Abstract
The production of the human papillomavirus type 16 (HPV-16) is intimately tied to the differentiation of the host epithelium that it infects. Infection occurs in the basal layer of the epithelium at a site of wounding, where the virus utilizes the host DNA replication machinery to establish itself as a low-copy-number episome. The productive stage of the HPV-16 life cycle occurs in the postmitotic suprabasal layers of the epithelium, where the virus amplifies its DNA to high copy number, synthesizes the capsid proteins (L1 and L2), encapsidates the HPV-16 genome, and releases virion particles as the upper layer of the epithelium is shed. Papillomaviruses are hypothesized to possess a mechanism to overcome the block in DNA synthesis that occurs in the differentiated epithelial cells, and the HPV-16 E7 oncoprotein has been suggested to play a role in this process. To determine whether E7 plays a role in the HPV-16 life cycle, an E7-deficient HPV-16 genome was created by inserting a translational termination linker (TTL) in the E7 gene of the full HPV-16 genome. This DNA was transfected into an immortalized human foreskin keratinocyte cell line shown previously to support the HPV-16 life cycle, and stable cell lines were obtained that harbored the E7-deficient HPV-16 genome episomally, the state of the genome found in normal infections. By culturing these cells under conditions which promote the differentiation of epithelial cells, we found E7 to be necessary for the productive stage of the HPV-16 life cycle. HPV-16 lacking E7 failed to amplify its DNA and expressed reduced amounts of the capsid protein L1, which is required for virus production. E7 appears to create a favorable environment for HPV-16 DNA synthesis by perturbing the keratinocyte differentiation program and inducing the host DNA replication machinery. These data demonstrate that E7 plays an essential role in the papillomavirus life cycle.
Human papillomaviruses (HPV) are small, double-stranded DNA viruses that infect epithelial cells and lead to the production of warts. A subset of HPVs infect the anogenital tract and can be placed into two categories, the low- and high-risk genotypes. While both low- and high-risk HPV genotypes lead to the production of warts, the high-risk genotypes are also associated with anogenital cancers including cervical cancer. HPV-16 is the genotype most commonly found in cervical cancer (31). The HPV life cycle is intimately tied to the differentiation of the host epithelium that it infects. The HPV life cycle begins in the basal layer of the epithelium, where the virus is thought to gain entry at a site of wounding. In this layer of the epithelium, the nonproductive stage of the HPV life cycle occurs, where the virus establishes itself as a low-copy-number episome by synthesizing its DNA on average once per cell cycle via a bidirectional theta mode (1a, 8, 12, 30). The productive stage of the HPV life cycle occurs in the suprabasal layers of the epithelium, where the virus amplifies its DNA to a high copy number. Here, the virus switches from a theta to a rolling-circle mode of DNA replication (8), synthesizes the capsid proteins, L1 and L2, and releases assembled virions (15). The study of the HPV-16 life cycle and the role of the various viral genes in the life cycle has been hindered by the lack of a cell culture system which supports the viral life cycle. Nevertheless, studying the life cycle of the virus is important, because understanding the life cycle is important for creating antiviral therapies that can stop the spread of HPV-16, which has been associated with the majority of cervical cancers.
An important feature of the HPV life cycle is that it depends on the host DNA replication machinery to synthesize its DNA, because the virus does not encode a DNA polymerase. HPV provides the viral proteins E1 and E2, which bind to the origin of HPV DNA replication and recruit the host factors necessary for viral DNA synthesis, including DNA polymerase α (4). The host DNA replication machinery is readily available in the proliferating basal layer of the epithelium where the nonproductive stage of the HPV life cycle occurs. However, host DNA replication machinery is thought to become limiting in the postmitotic, differentiated cells located in the suprabasal compartment of the epithelium (2, 9, 10). Paradoxically, it is in the suprabasal compartment of the epithelium where HPV amplifies its DNA to high copy number. Thus, the virus probably possesses a mechanism to make these cells permissive for DNA synthesis during the productive stage of the viral life cycle.
The viral oncogene E7 is hypothesized to cause the postmitotic suprabasal cells to become permissive for DNA synthesis (2, 6); however, the role of E7 in the HPV life cycle has not been elucidated. E7 in the context of the HPV-16 life cycle could behave differently from E7 alone, because its activities may be affected by other viral genes. For example, E6 and E7 affect different proteins involved in the cell cycle; E6 binds and degrades p53, while E7 binds to pRb (17). Previous studies in which E7 has been expressed in the absence of the rest of the HPV genome have demonstrated that E7 alone is sufficient to induce DNA synthesis in differentiated keratinocytes and that E7 alone induces factors of the host DNA replication machinery such as proliferating-cell nuclear antigen (PCNA) (2, 6). The induction of host DNA replication proteins by E7 alone is thought to occur through the ability of E7 to sequester pRb, liberating E2F and allowing it to induce the expression of genes encoding the DNA replication machinery. Other papillomavirus genes, notably E6, also can alter epithelial differentiation and induce epithelial cell hyperplasia (23). Therefore, it is unclear what role E7 specifically plays in the viral life cycle. Our study has focused on whether E7 creates a favorable environment for viral DNA synthesis in differentiated keratinocytes during the productive stage of the viral life cycle in the presence of other viral genes that may affect the activities of E7 or may have redundant activities.
To assess the role of E7 in the papillomavirus life cycle, we obtained cell lines that supported stable maintenance of the E7-deficient HPV-16 genome. This was possible using an immortalized human foreskin keratinocyte (HFK) cell line, BC-1-Ep/SL, which supports the full viral life cycle of HPV-16 (7). Using cells harboring the wild-type or E7-deficient HPV-16 genome episomally, we were able to determine the effects of the loss of E7 on the HPV-16 life cycle. The cells harboring wild-type or E7-deficient HPV-16 were grown using organotypic raft cultures and also suspended in methylcellulose to induce differentiation and thus promote the productive stage of the HPV-16 life cycle. The loss of E7 had a negative effect on the productive stage of the HPV life cycle as evidenced by the lack of viral DNA amplification and reduced L1 expression. Cells harboring the wild-type HPV-16 genome induced the host DNA replication machinery and inhibited the differentiation of keratinocytes in the suprabasal layer of the rafts. Both properties were dependent on the presence of an intact E7 gene. We hypothesize, therefore, that the perturbation of differentiation and induction of DNA synthesis by E7 contributes to its ability to create a favorable environment for viral DNA amplification during the productive stage of the HPV-16 life cycle.
MATERIALS AND METHODS
HPV DNA preparation for transfections.
As a source of HPV-16 DNA, plasmid pEFHPV-16W12E, derived from W12E cells, was used (GenBank accession no. AF125673) (7). To construct pEFHPV-16W12E/E7TTL, a translational termination linker (TTL), 5′-TTAGTTAACTAA-3′, was inserted at nucleotide 711 in the E7 gene of pEFHPV-16W12E. For transfections into BC-1-Ep/SL cells, the viral DNA sequences from pEFHPV-16W12E or pEFHPV-16W12E/E7TTL were excised from the pUC19 vector by digestion with BamHI. The HPV DNAs were gel purified by electroelution, ethanol precipitated, quantified, and ligated at low concentrations (50 ng/μl) to avoid the formation of multimers.
Cell culture.
Epithelial cells were cultured as described previously (8, 16). Briefly, cells were maintained at subconfluence on mitomycin C-treated m1 3T3 feeder cells in F medium (0.66 mM Ca2+) composed of 3 parts Ham's F12 medium to 1 part Dulbecco's modified Eagle's medium and supplemented with the following components: 5% fetal bovine serum (FBS), adenine (24 μg/ml), cholera toxin (8.4 ng/ml), epidermal growth factor (10 ng/ml), hydrocortisone (2.4 μg/ml), and insulin (5 μg/ml). When the epithelial cells reached subconfluence, the m1 3T3 feeder cells were removed with 0.02% EDTA and vigorous pipetting. The epithelial cells were removed from the tissue culture dishes by treatment with 0.1% trypsin–0.5 mM EDTA at 37°C.
Stable transfections.
