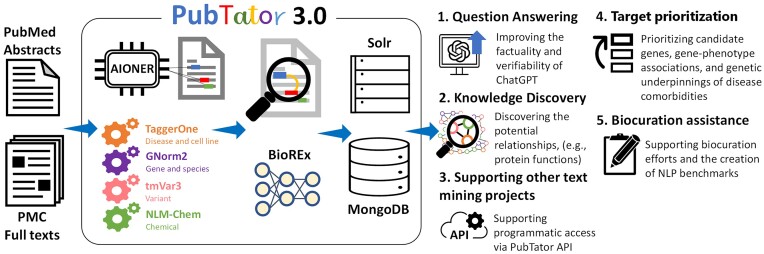
Abstract
PubTator 3.0 (https://www.ncbi.nlm.nih.gov/research/pubtator3/) is a biomedical literature resource using state-of-the-art AI techniques to offer semantic and relation searches for key concepts like proteins, genetic variants, diseases and chemicals. It currently provides over one billion entity and relation annotations across approximately 36 million PubMed abstracts and 6 million full-text articles from the PMC open access subset, updated weekly. PubTator 3.0's online interface and API utilize these precomputed entity relations and synonyms to provide advanced search capabilities and enable large-scale analyses, streamlining many complex information needs. We showcase the retrieval quality of PubTator 3.0 using a series of entity pair queries, demonstrating that PubTator 3.0 retrieves a greater number of articles than either PubMed or Google Scholar, with higher precision in the top 20 results. We further show that integrating ChatGPT (GPT-4) with PubTator APIs dramatically improves the factuality and verifiability of its responses. In summary, PubTator 3.0 offers a comprehensive set of features and tools that allow researchers to navigate the ever-expanding wealth of biomedical literature, expediting research and unlocking valuable insights for scientific discovery.
Graphical Abstract
Graphical Abstract.
Introduction
The biomedical literature is a primary resource to address information needs across the biological and clinical sciences (1), however the requirements for literature search vary widely. Activities such as formulating a research hypothesis require an exploratory approach, whereas tasks like interpreting the clinical significance of genetic variants are more focused.
Traditional keyword-based search methods have long formed the foundation of biomedical literature search (2). While generally effective for basic search, these methods also have significant limitations, such as missing relevant articles due to differing terminology or including irrelevant articles because surface-level term matches cannot adequately represent the required association between query terms. These limitations cost time and risk information needs remaining unmet.
Natural language processing (NLP) methods provide substantial value for creating bioinformatics resources (3–5), and may improve literature search by enabling semantic and relation search (6). In semantic search, users indicate specific concepts of interest (entities) for which the system has precomputed matches regardless of the terminology used. Relation search increases precision by allowing users to specify the type of relationship desired between entities, such as whether a chemical enhances or reduces expression of a gene. In this regard, we present PubTator 3.0, a novel resource engineered to support semantic and relation search in the biomedical literature. Its search capabilities allow users to explore automated entity annotations for six key biomedical entities: genes, diseases, chemicals, genetic variants, species, and cell lines. PubTator 3.0 also identifies and makes searchable 12 common types of relations between entities, enhancing its utility for both targeted and exploratory searches. Focusing on relations and entity types of interest across the biomedical sciences allows PubTator 3.0 to retrieve information precisely while providing broad utility (see detailed comparisons with its predecessor in Supplementary Table S1).
System overview
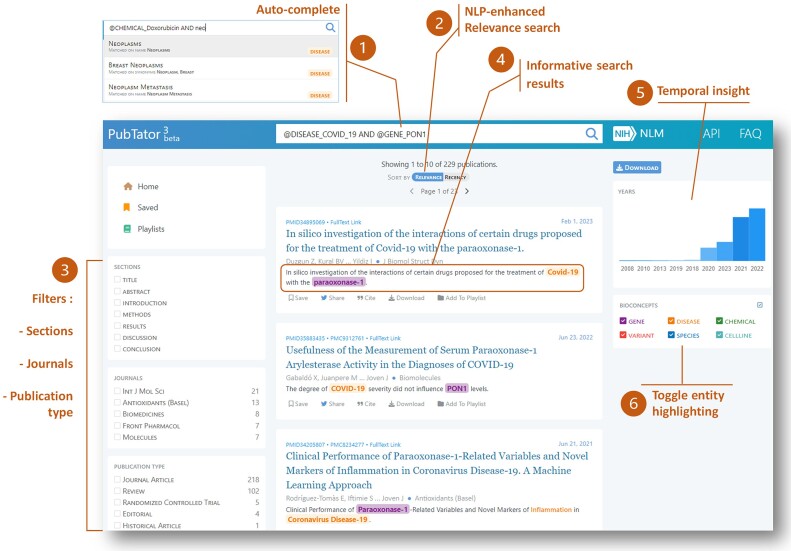
The PubTator 3.0 online interface, illustrated in Figure 1 and Supplementary Figure S1, is designed for interactive literature exploration, supporting semantic, relation, keyword, and Boolean queries. An auto-complete function provides semantic search suggestions to assist users with query formulation. For example, it automatically suggests replacing either ‘COVID-19″ or "SARS-CoV-2 infection’ with the semantic term ‘@DISEASE_COVID_19″. Relation queries – new to PubTator 3.0 – provide increased precision, allowing users to target articles which discuss specific relationships between entities.
