Abstract
We identified an interferon regulatory factor motif (IRF-E) upstream of an NF-κB binding site in the interleukin-15 (IL-15) promoter. Since these two motifs are part of the virus-inducible enhancer region of the beta interferon promoter, we speculated that there might be similar responses of these two genes to stimuli such as viruses. To test this hypothesis, L929 cells were infected with Newcastle disease virus (NDV), which led to the induction of IL-15 mRNA and protein expression. Using IL-15 promoter-reporter deletion constructs, a virus-inducible region, encompassing IRF-E, NF-κB, and a 13-nucleotide sequence flanked by these two motifs, was mapped to the −295-to-−243 position relative to the transcription initiation site. Using cotransfection studies, it was demonstrated that all three motifs were essential to achieve the maximum promoter activity induced by IRF-1 and NF-κB expression plasmids. The presence of a virus-inducible region in the IL-15 promoter suggests a role for IL-15 as a component of host antiviral defense mechanisms.
Interleukin-15 (IL-15) is a cytokine that utilizes the β and γ chains of the IL-2 receptor as well as its own private α chain (IL-15Rα) in T and natural killer (NK) cells. IL-15 stimulates T- and B-cell proliferation, lymphokine-activated killer cell induction, and B-cell differentiation (1, 8, 14). There is evidence for an essential role for IL-15, but not IL-2, in the effective development of NK cells (26, 29, 30). Unlike IL-2 mRNA, which is predominantly produced by activated T cells, IL-15 mRNA is constitutively expressed in a wide variety of nonlymphoid cells, including activated monocytes/macrophages (6, 9, 14), dendritic cells (16), epithelial cells (32), skeletal muscle cells (14, 31), astrocytes, and microglial cells (18). In addition to the constitutive nature of the IL-15 mRNA expression, we have demonstrated the induction of IL-15 mRNA expression upon lipopolysaccharide (LPS) and gamma interferon (IFN-γ) treatment in monocytes (6) and bone marrow cells (29). To study the transcriptional regulation of the IL-15 gene, we cloned its 5′ regulatory region and demonstrated that it had promoter activity by using reporter constructs. Further analysis of this region revealed the presence of two well-characterized motifs, namely, the IFN regulatory factor element (IRF-E) and nuclear factor-kappa B (NF-κB) binding site adjacent to each other. It was demonstrated that both of these two motifs were functionally active (4, 29).
There are many genes that are regulated by the IRF or the NF-κB family of transcription factors (28, 36). However, there are few known genes in which these two motifs reside at close proximity (10, 13, 27); one of them is IFN-β (11, 13). The IFN-β promoter region contains a virus-inducible enhancer element which consists of four positive regulatory domains (PRDI-to-IV). PRDI and PRDIII are IRF-E motifs, PRDII is a canonical NF-κB motif, and PRDIV is recognized by the heterodimer protein ATF-2–c-Jun (23). NF-κB defines a family of dimeric transcription factors composed of combinations of members of the Rel/NF-κB family such as p50, p52, p65 (RelA), c-Rel, and RelB. The predominant species is the p50-p65 complex, which is retained in the cytoplasm by its inhibitor IκB. Particular cell stimuli such as mitogens, cytokines, and viruses cause proteolytic degradation of the IκB subunits in the cytoplasm and subsequent translocation of the NF-κB proteins into the nucleus, which results in the activation of NF-κB target genes (5, 24, 36). IRF-E motif in the IFN-β is recognized by different members of the IRF family, including IRF-1, IRF-2, IRF-3, and IRF-7 (22, 41). IRF-1 and IRF-2 are expressed in a variety of cells. IRF-1 expression is induced by virus infection and also by IFN-α/β and IFN-γ. IRF-1 activates the IRF-E elements present in the IFN-β promoter region. In contrast to that, IRF-2 is an inhibitor of transcription (15, 37). IRF-3 is a recently described protein, which is constitutively expressed by variety of cells (2). Its expression does not increase after viral infection or treatment with IFNs. However, IRF-3 is phosphorylated after virus infection, which results in its translocation to the nucleus and activation of several genes (21, 34). In addition to IFN-α and -β promoters, the RANTES (regulated on activation normal T-cell expressed and secreted) chemokine promoters are activated by IRF-3 (20).
The presence of enhancer elements such as NF-κB and IRF-E in the promoter region of IL-15 gene raises the possibility of comparable responses to certain stimuli which activate the IFN-β gene. It has been documented that viruses induce the expression of both IRF and NF-κB proteins, which subsequently activate a number of cellular genes (36, 37). In particular, the IFN-β gene is induced by viruses and its protein is released from virus-infected cells, thereby conferring antiviral effects on neighboring cells (40). The resemblance of IL-15 and IFN-β gene promoters prompted us to study the response of IL-15 gene to viruses.
We utilized mouse fibroblast cell line L929 to study IL-15 gene transcription after virus infection. The Newcastle disease virus (NDV) has been shown to induce IFN-β mRNA in L929 cells (25). Here, we demonstrated that NDV also induced IL-15 mRNA and protein expression in these cells. We further identified a virus-inducible region in the mouse IL-15 promoter region. This region consists of IRF-E, NF-κB, and a 13-nucleotide spacer sequence which separates the IRF-E and NF-κB motifs. Using deletion and mutational analysis, we demonstrated that all the three elements of NF-κB, IRF-E, and the spacer sequence are essential for activation of the IL-15 promoter by viruses. Together, the data presented in this study demonstrate that IL-15 gene becomes activated by viruses through a virus-inducible region in its promoter.
MATERIALS AND METHODS
Cloning of the IL-15 promoter and plasmid construction.
The mouse IL-15 promoter was cloned as described previously (4) using two antisense primers from the mouse IL-15 exon 1 sequence (5′-CCTGACCTCTCTGAGCTGTTAGATGTGG-3′, 5′-CCAAACACAGCAGGATCCCGTCTTCG-3′). The amplified fragment was subcloned into the PCR2.1 vector (Invitrogen). The transcription initiation site was determined using S1 nuclease mapping (Ambion).
The −706/Luc, −295/Luc, −243/Luc, and −243(+NF-κB) deletion constructs were PCR amplified and then subcloned into the pGL3 Basic plasmid (Promega). The −243(+IRF-E)/Luc construct was generated using a primer which had the IRF-E motif from the mouse IL-15 promoter region (CTTTCTCTTTCACTTTTCT) and the −243/Luc construct as the template. The 123/Luc, 124/Luc, 129/Luc, and 99/Luc constructs were generated by cloning the corresponding double stranded oligonucleotides into the pGL3-promoter plasmid using KpnI and XhoI sites (Promega). The mutant constructs were generated using a site-directed mutagenesis kit (Stratagene). The construct −295(mtIRF-E)/Luc had four mutations in the following sites: CTCTTTCTCT(T→G)(T→C)CACTT(T→G)(T→C)CT. The construct −295(mtNF-(B)/Luc had two mutations in the following sites: G(G→A)(G→T)ACTCCCC. The construct −295(mtIRF-E+mtNF-(B)/Luc had the same mutations in both IRF-E and NF-κB sites.
The IRF-1 expression plasmid contains IRF-1 cDNA in pACT-1 (15), and the p50 and p65 cDNAs were subcloned into pMT2T plasmid (7). The IRF-3 expression plasmids, including wild-type IRF-3, the constitutively active form of IRF-3 or IRF-3(5D), or the nonfunctional mutant form of IRF-3 which lacks its DNA-binding domain orIRF-3(ΔN) (these plasmids were generous gifts from John Hiscott at The University of McGill) were generated in the CMVBL vector (19).
Reporter assays.
