SUMMARY
Although dysregulated cholesterol metabolism predisposes aging tissues to inflammation and a plethora of diseases, the underlying molecular mechanism remains poorly defined. Here, we show that metabolic and genotoxic stresses, convergently acting through liver X nuclear receptor, upregulate CD38 to promote lysosomal cholesterol efflux, leading to nicotinamide adenine dinucleotide (NAD+) depletion in macrophages. Cholesterol-mediated NAD+ depletion induces macrophage senescence, promoting key features of age-related macular degeneration (AMD), including subretinal lipid deposition and neurodegeneration. NAD+ augmentation reverses cellular senescence and macrophage dysfunction, preventing the development of AMD phenotype. Genetic and pharmacological senolysis protect against the development of AMD and neurodegeneration. Subretinal administration of healthy macrophages promotes the clearance of senescent macrophages, reversing the AMD disease burden. Thus, NAD+ deficit induced by excess intracellular cholesterol is the converging mechanism of macrophage senescence and a causal process underlying age-related neurodegeneration.
Graphical Abstract
In brief
Terao et al. demonstrated how dysregulated cholesterol metabolism drives macrophage senescence and the development of subretinal drusenoid deposits in mice. LXR/CD38 signaling activated by cholesterol reduces NAD+ availability and promotes macrophage senescence. Senolytic agents targeting NAD+ decline and senescent macrophages are a potential therapeutic option against early AMD.
INTRODUCTION
Discoveries that implicate cellular aging and senescence as a causal mechanism underlying systemic aging offer a potential therapeutic avenue to target a small subset of cells with the goal of delaying multi-organ deterioration.1 With age, ectopic senescent cells accumulate in multiple organs, leading to tissue dysfunction.2 Senescent cells release a set of cytokines, chemokines, and growth factors called senescence-associated secretory phenotype (SASP).3–5 Previously studies have identified tumor necrosis factor (TNF)-α, interleukin (IL)-1α, IL-1β, IL-6, CXCL1, MCP1, and interferon (IFN)-γ as commonly identified SASP factors across different cell types.6–8 SASPs promote the recruitment of immune cells and senescence induction in the surrounding tissue microenvironment, thereby causing chronic sterile inflammation.9 Targeting cellular aging through senotherapies, including elimination of senescent cells (senolytics) or suppression of SASP (senomorphics), has been shown to be effective in delaying age-related and metabolic disease manifestations.10–14 However, currently available senolytic agents are commonly derived from chemotherapeutic agents with adverse safety profiles, thereby limiting their utility. An increasing geriatric population and the high prevalence of age-related diseases demand a greater understanding of the biology of senescence induction and the development of targeted senotherapeutic interventions.
Metabolic derangement and obesity during aging promote tissue accumulation of senescent cells, thereby promoting tissue inflammation and dysfunction.15,16 Cholesterol is an essential component for maintaining cellular homeostatic functions as a precursor of bile acids, oxysterols, and steroid hormones.17 As such, intracellular cholesterol levels are tightly regulated by balancing influx and efflux.18 An imbalance in these processes leads to excess levels of intracellular cholesterol that promotes cytopathic and pathological processes through cholesterol crystallization and oxidation.19 Intracellular cholesterol is maintained by cholesterol efflux, primarily regulated by ATP binding cassette (ABC) subfamily A member 1 (ABCA1) and G1 (ABCG1), to prevent the excess accumulation of intracellular cholesterol.20,21 The age-associated decline in cholesterol efflux in myeloid cells caused by ABCA1/G1 deficiency has previously been shown to induce aging phenotypes in the eyes and cardiovascular system.22–24 These include features of age-related macular degeneration (AMD), the leading cause of blindness in people over 50 years of age in industrialized nations25; atherosclerosis, the precursor to cardiovascular disease-associated morbidity and mortality26; and Alzheimer’s disease, the leading cause of dementia.27 Additionally, a recent study reports that cholesterol promotes senescence-associated inflammation by promoting SASP expression.28 Thus, dysregulated cholesterol metabolism and cholesterol overload contribute to diverse age-associated diseases such as AMD, cardiovascular diseases, diabetes, and Alzheimer’s disease.29,30 Our previous study demonstrated that ABCA1/G1 decline in macrophages during aging.22
Abca1/g1-knockout bone-marrow-derived macrophages (BMDMs) deficient in cholesterol efflux accumulate intracellular cholesterol. Here, we demonstrate by gene expression analysis that CD38, a multi-faceted ectoenzyme with NADase activity, is substantially upregulated in Abca1/g1-knockout BMDMs. Cholesterol accumulation in BMDMs deficient in cholesterol efflux upregulates the transcription of CD38 through liver X receptor (LXR)-a activation, thereby promoting the degradation of NAD+ and eventually inducing the cellular senescence. Our findings delineate the molecular mechanism underlying NAD+ depletion in macrophages during metabolic dysregulation and aging and the cellular mechanism underlying the development of AMD. More importantly, we demonstrate the causal role of NAD+ depletion in senescence induction and the potential of NAD+ repletion as a senotherapeutic agent that may enhance the efficacy of existing disease interventions.
RESULTS
Cholesterol overload promotes macrophage NAD+ depletion by CD38 through LXR activation
ABC transporters play a critical role in cholesterol efflux by mobilizing cholesterol out of cells. Our previous study has found that the genetic deletion of Abca1 and Abcg1 in myeloid cells leads to aging phenotypes in the eye.31 In AMD, lipid-rich deposits form beneath the retina (subretinal drusenoid deposits or sub-retinal pigment epithelium [RPE] drusen) and genes involved in cholesterol metabolism, including Abca1, are strongly implicated in the development and progression of AMD.25,32–34 To further explore the biological roles of cholesterol efflux in macrophages during the aging process, we have isolated BMDMs from Abca1/g1f/f LysM-Cre (Abca1/g1-m/-m) and age- and sex-matched controls (Abca1/g1f/f). Shotgun lipidomics analysis using liquid chromatography-mass spectrometry (LC/MS) confirmed a significantly increased accumulation of cholesterol esters in BMDMs isolated from Abca1/g1-m/-m, without substantially affecting other lipid classes (Figures 1A, S1A, and S1B). The excess accumulation of cholesterol in macrophages induces membrane fluidity disruption, cytokine production, and endoplasmic reticulum (ER) stress, eventually leading to an inflammatory state and cell death.23,35,36 To characterize the molecular changes that are underlying macrophage-driven pathology and induced by the intra-cellular accumulation of cholesterol, we performed the transcriptomic analysis of BMDMs isolated from Abca1/g1-m/-m using RNA sequencing (RNA-seq). Unbiased differential gene expression analysis identified that genes associated with lipid metabolism (scd1 and apoc1), lysosomal enzyme (lyz1), cell adhesion (vascular cell adhesion protein 1), and inflammation (IL-1a) were upregulated (Figures 1B and S1C). Gene set enrichment analysis (GSEA) displayed enrichment in pathways such as leukocyte cell-cell adhesion, IFN-γ response, triglyceride metabolic process, positive regulation of angiogenesis, and response to hypoxia (Figure S1D). Interestingly, Cd38 was one of the highly upregulated genes in Abca1/g1-m/-m BMDMs. Consistently, immunofluorescence staining and real-time quantitative reverse-transcription polymerase chain reaction (RT-PCR) validation also demonstrated upregulation of CD38 in Abca1/g1- m/-m BMDMs (Figures 1C and 1E). CD38 has been characterized as a key nicotinamide adenine dinucleotide (NAD+)-consuming enzyme, which converts NAD+ to nicotinamide (NAM), adenosine diphosphate ribose (ADPR), cyclic ADPR (cADPR), and nicotinic acid adenine dinucleotide phophate (NAADP).37,38 CD38 is widely expressed in immune cells, including T cells, B cells, natural killer cells, neutrophils, and myeloid cells, and localizes in the plasma membrane with its catalytic domain.39–41 NAD+ is a universal cellular currency and a coenzyme that plays a critical role in redox reactions and metabolic homeostasis.42 NAD+ regulates energy metabolism, mitochondrial function, DNA repair, gene expression, and chromatin structure through NAD+-consuming enzymes such as sirtuins (SIRTs), SARM1, and poly (ADP-ribose) polymerase (PARP) (Figure 1D).43,44 However, increasing levels of CD38 deplete NAD+ levels in multiple tissues during aging.39,45 Additionally, a recent report demonstrated that accumulation of CD38-positive immune cells has been reported in multiple tissues, including the spleen, white adipose tissue, and liver, and that accumulation of CD38-expressing immune cells with age within tissues promotes NAD+ depletion leading to age-related tissue dysfunction.46 Given the key role of CD38 in NAD+ metabolism, we evaluated whether other genes involved in NAD+ biosynthesis and degradation are also affected. RT-qPCR analysis showed that none of the other major NAD+ biosynthetic enzymes (nicotinamide phosphoribosyltransferase [Nampt], nicotinamide mononucleotide adenylyltransferase [Nmnat1, Nmnat3]) were affected in Abca1/g1-m/-m BMDMs (Figure 1E). The expression of Nmnat2 was undetectable in BMDMs (data not shown).
Figure 1. Disruption of cholesterol metabolism reduces macrophage NAD+ availability via CD38 upregulation.
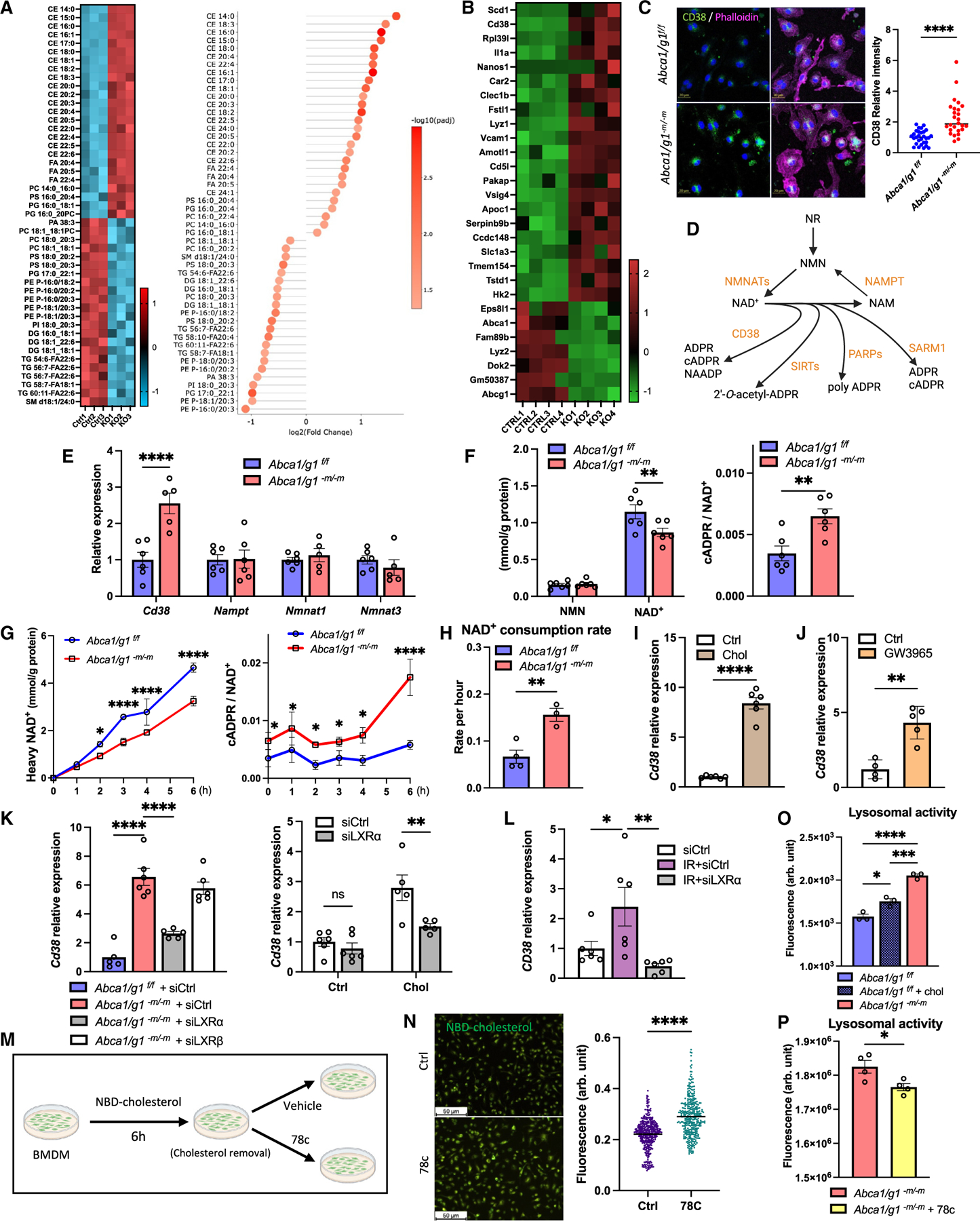
(A) LC/MS lipidomics data (heatmap and lollipop chart) comparing BMDMs isolated from Abca1/g1f/f (control [Ctrl]) and Abca1/g1-m/-m (knockout [KO]).
(B) Heatmap of upregulated and downregulated genes in bulk RNA-seq of BMDMs.
(C) Immunofluorescence images of BMDMs stained for CD38 and phalloidin and the quantification of CD38.
(D) Schematic representation of NAD+ metabolism.
(E) mRNA expression of genes synthesizing and consuming NAD+ in BMDMs.
(F) Intracellular NMN, NAD+, and cADPR/NAD+ ratio in BMDMs.
(G) NAD+ flux assay evaluating synthesized (heavy) NAD+ and cADPR/NAD+ ratio.
(H) The consumption rate of NAD+ in BMDMs calculated by NAD+ flux assay.
(I and J) (I) mRNA expression of CD38 in WT BMDMs treated with cholesterol and (J) an LXR agonist (GW3965).
(K) mRNA expression of CD38 in Abca1/g1-m/-m BMDMs transfected with Nr1h3 (siLXRα) and Nr1h2 (siLXRβ) siRNA and WT BMDMs treated with cholesterol while transfected with siLXRα.
(L) mRNA expression of Cd38 in BMDMs treated with irradiation under siRNA knockdown of LXRα.
(M) Experimental design of cholesterol flux assay using NBD cholesterol on BMDMs.
(N) Representative images of BMDMs treated with a selective CD38 inhibitor (78c) and the quantification of fluorescence intensity of each cell.
(O and P) (O) Lysosomal activity of BMDMs comparing among Abca1/g1f/f treated with/without cholesterol and Abca1/g1-m/-m and (P) between Abca1/g1-m/-m treated with/without 78c. NR, nicotinamide riboside; NMN, nicotinamide mononucleotide; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; cADPR, cyclic adenosine diphosphate ribose; NAADP, nicotinic acid adenine dinucleotide phosphate; ADPR, adenosine diphosphate ribose; NMNAT, nicotinamide mononucleotide adenylyltransferase; NAMPT, nicotinamide phosphoribosyltransferase; SIRT, sirtuin; PARP, poly ADP-ribose polymerase; SARM1, sterile α and TIR motif-containing 1. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, t test for comparison between two groups, one-way ANOVA followed by Bonferroni correction for multiple comparison, and two-way ANOVA followed by Bonferroni correction for comparison with multiple time points. Data are represented as mean ± SEM. The length of scale bar is indicated in each image. See also Figure S1.
To characterize the impact of CD38 upregulation on the NAD+ metabolism in macrophages, we next quantified intracellular NAD+ levels and flux. LC/MS analysis revealed significant reduction of steady-state NAD+ levels and an increase in cADPR/NAD+ ratio, a more sensitive measure of NAD+ degradation reflecting the baseline NAD+ changes,47 in Abca1/g1-m/-m BMDMs (Figure 1F). We then quantified the NAD+ metabolism by flux assay using isotopic-labeled NAM labeled with deuterium (2,4,5,6-deuterium NAM [D4-NAM]). Abca1/g1-m/-m BMDMs showed consistently lower accumulation of heavy NAD+ (Figure 1G). cADPR/NAD+ ratios were also significantly increased in Abca1/g1-m/-m BMDMs across all time points. Finally, specific quantification of NAD+ degradation by inhibiting NAD+ biosynthesis using NAMPT inhibitor, FK866, revealed significantly higher NAD+ degradation in Abca1/g1-m/-m BMDMs (Figure 1H). The NAD+ consumption rate based on the degradation assay in Abca1/g1-m/-m BMDMs was approximately 2.3-fold higher compared to the control. Our results indicate that the intracellular cholesterol accumulation decreases NAD+ levels in Abca1/g1-m/-m macrophages through the increased NAD+ degradation by CD38.
How does cholesterol promote CD38 expression in BMDMs? LXR is a nuclear receptor that is activated by intracellular oxidized cholesterols and regulates cholesterol homeostasis. Through the transcriptional response via the LXR response element (LXRE), oxidized cholesterols promote cholesterol metabolism and efflux in collaboration with LXR.48 Cholesterol loading significantly increased the expression of Cd38 in wild-type (WT) BMDMs (Figure 1I), suggesting that increased cholesterol influx in addition to decreased efflux upregulated CD38 in BMDMs. Moreover, an LXR agonist (GW3965) significantly increased the expression of Cd38 in WT BMDMs (Figure 1J), supporting that CD38 is upregulated through LXR activation. Conversely, small interfering RNA (siRNA) knockdown of LXRα (encoded by Nr1h3) significantly reduced the expression of Cd38 in both Abca1/g1-m/-m BMDMs and cholesterol-loaded WT BMDMs, while the knockdown of LXRβ (encoded by Nr1h2) did not (Figure 1K), suggesting that CD38 is upregulated by intracellular cholesterol through the activation of LXRα. Interestingly, irradiation employed as genotoxic mimetic of aging-related stress significantly increased the levels of cholesterol species and mRNA expression of Cd38 in WT BMDMs (Figures 1L and S1E). CD38 upregulation in response to irradiation was abrogated by LXRα knockdown (Figure 1L). These results revealed that both metabolic and genotoxic stress in BMDMs upregulates CD38 through LXRα activation and depletes NAD+ availability by its degradation.
