Abstract
Aims
Poly (ADP-ribose) polymerase (PARP) inhibitors (PARPIs) represent a standard of care for the clinical management of high-grade serous ovarian cancer (HGSOC). The recognition of homologous recombination deficiency (HRD) has emerged as a predictive biomarker of response for first-line PARPIs treatment in patients with HGOSC. On the other hand, this test is extremely complex and therefore it is often externalised. Regrettably, the reliability of outsourced HRD testing can be troubled by inconclusive results and high rejection rates. In this methodological study, we assessed the technical feasibility, interassay and interlaboratory reproducibility of in-house HRD testing using three different commercially available next-generation sequencing assays.
Methods
A total of n=20 epithelial ovarian cancer samples previously analysed with MyChoice CDx were subjected to HRD retesting using three different platforms in three different major pathology laboratories, that is, SOPHiA DDM HRD Solution, HRD focus and Oncomine homologous recombination repair pathway predesigned panel. Concordance was calculated by Cohen’s (dual) and Fleiss (triple) κ coefficients.
Results
In-house BRCA1/2 molecular testing yielded a concordance rate >90.0% among all participating centres. HRD scores were successfully calculated by each institution with a concordance rate of 76.5%. Concerning the external gold standard test, the overall percentage of agreement ranged from 80.0% to 90.0% with a positive percentage agreement ranging from 75.0% to 80.0% and a negative percentage agreement ranging from 80.0% to 100%.
Conclusions
In-house testing for HRD can be reliably performed with commercially available next-generation sequencing assays.
Keywords: ovarian neoplasms; point-of-care testing; pathology, molecular
WHAT IS ALREADY KNOWN ON THIS TOPIC
Homologous recombination deficiency (HRD) status has revolutionised the clinical management of patients with high-grade serous ovarian cancer.
WHAT THIS STUDY ADDS
In-house harmonised next-generation sequencing (NGS) procedures may represent a valid testing strategy for HRD evaluation.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
In-house harmonised NGS procedures for HRD testing lead an improvement of successful testing rate.
Introduction
Epithelial ovarian cancer (EOC) represents the leading cause of death among gynecological malignancies.1 Particularly, high-grade serous ovarian cancer (HGSOC) is characterised by a high mortality rate linked to advanced stage at diagnosis.2 The standard therapeutic schemes commonly rely on surgery combined with platinum-based chemotherapies.3 Lately, the overall survival of these patients has gradually improved thanks to novel and emerging therapeutic strategies.4 One of the most promising therapeutic approaches against HGSOC is the use of poly (ADP-ribose) polymerase (PARP) inhibitors (PARPIs). The mechanism of action of PARPIs is rather complex, involving the inhibition of PARP-mediated homologous recombination repair (HRR), upregulation of non-homologous end joining and PARP ‘trapping’.5–8
Accounting for 20%–25% of HGSOC cases, BRCA1/2 mutations hold a predictive value for the selection of platinum-sensitive patients with HGSOC eligible for PARPi treatment.9–11 However, other key actors are involved in the HRR system.12–14 In detail, functional defects in HRR, known as homologous recombination deficiency (HRD), promote the activation of error-prone DNA repair mechanisms that induce genomic instability and malignant cell transformation.15 16 The role of HRD status in the clinical selection of patients with HGSOC for PARPIs treatment has been proved by several clinical trials showing the remarkable clinical benefits of PARPi treatment against HRR-deficient compared with HRR-proficient tumours.16–19
Conventionally, HRD status is determined by analysing the signature approached with the evaluation of genomic instability (ie, genomic scars), including loss of heterozygosity (LOH), telomeric allelic imbalance (TAI) and large-scale state transitions (LST).20 21 In clinical trials, HRD-induced genomic scars are generally assessed by centralised next-generation sequencing (NGS)-based assays. This implies that HRD testing requires outsourcing analysis—a circumstance that may drastically impact on the success rate of molecular tests in clinical practice. Moreover, adding to the complexity of achieving accurate and reproducible results is the increasing number of different NGS-based assays commercially available for HRD status evaluation and the ensuing lack of standardised protocols and data interpretation.19 A possible solution to these issues could be the application of in-house testing for HRD. However, although in-house HRD testing should bridge the gap between molecular tests and diagnostic routine samples,22 standardising analytical pipelines and harmonising results remain challenging. Indeed, even in this case, simplification of complex technical procedures and harmonisation of diverse data analysis workflows are essential steps to implement in-house HRD testing in routine clinical practice. In this retrospective study, we tested the feasibility of in-house HRD testing to evaluate HRD status in patients with HGSOC. Here, we evaluated the interassay and interlaboratory reproducibility of HRD scores of three commercially available HRD assays used by three Italian referral institutions to determine HRD status in EOC tissue samples. All patients had been previously diagnosed with the MyChoice CDx Myriad HRD test (Myriad Genetics, Salt Lake City, USA).
Methods
Study design and samples
We retrospectively analysed a series of n=20 formalin-fixed paraffin-embedded (FFPE) tissue samples from patients with EOC. All samples, previously externalised for HRD testing with the Food and Drug Administration (FDA)-approved MyChoice CDx were retrieved from the institutional archives of the European Institute of Oncology, IRCCS, Milan, Italy. Each case was defined as HRD positive or negative according to the clinically validated cut-off Genomic Instability Score of 42 (table 1).
Table 1.
