Abstract
Immunoglobulin (Ig) E is central to the pathogenesis of allergic conditions, including allergic fungal rhinosinusitis. However, little is known about IgE antibody secreting cells (ASCs). We performed single-cell RNA sequencing from cluster of differentiation (CD)19+ and CD19− ASCs of nasal polyps from patients with allergic fungal rhinosinusitis (n = 3). Nasal polyps were highly enriched in CD19+ ASCs. Class-switched IgG and IgA ASCs were dominant (95.8%), whereas IgE ASCs were rare (2%) and found only in the CD19+ compartment. Through Ig gene repertoire analysis, IgE ASCs shared clones with IgD−CD27− “double-negative” B cells, IgD+CD27+ unswitched memory B cells, and IgD−CD27+ switched memory B cells, suggesting ontogeny from both IgD+ and memory B cells. Transcriptionally, mucosal IgE ASCs upregulate pathways related to antigen presentation, chemotaxis, B cell receptor stimulation, and survival compared with non-IgE ASCs. Additionally, IgE ASCs have a higher expression of genes encoding lysosomal-associated protein transmembrane 5 (LAPTM5) and CD23, as well as upregulation of CD74 (receptor for macrophage inhibitory factor), store-operated Calcium entry-associated regulatory factor (SARAF), and B cell activating factor receptor (BAFFR), which resemble an early minted ASC phenotype. Overall, these findings reinforce the paradigm that human ex vivo mucosal IgE ASCs have a more immature plasma cell phenotype than other class-switched mucosal ASCs and suggest unique functional roles for mucosal IgE ASCs in concert with Ig secretion.
INTRODUCTION
Chronic rhinosinusitis with nasal polyposis (CRSwNP) is an umbrella term for a heterogenous collection of diseases that is estimated to affect 4.9% of people in the United States1-4. Nasal polyps (NPs) in humans can be categorized as having biomarkers of type 2 inflammation (T2-high) or lacking evidence of type 2 inflammation (T2-low), with T2-high subsets including allergic fungal rhinosinusitis (AFRS) and aspirin-exacerbated respiratory disease3,5-8. AFRS is thought to result from a sustained T2 immune response at the mucosal membrane primarily directed at a variety of fungal antigens, including epitopes present on Aspergillus fumigatus. Interestingly, AFRS has been described as the upper airway correlate of allergic bronchopulmonary aspergillosis5-9, a disease resulting in high systemic immunoglobulin E (IgE) levels. Although IgE is a critical mediator of disease pathology in AFRS, little is known about the biology of human IgE antibody secreting cells (ASCs) in the respiratory mucosa.
Of all antibody isotypes, IgE has the lowest serum concentration and the shortest half-life10. IgE ASCs are extremely rare in peripheral blood and human bone marrow (BM) and are notoriously difficult to isolate, primarily because they lack unique surface markers10-12. Many naïve and transitional B cells differentially express CD23, the low-affinity IgE receptor, and therefore, selecting with anti-IgE antibodies can lead to false positives with fluorescence-activated cell sorting. Single-cell RNA sequencing (scRNA-seq) paired with Ig gene (VDJ) analysis is consequently required to ensure isolation of genuine IgE ASCs. Using this approach, Croote et al.13 sorted IgE-positive B cells in the peripheral blood of children with peanut allergy and found that only 89 of 973 cells were bona fide IgE ASCs. Compared with non-IgE ASCs, these cells upregulated CD23, major histocompatibility complex (MHC) class II genes, and LAPTM5, and downregulated LGALS1 and S100 genes. Although sorting for CD19+ IgE+ from peripheral blood cells results in B cells predominantly, the few ASCs that were isolated were skewed to an early ASC or plasmablast phenotype because they were isolated in the blood. Thus, it was not clear if the circulating blood IgE ASCs in food allergy are transcriptionally similar to naturally occurring mucosal IgE ASCs involved in allergic rhinitis, asthma, AFRS, or allergic bronchopulmonary aspergillosis.
To understand the transcriptional programs of mucosal IgE ASCs, which are critical for the pathogenesis of atopic diseases, we analyzed 5,907 single CD19+ and CD19− ASCs in the respiratory mucosa and compared VDJ repertoire and transcriptional profiles of IgE versus non-IgE ASCs in patients with AFRS as a model of allergen-driven mucosal disease. Of all mucosal ASCs, IgA and IgG accounted for over 90% of all plasma cells, with IgA2 and IgG2 comprising the major subclasses. A total of 116 IgE ASCs were identified and could only be found in the immature CD19+ compartment despite the substantial connectivity between the mature CD19− and CD19+ compartments found in ASCs from other isotypes. IgE ASCs featured both high and low heavy chain V region (IGHV) mutation frequencies with clonal connectivity shared with IgD+CD27− unswitched memory (USM), IgD−CD27− double-negative (DN) B cells, and IgD−CD27+ switched memory (SWM) B cells. The transcriptional analysis of IgE versus non-IgE ASCs revealed unique gene expression signatures related to antigen presentation, chemotaxis, B-cell receptor (BCR) stimulation, and ASC survival, which were reminiscent of early minted ASCs. Unsupervised clustering resulted in the majority of IgE ASCs located together in cluster 5, which contained the highest expression of CD19 and human leukocyte antigen (HLA) class I and II genes and the lowest CD138 expression. They also had the highest gene expression of LAPTM5, CD23, CD74 [the receptor for macrophage inhibitory factor (MIF)], SARAF, and BAFFR resembling an early minted ASC phenotype. Overall, we reveal characteristics of IgE ASCs, which suggest that these cells are more immature or newly minted than other mucosal class-switched ASCs within the NP.
RESULTS
NPs from patients with AFRS are enriched in ASCs
NPs from three adults (ages 23–38 years old) with AFRS were collected after endoscopic sinus surgery for the treatment of chronic rhinosinusitis. Tissue dissociation was performed as previously described to isolate ASCs and B cells14. The polyps weighed a mean of 1.2 g (Methods, Supplementary Table 1). None of the donors were taking oral corticosteroids or monoclonal antibodies targeting the mediators of T2 inflammation at the time of surgery. The cells were stained with a flow cytometry panel designed to enrich ASCs (Fig. 1A). Previously, we demonstrated that NPs contain higher proportions of IgE ASCs as a percentage of total CD19+ B cells; although, these cells are still quite rare14. We sorted up to 20,000 CD19+IgD−CD27hiCD38hi and CD19−CD38hiCD138+ ASCs per polyp and performed scRNA-seq using the 10X Genomics platform for paired 5’ VDJ and transcriptional libraries.
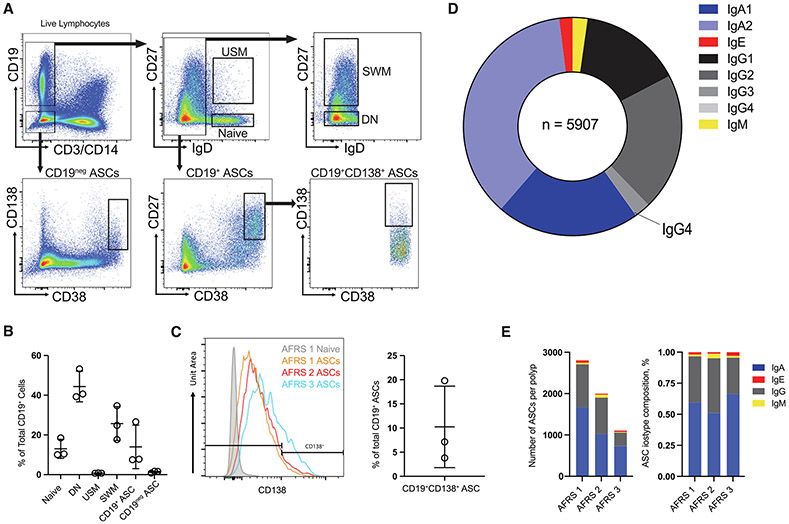
Fig. 1.
B cell and plasma cell composition of NPs. A, primary population gating strategy for fluorescence-activated cell sorted CD19+ ASCs, CD19− ASCs, and B cell subsets. One representative patient sample was selected for display (AFRS 1); B, B cell subset frequency of total CD19+ B cells in NPs; C, histograms of CD138 expression in naïve B cells from one donor (AFRS 1) and in ASCs from AFRS 1, 2, and 3 (left). CD19+CD138+ cell frequency of total CD19+ ASCs (right); D, relative proportions of immunoglobulin isotype subclasses as a percentage of total VDJ sequences isolated (n = 5907); E, stacked barplots showing the absolute number of ASCs per polyp (left) and the relative composition of each NP VDJ repertoire (right). AFRS = allergic fungal rhinosinusitis; ASC = antibody secreting cell; CD = cluster of differentiation; DN = double-negative B cells; Ig = immunoglobulin; NP = nasal polyp; SWM = switched memory; USM = unswitched memory; VDJ = immunoglobulin gene.
