Abstract
Background
The BCR::ABL1 is a hallmark of chronic myeloid leukemia (CML) and is also found in acute lymphoblastic leukemia (ALL). Most genomic breaks on the BCR side occur in two regions - Major and minor - leading to p210 and p190 fusion proteins, respectively.
Methods
By multiplex long-distance PCR or next-generation sequencing technology we characterized the BCR::ABL1 genomic fusion in 971 patients (adults and children, with CML and ALL: pediatric ALL: n = 353; pediatric CML: n = 197; adult ALL: n = 166; adult CML: n = 255 patients) and designed “Break-App” web tool to allow visualization and various analyses of the breakpoints. Pearson’s Chi-Squared test, Kolmogorov-Smirnov test and logistic regression were used for statistical analyses.
Results
Detailed analysis showed a non-random distribution of breaks in both BCR regions, whereas ABL1 breaks were distributed more evenly. However, we found a significant difference in the distribution of breaks between CML and ALL. We found no association of breakpoints with any type of interspersed repeats or DNA motifs. With a few exceptions, the primary structure of the fusions suggests non-homologous end joining being responsible for the BCR and ABL1 gene fusions. Analysis of reciprocal ABL1::BCR fusions in 453 patients showed mostly balanced translocations without major deletions or duplications.
Conclusions
Taken together, our data suggest that physical colocalization and chromatin accessibility, which change with the developmental stage of the cell (hence the difference between ALL and CML), are more critical factors influencing breakpoint localization than presence of specific DNA motifs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02053-4.
Keywords: Acute lymphoblastic leukemia, Chronic myeloid leukemia, Genomic breakpoints, BCR, ABL1
The BCR::ABL1 fusion gene is not only a hallmark of chronic myeloid leukemia (CML) but is also present in a proportion of patients (3–5% in children, 20–30% in adults) with acute lymphoblastic leukemia (ALL). The t(9;22)(q34;q11) translocation, recognized at the chromosomal level as “Philadelphia chromosome”, involves DNA breaks in large, mostly intronic regions of the BCR and ABL1 genes. The majority of breakpoints on the BCR side occur in two “breakpoint cluster regions” – “minor” (between exons 1 and 2; ~ 71.5 kilobase pairs [kbp], resulting in the p190 fusion protein, prevalent in ALL and scarce in CML) and “Major” (between exons 13 and 15; ~ 2.9 kbp, resulting in the p210 fusion protein, less frequent in ALL and almost exclusive in CML). On the ABL1 side, breakpoints are mostly localized between exon 1 and exon 2 (~ 140 kbp), however, breaks upstream of the ABL1 have also been described [1, 2].
Numerous studies have been published focusing on aberrant expression and downstream signaling of the BCR::ABL1 fusion gene/protein. However, due to the large intronic areas where the breakpoints occur, only a few papers have been published focusing on the primary structure of the BCR::ABL1 fusions. Moreover, to our knowledge, only one study included (a limited number of) patients with the minor form of BCR::ABL1 fusion (n = 25) and suggested RAG involvement in the double-strand break initiation in a subset of ALL [2]. Analysis focused on the localization of breakpoint sites in the Major BCR area showed a bimodal distribution of breakpoints [3, 4]. In contrast, the distribution of breakpoints in ABL1 was shown to be more uniform; however, some studies suggested subtle differences in distribution with respect to the gender or age of patients [3, 5].
Several analyses concerned the localization of breakpoints within BCR and ABL1 genes with respect to the presence of interspersed repeats (IR), recombination signal sequences for RAG-recombinase (RSS), or motifs known to mediate DNA breaks (e.g. cleavage sites for topoisomerases, immunoglobulin switch sequences, etc.) [1–4]. The primary structure of fusion sequences – mostly represented by short homologies, blunt-end connections or short insertions of random nucleotides – has led to the hypothesis that non-homologous end joining (NHEJ) is probably responsible for the BCR and ABL1 fusion [1, 4, 6].
In the present study, to definitively assess these factors and analyze genomic fusions in an extensive and fully representative cohort, we searched for and identified BCR::ABL1 genomic breakpoints in 971 patients with BCR::ABL1-positive ALL (n = 519) and CML (n = 452). Our cohort includes both pediatric (BCR::ABL1-positive ALL: n = 353; CML: n = 197) and adult (BCR::ABL1-positive ALL: n = 166; CML: n = 255) patients, with part of the sequences published previously [3, 7–9]. In general, all included patients were sent to the BCR::ABL1 genomic breakpoint identification after the presence of the fusion was already revealed by routine diagnostics - cytogenetics and/or reverse-transcriptase (RT-) PCR. Hence, our cohort might be slightly negatively biased towards very unusual fusions, missed by routine diagnostics – while our NGS approach with probes covering both BCR and ABL1 breakpoint regions is capable to detect also unusual breakpoints, such cases (e.g. micro-BCR::ABL1) might be underrepresented in our study.
