Abstract
In contrast to pathogenic human immunodeficiency virus and simian immunodeficiency virus (SIV) infections, chronic SIVagm infections in African green monkeys (AGMs) are characterized by persistently low peripheral and tissue viral loads that correlate with the lack of disease observed in these animals. We report here data on the dynamics of acute SIVagm infection in AGMs that exhibit remarkable similarities with viral replication patterns observed in peripheral blood during the first 2 weeks of pathogenic SIVmac infections. Plasma viremia was evident at day 3 postinfection (p.i.) in AGMs, and rapid viral replication led by days 7 to 10 to peak viremias characterized by high levels of antigenemia (1.2 to 5 ng of p27/ml of plasma), peripheral DNA viral load (104 to 105 DNA copies/106 peripheral blood mononuclear cells [PBMC]), and plasma RNA viral load (2 × 106 to 2 × 108 RNA copies/ml). The lymph node (LN) RNA and DNA viral load patterns were similar to those in blood, with peaks observed between day 7 and day 14. These values in LNs (ranging from 3 × 105 to 3 × 106 RNA copies/106 LN cell [LNC] and 103 to 104 DNA copies/106 LNC) were at no time point higher than those observed in the blood. Both in LNs and in blood, rapid and significant decreases were observed in all infected animals after this peak of viral replication. Within 3 to 4 weeks p.i., antigenemia was no longer detectable and peripheral viral loads decreased to values similar to those characteristic of the chronic phase of infection (102 to 103 DNA copies/106 PBMC and 2 × 103 to 2 × 105 RNA copies/ml of plasma). In LNs, viral loads declined to 5 × 101 to 103 DNA copies and 104 to 3 × 105 RNA copies per 106 LNC at day 28 p.i. and continued to decrease until day 84 p.i. (<10 to 3 × 104 RNA copies/106 LNC). Despite extensive viremia during primary infection, neither follicular hyperplasia nor CD8+ cell infiltration into LN germinal centers was detected. Altogether, these results indicate that the nonpathogenic outcome of SIVagm infection in its natural host is associated with a rapidly induced control of viral replication in response to SIVagm infection, rather than with a poorly replicating virus or a constitutive host genetic resistance to virus replication.
Studies of nonhuman primate models for AIDS are extremely useful in addressing the central issues in lentiviral pathogenesis, in particular the role of host factors early after viral exposure. Macaques infected by the simian immunodeficiency virus SIVmac experience a broad range of progression rates (rapid, normal, or slow progression) as is the case for human immunodeficiency virus type 1 (HIV-1)-infected individuals (49). Similar to the situation in humans (12, 15), continuous viral replication takes place in lymphoid tissues, such as lymph nodes (LNs), throughout the course of SIVmac infection (2, 27, 39). As also seen in humans (30), a strong correlation exists between the pattern of viral replication and the clinical course of the infection (10, 21). The level at which viremia stabilizes in macaques 6 weeks postinfection (p.i.) thus is highly predictive of the outcome of the infection. A low viral load in plasma (<104 RNA copies/ml) at this early time point predicts long-term survival, whereas a viral burden that remains above 105 RNA copies/ml is strongly correlated with a pathogenic outcome (44, 46, 49). It has also been shown that infection of macaques with attenuated SIVmac (SIVmacΔnef) is characterized by a low level of viral replication during all phases of infection (8, 9, 26, 50). Despite this low-replication level, SIVmacΔnef induces a strong and particularly early follicular hyperplasia in LNs, possibly indicating that attenuated SIVmac elicits a more rapid immune response than does pathogenic SIVmac (9).
Attenuated SIV infections in macaques, however, have been achieved only by using molecular clones. In contrast to SIV infections in macaques, the outcomes of SIV infections in their natural hosts are generally nonpathogenic. One of the most studied models of natural lentiviral infection is SIVagm infection of African green monkeys (AGMs). AGMs show a seroprevalence rate of about 45% in the wild (19, 24, 31, 36). Both naturally infected AGMs and AGMs experimentally infected with SIVagm isolates remain healthy without any biological or clinical signs of AIDS (4, 24, 36). It is not known why infections of AGMs by SIVagm do not result in clinical AIDS.
Previous studies with the AGM model were performed largely during the chronic phase of infection. It was demonstrated that SIVagm replicates continuously in chronically infected AGMs, with a rate similar to that of HIV-1 in humans (32). Despite this continuous replication, the amount of cell-associated viral DNA in peripheral blood remains very low (mean 28 copies/106 peripheral blood mononuclear cells [PBMC]) during the chronic phase of infection (5) and resembles that in individuals treated with highly active antiretroviral therapy (45). Furthermore, unlike the situation during progressive HIV-1 or SIVmac infections, the viral load observed in the LNs of long-term-infected AGMs is not higher than that in PBMC (5). This low viral load in LNs is associated with a lack of trapping of virus particles by follicular dendritic cells (FDC), as indicated by in situ hybridization studies performed during the chronic phase of infection (reference 5 and our unpublished observations). Contrary to nonprogressive infections with SIVmacΔnef in macaques, no follicular hyperplasia is observed in long-term SIVagm-infected AGMs. It is, however, not excluded that an early but transient development of LN germinal centers (GCs) occurs.
Host factors almost certainly play an important role in AGM resistance to disease progression, because SIVagm infection in another host (pig-tailed macaques) can lead to high viral loads, follicular hyperplasia and follicular depletion in LNs, and finally AIDS (20). As demonstrated in progressive HIV-1 and SIVmac infections, critical host responses are likely to occur within days after viral exposure (29). Until now, no precise immunological or virological data were available concerning the early phases of infection in AGMs or in any other natural host species of SIV.
Here, we have examined the viral load in peripheral blood and in LNs of AGMs (Cercopithecus sabaeus) and described histological features of the LNs during the early phases of SIVagm infection to provide clues helpful for understanding critical events of the initial lentivirus-host interactions. This study reveals an early and extensive SIVagm replication in its natural host. Viral replication is, however, rapidly and efficiently controlled. This control of viral replication is not associated with LN hyperplasia or CD8+ cell infiltration in LN GCs. These data provide evidence that host factors which exert their effects prior to the detection of humoral responses are able to rapidly curtail SIVagm replication in AGMs.
MATERIALS AND METHODS
Virus stocks.
To avoid in vitro culture selection, all inocula used in this study consisted of either PBMC plus plasma or plasma alone obtained directly from SIVagm-infected AGMs.
To constitute a sufficiently large stock of PBMC and plasma from a single naturally infected monkey, blood was sequentially (six times) collected from an adult AGM (animal 92018) that was seropositive for SIV at time of capture and simian T-cell leukemia virus (STLV) seronegative. PBMC were obtained by Ficoll-Hypaque density gradient centrifugation of whole blood collected in EDTA-K2 tubes. PBMC and plasma samples were aliquoted and frozen at −70°C.
To determine the infectious titer of the PBMC collected from the naturally infected AGM 92018, one PBMC vial (106 cells) of each harvest was thawed and the cells were pooled. The infectious titer of the pool was determined by a limiting-dilution assay. Briefly, serial fivefold dilutions of the PBMC pool, from 106 cells down to 64 cells, were added to 0.5 × 106 SupT1 cells (four wells per dilution). Virus production was measured by detection of SIV p27 core antigen in the supernatant of each well at days 14 and 28 of culture. The infectious titer of the PBMC pool was then expressed as the 50% tissue culture infectious dose (TCID50) determined by the Karber calculation method. The titer of the PBMC pool corresponded to 10 TCID50/106 PBMC. This PBMC pool constituted the cell-associated inoculum. It was constituted by thawing and pooling the PBMC of each of the AGM 92018 blood harvests immediately before inoculation of the animal.
