Abstract
Desertification is known to be a major threat to biodiversity, yet our understanding of the consequent decline in biodiversity remains insufficient. Here, we predicted climate change-induced range shifts and genetic diversity losses in three model dung beetles: Colobopterus erraticus, Cheironitis eumenes, and Gymnopleurus mopsus, distributed across the Gobi Desert and Mongolian Steppe, areas known for desertification. Phylogeographic analyses of mitochondrial COI sequences and species distribution modeling, based on extensive field investigations spanning 14 years, were performed. Species confined to a single biome were predicted to contract and shift their distribution in response to climate change, whereas widespread species was predicted to expand even if affected by range shifts. We indicated that all species are expected to experience significant haplotype losses, yet the presence of high singleton frequencies and low genetic divergence across geographic configurations and lineages mitigate loss of genetic diversity. Notably, Cheironitis eumenes, a desert species with low genetic diversity, appears to be the most vulnerable to climate change due to the extensive degradation in the Gobi Desert. This is the first study to predict the response of insects to desertification in the Gobi Desert. Our findings highlight that dung beetles in the Gobi Desert and Mongolian Steppe might experience high rates of occupancy turnover and genetic loss, which could reshuffle the species composition.
Keywords: Desertification, Dryland, Ecological niche model, Mongolia, Scarabaeidae, Vulnerability
Subject terms: Biodiversity, Climate-change ecology
Introduction
Global climate change is a major threat to biodiversity and has been proposed to profoundly affect species distribution, genetic diversity, behavior, and fitness1,2. Species confronted with adverse climate change have been reported to exhibit diverse responses, including range-shifting by migration and persistence mediated by either phenotypic plasticity or local adaptation1,3,4. However, climate change that occurs at a rate surpassing species’ adaptive capacity can reduce population sizes and cause the extinction of local populations, inevitably eroding genetic diversity at the species level3,5.
Climate change models forecast an increase in global temperatures and disruption of precipitation patterns, which pose various challenges to ecosystems at regional scales. Desertification refers to the permanent degradation of land accompanied by changes in soil properties and precipitation. This process is a major climate change issue in arid, semi-arid, and sub-humid regions6,7. By the end of the twenty-first century, climate change could lead to an expansion of dryland area by 10% in total7. East Asia is one of the key areas that highly suffered from different levels of desertification along with the expansion of the Gobi Desert8,9. The Gobi Desert, the second largest rain shadow desert covering 1.3 million km2, stretches across the southern regions Mongolia as well as northern China10. So far, only a few studies have investigated and documented high rates of range shift on plant11,12 and mammals13 in the Gobi Desert and nearby sub-arid steppe region, and the impact of climate change on insects in this region has not yet been investigated.
Species distribution models (SDMs) are powerful tools for estimating the relative suitability of habitat for specific species14. As a result of the simplicity of modeling and the accessibility of fine-scale climatic data, SDMs have been implemented extensively to predict species’ current distribution and changes in species’ distribution that could arise in response to climate change15,16. However, SDM approaches, which are based on limited environmental data, have been criticized as oversimplistic since they do not account for evolutionary processes and biological features that affect species’ distribution17,18. For instance, integrating SDMs and phylogeographic information would provide a powerful framework for inferring the effects of future climate change on target organisms5,19.
Phylogeography may provide insight into the contemporary and future responses of species to environmental changes. Indeed, genetic structure can provide insight into a species’ evolutionary history and into the dynamics of contemporary gene flow among populations, and genetic diversity can be used to evaluate species’ evolutionary potential and resilience to environmental change4,5. In addition, populations that suffer from unsuitable habitats or limited connectivity often harbor low genetic diversity as a result of bottlenecks or inbreeding depression and are, as a result, highly vulnerable to environmental change20. Therefore, investigating the genetic diversity and structure of populations across species’ distribution ranges is critical to evaluating the vulnerability and adaptive capacity of species to environmental change5,21.
Dung beetles are key organisms in terrestrial ecosystems because they contribute to various ecosystem processes, including nutrient cycling, parasite suppression, and secondary seed dispersal22. The group has been widely used as a bioindicator of biodiversity and ecosystem health because of their ecological importance, sensitivity to environmental factors, and established standardized sampling methods23,24. Serious concerns exist regarding the global decline in dung beetle diversity, and empirical data show that populations of many dung beetle species have significantly declined in response to habitat fragmentation25 and climate change26,27.
Mongolia spans a vast plateau with wide steppe and desert regions. More than 67 million livestock, including horses, cattle, sheep, goats, camels, and yaks, are farmed across the Gobi Desert and Mongolian Steppe, mostly through nomadic herding (National Statistics Office of Mongolia; https://www.en.nso.mn/, accessed on April 08, 2022). This environment, abundant in food resources throughout the country, dung beetles are essential for maintaining the ecosystem health. To date, 78 dung beetle species have been documented in Mongolia28. Research on the conservation of dung beetles in Mongolia has been primarily confined to a few national reserves29,30. However, Mongolia is a particularly well-suited region for investigating the effects of desertification through climate change on organisms. Encompassing two distinct climate regions and less affected by other anthropogenic disturbances such as habitat fragmentation, and chemical treatments, the country act as a natural laboratory to pinpoint and understand the direct implications of climate change.
Accordingly, this study explored how future climate change-induced range shifts and the loss of genetic diversity may differ among dung beetles in desert and sub-arid steppe region by incorporating SDMs and phylogeographic analyses. To achieve this, we investigated the genetic diversity, population structure, demographic history, and distribution of three model species from the Gobi Desert and Mongolian Steppe in Mongolia. We then predicted their future distribution and genetic diversity under climate change scenarios.
Results
Genetic diversity and haplotype network analyses
Mitochondrial COI sequences were generated from 173, 152, and 273 specimens of C. erraticus (12 locations), C. eumenes (10 locations), and G. mopsus (10 locations), respectively (Table 1). The C. erraticus sequences (836 bp) yielded 29 polymorphisms, including 18 parsimony-informative sites and 44 haplotypes (36 singletons; 81.8%). Meanwhile, the C. eumenes sequences (789 bp) yielded 29 polymorphisms, including 16 parsimony-informative sites and 26 haplotypes (18 singletons, 69.2%), and the G. mopsus sequences (658 bp) yielded 81 polymorphisms, including 51 parsimony-informative sites and 135 haplotypes (111 singletons; 82.2%).
Table 1.