Two micrograms of reporter or expression plasmids was transfected into p19 cells using Genepulser II (Bio-Rad; 250 V/950 μF) and into L929 cells using the DEAE-dextran method. The luciferase activity was measured 48 h after transfection (Promega). The total amount of the DNA transfected into the cells was equalized by adding irrelevant DNA (pUC19). Each assay was performed at least three times. The average value and the standard deviation for each transfection is demonstrated in each graph.
EMSA and footprinting assay.
Electrophoretic mobility shift assay (EMSA) for IRF-E was performed according to the protocol published previously (15) and CTTTCTCTTTCACTTTTCT as the probe. The supershift assay was performed using an anti-IRF-1 antibody (1 μg/ml) (a generous gift from Taniguchi and Taki, University of Tokyo, Tokyo, Japan). The NF-κB EMSA was performed using the NF-κB motif from the IL-15 promoter (TTGGGACTCCCCGG) and was compared to the consensus NF-κB (cNF-κB) motif from the human immunoglobulin κB gene (AGGGGACTTTCCCAG) as described previously (4). The COS p50-p65 transfected cell extract used as a positive control, and anti-p50 and anti-p65 antibodies used in the supershift assays were described previously and were generous gifts from U. Siebenlist (National Institute of Allergy and Infectious Diseases, National Institutes of Health). The EMSA was performed as described previously (4). All of the antibodies used in these assays (except the anti-IRF-1, -p50 and -p65 antibodies) were supershift-quality antibodies and were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.).
RPA.
L929 cells were infected with NDV as described by Watanabe and Kawade (40). At 1, 3, 6, and 12 h postinfection, total RNA was extracted from these cells. Then, 10 μg of the total RNA was used for an RNase protection assay (RPA) as described previously (4) using probes for mouse IRF-1 or IL-15, which were generated by cloning part of the IRF-1 or IL-15 cDNA, respectively, in pCR2.1 plasmid (Invitrogen). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe was purchased from a commercial source (Ambion).
Northern blot analysis.
A total of 20 μg of the same preparation of total RNA which was used for RPA was utilized in a Northern blot analysis. The blot was probed with the labeled IFN-β probe and subsequently with β-actin probe to monitor the mRNA loading.
Western blot analysis.
In order to detect the IL-15 protein produced by NDV-infected L929 cells, the cell pellet and supernatant of the NDV(−) and NDV(+) L929 cells were collected 24 h postinfection. The lysates were prepared with a modified radioimmunoprecipitation assay buffer and immunoprecipitated using an anti-mouse IL-15 monoclonal antibody (Pharmingen). Immunoblotting was performed using the same antibody with the enhanced chemiluminescence detection method according to the manufacturer's instructions (Amersham).
RESULTS
Molecular cloning of the mouse IL-15 promoter.
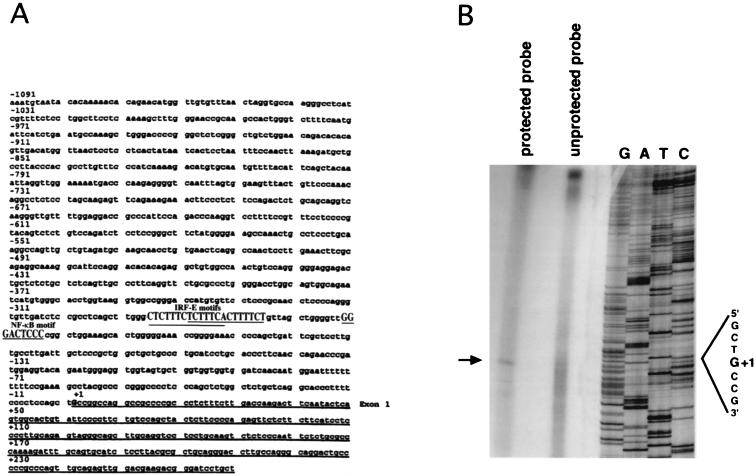
To study the transcriptional regulation of the IL-15 gene, we cloned the IL-15 5′ regulatory region using a mouse genomic library as described in the Materials and Methods (Fig. 1A). The transcription initiation site was determined by S1 nuclease mapping using mRNA isolated from the 10P12 cell line that constitutively expresses IL-15 mRNA (Fig. 1B). Upon sequence analysis, we discovered two overlapping IRF-E motifs (these two motifs as shown in A-rich sequence are GAGAAAGAGAAAGAGAAAAGA) and one NF-κB motif (GGGACTCCCC) at nucleotide positions −295 and −253, respectively, relative to the transcription initiation site. The IRF-E and NF-κB motifs were separated by 13 bp as shown in Fig. 1A.
FIG. 1.
(A) Nucleotide sequence of the mouse IL-15 5′-flanking region. These sequence data have been submitted to the GenBank database under accession number AF038164. The underlined portion represents the exon 1 sequence. Positions of the IRF-1 and NF-κB putative motifs (shown in uppercase) relative to the transcription initiation site (G, denoted as +1) are shown. (B) S1 mapping analysis of the mouse IL-15 gene. The transcription initiation site was determined using an S1 nuclease assay. The arrow indicates the protected fragment which comigrated with a C fragment in the DNA sequencing ladder (shown as G+1) that was generated using the antisense strand as a template.
Induction of IL-15 mRNA and protein in L929 cells by NDV.
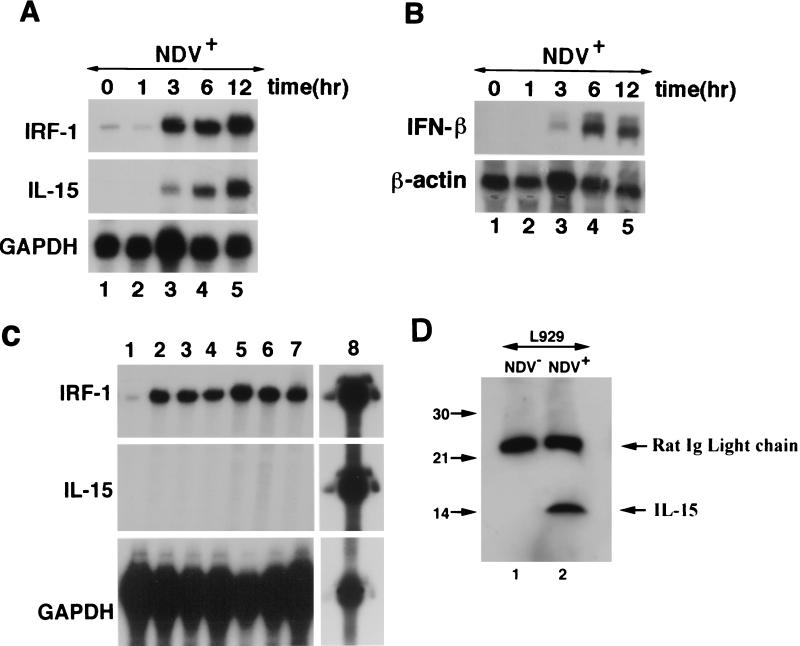
To study IL-15 transcription, the L929 cell line, a mouse fibroblast cell line, which has been used extensively in IFN regulatory pathway studies, was selected. It was reported that, when infected with NDV, the IFN-β gene was induced in L929 cells (13). To examine the effect of viral infection on IL-15 gene expression, L929 cells were treated with NDV. Total RNA was isolated from these cells which was used subsequently to monitor for IL-15 and IFN-β mRNA levels before and after infection. As shown in Fig. 2A, IL-15 mRNA was induced only after treating L929 cells with NDV as determined by an RPA. Since IRF-1 is one of the genes which becomes activated by viruses, its expression in NDV-infected L929 cells was examined. As shown in Fig. 2A, IRF-1 mRNA was also induced following NDV infection almost in parallel with that of IL-15. In this assay, GAPDH probe was included to monitor for the quality and loading of each RNA sample. To confirm that IFN-β mRNA was upregulated by NDV infection, a Northern blot analysis was performed. As shown in Fig. 2B, IFN-β mRNA was expressed only after NDV infection in L929 cells, and the kinetics of its expression were identical to that of IL-15. This blot was stripped and reprobed with β-actin to demonstrate the amount of the RNA loaded in each lane.