The BCR::ABL1 breakpoints were characterized originally by long-distance PCR (n = 427) and later by target enrichment NGS (n = 544) with custom-designed probes covering the following areas (according to GRCh38/hg38): minor BCR - chr22:23,180,958 − 23,254,000; Major BCR - chr22:23,289,491 − 23,292,664; micro BCR - chr22:23,311,732 − 23,313,035; (EXOSC2)/ABL1 - chr9:130,699,582 − 130,855,101. While breakpoint identification by PCR yielded some negative results (the proportion of unsuccessful attempts to obtain a genomic breakpoint was approximately 10–15%), the success rate by NGS was close to 100% (with rare failures attributable mainly to poor DNA quality or very low [< 5%] blast percentage). For more details see Additional methods.
Of the theoretically expected 1942 genomic breakpoints in BCR and ABL1 genes, we identified exact position for 1935 breakpoints (396, 575 and 964 breakpoints in the minor BCR, Major BCR and ABL1, respectively). In six patients examined in the early part of the study, the ABL1 breakpoints were not precisely characterized (three fusions with a breakpoint upstream of ABL1 and three fusions with a large ABL1 inversion) and one patient had a repetitive sequence that prevented reliable precise identification of the ABL1 breakpoint position. The breakpoints were mostly located in introns (n = 1852; 96% of all identified breakpoints); however, 40 patients (4%) had genomic breakpoints upstream of ABL1 (n = 8 in EXOSC2 gene; n = 32 in intergenic area between EXOSC2 and ABL1), and 45 patients had breakpoints in exons (ABL1 exon 1, n = 6; ABL1 exon 1b, n = 2; ABL1 exon 2, n = 1; BCR exon 2, n = 1; BCR exon 14, n = 25; BCR exon 15, n = 10). The breakpoint sites covered almost the complete breakpoint areas with only a few gaps larger than 1kbp, where no breakpoint was detected (minor BCR: n = 8; 1.1–3.3 kbp; ABL1: n = 7; 1.1–2.3 kbp). Where analyzed, the transcript variant (minor vs. Major) always corresponded to the genomic fusion localization. As already described, in some patients with Major BCR::ABL1 fusion, low levels of minor BCR::ABL1 transcript are also expressed due to alternative splicing; only the Major BCR::ABL1 genomic breakpoint was identified in all such cases. For all breakpoint positions and basic characteristics of the patients see Additional Table 1.
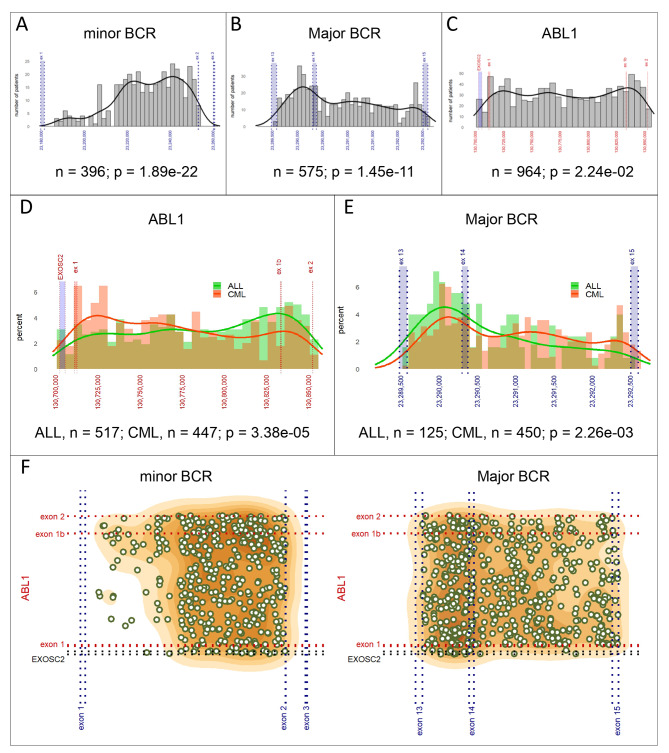
The uniformity test performed on breakpoint distribution within the minor (n = 396) and Major (n = 575) BCR areas revealed a non-random pattern (p = 1.89e-22 and p = 1.45e-11, respectively; see Fig. 1A, B) with breakpoints accumulating in the 3’ end area of the intron 1 (minor BCR) and in intron 13 (Major BCR). Within the ABL1 area, breakpoint sites were distributed more evenly (p = 2.24e-02; see Fig. 1C).
