Abstract
The increasing prevalence and incidence of colorectal cancer (CRC), particularly in young adults, underscore the imperative to comprehend its fundamental mechanisms, discover novel diagnostic and prognostic markers, and enhance therapeutic strategies. Here, we integrated multi-omics data, including gene expression, somatic mutation data and DNA methylation data, to unravel the intricacies of tumor microenvironment (TME) in CRC and search for novel prognostic markers. By calculating the immune score for each patient from the expression profile, we delineated the differential immune cell fraction, constructed an immune-related multi-omics atlas, and identified molecular characteristics. The entire colorectal dataset (n = 343) was randomly divided into training (n = 249) and testing datasets (n = 94). We screened 144 immune-related genes, 6 mutant genes, and 38 methylation probes associated with overall survival (OS). These makers were then incorporated into a 10-gene prognostic model using Lasso and Cox regression in the training dataset, and the model's performance was evaluated in an independent validation dataset. The model exhibited satisfactory results (average concordance index [C-index] = 0.77), with the average 1-year, 3-year, and 5-year AUCs being 0.79, 0.76, and 0.76 in the training dataset and 0.74, 0.80, and 0.90 in the testing dataset. Furthermore, the prognostic model demonstrated applicability in guiding chemotherapy for CRC patients and exhibited a degree of pan-cancer utility in risk stratification. In conclusion, our integrated analysis of multi-omics data revealed immune-related genetic and epigenetic characteristics of the TME. We propose an integrative prognostic model that can stratify risk and guide chemotherapy for CRC patients. The generalizability of the model in risk stratification across different cancer types was validated in Pan-Cancer cohort.
Keywords: Multi-omics, Tumor microenvironment, Immunotherapy, Prognostic model, Colorectal cancer
1. Introduction
Colorectal cancer (CRC) is the third most common cancer and exhibits the fourth highest mortality rate worldwide [1]. Early diagnosis and prompt treatment can improve the prognosis of CRC. Genetic changes are the main reason for the occurrence and progression of CRC [2]. Therefore, genetically related therapeutic strategies, including chemotherapy, precision therapy, and immunotherapy or some combination, can help prolonging the patient's survival [[3], [4], [5]]. Precision therapy requires specific gene mutations in CRC patients; i.e., the EGFR acts as a sensitive therapeutic target for the tyrosine kinase inhibitor (TKI), BRAFV600E, is responsible for the phosphorylation and activation of MEK1 and MEK2; KRAS, initiates the KRAS pathway, and HER2, activates tyrosine kinase activity [6,7]. Unfortunately, most of colorectal patients lack targetable mutations. Immunotherapy on immune checkpoint blockers (ICBs), such as nivolumab and ipilimumab, has shown strong activity as first-line treatments. However, the method is affected by the tumor microenvironment (TME), such as T cell abundance, tumor mutation burden, and regulatory relationships with other biomolecules, highlighting the importance and urgency of deeply understanding how the TME directs the therapeutic and prognostic outcomes [8].
The TME is composed of stromal cells, immune/inflammatory cells, blood vessels and cytokines, which affects the occurrence, development, metastasis, recurrence and drug resistance of tumors [9,10]. The promoting roles of the TME in the immunotherapeutic response, drug resistance and prognosis have been demonstrated in various cancer types [[11], [12], [13]]. For example, a consensus molecular subtype classification system consisting of four subtypes (CMS1–CMS4) [14], and its intersection with a coordinate immune response cluster of 28 immune genes, was found to reveal relatively poor immune infiltration (i.e., CD4+ T cells) and low inhibitory molecule (i.e., CTLA4, PDL1, PDL2, LAG3, and TIM3) expression [15]. Additionally, Chen et al. established a model comprising 5 differentially expressed immune genes as predictive signatures and attempted to determine their relationship with clinicopathological factors in colon cancer [16]. However, most CRC genetic prediction models are constructed by single-omics data and are not comprehensive enough to achieve satisfactory performance [[17], [18], [19], [20], [21], [22]]. Pankaj Ahluwalia et al. performed a gene expression prognostic model and identified four prognostic genes in 597 CRC patients, with a 1-year AUC of 0.590 [19].
The aim of this study was to estimate TME patterns in CRC patients based on the RNA-seq data, identify the multi-omics characteristics related to immunity, and integrate these genetic or epigenetic characteristics with prognostic significance in a predictive model. The results of this study are expected to provide a more comprehensive immune genome and epigenome landscape, identify better prognostic predictors, and provide guiding values for individualized treatment and chemotherapeutic drugs selection for patients with CRC.
2. Results
2.1. TME construction in colorectal
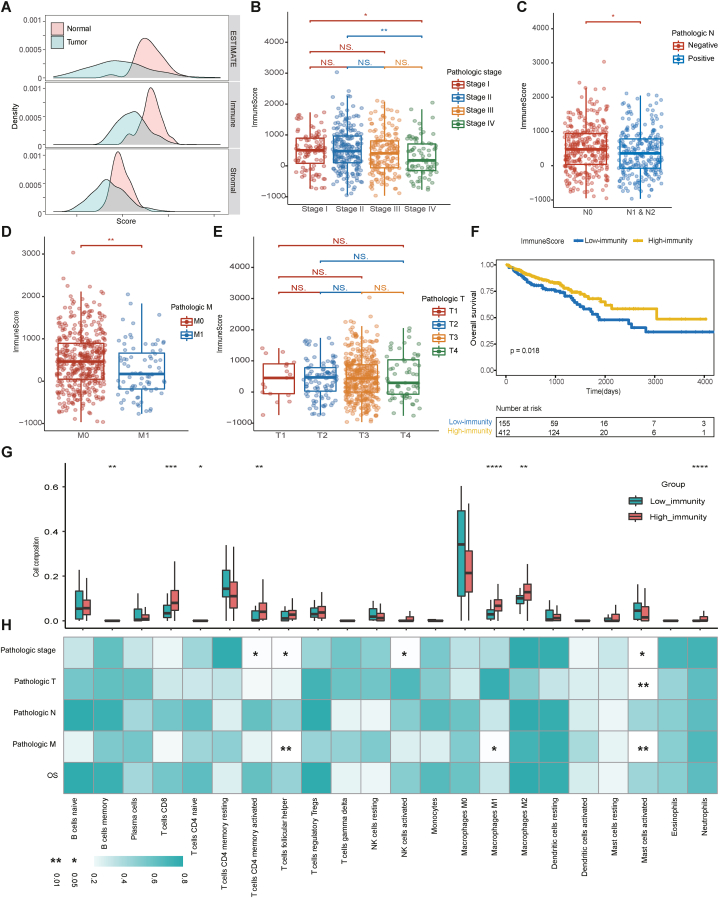
To predict the presence of stromal and immune cells in tumor tissue, we calculated the immune score, ESTIMATE score and stromal score from gene expression data in the dataset (n = 618) using the R package “estimate” (Table S1) [23]. The immune, ESTIMATE and stromal scores represent the infiltration level of the immune cells, tumor purity and tumor-associated stromal cells, respectively. The tumor samples had a significantly lower distribution of immune scores than the normal samples (Mann–Whitney U test, p < 0.0001), and so were the stromal scores (Mann–Whitney U test, p < 0.0001) and estimate scores (Mann–Whitney test, p < 0.0001), indicating that higher tumor purity existed in the colorectal samples (Fig. 1A). Then, we investigated the relationship between tumor purity and clinical characteristics, and no difference was found in estimate scores among pathological stage (Fig. S1A), TNM stage (Figure S1B, C and D) or prognosis (HR: 1.146; 95 % CI: 0.787–1.671; log-rank p = 0.48) (Figs. S1E and F).