FIG. 2.
(A) Parallel induction of IRF-1 and IL-15 mRNA in NDV+ L929 cells. An RPA was carried out in NDV-infected L929 cells at 0, 1, 3, 6, and 12 h postinfection. Both IRF-1 and IL-15 mRNA appeared in parallel 3 h after NDV infection of L929 cells. The GAPDH probe was included in this experiment to monitor for RNA quality and loading. (B) Induction of IFN-β mRNA after NDV infection with L929 cells. Northern blot analysis was performed using the same RNA which was used in the RPA shown in panel A. The blot was hybridized with IFN-β and subsequently with β-actin probes. Induction of the IFN-β mRNA occurs after 3 h, indicating similar kinetics when compared to IL-15 and IRF-1 mRNA induction. β-Actin bands in this figure show equal levels of RNA loaded in each lane. (C) Induction of IRF-1, but not IL-15 mRNA, by interferons in L929 cells. L929 cells were treated with media alone (lane 1), 100 ng of IFN-α per ml for 6 h (lane 2) and 12 h (lane 3), 100 ng of IFN-β per ml for 6 h (lane 4) and 12 h (lane 5), and 100 ng of IFN-γ per ml for 6 h (lane 6) and 12 h (lane 7). An RPA identical to that shown in panel A was used to analyze the RNA obtained from these cells for expression of IRF-1, IL-15, and GAPDH. To demonstrate the quality of the IL-15, IRF-1, and GAPDH probes used in this assay, the unprotected probe was loaded in lane 8. (D) Induction of the IL-15 protein in NDV-infected L929 cells. Western blot analysis was performed with the total cellular lysates from NDV-infected and noninfected L929 cells. IL-15 was detected only in the cellular lysates prepared from NDV-infected L929 cells. Immunoglobulin light-chain molecules of the anti-IL-15 antibody used for immunoprecipitation were present in both lanes migrating at 25 kDa.
Next, we examined whether IL-15 mRNA induction by NDV is mediated by induction of IFN-β mRNA. Since the kinetics of mRNA induction for both IL-15 and IFN-β are almost identical (Fig. 2), it is unlikely that IL-15 mRNA is induced by IFN-β. In contrast, the kinetics of induction of these two genes follow that of IRF-1 (IRF-1 mRNA is induced 3 h after NDV infection which proceeds by IL-15 and IFN-β mRNA induction that occurs 6 h after NDV infection). This suggests that the induction of IL-15 and IFN-β mRNA by NDV are parallel, but not consequential, events which may be mediated by common transcription factors. Nevertheless, to answer the question of whether IL-15 mRNA is induced by IFNs in L929 cells, we examined IL-15 mRNA expression after treating L929 cells with various doses of IFN-α, -β, and -γ using an RPA identical to that used in Fig. 2A. As shown in Fig. 2C, while IRF-1 mRNA was induced, under no circumstances was IL-15 mRNA induced. This confirms that IFN alone is not sufficient to induce IL-15 mRNA in L929 cells. Furthermore, it indicates that IRF-1 alone is not sufficient either for induction of IL-15 mRNA in these cells. Together, these results suggest that NDV infection of L929 cells provides a signal in addition to that provided by IFNs or IRF-1 which induces IL-15 mRNA.
To determine whether IL-15 mRNA induction resulted in IL-15 protein production, a Western blot analysis was performed using lysates from L929 cells before and after NDV infection. As shown in Fig. 2D, only NDV-infected L929 cells produced IL-15 protein, a finding which is in accord with IL-15 mRNA induction by this virus.
Identification of a virus-responsive element in the IL-15 promoter.
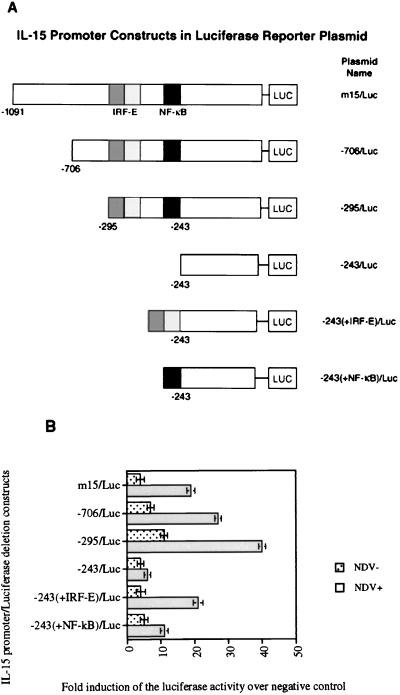
To define the region of the IL-15 promoter which responds to NDV infection, a set of deletion constructs spanning the IL-15 promoter was generated. Figure 3A shows a schematic representation of various IL-15 constructs in the pGL3-basic luciferase reporter plasmid. L929 cells were transfected with various IL-15 promoter-reporter constructs, and their luciferase activities were measured before and after NDV infection (Fig. 3B). The promoter activity of the full-length IL-15 promoter in the luciferase construct (m15/Luc), the −706/Luc construct, and the −295/Luc construct which contained the IRF-E and NF-κB motifs increased to the maximum of 40-fold after NDV infection. The luciferase activity of the −295/Luc construct after NDV infection was greater than those of the m15/Luc and −706/Luc constructs. This may be due to elimination of some inhibitory motifs which are present upstream of the −295 position. The luciferase activity of the IL-15 promoter after NDV infection was almost completely abrogated when the luciferase activity of the −295/Luc deletion construct was compared to that of the −243/Luc deletion construct, indicating that the region between positions −295 and −243 was crucial for IL-15 induction after virus infection. Further analysis revealed that this region includes both the IRF-E and NF-κB binding motifs. To assess the contribution of each of these transcription factors, reporter constructs were generated that included the IRF-E or NF-κB binding sequences added to the −243/Luc construct. The −243(+NF-κB)/Luc construct, which was generated by adding the NF-κB motif to the −243/Luc, exhibited a 10-fold increase in its reporter activity after NDV infection. The reporter activity of the construct −243(+IRF-E)/Luc that contained the IRF-E motif was increased 20-fold after NDV infection. As shown in Fig. 3B, the reporter activities of the −243(+NF-κB)/Luc and −243(+IRF-E)/Luc constructs were 75 and 50% lower, respectively, than that of the −295/Luc construct. These observations suggest that the region between positions −295 and −243, which includes IRF-E and NF-κB motifs, is essential for optimal NDV activation of the IL-15 promoter.
FIG. 3.
(A) Schematic representation of IL-15 promoter deletion constructs subcloned into pGL3-basic luciferase plasmid. (B) Induction of IL-15 promoter reporter activity in NDV(+) L929 cells. Reporter assays were carried out in NDV(+) and NDV(−) L929 cells. The luciferase activity is shown as the fold induction over the negative control, which is the luciferase plasmid with no promoter sequence (pGL3-basic). A maximum of 40-fold induction in the reporter activity was observed after NDV infection of L929 cells only when these cells were transfected with constructs bearing the IRF-E and NF-κB motifs (−706/Luc and −295/Luc). Deletion of this region containing IRF-E and NF-κB motifs in −243/Luc construct resulted in the loss of promoter activity after NDV infection, indicating that this region is essential for IL-15 promoter activation by NDV. Addition of NF-κB motif to the −243/Luc construct [−243(+NF-κB)/Luc] decreased the reporter activity about fourfold over that of −295/Luc after NDV infection. Addition of IRF-E motif to the −243/Luc construct [−243(+IRF-E)/Luc] caused an approximately twofold decrease in the promoter activity of the −295/Luc construct.
