Abstract
Adeno-associated virus (AAV) establishes latency in infected cells by integrating into the cellular genome, with a high preference for a unique region, called AAVS1, of the human chromosome 19. The AAV proteins Rep78 and -68 are postulated to initiate the site-specific integration process by binding to a Rep binding site (RBS) in AAVS1. We provide further evidence to corroborate this model by demonstrating that the AAVS1 RBS in human cell lines is located near a DNase I hypersensitive “open” chromatin region and therefore is potentially easily accessible to Rep proteins. This open conformation is maintained in transgenic rats which carry an AAVS1 3.5-kb DNA fragment and are proficient for Rep-mediated site-specific integration. Interestingly, the core of the DNAse I hypersensitive site in AAVS1 corresponds to a sequence displaying transcriptional enhancer-like properties, suggesting that AAVS1 constitutes a transcription-competent environment. The implications of our findings for AAV physiology and gene therapy are discussed.
The human adeno-associated virus (AAV) has evolved a complex strategy to propagate in its host cells: it replicates only in the presence of helper factors, provided either by coinfecting viruses, such as adenovirus (Ad) or herpesvirus, or by genotoxic stimuli (3, 25). In the absence of these stimulatory agents, the virus integrates stably and efficiently into a specific site (called AAVS1) of the human chromosome 19 (26, 27, 45, 46), from which it is rescued following helper virus superinfection (3, 25, 45).
AAV has a linear, single-stranded DNA genome of approximately 4.7 kb which consists of two open reading frames (ORFs), namely rep and cap, and has inverted terminal repeats (ITRs) at each end that fold into a hairpin structure and function as origins of replication (3, 25, 49). By using alternate promoters (p5 and p19) and splicing, the rep gene encodes four overlapping proteins: two larger (Rep78 and Rep68) and two smaller (Rep52 and Rep40). Rep78 and Rep68 are multifunctional proteins that differ at the C terminus, but have the same DNA binding, helicase, ATPase, and endonuclease activities (3, 20, 21, 49, 59, 62). Rep78 and -68 are essential for AAV replication as well as for its integration into AAVS1 (3, 10, 45). They bind a specific sequence (Rep binding site [RBS]) in the ITRs and cleave in a strand- and site-specific manner a nearby sequence called a terminal resolution site (TRS) (6, 10, 44, 48, 58); both binding and cleavage are crucial for AAV replication (3, 25, 49). An RBS flanked by a TRS is also present in AAVS1 (54, 57), and in vitro experiments have demonstrated that Rep78 and -68 can mediate the formation in vitro of a complex between this sequence of human chromosome 19 and the AAV ITRs (57). In addition, genetic analysis performed with an Epstein-Barr virus-based episomal vector containing various fragments of AAVS1 has demonstrated that the RBS and the TRS in AAVS1 are the two cis-acting signals required for AAV site-specific integration (17, 31, 32). This evidence has led to the hypothesis that the simultaneous binding of Rep78 and -68 to the RBSs present in the ITRs and in AAVS1 is the first step in the integration mechanism (25, 31, 32). The subsequent nicking of the flanking DNA cleavage sites would then initiate the actual integration process, which occurs at variable distances from the 3′ end of the RBS and probably proceeds through a replicative type of recombination (25, 31, 32).
We speculated that a corollary to this model of the AAV integration at chromosome 19 is that the RBS in AAVS1 should be present in a chromatin region maintained in an open conformation in order to be easily accessible to Rep78 and -68 proteins. We thus decided to test this hypothesis by assessing whether the RBS in AAVS1 maps near or at a DNAse I hypersensitive site (DHSs). DHSs constitute a minor (ca. 1%) fraction of the bulk genome and represent regions either which are nucleosome free or which do not carry canonical nucleosomes and to which trans-acting factors bind (5, 12, 18, 19, 38, 53). DHSs have been functionally associated mainly with processes such as transcription, but have also been associated with replication, recombination, and chromosome segregation (18, 43, 61). They can thus be considered as exposed chromatin regions that allow access of trans-acting factors to important cis-acting DNA sequences (9, 12, 18, 61).