Localities and genetic diversity indices for sampled populations of three species of dung beetles in Mongolia.
| Species | Sites | Regions | Coordinates | Number of individuals (N) | Number of Haplotypes (Nh) | Allelic richness (Ar) | Haplotype diversity (Hd) (SD) | Nucleotide diversity (π) (SD) | Tajima’s D | Fu’s Fs |
|---|---|---|---|---|---|---|---|---|---|---|
| Colobopterus erraticus | CER1402 | Selenge Aimag | 49° 38′ 46.26′′ N 106° 01′ 42.64′′ E | 14 | 6 | 1.778 | 0.7363 (0.1092) | 0.0013 (0.0010) | − 1.1150 | − 2.7635 |
| CER1502 | Töv Aimag | 47° 26′ 56.36′′ N 106° 46′ 42.80′′ E | 5 | 3 | 1.600 | 0.7000 (0.2184) | 0.0022 (0.0017) | − 0.4102 | 0.4690 | |
| CER16EX2 | Selenge Aimag | 50° 05′ 58.4′′ N 106° 12′ 47.7′′ E | 4 | 3 | 0.8333 (0.2224) | 0.0026 (0.0021) | − 0.0650 | 0.6080 | ||
| CER1621 | Bayankhongor Aimag | 46° 23′ 15.85′′ N 100° 48′ 48.63′′ E | 19 | 9 | 2.196 | 0.8421 (0.0670) | 0.0049 (0.0029) | − 0.4079 | − 0.8753 | |
| CER1626 | Arkhangai Aimag | 47° 23′ 07.53′′ N 102° 12′ 57.64′′ E | 8 | 5 | 2.214 | 0.8571 (0.1083) | 0.0021 (0.0015) | − 0.5034 | − 1.4953 | |
| CER1808 | Zavkhan Aimag | 47° 41′ 22.65′′ N 97° 15′ 30.93′′ E | 18 | 13 | 2.770 | 0.9608 (0.0301) | 0.0084 (0.0047) | 1.3475 | 0.9310 | |
| CER1822 | Khovd Aimag | 48° 00′ 59.70′′ N 91° 37′ 15.25′′ E | 20 | 6 | 1.842 | 0.0000 (0.0624) | 0.0016 (0.0002) | 0.4159 | − 1.5046 | |
| CER1005 | Khövsgöl Aimag | 50° 32′ 15.6′′ N 101° 30′ 55.6′′ E | 1 | 1 | 1.0000 (0.0000) | 0.0000 (0.0000) | 0.0000 | 0.0000 | ||
| CER2208 | Arkhangai Aimag | 47° 35′ 17.73′′ N 101° 12′ 51.48′′ E | 22 | 11 | 2.374 | 0.8831 (0.0471) | 0.0053 (0.0030) | 0.2825 | − 1.7727 | |
| CER2213 | Arkhangai Aimag | 48° 23′ 44.08′′ N 101° 18′ 46.40′′ E | 16 | 9 | 2.406 | 0.8917 (0.0543) | 0.0028 (0.0018) | − 1.1568 | − 3.5835 | |
| CER2218 | Khövsgöl Aimag | 49° 35′ 31.79′′ N 101° 59′ 14.78′′ E | 23 | 6 | 1.816 | 0.7668 (0.0489) | 0.0015 (0.0011) | − 0.2460 | − 1.3902 | |
| CER2222 | Bulgan Aimag | 48° 57′ 47.33′′ N 102° 47′ 27.31′′ E | 23 | 7 | 1.898 | 0.7826 (0.0533) | 0.0015 (0.0011) | − 1.0794 | − 2.4441 | |
| Total | 173 | 44 | 0.8370 (0.0180) | 0.0037 (0.0004) | − 1.1127 | − 26.2318 | ||||
| Cheironitis eumenes | CEU1609 | Umnugobi Aimag | 44° 09′ 38.65′′ N 104° 05′ 29.37′′ E | 22 | 7 | 1.729 | 0.5974 (0.1184) | 0.0015 (0.0011) | − 1.7885 | − 2.8194 |
| CEU1713 | Dornogobi Aimag | 45° 13′ 28.06′′ N 110° 8′ 46.33′′ E | 24 | 5 | 1.431 | 0.5400 (0.1089) | 0.0015 (0.0011) | − 1.4774 | − 0.5116 | |
| CEU1415 | Dornogobi Aimag | 46° 0′ 11.87′′ N 108° 50′ 7.61′′ E | 5 | 4 | 3.000 | 0.9000 (0.1610) | 0.0030 (0.0023) | − 1.1455 | − 0.7012 | |
| CEU1706 | Dornogobi Aimag | 45° 49′ 9.25′′ N 109° 18′ 11.57′′ E | 3 | 3 | 1.0000 (0.2722) | 0.0068 (0.0056) | 0.0000 | 0.4576 | ||
| CEU1607 | Umnugobi Aimag | 45° 49′ 9.25′′ N 109° 18′ 11.57′′ E | 2 | 1 | 0.0000 (0.0000) | 0.0000 (0.0000) | 0.0000 | 0.0000 | ||
| CEU1608 | Umnugobi Aimag | 44° 10′ 41.30′′ N 104° 22′ 13.87′′ E | 5 | 1 | 0.000 | 0.0000 (0.0000) | 0.0000 (0.0000) | 0.0000 | 0.0000 | |
| CEU22A4 | Umnugobi Aimag | 44° 21′ 18.26′′ N 105° 15′ 26.49′′ E | 24 | 9 | 2.172 | 0.7065 (0.0992)) | 0.0019 (0.0013) | − 1.4720 | − 4.0130 | |
| CEU22A6 | Umnugobi Aimag | 43° 56′ 56.6′′ N 104° 57′ 41.0′′ E | 28 | 6 | 1.281 | 0.4815 (0.1083) | 0.0014 (0.0010) | − 1.8656 | − 1.5132 | |
| CEU22A7 | Umnugobi Aimag | 43° 54′ 51.3′′ N 104° 55′ 14.4′′ E | 21 | 6 | 1.646 | 0.6000 (0.1104) | 0.0009 (0.0007) | − 1.4856 | − 3.3056 | |
| CEU22A8 | Umnugobi Aimag | 43° 41′ 12.7′′ N 104° 39′ 42.2′′ E | 18 | 5 | 1.111 | 0.4052 (0.1428) | 0.0013 (0.0010) | − 2.1893 | − 1.2567 | |