IRF-1 protein binds to the IRF-E motif in the IL-15 promoter region after NDV infection.
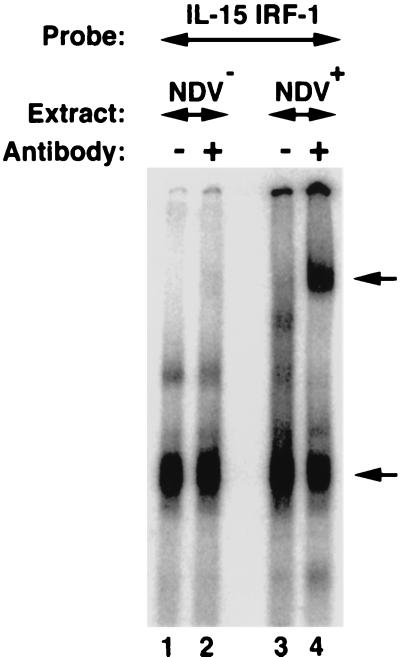
To determine whether any transcription factor binds to the IRF-E motif upon NDV infection, an EMSA experiment was performed. As shown in Fig. 4, the IRF-E motif forms a DNA-protein complex with cell extracts generated from L929 cells before and after NDV infection. Addition of the anti-IRF-1 antibody resulted in a mobility shift of a portion of the complex only in NDV-infected cells, thus confirming that NDV infection facilitated the complex formation between IRF-1 protein and the IRF-E element in L929 cells. The lower band indicated by an arrow was likely a nonspecific band since it was present in L929 cell extracts both before and after NDV infection. Furthermore, various antibodies against different members of the IRF family, including IRF-2, IRF-3, IRF-7, ICSBP, ICSAT, and ISGF3, were used in supershift assays in order to identify other IRF family members which may be present in NDV-infected L929 cells. However, no other IRF-related protein was identified. Nevertheless, we cannot exclude the possibility that these antibodies may not be able to recognize their cognate protein in a DNA-protein complex. These data support the view that the IRF-1 protein is induced only after NDV infection of L929 cells and is recruited to the already existing DNA-protein complex and thereby exerts its effect on the IL-15 promoter.
FIG. 4.
Formation of the IRF-E–IRF-1 complex in NDV-infected L929 cells. A radiolabeled IRF-E probe from the IL-15 promoter region (CTTTCTCTTTCACTTTCT) was incubated with total cellular extracts from NDV-infected (lanes 3 and 4) and noninfected (lanes 1 and 2) L929 cells. The resulting complex was analyzed on a nondenaturing polyacrylamide gel. When an antibody against IRF-1 was included in the reaction, it supershifted an IRF-E–IRF-1 complex only from NDV-infected L929 cells (lane 4; indicated by the upper arrow). Note the presence of a residual complex in lanes 1 to 4 that was not shifted by the addition of the antibody. This complex was likely to be a nonspecific band since it was present in L929 cell extract both before and after NDV infection. The lower arrow in NDV-infected L929 cells indicates a complex that at least contains IRF-1 protein bound to the IRF-E probe.
NDV infection induces p50 and p65 subunits of the NF-κB family in L929 cells.
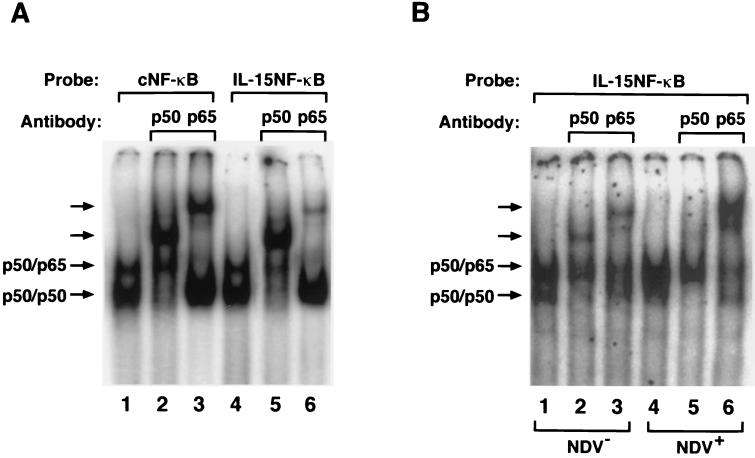
In order to evaluate the status of the NF-κB expression in the L929 cells before and after NDV infection, several EMSAs were performed. The NF-κB motif from the IL-15 promoter region was compared with the consensus NF-κB motif from the immunoglobulin light chain in its ability to interact with the p50 and p65 subunits of the NF-κB protein. As shown in Fig. 5A, p50 and p65 proteins of the NF-κB formed the p50-p50 homodimer and p50-p65 heterodimer with both the consensus NF-κB motif and the IL-15 NF-κB motif when extracts from the p50 and p65 transfected COS cells were used. The nature of these dimers was confirmed using antibodies against p50 or p65 proteins. These data demonstrate that p50 and p65 subunits of the NF-κB protein bind to the IL-15 NF-κB motif. In order to determine if the NF-κB proteins were present in the resting or NDV-infected L929 cells, we performed an EMSA using the extracts from the L929 cells before and after NDV infection. As shown in Fig. 5B, the p50-p50 homodimer and p50-p65 heterodimer were present in L929 cells before NDV infection, indicating that these proteins were constitutively produced by these cells, a phenomenon which may contribute to the constitutive expression of the IL-15 gene. However, after NDV infection of these cells, an additional induction of the p50-p65 heterocomplex was observed which was confirmed by a supershift assay using an anti-p65 antibody. This observation is in accord with previous studies demonstrating that the p65 protein is induced upon stimulation such as that observed with virus infection (12).
FIG. 5.
(A) The formation of the NF-κB motif–p50 and –p65 complexes before and after NDV infection in L929 cells. Binding of the p50 and p65 subunits of the NF-κB protein to the NF-κB motif from the IL-15 promoter region (IL-15 NF-κB) TTGGGACTCCCCGG and immunoglobulin κB promoter region as consensus NF-κB (cNF-κB) AGGGGACTTTCCCAG was compared in an EMSA using COS cell extract transfected with p50 and p65 expression constructs. Both cNF-κB and IL-15–NF-κB motifs bound to p50-p50 and p50-p65 complexes, as indicated by arrows (lanes 1 and 4). These complexes were further analyzed using antibodies against p50 and p65 proteins (lanes 2, 3, 5, and 6). (B) The induction of the NF-κB proteins after NDV infection. Both p50-p50 homodimer and p50-p65 heterodimer bound to the IL-15–NF-κB motif, as shown by arrows (lanes 1 and 4). However, the p65 protein was greatly induced after NDV infection of the L929 cells, as shown in lane 4 and in the supershifted complex using anti-p65 antibody (compare lanes 3 and 6).
IRF-E and NF-κB binding motifs are essential for activation of the IL-15 promoter.