Intracellular cholesterol accumulation promotes the expression of cholesterol efflux gene, Abca1, also through the activation of LXR in order to maintain cholesterol homeostasis in macrophages.24,49 While considering its role in cholesterol homeostasis, why does LXR also induce CD38, which is seemingly counterintuitive for maintaining effective macrophage function and cellular homeostasis during such metabolic stress? An earlier study has described that CD38 enhances lysosomal calcium release through synthesis of NAADP, thereby promoting cholesterol ester hydrolysis.50–52 Free cholesterol is then efflux by ABC transporters, ABCA1/G1, to extracellular high-density lipoprotein. Interestingly, Abca1 and Cd38 deficiency both have been reported to cause intracellular cholesterol accumulation, contributing to the development of atherosclerosis.53,54 Therefore, we hypothesize that the physiological role of CD38 is to regulate lysosomal function and that LXR upregulates CD38 in order to promote the lysosomal cholesterol processing and eventual efflux from the cell. To test this hypothesis, we evaluated the flux of fluorescently tagged cholesterol in BMDMs (Figure 1M). In fact, Cd38 knockdown and inhibition by CD38 selective inhibitor (78c) both resulted in significantly increased cholesterol accumulation in BMDMs (Figures S1F–S1K). Moreover, 78c significantly reduced the rate of decline in the fluorescence (Figure 1N), indicating that CD38 is necessary for efficient cholesterol efflux. Additionally, both cholesterol stimulation and deficient cholesterol efflux by Abca1/g1 deficiency promoted lysosomal activity and subsequent cholesterol processing (Figure 1O). Finally, CD38 selective inhibitor significantly decreased the lysosomal activity in Abca1/g1-m/-m BMDMs (Figure 1P). These findings indicate that LXR promotes CD38 expression in coordination with cholesterol efflux by ABCA1/G1 to efficiently unload intracellular cholesterol from macrophages by promoting lysosomal cholesterol processing.
NAD+ depletion induces macrophage senescence and dysfunction
A progressive decrease in NAD+ availability during aging and disease has been demonstrated in many tissue types, including the brain during age-related neurodegeneration and the retina.55,56 Recent studies have revealed that senescent cells promote the expression of CD38, leading to tissue NAD+ depletion. However, how NAD+ depletion in the macrophage affects its cellular aging remains unclear. Cellular senescence, characterized by cell-cycle arrest, increased senescence-associated β-galactosidase (SA-β-gal) activity, and upregulation of key senescence markers (p16 and p21) and SASP factors, is a key source of chronic tissue inflammation during aging and disease conditions.57–60 Cellular senescence is induced by several stresses, such as DNA damage, metabolic stress, and telomere dysfunction, as well as physiological aging.61 Cellular senescence in immune cells, also known as immunosenescence, often occurs in the setting of chronic inflammation.62 Senescence in macrophages has been shown to reduce their phagocytic and autophagic capacity and upregulates proinflammatory cytokines.63 Systemic removal of senescent cells in genetically engineered mice, INK-ATTAC mice, or by senolytic agents such as dasatinib and quercetin (D + Q) promotes healthy aging and ameliorates age-related tissue dysfunction.64–66 Senescent cells secrete SASP factors such as TNF-α and vascular endothelial growth factor (VEGF) that promote senescence in its surrounding tissue microenvironment and promote chronic inflammation.3–5
As the decreasing NAD+ level drives age-related diseases such as metabolic disorders and their complications,67 we evaluated the impact of NAD+ decline associated with cholesterol overload in macrophages on cellular senescence. RT-qPCR revealed that senescence markers (Cdkn1a mRNA encoding p21Cip1, and Cdkn2a mRNA encoding p16Ink4a) and the inflammatory cytokines that comprise SASP factors (Vegf-a and Tnf-α) were both significantly increased in Abca1/g1-m/-m BMDMs (Figure 2A). In situ hybridization confirmed the increased expression of p16 and p21. Abca1/g1-m/-m BMDMs also displayed markedly increased SA-β-Gal activity (Figure 2B). Immunofluorescence staining of Abca1/g1-m/-m BMDMs demonstrated significantly reduced expression of Ki67, a proliferation marker, indicating decline in cell cycling and proliferation capacity (Figures S2A and S2B). Moreover, cholesterol loading of WT macrophages also promoted the expression of senescence markers (Figure 2C), supporting the conclusion that intracellular cholesterol accumulation by both increased influx and decreased efflux induced macrophage senescence. To further delineate the molecular pathway underlying macrophage senescence in response to cholesterol overload, we assessed the impact of the individual components of the cholesterol/LXR/CD38/NAD+ pathway on macrophage senescence. Activation of LXR using GW3965, overexpression of CD38, and inhibition of NAD+ biosynthesis by FK866 all significantly promoted the expression of p16 and p21 in WT BMDMs (Figures 2D, 2E, and 2F). These data indicate that the cholesterol/LXR/CD38 pathway is the mediator of cellular senescence in macrophages through the depletion of NAD+ availability. To further confirm that NAD+ depletion triggers macrophage senescence, we evaluated whether NAD+ augmentation can reverse the senescence phenotype. NAD+ repletion by NAD+ intermediate, NMN, significantly reversed NAD+ and suppressed the expression of senescence markers and SASP factors (Vegf-a, Vcam1) in Abca1/g1-m/-m BMDMs (Figures 2G and S2C). Moreover, other senescence marker profiles defined in previous studies were also improved by NMN treatment (Figure S2D).68 Treatment of Abca1/g1-m/-m BMDMs with the CD38 selective inhibitor also significantly suppressed the NAD+ depletion and the expression of senescence markers, indicating that NAD+ repletion can reverse the senescence phenotype of Abca1/g1-m/-m BMDMs (Figures 2H and S2E).
Figure 2. Reduced NAD+ availability induces macrophage senescence and dysfunction.
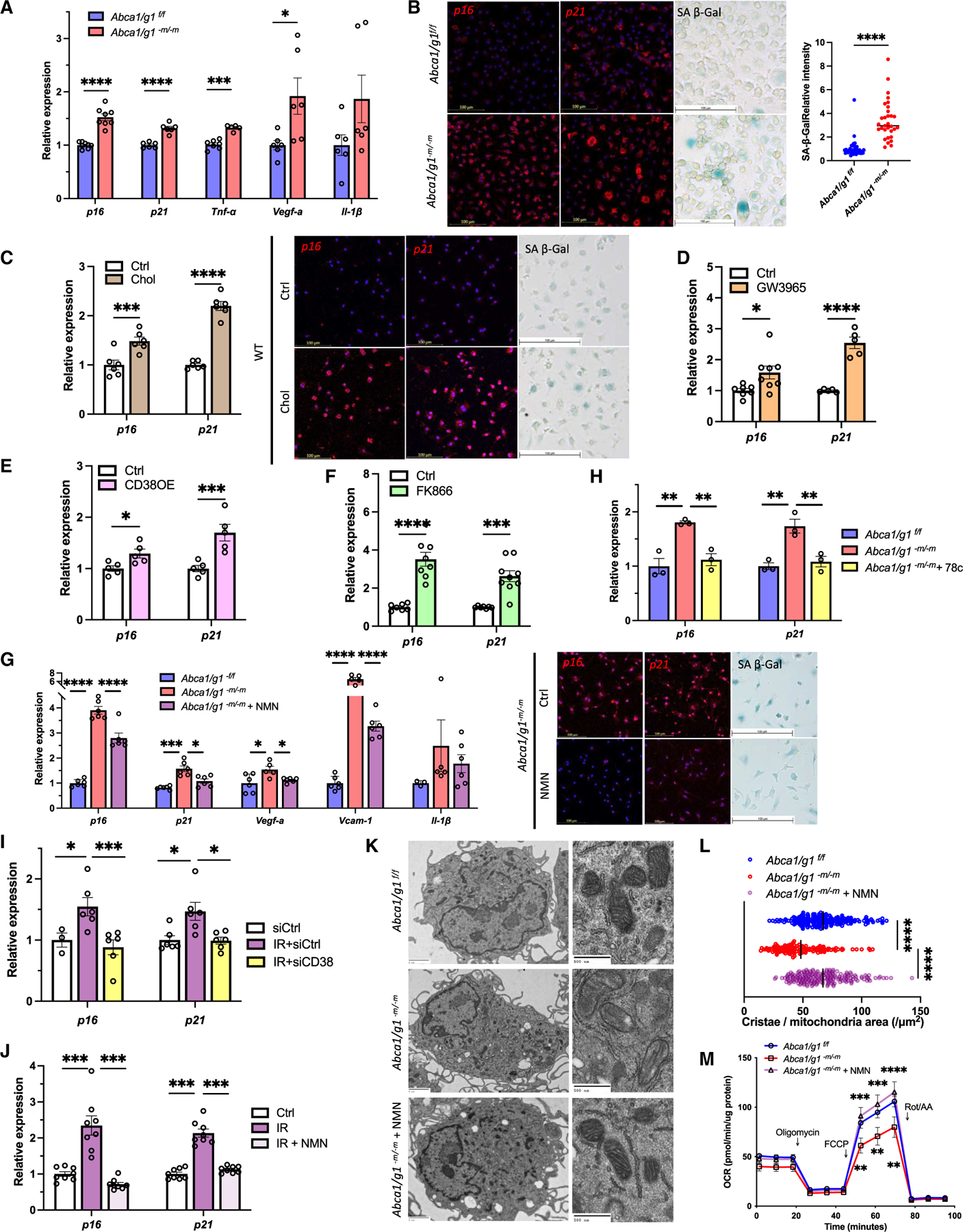
(A) mRNA expression of senescence markers (p16 and p21) and SASP factors (Tnf-α, Vegf-a, and Il-1β) in BMDMs isolated from Abca1/g1f/f and Abca1/g1-m/-m.
(B) Images of in situ hybridization (ISH; p16 and p21) and SA-β-Gal staining and the quantification of SA-β-Gal.
(C) mRNA expression and ISH of p16 and p21, and SA-β-Gal staining of WT BMDMs treated with cholesterol.
(D–F) (D) mRNA expression of p16 and p21 in WT BMDMs treated with an LXR agonist (GW3965), (E) CD38-overexpression, and (F) a selective NAMPT inhibitor (FK866).
(G) mRNA expression of senescence markers (p16 and p21) and SASP factors (Vegf-a, Vcam1, and Il-1β), representative images of ISH (p16 and p21), and SA-β-Gal staining of Abca1/g1-m/-m BMDMs treated with NMN.
(H) mRNA expression of p16 and p21 in Abca1/g1-m/-m BMDMs treated with the CD38 inhibitor (78c).
(J) (I and J) (I) mRNA expression of p16 and p21 in irradiation-induced senescent BMDMs transfected with siCD38 and (J) treated with NMN.
(K and L) (K) Representative EM images and (L) the quantification of the number of cristae in Abca1/g1f/f and Abca1/g1-m/-m BMDMs treated with or without NMN.
(M) Seahorse assay evaluating the oxygen consumption rate (OCR) of BMDMs. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, t test for comparison between two groups, one-way ANOVA followed by Bonferroni correction for multiple comparison, and two-way ANOVA followed by Bonferroni correction for comparison with multiple time points. Data are represented as mean ± SEM. The length of scale bar is indicated in each image. See also Figure S2.
In addition to metabolic stress, DNA damage triggers cellular senescence.69,70 Our data demonstrated that irradiation induces CD38 expression in BMDMs (Figure 1L) Consistently, irradiation reduced NAD+ levels in BMDMs (Figure S2F). Senescence markers induced by irradiation were significantly suppressed by siRNA knockdown of Cd38 (Figure 2I). Furthermore, NAD+ augmentation by NMN also suppressed the expression of senescence markers in macrophages treated with irradiation (Figure 2J). These findings reveal NAD+ depletion as a central mechanism underlying the induction of macrophage senescence and that NAD+ repletion can suppress senescence phenotypes in macrophages. To characterize the impact of NAD+ depletion and subsequent cellular senescence on macrophage function, we performed histological analysis using electron microscopy (EM). Mitochondrial dysfunction is a common feature of cellular senescence.71 Abca1/g1-m/-m BMDMs revealed significantly less mitochondrial cristae density, consistent with a previous report demonstrating that NAD+ depletion leads to impaired macrophage mitochondrial function (Figures 2K and 2L).72 Further characterization of mitochondrial function demonstrated that maximal respiratory capacity significantly decreased in Abca1/g1-m/-m BMDMs and was reversed by NMN supplementation (Figure 2M). Taken together, these data demonstrate that NAD+ depletion mediated by CD38 is a converging mechanism underlying senescence induction and causes cellular dysfunction in senescent macrophages.
Cholesterol efflux defect promotes ectopic accumulation of senescent macrophages in subretinal space through modulation of CD38
Myeloid-specific deletion of Abca1/g1 results in the development of lipid-rich extracellular subretinal drusenoid deposits (SDD) and retinal dysfunction, pathognomonic features of AMD (Figure 3A).31 In fact, genes involved in cholesterol metabolism, including Abca1, are strongly implicated in the development and progression of AMD.32–34 Consistently, the retinas of aged mice demonstrated significantly more cholesterol content compared to that of young mice (Figures 3B and S3A–S3D), suggesting an increasing cholesterol burden in the retina during aging. We hypothesized that the age- and genetic-associated increase in cholesterol burden induces macrophage senescence, thereby promoting the ectopic accumulation of inflammatory macrophages, tissue dysfunction, and neurodegeneration. Immunohistological evaluation revealed significant increases in the ectopic accumulation of IBA-1-positive myeloid cells in the subretinal space of Abca1/g1-m/-m mice (Figure 3C). Additionally, accumulated myeloid cells contained lipofuscin, a main component of drusen and SDD, suggesting that senescent myeloid cells are the source of the drusenoid deposits. Given that loss of cholesterol efflux in Abca1/g1-m/-m BMDMs induces senescence by NAD+ depletion, we evaluated the expression of CD38 and senescent markers in vivo. We detected strong expression of CD38 in the IBA-1-positive myeloid cells accumulated in Abca1/g1-m/-m mice (Figure 3D). In contrast, there were no cells enriched in CD38 expression in the age-matched control mice, indicating that macrophages expressing CD38 are highly enriched in lipofuscin and likely contribute to the pathogenesis of AMD.
Figure 3. Cholesterol efflux defect causes macrophage senescence and lipofuscin accumulation in eye.
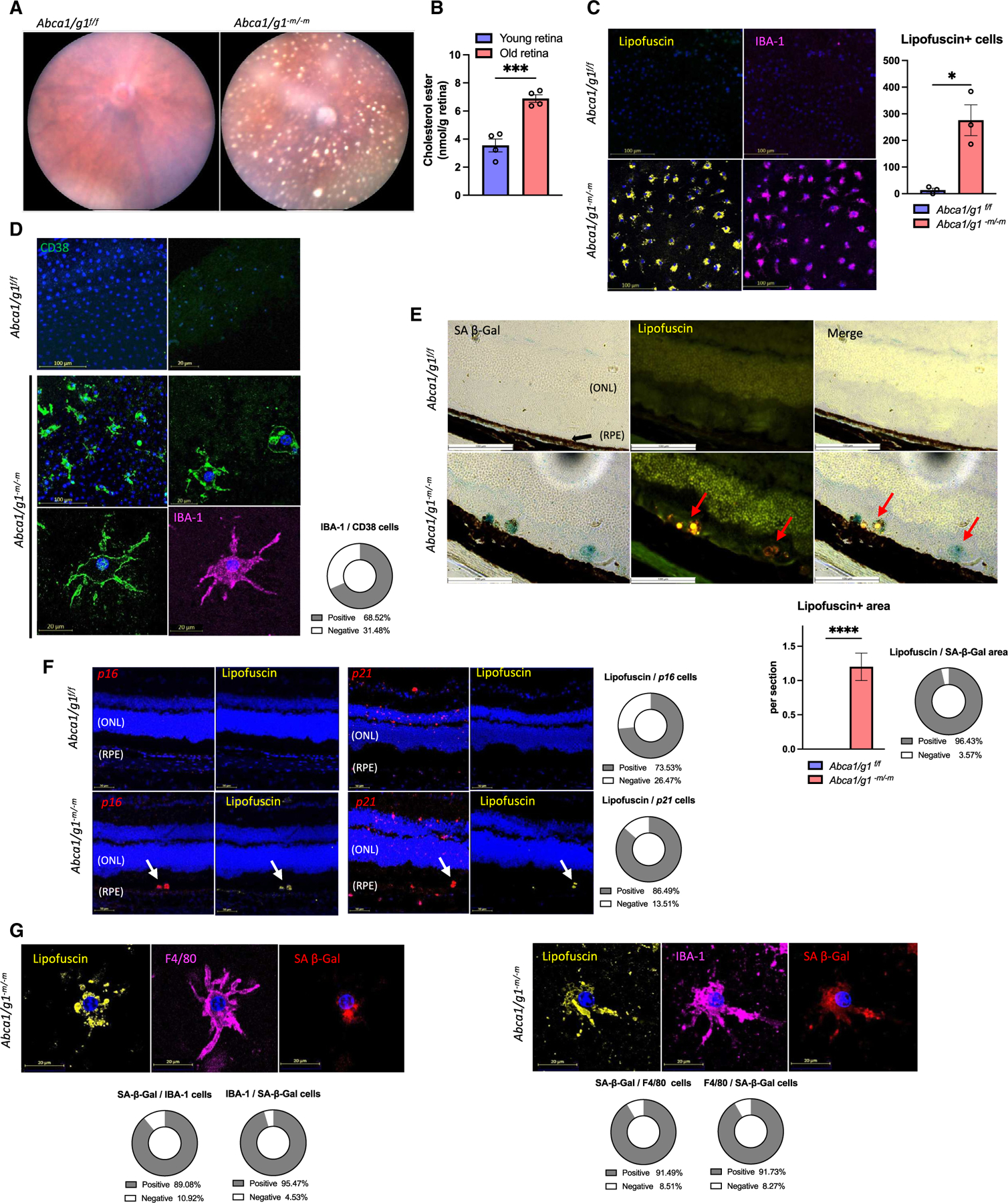
(A) Representative fundus photography images of Abca1/g1-m/-m presenting with SDD.