CD19+ ASCs were particularly prevalent, with an average of 14% of all CD19+ B cells (Fig. 1B). This frequency is much higher than would be expected in peripheral blood, where the frequency has been reported to be 1.4% and even lower in BM from healthy adults15,16. CD19− ASCs were less common, comprising a mean of 0.6% of total CD19− cells (1.4% of the total CD19+ cells), but still were more prevalent than in the peripheral blood15. On average, only 10.3% of the CD19+ ASCs coexpressed CD138 (Fig. 1C) compared with BM ASCs, whereas the frequency is typically higher than 50%. The majority of B cells found in the peripheral blood of healthy adults have a naïve phenotype. However, in the NPs we examined, DN (IgD−CD27−) B cell subsets were the most abundant, with a mean of 44.4% ± 7.7% of the total CD19+ cells. In peripheral blood, DN B cell frequencies are low in healthy adults. However, higher frequencies can be observed in patients with autoimmune disease and severe acute COVID-19, attributed to the strong extrafollicular differentiation of ASCs14,17-19. Other B cell subsets identified in the NP include SWM (25.7% ± 8.5%), USM (0.8% ± 0.1%), and naïve B cells (13% ± 4.8%, Fig. 1B).
Single-cell ASC isolation from AFRS NPs
We obtained high-confidence VDJ transcriptional data (see Methods) from 5,910 total cells (n = 3,724 CD19+, n = 2,187 CD19−). To arrive at this final number, we initially removed low-quality or multiple cells by removing cells that had less than 1,000 or greater than 5,000 genes, as well as those that had more than 25% of their transcripts consisting of mitochondrial genes. Because ASCs have increased Ig transcripts compared with B cells, we excluded cells with less than 25% of their transcripts based on Ig heavy and light chain genes (see Methods, Supplementary Figs. 2A-C). As previously described17-19, we included single-cell heavy chain sequences, even if paired light chain data were not available, if the cell still met the filtering criteria. Therefore, the number of heavy chain sequences included in repertoire analysis (n = 5,910) is slightly higher than the number of light chain sequences (n = 5,853). Interestingly, we did isolate VDJ sequence data from three IgD ASCs. Although the mucosal IgD ASCs had highly mutated VDJ sequences similar to what has been described by others20, no transcriptional data were available from these cells and we subsequently removed them from the final analysis, leaving 5,907 ASCs. After ensuring that there were no differences in VDJ data related to individuals (Supplementary Fig. 1A), we combined data from the three subjects to provide a robust comparison of IgE and non-IgE ASCs.
NP mucosal ASCs are dominated by IgA and IgG ASCs
The VDJ repertoire of the three polyps reflected the ASC composition of a mucosal site, as described in other studies, and was dominated by ASCs of IgG and IgA isotypes, with few (n = 131) IgM ASCs and 116 IgE ASCs (Fig. 1D). Among IgA ASCs (n = 3,420), the prevalence of IgA2 ASCs was roughly double that of IgA1 ASCs. Within the IgG compartment (n = 2,240), IgG1 (n = 893) and IgG2 (n = 1,206) ASCs were the most prevalent, with a small fraction of IgG3 (n = 138) and only three IgG4 ASCs. ASCs of all isotypes were found in higher frequencies in the CD19+ compartment than the CD19− compartment, with the notable exception of IgA2, which was more equally distributed between the two compartments (Supplementary Fig. 1B). Light chain analysis demonstrated an elevated κ/λ ratio in both the CD19+ (ratio = 2.1) and CD19− (ratio = 2.7) ASC compartments similar to what would be expected in peripheral blood21,22 (Supplementary Fig. 1C).
IgE ASCs have distinct VDJ repertoire characteristics
By VDJ analysis, we identified 116 IgE ASCs, which represented 2% of the total ASC sequences analyzed (Fig. 1D). All IgE ASCs were contained within the CD19+ compartment (Fig. 2A). Although a majority (n = 56) of IgE ASCs were isolated from a single donor, each NP had similar proportions of IgE ASCs relative to total ASCs (Fig. 1E). Akin to previous studies, which showed that IgE ASCs had a lower IGHV mutation frequency than non-IgE ASCs14, the IgE ASCs in our dataset had the second lowest median IGHV mutation frequency (7.1%), with a range of IGHV mutation frequencies (2.36%−14.9%) and had significantly lower median mutation frequency than IgA1 (8.7%), IgA2 (11.3%), and IgG2 (10.5%) ASCs (Fig. 2B). IgG4 ASCs had the lowest median V region mutation frequency (5.5%); although, the significance of this finding is unclear because only three VDJ sequences were isolated from IgG4 ASCs. Among the IGHV gene segments, IgE ASCs had relatively fewer mutations in the three framework regions (FR1—3) and complementarity determining regions (CDR 1-2) (Supplementary Figs. 1D-H). Additional analysis of selection pressures using a Bayesian estimation of antigen-driven section in Ig sequences (BASELINe) score showed that IgE ASCs experienced less selection pressures than any other isotype (Fig. 2C)23. Finally, there was no significant difference in the mutation frequency of IgE ASCs that were members of expanded lineages compared with IgE ASCs that were singlets in this dataset (data not shown).
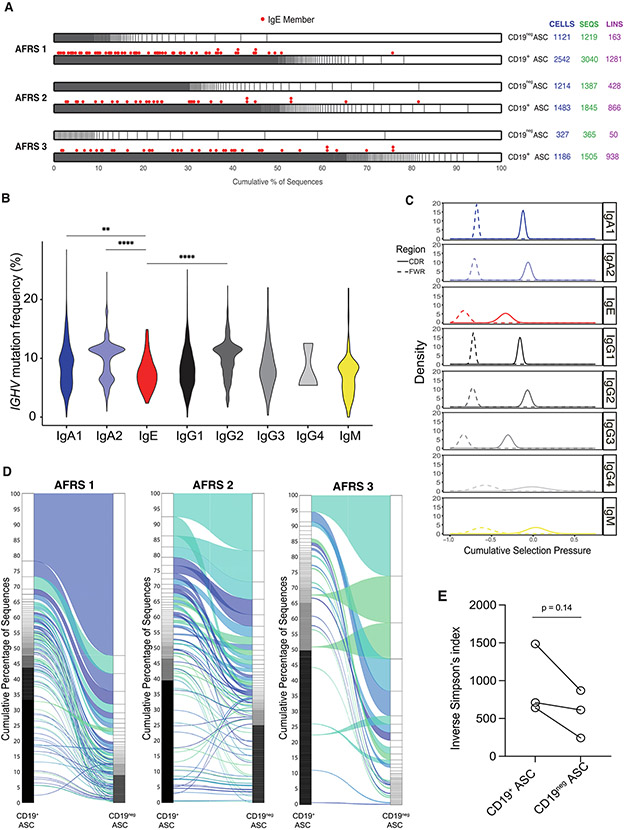
Fig. 2.
NP VDJ repertoire features. A, relative lineage size in the CD19+ and CD19− compartments, stratified by polyp. IgE members of lineages are denoted by red spots above their respective lineage; B, violin plot showing IGHV mutation frequencies across isotype subclasses; C, BASELINe analysis showing cumulative selection pressures on VDJ complementarity determining regions (solid line) and framework regions (dashed line) across isotype subclasses; D, alluvial plots showing connectivity between CD19+ ASCs and CD19− ASCs within each polyp; E, relative VDJ repertoire diversity within the CD19+ and CD19− compartments as measured by the inverse Simpson index. Statistical significance was determined using (B) Kruskal-Wallis testing with Dunn multiple comparisons testing between groups or (E) paired Student’s t test. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001. AFRS = allergic fungal rhinosinusitis; ASC = antibody secreting cell; CD = cluster of differentiation; Ig = immunoglobulin; IGHV = IgE ASCs featured both high and low heavy chain V region; lins = number of lineages; NP = nasal polyp; ns = not significant; Seqs = number of sequences; VDJ = immunoglobulin gene.
AFRS CD19+ ASCs contain an oligoclonal VDJ repertoire connected with CD19− ASCs
Due to the somatic hypermutation of expanded B cell clones, the lineages from the same clonal origin were defined as having the same V gene, J gene, CDR3 length, and 85% overall sequence homology24. Across all three polyps, oligoclonality was noted with large CD19+ lineages that were more expanded into the CD19− compartment, suggesting selection for a longer-lived, possibly tissue-resident, ASC phenotype (Fig. 2D). A trend toward significantly less diversity in the CD19− compartment was also observed, as measured by an inverse Simpson index; although, this may have simply reflected the low numbers of CD19− ASCs (Fig. 2E). Interestingly, the majority of IgE ASCs were singlets and did not belong to any ASC lineage. However, there were some, albeit few, expanded lineages, which contained IgE members. All included, these data suggest two developmental pathways for IgE ASC differentiation: the majority from extrafollicular IgD+ B cells and some from germinal center (GC)-derived memory B cells. In the models of human peanut allergy, antibody convergence has been observed13,25, suggesting IgE ASCs may involve public clones. Convergence is defined as VDJ sequences with identical V genes, J genes, and CDR3 sequences shared between two or more individuals in response to similar antigens and was not identified in this dataset. There was preferential utilization of the V3-15 allele among non-IgE ASCs (Supplementary Fig. 1I), but generally, IgE ASCs were not skewed toward a single V gene allele.