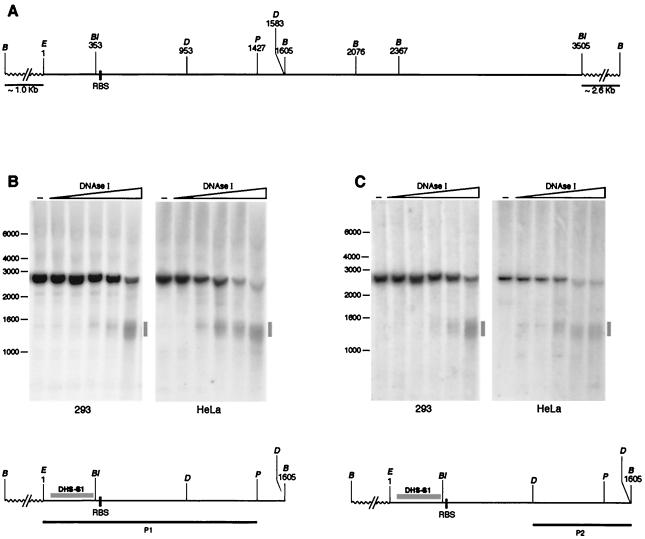
Figure 1A is a schematic representation of the region of human chromosome 19 containing the AAV integration site originally described by Kotin and colleagues (27). All Rep-mediated integrations documented so far (27, 28, 38–41, 50, 60) have been mapped within the 1.6-kb EcoRI-BamHI fragment containing the RBS (Fig. 1A), and we thus explored this region and its flanking sequences for the presence of DHSs. 293 and HeLa cells, in which Rep-mediated site-specific integration has been repeatedly reported, were selected as target cells (2, 28, 36, 39, 41, 47). DNAse I sensitivity assays were performed essentially as described previously (52, 61) with minor modifications. Briefly, 293 and HeLa cells were collected, resuspended in phosphate-buffered saline, and centrifuged for 10 min at 4°C. They were then resuspended at a density of 5 × 106 cells per ml of TNM buffer (10 mM Tris-Cl [pH 7.5], 10 mM NaCl, 3 mM MgCl2) and incubated for 10 min in ice. Swollen cells were broken in a tight-fitting Dounce homogenizer, and intact purified nuclei were recovered by centrifuging the broken cells for 10 min at 1,300 × g through a 0.25 M sucrose–TNM buffer cushion. Nuclei were then resuspended at a density of 2 × 107 per ml of digestion buffer (15 mM Tris-Cl [pH 7.5], 15 mM NaCl, 3 mM MgCl2, 60 mM KCl, 0.25 mM sucrose, 0.5 mM dithiothreitol, 1 mM, phenylmethylsulfonyl fluoride). Increasing concentrations of DNAse I were then added to aliquots (100 μl each) of the nuclear suspensions. After 10 min of incubation at 37°C, the reactions were stopped by adding 400 μl of stop solution (15 mM EDTA [pH 8.0], 0.6% sodium dodecyl sulfate). Forty micrograms of proteinase K was added to each suspension, and samples were incubated for 12 h at 37°C. Genomic DNA was purified by phenol extraction and ethanol precipitation. After treatment with RNase, DNA samples were digested to completion with the appropriate enzymes, electrophoresed on a 0.8% agarose gel, and blotted onto nylon membranes. First, we checked whether DHSs mapped in the 2.6-kb BamHI fragment containing the RBS (Fig. 1A). In the first experiment, BamHI-digested DNA was hybridized with probe P1 (EcoRI-PstI fragment): as shown in Fig. 1B, a DHS (called DHS-S1) was detectable in both cell lines as a smear centered at 1,400 bp and approximately spanning the 1,250- to 1,550-bp region. This approximately mapped the DHS near to the RBS, but since P1 was not located at either end of the 2.6-kb BamHI fragment, we could not discriminate whether DHS-S1 was mainly located 5′ or 3′ of the RBS. To further address this point, we performed a canonical indirect end-labelling experiment by hybridizing BamHI-digested DNA with a different probe (probe P2; DraIII-DraIII fragment) that adjoins exactly the 3′ end of the region analyzed (Fig. 1C). As shown in Fig. 1C, also in this case, a smear approximately spanning the 1,250- to 1,550-bp region was detected, while no signal was evident at a molecular weight higher than 1,600 bp. This pattern mapped the “core” of the DHS downstream of the EcoRI site and mainly, if not exclusively, 5′ to the RBS. Taken together, the results shown in Fig. 1B and C localize, within the sensitivity limits of the end-labelling technique, the core of DHS-S1 in an ∼300-bp-long region (spanning approximately nucleotides [nt] 50 to 350 of the AAVS1 sequence represented in Fig. 1A) located 5′ and in near proximity to the RBS (Fig. 1B and C). Notably, control DNase I digestions of the cloned and purified AAVS1 fragment failed to detect any DHS (data not shown), thus ruling out that the DHS-S1 sequence is hypersensitive to DNase I for reasons unrelated to chromatin structure.
FIG. 1.