(B) Quantification of cholesterol ester in retinal samples from young (2 months old) and old (27 months old) measured by LC/MS.
(C) Immunofluorescence of RPE flat mount showing lipofuscin- and IBA-1-positive cells and the quantification of lipofuscin-positive cells.
(D) Immunofluorescence images of RPE flat mount for CD38 and IBA-1. The circle graph shows the percentage of CD38-positive cells in IBA-1-positive cells.
(E) SA-β-Gal staining of retinal sections. Note that the SA-β-Gal-positive areas merge with lipofuscin (arrow) in Abca1/g1-m/-m retinal sections. The bar graph shows the number of lipofuscin-positive areas and the circle graph shows the percentage of SA-β-Gal-positive areas within lipofuscin-positive areas in the subretinal space.
(F) ISH (p16 and p21) of retinal sections and the quantification of the number of ISH-positive areas in lipofuscin-positive areas. Note that the lipofuscin-positive area merges with the p16/p21-expressing area (arrows in each image).
(G) Co-staining images of SPIDER-β-Gal and immunofluorescence (lipofuscin, F4/80, and IBA-1) in RPE flat mount. The circle graph shows the percentage of myeloid cell marker-positive cells in SPIDER-β-Gal-positive cells and SA-β-Gal-positive cells in myeloid cells. ONL, outer nuclear layer; RPE, retinal pigment epithelium. *p < 0.05; ***p < 0.001; ****p < 0.0001, t test for comparison between two groups. Data are represented as mean ± SEM. The length of scale bar is indicated in each image. See also Figure S3.
To further characterize the cellular states of macrophages accumulated in the subretinal space of Abca1/g1-m/-m mice, we evaluated the levels of senescence markers. Lipofuscin-rich cells of the subretinal layer in Abca1/g1-m/-m were SA-β-Gal positive (Figure 3E). Additionally, we detected increased mRNA expression of p16, specifically in lipofuscin-positive cells (Figure 3F). In contrast to the p16 localization, p21-positive cells were observed only in the inner layer of the control retina, while the retinas of Abca1/g1-m/-m also showed p21-positive cells that co-localize with lipofuscin in the subretinal layer (Figure 3F). Moreover, RPE flat-mount and sagittal-section staining of Abca1/g1-m/-m revealed that F4/80-positive or IBA-1-positive macrophages containing lipofuscin showed increased SA-β-Gal activity(Figure 3G). Additionally, p16-positive cells were IBA-1 and F4/80 (Figure S3E). These results demonstrate that subretinal macrophage in Abca1/g1-m/-m mice display lipofuscin enrichment, CD38 expression, and senescent markers and suggest that senescent macrophages contribute to the development of SDD.
Accumulation of subretinal senescent macrophages is directly responsible for drusenoid deposits and neurodegeneration in the AMD model
Macrophages are one of the most abundant immune cells throughout the body, contributing to host immune defense, tissue homeostasis, and repair.73 Therefore, a global cholesterol efflux defect in macrophages could lead to systemic metabolic derangement and inflammation, thereby contributing to the development of SDD and AMD phenotypes.31 Additionally, the LysM promoter, which we utilized for our conditional knockout, has been shown to express Cre-recombinase in a subset of microglia; therefore, we are unable to negate the impact of cholesterol efflux defect in retinal microglia on the AMD pathogenesis. To clarify whether infiltration of senescent macrophages is sufficient to induce AMD phenotypes, we examined whether the local injection of senescent BMDMs can induce SDD- and AMD-associated retinal dysfunction, by injecting Abca1/g1-m/-m BMDMs into the eyes of WT mice (Figure 4A). Two weeks after the subretinal injection, a substantial number of SDDs were observed only after subretinal injection of Abca1/g1-m/-m BMDMs and not after injection of Abca1/g1f/f control BMDMs (Figure 4B). In contrast, intravitreal injection of macrophages did not cause SDD. Subretinal deposits induced by the subretinal injection of Abca1/g1-m/-m BMDMs contained lipofuscin similar to the spontaneous SDD observed in Abca1/g1-m/-m mice (Figure 4C). Furthermore, subretinal injection of Abca1/g1-m/-m BMDMs led to impaired retinal function quantified by electroretinograms (ERGs) (Figure 4D) and photoreceptor degeneration (Figures 4E, S4A and S4B), indicative of retinal neurodegeneration. These data demonstrate that subretinal infiltration of cholesterol efflux-deficient BMDMs is responsible for dry AMD pheno-type including SDD and retinal neurodegeneration observed in Abca1/g1-m/-m mice. Moreover, to determine whether microglia contribute to the SDD generation, we evaluated the retina of microglia-specific Abca1 and Abcg1 knockout mice using Cx3cr1-CreERT2 or Tmem119-CreERT2 mice. Both Abca1/g1f/f;Cx3cr1-CreERT2 and Abca1/g1f/f;Tmem119-CreERT2 presented with minimal amount of SDDs (Figure S4C), supporting our conclusion that BMDMs are the dominant cell type responsible for the SDD biogenesis.
Figure 4. Accumulation of subretinal senescent macrophages is responsible for drusenoid deposits and neurodegeneration in an AMD model.
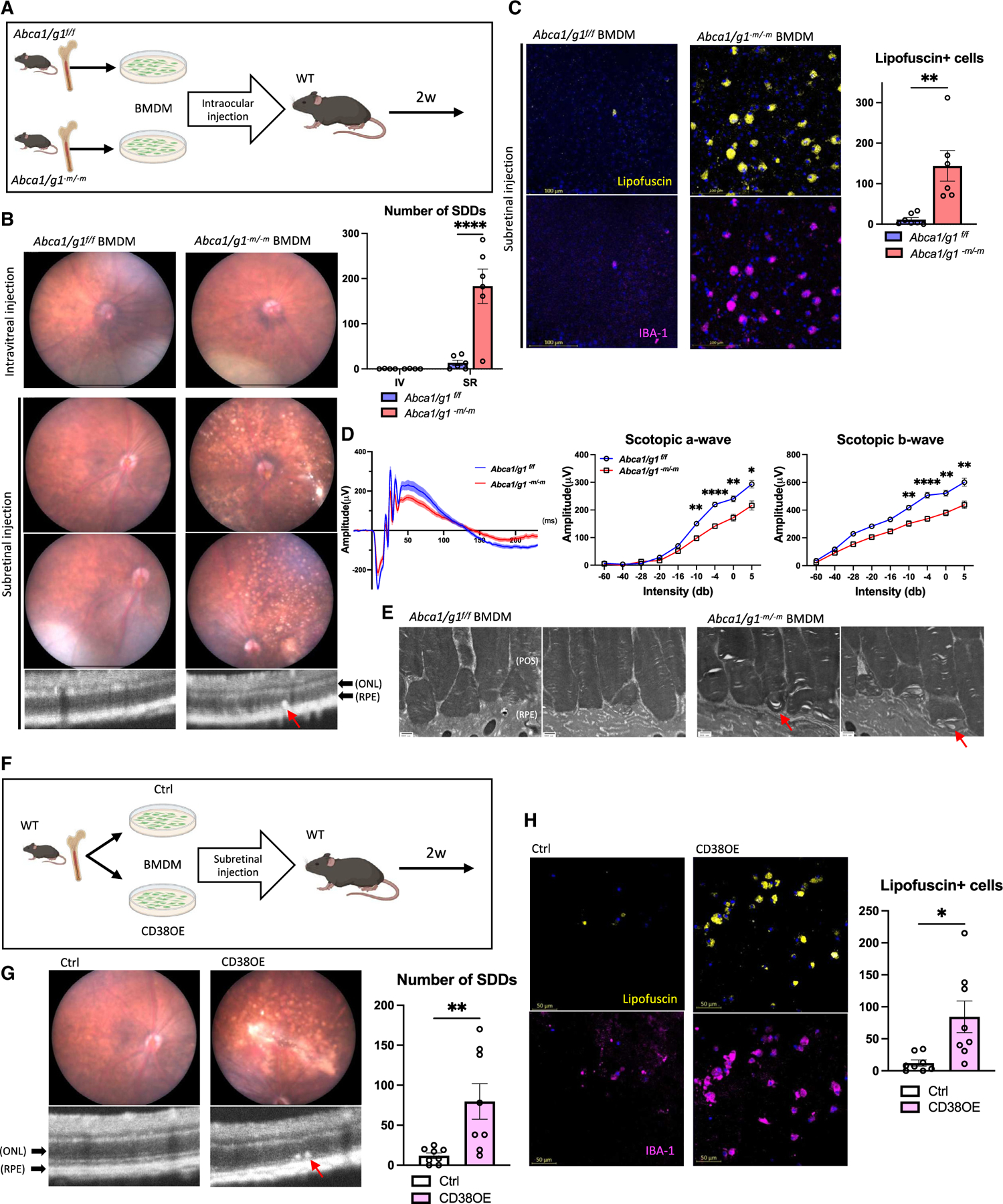
(A) Experimental design of the subretinal administration of BMDMs into eyes of WT mice.
(B) Fundus and optical coherence tomography (OCT) images and the quantification of SDD. Note a subretinal deposit in subretinal space of retina treated with subretinal injection of Abca1/g1-m/-m BMDMs (arrow).
(C) Representative images of RPE flat mount showing lipofuscin- and IBA-1-positive cells and the quantification of lipofuscin-positive cells.
(D) Electroretinography waveform of mice treated with Abca1/g1f/f and Abca1/g1-m/-m BMDMs, and the quantification of amplitude of scotopic a and b wave. Each waveform and graph indicate the mean ± SEM.
(E) Representative EM images of retinal sections. Note that outer segment disc membranes were disrupted in retinal sections treated with subretinal administration of Abca1/g1-m/-m BMDMs.
(F) Experimental design of the subretinal administration of CD38-overexpressing BMDMs into WT retina.
(G) Fundus and OCT images and the quantification of SDD. Note a subretinal deposit in subretinal space of retina treated with subretinal injection of CD38-overexpressing BMDMs (arrow).
(H) Representative images of RPE flat mount showing lipofuscin- and IBA-1-positive cells and the quantification of lipofuscin-positive cells. POS, photoreceptor outer segment. *p < 0.05; **p < 0.01; ****p < 0.0001, t test for comparison between two groups and two-way ANOVA followed by Bonferroni correction for comparison with multiple time points. Data are represented as mean ± SEM. The length of scale bar is indicated in each image. See also Figure S4.
To further determine the precise cellular process that is essential for the AMD phenotype, we asked whether the intracellular cholesterol accumulation or the senescence conversion of the macrophage is sufficient to induce SDD and subretinal macro phage accumulation. Given that Abca1/g1-m/-m BMDMs are both enriched in intracellular cholesterol and are also senescent, intracellular cholesterol accumulation may simply cause SDD formation by promoting local cholesterol deposition. Therefore, we performed the injection of Cd38-overexpressing BMDMs into subretinal space to determine whether the senescent macrophage without intracellular cholesterol accumulation is sufficient to promote SDD formation (Figure 4F). The injection of Cd38-overexpressing BMDMs induced SDD and subretinal accumulation of lipofuscin-positive macrophages (Figures 4G and 4H). Therefore, these results suggest that the CD38-induced senescence by NAD+ depletion promotes lipofuscinogenesis within macrophages, thereby promoting the dry AMD phenotype and neurodegeneration.
The clearance of senescent cells prevents the development of subretinal drusenoid deposits
To further establish that senescent macrophages are the pathological source of SDD material and neurodegeneration in vivo, we examined effects of senolysis by using both a genetic model and pharmacological senolysis in Abca1/g1-m/-m mice. INK-ATTAC mice are transgenic mice expressing FKBP-caspase 8 fusion protein under the INK4a promoter. p16Ink4a-positive senescent cells can be effectively ablated by the administration of AP20187, which dimerizes the fusion protein and selectively induces apoptosis of p16-positive senescent cells.64,65 Three months of senolysis by the administration of AP20187 in Abca1/g1-m/-m;INK-ATTAC mice (Figure 5A) almost completely prevented the formation of SDD (Figure 5B). Consistent with the lack of SDD, senolysis prevented subretinal accumulation of CD38-positive cells, SA-β-Gal-positive cells, and lipofuscinogenesis in Abca1/g1-m/-m;INK-ATTAC mice (Figures 5C and S5A). Macrophages identified in the subretinal space of Abca1/g1-m/-m;INK-ATTAC mice did not display SA-β-Gal staining or the accumulation of lipofuscin, unlike in Abca1/g1-m/-m mice, demonstrating the effective senolysis in INK-ATTAC mice. Transcriptomic analysis of the RPE/choroid complex samples isolated from Abca1/g1-m/-m;INK-ATTAC mice displayed a significant reduction in myeloid markers (Cd300e, Treml4), proinflammatory cytokines (Cxcl1), and prostaglandin biosynthetic enzymes (Prgs2) (Figures 5D and S5B), further supporting the microscopic findings. Intracellular lipid in RPE cells, and Bruch’s membrane thickening, which are key manifestations of AMD, were also repressed in Abca1/g1-m/-m;INK-ATTAC mice (Figure 5E). Furthermore, Abca1/g1-m/-m;INK-ATTAC mice displayed preserved retinal function characterized by higher ERG response on scotopic a and b wave (Figures 5F and S5C). Abca1/g1-m/-m; INK-ATTAC mice showed improved RPE phenotype, including higher dark-adapted ERG response, reduced RPE cell loss, and normalization of RPE morphology (Figures 5F and S5D), indicating preserved RPE function.
Figure 5. Senescence clearance prevents the development of subretinal drusenoid deposits.
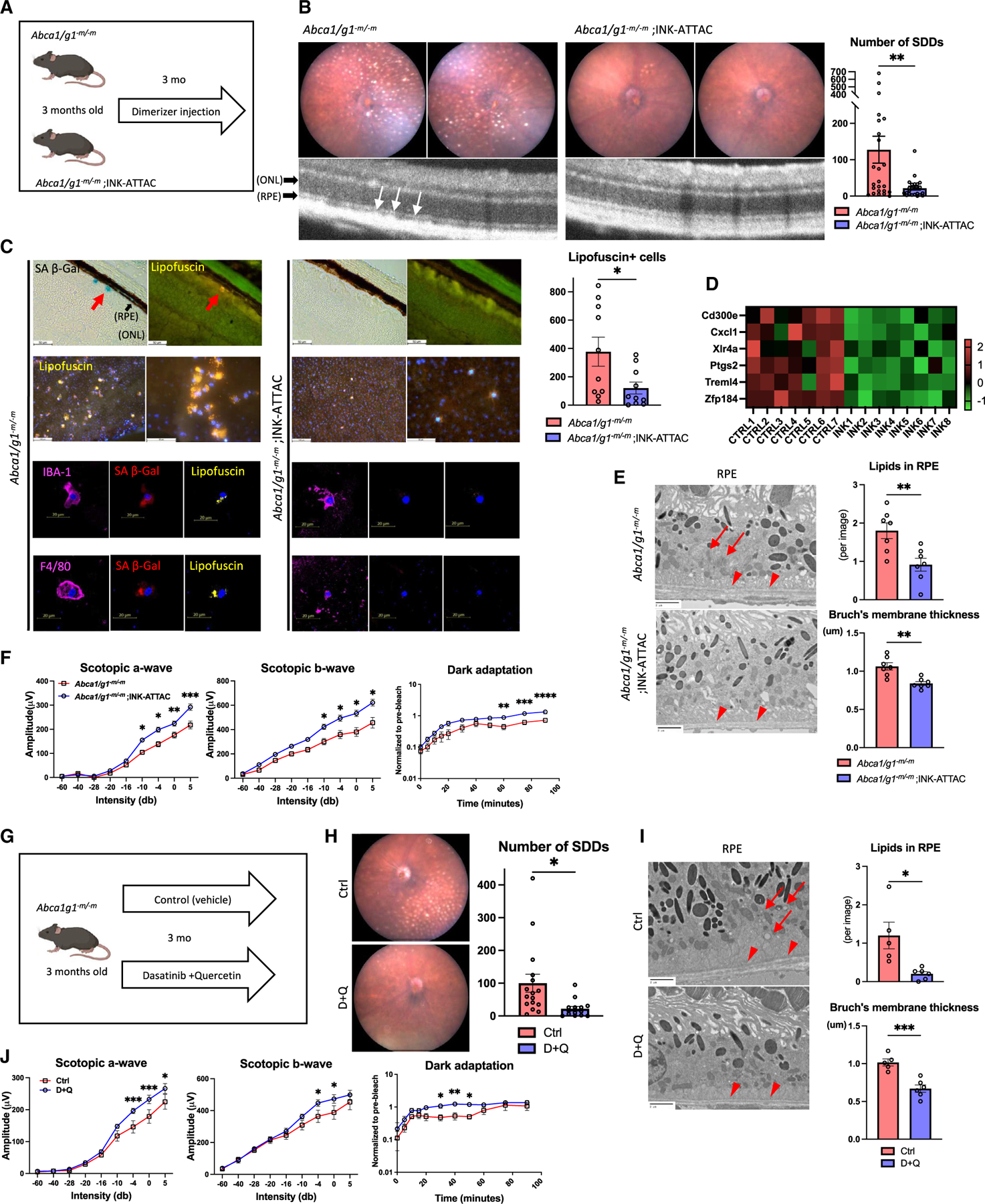
(A) Experimental design using Abca1/g1-m/-m;INK-ATTAC.
(B) Representative fundus images and OCT and the quantification of the number of SDD.
(C) Representative SA-β-Gal staining images of retinal section, immunofluorescence of RPE flat mount showing lipofuscin-, myeloid cell marker-, and SPiDER β-Gal-positive cells, and the quantification of lipofuscin-positive cells on RPE flat mount. Note SA-β-Gal- and lipofuscin-positive area (arrow) in Abca1/g1-m/-m indicating SDD.