Figure 1.
PubTator 3.0 system overview and search results page: 1. Query auto-complete enhances search accuracy and synonym matching. 2. Natural language processing (NLP)-enhanced relevance: Search results are prioritized according to the strength of the relationship between the entities queried. 3. Users can further refine results with facet filters—section, journal and type. 4. Search results include highlighted entity snippets explaining relevance. 5. Histogram visualizes number of results by publication year. 6. Entity highlighting can be switched on or off according to user preference.
PubTator 3.0 offers unified search results, simultaneously searching approximately 36 million PubMed abstracts and over 6 million full-text articles from the PMC Open Access Subset (PMC-OA), improving access to the substantial amount of relevant information present in the article full text (7). Search results are prioritized based on the depth of the relationship between the query terms: articles containing identifiable relations between semantic terms receive the highest priority, while articles where semantic or keyword terms co-occur nearby (e.g. within the same sentence) receive secondary priority. Search results are also prioritized based on the article section where the match appears (e.g. matches within the title receive higher priority). Users can further refine results by employing filters, narrowing articles returned to specific publication types, journals, or article sections.
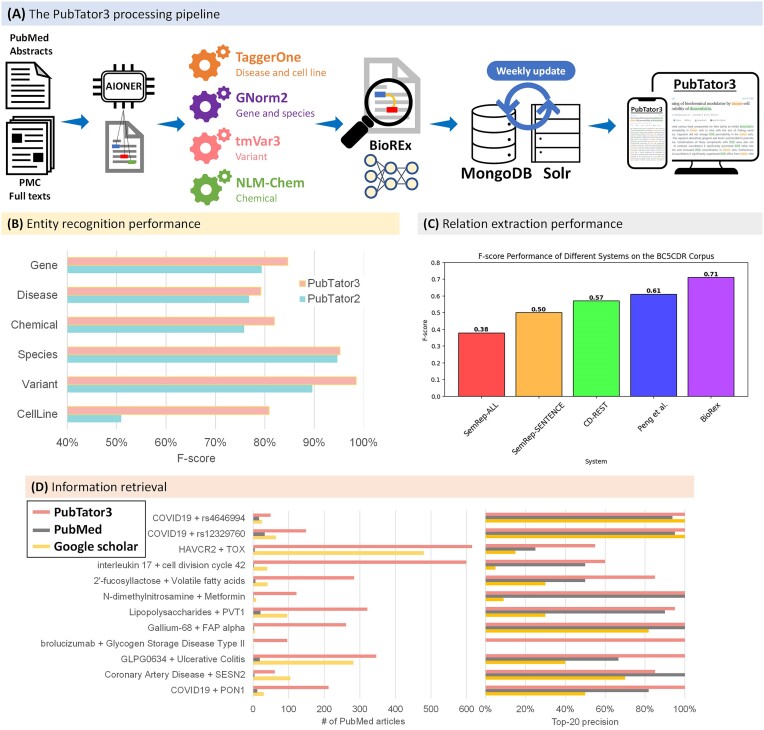
PubTator 3.0 is supported by an NLP pipeline, depicted in Figure 2A. This pipeline, run weekly, first identifies articles newly added to PubMed and PMC-OA. Articles are then processed through three major steps: (i) named entity recognition, provided by the recently developed deep-learning transformer model AIONER (8), (ii) identifier mapping and (iii) relation extraction, performed by BioREx (9) of 12 common types of relations (described in Supplementary Table S2).
Figure 2.