Fig. 1.
Construction of TME in patients with colorectal cancer. (A) The distributions of estimate scores, immune scores, and stromal scores between tumor and normal samples. (B–E) Immune scores on the (B) pathologic stage, (C) pathologic N stage, (D) pathologic M stage, and (E) pathologic T stage. (F) Kaplan–Meier curves show the independent relevance between OS time and immune scores. (G) The immune cell fractions between the high and low-immunity cohorts. (H) Correlations between clinical characteristics and immune cell fractions. The blue-to-white gradient indicates the significance level with * representing p < 0.05 and ** representing p < 0.01. (G) and (H) share the same x axis.
Furthermore, the relationship among stromal score, immune cells and clinical characteristics was discussed, as stromal cells play an important role in tumor growth, disease progression and drug resistance, and infiltrating immune cells, especially infiltrating T lymphocytes, are associated with tumor growth, invasion and metastasis [[24], [25], [26]]. A continuous decrease in the immune score was observed during the progression of the disease (stage I: Median = 502.80, IQR = 30.69 to 897.60; stage II: Median = 480.20, IQR = 97.51 to 971.50; stage III: Median = 404.30, IQR = −61.47 to 810.50; and stage IV: Median = 176.60, IQR = −164.40 to 722.10). Notably, the immune scores of stage I (Mann–Whitney test, p = 0.0205) and stage II (Mann–Whitney test, p = 0.0020) were significantly higher than that of stage IV (Fig. 1B). According to TNM stage, the immune scores showed significant differences among lymph nodes (Mann–Whitney test, p = 0.0312) (Fig. 1C) and distant metastasis (Mann–Whitney test, p = 0.0017) (Fig. 1D) but not in T stage (Fig. 1E). The stromal score showed a continuous increase during the progression of the disease (stage I: Median = −690.80, IQR = −1178.00 to −325.00; stage II: Median = −627.30, IQR = −1108.00 to 5.74; stage III: Median = −606.80, IQR = −977.60 to −61.27; and stage IV: Median = −563.80, IQR = −1127.00 to −36.08). The stromal score of stage III was significantly higher than that of stage I (Mann–Whitney, p = 0.0370) (Fig. S2A). Regarding TNM staging, the higher stromal scores were correlated to higher T stage (Fig. S2B) but had no relationship with lymph node metastasis (Mann–Whitney, p = 0.0158) (Fig. S2C) or distant metastasis (Mann–Whitney, p = 0.9649) (Fig. S2D). More importantly, higher immune scores were significantly associated with longer OS (HR: 0.623; 95%CI: 0.420–0.923; log-rank p = 0.018) (Fig. 1F), whereas ESTIMATE scores have no significant relevance with the prognosis (Figs. S1E and F) and higher stromal scores were related to impaired OS (HR: 1.532; 95%CI: 1.046–2.242; log-rank p = 0.028) (Figs. S2E and F). Meanwhile, multivariate Cox proportional hazards regression analysis revealed that the immune scores (HR: 0.519; 95 % CI: 0.287–0.938), pathologic T (T1, reference; T4, HR: 1.970, 95 % CI: 1.247–3.110), and age (HR: 1.041; 95 % CI: 1.022–1.061) were independent predictors of OS in CRC patient. The C-indexes was 0.791 (Table 1). As tumor-infiltrating immune cells might have a stronger clinical correlation and prognostic significance than stromal cells, the following multi-omics analyses mainly focused on the tumor immune microenvironment (TIME) and immunity-related genes.
Table 1.
Multivariate cox proportional hazards regression of the scores and clinical variables.
| Factors | HR | 95 % CI | P value |
|---|---|---|---|
| Stromal score | 1.151 | 0.5566–2.379 | 0.705 |
| ESTIMATE score | 1.325 | 0.6534–2.688 | 0.435 |
| Immune score | 0.519 | 0.2869–0.938 | 0.030c |
| Pathologic M | 1.832 | 0.7318–4.584 | 0.196 |
| Pathologic N | 1.508 | 0.9942–2.289 | 0.053. |
| Pathologic T | 1.969 | 1.2468–3.11 | 0.004b |
| Pathologic stage | 1.310 | 0.6639–2.585 | 0.436 |
| Age | 1.041 | 1.0219–1.061 | <0.001a |
| Gender | 0.805 | 0.5364–1.207 | 0.293 |
p ≤ 0.001.
p ≤ 0.01.
p ≤ 0.05. p ≤ 0.1.
2.2. Analysis of tumor infiltrating cell composition
The cell composition between the high and low-immunity cohorts was further explored using R the package “CIBERSORT”, and the relationship between the immune cell subsets and clinical characteristics was discovered using Spearman's rho rank correlation. A total of 255 samples with p values less than 0.05, as described in the methods section, were reserved and dissected into 22 types of immune cells. Of these samples, 231 were included in the high-immunity cohort and 24 in the low-immunity cohort. The high-immunity cohort had significantly larger fractions of CD8 T cells (Mann–Whitney test, p = 0.0005), memory activated CD4 T cells (Mann–Whitney test, p = 0.0026), M1 macrophages (Mann–Whitney test, p < 0.0001), and M2 macrophages (Mann–Whitney test, p = 0.0016) (Fig. 1G). These cell members have pro-immune functions [[27], [28], [29], [30], [31]]. Notably, some of the immune cell members had correlations with clinical characteristics. Activated mast cells had significant differences in tumor stage (Mann–Whitney test, p = 0.0009) and distant metastasis (Mann–Whitney test, p = 0.0106). Most of the activated memory CD4 T cells were distributed in the microenvironment of early-stage tumors (Mann–Whitney test, p = 0.0281), as were follicular helper T cells (Mann–Whitney test, p = 0.0163). The latter (Mann–Whitney test, p = 0.0028) and M1 macrophages (Mann–Whitney test, p = 0.0146) were also more common in patients who had no distant metastasis. Furthermore, although one type of cell member contributed little to OS in patients with CRC (Fig. 1H), the combination showed prognostic significance (Fig. 1F).