| Total | 152 | 26 | 0.5660 (0.0480) | 0.0016 (0.0002) | − 2.2122 | − 25.4402 | ||||
| Gymnopleurus mopsus | GM1403 | Selenge Aimag | 49° 45′ 25.43′′ N 106° 9′ 58.01′′ E | 34 | 20 | 6.925 | 0.9340 (0.0301) | 0.0082 (0.0045) | − 1.32765 | − 7.48445 |
| GM1417 | Dornogobi Aimag | 45° 09′ 19.52′′ N 109° 58′ 0.28′′ E | 34 | 28 | 8.398 | 0.9857 (0.0115) | 0.0081 (0.0045) | − 1.4744 | − 22.7255 | |
| GM1510 | Umnugobi Aimag | 44° 14′ 16.51′′ N 106° 53′ 59.43′′ E | 12 | 11 | 8.318 | 0.9848 (0.0403) | 0.0082 (0.0048) | − 1.30233 | − 5.19347 | |
| GM1604 | Dundgobi Aimag | 44° 54′ 47.99′′ N 105° 32′ 37.74′′ E | 28 | 28 | 9.000 | 1.0000 (0.0101) | 0.0091 (0.0050) | − 1.49167 | − 25.26433 | |
| GM1605 | Dundgobi Aimag | 43° 46′ 28.95′′ N 104° 47′ 29.96′′ E | 32 | 29 | 8.661 | 0.9919 (0.0110) | 0.0081 (0.0045) | − 1.33654 | − 25.38005 | |
| GM1611 | Umnugobi Aimag | 44° 22′ 33.54′′ N 104° 02′ 32.60′′ E | 28 | 27 | 8.881 | 0.9974 (0.0104) | 0.0091 (0.0050) | − 1.50788 | − 24.92409 | |
| GM1628 | Orkhon Aimag | 47° 21′ 02.14′′ N 103° 44′ 06.94′′ E | 34 | 19 | 6.722 | 0.9376 (0.0213) | 0.0091 (0.0049) | − 0.74768 | − 3.40479 | |
| GM1630 | Töv Aimag | 47° 44′ 27.8′′ N 105° 51′ 58.7′′ E | 37 | 10 | 5.289 | 0.8889 (0.0226) | 0.0091 (0.0049) | 0.28248 | 1.79525 | |
| GM162b | Gobisumber Aimag | 46° 06′ 26.39′′ N 108° 43′ 20.88′′ E | 11 | 11 | 9.000 | 1.0000 (0.0388) | 0.0098 (0.0057) | − 0.82427 | − 5.8677 | |
| GM2217 | Khövsgöl Aimag | 49° 27′ 32.61′′ N 101° 32′ 20.78′′ E | 23 | 7 | 4.077 | 0.7826 (0.0717) | 0.0072 (0.0041) | 0.30558 | 1.85054 | |
| Total | 273 | 135 | 0.9717 (0.0044) | 0.0088 (0.0003) | − 1.6712 | − 24.7955 | ||||
Significant values (P < 0.05, neutrality test) are indicated in bold.
Estimates of haplotype diversity for populations represented by at least five specimens (N ≥ 5) ranged from 0.7000 to 0.9608 (mean = 0.8370), from 0.0000 to 0.9000 (mean = 0.5660), and from 0.7826 to 1.0000 (mean = 0.9717) for C. erraticus, C. eumenes, and G. mopsus, respectively (Table 1). Meanwhile, nucleotide diversity estimates ranged from 0.0013 to 0.0084 (mean = 0.0037), from 0.0000 to 0.0030 (mean = 0.0016), and from 0.0072 to 0.0098 (mean = 0.0088) for C. erraticus, C. eumenes, and G. mopsus, respectively (Table 1). Most of the C. erraticus and C. eumenes localities yielded relatively low allelic richness (Ar), ranging from 1.600 to 2.770 (C. erraticus) and from 0.000 to 3.000 (C. eumenes). In contrast, the G. mopsus localities yielded relatively high allelic richness, ranging from 4.077 to 9.000.
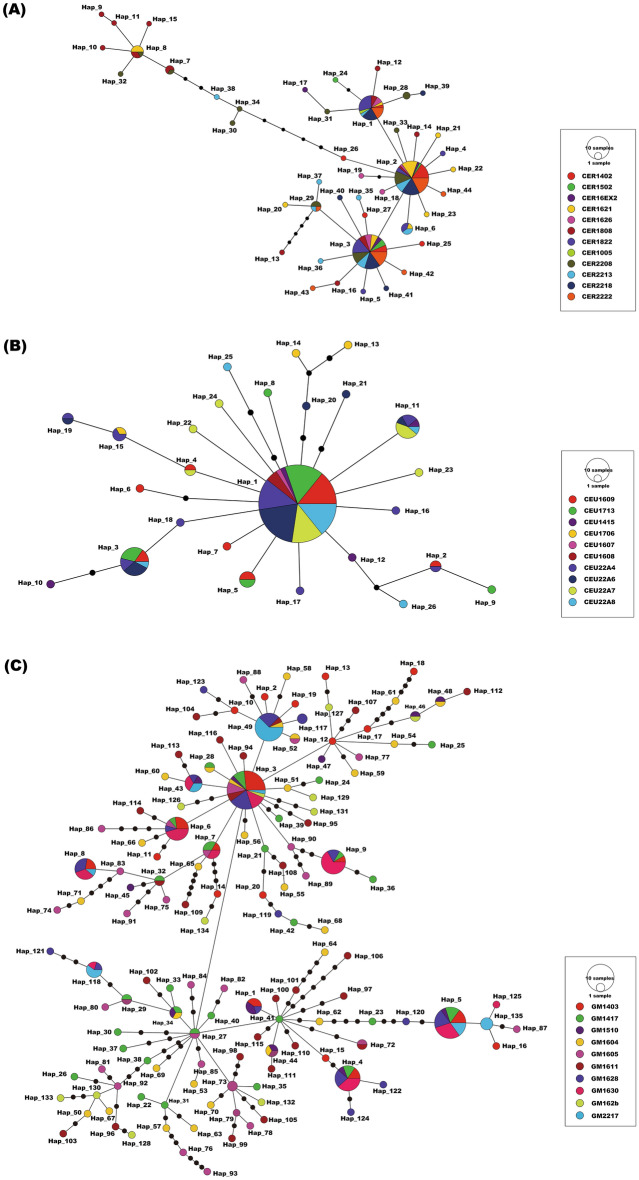
The haplotype networks of three species exhibited different topologies (Fig. 1). In the C. erraticus haplotype network, three dominant haplotypes (H1, H2, and H3), each one mutation step apart, accounted for 13.9% (H1, N = 24), 26.6% (H2, N = 46), and 27.2% (H3, N = 47), respectively. Haplotypes from CER1808, CER1621, and CER2213, clustered into haplogroups with approximately 10 mutational steps of H2 (Fig. 1A). In the C. eumenes haplotype network, the most dominant haplotype (H1) accounted for 65.1% (N = 99) of specimens and was located at the most internal position in the network, presumably to be an ancestral haplotype (Fig. 1B). In the G. mopsus haplotype network, the most dominant haplotype (H3) accounted for 11.0% (N = 30) of specimens and was located at an internal position in the network (Fig. 1C). This haplotype was detected in specimens from all the sampled localities, which indicated its extensive distribution.
