Abstract
In the past three decades, extraordinary advances have been made in the understanding of the pathogenesis of, and treatment options for, inflammatory arthritides, including rheumatoid arthritis and spondyloarthritis. The use of methotrexate and subsequently biologic therapies (such as TNF inhibitors, among others) and oral small molecules have substantially improved clinical outcomes for many patients with inflammatory arthritis; for others, however, these agents do not substantially improve their symptoms. The emerging field of pharmacomicrobiomics, which investigates the effect of variations within the human gut microbiome on drugs, has already provided important insights into these therapeutics. Pharmacomicrobiomic studies have demonstrated that human gut microorganisms and their enzymatic products can affect the bioavailability, clinical efficacy and toxicity of a wide array of drugs through direct and indirect mechanisms. This discipline promises to facilitate the advent of microbiome-based precision medicine approaches in inflammatory arthritis, including strategies for predicting response to treatment and for modulating the microbiome to improve response to therapy or reduce drug toxicity.
Inflammatory arthritides, including rheumatoid arthritis (RA), psoriatic arthritis (PsA) and ankylosing spondylitis (AS), are chronic, destructive, inflammatory disorders characterized by synovitis that can lead to accelerated morbidity, mortality and disability1-4. Over the past three decades, understanding of the immunological and molecular mechanisms in the pathogenesis of these disorders has advanced considerably, in particular with the discovery of TNF, IL-6 and other pro-inflammatory cytokines as important promoters of joint inflammation in RA, and of the role of TNF, IL-23 and IL-17 in spondyloarthritis (SpA; including PsA and AS) biology5. Furthermore, the use of methotrexate and, more recently, the advent of biologic therapies (such as those targeting TNF, IL-6 and the IL-23–IL-17 axis), as well as novel small molecules (for example, inhibitors of Janus kinase (JAK) or phosphodiesterase 4), has led to substantial improvements in clinical outcomes, ameliorating the quality of life for millions of patients with these forms of inflammatory arthritis. However, up to one half of patients with moderate or severe arthritis have no or suboptimal improvement in their symptoms with these treatments6-13. Therefore, insights into the underlying mechanisms that determine the pharmacokinetics and pharmacodynamics of anti-rheumatic drugs are urgently needed to maximize clinical response while eliminating patient frustration and wasteful health-care expenditure14,15. Multiple candidate biomarkers have been proposed for predicting (non)response to therapy, including clinical phenotype, host genetics, cytokines and autoantibodies, but they have either failed to be reproducible across cohorts or require lengthy treatment trials, during which joint damage could accrue.
Mounting evidence suggests that non-human genetic factors, most notably those derived from the trillions of microorganisms that live within and on the human body (the microbiota), might contribute to the development of RA and SpA in genetically susceptible individuals16-18. Although research examining intestinal communities as determinants of pathogenesis in inflammatory arthritis continues, the focus of study has also been expanded to include the mechanisms by which the aggregate genetic content of the gut microbiota (that is, the gut microbiome) encodes enzymatic machinery that modulates the pharmacokinetics of, and response to, immunomodulatory drugs19. The study of drug–microbiome interactions, termed pharmacomicrobiomics20-22, builds upon extensive research dating back to the 1930s on how microorganisms affect drug metabolism23,24. Novel sequencing technology enables researchers to dissect in ever more detail the constituent members of the gut microbiota and their genes and to investigate the effect of variations within the human gut microbiome on drugs; such research has already provided important insights into the effects of the microbiome on treatment response in autoimmunity and oncology, particularly those related to clinical outcomes of checkpoint inhibitors and biologic therapies19,25.
In this Review, we describe evidence from studies in animal models and humans characterizing the dynamic interactions between the gut microbiota and xenobiotics, with special emphasis on pharmaceuticals relevant to rheumatology. We also discuss the tools available to study pharmacomicrobiomics and describe relevant translational data in cancer and autoimmune diseases, as well as ongoing work in RA and SpA. Last, we discuss strategies to incorporate pharmacomicrobiomics into the realm of precision medicine in rheumatology, with an emphasis on the development of tools to predict treatment response and the development of microbiome-derived adjuvant therapies.
Gut microorganisms in drug metabolism
From the earliest stages of life, humans ingest a multitude of xenobiotics, including a variety of chemicals and medications26. Immediately after birth, humans are rapidly colonized by trillions of microorganisms (collectively referred to as the microbiota), many of which will ultimately inhabit their gastrointestinal tract27,28. The microbiota has a variety of critical roles in human physiology: supplementing host nutrition, aiding metabolism (for example, by catabolizing dietary and host-derived polysaccharides into short-chain fatty acids)29 and directly affecting maturation and development of the immune system and defence against pathogens30,31.
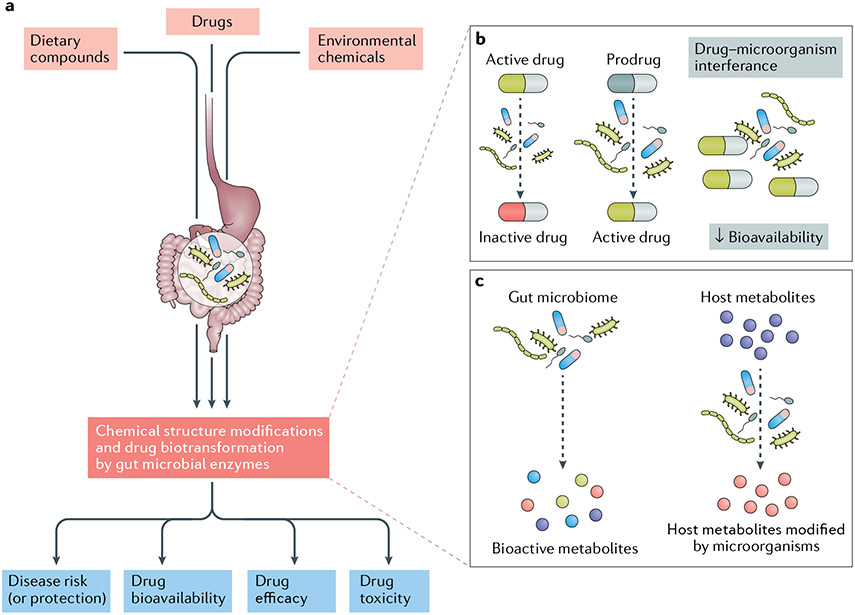
Intriguingly, variability between individuals in the composition and metabolic competence of their microbiomes has a unique role in determining the clinical efficacy of (and development of adverse events associated with) some medications. This variability arises because specific, direct modifications of the chemical structures of ingested drugs are dependent on the composition of gut microbial communities and their collective enzymatic activity, which can differentially modify the bioavailability of these medications and ultimately determine their biological fate and clinical effects32-34 (FIG. 1 a). Within the umbrella concept of precision medicine, the study of drug–microbiome interactions (that is, pharmacomicrobiomics) is gaining traction. A long-term goal of this research discipline is to manipulate complex host-associated microbial communities to improve drug efficacy, predict treatment outcomes and reduce the development of adverse events. This concept is not foreign to rheumatologists, who were arguably among the first clinicians to apply pharmacomicrobiomics; as we describe, classic examples include the prodrug sulfasalazine (which requires cleaving by the gut microbiome in order to become an active drug)35, as well as cyclophosphamide and methotrexate. Although fundamental insights into how gut microbiome-dependent biotransformations of xenobiotics affect human health are limited36, numerous studies have highlighted the extent to which microbial xenobiotic metabolism varies between individuals, the mechanisms by which these microbial activities influence human biology and how these reactions can be logically manipulated for therapeutic purposes37,38.