To constitute a cell-free inoculum, the plasma collected from the naturally infected AGM 92018 was analyzed similarly. Briefly, one plasma vial (1 ml) from each collected blood harvest was thawed, and the six plasma aliquots were pooled. Serial dilutions of the plasma pool were added to SupT1 cells. No virus production could be detected in the cultures. This is in agreement with another study that described a higher efficiency of virus isolation from PBMC (82%) than from plasma (24%) of chronically infected AGMs (18).
We therefore collected plasma from an acutely infected AGM. Briefly, a single SIV- and STLV-seronegative infant AGM (97026) was inoculated with 107 PBMC (corresponding to 100 TCID50 on SupT1 cells) of the cell-associated inoculum. Blood was collected during primary infection (day 8). The plasma obtained after Ficoll-Hypaque gradient was titrated as described above, and its infectious titer was 7,000 TCID50/ml.
Animals and infections.
All AGMs used in this study, including the animals 92018 and 97026, belong to the species C. sabaeus. They were wild born in Senegal, West Africa, and maintained in captivity at the Pasteur Institute facilities in Dakar, Senegal. Their ages were estimated according to physical and morphological criteria (16, 38). Animals with estimated ages between 18 months and 3 years were classified as juveniles, younger monkeys were classified as infants, and older animals were considered adults.
SIV and STLV seronegativity prior to inoculation was demonstrated by Western blotting (Lav blot II Diagnostics Pasteur; Diagnostics Biotechnology). For each intervention, animals were ketamine-HCl anesthetized. Four SIV-negative juvenile AGMs (designated 96001, 96008, 96011, and 96023, comprising two males and two females weighing approximately 2.5 kg each) were inoculated with 300 TCID50 of the cell-associated inoculum (immediately before inoculation, the cells were pooled in order to constitute the PBMC pool at 10 TCID50/106 PBMC as described above; 3 × 107 PBMC of this pool, resuspended in 1 ml of plasma from AGM 92018, corresponding to 300 TCID50 on SupT1 cells, were inoculated into each monkey).
Two juvenile monkeys were used as controls: one SIV- and STLV-negative monkey (AGM 96014) received the same amount of PBMC and plasma from an uninfected AGM; the other (AGM 96030) was inoculated with 300 TCID50 of the cell-free inoculum (42.5 μl of plasma from AGM 97026).
Specimen collection.
Whole blood was collected from all monkeys in EDTA-K2 tubes, twice weekly through the first 3 weeks and then at days 28, 35, 42, 84, and 340. PBMC were isolated by Ficoll-Hypaque gradient. PBMC and plasma aliquots were stored at −70°C and used to quantify viral load. Excisional axillar and inguinal LN biopsies were collected during primary infection at days 0, 3, 10, 17, and 28 for AGMs 96001 and 96008; at days 0, 7, 14, and 21 for AGMs 96011 and 96023; and at days 0, 7, 14, 21, and 28 for the control animal AGM 96014. Additional LN specimens were collected at 3 months p.i. for AGMs 96001, 96011, and 96023 and at 12 months p.i. for AGMs 96008 and 96023. Only plasma samples were collected for the control animal AGM 96030. For each LN specimen, one fragment was fixed overnight in 4% paraformaldehyde (PFA), dehydrated in graded ethanols, and then embedded in paraffin. A second fragment was cryopreserved by immersion in OCT compound (Tissue-Tek; Miles), snap frozen in liquid nitrogen, and stored at −70°C until use for quantification of viral load in LN cells (LNC) and for immunohistochemical analyses.
Determination of CD4 and CD8 lymphocyte subsets.
Lymphocyte subsets from AGMs were analyzed by fluorescence-activated cell sorter analysis (FACScan; Becton Dickinson, Heidelberg, Germany). For staining, 100 μl of cell suspension (5 × 105 PBMC) was mixed with 10 μl of monoclonal antibody (MAb) Leu2a-phycoerythrin (CD8; Becton Dickinson, Mountain View, Calif.) or 10 μl of unconjugated anti-CD4 (Sanofi Recherche, Marnes-La-Coquette, France) and incubated at 4°C for 30 min. After two washing steps with phosphate-buffered saline, 10 μl of fluorescein isothiocyanate-conjugated sheep anti-mouse was added to tubes containing unconjugated anti-CD4 and incubated in the dark at 4°C for 30 min. Following staining, the PBMC were washed and fixed with 1% PFA. An irrelevant anti-mouse immunoglobulin G1 MAb (Lagitre) was used as a negative control. The absolute numbers of cells were calculated from total white blood cell counts and percentages of lymphocytes (gating by scatter).
Histological and immunohistochemical analysis.
Five-micrometer sections of PFA-fixed paraffin-embedded tissues were used for histological analysis. After deparaffination in xylene, sections were rehydrated and stained with hematoxylin-eosin in order to search for elementary lesions.
Five-micrometer sections of either PFA-fixed paraffin-embedded or cryopreserved OCT-embedded tissues were also analyzed by immunohistochemistry to study the distribution of LNC subpopulations. Immunostaining was performed using the alkaline phosphatase (AP)–anti-AP reaction (11), with fast red TR (Sigma) as chromogen. The reaction was terminated using 0.1 M Tris, and the slides were counterstained with Harris hematoxylin. MAbs directed against human cell surface antigens CD4 (F101.69; Sanofi Recherche), CD8 (Leu-2a; Becton Dickinson), CD20 (pan-B; Dako, Copenhagen, Denmark), CD68 (KiM7; Valbiotech, France), and DRC-1 (Dako), which cross-react with AGM CD4, CD8, B lymphocytes, macrophages, and FDC, respectively, were used.
Antigen and antibody detection.
Plasma samples were assayed for p27 antigen by using an antigen capture enzyme-linked immunosorbent assay (ELISA) for SIV Gag p27 (Coulter Corporation, Hialeah, Fla.) with standards provided in the kit according to the manufacturer's guidelines. This ELISA kit was initially designed to detect p27 of SIVmac, but it was reported to detect also that of SIVagm (28). In addition, we confirmed its cross-reactivity for SIVagm.sab p27 by testing culture supernatants of cells infected by SIVagm.sab, SIVagm.ver, and SIVagm.tan as well as plasma samples from acutely infected sabaeus and vervet monkeys. Supernatants from noninfected cells and plasma from non-infected AGMs were used as negative controls (data not shown). Antibody responses to SIV were monitored with an HIV-1/HIV-2 ELISA (genelavia mixt; Sanofi-Pasteur) and confirmed by Western blotting (Lav blot II Diagnostics Pasteur).
Quantification of viral DNA copies in PBMC and LNs.
Frozen LN samples and PBMC pellets were disrupted in a refrigerated mortar containing lysis buffer. Genomic DNA was extracted using a QIAamp/DNA tissue kit (Qiagen, Courtaboeuf, France) according to the manufacturer's guidelines and stored at −20°C. The quality and concentration of the genomic DNA were determined by both a semiquantitative ethidium staining procedure (43) and measurements of optical density.
Quantification of viral DNA copy numbers was performed by limiting-dilution quantitative PCR in the SIVagm.sab env gene. For each sample of genomic DNA, eight dilutions ranging from 1 μg (approximately 105 cells) to 0.01 ng were subjected to PCR. Dilutions of an external standard corresponding to 106 copies down to 1 copy of a plasmid containing the entire genome of SIVagm.sab-1 (22) were subjected to PCR in parallel. To submit an equal amount of total DNA (1 μg) to each reaction, dilutions of genomic and standard DNA were adjusted with genomic DNA from an SIV-negative AGM.