Figure 1.
Haplotype networks of COI sequences of three model species. (A) Colobopterus erraticus, (B) Cheironitis eumenes, (C) Gymnopleurus mopsus.
Population structure and demographic history analyses
Populations of C. erraticus and G. mopsus exhibited significant pairwise genetic differentiation, whereas those of C. eumenes did not. The pairwise differences (FST) of C. erraticus populations ranged from − 0.066 to 0.367 (Supplementary Information, Table S1). However, the genetic differentiation between CER1808 and the other three sites was significant, which indicate that the local population is genetically isolated from the others. Meanwhile, the pairwise differences (FST) of C. eumenes ranged from − 0.087 to 0.081 (Table S2), with all FST values indicating low levels of genetic differentiation. Finally, the pairwise differences (FST) of G. mopsus populations ranged from − 0.014 to 0.078 (Table S3). As in C. erraticus, most of the FST values were relatively small, indicating a low inter-population differentiation level. Only four of the 45 comparisons among the 10 populations indicated significant differentiation.
In C. erraticus, AMOVA indicated significant spatial genetic differentiation between the Khangai Mountains group and other populations (Table 2). In G. mopsus, AMOVA indicated that significant spatial genetic structuring occurred under climate and geography, as reported by previous study31. However, no spatial genetic structuring was observed in the geographical groupings of C. eumenes.
Table 2.
Analysis of molecular variance (AMOVA) for sampled populations of three dung beetle species in Mongolia.
| Species | Hypothesis | d.f | Percent of Variation | F-statistics | P value |
|---|---|---|---|---|---|
| Colobopterus erraticus | Two groups (Khangai, Others) | 1 | 19.13 | 0.19132 | 0.00782 |
| 8 | 2.48 | 0.03073 | 0.06354 | ||
| 158 | 78.38 | 0.21616 | 0.000001 | ||
| Three groups (N, C, W) | 2 | − 0.3 | − 0.00299 | 0.39785 | |
| 7 | 13.78 | 0.13742 | 0.000001 | ||
| 158 | 86.52 | 0.13483 | 0.000001 | ||
| Cheironitis eumenes | Two groups (S, SE) | 1 | 0.86 | 0.00856 | 0.261 |
| 6 | − 0.18 | − 0.00182 | 0.49462 | ||
| 139 | 99.32 | 0.00675 | 0.4086 | ||
| Gymnopleurus mopsus | Two groups (Steppe, Desert) | 1 | 2.5 | 0.025 | 0.00978 |
| 8 | 1.19 | 0.01223 | 0.02248 | ||
| 263 | 96.31 | 0.03692 | 0.000001 | ||
| Three groups (N, C, S) | 2 | 2.02 | 0.02025 | 0.0088 | |
| 7 | 1.22 | 0.01249 | 0.02542 | ||
| 263 | 96.75 | 0.03248 | 0.000001 | ||
| Four groups (NC, NW, S, SE) | 3 | 2.34 | 0.02344 | 0.000001 | |
| 6 | 0.69 | 0.0071 | 0.15738 | ||
| 263 | 96.96 | 0.03037 | 0.000001 |
Significant values (P < 0.05) are indicated in bold.
The evaluation of the population expansion in C. eumenes and G. mopsus yielded significantly negative values when all populations were pooled in a single dataset (Table 1). Evaluation of C. erraticus yielded negative values for both indices (Tajima’s D = − 1.1127, P = 0.1230; Fu’s Fs = − 26.2318, P < 0.0001), and only Fu’s Fs was significant. A multimodal mismatch distribution of pairwise differences was observed for the C. erraticus populations, with significant statistics for both the sum of squared deviations (SSD) and raggedness index values (SSD = 0.0151, P < 0.0001, raggedness index = 0.0653, P < 0.0001), which indicated a constant population size under the neutral model (Supplementary Information, Fig. S1). A unimodal mismatch distribution was observed for C. eumenes (skewed) and G. mopsus. However, the SSD and raggedness index values were non-significant (C. eumenes: SSD = 0.0078, P = 0.66; raggedness index = 0.0629, P = 0.54; G. mopsus: SSD = 0.0005, P = 0.92; raggedness index = 0.0053, P = 0.91), which indicated historical population expansion (Fig. S1).