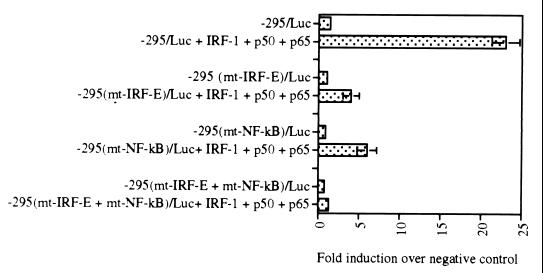
In order to assess the contribution of each IRF-E and NF-κB motif to the activity of the IL-15 promoter, several deletion constructs were generated in the −295/Luc construct. The −295/Luc construct was selected for introducing mutations since this construct showed the highest promoter activity after NDV infection of L929 cells (Fig. 3B). The −295(mtIRF-E)/Luc and −295(mtNF-κB)/Luc constructs have mutations in the IRF-E and NF-κB sites as described in Materials and Methods. The −295(mtIRF-E + NF-κB)/Luc construct bears mutations in both the IRF-E and NF-κB binding sites. Since both IRF-1 and NF-κB proteins were induced after NDV infection (Fig. 4 and 5), the mutant constructs were cotransfected with the IRF-1, p50, and p65 expression plasmids. Cotransfection studies were performed in p19 cell line, a cell line that is deficient in endogenous IRF-1 and has minimal levels of the NF-κB proteins as determined by EMSA (data not shown). As shown in Fig. 6, cotransfection of −295/Luc construct with IRF-1, p50, and p65 expression plasmids resulted in an approximately 25-fold increase of its reporter activity (fold increase is measured as luciferase activity of the construct over that of the pGL3 basic plasmid). However, cotransfection of the mutant construct −295(mtIRF-E)/Luc or −295(mtNF-κB)/Luc constructs with these expression plasmids resulted in only four- and sixfold increases, respectively. When the double mutant construct −295(mtIRF-E + mtNF-κB)/Luc construct was cotransfected with IRF-1, p50, and p65 expression plasmids, no induction of its luciferase activity was observed. These data indicate that both IRF-E and NF-κB sites are essential to confer maximum promoter activity to the IL-15 promoter.
FIG. 6.
Mutational analysis of IL-15 virus inducible element region. The −295/Luc construct contains both IRF-E and NF-κB motifs. This construct was activated almost 25-fold by IRF-1 and NF-κB p50 and p65 expression plasmids when it was cotransfected into p19 cells. Introducing mutation in IRF-E or NF-κB motifs in the −295/Luc construct resulted in a significant decrease in the luciferase activity of the −295(mtIRF-E)/Luc and −295(mtNF-κB)/Luc constructs in similar experiments. When the −295(mtIRF-E + mtNF-κB)/Luc construct, which bears mutations in both IRF-E and NF-κB motifs, was cotransfected with IRF-1, p50, and p65 expression plasmids, no luciferase activity was observed. These data indicate that both IRF-E and NF-κB motifs are essential for activity of IL-15 promoter region induced by IRF-E and NF-κB binding elements.
Cooperation between IRF-1 and NF-κB transcription factors in activating the virus-inducible region of the IL-15 promoter.
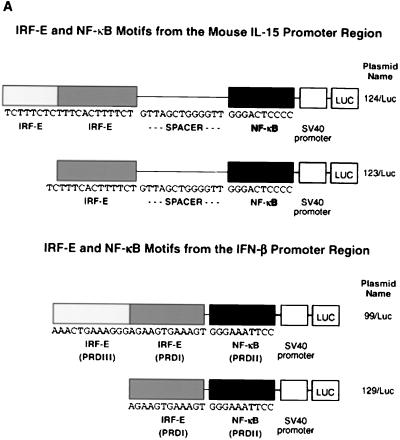
To further study the potential interaction between IRF-1 and NF-κB, the region of the IL-15 promoter at positions −295 to −243 nucleotide, which contained two IRF-E motifs and the NF-κB enhancer element, was cloned into the luciferase reporter plasmid (pGL3) with the simian virus 40 promoter sequence (124/Luc) (Fig. 7A). In order to assess the contribution of the second IRF-E motif, another luciferase construct which included only one of the IRF-E motifs of the 124/Luc plasmid was generated and named 123/Luc. Since IRF-E (PRDI and PRDIII) and NF-κB (PRDII) motifs were also in proximity in the IFN-β promoter, the region of the IFN-β promoter that contained these motifs was cloned into the luciferase plasmid (99/Luc and 129/Luc, respectively) (Fig. 7A). These constructs were used for transfection into p19 cells.
FIG. 7.
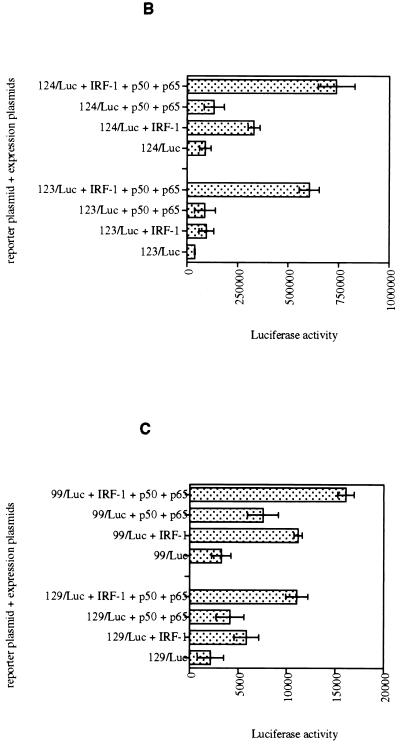
(A) Schematic representation of virus-inducible regions of IL-15 and INF-β promoters subcloned into the pGL3-promoter luciferase plasmid. (B) IL-15 virus-inducible constructs respond to IRF-1 and NF-κB proteins in p19 cells. The luciferase assay was carried out in p19 embryonic carcinoma cells that lack endogenous IRF-1 protein expression. IRF-1, p50, or p65 expression plasmids, singly or in combination, were cotransfected with the IL-15 virus-inducible reporter constructs (123/Luc and 124/Luc) as indicated in each graph. The promoter activity is shown as the luciferase activity. The IRF-1, p50, and p65 expression plasmids induced 123/Luc and 124/Luc constructs minimally. However, cotransfection of all three plasmids activated the reporter constructs about 10-fold. (C) PRDI-PRDIII regions from the IFN-β virus-inducible constructs respond to IRF-1 and NF-κB proteins in p19 cells, but to a lesser extent compared to those of IL-15. The same experiments were performed with IFN-β reporter constructs (99/Luc and 129/Luc constructs) as described in the Fig. 6 legend. Cotransfection of all three plasmids activated the reporter constructs about fivefold.