(A) The PubTator 3.0 processing pipeline: AIONER (8) identifies six types of entities in PubMed abstracts and PMC-OA full-text articles. Entity annotations are associated with database identifiers by specialized mappers and BioREx (9) identifies relations between entities. Extracted data is stored in MongoDB and made searchable using Solr. (B) Entity recognition performance for each entity type compared with PubTator2 (also known as PubTatorCentral) (13) on the BioRED corpus (15). (C) Relation extraction performance compared with SemRep (11) and notable previous best systems (12,13) on the BioCreative V Chemical-Disease Relation (14) corpus. (D) Comparison of information retrieval for PubTator 3.0, PubMed, and Google Scholar for entity pair queries, with respect to total article count and top-20 article precision.
In total, PubTator 3.0 contains over 1.6 billion entity annotations (4.6 million unique identifiers) and 33 million relations (8.8 million unique pairs). It provides enhanced entity recognition and normalization performance over its previous version, PubTator 2 (10), also known as PubTator Central (Figure 2B and Supplementary Table S3). We show the relation extraction performance of PubTator 3.0 in Figure 2C and its comparison results to the previous state-of-the-art systems (11–13) on the BioCreative V Chemical-Disease Relation (14) corpus, finding that PubTator 3.0 provided substantially higher accuracy. Moreover, when evaluating a randomized sample of entity pair queries compared to PubMed and Google Scholar, PubTator 3.0 consistently returns a greater number of articles with higher precision in the top 20 results (Figure 2D and Supplementary Table S4).
Materials and methods
Data sources and article processing
PubTator 3.0 downloads new articles weekly from the BioC PubMed API (https://www.ncbi.nlm.nih.gov/research/bionlp/APIs/BioC-PubMed/) and the BioC PMC API (https://www.ncbi.nlm.nih.gov/research/bionlp/APIs/BioC-PMC/) in BioC-XML format (16). Local abbreviations are identified using Ab3P (17). Article text and extracted data are stored internally using MongoDB and indexed for search with Solr, ensuring robust and scalable accessibility unconstrained by external dependencies such as the NCBI eUtils API.
Entity recognition and normalization/linking
PubTator 3.0 uses AIONER (8), a recently developed named entity recognition (NER) model, to recognize entities of six types: genes/proteins, chemicals, diseases, species, genetic variants, and cell lines. AIONER utilizes a flexible tagging scheme to integrate training data created separately into a single resource. These training datasets include NLM-Gene (18), NLM-Chem (19), NCBI-Disease (20), BC5CDR (14), tmVar3 (21), Species-800 (22), BioID (23) and BioRED (15). This consolidation creates a larger training set, improving the model's ability to generalize to unseen data. Furthermore, it enables recognizing multiple entity types simultaneously, enhancing efficiency and simplifying the challenge of distinguishing boundaries between entities that reference others, such as the disorder ‘Alpha-1 antitrypsin deficiency’ and the protein ‘Alpha-1 antitrypsin’. We previously evaluated the performance of AIONER on 14 benchmark datasets (8), including the test sets for the aforementioned training sets. This evaluation demonstrated that AIONER’s performance surpasses or matches previous state-of-the-art methods.
Entity mentions found by AIONER are normalized (linked) to a unique identifier in an appropriate entity database. Normalization is performed by a module designed for (or adapted to) each entity type, using the latest version. The recently-upgraded GNorm2 system (24) normalizes genes to NCBI Gene identifiers and species mentions to NCBI Taxonomy. tmVar3 (21), also recently upgraded, normalizes genetic variants; it uses dbSNP identifiers for variants listed in dbSNP and HGNV format otherwise. Chemicals are normalized by the NLM-Chem tagger (19) to MeSH identifiers (25). TaggerOne (26) normalizes diseases to MeSH and cell lines to Cellosaurus (27) using a new normalization-only mode. This mode only applies the normalization model, which converts both mentions and lexicon names into high-dimensional TF-IDF vectors and learns a mapping, as before. However, it now augments the training data by mapping each lexicon name to itself, resulting in a large performance improvement for names present in the lexicon but not in the annotated training data. These enhancements provide a significant overall improvement in entity normalization performance (Supplementary Table S3).
Relation extraction
Relations for PubTator 3.0 are extracted by the unified relation extraction model BioREx (9), designed to simultaneously extract 12 types of relations across eight entity type pairs: chemical–chemical, chemical–disease, chemical–gene, chemical–variant, disease–gene, disease–variant, gene–gene and variant–variant. Detailed definitions of these relation types and their corresponding entity pairs are presented in Supplementary Table S2. Deep-learning methods for relation extraction, such as BioREx, require ample training data. However, training data for relation extraction is fragmented into many datasets, often tailored to specific entity pairs. BioREx overcomes this limitation with a data-centric approach, reconciling discrepancies between disparate training datasets to construct a comprehensive, unified dataset.