List of 20 patients with ovarian cancer selected from internal archive of Pathology Division of European Institute of Oncology, Milan
| Sample ID | Histotype | Specimen type | Site | Cellularity (%) | Neoadjuvant therapy | BRCA1 status | BRCA2 status | HRD |
| ID_1 | HGSOC | Surgical resection | Adnexa uteri | 75 | 0 | WT | WT | NEG |
| ID_2 | HGSOC | Surgical resection | Fallopian tube | 40 | 1 | p.Ser288Argfs*10 | WT | POS |
| ID_3 | HGSOC | Biopsy | Omentum | 80 | 0 | WT | WT | POS |
| ID_4 | HGSOC | Surgical resection | Adnexa uteri | 60 | 0 | WT | WT | POS |
| ID_5 | HGSOC | Surgical resection | Ovary | 70 | 0 | WT | WT | NEG |
| ID_6 | EnOC | Surgical resection | Ovary | 60 | 0 | WT | WT | NEG |
| ID_7 | HGSOC | Surgical resection | Ovary | 70 | 0 | WT | WT | NEG |
| ID_8 | HGSOC | Surgical resection | Ovary | 60 | 0 | WT | WT | NEG |
| ID_9 | HGSOC | Surgical resection | Ovary | 60 | 0 | WT | WT | NEG |
| ID_10 | HGSOC | Surgical resection | Adnexa uteri | 60 | 1 | WT | WT | NEG |
| ID_11 | HGSOC | Biopsy | Peritoneum | 80 | 0 | WT | WT | POS |
| ID_12 | HGSOC | Surgical resection | Ovary | 80 | 0 | WT | WT | NEG |
| ID_13 | HGSOC | Surgical resection | Ovary | 60 | 0 | WT | WT | POS |
| ID_14 | Mixed OC | Surgical resection | Ovary | 60 | 0 | WT | WT | NEG |
| ID_15 | EnOC | Surgical resection | Ovary | 40 | 0 | WT | WT | NEG |
| ID_16 | HGSOC | Surgical resection | Ovary | 60 | 0 | WT | WT | POS |
| ID_17 | HGSOC | Surgical resection | Omentum | 60 | 0 | WT | WT | NEG |
| ID_18 | HGSOC | Surgical resection | Peritoneum | 50 | 1 | WT | WT | NEG |
| ID_19 | HGSOC | Surgical resection | Ovary | 60 | 0 | WT | WT | POS |
| ID_20 | HGSOC | Surgical resection | Fallopian tube | 70 | 0 | WT | WT | NEG |
BRCA, BReast CAncer associated gene; EnOC, endometrioid ovarian cancer; GSS, Genomic Scar Score; HGSOC, high-grade serous ovarian cancer; HRD, homologous recombination deficiency; NEG, negative; POS, positive; VUS, variant of uncertain significance; WT, wild-type.
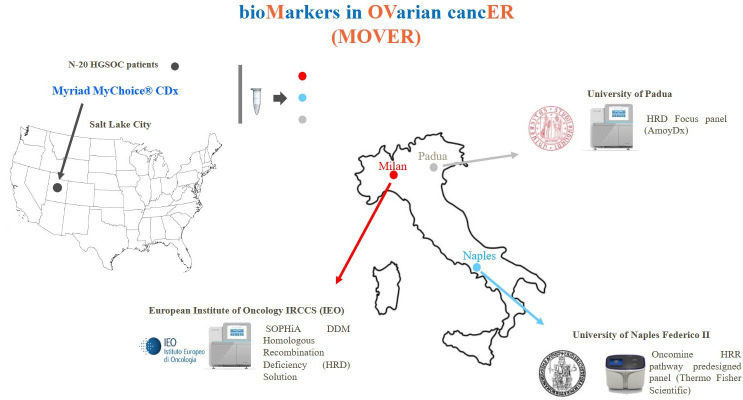
The study was conducted to evaluate in-house analysis of HRD status in three reference centres (Division of Pathology of the European Institute of Oncology, Milan; Department of Pathology of Federico II University, Naples; Department of Pathology, University of Padua, Padua, Italy) using three commercially available HRD panels (ie, SOPHiA DDM HRD Solution, SOPHiA Genetics, Saint-Sulpice, Switzerland; HRD focus panel, Amoy Diagnostics, Xiamen, Fujian, China; Oncomine HRR pathway predesigned panel, Thermo Fisher Scientific). In addition, a reference DNA sample (OncoSpan gDNA reference standard, Horizon Discovery, Cambridge, UK) containing 386 different variants across 152 cancer-related genes was also distributed to each participating centre to evaluate the technical reproducibility and concordance rate between each internal workflow (figure 1).
Figure 1.
Study design. HGSOC, high-grade serous ovarian cancer; HRR, homologous recombination repair.
The centres were anonymised as centre #1, centre #2 and centre #3 (online supplemental table 1).
jcp-2023-208852supp001.pdf (35KB, pdf)
Nucleic acids purification
Seven unstained slides from representative FFPE tissue blocks were cut into 4 μm thick sections. In brief, DNA was extracted with the Maxwell RSC DNA FFPE Kit (Promega, Madison, Wisconsin, USA) according to the manufacturer’s instructions and then quantified with the QuantiFluor ONE dsDNA System (Promega) on the Quantus Fluorometer (Promega).