A DHS maps near the RBS in AAVS1. (A) Schematic representation of the region of AAVS1 analyzed. The restriction map is limited to those sites that were relevant for our studies BamHI (B), BglI (Bl), EcoRI (E), PstI (P), and DraIII (D). The bold line represents part of the published AAVS1 sequence (27), with the nucleotide numbering starting at the indicated EcoRI site (position +1) and ending at the BglI site at position +3505. The 5′ and 3′ sequences flanking the 1- to -3505 region are represented by dotted lines. The BamHI sites present in these flanking regions are also indicated. (B and C) Nuclei isolated from 293 or HeLa cells were digested with increasing concentrations (1, 2, 10, 20, and 40 U/ml) of DNAse I (Boehringer Mannheim, catalog no. 776785). DNA was then isolated, digested with the indicated restriction enzymes, and incubated with probe P1 (B) or P2 (C). The two probes were derived from plasmid pRVK (gift of K. Berns, Cornell Medical School, Ithaca, N.Y.), spanning nt 1 to 3525 of AAVS1 (27). P1 was obtained as an EcoRI-PstI fragment and spans nt 1 to 1427; P2 was a DraIII fragment spanning nt 953 to 1583. The diagram below each autoradiogram shows the restriction fragment being analyzed, the identity and position of each hybridization probe, the position of the RBS, and the location of DHS-S1 as identified with each specific probe. The positions of the molecular weight standards (in kilobases) are shown to the left of each gel. −, lanes loaded with genomic DNA from untreated nuclei.
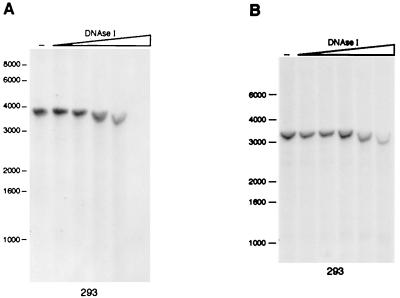
We next analyzed the AAVS1 segment from the BglI site at position 353 to the extreme 3′ BamHI site (Fig. 1A). In the first assay, DNAse I-treated genomic DNA of 293 cells was digested with BamHI restriction enzyme and hybridized with probe P3 (BamHI-BglI fragment; Fig. 2A), which detected the 3′-terminal 3.7 kb of the AAVS1 region (Fig. 1A). As shown in Fig. 2A, no DHS was detected. Secondly, DNase I-treated genomic DNA of 293 cells was digested with BglI restriction enzyme, which released a 3.2-kb fragment (Fig. 2B) that contains the RBS at a 20-bp distance from its 5′ extremity and overlaps with the 2.6-kb BamHI fragment analyzed in the experiments shown in Fig. 1B and C. When this DNA was hybridized with probe P4 (BglI-PstI fragment; Fig. 2B), no hypersensitive sites were detectable: this confirmed that, within the sensitivity limits of the assay, DHS-S1 is indeed centered in a region located 5′ to the RBS. In conclusion, in the 7.1-kb BamHI-BamHI region analyzed (Fig. 1A), only one DHS site (DHS-S1) was identified, and this mapped in close proximity to the RBS (Fig. 1B and C).
FIG. 2.
No DHSs are present in AAVS1 downstream of the RBS. Nuclei isolated from 293 cells were digested with DNAse I, purified, digested with the indicated restriction enzymes, and incubated with probe P3 (A) or P4 (B). P3 was a BamHI-BglI fragment spanning nt 2367 to 3505; P4 was a BglI-PstI fragment extending from nt 353 to 1427. The restriction fragment being analyzed and the identity and position of each hybridization probe are shown. The positions of the molecular weight standards (in kilobases) are shown to the left of each gel. −, lanes loaded with genomic DNA from untreated nuclei.
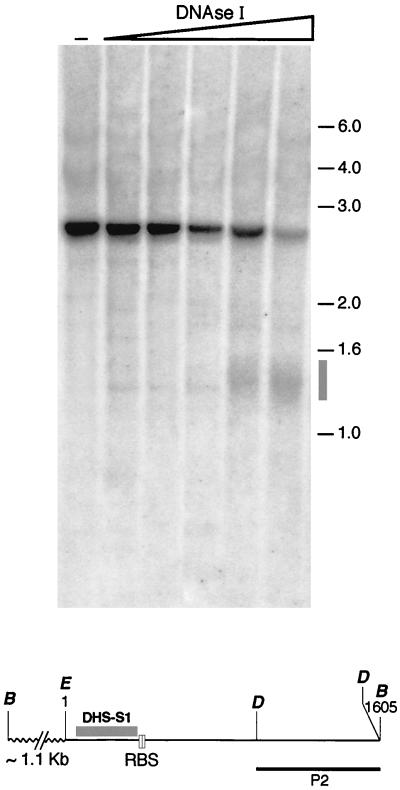
The chromatin configuration associated with functionally important DHSs is often dominant over the surrounding regions and is maintained in different chromosomal environments (11, 13, 15, 18, 35, 36, 51). The possibility to test whether this rule also applies to DHS-S1 was offered by recently generated transgenic rats which carry the 3.5-kb region of human AAVS1 and represent a valid model for studying AAV site-specific integration (42). Intact nuclei from transgenic primary rat fibroblasts, in which Rep-mediated integration at AAVS1 has been documented (42), were isolated and treated with increasing concentrations of DNase I. Southern blot analysis of BamHI-digested genomic DNA demonstrated that DHS-S1 at human AAVS1 was maintained even in the context of the rat genome (Fig. 3). This result strongly suggests that the DHS-S1 sequence imposes a position-independent chromatin configuration.