Connectivity of ASCs to DN, USM, and SWM B cell subsets in NPs
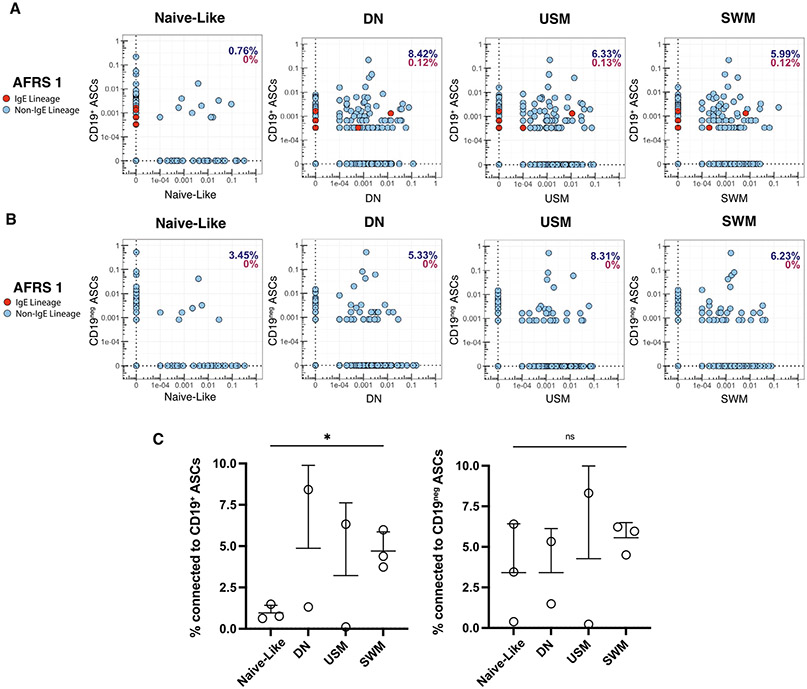
Using a separate pipeline for bulk VDJ sequencing data (see Methods), we evaluated the connectivity between mucosal sorted B-cell subsets and CD19+ and CD19− ASCs. In addition to sorted ASCs for the single-cell analysis, we also sorted four mucosal B-cell subsets (naïve, USM, SWM, and DN) based on canonical markers CD27 and IgD (Fig. 1A) for bulk VDJ sequencing in two of the three polyps (AFRS1 and AFRS2). Only naïve and SWM subsets were retrieved from AFRS 3 due to limited numbers. The majority of both CD19+ and CD19− ASC connectivity occurred mainly in the USM, SWM, and DN subsets compared with naïve-like B cells. However, due to the small sample size, only the connectivity between SWM and CD19+ ASCs was statistically significant (Figs. 3A and 3C). CD19− ASCs showed a similar connectivity trend but were not statistically significant (Figs. 3B and 3C). There was connectivity identified between IgE ASCs and other B cell subsets in only one donor (AFRS 1) and only within three subsets: DN, USM, and SWM (Fig. 3A).
Fig. 3.
Connectivity of V(D)J region of nasal polyp ASCs with sorted B cell populations. A, connectivity plots for CD19+ sorted ASCs from polyp AFRS 1 with bulk VDJ sequencing data from four sorted B cell subsets. Red dots denote IgE ASCs; B, connectivity plots for CD19− sorted ASCs from polyp AFRS 1 with bulk VDJ sequencing data from four sorted B cell subsets. Red dots denote IgE ASCs; C, connectivity of VDJ regions of CD19+ sorted ASCs (left) and CD19− ASCs (right) with four sorted B cell populations. Statistical significance was determined using paired Student’s t test. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001. AFRS = allergic fungal rhinosinusitis; ASC = antibody secreting cell; CD = cluster of differentiation; DN = double-negative B cells; Ig = immunoglobulin; ns = not significant; SWM = switched memory; USM = unswitched memory; VDJ = immunoglobulin gene.
Mucosal IgE ASCs are transcriptionally distinct in AFRS
To characterize mucosal-derived ASCs in NPs, single-cell transcriptomes were also analyzed. Isotype labels were assigned using available single-cell VDJ data (n = 4,705). In cases where high-confidence transcriptional data were available without paired VDJ data, an isotype label was assigned based on the highest heavy chain gene expression (n = 1,744, see Methods). After the exclusion of non-ASC contaminating cells, low-quality cells, dying cells, and doublets (see Methods, Supplementary Figs. 2A-C), the remaining cells were identified as ASCs, with the expression of genes encoding canonical ASC markers, including JCHAIN and transcription factors, IRF4, PRDM1 (BLIMP-1), and XBP126 (Fig. 4A, Supplementary Figs. 2D-F). We analyzed 6,449 total ASCs, including 113 IgE ASCs, which were clustered in an unsupervised manner.
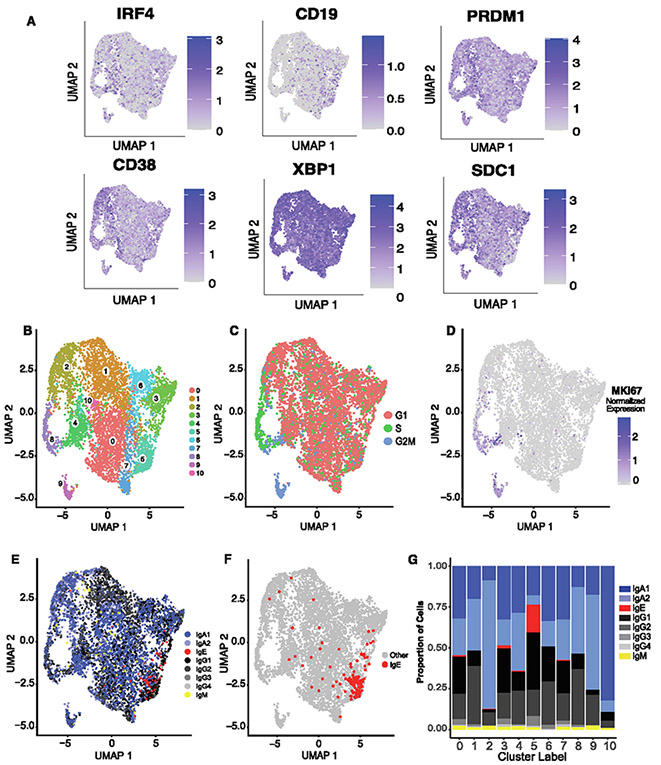
Fig. 4.
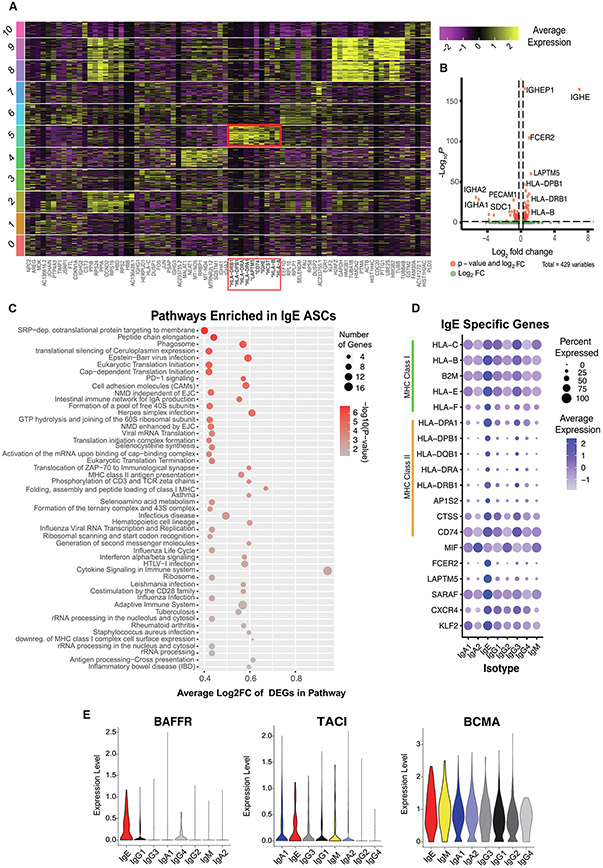
NP ASCs transcriptional features. A, gene expression of canonical gene markers of ASCs shown as log normalized counts; B, two-dimensional UMAP projection using scRNA-seq data of sorted ASCs from three nasal polyps; C, location of cell cycle phases of NP ASCs on a two-dimensional UMAP projection of scRNA-seq data of nasal polyp ASCs; D, location of MKI67 gene expression on a two-dimensional UMAP projections; E, location of each isotype overlaid onto two-dimensional UMAP projections; F, location of IgE ASCs, which reside primarily in cluster 5 portrayed onto the UMAP projection; G, stacked bar plot indicating the proportion of isotypes per cluster. ASC = antibody secreting cell; CD = cluster of differentiation; Ig = immunoglobulin; NP = nasal polyp; scRNA-seq = single-cell RNA sequence; UMAP = Uniform Manifold Approximation and Projection.