The recircularized HPV-16W12E DNA or HPV-16W12E/E7TTL DNA (2 to 3.2 μg) was cotransfected with 1.2 to 1.8 μg of pEGFPN1 (Clonetics), which encodes the green fluorescent protein and confers G418 resistance, into immortalized HFKs (BC-1-EP/SL cells). The DNA was transfected into the cells on a 6-cm dish in low-Ca2+ F medium supplemented with adenine (24 μg/ml), cholera toxin (8.4 ng/ml), epidermal growth factor (10 ng/ml), hydrocortisone (2.4 μg/ml), and insulin (5 μg/ml) by using LipofectACE (Gibco-BRL), LipofectAMINE (Gibco-BRL), or Superfect (Qiagen) as specified by the manufacturer. At 1 day posttransfection, the cells were trypsinized and plated in F medium (0.66 mM Ca2+) supplemented with 5% FBS, adenine (24 μg/ml), cholera toxin (8.4 ng/ml), hydrocortisone (2.4 μg/ml), and insulin (5 μg/ml) on 10-cm dishes containing m1 3T3 feeder cells. At 2 days posttransfection, 100 μg of G418 per ml was added to the medium. The level of G418 was reduced to 50 μg/ml 4 days after transfection. The cells were fed every other day until the untransfected control cells died, usually 5 to 6 days after selection began. The resulting G418-resistant colonies (2 to 10 colonies per 10-cm dish) were pooled and expanded for Southern analysis. This pool was referred to as a cell population. To generate subclones, 1,000 cells from the populations were plated on a 10-cm dish of m1 3T3 feeder cells. After 5 to 7 days, individual colonies were picked and expanded. Cell lines 41AS2 and 13-9 are subcloned populations. All other cell lines are cell populations.
Screening stable transfectants.
Hirt DNA (low-molecular-weight DNA) (14) was extracted from one 10-cm dish of each HPV-16 or HPV-16/E7TTL stably transfected cell population. Half of the resulting DNA from each cell population was linearized, while the other half remained undigested to determine the presence of open-circular and supercoiled viral DNA, indicators of episomal DNA as detected in productive HPV infections. Hirt DNA extracted from W12E cells was used as a positive control and a marker for open-circular, linear, and supercoiled HPV-16 DNA. The DNA was electrophoresed on a 0.8% agarose gel and transferred to a nitrocellulose membrane (Schleicher & Schuell). The blot was probed with a full-length HPV-16 probe generated by restriction enzyme digestion of pEFHPV-16W12E with BamHI and labeled with [α-32P]dCTP using a random-primer labeling kit (Amersham). To visualize HPV DNA, the blot was exposed to a PhosphorImager screen for 1 day or X-ray film for 2 to 5 days.
Raft cultures.
Transwell inserts (24 mm in diameter and 0.4 μm in pore size) (Costar) were coated with 1 ml of bovine tendon collagen type I (1 mg/ml) (Upstate Biotechnology, Inc.) (1, 7). The remaining collagen was impregnated with early-passage human foreskin fibroblasts (7.5 × 105 cells/ml) and plated on the collagen-coated Transwell inserts. The collagen was allowed to contract for 5 days in a 5% CO2 incubator at 37°C in F12 medium containing 10% FBS. After the collagen had contracted, 7 × 105 keratinocytes per 50 μl of keratinocyte plating medium (F medium [1.88 mM Ca2+] containing 0.5% FBS, adenine [24 μg/ml], cholera toxin [8.4 ng/ml], hydrocortisone [2.4 μg/ml], and insulin [5 μg/ml]) were plated on the collagen plug. Four days after the keratinocytes were plated, the rafts were placed on two 1-in2 cotton pads (Schleicher & Schuell) in a six-well tray (Organogenesis) to lift to the air-liquid interface. The rafts were fed from below the Transwell insert with cornification medium (keratinocyte plating medium containing 5% FBS) every third day. Ten days after being lifted to the air-liquid interface, the rafts were fed for 8 h with cornification medium containing 10 μM bromodeoxyuridine (BrdU). Subsequently, the rafts were fixed in 4% formalin at 4°C for 15 h, embedded in 2% agar–1% formalin followed by paraffin, and cut into 4-μm cross sections.
DNA in situ hybridization.
DNA in situ hybridization was performed on 4-μm cross sections of paraffin-embedded rafts using the Microprobe system (Fisher) (3). Briefly, slides were dewaxed in xylene-HemoDe (FisherBrand) at a 1:3 ratio. The slides were then hydrated in a graded series of alcohol washes, digested with 3 mg of pepsin per ml at 37°C for 20 min, neutralized with Tris-saline Brij (pH 7.5), and dehydrated with a graded series of alcohol washes. The following biotin-labeled DNA probes were applied to the slides at 1.5 μg/ml: full-length HPV-16, human placental DNA (positive control), and pBR322 (negative control). To denature the probes and target DNA, the slides were heated to 105°C for 18 min. Hybridization was carried out at 37°C for 2 h. Hybrids were detected by treatment with avidin-alkaline phosphatase conjugate (1:300) (Sigma) for 20 min (the condition found to detect amplified HPV DNA). Under less dilute conditions, HPV DNA can be found throughout the raft in cells harboring either HPV-16 or HPV-16/E7TTL episomally. For color development, the slides were incubated with McGadey reagent (nitroblue tetrazolium chloride [0.33 mg/ml] and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt [0.16 mg/ml] [both from Boehringer Mannheim Biochemicals]) for 1 h at 37°C. The sections were counterstained with nuclear fast red, mounted with Crystal Mount (Biomeda Corp., Foster City, Calif.), and postmounted with Permount (Fisher).
Immunohistochemistry.
Immunohistochemistry was performed on 4-μm cross sections of paraffin-embedded rafts using the Vectastain ABC kit (Vector). The slides were dewaxed in xylene and rehydrated in a graded series of alcohol washes. For L1, PCNA, keratin 10 (K10), filaggrin, and involucrin immunohistochemistry, the following conditions were used. After dewaxing, the slides were treated with 3 mg of pepsin per ml for 10 min, incubated with the blocking serum supplied in the Vectastain ABC kit, and incubated with the primary antibodies at room temperature. For L1 staining, the anti-L1 antibody (camvir-1) was used at a dilution of 1:50 for 3 h. PCNA staining was performed at a dilution of 1:200 for 3 h using the PCNA-specific antibody (clone 19F4 [Boehringer Mannheim]). For keratin 10 staining, the K10-specific antibody (clone Ck 8.60 [Sigma]) was used at a dilution of 1:200 for 2.5 h. For filaggrin staining, a monoclonal anti-human filaggrin antibody (Biomedical Technologies Inc.) was diluted 1:100 for 3 h. Involucrin staining was performed using the anti-involucrin antibody (clone SY5 [Sigma]) at a dilution of 1:200 for 3 h. DNA polymerase α (DNA polα), p53, mdm2, and p21 immunohistochemistry was performed under the following conditions. To expose the epitope, the slides were boiled in a microwave for 10 min in 10 mM sodium citrate buffer (pH 6.0). The slides were treated with blocking serum, as mentioned previously, followed by incubation with the primary antibody. DNA polα (clone CL-22-2-42B [PanVera]) and mdm2 (clone 2A10) (5) immunohistochemistry was performed at a dilution of 1:100, anti-human p21 (clone 6B6 [PharMingen]) was used at a dilution of 1:500, and anti-human p53 (clone DO-1) was used at a dilution of 1:1,000, all for 3 h. All antibodies were detected using the Vectastain ABC kit as specified by the manufacturer. Slides were counterstained with hematoxylin (Vector) for 2 min to reveal the tissue morphology and mounted with Cytoseal (Stephens Scientific). For detection of BrdU incorporation, the BrdU staining kit (Calbiochem) was used as specified by the manufacturer, except that incubation with the primary antibody was performed for 3 h. To quantify the percentage of positively stained cells presented in Table 2 for each sample, the number of positively stained cells in four fields (magnification, ×40) was averaged and divided by the average of total nucleated cells in four fields (×40).
TABLE 2.