We evaluated the relations extracted by BioREx using performance on manually annotated relation extraction datasets as well as a comparative analysis between BioREx and notable comparable systems. BioREx established a new performance benchmark on the BioRED corpus test set (15), elevating the performance from 74.4% (F-score) to 79.6%, and demonstrating higher performance than alternative models such as transfer learning (TL), multi-task learning (MTL), and state-of-the-art models trained on isolated datasets (9). For PubTator 3.0, we replaced its deep learning module, PubMedBERT (28), with LinkBERT (29), further increasing the performance to 82.0%. Furthermore, we conducted a comparative analysis between BioREx and SemRep (11), a widely used rule-based method for extracting diverse relations, the CD-REST (13) system, and the previous state-of-the-art system (12), using the BioCreative V Chemical Disease Relation corpus test set (14). Our evaluation demonstrated that PubTator 3.0 provided substantially higher F-score than previous methods.
Programmatic access and data formats
PubTator 3.0 offers programmatic access through its API and bulk download. The API (https://www.ncbi.nlm.nih.gov/research/pubtator3/) supports keyword, entity and relation search, and also supports exporting annotations in XML and JSON-based BioC (16) formats and tab-delimited free text. The PubTator 3.0 FTP site (https://ftp.ncbi.nlm.nih.gov/pub/lu/PubTator3) provides bulk downloads of annotated articles and extraction summaries for entities and relations. Programmatic access supports more flexible query options; for example, the information need ‘what chemicals reduce expression of JAK1?’ can be answered directly via API (e.g. https://www.ncbi.nlm.nih.gov/research/pubtator3-api/relations?e1=@GENE_JAK1&type=negative_correlate&e2=Chemical) or by filtering the bulk relations file. Additionally, the PubTator 3.0 API supports annotation of user-defined free text.
Case study I: entity relation queries
We analyzed the retrieval quality of PubTator 3.0 by preparing a series of 12 entity pairs to serve as case studies for comparison between PubTator 3.0, PubMed and Google Scholar. To provide an equal comparison, we filtered about 30% of the Google Scholar results for articles not present in PubMed. To ensure that the number of results would remain low enough to allow filtering Google Scholar results for articles not in PubMed, we identified entity pairs first discussed together in the literature in 2022 or later. We then randomly selected two entity pairs of each of the following types: disease/gene, chemical/disease, chemical/gene, chemical/chemical, gene/gene and disease/variant. None of the relation pairs selected appears in the training set. The comparison was performed with respect to a snapshot of the search results returned by all search engines on 19 May 2023. We manually evaluated the top 20 results for each system and each query; articles were judged to be relevant if they mentioned both entities in the query and supported a relationship between them. Two curators independently judged each article, and discrepancies were discussed until agreement. The curators were not blinded to the retrieval method but were required to record the text supporting the relationship, if relevant. This experiment evaluated the relevance of the top 20 results for each retrieval method, regardless of whether the article appeared in PubMed.
Our analysis is summarized in Figure 2D, and Supplementary Table S4 presents a detailed comparison of the quality of retrieved results between PubTator 3.0, PubMed and Google Scholar. Our results demonstrate that PubTator 3.0 retrieves a greater number of articles than the comparison systems and its precision is higher for the top 20 results. For instance, PubTator 3.0 returned 346 articles for the query ‘GLPG0634 + ulcerative colitis’, and manual review of the top 20 articles showed that all contained statements about an association between GLPG0634 and ulcerative colitis. In contrast, PubMed only returned a total of 18 articles, with only 12 mentioning an association. Moreover, when searching for ‘COVID19 + PON1’, PubTator 3.0 returns 212 articles in PubMed, surpassing the 43 articles obtained from Google Scholar, only 29 of which are sourced from PubMed. These disparities can be attributed to several factors: (i) PubTator 3.0's search includes full texts available in PMC-OA, resulting in significantly broader coverage of articles, (ii) entity normalization improves recall, for example, by matching ‘paraoxonase 1’ to ‘PON1’, (iii) PubTator 3.0 prioritizes articles containing relations between the query entities, (iv) Pubtator 3.0 prioritizes articles where the entities appear nearby, rather than distant paragraphs. Across the 12 information retrieval case studies, PubTator 3.0 demonstrated an overall precision of 90.0% for the top 20 articles (216 out of 240), which is significantly higher than PubMed's precision of 81.6% (84 out of 103) and Google Scholar's precision of 48.5% (98 out of 202).
