Abstract
The rapid and persistent emergence of drug-resistant bacteria poses a looming public health crisis. The possible task of developing new sets of antibiotics to replenish the existing ones is daunting to say the least. Searching for adjuvants that restore or even enhance the potency of existing antibiotics against drug-resistant strains of bacteria represent a practical and cost-effective approach. Herein we describe the discovery of potent adjuvants that extend the antimicrobial spectrum of existing antibiotics and restore their effectiveness towards drug-resistant strains including mcr-1-expressing strains. From a library of cationic compounds, MD-100, which has a diamidine core structure, was identified as a potent antibiotic adjuvant against Gram-negative bacteria. Further optimization efforts including the synthesis of ~20 compounds through medicinal chemistry work led to the discovery of a much more potent compound MD-124. MD-124 was shown to sensitize various Gram-negative bacterial species and strains, including multi-drug resistant (MDR) pathogens, towards existing antibiotics with diverse mechanisms of action. We further demonstrated the efficacy of MD-124 in an ex-vivo skin infection model and in an in vivo murine systemic infection model using both wild-type and drug-resistant E. coli strains. MD-124 functions through selective permeabilization of the outer membrane of Gram-negative bacteria. Importantly, bacteria exhibited low resistance frequency towards MD-124. In-depth computational investigations of MD-124 binding to the bacterial outer membrane using equilibrium and steered molecular dynamics simulations revealed key structural features for favorable interactions. The very potent nature of such adjuvants distinguishes them as very useful leads for future drug development in combating bacterial drug resistance.
Keywords: antibiotics adjuvants, effective in vivo, extending the antimicrobial spectrum, overcoming multi-drug resistance, LPS binding
Graphical Abstract:
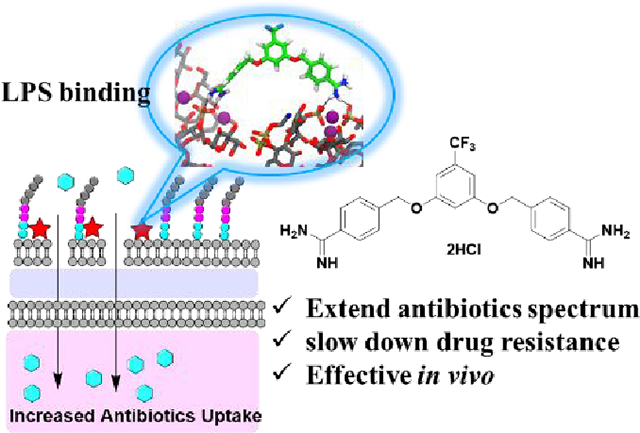
Introduction
The emergence of multi-drug resistance (MDR) in bacteria represents a major threat to public health.1–2 The so-called “super bugs” that are resistant to almost all current antibiotics are frequently isolated from clinics, leaving few therapeutic options available to handle the most difficult cases. To invent a whole new set of antibiotics to overcome such drug-resistant problems would be a huge undertaking to say the least. To compound this problem, there is a decreasing rate of new antibiotic discovery.3 Therefore, the field is in urgent need for new approaches to either attenuate the rate of emergence of drug resistance or directly tackle the issue of infection by drug-resistant bacteria. There are several major mechanisms through which bacteria develop resistance, including 1) reduced permeability for antibiotics and reduction of porin protein expression; 2) over-expression of efflux pumps and metabolizing enzymes for drug degradation; and 3) reduced affinity towards the drug target due to mutation of the target(s).4–5 While the latter one usually leads to resistance to certain types of antibiotics, the first two mechanisms often result in resistance to a wide range of antibiotics, and thus MDR.4 Indeed, the discovery of new antibiotics is still very important.6–17 In parallel, a somewhat different approach is the sensitization of bacteria towards existing antibiotics by adjuvants with well-defined efficacy and safety profiles.18–43 This is especially true in the case of MDR Gram-negative bacteria, which are often considered more difficult to overcome because of the existence of an outer membrane.44–46 Strategies that can permeabilize the outer membrane of Gram-negative bacteria and increase the uptake of antibiotics would add extra ammunition to the current arsenal of effective antibiotics without the need to develop new antibiotics as the sole solution.
Previous efforts in the search for compounds capable of sensitizing Gram-negative bacteria have focused on using polycationic peptides such as colistin and its analogs such as polymyxin B nonapeptide (PMBN), with very promising results.19, 22, 47–50 Recently, a linear antibacterial peptide was identified as a broad-spectrum antibiotic adjuvant.21 Similarly, cationic polymers have been shown to sensitize Gram-negative bacteria toward antibiotics.51–54 Recently, an FDA-approved antifungal drug, pentamidine (NebuPent® as the isethionate salt), was reported as a non-polypeptide sensitizer of Gram-negative bacteria including drug-resistant strains, with the ability to sensitize by ~30-fold at the dosage of ~50 μg/ml.55 Pentamidine disrupts bacterial outer membrane by binding to lipopolysaccharide (LPS). Subsequently, efforts have been made to probe the structure and activity relationships (SAR) by modifying the linker between the two amidine groups.56 Along this line, several other small-molecule bacterial sensitizers with diverse mechanisms such as LPS binding, outer membrane protein biogenesis impairment, and efflux inhibition have been discovered.41, 57–68 Such compounds serve as excellent examples of initial success, with much room for exploring improved potency, mechanistic understanding, establishing in vivo activity and examining broad chemical space for success.
We took an integrated approach combining mechanistic insights, medicinal chemistry efforts, bioassays, computational investigation, and animal model efficacy work. Herein, we report the discovery of novel bacterial sensitizers originating from a library of thousands of compounds and the study of sensitizer-outer membrane interactions using a combination of experimental efforts and sophisticated computational studies. We identified a new bacterial sensitizer scaffold, elucidated the mechanism of sensitization, and demonstrated its efficacy against various Gram-negative species, including several MDR strains. The efficacy of one such compound, MD-124, was demonstrated in an ex-vivo skin burn infection model, in which MD-124 enabled the use of novobiocin to treat Gram-negative bacterial skin infection. MD-124 also increased the efficacy of clindamycin in treating carbapenem-resistant E. coli in a skin burn infection model. Further, MD-124 was shown to sensitize mcr-1-expressing Gram-negative bacteria towards polymyxin B in a bacterial skin infection model. Finally, we validated the efficacy of MD-124 in a murine model of systemic infection using both wild-type and drug-resistant E. coli strains. Dramatically improved survival was observed in the treatment group with a combination of MD-124 and novobiocin, compared with novobiocin or MD-124 alone group.
Results
Library screen and chemical synthesis
Considering the grave threat from drug resistant Gram-negative bacteria, our initial efforts focused on the development of sensitizers of Gram-negative bacteria. The effort started with a library of ~ 2,500 mono-, di- and tri-cation (amine, amidine, guanidine, etc.) compounds collected over the years from the Boykin lab and references cited therein.69–71 We examined their structures and chose 150 cationic compounds with diverse chemical properties, including variations in linker length between the cationic groups, linker hydrophobicity/hydrophilicity, and rigidity. These compounds were tested for their ability to sensitize wild-type E. coli (ATCC 25922) towards rifampicin, which is generally considered to be ineffective against Gram-negative bacteria (Scheme 1). We found four compounds capable of 32-fold sensitization at a concentration equal to or below 10 μg/ml (Table S1). The sensitization fold is calculated with the following formula: MIC of antibiotic only /MIC of antibiotic with bacterial sensitizer. After assessment of their sensitization ability and cytotoxicity, the core structure MD-100 (Scheme 1 and Table S1) was chosen for medicinal chemistry modifications. MD-100 was found to sensitize E. coli towards rifampicin for 32-fold at 10 μg/mL (22 μM) and 8-fold at 4.5 μg/mL (10 μM) (Table S1 and scheme 2). To further optimize the activity of MD-100, we synthesized ~ 20 analogs (Scheme 2 and 3). Briefly, dinitrile compounds were synthesized first through method A (Scheme S1). Diamidine compounds were then synthesized from dinitrile compounds, either through method B or method C. Substituted amidine or cyclized amidine compounds were synthesized from dinitrile compounds through method D.
Scheme 1.
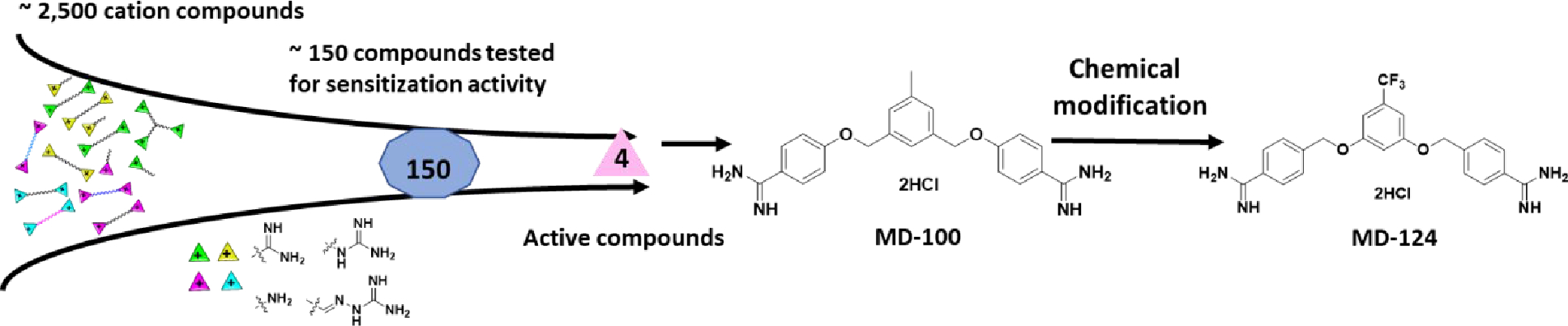
Discovery of MD-100 and MD-124.
Scheme 2.
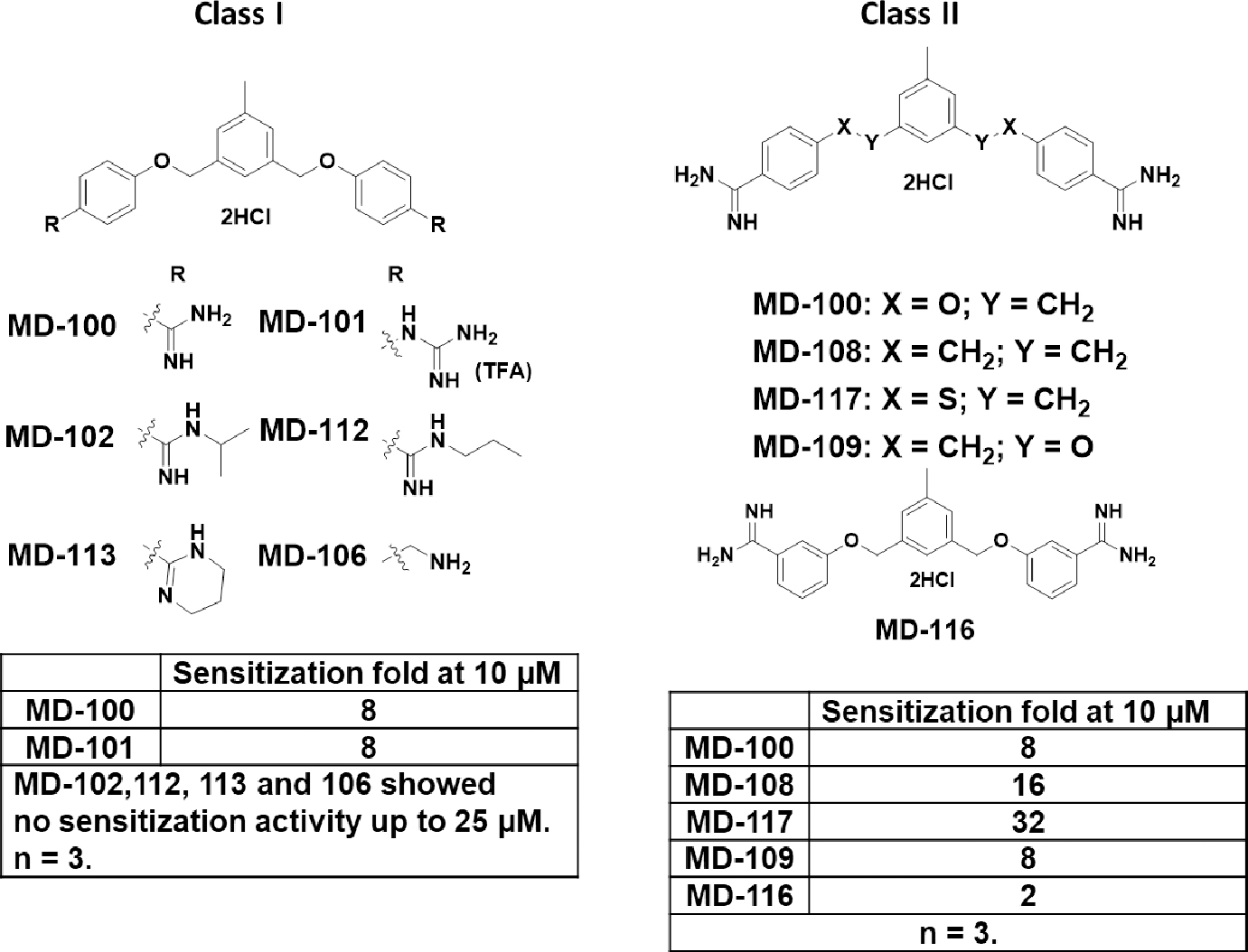
Class I and II analogs of MD-100.
Scheme 3.