A total of 11 clusters were obtained using a resolution of 0.5 and visualized using a Uniform Manifold Approximation and Projection (UMAP) embedding (Fig. 4B), with IgE ASCs primarily clustering in cluster 5. IgE ASCs appear to cluster more closely together than other isotypes (Figs. 4E-G, Supplementary Fig. 5), with 70% of IgE ASCs assigned to cluster 5 (Fig. 4G), suggesting that IgE ASCs are more transcriptionally homogenous relative to ASCs of other isotypes. To ensure that Ig genes were not driving IgE ASCs to cluster together, we confirmed that removing or including Ig genes did not affect the clustering of IgE ASCs nor the other notable clusters in a meaningful way (Supplementary Fig. 6, see Methods). Interestingly, IgA2 and IgG2 were predominant in clusters 9, 8, and 2, with IgA2 being most dominant in cluster 10. Although IgE ASCs were observed in clusters 0, 3, 4, 5, and 7, cluster 5 contained the majority. Clusters 8 and 9 were enriched in genes associated with S phase (cluster 8) and G2/M phase (cluster 9) of the cell cycle (Fig. 4C). Notably, MKI67 was not upregulated in cluster 5 (Fig. 4D), which confirms that IgE ASCs are not rapidly proliferating. The top 10 significantly upregulated differentially expressed genes (DEGs) were identified for each cluster, as shown in Fig. 5A, with cluster 5 notably upregulating the genes associated with the MHC. A gene ontology analysis identified upregulated DEGs in cluster 8 to be enriched in translational activity, oxidative phosphorylation, and DNA replication, whereas upregulated DEGs in cluster 9 showed similar translational and metabolic activity (Fig. 5A, Supplementary Fig. 4A).
Fig. 5.
IgE ASC-specific transcriptional features. A, heatmap showing the gene expression of the top 10 DEGs identified per cluster. The red box indicates the 10 DEGs upregulated in cluster 5; B, volcano plot of DEGs identified by comparing IgE ASCs against all other CD19+ ASCs; C, dot plot indicating the number of genes and average log2fold change of genes differentially expressed in pathways enriched in IgE ASCs. DEGs detected by comparing IgE versus all other CD19+ NP ASCs; D, dot plot showing IgE specific genes, including MHC Class I and MHC Class II genes, across sorted ASC NPs grouped by isotypes; E, sorted violin plots for the log normalized gene expression for genes encoding BAFFR, TACI and BCMA. ASC = antibody secreting cell; CD = cluster of differentiation; DEGs = differentially expressed genes; HLA = ; Ig = immunoglobulin; MHC = ; NP = nasal polyp; scRNA-seq = single-cell RNA sequence.
IgE ASCs retain characteristics of a B cell phenotype
Due to the heterogeneity of the other isotypes, a differential gene expression analysis to obtain IgE ASC-specific genes was performed based on isotype rather than cluster. As previously mentioned, IgE ASCs clustered together more strongly than the other isotypes, suggesting a strong transcriptional similarity across IgE ASCs. DEGs were obtained by comparing IgE ASCs (n = 113) to all other CD19+ ASC isotypes (IgM, IgG, IgA, n = 4,252) because IgE ASCs were found only in the CD19+ ASC subset by flow cytometric immunophenotyping. We identified 161 DEGs in IgE ASCs (Bonferroni corrected p value < 0.05), with 106 genes upregulated with respect to other isotypes and 55 genes downregulated (Fig. 5B). We performed a pathway enrichment analysis for IgE ASC-specific DEGs and found that enriched pathways were related to antigen processing and presentation, interferon-γ signaling, cell adhesion, and protein synthesis (Bonferroni corrected p value < 0.05) (Fig. 5C). The DEG analysis revealed 13 genes to be differentially upregulated in IgE ASCs related to antigen processing and presentation (Fig. 5D). Specifically, the genes involved in both MHC I (HLA-C, HLA-B, B2M, HLA-E, HLA-F) and MHC II antigen presentation (HLA-DPA1, HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DRB1, AP1S2, CD74, CTSS) were significantly upregulated compared with other ASC isotypes. Surface MHC II expression decreases as B cells differentiate into plasma cells27, suggesting that IgE cells are either more recently differentiated compared with other isotypes or may retain MHC class II genes. Nonetheless, these results suggest that in addition to Ig secretory function, IgE ASCs may be actively presenting antigen to local T cell subsets.
Surface expression of tumor necrosis factor superfamily receptors, including BAFFR, BCMA, and TACI, play critical roles in the survival of B cells and ASCs. BAFFR is known to downregulate as B cells differentiate into ASCs28. However, IgE ASCs demonstrate differentially upregulated TNFRSF13C (BAFFR) expression compared with ASCs of other isotypes. In contrast, there was a similarly high expression of TNFRSF17 (BCMA) and notable expression of TNFRSF13B (TACI) among IgE ASCs compared with non-IgE ASCs (Fig. 5E)26. These data provide evidence that IgE ASCs show transcriptional hallmarks of an early ASC phenotype, with the maintenance of features not typical of IgG ASCs.
Transcriptional features of IgE ASCs suggest recent activation and recruitment
CD23 (FCER2) is an Ig receptor in the C-type lectin superfamily that serves as the low-affinity receptor for IgE29,30. Although CD23 expression is present to varying degrees on multiple B cell subsets, this receptor increases in response to BCR stimulation in the presence of interleukin (IL)-431,32. However, in other models of B-cell activation, it appears that the expression of CD23 is decreased on mucosal ASCs14. FCER2 was the most significantly upregulated non-Ig gene found when comparing IgE ASCs with CD19+ non-IgE ASCs (Figs. 5B and 5D). Therefore, unique to IgE ASCs, perhaps CD23 is critical in regulating IgE synthesis with its negative feedback, whereas in ASCs of other isotypes, CD23 is downregulated because it plays no role in their function. We also observed the upregulation of LAPTM5 and SARAF, both of which are involved in BCR ligation (Fig. 5D). LAPTM5, which was previously shown to be upregulated on IgE ASCs13, is associated with lysosomal degradation of both pre-BCR in immature B cells33 and surface BCR after stimulation in mature B cells34. SARAF is associated with promoting slow inactivation of store-operated Ca2+ entry to prevent excess refill35. The upregulation of SARAF observed in the IgE ASCs in our dataset may prevent overstimulation and apoptosis by slow Ca2+ refill. The differential upregulation of these two genes in this context may imply protective responses to potential ongoing BCR stimulation of IgE ASCs.
CXCR4 and KLF2 were differentially upregulated in IgE ASCs (Fig. 5D). KLF2 is a transcription factor, that controls homeostasis and trafficking of activated B cells into peripheral blood36. KLF2 controls the regulation of sphingosine-1 phosphate receptor and potentially the migration of IgE ASCs. ASCs in the NP also universally express MIF, a chemokine that binds to CD74 (Fig. 5D). MIF expression by cells in the NP niche may promote the recruitment or retention of IgE ASCs to CXCL12 abundant regions by cooperative engagement of CXCR4 and CD7437, resulting in an increase of intracellular Ca2+ mobilization and F-actin polymerization.
Expanded ASC lineages and low IGHV mutation frequency IgE ASCs show specificity to fungal antigens
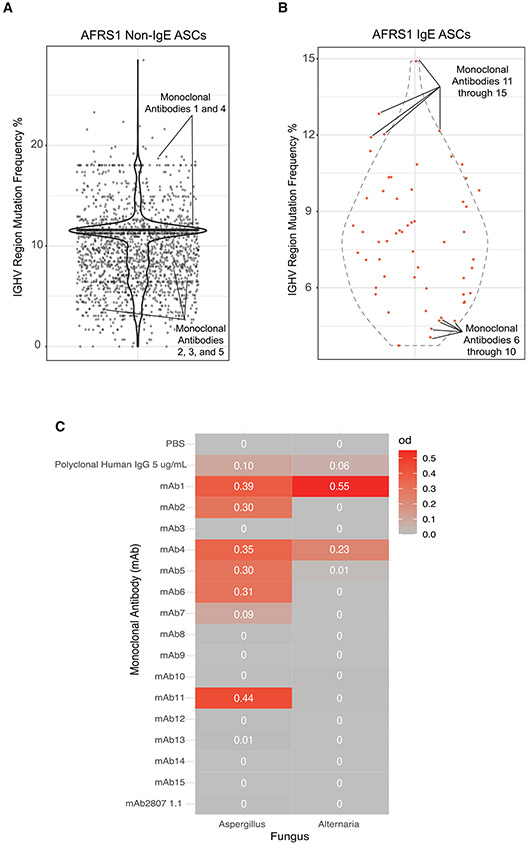
To test the hypothesis that NP IgE ASCs contribute to disease pathology in AFRS by producing IgE with fungal specificity, we selected 15 Ig gene sequences and synthesized monoclonal antibodies (mAbs). mAbs 1 through 5 were selected from non-IgE ASCs with expanded lineages (Fig. 6A, Supplementary Table 2). mAbs 1–5 had 150, 30, 20, 1,103, and 95 clonal members in its lineage, respectively. Two were highly mutated and three were less mutated (Fig. 6A). Even from the small numbers selected, four of five clones were specific to Aspergillus, demonstrating expanded lineages due to fungal antigens, resulting in mucosal ASC in AFRS patients. mAbs 1 and 4 also showed specificities to Alternaria antigens, which may have cross-reactive epitopes. mAbs in this group had both high (18%) and low (3.1%) mutation frequencies, suggesting origins from both GC-derived memory B cells and extrafollicular B cells. Notably, mAb 1 (IgA2) and 11 (IgE) were both members of the same lineage and showed specificities to Aspergillus fungal antigens, illustrating the expansion of both IgE and IgA2 antibodies (Figs. 6B and 6C).