Fig. 1 ∣. Gut microorganisms in drug metabolism and physiology.
a ∣ Bacteria and other microorganisms that inhabit the human gut can directly alter the chemical structures of many dietary components, environmental chemicals and pharmaceuticals. These biotransformations have the potential to affect drug bioavailability, pharmacokinetics, clinical efficacy and the development of adverse events. An accumulating body of evidence is clarifying the molecular mechanisms responsible for many of these biological changes in anti-inflammatory medications. b ∣ Microorganisms can directly alter a drug through inactivation, activation or direct physical interactions that alter the drug’s bioavailability. c ∣ Indirect mechanisms of drug biotransformation include the production of intermediate bioactive metabolites by gut microorganisms and the alteration of host gene regulation and expression in response to microbial interactions.
Notably, the collective gut microbiome-mediated modification of xenobiotics has a large metabolic component that is yet to be uncovered in its entirety39. The reasons for the vastness of this microbial enzymatic catalogue are multifactorial. The first reason relates to the greater abundance and diversity of bacterial cells relative to the more homogeneous host-intestinal cell population40. Equally important is the fact that gut bacteria are constantly subjected to evolutionary pressures exerted by the host and its ingested xenobiotics, which oblige the microorganisms to adapt to environmental fluctuations by altering their functional abilities and extracting vital nutrients for survival. These adaptations lead, in turn, to an extraordinary expansion of the number of xenobiotics that become subject to gut microbial metabolism21,41.
These biotransformations occur through two main mechanisms (FIG. 1 b,c). The first mechanism involves the direct interference of microbial enzymes with ingested medications, leading to the generation of end-products (or metabolites) that vary from the original prodrug. Several examples of this direct interference have been described42, including the bacterial production of bioactive compounds (as in the hydrolysis of hydroxy-cinnamate esters by microbial cinnamoyl esterases)43, microbial detoxification of drugs (for example, selected strains of the prevalent gut Actinobacterium Eggerthella lenta inactivate the cardiac drug digoxin)44-47, direct interaction between microbial cells and xenobiotics (for example, physical attachment of Helicobacter pylori adhesins to levodopa, which decreases the bioavailability of the drug)48 and interruption of the enterohepatic circulation and enteropathy of drugs (for example, inhibitors of luminal bacterial β-glucuronidase halt the hydrolysis of NSAID glucuronides and alleviate NSAID gut toxicity)49. The second mechanism involves indirect effects of host–microorganism interactions on drugs, including the alteration of host gene expression in response to microbial interactions50, production of intermediate metabolites by gut microorganisms (for example, dietary-derived phosphatidylcholine conversion by the intestinal bacteria to trimethylamine)51-53 and competition between bacterial metabolites and drugs for binding sites in host enzymes (as in the case of bromovinyl uracil, a metabolite of the anti-viral drug sorivudine that inhibits the degradation of 5-fluorouracil, resulting in its accumulation in the blood and a marked increase in its toxicity)54.
Tools to study pharmacomicrobiomics
Multiple methodologies are used to generate complementary lines of evidence implicating the microbiome in drug pharmacology. These approaches include the use of clinical studies, involving well-phenotyped cohorts with extensive clinical and demographic details, along with in vitro and ex vivo mechanistic experiments and studies in ‘humanized’ murine models55 (BOX 1). This integrative approach has been critical in the identification of the microbial strains, microbial consortia, genes and/or metabolites necessary for drug biotransformation. The use of these methods was pioneered in original work exploring how gut microorganisms metabolize drugs such as digoxin and irinotecan44,45,56,57.
Box 1 ∣. Methods and tools for studying pharmacomicrobiomics.
Metagenomics
The study of a microbial community by sequencing the aggregate genetic material from an environmental or clinical sample. Abundance of drug-related pathways and/or specific enzymes within a microbial community can provide insights into non-host-driven biotransformation processes.
Gnotobiotic animals
Animals in which the composition of all microorganisms present is known; the term ‘gnotobiotic’ derives from the Greek words ‘gnostos’ (meaning ‘known’) and ‘bios’ (‘life’).
Germ-free mice
Mice bred and raised under conditions to render them free from all microorganisms. Transplanting whole human faecal microbial communities (or specific taxa or consortia) into germ-free mice enables the study of biotransformations within a known taxonomic environment.
Human microbiota-associated (‘humanized’) mice
Mice in which human faecal microbiota is established in germ-free mice through the transplantation of fresh or frozen gut microbiota samples (that is, faecal microbial transplantation).
Metabolomics
The quantification of all metabolites of a biological system, commonly using high-throughput analytical platforms such as nuclear magnetic resonance spectroscopy, gas chromatography–mass spectrometry and liquid chromatography–mass spectrometry; in pharmacomicrobiomics, the focus is bacteria-derived and drug metabolites. Non-targeted metabolomics are optimized to cover as much of the metabolome as possible, whereas targeted metabolomics can accurately quantify a known set of metabolites.
Computational methods and machine learning
Integrative network analysis, pathway analysis and predictive models combine clinical phenotypic data, 16S rRNA gene sequencing data and metagenomic and metabolomic features to characterize interactions between drugs, the microbiome, metabolites and host factors and their effects on drug bioavailability and pharmacokinetics. These methods and models can then be used to predict clinical responses and the deleterious effects of medications of interest.
A prototypical clinical study would involve samples obtained from a human population of interest (that is, individuals with a specific disease or clinical phenotype) and interrogate the microbiome at various taxonomic and functional levels, including the relative abundance of bacteria (using 16S rRNA gene sequencing (16S-seq)), gene families (metagenomic sequencing), microbial gene expression (metatranscriptomics) and metabolites (targeted and untargeted metabolomics). The results would then be analysed to characterize whether the pretreatment features (alone or in combination) of the microbiome correspond to a particular clinical outcome, most commonly in the form of efficacy or toxicity. Machine learning methods, including random forest and related decision-tree algorithms, could then be applied to create a predictive tool25. These analyses not only have the ability to rapidly inform clinical practice but also generate hypotheses regarding the mechanisms by which microbial transformations of drugs change their pharmacokinetic properties or lead to compound inactivation or prodrug activation.
Although patient cohort studies are critical for identifying associations between microbial factors and drug response, additional methods are required to provide causal evidence of microbially mediated drug metabolism. One such experimental approach employs the quantification of drug concentrations and related metabolites following the ex vivo incubation of the compound of interest with stool samples, microbial communities or specific bacterial strains under anaerobic conditions. Several platforms are commonly employed in drug metabolism and pharmacokinetics studies, including the many variations of mass spectrometry, most commonly liquid (or gas) chromatography–mass spectrometry and nuclear magnetic resonance spectroscopy. The application of these methods to analysing samples from patients enables the characterization of inter-individual variation in the rate of drug metabolism by gut microorganisms, comparisons between categories of clinical response or adverse events and hypothesis-generating research in model systems.
Whereas ex vivo profiling of human samples provides evidence for microorganism-mediated metabolism, in vitro studies are required to identify the bacterial genes or operons responsible for drug biotransformation. The recognition of which specific genes are involved in these biological processes requires the incubation of the drug of interest with bacterial strains, followed by comparative genomics and heterologous expression or deletion of key genes; such studies are capable of providing mechanistic insights into the role of the microbiome in drug metabolism58.
A complementary in vivo strategy incorporates the use of gnotobiotic animals and humanized mouse models59 (BOX 1) to further investigate the direct role of the microbiota in modulating drug pharmacokinetics. These techniques enable the study of intestinal microorganism–host interactions in human physiology, pathogenesis and pharmacology60, while avoiding the confounding effects of commonplace variations such as host genotype and diet. In such studies, gnotobiotic mice are typically either germ-free animals or those colonized with defined microbiota61, and humanization is achieved by transplanting whole human faecal microbial communities into germ-free mice, in order to interrogate biotransformations within a representative taxonomic environment. Experiments using germ-free animals are of course subject to a number of limitations, perhaps the most relevant of which is that the gut physiology of these animals is altered in comparison with wild-type animals, which in turn decreases their potential enzymatic and metabolic capabilities. However, humanization experiments can certainly help to explore the physiological effects of bacteria on the activation, inactivation and bioavailability of drugs in wild-type animals19 or interrogate clinical outcomes in specific disorders14,38 by the use of humanized murine (mouse and rat) models of autoimmune disease or inflammatory arthritis.
