Nanosensors. Nano Lett. 2021, 21 (5), 2272–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gao S; Guisán JM; Rocha-Martin J Oriented Immobilization of Antibodies onto Sensing Platforms-a Critical Review. Analytica chimica acta 2022, 1189, No. 338907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.