Summary of immunohistochemical staining for DNA synthesis markers in raft cultures
| DNA synthesis marker and layer of raft culture | % of cells positive for marker in indicated layer of raft culture
|
||||
|---|---|---|---|---|---|
| BC-1-Ep/SL | BC-1-Ep/SL/HPV-16 (41AWT) | BC-1-Ep/SL/HPV-16 (11-1) | BC-1-Ep/SL/HPV-16/E7TTL (13-9) | W12E | |
| BrdU | |||||
| Basal layer | 13 | 24 | 20 | 19 | 26 |
| Suprabasal layer | 0 | 17 | 14 | 0 | 20 |
| PCNA | |||||
| Basal layer | 34 | 45 | 43 | 42 | 76 |
| Suprabasal layer | 0 | 12 | 10 | 0 | 31 |
| DNA polα | |||||
| Basal layer | 14 | 96 | 82 | 64 | 100 |
| Suprabasal layer | 0 | 54 | 53 | 14 | 55 |
| mdm2 | |||||
| Basal layer | 15 | 90 | 81 | 16 | 97 |
| Suprabasal layer | 16 | 52 | 73 | 11 | 48 |
HPV DNA levels in methylcellulose-cultured cells.
Epithelial cells grown to subconfluence on m1 3T3 feeder cells were trypsinized and counted. A total of 107 cells were suspended in a 50-ml conical tube (Falcon) containing 20 ml of F medium (0.66 mM Ca2+) composed of 1.68% methylcellulose, 20% FBS, adenine (24 μg/ml), cholera toxin (8.4 ng/ml), hydrocortisone (2.4 μg/ml), and insulin (5 μg/ml). To recover cells from suspension, the methylcellulose medium was diluted with serum-free F medium and centrifuged at 2,000 rpm for 20 min in a Beckman GPR centrifuge. After aspiration of the dilute methylcellulose, the resulting cell pellet was washed twice in 1× phosphate-buffered saline and centrifuged. Cells were counted to determine the number of cells recovered. Hirt DNA (14) was extracted from 5 × 105 cells and electrophoresed on a 0.8% agarose gel. Southern analysis was performed using an [α-32P]dCTP-labeled HPV-16 probe. The blots were exposed to a PhosphorImager screen, and the DNA was quantified using ImageQuant software.
Apoptosis.
DNA fragmentation was detected in 4-μm sections of paraffin-embedded raft cultures using the ApopTag kit (Intergen). Slides were dewaxed in xylene and rehydrated in a graded series of alcohol washes. Fragmented DNA was labeled in situ with digoxigenin using the terminal deoxynucleotidyltransferase (TdT) enzyme at 37°C for 1 h. Digoxigenin-labeled DNA was detected with an antidigoxigenin antibody conjugated to fluorescein. The slides were counterstained with fast green to reveal tissue morphology, mounted using 0.4% N-propyl gallate–50% glycerol in phosphate-buffered saline, and viewed by fluorescence microscopy.
RESULTS
The viral oncogene E7, in the context of the full episomal HPV-16 genome, extends the life span of HFKs.
Previously, we showed that HFKs can support the HPV-16 life cycle (7). To determine whether E7 plays a role in the HPV-16 life cycle, we introduced a TTL into the E7 gene of a plasmid containing a replication-competent HPV-16 genome (7). Early-passage HFKs were cotransfected with plasmid pEGFPN1 (which confers resistance to G418 and expresses the green fluorescent protein) and either the wild-type or E7 mutant HPV-16 genome. Following selection in G418, the resulting colonies were expanded and screened for the presence of episomal HPV DNA by Southern analysis. While 100% of the colonies arising from the transfection with wild-type HPV-16 could be screened for the presence of episomal viral DNA, analysis of the colonies arising from transfections with the E7-deficient HPV-16 genome was problematic. Cells transfected with this viral genome senesced before they could be expanded to sufficient numbers for analysis. Since cells transfected with plasmid pEGFPN1 alone also senesced, we interpret these data to indicate that wild-type HPV-16, but not E7-defective HPV-16, causes an extension of the life span of HFKs.
The viral oncogene E7 is not necessary for HPV-16 DNA replication during the nonproductive stage of the viral life cycle.
An immortalized HFK cell line, BC-1-Ep/SL (NIKS; see reference 1), can support the HPV life cycle (7). To circumvent the problem of senescence in early-passage HFKs, we used the BC-1-Ep/SL cells to determine the role of E7 in the HPV-16 life cycle. BC-1-Ep/SL cells were transfected as described above for the early-passage HFKs and cultured to maintain their basal cell-like properties, thereby providing a cellular environment supportive of the nonproductive stage of the HPV life cycle. We screened for the presence of episomal HPV DNA by isolating low-molecular-weight DNA and performing Southern analysis. Some examples of the cell populations harboring episomal HPV DNA are shown in Fig. 1. Supercoiled and open-circular DNA was present in all samples in Fig. 1, indicative of episomal HPV DNA as seen in HPV infections. The Southern blot shown in Fig. 1 contains examples of BC-1-Ep/SL cells stably harboring the wild-type HPV-16 genome episomally [BC16/E7(+) cells] (lanes 2 to 5) and examples of cells stably harboring the E7-deficient HPV-16 genome episomally [BC16/E7(−) cells] (lanes 6 to 11). The number of colonies screened and found to have episomal HPV DNA is summarized in Table 1. We found that comparable numbers of colonies harbored wild-type or E7-deficient HPV-16 episomally (Table 1). The copy numbers of the wild-type or E7-deficient HPV-16 genomes in the different populations were assessed by Southern analysis of total genomic DNA and found to range from 5 to 50 copies/cell. One population (13-9) harboring the E7TTL HPV-16 DNA had a copy number of several hundred. The similarity in copy number between wild-type and E7-deficient HPV-16 correlated with similarities in their efficiency of replication as assessed in transient-replication assays (data not shown). These results demonstrate that E7 is not needed for HPV-16 DNA synthesis in the nonproductive stage of the viral life cycle.
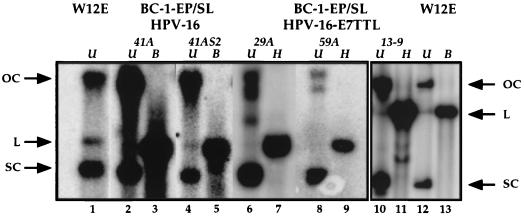
FIG. 1.
Analysis of HPV DNA isolated from BC-1-Ep/SL stably transfected populations. Shown is an autoradiograph of a Southern blot containing Hirt DNA extracted from BC-1-Ep/SL/HPV-16 [BC16/E7(+)] populations (41A and 41AS2), BC-1-Ep/SL/HPV-16/E7TTL [BC16/E7(−)] populations (29A, 59A, and 13-9), and W12E cells. The blot was hybridized to a full-length HPV-16 DNA probe. Undigested (U) Hirt DNA from the BC16/E7(+) cells (41A and 41AS2), the BC16/E7(−) cells (29A, 59A, and 13-9), and W12E cells contains open-circular (OC) and supercoiled (SC) HPV-16 DNA (lanes 1, 2, 4, 6, 8, 10, and 12). The DNA from the BC16/E7(+) cells (41A and 41AS2) was linearized (L) by digestion with BamHI (B) (lanes 3 and 5), and the DNA from BC16/E7(−) cells (29A, 59A, and 13-9) was linearized (L) by digestion with HpaI (H) (lanes 7, 9, and 11). Lanes 10 to 13 are from a different gel and are shown as a separate box. Arrows on either side of the autoradiographs indicate the migration of open-circular, linear, and supercoiled DNA.
TABLE 1.
Summary of stable transfectants harboring episomal HPV-16 DNA
| Cell line | HPV DNA | No. of transfections | Total no. of populations screened | No. with episomal DNA | % with episomal DNA |
|---|---|---|---|---|---|
| HFK | HPV-16 | 38 | 38 | 6 | 16 |
| HFK | HPV-16 E7TTL | 50 | NAa | NA | NA |
| BC-1-Ep/SL | HPV-16 | 117 | 117 | 9 | 7 |
| BC-1-Ep/SL | HPV-16 E7TTL | 79 | 79 | 7 | 9 |
NA, not available.
Loss of E7 leads to defects in the productive stage of the viral life cycle.