In-house HRD testing
DNA samples were handled according to the internal standardised diagnostic procedures of each laboratory; in-house HRD testing was evaluated according to the manufacturer’s instructions for each HRD panel. In detail, 1.25 ng/µL (corresponding to 50 ng in 40 µL) of DNA was used for manual library preparation with the SOPHiA DDM HRD solution panel. This assay is a hybridisation-based approach able to combine the Genomic Integrity Index (GII) with deleterious mutations in n=28 hours (including BRCA1/2) complex-related genes. In addition, HRD genomic scar was calculated by an optimised analytic pipeline based on deep learning algorithms for the analysis of low-pass whole genome sequencing data. Sequencing was performed on the Illumina NextSeq 550Dx platform (Illumina, San Diego, California, USA). HRD score was calculated by proprietary software. In particular, a negative or positive GII score was indicative of HRD negative or positive status, respectively.
Generally, the Oncomine HRR pathway predesigned panel allows laboratory clinicians to analyse the high functional impact of point mutations, indels and CNV (Copy Number Variation) found in n=27 hours related genes.23 In detail, an optimum of n=20 ng DNA input was achieved. Library preparation was manually performed as follows: 10 samples were amplified with the Ion AmpliSeq Library Kit Plus (Thermo Fisher Scientific, Waltham, Massachusetts, USA) under thermal conditions (25 cycles), as specified by the manufacturer’s instructions. After two purification steps, libraries were diluted at 50 pM and pooled for template generation on an automatic platform (Ion Chef system, Thermo Fisher Scientific). Overall, two sets of n=10 pooled libraries were loaded on the Ion Chef platform by adopting Ion 540 Kit-Chef (Thermo Fisher Scientific), according to the manufacturer’ s instructions. Sequencing was performed on Ion 540 chip. Data were processed by using signal processing and base calling default parameters on Torrent Suite (V.5.0.2). Coverage analysis and variant calling were carried out with a customised workflow based on optimised bed files on Torrent Suite. In detail, a median depth coverage of 2000X and a quality score ≥20 were used to filter variants at 5% of allele frequency, as indicated by manufacturer’s guidelines. Moreover, raw data were manually inspected on Golden Helix Genome Browser V.2.0.7 (Bozeman, Montana, USA) to confirm the molecular alterations automatically detected by a dedicated informatics pipeline. HRD score was calculated by a customised bioinformatics pipeline able to integrate LOH from referenced genes and CNV in BRCA1/2 genes. Particularly, HRD status was identified in accordance with ≥10 hotspot mutations in referenced genes and/or CNV-positive status. CNV was identified when ≥3 statistically significant unbalanced BRCA1/2 regions were detected in relation to normalised read counts.
Amoy HRD focus panel enables to detect SNVs (Single Nucleotide Variations) and indels in coding/non-coding regions from n=27 HRR-related genes and hotspot mutations in n=5 driver genes in solid tumours starting from tissue and liquid biopsy specimens.24 Moreover, this assay covers BRCA1/2 large rearrangements from peripheral blood samples. In brief, target selection is based on the Halo-shape Annealing and Defer-Ligation Enrichment system, which is an improved version of the Molecular Inversion Probe technology. A set of n=10 samples (n=9 samples plus internal control) were simultaneously processed in each run. Library construction was performed with high-sensitive probes that hybridised the target region. After DNA extension from the corresponding probe arm, circular DNA was obtained and repaired by DNA ligation. Moreover, circular DNA was cleaned by exonucleases, which cleave single and double DNA strands, and finally amplified with a universal primer to create the template for the sequencing process on a NextSeq 550 system. Data analysis was automatically carried out with proprietary software able to identify BRCA1/2 molecular alterations and calculate the Genomic Scar Score (GSS). Samples with a GSS ≥50 were considered HRD positive.
Statistical analysis
Interassay and interlaboratory reproducibility was measured as the percentage of agreement between two in-house HRD testing methods. The overall percentage of agreement (OPA) was calculated to assess the concordance between the three in-house HRD testing panels and the outsourced HRD assay (reference standard). HRD status was considered as a categorical variable (positive vs negative) according to the HRD score cut-off of each testing method (online supplemental table 1).
Results
Reference DNA samples were successfully analysed by each participating centre (online supplemental table 2). In detail, the total number of reads, percentage of aligned reads and uniformity of coverage were successfully achieved by each participating institution (online supplemental table 1). Overall, a concordance rate of 80.0% was observed for BRCA1/2 status. In particular, centres #1 and #3 detected four out of five (80.0%) clinically relevant BRCA1/2 alterations harboured by the OncoSpan gDNA. Instead, centre #2 identified three out of five (60.0%) BRCA1/2 referral alterations (table 2). Remarkably, all participating centres successfully calculated a negative HRD status for the reference DNA sample (table 2).
Table 2.
Schematic representation of BRCA1/2 clinically relevant molecular alterations and HRD score (pathogenetic and VUS alterations according to ClinVar database) detected by each participating institution on reference DNA sample (OncoSpan gDNA reference standard, Horizon Discovery, Cambridge, UK)
| Centre #1 | Centre #2 | Centre #3 | ||||
| Gene | Results | GIS SOPHiA |
Results | HRD score | Results | GSS Amoy |
| BRCA1 | c.4327C>T: p.(Arg1443*) (pathogenetic) |
−11.2 (NEG) | c.4327C>T: p.(Arg1443*) (pathogenetic) |
NEG | c.4327C>T: p.(Arg1443*) (pathogenetic) |
NEG |
| BRCA2 | c.8021dup: p.(Ile2675Aspfs*6) (pathogenetic) |
c.5073del: p.(Lys1691Asnfs*15) (VUS) |
c.8021dup: p.(Ile2675Aspfs*6) (pathogenetic) |
|||
| c.5073del: p.(Lys1691Asnfs*15) (VUS) |
c.5073del: p.(Lys1691Asnfs*15) (VUS) |
|||||
| c.5351del: p.(Asn1784Thrfs*7) (pathogenetic) | ||||||
| c.5351del: p.(Asn1784Thrfs*7) (pathogenetic) |
c.5351del: p.(Asn1784Thrfs*7) (pathogenetic) |
|||||
BRCA, BReast CAncer associated gene; GIS, Genomic Instability Score; GSS, Genomic Scar Score; HRD, homologous recombination deficiency; NEG, negative; VUS, variant of uncertain significance.
jcp-2023-208852supp002.pdf (30.4KB, pdf)
In particular, centres #1 and #2 successfully analysed BRCA1/2 status and HRD scores in all 20 samples (success rate 100%); centre #3 analysed 17 out of 20 cases (success rate: 85%). Remarkably, only centre #2 detected a pathogenic BRCA1 alteration in PD_6 (table 3).