Fig. 1.
BCR and ABL1 breakpoint distribution. Breakpoint distribution within the minor BCR (A), Major BCR (B) and ABL1 (C) breakpoint areas; comparison of breakpoints distribution between ALL and CML patients within ABL1 (D) and Major BCR (E) areas; overall distribution of breakpoints and the relationship between breakpoint location on the ABL1 side and in the minor and Major BCR regions. Gene coordinates are given according to GRCh38/hg38. The uniformity of breakpoint site distribution was tested using Pearson’s Chi-Squared test, comparison of breakpoint distribution between the groups was tested using Kolmogorov-Smirnov test. Images adapted from the “Break-App” web tool
Importantly, the breakpoint distribution within the ABL1 gene differed significantly between CML and ALL patients (p = 3.38e-05; see Fig. 1D), with a higher accumulation of breakpoints in the 5’ part of the ABL1 breakpoint area in CML and in the 3’ part of the area in ALL. This difference in breakpoint distribution was not driven by the type of fusion (minor vs. Major BCR) as the difference between CML and ALL was still apparent when only Major BCR::ABL1-positive patients were analyzed (445 CML and 123 ALL; p = 2.11e-02). Moreover, the different breakpoint distribution between CML and ALL patients was also evident in the Major BCR area (450 CML and 125 ALL; p = 2.26e-03, see Fig. 1E).
Using logistic regression, several tests were performed to evaluate the influence of sex, age and/or type of fusion (minor vs. Major BCR fusion [for ABL1 breakpoints in ALL patients]). Only age at diagnosis was found to moderately influence the breakpoint distribution within the minor BCR area in ALL (≤ 16 [n = 266] vs. > 16 [n = 127] years; p = 7.89e-03; see Additional Fig. 1). The overall distribution of the breakpoints is shown in Fig. 1F.
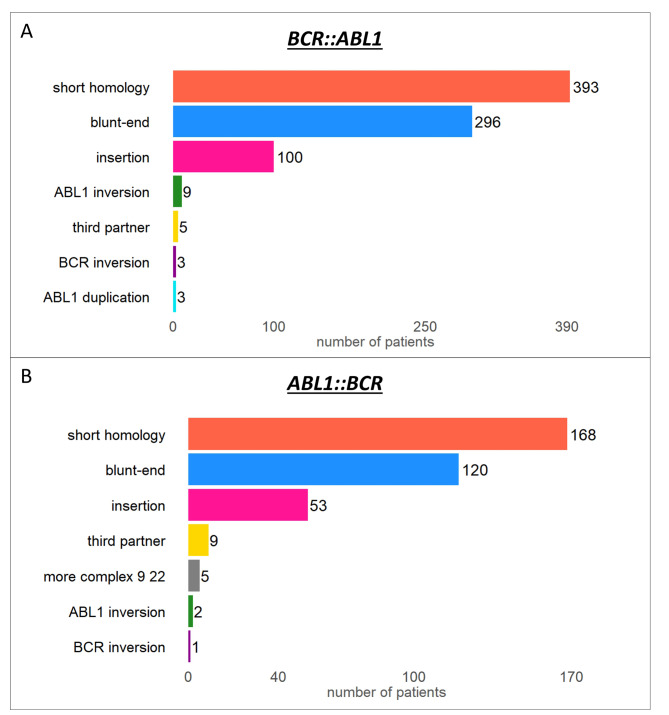
The detailed analysis of genomic breakpoints showed that fusions are mostly formed in loci with short homologies (48.6%; median length = 1 bp, range 1–71 bp), by blunt-end junctions (36.6%) or by a junction with the insertion of a few random nucleotides (12.4%; 1–42 bp, median length = 2.5 bp; see Fig. 2A). This primary structure suggests that non-homologous end joining (NHEJ) is responsible for the double-strand break repair, consistent with other findings [1, 4–6]. However, in some cases, BCR and ABL1 gene fusion may be a more complex process, as evidenced by insertions of part of chromosome 9 (including the ABL1 gene) into the BCR or insertions of sequences (up to 12.3 kbp) from another chromosome between BCR and ABL1. For more details regarding these few specific exceptions see Additional Results.
