Abstract
Methyl orange (MO) is a dye commonly used in the textile industry that harms aquatic life, soil and human health due to its potential as an environmental pollutant. The present study describes the dye degradation ability of Serratia marcescens strain ED1 isolated from textile effluent and characterized by 16S rRNA gene sequence analysis. The laccase property of bacterial isolate was confirmed qualitatively. The effects of various factors (pH, temperature, incubation time, and dye concentration) were evaluated using Response Surface Methodology (RSM). The maximum dye (MO) degradation was 81.02 % achieved at 37 °C temperature and 7.0 pH with 200 mg/L dye concentration after 48 h of incubation. The beef extract, ammonium nitrate and fructose supplementation showed better response during bioremediation among the different carbon and nitrogen sources. The degree of pathogenicity was confirmed through the simple plate-based method, and an antibiotic resistance profile was used to check the low-risk rate of antibiotic resistance. However, the fate and extinct of degraded MO products were analysed through UV–Vis spectroscopy, FT-IR, and GC-MS analysis to confirm the biodegradation potential of the bacterial strain ED1 and intermediate metabolites were identified to propose metabolic pathway. The phytotoxicity study on Vigna radiata L. seeds confirmed nontoxic effect of degraded MO metabolites and indicates promising degradation potential of S. marcescens strain ED1 to successfully remediate MO dye ecologically sustainably.
Keywords: Textile wastewater, 16S rRNA, AST, Nitrogen and carbon source, Laccase
1. Introduction
The industrial textile sector plays an essential role in the global economy and societal fabric, underlying critical facets of day-to-day life. However, its growth comes conjointly with considerable environmental risk, mainly demonstrated through large amounts of wastewater production. This wastewater is a complex mixture of diverse inorganic and organic pollutants, ranging synthetic dyes, heavy metals, aliphatic hydrocarbons, polycyclic aromatic hydrocarbons (PAHs), benzene, and endocrine-disrupting chemicals (EDCs) [1,2]. Of particular concern are synthetic dyes, the complex aromatic compounds acclaimed for their stability and recalcitrance to bioremediation. Their persistence raises substantial challenges to conventional degradation processes, requiring novel approaches for successful remediation.
Besides other groups of textile dyes, azo dyes are synthetic colorants widely used in the textile, food, and paper-pulp industries. They are preferred over natural dyes due to their superior fastness, lower cost, ease of application, and excellent stability against light, temperature, synthetic chemicals, and microbial destruction [3]. The discharge of azo dyes containing colorants into the atmosphere, on the other hand, can pose a significant danger to aquatic, soil life, and human health, causing inflammation, diarrhoea, and gastrointestinal illnesses. Furthermore, azo dyes and their decomposed compounds are poisonous, cancerous, and mutagenic [4]. Methyl orange (MO) is the most commonly used azo dye in the textile business, and it is readily discharged into the atmosphere via industrial waste. It is extremely water soluble and has a unique orange-red hue [5]. The discharge of MO dye into the atmosphere without sufficient treatment can have detrimental consequences on the water, soil, air, and human health. For example, MO dye can potentially contribute to the development of smog and ground-level ozone. These contaminants can negatively impact human health, increasing the risk of heart disease and causing respiratory issues. Additionally, it floats on land or water surfaces, where it may enter lakes, rivers, and other bodies of water. This harms aquatic life and makes water unsuitable for human consumption [6]. Additionally, MO dye contributes to climate change because it can combine with other atmospheric chemicals to create greenhouse gases like CO2 and CH4, which trap heat and promote global warming and chemical characteristics of MO dye is shown in Table S1. Many scientists are trying to discover ways to decompose such manufactured compounds to safeguard the ecosystem [7]. Various biological, chemical, and physiochemical treatment techniques have been used to handle industrial textile wastewater, including membrane separation, microbes, enzyme decomposition, adsorption, oxidation, filtering, coagulation, and precipitation [8].
In recent years, eliminating azo dyes and dye residues from the atmosphere has been successfully accomplished through microorganism decomposition. Numerous microorganisms, such as bacteria, fungi, and algae, can degrade and discolor azo dyes [9]. Two distinct processes—dye adsorption by microbial biomass and dye breakdown by microbial cells—are primarily involved in microbial degradation [10]. Although various microorganisms are shown to be effective in mineralizing and decolorizing textile pollutants, bacterial degradation is a preferable choice as bacteria are quick to grow, easily culturable, and better adaptable to various environmental circumstances with more flexible behaviour over biomass development on diverse substrates. Several bacterial species have been found to decompose textile effluent pollutants effectively; these include Bacillus sp., Thermus sp., Aeromonas sp., Pseudomonas sp., Serratia sp., Shewanella sp., Sphingomonas xenophaga BN6, Pigmentiphaga kullae K24, Caulobacter subvibrioides C7-D, Salinivibrio sp., Franconibacter sp.1MS, and Geobacillus thermoleovorans KNG 112 [11,12]. According to earlier study findings, S. marcescens exhibits disintegrating activity for lignin, palmarosa oil (green oil), oily culinary refuse, p-cresol, low-density polyethylene, chlorobenzenes and pentachlorophenol (trash from wood and paper mills) [[13], [14], [15], [16], [17], [18]]. Extracellular laccase, manganese peroxidase (MnP), NADH-DCIP reductase, MG reductase, and CV reductase enzyme of bacteria Serratia sp. was discovered during the decolorization of triphenylmethane (TPM) dyes of Malachite Green (MG) and Crystal Violet (CV) [19].
The degradation process of azo dyes is facilitated by bacterial enzymes, namely azoreductase and, laccase, which catalyse the cleavage of azo bonds (-N N-) present in the dye molecule. The resulting aromatic amines are oxidized and metabolised into less hazardous by-products [20]. An extraordinary source of antibiotic-resistant bacteria and genes in wastewater treatment plants (WWTPs). furthermore, WWTPs can provide an ideal environment for transferring antibiotic resistance genes between bacteria. As a result, to use these bacteria on a wide scale in treatment plants, it is essential to determine the antibiotic resistance profile of chosen bacteria [21]. The utilization of antimicrobial susceptibility testing (AST) in bioremediation is a crucial technique for determining the levels of antibiotic resistance present in bacterial isolates. Automated platforms, such as the Vitek® 2 compact system, can be utilised to conduct this testing, offering prompt and precise outcomes. The efficiency of microbial degradation of azo dyes may be impacted by several factors, such as pH, temperature, incubation duration, degradation timeframe, and concentration of the degraded dye [22]. The Response surface methodology (RSM) assists in optimizing these practical factors and improving the efficacy of bacterial decomposition [23]. Multiple techniques exist for evaluating the toxicity of dyes and their by-products, each characterised by unique sensitivity, resolution, and cost levels. In order to verify the deterioration of the dye and/or the existence of aromatic amines, it is possible to conduct analyses utilising techniques such as ultraviolet–visible spectrophotometry, Fourier-Transform infrared spectroscopy (FT-IR), and Gas Chromatography-Mass Spectrometer (GC-MS) [24].
The present investigation centres on the isolation of bacteria, evaluation of their AST profile, and basic plate-based pathogenicity, as well as the enhancement of biotransformation through utilising a central composite design (CCD) in response surface methodology. Furthermore, the assessment of MO dye degradation was conducted through the utilization of a UV–vis spectrophotometer, FT-IR, and GC-MS. The potential degradation pathway was analysed, and the toxicity of both the initial dye (MO) and the degraded metabolites was evaluated on certified mung bean (Vigna radiata L.) seeds. The utilization of microbial degradation presents a viable strategy for the remediation of industrial effluents that harbour such contaminants. This research endeavour aims to address, utilize, and provide foundational insights regarding said bacteria.
2. Material and methods
2.1. Materials
The analytical-grade dyes utilised in the present investigation were procured from the Central Drug House (P) Ltd. (CDH). Beef extract, peptone, tryptone, dextrose, sucrose, glucose, maltose, starch, fructose, mannitol, lactose, NH₄Cl, (NH₄) ₂C₂O₄, (NH₄) ₂SO₄, NH₄NO₃, KNO₃, NaNO₃ and nutrient media (in g/L: beef extract 5, peptone 3, sodium chloride 5, and agar-agar 20) were procured from Hi-media (Mumbai, India). Nutrient broth (NB) was used to isolate the bacterial strains. 2-2′-azino-bis- [3-ethyl benzthiazoline-6-sulfonic acid] (ABTS), Czapek–Dox Agar (CDA) was used for enzyme assay procured from Sigma-Aldrich in Canada.
2.2. Sample collection, decolorization, and quantitative screening
The wastewater samples were collected from a textile factory's effluent at SIDCUL (State Industrial Development Corporation of Uttarakhand), Haridwar, India. To identify the bacterial populations, a 1 mL effluent sample was mixed with 10 mL of nutritional broth in a 20 mL test container. The mixture was then incubated at 37 °C for 24 h 0.5 μL aliquot was withdrawn from the tube, then serially diluted to 10−7 concentration. Using the spread-plate approach, a loopful of the culture was spread on nutrient agar plates modified with MO dye and incubated for 24 h. The colonies grown with the clear zone were picked and transferred to fresh media to obtain a pure culture [25].
For further screening individual colonies of the selected decolorizing bacterial strain were inoculated in 100 mL of nutrient broth in a 250 mL flask and incubated at 37 °C for 24 h 5 mL of the bacterial suspension was added to 95 mL nutrient broth containing MO solution in 250 mL conical flasks, and kept at 37 °C on static incubation [26]. A Systronics UV–Vis Double Beam spectrophotometer 2201 was used to measure the optical density of the supernatant at 446 nm after 4 mL aliquots were periodically removed and centrifuged at 10,000 rpm for 8 min. Control was maintained as MO dye solution without bacteria and kept the same. Bacterial strain giving more decolorization efficiency was selected and screened for the dye concentration (50, 100, 200 and 300 mg/L) on dye degradation under the applied conditions. The experiments were carried out in triplicates. The decolorization was calculated using the following formula [27].
2.3. Biochemical characterization, enzymatic assay and molecular identification
Selecting the bacterial strain that produces the most decolorization through morphology and biochemistry involves a combination of visual observation of colony and cell morphology and biochemical tests to identify the specific strain. Observed morphological characteristics included cell size, shape, gram nature, motility, and endospore generation. Tests for catalase, oxidase, protease, and indole synthesis were part of the biochemical analysis [28]. The biochemical analysis was done using biochemical HiCarboTM testing kits (KB009 and KB003) and culture characterized according to Bergey's manual [29]. A qualitative laccase activity test was performed on bacterial stain ED1 by inoculating bacteria on a CDA plate enriched with 3 mM ABTS dye at 27 °C for 48 h. The appearance of the green colored zone around the bacterial colony indicates the presence of laccase activity in bacteria [30].
The bacterium genus was confirmed using Sanger sequencing and examination of the 16S rRNA signature. The 16S rRNA gene target sequences are amplified using universal primers (forward primer 5′- GGATGAGCCCGGCCTA-3′ and reverse primer 5′- CGGTGTACAAGGCCCGG-3′) prior to sequencing and homology creation using a ribosomal DNA library. The following conditions were used to conduct the PCR reactions. The template DNA was heated for 3 min at a pre-denaturation temperature of 94 °C to denature it. Following this, the experimental procedure involved 30 cycles of denaturation at a temperature of 94 °C for 1 min, followed by annealing for 1 min, elongation at 72 °C for 2 min, and a final extension at 72 °C for 7 min. The amplified products were separated in a 1.5 % agarose gel and resolved with the aid of 0.5X tris-acetate-EDTA (TAE) buffer. Subsequently, a gel documentation system was employed to visualise and capture an image of the gel. The 16S rRNA gene was analysed using the BLAST (blastn) similarity search tool provided by the National Center for Biotechnology Information (NCBI). The phylogenetic tree was constructed and evolutionary analyses were performed using the neighbour-joining method with MEGA XI [[31], [32], [33], [34]].