Species distribution modeling
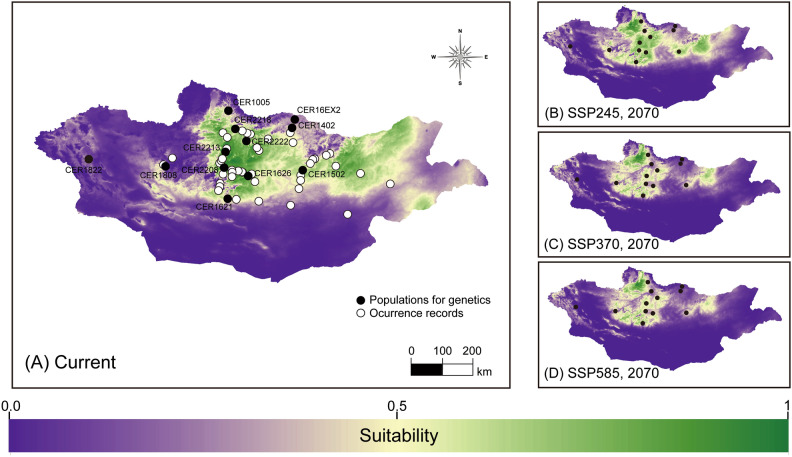
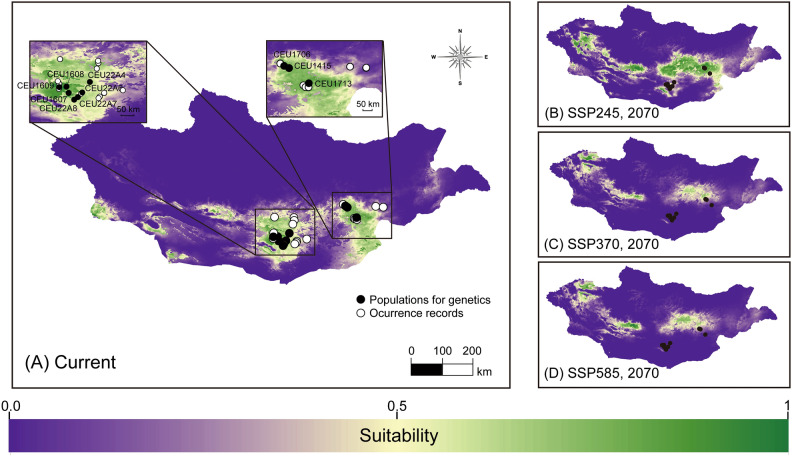
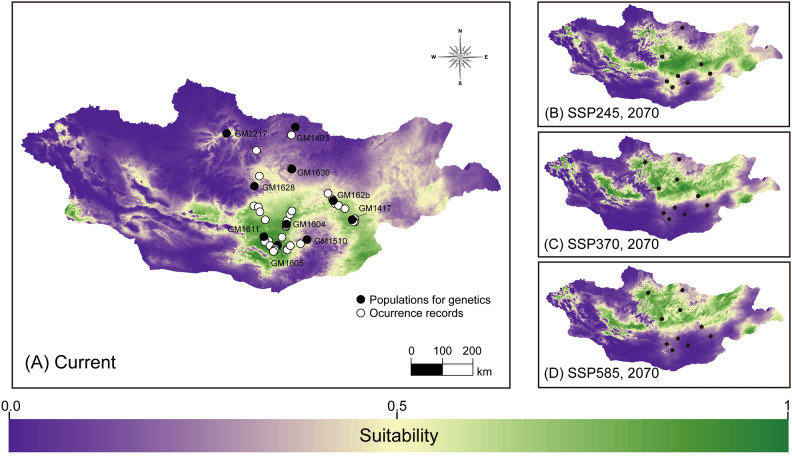
The optimal SDMs for each species yielded satisfactory predictive performance (C. erraticus: RM = 1, FC = LQ, AUCtest = 0.830, AUCdiff = 0.077, OR10 = 0.140, ΔAICc = 0; C. eumenes: RM = 0.5, FC = LQ, AUCtest = 0.953, AUCdiff = 0.033, OR10 = 0.100, ΔAICc = 0; G. mopsus: RM = 1.5, FC = LQHP, AUCtest = 0.860, AUCdiff = 0.071, OR10 = 0.105, ΔAICc = 17.350; Fig. S2). The models predicted that C. erraticus and C. eumenes were distributed in the northeastern steppe and southern desert, respectively, and predicted that G. mopsus was distributed among both steppe and desert biomes across central Mongolia (Figs. 2, 3, and 4, S3–S5). Presence–absence binary maps were created using 10% training logistic thresholds for each species (thresholds: C. erraticus, 0.2864; C. eumenes, 0.3427; and G. mopsus, 0.2931).
Figure 2.
Distribution and environmental suitability predictions of Colobopterus erraticus. (A) Current species distribution model with the occurrence records (white dots), and sites for population genetics (black dots); (B–D) Predicted future distributions in 2070 under the SSP245, SSP370, and SSP585 scenarios, respectively. The map of distribution model was modified using ArcGIS 10.8 (www.esri.com)67.
Figure 3.
Distribution and environmental suitability predictions of Cheironitis eumenes. (A) Current species distribution model with the occurrence records (white dots), and sites for population genetics (black dots); (B–D) Predicted future distributions in 2070 under the SSP245, SSP370, and SSP585 scenarios, respectively. The map of distribution model was modified using ArcGIS 10.8 (www.esri.com)67.
Figure 4.
Distribution and environmental suitability predictions of Gymnopleurus mopsus. (A) Current species distribution model with the occurrence records (white dots), and sites for population genetics (black dots); (B–D) Predicted future distributions in 2070 under the SSP245, SSP370, and SSP585 scenarios, respectively. The map of distribution model was modified using ArcGIS 10.8 (www.esri.com)67.
Predicting distribution and genetic diversity under climate change scenarios
Estimations of future C. erraticus distribution indicated a westward shift. The northeastern area of the species’ distribution contracted and shifted among the various time points and climate scenarios (Fig. 2). Under all climate scenarios, the distribution of C. erraticus exhibited a net reduction in area, ranging from − 18.55 to − 46.26% and from − 31.80 to − 63.23% in 2050 and 2070, respectively (Table 3). In contrast, the future projections of C. eumenes revealed a severe reduction in the southern desert region and a northward shift to central steppe region (Fig. 3). The distribution of C. eumenes was projected to either increase or decrease, with predicted changes in area ranging from − 79.01 to 19.98% and from − 28.02 to 58.42% in 2050 and 2070, respectively (Table 3). For G. mopsus, the species’ distribution was predicted to shift northward, with a net expansion in the northern steppe region (Fig. 4). Under all climate scenarios, the distribution of G. mopsus exhibited a net expansion in the area, ranging from 27.23 to 33.49% and from 21.63 to 43.44% in 2050 and 2070, respectively (Table 3).
Table 3.
Predicted future genetic diversity and distribution for sampled populations of three dung beetle species in Mongolia under current conditions and future climate change scenarios.