Fig. 2.
Primary structure of BCR::ABL1 (A) and ABL1::BCR (B) breakpoints
Images adapted from the “Break-App” web tool
We did not specifically look for the reciprocal ABL1::BCR fusion in all patients. However, the usage of NGS custom target enrichment enabled us to detect this breakpoint incidentally in some patients. In total, we characterized the ABL1::BCR junction in 415 (43%) patients (332 ALL, 83 CML) with a known BCR::ABL1 sequence. The distribution of reciprocal breakpoints was highly consistent with the BCR::ABL1 fusions, as the vast majority of the “forward” (BCR::ABL1) and “reciprocal” (ABL1::BCR) breakpoints in the given gene were located within a ± 100 bp window (81% in BCR and 80% in ABL1). In 42 patients (10%) the BCR::ABL1 and ABL1::BCR breakpoints were located within a ± 1 bp on both fusion partners; in 14 patients, a perfectly balanced translocation was detected (see Additional Fig. 2). Thus, in majority of cases the t(9;22) translocation was perfectly reciprocal or very close to it even at the nucleotide level. Large deletions (over 10 kbp) of chromosomes 22 and/or 9 (which can be identified in cases with both BCR::ABL1 and ABL1::BCR fusion available) were present relatively rarely (n = 20 [the largest deletion 2,171 kbp] and n = 21 [the largest deletion 1,005 kbp], respectively; i.e. 41/415 = 10% of patients). Duplications larger than 10 kbp were detected almost exclusively on chromosome 9 (n = 7; the largest duplication ~ 2,900 kbp; chromosome 22: n = 1; ~ 55 kbp). This analysis may be slightly biased as we did not specifically look for the reciprocal fusion in all patients and in some cases where the reciprocal fusion was not retrieved directly in the target enrichment NGS output, larger structural aberrations may have been involved, resulting in possible under-representation of more complex translocations. However, this bias would not dramatically impact our findings, since in a consecutive series of 404 patients analyzed by the same NGS approach, the reciprocal fusion was included in the NGS output in 87% of cases; the remaining 13% might comprise some further cases with more complex mechanism of fusion.
The primary structure of ABL1::BCR fusions corresponded to that of BCR::ABL1 counterparts, with the vast majority of fusion sequences involving short homologies, short insertions and blunt-end junctions (Fig. 2B). The overall comparison of BCR::ABL1 and ABL1::BCR fusions is listed in Additional Table 2.
We searched for RSS, specific motifs known to mediate DNA breaks (59 motifs, adopted from Ross et al., 2013) [4] and interspersed and other types of repeats within particular DNA areas (see Additional Methods). Despite the non-random pattern in both BCR regions, we did not find any significant association between the localization of breakpoints and any type of DNA motif or DNA sequences with specific chromatin structure or any evidence of breakpoint clustering within or in the neighborhood (± 10 bp) of any searched DNA or epigenetic motif. For further details see Additional Table 3 and Additional Results.
In our previous studies on BCR::ABL1-positive leukemias [7, 10–12], we defined “CML-like” leukemias, diagnosed as ALL but exhibiting BCR::ABL1 fusion in multipotent, not fully leukemic progenitors, biologically resembling CML in lymphoid blast crisis. It would be intriguing to compare whether the distribution of fusions in these CML-like leukemias differs from typical ALL and is closer to that of typical CML. Our data indeed suggest slightly more frequent breaks in the 5’ portion of ABL1 and downstream of intron 13 of BCR (more typical for CML) in CML-like leukemias than in typical ALL (data not shown), but the differences are relatively small and do not reach statistical significance, possibly due to the limited number of patients for this analysis (99 typical ALL vs. 53 CML-like leukemias).
To visualize our results and enable data browsing and various analyses of BCR::ABL1 breakpoint positions, we developed an interactive “Break-App” web tool (available at https://clip.lf2.cuni.cz/break-app). The web tool enables the analysis of breakpoint sites distribution in general or within/between particular subgroups (diagnosis, sex, age-specific, etc.). Furthermore, it comprises detailed information about (i) the primary structure of the breakpoints, (ii) ABL1::BCR breakpoints, (iii) breakpoint positions with respect to DNA motifs and chromatin structure; (iv) detailed information about KMT2A breakpoints (see Additional Results). We plan to update this tool regularly with new data. Not only can further BCR::ABL1 breakpoint positions be easily included, but, if desirable, the tool can also be adapted for other fusions or breakpoints.