FIG. 3.
DHS-S1 mapping in the genome of transgenic rats carrying the AAVS1 3.5-kb DNA fragment. Primary fibroblasts from ear biopsies of transgenic rats (line 15) were isolated as described previously (42). Isolated nuclei were incubated with increasing concentrations of DNAse I as described in the legend to Fig. 1. Genomic DNA was then purified, digested with restriction enzyme BamHI, transferred to nylon membranes, and hybridized with probe P2 (see also Fig. 1C). The fine structure of the human AAVS1 region integrated in the genome of transgenic rat line 15 has been previously described (42). At the bottom of the figure is a schematic representation of the BamHI restriction fragment recognized by the probe P2: the bold line represents part of the human AAVS1 sequence with the nucleotide numbering starting at the indicated EcoRI site (position +1) and ending at the BamHI site at position +1605 (27). The positions of the molecular weight standards (in kilobases) are indicated. −, genomic DNA from untreated nuclei.
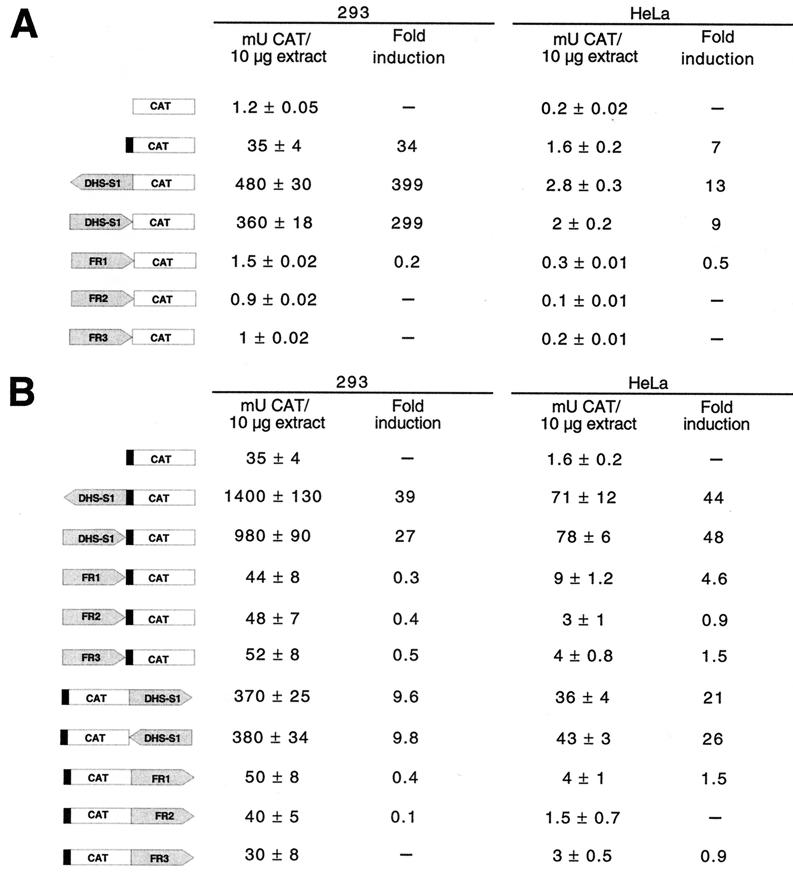
DHSs mainly reside at cis-acting DNA elements that function in gene regulation, such as promoters and enhancers (12, 18, 29, 53): we thus tested whether the identified DHS in AAVS1 had transcription regulatory properties. The core region of DHS-S1 was cloned in two opposite orientations upstream of a promoterless reporter chloramphenicol acetyltransferase (CAT) gene (Fig. 4A) and transfected in HeLa and 293 cells. A plasmid containing the CAT gene downstream of the simian virus 40 (SV40) promoter (pCAT 3-promoter vector; Promega) was used in control experiments. As shown in Fig. 4A, DHS-S1 stimulated transcription of the CAT gene in both cell lines. Activation was stronger in 293 cells (∼300- to -400-fold activation) than in HeLa cells (∼10- to 13-fold induction) and was mainly not orientation dependent. To verify that the transcriptional activity of DHS-S1 was not due to any artifact in the constructs, we tested the transcriptional properties of three randomly selected fragments of AAVS1 adjacent but not contiguous to DHS-S1. These three fragments, FR1, FR2, and FR3, spanning nt 1313 to 1616, 2077 to 2371, and 2633 to 2927, respectively, were cloned upstream of the promoterless CAT gene and analyzed in transient assays. As indicated in Fig. 4A, none of these three fragments stimulated transcription in 293 and HeLa cells, thus validating the results obtained with DHS-S1.