| Species | Scenarios/Time period | Haplotype retained (%) | Haplotype diversity (Hd) | Nucleotide diversity (π) | Change in distribution area from Current (%) |
|---|---|---|---|---|---|
| Colobopterus erraticus | Current | 0.837 | 0.004 | ||
| SSP245/2050 | 86.36 | 0.848 | 0.004 | − 18.55 | |
| SSP245/2070 | 86.36 | 0.848 | 0.004 | − 31.80 | |
| SSP370/2050 | 79.55 | 0.865 | 0.005 | − 37.70 | |
| SSP370/2070 | 65.91 | 0.862 | 0.005 | − 55.83 | |
| SSP585/2050 | 79.55 | 0.865 | 0.005 | − 46.26 | |
| SSP585/2070 | 45.55 | 0.827 | 0.003 | − 63.23 | |
| Cheironitis eumenes | Current | 0.566 | 0.002 | ||
| SSP245/2050 | 57.69 | 0.584 | 0.002 | + 8.49 | |
| SSP245/2070 | 80.77 | 0.562 | 0.002 | + 58.42 | |
| SSP370/2050 | 80.77 | 0.562 | 0.002 | + 19.98 | |
| SSP370/2070 | 11.54 | 1.000 | 0.007 | − 36.80 | |
| SSP585/2050 | 26.92 | 0.964 | 0.005 | − 79.01 | |
| SSP585/2070 | 0 | 0 | 0 | − 28.02 | |
| Gymnopleurus mopsus | Current | 0.971 | 0.009 | ||
| SSP245/2050 | 89.63 | 0.975 | 0.009 | + 29.46 | |
| SSP245/2070 | 89.63 | 0.975 | 0.009 | + 43.44 | |
| SSP370/2050 | 89.63 | 0.975 | 0.009 | + 33.49 | |
| SSP370/2070 | 31.11 | 0.937 | 0.009 | + 21.63 | |
| SSP585/2050 | 56.30 | 0.962 | 0.009 | + 27.23 | |
| SSP585/2070 | 31.11 | 0.937 | 0.009 | + 21.89 | |
The predicted loss of future genetic diversity in the overall population varied based on the climatic niche and phylogeography of each species. For example, six of the 12 C. erraticus populations were predicted to be affected by climate change (Table S4). The CER1402, CER16EX2, and CER1822 populations were predicted to be extirpated in every scenario, whereas CER1808 (currently exhibiting low suitability) increased in suitability under most of the future climatic scenarios (all but SSP585 in 2070). In addition, 24 (54.6%) of the 44 C. erraticus haplotypes were estimated to be affected by at least one climate change scenario, and six were predicted to be completely lost in response to climate change (Fig. S6). For C. eumenes, all populations were predicted to be affected by climate change (Table S4). The CEU22A4 and CEU22A8 populations were predicted to be extirpated in every scenario, and under the most severe scenario, local extinction was predicted for all populations by 2070. In addition, five (19.2%) of the C. eumenes 26 haplotypes were predicted to be lost in response to climate change (Fig. S7). For G. mopsus, six of the 10 populations were predicted to be affected by climate change (Table S4). The GM1510 population was predicted to be extirpated in every scenario, whereas GM1403 (currently exhibiting low suitability) was predicted to increase in suitability. In addition, 107 (79.3%) of the 135 G. mopsus haplotypes were predicted to be affected by at least one climate change scenario, and two haplotypes were predicted to be lost (Fig. S8). However, despite the predicted losses in the number of haplotypes, none of the three species were expected to undergo large reductions in haplotype or nucleotide diversity (Table 3). Similarly, there was no evident loss of phylogenetic diversity (e.g., loss of clades) in the phylogenetic tree of each species. Instead, the missing haplotypes were scattered throughout the tree.
Discussion
This study is the first to model the effects of climate change on insects from the Gobi Desert and Mongolian Steppe region. Given the serious concerns about desertification in the Gobi Desert and adjacent regions7,8, there have been few studies investigating its influences on organisms there11–13. This issue fundamentally stems from the lack of accumulated data in this region. Our results on the distribution and genetic diversity of Mongolian dung beetles can be valuable data for future studies on global-scale desertification as well as for the responses of organisms to desertification in the Gobi Desert.
Genetic diversity and structure can be used as indicators of the tolerance and adaptive potential of a population to environmental stress32. In this study, C. erraticus and G. mopsus exhibited high genetic diversity with high singleton frequency, which could reflect the species’ adaptive potential. In contrast, the desert-dwelling C. eumenes exhibited moderate genetic diversity, with a single haplotype accounting for 65% of specimens, which could indicate that the adaptive potential of this species is lower than that of either C. erraticus or G. mopsus.
Most of the FST values among the populations of the three species were relatively small, suggesting low levels of interpopulation genetic differentiation, which could be related to contemporary gene flow or demographic events such as bottlenecks, recent divergence, or selection. For example, there were no significant FST values for distant populations, C. erraticus populations at CER1822 and CER1502 (over 1100 km), C. eumenes populations at CEU1609 and CEU1713 (over 450 km), and G. mopsus populations at GM1417 and GM2207 (over 790 km; Tables S1–S3). Notably, significant differences were observed between the Khangai Mountains and other populations of C. erraticus (Table 2). These differences could be derived from historical demographic events involving geographic barriers, for instance, the founder effect or selection on historically small, isolated populations from genetically diverse population in the past33. This phenomenon was observed only in the Khangai Mountains populations, whereas the Altai Mountains population (CER1822) did not exhibit significant differences from the other populations (except for CER1808). The genetic structures in both geographic and climatic group for G. mopsus coincided with those reported by previous study31.
The demographic histories of the three dung beetle species were observed to differ. The haplotype networks of all three species showed major haplotypes surrounded by many unique haplotypes. For C. eumenes, a single major haplotype was identified, while multiple major haplotypes were observed in the other two species. Our demographic history analyses indicated that C. eumenes and G. mopsus have undergone recent rapid expansion. A previous study proposed the domestication-driven expansion hypothesis to explain this recent expansion of Mongolian dung beetles31. However, Tajima’s D analysis and the mismatched distribution of the overall population revealed that C. erraticus maintained a constant-sized population rather than undergoing a recent expansion. One plausible explanation for this result could be the difference in food ranges among the three species. C. eumenes and G. mopsus prefer the feces of large herbivores, primarily livestock, whereas C. erraticus has a broader food range that includes the feces of small mammals. The presence of C. erraticus inside the burrows of rodents has been reported, indicating that the species can utilize the dung or residues found within burrows34. Therefore, C. erraticus may have been less affected by domestication events than the other two species. Another hypothesis is that the demographic histories of dung beetles in Mongolia were influenced more by late Quaternary climate change than by domestication events. Several previous studies have reported the influence of the late Quaternary climate oscillation, usually the last glacial maximum, on the population demography of species in Mongolia35,36.
The SDMs used in this study suggest that the distributions of dung beetles inhabiting a single biome type will contract and shift substantially in response to climate change. In contrast, the distributions of species inhabiting multiple biomes will expand, even if affected by range shifts. These trends are consistent with the idea that generalists and specialists respond differently to rapid environmental change. Species with broad tolerance exhibit range expansion, whereas species with restricted tolerance undergo range contraction37,38.