2.3. Identification of the immune-related differentially expressed genes
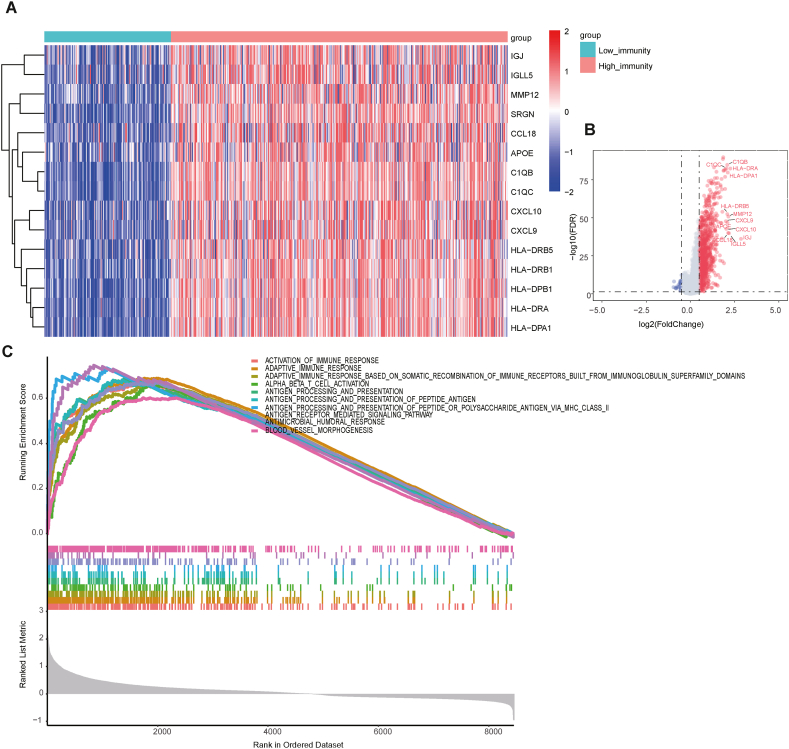
To explore the difference between the high and low-immunity cohorts at the level of the expression profile, the RNA-seq data from the patients with CRC portal were used for differentially expressed genes. The genes with |log2(fold change)| > 0.5 and adjusted p value < 0.05 were considered to be differentially expressed, of which the 15 most significantly upregulated genes are shown in Fig. 2A. A total of 940 and 28 genes were significantly upregulated and downregulated in the high-immunity cohort, respectively (Table S2). The most significantly upregulated twelve genes with log2(fold change) > 2 and adjusted p value < 0.05 were labeled (Fig. 2B). Significantly upregulated genes, such as CXCL9, CXCL10, and CCL18, are responsible for recruiting different types of immune cells [32]. To determine the potential functional mechanism of high-immunity cohort, the differentially expressed genes (DEGs) were submitted for Gene Ontology (GO) analysis using clusterProfiler [33]. As expected, the upregulated genes in the high-immunity cohort were enriched in immune-related biological processes such as activation of the immune response and antigen processing and presentation (Fig. 2C), indicating a positive role in the enhancement of tumor-associated immunity. Furthermore, several upregulated genes, such as IGJ, IGLL5 [34], MMP12 [35], and APOE [36], have been experimentally verified to regulate T cell activation and tumor infiltration. C1QB and C1QC are involved in clearing of apoptotic cells and producing anti-inflammatory cytokines [37], while HLA−DRB5 and HLA−DRA participate in HLA class II composition and enhance antigen affinity in antigen presentation [38,39].
Fig. 2.
Identification of the Immune Infiltration-Related Expression Genes. (A) Heatmap displaying the top 15 immune-related differentially expressed genes, including IGJ, IGLL5, MMP12, SRGN, CCL18, APOE, C1QB, C1QC, CXCL10, CXCL9, HLA−DRB5, HLA−DRB1, HLA−DPB1, HLA−DRA, HLA−DPA1. The top bar represents the grouping information, and the central plot represents the gradient of normalized log2FPKM (fragments per kilobase of transcript per million mapped reads). (B) Volcano plot showing the differentially upregulated (red nodes) and downregulated genes (blue nodes) with log2(fold change) > 2 and adjusted p value < 0.05 were labeled. (C) Gene set enrichment analysis (GSEA) shows the significant enrichment of 10 immune-related biological processes.
2.4. Identification of immune-related differentially somatic mutations
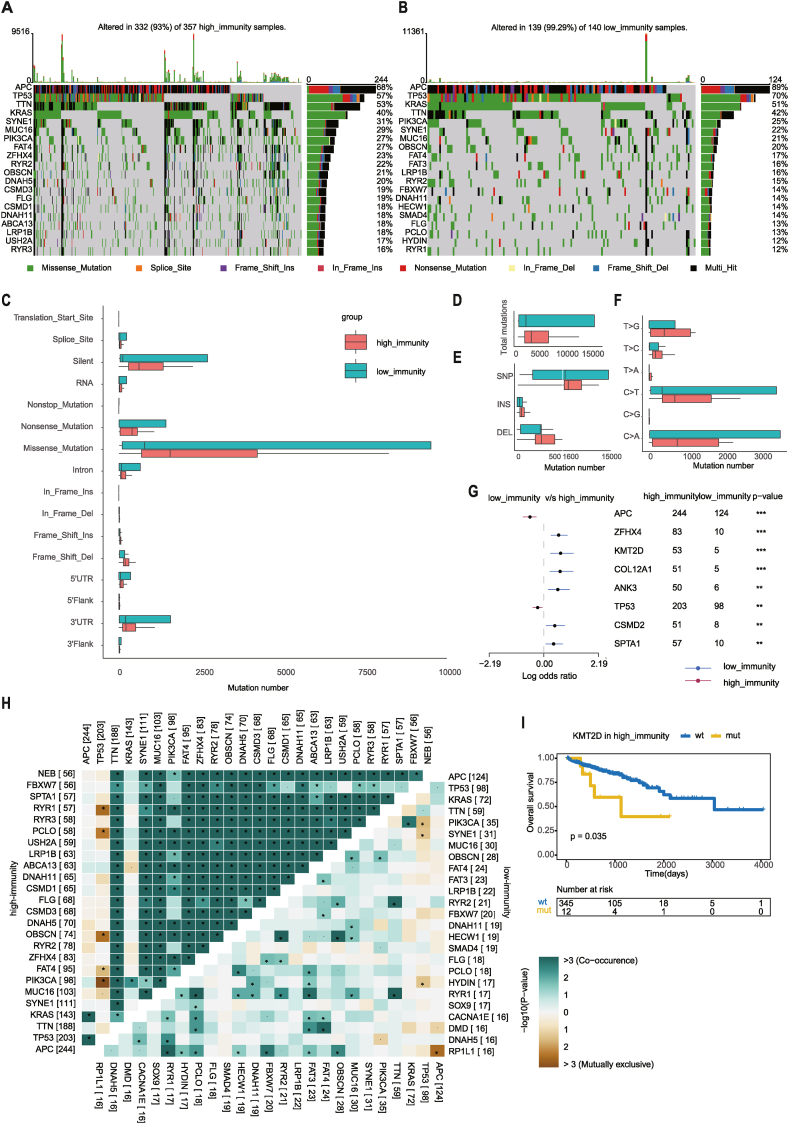
To explain the difference between high and low-immunity cohorts at the somatic mutation level, whole exome sequencing (WES) data including the single-nucleotide variant (SNV), single-nucleotide polymorphism (SNP), insertion (INS), and deletion (DEL) were analyzed and visualized using the R package “maftools” [23]. Table S3A lists a total of 964 differentially mutated genes (DMGs), and Fig. 3A and B shows the top 20 most frequently mutated genes in the high and low-immunity cohorts. APC, TP53, TTN, and KRAS occupied the top four positions in both cohorts, and three of them have previously been shown to be related to the occurrence and development of CRC [2]. This result suggested that the genes mainly play a role in tumor progression rather than immune invasion. Surprisingly, FBXW7 and SMAD4 were ranked in the top 20 in the low-immunity cohort, but not in the high-immunity cohort. FBXW7 promotes innate antiviral immunity by mediating the ubiquitination of SHP2 [40], and participates in the inflammatory response by degrading EZH2 and increasing the expression of CCL2 and CCL7 in CX3CR1hi macrophages [41]. Gu et al. demonstrated that SMAD4 promotes T cell proliferation by targeting Myc, and therefore affects autoimmunity and antitumor immunity by constructing a genetic model of SMAD4-deleted mice [42]. As shown in Fig. 3C, missense mutations are the most common mutations in both the high (n = 357) and low-immunity (n = 140) cohorts. Therefore, we further analyzed the mutation types to reveal the possible mutation mechanism. Overall, no difference in the counts of mutations was between the high (median = 168, IQR: 123–265) and low-immunity (median = 157.5, IQR: 128–201.8) cohorts (Mann–Whitney U test, p = 0.1504) (Fig. 3D), but the mutation frequency of the low-immunity (median = 0.57, IQR: 0.34–0.76) (Mann–Whitney U test, p < 0.0001) cohort was significantly higher than that of the high-immunity (median = 0.36, IQR: 0.21–0.54) cohort. High mutation frequency has been proved to be associated with high tumor purity and low heterogeneity, which may cause tumor progression and poor prognosis [43,44]. For