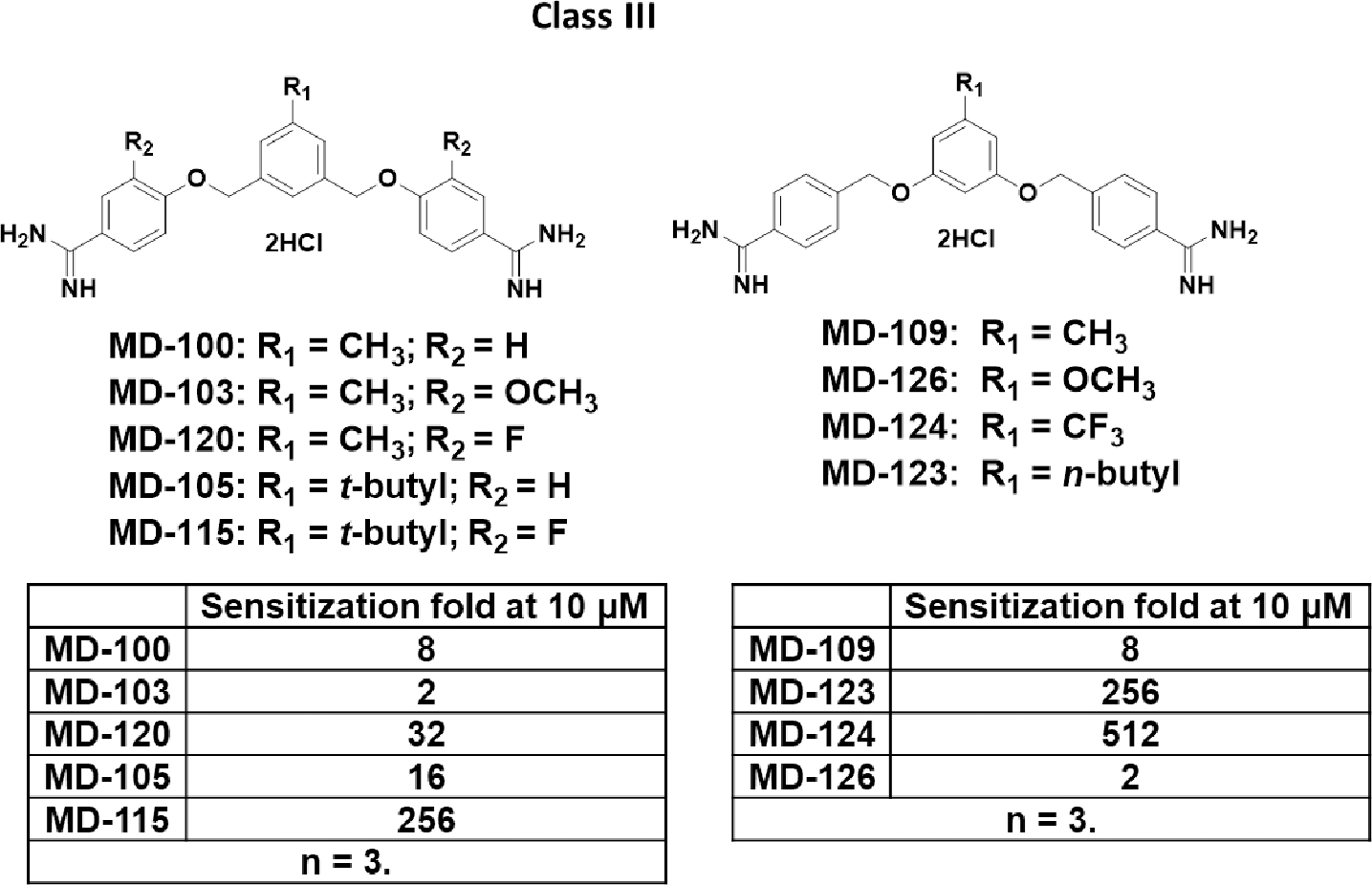
Class III analogs of MD-100.
We tested their ability to sensitize E. coli towards rifampicin and utilized the sensitization fold at 10 μM as the potency index. It is well known that the cationic property of these bacterial sensitizers plays a key role in their activity.47, 50 As a result, we firstly focused on modifying the amidine group (Class I in Scheme 2). A guanidine analog MD-101 (trifluoroacetic acid salt form) was synthesized and it showed similar activity with MD-100. Substitutions on the amidine group almost completely abolished the sensitization activity. MD-102, 112 and 113, which have the amidine alkylated or being part of a ring failed to sensitize E. coli toward rifampicin at up to 25 μM. Changing the amidine group to an amine group (MD-106) led to the abolishment of the activity.
The hydrophobicity/hydrophilicity of linker between the two diamidine groups has been reported to affect sensitization ability.55 We then modified the linker between the middle and flanking phenyl rings and synthesized three analogs (class II in Scheme 2). A less polar linker between the middle and flanking phenyl rings contributed to an increase in sensitization magnitude. When X in class II was changed from oxygen (MD-100) to methylene (MD-108) or to sulfur (MD-117), potency increased. For example, 10 μM MD-100 sensitize E. coli towards rifampicin for 8-fold; while for MD-108 and MD-117, the sensitization fold was 16 and 32, respectively. Interestingly, switching the oxygen and methylene group between the middle and flanking phenyl rings did not affect the activity (MD-100 vs. MD-109). Shortening the distance between the two amidine groups by changing the amidine from para to meta positions (MD-116) significantly decreased the activity.
We then focused on modifying the substituents on the middle and flanking phenyl rings and synthesized class III analogs (Scheme 3). Introduction of an electron-withdrawing group at the R2 position increased the activity while an electron-donating group decreased the activity. For example, changing the hydrogen atom in the R2 position in MD-100 to a fluoro group (MD-120) increased the sensitization fold from 8 to 32-fold, while introduction of a methoxy group at the same position (MD-103) decreased the activity. The comparison between MD-105 and MD-115 supported the same trend. Hydrophobic substituents at the R1 position significantly increased the activity. For example, changing the methyl group in MD-109 to n-butyl group (MD-123) increased the sensitization ability from 8 to 256-fold. The comparison between MD-100 and MD-105 revealed a similar trend, in which the methyl group was changed to a t-butyl group. Introduction of an electron-withdrawing group at the R1 position significantly increased the potency while an electron-donating group decreased the potency. For instance, changing the methyl group in MD-109 to the trifluoromethyl group (MD-124) increased the sensitization ability from 8 to 512-fold. The introduction of a methoxy group (MD-126) in the R1 position almost completely abolished the activity.
Identification of MD-124 as a potent sensitizer of Gram-negative bacteria
Among all the compounds prepared in the subsequent optimization efforts, MD-124 showed superior bacterial sensitization activities and was chosen for further biological evaluation (Scheme 1). MD-124 at 5 μg/ml (~ 10 μM) was able to sensitize E. coli towards rifampicin by 512-fold, lowering the minimum inhibitory concentration (MIC) of rifampicin from 10 to 0.02 μg/ml (Fig. 1A). MD-124 itself at the same concentration showed no effect on bacterial growth (Fig. 1B). A checkerboard assay showed that MD-124 sensitized E. coli towards rifampicin in a concentration-dependent manner, reaching over 16,000-fold when 12 μg/ml of MD-124 was used, with the MIC of rifampicin being 0.6 ng/ml (Fig. 1C, blue represents MIC). The fractional inhibitory concentration (FIC) index was calculated to be 0.09 based on the checkerboard assay (The MIC of MD-124 itself on E. coli is 50 μg/ml), indicating a strong synergistic effect between MD-124 and rifampicin.72 We observed that the sensitization ability of MD-124 changed drastically in the range of 3 to 12 μg/ml. To investigate this further, a checkerboard assay of MD-124 in the concentration range of 0 to 10 μg/ml was performed (Fig. 1D). It is worth mentioning that 5 to 10 μg/ml MD-124 showed no direct bacteriostatic or bactericidal effects when used in this concentration range. Therefore, MD-124 is considered as an adjuvant at this concentration range. MD-124 at 5 μg/ml sensitized E. coli towards a broad range of existing antibiotics with diverse targets and mechanisms of action such as clarithromycin (256-fold), erythromycin (128-fold), novobiocin (64-fold), trovafloxacin (32-fold), polymyxin B (32-fold) and chloramphenicol (8-fold) (Fig. 1G and Table S2A). A greater degree of sensitization was achieved when MD-124 was combined with antibiotics that tend to possess characteristics unfavorable for membrane permeation. This indicates that the sensitization ability of MD-124 varies depending on the antibiotics used in combination. When combined with MD-124, the MIC of various antibiotics that are viewed as relatively narrow-spectrum ones was lowered to around 1 μg/ml or below (Fig. 1G and Table S2). Together, those results demonstrated that MD-124 could potentiate antibiotics with diverse targets and mechanisms of action at sub-MIC levels. Pentamidine and PMBN have been shown as potent antimicrobial adjuvants capable of overcoming drug-resistance.48, 55 Compared with them, MD-124 showed much improved potency (Fig. 1F). For example, 30 μg/ml pentamidine and 20 μg/ml PMBN were shown to sensitize E. coli towards rifampicin by 8 and 16 folds respectively, while 5 μg/ml MD-124 was able to sensitize E. coli by 512-fold. The above tests were performed using reference strain E. coli 25922 from American Type Culture Collection (ATCC). To test if the observed effects are translatable to other E. coli strains, we also tested MD-124 on another reference strain (ATCC 10536) and found similar sensitization activity (Table S2B). Besides E. coli, MD-124 also showed sensitization effect on various other wild-type Gram-negative bacterial species such as Acinetobacter baumannii (ATCC 17978), Klebsiella pneumoniae (ATCC 43816) and Salmonella enterica Typhimurium (ATCC 14028), the resistant strains of which are listed as WHO Priority I pathogens in urgent need of new antibiotics (Fig. 1E).46
Fig. 1. MD-124 sensitizes Gram-negative bacteria towards various antibiotics.
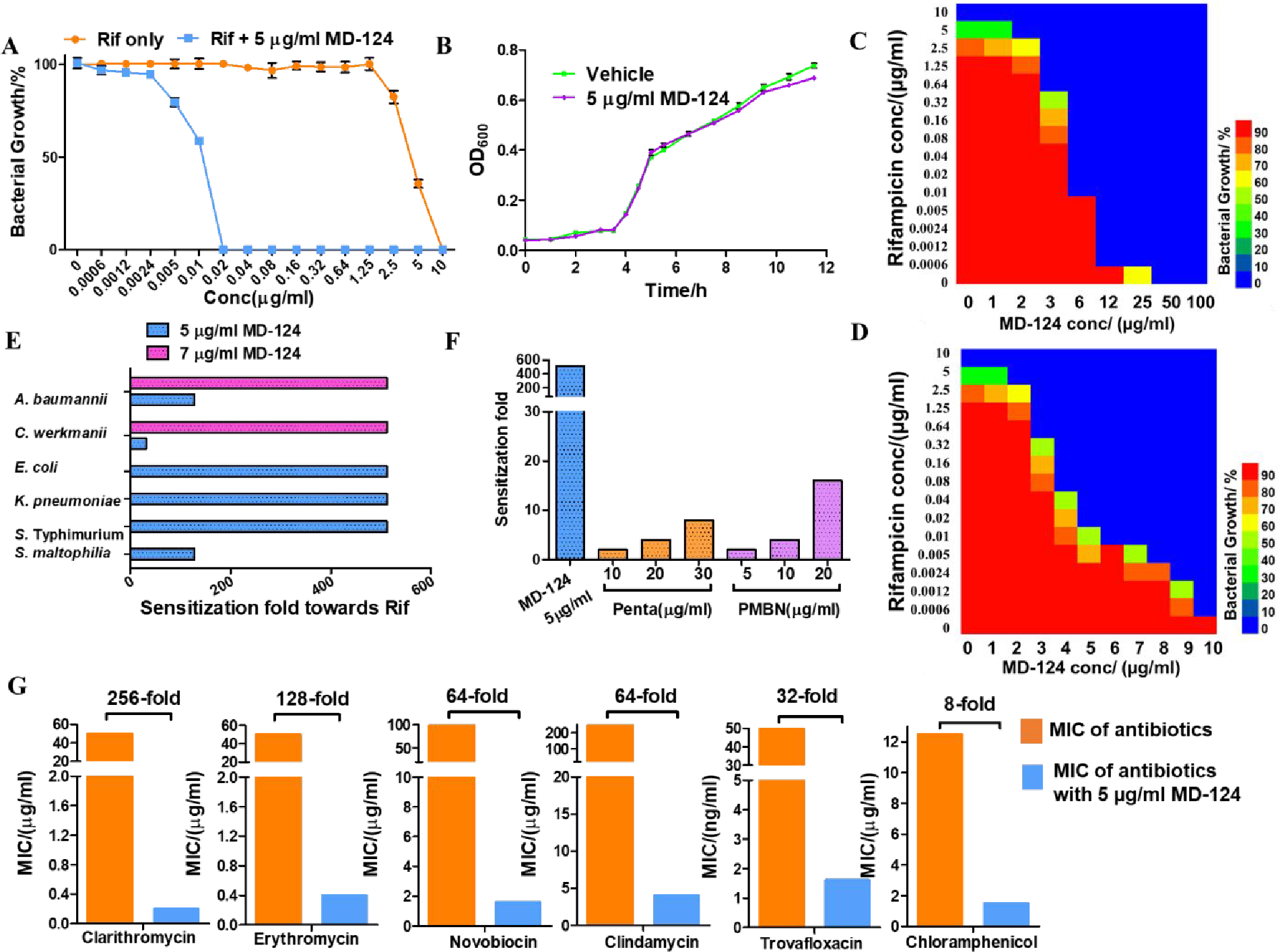
(A) 5 μg/ml MD-124 sensitized E. coli towards rifampicin (Rif). E. coli was cultured with rifampicin at various concentrations in the presence or absence of 5 μg/ml MD-124 for 24 h at 37 °C. Then bacterial growth density was determined by measuring OD600. Values are means ± SD. n = 3. (B) 5 μg/ml MD-124 itself showed no inhibition effect on bacterial growth. E. coli (5 × 105 CFUs/ml) was cultured in the absence or presence of 5 μg/ml MD-124 and OD600 values at different time points were recorded. Values are means ± SD. n = 3. (C) Checkerboard assay of MD-124 (0 to 100 μg/ml) and rifampicin on E. coli. From red bar to blue bar: decreasing bacterial growth density. (D) Checkerboard assay of MD-124 (0 to 10 μg/ml) and rifampicin on E. coli. (E) MD-124 sensitizes various Gram-negative bacterial species towards rifampicin (Rif). Sensitization fold = MIC of rifampicin only /MIC of rifampicin with MD-124. (F) Comparison among MD-124, pentamidine and PMBN for their sensitization effects on E. coli towards rifampicin. (G) 5 μg/ml MD-124 sensitizes E. coli towards a broad range of antibiotics. All results shown represent biologically independent triplicates.
Since cytotoxicity is an essential factor to be considered in drug discovery, we tested the cytotoxicity of MD-124 on mammalian cells in the cell culture medium supplemented with 10% fetal bovine serum (FBS). The IC50 of MD-124 on mammalian cells such as HEK293 and NIH3T3 was found to be ~100 μM (Fig. S1). Drug molecules can bind to serum proteins, leading to a decreased concentration of free molecules and potentially a decreased efficacy.73–74 To test if serum protein binding would affect the sensitization activity of MD-124, we then tested the sensitization ability of MD-124 on E. coli towards rifampicin in the absence and presence of 10% FBS, the same serum concentration used for mammalian cell culture. We observed that 5 μg/ml MD-124 was able to sensitize E. coli towards rifampicin by 512-fold under both conditions, indicating the neglectable influence of serum protein under such experimental conditions.