One microgram of total adjusted DNA was then added to the PCR mixture containing 200 μM each deoxynucleoside triphosphate, 2 U of Taq polymerase (Appligene), and 750 nM (first PCR) or 250 nM (nested PCR) each primer. The following primer pair, slightly modified from previously published primers EnvA-EnvB (22) in order to be specific for SIVagm.sab, was used to amplify, a 1,150-bp env fragment: EnvAsab (5′ GAG GCT TGT GAT AAA ACT TAT TGG GAT 3′)-EnvBsab (5′ AGA GCA GTG ACG CGG GCA TTG AGG 3′). The PCR protocol consisted of 1 cycle of denaturation (94°C, 5 min), hybridization (55°C, 1 min), and extension (72°C, 1 min), 38 cycles of denaturation (94°C, 1 min), hybridization (55°C, 1 min), and extension (72°C, 1 min), and a final cycle of extension (72°C, 10 min). The limit of detection of this PCR was 103 copies of SIVagm DNA. A nested PCR was used to detect very low copy numbers. Five microliters of each product from the first PCR was mixed with the PCR buffer described above. The cycling conditions were the same. The following primer pair, adapted from previously published NS3s-NS3as (31), was used: NS3s sab (5′ TGT AGA AGA CCA GGA AAT AAG ACA 3′)-NS3as sab (5′ AAG CCT AGG AAC CCT AGC ACG AA 3′). The limit of detection of this nested PCR was one copy of SIVagm.sab DNA.
A fragment of the β-actin gene was always amplified in parallel to monitor the quantity and the quality of each DNA extract amplified by PCR. The dilutions corresponding to 1 and 0.01 ng of genomic DNA were used for PCR, because the signal of the amplified β-actin fragment is not saturated at these concentrations. Conditions for this PCR were as above except that β-actin primers (β1 [5′ GTG GGG CGC CCC AGG CAC CA3′] and β2 [5′ CTC CTT AAT GTC ACG CAC GAT TTC3′]) were used at a final concentration of 1 μM.
Quantification of viral RNA copies in plasma and LNs.
RNA was extracted from plasma using a Qiagen viral RNA extraction kit. Frozen LNs were disrupted in refrigerated mortars, and RNA was extracted using a Qiagen RNA/DNA extraction kit. For plasma viral load quantification, seven 10-fold serial dilutions of RNA were subjected to limiting-dilution reverse transcription (RT)-PCR in the SIVagm.sab env gene. For RNAs extracted from LNs, concentrations were determined by using a spectrophotometric operation system (Gene QuantII; Pharmacia Biotech) that allows a highly sensitive and precise measurement in a broad range (from 4 × 100 to 2 × 103 μg of RNA/ml). Five 10-fold serial dilutions of each LN RNA extract starting from 100 ng were subjected to RT-PCR. Plasma and LN RNA copy numbers were quantified by using a standard RNA. The latter standard was obtained by amplification of an env fragment of SIVagm.sab-1 (22) with the EnvAsab-EnvBsab primer pair. The env PCR product was then inserted into an expression vector using a TopoTA cloning kit (Invitrogen, Groningen, The Netherlands); the plasmid obtained was linearized with BamHI downstream the env insert, and the env fragment was transcribed in vitro by the MEGAScript T7 method (Ambion, Austin, Tex.). The 1.2-kb env transcript was purified on Qiagen RNeasy columns and quantified by the highly sensitive and specific Gene QuantII spectrophotometric operation system. To ensure that the transcripts were full length, we verified the size on a denaturing polyacrylamide gel. Aliquots were stored at −80°C until use. Immediately before use, one aliquot of RNA was thawed, and serial dilutions from 1012 down to 1 copy were used as an external standard in each experiment. For all RNAs, a one-step RT-PCR was first performed with a Superscript one-step RT-PCR kit (Gibco BRL) followed by a nested PCR in order to increase the sensitivity of the assay (100 copies of standard RNA). The sensitivity of the assay was the same whether we added a carrier RNA (1 μg of rRNA) or not to the target and to the standard RNA. Primers and PCR conditions were the same as those described for viral DNA quantification.
As a positive control RNA, we used SIVagm.sab-1 derived from Cos-7 cell transfection. Briefly, Cos-7 cells were transfected with plasmid psab-1 (22) by the Superfect reagent method (Qiagen); supernatants were collected and submitted to RT-PCR.
The absence of contamination by genomic DNA was verified by subjecting 100 ng of RNA to PCR without the RT step. To monitor the recovery of intact RNA and the uniform efficiency of each RT-PCR assay, two dilutions of each LN RNA extract were also analyzed by RT-PCR using the β-actin-specific primers described above; these two dilutions (1 and 0.01 ng of RNA) were used because they avoid saturation of the β-actin PCR signal. To assess the presence of putative RT-PCR inhibitors in the plasma RNA samples, a known concentration of the standard RNA was added to plasma collected at day 0 from three AGMs, followed by the RNA extraction step described above; the three RNA extracts were then diluted and subjected to RT-PCR, and sensitivities of these RT-PCR reactions were compared with that obtained with the standard RNA alone.
To control the sensitivity of the env primers, we also developed a limiting-dilution RT-PCR assay for the gag gene of SIVagm.sab. The primers C1S (5′-AAG TAT AAG TTA AGA CAT CTA ITA TGG GCA-3′) and C4S (5′-GCA TTC TGG ATC AAC AGA GAC TGI GTC ATC CA-3′) were used for the first PCR; C2S (5′-GTC ACG CAG AAA TIA AAG TGA AA-3′) and C3S (5′-TCC TCA ATC ACT TTT ACC CA-3′) were used for nested PCR under the same conditions as used for quantification in env.
To validate the limiting-dilution PCR assay, randomly selected plasma and LN RNA samples were also quantified by a real-time PCR assay. Briefly, total RNA was transcribed using a TaqMan Gold RT-PCR kit and random hexamers (PE Applied Systems). PCRs were carried out in a spectrofluorometric thermal cycler (ABI PRISM 7700). cDNA was added to the universal master mix (Perkin-Elmer), and 10 μM each primer and 10 μM probe were added. The primers (J15S [5′-CTG GGT GTT CTC TGG TAA G-3′] and 5′ J15S [5′-CAA GAC TTT ATT GAG GCA AT-3′]) and a TaqMan probe (J15P [6FAM-CGA ACA CCC AGG CTC AAG CTG G-6TAMRA]), hybridizing to conserved regions of the SIVagm.sab long terminal repeat (LTR), were designed by Althea Technologies Inc. (San Diego, Calif.). They allowed amplification of a 180-bp LTR fragment. A first cycle of denaturation (95°C, 10 min) was followed by 45 cycles of denaturation (95°C, 10 s), hybridization (50°C, 30 s), and extension (72°C, 30 s). A SIVagm.sab LTR standard RNA was constructed according to the same protocol as described for the env standard except that it was generated by in vitro transcription of an LTR fragment corresponding to a PCR product of plasmid psab-1 (obtained with the LTR primers LTR2A [5′-AAC TAA GGC AAG ACT TTA TTG AGG-3′] and LTR4S [5′-ACT GGG CGG TAC TGG GAG TGG CTT-3′]) and inserted into the pCR2.1 vector. Known amounts of this SIVagm.sab LTR standard RNA were used to determine the target copy numbers.
Interassay variations of the DNA and RNA viral load assays.
To assess the interassay variability of the limiting-dilution PCR assay for DNA quantification, a single DNA sample was repeatedly tested 10 times (each test was performed on a separate day). The resulting coefficient of variation (CV) was 75%.