variant type analysis, no difference in the number of SNVs was found between the high and low-immunity cohorts (Mann–Whitney U test, p = 0.1492) (Fig. 3E); however, T > A (Mann–Whitney U test, p = 0.0003) and T > G (Mann–Whitney U test, p = 0.0098) were more common in the high-immunity cohort than in the low-immunity cohort (Fig. 3F). Significantly, samples in the high-immunity cohort had more numbers of INSs (Mann–Whitney U test, p = 0.0118) and DELs (Mann–Whitney U test, p = 0.047) than those in the low-immunity cohort (Fig. 3E). Next, we investigated the mutation frequencies between the high and low-immunity cohorts and identified the genes with differential mutation frequencies (Fisher's exact test, p < 0.01) (Fig. 3G). Symbiosis or exclusivity is a characteristic of many oncogene mutation patterns, which can determine drug selection and predict the therapeutic effect. Therefore, we detected the important mutually exclusive or cooccurrence gene pairs among top 25 most frequently mutated genes using paired Fisher's exact test. Notably, compared with the low-immunity cohort, the high-immunity cohort had a common cooccurrence landscape, except TP53, APC and KRAS, which were exclusive to most other genes in both of the two cohorts. TP53 was found to be mutually exclusive to RYR1, PCLO, OBSCN, PIK3CA, and FAT4 in the high-immunity cohort, and to HYDIN, SYNE1, and PIK3CA in the low-immunity cohort (Fig. 3H). This result indicates a redundancy effect in the pathway between them, and the selection advantage retains multiple mutations. Furthermore, KMT2D is another typical example illustrating the different mutation spots between two cohorts (Fig. S3A) and the plausible chain reaction of the differences in prognostic impact (Fig. 3I and Fig. S3B).
Fig. 3.
Landscape of Somatic Mutation in High and Low-Immunity Cohorts. (A–B) Oncoplots show the mutation distribution of the top 20 most frequently mutated genes. The central panel shows the types of mutations in each colorectal sample. The upper panel shows the mutation numbers of each sample. The bar plots on the right side show the frequency and mutation type of genes mutated in the (A) high and (B) low-immunity cohorts, respectively. (C–F) Boxplots showing the mutation numbers of (C) mutation type classified by effects, (D) total mutations, (E) INDEL and SNP, and (F) SNV between high and low-immunity cohorts. (G) Forest plot displaying the top 8 most significantly differentially mutated genes between the high and low-immunity cohorts. (H) The heatmap describes the mutually cooccurring and exclusive mutations of the top 25 frequently mutated genes. The color and symbol in each cell represent the statistical significance of the exclusivity or cooccurrence for each pair of genes. (I) Kaplan–Meier curve shows the independent relevance between OS time and KMT2D mutation in high-immunity cohort.
2.5. Identification of immune-related differentially methylated probes
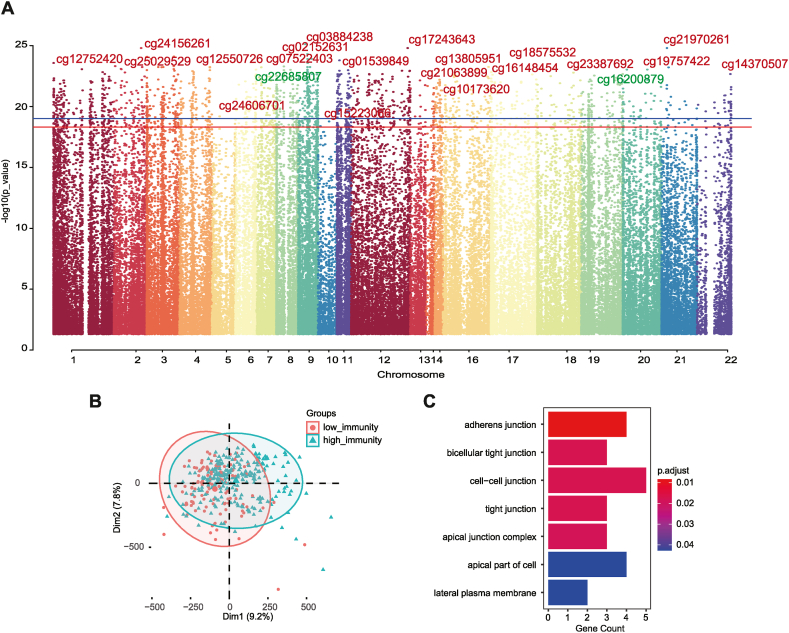
To explain the difference between the high and low-immunity cohorts based on DNA methylation patterns, the Illumina 450k DNA methylation data from the patients with CRC were analyzed for differentially expressed genes using the R package “ChAMP”. Aberrant methylation can cause tumor formation and progression [45,46]. In 443 samples with no missing beta values used to detect the differential methylation probes (DMPs), a total of 1830 immune-related DMPs were identified according to the standard of |log2(fold change)| > 0.15 and adjusted p-value <0.05 between high and low-immunity cohorts (Fig. 4A and B; Table S4). A total of 1716 (93.77 %) hypermethylated probes across 944 genes and 114 (6.23 %) hypomethylated probes involving 88 genes were identified in the high-immunity cohort, compared with the low-immunity cohort. The functions of DMP-related genes upregulated in high-immune cohorts were enriched based on GO analysis. The 7 enriched GO biological processes showed that they had a potential role in cell signal transduction (Fig. 4C). On the one hand, they upregulated the cell junction pathway; on the other hand, the lateral plasma membrane and apical complex were downregulated, indicating that the difference in prognosis of the tumor immune microenvironment may be partly due to abnormal methylation affecting cell recognition and connection.
Fig. 4.
Differential Methylation Pattern in TIME. (A) Manhattan plot of the genome-wide differential methylation probes in high and low-immunity cohorts. (B) Principal component analysis showed the distribution of differentially methylated probes in the high and low-immunity cohorts. (C) Bar plot showing the results of GO biological process (GOBP) enrichment analyses on DMP-associated genes using R package “clusterProfiler”.