FIG. 4.
Transcriptional activity of DHS-S1. (A) Structure and activity of constructs containing DHS-S1 cloned upstream of the CAT gene. The whole DHS-S1 was excised as an SmaI-SmaI fragment (nt 18 to 353 of AAVS1) from plasmid pRVK and cloned in both orientations (indicated by the direction of the arrows) upstream of the intron and the CAT gene into the SmaI site of pCAT 3-basic vector (Promega). Black boxes represent the SV40 promoter. The promoterless plasmid is the pCAT 3-basic vector (Promega), and the control plasmid containing the SV40 promoter is the pCAT 3-promoter vector (Promega). Regions FR1 (nt 1313 to 1616 of AAVS1) and FR3 (nt 2633 to 2927 of AAVS1) were amplified by PCR with plasmid pRVK as a substrate and oligonucleotides carrying SmaI sites at either end. The resulting fragments were digested with restriction enzyme SmaI and cloned into the SmaI site of the pCAT 3-basic vector. Region FR2 (nt 2077 to 2371 of AAVS1) was excised from plasmid pRVK as a BamHI-flanked fragment, filled in by treatment with Klenow enzyme, and inserted into the SmaI site of pCAT 3-basic vector. Ten micrograms of each plasmid was transfected as described previously by calcium phosphate precipitation into 293 and HeLa cells (1). At 15 h posttransfection, the medium was changed: after an additional 24 h, cells were collected and CAT activity in cell extracts was measured by using the Quan-T-CAT assay system (Amersham, Pharmacia). CAT activities were normalized against the luciferase activity derived from a cotransfected cytomegalovirus-luciferase plasmid (1). Results are expressed as milliunits of CAT enzyme present in 10 μg of cell extracts and as fold induction with respect to the promoterless pCAT 3-basic vector. The data are means ± standard deviations of two independent experiments in which two separate plasmid preparations were used. (B) Enhancer activity of DHS-S1 in 293 and HeLa cells. DHS-S1 was cloned in both orientations (represented by the direction of the arrows) into the pCAT 3-promoter vector either into the SmaI site upstream of the SV40 promoter or into the BamHI site downstream of the CAT gene. Similarly, FR1, FR2, and FR3 were cloned in one orientation either into the SmaI site or into the BamHI site of pCAT 3-vector. Transfections were performed, and CAT activities were measured and normalized as described in the legend to Fig. 4A. Results are expressed as milliunits of CAT enzyme in 10 μg of cell extracts; fold induction was calculated with respect to that of the pCAT 3-promoter vector. The data are means ± standard deviations of three independent experiments in which at least two different plasmid preparations were used.
The finding that DHS-S1 acted essentially in an orientation-independent manner suggested that this region could be a transcriptional enhancer. To check this point, DHS-S1 was cloned into pCAT 3-promoter vector either 5′ to the SV40 promoter or 3′ to the polyadenylation site of the CAT gene. Control plasmids were generated by using FR1, FR2, and FR3 fragments. The various constructs are represented in Fig. 4B, which also shows their behavior in transfected 293 and HeLa cells. In both cells, DHS-S1 markedly enhanced the SV40 promoter activity when cloned either upstream of the SV40 promoter or downstream of the CAT gene, although the stimulation was greater from an upstream position (20- to 50-fold induction) than from a downstream position (10- to 25-fold induction) (Fig. 4B). The enhancer effect was not orientation dependent (Fig. 4B) and not artifactual, in that control fragments FR1, FR2, and FR3 did not behave as transcriptional enhancers (Fig. 4B). In conclusion, DHS-S1 exhibited the properties of a true enhancer in that it was effective in both orientations and activated the SV40 promoter either from a proximal site (upstream of the promoter) or from a remote site (downstream of the CAT gene) (4).