Fig. 6.
IGHV mutational frequencies of nasal polyp ASCs. A, violin plot of IGHV region mutation frequencies for AFRS1 non-IgE ASCs with mAbs annotated by solid lines; B, violin plot of IGHV region mutation frequencies for AFRS1 IgE ASCs with mAbs annotated by solid lines; C, monoclonal antibody reactivity against Aspergillus and Alternaria fungal antigens. AFRS = allergic fungal rhinosinusitis; ASC = antibody secreting cell; Ig = immunoglobulin; IGHV = IgE ASCs featured both high and low heavy chain V region; mAb = monoclonal antibody; PBS = .
mAbs 6 through 15 were synthesized from sequences derived from IgE ASCs, with the five lowest (mAb 6—10) and five highest (mAb 11—15) mutation frequencies (Fig. 6B, Supplementary Table 2). From this group, only mAbs 6, 7, and 11 demonstrated specificities to fungal antigens. Of the mAbs generated from IgE singletons, only mAbs 6 and 7 showed specificity against Aspergillus fungal antigens (Fig. 6C). The majority of mAbs from single IgE ASC clones did not react against fungal Alternaria antigens; although, the non-IgE ASCs were positive. In all, the majority of expanded non-IgE ASCs were fungal-specific in the NPs of patients with AFRS, and fungal-specific IgE arose from both IgE ASC singletons and expanded clones.
DISCUSSION
AFRS is a challenging clinical entity that involves polyp recurrence, symptom exacerbations, and revision surgery when disease is uncontrolled6. At present, a definitive treatment for this condition has yet to be identified and many patients will require sinus surgery despite aggressive non-invasive interventions. Previous investigation into the pathophysiology of AFRS has led to a model in which a sustained local T2 immune response leads to progressive symptoms of this disease. Central to this paradigm of sustained T2 inflammation is the production of IgE, which sensitizes mast cells, basophils, and upstream antigen presenting cells to endemic fungal antigens. Yet, despite the recent advances in the characterization of IgE ASCs, many questions remain about their B-cell ontogeny and role on the mucosal surface. Here, we report that the majority of IgE ASCs isolated from NPs in AFRS appear to have an immature, early ASC phenotype, with features reminiscent of early blood ASCs or plasmablasts. Their origins from antigen-experienced B cells (USM, SWM, and DN) suggest a fungal-specific mucosal B cell reservoir. Interestingly, only some IgE ASCs were directed against common fungal antigens, providing potential mechanisms of epitope spreading through CD23. Finally, these B cell features on IgE ASC maybe a sign of early minted ASC, implicating a short half-life and continuous generation of IgE ASC to perpetuate the local mucosal T2 response.
IgE is difficult to study because it has the shortest half-life in serum of all antibody isotypes. Moreover, IgE ASCs are non-proliferative, extremely rare in frequency, and there is controversy about a long-lived reservoir. Thus, it would seem that IgE ASCs are replenished rapidly upon re-exposure to fungal aeroallergens in upper airway diseases, such as AFRS. Interestingly enough, human IgE memory B cells have not been definitively isolated and their existence remains controversial38-41 because IgE class-switch recombination rapidly leads to ASC differentiation. Hence, B cell reservoirs for IgE ASC in AFRS may arise from several sources. First, GC-derived IgA or IgG B cells implicated in mucosal defense may undergo local sequential class switch recombination (CSR) into IgE ASCs in the setting of a dysregulated T2 immune response. In this scenario, fungal-specific SWM B cells would clonally expand and differentiate into IgE ASCs locally to produce antibodies, such as with mAb 11, which is highly mutated. The lack of bona fide IgE memory B cells and evidence of clonal connectivity of IgG memory B cells support this hypothesis38,39. The second source of IgE ASCs maybe of B cells arising through an extrafollicular pathway, as shown by fungal-specific antibodies (mAbs 6, 7) with low mutation frequencies. These IgE ASC likely originated from naïve-like or USM B cells. Ultimately, the source of IgE ASCs may depend on the primary versus repeat exposures and the fungal antigens. Initially, the fungal antigens may predispose to a rapid differentiation through extrafollicular and possibly GC pathways, whereas seasonal re-exposure to antigens may occur through differentiation from SWM or DN B cells in inducible nasal-associated lymphoid tissue.
In AFRS, IgE ASCs were only found within the CD19+ ASC compartment and had the lowest IGHV mutation frequency and lowest VDJ selection pressures. Yet, they were not connected to naïve-like subsets in the NP mucosa, which was shown in our previous study14. However, IgE ASC connectivity with the USM and DN subsets were readily observed. The reasons for the lack of IgE ASC connectivity with naïve-like B cells origins may be several-fold. AFRS is a special subset of nasal polyposis driven by fungal exposures and therefore may have different pathophysiologic mechanisms compared with allergic rhinitis. Repeat fungal exposures may have expanded a population of antigen-experienced USM and DN B cells as a source of continuous IgE ASC differentiation. Another reason could have been the gating strategies of the IgD+ naïve-like B cell subset, which may have inadvertently included the USM (IgD+CD27+) B cell subset in the first study. Lastly, the small sample size may have missed connectivity with naïve-like B cells. Nonetheless, both studies support the model of local extrafollicular IgE ASC generation from two separate mucosal B cell origins: mucosal naïve-like and antigen-experienced B cell subsets.
Previous studies identified a large expansion of DN B cells likely originating from extrafollicular naïve B-cell activation in patients with autoimmune disease, thus resulting in lower VH mutation rates than conventional SWM18. Similar expansions of DN B cell in the nasal mucosa in AFRS parallel this model of extrafollicular B cell responses, which may be critical to maintain an antigen-experienced B cell origin of IgE ASC in allergic diseases. In this model, IgE ASCs would be predicted to respond to fungal antigens present in the NP inflammatory milieu. Although fungal-specific IgE antibodies were identified, our study did not further elucidate the specificities of the majority of the highly mutated IgE mAbs selected. Thus, it will be important for future studies to determine if highly mutated IgE ASCs truly react against non-fungal targets in AFRS and whether this has an impact on disease pathogenesis.
Interestingly, MHC class II molecules and FCER2 (CD23) were differentially upregulated by IgE ASCs, a finding which was also shown in a recent study by Croote et al.13. Similar to CD19, MHC class II molecules are downregulated as plasmablasts mature into plasma cells16. Therefore, differential upregulation of MHC class II proteins in IgE ASCs implicate more recent activation and differentiation than ASC of other isotypes. However, an alternative hypothesis may be that CD23 and MHC class II surface expression in fact may be related. CD23 can bind to MHC class II molecules on the surface of B cells and facilitate antigen presentation42. In some plasma cell dyscrasias, antigen presentation has been described supporting this model43. Although we thought ASCs of all isotypes matured similarly by downregulating MHC class II, IgE ASCs appear to retain MHC class II and upregulate CD23 (FCER2) suggesting novel mechanisms of possible epitope spreading during antigen presentation, perhaps to perpetuate T2 local immune response30.
IgE ASCs also differentially express CXCR4, a chemokine receptor for CXCL12. CXCR4 has been shown for BM homing or migrate to the GC dark zones during affinity maturation44,45. Due of the paucity of IgE ASCs found in the human BM, even in patients with allergic disease, it is unlikely that they are primed to migrate to the BM; although, BM-derived IgE ASCs in humans have been described11,12. For mucosal IgE ASC, CXCR4 may promote homing or retention in tissues sites with high CXCL12 concentrations, such as the inflamed respiratory mucosa.
The universal expression of MIF on NP ASCs suggests potential recruitment of IgE ASCs through its receptors CD74 and CXCR4. MIF has been shown to play a role in survival of diseased B cells through CD74 and upregulation of BCL2 and to be important in differentiation and activation of transitional and activated SWM B cells46,47. However, its role on ASCs is not entirely clear. In the 1980’s, a soluble glycosylation inhibitory factor, which was originally described as a suppressor of IgE synthesis on B cells, was later found to be a post-translational modified MIF48,49. Thus, the higher expression of MIF on mucosal IgE ASC may suggest a role in IgE synthesis or survival. MIF has also been shown to stimulate intracellular Ca2+ to promote migration. We observed SARAF to be upregulated in IgE ASCs in the NP, which is known to prevent Ca2+ influx or refill when stores are depleted. Thus, SARAF may limit ongoing B cell activation on an IgE ASC with high major histocompatibility complex (MHC) class II expression to prevent apoptosis.