Drug biotransformation in mice
The pharmacomicrobiomics of several anti-rheumatic and immunosuppressive drugs have been studied in gnotobiotic experiments over the past few decades.
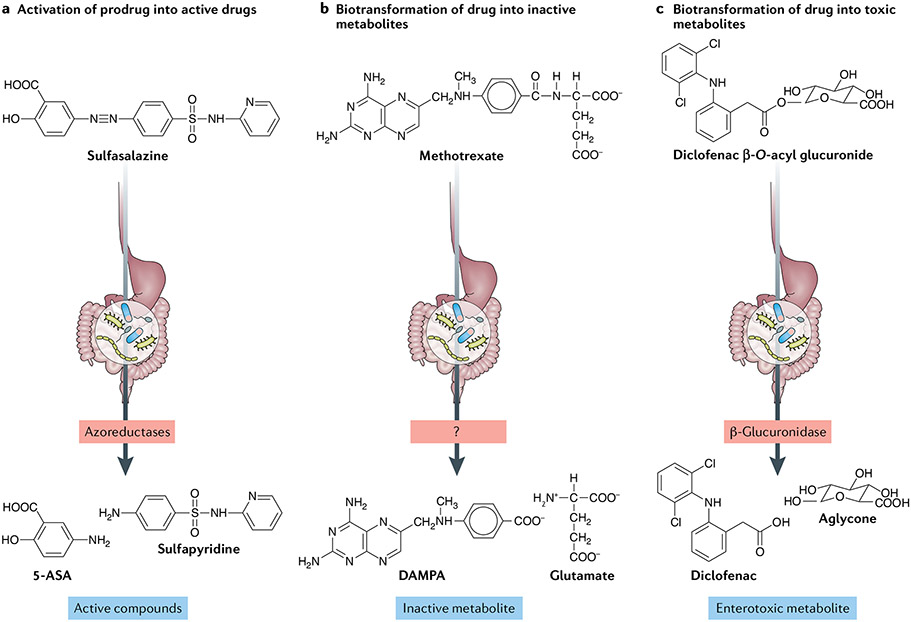
The prodrug sulfasalazine is considered the first rationally engineered medication for RA62 and, curiously, it was developed synthetically to combine an antibiotic, sulfapyridine, with an anti-inflammatory 5-aminosalicylic acid (5-ASA) molecule63 through an azo double bond. Sulfasalazine reaches the large intestine in its inactive form, where azoreductases encoded by the gut microbiome cleave the azo bond to release sulfapyridine and 5-ASA (FIG. 2a.). Sulfapyridine is then almost completely absorbed to promote anti-arthritic effects, whereas nearly all of the 5-ASA is excreted and becomes the active compound for the treatment of ulcerative colitis64. The role of the intestinal microbiome in sulfasalazine metabolism was demonstrated in classic gnotobiotic studies in the 1970s, in which conventionally raised rats fully converted sulfasalazine into its constituent molecules, whereas antibiotics-treated or germ-free animals excreted mostly the prodrug35. Importantly, a consortium of four gut microbiota-derived bacterial strains was sufficient to re-establish sulfasalazine metabolism in these animals35. These findings were later confirmed by experiments utilizing ex vivo incubation of sulfasalazine with human faecal samples65.
Fig. 2 ∣. Mechanisms of gut microbiome modulation of anti-rheumatic drug disposition and response.
The microbial metabolism of anti-rheumatic drugs can lead to their activation or inactivation, or result in the production of toxic compounds. a ∣ Activation is the conversion of a prodrug into its bioactive form, thus contributing to therapeutic concentrations. For example, biotransformation of sulfasalazine produces 5-aminosalicylic acid (5-ASA) and sulfapyridine (the active form of the prodrug in rheumatoid arthritis), b ∣ Inactivation is the conversion of an active metabolite into a less bioactive metabolite. For example, methotrexate is converted into 2,4-diamino-N10-methylpteroic acid (DAMPA) through the action of an (as yet uncharacterized) microbial enzyme. c ∣ Toxicity results from the production of bacterial metabolites that are deleterious to the host, for example, through the hydrolysis of glucuronidated NSAIDs.
The fact that the murine gut microbiome alters methotrexate metabolism has been known for decades66,67; remarkably, the gut microbiome of mice can mediate the metabolism of methotrexate, producing glutamate and the inactive metabolite 2,4-diamino-N10-methylpteroic acid (DAMPA) (FIG. 2b). Studies showed that, although methotrexate metabolites can be excreted and quantified in the faeces of conventionally reared animals, DAMPA is not detected in germ-free or antibiotics-treated mice, suggesting that the gut microbiome is necessary for this biotransformation.
The intestinal microbiome was also found to modulate the immunosuppressive effects of cyclophosphamide, a drug used for treating both arthritis and cancer68. Cyclophosphamide promotes a microbiota perturbation in the small intestine of cancer-bearing mice and induces the translocation of Gram-positive bacteria to secondary lymphoid organs, where they activate immune responses driven by pathogenic T helper 17 (TH17) cells and memory T helper 1 (TH1) cells68. However, under germ-free conditions (or after depletion of Gram-positive bacteria with antibiotics), these mice show decreased TH17 responses and their cancer becomes resistant to cyclophosphamide, suggesting that the gut microbiota can help to shape the anti-cancer (and potentially anti-rheumatic) immune response to this drug and related compounds69. Although informative, these proof-of-principle, mechanistic studies were performed in mouse models, in which the microbiome composition differs substantially from that of humans. With this limitation in mind, subsequent work has looked at the generalizability of microbiome-mediated biotransformations in patients with rheumatic or oncological diseases.
Drug modulation by human gut microorganisms
Many of the initial studies in modern human pharmacomicrobiomics have been in immuno-oncology. These studies are of interest to the rheumatology field, as many drugs used in the treatment of cancer are either also used in rheumatology (for example, methotrexate and cyclophosphamide) or known to cause autoimmune-like syndromes (such as checkpoint inhibitor-induced inflammatory colitis or arthritis). Several examples elegantly illustrate how the gut microbiome can modulate response to therapy in human disease. A pivotal study in 2016 analysed outcomes in patients with metastatic melanoma undergoing treatment with the checkpoint inhibitor ipilimumab, a monoclonal antibody that blocks cytotoxic T lymphocyte antigen 4 (CTLA4), and correlated the pretreatment composition of the patients’ microbiota with the development of colitis after treatment70. Baseline gut microbiota composition also predicted colitis in a subsequent ipilimumab study71, suggesting the possibility that microbial biomarkers might enable interventions to reduce the risk of inflammatory complications following immunotherapy.
Other work in the field of pharmacomicrobiomics has revealed that the baseline gut microbiome of patients with metastatic melanoma and other tumours can predict the outcomes of treatment with anti-programmed cell death protein 1 and anti-CTLA4 immunotherapies25,72-74. Importantly, modulation of the gut microbiome of germ-free mice via faecal microbiota transplantation (FMT) using samples from patients who responded to immunotherapy with ipilimumab could alter antitumour immunity and improve therapeutic response in the recipient mice75. Perhaps most intriguing is a 2018 report describing the successful implementation of FMT using samples from a single healthy unrelated donor to treat two patients with refractory immune checkpoint inhibitor-associated colitis; following FMT and gut microbiome reconstitution in both patients, the proportion of regulatory T cells increased within the colonic mucosa and clinical symptoms of colitis resolved76.
A 2019 study42 expanded our knowledge on the capacity of the human intestinal microbiome to biotransform oral medications prescribed for a wide range of clinical purposes, by combining the use of high-throughput functional genomic analyses and mass spectrometry to systematically identify human gut microorganisms and their gene products that metabolize drugs. Intriguingly, the results show that a large variety of human gut bacteria can metabolize a wide array of drugs, including anti-fungal, anti-hypertensive, anti-viral and hormone replacement medications; indeed, more than two thirds (176 of 271) of the tested medications were ultimately biotransformed42. However, the screening platform used in this study lacked controls, making the results and cut-off levels (that is, at what level a drug would be considered ‘metabolized’) challenging to interpret. Further work will be required to validate this approach.