2.4. AST and identification
The VITEK 2 compact systems in microbiology employ a growth-based approach intended for the prompt detection and assessment of bacterial and fungal organisms' antibiotic susceptibility. The provision of colorimetric reagent cards that yield digitally decoded outcomes for bacterial colonies presents several benefits over conventional AST techniques. These advantages include expedited turnaround time, increased throughput, and enhanced precision and reproducibility [35]. In 0.45 % saline, the 0.5 McFarland standard of the bacterial solution was diluted to 1.5 ꓫ 107 CFU/mL. The VITEK 2 apparatus automatically filled, sealed, and loaded with cards for incubation and readout. The system utilised Vitek® 2 GN and Vitek® 2 AST- N281 cards for Enterobacteriaceae, which contained ticarcillin/clavulanic acid, piperacillin/tazobactam, ceftazidime, cefoperazone/sulbactam, cefepime, aztreonam, doripenem, imlpenem, meropenem, amikacin, gentamicin, ciprofloxacin. By comparing the growth of the isolate with the known MICs, the minimum inhibitory concentration (MIC) was ascertained [36,37].
2.5. Plate-based pathogenicity of isolated strain ED1
Although the isolated bacterium considered of low virulence but it is possible to became virulent under different environmental conditions. Bacterial virulence is complex process that involves the interplay between the bacterium and its host environment, and its interplay can be influenced by wide range of factors such as temperature, pH, nutrient availability, and the presence of other microorganisms [38,39]. The assessment of degree of pathogenicity was conducted through the examination of bacterial growth patterns on Blood Agar Plate (BAP) and MacConkey Agar Plate (MAP) accordingly (CDH JO 2008F). The utilization of lactose fermentation is employed in the selective identification of enteric and Gram-negative bacteria via using the MAP. The haemolytic properties of bacterial colonies were evaluated on BAP by subjecting them to incubation at 37 °C for a duration of 18–24 h (extended for an additional 24 h when haemolysis was not observed) and the three types of hemolysis, namely α, β, and γ, were assessed.
The study observed the use of MAP, wherein the formation of violet-colored media on bacterial plates after 24 h of incubation was utilised to aid in the identification of lactose-fermenting bacteria. The presence of greenish clear zones in blood media may suggest the potential existence of pathogenic bacteria that could threaten the host. BAP and MAP offer insight into the potential for pathogenicity but do not serve as conclusive assessments. Subsequently, an investigation was carried out on BAP and MAP utilising a methodology based on scoring [40].
2.6. Carbon and nitrogen sources effects on decolorization
Seven carbon sources (Sucrose, Glucose, Maltose, Starch, Fructose, Mannitol, and Lactose) and nine nitrogen sources (organic: Peptone, Beef extract, and Tryptone and inorganic: NH₄Cl, (NH₄)₂C₂O₄, (NH₄)₂SO₄, KNO₃, NaNO₃) were used to test the effect of carbon and nitrogen (1g/100 mL) supplements on the bacterial growth medium enriched with MO dye. The negative control contains MO dye (200 mg/L) solution without bacterial inoculum or organic or mineral media supplements [41]. For isolate ED1, the maximum growth rate was observed at regular intervals for 5 days at an optimized pH 7.0 and temperature 37 °C.
2.7. Statistical optimization and experimental design
RSM is a mathematical model that allows the statistical design of a response surface 2-D and 3-D plots, which can be used to predict the optimal values of the process parameters that impart maximum dye degradation results [42]. For the effective decolorization of MO dye, central composite design (CCD) was used, and four different factors, i.e., pH, temperature, incubation time, and dye concentration, were considered for experimentation and the relationship between factors [43]. The four factors were divided into five levels (-α, −1, 0, +1, +α), and their relationship was determined by fitting the second-order polynomial equation [44]. Table 1 contains a list of the combinations of uncoded independent variables.
| (equation 1) |
Table 1.
Actual levels and range of independent variables for CCD.
| Factors | -α | −1 | 0 | +1 | + α |
|---|---|---|---|---|---|
| pH | 3 | 5 | 7 | 9 | 11 |
| Temperature, ⁰C | 23 | 30 | 37 | 44 | 51 |
| Incubation time, hours | 32 | 40 | 48 | 56 | 65 |
| Dye concentration, mg/L | 100 | 150 | 200 | 250 | 300 |
The variables Xi and Xj represent dimensionless coded variables, while Y denotes the response variable, which signifies the percentage of dye decolorization. The model constant value is designated by β0, the linear coefficient by βi, the square coefficient by βii, and the cross-product coefficient by βij [44].
The coefficient of regression (R2) and analysis of variance (ANOVA) were computed to assess the model's fit quality. The significant impacts of governing factors on decolorization were also investigated using the F-test and P-value. Only variables with P-values lower than 0.05 were considered to be necessary. Three-dimensional (3D) response surfaces and counterplots were drawn to show the variables' individual and cumulative impacts on the reaction. The decolorization was measured in duplicate, and the precision of the design was determined by using the mean.
A total of 30 runs were conducted using a full factorial CCD (4 factors at 5 levels) [45].
| (equation 2) |
The variable N represents the overall quantity of experiments conducted, while n denotes the quantity of factors involved. A regression model was developed utilising Design Expert software (Version 7.0.0) [45].
2.8. Analysis of MO dye degraded product
2.8.1. UV–visible spectroscopy analysis
The alteration in the decolorization of MO dye was evaluated by utilising a UV–Vis spectrophotometer (Systronics UV–Vis Double Beam spectrophotometer 2201) to examine the modifications in the spectra. In the decolorization study, a 4 mL (v/v) decolourized sample was obtained from flasks at various intervals and subjected to centrifugation at 8000 rpm for 15 min to eliminate cellular biomass. The acquired supernatant was utilised to observe the decolorization process within the wavelength spectrum of 200–700 nm [46].
2.8.2. FT-IR analysis
The identification of various functional groups in both MO dye and its degraded product was accomplished through FT-IR analysis. After decolorization, a 20 mL aliquot was extracted from the flasks and centrifuged. The metabolite was then extracted using an equal volume of ethyl acetate dried with Na2SO4 and further concentrated and dried using a rotatory evaporator. Subsequently, a specimen was deposited onto a thin KBr pellet prepared under vacuum conditions using a PCI hydraulic press (15 tonnes) and secured in a sample holder. The FT-IR analysis was conducted utilising the Nicolet™ 6700 Thermo Scientific, USA spectrometer, spanning a range of 400–4000 cm−1 at a 16-scan speed [47].
2.8.3. GC- MS analysis
The biodegradation products of MO dye were identified through GC-MS analysis, both before and after bacterial treatment, using a selected strain of bacteria. The collected samples were centrifuged for 10 min at room temperature. The supernatant collected was extracted thrice with an equal volume of ethyl acetate, dried with Na2SO4 and further concentrated in the rotatory evaporator [48]. The control group, prior to treatment, and the extracted degraded product were analysed using a GC-MS equipped with an AOC-20i Auto-injector (Model: GC-MS-TQ8040). The conditions under which the operation was conducted are as follows: The front inlet injection mode was maintained in splitless mode, with an initial temperature of 280 °C. The samples were subjected to separation using an SH-Rxi-5Sil MS capillary column consisting of 5 % biphenyl and 95 % dimethyl polysiloxane. The column had a nominal film thickness of 0.25 μm df, an internal diameter of 0.25 mm, and a length of 30 m. The temperature of the column was initially set to 80 °C and gradually increased to 260 °C at a rate of 5 °C per minute, where it was maintained for a duration of 15 min. The mass spectrometry data was obtained by scanning within 30–490 m/z. Identifying potential degradation products was predicated on their correspondence with the standard mass spectrum found within the GC-MS library database [49]. The chromatogram's peaks were identified through a comparative analysis of the National Institute of Standards and Technology (NIST) and Wiley online libraries. The reaction intermediates were identified through spectral comparison with established standards [50].
2.8.3.1. Possible pathway of MO dye degradation
In this study, the degradation pathway of MO dye has been proposed by utilising the results obtained from GC-MS analysis. MO dye metabolites formed after the treatment were further identified by GC- MS analysis. Based on the parent structure of MO dye and metabolites produced during the treatment process, the possible degradation pathway of MO dye was drawn. All structures were drawn in the ChemDraw programme [51].
2.9. Phytotoxicity study
Toxicological studies of untreated (UT) and bacterial-treated (BT) MO were carried out by phytotoxicity studies. A study was conducted to assess phytotoxicity in Vigna radiata L. (mung Var.IPM3) seeds through a seed germination test that evaluated seed germination and early seedling growth parameters. The acquisition of mung seeds was made from Uttarakhand Seeds and Tarai Development Corporation Limited (USTDCL), accompanied by the identification tag number C33214320004. A test on seed germination was carried out using Petri - plates. Product obtained after 48 h was extracted in the ethyl acetate and dissolved in the water to form a final concentration of 200 mg/L for phytotoxicity studies. The UT and BT MO dye samples were added to distilled water at a concentration of 200 mg/L. Distilled water was utilised to prepare UT and BT MO dye four test solutions with varying concentrations of 25 %, 50 %, 75 %, and 100 % v/v. Ten viable seeds were subjected to sterilisation using a 2.0 % w/v mercuric chloride solution for a duration of 2 min [20]. The experimental procedure involved the placement of seeds between Whatman No. 1 filter papers on Petri - plates, followed by irrigation with 4 mL of distilled water in the control group, as well as in the UT and BT MO dye solution groups. The Petri - plates were placed in a BOD incubator at 28 ± 1 °C. The number of seeds germinated was observed after 48 h. Following a period of 5 days, a sample of three seedlings were selected at random from each Petri plate for the purpose of measuring their respective root lengths, shoot lengths, and biomass. The experimentation involved conducting toxicity tests in three sets, with the percentage of germination and inhibition documented according to the following formula [52].
2.10. Statistical analysis
All the experiments were carried out in triplicates and data are represented as mean ± SE. All graphs created in this paper were plotted using OriginPro 2024 (64-bit) SR1 software for data visualization and analysis.
3. Results
3.1. Screening and characterization of bacteria strain
From the effluent sample, eight morphologically different bacterial strains- ED1, ED2, ED5, ED9, ED12, ED13, ED18, and ED24 were selected primarily based on decolorization zone (≈10 mm) on MO dye-supplemented plate and further screened for their potential to degrade MO dye. Further screening of selected strains showed a decolorization rate obtuse after 72 h by ED2, ED5, ED9, ED12, ED13, ED18, and ED24 for MO dye. Strain ED1 was exposed to show the eminent decolorization (more than 80 %) of MO in 48 h. After the selection of efficient bacterial strain ED1, effect of dye concentration was also screened and highest degradation potential observed for 50 mg/L MO dye solution which is about 92 % and on highest concentration where isolated bacterial strain ED1 can survive and degraded MO dye solution was 200 mg/L as depicted in Fig. 2D. MO dye concentration on 300 mg/L dose not shows any degradation after 42 h of incubation.
Fig. 2.
Optimization of MO dye decolorization by S. marcescens isolate using different A. organic sources, B. inorganic nitrogen sources and, C. using different carbon sources, D. Decolorization of MO dye on different concentration.