In this study, there were several limitations. The first includes the use of nasal steroids by all donors, which certainly can affect the local mucosal microenvironment. However, it may have less of an effect on the biology of ASC than B cells because corticosteroids result in lymphopenia. Perhaps, due to the low doses of inhaled steroids, we were still able to isolate enough mucosal B cell and ASC for the single-cell analysis. A second limitation was the low numbers of IgE ASCs (116 IgE ASC), which may have constrained the power of our analysis; however, interestingly, Croote et al.13 identified only 89 bona fide IgE ASC, suggesting that these cells are extremely rare. Although the proportion of IgE ASCs was low (2% of all isotypes by VDJ), our approach of isolating all mucosal CD27+CD38+ASC and phenotyping by VDJ analysis was essential to identify a bona fide IgE ASCs because they do not express any unique surface markers. In all, the abundance of IgE ASC in the mucosa of AFRS probably afforded feasibility to analyze 116 bona fide IgE ASC. For future studies, using a combination of HLA-DR, CXCR4, and CD23 to enrich for bona fide IgE ASC may also prove useful.
Our findings in this manuscript may have direct clinical relevance because the case reports describing the use of a monoclonal antibody, which inhibits IL-4 and IL-13 signaling, dupilumab, has shown promise in the treatment of AFRS. More recently, two clinical trials will investigate the effectiveness of dupilumab in AFRS (NCT05545072, NCT04684524). Whether B-cell reservoirs, such as USM and DN, of IgE ASC remain after local inhibition of IgE class-switching would be important to understand. Similarly, ability to measure the longevity of an IgE mucosal ASC when IL-4-mediated ASC differentiation is halted would also be valuable.
In summary, this study demonstrates the first single-cell analysis of a bona fide mucosal IgE ASC, with features revealing a newly minted ASC due to the high expression of CD19 and HLA class I and II genes. Whether this phenotype merely signifies early minted ASC immaturity or is also indicative of additional functions of IgE ASC, such as antigen presentation, will need further study. In all, this study provides single-cell methods to study bona fide mucosal IgE ASC to reveal novel molecular mechanisms.
METHODS
Human subjects
Polyps AFRS1 through 3 (n = 3) were obtained through the discarded tissue protocol from patients undergoing endoscopic sinus surgery at the Ambulatory Surgery Center of Emory University Hospital in Atlanta, Georgia, USA. All research was approved by the Emory Institutional Review Board (Emory IRB 66294) and performed in accordance with all relevant guidelines/regulations. All patients were undergoing functional endoscopic sinus surgery for symptomatic CRSwNP at the discretion of their treating physicians. Patients were diagnosed with CRSwNP on the basis of established guidelines50,51. Polypectomy specimens were placed in chilled Roswell Park Memorial Institute (RPMI) medium and transported at room temperature to the laboratory for processing. Baseline subject demographics are included in Supplementary Table 1. Notably, all research subjects identified as Black or African American, which may reflect the epidemiology of this disease51-54. Patient sex was self-reported and confirmed by treating physicians. All three NP specimens were obtained from patients with AFRS diagnosed using the Bent and Kuhn criteria55; these patients were not taking oral corticosteroids or any monoclonal antibody therapy for asthma at the time of surgery. All patients were using steroid nasal rinses at the time of surgery, per standard of care.
NP mechanical and enzymatic digestion
To isolate ASCs, the NP tissue specimens were processed, as described previously14. Briefly, NPs were extracted from media, transferred to a petri dish, weighed, and then finely minced using sterile forceps and a scalpel. After mechanical separation, Dulbecco's Modified Eagle Medium (DMEM) was added to the petri dish and the specimens were triturated using a modified sterile P1000 pipette tip and washed with additional DMEM before being placed in a 50-ml conical tube. A digestion mix was then added containing 10 ml DMEM, supplemented with antibiotic/antimycotic at a concentration of 100 ug/ml (Gibco), DNAse I at a concentration of 100 ug/ml (Sigma), and collagenase/protease mixture (Liberase TL Research Grade, Millipore Sigma) at a concentration of 150 ug/ml. Once the digestion mix was added to the 50-ml conical tube, the mixture was vortexed for 30 seconds, and the cells were incubated for a total of 45 minutes, vortexing every 15 minutes. After the final vortex step, the samples were filtered through a 40-μm cell strainer, rinsed with 10 ml RPMI, and centrifuged at 500 g for 10 minutes. The supernatant was then removed, and the cells were resuspended in 5 ml red blood cell lysis buffer for 3 minutes at 4°C. The cells were washed twice with RPMI, resuspended in complete RPMI medium with 10% fetal calf serum (R10), and were counted with an automated hemocytometer.
Flow cytometric immunophenotyping and fluorescence-activated cell sorting
Cells were centrifuged again at 500 g for 5 minutes, the supernatant was removed, and the cells were resuspended in 50 μl of R10. An antibody cocktail was then added to the single-cell suspension, which included the following: normal mouse serum, FITC anti-human IgD (BD, clone IA6-2), Brilliant Violet 711 anti-human CD3 (Biolegend, clone OKT3), Brilliant Violet 711 anti-human CD14 (Biolegend, clone M5E2), PE/Cy7 anti-human CD19 (BD, clone SJ25C1), V450 anti-human CD38 (BD, clone HIT2), APC anti-human CD138 (Miltenyi Biotec, clone 44F9), APC-eFluor 780 anti-human CD27 (eBiosciences, clone O323), and LiveDead Aqua (Invitrogen). Cells were stained for 20 minutes at 4°C in staining buffer and then washed for immediate sorting. Cells were sorted on a BD FACSAria II cell sorter using BD FACSDiva software. Up to 20,000 CD19+ ASCs and 20,000 CD19− ASCs were sorted into 500 μl of complete RPMI medium with 5% fetal calf serum (R5) and were taken directly for scRNA-seq. Additional populations, including naïve, DN, SWM, and USM, were also sorted into 350 μl of RLT buffer (Qiagen) and were frozen at −80°C until they were taken for bulk VDJ sequencing. FlowJo v10 (Treestar) was used to generate flow cytometry plots.
Single-cell VDJ sequencing and data processing
The sorted cells were then centrifuged at 500 g for 10 minutes at 4°C and then washed with 1000 μl of 0.04% bovine serum albumin (BSA) (non-acetylated) in phosphate buffered saline (PBS) three times. After the final wash, the media was carefully removed until roughly 20 μl of media remained. Cells were resuspended and the volume was measured using a pipette. The cell concentration was calculated and, using the cell suspension volume calculator table provided by 10X Genomics, nuclease-free water was added. The cell suspension and nuclease-free water mixture was added to the master mix and was loaded onto the 10X Genomics Chromium, according to the manufacturer’s standard protocol [Chromium Next GEM Single-Cell V(D)J Reagent Kits, v1.1]. 5’ VDJ libraries were sequenced on an Illumina NovaSeq using 150bp × 2 paired end sequencing. The resultant sequences were demultiplexed using Cell Ranger software (v3.0.2) and aligned using IMGT HighV-QUEST tool56. Further analysis was conducted using inhouse developed software, including the IgSeq program for clonal clustering, isotype, and mutation analysis17.
Bulk VDJ sequencing and data processing
Bulk, heavy chain VDJ raw sequence data were processed as previously described17. Briefly, paired end reads were joined and then filtered based on read length (excluding reads <200 bp) and base quality score metrics. Isotype was determined through alignment of the constant region, and then reads were annotated using the IMGT/HighV-QUEST tool56. Sequences were assigned to groups characterized by the same V gene, J gene, and CDR3 length, and subsequently, further clustered into lineages based on 85% CDR3 nucleotide sequence similarity within each group. The sequences from different populations were clustered together to assess clonal relatedness. IMGT output, cell population, and lineage unique identifiers were written to SQLite databases and accessed by custom analysis and visualization tools written in MATLAB (The Mathworks, Natick, Massachusetts, USA www.mathworks.com) for further analysis.
scRNA-seq gene expression sequencing and data processing
In parallel to the single-cell VDJ sequencing, 5’ gene expression libraries were constructing according to the manufacturer’s standard protocol (Chromium Next GEM Single-Cell V(D)J Reagent Kits, v1.1). 5’ gene expression libraries were sequenced on an Illumina NovaSeq. The resultant sequences were demultiplexed and converted to fastq files using CellRanger v3.1.0 from 10x Genomics57. The sequencing results were mapped to GRCh38 reference genome from the 10x Genomics website and quantified using CellRanger v3.1.0. A counts matrix was generated with filtered feature barcodes used as input for Seurat v4.058. The total number of cells identified was 15,942. The mean number of genes detected per cell was 1709 and the mean unique molecular identifiers per cell was 18,209. Data analysis was performed in R using Seurat v4.0. Unless specified, the quantitative parameters were set to default values.
To remove possible empty droplets, low-quality cells, and possible multiplets, cells with less than 1,000 or more than 5,000 genes detected were excluded from the analysis. In addition, to remove possible dying, dead, or uncharacteristically stressed cells, cells having more than 25% of their transcripts being mitochondrial genes were excluded from the analysis. Lastly, to ensure that antibody secreting B cells were captured, an additional filter was implemented based on the amount of Ig heavy and light chains (λ and κ genes)59. Cells with less than 25% of their transcripts belonging to these genes were excluded (Supplementary Fig. 2C). After a preliminary unsupervised clustering analysis, the marker genes for potentially contaminating cells were used to detect and remove cells that were not ASCs (Supplementary Figs. 2D-F). After the removal of low-quality and contaminating cells, a total of 6449 high-confidence ASCs were used for the analysis.