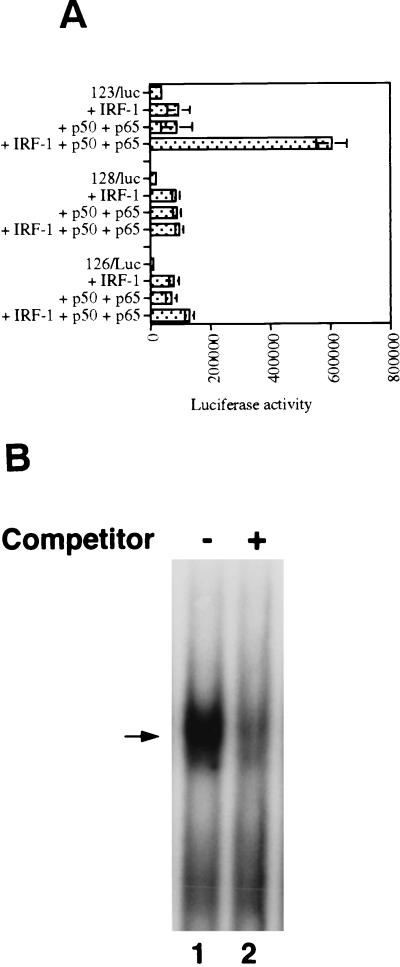
As shown in Fig. 7B, cotransfection of IRF-1 expression plasmid with the IL-15 reporter-promoter constructs, 123/Luc and 124/Luc, resulted in a minimum increase in their luciferase activities (about two- and fourfold, respectively). Similarly, cotransfection of these two constructs with NF-κB p50 and p65 expression plasmids also increased the luciferase activities of the 123/Luc and 124/Luc constructs only about twofold. However, cotransfection of IRF-1, p50, and p65 expression plasmids together with 123/Luc or 124/Luc constructs led to a 10-fold increase of the promoter activities of both reporter constructs, suggesting that NF-κB and IRF-1 are both needed for meaningful induction of 123/Luc and 124/Luc reporter activities. Comparing luciferase activities of 123/Luc (with one IRF-E motif) and 124/Luc (with two IRF-E motifs) constructs in cotransfection studies reveals that the second IRF-E motif increases the activity of the enhancer region in response to the IRF-1 protein about twofold. However, it does not contribute to the luciferase activity of these constructs when cotransfected with IRF-1, p50, and p65 plasmids, since they both showed a 10-fold increase in their luciferase activities. This suggests that the necessary interactions between IRF-1 and NF-κB proteins can be adequately formed with one IRF-E motif.
In a similar experiment, IRF-1, p50, and p65 expression plasmids were cotransfected with 99/Luc and 129/Luc plasmids (from IFN-β promoter). As shown in Fig. 7C, cotransfection of 129/Luc and 99/Luc constructs with IRF-1 expression plasmids yielded a minimum increase in the promoter activities of each construct (about two- and fourfold, respectively). Additionally, cotransfection of 129/Luc or 99/Luc constructs with NF-κB p50 and p65 expression plasmids resulted in only 2- and 1.5-fold increases in 129/Luc and 99/Luc activities, respectively. However, cotransfection of IRF-1, p50, and p65 expression plasmids with 129/Luc and 99/Luc constructs resulted in a fivefold increase in the promoter activity of each construct. These data suggest that IRF-1 and NF-κB cooperate in activating the transcription of both IL-15 and IFN-β promoters.
The spacer sequence is essential for activity of the IL-15 virus-inducible region.
As shown in Fig. 7A, the virus-inducible region of IL-15 promoter consists of IRF-E, NF-κB, and a 13-bp sequence separating these two motifs. Both IRF-E and NF-κB motifs have been shown to be essential in activating IL-15 promoter. In order to study the role and significance of the spacer sequence, we generated several constructs in which the spacer sequence was either removed or replaced by a mutant sequence. In the 128/Luc construct the spacer sequence was removed from the IL-15 reporter construct (123/Luc) and in the 126/Luc construct the spacer sequence was replaced by an irrelevant sequence with the same number of nucleotides (GACTCTGAGCTCA). These reporter constructs contained only one IRF-E enhancer motif, since addition of the second IRF-E motif in cotransfection studies did not contribute to the overall promoter activity of the 123/Luc construct (Fig. 7B). As shown in Fig. 8A, both 126/Luc and the 128/Luc constructs showed lower promoter activities in response to IRF-1, p50, and p65 cotransfection compared to that of the 123/Luc construct. These data indicate that the spacer sequence had a positive effect on the transcription of IL-15 virus-inducible region reporter construct when it was cotransfected with IRF-1 and NF-κB expression plasmids. This positive effect seems to be due to the specific sequence of the spacer region and not merely due to the presence of a particular distance (13 nucleotides) between the IRF-E and NF-κB motifs. This raises the possibility of the spacer sequence acting as a binding and recognition site for some proteins. In order to examine this possibility, an EMSA was performed to examine whether the spacer sequence is recognized by a protein in the p19 cell extract. Since all the cotransfection studies were performed in uninfected cells, we used uninfected p19 cell lysates in gel shift assays to assess the basal expression of a potential DNA-binding protein which recognizes this sequence. A DNA-protein complex was generated, and its formation was competed by adding 50× molar excess of the unlabeled spacer probe which showed the specificity of this binding (Fig. 8B). This suggests that the spacer sequence may serve as a motif for a protein which has a basal level of expression in the p19 cells. It is possible that the expression of this protein increases or gets modified after virus infection. This protein may contribute to the inducibility of the IL-15 virus-inducible region.
FIG. 8.
The spacer sequence contributes to the IL-15 virus-inducible reporter activity. (A) The 128/Luc construct with the native spacer sequence is activated about 10-fold when cotransfected with IRF-1, p50, and p65 expression plasmids into the p19 cells. However, when the spacer sequence was removed in 128/Luc, no reporter activity was observed in a similar experiment. Replacement of the native spacer sequence with an irrelevant sequence with the same number of nucleotides in the 126/Luc construct did not increase the promoter activity of this construct when it was cotransfected with IRF-1, p50, and p65 expression plasmids. This indicates the importance of the spacer sequence in the activity of the virus-inducible region. (B) The spacer sequence forms a DNA-protein complex with p19 cell extracts. Total cell extracts were prepared and used in an EMSA with the spacer as a probe (CTGTTAGCTGGGGTT). The arrow indicates the position of the DNA-protein complex. The spacer unlabeled probe was added at 50× excess, as indicated in this figure.
IRF-3 activates IL-15 promoter-reporter constructs.
It was shown recently that IRF-3 is an essential factor for virus-induced activation of IFN-β gene (17, 41). IRF-3 is constitutively expressed in a variety of cells, and its mRNA expression is not induced after NDV infection. However, IRF-3 is phosphorylated after virus infection, which is followed by its subsequent translocation to the nucleus. The phosphorylated form of IRF-3 is thought to be the transcriptionally active form of this molecule (19). Since L929 cells are fibroblast cells and IRF-3 is expressed in these cells, we examined the role of IRF-3 transcription factor in activating the IL-15 promoter.
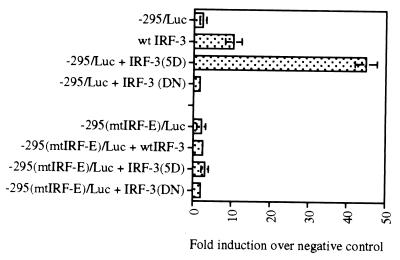
In order to examine the effect of the IRF-3 on activation of the IL-15 promoter, cotransfection studies were performed using IL-15 promoter-reporter constructs and IRF-3 expression plasmids. The −295/Luc construct was used for transfection studies since this construct produced the highest reporter activity after virus infection (Fig. 3B). The −295/Luc construct was cotransfected with a wild-type IRF-3 expression plasmid. In order to resemble the phosphorylated form of the IRF-3 in these cotransfection studies, an active mutant of IRF-3, namely IRF-3(5D), which is capable of constitutively activating the IFN-β promoter-reporter was used (19). As a control, a mutant form of IRF-3, IRF-3(ΔN) which lacks its DNA-binding domain, was also used in similar experiments. As shown in Fig. 9, the luciferase activity of the −295/Luc construct increased when it was cotransfected with an active form of IRF-3(5D) expression plasmid about 45-fold. The wild-type form of IRF-3 expression plasmid did not increase the luciferase activity of the −295/Luc construct significantly. The IRF-3(ΔN) mutant construct did not induce the −295/Luc construct luciferase activity either. These data suggest that IRF-3 transcription factor in its active (phosphorylated) form can activate IL-15 promoter. The IRF-3 or IRF-3(5D) plasmids was not able to induce promoter activity in the −295(mtIRF-E)/Luc construct which bears mutations in its IRF-E motif in the same experiment. This clearly indicates that IRF-3 transcription factor confers promoter activity to the IL-15 promoter via the IRF-E binding site. Upon virus infection, the IRF-3 protein becomes phosphorylated and can activate transcription of several genes, including that for IL-15.