Taken together, pharmacomicrobiomic data provide evidence that the gut microbiome can modulate the effects of parenteral immunotherapies and metabolize a sizable selection of oral medications (including anti-inflammatory drugs), with potential implications for the treatment of chronic inflammatory and autoimmune disorders77,78.
Pharmacomicrobiomics in autoimmunity
Research groups investigating human autoimmune diseases have utilized pharmacomicrobiomics methods in the analysis of the intestinal microbiome and/or its genetically encoded functions as predictors of response to biologic therapies (TABLE 1). Three prospective studies using samples from patients with inflammatory bowel disease (IBD)79 investigated associations between features of the microbiome and response to TNF inhibitors in biologic-naive patients with ulcerative colitis80, the α4β7 integrin blocker vedolizumab in patients with IBD81 and the IL-12–IL-23 blocker ustekinumab in patients with Crohn’s disease82. Pharmacomicrobiomics has also been applied to the study of drugs used for the treatment of human rheumatic diseases20,21. For example, the metabolic fate of paracetamol (also known as acetaminophen) was shown to be markedly associated with an individual’s pretreatment urinary concentration of p-cresol sulfate, a co-metabolite derived from the human gut microbiota83,84. As discussed, azo-bonded prodrugs used in the treatment of IBD and inflammatory arthritis (including sulfasalazine) rely on colonic bacteria for cleavage of the azo bonds via microbial azoreductases, which releases the biologically active compound in the large intestine. These enzymes are ubiquitous across the human gut microbiome85,86 and each azoreductase can bind multiple substrates87,88. However, the rate at which azo compounds are metabolized is substrate dependent. Moreover, the gut microbiota can metabolize the downstream metabolites of these azo reductions; for example, 5-ASA is inactivated by bacterial arylamine N-acetyltransferases89. Importantly, the activity of azoreductases has a high inter-individual variability89,90, further underscoring the need to incorporate gut microbiome analysis and metabolomics when studying clinical disparities in drug efficacy. This need was exemplified in a study using an in vitro colonic simulator to determine the rates of metabolism of sulfasalazine and other azo-bonded prodrugs in the presence of human-derived colonic bacteria65.
Table 1 ∣.
Pharmacomicrobiomic studies in autoimmune and rheumatic diseases
| Disease | Study design | Intervention | Result | Ref. |
|---|---|---|---|---|
| Ulcerative colitis | Prospective | TNF inhibitors | Non-responders characterized by high dysbiosis indices and a lower abundance of Faecalibacterium prausnitzii at baseline | 80 |
| Ulcerative colitis and Crohn’s disease | Prospective | Vedolizumab | High microbial diversity at baseline, specific taxa (e.g. Roseburia inulinivorans) and several microbial pathways enriched in patients achieving remission | 81 |
| Crohn’s disease | Prospective | Ustekinumab | Patients achieving remission had high microbial diversity and enrichment of specific taxa at baseline | 82 |
| Axial SpA | Prospective | TNF inhibitors | High relative abundance of the order Burkholderiales at baseline was modestly predictive of future response | 92 |
| PsA and SpA | Prospective | TNF and IL-17A inhibitors | Abundance of several specific taxa (e.g. Clostridiales) shifted after treatment with IL-17 blockade (as compared with TNF inhibition); Candida albicans was expanded in a subset of patients following IL-17 blockade | 94 |
| Treatment-naive, chronic RA | Prospective | Herbal remedies with or without methotrexate | Oral microbiome (and to a lesser degree the gut microbiome) distinguished responders from non-responders | 126 |
| Treatment-naive, new-onset RA | Prospective | Methotrexate | Gut metagenome at baseline could differentiate methotrexate responders from non-responders; ex vivo incubation with methotrexate of samples from patients with treatment-naive, new-onset RA correlated with the magnitude of future clinical response | 129 |
PsA, psoriatic arthritis; RA rheumatic arthritis; SpA, spondyloarthritis.
The intestinal microbiome has also been explored as a modulator of clinical outcome of treatment with monoclonal antibody therapies for inflammatory arthritis (TABLE 1). In 2018, a pilot study investigated whether baseline gut microbiota of patients with axial SpA predicted response to TNF inhibition91. Evaluation of stool samples from 19 patients using 16S-seq before and 3 months after anti-TNF treatment coupled with assessments of SpA disease activity suggested that a high relative abundance of the order Burkholderiales prior to initiation of anti-TNF therapy was modestly predictive of future response, although these results were not statistically significant after correction for multiple comparisons92.
An intriguing study in the β-1,3-glucan (curdlan)-triggered SKG mouse model of SpA revealed that treatment of SKG mice with anti-IL-23 monoclonal antibodies before curdlan injection not only suppressed SpA development but also shifted the faecal microbiota composition (with an increase in the relative abundance of the families Clostridiales and Lactobacillaceae) and prevented the outgrowth of SpA-associated pathobionts93. These results suggest that the interplay between host IL-23 and gut bacteria might promote the emergence of clinically evident SpA in genetically predisposed individuals.
The gut microbiota is also perturbed in patients with new-onset PsA, with dysbiosis resembling that seen in patients with IBD18. Treatment with either IL-17 blockade or TNF blockade affects the gut bacterial and fungal microbiota of patients with PsA and SpA too94. The relative abundance of several specific bacterial taxa, particularly Clostridiales, shifted after both treatments, with the changes more prominent with IL-17 blockade compared with TNF blockade. Intriguingly, in a subgroup of patients, initiation of IL-17A blockade was associated with a perturbation of intestinal fungal taxa, most notably Candida albicans. These results are not unexpected, as most clinical trials have reported occurrences of oropharyngeal candidiasis after IL-17A blockade95. However, intestinal candidiasis could help to explain why this treatment strategy failed in IBD96 and could potentially predict which (small subset of) patients with SpA treated with these biologics will develop (sub) clinical IBD96,97.
Predicting response to methotrexate
Despite remarkable advances in understanding the pathogenesis of RA and the discovery of numerous new therapies, oral methotrexate remains the anchor drug for the treatment of RA and related autoimmune conditions worldwide98. An accumulating body of literature suggests that early and aggressive intervention with methotrexate results in low disease activity, slow radiographic progression and can even lead to remission in some patients with RA99. This principle is now reflected in current treatment guidelines for RA, most notably those from the ACR (published in 2015) and EULAR (2020), which recommend the use of methotrexate in all patients with early RA100,101. However, more than half of patients with moderate or severe RA show no or suboptimal improvement in their symptoms in response to methotrexate therapy8,102-104, and bioavailability of the drug is known to be highly variable between individuals105-107. The reasons for these disparities remain unclear, and despite decades of study, differences in clinical response to methotrexate cannot be accurately predicted by host genetic factors or other established biomarkers108. An initial effort using concentrations of red blood cell methotrexate polyglutamates explained <20% of the variation in drug response109,110 and required a lengthy trial of methotrexate treatment, but the findings have not been consistently reproduced in other cohorts111,112. Other factors explored as potential determinants of methotrexate efficacy have included serum or plasma concentrations of methotrexate106,113, clinical factors such as sex and disease activity114-117 and circulating CD39+ regulatory T cells117,118. More than 70 genetic studies have also explored polymorphisms in candidate genes as predictors of methotrexate response, but no genetic marker has yet been sufficiently validated119.
A handful of cohort studies have integrated clinical, demographic and host-genomic factors into models to predict (lack of) responsiveness to methotrexate120,121. More than a decade ago, pivotal work led to the first clinical–pharmacogenetic model (that is, combining risk alleles with sex, smoking and the presence of rheumatoid factor) to predict the efficacy of methotrexate monotherapy in patients with recent-onset RA (defined as disease duration <2 years)122. Although this tool has improved the original genetics-based models by integrating multiple variables, its accuracy remains imperfect, and the clinical application of this model is not generalizable across populations123-125.