Table 3.
List of BRCA1/2 pathogenetic variations and HRD score on a retrospective series of n=20 patients with HGSOC inspected with commercially available NGS assays
| Sample ID | Gene | Centre #1 | Centre #2 | Centre #3 | ||||
| Results | HRD SOPHiA | GSS Myriad | Results | HRD score | Results | GSS | ||
| ID_1 | BRCA1/2 | WT | 3.4 | 40 | WT | LOH* | WT | 77.7 |
| ID_2 | BRCA1/2 | WT | 16.8 | 60 | WT | NEG | WT | 98.7 |
| ID_3 | BRCA1/2 | WT | 5.3 | 53 | WT | LOH | WT | 92.7 |
| ID_4 | BRCA1/2 | WT | −1.6 | 47 | WT | LOH | WT | 88.3 |
| ID_5 | BRCA1/2 | WT | −3.3 | 28 | WT | NEG | WT | 17.1 |
| ID_6 | BRCA1/2 | WT | −11.4 | 3 | p.Glu577AsnfsTer11 | NEG | WT | 4.1 |
| ID_7 | BRCA1/2 | WT | −18.8 | 18 | WT | NEG | WT | 11.8 |
| ID_8 | BRCA1/2 | WT | −10.7 | 16 | WT | NEG | WT | 7.2 |
| ID_9 | BRCA1/2 | WT | −11 | 15 | WT | NEG | WT | 19 |
| ID_10 | BRCA1/2 | WT | −1.4 | 23 | WT | NEG | NA | NA |
| ID_11 | BRCA1/2 | WT | 11.9 | 75 | WT | NEG | NA | NA |
| ID_12 | BRCA1/2 | WT | −5.4 | 15 | WT | NEG | WT | 6.5 |
| ID_13 | BRCA1/2 | WT | 12.6 | 56 | WT | NEG | WT | 99.3 |
| ID_14 | BRCA1/2 | WT | −15.5 | 9 | WT | NEG | WT | 7.9 |
| ID_15 | BRCA1/2 | WT | −21.3 | 2 | WT | NEG | WT | 0.7 |
| ID_16 | BRCA1/2 | WT | 14.9 | 55 | WT | LOH | WT | 89.7 |
| ID_17 | BRCA1/2 | WT | −5.7 | 34 | WT | NEG | WT | 50.1* |
| ID_18 | BRCA1/2 | WT | −12.4 | 19 | WT | NEG | NA | NA |
| ID_19 | BRCA1/2 | WT | 8.1 | 56 | WT | LOH | WT | 97.2 |
| ID_20 | BRCA1/2 | WT | −9.6 | 25 | WT | NEG | WT | 19.3 |
HRD positive (green) and negative (red) status is shown.
*Borderline results.
BRCA, BReast CAncer associated gene; GSS, Genomic Scar Score; HRD, homologous recombination deficiency; ID, identify number; LOH, loss of heterozygosis; NA, not assessed; NEG, negative; NGS, next-generation sequencing; WT, wild-type.
All participating centres successfully calculated HRD scores in all the analysed ovarian cancer samples (table 4). Overall, a concordance rate in HRD status of 76.5% (13 out of 17) was detected by the three assays used in the three participating centres. Centres #1, #2 and #3 detected a positive HRD score in 7 out of 20 cases (35.0%), in 5 out of 20 cases (25.0%) and in 8 out of 17 cases (47.0%), respectively (tables 3 and 4). The Amoy HRD focus panel and SOPHiA DDM HRD Solution assay showed a concordance rate of 88.2% (15 out of 17 cases) for the evaluation of the HRD score. Similarly, the Amoy HRD focus panel and Oncomine HRR pathway predesigned panel showed a concordance rate of 82.4% (14 out of 17). Finally, the Oncomine HRR pathway predesigned panel and the SOPHiA DDM HRD Solution assay showed a concordance rate of 80.0% (16 out of 20 cases).
Table 4.
Schematic representation of HRD score evaluated by each participating institution by using commercially available NGS assays on a retrospective series of diagnostic HGSOC specimens
| Sample ID | Centre #1 | Centre #2 | Centre #3 | |
| Illumina NextSeq 550Dx | Illumina HiSeq 2500 | Ion Gene Studio S5 Plus System | Illumina NextSeq 550 platform | |
| SOPHiA DDM HRD solution panel | MyChoice CDx Myriad HRD | Oncomine HRR pathway panel | Amoy HRD focus panel | |
| ID_1 | POS | NEG | POS* | POS |
| ID_2 | POS | POS | NEG | POS |
| ID_3 | POS | POS | POS | POS |
| ID_4 | NEG | POS | POS | POS |
| ID_5 | NEG | NEG | NEG | NEG |
| ID_6 | NEG | NEG | NEG | NEG |
| ID_7 | NEG | NEG | NEG | NEG |
| ID_8 | NEG | NEG | NEG | NEG |
| ID_9 | NEG | NEG | NEG | NEG |
| ID_10 | NEG | NEG | NEG | NA |
| ID_11 | POS | POS | NEG | NA |
| ID_12 | NEG | NEG | NEG | NEG |
| ID_13 | POS | POS | NEG | POS |
| ID_14 | NEG | NEG | NEG | NEG |
| ID_15 | NEG | NEG | NEG | NEG |
| ID_16 | POS | POS | POS | POS |
| ID_17 | NEG | NEG | NEG | POS* |
| ID_18 | NEG | NEG | NEG | NA |
| ID_19 | POS | POS | POS | POS |
| ID_20 | NEG | NEG | NEG | NEG |
*Borderline results.