The bacterial colonies' growth on nutrient agar plates indicated significant diversity in cell morphology and other aspects of colonial morphology among all the isolates. The bacterial strain ED1 was identified as a gram-negative, rod-shaped bacteria that exhibits a distinct pink or red hue due to a red pigment as depicted tin Fig. 1A. The biochemical tests were performed to determine the presence or absence of specific enzymes or metabolic pathways in the isolated bacteria. The organism exhibits glucose utilization, saccharose utilization, esculin hydrolysis, methyl red, voges-proskauer, citrate utilization, urease, catalase, nitrate reduction, ornithine, lysine, adanitol, trehalose biochemical activity and with absence activity of endospore formation, lactose utilization, indole production, H2S production and tryptophan deaminase. Table 2 displays the outcomes of the morphological and biochemical examinations.
Fig. 1.
A. Pure isolate of ED1 strain, B. The Isolate ED1 on MAP media plate, C. The Isolate ED1 on BAP media plate, D. Phylogenetic tree of S. marcescens based on the 16S rRNA gene sequence.
Table 2.
Biochemical characters of isolate ED1.
| Tests | ED1 |
|---|---|
| Gram's staining | -ve |
| Shape | Rod |
| Cultural characteristics | Red, smooth, convex, entire and round colonies |
| Motility | Motile |
| Endospore formation | -ve |
| Lactose utilization | -ve |
| Glucose utilization | +ve |
| Saccharose utilization | +ve |
| Esculin hydrolysis | +ve |
| Indole production | -ve |
| Methyl red | +ve |
| Voges-Proskauer | +ve |
| Citrate utilization | +ve |
| Urease activity | +ve |
| Catalase activity | +ve |
| H2S production | -ve |
| Nitrate reduction | +ve |
| Ornithine | +ve |
| Lysine | +ve |
| Adanitol | +ve |
| Trehalose | +ve |
| TDA | -ve |
+ = Positive, - = Negative, TDA = Tryptophan Deaminase.
In this study, the bacterial strain ED1 were assayed for laccase activity by qualitative ABTS assay that showed green colored zones around each bacterial colony in an agar plate after 48 h incubation period as shown in Fig. S2.
3.2. AST and identification
AST and bacterial identification by Food and Drug Administration (FDA) approved following generation identification tool based on the prior standard library and reference MIC (minimum inhibitory concentration for preventing the growth of isolate) data offered a modern VITEK 2 (bioMerieux) compact system that resulted in the identification of the isolate. Serratia marcescens with the bio number 2025711451004230 (Lab ID: ED123082022) with 97 % probability was successfully identified by the VITEK 2 system through the 9.02 h processing period. Interestingly, S. marcescens isolate was susceptible to most antibiotic drugs except colistin, which displayed resistance due to equal or greater than 16.03 ± 0.03 μg/mL MIC value in the AST profile, as shown in Table 3; all other antibiotics displayed susceptibility for the specified MIC. Trimethoprim/Sulfamethoxazole showed the lowest susceptible for MIC of 20 ± 0.03 μg/mL and the highest susceptible for MIC of 0.12 ± 0.003 μg/mL for levofloxacin and doripenem. In all 17 used antibiotics, only colistin showed resistance and two antibiotics piperacillin/tazobactam and imlpenem was not shown by this system for S. marcescens isolate. The susceptibility and resistance of each antibiotics under different MIC value were interpreted by following guidelines from Clinical and Laboratory Standards Institute (CLSI).
Table 3.
Antibiotic-bacterial isolate relationship through the VITEK 2 (bioMerieux) compact system.
| Antibiotics | MIC (μg/mL) | Interpretation |
|---|---|---|
| Ticarcillin/Clavulanic acid | 8.00 ± 0.06 | S |
| Piperacillin/Tazobactam | – | – |
| Ceftazidime | 1.03 ± 0.09 | S |
| Cefoperazone/Sulbactam | 8.10 ± 0.06 | S |
| Cefepime | 1.07 ± 0.09 | S |
| Aztreonam | 1.03 ± 0.04 | S |
| Doripenem | 0.12 ± 0.003 | S |
| Imlpenem | – | – |
| Meropenem | 0.25 ± 0.005 | S |
| Amikacin | 2.03 ± 0.03 | S |
| Gentamicin | 1.17 ± 0.17 | S |
| Ciprofloxacin | 0.25 ± 0.003 | S |
| Levofloxacin | 0.12 ± 0.003 | S |
| Minocycline | 2.16 ± 0.17 | S |
| Tigecycline | 1.03 ± 0.03 | S |
| Colistin | <16.03 ± 0.03 | R |
| Trimethoprim/Sulfamethoxazole | 20 ± 0.03 | S |
R, resistant; I, intermediate; S, susceptible; -, Not detected.
3.3. Sequence alignment and phylogenetic analysis
Molecular techniques reconfirmed the Vitek 2 results. After performing alignment and sequencing procedures, the obtained sequence data were compared with 16S rRNA gene sequences of other organisms previously deposited in the NCBI Gene bank database, with the accession number ON 196501. Its genomic DNA gel electrophoresis image depicted by Fig. S1. The phylogenetic tree of the isolate, which was presented in Fig. 1D, indicates a 97 % similarity with Serratia marcescens. Because it possessed the decolorization and degradation qualities required for the biotransformation of the MO dye, this strain was selected for the degradation experiments.
3.4. Pathogenicity of isolated bacteria
The simple plate-based pathogenicity test on a blood agar plate can provide helpful information about the pathogenic properties of a bacterium. Bacteria can produce enzymes or toxins that can lyse (break down) red blood cells or create a transparent or opaque zone around bacterial growth. This indicates that the bacterium has haemolytic properties. A haemolysis test through simple plate assay confirmed that the bacterial isolate did not exhibit any haemolytic patterns/clear zones on BAP media plates. MacConky agar is a widely employed medium in microbiology that isolates and distinguishes gram-negative bacteria based on their selective and differential properties. The bacterial plate was subjected to MAP analysis, which confirmed the bacteria's non-lactose fermenting, gram-negative characteristics due to the absence of violet-color colonies on the MAP. The plates are displayed in Fig. 1 B and C.
3.5. Effect of various carbon and nitrogen sources on decolorization
The present study investigated the impact of diverse organic and inorganic nitrogen sources (1 g/L), and the outcomes were graphically represented in Fig. 2. Among all used, three organics (beef extract, peptone, tryptone) and six inorganic nitrogen (ammonium chloride, oxalate, sulfate, and nitrate with potassium and sodium nitrate) sources showed rapid bacterial growth and best performance in beef extract and ammonium nitrate, respectively as shown in Fig. 2A and B. The results revealed that the carbon source, lactose and fructose confront enhancement in MO dye decolorization up to 82.89 and 90 % respectively, within 120 h due to nutrition available for bacteria through absorption and metabolization of available carbon sources as shown in Fig. 2C. Furthermore, as compare to other nitrogen sources, peptone, beef extract and ammonium nitrate displayed high performance of about 81.45, 81.69 and 89 % respectively, degradation efficacy of MO dye by S. marcescens ED1.
3.6. Estimation of a response surface for maximal degradation and statistical validation
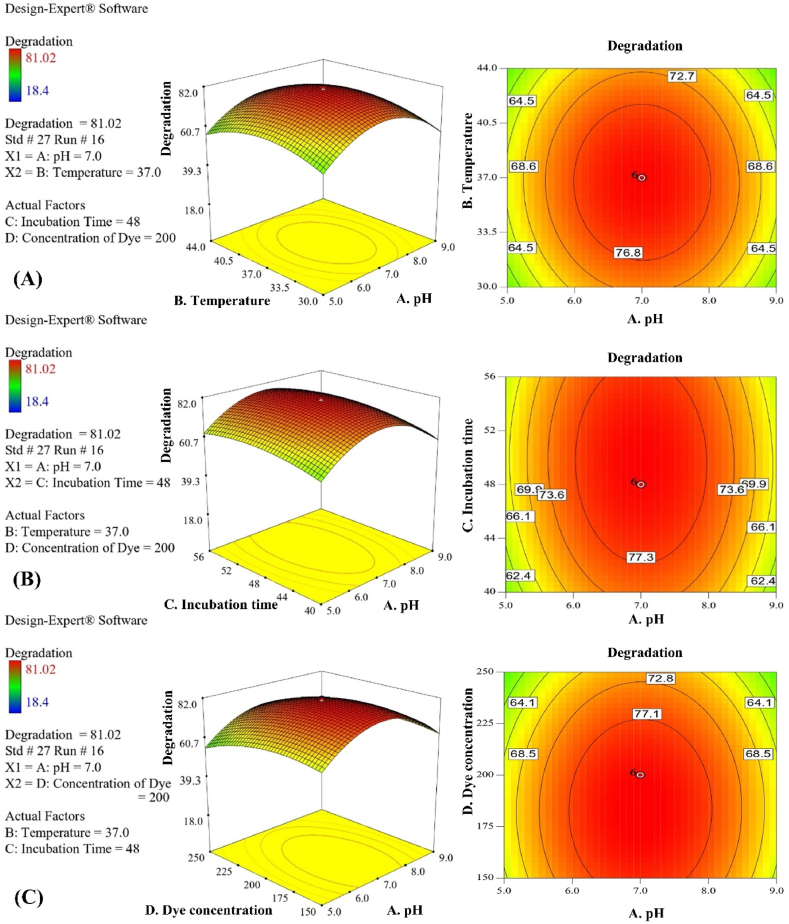
An experimental investigation was carried out in optimal circumstances to validate the model's analysis derived from statistical analysis of CCD. The comparison between actual and predicted responses was performed. The results indicate that the maximum degradation rate was observed at a pH of 7.0, a temperature of 37 °C, an incubation time of 48 h, and a dye concentration of 200 mg/L, as illustrated in the 2D and 3D response surface plots presented in Fig. 4(A-F) response.
Fig. 4.
Response surface plots and contour plots, A. pH and temperature, B. pH and incubation time, C. MO dye concentration and pH, D. incubation time and temperature, E. the concentration of MO dye and temperature, F. MO dye concentration and incubation time. This experiment focused on the examination of two specific factors, while the remaining factors were held constant at a value of zero.
The following was the final model-based polynomial equation (second order) for the degradation in terms of actual factors:
| Degradation % (Y) = −620.18943 + 56.54974A + 12.23346B + 7.51871C + 1.00607D −0.01593AB - 0.022070AC + 0.00199AD + 0.0017075BC - 0.0003339BD - 0.003560CD - 3.94622A2 - 0.16530B2 - 0.067674C2 - 0.0035609D2 | (3) |
The statistical analysis of variance (ANOVA) yielded significant results, as evidenced by a large F-value of 1421.127 and a statistically significant P-value of less than 0.05 (<0.0001), as presented in Table 4. The "Pred R-Squared" value of 0.9961 exhibits a satisfactory level of concurrence with the "Adj R-Squared" value of 0.9985. Additionally, the R Squared value of 0.9992, which is in close proximity to 1, implies that the model employed to traverse the design space is effective (Fig. 3A and B). The absence of a significant Lack of Fit (LOF) value indicates that the model is statistically appropriate for the provided data.
Table 4.