2.6. Construction of the prognostic model by integrated multi-omics data
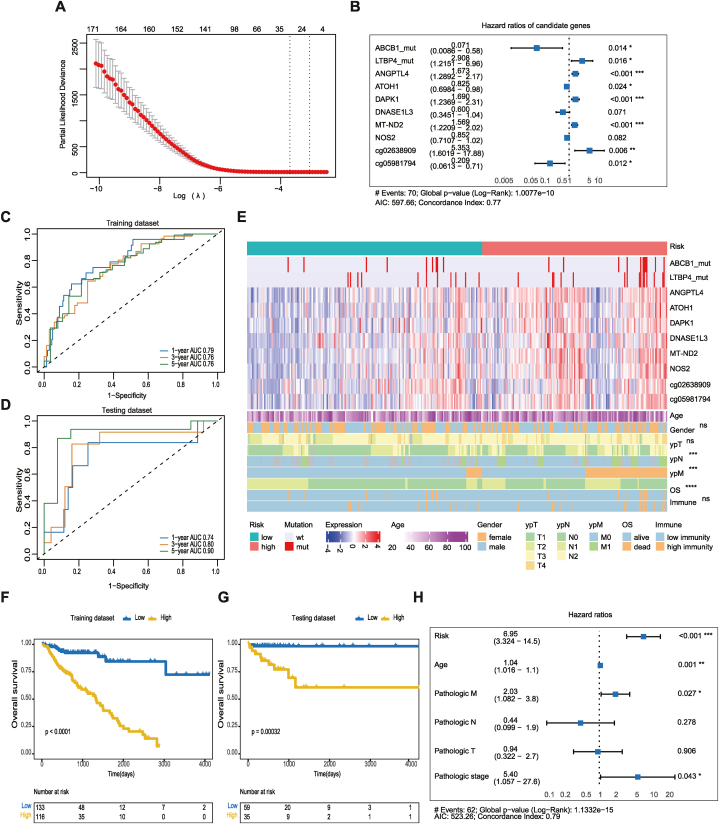
By analyzing the multi-omics data above, we identified the immune-related molecular features of expression, mutation and methylation. A total of 968 differentially expressed genes, 298 frequently mutated genes, and 1830 differentially methylated probes were integrated for subsequent analysis. Lasso regression and Cox proportional hazards regression were accepted to determine immune-related prognostic features. Meanwhile, the joint and separate effects of features were investigated to determine which one had the best performance. For joint effects, all candidate genetic features were combined in the training dataset using the stepwise method (Fig. 5A), of which 144 events composed of DEGs, 6 frequent mutations, and 38 DMPs were identified to significantly and independently influence OS using a univariate Cox proportional hazards model (Table S5). Next, a 10-gene prognostic model using the Lasso and Cox regression analysis with a concordance index (C-index) of 0.77 was established, including 2 DMGs (ABCB1, LTBP4), 6 DEGs (ANGPTL4, ATOH1, DAPK1, DNASE1L3, MT-ND2, NOS2), and 2 DMPs (cg02638909, cg05981794) (Fig. 5B). In brief, the expression levels of ANGPTL4, DAPK1, and MT-ND2, the mutation of LTBP4, and the methylation levels of the cg02638909 probe had significantly positive contributions to a poorer prognosis, while the expression levels of ATOH1, the mutation of ABCB1, and the methylation levels of the cg05981794 probe played contrary roles. Then, the risk scores related to OS status, were calculated for each sample in the training and testing datasets based on the prognostic models. The prediction ability of the model was verified by timeROC analysis, and the AUC values at 1 year, 3 years, and 5 years were 0.79, 0.76, and 0.76, respectively, on the training datasets (Fig. 5C). On the testing datasets, the performance remained excellent, with AUC values of 1 year, 3 years, and 5 years of 0.74, 0.80, and 0.90, respectively (Fig. 5D).
Fig. 5.
Establishment of the prognostic model using immune-related multi-omics data through colorectal datasets. (A) Identification of the optimal penalization coefficient lambda in the Lasso regression model. (B) Forest plot of the prognostic impact of 10 candidate variables. (C–D) ROC curves of the risk score for predicting 1-year, 3-year, and 5-year OS in the (C) training and (D) testing datasets. (E) There are three parts in the heatmap. The top bar shows the risk score. The middle part represents the mutation status (0 for non-mutated and 1 for mutated), normalized values of log2FPKM and log2 beta. The bottom part shows the demographic and clinical features (age, gender, pathological stage, and OS) and immune scores. (F–G) Kaplan–Meier curves show the independent relevance between OS time and risk scores in the (F) training and (G) testing datasets. (H) Forest plot of the prognostic impact of risk score and clinical features.
Moreover, the samples were classified into high and low-risk groups according to the optimal cutoff point for each risk score using ROC curves. As shown in Fig. 5E, the mutation status, expression, methylation probe level, clinical characteristics and immune score were displayed according to the high and low-risk groups mentioned above. The patients classified in the high-risk group were more likely to have lymph node metastasis (Fisher's exact test, p = 0.0007), distal metastasis (Fisher's exact test, p = 0.0003) or die (Fisher's exact test, p < 0.0001). Patients classified in the high-risk group had poorer OS than those classified in the low-risk group in both the training dataset (HR: 2.642; 95 % CI: 1.425–4.897; log-rank p < 0.0001) (Fig. 5F) and testing dataset (HR: 2.567; 95 % CI: 1.181–5.576; log-rank p = 0.00032) (Fig. 5G). The risk remained an independent prognostic predictor when incorporated into a multivariable cox regression model including traditional predictors (HR: 6.950; 95 % CI, 3.320–14.500; log-rank p < 0.001) (Fig. 5H)
For separate effects, on the one hand, we reproduced the same strategy of modeling (Figs. S4A and B) and verification methods (Figs. S4C and D) as mentioned above in differential expression (left), somatic mutation (middle) and methylation (right) profiles, and found that none of the single-omics models could provide a powerful enough prognostic prediction compared to the multi-omics model. On the other hand, 10 genes enrolled in the prognostic model and risk score above were adopted for Kaplan–Meier survival analyses separately (Fig. S5). Among gene derived from DEGs, somatic mutant profiles, and DMPs, ABCB1, LTBP4, ATOH1, DAPK1, DNASEIL3, NOS2, and cg02638909 can be used as an independent risk factor with a significant effect on OS. ATOH1 (HR: 0.430; 95 % CI: 0.266–0.693; log-rank p = 0.00037) was the best in risk classification among the remaining 9 genes. The HR of the risk score was higher than that of ATOH1, meaning that the classification factor of multi-omics analysis (HR: 7.900; 95 % CI: 4.337–14.390; log-rank p < 0.0001) can more accurately distinguish patients at high risk from patients at low risk than that of single-omics analysis.
2.7. Chemotherapeutic drug response of the prognostic model
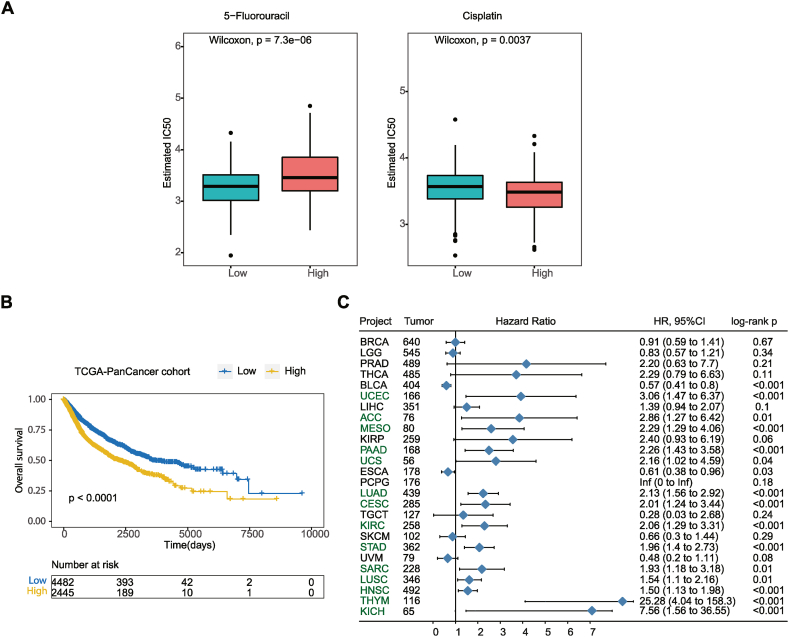
To explore the clinical application of the prognostic model, we evaluated the response to 5-fluorouracil and cisplatin treatment between high and low-risk groups, considering that chemotherapy is the traditional strategy for the treatment of CRC. In the dataset, patients in the low-risk group were more sensitive to 5-fluorouracil (Wilcoxon, p < 0.0001), while those in the high-risk group were more sensitive to cisplatin (Wilcoxon, p = 0.0037) (Fig. 6A). This result suggests that risk scores based on the 10-gene prognostic models can be used for guiding personalized treatment of patients with CRC.