MD-124 is effective against drug-resistant Gram-negative bacterial strains
We then focused on examining the sensitization effect of MD-124 on various clinically relevant drug-resistant strains, especially Gram-negative bacteria belonging to the ESKAPE category of pathogens.75 Checkerboard assays revealed MD-124 was able to sensitize A. baumannii (wild-type, ATCC 17978) towards rifampicin, decreasing the MIC from 5 μg/ml to 0.04 and 0.01 μg/ml, in the presence of 5 and 7 μg/ml of the sensitizer, respectively (Fig. 2A). The FIC index between MD-124 and rifampicin was calculated to be 0.14 (Table S3). A similar sensitization effect on A. baumannii was also observed with novobiocin through a checkerboard assay and the FIC index between MD-124 and novobiocin was determined to be 0.20 (Fig. 2B). MD-124 at 5 μg/ml also sensitized A. baumannii towards a variety of other antibiotics such as clarithromycin, fusidic acid and clindamycin, achieving 64-, 128- and 8-fold sensitizations, respectively (Table S4). Another ESKAPE pathogen species, K. pneumoniae (wild-type, ATCC 43816), also showed susceptibility to MD-124. As shown in Fig. 2C and 2D, MD-124 sensitized K. pneumoniae towards rifampicin and clarithromycin by 512-fold or more; and at 10 μg/ml it brought the MIC of rifampicin and clarithromycin from more than 20 μg/ml to 0.2 μg/ml or less. The FIC index between MD-124 and rifampicin or clarithromycin was determined to be 0.15 and 0.16, respectively. Many other antibiotics such as clindamycin, chloramphenicol, fusidic acid and novobiocin also showed synergistic effect when combined with 5 μg/ml MD-124 with sensitization of 4–64 fold in inhibiting K. pneumoniae growth (Table S5).
Fig. 2. MD-124 sensitization of wild-type A. baumannii, K. pneumoniae and drug-resistant Gram-negative strains towards existing antibiotics.
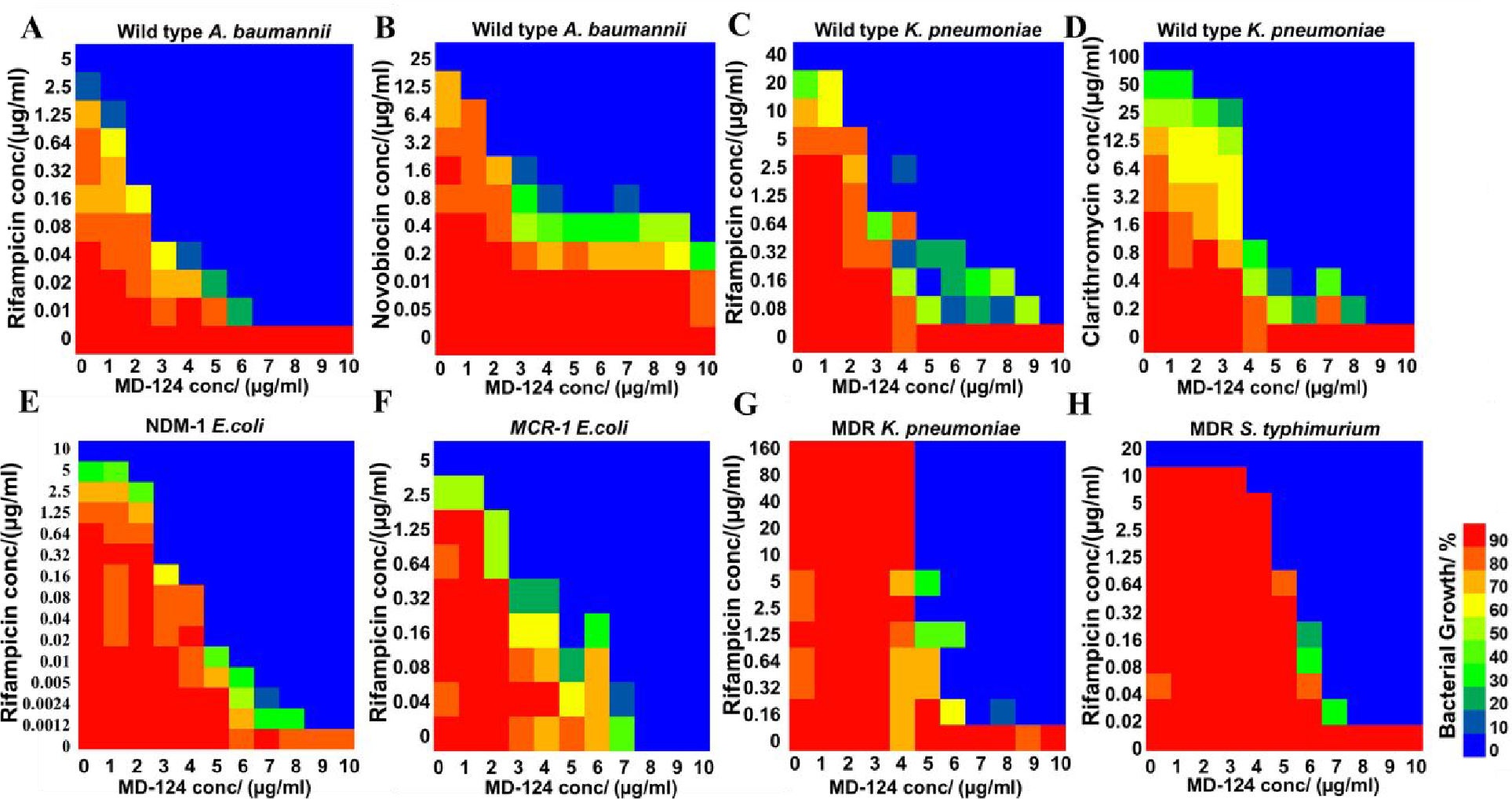
(A and B) Checkerboard assays showed sensitization of A. baumannii by MD-124 towards rifampicin and novobiocin. (C and D) Checkerboard assays showed sensitization of K. pneumoniae by MD-124 towards rifampicin and clarithromycin. (E) Sensitization of an NDM-1-expressing strain of E. coli by MD-124 towards rifampicin. (F) Sensitization of an mcr-1-expressing strain of E. coli by MD-124 towards rifampicin. (G) Sensitization of MDR K. pneumoniae by MD-124 towards rifampicin. (H) Sensitization of MDR S. Typhimurium by MD-124 towards rifampicin. All experiments were done at least in biologically independent duplicate.
The emergence and the spread of carbapenem-resistant Enterobacteriaceae are causing rising concerns. This class is listed as Priority 1 pathogens by the WHO for developing new antibiotics.76–77 To test the effect of MD-124 on carbapenem-resistant bacteria, an NDM-1 (New Delhi Metallo-beta-lactamase 1)-expressing E. coli strain was constructed to induce carbapenem-resistance.78 Compared with wild-type E. coli, the NDM-1-expressing strain showed a 30- to 100-fold increase in MIC towards β-lactam antibiotics such as ampicillin, ceftazidime and a carbapenem antibiotic, meropenem (Table S6). We found that MD-124 was able to sensitize the NDM-1-expressing strain of E. coli towards various antibiotics. Specifically, MD-124 sensitized the NDM-1-expressing strain towards rifampicin in a concentration-dependent manner, decreasing the MIC from 10 to 0.02 and 0.005 μg/ml, when 5 and 7 μg/ml MD-124 were used, respectively (Fig. 2E). MD-124 also showed synergistic effects with rifampicin with an FIC index of 0.09 (Table S3) and sensitized carbapenem-resistant E. coli towards various antibiotics with distinct mechanisms of action (Table S7).
Polymyxin- and colistin-resistant strains (harboring the mcr-1 gene) are threatening the last line of defense of antimicrobial treatment.79–80 Bacteria expressing mcr-1 can modify the phosphate group on lipid A, resulting in decreased negative charge density on the Gram-negative bacteria outer membrane, and subsequent resistance to colistin.79, 81 We observed that MD-124 sensitized mcr-1-expressing E. coli towards rifampicin and several other antibiotics (Fig. 2F, Table S8). MD-124 showed synergistic effects with rifampicin on mcr-1-expressing E. coli with a FIC index of 0.37 (Table S3). Interestingly, MD-124 also sensitized this strain towards polymyxin B, bringing its MIC from 30 to 0.9 μg/ml. Such results suggest that MD-124 is able to convert the polymyxin B-resistant E. coli strain to a polymyxin B-sensitive strain (Fig. S2).
To further evaluate the role of MD-124 in antimicrobial therapy, we then assessed if MD-124 can sensitize MDR Gram-negative strains. Thus, MD-124 was tested on one MDR K. pneumoniae strain (ATCC BAA-2472) and one MDR S. Typhimurium strain (ATCC 14028), both of which are resistant to almost all antibiotics. Specifically, they are resistant to a wide range of antibiotics including colistin, aminoglycoside, fluoroquinolone, tetracycline, β-lactam families of antibiotics and other class of antibiotics (Table S9). These MDR strains represent the most severe challenges in antimicrobial treatment. The MIC of rifampicin on this MDR K. pneumoniae strain is >320 μg/ml. MD-124 decreased the MIC of rifampicin to 10 μg/ml and below 0.2 μg/ml at a dose of 5 and 7 μg/ml, respectively. (Fig. 2G, Table S3). A similar phenomenon was observed on the MDR S. Typhimurium strain. While the MIC of rifampicin itself is 40 μg/ml, this was brought down to below 0.05 μg/ml when combined with 7 μg/ml of MD-124 (Fig. 2H, Table S3). As summarized in Table S3, MD-124 potentiated rifampicin against drug-resistant strains, including MDR strains of Gram-negative bacteria.
Bacteria exhibit low resistance frequency towards MD-124
Because bacteria can quickly develop resistance to new antimicrobial agents, we then evaluated the frequency of resistance development of E. coli towards MD-124 using a literature reported method.55 Briefly, E. coli (ATCC 25922) was cultured with antibiotic and 10 μg/ml MD-124. For each antibiotic, the concentration ranged from 2 to 7-fold of its respective MIC. We chose clindamycin, novobiocin, and trovafloxacin for this study to include antibiotics with different mechanisms of action. Colonies that were resistant to the MD-124-antibiotic combination were studied further to determine whether the observed resistance phenotype was due to resistance against MD-124 or against the antibiotic used in the combination. Therefore, the corresponding resistant colonies were subjected to another antibiotic combination: MD-124 and rifampicin. Our results indicated that MD-124 was still effective in sensitizing those strains towards rifampicin, with the same potency as on the wild-type E. coli. Hence such results strongly suggest that these resistant strains are not resistant to MD-124. No strain was found to be resistant to 10 μg/ml MD-124 out of 2.7 × 1010 CFUs, giving a resistant frequency of less than 3.7 ×10−11. It is worth noting that the resistance frequency here is not defined based on direct inhibition/bactericidal effect as it is usually the case for traditional antibiotics; rather, it is based on the sensitization effect of MD-124 at a concentration far below its MIC (the MIC of MD-124 itself on E. coli was determined to be 50 μg/ml) since MD-124 serves as an adjuvant here.
MD-124 sensitizes Gram-negative bacteria through binding to a key component of LPS: lipid A
We next investigated the mechanism of action for MD-124. The fact that MD-124 can sensitize Gram-negative bacteria towards a broad range of antibiotics and that the outer membrane is the major barrier of antibiotic uptake led us to examine the disruption of bacterial outer membrane as a possible mechanism (Fig. 3A). Further, polymyxin analogs and pentamidine are also known to interact with the outer membrane. MD-124 and our initial lead compound, MD-100, were used for this study. MD-124 showed compromised ability to sensitize an outer membrane “leaky” mutant of E. coli, strain NR698 (Fig. 3B), which is already susceptible to antibiotics such as clarithromycin and erythromycin.82 MD-124 at 5 μg/ml was shown to sensitize wild-type E. coli towards clarithromycin by 256-fold (Fig. 1G), and NR698 by only 4-fold. We also observed the same phenomenon when MD-100 was used on both E. coli wild-type and NR698 mutant strains (Fig. S3A). Such results support the involvement of the outer membrane integrity in the sensitization ability of MD-124 and MD-100. We then directly probed the disruption of the bacterial outer membrane by MD-124 using a lysozyme assay on E.coli (ATCC 25922).83 Briefly, lysozyme can break down peptidoglycan and lead to bacterial lysis. While lysozyme cannot naturally penetrate intact Gram-negative bacterial outer membrane, disruption of the outer membrane would result in increased lysozyme efficacy, leading to bacterial lysis. We found that lysozyme caused quick bacterial lysis in the presence of 25 μg/ml of MD-124 or 50 μg/ml of MD-100 and no lysis in the absence of the sensitizer (Fig. 3C, blue bar). MD-124 and MD-100 themselves showed no lytic effects (Fig. 3C, pink bar). These results indicate that MD-124 was able to disrupt outer membrane integrity. In comparison, polymyxin B at 25 μg/ml and pentamidine at 50 μg/ml also caused bacterial lysis when incubated together with lysozyme. Collectively, these results support the notion that the sensitizers function by permeabilizing membrane and allowing for increased antibiotic entry.
Fig. 3. Mechanistic studies revealed that MD-100 and MD-124 sensitize E. coli by disrupting the outer membrane and increasing antibiotics uptake through binding to LPS.