Each cell’s gene expression measurement was normalized using Seurat “LogNormalize” function, which normalized each cell by their total expression, multiplied by a scale factor of 10,000, and log-transformed the result. After identifying 2750 highly variable features using variance-stabilizing transformation, integration was used by identifying corresponding anchors between the three donors’ samples using 30 dimensions to remove the donor-specific differences. Next, a linear transformation was used to scale the data. The expression of each gene was shifted so the mean expression across all cells is 0 and the variance is 1 across all cells. The principle components were computed, and an elbow plot was used to choose components 2 through 12 for subsequent clustering, which captured the majority of variance across the cells. Clusters were obtained using the Seurat v4.0 unsupervised, graph-based approach, with the resolution parameter set to 0.5 and visualized using a two-dimensional UMAP embedding. To determine which phase of the cell cycle each cluster resided, the cell cycle scores were computed for each cell using the Seurat CellCycleScoring function. Unless specified, the quantitative parameters were set to default values.
Because Ig genes have been observed to contribute to a substantial number of genes expressed in ASCs, we sought to determine whether IgE cells clustering together could be influenced by Ig genes. As a comparison, a total of 206 known Ig genes were removed before clustering, with the same clustering parameters used, as previously described. The Rand index was computed using the R package Dune between the working cluster set and the cluster set obtained after removing Ig genes with a value of 0.43 retention. Supplementary Fig. 6 depicts which clusters have the highest and lowest retention, with clusters 2, 5, 8, and 9 having >80% of cells clustering together without Ig genes. This indicates that IgE cells, which reside in cluster 5, possess a unique transcriptional program to drive their clustering rather than being clustered by the influence of Ig genes.
To evaluate the DEGs between IgE ASCs versus other isotypes, the isotype labels were obtained by one of two different means. VDJ data with isotype labels from IMGT’s HighV-QUEST tool were available for 4705 cells, which left the remaining 1,744 cells that did not have VDJ data without an isotype label. Isotype labels were given to these remaining cells by assessing which Ig heavy chain gene was expressed the most according to the normalized gene expression. To evaluate the accuracy of this prediction, the 4,705 cells with high-confidence isotype labels from IMGT HighV-QUEST were given predicted labels based on which Ig heavy chain gene was expressed the most according to the normalized gene expression. The R package rfUtilities v.2.1-560 accuracy function was used to determine the gene expression isotype label accuracy by measuring the percent of cells correctly classified and computing a Cohen κ value to measure the agreement of isotype labels. For the 4,705 cells with VDJ based isotype labels, 94.5% were correctly classified by assessing which Ig heavy chain gene was expressed the most according to the normalized gene expression, with a Cohen κ yielding a value of 0.93. This provided enough confidence to predict isotype labels for the 1,744 cells that did not have VDJ data using this method. The correlation between the isotype labels and gene expression is displayed in Supplementary Fig. 3.
scRNA-seq comparative analysis
To identify the DEGs for IgE ASCs (n = 113 cells) compared with CD19+ cells of other isotypes (n = 4,252), Seurat FindMarkers was implemented using a Wilcoxon Rank Sum test implemented with Seurat v4.0 on the normalized gene expression from the RNA assay. The tested genes were required to be detected at a minimum of 10% cells, and only genes with an absolute average log2 fold change greater than 0.25 and Bonferroni corrected p value below 0.05 were considered differentially expressed. A total of 161 genes were differentially expressed in IgE ASCs, 106 of which were found to be upregulated with respect to other isotypes and 55 found to be downregulated. These genes were used subsequently for functional analysis of IgE ASCs.
scRNA-seq pathway enrichment analysis
To identify the pathways enriched in IgE ASC-specific genes, we performed a pathway enrichment analysis using ToppFun, which is part of the ToppGene Suite61. This analysis was performed using the DEGs obtained from IgE ASCs versus CD19+ other isotypes to evaluate enrichment for functional annotation and belonging to protein networks. The potentially upregulated pathways were identified using DEGs with log2 fold change greater than 0.25, whereas the potentially down regulated pathways were identified using DEGs with log2 fold change less than −0.25. Multiple databases were queried against, including BioCyc, Reactome, KEGG, Pathway Interaction Database, GenMAPP, Panther DB, Pathway Ontology, SMPDB, and MSigDB C2 Biocarta (v7.1). Bonferroni was used to correct for multiple corrections, with a 0.05 significance threshold. A total of 74 pathways were found to be enriched in upregulated DEGs in IgE ASCs, and seven pathways were found to be enriched in downregulated DEGs.
scRNA-seq gene ontology analysis
To elucidate functions based on genes unique to each cluster, a DEG analysis was conducted, followed by a gene ontology analysis. For each cluster, the Seurat FindMarkers function was used to find DEGs for each cluster versus all other clusters. The resulting DEGs were obtained using the same criteria as identifying DEGs for IgE ASCs described previously. The DEGs with a positive log2 fold change value were queried separately from DEGs with a negative log2 fold change value. The R package pathfindR was used to query DEGs in the Kegg database for the gene ontology analysis. The DEGs obtained for each cluster along with their log2 fold change in addition to their adjusted p values were used to query Kegg and obtain insight into each cluster’s functionality.
Monoclonal antibody synthesis
After the antibodies of interest were selected, the DNA sequences of paired heavy and light chains were optimized and sent to GenScript for synthesis. In brief, the complete DNA sequences were sub-cloned into a pcDNA3.4 vector. Transfection grade plasmids were maxi-prepared for HD 293F cell expression, whereas, in parallel, cells were cultured at 37°C with 8% CO2 on a shaker in Erlenmeyer flasks (Corning Inc.). Cells were seeded at appropriate density in flasks 1 day before transfection. Transfection reagent and DNA were mixed at an optimal ratio on the day of transfection and then added to the flask containing cells. The recombinant plasmids encoding target antibodies were transiently co-transfected into suspension HD 2,93F cell cultures. The cell culture broth was then centrifuged and filtered. The filtered supernatant was loaded onto an affinity purification column at an appropriate flowrate and after washing and elution, the eluted fractions were pooled and the buffer was exchanged to the final formulation buffer. Purified antibodies were analyzed by SDS-PAGE and HPLC to determine the molecular weight and purity. The final antibody concentrations were determined by the A280 method.
Fungal enzyme-linked immunosorbent assay
Soluble fungal lysates for Aspergillus f. and Alternaria alternans were purchased (Greer). After extensive optimization, fungal lysates were diluted to a final concentration of 40 μg/ml of fungal lysate in bicarbonate/carbonate antigen coating buffer (100 mM NaHCO3 in deionized water, pH adjusted to 9.6). A total of 100 μl of diluted antigen in coating buffer was plated onto a flat-bottom 96-well plate (ThermoFisher) and incubated at 4°C overnight and was then blocked with 200 μl of blocking buffer (SuperBlock Blocking Reagent, ThermoFisher) for 2 hours at 37°C. After the addition of standards (polyclonal rabbit anti-Aspergillus IgG, Bio-Rad) and polyclonal rabbit anti-Alt a1 IgG, MyBioSource) and the addition of diluent for controls, the primary monoclonal antibodies were diluted and added to the plate. Antibodies were then incubated for 1 hour at room temperature on a shaker. Secondary antibodies for the standards (monoclonal anti-rabbit IgG/alkaline phosphatase, Millipore Sigma, 1:15,000) and for the monoclonal antibodies (monoclonal goat anti-human IgG/alkaline phosphatase, Millipore Sigma, 1:15,000) were added to the appropriate wells for 1 hour at room temperature on a shaker, and then 100 μl of substrate solution (KPL) was added to each well. After sufficient color development, the plate was placed on a microplate reader (Synergy 2 MultiMode Microplate Reader, Biotek) and absorbance was read at 620 nm. Plates were washed three times with wash buffer (1× PBS containing 0.1% Tween 20) after each step.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the contributions of Dr Alessia Corrado, Dr Chester J. Joyner, Mindy Hernandez, Dr Vanessa Engineer, and Sang Le for their time and expertise. The authors are grateful for the efforts of the Flow Cytometry Core at the Emory Vaccine Center, which made this work possible.
FUNDING
This work was supported by National Institutes of Health grants T32-HL007563-33 (RPR), R01AI121252 (FEL), P01-AI125180 (IS, FEL), U01AI141993 (FEL, IS, GG), T32-HL116271 (RPR), Genentech Award ML42271 (FEL, IS, GG).
Footnotes
DECLARATION OF COMPETING INTEREST
FEL is the founder of MicroB-plex, Inc, serves on the scientific board of Be Bio Pharma, and is the recipient of research grants from the Bill and Melinda Gates Foundation and Genentech. FEL has also served as a consultant for AstraZeneca. JML serves on the scientific advisory board for GSK, Regeneron Pharma, and Honeywell International. JML has also received research grants from Sanofi. SKW consults for Stryker and NeurENT and is on the scientific board of OptiNose and SinopSys Surgical. RPR, MB, MCW, JL, JD, MD, CLS, SK, KSC, JH, AR, CMT, IS, and GG have no competing interests to declare.