FIG. 9.
IRF-3 activates the promoter activity of the IL-15 promoter-reporter constructs. The −295/Luc construct of IL-15 promoter was cotransfected with the wild-type IRF-3 or its active form IRF-3(5D) into p19 cells. The IRF-3(5D) expression plasmid conferred about a 45-fold increase in the luciferase activity over that of the basic luciferase (pGL3) plasmid. In contrast, the IRF-3 dominant-negative IRF-3(DN) did not activate the −295/Luc construct. In a similar experiment, no promoter activity was observed when −295(mtIRF-E)/Luc reporter construct was used which bears mutant IRF-E motif.
DISCUSSION
In this study, we demonstrated that the cloned IL-15 promoter region was a functional promoter which was activated after NDV infection of L929 cells. The region responsive to virus infection in the IL-15 promoter region was mapped to be between positions −295 and −243 relative to the transcription start site. This region includes two overlapping IRF-E motifs, one NF-κB binding motif, and a 13-nucleotide spacer sequence which separates these IRF-E and NF-κB motifs. By using IL-15 promoter-reporter mutation and deletion constructs in transfection studies, it was demonstrated that both IRF-E and NF-κB motifs are essential elements of the virus-inducible region of the IL-15 promoter. As demonstrated in cotransfection studies, IRF-1 or NF-κB (p50 and p65) expression plasmids are able to activate the virus-inducible region of the IL-15 promoter-luciferase constructs which bear IRF-E and NF-κB motifs. However, the maximum luciferase activity was achieved when all three plasmids were cotransfected. These data indicate that there is a cooperation between IRF-1 and NF-κB p50 and p65 subunits in activating the IL-15 promoter. These data are in accord with previous reports indicating cooperation between IRF-1 and NF-κB proteins in activating the promoter of other genes, including those of IFN-β, major histocompatibility complex class I, VCAM-I, and iNO synthase (10, 27, 35, 38).
Furthermore, it seems that the spacer sequence separating IRF-E and NF-κB motifs in the IL-15 promoter region plays an important role in activating the IL-15 promoter virus-inducible region. Its removal or substitution with an irrelevant sequence abolished the promoter activity of the IL-15 virus-inducible region reporter construct when it was cotransfected with IRF-1, p50, and p65 expression plasmids. This indicates that not only the distance between IRF-E and NF-κB binding motifs but also its sequence content is crucial for the activity of the virus-inducible region of IL-15 promoter. The spacer sequence was recognized by a protein(s) present in the extracts of p19 cells, as shown by a gel shift assay. This protein(s) may serve as a transcription factor or coactivator of transcription. However, it did not seem that this sequence can confer promoter activity alone, since three tandem repeats of this sequence in the luciferase reporter plasmid did not induce any promoter activity when it was transfected into p19 cells. It is possible that this protein(s) acts as a glue factor, bringing the IRF-1 and NF-κB subunits together, and provides the necessary interactions between these elements. It has been demonstrated that the presence of the high-mobility group, HMGI(Y), is required to fully activate the transcription of the virus-inducible element in the IFN-β promoter (11, 39). HMGI(Y) proteins bind to the AT-rich sequence in the PDRII and to the flanking AT sites of the PRDIV. It will be interesting to see if the protein(s) which binds to the spacer region in the IL-15 promoter is acting on this enhancer region with a function similar to that of HMGI(Y).
As shown in this study, IRF-3 and, more significantly, its constitutively active mutant IRF-3(5D) is capable of activating IL-15 promoter in transient assays. It has been shown in several reports that IRF-3 is a crucial factor in activating IFN-β gene transcription (17, 41). This indicates that in addition to IRF-1 and NF-κB transcription factors, another element, namely, IRF-3, is utilized by both virus-inducible promoter regions of IL-15 and IFN-β. This, in turn, results in a similar induction pattern of IL-15 and IFN-β genes in response to NDV infection. As mentioned before, IRF-3 is a transcription factor which is constitutively expressed in variety of cells and is located in the cytoplasm in a latent state. After viral infection, IRF-3 becomes phosphorylated at multiple serine and threonine residues which are located at the carboxy terminus of this molecule. Phosphorylation of IRF-3 alters its protein conformation, which allows its translocation into the nucleus and its association with transcriptional partners. One of these partners is the CREB binding protein CBP (also called p300) coactivator (19). Interaction between IRF-3 and CBP may help to bring this coactivator to the virus-inducible enhancer element of the (PRDI-PRDIV) in the IFN-β promoter region. It would be curious to examine the presence of CBP coactivator in the IL-15 virus-inducible region and to study its role in regulation of the IL-15 transcription.
In contrast to IRF-3, IRF-7 is predominantly expressed by lymphoid cells (3). Its mRNA expression is induced after virus infection, and its protein is translocated into the nucleus. It has been shown that IRF-7 protein exists in a phosphorylated state in the nucleus after virus infection (33, 42). IRF-7 seems to have a preferential effect on transcription of IFN-α gene promoters, which are mostly expressed in cells of lymphoid origin. It would be of interest to examine the virus inducibility of the IL-15 gene in a variety of lymphoid cells. Although IL-15 mRNA is constitutively expressed by a variety of cells, including T cells, it has been difficult to induce IL-15 mRNA expression in these cells. We previously reported the overexpression of IL-15 mRNA in T cells infected with human T lymphotrophic virus type 1 which occurred through induction of NF-κB proteins (4). It would be interesting to study the effect of IRF-7 in regulating the transcription of IL-15 gene by other viruses in T cells or other cells of lymphoid origin.
Together, the data indicate that IL-15 mRNA is induced after NDV infection in L929 cells and that this induction is through the action of several enhancer elements in the virus-inducible region of the IL-15 promoter, which includes IRF-E, a spacer sequence, and NF-κB. Although each of these elements seems to be necessary for induction of IL-15 mRNA, none of them alone is sufficient to activate IL-15 gene expression. L929 cells were treated with agents to induce IRF-1, including IFN-α, IFN-β, and IFN-γ (Fig. 2C). Although IRF-1 mRNA was induced, IL-15 mRNA was not expressed in any of the conditions applied. This suggests that IRF-1 alone does not provide a sufficient signal to drive IL-15 transcription in these cells. This may be explained by our findings in this study that IRF-3(5D), which mimics the phosphorylated and active form of the IRF-3, can significantly activate IL-15 promoter. Treatment of L929 cells with IFNs does not seem to cause phosphorylation and activation of IRF-3 and thereby activation of IL-15 promoter and induction of its mRNA. Furthermore, it rules out the possibility that IL-15 induction in L929 cells by NDV is through induction of IFN genes. Interestingly, addition of LPS (an NF-κB inducer) alone or in combination with IFN-γ did not induce IL-15 mRNA expression either (data not shown). These data demonstrate that NDV infection provides a unique signal, independent from those of IFNs or LPS, which results in IL-15 induction in L929 cells. However, it has been demonstrated before that IL-15 mRNA can be induced in freshly isolated monocytes after treatment of these cells with LPS and IFN-γ (6). This suggests that there are cell-specific controls over induction of IL-15 mRNA. The agents which are capable of inducing IL-15 mRNA in some cells may not be able to induce it in another cells. The biological and physiological significance of this preferential induction of IL-15 in various cells by common agents requires further scrutiny.