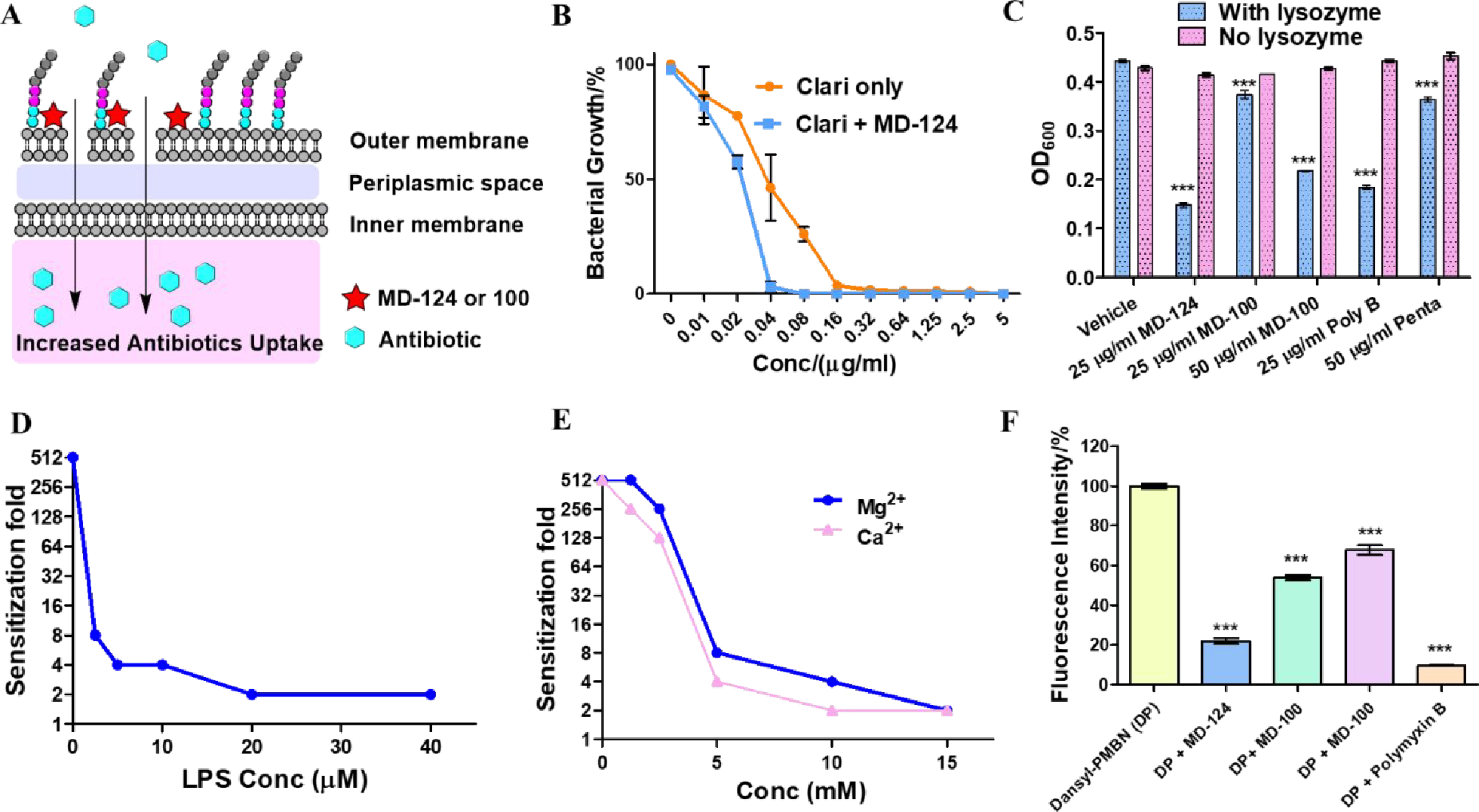
(A) Proposed sensitization mechanism of MD-124 and MD-100. (B) MD-124 showed decreased ability to sensitize E. coli strain NR698 (a mutant with a “leaky” outer membrane) towards clarithromycin. Values are means ± SD. n = 3. (C) Bacterial sensitizers facilitated E. coli lysis by lysozyme. Poly B and Penta are short for polymyxin B and pentamidine, respectively. As control, E. coli was incubated with various bacterial sensitizers at the same concentration in the absence of lysozyme (pink bar). Values are means ± SD. n = 3. P values were determined using unpaired two-tailed Student’s t-tests. ***: P < 0.001 compared with vehicle group. (D) LPS decreased the sensitization ability of MD-124 on E. coli in a concentration-dependent manner. (E) Mg2+ and Ca2+ decreased the sensitization ability of MD-124 in a concentration-dependent manner. (F) Dansyl-PMBN (DP) displacement assay. 10 μM Dansyl-PMBN was added to E. coli (OD600 = 0.3) in HEPES buffer (pH = 7.4), and the fluorescent intensity was recorded as F1 (Ex = 340 nm; Em = 520 nm). Then 200 μM compounds were added, and the fluorescent intensity was recorded as Fx. Fluorescent intensity/% = (F1-Fx)/F1. The fluorescent intensity of 10 μM Dansyl-PMBN with E. coli (F1) was marked as 100 %. Values are means ± SD. n = 3. P values were determined using unpaired two-tailed Student’s t-tests. ***: P < 0.001 compared with vehicle group.
We then conducted various experiments to determine the molecular target(s). By adding LPS to the growth media, we were able to abolish the sensitization effect of MD-124 and MD-100 (Fig. 3D and Fig. S3B). This finding brought lipid A, an essential component of LPS, which is responsible for maintaining the outer membrane integrity of Gram-negative bacteria, to the forefront of our investigation.84–85 Mg2+ and Ca2+ were able to dampen the bacterial sensitization activity of MD-124 on E. coli in a concentration-dependent manner (Fig. 3E). Divalent cations such as Mg2+ and Ca2+ are known to bridge between adjacent lipid A and enhance membrane integrity.86–87 Therefore, high concentrations of divalent cations may displace the diamidine and thus exert antagonistic effects against MD-124. This phenomenon was also observed on MD-100 (Fig. S3B). It is worth noting that the growth medium for E. coli is Mueller Hinton Broth II, a cation-adjusted broth that already contains approximately 0.5 mM Mg2+ and 0.5 mM Ca2+. Therefore, the Mg2+ and Ca2+ concentrations in Fig. 3E reflect the exogenous amounts and not the total amounts in the culture medium. The free magnesium and calcium concentration in human blood is between 0.55 to 0.75 mM and 1.1 to1.3 mM, respectively.88–89 Based on the high concentrations of cation needed to antagonize the effect of MD-124, as observed in the current study, one can predict that physiological concentrations of Mg2+ or Ca2+ will have a very limited effect on the sensitization ability of MD-124. A fluorescent conjugate of a polymyxin-B nonapeptide derivative, Dansyl-PMBN (Scheme S2) has been reported to increase fluorescent intensity after binding to the lipid A part of LPS.90 Therefore, we chose Danzyl-PMBN (PB) as an indicator agent in a displacement assay. Addition of 200 μM MD-124 to a mixture of E. coli and 10 μM Dansyl-PMBN led to a decrease of the fluorescent intensity of Dansyl-PMBN, indicating binding of MD-124 to lipid A and displacement of Dansyl-PMBN (Fig. 3F). Pentamidine and MD-100 were also able to displace Dansyl-PMBN, although not as much as MD-124. Another piece of evidence that supports that lipid A is the molecular target of MD-124 was obtained through the sensitization activity comparison between wild-type and mcr-1-expressing E. coli. Bacteria harboring mcr-1 can modify the phosphate group on lipid A and lead to decreased negative charge density,79 which might decrease the sensitization activity of MD-124. Indeed, MD-124 showed slightly decreased sensitization activity on mcr-1-expressing E. coli than on wild-type strain. For example, it required 3 μg/ml and 5 μg/ml of MD-124 to achieve a 32-fold sensitization towards rifampicin on wild-type E. coli and mcr-1-expressing E. coli, respectively (Fig. 1D and Fig. 2F). It is worth mentioning that this resistance towards MD-124 by mcr-1 is moderate (less than 2-fold). Collectively, these results support LPS as the molecular target of MD-124.
Computational investigations of sensitizer-membrane interactions
To further understand the sensitizer-membrane interactions, we performed molecular dynamics (MD) simulations of a model E. coli OM in the presence of MD-124 using previously our published methods.87, 91–92 The simulations revealed that MD-124 quickly adsorbs to the LPS molecules in the outer leaflet of the E. coli OM when it is placed 20 Å above the top of the LPS (Fig. 4A). Similar behavior was observed for simulations of the antimicrobial peptide LL-37 with a Salmonella enterica OM.93 Adsorption is followed by the formation of stabilizing interactions, such as salt bridges between the positively charged amidine groups of MD-124 and the negatively charged phosphate groups of LPS. Normally, these phosphate groups are bridged by divalent cations (Ca2+ and Mg2+), thus forming a tight network.87, 92 Disruption of hydrogen bonds between the phosphates and the neighboring sugars due to interaction with adjuvants has been observed. We also observed MD-124 to interact with ion-bridged phosphates; while we did not see the ions displaced during the 1.3-μs simulation, the interactions between ions and phosphates appear weakened. For comparison, we also performed MD simulations of a model E. coli OM in the presence of a very weak sensitizer, MD-126. MD-124 and 126 were found to adopt similar geometries in water (Fig. 4C), as measured by the distance between the two amidine groups within each molecule (d1, Fig. 4B); this was expected given their similar structures. However, when they directly interacted with the LPS layer of the OM, the distance (d1) of MD-126 became much smaller than in water (Fig. 4D). In contrast, the average distance (d1) between the positively charged ends of MD-124 remained very large, leading us to the working hypothesis that a larger separation between the ends contributes to the effectiveness of sensitizers. To better quantify the connection between d1 and membrane interaction for each sensitizer, we performed steered molecular dynamics (SMD) simulations in which we slowly pull (0.25 Å/ns) each sensitizer through the model OM. Our analysis shows large differences in the d1 values between MD-124 and MD-126 in the hydrophilic part (Lipid A sugars and core sugars) of the LPS layer (Fig. 4, E and F), with larger values for MD-124, just as was observed in the equilibrium simulations. Overall, our simulations show that MD-124 and MD-126 behave similarly in water, in the hydrophobic part of the LPS layer, and in the lower leaflet of the OM. Differences between the two sensitizers were only observed in the hydrophilic part of the LPS layer, suggesting that this is the region in which they are active.
Fig. 4. Molecular dynamics simulations of the interactions between bacterial sensitizers and E. coli outer membrane (OM).
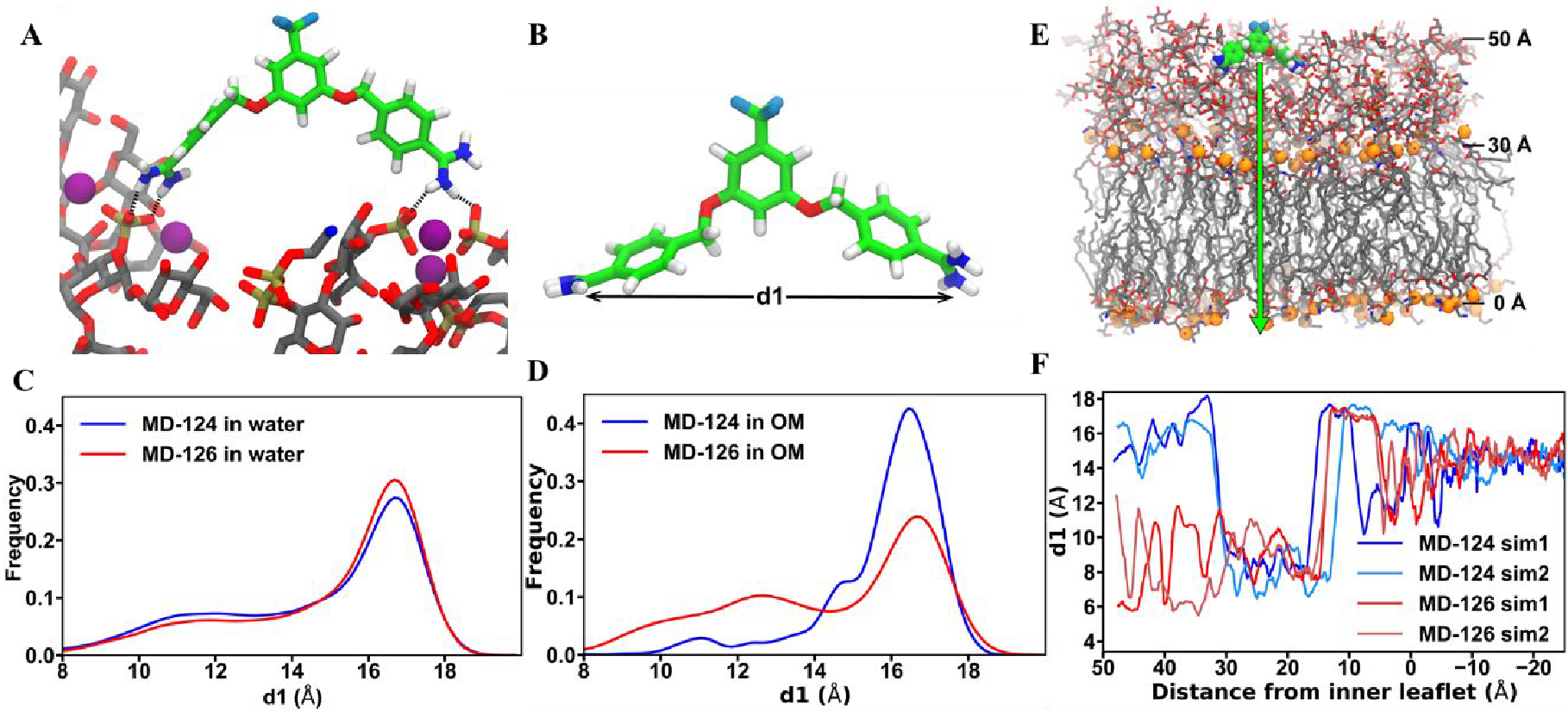
(A) MD-124 interacts with phosphate groups of LPS. A representative state for one of the two copies of MD-124 present in the E. coli OM simulation. Hydrogen bonds are indicated by dotted lines. Purple spheres are Ca2+ ions. (B) d1 is defined as the distance between the two positively charged diamidine groups. (C) MD-124 and MD-126 adopt similar geometries in water in terms of the distance between the two amidine groups (d1). (D) MD-126 diamidine groups come closer together than those of MD-124 when they directly interact with the LPS layer of the OM. (E and F) Steered molecular dynamics (SMD) simulations (pulling speed of 0.25 Å/ns) show large differences in the d1 values between MD-124 and MD-126 in the hydrophilic part (Lipid A sugars and core sugars) of the LPS layer. Sim1 and Sim2 are short for simulation 1 and simulation 2.