Fig. 6.
Development of the chemotherapeutic drugs and verification of Pan-cancer performance. (A) Distribution of the estimated IC50 of 5-Fluorouracil and Cisplatin among the high and low-risk groups in patients with colorectal cancer. (B) Kaplan–Meier curves of OS among 26 pan cancer species. (C) The forest map shows the effect of risk score on prognosis in 26 pan cancer species.
2.8. The generalization of prognostic models
To verify whether the 10-gene prognosis model is general useable, other cohorts from Pan-Cancer was download as external validation. In Pan-cancer species, we obtained the risk scores of 26 cancer species, and stratified the patients into high and low-risk groups according to the risk scores of each cancer species. As shown in Fig. 6B, the risk score, in pan-cancer cohorts, showed a statistically significant risk stratification ability in OS (HR: 1.586; 95%CI: 1.446–1.739; log-rank p < 0.0001). Fig. 6C shows the distribution of Hazard Ration values in 26 cancer species (Table S6). Among the 14 cancer species (HNSC, LUAD, STAD, LUSC, CESC, KIRC, SARC, PAAD, UCEC, THYM, MESO, ACC, KICH, and UCS), the OS of patients in the high-risk group was significantly worse than that in the low-risk group (Fig. S6), which was completely consistent with the results we found in patients with CRC. Especially in THYM, the recurrence risk of patients in the high-risk group was 25.28 times higher than that in the low-risk group (log-rank p < 0.0001). Nevertheless, there were cases where the 10-gene prognostic model fails to make a meaningful risk stratification of patients in cancer types, such as BRCA, LGG, PRAD, THCA, LIHC, KIRP, PCPG, TGCT, SKCM, UVM, ESCA, and BLCA (Fig. S7).
3. Discussion
In exploring the complex mechanisms of CRC, our study aims to address the current gaps in understanding the impact of the TME on targeted therapy and prognosis, particularly in the context of unresolved clinical challenges such as the selection and dose control of chemotherapeutic drugs. To construct the TME of CRC patients, we utilized a gene expression profile unraveling the immune-infiltration landscape. Notably, significant differences were observed in immune cells between high and low-immunity cohorts. CD8+ T cells, with their specific antitumor effects, were found to be elevated in the high-immunity cohort, correlating negatively with pathological stage and distal metastasis in CRC patients. Moreover, the high proportion of M1 and M2 macrophages in the high-immunity cohort indicated both tumor inhibition and promotion of fibroblast proliferation, respectively [47]. Importantly, our study demonstrated that the composition of immune cells in the TME was not a binary “antitumor” or “protumor” environment but rather an interaction between these immune and nonimmune cells [48]. Then, we explored the potential function of the TME based on genetic or epigenetic characteristics under different immune levels. Upregulated genes in the high-immune cohort were associated with the immune response and participating in antigen presentation. Differentially methylated genes were implicated in facilitating cell-to-cell interactions, immune cell adhesion to tumors, and further promoting CRC progression [49]. Finally, we integrated genetic or epigenetic features with prognostic significance to construct a multi-omics prognostic model. The model exhibited superior prediction performance and a higher AUC value compared to previous single-omics model [16]. Notably, the model's stability in predicting high AUC values in the testing dataset underscored its robustness. The inclusion of independent genetic and epigenetic features known for their prognosis roles in prior studies further validated the model's clinical relevance [[50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63]]. Importantly, our prognostic model effectively stratified patients into high and low-risk groups in both training and validation cohorts. This personalized risk stratification holds the potential to guide treatment strategies, avoiding overtreatment or ineffective interventions. The model's application in predicting the sensitivity of patients to common chemotherapeutic drugs, 5-Fluorouracil and Cisplatin, demonstrated its utility in guiding chemotherapy. This approach aligns with the broader goal of enhancing OS and developing novel treatment strategies for CRC patients. Additionally, our study extended its impact by applying the prognostic model to 26 Pan-Cancer cohorts, reaffirming its efficacy in risk stratification. While some individual cancer species did not show significant risk stratification, the overall performance across diverse cancers, such as HNSC, LUAD, STAD, LUSC, CESC, KIRC, SARC, PAAD, UCEC, THYM, MESO, ACC, KICH, and UCS, showcased the model's pan-cancer potential. However, limitations exist, including the high cost and complexity of multi-omics sequencing data, necessitating further validation through external datasets and clinical trials. Although, with the development of sequencing technology and the reduction of sequencing cost, the model is expected to produce a three-in-one toolkit, opening up a way for its implementation and promotion. Despite these limitations, our study provided valuable insights into the tumor microenvironment of CRC patients, offering a powerful prognostic model with pan-cancer applicability and guiding potential for chemotherapeutic interventions. Further developments in sequencing technology and cost reduction may pave the way for wider implementation and promotion of our model.
4. Materials and methods
4.1. Data collection and processing
The data of 567 colorectal RNA-seq samples with OS information and 51 normal samples, 497 WES (whole exome sequencing) data samples, and 366 Illumina 450k DNA methylation array samples were collected from the colorectal dataset (https://xenabrowser.net/datapages/) (Table S7). GTF files were downloaded from Ensemble (http://asia.ensembl.org) for mRNA annotation. The 32 cancer species applied for external validation with the gene expression RNA-seq, the somatic mutation (SNP and INDEL), and the Methylation450k datasets were downloaded.
4.2. Immune score construction
The immune cell fraction, including stromal score, immune score and estimate score in RNA expression profiles was estimated by the R package “ESTIMATE” [23]. Then, the patients with CRC expression profiles were divided into a high-immunity (above the optimal cutoff value, n = 412) and low-immunity (below the optimal cutoff value, n = 155) cohorts by the optimal cutoff value of the immune score, which was determined based on the “surv_cutpoint” function of the R package “survminer”. The infiltration level and immune cell distribution levels are strongly associated with disease progression and OS outcomes [10]. 22 immune cell types in samples with a p value less than 0.05 was used to ensure the quality of given tumor microenvironment results. The p-values were obtained by CoreAlg based on the Permutation Test method of the R package “CIBERSORT” [64]. The comparison of immune cell distribution levels between the high and low-immunity cohorts was analyzed using the Mann–Whitney test. The p value of the correlation between immune cells and clinical features of tumors was obtained by Pearson analysis. For the WES and methylation profiles, we constructed high and low-immunity cohorts by mapping the sample IDs of the RNA-seq profiles.