HRD, homologous recombination deficiency; HRR, homologous recombination repair; ID, identify number; NA, not assessed; NEG, negative; NGS, next-generation sequencing; POS, positive.
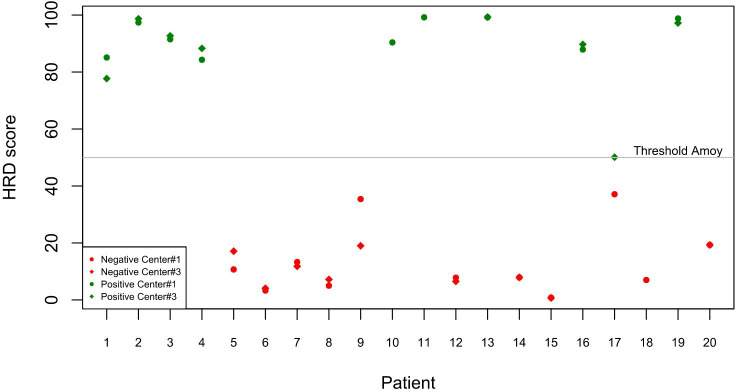
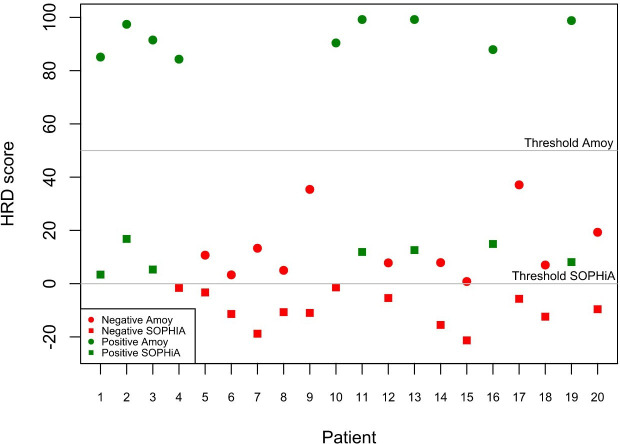
To investigate interlaboratory and interassay variability separately, we also evaluated the HRD status of the 20 EOC samples by using the same assay in two participating centres and two in-house HRD panels performed in a single institution. In detail, the use of the Amoy HRD focus panel yielded a concordance rate of 94.1% (16 out of 17 cases) between centre #1 and centre #3 (figure 2 and online supplemental table 2). Moreover, the use of both the Amoy HRD focus panel and the SOPHiA DDM HRD Solution assay by centre #1 yielded concordant HRD status results in 18 out of 20 cases, thereby showing a concordance rate of 90.0% (figure 3 and online supplemental table 3).
Figure 2.
Concordance rate between SOPHiA DDM HRD Solution assay and Amoy HRD focus between center #1 and center #3. HRD, homologous recombination deficiency.
Figure 3.
Concordance rate of center #1 by using SOPHiA DDM HRD Solution assay and Amoy HRD focus assay. HRD, homologous recombination deficiency.
jcp-2023-208852supp003.pdf (35.4KB, pdf)
High concordance rates were also seen in the classification of HRD status between the reference MyChoice CDx Myriad HRD test and the other in-house panels. In particular, we observed an OPA of 88.2% (15 out of 17) between the reference standard and the Amoy HRD focus panel; an OPA of 80.0% (16 out of 20) between the reference standard and the Oncomine HRR pathway predesigned panel and an OPA of 90% (18 out of 20 cases) between the reference standard test and the SOPHiA DDM HRD Solution assay (table 3). In addition, positive percentage agreement (PPA) and negative percentage agreement (NPA) were also evaluated between the MyChoice CDx Myriad HRD panel and the other NGS assays used to calculate HRD scores. Interestingly, a PPA and an NPA of 85.7% and 92.3%, 80.0% and 80.0%, 75.0% and 100.0% were observed between the MyChoice CDx Myriad HRD panel and the SOPHiA DDM HRD Solution assay, the Oncomine HRR pathway predesigned panel and Amoy HRD focus panel, respectively (figure 4).
Figure 4.
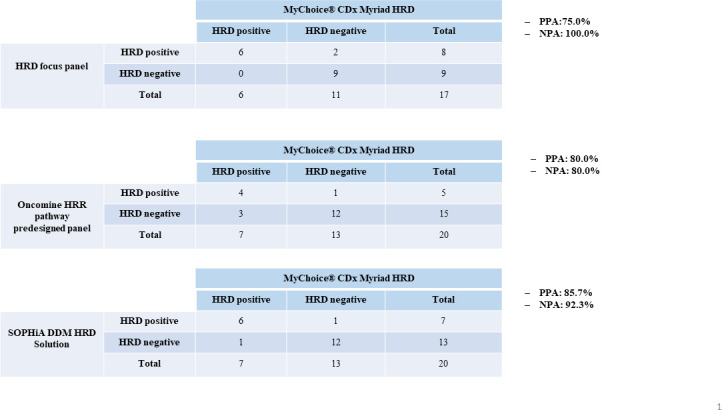
Negative percentage agreement (NPA) and positive percentage agreement (PPA) evaluated for each technical assay in comparison with gold standard externalised next-generation sequencing assay. HRD, homologous recombination deficiency; HRR, homologous recombination repair.