ANOVA results for quadratic model generated for the degradation of MO dye.
| Source | Sum of Squares | Degree of freedom | Mean Square |
F Value |
p-value Prob > F |
Result |
|---|---|---|---|---|---|---|
| Model | 8497.92 | 14 | 606.99 | 1421.13 | < 0.0001 | Significant |
| A-pH | 0.26 | 1 | 0.26 | 0.61 | 0.4453 | |
| B-Temperature | 10.71 | 1 | 10.71 | 25.07 | 0.0002 | |
| C-Incubation Time | 73.33 | 1 | 73.33 | 171.67 | <0.0001 | |
| D-Concentration of Dye | 345.12 | 1 | 345.12 | 808.00 | <0.0001 | |
| AB | 0.80 | 1 | 0.80 | 1.87 | 0.1922 | |
| AC | 1.96 | 1 | 1.99 | 4.67 | 0.0473 | |
| AD | 0.64 | 1 | 0.64 | 1.49 | 0.2412 | |
| BC | 0.15 | 1 | 0.15 | 0.34 | 0.5671 | |
| BD | 0.22 | 1 | 0.22 | 0.51 | 0.4854 | |
| CD | 32.46 | 1 | 32.46 | 76.00 | <0.0001 | |
| A2 | 6834.18 | 1 | 6834.18 | 16000.55 | <0.0001 | |
| B2 | 1799.54 | 1 | 1799.54 | 4213.18 | <0.0001 | |
| C2 | 514.53 | 1 | 514.53 | 1204.64 | <0.0001 | |
| D2 | 892.29 | 1 | 892.29 | 2089.09 | <0.0001 | |
| Residual | 6.41 | 15 | 0.43 | |||
| Lack of Fit | 5.57 | 10 | 0.56 | 3.34 | 0.096 | Not significant |
| Pure Error | 0.83 | 5 | 0.17 | |||
| Cor Total | 8504.32 | 29 |
Fig. 3.
Regression plots of decolorization of MO dye, A. Plot represent normal plot of residuals, B. Plot the projection made by the algorithm versus the actual amount of decolorization.
The Lack of Fit value is typically assessed using an F-test. If the Lack of Fit F-value is not significant (i.e., if the p-value associated with the F-value is greater than a chosen significance level, often 0.05), it suggests that the model fits the data adequately, and there is no significant lack of fit. Conversely, if the LOF, F-value is significant, it indicates that there is a lack of fit, and the model does not adequately explain the variation in the data.
The response variable (Degradation %) is affected by all parameters, and their influencing order can be ranked according to F- values as Dye Concentration (808.00) > Incubation Time (171.67) > Temperature (25.07) > pH (0.61), elucidating dye concentration has the most influencing effect in this study.
3.7. Analysis of MO dye degraded product
3.7.1. UV–visible spectroscopy analysis
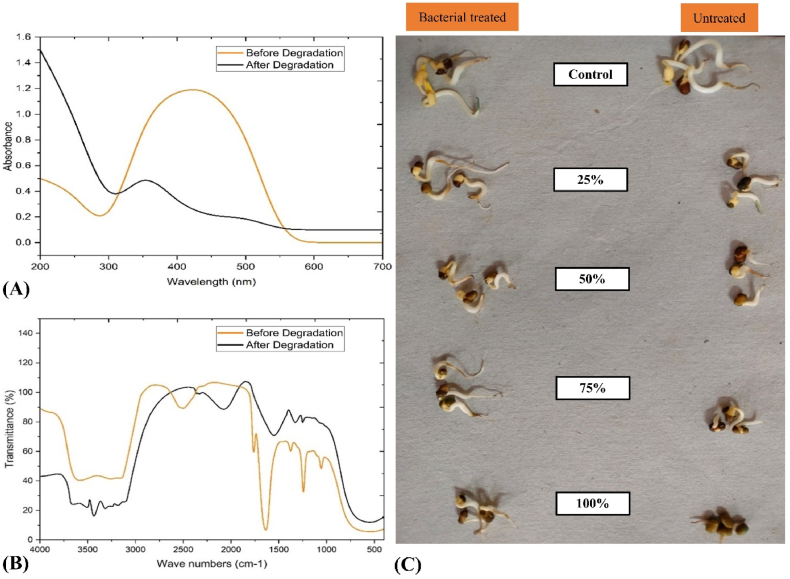
UV–Vis spectrophotometric analysis showed a characteristic λmax of MO dye at 446 nm wavelength that was absent in the degraded product, indicating structurally dissipation of MO dye. The process of dye decolorization may be linked to its degradation. The absorption peak that was observed exhibited disappearance within 24–48 h after incubation. Fig. 5 A shows the disappearance of a particular peak, indicating the elimination of MO dye through the degradation of azo bond and aromatic rings by the S. marcescens isolate found in the industrial effluent from the textile industry. The primary cause of MO dye decolorization by this bacterium is widely attributed to biodegradation.
Fig. 5.
A. Observations of the UV–Vis wavelengths of MO dye both before and after 48 h of degradation, B. FTIR spectra of MO dye before and after 48 h of degradation, C. On mung beans, the toxicity profile of untreated and bacterially-treated MO dye at various concentrations (25 %, 50 %, 75 %, and 100 %).
3.7.2. FT- IR analysis
Fourier-transform infrared spectroscopy (FT-IR) is a fundamental technique employed to verify the presence of MO dye and its degraded by-product. It can be used to determine if the product has been entirely biodegraded [53]. Functional groups indicate that the dye has been degraded into more straightforward, biodegradable molecules. The comparison of FT-IR spectra between the original dye (MO) and its degraded metabolites within a 48-h incubation period indicates the biodegradation of MO dye by bacterial isolate, as depicted in Fig. 5B. The FT-IR spectra for the unaltered MO dye (prior to degradation) exhibited an absorption peak at 34095.962 cm−1 and 3186.153 cm−1, signifying the stretching vibration of the N–H group of amines [54]. The appearance of a band around 3135.923 cm−1 indicated the stretching of the aromatic C–H group. The peak associated at 2514.093 cm−1 corresponded to the stretching of the C–H group. Additionally, the peak was recorded at 1055.969 cm−1 for stretching vibration of an ether group (-C-O-) [55]. The peaks at 1637.261 cm−1 for N N and 1243.007 cm−1 for C–N coniform the azo nature of the dye [22,56]. Peak 1375.235 cm−1 represented and confirmed the dye's sulphonic nature (S O) [9].
In comparison to the control MO dye, the spectra of the treated MO dye exhibited an absorption peak at 3680.270 cm-1, corresponding to the –OH group, and a new peak formed at 3434.122 cm−1 and 3320.946 cm−1 after the breakage of 34095.9619 cm−1 and 3186.153 cm−1. This is because the hydroxyl group typically exhibits a broad and robust peak in the range of 3600-3200 cm−1 due to the stretching of the O–H bond. The peak at 3434.122 cm−1 and 3320.946 cm−1 is typically associated with the stretching vibration of the N–H bend [48]. However, the peak at 34095.962 cm−1 and 3186.153 cm−1 for N–H stretching vibration widened or shifted the band locations. A peak confirmed a new C C group of stretching vibration at 2077.923 cm−1, a newly generated peak in the treated sample that was not present in the untreated sample. The peak represents 1553.373 cm−1,1326.386 cm−1, and 1255.724 cm−1, which indicate the presence of the C–O group, benzene ring mixed with the CH in-plane bending from the phenyl ring and the ethylene bridge, and amide III bands arise from C–N stretching respectively [54]. The degenerated sample's missing peak showed that the group had been eliminated during the decolorization procedure. The observed absence of spectra in the treated samples can be attributed to the comprehensive degradation of the dye molecule, which resulted in a shift in functional group peaks when compared to the control [57].
3.7.3. GC- MS analysis and pathway of degradation
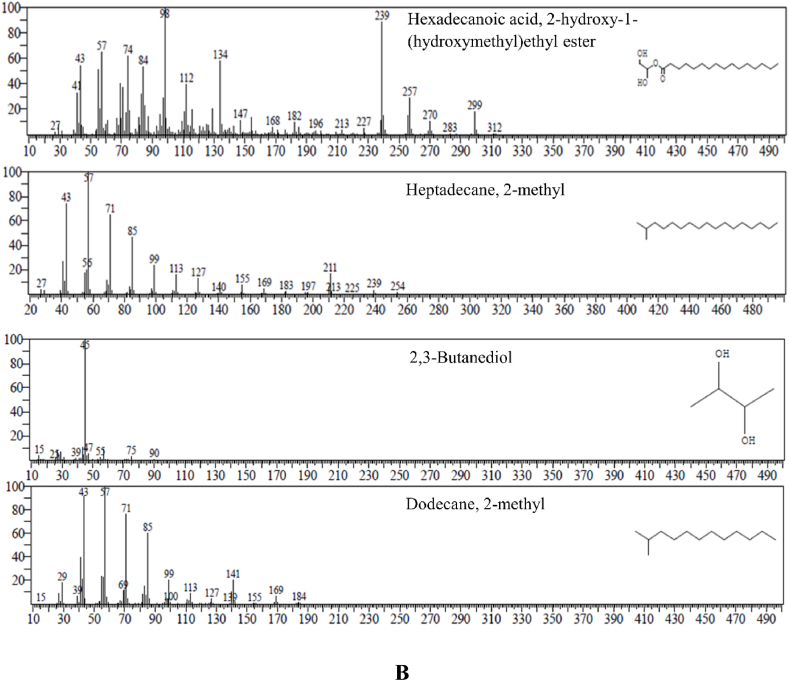
The GC-MS findings of the different dye-degraded metabolite products are shown in Table 5. In the present investigation, GC-MS analysis confirms that MO dye undergoes various transformations after being degraded by isolating ED1 bacteria. Fig. 6A, B depicts chemical identification by GC-MS spectra and chromatograms of extracted metabolites. Initially, S. marcescens ED1 was observed to asymmetrically cleave azo bond of MO dye through laccase enzyme activity and simultaneously convert into degraded metabolite, i.e. naphthalene, 2-methoxy (R-Time = 17.891; m/z = 115.05) with additional dimethyl sulfoxide (R-Time = 3.726 m/z = 63.00), and 2-oxopropinamide (R-Time = 4.831; m/z = 87.15) by the deamination, desulfonation, and carboxylation processes. Sequentially, via decarboxylation and oxidation, another intermediate metabolite product called dibutyl phthalate (R-Time = 14.884; m/z = 149.00) was isomerized into 1,2-benzenedicarboxylic acid, bis(2-methylpropyl) ester (R-Time = 17.891; m/z = 149.00). This product undergoes stepwise decarboxylation and dearomatization to yield hexadecanoic acid, 2- hydroxyl-1- (hydroxymethyl) ethyl ester (R-Time = 23.740; m/z = 98.10), which subsequently converted into common dye degraded metabolite product 2,3-butanediol (R-Time = 2.941; m/z = 45.05) and simple hydrocarbon chain metabolite product heptadecane- 2- methyl (R-Time = 16.177; m/z = 57.05). The product further undergoes decarboxylation to yield small hydrocarbon chain metabolite, i.e. dodecane, 2-methyl (R- Time = 15.756; m/z = 57.05), that ultimately yields simple inorganic compounds after 48 h. Alternatively, these intermediate metabolites may undergo directly or indirectly enter into fatty acid-oxidation reactions to produce NADH2 and FADH2. These reduced molecules may be utilised in the production of ATP. The suggested method for using isolated bacteria ED1, i.e., S. marcescens, to reduce sulfonated MO dye is depicted in Fig. 7. A pathway is proposed for the degradation of MO dye, illustrating the numerous phases involved with the confirmation of degradation into various end products. Additionally, additional research is required to clarify and validate these processes.
Table 5.