4.3. Differential multi-omics data analysis
For gene expression, the RNA-seq data were analyzed for DEGs between the high (n = 412) and low-immunity cohorts (n = 155), with the expression log2(fold change) greater than 0.5 and adjusted p value lower than 0.05 (Benjamini and Hochberg method) using the R package “limma” [65]. For somatic mutations, WES data was analyzed between high (n = 412) and low-immunity cohorts (n = 155) using the R package “maftools”. The differential mutation pattern was identified using Fisher's exact test and side-by-side comparisons by oncoplot. In addition, the differentially mutated genes were found using Fisher's test on 2x2 contingency table with a p value less than 0.05, and the forestplot is shown using R package “forestplot”. Mutually exclusive or cooccurring events were assessed using pairwise Fisher's exact test. For DNA methylation, the Illumina Infinium 450k DNA methylation array data were analyzed using the R package “ChAMP” [66,67]. The probes with at least one missing value were removed, and 371,495 probes and 443 samples were eligible for further analysis. Differentially methylated probes (DMPs) were achieved through the standard process of ChAMP with an arraytype of 450k. Probes with both adjusted p value < 0.05 (Benjamini and Hochberg method) and log2(fold change) > 0.15 were considered as DMPs.
4.4. Establishing and evaluating the prognosis prediction model
To establish the prognostic prediction model, several steps were taken into consideration. First, 343 samples of significantly expressed genes, mutated genes, and methylation probes with OS information were integrated. Second, the prognostic features with p values less than 0.05 in univariate Cox proportional hazards regression were retained for subsequent analysis using the R package “survival”. Third, less informative features were filtered in Lasso-Cox regression model using R package “glmnet”. Finally, the entire datasets were randomly divided into training and testing datasets at a ratio of 7:3. Multivariate cox proportional hazards regression with 10-fold cross validation and stepwise bidirectional analysis was applied for the multi-omics prognostic model in the training dataset (n = 249) using R package “glmnet”. In the construction, the dataset was randomly divided into an internal training set and an internal validation set with 10 repeats, resulting in 10 independent internal training sets and internal validation sets. The internal training set was to obtain the optimal variable combination and the internal validation set is for tuning. The testing dataset (n = 94) was to prove the generalization ability of the tuned model. The risk scores generated by the prognostic model were further combined with several clinical characteristics using a multivariate Cox proportional hazards regression model for predicting OS. Then, patients were divided into a high and low-risk groups by the risk score. The AUC values of 1-year, 3- year, and 5-year receiver operating characteristic (ROC) curves were used to represent the predictive effect of the prognostic model using the R package “timeROC”.
4.5. Predicting the clinical chemotherapeutic response
The chemotherapy response to 5-fluorouracil and cisplatin in the colorectal cohort was estimated using the R package “pRRophetic” [68]. The method fitted a ridge regression model to predict clinical response to chemotherapy according to gene expression and drug sensitivity of cgp2016 cell lines [69]. The drug sensitivity was quantified based on 50 % of cellular growth inhibition (IC50).
4.6. External validation of prognostic model in pan-cancer cohort
Considering that a small sample cohort may cause analysis errors, we excluded 4 cancer types with less than 50 tumor samples (GBM, n = 49; DLBC, n = 37; CHOL, n = 36; OV, n = 9), and utilized the remaining 26 solid tumor cohorts (except for COAD and READ) for subsequent validation. The risk scores of each cohort were obtained according to the 10-gene prognostic model. Then, Kaplan–Meier curves and the log-rank test were used to evaluate the risk stratification ability of 10-gene prognostic model.
4.7. Statistical analysis
Statistical analysis was performed using Fisher's exact test for categorical variables, and the Wilcoxon test or Mann–Whitney U test for continuous variables. We estimated the mutual exclusivity of mutations by Monte Carlo simulation. We calculated the relevance and p value between continuous variables using Pearson's correlation or Spearman's correlation based on the “rcorr” function of the R package “Hmisc”. GSEA analyses of DEGs and GO analyses of the DMPs were performed using the R package “clusterProfiler” [33]. Kaplan–Meier curves and the log-rank test were used for the OS analysis. Statistical tests were performed in R (version 3.2.0). A two-tailed p value < 0.05 was regarded as statistically significant.
Funding statement
This study was supported by National High Level Hospital Clinical Research Funding (BJ-2022-151) and the Fundamental Research Funds for the Central Universities (3332019120).
Ethical declarations
Review and/or approval by an ethics committee was waived for this study as TCGA data are publicly available. Ethical approval has already been granted for all the patients included in the database. Users can download data for free for research and academic publishing. All the authors have no conflicts of interest to declare.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article or its supplementary materials.
CRediT authorship contribution statement
Yifei Li: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Conceptualization. Hexin Li: Validation, Investigation, Data curation. Gaoyuan Sun: Validation, Investigation, Data curation. Siyuan Xu: Validation, Investigation, Data curation. Xiaokun Tang: Validation, Investigation, Data curation. Lanxin Zhang: Writing – original draft, Investigation. Li Wan: Writing – original draft, Investigation. Lili Zhang: Writing – original draft, Investigation. Min Tang: Writing – review & editing, Methodology, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32744.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Bert Vogelstein N.P., Velculescu Victor E., Zhou Shibin, Diaz Luis A., Jr., Kenneth W. Kinzler*, cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merker Jason D., G R.O., Compton Carolyn, Diehn Maximilian, Hurley Patricia, Lazar Alexander J., Lindeman Neal, Lockwood Christina M. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J. Clin. Oncol. 2017 doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 4.Stintzing S., et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019;30(11):1796–1803. doi: 10.1093/annonc/mdz387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dienstmann R., et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 2017;17(2):79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 6.Sartore-Bianchi A., et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 7.Liao W., et al. KRAS-IRF2 Axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell. 2019;35(4):559–572 e7. doi: 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma P., et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binnewies M., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coussens Lisa M., L Z., Karolina Palucka A. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;(339):286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K., Hwang H., Nam K.T. Immune response and the tumor microenvironment: how they communicate to regulate gastric cancer. Gut Liver. 2014;8(2):131–139. doi: 10.5009/gnl.2014.8.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariathasan S., et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roumenina L.T., et al. Context-dependent roles of complement in cancer. Nat. Rev. Cancer. 2019;19(12):698–715. doi: 10.1038/s41568-019-0210-0. [DOI] [PubMed] [Google Scholar]