(D) Heatmap of the bulk RNA-seq representing downregulated expression in RPE/choroid complex samples.
(E) Representative EM images showing RPE cells and Bruch’s membrane, and the quantification of the number of intracellular lipids in RPE cells and Bruch’s membrane thickness. Note intracellular lipids in RPE cells (arrow) and thickened Bruch’s membrane (arrowhead) in Abca1/g1-m/-m.
(F) The quantification of electroretinography (scotopic a-wave, b-wave, and dark-adaptation electroretinography) amplitudes.
(G) Experimental design of the experiment using senolytic drugs (dasatinib and quercetin [D + Q]) for Abca1/g1-m/-m.
(H) Representative fundus images and the quantification of SDD.
(I) Representative EM images showing RPE cells and Bruch’s membrane and the quantification of the number of intracellular lipids in RPE cells and Bruch’s membrane thickness. Note intracellular lipids in RPE cells (arrow) and thickened Bruch’s membrane (arrowhead) in Abca1/g1-m/-m treated with vehicle.
(J) The quantification of electroretinography amplitudes. *p < 0.05; **p < 0.01; ****p < 0.0001, t test for comparison between two groups and two-way ANOVA followed by Bonferroni correction for comparison with multiple time points. Data are represented as mean ± SEM. The length of scale bar is indicated in each image. See also Figure S5.
We additionally examined effects of pharmacological senolysis (D + Q) on their phenotypes in Abca1/g1-m/-m mice (Figure 5G). Half-maximal inhibitory concentration (IC50) experiments revealed the higher susceptibility of Abca1/g1-m/-m BMDMs to D + Q-induced apoptosis (Figures S5E and S5F). D + Q administration provided similar effects on the AMD phenotype to the senolysis in INK-ATTAC mice, including the suppression of SDD formation (Figure 5H), Bruch’s membrane thickening and intracellular lipid accumulation in RPE cells (Figure 5I), and retinal and RPE dysfunction (Figures 5J and S5G). Taken together, senescent macrophage is responsible for the formation of SDD and subsequent retinal dysfunction in Abca1/g1-m/-m mice, and senolytics are an effective therapeutic approach to prevent the development of AMD. Moreover, given that elimination of senescent macrophages is sufficient to reverse histopathological findings and functional deficits in RPE, RPE dysfunction is likely the consequence of the subretinal accumulation of senescent macrophages.
NAD+ augmentation suppresses accumulation of subretinal drusenoid deposits
Given that cholesterol-induced senescence can be reversed by NAD+ repletion, we further speculated that NAD+ repletion by NMN could ameliorate the AMD phenotype in Abca1/g1-m/-m mice (Figure 6A). Administration of NMN significantly reduced the number of SDDs in Abca1/g1-m/-m (Figure 6B). NMN administration also reduced the accumulation of lipofuscin-positive macrophages (Figure 6C), demonstrating that NAD+ augmentation suppresses senescence states and promotes the clearance of SDDs. Furthermore, retinal and RPE dysfunction detected by ERG, intracellular lipid in RPE cells, and Bruch’s membrane thickening were also repressed in Abca1/g1-m/-m; INK-ATTAC mice (Figures 6D and 6E). Consistently, systemic administration of NMN and 78c suppressed SDD and the accumulation of lipofuscin induced by subretinal injection of Abca1/g1-m/-m BMDMs (Figures S6A–S6E), demonstrating that NAD+ augmentation promotes the clearance of senescent macrophages. We cannot demonstrate the reversion of macrophage senescence phenotype in vivo given subretinal macrophages are eliminated with NMN treatment. However, these data demonstrate that NAD+ augmentation promotes the clearance of subretinal macrophages and prevents SDD formation. Therefore, NAD+ repletion strategy is an effective approach to treat dry AMD.
Figure 6. NAD+ augmentation promotes the clearance of subretinal senescent macrophages and drusenoid deposits.
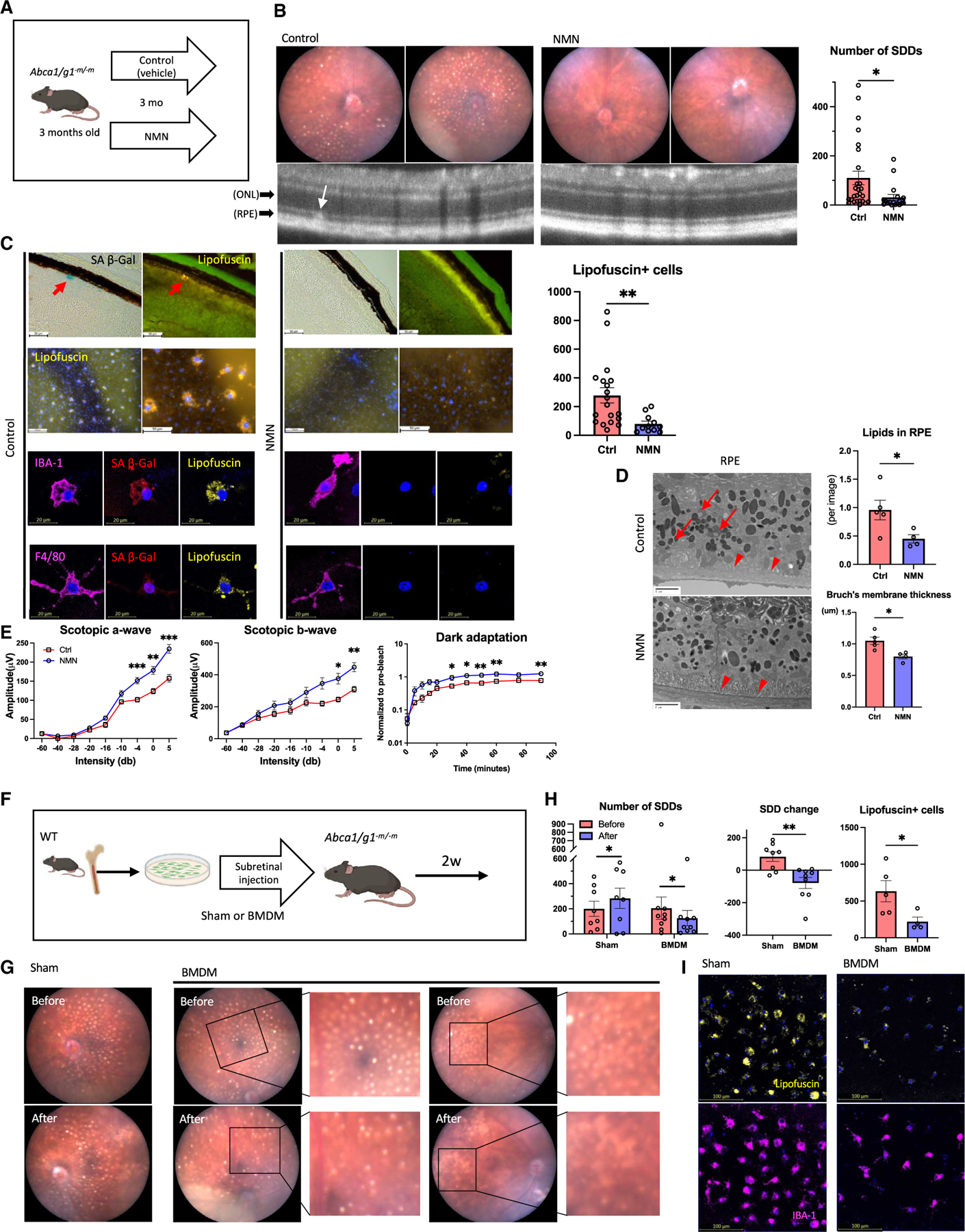
(A) Experimental design for the treatment with NMN.
(B) Representative fundus images and OCT images of eyes the treatment with NMN and the quantification of SDD.
(C) Representative SA-β-Gal staining images of retinal section, immunofluorescence of RPE flat mount showing lipofuscin-, myeloid cell marker-, and SPiDER β-Gal-positive cells, and the quantification of lipofuscin-positive cells on RPE flat mount. Note SA-β-Gal- and lipofuscin-positive area (arrow) in Abca1/g1-m/-m indicating SDD.
(D) Representative EM images showing RPE cells and Bruch’s membrane, and the quantification of the number of intracellular lipids in RPE cells and Bruch’s membrane thickness. Note intracellular lipids in RPE cells (arrow) and thickened Bruch’s membrane (arrowhead) in Abca1/g1-m/-m.
(E) The quantification of electroretinography (scotopic a-wave, b-wave, and dark-adaptation electroretinography) amplitudes.
(F) Experimental design of subretinal administration of healthy BMDMs into Abca1/g1-m/-m mice with SDD.
(G) Representative fundus images of eyes before and after subretinal injection of BMDMs or vehicle. Before and after images were taken in identical eyes.
(H) The quantification of SDD and lipofuscin-positive cells.
(I) Representative images of RPE flat mount showing lipofuscin-positive cells. *p < 0.05; **p < 0.01, t test for comparison between two groups. Data are represented as mean ± SEM. The length of scale bar is indicated in each image.
Subretinal injection of healthy macrophages promotes senescence clearance and reverses dry AMD phenotype
Whether the subretinal infiltration of BMDMs is simply a pathogenic consequence of diseased macrophages or the homeostatic process of subretinal cholesterol clearance is unclear to date. During AMD progression, myeloid cells can be recruited to the subretinal space by local upregulation of inflammatory cytokines, leading to the aberrant accumulation of senescent macrophages. Alternatively, BMDMs may periodically survey the subretinal space to maintain tissue homeostasis and prevent accumulation of waste products. We hypothesized that if BMDMs are recruited to the subretinal space by local inflammation, subretinal administration of WT BMDMs may exacerbate the AMD phenotype. To address this question, we performed subretinal administration of WT BMDMs into 6-month-old Abca1/g1-m/-m mice that already demonstrated high SDD burden (Figure 6F). Surprisingly, subretinal administration of WT BMDMs significantly reduced the number of SDDs compared to that before the administration and sham control mice (Figures 6G and 6H). Furthermore, the numbers of subretinal senescent lipofuscin-positive macrophages were significantly reduced (Figures 6H and 6I), indicating that the local administration of healthy BMDMs promotes the clearance of senescent macrophages. Co-incubation of Abca1/g1-m/-m BMDMs and WT BMDMs labeled with lipophilic dyes of different colors demonstrated their co-localization, suggesting that healthy macrophages clear senescent macrophages by phagocytosis (Figure S6F). These results demonstrate that healthy BMDMs have the potential to improve the cholesterol clearance, suppress the accumulation of senescent macrophages, and reverse the dry AMD phenotypes in the already diseased retina. Given that there are no effective clinical treatment options to date to reverse dry AMD at its early stages prior to vision loss, cell-mediated clearance is an innovative approach for the potential treatment of pre-existing dry AMD to prevent further neurodegeneration and vision loss.
DISCUSSION
During aging, the body encounters genotoxic and metabolic stress, causing aberrant immune activation and dysregulation.15,74,75 A large body of literature has associated the inflammatory macrophage with tissue inflammation, organ dysfunction, and the development of age-related diseases.76 These inflammatory macrophages induce cellular senescence in the local tissue, further exacerbating organ dysfunction and systemic inflammation.77 Senolysis by genetic models and repurposed chemotherapeutic agents have been observed to reduce the accumulation of senescent cells and are now proposed to be effective against the progression of age-related diseases. Such observations have heightened interest in selective and potent targeting of mechanisms of senescence induction and senescent cells themselves. In this study, senolysis improved the phenotypic hallmarks of dry AMD in a murine model. We have shown that senolysis and NAD repletion efficiently removed senescent CD38-positive macrophages in subretinal space. However, there remains a possibility that systemic elimination of senescence by senolysis and NAD+ augmentation acts on other cell types besides macrophages. Indeed, senescence in other retinal cell types such as RPE and photoreceptors has been observed in human AMD.78,79 Although further human studies are necessary to identify the cell type responsible for the disease pathology, our study demonstrated senescent macrophages by the NAD+ decline drive phenotypes of an AMD model in mice. Moreover, SASP factors (TNF-α, VEGF) expressed in AMD patients are upregulated in the senescent macrophages in our study, leaving the possibility that SASP may also contribute to drusen biogenesis and neurodegeneration in retina.80
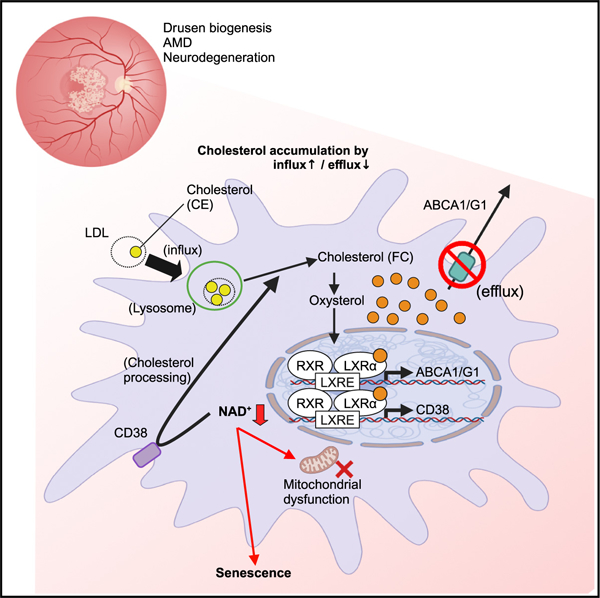
Our data describing that LXR/CD38 activation mediates aging, genotoxicity, and cholesterol-induced macrophage senescence, thereby driving age-related disease, constitute a biologic basis for testing NAD+ repletion as a senotherapeutic agent (Figure 7). Given that CD38 induces cellular senescence, it seems paradoxical for macrophages to co-regulate CD38 and ABCA1 under LXRE in response to cholesterol overload. Although the exact mechanism of how CD38 promotes cholesterol processing in macrophages remains unclear, our data suggest that CD38 and ABCA1/G1 both enhance cholesterol homeostasis by coordinating the cholesterol processing and efflux while CD38 halts cellular metabolism. Indeed our previous work revealed that whole-body knockout of LXRα leads to AMD pheno-types such as visual dysfunction and the accumulation of lipid droplets in RPE cells, demonstrating the essential role of LXR in cholesterol homeostasis and visual function.81 Additionally, a previous study of CD38 in pathogen response demonstrated its essential role in host defense.48 Taken together, these data suggest that CD38-induced senescence is a byproduct of physiological response and likely mediated through chronic metabolic and inflammatory stress that was unexpected during evolution. Additionally, LXR-mediated senescence may enhance the immune clearance of macrophages that are totally over-whelmed with intracellular cholesterol overload or have dysfunctional cholesterol clearance. Consistent with this idea, administration of healthy macrophages promoted the clearance of senescent macrophages. Interestingly, a recent study suggested that induction of cellular senescence enhances cholesterol metabolism by ABCA1 upregulation.28 In fact, we also demonstrated that LXR induces CD38 expression in response to diverse triggers of cellular senescence, demonstrating that cholesterol-mediated LXR activation is a common mechanism during senescence induction. However, our data on the reversal of senescence by NAD+ augmentation suggest that cholesterol-mediated LXR/CD38 activation and subsequent NAD+ degradation induce cellular senescence at least in macrophage, rather than the reverse scenario of the senescence conversion inducing the cholesterol accumulation and enhancement of cholesterol metabolism as a consequence.28
Figure 7. Models of NAD+-mediated induction of senescence and cholesterol metabolism.
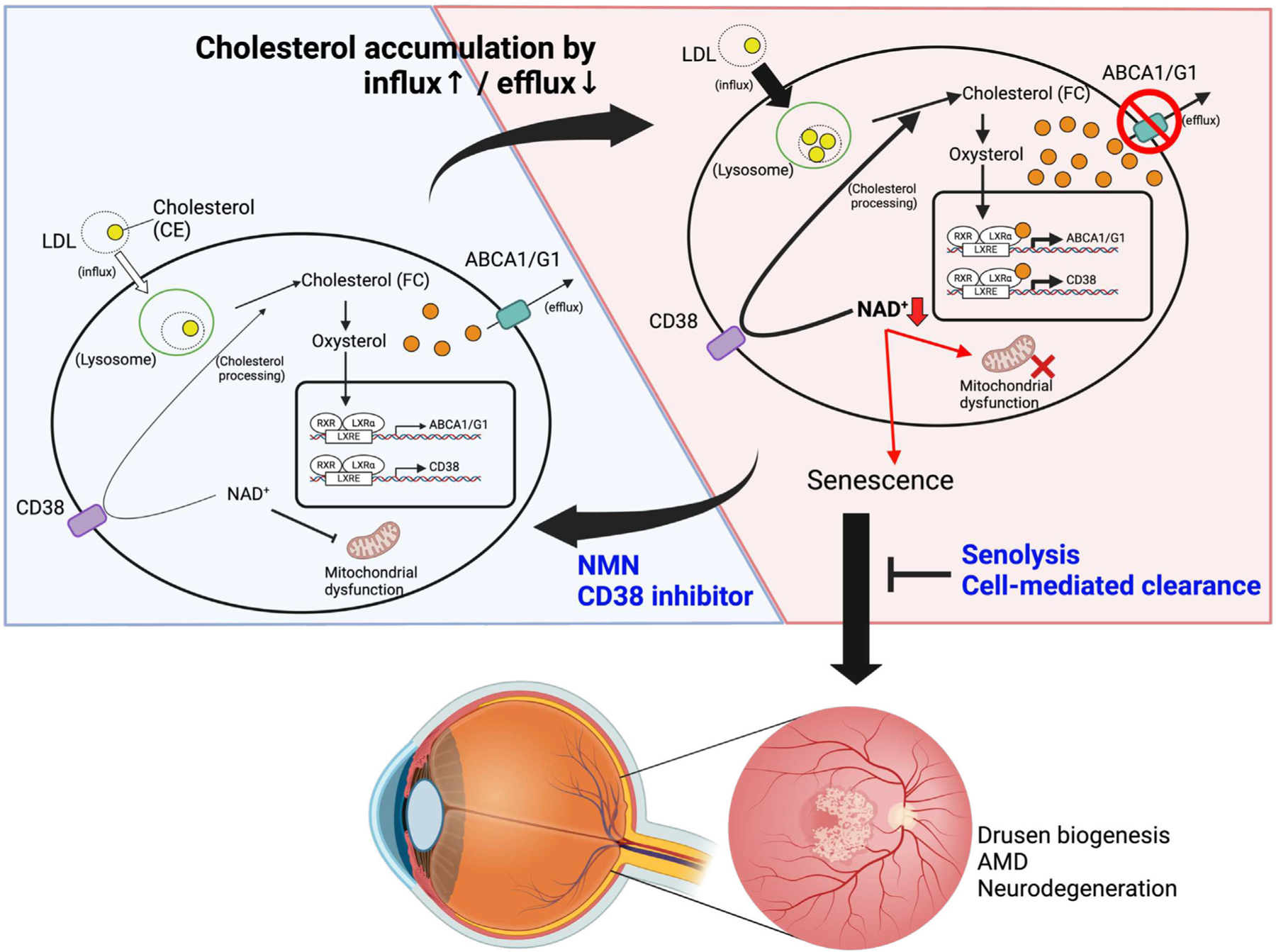
Intracellular cholesterol accumulation activates LXR-mediated expression of ABCA1/G1 and CD38 to maintain cholesterol homeostasis. During aging and in patients with genetic susceptibility, excessive intracellular cholesterol levels drive the cholesterol/LXR/CD38 axis, thereby inducing macrophage senescence by NAD+ depletion. Accumulation of subretinal senescent macrophages with lipofuscin leads to the pathogenic SDD phenotype and the development of dry AMD. Senolysis and cell-mediated clearance of senescent macrophages prevent or reverse AMD pathology. NAD+ repletion by NMN and CD38 inhibition suppresses macrophage senescence and is a viable therapeutic approach against age-related neurodegeneration.