Discussion
HRD testing has become an essential predictive biomarker for all patients with HGSOC who may benefit from PARPi therapies.24 However, the clinical need for widespread routine HRD testing remains challenging in Italian molecular diagnostic laboratories.23 24 Although external clinically validated assays are available for routine HRD status analysis, not all patients can benefit from this approach mainly because it is costly and not reimbursable by the Italian Healthcare System. Moreover, centralised approaches show a significant rejection rate (30%–40%) due to the strict quality requirements of externalised procedures.23 In addition, external analyses require increased turnaround time, which sometimes does not reflect the actual clinical need.25 For these reasons, the identification of an alternative low-cost, highly reproducible, in-house assay for the evaluation of the HRD status in clinical samples would be an ideal alternative to guarantee this service to the highest possible number of patients. Several in-house HRD-testing strategies have been implemented, mainly involving different gene panels, bioinformatic pipelines, HRD score cut-off and sequencing platforms.26
In this retrospective study, we evaluated the HRD status in FFPE ovarian cancer samples using three commercially available in-house NGS-based assays performed in three laboratories. We observed concordance rates from 80% to 90% between in-house testing and the outsourced FDA-approved MyChoice CDx Myriad HRD test. The interassay and laboratory agreement rates were also encouraging, ranging from 71% to 94%. In addition, a reference standard DNA was also analysed by each participating centre to assess the technical validity of each in-house assay. Five out of six BRCA1/2 pathogenetic alterations covered by the DNA reference standard were detected by the SOPHiA DDM HRD Solution assay and Amoy HRD focus panel. Only the Oncomine HRR pathway predesigned panel failed to detect one of these alterations. We speculate that this discrepancy was probably due to the fact that the low quality of the homopolymeric region was not adequately covered by the bioinformatics mutation calling pipeline of the amplicon-based and semiconductor NGS platforms.26–28
Remarkably, HRD status was successfully evaluated in 100% of cases analysed with SOPHiA DDM HRD Solution assay and the Oncomine HRR pathway predesigned panel, and in 85% of cases analysed with the Amoy HRD focus panel. Three cases were excluded from the molecular analysis performed by centre #1 because of the technical capability of multiplexing of sequencing cartridges (n=9 specimens plus internal run control). The overall agreement between the three in-house HRD testing assays was of 76.5% (13 out of 17 tested samples). It was observed that all technical approaches were concordant to successfully detect 4 out of 13 (30.8%) and 9 out of 13 (69.2%) HRD-positive and HRD-negative cases, respectively (table 4). Overall agreement is strongly dependent from the difference in terms of reference range between hybridisation-based assays and Oncomine HRR pathway predesigned panel.29 30 Indeed, the technical performance of the Oncomine HRR pathway predesigned panel was slightly lower than that of the other two assays (concordance rate of 80.0%/82.4% vs 88.2%), especially for HRD-positive cases (only 20%). This difference was expected since the Oncomine HRR pathway assay is designed to detect only one (LOH) of the three genomic scars (LOH, LST, TAI), which instead are covered by the other HRD panels.29 At the sight of this critical issue, a non-negligible percentage of patients with HGSOC should be clinically administrated on the basis of false negative molecular results decreasing the population of patients with tumour that could benefit from PARPi administration. Moreover, whereas the HRD score was automatically calculated by proprietary software with the SOPHiA DDM HRD Solution and Amoy HRD focus assays, a customised analytical pipeline was implemented to define the HRD scores with the Oncomine HRR pathway predesigned panel. As of today, an automatised bioinformatics pipeline focused on the molecular fingerprint covered by NGS panels still remains an open challenge. However, some efforts are being made in this regard. For instance, Thermo Fisher recently announced that a genomic instability metric, able to calculate an HRD score in patients with HGSOC, is currently being investigated to optimise HRD calling in diagnostic specimens. Moreover, a comparative study will be performed to identify the most appropriate cut-off values for the clinical application of this approach.30
Overall, the Amoy HRD focus panel and SOPHiA DDM HRD Solution panel detected a positive HRD score in a substantial percentage of patients (from 30.0% to 45.0%). These data were consistent with the percentage of HRD-positive HGSOC evaluated in clinical trials.7 17 A high concordance rate was also observed between HRD testing performed in a single laboratory using both the Amoy HRD focus and SOPHiA DDM HRD Solution panels (90%), and in two laboratories using the same panel (94.1%). These results highlight that in-house methods display a high interassay and interlaboratory reproducibility. Moreover, the comparison with a clinically validated HRD testing assay (MyChoice Cdx Myriad HRD test) revealed a high percentage of agreement for both the Amoy HRD focus panel (88.2%) and SOPHiA DDM HRD Solution panel (90%). The majority of the interassay and interlaboratory discordances were observed in cases with an HRD score close to the predefined cut-off value of the in-house assay. This aspect may be related to the technical variability observed in real-world practice.31 Interestingly, the number of HRD-positive cases and overall agreement scores observed between in-house HRD testing and MyChoice Cdx Myriad HRD test were consistent with the percentages reported in previous studies.32 33
Although this study provides encouraging preliminary evidence for the possibility of implementing of in-house NGS-based HRD testing in routine clinical practice, it does have some main limitations. The first is that being a retrospective study based on a small number of selected cases, it did not allow us to compare the failure rate between in-house and outsourced HRD testing. The second limitation is that the small number of samples analysed yielded only preliminary results that will necessarily require further validation. Finally, we were unable to assess the clinical validity and utility of in-house HRD evaluation that was beyond the scope of this work aiming to verify the analytical reproducibility of in-house HRD testing. Therefore, building on these preliminary insights, we hope that future research will validate the clinical benefits that routine in-house HRD testing could have on patients with HGSOC.