To determine the effect of the loss of E7 on virion production, we induced differentiation of the cells harboring either the wild-type or the E7-deficient HPV-16 genome episomally by suspension in methylcellulose or use of raft cultures. Using these cultures, viral DNA amplification, a hallmark of the productive stage of the HPV life cycle, was monitored. Evidence for viral DNA amplification was determined by comparing the level of HPV-16 DNA produced during the nonproductive stage of the life cycle to the level of HPV-16 DNA produced in the productive stage of the life cycle. To mimic the nonproductive stage of the viral life cycle, cells were cultured under conditions in which they remained primarily undifferentiated. To induce the productive stage of the viral life cycle, cells were induced to differentiate by suspension in semisolid methylcellulose medium for 1 and 2 days (8, 13, 21). Low-molecular-weight DNA was extracted from an equivalent number of undifferentiated and differentiated cells and subjected to Southern analysis. After 1 day in methylcellulose, the amount of wild-type HPV-16 DNA increased dramatically and reproducibly (Fig. 2A, lanes 3 and 4 and lanes 6 and 7). Two days after suspension in methylcellulose, the amount of wild-type HPV-16 DNA was larger than the amount present in the undifferentiated cells but was reduced compared to the amount present after 1 day in methylcellulose (lanes 3 to 8). This reduction probably reflects the induction of apoptosis in keratinocytes upon suspension in methylcellulose (22). In contrast, cells harboring the E7-deficient HPV-16 genome reproducibly failed to exhibit any increase in viral DNA production after suspension in methylcellulose; rather, they showed a decreased amount of viral DNA regardless of the HPV DNA copy number (Fig. 2A, lanes 9 to 17; Fig. 2B, lanes 3 to 5). Again, this decrease probably is attributable to the induction of apoptosis by suspension of cells in methylcellulose. These initial results provide evidence that E7 is necessary for viral DNA amplification.
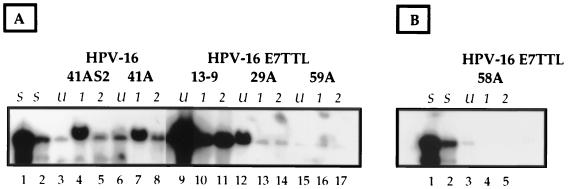
FIG. 2.
Southern analysis of HPV DNA extracted from stably transfected BC-1-Ep/SL cells cultured in methylcellulose. Shown are autoradiographs of Southern blots containing Hirt DNA extracted from 2 × 105 cells of BC-1-Ep/SL/HPV-16 [BC16/E7(+)] populations (41A and 41AS2) and BC-1-Ep/SL/HPV-16/E7TTL [BC16/E7(−)] populations (29A, 59A, and 13-9) (A) and a BC-1-Ep/SL/HPV-16/E7TTL population (58A) (B) cultured under various conditions. The resulting DNA was linearized with BamHI. These cells were cultured under conditions so that they remained primarily undifferentiated (U) (lanes 3, 6, 9, 12, and 15 [A] and lane 3 [B]), suspended in methylcellulose for 1 day (1) (lanes 4, 7, 10, 13, and 16 [A] and lane 4 [B]), and suspended in methylcellulose for 2 days (2) (lanes 5, 8, 11, 14, and 17 [A] and lane 5 [B]). Two amounts of standards (S) of linearized HPV-16 DNA were run, 1,000 pg (lanes 1) and 100 pg (lanes 2). The blot was hybridized to a full-length HPV-16 DNA probe. Note that in all of the cell populations used in these experiments, only episomal HPV-16 genomes could be detected by Southern analyses (Fig. 1 and data not shown).
The effect of the loss of E7 on viral DNA amplification was also monitored in raft cultures containing cells harboring either the wild-type or the E7-deficient HPV-16 genome. The W12E raft cultures were used as a positive control for productive HPV-16 infections throughout this study because W12E cells were derived from an HPV-16-infected patient and harbor HPV-16 episomally (16, 24, 25). Paraffin-embedded sections of the raft cultures were stained with hematoxylin and eosin to reveal stratification of the keratinocytes (Fig. 3A to D). Viral DNA amplification was monitored by in situ hybridization using a biotin-labeled HPV-16 DNA probe. Using conditions that allowed the detection only of cells with highly amplified HPV-16 DNA, we detected amplified HPV-16 in the suprabasal layers of BC16/E7(+) rafts (Fig. 3F) and W12E rafts (Fig. 3G) but not in raft cultures composed of the parental BC-1-Ep/SL cells (Fig. 3E) or BC16/E7(−) (Fig. 3H) cells. Using in situ hybridization conditions that allowed the detection of low levels of HPV DNA, we found that all cells in the HPV+ rafts, including the BC16/E7(−) rafts, contained HPV-16 DNA (data not shown). This latter result confirms that BC16/E7(−) rafts had appropriate reservoirs of cells in which amplification should have been detectable, if it had occurred. These results provide additional evidence that E7 is essential for HPV-16 DNA amplification.
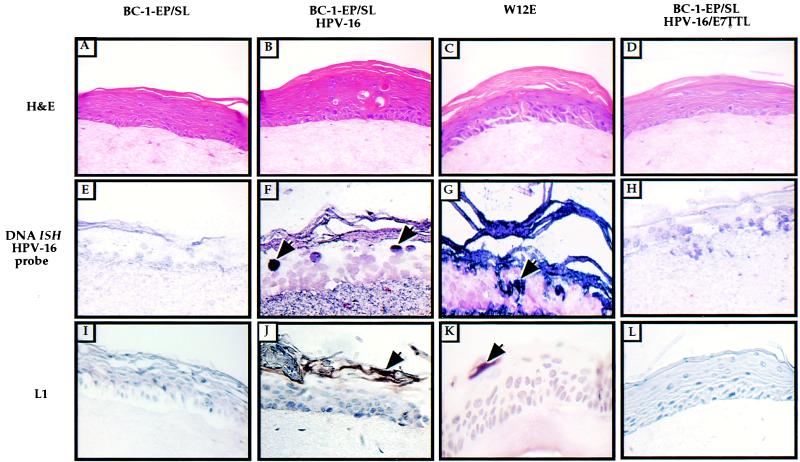
FIG. 3.
HPV-16 DNA in situ hybridization and L1 immunohistochemical analysis of organotypic raft cultures. Shown are epithelial organotypic raft cultures of BC-1-Ep/SL cells, BC-1-Ep/SL/HPV-16 cells [BC16/E7(+)] (41A), W12E cells, and BC-1-Ep/SL/HPV-16/E7TTL cells [BC16/E7(−)] (13-9) which were maintained on a dermal equivalent of collagen impregnated with human foreskin fibroblasts. The rafts were lifted to the air-liquid interface after 4 days in culture and harvested 10 days later. The rafts were fixed in 4% formalin, embedded in paraffin, and cut into 4-μm serial sections. (A to D) Cross sections from each sample stained with hematoxylin and eosin are shown: BC-1-Ep/SL (A), BC-1-Ep/SL/HPV-16 (B), W12E (C), and BC-1-Ep/SL/HPV-16/E7TTL (D). (E to H) For DNA in situ hybridization (DNA ISH), cross sections from each sample were hybridized with a biotin-labeled HPV-16 DNA probe and detected by treating the slides with an avidin-alkaline phosphatase conjugate followed by NBT-BCIP. Positive nuclei are stained dark purple and are indicated by arrows. Many positive nuclei are present in the BC-1-Ep/SL/HPV-16 rafts (F) and in the W12E rafts (G); no nuclei stained positively in the BC-1-Ep/SL/HPV-16/E7TTL rafts (H) or the BC-1-Ep/SL rafts (E). (I to L) For L1 immunohistochemistry, cross sections from each sample were incubated with an anti-L1 antibody and detected using the Vectastain ABC kit. Positive cells are stained brown and are indicated by arrows. Positive cells were detected in BC-1-EP/SL/HPV-16 rafts (J) and W12E rafts (K) but not BC-1-EP/SL rafts (I), and a few weakly positive cells were detected in BC-1-Ep/SL/HPV-16/E7TTL rafts (L).
We monitored another hallmark of the productive stage of the HPV-16 life cycle, the expression of the late capsid protein L1. The expression of this protein was monitored in raft cultures by immunohistochemistry using an antibody against HPV-16 L1 (camvir-1) (Fig. 3). Since L1 is synthesized during the productive stage of the HPV life cycle, it serves as a marker for the productive stage of the viral life cycle. L1-positive cells were found in the granular layer of the raft cultures containing BC16/E7(+) cells (Fig. 3J) and W12E cells (Fig. 3K) but not BC-1-Ep/SL cells (Fig. 3I). L1-positive cells were detected less frequently in the rafts containing BC16/E7(−) cells (Fig. 3L) than in those containing the BC16/E7(+) cells, indicating that the loss of E7 has a negative effect on the production of the viral capsid protein L1.