Case study II: retrieval-augmented generation
In the era of large language models (LLMs), PubTator 3.0 can also enhance their factual accuracy via retrieval augmented generation. Despite their strong language ability, LLMs are prone to generating incorrect assertions, sometimes known as hallucinations (30,31). For example, when requested to cite sources for questions such as ‘which diseases can doxorubicin treat’, GPT-4 frequently provides seemingly plausible but nonexistent references. Augmenting GPT-4 with PubTator 3.0 APIs can anchor the model's response to verifiable references via the extracted relations, significantly reducing hallucinations.
We assessed the citation accuracy of responses from three GPT-4 variations: PubTator-augmented GPT-4, PubMed-augmented GPT-4 and standard GPT-4. We performed a qualitative evaluation based on eight questions selected as follows. We identified entities mentioned in the PubMed query logs and randomly selected from entities searched both frequently and rarely. We then identified the common queries for each entity that request relational information and adapted one into a natural language question. Each question is therefore grounded on common information needs of real PubMed users. For example, the questions ‘What can be caused by tocilizumab?’ and ‘What can be treated by doxorubicin?’ are adapted from the user queries ‘tocilizumab side effects’ and ‘doxorubicin treatment’ respectively. Such questions typically require extracting information from multiple articles and an understanding of biomedical entities and relationship descriptions. Supplementary Table S5 lists the questions chosen.
We augmented the GPT-4 large language model (LLM) with PubTator 3.0 via the function calling mechanism of the OpenAI ChatCompletion API. This integration involved prompting GPT-4 with descriptions of three PubTator APIs: (i) find entity ID, which retrieves PubTator entity identifiers; (ii) find related entities, which identifies related entities based on an input entity and specified relations and (iii) export relevant search results, which returns PubMed article identifiers containing textual evidence for specific entity relationships. Our instructions prompted GPT-4 to decompose user questions into sub-questions addressable by these APIs, execute the function calls, and synthesize the responses into a coherent final answer. Our prompt promoted a summarized response by instructing GPT-4 to start its message with ‘Summary:’ and requested the response include citations to the articles providing evidence. The PubMed augmentation experiments provided GPT-4 with access to PubMed database search via the National Center for Biotechnology Information (NCBI) E-utils APIs (32). We used Azure OpenAI Services (version 2023-07-01-preview) and GPT-4 (version 2023-06-13) and set the decoding temperature to zero to obtain deterministic outputs. The full prompts are provided in Supplementary Table S6.
PubTator-augmented GPT-4 generally processed the questions in three steps: (i) finding the standard entity identifiers, (ii) finding its related entity identifiers and (iii) searching PubMed articles. For example, to answer ‘What drugs can treat breast cancer?’, GPT-4 first found the PubTator entity identifier for breast cancer (@DISEASE_Breast_Cancer) using the Find Entity ID API. It then used the Find Related Entities API to identify entities related to @DISEASE_Breast_Cancer through a ‘treat’ relation. For demonstration purposes, we limited the maximum number of output entities to five. Finally, GPT-4 called the Export Relevant Search Results API for the PubMed article identifiers containing evidence for these relationships. The raw responses to each prompt for each method are provided in Supplementary Table S6.
We manually evaluated the accuracy of the citations in the responses by reviewing each PubMed article and verifying whether each PubMed article cited supported the stated relationship (e.g. Tamoxifen treating breast cancer). Supplementary Table S5 reports the proportion of the cited articles with valid supporting evidence for each method. GPT-4 frequently generated fabricated citations, widely known as the hallucination issue. While PubMed-augmented GPT-4 showed a higher proportion of accurate citations, some articles cited did not support the relation claims. This is likely because PubMed is based on keyword and Boolean search and does not support queries for specific relationships. Responses generated by PubTator-augmented GPT-4 demonstrated the highest level of citation accuracy, underscoring the potential of PubTator 3.0 as a high-quality knowledge source for addressing biomedical information needs through retrieval-augmented generation with LLMs such as GPT-4. In our experiment, using Azure for ChatGPT, the cost was approximately $1 for two questions with GPT-4-Turbo, or 40 questions when downgraded to GPT-3.5-Turbo, including the cost of input/output tokens.