LXRs are nuclear receptors that are activated by oxysterol and essential to maintain lipid homeostasis and immune responses. Oxysterols exhibit deleterious effects on inflammation, oxidative stress, and cell survival and have been suggested to contribute to age-related tissue dysfunction and diseases.82–85 In the context of AMD, oxysterol signatures can distinguish AMD from physiologic aging.86 A recent study demonstrated that LXR activation also promoted the development of myeloid-derived immune-suppressive cells in both cancer and chronic inflammatory states such as autoimmune hepatitis.87,88 However, conflicting roles of LXR as both promotors and suppressors of immune responses and tissue homeostasis suggest its high context, tissue, and cell-type dependency. Given that the impact of senescence in tissue homeostasis and repair is also context and cell-type dependent, it is plausible that activation of LXR induces senescent cells in many of these chronic and inflammatory conditions. Additionally, previous studies have shown that CD38 expression can be induced by other members of the nuclear receptor family, including retinoid acid receptor alpha.89,90 Although our result demonstrated that LXR is responsible for the genotoxic and cholesterol-induced macrophage senescence, we cannot discard the possibility of an alternative pathway that promotes cellular senescence through NAD+ depletion by CD38.
CD38 is a multifunctional enzyme that plays a crucial role in various cellular processes, including calcium signaling, immune function, and cell survival. Several studies have implicated CD38 in age-related metabolic dysfunction, tissue inflammation, immune dysfunction, and tumor development through the disruption of NAD+ homeostasis.45,91–93 Loss of CD38 may ameliorate metabolic functions including glucose tolerance and mitochondrial energy generation that occur during aging.94 CD38 is induced by factors secreted from senescent cells providing a reciprocal link between senescence and age-related cellular NAD+ decline, which decreases to 40%–70% by CD38 upregulation with age or several stimuli.46,93,95,96 In contrast, NAD+ augmentation by NMN and NR has a protective effect against age-associated metabolic dysfunction such as mitochondrial dysfunction and glucose intolerance.94,97 Our previous study of long-term NMN administration demonstrated that NMN ameliorated age-related retinal dysfunction, consistent with our current findings.98 Interestingly, we demonstrate that, upon cholesterol overload, CD38 is induced by LXR to promote cholesterol efflux in collaboration with ABCA1/G1, demonstrating the physiological rationale underlying its upregulation. Given that, in addition to cholesterol overload, genotoxic stress mediated by irradiation also upregulates CD38 through LXR, cholesterol accumulation seems to be the initiating event in diverse triggers of senescence. However, although genotoxic stress induces intracellular cholesterol accumulation,28 the molecular mechanism underlying cholesterol accumulation during cellular senescence by the genotoxic insults remains unclear and requires further investigation. Additionally, while our study demonstrates that NAD+ depletion plays a central role in senescence induction by various insults, given its pleiotropic role in cellular processes, decreased activity of which NAD+-dependent enzyme is responsible for the induction of senescence requires further exploration.
Our data suggest that modulation of the LXR/CD38/NAD+ pathway is an effective senotherapeutic intervention at the cellular level. Given that the safety of the NAD+ augmentation approach has been demonstrated in human trials, it may enhance the treatment efficacy of existing therapies against various age-associated diseases.99 Future studies are warranted to study the potential role of myeloid senescence in the etiology of other age-related diseases to further explore the application of NAD+-based senoreversal intervention. In summary, our findings uncover cholesterol-LXR-CD38 as a converging mechanism underlying senescence induction and that NAD+ augmentation is an effective approach to reverse macrophage senescence and age-related diseases.
Limitations of the study
Although our study demonstrates that CD38 upregulation and subsequent NAD+ degradation induces macrophage senescence, further exploration is needed to clarify whether the induction of cellular senescence is mediated by the depleted availability of NAD+ or can also be modulated by the metabolic byproduct(s) of NAD+ degradation. Given the essential role of NAADP in the hydrolysis of cholesterol ester and cADPR as a key cellular messenger in calcium signaling, it is imperative to investigate their biological roles in cholesterol processing and cellular senescence. Additionally, if NAD+ depletion is indeed the causal mechanism of macrophage senescence as our data suggests, further exploration of downstream pathways such as SIRTs and PARPs is necessary to elucidate the precise mechanism of macrophage senescence induction. Furthermore, how macrophages enter and survey the subretinal space needs to be explored as the eye is classically considered an immune-privileged site. Lastly, the generalizability of our findings in mice to human AMD pathogenesis needs to be assessed for potential translation into therapy.
STAR☆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rajendra Apte (apte@wustl.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
RNA-seq data have been deposited at GEO and lipidomics data have been deposited at Mendeley Data (codes are listed in Key Resources Table). These data are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-Iba1 | Wako | Cat#019-19741; RRID:AB_839504 |
| Rat monoclonal anti-F4/80 | abcam | Cat#ab6640; RRID:AB_1140040 |
| Sheep polyclonal anti-CD38 | R&D Systems | Cat#AF4947; RRID:AB_1241945 |
| Rabbit polyclonal anti-Ki67 | abcam | Cat#abcam15580; RRID;AB_443209 |
| Phalloidin-iFluor 647 | abcam | Cat#ab176759 |
| Alexa Fluor 647 donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711-605-152 |
| Alexa Fluor 647 donkey anti-rat IgG | Jackson ImmunoResearch | Cat# 712-605-153 |
| Alexa Fluor 488 donkey anti-sheep IgG | Jackson ImmunoResearch | Cat#713-545-147 |
| Goat polyclonal antibody to Biotin (FITC) | abcam | Cat#ab6550 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| pCMV6-XL6 | OriGene Technologies | Cat#PCMV6XL6 |
| pCMV6-AC-CD38-GFP | OriGene Technologies | Cat#MG204265 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DMEM | gibco | Cat#11965-084 |
| L-glutamate | gibco | Cat#25030-081 |
| Penicillin/streptomycin (P/S) | gibco | Cat#15140-122 |
| Accutase | Sigma-Aldrich | Cat#A6964 |
| Cholesterol | Sigma-Aldrich | Cat#C4951 |
| DNase | Sigma-Aldrich | Cat#D4527 |
| DNA digestion buffer | ZYMO RESEARCH | Cat#E1010-1 |
| High Capacity cDNA Reverse Transcription Kit | Applied biosystems | Cat#4368813 |
| TaqMan Fast Advanced Master Mix | Applied biosystems | Cat#4444554 |
| Nicotinamide-2,4,5,6-d4 | C/D/N Isotopes | Cat#D-3457 |
| Lipofectamine RNAiMAX transfection reagent | Thermo Fisher Scientific | Cat#13778150 |
| Lipofectamine 3000 transfection reagent | Thermo Fisher Scientific | Cat#L3000-008 |
| Opti-MEM | gibco | Cat#31895-070 |
| FK866 | Chem Cruz | Cat#sc-205325A |
| DMEM, high glucose | Sigma-Aldrich | Cat#D1145 |
| β-nicotinamide mononucleotide | Sigma-Aldrich | Cat#N3501 |
| GW3965 | Sigma-Aldrich | Cat#G6295 |
| 78c | MedChemExpress | Cat#HY-123999 |
| AP-20187 | APExBIO | Cat#B1274 |
| Dasatinib | Sigma-Aldrich | Cat#SML2589 |
| Quercetin | Sigma-Aldrich | Cat#Q4951 |
| Seahorse XF DMEM Medium | Agilent Technologies | Cat#103575-100 |
| Seahorse XF Cell Mito Stress Test kit | Agilent Technologies | Cat#103015-100 |
| Seahorse XF24 V7 PS Cell Culture Microplate | Agilent Technologies | Cat#100777-004 |
| Co-detection blocker | Advanced Cell Diagnostics | Cat#323170 |
| Co-detection antibody diluent | Advanced Cell Diagnostics | Cat#323160 |
| RNAscope probe Mm-Cdkn1a | Advanced Cell Diagnostics | Cat#408551 |
| RNAscope probe Mm-Cdkn2a | Advanced Cell Diagnostics | Cat#411011 |
| RNAscope 2.5 HD detection reagents- red | Advanced Cell Diagnostics | Cat#322360 |
| SenTraGor Cell Senescence Reagent | Cayman chemical | Cat#35568 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat#23225 |
| NBD Cholesterol Staining Dye Kit | abcam | Cat#ab269448 |
| Senescence β-Galactosidase staining kit | Cell Signaling | Cat#9860 |
| SPiDER β -galactosidase staining kit | DOJINDO | Cat#SG02 |
| Lysosome intracellular activity assay kit | abcam | Cat#ab234622 |
| TaqMan array standard plates | Thermo Scientific | Cat#4391524 |
| Click-iT™ plus TUNEL Assay kits for in situ apoptosis eetection | Invitrogen | Cat#C10618 |
| Fluorescent cell linker kit for general cell membrane labeling | Sigma-Aldrich | Cat# PKH26, PKH67 |
|
| ||
| Deposited data | ||
|
| ||
| The lipidomics data of BMDMs from Abca1/g1 -m/-m and Abca1/g1 f/f | This study | Mendeley Data: https://doi.org/10.17632/gd4mprzbvh.1 |
| The RNA sequence data for BMDMs from Abca1/g1 -m/-m and Abca1/g1 f/f | This study | GEO: GSE234507 |
| The lipidomics data of retina isolated from young and old mice | This study | Mendeley Data: https://doi.org/10.17632/j9gv8x9td7.1 |
| The RNA sequence data for retinal pigment epithelium/choroid tissue of Abca1/g1 -m/-m and INK-ATTAC-Abca1/g1 -m/-m | This study | GEO: GSE234641 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: Abca1fl Abcg1fl: B6.Cg-Abca1tm1Jp Abcg1tm1Tall/J | The Jackson Laboratory | RRID: IMSR_JAX:021067 |
| Mouse: LysM Cre: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | RRID: IMSR_JAX:004781 |
| Mouse: INK-ATTAC | Unity Biotechnology | N/A |
| Mouse: Cx3cr1 CreERT2: B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ | The Jackson Laboratory | RRID: IMSR_JAX:021160 |
| Mouse: Tmem119 Cre ERT2: C57BL/6-Tmem119em1(cre/ERT2)Gfng/J | The Jackson Laboratory | RRID: IMSR_JAX:031820 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for qPCR, see Table S1 | Thermo Scientific | N/A |
| AllStars Negative Control siRNA | QIAGEN | Cat#1027281 |
| siRNA targeting Nr1h3 (NM_013839) | QIAGEN | Cat#SI02740339 |
| siRNA targeting Nr1h2 (NM_009473) | QIAGEN | Cat#SI00185241 |
| siRNA targeting Cd38 (NM_007646) | QIAGEN | Cat#SI02666398 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LipidSig | N/A | http://chenglab.cmu.edu.tw/lipidsig/ |
| CellProfiler | https://cellprofiler.org | |
| MassHunter Quantitative Analysis | Agilent Technologies | https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software/ |
| FlowJo | BD Biosciences | https://www.flowjo.com/solutions/flowjo |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/download.html |
| Sample Size Calculator | ClinCalc | https://clincalc.com/stats/samplesize.aspx. |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
All animal experiments were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and Washington University School of Medicine in St. Louis Animal Care and Use guidelines and after approval by the Institutional Animal Care and Use Committee. Mice (2 months old-27 months old) of both sexes were used in this study. Mice were housed in a 12-h light/dark cycle with free access to food and water. Wild-type C57BL/6J mice were purchased from the Jackson Laboratory (000664). Abca1flox/flox/Abcg1flox/flox (Abca1/g1f/f), LysM-Cre, Cx3cr1-CreERT2, Tmem119-CreERT2 mice were purchased from the Jackson Laboratory as previously described (021067, 004781,021160, and 031820, respectively).31 To activate Cre-ERT2 fusion protein, tamoxifen-infused chow (500 mg/kg chow) was provided to mice expressing CreERT2. Phenotypes were evaluated 3 months after tamoxifen-infused chow feeding for 3 weeks on a 6 days-on/1 day-off weekly. INK-ATTAC transgene mice were provided by Unity Biotechnology and crossed with Abca1/g1-m/-m to generate Abca1/g1-m/-m; INK-ATTAC.
Primary cell cultures
Bone-marrow derived macrophages (BMDMs) were isolated as previously described.100 After 2-month-old wild-type or indicated mutant mice were euthanized by carbon dioxide (CO2) incubation, hindlimb bones were harvested. Bone marrow cells were isolated from limb bones by flushing with DMEM media (gibco, 11965–084) and cultured in a humidified incubator at 37◦C with 5% CO2 with DMEM media containing 10% fetal bovine serum (FBS), 10% conditioned media collected from CMG 14–12 cells containing macrophage colony stimulating factor (CMG solution), 1% L-glutamate (gibco, 25030–081), and 1% penicillin-streptomycin (P/S; gibco, 15140–122). Culture media were exchanged on day 3. Six days after the incubation, adherent cells were harvested using Accutase (Sigma-Aldrich, A6964) and seeded with DMEM media containing 10% FBS, 2% CMG solution, 1% L-glutamate, and 1% P/S (BMDM culture media) for further experiments.
Cell transfection
24 h after cells were seeded, the transfection of siRNA and DNA plasmid was performed. Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific, 13778150) and lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, L3000-008) diluted by Opti-MEM (gibco, 31895–070) was used for the transfection of siRNA and DNA plasmid respectively in accordance with manufacture’s protocol. 24 h after the siRNA transfection, cells were harvested or used for further experiments.
For DNA plasmid transfection, 1.5 μg DNA plasmid was added to each well. Cells were harvested or used for further experiments 72 h after the transfection.
Cholesterol loading and cell treatments
For cholesterol loading, cells were serum-starved overnight by replacing with BMDM culture media without FBS and CMG solution. The next day cholesterol solution with methyl-β-cyclodextrin (Sigma-Aldrich, C4951; 50 μM) was added to each well. When indicated, FK866 (Chem Cruz, sc-205325A; 100 μM), GW3965(Sigma-Aldrich, G6295; 1μM), 78c (MedChemExpress, HY-123999; 200nM), and nicotinamide mononucleotide (NMN; Sigma-Aldrich, N3501; 500 μM) were used following serum-starvation.
To induce senescence, Ionizing radiation (IR) was utilized. After BMDMs were exposed to 5 Gy of IR using X-ray irradiator (X-RAD 320, Precision X-ray), BMDM culture media was replaced every other day until harvest.
Intravitreal and subretinal injection of bone marrow derived macrophages (BMDMs)
Mice were anesthetized with intraperitoneal injection of ketamine (81.8 mg/kg BW) and xylazine (9.4 mg/kg BW). Pupils were dilated using an eye drop solution of 1% tropicamide (Somerset Therapeutics). BMDMs (10,000 cells/μl, 1 μL) in DMEM were injected into the vitreous cavity or subretinal space using 33-gauge Hamilton syringe. Phenotypes were evaluated 2 weeks after the intraocular injection.
Drug treatments in vivo
To ablate p16Ink4a-positive cells, Abca1/g1f/f LysM-Cre (Abca1/g1-m/-m); INK-ATTAC mice were treated with intraperitoneal injection of AP20187 (APExBIO, B1274; 10 mg/kg BW) every other day for 3 months from the age of 3 months. AP20187 reconstituted in ethanol (50 mg/mL) was diluted by PEG 400, tween 80, and H2O. For senolytic drugs experiments, Abca1/g1-m/-m mice were treated with the combination of dasatinib (Sigma-Aldrich, SML2589; 5 mg/kg BW) and quercetin (Sigma-Aldrich, Q4951; 50 mg/kg BW) dissolved in dimethyl sulfoxide (DMSO), tween 20, polyethylene glycol (PEG) 400, H2O or vehicle in 5 consecutive days every 2 weeks via oral gavage for 3 months from the age of 3 months. Two-month-old WT mice which received the subretinal injection of Abca1/g1-m/-m BMDMs were treated with NMN (300 mg/kg BW) dissolved in PBS, 78c (10 mg/kg BW) dissolved in DMSO, or vehicle intraperitoneally every day for 2 weeks.