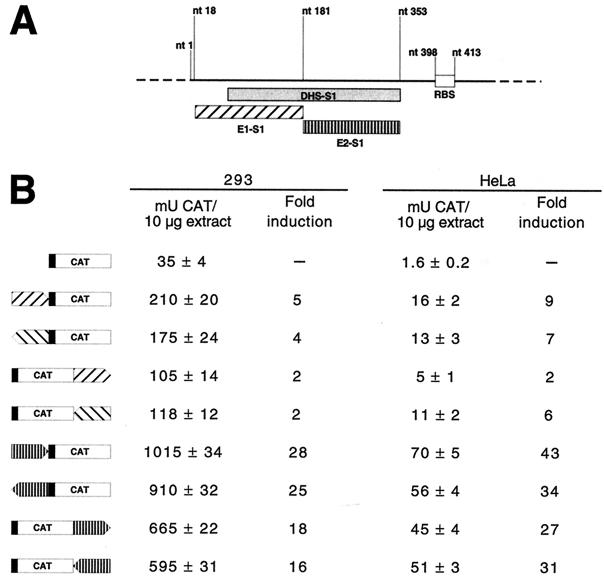
To further characterize DHS-S1, we also analyzed the enhancer properties of two deletion mutants: E1-S1, spanning nt 18 to 181 of the published AAVS1 sequence (27), and E2-S1, which runs from nt 181 to 353 (Fig. 5A). Each fragment was cloned in both possible orientations in the plasmid pCAT 3-promoter—either 5′ or 3′ to the CAT gene—and the various constructs were transfected into 293 and HeLa cells (Fig. 5B). As shown in Fig. 5B, E2-S1 displayed almost the same enhancer activity as the full-length DHS-S1 (compare Fig. 4B and Fig. 5B): however, E1-S1 also was still active, thus suggesting that cis-acting signals are at least partially redundant in DHS-S1. Taken together, these results correlate well with the presence of potential binding sites for several transcription factors (such as AP-1, AP-2, Sp1, and CREB) scattered along the entire hypersensitive site, but mainly concentrated (in particular the Sp1 sites) (27) in the 3′ half of DHS-S1, as revealed by DHS-S1 sequence analysis using the MAP program and the Transcription Factor Database (Wisconsin Package, version 10.0; Genetics Computer Group, Madison, Wisc.) (16).
FIG. 5.
Enhancer activity of DHS-S1 is mainly located in the 181 to 353 region. (A) Graphic representation of the two DHS-S1 subregions, E1-S1 (nt 18 to 181) and E2-S1 (nt 181 to 353), whose transcriptional activity was tested in transient transfection assays. (B) Enhancer activity of E1-S1 and E2-S1 in 293 and HeLa cells. E1-S1 and E2-S1 were obtained as PCR fragments by using plasmid pRVK (see legend to Fig. 1) as a template, and cloned in both orientations (indicated by the direction of the arrows) into plasmid pCAT 3-promoter either upstream of the SV40 promoter or downstream of the CAT gene (see the legend to Fig. 4). Transfections, CAT assays, and normalization of the results were performed as described in the legend to Fig. 4. Results are expressed as milliunits of CAT per 10 μg of cell extracts and as fold induction with respect to the pCAT 3-promoter vector. The data are means ± standard deviations of four independent experiments in which two different plasmid preparations were used.
In this report, we have demonstrated that AAV site-specific integration into chromosome 19 takes place near a DHS and in an apparently transcriptionally active region. It is known that DHSs associated with enhancer functions confer chromatin accessibility to proximal DNA regions (12, 22, 23). Therefore, the finding that DHS-S1 maps in near proximity to the RBS (at less than 1 nucleosomal DNA unit length from the mapped 3′ end of the DHS) strongly suggests that the RBS is indeed maintained in an open chromatin conformation and hence is easily accessible to Rep proteins in vivo (9, 12, 53, 61). This further corroborates the widely accepted model for Rep-mediated site-specific integration: the specificity of AAV integration might thus be dictated by the DNA sequence at AAVS1 and facilitated by the chromatin configuration in that region (17, 31, 32, 57). This hypothesis is further supported by results obtained by Kotin and colleagues using episomal integration substrates: in that case, the addition of an AAVS1 segment corresponding to DHS-S1 led to an integration frequency threefold higher than that observed with a substrate containing only the RBS and the TRS (31, 32). Interestingly, one region (from nt 209 to 326 of AAVS1) overlapping with DHS-S1, and in particular with the E2-S1 segment, has been associated with episomal DNA rearrangements by Linden et al. (31, 32). This region carries the motif M26, which has been characterized as an enhancer of meiotic recombination in fission yeast (14, 24, 55). A role for this sequence in AAV integration has only been hypothesized, but our finding that it co-maps with a nuclease hypersensitive site, much as has been observed in Schizosaccharomyces pombe, is of interest and supports this conjecture (33, 34).