Discussion
Previous versions of PubTator have fulfilled over one billion API requests since 2015, supporting a wide range of research applications. Numerous studies have harnessed PubTator annotations for disease-specific gene research, including efforts to prioritize candidate genes (33), determine gene–phenotype associations (34), and identify the genetic underpinnings of disease comorbidities (35). Several projects have used PubTator to create gene and genetic variant resources (36,37) or to enrich disease knowledge graphs (38,39). Moreover, PubTator has supported biocuration efforts (40,41) and the creation of NLP benchmarks (42). With enhanced accuracy, PubTator 3.0 will better support these use cases.
Introducing relation annotations to PubTator 3.0 opens novel avenues for expanded use scenarios. With relations precomputed from the literature, complex research questions can often be answered directly. Drug repurposing, for example, can be formulated as identifying chemicals which target specific genes. Conversely, determining the genetic targets of a chemical can be achieved by querying the same chemical/gene relations. Clinicians evaluating genetic variants, e.g. for rare diseases or personalized medicine, may explore the relationships between specific genetic variants and disease. Biologists, on the other hand, may utilize interactions between multiple genes to assemble complex molecular pathways.
There are several notable limitations for PubTator 3.0. Although it is capable of extracting relations from full-text articles, this feature is currently restricted to abstracts due to computational constraints. However, the system has been designed to support full-text relation extraction in a future enhancement. The current system only extracts 12 relation types, though these represent common uses. Finally, entity annotation and relation extraction are automated; though these systems exhibit high performance, their accuracy remains imperfect.
Conclusion
PubTator 3.0 offers a comprehensive set of features and tools that allow researchers to navigate the ever-expanding wealth of biomedical literature, expediting research and unlocking valuable insights for scientific discovery. The PubTator 3.0 interface, API, and bulk file downloads are available at https://www.ncbi.nlm.nih.gov/research/pubtator3/.
Supplementary Material
Notes
Present address: Alexis Allot, The Neuro (Montreal Neurological Institute-Hospital), McGill University, Montreal, Quebec H3A 2B4, Canada.
Present address: Ling Luo, School of Computer Science and Technology, Dalian University of Technology, 116024 Dalian, China.
Present address: Qingyu Chen, Biomedical Informatics and Data Science, Yale School of Medicine, New Haven, CT 06510, USA.
Contributor Information
Chih-Hsuan Wei, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Alexis Allot, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Po-Ting Lai, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Robert Leaman, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Shubo Tian, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Ling Luo, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Qiao Jin, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Zhizheng Wang, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Qingyu Chen, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Zhiyong Lu, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Data availability
Data is available through the online interface at https://www.ncbi.nlm.nih.gov/research/pubtator3/, through the API at https://www.ncbi.nlm.nih.gov/research/pubtator3/api or bulk FTP download at https://ftp.ncbi.nlm.nih.gov/pub/lu/PubTator3/.
The source code for each component of PubTator 3.0 is openly accessible. The AIONER named entity recognizer is available at https://github.com/ncbi/AIONER. GNorm2, for gene name normalization, is available at https://github.com/ncbi/GNorm2. The tmVar3 variant name normalizer is available at https://github.com/ncbi/tmVar3. The NLM-Chem Tagger, for chemical name normalization, is available at https://ftp.ncbi.nlm.nih.gov/pub/lu/NLMChem. The TaggerOne system, for disease and cell line normalization, is available at https://www.ncbi.nlm.nih.gov/research/bionlp/Tools/taggerone. The BioREx relation extraction system is available at https://github.com/ncbi/BioREx. The code for customizing ChatGPT with the PubTator 3.0 API is available at https://github.com/ncbi-nlp/pubtator-gpt. The details of the applications, performance, evaluation data, and citations for each tool are shown in Supplementary Table S7. All source code is also available at https://doi.org/10.5281/zenodo.10839630.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Intramural Research Program of the National Library of Medicine (NLM), National Institutes of Health; ODSS Support of the Exploration of Cloud in NIH Intramural Research. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
References