The failure of other factors to account for differences in the response to methotrexate raises the possibility that this variability could be driven, at least partially, by inter-individual disparities in the composition and function of the gut microbiome. As discussed earlier, work in germ-free and antibiotics-treated mice demonstrated decreased intestinal absorption and metabolism of methotrexate in these mice relative to wild-type mice66,67, suggesting a critical role for the gut microbiome in the biotransformation of this drug. Moreover, the gut microbiomes of patients with untreated, new-onset RA have been found to vary in bacteria-derived purine metabolic pathways, including biosynthesis of tetrahydrofolate (and other purines)17, which could modulate the absorption, bioavailability and downstream therapeutic effects of oral methotrexate.
A 2015 study found that the oral microbiome (and to a significantly lesser extent the gut microbiome) distinguished individuals with RA from healthy controls, and that microbiome alterations correlated with clinical indices and response to therapy, suggesting potential diagnostic and prognostic value126. However, this study focused primarily on patients with longstanding, established RA, who are known to harbour a markedly distinct gut microbiome relative to patients with new-onset RA17. In addition, response to methotrexate was predicted on the basis of the abundance of metagenomics-catalogued species rather than specific gene orthologues, thus precluding a detailed functional analysis.
A study using 16S-seq demonstrated that, over time, oral methotrexate at doses conventionally used in RA does not lead to consistent perturbations in gut microbial ecology127. However, applying in vitro and gnotobiotic methods, methotrexate can be observed to affect the composition of the gut microbiota of humanized mice in a dose-dependent manner and to directly inhibit the growth of some human gut bacteria128. Taken together, these data suggest that methotrexate, by altering bacterial physiology, might exert its anti-inflammatory effects in part by modulating the gut microbiome of patients with RA. Intriguingly, ongoing studies have demonstrated that the pretreatment microbiomes of patients with new-onset RA can be used to differentiate methotrexate responders from non-responders129. Moreover, use of machine learning techniques resulted in a robust predictive model, and remaining concentrations of methotrexate after ex vivo incubation with pretreatment samples from patients with new-onset RA correlated with the magnitude of future clinical response, suggesting a direct effect of the gut microbiome on methotrexate bioavailability and response to therapy129. Together, these results provide the first step towards the use of the gut microbiome to predict response to oral methotrexate therapy in patients with new onset RA and perhaps even its use as a target for manipulation in the treatment of rheumatic and autoimmune disease. Work is ongoing to understand if parenteral administration of methotrexate (and biologic therapies) can also be affected by the gut microbiome and whether using the microbiome as a predictor of response can be applied to other oral anti-rheumatic drugs (for example, JAK inhibitors).
Applications for precision medicine
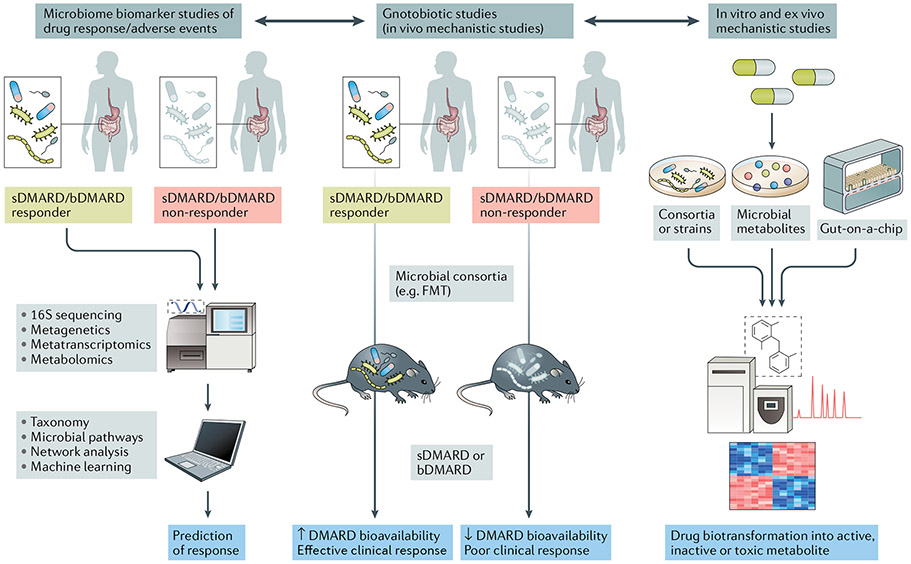
Advancing our knowledge and the translational applicability of pharmacomicrobiomics is highly relevant to our understanding of drug efficacy and adverse reactions to medications routinely prescribed in rheumatology (FIG. 3). Because the magnitude of response to drugs such as methotrexate, sulfasalazine and other synthetic and biologic DMARDs is known to have a high and unpredictable interindividual variability, the incorporation of precision medicine strategies based on features of the gut microbiome could help to guide a more rational use of these treatments (BOX 2).
Fig. 3 ∣. Translational implications of pharmacomicrobiomic studies in rheumatic diseases.
In clinical studies with deeply phenotyped patient populations and known outcomes of synthetic (sDMARDs) or biologic DMARDs (bDMARDs) (such as efficacy and adverse events) (left panel), microbial features can be integrated with established biomarkers of response (for example, host genetics or immune cell profiles) via machine learning and network analyses to develop predictive tools. Mechanistic studies applying in vivo methods (middle panel) and in vitro or ex vivo methods (right panel) can complement and expand the understanding of drug biotransformation by the human gut microbiome, including activation, inactivation, conversion into toxic metabolites and bioavailability. FMT, faecal microbiota transplantation.
Box 2 ∣. Potential applications of pharmacomicrobiomics in precision medicine.
Intestinal microbiome as a biomarker of response
Microbial community composition, the relative abundance of specific taxa, microbial pathways or metabolites could be measured to predict the efficacy and/or toxicity of synthetic and biologic DMARDs and other commonly used anti-rheumatic medications. This information could help to guide clinical decision-making and the initiation of early and effective treatments in rheumatoid arthritis, psoriatic arthritis and other related diseases.
Microbiome-modulating strategies
Taxonomic, metagenomic and metabolomic approaches enable the identification of microbial communities, strains and/or metabolites that can modulate drug bioavailability and improve clinical efficacy (or decrease the occurrence of adverse events). Strategies to modulate the microbiome include adjuvant therapies that either introduce communities or consortia (for example, via faecal microbiota transplantation or probiotics) or the modification of microbial composition through natural or engineered products (for example, probiotics).
Inhibition of gut microbial enzymes
Small molecules can be designed to inhibit the activity of bacterial functional pathways involved in the biotransformation of drugs into toxic metabolites (for example, the inhibition of β-glucuronidase to prevent NSAID-associated enteropathy).
From a diagnostic perspective, it is possible to envision the application of pharmacomicrobiomics in rheumatology through the measurement of microbial species, genes, transcripts and/or proteins that affect drug metabolism, small-molecule transport or immunoprotective responses19. This information could empower both clinicians and patients to adopt the best course of therapeutic action, on the basis of pretreatment gut microbial features (FIG. 3). In turn, this information can guide decision-making by either avoiding medications that are likely to fail to achieve meaningful clinical outcomes or engineer new avenues of microbiome-modulating strategies (sequential or adjuvant) that can lead to a desirable composition of microorganisms or genes to improve drug bioavailability and symptom amelioration. As discussed, these approaches have already proven successful in oncology (for example, the use of baseline gut microbiota as a predictor of clinical response and the development of colitis in checkpoint inhibitor trials72-74, as well as the use of FMT for the treatment of colitis70,71,130,131), and they are now being employed in human inflammatory arthritis. One relevant example is the FLORA study132, an ongoing randomized, placebo-controlled trial of FMT in patients with active PsA who have an inadequate response to methotrexate.