Loss of E7 results in reduced host DNA synthesis and reduced expression of the DNA replication machinery in the suprabasal layers of raft cultures during the productive stage of the HPV-16 life cycle.
E7 alone induces host DNA synthesis and factors of the host DNA replication machinery such as PCNA in the suprabasal layers of human keratinocyte raft cultures (2, 6). This property of E7 probably contributes to its above-demonstrated role in the productive stage of the viral life cycle. However, E6 also can induce hyperplasia in vivo, leading to the presence of DNA synthesis-competent cells in the suprabasal compartment in the mouse epidermis (23). To assess whether E7 is necessary for creating a favorable environment for the productive stage of the viral life cycle, immunohistochemistry was performed on BC16/E7(+) rafts or on BC16/E7(−) rafts by using antibodies against different markers of DNA synthesis (BrdU, PCNA, and DNA polα).
To determine whether DNA synthesis occurred in the suprabasal layers of the E7-deficient HPV-16 raft cultures, cells were labeled with the thymidine analog BrdU for 8 h. BrdU incorporation was detected in the basal and suprabasal layers of the BC16/E7(+) rafts (Fig. 4B) and the W12E rafts (Fig. 4C). In contrast, the BC16/E7(−) rafts (Fig. 4D), like the BC-1-Ep/SL rafts (Fig. 4A), contained BrdU incorporated in the basal layer only. Quantitation of BrdU immunohistochemistry is provided in Table 2. These data indicate that E7 in the context of the full HPV-16 genome is required for HPV-16 to induce DNA synthesis in the suprabasal compartment of the epithelia.
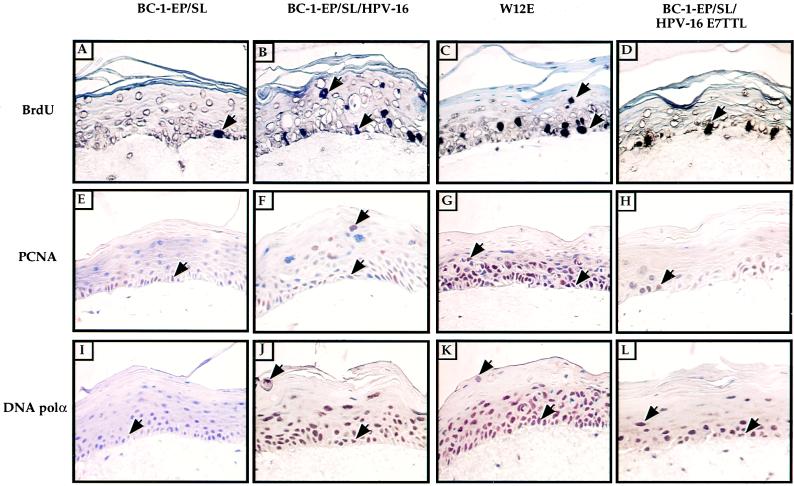
FIG. 4.
Immunohistochemical staining for indicators of DNA synthesis in organotypic raft cultures. Shown are epithelial organotypic raft cultures of BC-1-Ep/SL cells, BC-1-Ep/SL/HPV-16 [BC16/E7(+)] cells (41A), BC-1-Ep/SL/HPV-16/E7TTL [BC16/E7(−)] cells (13-9), and W12E cells. (A to D) For BrdU detection, rafts were labeled with BrdU for 8 h before being fixed. BrdU incorporation was detected by immunohistochemical staining using a biotin-conjugated BrdU antibody available in the BrdU staining kit (Calbiochem). BrdU-positive cells were detected in the basal and suprabasal layers of BC-1-Ep/SL/HPV-16 rafts (B) and W12E rafts (C) but only in the basal layer of BC-1-Ep/SL rafts (A) and BC-1-Ep/SL/HPV-16/E7TTL rafts (D). (E to H) PCNA was detected by immunohistochemical staining using an antibody against PCNA (clone 19F4). PCNA-positive cells were detected in the basal and suprabasal layers of BC-1-Ep/SL/HPV-16 rafts (F) and W12E rafts (G) but only in the basal layer of BC-1-Ep/SL rafts (E) and BC-1-Ep/SL/E7TTL rafts (H). (I to L) DNA polα was detected by immunohistochemical staining using an antibody for DNA polα (clone CL-22-2-42B). Many DNA polα-positive cells were found in the basal and suprabasal layers of BC-1-Ep/SL/HPV-16 rafts (J) and W12E rafts (K) and were found as high as the granular layer; DNA polα was found in the basal and suprabasal layers of BC-1-Ep/SL/HPV-16/E7TTL rafts (L); there were fewer positive cells in BC-1-Ep/SL/HPV-16/E7TTL rafts than in rafts containing wild-type HPV-16, and no positive cells were found in the granular layer. DNA polα was found only in the basal layer of BC-1-Ep/SL rafts (I). All BrdU-, PCNA-, and DNA polα-positive cells are brown, and examples of positive cells are indicated by arrows. Note that most cells which are positive are not indicated by arrows.
To determine the effect that the loss of E7, in the context of the HPV-16 life cycle, had on the host DNA replication machinery, immunohistochemistry was performed on paraffin-embedded cross sections of raft cultures using antibodies against PCNA and DNA polα. PCNA was found in the basal and suprabasal layers of all HPV-16-positive rafts tested (Fig. 4F and G; Table 2). In contrast, PCNA was detected only in the basal layer of BC16/E7(−) rafts (Fig. 4H) and BC-1-Ep/SL rafts (Fig. 4E). This result indicates that during the HPV-16 viral life cycle, E7 is necessary for the induction of PCNA. DNA polα, an important component of the DNA replication machinery, was found in many cells in the basal and suprabasal layers of the BC16/E7(+) rafts (Fig. 4J) and in W12E rafts up to and including the granular layer (Fig. 4K; Table 2). The BC16/E7(−) rafts contained fewer DNA polα-positive nuclei in the basal and suprabasal layers (Fig. 4L). The BC-1-Ep/SL rafts contained weakly positive cells only in the basal layer (Fig. 4I), indicating that while E7 may induce DNA polα, other HPV-16 genes may also contribute to this induction.
Loss of E7 results in reduced expression of the cellular proteins p53, mdm2, and p21.
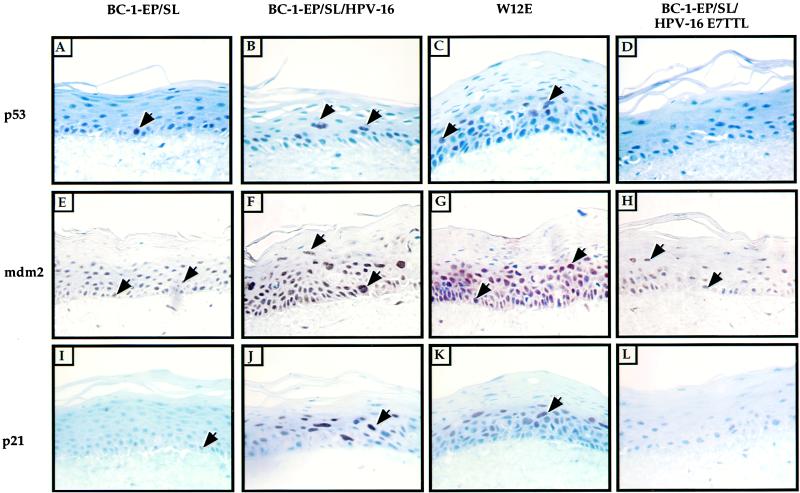
Levels of the cellular proteins p53, mdm2, and p21 have been shown previously to be increased by the viral oncogene E7 (22a, 28). We wanted to determine whether the expression of these proteins in an HPV-16 infection is altered. To address this question, immunohistochemistry was performed using antibodies against the cellular proteins p53, mdm2, and p21. p53 was detected in the suprabasal layers of rafts containing cells harboring the wild-type HPV-16 genome episomally, BC16/E7(+) rafts (Fig. 5B), and W12E rafts (Fig. 5C). In contrast, rafts composed of BC16/E7(−) cells contained few p53-positive cells (Fig. 5D) and BC-1-Ep/SL rafts contained p53-positive cells in the basal layer only (Fig. 5A). These results indicate that E7 increased the levels of p53 in the suprabasal layers of the rafts even in the presence of an intact E6 gene. This was only true in the suprabasal layer of the raft. In the basal layer of BC16/E7(+) rafts, p53 was not present, indicating that E6 is performing its expected role of targeting p53 for degradation there.