- 14.Guinney J., et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lal N., et al. An immunogenomic stratification of colorectal cancer: implications for development of targeted immunotherapy. OncoImmunology. 2015;4(3) doi: 10.4161/2162402X.2014.976052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H., Luo J., Guo J. Development and validation of a five-immune gene prognostic risk model in colon cancer. BMC Cancer. 2020;20(1):395. doi: 10.1186/s12885-020-06799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagès F., et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z., et al. Genomics and prognosis analysis of epithelial-mesenchymal transition in colorectal cancer patients. BMC Cancer. 2020;20(1):1135. doi: 10.1186/s12885-020-07615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahluwalia P., et al. Identification and clinical validation of a novel 4 gene-signature with prognostic utility in colorectal cancer. Int. J. Mol. Sci. 2019;20(15) doi: 10.3390/ijms20153818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Romero J., et al. Survival marker genes of colorectal cancer derived from consistent transcriptomic profiling. BMC Genom. 2018;19(Suppl 8):857. doi: 10.1186/s12864-018-5193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang Y., et al. Multi gene mutation signatures in colorectal cancer patients: predict for the diagnosis, pathological classification, staging and prognosis. BMC Cancer. 2021;21(1):380. doi: 10.1186/s12885-021-08108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng Y., et al. A methylation-driven gene panel predicts survival in patients with colon cancer. FEBS Open Bio. 2021;11(9):2490–2506. doi: 10.1002/2211-5463.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Zhang J.R.C.G., et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003;(348):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 25.Franck Pagès A.B., Camus Matthieu, M Sc, Fatima Sanchez-Cabo A.C., B S., Molidor Robert, Mlecnik Bernhard. Effector memory T cells, early metastasis, and survival in colorectal cancer. N. Engl. J. Med. 2005;(353):2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 26.Mlecnik B., et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J. Clin. Oncol. 2011;29(6):610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 27.Selitsky S.R., et al. Prognostic value of B cells in cutaneous melanoma. Genome Med. 2019;11(1):36. doi: 10.1186/s13073-019-0647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navegantes K.C., et al. Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J. Transl. Med. 2017;15(1):36. doi: 10.1186/s12967-017-1141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Yang D.P.C. Tumor–host immune interactions and dendritic cell dysfunction. Adv. Cancer Res. 2004 doi: 10.1016/S0065-230X(04)92002-7. [DOI] [PubMed] [Google Scholar]
- 30.Ryzhov S., et al. Adenosine-activated mast cells induce IgE synthesis by B lymphocytes: an A2B-mediated process involving Th2 cytokines IL-4 and IL-13 with implications for asthma. J. Immunol. 2004;172(12):7726–7733. doi: 10.4049/jimmunol.172.12.7726. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg A.S., et al. A role for plasma cell targeting agents in immune tolerance induction in autoimmune disease and antibody responses to therapeutic proteins. Clin. Immunol. 2016;165:55–59. doi: 10.1016/j.clim.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Zheng X., Hu Y., Yao C. The paradoxical role of tumor-infiltrating immune cells in lung cancer. Intractable Rare Dis Res. 2017;6(4):234–241. doi: 10.5582/irdr.2017.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson C., et al. Prognostic implications of the expression levels of different immunoglobulin heavy chain-encoding RNAs in early breast cancer. NPJ Breast Cancer. 2020;6:28. doi: 10.1038/s41523-020-0170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y., et al. Three genes predict prognosis in microenvironment of ovarian cancer. Front. Genet. 2020;11:990. doi: 10.3389/fgene.2020.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavazoie M.F., et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell. 2018;172(4):825–840 e18. doi: 10.1016/j.cell.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukundan L., et al. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 2009;15(11):1266–1272. doi: 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khodadoust M.S., et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543(7647):723–727. doi: 10.1038/nature21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godfrey D.I., et al. Unconventional T cell targets for cancer immunotherapy. Immunity. 2018;48(3):453–473. doi: 10.1016/j.immuni.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Song Y., et al. E3 ligase FBXW7 is critical for RIG-I stabilization during antiviral responses. Nat. Commun. 2017;8 doi: 10.1038/ncomms14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He J., et al. Fbxw7 increases CCL2/7 in CX3CR1hi macrophages to promote intestinal inflammation. J. Clin. Invest. 2019;129(9):3877–3893. doi: 10.1172/JCI123374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu A.D., et al. A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor beta receptor signaling. Immunity. 2015;42(1):68–79. doi: 10.1016/j.immuni.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin H.T., et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat. Commun. 2017;8(1):1377. doi: 10.1038/s41467-017-01470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sallman D.A., Padron E. Integrating mutation variant allele frequency into clinical practice in myeloid malignancies. Hematol Oncol Stem Cell Ther. 2016;9(3):89–95. doi: 10.1016/j.hemonc.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Koch A., et al. Analysis of DNA methylation in cancer: location revisited. Nat. Rev. Clin. Oncol. 2018;15(7):459–466. doi: 10.1038/s41571-018-0004-4. [DOI] [PubMed] [Google Scholar]
- 46.Soozangar N., et al. Comparison of genome-wide analysis techniques to DNA methylation analysis in human cancer. J. Cell. Physiol. 2018;233(5):3968–3981. doi: 10.1002/jcp.26176. [DOI] [PubMed] [Google Scholar]
- 47.Mantovani A., et al. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giraldo N.A., et al. The clinical role of the TME in solid cancer. Br. J. Cancer. 2019;120(1):45–53. doi: 10.1038/s41416-018-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alan G., Baxter P.D.H. Activation rules: the two-signal theories of immune activation. Nat. Rev. Immunol. 2002;2(6):439–446. doi: 10.1038/nri823. [DOI] [PubMed] [Google Scholar]
- 50.O'Connor G. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. JAMA Oncol. 2011;306(16):1765–1774. doi: 10.1001/jama.2011.1529. [DOI] [PubMed] [Google Scholar]
- 51.Soldevilla B., et al. Prognostic impact of DeltaTAp73 isoform levels and their target genes in colon cancer patients. Clin. Cancer Res. 2011;17(18):6029–6039. doi: 10.1158/1078-0432.CCR-10-2388. [DOI] [PubMed] [Google Scholar]
- 52.Bogdan Badic S.D., El Khoury Flaria, De La Grange Pierre, et al. Prognostic impact of cancer stem cell markers ABCB1, NEO1 and HIST1H2AE in colorectal cancer. Am J Transl Res. 2020;12(9):5797–5807. [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou S., et al. High expression of angiopoietin-like protein 4 in advanced colorectal cancer and its association with regulatory T cells and M2 macrophages. Pathol. Oncol. Res. 2020;26(2):1269–1278. doi: 10.1007/s12253-019-00695-0. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J., et al. ANGPTL4 overexpression is associated with progression and poor prognosis in breast cancer. Oncol. Lett. 2020;20(3):2499–2505. doi: 10.3892/ol.2020.11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tak-Pan Ng Kevin, A X., Qiao Cheng, Guo Dong Yong, Lim Zophia Xue-Hui, Sun Chris Kin-Wai, Fung J.H.-S. Clinical relevance and therapeutic potential of angiopoietin-like protein 4 in hepatocellular carcinoma. Mol. Cancer. 2014;13(196) doi: 10.1186/1476-4598-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Zhang Z., Yu Z. Identification of a novel glycolysis-related gene signature for predicting metastasis and survival in patients with lung adenocarcinoma. J. Transl. Med. 2019;17(1):423. doi: 10.1186/s12967-019-02173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verine J., et al. Determination of angptl4 mRNA as a diagnostic marker of primary and metastatic clear cell renal-cell carcinoma. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curiel T.J., et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 59.Ivanovska J., et al. DAPK loss in colon cancer tumor buds: implications for migration capacity of disseminating tumor cells. Oncotarget. 2015;6(34):36774–36788. doi: 10.18632/oncotarget.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H.Y., Lee Y.R., Chen R.H. The functions and regulations of DAPK in cancer metastasis. Apoptosis. 2014;19(2):364–370. doi: 10.1007/s10495-013-0923-6. [DOI] [PubMed] [Google Scholar]
- 61.Gozuacik D., et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15(12):1875–1886. doi: 10.1038/cdd.2008.121. [DOI] [PubMed] [Google Scholar]
- 62.Yuan W., et al. Downregulation of DAPK1 promotes the stemness of cancer stem cells and EMT process by activating ZEB1 in colorectal cancer. J. Mol. Med. (Berl.) 2019;97(1):89–102. doi: 10.1007/s00109-018-1716-8. [DOI] [PubMed] [Google Scholar]
- 63.Yang X., et al. MicroRNA-613 promotes colon cancer cell proliferation, invasion and migration by targeting ATOH1. Biochem. Biophys. Res. Commun. 2018;504(4):827–833. doi: 10.1016/j.bbrc.2018.09.054. [DOI] [PubMed] [Google Scholar]
- 64.Newman A.M., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ritchie M.E., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian Y., et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33(24):3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morris T.J., et al. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014;30(3):428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paul Geeleher N.J.C., Huang R Stephanie. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15(3):R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article or its supplementary materials.