METHOD DETAILS
Shotgun lipidomics analysis
Retinal samples from young (2 months old) and old (27 months old) mice were collected for lipidomics analysis. For homogenized tissue, each whole retina was collected in a 2 mL homogenizer tube pre-loaded with 2.8 mm ceramic beads (OMNI INTERNATIONAL, #19–628). 0.75 mL PBS was added to the tube and homogenized in the Omni Bead Ruptor Elite (3 cycles of 10 s at 5 m/s with a 10 s dwell time). Homogenate containing 2–6mg of original tissue was transferred to a glass tube for extraction. To evaluate the lipid profile in BMDMs, differentiated BMDMs were collected after 24-h serum starvation. Culture media was removed and replaced by 500mL PBS. BMDMs were scraped and harvested into glass tubes for extraction. A modified Bligh and Dyer extraction was carried out on all samples.101 Prior to biphasic extraction, an internal standard mixture consisting of 70 lipid standards across 17 subclasses was added to each sample (AB Sciex 5040156, Avanti 330827, Avanti 330830, Avanti 330828, Avanti 791642). Following two successive extractions, pooled organic layers were dried down in a SpeedVac SPD300DDA (Thermo Fisher Scientific) using ramp setting 4 at 35◦C for 45 min with a total run time of 90 min. Lipid samples were resuspended in 1:1 methanol/dichloromethane with 10mM Ammonium Acetate and transferred to robovials (Thermo Fisher Scientific, 10800107) for analysis. Samples were analyzed on the Sciex 5500 with DMS device (Lipidyzer Platform) with an expanded targeted acquisition list consisting of 1450 lipid species across 17 subclasses. Differential Mobility Device on Lipidyzer was tuned with EquiSPLASH LIPIDOMIX (Avanti, 330731). Data analysis was performed on an in-house data analysis platform comparable to the Lipidyzer Workflow Manager.102 Quantitative values were normalized to the number of cells or mg of tissue. Differential expression analysis was performed using LipidSig.
RNA isolation, qPCR, and senescence panel assay
Samples were collected in TRIzol reagent. mRNA was obtained using NucleoSpin RNA columns (Takara, 740955) according to the manufacturer’s instructions. gDNA was removed using DNase (Sigma-Aldrich, D4527) diluted with DNA digestion buffer (ZYMO RESEARCH, E1010-1). cDNA was obtained using a cDNA reverse transcription kit (applied biosystems, 4368813). TaqMan array standard plates were purchased from Thermo (4391524) and customized to evaluate to senescence markers identified in a previous study.68 qPCR was performed using probes for each target gene and TaqMan Fast Advanced Master Mix (applied biosystems, 4444554). β-actin was utilized as the internal control. Relative expression of target genes normalized to an internal control was calculated using the ΔΔCt method. mRNA transcript levels were measured using StepOnePlus Real-Time PCR System (Thermo Fisher Scientific).
RNA sequencing and analysis
Samples were prepared according to the library kit manufacturer’s protocol, indexed, pooled, and sequenced on an Illumina NovaSeq 6000. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq2 software. RNA-seq reads were then aligned and quantitated to the Ensembl release 101 primary assembly with an Illumina DRAGEN Bio-IT on-premise server running version 3.9.3–8 software.
All gene counts were then imported into the R/Bioconductor package EdgeR103 and TMM normalization size factors were calculated to adjust for samples for differences in library size. Ribosomal genes and genes not expressed in the smallest group size minus one sample greater than one count-per-million were excluded from further analysis. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package Limma.104 Weighted likelihoods based on the observed mean-variance relationship of every gene and sample were then calculated for all samples and the count matrix was transformed to moderated log 2 counts-per-million with Limma’s voomWithQualityWeights.105 The performance of all genes was assessed with plots of the residual standard deviation of every gene to their average log-count with a robustly fitted trend line of the residuals. Differential expression analysis was then performed to analyze for differences between conditions and the results were filtered for only those genes with Benjamini-Hochberg false-discovery rate adjusted pp-values less than or equal to 0.05.
For each contrast extracted with Limma, global perturbations in known Gene Ontology (GO) terms, MSigDb, and KEGG pathways were detected using the R/Bioconductor package GAGE106 to test for changes in expression of the reported log 2 fold-changes reported by Limma in each term versus the background log 2 fold-changes of all genes found outside the respective term. Perturbed KEGG pathways where the observed log 2 fold-changes of genes within the term were significantly perturbed in a single-direction versus background or in any direction compared to other genes within a given term with pp-values less than or equal to 0.05 were rendered as annotated KEGG graphs with the R/Bioconductor package Pathview.107
Immunofluorescence
Slides or flatmount samples were fixed with 10% formalin for 15 min. Blocking and hyperpermeabilization were performed through PBS containing 5% bovine serum albumin (BSA) and 0.1% Triton X-100 for 30–60 min. Samples were incubated with rabbit polyclonal anti-IBA-1, rat monoclonal anti-F4/80, sheep polyclonal anti-CD38, and rabbit polyclonal anti-Ki67 as primary antibodies overnight at 4◦C. On the following day, samples were washed with PBS and incubated with Alexa Fluor-conjugated corresponding secondary antibodies at room temperature for 1 h. When indicated, fluorescent phalloidin conjugate (abcam, ab176759) was used along with secondary antibodies. Nuclei were stained with DAPI solution (5 μg/mL). In order to quench the autofluorescence and avoid the bleed-through from lipofuscin in subretinal drusenoid deposits, SenTraGor Cell Senescence Reagent (Cayman chemical, 35568), a biotinylated analog of Sudan Black B, was used according to the manufacturer’s protocol. In the detection of lipofuscin in immuno-fluorescence images, fluorescein-conjugated anti-biotin antibody (abcam, ab6550) was used after quenching. For co-staining with in situ hybridization (ISH), co-detection blocker (Advanced Cell Diagnostics, 323170) was used for blocking and antibodies were diluted in co-detection antibody diluent (Advanced Cell Diagnostics, 323160). ISH described below was performed after the incubation with primary antibodies overnight, followed by the incubation with secondary antibodies. Images were taken using a Zeiss LSM 800 microscope.
LC/MS measurement for NAD+ metabolites
Lyophilized samples were reconstituted with 30 μL of 5 mM ammonium formate and centrifuged at 12,000 g for 10 min. Cleared supernatant were transferred to the 96-well sample tray. Serial dilutions of standards for each metabolite in 5 mM ammonium formate were used for calibration. Liquid chromatography was performed by HPLC (1290; Agilent Technologies) with Atlantis T3 (LC 2.1 3 150 mm, 3 μm; Waters)108 and 10 μL of samples or standards were injected at a flow rate of 0.15 mL/min with 5 mM ammonium formate for mobile phase A and 100% methanol for mobile phase B. Metabolites were eluted with gradients of 0–10 min, 0–70% B; 10–15 min, 70% B; 16–20 min, 0% B. The metabolites were detected with a triple quadrupole mass spectrometer (6460 MassHunter; Agilent Technologies) under positive ESI multiple reaction monitoring (MRM). Metabolites were quantified by MassHunter quantitative analysis tool (Agilent).
NAD+ flux assay
The net accumulation of heavy NAD+ were measured in BMDMs incubated with D4-Nam (320 μM: 2,3,4,5 deuterium Nam). BMDMs were incubated with D4-Nam over 6 h and cellular metabolites were collected as described.109 Briefly, after BMDMs were incubated with D4-Nam, cells were washed with 0.9% NaCl and incubated with 50% methanol for 5 min, followed by chloroform extraction. Samples were lyophilized and stored at —20◦C for LC/MS measurement. Each concentration of metabolites was normalized to the amount of total protein measured using Pierce BCA Protein Assay Kit (Thermo Scientific, 23225). The total amount of D3-NAD+ and D4-NAD+ was defined as the labeled (heavy) NAD+ due to the replacement of deuterium at C4 position with non-labeled proton during NAD+ - NADH cycling.110 This net NAD+ accumulation of heavy NAD+ accounts for the amount of labeled NAD+ remaning after NAD+ produced subtracted by the NAD+ consumed. For quantifying the specific NAD+ consumption rate, FK866 (100 μM) was added to each well 3 h after labeling NAD+ with D4-Nam. The consumption rate was calculated as a rate of heavy NAD+ decline between 1 h and 3 h post FK866 addition.
NBD-cholesterol flux assay
In order to measure the intracellular concentration of fluorescence-tagged cholesterol, BMDMs were incubated with NBD Cholesterol Staining Dye Kit (abcam, ab269448; 1:20) diluted by DMEM, high glucose (Sigma-Aldrich, D1145) containing 1% L-glutamate and 1% P/S, when indicated, along with the treatment with 78c or DMSO. Cells were incubated for 2.5 h and images were obtained using DMi8 (Leica). In the evaluation of cholesterol processing and efflux in BMDMs, BMDMs were treated with NBD-cholesterol diluted by media described above. After 6 h, NBD-tagged cholesterol in culture media was removed by replacing with culture madia containing 78c or DMSO. Cells were incubated for 20 h and images were obtained using DMi8 (Leica). The fluorescence intensity in each cell was measured using CellProfiler.
Organelle function assays
Oxygen consumption rate (OCR) measurement to evaluate mitochondrial respiration was measured using the Seahorse XF Cell Mito Stress Test kit (Agilent Technologies, 103015-100) on a XFe24 Extracellular Flux Analyzer (Agilent Technologies). BMDMs were plated in Seahorse XF24 V7 PS Cell Culture microplates (Agilent Technologies, 100777–004) at 100,000 cells per well in BMDM culture media. On the night before the experiment, the Seahorse XFe24 sensor cartridge was hydrated with Seahorse XF Calibrant solution (1mL per well) and placed in a non-CO2 incubator at 37◦C. One hour prior to the experiment, standard medium was removed and replaced with 500uL pre-warmed Seahorse XF DMEM medium (Agilent Technologies, 103575–100) supplemented with 1mM sodium pyruvate, 2mM glutamine, and 10mM glucose, pH 7.4, and placed in a non-CO2 incubator at 37◦C. Chemicals from the Mito Stress Test kit were loaded into the sensor cartridge to provide the following final in-well concentrations: 1.5μM oligomycin, 2.5μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and 1 μM rotenone/antimycin A (rot/AA). Following calibration of the censor cartridge in the XFe24 Extracellular Flux Analyzer, the cell plate was inserted and OCR measured. Each cycle consisted of 3 min of mixing and a 2-min pause, followed by a 3-min measurement period. Each cycle repeated 3 times. Total protein content of each well was measured using Pierce BCA Protein Assay Kit and used to normalize OCR readings.
Lysosome intracellular activity assay (abcam, ab234622) was used to evaluate the capability of degradation by lysosome. The kit was used according to the manufacturer’s instructions. A negative control was pre-treated with bafilomycin A1 at 37◦C for 2 h. BMDMs were treated with a self-quenched substrate at 37◦C for 1 h. Cells were washed twice with ice-cold assay buffer and used for further measurement using a flow cytometer.
Electroretinogram (ERG)
Mice were dark adapted overnight. All procedures during ERG were performed under dark red light illumination. Mice were anesthetized with intraperitoneal injection of 122 mg/kg ketamine and 14.1 mg/kg xylazine. Pupils were dilated with the 1% tropicamide. ERG was performed using UTAS BigShot System (LKC Technologies Inc.), maintaining body temperature at 37◦C with a heating pad.Scotopic ERG was taken using contact lens electrodes placed bilaterally. The sequence of white light flash (10 μs) was used as full-field flash stimuli. A-wave was defined as the amplitude from the baseline to the lowest negative point after the stimuli. B-wave was measured from a-wave to the highest peak. To evaluate RPE function, dark adaptation was measured as previously described.31 After taking the photoresponse as the baseline, both eyes were exposed to LED light (150,000 lux) for 15 s to photobleach the visual pigment. The recovery of ERG response was measured at fixed post-bleach time points within 90 min after the bleach. A-wave amplitudes at each point were normalized to the baseline.
Live retinal imaging
Mice were anesthetized with intraperitoneal injection of ketamine and xylazine as described above. Pupils were dilated by the 1% tropicamide. Fundus photography was taken using Micron III (Phoenix) with optical coherence tomography (Phoenix, OCT2). The number of subretinal drusenoid deposits were evaluated using images of central area as well as peripheral area in each mouse.
In order to measure thickness of the outer nuclear layer (ONL), OCT images were processed by ImageJ software (National Institutes of Health) and ONL thickness was measured every 100μm from optic nerve.
Transmission electron microscopy
For ultrastructural analyses, BMDMs were fixed in 2% paraformaldehyde/2.5% glutaraldehyde (Ted Pella Inc., Redding, CA) in 0.1 M sodium cacodylate buffer for 2 h at room temperature. Enucleated eyes were dissected out and anterior chamber including cornea and lens were removed. Eye cups were fixed in the same fixative buffer for 2 h at room temperature and then overnight at 4◦C to get better preservation of retinal layers. Samples were washed in sodium cacodylate buffer and postfixed in 1% osmium tetroxide (Ted Pella Inc.) for 1 h at room temperature. After three washes in distilled water, samples were en bloc stained in 1% aqueous uranyl acetate (Electron Microscopy Sciences, Hatfield, PA) for 1 h. Samples were then rinsed in distilled water, dehydrated in a graded series of ethanol, and embedded in Eponate 12 resin (Ted Pella Inc). Ultrathin sections of 95 nm sections were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems, Bannockburn, IL), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc., Peabody, MA) equipped with an AMT 8 megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques, Woburn, MA). The number of intracellular lipids in retinal pigment epithelium and Bruch’s membrane thickness in each sample were evaluated by calculating the average of randomly taken images.
SA-β-gal staining and SPiDER β-gal staining
Senescence β-Galactosidase staining kit (Cell Signaling, 9860) was utilized to visualize senescent cells in retinal sagittal sections and BMDMs. Briefly, β-galactosidase activity was evaluated at pH 6. Sagittal sections and BMDMs were incubated with 5-Bromo-4-Chloro-3-Indolyl β-D-Galactopyranoside at 37◦C for 48 h and 6 h, respectively. Lipofuscin was detected as the yellow-gold autofluorescence (Ex 475 nm).111,112 Images were obtained with DMi8 (Leica).
SPiDER β-Gal staining kit (DOJINDO, SG02) was used in order to visualize senescent cells in RPE-flatmount. Samples were incubated with SPiDER β-Gal at 37◦C for 2 h in line with the manufacturer’s instructions. In order to prevent the bleed-through by auto-fluorescence from lipofuscin into other channels, lipofuscin was visualized using SenTraGor Cell Senescence Reagent for co-staining as described in Immunofluorescence section. Images were obtained with LSM 800 (Zeiss).
In situ hybridization
Formalin-fixed and dehydrated slides were pretreated with RNAscope protease reagents (Advanced Cell Diagnostics, 322381). Cdkn1a (Advanced Cell Diagnostics, 408551) and Cdkn2a (Advanced Cell Diagnostics, 411011) probes were added on each slide and incubated at 40◦C for 2 h in a HybEZ Oven (Advanced Cell Diagnostics, 241000ACD). Amplification steps and color development using RNAscope 2.5 HD detection reagents (Advanced Cell Diagnostics, 322360) were performed in accordance with manufacturer’s instructions. After DAPI staining (5 mg/mL) for 5 min, coverslips were place on slides. Images were taken by LSM 800.
TUNEL assay
After BMDMs were treated with D + Q for 24 h, cells were fixed with 10% formalin for 15 min. Click-iT Plus TUNEL Assay Kits for In Situ Apoptosis Detection (Invitrgen, C10618) was used in order to detect apoptotic cells. Images were taken by DMi8.
Endocytosis assay
Fluorescent cell linker kits for general cell membrane labeling (Sigma-Aldrich, PKH26, PKH67) were used based on the manufacture’s protocol. Cytomembranes of BMDMs were stained by dye solution containing the lipophilic dye (1μM) provided in the kit. After the incubation for 5 min, dye solution was removed and BMDMs were resuspended with culture media. BMDMs were plated on chamber slides and incubated for 24 h.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments except for sequence study and lipidomics measurement were performed more than 2 times for replicates. In experimental groups of mice, littermates of the same sex were randomly assigned. Sample size was estimated by Sample Size Calculator (ClinCalc). Lipidomics data was analyzed using LipidSig. Data obtained from FACS was analyzed using FlowJo. Statistical analysis was performed using GraphPad Prism. A two-tailed, unpaired student’s t-test was utilized for a comparison between two groups. To compare three or more means, a two-tailed, unpaired student’s t-test and a two-way ANOVA with the Bonferroni post-hoc test was used to compare two or more means, respectively. Statistical analysis of lipidomics data was performed by Benjamini & Hochberg method equipped in LipidSig. Statistically significant outliers were excluded from analysis. p < 0.05 was considered statistically significant. Data were presented as bar or line graphs (mean ± standard error of the mean). When indicated, dot plots show each value.
Supplementary Material
Highlights.
Cholesterol promotes macrophage senescence by NAD depletion via LXR/CD38 signaling
Senescent macrophages accumulate in the subretinal space in AMD model mice
NAD augmentation and senolytics can suppress subretinal drusenoid deposits
Cell-mediated clearance has the capacity to remove subretinal deposits
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants R01 EY019287 (R.S.A.) and P30 EY02687 (Vision Core Grant); Jeffrey T. Fort Innovation Fund (R.S.A.); Starr Foundation AMD Research Fund (R.S.A.); Siteman Retina Research Fund (R.S.A.); Carl Marshall and Mildred Almen Reeves Foundation; Retina Associates of St. Louis Research Fund (R.S.A.); an unrestricted grant from Research to Prevent Blindness to the John F. Hardesty, MD Department of Ophthalmology and Visual Sciences at Washington University School of Medicine in St. Louis; Williams D. Owens Anesthesiology Research Fellowship Award (M.Y.); Bayer Retina Award in Japan (R.T.); the International Retinal Research Foundation (R.T.); and the Japan Society for the Promotion of Science Overseas Research Fellowship (R.T.). T.L. was supported by NIH Training Grant (1T32GM1397740-1). C.W.P. was supported by the Washington University in St. Louis Vision Science Training grant (T32 EY013360), Vitreoretinal Surgery Foundation Fellowship (VGR0023118), and McDonnell Center for Cellular and Molecular Neurobiology Small Grants Program (GF0011083). J.B.L. was supported by the NIH grant (F30DK130282) and the Washington University in St. Louis Medical Scientist Training Program (NIH grant T32GM07200). Y.S. was supported by NIH grant RF1 AG013730 (to Jeffrey Milbrandt).