FIG. 5.
Immunohistochemical staining for the cellular proteins p53, mdm2, and p21. Shown are epithelial organotypic raft cultures of BC-1-Ep/SL cells, BC-1-Ep/SL/HPV-16 [BC16/E7(+)] cells (41A), BC-1-Ep/SL/HPV-16/E7TTL [BC16/E7(−)] cells (13-9), and W12E cells. p53 was detected by immunohistochemical staining using an antibody against human p53 (clone DO-1). (A to D) p53-positive cells were detected in the suprabasal layers of BC-1-Ep/SL/HPV-16 rafts (B) and W12E rafts (C) but only in the basal layer of BC-1-Ep/SL rafts (A); little p53 staining could be seen in BC-1-Ep/SL/HPV-16/E7TTL rafts (D). (E to H) mdm2 was detected by immunohistochemical staining using an antibody against mdm2 (clone 2A10). Many positive cells were detected in the basal and suprabasal layers of BC-1-Ep/SL/HPV-16 rafts (F) and W12E rafts (G); fewer positive cells were detected in BC-1-Ep/SL rafts (E) and BC-1-Ep/SL/HPV-16/E7TTL rafts (H). (I to L) p21 was detected by immunohistochemical staining using an antibody against human p21 (clone 6B6). Positive cells were detected in the suprabasal layer of BC-1-Ep/SL/HPV-16 rafts (J) and W12E rafts (K); weakly positive cells were detected in the basal layer of BC-1-Ep/SL rafts (I) and BC-1-Ep/SL/HPV-16/E7TTL rafts (L). p53-, mdm2-, and p21-positive cells are brown, and examples of positive cells are indicated by arrows. Note that most cells which are positive are not indicated by arrows.
It has recently been shown that E7 disrupts the interaction between mdm2 and p53 (22a). Additionally, levels of mdm2 are elevated in E7-expressing cells (28). This upregulation of mdm2 by E7 is dependent on p53, and E6 downregulates this effect by E7 (22a). A potentially important feature of mdm2 is that it stimulates E2F-responsive transcription, which is essential in the transactivation of the host DNA replication machinery (19, 26). To determine whether E7 upregulates mdm2 in the context of the HPV-16 life cycle where E6 is also present, immunohistochemistry was performed using an anti-mdm2 antibody. The number of mdm2-positive cells in the basal layer of BC16/E7(+) rafts (Fig. 5F) and W12E rafts (Fig. 5G) (81 to 97%) was larger than the number of positive cells in the basal layer of BC16/E7(−) rafts (Fig. 5H) (16%) or BC-1-Ep/SL rafts (Fig. 5E) (15%) (Table 2). mdm2 was also detected at different levels in the suprabasal layers of all the rafts. The number of mdm2-positive cells in the suprabasal layers of BC16/E7(+) and W12E rafts (48 to 73%) was larger than that in the BC16/E7(−) rafts (11%) or the BC-1-Ep/SL rafts (16%). Thus, E7 can induce mdm2 in the context of the HPV-16 life cycle.
p21, an inhibitor of PCNA, is upregulated in E7-positive cells and abrogated in its cdk inhibition activity by E7 (11, 17). Like mdm2, p21 is a p53-responsive gene. Therefore, we predicted that we would find elevated levels of p21, given the elevated levels of p53 in the suprabasal compartment of the HPV-16-positive rafts. Immunohistochemistry using an antibody for human p21 was performed on W12E, BC16/E7(+), and BC16/E7(−) rafts. In control BC-1-Ep/SL rafts, there were weakly positive cells in the basal and parabasal layers of the raft (Fig. 5I). Rafts containing cells harboring the full-length HPV-16 genome episomally, BC16/E7(+) and W12E, contained strongly positive cells in the suprabasal layers (Fig. 5J and K). The p21 staining pattern in the BC16/E7(−) rafts (Fig. 5L) was similar to that seen in the BC-1-Ep/SL rafts; cells in these rafts were only weakly positive for p21. Thus, two p53-responsive proteins, mdm2 and p21, are present at elevated levels in the suprabasal compartment of HPV-16-positive rafts, depending on the presence of E7.
Perturbation of the normal program of keratinocyte differentiation in raft cultures by HPV-16 is dependent on E7.
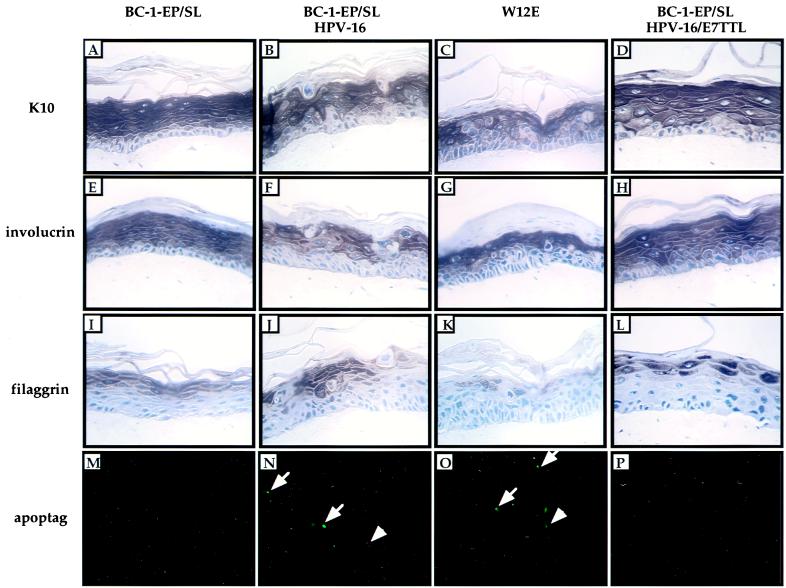
The viral oncogenes E6 and E7 each perturb the differentiation program of early-passage HFKs (20, 29) and in the mouse epidermis (23). The full-length HPV-16/W12E genome harbored episomally in BC-1-Ep/SL cells also can disrupt the differentiation program of the cells (7). Differentiated keratinocytes are postmitotic; thus, E7 may inhibit differentiation to allow DNA synthesis to occur in the suprabasal layer of the epithelium. To determine the effect that the loss of E7 in the full, episomal HPV-16 genome had on the differentiation program of BC-1-Ep/SL cells, immunohistochemistry was performed on raft cultures using antibodies for markers of epithelial differentiation, K10, involucrin, and filaggrin. K10 and involucrin are expressed in the spinous layer of normal epithelia, and filaggrin is expressed in the granular layer. The BC-1-Ep/SL raft stained uniformly for K10 and involucrin in the spinous layer and filaggrin in the granular layer (Fig. 6A, E and I). While the BC16/E7(+) (Fig. 6B, F, and J) and W12E (Fig. 6C, G, and K) rafts were positive for K10 and involucrin in the spinous layer and filaggrin in the granular layer, the staining was not uniform. Large dysplastic cells with enlarged nuclei existed within these layers and did not stain positively for these differentiation markers. In contrast, the BC16/E7(−) rafts did not contain dysplastic cells and exhibited a more uniform staining for all three differentiation markers (Fig. 6D, H, and L), as seen in the BC-1-Ep/SL rafts. These data indicate that in the context of the HPV-16 life cycle, E7 perturbs the differentiation program of keratinocytes.
FIG. 6.