Similar to LPS in Gram-negative bacteria, various phospholipids serve structural roles in eukaryotic cells. Since the amidine group is known to bind to the phosphate group, potential cytotoxicity of MD-124 could arise in eukaryotic cells.94 To address this potential limitation, we conducted additional experiments. While adding extra LPS was found to effectively dampen or even abolish the activity of MD-124 (Fig. 3D), exogenous phospholipid from eukaryotic cells such as phosphatidylcholine was much less potent in neutralizing the sensitization ability of MD-124 (Fig. S4). For example, 10 μM LPS was able to almost completely abolish the activity of MD-124 (5 μg/ml), while 160 μM phosphatidylcholine did not alter the sensitization effect of MD-124. These results suggest that MD-124 is selective towards LPS over phospholipids from eukaryotic cells, such as phosphatidylcholine. Such findings are expected because the structures of lipid A and phospholipids from eukaryotic cells are quite different, especially in that the phosphate groups in lipid A are doubly charged while in phospholipids, the phosphate group is singly charged.95
MD-124-antibiotic combinations show potent antimicrobial effects an ex-vivo skin infection model
We examined the effect of MD-124 on E. coli in an ex-vivo skin-burn infection model.96 To achieve topical application, compounds were formulated as hypromellose gel.97–98 Briefly, human skin (about 1 cm × 1 cm) was burnt with a soldering iron (95 °C) for 10 s; then 105 CFUs of bacteria were inoculated to the burnt skin and was cultured at 37 °C for 1 h. Then the skin was loaded with hypromellose gel containing different antibiotics and cultured at 37 °C. After 24 h of incubation, the skin sample was homogenized and the supernatants were serially diluted and plated on agar plates, and bacterial counts were carried out. We firstly tested the combination of MD-124 with novobiocin because MD-124 showed good sensitization activity with novobiocin in vitro and the good water solubility of novobiocin (sodium salt form) for easy formulation. While neither 4‰ (w/w) novobiocin nor 1.5‰ MD-124 alone achieved significant inhibition of wild-type (WT) E. coli (ATCC 25922) growth, the combination of novobiocin and MD-124 effectively decreased the bacterial load compared with the vehicle group (Fig. 5A). For example, while novobiocin or MD-124 alone each led to approximately 108 CFUs, their combination resulted in an approximately 3-log decrease (105 CFUs); 1‰ polymyxin B hypromellose gel was used as the positive control. To further investigate the scope and effectiveness of MD-124, we then tested the combination of MD-124 and clindamycin on drug-resistant Gram-negative bacteria. MD-124 and clindamycin also effectively treated skin infection caused by NDM-1-expressing E. coli, leading to a reduction of the bacterial load by more than 2,000-fold over clindamycin alone (Fig. 5B). The dosage used was comparable to or lower than the commonly used ointment preparations, containing 5 to 20‰ antibiotics. Those results showed MD-124 potentiated the activity of novobiocin and clindamycin against E. coli; both antibiotics are typically considered ineffective against E. coli. Besides extending the spectrum of antibiotics which are ineffective in treating Gram-negative bacterial infection, restoring the effectiveness of antibiotics towards drug-resistant strains is another important property of bacterial sensitizers. As described earlier, MD-124 restored the effectiveness of polymyxin B towards mcr-1-expressing E. coli (Fig. S2). Thus, we investigated whether MD-124-polymyxin B combination would be effective in treating infection caused by mcr-1-expressing bacteria in this skin-burn model. Treatment with polymyxin B alone (3‰, w/w) showed minor effect, compared with the vehicle group (Fig. 5C). In contrast, the MD-124-polymyxin B combination effectively reduced the bacterial load (Fig. 5C).
Fig. 5. Validation of MD-124 efficacy in an ex-vivo skin infection model (A, B, C) and in-vivo systemic infection model in mice (D, E, F).
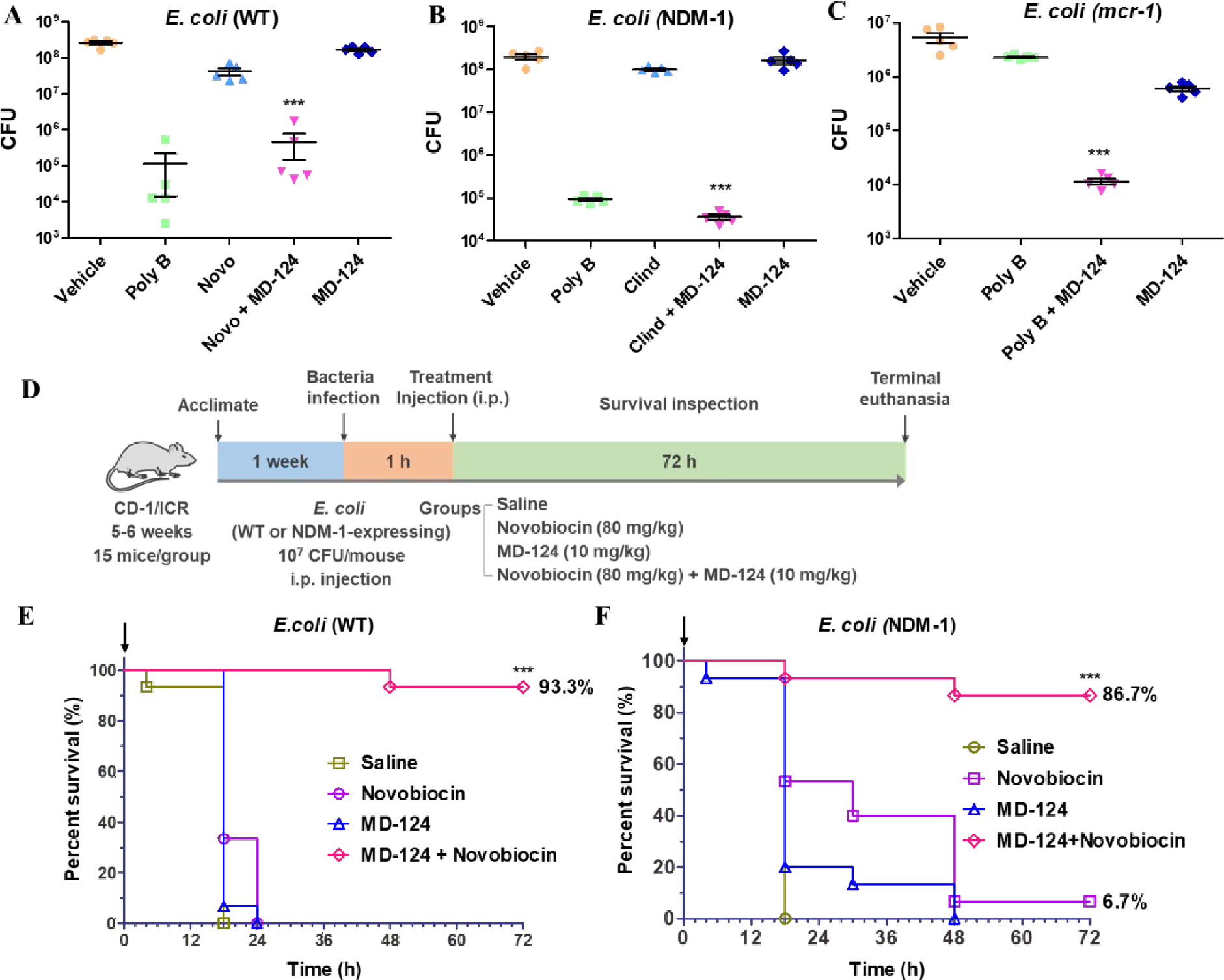
(A) Combination of MD-124 and novobiocin inhibited wild-type (WT) E. coli growth. Concentration (w/w) for polymyxin B (poly B), novobiocin (Novo) and MD-124 were 1‰, 4‰ and 1.5‰. The same concentration was used for novobiocin and MD-124 in the combination treatment groups (Novo + MD-124). (B) Combination of MD-124 and clindamycin inhibited the growth of NDM-1-expressing E. coli. Concentration (w/w) for polymyxin B (poly B), clindamycin (Clind) and MD-124 were 1‰, 3‰ and 1.5‰. The same concentration was used for clindamycin and MD-124 in the combination groups (Clind + MD-124). (C) Combination of MD-124 and polymyxin B inhibited the growth of mcr-1-expressing E. coli. Concentration (w/w) for polymyxin B (poly B) and MD-124 were 3‰ and 1.5‰. The same concentration was used for polymyxin B and MD-124 in the combination groups (Poly B + MD-124). For Fig. A, B, C, values are means ± SEM. n = 5, P values were determined using unpaired two-tailed Student’s t-tests. ***P < 0.001 vs antibiotics or MD-124 alone group. (D) Schematic illustration of the experimental procedures of a systemic infection model in mice. (E and F) MD-124 and novobiocin combinations significantly increased the survival rates of mice after infection by WT E. coli (E) and NDM-1-expressing E. coli (F). Arrows in Fig. E and F indicate the treatment time. The concentration for MD-124 and novobiocin is 10 mg/kg and 80 mg/kg respectively. The same concentration was used for novobiocin and MD-124 in the combination treatment groups. For Fig. E and F, n = 15 biologically independent animals per group. ***P < 0.001 vs antibiotics or MD-124 alone group. Statistical analysis using Log-rank (Mantel-Cox) test.
MD-124-antibiotic combinations show potent antimicrobial effects in a systemic infection model in mice
We then tested the in vivo efficacy of MD-124 in a systemic infection model in mice following literature procedures.19, 21, 55, 59 Briefly, each mouse was infected with 107 CFUs of E. coli (WT or NDM-1-expressing) intraperitoneally (i.p.). One hour post infection, the mice were subjected to different treatments through i.p. injection. Survival of the mice in each group was monitored for 72 h (Fig. 5D). As shown in Fig. 5E, all the mice treated with saline died from infection within 18 h post-infection. Novobiocin was chosen to pair with MD-124 for the same reason as in the ex vivo model. Single dose of combination of MD-124 (10 mg/kg) and novobiocin (80 mg/kg) treatment increased survival rate from 0% (novobiocin or MD-124 only) to 93% in the case of WT E. coli (ATCC 25922) infection. Encouraged by those results, we then tested if MD-124 was effective against drug-resistant strains. As described earlier, MD-124 sensitized NDM-1-expressing E. coli towards a variety of antibiotics (Table S7). We tested MD-124 in the mice infected with NDM-1-expressing E. coli and observed a similar effect (Fig. 5F). MD-124 (10 mg/kg) and novobiocin (80 mg/kg) combination group achieved a survival rate of 87%, while the MD-124 only and novobiocin only group has a survival rate of 0% and 6.7%, respectively. Mice that survived after 72 h in the MD-124 and novobiocin combination group showed normal body temperature, recovered body weight, and normal vibrant behavior, indicating a cure from the otherwise lethal infection. Taken together, these results showed MD-124 extended the spectrum of novobiocin and potentiated novobiocin for treating drug-resistant Gram-negative bacterial infection in vivo and greatly increased the efficacy of the tested antibiotic.
Discussion
In the combat against existing and emerging drug-resistant Gram-negative bacterial strains, there is a need for innovative approaches to develop therapeutics. There are ongoing efforts in many directions, including targeting new mechanism(s) of actions and regulating bacterial quorum sensing.6, 8, 42, 99 One “economical” approach is to salvage existing antibiotics against drug-resistant strains by re-sensitizing the bacteria towards these antibiotics. This way, the entire arsenal of antibiotics will still be available for clinical use. With this goal in mind, we discovered a new chemical moiety, MD-124, which can significantly sensitize drug-resistant Gram-negative bacteria, including carbapenem-resistant, colistin-resistant, and other MDR strains, towards existing antibiotics. MD-124 can sensitize Gram-negative bacteria, including MDR strains, to the point that it allows the use of some narrow-spectrum antibiotics to be effective treatment options. This sensitization effect of MD-124 works on antibiotics with diverse targets and mechanisms of action, bringing the MIC of various antibiotics on Gram-negative bacteria below 1 μg/ml. Compared with previously discovered sensitizers such as PMBN and pentamidine, MD-124 showed much improved potency (Fig. 1F). For each bacterial species tested herein, including E. coli, K. pneumoniae, and S. Typhimurium, two reference strains were tested for cross-validation (Table S2, S3, and S5, Fig. 1E, 2).
Besides its potent sensitization effect, MD-124 also exhibited a very low propensity to induce bacterial resistance. A bacterial sensitizer should ideally 1) extend the spectrum of antibiotics; and 2) restore the effectiveness of existing antibiotics towards drug-resistant strains. The second point might arguably be more clinically relevant. We believe the MD-124, as described in this study, was able to fulfill both objectives. Indeed, MD-124 enabled narrow-spectrum antibiotics to treat Gram-negative bacterial infections (Fig. 5, A, B, C, and D) and restored the effectiveness of polymyxin B towards mcr-1-expressing strain (Fig. 5C) at the same time.
Finally, MD-124 sensitizes Gram-negative bacteria by binding to LPS, disrupting the outer membrane integrity and increasing the antibiotic uptake. MD-124 has been shown to be effective against all the Gram-negative strains of bacteria studied herein (Fig. 1E and 2), presumably because they share very similar LPS structures, especially the lipid A part. Bacteria can express the mcr-1 gene, which confers colistin resistance by modifying LPS.80, 100 We found mcr-1-expressing E. coli only showed moderately increased effective dose of MD-124 (3 to 5 mg/ml) compared with the wild-type (Fig. 1D and 2F). Very importantly, MD-124 can sensitize mcr-1-expressing E. coli towards various antibiotics, including polymyxin B (Table S8, Fig. S2), which also targets the outer membrane.
We recognize that drug development is a complex, costly (Averaging over $1B),101 and lengthy undertaking.74, 102 The promising results described only constitute the first step in potentially translating the initial success into clinical therapeutics. Much more work is needed in areas of toxicology, pharmacokinetics, metabolism, and efficacy beyond rodents. It is our hope that the compounds described can serve either as leads for future development or inspirations for other efforts in the same area so as to facilitate the development of potent adjuvants to overcome bacterial drug resistant problems.
In summary, we have described the discovery of a group of small molecules that can significantly sensitize Gram-negative bacteria towards existing antibiotics and extend the spectrum of antibiotics. One such compound, MD-124, was shown to achieve antimicrobial effects through combinations with various antibiotics. Remarkably, the combination was shown to be effective against some pathogens of the highest priority for antibiotic development, including those with mcr-1 and NDM-1 mediated drug-resistance and MDR strains of Gram-negative bacteria. Further, the bacterial resistance rate for MD-124 was low, which further signifies its therapeutic values. MD-124 enables the use of antibiotics like novobiocin and clindamycin to treat not only wild-type but also carbapenem-resistant Gram-negative bacteria in a skin infection model. MD-124 also enables the use of polymyxin B in an ex-vivo skin infection model for the treatment of infection by bacteria with mcr-1 induced resistance to the polymyxin family of antibiotics. Finally, MD-124, when combined with novobiocin, significantly increased the survival rates of mice infected with either WT or drug-resistant (NDM-1-expressing) E. coli. Extensive computational efforts also led to significant insight into how MD-124 interacts with the bacterial outer membrane.
Experimental section
General Information.