Footnotes
Handling editor: Runjan Chetty.
@UmbertoMalapel1
MB and AI contributed equally.
Contributors: Conceptualisation: FP, UM, GT, MB and AI. Methodology: all authors. Software: all authors. Validation: all authors. Formal analysis: all authors. Investigation: all authors. Resources: all authors. Data curation: all authors. Writing—original draft preparation: FP, EG-R and NF. Writing—review and editing: all authors. Visualisation: all authors. Supervision: UM, MB, GT and AI. Project administration: UM and GT. Funding acquisition: GT. Guarantor: UM.
Funding: This study was funded by the University of Naples Federico II.
Competing interests: EG-R has relevant relationship (advisory fees, honoraria, travel accommodation and expenses, grants and non-financial support) with AstraZeneca, Exact Sciences, GlaxoSmithKline (GSK), Novartis, Roche, Thermo Fisher Scientific unrelated to the current work. MF reports research funding (to Institution) from QED, Macrophage pharma, Astellas, Diaceutics; personal honoraria as invited speaker from Roche, Astellas, AstraZeneca, Incyte, Bristol Myers Squibb (BMS), Merck Serono, Pierre Fabre, GSK, Novartis, Amgen; participation in advisory board for Amgen, Astellas, Roche, Merck Serono, GSK, Novartis, Janssen unrelated to the current work. NF reports honoraria from Merck Sharp and Dohme (MSD), Boehringer Ingelheim, Novartis, AstraZeneca and Daiichi Sankyo unrelated to the current work. CDA Consulting/Advisor: Roche, AstraZeneca, Lilly, GSK, Novartis, Pfizer, Seagen Honoraria: Novartis, Pfizer, Lilly. Research funding to the institution: Novartis, Daiichi Sankyo. Travel, accommodation, expenses: Roche, AstraZeneca, Lilly, GSK, Novartis, Celgene, Pfizer unrelated to the current work. GV reports speaking honoraria from PharmaMar, AstraZeneca, Roche, Clovis and GSK-Tesaro and travel grants from GSK-Tesaro and PharmaMar and has been part of the advisory boards of Tesaro, Amgen and PharmaMar outside of the submitted work. UM has received personal fees (as consultant and/or speaker bureau) from Boehringer Ingelheim, Roche, MSD, Amgen, Thermo Fisher Scientific, Eli Lilly, Diaceutics, GSK, Merck and AstraZeneca, Janssen, Diatech, Novartis and Hedera unrelated to the current work. GT reports personal fees (as speaker bureau or advisor) from Roche, MSD, Pfizer, Boehringer Ingelheim, Eli Lilly, BMS, GSK, Menarini, AstraZeneca, Amgen and Bayer, unrelated to the current work. MB has received honoraria for consulting, advisory role, speakers’ bureau, travel, accommodation, expenses from MSD Oncology, Roche/Genetech, AstraZeneca, Thermo Fisher Scientific, GSK and Illumina unrelated to the current work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on request to the corresponding author. All data relevant to the study are included in the article or uploaded as supplementary information All data that are publicly available and used in the writing of this article in the text and the reference list.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
The study received the European Institute Review Board approval: UID 2386. Written informed consent was acquired from all patients and documented according to 'The Italian Data Protection Authority' (http://www.garanteprivacy.it/web/guest/home/docweb/-/docwebdisplay/export/2485392). All information regarding human material was managed using anonymous numerical codes and all samples were handled in compliance with the Helsinki Declaration (http://www.wma.net/en/30publications/10policies/b3/).