Immunohistochemical staining for terminal differentiation markers of organotypic raft cultures. Shown are epithelial organotypic raft cultures of BC-1-Ep/SL cells, BC-1-Ep/SL/HPV-16 [BC16/E7(+)] cells (41A), BC-1-Ep/SL/HPV-16/E7TTL [BC16/E7(−)] cells (13-9), and W12E cells. (A to D) K10 was detected by immunohistochemical staining using an anti-K10 antibody (clone Ck 8.60). Positive cells are brown and were detected in the suprabasal layers of BC-1-Ep/SL rafts (A), BC-1-Ep/SL/HPV-16 rafts (B), W12E rafts (C), and BC-1-Ep/SL/HPV-16/E7TTL rafts (D). (E to H) Involucrin was detected by immunohistochemical staining using an anti-involucrin antibody (clone SY5). Positive cells are brown and are present in the suprabasal layers of BC-1-Ep/SL rafts (E), BC-1-Ep/SL/HPV-16 rafts (F), W12E rafts (G), and BC-1-Ep/SL/HPV-16/E7TTL rafts (H). (I to L) Filaggrin was detected with an antifilaggrin antibody. Positive cells are stained brown and were detected in the granular layer of BC-1-Ep/SL rafts (I), BC-1-Ep/SL/HPV-16 rafts (J), W12E rafts (K), and BC-1-Ep/SL/HPV-16/E7TTL rafts (L). (M to P) DNA fragmentation was detected in situ using the Apoptag kit. DNA fragmentation was detected in the suprabasal layers of BC-1-Ep/SL/HPV-16 rafts (N) and W12E rafts (O) but not in BC-1-Ep/SL rafts (M) or BC-1-Ep/SL/HPV-16/E7TTL rafts (P). Cells undergoing apoptosis are green. Examples of positive cells are indicated by white arrows.
E7 induces apoptosis in the context of the full HPV-16 life cycle.
E7 alone has previously been demonstrated to induce programmed cell death (apoptosis) in transgenic mice (16, 23). Additionally, E6 has been shown to counteract the effect of E7 by inhibiting apoptosis (23). Because these viral oncogenes have opposing effects on apoptosis, we wanted to determine whether apoptosis occurred during the full HPV-16 life cycle in the presence of both genes. We assayed for fragmented DNA, a hallmark of apoptosis, using the TdT-mediated dUTP-biotin nick end labeling (TUNEL) assay. The presence of fluoroscein-positive cells, indicative of fragmented DNA, was not detected in BC-1-Ep/SL rafts (Fig. 6M). Many fluorescein-positive cells were detected in the suprabasal layers of BC16/E7(+) (Fig. 6N) and W12E (Fig. 6O) rafts. Therefore, HPV-16 induces apoptosis. In contrast, fluorescein-positive cells could not be detected in the BC16/E7(−) rafts (Fig. 6P), indicating that E7 is necessary for this induction of apoptosis in the context of the HPV-16 life cycle.
DISCUSSION
Requirement for E7 in the HPV-16 life cycle.
In this study, we have determined that E7 plays a critical role in the productive stage of the HPV-16 life cycle. We found that cells harboring the E7-defective HPV-16 genome failed to amplify the viral DNA and had reduced L1 expression compared to cells harboring the wild-type HPV-16 genome. Only cells containing the wild-type HPV-16 genome supported DNA synthesis and overexpressed host factors implicated in DNA synthesis in the suprabasal layers of rafts. DNA synthesis did not occur in the suprabasal layers of rafts harboring the E7-defective HPV-16 genome, and the host DNA replication machinery was present at reduced levels compared to those in wild-type HPV-16 rafts. Lastly, E7 in the context of the full-length HPV-16 genome perturbed the program of keratinocyte differentiation. We hypothesize that some or all of these activities of E7 contribute to its creating a more favorable environment for HPV DNA synthesis and virus production in the suprabasal layers of the epithelium.
Our ability to identify a role for E7 in the productive stage of the papillomavirus life cycle relied on our use of the BC-1-Ep/SL immortalized HFK cell line. With these cells, we could expand cell populations harboring the E7-defective genome, a feat that was not possible using early-passage HFKs. Other investigators have tried to analyze E7 function in the HPV life cycle by using early-passage HFKs. As in our experience with E7-defective HPV-16 genomes, E7-defective HPV-18 and HPV-31 genomes were found not to extend the life span of the HFKs. As a consequence, full viral life cycle studies cannot be performed with HFKs, as was our experience. However, these investigators were able to expand cell populations sufficiently to monitor the genomic state of the E7-defective HPV-18 and HPV-31 genomes under conditions supporting the nonproductive stage of the viral life cycle. E7-defective HPV-18 was found to be maintained as an episome in HFKs (C. Meyers, personal communication), as was E7-defective HPV-16 in BC-1-Ep/SL cells (Fig. 1); however, the E7-defective HPV-31, although able to replicate as an episome transiently, could not be detected as a stable replicon (27). This difference between the behavior of HPV-31 and that of HPV-16 or HPV-18 may point to genotype-specific differences in the requirement for E7 in the nonproductive stage of the papillomavirus life cycle.
Effects of the loss of E7 on the host DNA replication machinery.
In this study, we found that wild-type HPV-16 induced DNA synthesis in the suprabasal layers of the epithelium in raft cultures and that this induction coincided with viral DNA amplification. In contrast, E7-deficient HPV-16 failed to induce DNA synthesis in the suprabasal layers of the epithelium, which coincided with an absence of viral DNA amplification. Thus, E7 is necessary to induce DNA synthesis and viral DNA amplification in the productive stage of the viral life cycle. E7 alone has been shown to induce the host DNA replication machinery, presumably through its interaction with pRb, which liberates E2F, allowing transactivation of the host DNA replication machinery (2). In this study, we demonstrated that during the HPV-16 life cycle, the loss of E7 results in the loss of the induction of PCNA and of DNA polα, both of which are components of the host DNA replication machinery and are necessary for viral DNA synthesis, in suprabasal epithelial cells. E7, by inactivating pRB, may indirectly cause increased expression of PCNA and DNA polα, which are encoded by two E2F-responsive genes.
HPV-16 infection induces p53 expression in the suprabasal layer of the epithelium.
The viral oncogene E7, when expressed alone, increases the levels of p53 in mouse embryo fibroblasts (22a) and in undifferentiated keratinocytes (22a). Additionally, p53 levels were shown to decrease in cells expressing both of the viral oncogenes E6 and E7 (22a, 28). This decrease in the levels of p53 presumably occurs through the ability of E6 to degrade p53. In our study, we were able to detect p53 in the basal layer of the BC-1-Ep/SL rafts only but not in the basal layer of rafts containing cells harboring HPV-16 episomally [BC16/E7(+) and W12E cells]. Thus, E6 appears to be proficient in degrading p53 in the basal layer of these rafts. The p53 expression pattern detected in the suprabasal layers of the rafts composed of differentiated keratinocytes was quite surprising. p53 levels were elevated in the suprabasal layers of rafts harboring wild-type HPV-16 episomally [BC16/E7(+) and W12E rafts], in which both the E6 and E7 genes are intact. This elevated level of p53 was dependent on the presence of E7, since it was not observed in BC16(E7−) rafts. These results indicate that in a productive HPV-16 infection E7 must at least in part override the effect of E6 on p53 in the suprabasal layers of the epithelium. This could be due to the inability of E6 to degrade efficiently the amount of p53 induced by E7 in the suprabasal compartment. Alternatively, E6 may not be present or functional in this compartment.
The expression patterns of two p53-responsive genes, p21 and mdm2, were characterized. p21 levels were elevated in cells harboring wild-type but not E7-defective HPV-16. Increased p21 levels would be predicted to suppress DNA synthesis through its inhibition of PCNA and cyclin-dependent kinase activity; however, this potentially negative effect of p21 may be counterbalanced by the capacity of E7 to bind and inactivate p21. Given the role of p21 as a cyclin-dependent kinase inhibitor, its induction would seem to be counterproductive to the viral life cycle and may represent a host defense response to unscheduled DNA synthesis in the suprabasal compartment of the HPV-16-positive rafts. However, it is also possible that elevated levels of p21 may contribute to the positive role of E7 in the productive stage of the viral life cycle, given the role of p21 as a scaffolding factor for the assembly of cyclin–cyclin-dependent kinase complexes. mdm2, another p53-responsive gene, also was elevated in cells harboring wildtype but not E7-defective HPV-16 DNA. Given that mdm2 can increase E2F-responsive transcription and induce S phase without mitosis in mammary epithelial cells (18), induced levels of mdm2 may contribute to unscheduled DNA synthesis in the suprabasal layers of the HPV-16-positive raft culture. That mdm2 and p53 were both elevated in the same epithelial compartment of the HPV-16-positive rafts suggests that the normal autoregulatory circuit in which mdm2 induced the degradation of p53 must be compromised, consistent with recent studies performed with human fibroblasts expressing HPV-16 E7 (22a).