All solvents were of reagent grade and were purchased from Fisher Scientific and Aldrich. Reagents and antibiotics were purchased from Aldrich, Oakwood, or VWR. The stationary phase of chromatographic purification is silica (230 × 400 mesh, Sorbtech). Silica gel TLC plate was purchased from Sorbtech. 1H-NMR (400 MHz) and 13C-NMR (100 MHz) spectra were recorded on a Bruker Avance 400 MHz NMR spectrometer. All compounds are >95% pure by HPLC. Mass spectral analyses were performed on an ABI API 3200 (ESI-Triple Quadruple). HPLC was performed on a Shimadzu Prominence UFLC (column: Waters C18 3.5 μM, 4.6×100 mm) or Agilent 1100 HPLC system (Column: Kromasil C18 5μm, 4.6 × 150 mm). OD600 and fluorescence intensity were recorded on a Perkin Elmer Enspire UV/ Vis/Fluorescence plate reader.
Safety statement
No unexpected or unusually high safety hazards were encountered.
Bacterial strains
Escherichia coli (ATCC 25922 or ATCC 10536), Acinetobacter baumannii (ATCC 17978), Klebsiella pneumoniae (ATCC 43816), Stenotrophomonas maltophilia (ATCC 31559), Salmonella enterica Typhimurium (ATCC 14028), Citrobacter werkmanii (ATCC 51114), MDR Klebsiella pneumoniae (ATCC BAA-2472) and MDR Salmonella enterica serovar Typhimurium (ATCC 700408) were also purchased from ATCC. E. coli strain NR698 was generously provided by Dr. Thomas J. Silhavy (Princeton University, Princeton, NJ).
Bacteria culture and MICs determination
All bacterial species were cultured in Mueller Hinton II (MH- II) Broth II (cation-adjusted). MICs of antibiotics only (no bacterial sensitizer) were determined by performing two-fold serial dilutions of antibiotics. MICs of antibiotics in the presence of bacterial sensitizers were determined by performing two-fold serial dilutions of antibiotics with or without a constant concentration of bacterial sensitizers. The MICs tests were performed on 96-well plates with a final volume of 200 μl Mueller Hinton Broth II (cation-adjusted) in each well. Each well was inoculated with 5 × 105 CFU/ml and plates were incubated for 24 h at 37 °C with continuous shaking of 200 rpm. The bacterial density was determined by OD600. The percentage (%) of bacterial growth is calculated with the following formula:(OD600 of antibiotic treatment group/OD600 of the non-treatment group) × 100. The MIC is defined as the concentration that inhibits more than 90% of bacterial growth.
Sensitization fold determination and fractional inhibitory concentration (FIC) index calculations.
The sensitization fold is calculated with the following formula: MIC of antibiotic only /MIC of antibiotic with bacterial sensitizer. For example, given a 10 μg/ml MIC of rifampicin for E.coli, and a 0.019 μg/ml MIC of rifampicin in the presence of 5 μg/ml MD-124, the calculated sensitization fold is 10 /0.019 = 512-fold.
The FIC index between compound a (e.g., antibiotic) and b (e.g., bacterial sensitizer) was calculated according to the formula below:72
FIC ≤ 0.5: Synergy; FIC between 0.5 and 1: Additive; FIC between 1 and 2: no interaction; FIC >4: antagonism.
Bacterial resistance frequency study
E. coli (ATCC 25922) was grown overnight in MH-II broth and resuspended to approximately 5 × 109 cells/ml in MH-II. A 200 μl volume of this suspension (approximately 1 × 109 cells) was transferred onto solid MH agar plates (100 mm Petri dishes) containing antibiotic with concentrations ranging from 2- to 7 -times of the MIC and 10 μg/ml MD-124. Three antibiotics including clindamycin, novobiocin or trovafloxacin was used separately with MD-124 in this study. Approximately 2.7 × 1010 CFUs were tested in the MD-124 and antibiotics combination. Plates were incubated at 37 °C for 48 h, after which colonies formed on the plate were counted. Those colonies were resistant to MD-124 and antibiotic combination. To find out whether those resistant strains are resistant to MD-124 or antibiotic, those colonies were then subjected to another antibiotic combination: MD-124 and rifampicin. It turned out that MD-124 was still able to sensitize those resistant strains towards rifampicin with the same potency on wild-type E. coli, which demonstrated those combinational therapy resistant strains were not resistant to MD-124. No strain was found to be resistant to 10 μg/ml MD-124 out of 2.7 × 1010 CFUs.
The resistance frequency towards MD-124 was calculated to be:
Lysozyme sensitivity assays
To evaluate the disruption of Gram-negative bacteria outer membranes by bacterial sensitizers, we performed lysozyme assays, as previously described.83 Briefly, E. coli (ATCC 25922) cells were grown in MH-II broth overnight, centrifuged at 3,000 X g for 10 min, washed with HEPES buffer (pH 7.2), and gently resuspended in HEPES buffer supplemented with 5 mM NaCl to obtain an E. coli suspension of OD600 0.8–1.0. Next, 100 μl of the bacterial suspension was added to 96-well plates containing 100 μl of lysozyme solution (100 μg/ml), and either 50 or 100 μg/ml of bacterial sensitizer prepared in HEPES buffer (final concentration of lysozyme:50 μg/ml). Plates were incubated at room temperature for 10 min and the OD600 was recorded. E. coli was also incubated with bacterial sensitizers in the absence of lysozyme to test if bacterial sensitizers would directly induce bacterial lysis. E. coli that was incubated with lysozyme alone or E. coli without any treatment were also included as negative controls.
Evaluation of the influence of exogenous LPS and Mg2+, Ca2+ on bacterial sensitizers
The MICs of bacterial sensitizer and antibiotic combination in the presence of LPS (ranging from 0 to 40 μM ) or Mg2+ or Ca2+ (ranging from 0 to 20 mM) were determined as described above (Bacteria culture and MICs determination section). E. coli (ATCC 25922) was used for this study.
Construction of MCR-1 and NDM-1 expressing E. coli strain
E. coli (ATCC 25922) was transformed with pGDP2 MCR-1 (Addgene, pGDP2 MCR-1, plasmid #118404) to generate colistin-resistant strains. E. coli (ATCC 25922) was transformed with pGDP1 NDM-1 (Addgene, plasmid # 112883, pGDP1 NDM-1) to generate strains resistant to a broad range of β-lactam antibiotics.103 The successful construction of the MCR-1 and NDM-1 expressing E.coli strains were validated by their increased resistance towards polymyxin B and β-lactam antibiotics (ampicillin, ceftazidime and meropenem) respectively.
Dansyl-PMBN displacement assay
Dansyl-PMBN was synthesized following a previously described protocol.90 Dansyl-PMBN was dissolved in H2O to make 500 μM stock. Bacterial sensitizers were dissolved in ethanol to make 10 mM stock solutions. E. coli (ATCC 25922) was cultured in MH-II overnight at 37 °C, centrifuged at 3,000 g for 10 min, washed with HEPES buffer (pH 7.2), and finally resuspended in HEPES buffer, to obtain a E. coli suspension of OD600 0.3. Using 96-well plates, 10 μM Dansyl-PMBN was added to a volume of 100 μl of the bacterial suspension and the fluorescence intensity was recorded (Ex = 340 nm; Em = 520 nm). Bacterial sensitizer was then added to achieve a final concentration of 200 μM. Then the mixture was incubated at room temperature for 3 min and the fluorescence intensity was recorded (Ex = 340 nm; Em = 520 nm).
Cytotoxicity test of bacterial sensitizers on mammalian cells
Cell viability was assessed by using Cell Counting Kit-8 (CCK-8, Dojindo, Japan). HEK293 or NIH3T3 cells were cultured in DMEM (Dulbecco’s Modified Eagle’s Medium) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Sigma-Aldrich; P4333) at 37 °C with 5% CO2. Cells were seeded in a 96-well plate one day before the experiment. Cells were then incubated with various concentrations of bacterial sensitizers at 37 °C in incubators with an atmosphere of 5% CO2 for 24 h, then 10 μL of CCK-8 solution was added to each well, and the plate was incubated for an additional 2 h at 37 °C with an atmosphere of 5% CO2. The optical density at 450 nm was recorded by plate reader, and the results were calculated as a percentage of viability compared with the untreated control.
Computational simulations
Initial force-field parameters for MD-124 and MD-126 were obtained from the CGenFF webserver at cgenff.umaryland.edu.104–105 Partial charges on the MD-124 and MD-126 central rings were further modified using the Force Field Toolkit (ffTK).106 Additionally, one dihedral angle parameter (with penalty of ~33 in both molecules) was improved using ffTK by fitting to a quantum mechanical dihedral scan at 6–31G*/MP2 level of theory. The CHARMM36 force field was used for all other components.107
We built the outer membrane (OM) using CHARMM-GUI.108–109 The OM comprised 36 lipopolysaccharides (LPS) and 105 phospholipids in the upper and lower leaflets, respectively. The phospholipid layer (inner leaflet) is composed of 90 PPPE (charge = 0), 11 PVPG (charge = −1), and 4 PVCL2 (charge = −2). The compositions and structures of the OM were based upon previous studies.87 Lipid A and core sugars of the LPS were neutralized by adding Ca2+ ions. After that, we added Na+ and Cl- ions in a bulk solvation box at a 0.15 M NaCl concentration. Equilibration of each system was performed using NAMD,110 followed by production simulations using Amber111 at a constant temperature of 310 K and pressure of 1 atm. Equilibration involved multiple steps: (i) We melted the lipid tails for 1 ns. (ii) We equilibrated for another 1 ns where everything was allowed to relax. (iii) We simulated the membrane for another 200 ns to obtain a converged area-per-lipid. Using the resulting membrane system, we added two copies of a given adjuvant molecule on top of the membranes followed by the removal of four Na+ ions to keep the overall system neutral. Furthermore, we also removed any clashing water molecules that were within 2 Å of the added molecules. Next, the combined system was equilibrated for 11 ns followed by production simulations with Amber. Hydrogen mass repartitioning (HMR) was employed to accelerate the simulations by allowing for a 4-fs time step.112–113 The Berendsen barostat was used for pressure control and Langevin thermostat for temperature control. Van der Waals interactions were cut off at 12 Å with a switching function starting at 10 Å. To prevent the molecules from leaving the upper bulk solvation box above the LPS, which is possible due to the use of periodic boundary conditions, we added a wall potential 40 Å from the lipid A phosphates in equilibrium simulations.
For steered MD (SMD) simulations, we added one molecule of an adjuvant on top of the equilibrated membrane followed by the removal of two Na+ ions and clashing water molecules. In addition, we prepared another system with a different initial orientation of the same molecule. We performed two SMD simulations for each molecule after equilibrating each system for 1 ns. In the SMD simulations, we pull a molecule slowly (0.25 Å/ns) towards the lipid head groups of the lower leaflet. Therefore, we capture the behavior of the molecules as it traverses the OM. These simulations were run for nearly 300 ns each with a 2-fs time step and without HMR.
Skin-burn infection model
Evaluation of MD-124-antibiotic combinations was conducted in a skin-burn infection model following published literature.96–97 E.coli wild-type (ATCC 25922) or MCR-1 and NDM-1 expressing E. coli strains were used in this study. Full Thickness human skin was purchase from ZenBio. Skin was cut to 1cm × 1 cm before use. MD-124 and antibiotics were formulated as water based hypromellose gel.98 2 g Hypromellose 4000 was suspended with 15 ml propylene glycol to prepare the gel base. 0.8 ml MD-124 and antibiotic in autoclaved H2O was mixed with 0.2 ml gel base to prepare MD-124-antibiotic combination gel for topical applications. 0.8 ml MD-124 or 0.8 ml antibiotic in H2O were also mixed with 0.2 ml gel base to prepare gel containing MD-124 or antibiotics only. Bacteria were grown in MH-II broth overnight, centrifuged at 3,000 g for 10 min, washed with PBS buffer (pH 7.4), and gently resuspended in PBS buffer to reach 1 × 107 CFU/ml. Human skin was burnt with a soldering iron (95 °C) for 10 s; then 105 CFUs bacteria (10 μl of 1× 107 CFU/ml bacteria stock) were inoculated to the burnt skin and was cultured at 37 °C for 1 h. Then the skin was loaded with hypromellose gel (~50 μl gel for each piece of skin) containing different compounds and cultured at 37 °C. After 24 h of incubation, each skin sample was transferred to a 1.5 ml tube with 1 ml PBS, homogenized (4,000 rpm with beads, 2 min) and the supernatants were serially diluted and plated on agar plates, and bacterial counts were carried out. The homogenization process did not affect bacterial viability.
Mouse infection models
This study was carried out in accordance with the guide for the Care and Use of Laboratory Animals and under the Georgia State’s Animal Welfare Assurance in accordance with the Public Health Service (PHS) Policy for Humane Care and Use of Laboratory Animals with assurance number D16–00527(A3914–01). The animal study protocol (A21045) was approved by the Georgia State’s Institutional Animal Care and Use Committee (IACUC). CD-1/ICR mice (female, 5–6 weeks) were purchased from Charles Rivers Laboratories and raised under standard sterile housing with free access to food and water. After acclimation, the average body weight was 26.2 g for wild-type (WT) E. coli (ATCC 25922) and 25.8 g for NDM-1 E. coli infection experiment. For the mouse studies, overnight cultures of E. coli grown in LB were diluted to an OD600 of 0.05 with fresh LB and allowed to grow at 37°C with shacking until reaching an OD600 of about 1.0 (about 2.5 hours). The bacteria were enumerated by microscopic counts (Zeiss, Germany) using a Hauser counting chamber and diluted with 5% mucin in normal saline (autoclaved) to 108 CFU/ml. The mouse was infected with E. coli (WT or NDM-1 expressing) by intraperitoneal (i.p.) injection of 0.1 ml of the 108 CFU/ml bacteria suspension (107 CFU/mouse). One hour post infection, treatments were administered intraperitoneally as described. After treatment, the animal comfort level was determined by observing their appearance and behavior. Bodyweight and temperature (monitored by infrared body temperature meter) were monitored 5 hours post-infection and on day 2 and day 3. Mice that showed signs of terminal systemic infection such as lethargy, head tilt, paralysis, labored breathing, or body temperature lower than 30℃ were euthanized and considered to reach clinical endpoint. Experimental endpoint was defined as 3 days post-infection for mice not reaching clinical endpoint. The survival data was collected and analyzed by Graphpad Prism 9 to generate the survival curve and statistical analysis using Log-rank (Mantel-Cox) test.
Supplementary Material
Acknowledgments
We acknowledge Dr. Thomas J. Silhavy (Princeton University, Princeton, NJ) for providing E. coli strain NR698.