To evaluate the interassay variability for RNA quantification by the limiting-dilution RT-PCR assay, six single RNA samples were each submitted to five distinct RT reactions, and each reverse-transcribed RNA was quantified by a distinct PCR performed on a separate day. The mean RNA copy numbers of the six samples ranged from 2.6 × 102 to 4.2 × 105, and the corresponding standard deviations ranged from 1.7 × 102 to 5.3 × 105. The CV ranged from 65 to 194% (mean, 133%). The variation of RNA quantification by the real-time PCR assay was assessed by calculating the mean CV as follows. Thirty-four RNA samples were each submitted to two distinct RT reactions. Each reverse-transcribed RNA was then quantified in duplicate by four distinct PCRs, each performed on a separate day. The average CV corresponded to 71%, a value within the range reported for a commercial RNA load assay (52 to 82%; Roche Molecular Systems) (47).
RESULTS
In this study we examined early virological and immunological events associated with nonpathogenic SIVagm infection in its natural host. Five AGMs were experimentally infected with wild-type SIVagm and followed during acute and postacute phases of infection.
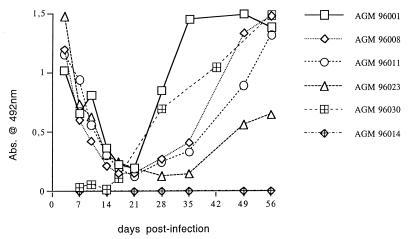
All animals, inoculated either with cell-associated or cell-free virus, seroconverted between days 28 and 42 (Fig. 1), as also demonstrated by immunoblotting and maintained a sustained antibody response (data not shown). Because the inoculum for four AGMs (96001, 96008, 96011, and 96023) consisted of a pool of PBMC and plasma from a chronically infected AGM, passively transferred anti-SIVagm antibodies were detectable at high levels at 3 days p.i. and declined subsequently (Fig. 1). The animal that did not receive passive antibodies (96030) showed a similar pattern of infection and seroconversion (Fig. 1; see also below).
FIG. 1.
Antibody responses in individual AGMs experimentally infected with wild-type SIVagm.sab92018. Absorbance values (Abs.) of sequential plasma samples are depicted for animals 96001, 96008, 96011, and 96023, which received cell-associated virus (plasma and PBMC), for animal 96030, which received cell-free virus alone, and for the negative control (96014).
After 2 years, none of the infected animals showed signs of AIDS-like clinical or biological symptoms. Transient variations in the percentages of CD4+ cells and CD8+ cells were observed (data not shown), but the absolute numbers of these lymphocyte subsets remained within the normal range (700 to 2,300 CD4+ cells/μl and 1,300 to 4,300 CD8+ cells/μl, respectively).
Extensive but transient viremia during the acute phase of SIVagm infection.
The kinetics of antigenemia during primary infection are shown in Fig. 2a. By day 7 p.i., those animals inoculated with cell-associated virus (96001, 96008, 96011, and 96023) had quantifiable levels of p27 SIV core antigen. Plasma antigenemia then rose to peak values by day 10, ranging from 1.2 to 5.0 ng/ml. AGM 96030, inoculated with cell-free virus, exhibited values in a similar range (2.26 ng/ml at day 10 p.i.), while plasma antigenemia in the mock-infected AGM 96014 remained negative (Fig. 2a). These results indicate that the peak of antigenemia observed in infected monkeys is due to early high levels of SIVagm replication in host cells rather than to virus replication in the allogenic cells from the inoculum or to stimulation of the host immune system by the allogenic cells. The extent of this early antigenemia varied up to fourfold among animals that had received identical inocula. Antigenemia then declined to undetectable levels that were reached between days 17 and 28 p.i. (Fig. 2a), before the detection of specific anti-SIVagm antibodies (Fig. 1). We cannot exclude a lower sensitivity of the p27 core antigen assay for SIVagm.sab with regard to SIVmac. The antigenemia levels would in this case be underestimated. The amount of p27 that we detected at the peak was, however, quite high and within the range of those (0.5 to 7 ng of p27/ml that have been reported in several studies for macaques infected by pathogenic SIVmac (9, 39, 50). In addition, the p27 levels in AGMs are consistent with their RNA viral loads in the blood (see below).
FIG. 2.
Viral load in peripheral blood from AGMs described in the legend to Fig. 1 (a) Quantification of plasma antigenemia. The mock-infected animal (96014) remained negative. (b) Quantification of SIVagm RNA in plasma by limiting-dilution RT-PCR. The dashed line marks the limit of PCR signal detection. (c) Quantification of SIVagm DNA in PBMC by limiting-dilution PCR. The dashed line marks the limit of PCR signal detection.
To further characterize the extensive viral replication taking place in SIVagm-infected AGMs soon after infection and to compare SIVagm replication dynamics to those of pathogenic SIV in macaques, we also evaluated RNA and DNA viral loads during the acute phase of infection. We developed highly sensitive limiting-dilution quantitative PCR and RT-PCR assays to evaluate the viral burden in AGMs. The reliability of the RT-PCR assay was tested in the following ways. As a control to determine whether the sensitivity of the RT-PCR could be affected by the presence of putative, naturally occurring inhibitory molecules in plasma, we mixed individual plasma samples, obtained before infection of animals 96008, 96011, and 96023, with SIVagm.sab standard RNA, extracted the RNA as usual, and subjected the extracts to limiting dilution RT-PCR. No changes in the sensitivity of the RT-PCRs were observed when the standard RNA was previously mixed with plasma in comparison to standard RNA alone (data not shown). Since viral load assays for SIV and HIV are often based on the gag gene, and in order to exclude a lowered sensitivity of the assay due to possible mismatches between the env primers and the target RNA, we also developed a limiting-dilution RT-PCR assay for the SIVagm.sab gag gene. Four randomly selected RNA samples (plasma and LN RNAs derived from animals 96001 and 96008 at day 10 p.i.) were quantified in parallel, using primers located in the env or gag gene. No significant differences in viral load estimates were observed (data not shown).
To further validate our limiting-dilution RT-PCR, we determined viral loads by real-time PCR. Randomly selected RNA samples (for AGM 96023, plasmas of days 7 and day 10, LNs of days 7 and day 14; for AGM 96014, plasma and LN of day 0) and a positive control RNA (supernatant of psab-1-transfected Cos cells) were quantified in parallel by the limiting-dilution RT-PCR assay and by real-time PCR. The two assays gave similar results (data not shown).
We then quantified RNA levels in the plasmas from all AGMs included in the study during the acute and postacute phases of SIVagm infection by the limiting-dilution RT-PCR assay (Fig. 2b). The data revealed a peak of plasma viral RNA, starting from day 7 or 10, that ranged from 2 × 106 to 2 × 108 copies per ml of plasma. The positive control animal AGM 96030, inoculated with cell-free virus derived from an animal in the acute phase of infection, showed a particularly rapid and elevated peak of plasma RNA (Fig. 2b). Those animals that showed the highest RNA values were, however, not necessarily those with the highest antigenemia (Fig. 2a and b). As was the case with antigenemia, viral RNA in plasma then declined rapidly; a decline of 10- to 1,000-fold was observed for all animals before day 35 (Fig. 2b). For three animals (96001, 96008, and 96030), the time point at which viral load stabilized (viral set point) was already reached between days 17 and 24. Between days 35 and 84, no significant variations in plasma viral RNA loads were noted, except for AGM 96011, which showed a further 10-fold decrease, and plasma viral loads were relatively low (2 × 103 to 2 × 105 RNA copies/ml). The naturally infected AGM 92018, which served as a donor animal, had an RNA plasma viral load in a similar range (3 × 105 copies per ml of plasma), suggesting that the experimentally induced infections reproduced the features of natural infection.
As shown in Fig. 2c, the kinetics of DNA viral loads in PBMC correlate with the profiles of antigenemia and RNA copy numbers in the plasmas of infected AGMs described above. DNA copy numbers peaked by day 7 or day 10 p.i. (104 to 105 copies/106 PBMC). The positive control animal AGM 96030 also showed a high viral DNA load (104 copies/106 PBMC). The DNA load then rapidly declined and reached values characteristic of the chronic phase of infection (102 to 103 DNA copies/106 PBMC) by as early as days 14 to 21 (Fig. 2c). Cell-associated DNA viral load was also determined in the naturally infected donor AGM (92018) at the time of sampling and was in the same range (102 DNA copies/106 PBMC) as the DNA viral loads measured in the chronic phase of the experimentally induced infections.