- 1. Lindberg D.A., Humphreys B.L. Rising expectations: access to biomedical information. Yearb Med. Inform. 2008; 3:165–172. [PMC free article] [PubMed] [Google Scholar]
- 2. Jin Q., Leaman R., Lu Z. PubMed and beyond: biomedical literature search in the age of artificial intelligence. EBioMedicine. 2024; 100:104988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rzhetsky A., Seringhaus M., Gerstein M. Seeking a new biology through text mining. Cell. 2008; 134:9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mayers M., Li T.S., Queralt-Rosinach N., Su A.I. Time-resolved evaluation of compound repositioning predictions on a text-mined knowledge network. BMC Bioinf. 2019; 20:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao S., Su C., Lu Z., Wang F. Recent advances in biomedical literature mining. Brief Bioinform. 2021; 22:bbaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li P.-H., Chen T.-F., Yu J.-Y., Shih S.-H., Su C.-H., Lin Y.-H., Tsai H.-K., Juan H.-F., Chen C.-Y., Huang J.-H. pubmedKB: an interactive web server for exploring biomedical entity relations in the biomedical literature. NucleicAcids Res. 2022; 50:W616–W622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Westergaard D., Staerfeldt H.H., Tonsberg C., Jensen L.J., Brunak S. A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLoS Comput. Biol. 2018; 14:e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo L., Wei C.-H., Lai P.-T., Leaman R., Chen Q., Lu Z. AIONER: all-in-one scheme-based biomedical named entity recognition using deep learning. Bioinformatics. 2023; 39:btad310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lai P.T., Wei C.H., Luo L., Chen Q., Lu Z. BioREx: improving biomedical relation extraction by leveraging heterogeneous datasets. J. Biomed. Inform. 2023; 146:104487. [DOI] [PubMed] [Google Scholar]
- 10. Wei C.-H., Allot A., Leaman R., Lu Z. PubTator central: automated concept annotation for biomedical full text articles. NucleicAcids Res. 2019; 47:W587–W593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kilicoglu H., Rosemblat G., Fiszman M., Shin D Broad-coverage biomedical relation extraction with SemRep. BMC Bioinf. 2020; 21:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng Y., Wei C.-H., Lu Z. Improving chemical disease relation extraction with rich features and weakly labeled data. J. Cheminformatics. 2016; 8:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu J., Wu Y., Zhang Y., Wang J., Lee H.J., Xu H. CD-REST: a system for extracting chemical-induced disease relation in literature. Database. 2016; 2016:baw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J., Sun Y., Johnson R.J., Sciaky D., Wei C.-H., Leaman R., Davis A.P., Mattingly C.J., Wiegers T.C., Lu Z. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database. 2016; 2016:baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luo L., Lai P.-T., Wei C.-H., Arighi C.N., Lu Z. BioRED: a Rich Biomedical Relation Extraction Dataset. Brief. Bioinf. 2022; 23:bbac282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Comeau D.C., Islamaj Doğan R., Ciccarese P., Cohen K.B., Krallinger M., Leitner F., Lu Z., Peng Y., Rinaldi F., Torii M.J.D. BioC: a minimalist approach to interoperability for biomedical text processing. Database. 2013; 2013:bat064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sohn S., Comeau D.C., Kim W., Wilbur W.J. Abbreviation definition identification based on automatic precision estimates. BMC Bioinf. 2008; 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Islamaj R., Wei C.-H., Cissel D., Miliaras N., Printseva O., Rodionov O., Sekiya K., Ward J., Lu Z. NLM-Gene, a richly annotated gold standard dataset for gene entities that addresses ambiguity and multi-species gene recognition. Sci. Data. 2021; 118:103779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Islamaj R., Leaman R., Kim S., Kwon D., Wei C.-H., Comeau D.C., Peng Y., Cissel D., Coss C., Fisher C. NLM-Chem, a new resource for chemical entity recognition in PubMed full text literature. Sci. Data. 2021; 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doğan R.I., Leaman R., Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014; 47:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei C.-H., Allot A., Riehle K., Milosavljevic A., Lu Z. tmVar 3.0: an improved variant concept recognition and normalization tool. Bioinformatics. 2022; 38:4449–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pafilis E., Frankild S.P., Fanini L., Faulwetter S., Pavloudi C., Vasileiadou A., Arvanitidis C., Jensen L.J. The SPECIES and ORGANISMS resources for fast and accurate identification of taxonomic names in text. PLoS One. 2013; 8:e65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arighi C., Hirschman L., Lemberger T., Bayer S., Liechti R., Comeau D., Wu C. Bio-ID track overview. BioCreative VI Challenge Evaluation Workshop. 2017; 14–19. [Google Scholar]