Kotin et al. reported the presence of an open ORF which maps in the AAVS1 sequence from nt 1620 to nt 2318, potentially codes for a 95-amino-acid-long polypeptide (no match with known protein sequences), and is expressed in human foreskin fibroblasts at low levels detectable only by reverse transcription-PCR (27). So far, this is the only report describing a transcriptional activity in AAVS1. At this stage, we do not know whether the DHS-1 is involved in the transcriptional regulation of this potential ORF or of any other gene (4, 7, 12). However, the presence of DHS-S1, as well as its enhancer-like properties, suggests, although it does not prove, that AAVS1 constitutes a transcriptionally competent environment (12, 29, 53, 56). AAV perpetuates its genetic information by establishing latency in cells: genotoxic agents or helper virus infections activate transcription of integrated AAV genes, and this leads to recovery and replication of the AAV genome (3). It is thus tempting to hypothesize that AAVS1 has evolved as a preferential site for AAV integration, also because it locates in a region of the human genome which is proficient for transcription and allows stably integrated exogenous genes to be reactivated. On the other side, the presence of DHS-S1 must be reconciled with the latency of AAV, whose genome is currently believed to be transcriptionally silent when integrated at AAVS1 (3). More work will thus be required to clarify whether and how DHS-S1 affects transcription from the integrated AAV genome during the different phases of its life cycle.
Our results are also of interest for gene therapy. It has been recently demonstrated that either Rep78 or Rep68 provided in trans promotes preferential integration at AAVS1 of a transgene flanked by the AAV ITRs (2, 39, 47, 50). Several types of viral and nonviral delivery vectors which incorporate Rep78 and -68 and ITR-flanked transgenes are being designed and tested (28, 37, 40, 41). In this connection, the identification of a DHS in AAVS1 represents a further step toward the functional characterization of AAVS1. In particular, the presence of DHS-S1 indicates that AAVS1 should indeed constitute a proper environment for transgene expression: interestingly, since DHS-S1 maps 5′ to the hot-spot integration sites in AAVS1, transgene integration should not lead to disruption of the DHS-S1 sequence (27, 28, 38–41, 50, 60). This point will clearly need to be addressed in consideration of DHS-S1 possibly playing an important role in cell physiology, especially because rearrangements at AAVS1 have been frequently associated with Rep-mediated integration (2, 37, 39, 41, 50).
Acknowledgments
We thank Gennaro Ciliberto for critically reading the manuscript. We also gratefully acknowledge Janet Clench for editing the manuscript and Manuela Emili for work with the graphics.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Balagué C, Kalla M, Zhang W-W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I, Linden R M. The cryptic life style of adeno-associated virus. Bioessays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood E, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 5.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 6.Brister J R, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulger M, Groudine M. Looping versus linking: toward a model of long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 8.Bungert J, Tanimoto K, Patel S, Liu Q, Fear M, Engel J D. Hypersensitive site 2 specifies a unique function within the human β-globin locus control region to stimulate globin gene transcription. Mol Cell Biol. 1999;19:3062–3072. doi: 10.1128/mcb.19.4.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartwright I L, Cryderman D E, Gilmour D S, Pile L A, Wallrath L L, Weber J A, Elgin S C R. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- 10.Chiorini J A, Wiener S M, Owens R A, Kyöstiö S R M, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis J, Tan-Un K C, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position variegation effect. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 14.Fox M E, Virgin J B, Metzger J, Smith G R. Position- and orientation-independent activity of the Schizosaccharomyces pombe meiotic recombination hot spot M26. Proc Natl Acad Sci USA. 1997;94:7446–7451. doi: 10.1073/pnas.94.14.7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganss R, Montoliu L, Monaghan A P, Schutz G. A cell specific enhancer far upstream of the mouse tyrosinase gene confers high level and copy number-related expression in transgenic mice. EMBO J. 1994;13:3083–3093. doi: 10.1002/j.1460-2075.1994.tb06607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh D. Status of the transcription factor database (TFD) Nucleic Acids Res. 1993;21:3117–3118. doi: 10.1093/nar/21.13.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross D S, Garrard W T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 19.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core hystone hyperacetylation co-maps with generalized DNAse I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im D-S, Muzyczka N. The AAV origin-binding protein Rep68 is an ATP dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 21.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenuwein T, Forrester W C, Qiu R-G, Grosschedl R. The immunoglobulin μ enhancer core establishes local factor access in nuclear protein independent of transcriptional stimulation. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T, Forrester W C, Fernández-Herrero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 24.Kon N, Krawchuk M D, Warren B G, Smith G R, Wahls W P. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotin R M. Prospects for the use of adeno-associated virus as a vector for human gene therapy. Hum Gene Ther. 1994;5:793–891. doi: 10.1089/hum.1994.5.7-793. [DOI] [PubMed] [Google Scholar]