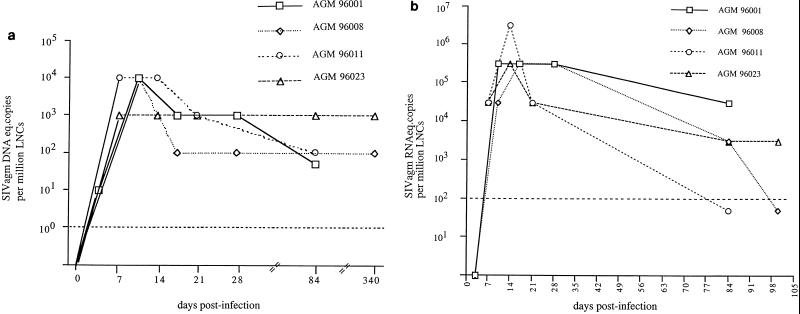
Early control of viral burden in LNs of AGMs.
In HIV and SIV infections, lymphoid organs are major replication sites for the virus (2, 12, 15). To evaluate viral replication dynamics in lymphoid organs of acutely SIVagm-infected AGMs, DNA and RNA viral copy numbers were determined in sequentially recovered LN biopsies. The pattern of cell-associated viral DNA in LNs is represented in Fig. 3a. The day of the peak could not be pinpointed in LNs as precisely as in blood samples, because LN biopsies were performed less frequently (every 7 days). However, for all animals, the peak was observed, as before, at approximately day 10 (between days 7 and 14). The viral DNA loads observed in LNs (103 to 104 DNA copies/106 LNC) were very similar to those measured in PBMC at the same time (Fig. 2c and 3a). At days 7 and 10, for example, identical copy numbers were observed in LNC and PBMC of all animals (103 copies for AGM 96008 at day 7; 104 copies for AGM 96023 at day 7 and for AGM 96001 and 96008 at day 10). At no time point after infection were the DNA copy numbers higher in LNs than in blood. This latter finding correlates with the data reported on the chronic phase of SIVagm infection (5). Values above 103 or 104 DNA copies/106 LNC cannot be excluded, however, in particular at day 10 (for AGMs 96011 and 96023) and at day 7 (for AGMS 96001 and 96008) when viral DNA loads peaked in PBMC.
FIG. 3.
Viral load in LNs from AGMs described in the legend to Fig. 1. The dashed line marks the limit of detection. (a) Quantification of SIVagm DNA in LNC by limiting-dilution PCR; (b) quantification of SIVagm RNA in LNC by limiting-dilution RT-PCR. The RNA levels below the detection limit were arbitrarily assigned the value of 50 copies.
A decrease of the DNA viral load then proceeded in the four animals (Fig. 3a). The viral DNA setpoint was reached at days 14 and 17 for AGMs 96023 and 96008, respectively, and between days 17 and 84 for AGMs 96001 and 96011. The DNA viral loads in LNs during the chronic phases of infection corresponded to 102 copies/106 LNC in three animals and to 103 copies/106 LNC in the fourth animal.
RNA viral loads in LNC are presented in Fig. 3b. Peak values were observed at day 10 for AGMs 96001 and 96008 (4 × 105 and 3 × 106 RNA copies/106 LNC, respectively) and at day 14 for AGMs 96011 and 96023 (3 × 106 and 3 × 105 RNA copies/106 LNC, respectively). As was the case for DNA viral loads, higher RNA copy numbers at days 7 (for AGMs 96001 and 96008) and 10 (for AGMs 96011 and 96023) cannot, however, be excluded. For three out of four animals, RNA loads declined rapidly by 10- to 100-fold by day 21 (Fig. 3b). After day 21, all animals showed a further decline in viral RNA burden of the same magnitude. At 3 months p.i., the RNA copy numbers in LNs varied from <10 to 4 × 104 copies/106 LNC among the animals. These low numbers correlate with in situ hybridization data showing only a few cells positive for viral RNA in chronically infected AGMs (reference 5 and our unpublished data).
Lack of morphology and cellular distribution changes in AGM LNs during primary infection.
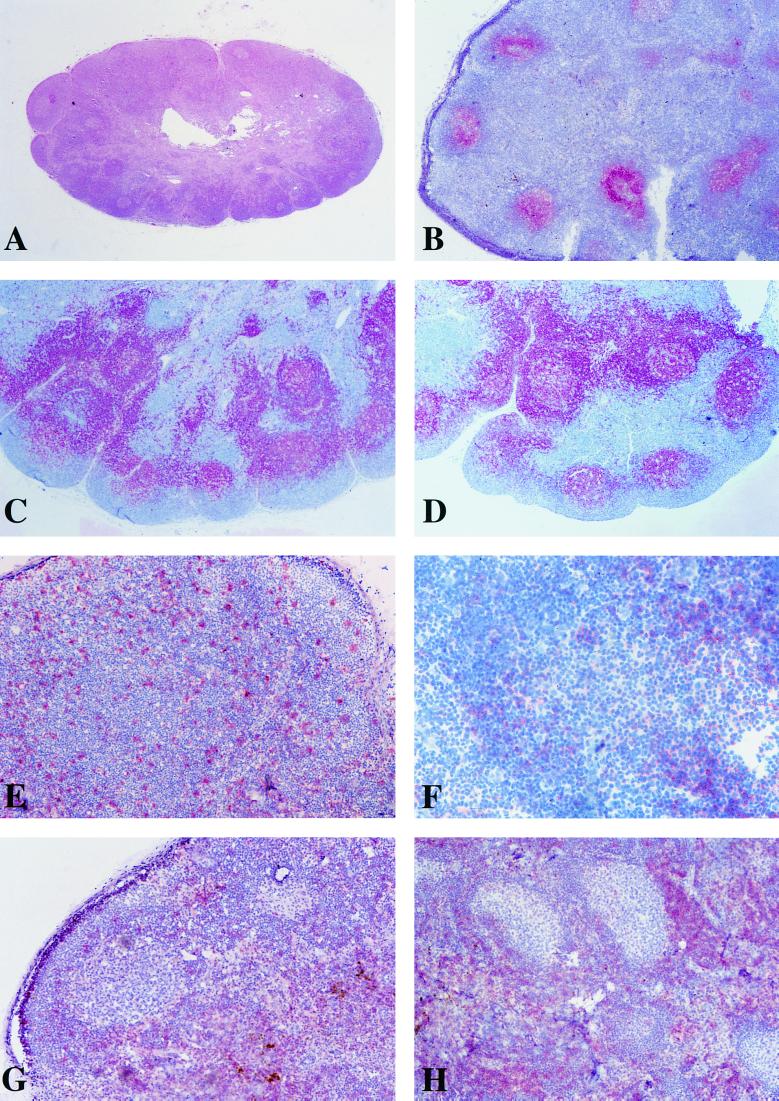
Early virus-host interactions in lymphoid organs, such as LNs, probably play a determinative role in the outcome of infection. We therefore looked for immunohistochemical changes in LNs during the acute phase of SIVagm infection. Axillary and inguinal LNs recovered from infected animals between days 0 and 28 p.i. showed well-conserved morphologies. As was the case for noninfected AGMs, the GCs were well demarcated, and mantle zones were intact, with no evidence of either lymphoid depletion or involution (Fig. 4A and B). Staining with the DRC-1 MAb demonstrated that the FDC network was normal and largely confined to the follicular zone (Fig. 4B), while staining of B lymphocytes showed the confines of follicular zones (Fig. 4C and D). In most animals, we observed a mild follicular hyperplasia (Fig. 4A). Such hyperplasia was, however, observed at every time point studied, including day 0 (Fig. 4C and D), and in the negative control AGM 96014 (data not shown), and is probably a consequence of exposure to other infectious agents in the originally wild animals.