Consequence of perturbing the program of keratinocyte differentiation.
The wild-type-HPV-16-containing rafts exhibited perturbations in the keratinocyte differentiation program. Large, dysplastic cells with enlarged nuclei were apparent in the spinous and granular layers of the rafts composed of cells harboring the wild-type HPV-16 genome episomally [BC16/E7(+) and W12E cells]. These large cells did not stain for the markers of keratinocyte differentiation: K10, involucrin, or filaggrin. Interestingly, these cells did not stain positively for K14, which is a marker normally expressed in the basal layer of the epithelium (data not shown). In contrast, the rafts composed of cells harboring the E7-deficient HPV-16 genome exhibited a uniform staining pattern for K10, involucrin, and filaggrin, as seen in BC-1-Ep/SL rafts. Since these large dysplastic cells were not present in the E7-deficient HPV-16 rafts, they may represent the cells which support the viral DNA amplification.
Rafts with cells harboring wild-type HPV-16 [BC16/E7(+) and W12E] contained numerous cells undergoing apoptosis in the suprabasal layers. This increased apoptosis correlated with the increased expression of the proapoptotic factor p53. That this cell death reflects a host defense response to the unscheduled DNA synthesis in the suprabasal epithelial compartment is supported by the fact that apoptotic cells were absent in the BC16/E7(−) rafts. However, it remains possible that induction of apoptosis may contribute to the viral life cycle, perhaps by facilitating virion release, as has been suggested for other viruses.
E6 and E7 share similar biological properties, including their independent abilities to induce epithelial hyperplasia and inhibit epithelial differentiation. Therefore, we were rather surprised to find that the loss of E7 led to a gross disruption of the productive stage of the papillomavirus life cycle, including the loss of a DNA synthesis-competent environment permissive for viral DNA amplification in the suprabasal compartment. In addition, we found that whereas p53 levels were suppressed in the basal compartment, they were elevated in the suprabasal compartment of HPV-positive rafts, and that this correlated with elevated levels of p53-responsive genes and induction of apoptosis. Taken together, these results imply that E6 is primarily expressed and functional in the nonproductive stage of the viral life cycle and is absent or plays a reduced role in the productive stage. A role for E6 in the nonproductive stage has been demonstrated recently by us and others based on the observation that E6-deficient HPVs are defective for episomal DNA maintenance in undifferentiated, early-passaged HFKs (27) and in BC1-EP/SL cells (E. Flores and P. F. Lambert, unpublished results). Further experiments are necessary to assess whether E6 plays any role in the productive stage of the viral life cycle.
ACKNOWLEDGMENTS
We acknowledge Harlene Edwards and Jane Weeks for their expertise in preparing histological sections of the organotypic raft cultures, Joshua Nelson for preparing tissue culture reagents, Cathy Ivarie for sharing her knowledge of the organotypic raft culture system, and Elizabeth Unger for helpful discussions regarding DNA in situ hybridization. We also thank Mary Ellen Perry for sharing her expertise on p53 and mdm2 and for providing the p53 and mdm2 antibodies used in this study. We gratefully acknowledge Bill Sugden and Mary Ellen Perry for critical reading of the manuscript.
This study was supported by a grant from the American Cancer Society (VM164) and NIH grants CA22443 and CA07175.
REFERENCES
- 1.Allen-Hoffmann B L, Schlosser S J, Ivarie C A, Sattler C A, Meissner L F, O'Connor S L. Normal growth and differentiation in a spontaneously immortalized near-diploid keratinocyte cell line, NIKS. J Investig Dermatol. 2000;114:444–455. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 1a.Auborn K J, Little R D, Platt T H, Vaccariello M A, Schildkraut C L. Replicative intermediates of human papillomavirus type 11 in laryngeal papillomas: site of replication initiation and direction of replication. Proc Natl Acad Sci USA. 1994;91:7340–7344. doi: 10.1073/pnas.91.15.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng S, Schmidt-Grimminger D-C, Murant T, Broker T R, Chow L T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 3.Chengiss M L, Unger E R. Application of a manual capillary action workstation to colorimetric in situ hybridization. J Histotechnol. 1993;16:33–38. [Google Scholar]
- 4.Chow L T, Broker T R. Papillomavirus DNA replication. Intervirology. 1994;37:150–158. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 5.Dazard J-E, Augias D, Neel H, Mils V, Marechal V, Basset-Séquin N, Piette J. MDM-2 protein is expressed in different layers of normal human skin. Oncogene. 1997;14:1123–1128. doi: 10.1038/sj.onc.1200922. [DOI] [PubMed] [Google Scholar]
- 6.Demeter L M, Stoler M H, Broker T R, Chow L T. Induction of proliferating cell nuclear antigen in differentiated keratinocytes of human papillomavirus-infected lesions. Hum Pathol. 1994;25:343–348. doi: 10.1016/0046-8177(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 7.Flores E R, Allen-Hoffmann B L, Lee D, Sattler C A, Lambert P F. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262:344–354. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- 8.Flores E R, Lambert P F. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs E. Epidermal differentiation and keratin gene expression. J Cell Sci. 1993;17:197–208. doi: 10.1242/jcs.1993.supplement_17.28. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs E. Epidermal differentiation: the bare essentials. J Cell Biol. 1990;111:2807–2814. doi: 10.1083/jcb.111.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert D M, Cohen S N. Bovine papillomavirus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell. 1987;50:59–68. doi: 10.1016/0092-8674(87)90662-3. [DOI] [PubMed] [Google Scholar]
- 13.Green H. Terminal differentiation of cultured human epidermal cells. Cell. 1977;11:405–416. doi: 10.1016/0092-8674(77)90058-7. [DOI] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infected cell cultures. J Mol Biol. 1967;36:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2045–2076. [Google Scholar]
- 16.Jeon S, Allen-Hoffmann B L, Lambert P F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones D L, Alani R M, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundgren K, Montes de Oca Luna R, McNeill Y B, Emerick E P, Spencer B, Barfield C R, Lozano G, Rosnberg M P, Finlay C A. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- 19.Martin K, Trouche D, Hagemeier C, Sorensen T S, La Thangue N B, Kouzarides T. Stimulation of E2F/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 20.McCance D J, R K, Fuchs E, Laimins L A. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc Natl Acad Sci USA. 1988;85:7169–7173. doi: 10.1073/pnas.85.19.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene expression and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachsenmeier K F, Sheibani N, Schlosser S J, Allen-Hoffmann B L. Transforming growth factor-B1 inhibits nucleosomal fragmentation in human keratinocytes following loss of adhesion. J Biol Chem. 1996;271:5–8. doi: 10.1074/jbc.271.1.5. [DOI] [PubMed] [Google Scholar]
- 22a.Seavey S E, Holubar M, Saucedo L J, Perry M E. The E7 oncoprotein of human papillomavirus type 16 stabilizes p53 through a mechanism independent of p19ARF. J Virol. 1999;73:7590–7598. doi: 10.1128/jvi.73.9.7590-7598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song S, Pitot H C, Lambert P F. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley M A, Browne H M, Appleby M, Minson A C. Properties of a non-tumorgenic human cervical keratinocyte cell line. Int J Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 25.Sterling J, Stanley M, Gatward G, Minson T. Production of human papillomavirus type 16 virions in a keratinocyte cell line. J Virol. 1990;64:6305–6307. doi: 10.1128/jvi.64.12.6305-6307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teoh G, Urashima M, Ogata A, Chauhan D, DeCaprio J A, Treon S P, Schlossman R L, Anderson K C. MDM2 protein overexpression promotes proliferation and survival of multiple myeloma cells. Blood. 1997;90:1982–1992. [PubMed] [Google Scholar]
- 27.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas J T, Laimins L A. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J Virol. 1998;72:1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodworth C D, Cheng S, Simpson S, Hamacher L, Chow L T, Broker T R, DiPaolo J A. Recombinant retroviruses encoding human papillomavirus type 18 E6 and E7 genes stimulate proliferation and delay differentiation of human keratinocytes early after infection. Oncogene. 1992;7:619–626. [PubMed] [Google Scholar]
- 30.Yang L, Botchan M. Replication of bovine papillomavirus type 1 DNA initiates within an E2-responsive enhancer element. J Virol. 1990;64:5903–5911. doi: 10.1128/jvi.64.12.5903-5911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]