Characterizing primary aberrations, including gene fusions, at the DNA level aids in understanding their origin and mechanisms, with practical applications such as obtaining patient-specific, highly sensitive targets for detecting residual leukemic cells [7, 11–15]. Advances in molecular techniques, particularly massively parallel sequencing, have significantly enhanced the feasibility of detecting genomic fusions, enabling efficient identification of breakpoint sites in a shorter time, regardless of the length and complexity of breakpoint regions in fusion partners. Previous publications on BCR::ABL1-positive leukemia have primarily focused on the Major form of the translocation due to technical challenges in determining breakpoints in the substantially larger minor BCR region (~ 2.9 vs. ~71.5 kbp).
In conclusion, our study of BCR::ABL1 fusions is based on the largest and most complex cohort of patients with BCR::ABL1 fusion identified at the DNA level to date. The complexity of our cohort allowed for the first time comparison of breakpoint distribution in CML vs. BCR::ABL1-positive ALL, revealing significant differences in both ABL1 and Major BCR loci. Importantly, our data are not biased towards only ‘canonical’ fusions, as the NGS approach allowed us to characterize genomic breakpoints in all patients where at least one side of the fusion is located in the area covered by our probes. No DNA or epigenetic motif responsible for the non-random distribution was found. Taken together, our data suggest that physical colocalization and chromatin accessibility, which change with the developmental stage of the cell (hence the difference between ALL that arises in a committed lymphoid progenitor, and CML that arises in a stem/multipotent cell), are more critical factors influencing breakpoint localization than the presence of specific DNA motifs. While we offer here a detailed insight and analysis of the genomic breakpoints, the cause and exact molecular mechanism underlying the origin of double-strand breaks in BCR and ABL1 genes and their fusion remains to be resolved.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Marco Beldinanzi for his help with samples preparation, Robert Ivanek for his help and suggestions on using the Gviz package, and Radek Senfeld for help in placing our “Break-App” web tool on the web.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- CML
Chronic myeloid leukemia
- NGS
Next generation sequencing
- NHEJ
Non–homologous end joining
- PCR
Polymerase chain reaction
Author contributions
LH and JZ designed the study; LH, LW, JSk, MK, AB, VP, JA, MB, CM, RK, TNT, EC, SC, MH, RS, LS, JSt, KMP, RM, MM, GCaz, GCar, JT, MKZ and JZ analyzed samples and provided data; all authors participated on data integration, interpretation, and presentation; LH and JZ designed the Break-App web tool, wrote the draft; and all authors revised the draft and contributed to the final manuscript.
Funding
Supported by grants from the Czech Health Research Council (NU21-03-00128), Charles University (GAUK 327322), by the project from Ministry of Health, Czech Republic 00064203 (University Hospital Motol, Prague, Czech Republic), MH CZ – DRO (IHBT, 00023736), by the project National Institute for Cancer Research (Program EXCELES, ID Project No. LX22NPO5102), and Cancer Australia PdCCRS1128727 (RS).
Data availability
The datasets generated during and/or analyzed during the current study are included in this published article and its Additional files and/or are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The project was approved by the Institutional Review Board of University Hospital Motol (Czech Republic) and Hunter Human Research Ethics Committee HNE 2019/ETH01219 (multisite, Australia). Informed consent was obtained following the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Burmeister T, Groger D, Kuhn A, Hoelzer D, Thiel E, Reinhardt R. Fine structure of translocation breakpoints within the major breakpoint region in BCR-ABL1-positive leukemias. DNA Repair (Amst) 2011;10(11):1131–7. doi: 10.1016/j.dnarep.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Score J, Calasanz MJ, Ottman O, Pane F, Yeh RF, Sobrinho-Simoes MA, et al. Analysis of genomic breakpoints in p190 and p210 BCR-ABL indicate distinct mechanisms of formation. Leukemia. 2010;24(10):1742–50. doi: 10.1038/leu.2010.174. [DOI] [PubMed] [Google Scholar]