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2. Seidman JD, Horkayne-Szakaly I, Haiba M, et al. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol 2004;23:41–4. 10.1097/01.pgp.0000101080.35393.16 [DOI] [PubMed] [Google Scholar]
- 3. Tsonis O, Gkrozou F, Vlachos K, et al. Upfront debulking surgery for high-grade serous ovarian carcinoma: current evidence. Ann Transl Med 2020;8:1707. 10.21037/atm-20-1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller DS, Blessing JA, Krasner CN, et al. Phase II evaluation of pemetrexed in the treatment of recurrent or persistent platinum-resistant ovarian or primary peritoneal carcinoma: a study of the gynecologic Oncology Group. J Clin Oncol 2009;27:2686–91. 10.1200/JCO.2008.19.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018;379:2495–505. 10.1056/NEJMoa1810858 [DOI] [PubMed] [Google Scholar]
- 6. Pujade-Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a brca1/2 mutation (solo2/engot-ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:1274–84. 10.1016/S1470-2045(17)30469-2 [DOI] [PubMed] [Google Scholar]
- 7. Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ariel3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949–61. 10.1016/S0140-6736(17)32440-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang S, Royer R, Li S, et al. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. Gynecol Oncol 2011;121:353–7. 10.1016/j.ygyno.2011.01.020 [DOI] [PubMed] [Google Scholar]
- 10. Dann RB, DeLoia JA, Timms KM, et al. Brca1/2 mutations and expression: response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. Gynecol Oncol 2012;125:677–82. 10.1016/j.ygyno.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 11. Ngoi NYL, Tan DSP. The role of homologous recombination deficiency testing in ovarian cancer and its clinical implications: do we need it? ESMO Open 2021;6:100144. 10.1016/j.esmoop.2021.100144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frey MK, Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecol Oncol Res Pract 2017;4:4. 10.1186/s40661-017-0039-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang M, Wu W, Wu W, et al. Parp-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 2006;34:6170–82. 10.1093/nar/gkl840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature 2001;411:366–74. 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- 15. Banerjee S, Moore KN, Colombo N, et al. 811MO maintenance olaparib for patients (PTS) with newly diagnosed, advanced ovarian cancer (OC) and a BRCA mutation (brcam): 5-year (Y) follow-up (f/u) from SOLO1. Annals of Oncology 2020;31:S613. 10.1016/j.annonc.2020.08.950 Available: 10.1016/j.annonc.2020.08.950 [DOI] [Google Scholar]
- 16. Oaknin A, Moore K, Colombo N, et al. Time to second progression (PFS2) and second subsequent therapy (TSST) for patients (PTS) with newly diagnosed, advanced ovarian cancer (OC) and a BRCA mutation (BRCAm) treated with maintenance (mt) olaparib (ola): phase III SOLO1 trial. Annals of Oncology 2019;30(suppl 5):v405. 10.1093/annonc/mdz250.003 [DOI] [Google Scholar]
- 17. Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. 10.1056/NEJMoa1911361 [DOI] [PubMed] [Google Scholar]
- 18. Timms KM, Mills GB, Perry M, et al. Comparison of genomic instability test scores used for predicting PARP activity in ovarian cancer. JCO 2020;38:1586. 10.1200/JCO.2020.38.15_suppl.1586 [DOI] [Google Scholar]
- 19. Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012;107:1776–82. 10.1038/bjc.2012.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Birkbak NJ, Wang ZC, Kim J-Y, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012;2:366–75. 10.1158/2159-8290.CD-11-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagener-Ryczek S, Merkelbach-Bruse S, Siemanowski J. Biomarkers for homologous recombination deficiency in cancer. J Pers Med 2021;11:612. 10.3390/jpm11070612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Denkert C, Romey M, Swedlund B, et al. Homologous recombination deficiency as an ovarian cancer biomarker in a real-world cohort: validation of decentralized genomic profiling. J Mol Diagn 2022;24:1254–63. 10.1016/j.jmoldx.2022.09.004 [DOI] [PubMed] [Google Scholar]
- 23. Fumagalli C, Betella I, Ranghiero A, et al. In-house testing for homologous recombination repair deficiency (HRD) testing in ovarian carcinoma: a feasibility study comparing amoydx hrd focus panel with myriad mychoicecdx assay. Pathologica 2022;114:288–94. 10.32074/1591-951X-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kohn EC, Lee JM, Ivy SP. The HRD decision--which PARP inhibitor to use for whom and when. Clin Cancer Res 2017;23:7155–7. 10.1158/1078-0432.CCR-17-2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pruneri G, De Braud F, Sapino A, et al. Next-generation sequencing in clinical practice: is it a cost-saving alternative to a single-gene testing approach? Pharmacoecon Open 2021;5:285–98. 10.1007/s41669-020-00249-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fumagalli C, Tomao F, Betella I, et al. Tumor brca test for patients with epithelial ovarian cancer: the role of molecular pathology in the era of parp inhibitor therapy. Cancers (Basel) 2019;11:1641. 10.3390/cancers11111641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loman NJ, Misra RV, Dallman TJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012;30:434–9. 10.1038/nbt.2198 [DOI] [PubMed] [Google Scholar]
- 28. Bragg LM, Stone G, Butler MK, et al. Shining a light on dark sequencing: characterising errors in ion torrent pgm data. PLoS Comput Biol 2013;9:e1003031. 10.1371/journal.pcbi.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeo ZX, Wong JCL, Rozen SG, et al. Evaluation and optimisation of indel detection workflows for ion torrent sequencing of the brca1 and brca2 genes. BMC Genomics 2014;15:516. 10.1186/1471-2164-15-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoppe MM, Sundar R, Tan DSP, et al. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst 2018;110:704–13. 10.1093/jnci/djy085 [DOI] [PubMed] [Google Scholar]
- 31. Thermo fisher introduces homologous recombination deficiency score for cancer profiling assay. n.d. Available: https://www.precisiononcologynews.com/sequencing/thermo-fisher-introduces-homologous-recombination-deficiency-score-cancer-profiling
- 32. Matthijs G, Souche E, Alders M, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet 2016;24:2–5. 10.1038/ejhg.2015.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fumagalli C, Betella I, Ranghiero A, et al. In-house testing for homologous recombination repair deficiency (hrd) testing in ovarian carcinoma: a feasibility study comparing amoydx hrd focus panel with myriad mychoicecdx assay. Pathologica 2022;114:288–94. 10.32074/1591-951X-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jcp-2023-208852supp001.pdf (35KB, pdf)
jcp-2023-208852supp002.pdf (30.4KB, pdf)
jcp-2023-208852supp003.pdf (35.4KB, pdf)
Data Availability Statement
Data are available on request to the corresponding author. All data relevant to the study are included in the article or uploaded as supplementary information All data that are publicly available and used in the writing of this article in the text and the reference list.