Funding:
Financial support from the GSU Brains and Behaviors Fellowship Program to B.Y. and the Molecular Basis of Disease program to M.R.C. is gratefully acknowledged. We also acknowledge the financial support of the Georgia Research Alliance through a GRA Eminent Scholar endowment to B.W. J.C.G. acknowledges support from the National Institutes of Health (R01-GM123169). Computational resources were provided through the Extreme Science and Engineering Discovery Environment (XSEDE; TG-MCB130173), which is supported by National Science Foundation (NSF; ACI-1548562). This work also used the Hive cluster, which is supported by the NSF under grant number 1828187 and is managed by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology.
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability:
All data are available in the main text or the supplementary materials. Any additional data can be requested from the corresponding author.
References
- 1.Davies SC;Fowler T;Watson J;Livermore DM;Walker D Annual Report of the Chief Medical Officer: infection and the rise of antimicrobial resistance. Lancet 2013, 381 (9878), 1606–1609. [DOI] [PubMed] [Google Scholar]
- 2.Laxminarayan R;Sridhar D;Blaser M;Wang M;Woolhouse M Achieving global targets for antimicrobial resistance. Science 2016, 353 (6302), 874. [DOI] [PubMed] [Google Scholar]
- 3.Ventola CL The antibiotic resistance crisis: part 1: causes and threats. Pharm. Ther. 2015, 40 (4), 277–283. [PMC free article] [PubMed] [Google Scholar]
- 4.Blair JMA;Webber MA;Baylay AJ;Ogbolu DO;Piddock LJV Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2014, 13, 42–51. [DOI] [PubMed] [Google Scholar]
- 5.Li X-Z;Plésiat P;Nikaido H The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28 (2), 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai Y;Meyer KJ;Iinishi A;Favre-Godal Q;Green R;Manuse S;Caboni M;Mori M;Niles S;Ghiglieri M;Honrao C;Ma X;Guo JJ;Makriyannis A;Linares-Otoya L;Böhringer N;Wuisan ZG;Kaur H;Wu R;Mateus A;Typas A;Savitski MM;Espinoza JL;O’Rourke A;Nelson KE;Hiller S;Noinaj N;Schäberle TF;D’Onofrio A;Lewis K A new antibiotic selectively kills Gram-negative pathogens. Nature 2019, 576 (7787), 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith PA;Koehler MFT;Girgis HS;Yan D;Chen Y;Chen Y;Crawford JJ;Durk MR;Higuchi RI;Kang J;Murray J;Paraselli P;Park S;Phung W;Quinn JG;Roberts TC;Rougé L;Schwarz JB;Skippington E;Wai J;Xu M;Yu Z;Zhang H;Tan M-W;Heise CE Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 2018, 561 (7722), 189–194. [DOI] [PubMed] [Google Scholar]
- 8.Martin JK;Sheehan JP;Bratton BP;Moore GM;Mateus A;Li SH-J;Kim H;Rabinowitz JD;Typas A;Savitski MM;Wilson MZ;Gitai Z A Dual-Mechanism Antibiotic Kills Gram-Negative Bacteria and Avoids Drug Resistance. Cell 2020, 181 (7), 1518–1532.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benoit SL;Maier RJ;Sawers RG;Greening C Molecular Hydrogen Metabolism: a Widespread Trait of Pathogenic Bacteria and Protists. Microbiol. Mol. Biol. Rev. 2020, 84 (1), e00092–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De La Cruz LKC;Benoit SL;Pan Z;Yu B;Maier RJ;Ji X;Wang B Click, Release, and Fluoresce: A Chemical Strategy for a Cascade Prodrug System for Codelivery of Carbon Monoxide, a Drug Payload, and a Fluorescent Reporter. Org. Lett. 2018, 20 (4), 897–900. [DOI] [PubMed] [Google Scholar]
- 11.Schmalstig AA;Benoit SL;Misra SK;Sharp JS;Maier RJ Noncatalytic Antioxidant Role for Helicobacter pylori Urease. J. Bacteriol. 2018, 200 (17), e00124–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skaar EP;Zackular JP Editorial overview: Microbe–microbe interactions: the enemy of my enemy is my friend. Curr. Opin. Microbiol. 2020, 53, iii–v. [DOI] [PubMed] [Google Scholar]
- 13.Hopper CP;De La Cruz LK;Lyles KV;Wareham LK;Gilbert JA;Eichenbaum Z;Magierowski M;Poole RK;Wollborn J;Wang B Role of Carbon Monoxide in Host–Gut Microbiome Communication. Chem. Rev. 2020, 120 (24), 13273–13311. [DOI] [PubMed] [Google Scholar]
- 14.Cui AL;Hu X-X;Gao Y;Jin J;Yi H;Wang X-K;Nie T-Y;Chen Y;He Q-Y;Guo H-F;Jiang J-D;You X-F;Li Z-R Synthesis and Bioactivity Investigation of the Individual Components of Cyclic Lipopeptide Antibiotics. J. Med. Chem. 2018, 61 (5), 1845–1857. [DOI] [PubMed] [Google Scholar]
- 15.Alexander EM;Kreitler DF;Guidolin V;Hurben AK;Drake E;Villalta PW;Balbo S;Gulick AM;Aldrich CC Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic Pathogen Klebsiella oxytoca. ACS Infect. Dis. 2020, 6 (7), 1976–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuegg J;Hansford KA;Elliott AG;Cooper MA;Blaskovich MAT. How to Stimulate and Facilitate Early Stage Antibiotic Discovery. ACS Infect. Dis. 2020, 6 (6), 1302–1304. [DOI] [PubMed] [Google Scholar]
- 17.Si Y;Basak S;Li Y;Merino J;Iuliano JN;Walker SG;Tonge PJ Antibacterial Activity and Mode of Action of a Sulfonamide-Based Class of Oxaborole Leucyl-tRNA-Synthetase Inhibitors. ACS Infect. Dis. 2019, 5 (7), 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown ED;Wright GD Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [DOI] [PubMed] [Google Scholar]
- 19.MacNair CR;Stokes JM;Carfrae LA;Fiebig-Comyn AA;Coombes BK;Mulvey MR;Brown ED Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 2018, 9 (1), 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King AM;Reid-Yu SA;Wang W;King DT;De Pascale G;Strynadka NC;Walsh TR;Coombes BK;Wright GD Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510 (7506), 503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song M;Liu Y;Huang X;Ding S;Wang Y;Shen J;Zhu K A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020, 5 (8), 1040–1050. [DOI] [PubMed] [Google Scholar]
- 22.Corbett D;Wise A;Langley T;Skinner K;Trimby E;Birchall S;Dorali A;Sandiford S;Williams J;Warn P;Vaara M;Lister T Potentiation of Antibiotic Activity by a Novel Cationic Peptide: Potency and Spectrum of Activity of SPR741. Antimicrob. Agents Chemother. 2017, 61 (8), e00200–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brochado AR;Telzerow A;Bobonis J;Banzhaf M;Mateus A;Selkrig J;Huth E;Bassler S;Zamarreño Beas J;Zietek M;Ng N;Foerster S;Ezraty B;Py B;Barras F;Savitski MM;Bork P;Göttig S;Typas A Species-specific activity of antibacterial drug combinations. Nature 2018, 559 (7713), 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaara M Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992, 56 (3), 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock RE;Wong PG Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 1984, 26 (1), 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savage PB Multidrug-resistant bacteria: overcoming antibiotic permeability barriers of Gram-negative bacteria. Ann. Med. 2001, 33 (3), 167–171. [DOI] [PubMed] [Google Scholar]
- 27.Savage PB;Li C;Taotafa U;Ding B;Guan Q Antibacterial properties of cationic steroid antibiotics. FEMS Microbiol. Lett. 2002, 217 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen HT;Venter H;Veltman T;Williams R;O’Donovan LA;Russell CC;McCluskey A;Page SW;Ogunniyi AD;Trott DJ In vitro synergistic activity of NCL195 in combination with colistin against Gram-negative bacterial pathogens. Int. J. Antimicrob. 2021, 57 (5), 106323. [DOI] [PubMed] [Google Scholar]
- 29.Blondiaux N;Moune M;Desroses M;Frita R;Flipo M;Mathys V;Soetaert K;Kiass M;Delorme V;Djaout K;Trebosc V;Kemmer C;Wintjens R;Wohlkönig A;Antoine R;Huot L;Hot D;Coscolla M;Feldmann J;Gagneux S;Locht C;Brodin P;Gitzinger M;Déprez B;Willand N;Baulard Alain R Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420. Science 2017, 355 (6330), 1206–1211. [DOI] [PubMed] [Google Scholar]
- 30.Douafer H;Andrieu V;Phanstiel O;Brunel JM Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019, 62 (19), 8665–8681. [DOI] [PubMed] [Google Scholar]
- 31.Allen Richard C;Brown Sam P;Levin Bruce R Modified Antibiotic Adjuvant Ratios Can Slow and Steer the Evolution of Resistance: Co-amoxiclav as a Case Study. mBio 2019, 10 (5), e01831–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorityala BK;Guchhait G;Fernando DM;Deo S;McKenna SA;Zhanel GG;Kumar A;Schweizer F Adjuvants Based on Hybrid Antibiotics Overcome Resistance in Pseudomonas aeruginosa and Enhance Fluoroquinolone Efficacy. Angew. Chem. Int. Ed. 2016, 55 (2), 555–559. [DOI] [PubMed] [Google Scholar]
- 33.García-Fernández E;Koch G;Wagner RM;Fekete A;Stengel ST;Schneider J;Mielich-Süss B;Geibel S;Markert SM;Stigloher C;Lopez D Membrane Microdomain Disassembly Inhibits MRSA Antibiotic Resistance. Cell 2017, 171 (6), 1354–1367.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim C;Kassu M;Smith KP;Kirby JE;Manetsch R Pyrazole-Thiazole Core-Containing Analogs Exhibit Adjunctive Activity with Meropenem against Carbapenem-Resistant Enterobacteriaceae (CRE). ChemMedChem 2021, 16 (18), 2775–2780. [DOI] [PubMed] [Google Scholar]
- 35.Heesterbeek DAC;Muts RM;van Hensbergen VP;de Saint Aulaire P;Wennekes T;Bardoel BW;van Sorge NM;Rooijakkers SHM Outer membrane permeabilization by the membrane attack complex sensitizes Gram-negative bacteria to antimicrobial proteins in serum and phagocytes. PLoS Pathog. 2021, 17 (1), e1009227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q;Chen S;Liu X;Lin W;Zhu K Equisetin Restores Colistin Sensitivity against Multi-Drug Resistant Gram-Negative Bacteria. Antibiotics 2021, 10 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cebrián R;Xu C;Xia Y;Wu W;Kuipers OP The cathelicidin-derived close-to-nature peptide D-11 sensitises Klebsiella pneumoniae to a range of antibiotics in vitro, ex vivo and in vivo. Int. J. Antimicrob. 2021, 58 (5), 106434. [DOI] [PubMed] [Google Scholar]
- 38.Chen T;Xu W;Yu K;Zeng W;Xu C;Cao J;Zhou T In Vitro Activity of Ceftazidime-Avibactam Alone and in Combination with Amikacin Against Colistin-Resistant Gram-Negative Pathogens. Microb. Drug Resist. 2020, 27 (3), 401–409. [DOI] [PubMed] [Google Scholar]
- 39.Chen L;Yu K;Chen L;Zheng X;Huang N;Lin Y;Jia H;Liao W;Cao J;Zhou T Synergistic Activity and Biofilm Formation Effect of Colistin Combined with PFK-158 Against Colistin-Resistant Gram-Negative Bacteria. Infect. Drug Resist. 2021, 14, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y;Jia Y;Yang K;Li R;Xiao X;Zhu K;Wang Z Metformin Restores Tetracyclines Susceptibility against Multidrug Resistant Bacteria. Adv. Sci. 2020, 7 (12), 1902227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart EM;Mitchell AM;Konovalova A;Grabowicz M;Sheng J;Han X;Rodriguez-Rivera FP;Schwaid AG;Malinverni JC;Balibar CJ;Bodea S;Si Q;Wang H;Homsher MF;Painter RE;Ogawa AK;Sutterlin H;Roemer T;Black TA;Rothman DM;Walker SS;Silhavy TJ A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl. Acad. Sci. U.S.A. 2019, 116 (43), 21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ni N;Li M;Wang J;Wang B Inhibitors and antagonists of bacterial quorum sensing. Med. Res. Rev. 2009, 29 (1), 65–124. [DOI] [PubMed] [Google Scholar]
- 43.Abdali N;Parks JM;Haynes KM;Chaney JL;Green AT;Wolloscheck D;Walker JK;Rybenkov VV;Baudry J;Smith JC;Zgurskaya HI Reviving Antibiotics: Efflux Pump Inhibitors That Interact with AcrA, a Membrane Fusion Protein of the AcrAB-TolC Multidrug Efflux Pump. ACS Infect. Dis. 2017, 3 (1), 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May KL;Grabowicz M The bacterial outer membrane is an evolving antibiotic barrier. Proc. Natl. Acad. Sci. U.S.A. 2018, 115 (36), 8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koebnik R;Locher KP;Van Gelder P Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 2000, 37 (2), 239–253. [DOI] [PubMed] [Google Scholar]
- 46.WHO. Media Centre. News Release. WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
- 47.Tsubery H;Ofek I;Cohen S;Fridkin M Structure−Function Studies of Polymyxin B Nonapeptide: Implications to Sensitization of Gram-Negative Bacteria. J. Med. Chem. 2000, 43 (16), 3085–3092. [DOI] [PubMed] [Google Scholar]
- 48.Vaara M Polymyxin Derivatives that Sensitize Gram-Negative Bacteria to Other Antibiotics. Molecules (Basel, Switzerland) 2019, 24 (2), 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaara M New polymyxin derivatives that display improved efficacy in animal infection models as compared to polymyxin B and colistin. Med. Res. Rev. 2018, 38 (5), 1661–1673. [DOI] [PubMed] [Google Scholar]
- 50.Rabanal F;Cajal Y Recent advances and perspectives in the design and development of polymyxins. Nat. Prod. Rep. 2017, 34 (7), 886–908. [DOI] [PubMed] [Google Scholar]
- 51.Si Z;Hou Z;Vikhe YS;Thappeta KRV;Marimuthu K;De PP;Ng OT;Li P;Zhu Y;Pethe K;Chan-Park MB Antimicrobial Effect of a Novel Chitosan Derivative and Its Synergistic Effect with Antibiotics. ACS Appl. Mater. Interfaces 2021, 13 (2), 3237–3245. [DOI] [PubMed] [Google Scholar]
- 52.Namivandi-Zangeneh R;Sadrearhami Z;Dutta D;Willcox M;Wong EHH;Boyer C Synergy between Synthetic Antimicrobial Polymer and Antibiotics: A Promising Platform To Combat Multidrug-Resistant Bacteria. ACS Infect. Dis. 2019, 5 (8), 1357–1365. [DOI] [PubMed] [Google Scholar]