- 26.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, Laughlin S M, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamartina S, Roscilli G, Rinaudo D, Delmastro P, Toniatti C. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Harju S, Peterson K R. Locus control regions: coming of age at a decade plus. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 30.Linden R M, Berns K I. Site-specific integration by adeno-associated virus: a basis for a potential gene-therapy vector. Gene Ther. 1997;4:4–5. doi: 10.1038/sj.gt.3300357. [DOI] [PubMed] [Google Scholar]
- 31.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno K I, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas A. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc Natl Acad Sci USA. 1998;95:87–89. doi: 10.1073/pnas.95.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz B D, Cado D, Chen V, Diaz P W, Winoto A. Adjacent DNA elements dominantly restrict the ubiquitous activity of a novel chromatin-opening region to specific tissues. EMBO J. 1997;16:5037–5045. doi: 10.1093/emboj/16.16.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortiz B D, Cado D, Winoto A. A new element within the T-cell receptor α locus required for tissue-specific locus control region activity. Mol Cell Biol. 1999;19:1901–1909. doi: 10.1128/mcb.19.3.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palombo F, Monciotti A, Recchia A, Cortese R, Ciliberto G, La Monica N. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus–adeno-associated virus vector. J Virol. 1998;72:5025–5034. doi: 10.1128/jvi.72.6.5025-5034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 39.Pieroni L, Fipaldini C, Monciotti A, Cimini D, Sgura A, Fattori E, Epifano O, Cortese R, Falombo F, La Monica N. Targeted integration of adeno-associated virus-derived plasmid in transfected human cells. Virology. 1998;249:249–259. doi: 10.1006/viro.1998.9332. [DOI] [PubMed] [Google Scholar]
- 40.Recchia A, Parks R J, Lamartina S, Toniatti C, Pieroni L, Palombo F, Ciliberto G, Graham F L, Cortese R, La Monica N, Colloca S. Site-specific integration mediated by a hybrid adenovirus/adenoassociated virus vector. Proc Natl Acad Sci USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinaudo D, Lamartina S, Roscilli G, Ciliberto G, Toniatti C. Conditional site-specific integration into human chromosome 19 using a ligand-dependent chimeric AAV/rep protein. J Virol. 1999;74:281–294. doi: 10.1128/jvi.74.1.281-294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzuto G, Gorgoni B, Cappelletti M, Lazzaro D, Gloaguen I, Poli V, Sgura A, Cimini D, Ciliberto G, Cortese R, Fattori E, La Monica N. Development of animal models for adeno-associated virus site-specific integration. J Virol. 1999;73:2517–2526. doi: 10.1128/jvi.73.3.2517-2526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 44.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samulski R J. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Genet Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 46.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. . (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 48.Snyder R O, Im D-S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanimoto K, Liu Q, Bungert J, Engel J D. The polyoma virus enhancer cannot substitute for DNAseI core hypersensitive sites 2–4 in the human beta-globin region. Nucleic Acids Res. 1999;27:3130–3137. doi: 10.1093/nar/27.15.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuan D, London I M. Mapping of DNAse I-hypersensitive sites in the upstream DNA of human embryonic ɛ-globin gene in K562 leukemia cells. Proc Natl Acad Sci USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Udavrdy A. Dividing the empire: boundary chomatin elements delimit the territory of enhancers. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wahls W P, Smith G R. A heterodimeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 56.Walters M C, Fierung S, Eidemiller J, Magis W, Groudine M, Martin D I K. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weitzman M D, Kyöstiö S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weitzman M D, Kyöstiö S R M, Carter B J, Owens R A. Interaction of wild-type and mutant adeno-associated virus (AAV) Rep proteins on AAV hairpin DNA. J Virol. 1996;70:2440–2448. doi: 10.1128/jvi.70.4.2440-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1996;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaret K. In vivo analysis of chromatin structure. Methods Enzymol. 1999;304:612–626. doi: 10.1016/s0076-6879(99)04036-7. [DOI] [PubMed] [Google Scholar]
- 62.Zhou X, Zolotukhin I, Im D-S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activity. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]