The authors acknowledge Wandy L. Beatty, Molecular Microbiology Imaging Facility in Washington University School of Medicine, for EM analyses; the University of California, Alicia Neiner, Department of Genetics in Washington University School of Medicine, for technical support; and Los Angeles lipidomics lab for the lipidomics measurement service. We also thank Washington University Diabetes Research Center Cores (NIH grant #5 P30 DK020579) for the use of XFe24 Extracellular Flux Analyzer, and Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis for the use of the Siteman Flow Cytometry, which provided FACS service. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA091842. We acknowledge the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. We thank Dr. Daniel Saban at Duke University for his advice regarding our myeloid cell studies.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114102.
DECLARATION OF INTERESTS
R.S.A. is a co-founder of Metro Midwest Biotech. M.Y. receives patent-licensing fees from Institute for Research on Productive Aging.
REFERENCES
- 1.Zhang L, Pitcher LE, Yousefzadeh MJ, Niedernhofer LJ, Robbins PD, and Zhu Y (2022). Cellular senescence: a key therapeutic target in aging and diseases. J. Clin. Invest. 132, e158450. 10.1172/JCI158450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zindy F, Quelle DE, Roussel MF, and Sherr CJ (1997). Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15, 203–211. 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 3.Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez PY, and Campisi J (2008). Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868. 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krtolica A, Parrinello S, Lockett S, Desprez PY, and Campisi J (2001). Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. USA. 98, 12072–12077. 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krtolica A, and Campisi J (2002). Cancer and aging: a model for the cancer promoting effects of the aging stroma. Int. J. Biochem. Cell Biol. 34, 1401–1414. 10.1016/s1357-2725(02)00053-5. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Yue X, Sun Z, Hambright WS, Feng Q, Cui Y, Huard J, Robbins PD, Wang Z, and Mu X (2022). Senolytic elimination of senescent macrophages restores muscle stem cell function in severely dystrophic muscle. Aging (Albany NY) 14, 7650–7661. 10.18632/aging.204275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oubaha M, Miloudi K, Dejda A, Guber V, Mawambo G, Germain MA, Bourdel G, Popovic N, Rezende FA, Kaufman RJ, et al. (2016). Senescence-associated secretory phenotype contributes to pathological angiogenesis in retinopathy. Sci. Transl. Med. 8, 362ra144. 10.1126/scitranslmed.aaf9440. [DOI] [PubMed] [Google Scholar]
- 8.Crespo-Garcia S, Tsuruda PR, Dejda A, Ryan RD, Fournier F, Chaney SY, Pilon F, Dogan T, Cagnone G, Patel P, et al. (2021). Pathological angiogenesis in retinopathy engages cellular senescence and is amenable to therapeutic elimination via BCL-xL inhibition. Cell Metabol. 33, 818–832.e7. 10.1016/j.cmet.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Hickson LJ, Eirin A, Kirkland JL, and Lerman LO (2022). Cellular senescence: the good, the bad and the unknown. Nat. Rev. Nephrol. 18, 611–627. 10.1038/s41581-022-00601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Pitcher LE, Prahalad V, Niedernhofer LJ, and Robbins PD (2023). Targeting cellular senescence with senotherapeutics: senolytics and senomorphics. FEBS J. 290, 1362–1383. 10.1111/febs.16350. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Tchkonia T, Pirtskhalava T, Gower AC, Ding H, Giorgadze N, Palmer AK, Ikeno Y, Hubbard GB, Lenburg M, et al. (2015). The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658. 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baar MP, Brandt RMC, Putavet DA, Klein JDD, Derks KWJ, Bourgeois BRM, Stryeck S, Rijksen Y, van Willigenburg H, Feijtel DA, et al. (2017). Targeted Apoptosis of Senescent Cells Restores Tissue Homeostasis in Response to Chemotoxicity and Aging. Cell 169, 132–147.e16. 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moiseeva O, Deschênes-Simard X, St-Germain E, Igelmann S, Huot G, Cadar AE, Bourdeau V, Pollak MN, and Ferbeyre G (2013). Metformin inhibits the senescence-associated secretory phenotype by interfering with IKK/NF-kappaB activation. Aging Cell 12, 489–498. 10.1111/acel.12075. [DOI] [PubMed] [Google Scholar]
- 14.Alimbetov D, Davis T, Brook AJC, Cox LS, Faragher RGA, Nurgozhin T, Zhumadilov Z, and Kipling D (2016). Suppression of the senescence-associated secretory phenotype (SASP) in human fibroblasts using small molecule inhibitors of p38 MAP kinase and MK2. Biogerontology 17, 305–315. 10.1007/s10522-015-9610-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Otín C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer AK, Tchkonia T, and Kirkland JL (2022). Targeting cellular senescence in metabolic disease. Mol. Metabol. 66, 101601. 10.1016/j.molmet.2022.101601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan AE, Mooney KM, Wilkinson SJ, Pickles NA, and Mc Auley MT (2016). Cholesterol metabolism: A review of how ageing disrupts the biological mechanisms responsible for its regulation. Ageing Res. Rev. 27, 108–124. 10.1016/j.arr.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Warner GJ, Stoudt G, Bamberger M, Johnson WJ, and Rothblat GH (1995). Cell toxicity induced by inhibition of acyl coenzyme A:cholesterol acyltransferase and accumulation of unesterified cholesterol. J. Biol. Chem. 270, 5772–5778. 10.1074/jbc.270.11.5772. [DOI] [PubMed] [Google Scholar]
- 19.Geng YJ, Phillips JE, Mason RP, and Casscells SW (2003). Cholesterol crystallization and macrophage apoptosis: implication for atherosclerotic plaque instability and rupture. Biochem. Pharmacol. 66, 1485–1492. 10.1016/s0006-2952(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 20.Langmann T, Klucken J, Reil M, Liebisch G, Luciani MF, Chimini G, Kaminski WE, and Schmitz G (1999). Molecular cloning of the human ATP-binding cassette transporter 1 (hABC1): evidence for sterol-dependent regulation in macrophages. Biochem. Biophys. Res. Commun. 257, 29–33. 10.1006/bbrc.1999.0406. [DOI] [PubMed] [Google Scholar]
- 21.Klucken J, Büchler C, Orsó E, Kaminski WE, Porsch-Ozcürümez M, Liebisch G, Kapinsky M, Diederich W, Drobnik W, Dean M, et al. (2000). ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc. Natl. Acad. Sci. USA. 97, 817–822. 10.1073/pnas.97.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sene A, Khan AA, Cox D, Nakamura REI, Santeford A, Kim BM, Sidhu R, Onken MD, Harbour JW, Hagbi-Levi S, et al. (2013). Impaired cholesterol efflux in senescent macrophages promotes age-related macular degeneration. Cell Metabol. 17, 549–561. 10.1016/j.cmet.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sene A, and Apte RS (2014). Eyeballing cholesterol efflux and macrophage function in disease pathogenesis. Trends Endocrinol. Metabol. 25, 107–114. 10.1016/j.tem.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuo M (2022). ABCA1 and ABCG1 as potential therapeutic targets for the prevention of atherosclerosis. J. Pharmacol. Sci. 148, 197–203. 10.1016/j.jphs.2021.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Apte RS (2021). Age-Related Macular Degeneration. N. Engl. J. Med. 385, 539–547. 10.1056/NEJMcp2102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Björkegren JLM, and Lusis AJ (2022). Atherosclerosis: Recent developments. Cell 185, 1630–1645. 10.1016/j.cell.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long JM, and Holtzman DM (2019). Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 179, 312–339. 10.1016/j.cell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roh K, Noh J, Kim Y, Jang Y, Kim J, Choi H, Lee Y, Ji M, Kang D, Kim MS, et al. (2023). Lysosomal control of senescence and inflammation through cholesterol partitioning. Nat. Metab. 5, 398–413. 10.1038/s42255-023-00747-5. [DOI] [PubMed] [Google Scholar]
- 29.Martins IJ, Hone E, Foster JK, Sünram-Lea SI, Gnjec A, Fuller SJ, Nolan D, Gandy SE, and Martins RN (2006). Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer’s disease and cardiovascular disease. Mol. Psychiatr. 11, 721–736. 10.1038/sj.mp.4001854. [DOI] [PubMed] [Google Scholar]
- 30.Cheung CMG, Gan A, Fan Q, Chee ML, Apte RS, Khor CC, Yeo I, Mathur R, Cheng CY, Wong TY, and Tai ES (2017). Plasma lipoprotein subfraction concentrations are associated with lipid metabolism and age-related macular degeneration. J. Lipid Res. 58, 1785–1796. 10.1194/jlr.M073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ban N, Lee TJ, Sene A, Choudhary M, Lekwuwa M, Dong Z, Santeford A, Lin JB, Malek G, Ory DS, and Apte RS (2018). Impaired monocyte cholesterol clearance initiates age-related retinal degeneration and vision loss. JCI Insight 3, e120824. 10.1172/jci.insight.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler TW, Grassmann F, Brandl C, Kiel C, Günther F, Strunz T, Weidner L, Zimmermann ME, Korb CA, Poplawski A, et al. (2020). Genome-wide association meta-analysis for early age-related macular degeneration highlights novel loci and insights for advanced disease. BMC Med. Genom. 13, 120. 10.1186/s12920-020-00760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, Raychaudhuri S, Tan PL, Oh EC, Merriam JE, Souied E, et al. (2010). Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc. Natl. Acad. Sci. USA. 107, 7395–7400. 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, et al. (2010). Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 107, 7401–7406. 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang N, and Westerterp M (2020). ABC Transporters, Cholesterol Efflux, and Implications for Cardiovascular Diseases. Adv. Exp. Med. Biol. 1276, 67–83. 10.1007/978-981-15-6082-8_6. [DOI] [PubMed] [Google Scholar]
- 36.Zahid MDK, Rogowski M, Ponce C, Choudhury M, Moustaid-Moussa N, and Rahman SM (2020). CCAAT/enhancer-binding protein beta (C/EBPbeta) knockdown reduces inflammation, ER stress, and apoptosis, and promotes autophagy in oxLDL-treated RAW264.7 macrophage cells. Mol. Cell. Biochem. 463, 211–223. 10.1007/s11010-019-03642-4. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q, Kriksunov IA, Graeff R, Lee HC, and Hao Q (2007). Structural basis for formation and hydrolysis of the calcium messenger cyclic ADP-ribose by human CD38. J. Biol. Chem. 282, 5853–5861. 10.1074/jbc.M609093200. [DOI] [PubMed] [Google Scholar]
- 38.Palade P (2007). The hunt for an alternate way to generate NAADP. Focus on “NAADP as a second messenger: neither CD38 nor base-exchange reaction are necessary for in vivo generation of NAADP in myometrial cells. Am. J. Physiol. Cell Physiol. 292, C4–C7. 10.1152/ajpcell.00390.2006. [DOI] [PubMed] [Google Scholar]
- 39.Zeidler JD, Hogan KA, Agorrody G, Peclat TR, Kashyap S, Kanamori KS, Gomez LS, Mazdeh DZ, Warner GM, Thompson KL, et al. (2022). The CD38 glycohydrolase and the NAD sink: implications for pathological conditions. Am. J. Physiol. Cell Physiol. 322, C521–C545. 10.1152/ajpcell.00451.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morra M, Zubiaur M, Terhorst C, Sancho J, and Malavasi F (1998). CD38 is functionally dependent on the TCR/CD3 complex in human T cells. Faseb. J. 12, 581–592. 10.1096/fasebj.12.7.581. [DOI] [PubMed] [Google Scholar]
- 41.Jackson DG, and Bell JI (1990). Isolation of a cDNA encoding the human CD38 (T10) molecule, a cell surface glycoprotein with an unusual discontinuous pattern of expression during lymphocyte differentiation. J. Immunol. 144, 2811–2815. [PubMed] [Google Scholar]
- 42.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. (2013). The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 154, 430–441. 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zapata-Pérez R, Wanders RJA, van Karnebeek CDM, and Houtkooper RH (2021). NAD(+) homeostasis in human health and disease. EMBO Mol. Med. 13, e13943. 10.15252/emmm.202113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masutani M, Miwa M, and Poltronieri P (2023). NAD(+) Consuming Enzymes: Involvement in Therapies and Prevention of Human Diseases. Anti Cancer Agents Med. Chem. 23, 1351–1354. 10.2174/1871520623666230320153757. [DOI] [PubMed] [Google Scholar]
- 45.Tarragó MG, Chini CCS, Kanamori KS, Warner GM, Caride A, de Oliveira GC, Rud M, Samani A, Hein KZ, Huang R, et al. (2018). A Potent and Specific CD38 Inhibitor Ameliorates Age-Related Metabolic Dysfunction by Reversing Tissue NAD(+) Decline. Cell Metabol. 27, 1081–1095.e10. 10.1016/j.cmet.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chini CCS, Peclat TR, Warner GM, Kashyap S, Espindola-Netto JM, de Oliveira GC, Gomez LS, Hogan KA, Tarragó MG, Puranik AS, et al. (2020). CD38 ecto-enzyme in immune cells is induced during aging and regulates NAD(+) and NMN levels. Nat. Metab. 2, 1284–1304. 10.1038/s42255-020-00298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexandris AS, Ryu J, Rajbhandari L, Harlan R, McKenney J, Wang Y, Aja S, Graham D, Venkatesan A, and Koliatsos VE (2022). Protective effects of NAMPT or MAPK inhibitors and NaR on Wallerian degeneration of mammalian axons. Neurobiol. Dis. 171, 105808. 10.1016/j.nbd.2022.105808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matalonga J, Glaria E, Bresque M, Escande C, Carbó JM, Kiefer K, Vicente R, León TE, Beceiro S, Pascual-García M, et al. (2017). The Nuclear Receptor LXR Limits Bacterial Infection of Host Macrophages through a Mechanism that Impacts Cellular NAD Metabolism. Cell Rep. 18, 1241–1255. 10.1016/j.celrep.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Costet P, Luo Y, Wang N, and Tall AR (2000). Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 275, 28240–28245. 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 50.Yang S, Zhang F, Li Q, and Li Q (2022). Niacin promotes the efflux of lysosomal cholesterol from macrophages via the CD38/NAADP signaling pathway. Exp. Biol. Med. 247, 1047–1054. 10.1177/15353702221084632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nabar NR, Heijjer CN, Shi CS, Hwang IY, Ganesan S, Karlsson MCI, and Kehrl JH (2022). LRRK2 is required for CD38-mediated NAADP-Ca(2+) signaling and the downstream activation of TFEB (transcription factor EB) in immune cells. Autophagy 18, 204–222. 10.1080/15548627.2021.1954779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang C, Li T, Li Y, Xu GJ, Deng QW, Chen YJ, Hou YN, Lee HC, and Zhao YJ (2018). CD38 produces nicotinic acid adenosine dinucleotide phosphate in the lysosome. J. Biol. Chem. 293, 8151–8160. 10.1074/jbc.RA118.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X, Yuan X, Li N, Dewey WL, Li PL, and Zhang F (2016). Lysosomal cholesterol accumulation in macrophages leading to coronary atherosclerosis in CD38(−/−) mice. J. Cell Mol. Med. 20, 1001–1013. 10.1111/jcmm.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westerterp M, Murphy AJ, Wang M, Pagler TA, Vengrenyuk Y, Kappus MS, Gorman DJ, Nagareddy PR, Zhu X, Abramowicz S, et al. (2013). Deficiency of ATP-binding cassette transporters A1 and G1 in macrophages increases inflammation and accelerates atherosclerosis in mice. Circ. Res. 112, 1456–1465. 10.1161/CIRCRESAHA.113.301086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lautrup S, Sinclair DA, Mattson MP, and Fang EF (2019). NAD(+) in Brain Aging and Neurodegenerative Disorders. Cell Metabol. 30, 630–655. 10.1016/j.cmet.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida M, Satoh A, Lin JB, Mills KF, Sasaki Y, Rensing N, Wong M, Apte RS, and Imai SI (2019). Extracellular Vesicle-Contained eNAMPT Delays Aging and Extends Lifespan in Mice. Cell Metabol. 30, 329–342.e5. 10.1016/j.cmet.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Childs BG, Durik M, Baker DJ, and van Deursen JM (2015). Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435. 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kao YC, Ho PC, Tu YK, Jou IM, and Tsai KJ (2020). Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 21, 1505. 10.3390/ijms21041505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldstein JL, and Brown MS (2015). A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161, 161–172. 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chrispin J, Martin SS, Hasan RK, Joshi PH, Minder CM, McEvoy JW, Kohli P, Johnson AE, Wang L, Blaha MJ, and Blumenthal RS (2013). Landmark lipid-lowering trials in the primary prevention of cardiovascular disease. Clin. Cardiol. 36, 516–523. 10.1002/clc.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Micco R, Krizhanovsky V, Baker D, and d’Adda di Fagagna F (2021). Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95. 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lian J, Yue Y, Yu W, and Zhang Y (2020). Immunosenescence: a key player in cancer development. J. Hematol. Oncol. 13, 151. 10.1186/s13045-020-00986-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, and De Benedictis G (2000). Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 908, 244–254. 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 64.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, and van Deursen JM (2011). Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236. 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, et al. (2016). Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530, 184–189. 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, et al. (2018). Senolytics improve physical function and increase lifespan in old age. Nat. Med. 24, 1246–1256. 10.1038/s41591-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okabe K, Yaku K, Tobe K, and Nakagawa T (2019). Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 26, 34. 10.1186/s12929-019-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hernandez-Segura A, de Jong TV, Melov S, Guryev V, Campisi J, and Demaria M (2017). Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr. Biol. 27, 2652–2660.e4. 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodier F, Coppé JP, Patil CK, Hoeijmakers WAM, Muñoz DP, Raza SR, Freund A, Campeau E, Davalos AR, and Campisi J (2009). Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979. 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng X, Wu Y, Brouwer U, van Vliet T, Wang B, Demaria M, Barazzuol L, and Coppes RP (2020). Cellular senescence contributes to radiation-induced hyposalivation by affecting the stem/progenitor cell niche. Cell Death Dis. 11, 854. 10.1038/s41419-020-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miwa S, Kashyap S, Chini E, and von Zglinicki T (2022). Mitochondrial dysfunction in cell senescence and aging. J. Clin. Invest. 132, e158447. 10.1172/JCI158447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minhas PS, Liu L, Moon PK, Joshi AU, Dove C, Mhatre S, Contrepois K, Wang Q, Lee BA, Coronado M, et al. (2019). Macrophage de novo NAD(+) synthesis specifies immune function in aging and inflammation. Nat. Immunol. 20, 50–63. 10.1038/s41590-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, Liu Y, Gao Y, Shou S, and Chai Y (2021). The roles of macrophage polarization in the host immune response to sepsis. Int. Immunopharm. 96, 107791. 10.1016/j.intimp.2021.107791. [DOI] [PubMed] [Google Scholar]