- 53.Bai S;Wang J;Yang K;Zhou C;Xu Y;Song J;Gu Y;Chen Z;Wang M;Shoen C;Andrade B;Cynamon M;Zhou K;Wang H;Cai Q;Oldfield E;Zimmerman Steven C;Bai Y;Feng X A polymeric approach toward resistance-resistant antimicrobial agent with dual-selective mechanisms of action. Sci. Adv. 7 (5), eabc9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tantisuwanno C;Dang F;Bender K;Spencer JD;Jennings ME;Barton HA;Joy A Synergism between Rifampicin and Cationic Polyurethanes Overcomes Intrinsic Resistance of Escherichia coli. Biomacromolecules 2021, 22 (7), 2910–2920. [DOI] [PubMed] [Google Scholar]
- 55.Stokes JM;MacNair CR;Ilyas B;French S;Côté J-P;Bouwman C;Farha MA;Sieron AO;Whitfield C;Coombes BK;Brown ED Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 17028–17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wesseling CMJ;Slingerland CJ;Veraar S;Lok S;Martin NI Structure–Activity Studies with Bis-Amidines That Potentiate Gram-Positive Specific Antibiotics against Gram-Negative Pathogens. ACS Infect. Dis. 2021, 7 (12), 3314–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin SE;Melander RJ;Brackett CM;Scott AJ;Chandler CE;Nguyen CM;Minrovic BM;Harrill SE;Ernst RK;Manoil C;Melander C Small Molecule Potentiation of Gram-Positive Selective Antibiotics against Acinetobacter baumannii. ACS Infect. Dis. 2019, 5 (7), 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacNair CR;Tsai CN;Brown ED Creative targeting of the Gram-negative outer membrane in antibiotic discovery. Ann. N. Y. Acad. Sci. 2020, 1459 (1), 69–85. [DOI] [PubMed] [Google Scholar]
- 59.Sun H;Zhang Q;Wang R;Wang H;Wong Y-T;Wang M;Hao Q;Yan A;Kao RYT;Ho P-L;Li H Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin. Nat. Commun. 2020, 11 (1), 5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Idowu T;Ammeter D;Rossong H;Zhanel GG;Schweizer F Homodimeric Tobramycin Adjuvant Repurposes Novobiocin as an Effective Antibacterial Agent against Gram-Negative Bacteria. J. Med. Chem. 2019, 62 (20), 9103–9115. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerman SM;Lafontaine A-AJ;Herrera CM;McLean AB;Trent MS A Whole-Cell Screen Identifies Small Bioactives That Synergize with Polymyxin and Exhibit Antimicrobial Activities against Multidrug-Resistant Bacteria. Antimicrob. Agents Chemother. 2020, 64 (3), e01677–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattingly AE;Cox KE;Smith R;Melander RJ;Ernst RK;Melander C Screening an Established Natural Product Library Identifies Secondary Metabolites That Potentiate Conventional Antibiotics. ACS Infect. Dis. 2020, 6 (10), 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blankson G;Parhi AK;Kaul M;Pilch DS;LaVoie EJ Structure-activity relationships of potentiators of the antibiotic activity of clarithromycin against Escherichia coli. Eur. J. Med. Chem. 2019, 178, 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cadelis MM;Li SA;Bourguet-Kondracki M-L;Blanchet M;Douafer H;Brunel JM;Copp BR Spermine Derivatives of Indole-3-carboxylic Acid, Indole-3-acetic Acid and Indole-3-acrylic Acid as Gram-Negative Antibiotic Adjuvants. ChemMedChem 2021, 16 (3), 513–523. [DOI] [PubMed] [Google Scholar]
- 65.Konai MM;Haldar J Lysine-Based Small Molecule Sensitizes Rifampicin and Tetracycline against Multidrug-Resistant Acinetobacter baumannii and Pseudomonas aeruginosa. ACS Infect. Dis. 2020, 6 (1), 91–99. [DOI] [PubMed] [Google Scholar]
- 66.García-Quintanilla M;Caro-Vega José M;Pulido Marina R;Moreno-Martínez P;Pachón J;McConnell Michael J Inhibition of LpxC Increases Antibiotic Susceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother. 60 (8), 5076–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muheim C;Götzke H;Eriksson AU;Lindberg S;Lauritsen I;Nørholm MHH;Daley DO Increasing the permeability of Escherichia coli using MAC13243. Sci. Rep. 2017, 7 (1), 17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barker CA;Allison SE;Zlitni S;Nguyen ND;Das R;Melacini G;Capretta AA;Brown ED Degradation of MAC13243 and studies of the interaction of resulting thiourea compounds with the lipoprotein targeting chaperone LolA. Bioorg. Med. Chem. Lett 2013, 23 (8), 2426–2431. [DOI] [PubMed] [Google Scholar]
- 69.Soeiro MNC;Werbovetz K;Boykin DW;Wilson WD;Wang MZ;Hemphill A Novel amidines and analogues as promising agents against intracellular parasites: a systematic review. Parasitology 2013, 140 (8), 929–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paul A;Guo P;Boykin DW;Wilson WD A New Generation of Minor-Groove-Binding-Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context. Molecules (Basel, Switzerland) 2019, 24 (5), 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo P;Farahat AA;Paul A;Harika NK;Boykin DW;Wilson WD Compound Shape Effects in Minor Groove Binding Affinity and Specificity for Mixed Sequence DNA. J. Am. Chem. Soc. 2018, 140 (44), 14761–14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall MJ;Middleton RF;Westmacott D The fractional inhibitory concentration (FIC) index as a measure of synergy. J. Antimicrob. Chemother. 1983, 11 (5), 427–433. [DOI] [PubMed] [Google Scholar]
- 73.Zhivkova ZD Studies on drug-human serum albumin binding: the current state of the matter. Curr. Pharm. Des. 2015, 21 (14), 1817–30. [DOI] [PubMed] [Google Scholar]
- 74.Han C;Wang B Factors that Impact the Developability of Drug Candidates. In Drug Delivery: Principles and Applications, 2nd edition; Wang B.; Hu L.; Siahaan TJ., Eds.; Wiley: 2016; pp 1–18. [Google Scholar]
- 75.Rice LB Progress and Challenges in Implementing the Research on ESKAPE Pathogens. Infect. Control Hosp. Epidemiol. 2010, 31 (S1), S7–S10. [DOI] [PubMed] [Google Scholar]
- 76.Codjoe FS;Donkor ES Carbapenem Resistance: A Review. Med. Sci. 2017, 6 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Asokan GV;Ramadhan T;Ahmed E;Sanad H WHO Global Priority Pathogens List: A Bibliometric Analysis of Medline-PubMed for Knowledge Mobilization to Infection Prevention and Control Practices in Bahrain. Oman Med. J. 2019, 34 (3), 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moellering RC NDM-1 — A Cause for Worldwide Concern. N. Engl. J. Med. 2010, 363 (25), 2377–2379. [DOI] [PubMed] [Google Scholar]
- 79.Liu Y-Y;Wang Y;Walsh TR;Yi L-X;Zhang R;Spencer J;Doi Y;Tian G;Dong B;Huang X;Yu L-F;Gu D;Ren H;Chen X;Lv L;He D;Zhou H;Liang Z;Liu J-H;Shen J Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16 (2), 161–168. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y;Tian G-B;Zhang R;Shen Y;Tyrrell JM;Huang X;Zhou H;Lei L;Li H-Y;Doi Y;Fang Y;Ren H;Zhong L-L;Shen Z;Zeng K-J;Wang S;Liu J-H;Wu C;Walsh TR;Shen J Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 2017, 17 (4), 390–399. [DOI] [PubMed] [Google Scholar]
- 81.Stoesser N;Mathers AJ;Moore CE;Day NPJ;Crook DW Colistin resistance gene mcr-1and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect. Dis. 2016, 16 (3), 285–286. [DOI] [PubMed] [Google Scholar]
- 82.Ruiz N;Falcone B;Kahne D;Silhavy TJ Chemical Conditionality: A GeneticStrategy to Probe Organelle Assembly. Cell 2005, 121 (2), 307–317. [DOI] [PubMed] [Google Scholar]
- 83.Fidai S;Farmer SW;Hancock REW. Interaction of Cationic Peptides with Bacterial Membranes. In Methods in Molecular Biology: antibacterial peptide protocols; Schafer WM., Ed. Humana Press,Totowa, NJ: 1997; Vol. 78, pp 187–204. [DOI] [PubMed] [Google Scholar]
- 84.Silhavy TJ;Kahne D;Walker S The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2 (5), a000414–a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steimle A;Autenrieth IB;Frick J-S Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 2016, 306 (5), 290–301. [DOI] [PubMed] [Google Scholar]
- 86.Clifton LA;Skoda MWA;Le Brun AP;Ciesielski F;Kuzmenko I;Holt SA;Lakey JH Effect of Divalent Cation Removal on the Structure of Gram-Negative Bacterial Outer Membrane Models. Langmuir 2015, 31 (1), 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balusek C;Gumbart, James C. Role of the Native Outer-Membrane Environment on the Transporter BtuB. Biophys. J. 2016, 111 (7), 1409–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jahnen-Dechent W;Ketteler M Magnesium basics. Clin. Kidney J. 2012, 5 (Suppl 1), i3–i14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bazydlo LAL;Needham M;Harris NS Calcium, Magnesium, and Phosphate. Lab. Med. 2014, 45 (1), e44–e50. [Google Scholar]
- 90.Moison E;Xie R;Zhang G;Lebar MD;Meredith TC;Kahne D A Fluorescent Probe Distinguishes between Inhibition of Early and Late Steps of Lipopolysaccharide Biogenesis in Whole Cells. ACS Chem. Biol. 2017, 12 (4), 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Domínguez-Medina CC;Pérez-Toledo M;Schager AE;Marshall JL;Cook CN;Bobat S;Hwang H;Chun BJ;Logan E;Bryant JA;Channell WM;Morris FC;Jossi SE;Alshayea A;Rossiter AE;Barrow PA;Horsnell WG;MacLennan CA;Henderson IR;Lakey JH;Gumbart JC;López-Macías C;Bavro VN;Cunningham AF Outer membrane protein size and LPS O-antigen define protective antibody targeting to the Salmonella surface. Nat. Commun. 2020, 11 (1), 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pavlova A;Hwang H;Lundquist K;Balusek C;Gumbart JC Living on the edge: Simulations of bacterial outer-membrane proteins. Biochim. Biophys. Acta. Biomembr. 2016, 1858 (7, Part B), 1753–1759. [DOI] [PubMed] [Google Scholar]
- 93.Martynowycz MW;Rice A;Andreev K;Nobre TM;Kuzmenko I;Wereszczynski J;Gidalevitz D Salmonella Membrane Structural Remodeling Increases Resistance to Antimicrobial Peptide LL-37. ACS Infect. Dis. 2019, 5 (7), 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fischer D;Li Y;Ahlemeyer B;Krieglstein J;Kissel T In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24 (7), 1121–1131. [DOI] [PubMed] [Google Scholar]
- 95.van Meer G;Voelker DR;Feigenson GW Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 2008, 9 (2), 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boekema BKHL;Pool L;Ulrich MMW The effect of a honey based gel and silver sulphadiazine on bacterial infections of in vitro burn wounds. Burns 2013, 39 (4), 754–759. [DOI] [PubMed] [Google Scholar]
- 97.de Breij A;Riool M;Cordfunke RA;Malanovic N;de Boer L;Koning RI;Ravensbergen E;Franken M;van der Heijde T;Boekema BK;Kwakman PHS;Kamp N;El Ghalbzouri A;Lohner K;Zaat SAJ;Drijfhout JW;Nibbering PH The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10 (423), eaan4044. [DOI] [PubMed] [Google Scholar]
- 98.Haisma EM;Göblyös A;Ravensbergen B;Adriaans AE;Cordfunke RA;Schrumpf J;Limpens RWAL;Schimmel KJM;den Hartigh J;Hiemstra PS;Drijfhout JW;El Ghalbzouri A;Nibbering PH Antimicrobial Peptide P60.4Ac-Containing Creams and Gel for Eradication of Methicillin-Resistant Staphylococcus aureus from Cultured Skin and Airway Epithelial Surfaces. Antimicrob. Agents Chemother. 2016, 60 (7), 4063–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rutherford ST;Bassler BL Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2 (11), a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chew KL;La M-V;Lin RTP;Teo JWP Colistin and Polymyxin B Susceptibility Testing for Carbapenem-Resistant and mcr-Positive Enterobacteriacea: Comparison of Sensititre, MicroScan, Vitek 2, and Etest with Broth Microdilution. J. Clin. Microbiol. 2017, 55 (9), 2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wouters OJ;McKee M;Luyten J Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009–2018. JAMA 2020, 323 (9), 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pharmaceutical Profiling in Drug Discovery for Lead Selection. AAPS Press, Arlington, VA: 2004. [Google Scholar]
- 103.Cox G;Sieron A;King AM;De Pascale G;Pawlowski AC;Koteva K;Wright GD A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chem. Biol. 2017, 24 (1), 98–109. [DOI] [PubMed] [Google Scholar]
- 104.Vanommeslaeghe K;MacKerell AD Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vanommeslaeghe K;Raman EP;MacKerell AD Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mayne CG;Saam J;Schulten K;Tajkhorshid E;Gumbart JC Rapid parameterization of small molecules using the Force Field Toolkit. J. Comput. Chem. 2013, 34, 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klauda JB;Venable RM;Freites JA;O’Connor JW;Tobias DJ;Mondragon-Ramirez C;Vorobyov I;MacKerell AD Jr.;Pastor RW Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu EL;Cheng X;Jo S;Rui H;Song KC;Dávila-Contreras EM;Qi Y;Lee J;Monje-Galvan V;Venable RM;Klauda JB;Im W CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jo S;Kim T;Iyer VG;Im W CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [DOI] [PubMed] [Google Scholar]
- 110.Phillips JC;Braun R;Wang W;Gumbart J;Tajkhorshid E;Villa E;Chipot C;Skeel D, R.;Kale, L.;Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Case DA;Cheatham III TE;Darden T;Gohlke H;Luo R;Merz KM Jr.;Onufriev A;Simmerling C;Wang B;Woods RJ The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Balusek C;Hwang H;Lau CH;Lundquist K;Hazel A;Pavlova A;Lynch DL;Reggio PH;Wang Y;Gumbart JC Accelerating Membrane Simulations with Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2019, 15, 4673–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hopkins CW;Le Grand S;Walker RC;Roitberg AE Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Any additional data can be requested from the corresponding author.