We revealed that the number of haplotypes would decrease following the extinction of local populations of all three species (Table 3). Under the most severe climate scenario (SSP585), C. erraticus and G. mopsus were predicted to experience haplotype losses of 54.5 and 68.9%, respectively, by 2070. Meanwhile, C. eumenes was predicted to have lost all extant haplotypes from the 10 sites investigated in this study. A reduction in the absolute number of haplotypes within the overall gene pool of a species is likely to reduce its evolutionary potential and resilience. However, despite reduced haplotype richness, the dung beetles modeled in this study were predicted to maintain genetic diversity (Table 3). These unexpected results may be attributed to the low mtDNA divergence of the species in both geographic configurations and lineages. In the phylogenetic trees of these species, unique haplotypes were scattered without geographic configuration or deep phylogenetic divergence. Therefore, haplotype loss due to local extinction would be significant under climate change scenarios, while low genetic divergence and a high singleton frequency could buffer the effects of climate change on genetic diversity and phylogeographic structure. Similar predictions have been reported by previous studies19,39.
Our predictions of future genetic diversity and distribution should be interpreted cautiously. First, the study could not address the extent to which dispersal might mitigate genetic diversity loss, and it was conservatively assumed that dispersal was restricted. In this study, local populations were assumed to disappear as habitats became climatically unsuitable, even when they were close to areas projected to persist or become climatically suitable. This assumption can enhance the prediction of potential genetic diversity loss, thereby contributing to effective conservation planning. However, it can reduce the prediction accuracy. For instance, the range of each species could shift due to enhanced fitness in locations at the leading edge and reduced fitness in locations at the trailing edge. This process tends to lead to a gradient in genetic diversity that increases toward the leading edge32,40. Second, the binary decision of presence or absence in the model neglected phenotypic plasticity and local adaptation. Several studies have investigated the local adaptation and phenotypic plasticity of dung beetles in terms of climatic differences. For example, Lim et al. (2020) reported variations in the morphology (body shape and size) of G. mopsus from steppe and desert in Mongolia41. Furthermore, Cuesta et al.42 reported the phenological plasticity of Iberian dung beetles, and Macagno et al. (2018) reported the behavioral plasticity of the model species Onthophagus taurus, which was observed to mitigate the negative effects of temperature increases43. This study indicates that C. erraticus and G. mopsus inhabit climatically unsuitable localities (C. erraticus: CER1822 and CER1005; G. mopsus: GM1403, GM1510, and GM1630) with relatively high genetic diversity (except for CER1005, which included only one individual), which might suggest that, in Mongolia, the response of dung beetles to climatic stress involves either local adaptation or phenotypic plasticity.
Researchers have recently attempted to incorporate evolutionary factors into SDMs44,45. For example, Benito-Garzόn et al.46 described a newly developed SDM that can incorporate fitness-related traits to estimate the sensitivity of species to environmental change. Abreu‐Jardim et al.39 forecasted the effects of climate change on the phylogeographic diversity of species. Nevertheless, such efforts remain in the early stages of development and have been applied to a few well-documented species. Determining the actual levels of genetic parameters required for responding to environmental change is challenging. Nonetheless, it’s essential to conserve a wide range of genotypes across both phylogeny and geography to retain a meaningful subset of this diversity21. Our results highlight that the spatial arrangement of haplotypes and lineages, reflecting the distinct phylogeographic histories of each species, offers valuable insights into their resistance against potential genetic diversity loss owing to climate change.
As a result of climate change, the distribution of steppe-inhabiting dung beetles was expected to shift toward the mountainous steppe. In contrast, the distributions of desert-inhabiting dung beetles were expected to shift toward the central region and to expand northward toward the Gobi Desert (Fig. 5). However, contrary to our expectations, the desert-inhabiting species C. eumenes experienced the most severe range contraction. This suggests that the modeled climate change may have exceeded the thermal and drought tolerance of dung beetles in the Gobi Desert. Widespread dung beetles were predicted to lose their current habitat in the southern desert region and to expand their distribution into the northern-central region, which was predicted to develop into a more suitable habitat (i.e., semi-desert habitat). Therefore, it is likely that the central region of Mongolia, which is an intermediate region between desert and steppe, will become a key region for dung beetle diversity conservation over the next 50 years (Fig. 5). National parks in central Mongolia, such as Hustai, Gorkhi-Terelj, and Khugu Tarna, serve as climate refuges and corridors for Mongolian dung beetles. These national parks will likely become the leading edge of the range-shifting populations of C. eumenes and the trailing edge (or refuge) and center (or corridor) populations of C. erraticus and G. mopsus, respectively. Therefore, conservation management in the central region of Mongolia is crucial for conserving biodiversity of Mongolian dung beetles.
Figure 5.
Future distribution of three model dung beetles in 2070 under the SSP585 scenario with schematic illustration of climate change and species response in the Gobi Desert and Mongolian Steppe. The map of distribution model was modified using ArcGIS 10.8 (www.esri.com)67.
In conclusion, we provide novel insights into the potential range shifts and genetic diversity losses of dung beetles from the Gobi Desert and Mongolian Steppe in response to climate change. Contrary to general expectations, desert species appear to be the most vulnerable to climate change due to the extensive degradation in the Gobi Desert. Both steppe and widespread species are likely to experience high rates of occupancy turnover and genetic loss, which could reshuffle the dung beetles community. Climate change-induced desertification should be considered an emerging threat to dung beetles in drylands worldwide.
Materials and methods
Study species, sampling, and DNA sequencing
Three dung beetle species that represent the typical biogeographic patterns of dung beetles in Mongolia were selected for this study: Colobopterus erraticus (Linnaeus, 1758), Cheironitis eumenes (Motschulsky, 1859), and Gymnopleurus mopsus (Pallas, 1781). These species generally inhabit the steppe, desert, and both biomes, respectively (Fig. 5). Adult dung beetles were collected either directly from dung pats of livestock (horses, cattle, sheep, goats, camels, and yaks) or using pit-fall traps baited with fresh horse dung. Specific preference for dung type was not observed for any species during field sampling. Specimens were preserved in 95% ethyl alcohol for subsequent DNA analysis.