FIG. 4.
Morphological analysis and immunohistochemical staining of LN sections harvested between days 0 and 28 p.i. from AGMs described in the legend to Fig. 1. (A) A 5-μm section of PFA-fixed paraffin-embedded LN, stained with hematoxylin-eosin; (B to H) 5-μm sections of PFA-fixed paraffin-embedded or cryopreserved OCT-embedded LN, stained by the AP–anti-AP method (11) and counterstained with Harris hematoxylin. No significant differences were observed between acutely infected and noninfected animals or within the same animal before and after infection. Representative sections are shown. (A) Classical histopathologic pattern of LN sections from AGM acutely infected with SIVagm (animal 96001, 10 days p.i., inguinal LN). (B) Typical image of the FDC network in LN from acutely SIVagm-infected AGM (animal 96001, 17 days p.i.). This frozen section of inguinal LN was stained with an anti-FDC MAb (DRC-1; Dako). (C) (D) Paraffin-embedded LN sections (animal 96001, inguinal LN) taken at days 0 and day 28 p.i., respectively, stained with a pan-B MAb (Dako). (E) Typical image of CD68+ cells distribution in LNs of infected and noninfected AGMs. Animal 96023 (day 0) is shown in this frozen section of inguinal LN stained with an anti-CD68 MAb (KiM7; Valbiotech). (F) Representative frozen LN section from acutely SIVagm-infected AGM (animal 96001, 10 days p.i., inguinal LN), stained with an anti-CD4 MAb (F101.69; Sanofi Recherche). (G and H) Frozen LN sections of animals 96014 (day 0, axillary LN) and 96011 (day 14 p.i., axillary LN), respectively, stained with an anti-CD8 MAb (Leu-2a; Becton Dickinson).
MAb directed to CD4+ cells, CD8+ cells, or macrophages did not reveal any specific differences in cellular distributions in the animals before and after inoculation, nor were any changes noted between infected and control animals (Fig. 4E to H). In each animal, and at every time tested before or after experimental infection, CD8+ cells were mainly localized in the extrafollicular zones (Fig. 4G and H). It is important to note that in every monkey, including the negative control animal, only very few CD8+ cells were found in GCs, indicating an absence of infiltration by these cells from extrafollicular zones. CD4+ cells shared the same localization as CD8+ cells, but it was not unusual to find a few of the former in follicular zones as well, typical for normal LNs (Fig. 4F). On the other hand, we did not find any such preferential localization of macrophages, which are present both in follicular and extrafollicular zones (Fig. 4E). In summary, no significant changes in morphology or cellular localization were observed in the LNs of SIVagm-infected AGMs within the first month p.i.
DISCUSSION
SIV infections in their natural hosts are generally asymptomatic, and their study provide clues concerning the basis of apathogenic lentivirus infection. This report describes for the first time the main features associated with the early stages of nonpathogenic SIV infection in the natural host species using the AGM nonhuman primate model. We demonstrate that primary infection by SIVagm in AGMs is characterized initially by high levels of viral replication by approximately day 10 p.i., both in the blood and in LNs (Fig. 2 and 3). This peak of viral replication is then consistently followed by a rapid and significant decrease of peripheral and tissue viral load. The viral DNA and RNA copy numbers in blood stabilized at the latest by day 35 in all studied AGMs (between days 14 and 35). In the chronic phase of infection (studied between days 35 and 350 p.i.), the blood and LN viral loads were comparable to the previously reported low levels of viral burden in other naturally or experimentally chronically infected AGMs (5, 18).
The profiles of viral replication, as measured by antigenemia or by RNA and DNA viral loads either in blood or in LNs, were very similar to each other despite the variability inherent to the viral load assays. The viral dynamics were also remarkably similar in each animal studied, although the animals were not inbred. In addition, the peak of antigenemia at approximately day 10 was repeatedly observed in five other wild-born AGMs exposed either to lower doses of the same cell-associated virus stock (30 and 100 TCID50) or to another SIVagm wild-type strain (our unpublished data). The overall consistency of the pattern of viral replication in these AGMs indicates that the peak of viral replication around day 10 and the subsequent significant decrease in viral burden are representative of SIVagm primary infections.
Interestingly, the RNA levels in plasma stabilized at 2 × 103 to 2 × 105 copies/ml in the AGMs studied. These values are below or very close to the minimum pathogenic threshold value (>105 RNA copies/ml) previously defined in SIVmac-infected macaques (44, 46). Early control at approximately these levels is also characteristic for HIV-1-infected long-term nonprogressors (1, 7, 30). These similarities between nonpathogenic SIVagm infection and long-term nonprogressive infections in humans may lead to more informative conclusions about the general mechanisms responsible for disease progression in humans.
There are, in theory, at least three main mechanisms that could, either separately or in combination, be responsible for a lack of disease in infected AGMs: (i) SIVagm could be attenuated in vivo, similarly to SIVmacΔnef (8, 9, 26); (ii) virus reservoirs could be distinct in AGMs, and/or (iii) the lack of disease might be associated with the induction of protective host immune responses. In each case, both quantitative and qualitative differences in virus-host interactions must be considered in assessing those factors which are critical for variations in pathogenesis, potentially complicating the interpretation of individual results.
The present data indicate that SIVagm is not likely to be attenuated for replication in its natural host, at least in the initial phases of infection. Indeed, the levels of antigenemia and peripheral DNA and RNA viral loads during the first 10 days following SIVagm infection in AGMs (Fig. 2 and 3) are similar to those reported for pathogenic SIVmac infections (9, 21, 39, 44, 46, 49, 50). These findings correlate with the capacity of at least one SIVagm strain to induce both high viral loads and disease in another monkey species, the pig-tailed macaque (20).
In addition, the kinetics of viremia were not significantly different between the four AGMs that were inoculated with a virus derived from a chronically infected AGM and AGM 96030, which was exposed to a virus isolated at day 8 p.i. (Fig. 2). These findings correlate with the constant and rapid replication, albeit at a low level, of SIVagm during the chronic phase of infection (32). They suggest that there is no selection for attenuated virus in individual monkeys at later stages of infection. Like SIVagm, another virus (SIVsm) also replicates actively in its natural host, although to a higher level than SIVagm in AGMs, at least during the chronic phase of infection (40). The capacity for efficient replication of SIVs in their natural hosts might thus be a common feature among naturally occurring SIVs and a consequence of long-term coevolution of the viruses and their hosts (3, 17, 31).
Since SIVagm replicates to high levels early in infection, the existence of a constitutive genetic host restriction of viral replication is not supported by our study. These findings correlate with our previously published data that indicated no correlation between mutations in the CCR5 coding region in AGMs and resistance to SIV infection (33). However, one cannot exclude the existence of either an inducible restriction of viral replication in the AGM host or the presence of an unknown subset of genetically resistant cells whose infection is required for disease.
If a restriction of viral replication is rapidly induced in response to viral infection, early host immune responses would be likely candidates for being involved in such an induction. Such a hypothesis would correlate with the observation that unspecific activation of peripheral AGM CD4+ cells in vitro induces a phenotypic conversion from the CD4+ cells to CD4− cells, rendering the latter resistant to SIVagm infection in vitro (34).