Although much will be learned from these proof-of-principle studies, other, less cumbersome, microbiome-regulating modalities are being tested, including adjuvant prebiotic and probiotic approaches that can potentially achieve similar results without the challenges and barriers of FMT (for example, risks inherent to the procedure, lack of clinical practicality and the potential to introduce pathogens into the recipient). Novel technologies, such as organs-on-chips (for example, gut-on-a-chip)133-135 and bacterial culturomics21,136, promise to aid in the understanding of the mechanisms underlying pharmacomicrobiomics by attempting to mimic the intestinal environment and to recapitulate physiological host–microorganism interactions. Drugs of interest can then be incubated in these systems to assess their effect on bacterial growth and metabolism137, as well as the mechanisms by which bacteria biotransform medications (FIG. 3).
Conclusions
Numerous studies have demonstrated that the absorption, distribution, metabolism and excretion of drugs and other xenobiotics require multistep, effective interactions between host and microbial pathways58. Therefore, the integration of clinical factors, host genomics and pharmacomicrobiomics in a rigorous and validated manner, and their application in extensively phenotyped cohorts, will establish the basic knowledge for major advances in personalized medicine in rheumatology. Further discoveries of drug–microbiome–host interactions will require the application of innovative bioinformatic and machine learning tools coupled with ex vivo, in vitro and gnotobiotic models.
Key points.
Culture-independent, high-throughput DNA and RNA sequencing technologies—coupled with deeper insight into host mucosal immunology — have substantially advanced our understanding of the role of microorganisms in modulating health and disease.
Pharmacomicrobiomics, an emerging field that describes the complex interaction of drugs with the microbiome, is increasingly considered an important factor in the prediction of therapeutic responses in many medical subspecialties.
Multiple tools, including ex vivo cultures, metabolomics and gnotobiotic experiments, have enabled a deeper mechanistic understanding of host–microbial interactions in the pharmacokinetics of many available drugs.
Emerging evidence supports the notion that the bioavailability, clinical efficacy and toxicity of several drugs used to treat human inflammatory arthritis can be modulated by human gut microorganisms and their enzymatic products.
Pharmacomicrobiomics could potentially be incorporated into precision medicine approaches in rheumatology.
Acknowledgements
The authors are supported by the NIH (grants R03AR072182 and R01AR074500 to J.U.S.; R01HL122593 and R01AR074500 to P.J.T.; K08AR073930 to R.N.). J.U.S. is further supported by The Riley Family Foundation, The Beatriz Snyder Foundation, the Rheumatology Research Foundation, the National Psoriasis Foundation and The Judith and Stewart Colton Center for Autoimmunity. P.J.T. is a Chan Zuckerberg Biohub investigator and a Nadia’s Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42-16) and the Searle Scholars Program (SSP-2016-1352). C.U. is supported by MINECO (SAF2017-90083-R).
Glossary
- Pharmacokinetics
The study of how an organism affects a drug, including absorption, distribution, bioavailability, metabolism and excretion.
- Pharmacodynamics
The study of the biochemical, physiological and molecular effects Of drugs on the body, including receptor binding, Post-receptor effects and chemical interactions.
- Xenobiotics
Chemical compounds (for example, drugs or pollutants) found within but not produced by living Organisms.
- Biotransformations
The processes by which a compound (for example, a drug) is transformed from one form to another by a chemical reaction within the body.
- Microbial consortia
Two or more microbial groups living symbiotically.
- Random forest
A data construct classifier applied to machine learning that develops large numbers of random decision trees that analyse multiple sets of variables.
- Operons
Genetic regulatory systems found in bacteria and their viruses in which genes encoding functionally related proteins are clustered along the DNA.
- Prebiotic
Non-digestible supplement that induces the growth (and/or activity) of commensal microorganisms.
- Probiotic
Supplement containing live microorganisms that can alter the composition of microbiota and are supposed to provide health benefits to the host.
- Bacterial culturomics
A method that allows for the description of the microbial composition by high-throughput culture platforms.
Footnotes
Competing interests
J.U.S. declares that he has served as a consultant for Amgen, BMS, Janssen, Novartis, Sanofi and UCB, and has received funds from Novartis to NYU School of Medicine to conduct investigator-initiated studies. J.U.S. and S.B.A. have been granted USPTO patent no. 10011883 (“Causative agents and diagnostic methods relating to rheumatoid arthritis”). P.J.T. declares he is on the scientific advisory boards for Kaleido, Seres, SNIPRbiome, uBiome, and WholeBiome; there is no direct overlap between the current article and these consulting duties. R.R.N. and C.U. declare no competing interests.
References
- 1.McInnes IB & Schett G The pathogenesis of rheumatoid arthritis. N. Engl. J. Med 365, 2205–2219 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Gladman DD, Antoni C, Mease P, Clegg DO & Nash P Psoriatic arthritis: epidemiology, clinical features, course, and outcome. Ann. Rheum. Dis 64, ii14–ii17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnas JL & Ritchlin CT Etiology and pathogenesis of psoriatic arthritis. Rheum. Dis. Clin. North Am 41, 643–663 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Taurog JD, Chhabra A & Colbert RA Ankylosing spondylitis and axial spondyloarthritis. N. Engl. J. Med 374, 2563–2574 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Keffer J. et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 10, 4025–4031 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McInnes IB et al. Secukinumab, a human anti-interleukin-17A monoclonal antibody, in patients with psoriatic arthritis (FUTURE 2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 386, 1137–1146 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Mease PJ et al. Adalimumab for the treatment of patients with moderately to severely active psoriatic arthritis: results of a double-blind, randomized, placebo-controlled trial. Arthritis Rheum. 52, 3279–3289 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Emery P. et al. Comparison of methotrexate monotherapy with a combination of methotrexate and etanercept in active, early, moderate to severe rheumatoid arthritis (COMET): a randomised, double-blind, parallel treatment trial. Lancet 372, 375–382 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Liu W. et al. Efficacy and safety of TNF-alpha inhibitors for active ankylosing spondylitis patients: multiple treatment comparisons in a network meta-analysis. Sci. Rep 6, 32768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baeten D. et al. Secukinumab, an interleukin-17A inhibitor, in ankylosing spondylitis. N. Engl. J. Med 373, 2534–2548 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Lee EB et al. Tofacitinib versus methotrexate in rheumatoid arthritis. N. Engl. J. Med 370, 2377–2386 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Gladman D. et al. Tofacitinib for psoriatic arthritis in patients with an inadequate response to TNF inhibitors. N. Engl. J. Med 377, 1525–1536 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Kavanaugh A. et al. Treatment of psoriatic arthritis in a phase 3 randomised, placebo-controlled trial with apremilast, an oral phosphodiesterase 4 inhibitor. Ann. Rheum. Dis 73, 1020–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdollahi-Roodsaz S, Abramson SB & Scher JU The metabolic role of the gut microbiota in health and rheumatic disease: mechanisms and interventions. Nat. Rev. Rheumatol 12, 446–455 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Curtis JR et al. Use of a validated algorithm to estimate the annual cost of effective biologic treatment for rheumatoid arthritis. J. Med. Econ 17, 555–566 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Scher JU & Abramson SB The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol 7, 569–578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scher JU et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher JU et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 67 128–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koppel N, Maini Rekdal V & Balskus EP Chemical transformation of xenobiotics by the human gut microbiota. Science 356, eaag2770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saad R, Rizkallah MR & Aziz RK Gut pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 4, 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doestzada M. et al. Pharmacomicrobiomics: a novel route towards personalized medicine? Protein Cell 9, 432–445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizkallah MSR & Aziz RK The Human Microbiome Project, personalized medicine and the birth of pharmacomicrobiomics. Curr. Pharmacogenomics Person. Med 8, 182–193 (2010). [Google Scholar]
- 23.Je T, Tréfouël, Nitti F & Bovet D Activité du p-aminophénylsulfamide surl’infection streptococcique expérimentar de la souris et du lapin. CR Soc. Biol 120, 23 (1935). [Google Scholar]
- 24.Butler V, Neu H & Lindenbaum J Digoxin-inactivating bacteria: identification in human gut flora. Science 220, 325–327 (1983). [DOI] [PubMed] [Google Scholar]