APPENDIX A. SUPPLEMENTARY DATA
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mucimm.2023.02.008.
DATA AVAILABILITY
The scRNA-seq data that support the findings in this paper are publicly available in the National Institutes of Health SRA database under the BioProject number PRJNA898288.
REFERENCES
- 1.Pleis JR, Lucas JW & Ward BW Summary health statistics for U.S. adults: National Health Interview Survey, 2008. Vital Health Stat. 10(242), 1–157 (2009). [PubMed] [Google Scholar]
- 2.Fokkens WJ et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology 50, 1–12 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Hopkins C. Chronic rhinosinusitis with nasal polyps. N. Engl. J. Med 381, 55–63 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya N. Incremental health care utilization and expenditures for chronic rhinosinusitis in the United States. Ann. Otol. Rhinol. Laryngol 120, 423–427 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Bent JP 3rd & Kuhn FA Allergic fungal sinusitis/polyposis. Allergy Asthma Proc. 17, 259–268 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Dykewicz MS, Rodrigues JM & Slavin RG Allergic fungal rhinosinusitis. J. Allergy Clin. Immunol 142, 341–351 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Glass D & Amedee RG Allergic fungal rhinosinusitis: a review. Ochsner J. 11, 271–275 (2011). [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilos DL Chronic rhinosinusitis: epidemiology and medical management. J. Allergy Clin. Immunol 128, 693–707 (2011). quiz 708–699. [DOI] [PubMed] [Google Scholar]
- 9.Marple BF Allergic fungal rhinosinusitis: current theories and management strategies. Laryngoscope 111, 1006–1019 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Dullaers M. et al. The who, where, and when of IgE in allergic airway disease. J. Allergy Clin. Immunol 129, 635–645 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Asrat S. et al. Chronic allergen exposure drives accumulation of long-lived IgE plasma cells in the bone marrow, giving rise to serological memory. Sci. Immunol 5, eaav8402 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Bellou A, Kanny G, Fremont S & Moneret-Vautrin DA Transfer of atopy following bone marrow transplantation. Ann. Allergy Asthma Immunol 78, 513–516 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Croote D, Darmanis S, Nadeau KC & Quake SR High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science 362, 1306–1309 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Corrado A. et al. Extrafollicular IgD+ B cells generate IgE antibody secreting cells in the nasal mucosa. Mucosal Immunol. 14, 1144–1159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garimalla S. et al. Differential transcriptome and development of human peripheral plasma cell subsets. JCI Insight 4, e126732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halliley JL et al. Long-lived plasma cells are contained within the CD19(−) CD38(hi)CD138(+) subset in human bone marrow. Immunity 43, 132–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tipton CM et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat. Immunol 16, 755–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenks SA et al. Distinct effector B cells induced by unregulated toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity 49, 725–739.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodruff MC et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol 21, 1506–1516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutzeit C, Chen K & Cerutti A The enigmatic function of IgD: some answers at last. Eur. J. Immunol 48, 1101–1113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzmann JA et al. Serum reference intervals and diagnostic ranges for free kappa and free lambda immunoglobulin light chains: relative sensitivity for detection of monoclonal light chains. Clin. Chem 48, 1437–1444 (2002). [PubMed] [Google Scholar]
- 22.Hutchison CA et al. Quantitative assessment of serum and urinary polyclonal free light chains in patients with chronic kidney disease. Clin. J. Am. Soc. Nephrol 3, 1684–1690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta NT et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershberg U & Luning Prak ET The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos. Trans. R. Soc. Lond. B Biol. Sci 370, 20140239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoh RA et al. Origins and clonal convergence of gastrointestinal IgE + B cells in human peanut allergy. Sci. Immunol 5, eaay4209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen DC, Joyner CJ, Sanz I & Lee FE Factors affecting early antibody secreting cell maturation into long-lived plasma cells. Front. Immunol 10, 2138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson ST et al. Partial plasma cell differentiation as a mechanism of lost major histocompatibility complex class II expression in diffuse large B-cell lymphoma. Blood 119, 1459–1467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smulski CR & Eibel H BAFF and BAFF-receptor in B cell selection and survival. Front. Immunol 9, 2285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz I. et al. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Front. Immunol 10, 2458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould HJ & Sutton BJ IgE in allergy and asthma today. Nat. Rev. Immunol 8, 205–217 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Yokota A. et al. Two species of human Fc epsilon receptor II (Fc epsilon RII/ CD23): tissue-specific and IL-4-specific regulation of gene expression. Cell 55, 611–618 (1988). [DOI] [PubMed] [Google Scholar]
- 32.Fournier S, Rubio M, Delespesse G & Sarfati M Role for low-affinity receptor for IgE (CD23) in normal and leukemic B-cell proliferation. Blood 84, 1881–1886 (1994). [PubMed] [Google Scholar]
- 33.Kawano Y. et al. A novel mechanism for the autonomous termination of pre-B cell receptor expression via induction of lysosome-associated protein transmembrane 5. Mol. Cell. Biol 32, 4462–4471 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouchida R, Kurosaki T & Wang JY A role for lysosomal-associated protein transmembrane 5 in the negative regulation of surface B cell receptor levels and B cell activation. J. Immunol 185, 294–301 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Baba Y & Kurosaki T Role of calcium signaling in B cell activation and biology. Curr. Top. Microbiol. Immunol 393, 143–174 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Winkelmann R. et al. B cell homeostasis and plasma cell homing controlled by Kruppel-like factor 2. Proc. Natl Acad. Sci. U. S. A 108, 710–715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klasen C. et al. MIF promotes B cell chemotaxis through the receptors CXCR4 and CD74 and ZAP-70 signaling. J. Immunol 192, 5273–5284 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Hoof I. et al. Allergen-specific IgG+ memory B cells are temporally linked to IgE memory responses. J. Allergy Clin. Immunol 146, 180–191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez-Saiz R. et al. Human BCR analysis of single-sorted, putative IgE(+) memory B cells in food allergy. J. Allergy Clin. Immunol 144, 336–339.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haniuda K, Fukao S, Kodama T, Hasegawa H & Kitamura D Autonomous membrane IgE signaling prevents IgE-memory formation. Nat. Immunol 17, 1109–1117 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Wade-Vallance AK & Allen CDC Intrinsic and extrinsic regulation of IgE B cell responses. Curr. Opin. Immunol 72, 221–229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karagiannis SN et al. Endocytosis and recycling of the complex between CD23 and HLA-DR in human B cells. Immunology 103, 319–331 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi Q, Dabadghao S, Osterborg A, Bergenbrant S & Holm G Myeloma bone marrow plasma cells: evidence for their capacity as antigen-presenting cells. Blood 90, 1960–1967 (1997). [PubMed] [Google Scholar]
- 44.Nie Y. et al. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med 200, 1145–1156 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mesin L, Ersching J & Victora GD Germinal Center B cell dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rijvers L. et al. The macrophage migration inhibitory factor pathway in human B cells is tightly controlled and dysregulated in multiple sclerosis. Eur. J. Immunol 48, 1861–1871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Binsky I. et al. TAp63 regulates VLA-4 expression and chronic lymphocytic leukemia cell migration to the bone marrow in a CD74-dependent manner. J. Immunol 184, 4761–769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watarai H. et al. Posttranslational modification of the glycosylation inhibiting factor (GIF) gene product generates bioactive GIF. Proc. Natl Acad. Sci. U. S. A 97, 13251–13256 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu YC, Nakano T, Elly C & Ishizaka K Requirement of posttranslational modifications for the generation of biologic activity of glycosylation-inhibiting factor. Proc. Natl Acad. Sci. U. S. A 91, 11227–11231 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fokkens WJ et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology 58, 1–64 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Orlandi RR et al. International consensus statement on allergy and rhinology: rhinosinusitis 2021. Int. Forum Allergy Rhinol 11, 213–739 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Wise SK, Ghegan MD, Gorham E & Schlosser RJ Socioeconomic factors in the diagnosis of allergic fungal rhinosinusitis. Otolaryngol. Head Neck Surg 138, 38–42 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Ghegan MD, Wise SK, Gorham E & Schlosser RJ Socioeconomic factors in allergic fungal rhinosinusitis with bone erosion. Am. J. Rhinol 21, 560–563 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Miller JD et al. Markers of disease severity and socioeconomic factors in allergic fungal rhinosinusitis. Int. Forum Allergy Rhinol 4, 272–279 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Bent JP 3rd & Kuhn FA Diagnosis of allergic fungal sinusitis. Otolaryngol. Head Neck Surg 111, 580–588 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Alamyar E, Duroux P, Lefranc MP & Giudicelli V IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol 882, 569–604 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tellier J. et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat. Immunol 17, 323–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy MA, Evans JS & Storfer A Quantifying Bufo Boreas connectivity in Yellowstone National Park with landscape genetics. Ecology 91, 252–261 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Chen J, Bardes EE, Aronow BJ & Jegga AG ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data that support the findings in this paper are publicly available in the National Institutes of Health SRA database under the BioProject number PRJNA898288.