Degraded products of MO dye after 48 h of incubation through GC- MS analysis.
| S.No. | Compound | Chemical formula | R- Time | Structure |
|---|---|---|---|---|
| 1 | 1,2-Benzenedicarboxylic acid, bis (2-methylpropyl) ester | C16H22O4 | 17.891 | 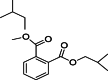 |
| 2 | Dodecane, 2-methyl | C13H28 | 15.756 | |
| 3 | 2,3-Butanediol | C4H10O2 | 2.941 |  |
| 4 | 2-Oxopropionamide, | C3H5NO2 | 4.831 | 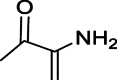 |
| 5. | Napthalene, 2- methoxy | C11H10O | 13.285 |  |
| 6 | Isoborneol | C10H18O | 9.210 |  |
| 7 | Heptadecane -2- methyl | C17H36 | 16.177 | |
| 8 | Hexadecanoic acid, 2- hydroxyl-1- (hydroxymethyl) ethyl ester | C19H38O4 | 23.740 | |
| 9 | Dibutyl phthalate, | C16H22O4 | 14.884 |  |
| 10 | Dimethyl Sulfoxide | C2H6OS | 3.726 | 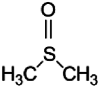 |
Fig. 6.
Chemical identification by GC-MS spectra, A. Chromatograms of Dimethyl Sulfoxide; Naphthalene, 2-methoxy; 2-Oxopropionamide; Dibutyl phthalate; 1,2-Benzenedicarboxylic acid, bis(2-methylpropyl) ester extracted metabolites of S. marcescens ED1 decolourized MO dye sample through GC-MS analysis, B. Chromatograms of Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl) ethyl ester; Heptadecane; 2-methyl, 2,3-Butanediol; Dodecane, 2-methyl extracted metabolites of S. marcescens ED1 decolourized MO dye sample through GC-MS analysis.
Fig. 7.
Proposed pathway for biodegradation of MO dye by S. marcescens ED1.
3.8. Toxicity of MO dye before and after bacterial treatment
Researchers frequently use the delicate mung bean plant as an example to assess the toxicity of contaminants [20]. In the current study, separately of UT (untreated) and BT (bacterially treated) MO dye in varying concentrations (25, 50, 75, and 100 %) of 200 mg/L was created and tested for their toxicity to the mung bean plant. The seedlings watered with 25 % and 50 % UT MO dye exhibited 100 % germination and the most significant root length and biomass. However, as shown in Fig. 5C, the seeds watered with a 75 % dose of unprocessed dye demonstrated a 30 % reduction in seed germination and adverse effects on root length, stalk length, and biomass output compared to the control. As shown in Table 6, the seed viability, root length, stalk length, and biomass output of the seedlings watered with 100 % concentration of UT dye were all inhibited by 40 %, 99.40 %, 92.3 %, and 83.94 %, respectively, when compared to the control.
Table 6.
Toxicity effects of untreated and treated MO dye seed on mung beans (Vigna radiata L.) germination.
| Conc. of MO dye |
No. of Seeds Sown |
No. of Seeds Germinated |
% Germination |
Root length (mm) |
Shoot length (mm) |
Biomass (mg) |
Root and shoot ratio |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UT MO dye | BT MO dye | UT MO dye | BT MO dye | UT MO dye | BT MO dye | UT MO dye | BT MO dye | UT MO dye | BT MO dye | UT MO dye |
BT MO dye |
||
| 25 % | 10 | 10 | 10 | 100 % | 100 % | 2.53 ± 0.029 | 17.76 ± 0.092 | 14.5 ± 0.058 | 62.46 ± 0.029 | 16.4 ± 0.058 | 27.86 ± 0.029 | 0.16 ± 0.006 | 0.28 ± 0.006 |
| 50 % | 10 | 9 | 10 | 90 % | 100 % | 1.63± 0.081 |
13.87 ± 0.029 | 12.7 ± 0.087 | 48.56 ± 0.087 | 8.56 ± 0.115 | 22.7 ± 0.098 | 0.133 ± 0.006 | 0.28 ± 0.006 |
| 75 % | 10 | 7 | 10 | 70 % | 100 % | 2.83 ± 0.029 | 13.7± 0.058 |
9.63 ± 0.029 | 42.8 ± 0.058 | 6.5 ± 0.058 | 17.5 ± 0.058 | 0.33 ± 0.017 | 0.32 ± 0.006 |
| 100 % | 10 | 6 | 10 | 60 % | 100 % | 0.23 ± 0.012 | 11.67 ± 0.087 | 5.23 ± 0.087 | 38.26 ± 0.087 | 4.5 ± 0.058 | 16.46 ± 0.173 | 0.03 ± 0.006 | 0.30 ± 0.009 |
| Control | 10 | 10 | 10 | 100 % | 100 % | 38.75 ± 0.023 | 38.75 ± 0.023 | 68.06 ± 0.063 | 68.06 ± 0.063 | 28.03± 0.029 |
28.03± 0.029 |
0.58 ± 0.006 | 0.58 ± 0.006 |
All the values are means of triplicates (n = 3) ± SE (Standard Error).
P < 0.05, F statistical value = 100870.07266.
*MO; methyl orange: Conc.: concentration; UT: untreated; BT: bacterial treated.
However, compared to nonbacterial treated MO dye, the seedlings watered with 25–75 % concentration of bacterially treated MO dye demonstrated 100 % germination and perpetual root length, stalk length, and biomass output, as shown in Fig. 5C. As opposed to the seeds watered with nonbacterial treated MO dye (100 %), the seeds rinsed with 100 % concentration of bacterially treated MO dye exhibited 100 % seed germination, 30.11 % root length, 56.21 % shoot length, and 58.72 % biomass output. According to this research, the bacteria-treated MO dye solution was less or entirely non-toxic for plants than the unprocessed MO dye.
4. Discussion
The demand from various end-use industries, including textiles, paper, leather, plastics, and printing inks, drives the consumption of dyes in globally [58]. The textile industry is India's largest dyes consumer, accounting for around 60 % of the total demand. During the textile manufacturing process of dyeing, printing, washing, and finishing, a large amount of effluent containing toxic chemicals, dyes, heavy metals, etc., is generated, which can harm the environment and human health if not treated appropriately [59]. To mitigate the harmful effects of textile effluent, treating it before discharge into the environment is crucial. Many bacteria can biodegrade and are used in bioremediation processes to clean up contaminated sites. Bacterial biodegradation is the process by which microorganisms break down organic compounds, such as pollutants, into simpler compounds that are less harmful to the environment. Microbial degradation offers a promising approach to treating industrial effluents containing toxic pollutants [60].
This given study mainly focuses on isolated Serratia marcescens strain ED1 dye degradation ability particularly targeting the degradation of MO. Our study extensively employed spectroscopic for monitoring dye removal efficiency and chromatographic for identifying intermediate metabolites within the proposed metabolic pathway to comprehensively characterize the degradation process. The chosen analytical methods were selected based on their sensitivity to detect low concentrations of dye and degradation products, specificity in distinguishing between different compounds, and compatibility with bacterial culture conditions. Optimizing parameter on the basis of RSM gave the maximize dye degradation efficiency of MO (81.02 %) with cost -effectiveness manner due to optimization of reagent usage to reduce the cost without affecting data quality. Validation procedures were done through pathogenicity of strain via the plate-based method and also asses the modern antibiotics with their MIC values for assessment of low risk rate of antibiotic resistant conducted to ensure the accuracy and reliability of our analytical methods.
The bacterial species belonging to the genus Serratia, a gram-negative facultative anaerobic bacterium in the family Enterobacteriaceae, has been recognised for its numerous roles in biodegradation. According to earlier study findings, S. marcescens exhibits disintegrating activity for lignin, chlorobenzenes, palmarosa oil (green oil), oily culinary refuse, p-cresol, low-density polyethylene, and pentachlorophenol (trash from wood and paper mills) [[13], [14], [15], [16], [17], [18]]. The 200 mg/L MO dye was found to be decomposed by S. marcescens for the first time in this research, to the best of our understanding and the literature survey. The MO pigment degraded by over 80 % after 48 h at 37 °C and pH 7.0 under steady circumstances. It shows that high concentration more than 200 mg/L of the selected MO dye is toxic for the selected bacterial strain. The study found a concentration above 200 mg/L was toxic for the isolated strain ED1. Therefore, in this study, a concentration of 200 mg/L dye was preferred for studying the MO dye degradation by strain ED1. This concentration favored implications in the real world, where the dye percentage in industrial effluent was high. The result was supported by Ref. [61], who reported that higher dye concentrations have a toxic effect on bacterial cell growth and may be due to azoreductase enzyme active sites interfering with dye molecules. Thus, lower dye concentrations show higher degradation efficiency than higher dye concentrations. Extracellular laccase and manganese peroxidase (MnP) activity were discovered during the decolorization of Ranocid Rapid Blue (RFB) and Procion Brilliant Blue-H-GR (PBB-HGR), according to Verma and Madamwar [19].
The biochemical and molecular characterization of isolated bacteria is a crucial aspect of microbiology that facilitates identifying and classifying bacteria based on their metabolic characteristics. Pure separated bacterial colonies on the nutrient agar plate screen revealed appreciable variance in colony appearance, including cell form, between all strains. The colony displayed gram-negative, rod-shaped, and motile bacteria, which are essential as the first piece of information in identifying microorganisms. The 16S rRNA gene is widely distributed and exhibits a high degree of conservation, rendering it a suitable molecular marker for taxonomic categorization and phylogenetic investigation [26]. The most efficient and dependable way of categorizing and recognizing bacteria is to align and compare the genome sequences of isolates and their close cousins. The variety of 16S rRNA gene sequences, their categorization, and their connection to the target species were all disclosed by the findings of the BLAST analysis. The 16S rRNA gene sequence is a technique commonly used for identifying and analyzing species because it is the most ideal, preserved, and steady code during microbial development. Using behavioural (morphological and molecular) and genotypic (16S rRNA gene sequence similarity) profiling techniques, 97 % identity with Serratia marcescens was discovered. Because it possessed the decolorization and degradation qualities required for the biotransformation of the MO dye, this strain was selected for the degradation experiments. A study done by Pathak and Kumar [29] also isolated four bacteria, Bacillus sp., Paracoccus sp., Pseudomonas sp., and Acinetobacter sp., from waste disposal sites showed positive results of polymer-degrading microorganisms [62]. studied the degradation of malachite green dye by a single isolated bacterium, Pseudomonas sp. YB2 from the sludge site decolorizes 90.40 % MG dye at a high concentration.
Although S. marcescens is generally considered a bacterium of low virulence, it can still cause infections, particularly in immunocompromised individuals or those with underlying medical conditions so that it can pose a risk to human health [63]. If a pathogenic bacterium is used to biodegrade a pollutant, there is a risk that it could escape into the surrounding environment and cause harm [64]. Therefore, it is essential to understand the pathogenicity of bacteria and carefully select non-pathogenic strains for use in the biodegradation process. This can help ensure that biodegradation's benefits are realized without adversely affecting human health or the environment. In this study, a plate-based assay of the isolated ED1 strain showed no hemolytic pattern on BAP, and neither lactose fermenting property on MAP suggested the non-pathogenic nature of isolated strain ED1. A similar study done by Darmawati et al. [40] conducted comparable plate-based pathogenicity on BAP, MAP, and CAP (Chocolate agar plate) for the selection of bacterial consortium made as a bioremediation agent of hospital effluent in central Java. They found that isolating bacteria that did not produce a hemolytic zone on the BAP media plate was adequate for this purpose.