The mechanism or group of mechanisms that most likely determine the level of pathogenesis of SIVagm in AGMs may indeed be related to the complex interactions between the multiple virus parameters and the various immune responses of the host. It seems reasonable to assume that normal or enhanced host immune responses contribute directly to the early control of viral replication. AGMs generally exhibit significantly higher percentages of CD8+ cells than do macaques or humans (6, 34), and it was demonstrated that these cells secrete soluble antiviral factors capable of inhibiting SIVagm replication in vitro (13). Depletion of CD8+ cells during the acute and chronic phases of SIVmac infection in macaques favors viral spreading (23, 42). In HIV-1-infected humans, moreover, the mobilization of a broader cytotoxic T-lymphocyte repertoire during primary infection seems to confer better protection (37). Virus-specific CD4 T helper responses, which are necessary to induce efficient CD8+ cell responses, appear to be associated with the control of viremia in some untreated long-term nonprogressors as well as in acutely HIV-1-infected individuals whose viremia is reduced by highly active antiretroviral therapy when started before seroconversion (41).
However, our data do not indicate a major role for quantitatively strong immune responses in SIVagm-infected AGMs. In fact, primary SIVagm infection was characterized by a lack of follicular hyperplasia (Fig. 4), in contrast to pathogenic and attenuated SIVmac infection in macaques (8). Absence of follicular hyperplasia has also been reported in later stages of SIVagm infection in AGMs (5) and for SIVsm infection in sooty mangabeys, despite significant high levels of viral DNA loads (5 × 103 to 2 × 104 copies/106 LNC) in the latter (40). The lack of any signs of CD8+ cell infiltrations into GCs (Fig. 4) also contrasts with pathogenic SIVmac infections (2, 39). Our data thus support the hypothesis that at least certain kinds of immune activation are less pronounced in SIV infections in their natural hosts than in infections with a pathogenic outcome (25, 48). This hypothesis correlates with our previous findings on the absence of an abnormal rate of peripheral CD4+ lymphocytes from chronically infected AGMs that are undergoing apoptosis, in contrast to pathogenic HIV-1 and SIVmac infections (14). The apparently quantitatively low levels of cytotoxic T-lymphocyte responses in chronically infected AGMs and sooty mangabeys in their natural hosts (25, 35) also support this hypothesis. The lack of disease progression in SIVagm-infected AGMs may thus be attributed to the development of essentially protective immune responses owing to a lack of abnormal activation and/or of dysfunction of immune cells in response to the infection.
In summary, our study demonstrates that SIVagm is not attenuated for replication in its natural host. Data on the dynamics of viral replication indicate that the constant low level of viral burden during chronic infection in AGMs is the result of a rapid induction of protective host responses rather than to a constitutive genetic restriction of virus replication in the host. Our study is consistent with the existence of a delicate balance between viral replication and host immune responses. Finally, the viral dynamics observed in these experiments are very similar to those found in human long-term survivors, and further studies in AGMs might yield clues about the host responses that are essentially protective and not damaging to the host.
ACKNOWLEDGMENTS
We thank M. Yaya and X. Pourrut for expert assistance with LN biopsies, M. Ndiaye and M. Touré for assistance with animal care, and P. Versmisse for excellent technical help. We are grateful to B. Hahn for the gift of the SIVagm.sab-1 plasmid and to R. Le Grand, B. Hurtrel, E. Khatissian, and V. Monceaux for providing control plasma samples. We thank Bob Bassin for critical reading of the manuscript.
This work was supported by grants from the French Agency for AIDS Research (ANRS). A.G., M.D.-T., M.C.M.-T., and C.K. were recipients of fellowships from the French Ministère de la Cooperation, the Bresilian Ministério da Educaçao, the French Foundation for Medical Research (Sidaction), and the Daimler Benz Foundation, respectively.
REFERENCES
- 1.Balotta C, Bagnarelli P, Riva C, Valenza A, Antinori S, Colombo M C, Sampaolesi R, Violin M, de Pasquale M P, Moroni L, Clementi M, Galli M. Comparable biological and molecular determinants in HIV type 1-infected long-term non progressors and recently infected individuals. AIDS Res Hum Retroviruses. 1997;13:337–341. doi: 10.1089/aid.1997.13.337. [DOI] [PubMed] [Google Scholar]
- 2.Baskin G, Martin L N, Murphey-Corb M, Hu F-S, Kuebler D, Davison B. Distribution of SIV in lymph nodes of serially sacrificed rhesus monkeys. AIDS Res Hum Retroviruses. 1995;11:273–285. doi: 10.1089/aid.1995.11.273. [DOI] [PubMed] [Google Scholar]
- 3.Beer B, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. doi: 10.1128/jvi.73.9.7734-7744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer B, Denner J, Brown C R, Norley S, Megede J Z, Coulibaly C, Plesker L, Holzhammer S, Baier M, Hirsch V, Kurth R. Simian immunodeficiency virus of African green monkey is apathogenic in the newborn natural host. J AIDS Hum Retrovirol. 1998;18:210–220. doi: 10.1097/00042560-199807010-00003. [DOI] [PubMed] [Google Scholar]
- 5.Beer B, Scherer J, Megede J, Norley S, Baier M, Kurth R. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long term simian immunodeficiency virus infection of African green monkeys. Virology. 1996;219:367–375. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 6.Buijs L, Bogers W M, Eichberg J W, Heeney J L. CD8+ cell-mediated immune responses: relation to disease resistance and susceptibility in lentivirus-infected primates. J Med Primatol. 1997;26:129–138. doi: 10.1111/j.1600-0684.1997.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Qin L, Zhang L, Safriti J, Ho D D. Virologic and immunologic characterisation of long-term survivors of human immunodeficiency virus type I infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarti L, Baptiste V, Cumont M-C, Khatissian E, Montagnier L, Hurtrel B. Comparison of primary infection in lymph nodes of macaques inoculated with pathogenic and attenuated SIV. J Med Primatol. 1994;23:229. [Google Scholar]
- 9.Chakrabarti L, Baptiste V, Khatissian E, Cumont M C, Aubertin A M, Montagnier L, Hurtrel B. Limited viral spread and rapid immune response in lymph nodes of macaques inoculated with attenuated simian immunodeficiency virus. Virology. 1995;213:535–548. doi: 10.1006/viro.1995.0026. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti L, Cumont M-C, Montagnier L, Hurtrel B. Variable course of primary simian immunodeficiency virus infection in lymph nodes: relation to disease progression. J Virol. 1994;68:6634–6643. doi: 10.1128/jvi.68.10.6634-6643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordell J L, Falini B, Erber W N, Ghosh A K, Abdulaziz Z, MacDonald S, Pulford K A, Stein H, Mason D Y. Immunoenzymatic labeling of monoclonal antibodies using immune complexes of alkaline phosphatase and monoclonal anti-alkaline phosphatase (APAAP complexes) J Histochem Cytochem. 1984;32:219–229. doi: 10.1177/32.2.6198355. [DOI] [PubMed] [Google Scholar]