- 74.Vijg J, and Dong X (2020). Pathogenic Mechanisms of Somatic Mutation and Genome Mosaicism in Aging. Cell 182, 12–23. 10.1016/j.cell.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.López-Otín C, Galluzzi L, Freije JMP, Madeo F, and Kroemer G (2016). Metabolic Control of Longevity. Cell 166, 802–821. 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 76.Albright JM, Dunn RC, Shults JA, Boe DM, Afshar M, and Kovacs EJ (2016). Advanced Age Alters Monocyte and Macrophage Responses. Antioxidants Redox Signal. 25, 805–815. 10.1089/ars.2016.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaw AC, Goldstein DR, and Montgomery RR (2013). Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol. 13, 875–887. 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Terao R, Ahmed T, Suzumura A, and Terasaki H (2022). Oxidative Stress-Induced Cellular Senescence in Aging Retina and Age-Related Macular Degeneration. Antioxidants 11, 2189. 10.3390/antiox11112189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blasiak J (2020). Senescence in the pathogenesis of age-related macular degeneration. Cell. Mol. Life Sci. 77, 789–805. 10.1007/s00018-019-03420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee KS, Lin S, Copland DA, Dick AD, and Liu J (2021). Cellular senescence in the aging retina and developments of senotherapies for age-related macular degeneration. J. Neuroinflammation 18, 32. 10.1186/s12974-021-02088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choudhary M, Ismail EN, Yao PL, Tayyari F, Radu RA, Nusinowitz S, Boulton ME, Apte RS, Ruberti JW, Handa JT, et al. (2020). LXRs regulate features of age-related macular degeneration and may be a potential therapeutic target. JCI Insight 5, e131928. 10.1172/jci.insight.131928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tesoriere L, Attanzio A, Allegra M, Cilla A, Gentile C, and Livrea MA (2014). Oxysterol mixture in hypercholesterolemia-relevant proportion causes oxidative stress-dependent eryptosis. Cell. Physiol. Biochem. 34, 1075–1089. 10.1159/000366322. [DOI] [PubMed] [Google Scholar]
- 83.Dasari B, Prasanthi JRP, Marwarha G, Singh BB, and Ghribi O (2010). The oxysterol 27-hydroxycholesterol increases beta-amyloid and oxidative stress in retinal pigment epithelial cells. BMC Ophthalmol. 10, 22. 10.1186/1471-2415-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ravi S, Duraisamy P, Krishnan M, Martin LC, Manikandan B, Raman T, Sundaram J, Arumugam M, and Ramar M (2021). An insight on 7-ketocholesterol mediated inflammation in atherosclerosis and potential therapeutics. Steroids 172, 108854. 10.1016/j.steroids.2021.108854. [DOI] [PubMed] [Google Scholar]
- 85.de Medina P, Silvente-Poirot S, and Poirot M (2022). Oxysterols are potential physiological regulators of ageing. Ageing Res. Rev. 77, 101615. 10.1016/j.arr.2022.101615. [DOI] [PubMed] [Google Scholar]
- 86.Lin JB, Sene A, Santeford A, Fujiwara H, Sidhu R, Ligon MM, Shankar VA, Ban N, Mysorekar IU, Ory DS, and Apte RS (2018). Oxysterol Signatures Distinguish Age-Related Macular Degeneration from Physiologic Aging. EBioMedicine 32, 9–20. 10.1016/j.ebiom.2018.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tavazoie MF, Pollack I, Tanqueco R, Ostendorf BN, Reis BS, Gonsalves FC, Kurth I, Andreu-Agullo C, Derbyshire ML, Posada J, et al. (2018). LXR/ApoE Activation Restricts Innate Immune Suppression in Cancer. Cell 172, 825–840.e18. 10.1016/j.cell.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li B, Lian M, Li Y, Qian Q, Zhang J, Liu Q, Tang R, and Ma X (2021). Myeloid-Derived Suppressive Cells Deficient in Liver X Receptor alpha Protected From Autoimmune Hepatitis. Front. Immunol. 12, 732102. 10.3389/fimmu.2021.732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drach J, McQueen T, Engel H, Andreeff M, Robertson KA, Collins SJ, Malavasi F, and Mehta K (1994). Retinoic acid-induced expression of CD38 antigen in myeloid cells is mediated through retinoic acid receptor-alpha. Cancer Res. 54, 1746–1752. [PubMed] [Google Scholar]
- 90.Mehta K, McQueen T, Manshouri T, Andreeff M, Collins S, and Albitar M (1997). Involvement of retinoic acid receptor-alpha-mediated signaling pathway in induction of CD38 cell-surface antigen. Blood 89, 3607–3614. [PubMed] [Google Scholar]
- 91.Hogan KA, Chini CCS, and Chini EN (2019). The Multi-faceted Ecto-enzyme CD38: Roles in Immunomodulation, Cancer, Aging, and Metabolic Diseases. Front. Immunol. 10, 1187. 10.3389/fimmu.2019.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chini EN, Chini CCS, Espindola Netto JM, de Oliveira GC, and van Schooten W (2018). The Pharmacology of CD38/NADase: An Emerging Target in Cancer and Diseases of Aging. Trends Pharmacol. Sci. 39, 424–436. 10.1016/j.tips.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cui H, Xie N, Banerjee S, Dey T, Liu RM, Antony VB, Sanders YY, Adams TS, Gomez JL, Thannickal VJ, et al. (2022). CD38 Mediates Lung Fibrosis by Promoting Alveolar Epithelial Cell Aging. Am. J. Respir. Crit. Care Med. 206, 459–475. 10.1164/rccm.202109-2151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Camacho-Pereira J, Tarragó MG, Chini CCS, Nin V, Escande C, Warner GM, Puranik AS, Schoon RA, Reid JM, Galina A, and Chini EN (2016). CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. Cell Metabol. 23, 1127–1139. 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Covarrubias AJ, Kale A, Perrone R, Lopez-Dominguez JA, Pisco AO, Kasler HG, Schmidt MS, Heckenbach I, Kwok R, Wiley CD, et al. (2020). Senescent cells promote tissue NAD(+) decline during ageing via the activation of CD38(+) macrophages. Nat. Metab. 2, 1265–1283. 10.1038/s42255-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Q, Chen W, Cong L, Wang M, Li H, Wang H, Luo X, Zhu J, Zeng X, Zhu Z, et al. (2024). NADase CD38 is a key determinant of ovarian aging. Nat. Aging 4, 110–128. 10.1038/s43587-023-00532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoshino J, Baur JA, and Imai SI (2018). NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metabol. 27, 513–528. 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y, Redpath P, Migaud ME, Apte RS, Uchida K, et al. (2016). Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metabol. 24, 795–806. 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pencina KM, Valderrabano R, Wipper B, Orkaby AR, Reid KF, Storer T, Lin AP, Merugumala S, Wilson L, Latham N, et al. (2023). Nicotinamide Adenine Dinucleotide Augmentation in Overweight or Obese Middle-Aged and Older Adults: A Physiologic Study. J. Clin. Endocrinol. Metab. 108, 1968–1980. 10.1210/clinem/dgad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao H, Roychoudhury J, Doggett TA, Apte RS, and Ferguson TA (2013). Age-dependent changes in FasL (CD95L) modulate macrophage function in a model of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 54, 5321–5331. 10.1167/iovs.13-12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hsieh WY, Williams KJ, Su B, and Bensinger SJ (2021). Profiling of mouse macrophage lipidome using direct infusion shotgun mass spec-trometry. STAR Protoc. 2, 100235. 10.1016/j.xpro.2020.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Su B, Bettcher LF, Hsieh WY, Hornburg D, Pearson MJ, Blomberg N, Giera M, Snyder MP, Raftery D, Bensinger SJ, and Williams KJ (2021). A DMS Shotgun Lipidomics Workflow Application to Facilitate High-Throughput, Comprehensive Lipidomics. J. Am. Soc. Mass Spectrom. 32, 2655–2663. 10.1021/jasms.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, Blewitt ME, Asselin-Labat ML, Smyth GK, and Ritchie ME (2015). Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 43, e97. 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luo W, Friedman MS, Shedden K, Hankenson KD, and Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinf. 10, 161. 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luo W, and Brouwer C (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hikosaka K, Ikutani M, Shito M, Kazuma K, Gulshan M, Nagai Y, Takatsu K, Konno K, Tobe K, Kanno H, and Nakagawa T (2014). Deficiency of nicotinamide mononucleotide adenylyltransferase 3 (nmnat3) causes hemolytic anemia by altering the glycolytic flow in mature erythrocytes. J. Biol. Chem. 289, 14796–14811. 10.1074/jbc.M114.554378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sasaki Y, Nakagawa T, Mao X, DiAntonio A, and Milbrandt J (2016). NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD(+) depletion. Elife 5, e19749. 10.7554/eLife.19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hara N, Shibata T, Osago H, Yamada K, and Tsuchiya M (2014). Formation of [nicotinamide-(2)H(3)]NAD(+) from [(2)H(4)]nicotinamide and [(2)H(4)]nicotinic acid in human HepG2N cells and involvement of (2)H/(1)H exchange at the redox site of NAD(+)/NADH. J. Nutr. Sci. Vitaminol. 60, 17–21. 10.3177/jnsv.60.17. [DOI] [PubMed] [Google Scholar]
- 111.Katz ML, and Robison WG Jr. (2002). What is lipofuscin? Defining characteristics and differentiation from other autofluorescent lysosomal storage bodies. Arch. Gerontol. Geriatr. 34, 169–184. 10.1016/s0167-4943(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 112.Pan C, Banerjee K, Lehmann GL, Almeida D, Hajjar KA, Benedicto I, Jiang Z, Radu RA, Thompson DH, Rodriguez-Boulan E, and Nociari MM (2021). Lipofuscin causes atypical necroptosis through lysosomal membrane permeabilization. Proc. Natl. Acad. Sci. USA. 118, e2100122118. 10.1073/pnas.2100122118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and lipidomics data have been deposited at Mendeley Data (codes are listed in Key Resources Table). These data are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-Iba1 | Wako | Cat#019-19741; RRID:AB_839504 |
| Rat monoclonal anti-F4/80 | abcam | Cat#ab6640; RRID:AB_1140040 |
| Sheep polyclonal anti-CD38 | R&D Systems | Cat#AF4947; RRID:AB_1241945 |
| Rabbit polyclonal anti-Ki67 | abcam | Cat#abcam15580; RRID;AB_443209 |
| Phalloidin-iFluor 647 | abcam | Cat#ab176759 |
| Alexa Fluor 647 donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711-605-152 |
| Alexa Fluor 647 donkey anti-rat IgG | Jackson ImmunoResearch | Cat# 712-605-153 |
| Alexa Fluor 488 donkey anti-sheep IgG | Jackson ImmunoResearch | Cat#713-545-147 |
| Goat polyclonal antibody to Biotin (FITC) | abcam | Cat#ab6550 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| pCMV6-XL6 | OriGene Technologies | Cat#PCMV6XL6 |
| pCMV6-AC-CD38-GFP | OriGene Technologies | Cat#MG204265 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DMEM | gibco | Cat#11965-084 |
| L-glutamate | gibco | Cat#25030-081 |
| Penicillin/streptomycin (P/S) | gibco | Cat#15140-122 |
| Accutase | Sigma-Aldrich | Cat#A6964 |
| Cholesterol | Sigma-Aldrich | Cat#C4951 |
| DNase | Sigma-Aldrich | Cat#D4527 |
| DNA digestion buffer | ZYMO RESEARCH | Cat#E1010-1 |
| High Capacity cDNA Reverse Transcription Kit | Applied biosystems | Cat#4368813 |
| TaqMan Fast Advanced Master Mix | Applied biosystems | Cat#4444554 |
| Nicotinamide-2,4,5,6-d4 | C/D/N Isotopes | Cat#D-3457 |
| Lipofectamine RNAiMAX transfection reagent | Thermo Fisher Scientific | Cat#13778150 |
| Lipofectamine 3000 transfection reagent | Thermo Fisher Scientific | Cat#L3000-008 |
| Opti-MEM | gibco | Cat#31895-070 |
| FK866 | Chem Cruz | Cat#sc-205325A |
| DMEM, high glucose | Sigma-Aldrich | Cat#D1145 |
| β-nicotinamide mononucleotide | Sigma-Aldrich | Cat#N3501 |
| GW3965 | Sigma-Aldrich | Cat#G6295 |
| 78c | MedChemExpress | Cat#HY-123999 |
| AP-20187 | APExBIO | Cat#B1274 |
| Dasatinib | Sigma-Aldrich | Cat#SML2589 |
| Quercetin | Sigma-Aldrich | Cat#Q4951 |
| Seahorse XF DMEM Medium | Agilent Technologies | Cat#103575-100 |
| Seahorse XF Cell Mito Stress Test kit | Agilent Technologies | Cat#103015-100 |
| Seahorse XF24 V7 PS Cell Culture Microplate | Agilent Technologies | Cat#100777-004 |
| Co-detection blocker | Advanced Cell Diagnostics | Cat#323170 |
| Co-detection antibody diluent | Advanced Cell Diagnostics | Cat#323160 |
| RNAscope probe Mm-Cdkn1a | Advanced Cell Diagnostics | Cat#408551 |
| RNAscope probe Mm-Cdkn2a | Advanced Cell Diagnostics | Cat#411011 |
| RNAscope 2.5 HD detection reagents- red | Advanced Cell Diagnostics | Cat#322360 |
| SenTraGor Cell Senescence Reagent | Cayman chemical | Cat#35568 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat#23225 |
| NBD Cholesterol Staining Dye Kit | abcam | Cat#ab269448 |
| Senescence β-Galactosidase staining kit | Cell Signaling | Cat#9860 |
| SPiDER β -galactosidase staining kit | DOJINDO | Cat#SG02 |
| Lysosome intracellular activity assay kit | abcam | Cat#ab234622 |
| TaqMan array standard plates | Thermo Scientific | Cat#4391524 |
| Click-iT™ plus TUNEL Assay kits for in situ apoptosis eetection | Invitrogen | Cat#C10618 |
| Fluorescent cell linker kit for general cell membrane labeling | Sigma-Aldrich | Cat# PKH26, PKH67 |
|
| ||
| Deposited data | ||
|
| ||
| The lipidomics data of BMDMs from Abca1/g1 -m/-m and Abca1/g1 f/f | This study | Mendeley Data: https://doi.org/10.17632/gd4mprzbvh.1 |
| The RNA sequence data for BMDMs from Abca1/g1 -m/-m and Abca1/g1 f/f | This study | GEO: GSE234507 |
| The lipidomics data of retina isolated from young and old mice | This study | Mendeley Data: https://doi.org/10.17632/j9gv8x9td7.1 |
| The RNA sequence data for retinal pigment epithelium/choroid tissue of Abca1/g1 -m/-m and INK-ATTAC-Abca1/g1 -m/-m | This study | GEO: GSE234641 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: Abca1fl Abcg1fl: B6.Cg-Abca1tm1Jp Abcg1tm1Tall/J | The Jackson Laboratory | RRID: IMSR_JAX:021067 |
| Mouse: LysM Cre: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | RRID: IMSR_JAX:004781 |
| Mouse: INK-ATTAC | Unity Biotechnology | N/A |
| Mouse: Cx3cr1 CreERT2: B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ | The Jackson Laboratory | RRID: IMSR_JAX:021160 |
| Mouse: Tmem119 Cre ERT2: C57BL/6-Tmem119em1(cre/ERT2)Gfng/J | The Jackson Laboratory | RRID: IMSR_JAX:031820 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for qPCR, see Table S1 | Thermo Scientific | N/A |
| AllStars Negative Control siRNA | QIAGEN | Cat#1027281 |
| siRNA targeting Nr1h3 (NM_013839) | QIAGEN | Cat#SI02740339 |
| siRNA targeting Nr1h2 (NM_009473) | QIAGEN | Cat#SI00185241 |
| siRNA targeting Cd38 (NM_007646) | QIAGEN | Cat#SI02666398 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LipidSig | N/A | http://chenglab.cmu.edu.tw/lipidsig/ |
| CellProfiler | https://cellprofiler.org | |
| MassHunter Quantitative Analysis | Agilent Technologies | https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software/ |
| FlowJo | BD Biosciences | https://www.flowjo.com/solutions/flowjo |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/download.html |
| Sample Size Calculator | ClinCalc | https://clincalc.com/stats/samplesize.aspx. |