Wastewater treatment plants (WWTPs) can be remarkable sources of antibiotic-resistant bacteria (ARB) and genes [65]. These can then be released into the environment through effluent discharge or biosolids as fertilizer. This can pose a risk to human and animal health and the environment by promoting the spread of antibiotic resistance. Knowing the sensitivity of bacteria to antibiotics is crucial for effectively treating bacterial infections, preventing antibiotic resistance, public health surveillance, and research. It can help identify emerging trends in antibiotics [66]. The study done by Roy et al. [21] on the bioremediation of malachite green dye by two bacterial strains isolated from textile effluents showed both isolate CV-S1 and CM-S1 were found sensitive to cefriaxone, trimethoprim/sulphamethoxazole, azithromycin, and tetracycline, median sensitive to neomycin and ampicilin, doxycycline and resistant to bacitracin, erythromycin, and cephradine. In this study, we used the Vitek 2 system, an automated microbiology system commonly used in clinical laboratories to identify microorganisms and determine their susceptibility to antibiotics; it offers a reliable and efficient method for performing AST. Higher MIC values in microbiological susceptibility tests often indicate lower susceptibility of the bacterium to the antibiotic. In this study MIC is 16.03 ± 0.03 μg/mL or above, it indicates that a high concentration of the antibiotic is required for inhibition of the growth of the bacteria. This suggests that the bacteria are resistant to the antibiotic at typical therapeutic doses. Interpreting MIC readings usually relies on predefined breakpoints set by organizations like the Clinical and Laboratory Standards Institute (CLSI) or the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Microorganisms are classified as susceptible, intermediate, or resistant to certain antibiotics according to their MIC values. This information can help design biodegradation strategies that minimize the risk of antibiotic resistance development and ensure the continued effectiveness of the biodegradation process. This approach was used in the study of AST through the Viteck 2 system of isolated bacteria used in the bioremediation process reported the first time.
The RSM method was used for analysis to isolate the impact of the two variables and hold all others constant. This method can reduce the energy and time spent refining, leading to more productive biodegradation procedures [67]. We performed 30 separate trials by varying the following four factors: pH, temperature, MO dye content, and breakdown duration. This statistical approach identifies the most favourable conditions for maximum MO dye degradation efficiency as 81.02 % at pH 7.0, temperature 37 °C with an incubation time of 48 h, and dye concentration 200 mg/L. Sandesh et al. [7] followed a similar method and procedure to optimize direct blue-14 dye degradation by Bacillus fermus to achieve 92.76 % at 68.78 ppm dye concentration in 72 h degradation time with 1 g of sucrose and 2.5 % (v/v) of inoculum.
Incorporating additional carbon and nitrogen sources has been found to augment the degradation of dyes due to their crucial functions in promoting the proliferation, metabolic activity, and enzyme synthesis of microorganisms engaged in the degradation mechanism. The carbon source, fructose, sustains and enhances the biodegradation process by up to 90 % due to enhanced laccase production by bacterial species [68]. The observed results are supported by a previous study where supplemented fructose functions as a co-metabolite that simulates gene expression of many functional proteins, i.e. oxidoreductases like azoreductase, Lacasse and ketol-acid reductoisomerase (NADP (+)), which generates reductive power to degrade azo bond of MO dye [69]. Additionally, the efficient nitrogen sources, beef extract and ammonium nitrate also improve the degradation of MO dye by promoting specific bacterial growth, stimulating enzyme production, facilitating metabolic activity, and creating a more favourable environment for the breakdown of complex dye molecules. The outcomes are also supported by a decolorization study conducted by Modi et al. [70], where certain organic compounds, such as peptone, significantly improved azo dye decolorization.
Characterizing the degraded product metabolites of dye utilising UV–Vis spectroscopy, FT-IR, and GC-MS is essential for understanding degradation pathways, ensuring product stability, complying with regulations, assessing environmental impact, and advancing research and development efforts in the dye industry [20,56]. UV–visible spectroscopy is a significant analytical technique employed for the analysis of dye decolorization. The process of dye decolorization may be linked to its degradation. In this study, the primary absorption peak of the MO dye is eliminated or a new absorption peak emerges, suggesting that the dye has undergone complete degradation or a new metabolite has been generated, respectively. Fourier Transform Infrared Spectroscopy (FT-IR) can analyze a sample's functional groups and chemical bonds. In the case of biodegradation products from a dye, FT-IR can be used to determine if the product has been wholly biodegraded [53]. Functional groups indicate that the dye has been degraded into more straightforward, biodegradable molecules. The formation of a new peak at 3680.270 cm−1 corresponds to the –OH group, and new peaks formed at 3434.122 cm−1 and 3320.946 cm−1 after the breakage of 34095.9619 cm−1 and 3186.153 cm−1 showed the N–H bend vibration. However, the peak at 34095.9619 cm−1 and 3186.153 cm−1 for N–H stretching vibration widened or shifted the band locations. C C group at 2077.923 cm−1 was a newly generated peak in the treated sample, which was not present in the untreated sample. The peak at 1553.373 cm−1,1326.386 cm−1, 1255.724 cm−1 in a sample of treated MO dye showed that changes occurred and, which confirmed the degradation process because these peaks were not present in the untreated sample. Khan et al. [71] also performed UV–Vis spectroscopy and FT-IR analysis of degraded products for conformation of an absence of functional groups in dye-treated samples. Various workers also investigated the confirmation of dye degradation through FT-IR analysis by the disappearance of the characteristic absorption peak, the rapid degradation, and the breakdown of aromatic rings collectively suggested that the isolated bacterium is actively involved in biodegrading the dye [51,[72], [73], [74]].
In the present study, GC-MS analysis has verified that MO dye undergoes degradation into various end products that produce energy-yielding processes. This study identified products during MO dye degradation that align with previous reports. The biodegradation of MO dye begins probably with a laccase-driven asymmetrical azo bond cleavage by deamination, simultaneously with desulfonication and carboxylation reactions that gave rise to Naphthalene, 2- methoxy, 2-oxopropinomide, and Dimethyl sulfoxide. These findings were also consistent with previous study carried out by Baena-Baldiris et al. [57], who reported formation of Naphthalene as a product during biodegradation of MO dye by azo bond cleavage through azoreductase and lignin peroxidase enzyme present in gold-mining Franconibacter sp., 1MS bacterium. Subsequently, these intermediate products are decarboxylated or oxidized to form Dibutyl phthalate (DBP) by some aromatic oxygenases, as reported by Tan et al. [75]. Further, the DBP product undergoes isomerization to rearrange into phthalic acid derivatives (i.e. 1,2- Benzenedicarboxylic acid, bis (2- methylpropyl) ester) that oxidized to form Butanediol/benzoic acid and simple methylated hydrocarbons like Heptadecane or Dodecane. The simple large-chain intermediate metabolites were detected in GC-MS due to slow bacterial metabolism. In this investigation, more simple products were produced due to rapid bacterial laccase reaction [76]. Alternatively, these intermediate compounds like phthalate, fatty acids, and aldehydes can transformed into acetyl-CoA through β-oxidation that undergoes the kerb cycle to generate reductive power NADH2 and FADH2. Mineralization degrades simple, intermediate compounds into carbon dioxide, water, and other inorganic compounds released into the environment.
The kind of cleavage, whether symmetric or asymmetric, is determined by the dye structure and the specific enzyme used. Bacterial laccases with low redox potential may not have the ability to break the azo bond [76]. Our study supports previous research showing laccase activity in Serratia bacteria, namely in the strain Serratia proteamaculans AORB19, which is well-known for its strong laccase production [30]. The current study also validates the effectiveness of laccase from S. marcescens ED1 in degrading azo dyes, as shown by the substantial degradation of MO dye. The results are consistent with the reported mechanism of laccase-mediated azo dye degradation, which includes asymmetric azo bond breaking, oxidative cleavage, dihydroxylation, deamination, demethylation, desulfonation, and subsequent chemical changes [77]. Furthermore, molecular docking studies documented in the literature have significantly contributed to understanding laccase's potential to interact with azo dyes and aid in their degradation. Such investigations found laccase to successfully degrade azo dyes, such as pigments red 23, fuchsin base, and Sudan IV, by formation of stable enzyme-substrate complex as confirmed by MD simulations [78].
Plant growth and reproduction depend on seeds; any harm to their sprouting or development can have severe biological and economic repercussions [79]. To manage environmental risk, it is essential to consider the seed toxicity of deteriorated goods. Determining the environmental effects of biodegradation processes and creating successful clean-up plans for polluted locations also require a knowledge of the seed toxicity of deteriorated goods [80]. Prior research by Sarkar et al. [81] revealed that Congo red (CR) (bacteria-degraded water) biodegraded products were non-toxic and had no detrimental effects on seed viability or plant development. As a result, comparable findings in our research supported the finding that the dye solution treated by bacteria was less harmful and demonstrated 100 % seed viability and excellent biomass output.
Due to their susceptibility to external stresses, mung legumes are frequently used as seeds in poisoning research. In the near future, plants may be fed with water or minerals from the bacteria-degraded intermediates for plant development.
Our methods offer flexibility in adapting to different bacterial isolates, as demonstrated by the successful degradation of MO by S. marcescens strain ED1 under optimized conditions. In future this study can contribute to ongoing efforts for scaling-up the process from laboratory-scale to industrial-scale, enzymes with potential applications in bioremediation, employing metagenomic and transcriptomic approaches for discovery of new enzymes and degradation mechanisms, designing and optimizing bioreactor systems with help of metallic nanocomposite for large-scale dye degradation, biosensors for detecting dye pollution, bioremediation strategies for contaminated sites, or value-added products from degradation by-products and also formulation of effective adsorbent with the help of bacterial enzyme and nanoparticles for adsorption of dye pollution.
5. Conclusion
The findings support, provide the feasibility and offer sustainable method for the recycling of effluent water. The present investigation represents the initial documentation of the superior MO degradation capacity of the isolated strain, owing to its non-hemolytic and non-antibiotic resistant nature. To the best of our knowledge and the literature survey, the 200 mg/L MO dye concentration is to be remediated by the S. marcescens ED1 strain marks the first instance in this research. The presence of beef extract, ammonium nitrate, and fructose enhanced the decolorization efficacy of the organism. The intermediates produced during the biodegradation of MO dye did not adversely affect the mung bean seed's germination, growth, and biomass. The primary objective of this investigation was to examine the effective utilization of bioresources and potential avenues for mitigating the ecological consequences of industrial operations. Overall, the results suggested that strain ED1 exhibits promise as a proficient decolorizer of azo dyes and has excellent potential for wastewater treatment.
Data availability statement
The data used to support the findings of this study were included in the article.