- 25.Jobin C. Precision medicine using microbiota. Science 359, 32–34 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Kuntz TM & Gilbert JA Introducing the microbiome into precision medicine. Trends Pharmacol. Sci 38, 81–91 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Yatsunenko T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gensollen T, Iyer SS, Kasper DL & Blumberg RS How colonization by microbiota in early life shapes the immune system. Science 352, 539–544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson JK et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Honda K & Littman DR The microbiome in infectious disease and inflammation. Annu. Rev. Immunol 30, 759–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sousa T. et al. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm 363, 1–25 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Birer C & Wright ES Capturing the complex interplay between drugs and the intestinal microbiome. Clin. Pharmacol. Ther 106, 501–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ & Balskus EP Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 364, eaau6323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peppercorn MA & Goldman P The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J. Pharmacol. Exp. Ther 181, 555–562 (1972). [PubMed] [Google Scholar]
- 36.Nayak RR & Turnbaugh PJ Mirror, mirror on the wall: which microbiomes will help heal them all? BMC Med. 14, 72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma AK, Jaiswal SK, Chaudhary N & Sharma VK A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota. Sci. Rep 7, 9751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spanogiannopoulos P, Bess EN, Carmody RN & Turnbaugh PJ The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol 14, 273–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gill SR et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Human Microbiome Project. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes E. et al. Therapeutic modulation of microbiota-host metabolic interactions. Sci. Transl Med 4, 137rv136 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R & Goodman AL Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couteau D, McCartney AL, Gibson GR, Williamson G & Faulds CB Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid. J. Appl. Microbiol 90, 873–881 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Lindenbaum J, Rund DG, Butler VP Jr., Tse-Eng D & Saha JR Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N. Engl. J. Med 305, 789–794 (1981). [DOI] [PubMed] [Google Scholar]
- 45.Haiser HJ et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haiser HJ, Seim KL, Balskus EP & Turnbaugh PJ Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 5, 233–238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koppel N, Bisanz JE, Pandelia ME, Turnbaugh PJ & Balskus EP Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. eLife 7, e33953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niehues M & Hensel A In-vitro interaction of L-dopa with bacterial adhesins of Helicobacter pylori: an explanation for clinicial differences in bioavailability? J. Pharm. Pharmacol 61, 1303–1307 (2009). [DOI] [PubMed] [Google Scholar]
- 49.LoGuidice A, Wallace BD, Bendel L, Redinbo MR & Boelsterli UA Pharmacologic targeting of bacterial beta-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther 341, 447–454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bjorkholm B. et al. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4, e6958 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang WH et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J 368, 1575–1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Craciun S & Balskus EP Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl Acad. Sci. USA 109, 21307–21312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakayama H. et al. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics 7, 35–43 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Aziz RK, Hegazy SM, Yasser R, Rizkallah MR & ElRakaiby MT Drug pharmacomicrobiomics and toxicomicrobiomics: from scattered reports to systematic studies of drug-microbiome interactions. Expert. Opin. Drug. Metab. Toxicol 14, 1043–10553 (1918). [DOI] [PubMed] [Google Scholar]
- 56.Wallace BD et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–8353 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saha JR, Butler VP Jr., Neu HC & Lindenbaum J Digoxin-inactivating bacteria: identification in human gut flora. Science 220, 325–3273 (1983). [DOI] [PubMed] [Google Scholar]
- 58.Bisanz JE, Spanogiannopoulos P, Pieper LM, Bustion AE & Turnbaugh PJ How to determine the role of the microbiome in drug disposition. Drug. Metab. Dispos 46, 1588–1595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen TL, Vieira-Silva S, Liston A & Raes J How informative is the mouse for human gut microbiota research? Dis. Model. Mech 8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faith JJ et al. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 4, 1094–1098 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turnbaugh PJ et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Svartz N. Treatment of rheumatoid arthritis with salicylazosulfapyridine. Acta Med. Scand. Suppl 341, 247–254 (1958). [DOI] [PubMed] [Google Scholar]
- 63.Svartz N. The treatment of rheumatic polyarthritis with acid azo compounds. Rheumatism 4, 180–185 (1948). [PubMed] [Google Scholar]
- 64.Das KM & Dubin R Clinical pharmacokinetics of sulphasalazine. Clin. Pharmacokinet 1, 406–425 (1976). [DOI] [PubMed] [Google Scholar]
- 65.Sousa T. et al. On the colonic bacterial metabolism of azo-bonded prodrugs of 5-aminosalicylic acid. J. Pharm. Sci 103, 3171–3175 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Valerino DM, Johns DG, Zaharko DS & Oliverio VT Studies of the metabolism of methotrexate by intestinal flora—I: Identification and study of biological properties of the metabolite 4-amino-4-deoxy-N10-methylpteroic acid. Biochem. Pharmacol 21, 821–831 (1972). [DOI] [PubMed] [Google Scholar]
- 67.Zaharko DS, Bruckner H & Oliverio VT Antibiotics alter methotrexate metabolism and excretion. Science 166, 887–888 (1969). [DOI] [PubMed] [Google Scholar]
- 68.Viaud S. et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iida N. et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubin K. et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun 7, 10391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaput N. et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol 28, 1368–1379 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Gopalakrishnan V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Routy B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Matson V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vetizou M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y. et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med 24, 1804–1808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith MH & Bass AR Arthritis after cancer immunotherapy: symptom duration and treatment response. Arthritis Care Res. 71, 362–366 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Cappelli LC et al. Clinical presentation of immune checkpoint inhibitor-induced inflammatory arthritis differs by immunotherapy regimen. Semin. Arthritis Rheum 48, 553–557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paramsothy S, Rosenstein AK, Mehandru S & Colombel JF The current state of the art for biological therapies and new small molecules in inflammatory bowel disease. Mucosal Immunol. 11, 1558–1570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Magnusson MK et al. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J. Crohns Colitis 10, 943–952 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Ananthakrishnan AN et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 21, 603–610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandborn WJ et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N. Engl. J. Med 367, 1519–1528 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Clayton TA, Baker D, Lindon JC, Everett JR & Nicholson JK Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilson ID Drugs, bugs, and personalized medicine: pharmacometabonomics enters the ring. Proc. Natl Acad. Sci. USA 106, 14187–14188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haiser HJ & Turnbaugh PJ Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res 69, 21–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ryan A. Azoreductases in drug metabolism. Br. J. Pharmacol 174, 2161–2173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morrison JM, Wright CM & John GH Identification, isolation and characterization of a novel azoreductase from clostridium perfringens. Anaerobe 18, 229–234 (2012). [DOI] [PubMed] [Google Scholar]