- 12.Embretson J, Zupancic M, Ribas J L, Burke A, Racz P, Tenner-Racz K, Haase A T. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 13.Ennen J, Findeklee H, Dittmar M T, Norley S, Ernst M, Kurth R. CD8+ T lymphocytes of African green monkeys secrete an immunodeficiency virus-suppressing lymphokine. Proc Natl Acad Sci USA. 1994;91:7207–7211. doi: 10.1073/pnas.91.15.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estaquier J, Idziorek T, De Bels F, Barré-Sinoussi F, Hurtrel B, Aubertin A-M, Venet A, Mehtali M, Muchmore E, Michel P, Mouton Y, Girard M, Ameisen J-C. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc Natl Acad Sci USA. 1994;91:9431–9435. doi: 10.1073/pnas.91.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox C H, Tenner-Racz K, Racz P, Firpo A, Pizzo P A, Fauci A S. Lymphoid germinal centers are reservoirs of human immunodeficiency virus type 1 RNA. J Infect Dis. 1991;164:1051–1057. doi: 10.1093/infdis/164.6.1051. [DOI] [PubMed] [Google Scholar]
- 16.Galat G. Thèse de Doctorat d'Etat. Paris, France: Université Pierre et Marie Curie; 1983. [Google Scholar]
- 17.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur I O, Peeters M, Shaw G, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–440. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 18.Hartung S, Boller K, Cichutek K, Norley S G, Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendry R M, Wells M A, Phelan M A, Schneider A L, Epstein J S, Quinnan G V. Antibodies to simian immunodeficiency virus in African green monkeys in Africa in 1957–1962. Lancet. 1986;ii:455. doi: 10.1016/s0140-6736(86)92156-2. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch V, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, Fuerst T R, Sutter G, Carrol M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin M J, Hui H, Robertson D L, Müller M C, Barré-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin X, Bauer D, Tuttleton S, Lewin S, Gettie A, Blanchard J, Irwin C, Safrit J, Mittler J, Weinberger L, Kostrikis L, Zhang L, Perelson A, Ho D. Dramatic rise in plasma viremia after CD8 (+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanki P J, Kurth R, Becker W, Dreesman G, McLane M F, Essex M. Antibodies to simian T-lymphotropic retrovirus type III in African green monkeys and recognition of STLV-III viral proteins by AIDS and related sera. Lancet. 1985;i:1330–1332. doi: 10.1016/s0140-6736(85)92818-1. [DOI] [PubMed] [Google Scholar]
- 25.Kaur A, Grant R M, Means R E, McClure H, Feinberg M, Johnson R P. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J Virol. 1998;72:9597–9611. doi: 10.1128/jvi.72.12.9597-9611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kestler H W, Ringler D J, Mori K, Panicall D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 27.Lackner A A. Pathology of simian immunodeficiency virus induced disease. Curr Top Microbiol Immunol. 1994;188:35–76. doi: 10.1007/978-3-642-78536-8_3. [DOI] [PubMed] [Google Scholar]
- 28.Lairmore M D, HofHeinz D E, Letvin N L, Stoner C S, Pearlman S, Toedter G P. Detection of simian immunodeficiency virus and human immunodeficiency virus type 2 capsid antigens by a monoclonal antibody-based antigen capture assay. AIDS Res Hum Retroviruses. 1993;9:565–575. doi: 10.1089/aid.1993.9.565. [DOI] [PubMed] [Google Scholar]
- 29.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 31.Müller M C, Saksena N K, Nerrienet E, Chappey C, Hervé V M A, Durand J-P, Legal-Campodonico P, Lang M-C, Digoutte J-P, Georges A J, Georges-Courbot M-C, Sonigo P, Barré-Sinoussi F. Simian immunodeficiency viruses from Central and Western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller-Trutwin M C, Corbet S, Dias Tavares M, Hervé V M A, Nerrienet E, Georges-Courbot M-C, Saurin W, Sonigo P, Barré-Sinoussi F. The evolutionary rate of nonpathogenic simian immunodeficiency viruses (SIVagm) indicates a rapid and continuous replication in vivo. Virology. 1996;223:89–102. doi: 10.1006/viro.1996.0458. [DOI] [PubMed] [Google Scholar]
- 33.Müller-Trutwin M C, Corbet S, Hansen J, Georges-Courbot M C, Diop O, Rigoulet J, Barré-Sinoussi F, Fomsgaard A. Mutations in CCR5-coding sequences are not associated with SIV carrier status in African nonhuman primates. AIDS Res Hum Retroviruses. 1999;15:931–939. doi: 10.1089/088922299310647. [DOI] [PubMed] [Google Scholar]
- 34.Murayama Y, Amano A, Mukai R, Shibata H, Matsunaga S, Takahashi H, Yoshikawa Y, Hayami M, Noguchi A. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int Immunol. 1997;9:843–851. doi: 10.1093/intimm/9.6.843. [DOI] [PubMed] [Google Scholar]
- 35.Norley S G, Kraus G, Ennen J, Bonilla J, König H, Kurth R. Immunological studies of the basis for the apathogenicity of simian immunodeficiency virus from African green monkeys. Proc Natl Acad Sci USA. 1990;87:9067–9071. doi: 10.1073/pnas.87.22.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohta Y, Masuda T, Tsujimoto H, Ishikawa K, Kodama T, Morikawa S, Nakai M, Honjo S, Hayami M. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiological survey of the virus in various nonhuman primates. Int J Cancer. 1988;41:115–122. doi: 10.1002/ijc.2910410121. [DOI] [PubMed] [Google Scholar]
- 37.Pantaleo G, Demarest J F, Schacker T, Vaccarezza M, Cohen O J, Daucher M, Graziosi C, Schnittman S S, Quinn T C, Shaw G M, Perrin L, Tambussi G, Lazzarin A, Sekaly R P, Soudeyns H, Corey L, Fauci A S. The qualitative nature of the primary immune response to HIV infection is a prognosticator of disease progression independent of the initial level of plasma viremia. Proc Natl Acad Sci USA. 1997;94:254–258. doi: 10.1073/pnas.94.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips-Conroy J E, Jolly C J, Petros B, Allan J S, Desrosiers R C. Sexual transmission of SIVagm in wild grivet monkeys. J Med Primatol. 1994;23:1–7. doi: 10.1111/j.1600-0684.1994.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 39.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey-Cuille M A, Berthier J L, Bomsel-Demontoy M C, Chaduc Y, Montagnier L, Hovanessian A G, Chakrabarti L A. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol. 1998;72:3872–3886. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz J, Kuroda M, Santra S, Sasseville V, Simon M, Lifton M, Racz P, Tenner-Racz K, Dalesandro M, Scallon B, Ghrayeb J, Forman M, Montefiori D, Rieber E, Letvin N, Reimann K. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 43.Spina C A, Guatelli J, Richman D. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J Virol. 1995;69:2977–2988. doi: 10.1128/jvi.69.5.2977-2988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staprans S I, Dailey P J, Rosenthal A, Horton C, Grant R M, Lerche N, Feinberg M B. Simian immunodeficiency virus disease course is predicted by the extent of virus replication during primary infection. J Virol. 1999;73:4829–4839. doi: 10.1128/jvi.73.6.4829-4839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamalet C, Lafeuillade A, Fantini J, Poggi C, Yahi N. Quantification of HIV-1 viral load in lymphoid and blood cells: assessment during four-drug combination therapy. AIDS. 1997;11:895–901. doi: 10.1097/00002030-199707000-00009. [DOI] [PubMed] [Google Scholar]
- 46.ten Haaft P J F, Verstrepen B E, Überla K, Rosenwirth B, Heeney J L. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todd J, Pachl C, White R, Yeghiazarian T, Johnson P, Taylor B, Holodniy M, Kern D, Hamren S, Chernoff D, et al. Performance characteristics for the quantitation of plasma HIV-1 RNA using branched DNA signal amplification technology. J Acquir Immune Defic Syndr. 1995;10:S35–S44. [PubMed] [Google Scholar]
- 48.Villinger F, Folks T M, Lauro S, Powell J D, Sundstrom J B, Mayne A, Ansari A A. Immunological and virological studies of natural SIV infection of disease-resistant nonhuman primates. Immunol Lett. 1996;51:59–68. doi: 10.1016/0165-2478(96)02556-4. [DOI] [PubMed] [Google Scholar]
- 49.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wykrzykowska J J, Rosenzweig M, Veazy R, Simon M A, Halvorsen K, Desrosiers R, Johnson R P, Lackner A. Early regeneration of thymic progenitors in rhesus macaques infected with SIV. J Exp Med. 1998;187:1767–1778. doi: 10.1084/jem.187.11.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]