CRediT authorship contribution statement
Akanksha Pandey: Writing – review & editing, Writing – original draft, Methodology, Data curation, Conceptualization. Vinay Mohan Pathak: Writing – review & editing, Formal analysis, Data curation. Navneet: Supervision. Minakshi Rajput: Validation, Investigation, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are thankful to Ravi Diagnostic laboratory for providing me with antimicrobial susceptibility testing (AST) and identification. The authors wish to thank the Head, Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar (India) for providing necessary facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32339.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bankole P.O., Omoni V.T., Tennison-Omovoh C.A., Adebajo S.O., Mulla S.I. Enhanced removal of dibutyl phthalate in a laccase-mediator system: optimized process parameters, kinetics, and environmental impact. J. Environ. Manag. 2023;348 doi: 10.1016/j.jenvman.2023.119227. [DOI] [PubMed] [Google Scholar]
- 2.Mishra P., Kiran N.S., Ferreira L.F.R., Yadav K.K., Mulla S.I. New insights into the bioremediation of petroleum contaminants: a systematic review. Chemosphere. 2023;326 doi: 10.1016/j.chemosphere.2023.138391. [DOI] [PubMed] [Google Scholar]
- 3.Santhanarajan A.E., Rhee C., Sul W.J., Yoo K., Seong H.J., Kim H.G., Koh S.C. Transcriptomic analysis of degradative pathways for azo dye acid blue 113 in Sphingomonas melonis B-2 from the dye wastewater treatment process. Microorganisms. 2022;10(2):438. doi: 10.3390/microorganisms10020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouthami K., Lakshminarayana L., Veeraraghavan V., Bilal M., Bharagava R.N., Ferreira L.F., Rahdar A., Bankole O.P., Américo-Pinheiro H.-P.J., Mulla S.I. Application of microbes in dye decolorization. Microb. Biotechnol.: Role in Ecological Sustainability and Research. 2022:237–254. doi: 10.1002/9781119834489.ch13. [DOI] [Google Scholar]
- 5.Amin S., Rastogi R.P., Chaubey M.G., Jain K., Divecha J., Desai C., Madamwar D. Degradation and toxicity analysis of a reactive textile diazo dye-Direct Red 81 by newly isolated Bacillus sp. DMS2. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.576680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan M.D., Singh A., Khan M.Z., Tabraiz S., Sheikh J. Current perspectives, recent advancements, and efficiencies of various dye-containing wastewater treatment technologies. J. Water Process Eng. 2023;53 doi: 10.1016/j.jwpe.2023.103579. [DOI] [Google Scholar]
- 7.Sandesh K., Kumar G., Chidananda B., Ujwal P. Optimization of Direct Blue-14 dye degradation by Bacillus fermus (Kx898362) an alkaliphilic plant endophyte and assessment of degraded metabolite toxicity. J. Hazard Mater. 2021;364:742–751. doi: 10.1016/j.jhazmat.2018.10.074. [DOI] [PubMed] [Google Scholar]
- 8.Abd El-Rahim W.M., Moawad H., A Azeiz A.Z., Sadowsky M.J. Biodegradation of azo dyes by bacterial or fungal consortium and identification of the biodegradation products. Egypt. J. Aquat. Res. 2021;47(3):269–276. doi: 10.1016/j.ejar.2021.06.002. [DOI] [Google Scholar]
- 9.Akansha K., Chakraborty D., Sachan S.G. Decolorization and degradation of methyl orange by Bacillus stratosphericus SCA1007. Biocatal. Agric. Biotechnol. 2019;18 doi: 10.1016/j.bcab.2019.101044. [DOI] [Google Scholar]
- 10.Benkhaya S., M'rabet S., El Harfi A. Classifications, properties, recent synthesis, and applications of azo dyes. Heliyon. 2020;6(1) doi: 10.1016/j.heliyon.2020.e03271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyedi Z.S., Zahraei Z., Jookar Kashi F. Decolorization of reactive black 5 and reactive red 152 Azo dyes by new haloalkaliphilic bacteria isolated from the textile wastewater. Curr. Microbiol. 2020;77(9):2084–2092. doi: 10.1007/s00284-020-02039-7. [DOI] [PubMed] [Google Scholar]
- 12.Daphedar A.B., Kakkalameli S., Faniband B., Bilal M., Bhargava R.N., Ferreira L.F.R., Mulla S.I. Decolorization of various dyes by microorganisms and green-synthesized nanoparticles: current and future perspective. Environ. Sci. Pollut. Res. 2022:1–16. doi: 10.1007/s11356-022-21196-9. [DOI] [PubMed] [Google Scholar]
- 13.Xing Zhilin, Chen Shangjie, Fuqing Xu. Authorea; 2023. Kinetic Analysis and Dual Biodegradation Pathway for Chlorobenzenes Removal by Serratia marcescens Strain TF-1 and its Performance in Contaminated Soil. [DOI] [Google Scholar]
- 14.Mohanan S., Maruthamuthu S., Muthukumar N., Rajasekar A., Palaniswamy N. Biodegradation of palmarosa oil (green oil) by Serratia marcescens. Int. J. Environ. Sci. Technol. 2007;4:279–283. doi: 10.1007/BF03326285. [DOI] [Google Scholar]
- 15.Singh T., Srivastava N., Bhatiya A.K., Mishra P.K. Analytical study of effective biodegradation of p-cresol using Serratia marcescens ABHI001: application in bioremediation. 3 Biotech. 2017;7:1–8. doi: 10.1007/s13205-017-1006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azeko S.T., Etuk-Udo G.A., Odusanya O.S., Malatesta K., Anuku N., Soboyejo W.O. Biodegradation of linear low-density polyethylene by Serratia marcescens subsp. marcescens and its cell free extracts. Waste biomass valori. 2015;6:1047–1057. doi: 10.1007/s12649-015-9421-0. [DOI] [Google Scholar]
- 17.Rajasekar A., Babu T.G., Pandian S.T.K., Maruthamuthu S., Palaniswamy N., Rajendran A. Role of Serratia marcescens ACE2 on diesel degradation and its influence on corrosion. J. Ind. Microbiol. Biotechnol. 2007;34(9):589–598. doi: 10.1007/s10295-007-0225-5. [DOI] [PubMed] [Google Scholar]
- 18.Singh S., Chandra R., Patel D.K., Rai V. Isolation and characterization of novel Serratia marcescens (AY927692) for pentachlorophenol degradation from pulp and paper mill waste. World J. Microbiol. Biotechnol. 2007;23:1747–1754. doi: 10.1007/s11274-007-9424-5. [DOI] [PubMed] [Google Scholar]
- 19.Du L., Li H., Wu G., Wei Y., Wang F., Xu L., Dong X. Efficient degradation and decolorization of triphenylmethane dyes by Serratia sp. WKD under extreme environmental conditions and the mechanism. Int. Biodeterior. Biodegrad. 2023;179 doi: 10.1016/j.ibiod.2023.105565. [DOI] [Google Scholar]
- 20.Kishor R., Purchase D., Saratale G.D., Ferreira L.F.R., Hussain C.M., Mulla S.I., Bharagava R.N. Degradation mechanism and toxicity reduction of methyl orange dye by a newly isolated bacterium Pseudomonas aeruginosa MZ520730. J. Water Process Eng. 2021;43 doi: 10.1016/j.jwpe.2021.102300. [DOI] [Google Scholar]
- 21.Roy D.C., Biswas S.K., Sheam M.M., Hasan M.R., Saha A.K., Roy A.K., Tang S.S. Bioremediation of malachite green dye by two bacterial strains isolated from textile effluents. Microbiol. Curr. Res. 2020;1:37–43. doi: 10.1016/j.crmicr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayed L., Khelifi E., Jannet H.B., Miladi H., Cheref A., Achour S., Bakhrouf A. Response surface methodology for decolorization of azo dye Methyl Orange by bacterial consortium: produced enzymes and metabolites characterization. J. Chem. Eng. 2010;165(1):200–208. doi: 10.1016/j.cej.2010.09.018. [DOI] [Google Scholar]
- 23.Siddiqui V.U., Ansari A., Ansari M.T., Akram M.K., Siddiqi W.A. Fabrication of a zinc oxide/alginate (ZnO/Alg) bionanocomposite for enhanced dye degradation and its optimization study. RSC Adv. 2022;12(12):7210–7228. doi: 10.1039/D1RA08991A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawat D., Sharma R.S., Karmakar S., Arora L.S., Mishra V. Ecotoxic potential of a Presumably non-toxic azo dye. Ecotoxicol. Environ. Saf. 2018;148:528–537. doi: 10.1016/j.ecoenv.2017.10.049. [DOI] [PubMed] [Google Scholar]
- 25.Barua S., Miah S., Mahmud M.N., Rahman I.M. Isolation and identification of naturally occurring textile effluent-degrading bacteria and evaluation of their ability to inhibit potentially toxic elements. Results Eng. 2023;17 doi: 10.1016/j.rineng.2023.100967. [DOI] [Google Scholar]
- 26.Wang Z.W., Liang J.S., Liang Y. Decolorization of Reactive Black 5 by a newly isolated bacterium Bacillus sp. YZU1. Int. Biodeterior. Biodegrad. 2013;76:41–48. doi: 10.1016/j.ibiod.2012.06.023. [DOI] [Google Scholar]
- 27.Cui D., Zhang H., He R., Zhao M. The comparative study on the rapid decolorization of azo, anthraquinone and triphenylmethane dyes by anaerobic sludge. Int. J. Environ. Res. Publ. Health. 2016;13(11):1053. doi: 10.3390/ijerph13111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rima S.A.J., Paul G.K., Islam S., Akhtar-E-Ekram M., Zaman S., Saleh M.A., Uddin M.S. Efficacy of Pseudomonas sp. and Bacillus sp. in textile dye degradation: a combined study on molecular identification, growth optimization, and comparative degradation. J Hazard Mater Lett. 2022;3 doi: 10.1016/j.hazl.2022.100068. [DOI] [Google Scholar]
- 29.Pathak V.M., Kumar N. Implications of SiO2 nanoparticles for in vitro biodegradation of low-density polyethylene with potential isolates of Bacillus, Pseudomonas, and their synergistic effect on Vigna mungo growth. Energy Ecol. Environ. 2017;2:418–427. doi: 10.1007/s40974-017-0068-5. [DOI] [Google Scholar]
- 30.Ali N.S., Huang F., Qin W., Yang T.C. Identification and characterization of a new Serratia proteamaculans strain that naturally produces significant amount of extracellular laccase. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.878360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 35.Ligozzi M., Bernini C., Bonora M.G., De Fatima M., Zuliani J., Fontana R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 2002;40(5):1681–1686. doi: 10.1128/JCM.40.5.1681-1686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bobenchik A.M., Hindler J.A., Giltner C.L., Saeki S., Humphries R.M. Performance of Vitek 2 for antimicrobial susceptibility testing of Staphylococcus spp. and Enterococcus spp. J. Clin. Microbiol. 2014;52(2):392–397. doi: 10.1128/JCM.02432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tshabuse F., Buthelezi N., Folami A.M., Donnelly L., Swalaha F.M. Rapid detection of drug-resistant Escherichia coli by Vitek 2 compact system. WaterSA. 2022;48(4):450–456. doi: 10.17159/wsa/2022.v48.i4.3941. [DOI] [Google Scholar]
- 38.Kurz C.L., Chauvet S., Andrès E., Aurouze M., Vallet I., Michel G.P., Ewbank J.J. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003;22(7):1451–1460. doi: 10.1093/emboj/cdg159. 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diard M., Hardt W.D. Evolution of bacterial virulence. FEMS Microbiol. Rev. 2017;41(5):679–697. doi: 10.1093/femsre/fux023. [DOI] [PubMed] [Google Scholar]
- 40.Darmawati S., Muchlissin S.I., Ernanto A.R., Sulistyaningtyas A.R., Fuad H., Rahman K.M.Z., Ethica S.N. Pathogenicity scoring system for selection of bacterial consortium formulated as bioremediation agent of hospital wastewater in central Java. IOP Conf. Ser. Earth Environ. Sci. 2021;707(1) doi: 10.1088/1755-1315/707/1/012003. [DOI] [Google Scholar]