- 3.Krumbholz M, Goerlitz K, Albert C, Lawlor J, Suttorp M, Metzler M. Large amplicon droplet digital PCR for DNA-based monitoring of pediatric chronic myeloid leukaemia. J Cell Mol Med. 2019;14(10):14321. doi: 10.1111/jcmm.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross DM, O’Hely M, Bartley PA, Dang P, Score J, Goyne JM, et al. Distribution of genomic breakpoints in chronic myeloid leukemia: analysis of 308 patients. Leukemia. 2013;27(10):2105–7. doi: 10.1038/leu.2013.116. [DOI] [PubMed] [Google Scholar]
- 5.Krumbholz M, Karl M, Tauer JT, Thiede C, Rascher W, Suttorp M, et al. Genomic BCR-ABL1 breakpoints in pediatric chronic myeloid leukemia. Genes Chromosomes Cancer. 2012;51(11):1045–53. doi: 10.1002/gcc.21989. [DOI] [PubMed] [Google Scholar]
- 6.Mattarucchi E, Guerini V, Rambaldi A, Campiotti L, Venco A, Pasquali F, et al. Microhomologies and interspersed repeat elements at genomic breakpoints in chronic myeloid leukemia. Genes Chromosomes Cancer. 2008;47(7):625–32. doi: 10.1002/gcc.20568. [DOI] [PubMed] [Google Scholar]
- 7.Hovorkova L, Zaliova M, Venn NC, Bleckmann K, Trkova M, Potuckova E, et al. Monitoring of childhood ALL using BCR-ABL1 genomic breakpoints identifies a subgroup with CML-like biology. Blood. 2017;129(20):2771–81. doi: 10.1182/blood-2016-11-749978. [DOI] [PubMed] [Google Scholar]
- 8.Linhartova J, Hovorkova L, Soverini S, Benesova A, Jaruskova M, Klamova H, et al. Characterization of 46 patient-specific BCR-ABL1 fusions and detection of SNPs upstream and downstream the breakpoints in chronic myeloid leukemia using next generation sequencing. Mol Cancer. 2015;14:89. doi: 10.1186/s12943-015-0363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machova Polakova K, Zizkova H, Zuna J, Motlova E, Hovorkova L, Gottschalk A, et al. Analysis of chronic myeloid leukaemia during deep molecular response by genomic PCR: a traffic light stratification model with impact on treatment-free remission. Leukemia. 2020;34(8):2113–24. doi: 10.1038/s41375-020-0882-1. [DOI] [PubMed] [Google Scholar]
- 10.Zaliova M, Fronkova E, Krejcikova K, Muzikova K, Mejstrikova E, Stary J, et al. Quantification of fusion transcript reveals a subgroup with distinct biological properties and predicts relapse in BCR/ABL-positive ALL: implications for residual disease monitoring. Leukemia. 2009;23(5):944–51. doi: 10.1038/leu.2008.386. [DOI] [PubMed] [Google Scholar]
- 11.Zuna J, Hovorkova L, Krotka J, Koehrmann A, Bardini M, Winkowska L, et al. Minimal residual disease in BCR::ABL1-positive acute lymphoblastic leukemia: different significance in typical ALL and in CML-like disease. Leukemia. 2022;36(12):2793–801. doi: 10.1038/s41375-022-01668-0. [DOI] [PubMed] [Google Scholar]
- 12.Zuna J, Hovorkova L, Krotka J, Winkowska L, Novak Z, Sramkova L, et al. Posttreatment positivity of BCR::ABL1 in acute lymphoblastic leukemia: should we keep track? Am J Hematol. 2023;98(10):E269–71. doi: 10.1002/ajh.27022. [DOI] [PubMed] [Google Scholar]
- 13.Lukes J Jr., Winkowska L, Zwyrtkova M, Starkova J, Sramkova L, Stary J et al. Identification of Fusion Gene breakpoints is feasible and facilitates accurate sensitive minimal residual disease monitoring on genomic level in patients with PML-RARA, CBFB-MYH11, and RUNX1-RUNX1T1. Hemasphere. 2020;4(6). [DOI] [PMC free article] [PubMed]
- 14.Zaliova M, Zuna J, Winkowska L, Janotova I, Skorepova J, Lukes J, et al. Genomic DNA-based measurable residual disease monitoring in pediatric acute myeloid leukemia: unselected consecutive cohort study. Leukemia. 2024;38(1):21–30. doi: 10.1038/s41375-023-02083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venn NC, Huang L, Hovorkova L, Muskovic W, Wong M, Law T, et al. Measurable residual disease analysis in paediatric acute lymphoblastic leukaemia patients with ABL-class fusions. Br J Cancer. 2022;127(5):908–15. doi: 10.1038/s41416-022-01806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are included in this published article and its Additional files and/or are available from the corresponding author on reasonable request.