It appears that NDV infection induces a set of transcriptional signals which trigger the expression of IL-15 and IFN-β genes independently. The antiviral effects of IFNs on neighboring cells have been known. However, we have not been able to demonstrate a direct antiviral effect for IL-15 in these cells. There are several reports suggesting a role for IL-15 as a chemoattractant for T cells, NK cells, and macrophages/monocytes (16). These cells migrate to the site of infection to eliminate virus-infected cells. Furthermore, it has been demonstrated that IL-15 plays a critical role in stimulation of CD8+ memory phenotype T cells (43). Production of IL-15 by virus-infected cells may help generate and maintain CD8+ memory cells which will build the immunity against this pathogen. Altogether, the data suggest that IL-15 may be a player in the antiviral defense mechanism.
ACKNOWLEDGMENT
We thank Colin Duckett (National Cancer Institute, National Institutes of Health) for his critical review of the manuscript.
REFERENCES
- 1.Armitage R J, Macduff B M, Eisenman J, Paxton R, Grabstein K H. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J Immunol. 1995;154:483–490. [PubMed] [Google Scholar]
- 2.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au W-C, Moore P A, Lafleur D W, Tombal B, Pitha P M. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 4.Azimi N, Brown K, Bamford R N, Tagaya Y, Siebenlist U, Waldmann T A. Human T cell lymphotropic virus type I Tax protein trans-activates interleukin 15 gene transcription through an NF-kappa B site. Proc Natl Acad Sci USA. 1998;95:2452–2457. doi: 10.1073/pnas.95.5.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 6.Bamford R N, Battiata A P, Burton J D, Sharma H, Waldmann T A. Interleukin (IL) 15/IL-T production by the adult T-cell leukemia cell line HuT-102 is associated with a human T-cell lymphotropic virus type I R region/IL-15 fusion message that lacks many upstream AUGs that normally attenuate IL-15 mRNA translation. Proc Natl Acad Sci USA. 1996;93:2897–2902. doi: 10.1073/pnas.93.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bours V, Burd P R, Brown K, Villalobos J, Park S, Ryseck R P, Bravo R, Kelly K, Siebenlist U. A novel mitogen-inducible gene product related to p50/p105-NF-κB participates in transactivation through a κB site. Mol Cell Biol. 1992;12:685–695. doi: 10.1128/mcb.12.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton J D, Bamford R N, Peters C, Grant A J, Kurys G, Goldman C K, Brennan J, Roessler E, Waldmann T A. A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci USA. 1994;91:4935–4939. doi: 10.1073/pnas.91.11.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doherty T M, Seder A S, Sher A. Induction and regulation of IL-15 expression in murine macrophages. J Immunol. 1996;156:735–741. [PubMed] [Google Scholar]
- 10.Drew P D, Franzoso G, Becker K G, Bours V, Carlson L M, Siebenlist U, Ozato K. NF-κB and interferon regulatory factor 1 physically interact and synergistically induce major histocompatibility class I gene expression. J Interferon Cytokine Res. 1995;15:1037–1045. doi: 10.1089/jir.1995.15.1037. [DOI] [PubMed] [Google Scholar]
- 11.Du W, Dimitris W, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 12.Fujita T, Ohno S, Yasumitsu H, Taniguchi T. Delimitation and properties of DNA sequences required for the regulated expression of human interferon-beta gene. Cell. 1985;41:489. doi: 10.1016/s0092-8674(85)80022-2. [DOI] [PubMed] [Google Scholar]
- 13.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, Roulston A, Lacoste J, Hiscott J. Viral induction of the human beta interferon promoter: modulation of transcription by NF-κB/rel proteins and interferon regulatory factors. J Virol. 1994;68:4707–4715. doi: 10.1128/jvi.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grabstein K H, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn M A, Ahdieh M, Johnson L, Alderson M R, Eatson J D, Anderson D M, Giri J G. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- 15.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 16.Jonuleit H, Wiedemann K, Müller G, Degwert J, Hoppe U, Knop J, Enk A H. Induction of IL-15 messenger RNA and protein in human blood-derived dendritic cells: a role for IL-15 in attraction of T-cells. J Immunol. 1997;158:2610–2615. [PubMed] [Google Scholar]
- 17.Juang Y-T, Lowther W, Kellum M, Au W-C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y B, Satoh J-L, Walker D G, Seung U K. Interleukin 15 gene expression in human astrocytes and microglia in culture. NeuroReport. 1996;7:1062–1066. doi: 10.1097/00001756-199604100-00022. [DOI] [PubMed] [Google Scholar]
- 19.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteosome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin R, Heylbroeck C, Genin P, Pitha P M, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis T, Whittemore L-A, Du W, Fan C-M, Keller A D, Palombella V J, Thanos D. Positive and negative control of human interferon-β gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.May J M, Ghosh S. Signal transduction through NF-κB. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 26.Mrozek E, Anderson P, Caligiuri M A. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- 27.Neish A S, Read M A, Thanos D, Pine R, Maniatis T, Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-κB as a transcriptional activator of vascular cell adhesion molecule-1. Mol Cell Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 29.Ogasawara K, Hida S, Azimi N, Tagaya Y, Sato T, Yokochi-fukuda T, Waldmann T A, Taniguchi T, Taki S. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature. 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 30.Puzanov I J, Williams N S, Schatzle J, Sivakumar P V, Bennett M, Kumar V. Ontogeny of NK cells and the bone marrow microenvironment: where does IL-15 fit in? Res Immunol. 1997;148:198–201. doi: 10.1016/s0923-2494(97)84225-3. [DOI] [PubMed] [Google Scholar]
- 31.Quinn L S, Haugk K L, Grabstein K H. Interleukin-15: a novel anabolic cytokine for skeletal muscle. Endocrinology. 1995;136:3669–3672. doi: 10.1210/endo.136.8.7628408. [DOI] [PubMed] [Google Scholar]
- 32.Reinecker H-C, MacDermott R, Mirau S, Dignass A, Podolsky D. Intestinal epithelial cells both express and respond to interleukin 15. Gastroentology. 1996;111:1706–1713. doi: 10.1016/s0016-5085(96)70036-7. [DOI] [PubMed] [Google Scholar]
- 33.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 34.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of IRF family transcription factor IRF-3 in virus-induced activation of the IFN-β gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 35.Saura M, Zaragoza C, Bao C, McMillan A, Lowenstein C J. Interaction of interferon regulatory factor-1 and nuclear factor kappaB during activation of inducible nitric oxide synthase transcription. J Mol Biol. 1999;289:259–471. doi: 10.1006/jmbi.1999.2752. [DOI] [PubMed] [Google Scholar]
- 36.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka N, Taniguchi T. Cytokine gene regulation: regulatory cis-elements and DNA binding factors involved in the interferon system. Adv Immunol. 1992;52:263–281. doi: 10.1016/s0065-2776(08)60877-9. [DOI] [PubMed] [Google Scholar]
- 38.Thanos D, Maniatis T. Virus induction of IFN-β gene expression requires the assembly of an enhancesome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 39.Thanos D, Maniatis T. The high mobility group protein HMGI(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe Y, Kawade Y. Induction, production and purification of natural mouse IFN-α and -β. In: Clemens M J, Morris A G, Gearing A J H, editors. Lymphokines and interferons: a practical approach. Oxford, England: IRL Press; 1987. pp. 1–14. [Google Scholar]
- 41.Wathelet M G, Lin C H, Parekh B S, Ronoco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 42.Yeow W S, Au W-C, Juang Y-T, Fields C D, Dent C L, Gewert D R, Pitha P M. Reconstitution of virus-mediated expression interferon α genes in human fibroblast cells by ectopic interferon regulatory factor-7. J Biol Chem. 2000;275:6313–6320. doi: 10.1074/jbc.275.9.6313. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Siquan S, Hwang I, Tough D F, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]