- 41.Biswas S., Kahali T., Mukherjee A., Chakraborty D., Guha C., Adhikary T., Basak P. Enzymes analysis, degradation kinetics, response surface optimization, and heavy metal tolerance of the biodegradation of malachite green by Stenotrophomonas koreensis. Ann. Finance. 2022:1–23. doi: 10.1080/10889868.2022.2143472. [DOI] [Google Scholar]
- 42.Pinheiro L.R.S., Gradissimo D.G., Xavier L.P., Santos A.V. Degradation of azo dyes: bacterial potential for bioremediation. Sustainability. 2022;14(3):1510–1532. doi: 10.3390/su14031510. [DOI] [Google Scholar]
- 43.Pathak V.M., Navneet Exploitation of bacterial strains for microplastics (LDPE) biodegradation. Chemosphere. 2023 doi: 10.1016/j.chemosphere.2023.137845. [DOI] [PubMed] [Google Scholar]
- 44.Verma M., Ekka A., Mohapatra T., Ghosh P. Optimization of kraft lignin decolorization and degradation by bacterial strain Bacillus velezensis using response surface methodology. J. Environ. Chem. Eng. 2020;8(5) doi: 10.1016/j.jece.2020.104270. [DOI] [Google Scholar]
- 45.Nouren S., Bhatti H.N., Iqbal M., Bibi I., Kamal S., Sadaf S., Safa Y. By-product identification and phytotoxicity of biodegraded Direct Yellow 4 dye. Chemosphere. 2017;169:474–484. doi: 10.1016/j.chemosphere.2016.11.080. [DOI] [PubMed] [Google Scholar]
- 46.Ibrahim A., El-Fakharany E.M., Abu-Serie M.M., ElKady M.F., Eltarahony M. M. Methyl orange biodegradation by immobilized consortium microspheres: experimental design approach, toxicity study and bioaugmentation potential. Biology. 2022;11(1):76. doi: 10.3390/biology11010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carolin C.F., Kumar P.S., Joshiba G.J. Sustainable approach to decolorize methyl orange dye from aqueous solution using novel bacterial strain and its metabolites characterization. Clean Technol. Environ. Policy. 2021;23(1):173–181. doi: 10.1007/s10098-020-01934. [DOI] [Google Scholar]
- 48.Thao T.T., Nguyen-Thi M.L., Chung N.D., Ooi C.W., Park S.M., Lan T.T. Microbial biodegradation of recalcitrant synthetic dyes from textile-enriched wastewater by Fusarium oxysporum. Chemosphere. 2023;325 doi: 10.1016/j.chemosphere.2023.138392. [DOI] [PubMed] [Google Scholar]
- 49.Rajput M., Bithel N. Phytochemical characterization and evaluation of antioxidant, antimicrobial, antibiofilm and anticancer activities of ethyl acetate seed extract of Hydnocarpus laurifolia (Dennst) Sleummer. 3 Biotech. 2022;12(9):215. doi: 10.1007/s13205-022-03267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao X., Wang H., Zhang S., Nishimura O., Li X. Azo dye degradation pathway and bacterial community structure in biofilm electrode reactors. Chemosphere. 2018;208:219–225. doi: 10.1016/j.chemosphere.2018.05.190. [DOI] [PubMed] [Google Scholar]
- 51.Ajaz M., Rehman A., Khan Z., Nisar M.A., Hussain S. Degradation of azo dyes by Alcaligenes aquatilis 3c and its potential use in the wastewater treatment. Amb. Express. 2019;9:1–12. doi: 10.1186/s13568-019-0788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Othman B., Haddad D., Tabbache S. Allelopathic effects of Sorghum halepense (L.) pers. And Avena sterilis L. Water extracts on early seedling growth of Portulacca oleracea L. and Medicago sativa L. Int. J. Med. Sci. 2018;5(10):7–12. [Google Scholar]
- 53.D'Souza E., Fulke A.B., Mulani N., Ram A., Asodekar M., Narkhede N., Gajbhiye S.N. Decolorization of Congo red mediated by marine Alcaligenes species isolated from Indian West coast sediments. Environ. Earth Sci. 2017;76(20):721. doi: 10.1007/s12665-017-7077-8. [DOI] [Google Scholar]
- 54.Movasaghi Z., Rehman S., ur Rehman D.I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008;43(2):134–179. doi: 10.1080/05704920701829043. [DOI] [Google Scholar]
- 55.Mishra S., Nayak J.K., Maiti A. A. Bacteria-mediated bio-degradation of reactive azo dyes coupled with bio-energy generation from model wastewater. Clean Technol. Environ. Policy. 2020;22:651–667. doi: 10.1007/s10098-020-01809-y. [DOI] [Google Scholar]
- 56.Haque M.M., Haque M.A., Mosharaf M.K., Marcus P.K. Decolorization, degradation, and detoxification of carcinogenic sulfonated azo dye methyl orange by newly developed biofilm consortia. Saudi J. Biol. Sci. 2021;28(1):793–804. doi: 10.1016/j.sjbs.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baena-Baldiris D., Montes-Robledo A., Baldiris-Avila R. Franconibacter sp., 1MS: a new strain in decolorization and degradation of azo dyes ponceau s red and methyl orange. ACS Omega. 2020;5(43):28146–28157. doi: 10.1021/acsomega.0c03786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alam S., Khan M.S., Umar A., Khattak R., Rahman N.U., Zekker I., Zahoor M. Preparation of Pd–Ni nanoparticles supported on activated carbon for efficient removal of basic blue 3 from water. Water. 2021;13(9):1211. doi: 10.3390/w13091211. [DOI] [Google Scholar]
- 59.Yaseen D.A., Scholz M. Textile dye wastewater characteristics and constituents of synthetic effluents: a critical review. Int. J. Environ. Sci. Technol. 2019;16:1193–1226. doi: 10.1007/s13762-018-2130-z. [DOI] [Google Scholar]
- 60.Jamee R., Siddique R. Biodegradation of synthetic dyes of textile effluent by microorganisms: an environmentally and economically sustainable approach. Eur. J. Microbiol. Immunol. 2019;9(4):114–118. doi: 10.1556/1886.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan A.U., Rehman M.U., Zahoor M., Shah A.B., Zekker I. Biodegradation of brown 706 dye by bacterial strain Pseudomonas aeruginosa. Water. 2021;13(21):2959. doi: 10.3390/w13212959. [DOI] [Google Scholar]
- 62.Tao Y., Wang F., Meng L., Guo Y., Han M., Li J., Wang S. Biological decolorization and degradation of malachite green by Pseudomonas sp. YB2: process optimization and biodegradation pathway. Curr. Microbiol. 2017;74:1210–1215. doi: 10.1007/s00284-017-1306-y. [DOI] [PubMed] [Google Scholar]
- 63.Genera F., Yousefian F., Danosos G., Friedler S., Martinez V. Multifocal Serratia marcescens infection in a healthy adult. JAAD Case Reports. 2022;30:48–50. doi: 10.1016/j.jdcr.2022.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dvorak P., Nikel P.I., Damborsky J., de Lorenzo V. Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 2017;35(7):845–866. doi: 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 65.Zahoor M., Ullah A., Alam S., Muhammad M., Hendroko Setyobudi R., Zekker I., Sohail A. Novel magnetite nanocomposites (Fe3O4/C) for efficient immobilization of ciprofloxacin from aqueous solutions through adsorption pretreatment and membrane processes. Water. 2022;14(5):724. doi: 10.3390/w14050724. [DOI] [Google Scholar]
- 66.Wikler M.A. vol. 26. 2006. pp. M7–A7. (Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard, Clsi (Nccls)). [Google Scholar]
- 67.Baliyan N., Dhiman S., Dheeman S., Kumar S., Maheshwari D.K. Optimization of indole-3-acetic acid using response surface methodology and its effect on vegetative growth of chickpea. Rhizosphere. 2021;17 doi: 10.1016/j.rhisph.2021.100321. [DOI] [Google Scholar]
- 68.Ayodeji F.D., Shava B., Iqbal H.M., Ashraf S.S., Cui J., Franco M., Bilal M. Biocatalytic versatilities and biotechnological prospects of laccase for a sustainable industry. Catal. Lett. 2023;153(7):1932–1956. doi: 10.1007/s10562-022-04134-9. [DOI] [Google Scholar]
- 69.Zhang Q., Xie X., Liu Y., Zheng X., Wang Y., Cong J., Sand W. Fructose as an additional co-metabolite promotes refractory dye degradation: performance and mechanism. Bioresour. Technol. 2019;280:430–440. doi: 10.1016/j.biortech.2019.02.046. [DOI] [PubMed] [Google Scholar]
- 70.Modi H.A., Rajput G., Ambasana C. Decolorization of water-soluble azo dyes by bacterial cultures, isolated from dye house effluent. Bioresour. Technol. 2010;101(16):6580–6583. doi: 10.1016/j.biortech.2010.03.067. [DOI] [PubMed] [Google Scholar]
- 71.Khan Z., Jain K., Soni A., Madamwar D. Microaerophilic degradation of sulphonated azo dye - reactive Red 195 by bacterial consortium AR1 through co-metabolism. Int. Biodeterior. Biodegrad. 2014;94:167–175. doi: 10.1016/j.ibiod.2014.07.002. [DOI] [Google Scholar]
- 72.Kabeer F.A., John N., Abdulla M.H. Biodegradation of malachite green by a newly isolated Bacillus vietnamensis sp. MSB17 from continental slope of the Eastern Arabian Sea: enzyme analysis, degradation pathway and toxicity studies. Bioremediat J. 2019;23(4):334–342. doi: 10.1080/10889868.2019.1671790. [DOI] [Google Scholar]
- 73.Ramesh P., Rajendran A. A. Photocatalytic dye degradation activities of green synthesis of cuprous oxide nanoparticles from Sargassum wightii extract. Chem. Phys. Impact. 2023;6 doi: 10.1016/j.chphi.2023.100208. [DOI] [Google Scholar]
- 74.Kumar S., Kumar S., Mir M.A., Vishnoi V.K., Pandey A., Pandey A. Bioefficacy of Sida cordifolia L. phytoextract against foodborne bacteria: optimization and bioactive compound analysis. Future Microbiol. 2023;18(17):1235–1249. doi: 10.2217/fmb-2023-0064. 2023. [DOI] [PubMed] [Google Scholar]
- 75.Tan F., Cheng J., Zhang Y., Jiang X., Liu Y. Genomics analysis and degradation characteristics of lignin by Streptomyces thermocarboxydus strain DF3-3. Biotechnol. biofuels bioprod. 2022;15(1):78. doi: 10.1186/s13068-022-02175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mani P., Fidal V.T., Bowman K., Breheny M., Chandra T.S., Keshavarz T., Kyazze G. Degradation of azo dye (acid orange 7) in a microbial fuel cell: comparison between anodic microbial-mediated reduction and cathodic laccase-mediated oxidation. Front. Energy Res. 2019;7:101. doi: 10.3389/fenrg.2019.00101. [DOI] [Google Scholar]
- 77.Legerská B., Chmelová D., Ondrejovic M. Degradation of synthetic dyes by laccases–a mini-review. Nova Biotechnol Chim. 2016;15(1):90–106. doi: 10.1515/nbec-2016-0010. [DOI] [Google Scholar]
- 78.Pande V., Joshi T., Pandey S.C., Sati D., Mathpal S., Pande V., Samant M. Molecular docking and molecular dynamics simulation approaches for evaluation of laccase-mediated biodegradation of various industrial dyes. J. Biomol. Struct. Dyn. 2022;40(23):12461–12471. doi: 10.1080/07391102.2021.1971564. [DOI] [PubMed] [Google Scholar]
- 79.Masarbo R.S., Niranjana S.R., Monisha T.R., Nayak A.S., Karegoudar T.B. Efficient decolorization and detoxification of sulphonated azo dye Ponceau 4R by using single and mixed bacterial consortia. Biocatal. Biotransform. 2019;37(5):367–376. doi: 10.1080/10242422.2019.1568414. [DOI] [Google Scholar]
- 80.Nawaz N., Ali S., Shabir G., Rizwan M., Shakoor M.B., Shahid M.J., Afzal M., Arsalan M., Hashem A., Alyemeni M.N., Ahmad P. Bacterial augmented floating treatment wetlands for efficient treatment of synthetic textile dye wastewater. Sustainability. 2020;12(9):3731. doi: 10.3390/su12093731. 2020. [DOI] [Google Scholar]
- 81.Sarkar S., Echeverría-Vega A., Banerjee A., Bandopadhyay R. Decolorisation and biodegradation of textile di-azo dye Congo red by Chryseobacterium geocarposphaerae DD3. Sustainability. 2021;13(19) doi: 10.3390/su131910850. 2021. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study were included in the article.