- 24. Wei C.H., Luo L., Islamaj R., Lai P.T., Lu Z. GNorm2: an improved gene name recognition and normalization system. Bioinformatics. 2023; 39:btad599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lipscomb C.E. Medical subject headings (MeSH). Bull. Med. Libr. Assoc. 2000; 88:265. [PMC free article] [PubMed] [Google Scholar]
- 26. Leaman R., Lu Z. TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Bioinformatics. 2016; 32:2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bairoch A. The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gu Y., Tinn R., Cheng H., Lucas M., Usuyama N., Liu X., Naumann T., Gao J., Poon H. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthcare. 2021; 3:1–23. [Google Scholar]
- 29. Yasunaga M., Leskovec J., Liang P. LinkBERT: Pretraining Language Models with Document Links. Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). 2022; 8003–8016. [Google Scholar]
- 30. Jin Q., Leaman R., Lu Z. Retrieve, summarize, and verify: how will ChatGPT affect information seeking from the medical literature?. J. Am. Soc. Nephrol. 2023; 34:1302–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tian S., Jin Q., Yeganova L., Lai P.T., Zhu Q., Chen X., Yang Y., Chen Q., Kim W., Comeau D.C. et al. Opportunities and challenges for ChatGPT and large language models in biomedicine and health. Brief Bioinform. 2023; 25:bbad493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sayers E. Entrez Programming Utilities Help. 2010; Bethesda, MD: National Center for Biotechnology Information (US). [Google Scholar]
- 33. Lieberwirth J.K., Buttner B., Klockner C., Platzer K., Popp B., Abou Jamra R. AutoCaSc: prioritizing candidate genes for neurodevelopmental disorders. Hum. Mutat. 2022; 43:1795–1807. [DOI] [PubMed] [Google Scholar]
- 34. Buch A.M., Vertes P.E., Seidlitz J., Kim S.H., Grosenick L., Liston C. Molecular and network-level mechanisms explaining individual differences in autism spectrum disorder. Nat. Neurosci. 2023; 26:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinto B.G.G., Oliveira A.E.R., Singh Y., Jimenez L., Goncalves A.N.A., Ogava R.L.T., Creighton R., Schatzmann Peron J.P., Nakaya H.I. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 2020; 222:556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitsuhashi N., Toyo-Oka L., Katayama T., Kawashima M., Kawashima S., Miyazaki K., Takagi T. TogoVar: a comprehensive Japanese genetic variation database. Hum. Genome Var. 2022; 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang J., Yuan J., Hu Z., Zhang Y., Zhang T., Xu M., Long M., Fan Y., Tanyi J.L., Montone K.T. et al. Systematic illumination of druggable genes in cancer genomes. Cell Rep. 2022; 38:110400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pu Y., Beck D., Verspoor K. Graph embedding-based link prediction for literature-based discovery in Alzheimer's disease. J. Biomed. Inform. 2023; 145:104464. [DOI] [PubMed] [Google Scholar]
- 39. Chen C., Ross K.E., Gavali S., Cowart J.E., Wu C.H. COVID-19 Knowledge Graph from semantic integration of biomedical literature and databases. Bioinformatics. 2021; 37:4597–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lou P., Jimeno Yepes A., Zhang Z., Zheng Q., Zhang X., Li C. BioNorm: deep learning-based event normalization for the curation of reaction databases. Bioinformatics. 2020; 36:611–620. [DOI] [PubMed] [Google Scholar]
- 41. Percha B., Altman R.B. A global network of biomedical relationships derived from text. Bioinformatics. 2018; 34:2614–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Legrand J., Gogdemir R., Bousquet C., Dalleau K., Devignes M.-D., Digan W., Lee C.-J., Ndiaye N.-C., Petitpain N., Ringot P. PGxCorpus, a manually annotated corpus for pharmacogenomics. Sci. Data. 2020; 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available through the online interface at https://www.ncbi.nlm.nih.gov/research/pubtator3/, through the API at https://www.ncbi.nlm.nih.gov/research/pubtator3/api or bulk FTP download at https://ftp.ncbi.nlm.nih.gov/pub/lu/PubTator3/.
The source code for each component of PubTator 3.0 is openly accessible. The AIONER named entity recognizer is available at https://github.com/ncbi/AIONER. GNorm2, for gene name normalization, is available at https://github.com/ncbi/GNorm2. The tmVar3 variant name normalizer is available at https://github.com/ncbi/tmVar3. The NLM-Chem Tagger, for chemical name normalization, is available at https://ftp.ncbi.nlm.nih.gov/pub/lu/NLMChem. The TaggerOne system, for disease and cell line normalization, is available at https://www.ncbi.nlm.nih.gov/research/bionlp/Tools/taggerone. The BioREx relation extraction system is available at https://github.com/ncbi/BioREx. The code for customizing ChatGPT with the PubTator 3.0 API is available at https://github.com/ncbi-nlp/pubtator-gpt. The details of the applications, performance, evaluation data, and citations for each tool are shown in Supplementary Table S7. All source code is also available at https://doi.org/10.5281/zenodo.10839630.