- 88.Chen H, Wang RF & Cerniglia CE Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr. Purif 34, 302–310 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delomenie C. et al. Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: evidence for highly selective acetylation of 5-aminosalicylic acid. J. Bacteriol 183, 3417–3427 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sim E, Abuhammad A & Ryan A Arylamine N-acetyltransferases: from drug metabolism and pharmacogenetics to drug discovery. Br. J. Pharmacol 171, 2705–2725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bazin T. et al. Microbiota composition may predict anti-TNF alpha response in spondyloarthritis patients: an exploratory study. Sci. Rep 8, 5446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim D. et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 5, 52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rehaume LM et al. IL-23 favours outgrowth of spondyloarthritis-associated pathobionts and suppresses host support for homeostatic microbiota. Ann. Rheum. Dis 78, 494–503 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Manasson J. et al. IL-17 inhibition in spondyloarthritis associates with subclinical gut microbiome perturbations and a distinctive IL-25-driven intestinal inflammation. Arthritis Rheumatol. 10.1002/art.41169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saunte DM, Mrowietz U, Puig L & Zachariae C Candida infections in patients with psoriasis and psoriatic arthritis treated with interleukin-17 inhibitors and their practical management. Br. J. Dermatol 177, 47–62 (2017). [DOI] [PubMed] [Google Scholar]
- 96.Gerard R, Sendid B, Colombel JF, Poulain D & Jouault T An immunological link between Candida albicans colonization and Crohn’s disease. Crit. Rev. Microbiol 41, 135–139 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Colombel JF, Sendid B, Jouault T & Poulain D Secukinumab failure in Crohn’s disease: the yeast connection? Gut 62, 800–801 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Favalli EG, Biggioggero M & Meroni PL Methotrexate for the treatment of rheumatoid arthritis in the biologic era: still an “anchor” drug? Autoimmun. Rev 13, 1102–1108 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Aletaha D & Smolen JS Diagnosis and management of rheumatoid arthritis: a review. JAMA 320, 1360–1372 (2018). [DOI] [PubMed] [Google Scholar]
- 100.Singh JA et al. 2015 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Rheumatol. 68, 1–26 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Smolen JS et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheum. Dis 10.1136/annrheumdis-2019-216655 (2020). [DOI] [PubMed] [Google Scholar]
- 102.Detert J. et al. Induction therapy with adalimumab plus methotrexate for 24 weeks followed by methotrexate monotherapy up to week 48 versus methotrexate therapy alone for DMARD-naive patients with early rheumatoid arthritis: HIT HARD, an investigator-initiated study. Ann. Rheum. Dis 72, 844–850 (2013). [DOI] [PubMed] [Google Scholar]
- 103.Hughes CD, Scott DL & Ibrahim F Titrate Programme Investigators. Intensive therapy and remissions in rheumatoid arthritis: a systematic review. BMC Musculoskelet. Disord 19, 389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smolen JS et al. Adjustment of therapy in rheumatoid arthritis on the basis of achievement of stable low disease activity with adalimumab plus methotrexate or methotrexate alone: the randomised controlled OPTIMA trial. Lancet 383, 321–332 (2014). [DOI] [PubMed] [Google Scholar]
- 105.Goodman SM, Cronstein BN & Bykerk VP Outcomes related to methotrexate dose and route of administration in patients with rheumatoid arthritis: a systematic literature review. Clin. Exp. Rheumatol 33, 272–278 (2015). [PMC free article] [PubMed] [Google Scholar]
- 106.Lebbe C, Beyeler C, Gerber NJ & Reichen J Intraindividual variability of the bioavailability of low dose methotrexate after oral administration in rheumatoid arthritis. Ann. Rheum. Dis 53, 475–477 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Roon EN & van de Laar MA Methotrexate bioavailability. Clin. Exp. Rheumatol 28, S27–32 (2010). [PubMed] [Google Scholar]
- 108.Halilova KI et al. Markers of treatment response to methotrexate in rheumatoid arthritis: where do we stand? Int. J. Rheumatol 2012, 978396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Angelis-Stoforidis P, Vajda FJ & Christophidis N, Methotrexate polyglutamate levels in circulating erythrocytes and polymorphs correlate with clinical efficacy in rheumatoid arthritis. Clin. Exp. Rheumatol 17, 313–320 (1999). [PubMed] [Google Scholar]
- 110.Dervieux T. et al. Polyglutamation of methotrexate with common polymorphisms in reduced folate carrier, aminoimidazole carboxamide ribonucleotide transformylase, and thymidylate synthase are associated with methotrexate effects in rheumatoid arthritis. Arthritis Rheum. 50, 2766–2774 (2004). [DOI] [PubMed] [Google Scholar]
- 111.Danila MI et al. Measurement of erythrocyte methotrexate polyglutamate levels: ready for clinical use in rheumatoid arthritis? Curr. Rheumatol. Rep 12, 342–347 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stamp LK et al. Methotrexate polyglutamate concentrations are not associated with disease control in rheumatoid arthritis patients receiving long-term methotrexate therapy. Arthritis Rheum. 62, 359–368 (2010). [DOI] [PubMed] [Google Scholar]
- 113.Hornung N, Ellingsen T, Attermann J, Stengaard-Pedersen K & Poulsen JH Patients with rheumatoid arthritis treated with methotrexate (methotrexate): concentrations of steady-state erythrocyte methotrexate correlate to plasma concentrations and clinical efficacy. J. Rheumatol 35, 1709–1715 (2008). [PubMed] [Google Scholar]
- 114.Bluett J. et al. Risk factors for oral methotrexate failure in patients with inflammatory polyarthritis: results from a UK prospective cohort study. Arthritis Res. Ther 20, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dekkers JS et al. Autoantibody status is not associated with early treatment response to first-line methotrexate in patients with early rheumatoid arthritis. Rheumatology 58, 149–153 (2019). [DOI] [PubMed] [Google Scholar]
- 116.Hider SL et al. Can clinical factors at presentation be used to predict outcome of treatment with methotrexate in patients with early inflammatory polyarthritis? Ann. Rheum. Dis 68, 57–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sergeant JC et al. Prediction of primary non-response to methotrexate therapy using demographic, clinical and psychosocial variables: results from the UK Rheumatoid Arthritis Medication Study (RAMS). Arthritis Res. Ther 20, 147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gupta V, Katiyar S, Singh A, Misra R & Aggarwal A CD39 positive regulatory T cell frequency as a biomarker of treatment response to methotrexate in rheumatoid arthritis. Int. J. Rheum. Dis 21, 1548–1556 (2018). [DOI] [PubMed] [Google Scholar]
- 119.Lopez-Rodriguez R. et al. Replication study of polymorphisms associated with response to methotrexate in patients with rheumatoid arthritis. Sci. Rep 8, 7342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Rotte M. et al. Development and validation of a prognostic multivariable model to predict insufficient clinical response to methotrexate in rheumatoid arthritis. PLoS One 13, e0208534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Plant D. et al. Gene expression profiling identifies classifier of methotrexate non-response in patients with rheumatoid arthritis. Arthritis Rheum. 10.1002/art.40810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wessels JA et al. A clinical pharmacogenetic model to predict the efficacy of methotrexate monotherapy in recent-onset rheumatoid arthritis. Arthritis Rheum. 56, 1765–1775 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Eektimmerman F et al. Validation of a clinical pharmacogenetic model to predict methotrexate nonresponse in rheumatoid arthritis patients. Pharmacogenomics 20, 85–93 (2019). [DOI] [PubMed] [Google Scholar]
- 124.Jenko B. et al. Clinical pharmacogenetic models of treatment response to methotrexate monotherapy in Slovenian and Serbian rheumatoid arthritis patients: differences in patient’s management may preclude generalization of the models. Front. Pharmacol 9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lopez-Rodriguez R. et al. Evaluation of a clinical pharmacogenetics model to predict methotrexate response in patients with rheumatoid arthritis. Pharmacogenomics J. 18, 539–545 (2018). [DOI] [PubMed] [Google Scholar]
- 126.Zhang X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med 21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Isaac S. et al. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J. Antimicrob. Chemother 72, 128–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nayak RR et al. Perturbation of the human gut microbiome by a non-antibiotic drug contributes to the resolution of autoimmune disease. bioRxiv 10.1101/600155 (2019). [DOI] [Google Scholar]
- 129.Isaac S. et al. The pre-treatment gut microbiome predicts early response to methotrexate in rheumatoid arthritis [abstract]. Arthritis Rheumatol. 71, 2769 (2019). [Google Scholar]
- 130.Costello SP et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA 321, 156–164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paramsothy S. et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 132.Kragsnaes MS et al. Efficacy and safety of faecal microbiota transplantation in patients with psoriatic arthritis: protocol for a 6-month, double-blind, randomised, placebo-controlled trial. BMJ Open 8, e019231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gjorevski N, Ranga A & Lutolf MP Bioengineering approaches to guide stem cell-based organogenesis. Development 141, 1794–1804 (2014). [DOI] [PubMed] [Google Scholar]
- 134.Trietsch SJ et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun 8, 262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bhatia SN & Ingber DE Microfluidic organs-on-chips. Nat. Biotechnol 32, 760–772 (2014). [DOI] [PubMed] [Google Scholar]
- 136.Lagier JC et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol 1, 16203 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Maier L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]