Genomic DNA was extracted from leg and thoracic tissues using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germanton, NC, USA) following the manufacturer’s protocol. A fragment of the mitochondrial cytochrome oxidase I gene (COI) was amplified using polymerase chain reaction (PCR) in 20 µL reaction volumes using coleopteran-specific primers (forward: C1-J-2183, 5′-CAA CAT TTA TTT TGA TTT TTT GG-3′; reverse: TL2-N-3014, 5′-TCC AAT GCA CTA ATC TGC CAT ATT A-3′)47, universal primers (forward: LCO1490, 5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′; reverse: HCO2198, 5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′)48, AccuPower PCR PreMix (Bioneer, Daejeon, South Korea), and the following conditions: initial denaturation at 94 ℃ for 3 min; followed by 36 cycles of 94 °C for 30 s, 47–52 °C for 30 s, and 72 °C for 90 s; and a final extension step at 72 °C for 5 min. Amplification was verified using electrophoresis on 1.5% agarose gels and UV visualization. The verified PCR products were purified enzymatically by Exonuclease I and Shrimp Alkaline Phosphatase (New England BioLabs, Ipswich, MA, USA) and then sequenced by Macrogen Inc. (Seoul, South Korea) using an ABI Prism 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). All sequences were aligned using the Clustal W49 algorithm, implemented in MEGA X v.10.2.650, and were checked manually to confirm alignment. Some of COI sequences for G. mopsus were obtained from GenBank (MF674025–MF674381)31. The newly obtained sequences were deposited in GenBank under accession numbers OR363454–OR363626 (C. erraticus), OR363219–363370 (C. eumenes), and OR364414–OR364436 (G. mopsus).
Genetic diversity and haplotype network analyses
The number of haplotypes (Nh), haplotype diversity (Hd), and nucleotide diversity (π) of each population were estimated for each species using DnaSP v.6.12.0351. Allelic richness (Ar) was calculated using the rarefaction method in CONTRIB v.1.02552, that corrects for unequal sample sizes among populations. Haplotype networks for each species were constructed using minimum-spanning tree algorithms in HapStar v.0.753 and optimized using PopART v.1.754. Only populations from which at least five individuals (N ≥ 5) were sampled were included in subsequent genetic analyses, except for demographic history and intraspecific phylogenetic analyses.
Population structure and demographic history analyses
To infer the genetic differentiation and spatial genetic structure of the three species, ARLEQUIN v.3.555 was used to calculate fixation index (FST) values for pairwise population comparisons and to conduct hierarchical analysis of molecular variance (AMOVA, 1000 permutations). Each population of the three species was assigned to a geographic or climatic region, depending on the species’ distribution pattern. The C. erraticus populations were assigned to (1) two geographic groups: the Khangai Mountains (CER1621, 1808, and 2208) and others (CER1402, 1502, 1626, 1822, 2213, 2218, and 2222) and (2) three geographic groups: North (CER1402 and 2218), Central (CER1502, 1621, 1626, 2208, 2213, and 2222), and West (CER1808 and 1821). The C. eumenes populations were assigned to two geographic groups: southern (CEU1608, 1609, 22A4, 22A6, 22A7, and 22A8) and southeastern (CEU1415 and 1713). The G. mopsus populations were assigned to (1) two climatic groups: steppe (GM1403, 1628, 1630, and 2217) and desert (GM1417, 1510, 1604, 1605, 1611, and 162b); (2) three geographic groups: North (GM1403 and 2217), Central (GM1628 and 1630), South (GM1417, 1510, 1604, 1605, 1611, and 162b); and (3) four geographic groups based on major road networks: Northcentral (GM1403 and 1630), Northwest (GM1628 and 2217), South (GM1510, 1604, 1605, and 1611), and Southeast (GM1417 and 162b). The total molecular variance was partitioned among the groups (Φct = ′′inter-group′′ genetic variation), within the groups (Φsc = ′′intra-group′′ genetic variation), and within populations regardless of the grouping (Φst = ′′inter-population′′ genetic variation).
To infer demographic history, the past demographic expansion of the population of the three species was tested Tajima’s D56, and Fu’s Fs57 neutrality tests with 1000 random samples in ARLEQUIN v3.5. Mismatch distribution (distribution of pairwise sequence difference) analysis was performed in DnaSP v.6.12.03 and sum of squared deviations (SSD) and raggedness index values were calculated to assess the goodness of fit between the observed and expected distributions of demographic expansion model.
Intraspecific phylogeny
Phylogenetic trees of the three species were reconstructed using Bayesian inference (BI) and Maximum likelihood (ML). BI trees were reconstructed using Markov Chain Monte Carlo (MCMC) methods, assuming a constant coalescent size for the tree prior, in BEAST v.10.458. The best-fit evolutionary substitution models were selected using the Akaike information criterion (AICc) in jModelTest v.2.1.759 as follows: TrN + I for C. erraticus and C. eumenes; GTR + G + I for G. mopsus. Strict clock model was selected for three species based on calculation of the Bayes factor58,60. A COI substitution rate of 0.0075 (substitutions/site/million years) was based on previous phylogenetic study on dung beetles61. The MCMC run was performed using 10–100 million generations for each species, with sampling every 1000 generations, and the first 10 or 20% of generations were discarded as burn-in (Table S5). The run was examined using Tracer v.1.662 to confirm that stationarity and convergence had been reached (effective sample size [ESS] > 200). The Maximum Clade Credibility (MCC) tree was evaluated using TreeAnnotator v.10.458. ML analyses were performed using IQ-TREE63 with 1000 replicates in ultrafast bootstrap64. Intraspecific coalescent trees were visualized using FigTree v.1.4.465.
Species distribution modeling
From 2009 to 2022, field investigations were conducted at 164 sites distributed across Mongolia, ranging from 50° N to 43° N and 91° E to 112° E. Many SDM studies rely on distribution records from open-access databases, such as the Global Biodiversity Information Facility (GBIF). However, such databases often lack data for neglected regions and species, such as dung beetles in the Gobi Desert and Mongolian Steppe. To address this challenge, this study used data collected during our extensive field investigations conducted over the past 14 years (Table S6). Duplicate occurrence records were eliminated and rarefied to 5 km, resulting in 56, 28, and 45 occurrence records for C. erraticus, C. eumenes, and G. mopsus, respectively.
Bioclimatic variables for modeling (Table S7) were obtained from WorldClim66 at a spatial resolution of 30 arcsec (ca. 1 km2) using ArcGIS Desktop 10.867. For future climate projections to 2050 and 2070, three different Shared Socioeconomic Pathways (SSPs), including two moderate stabilization scenarios (SSP245 and SSP370) and one high baseline emission scenario (SSP585), were derived from the CMIP6 global climate model (BCC-CSM)68. The limitation of food resources is a crucial factor in determining species’ distribution69. However, in Mongolia, the tremendous number of livestock widely distributed through nomadic herding provides sufficient food resources across the landscape (National Statistics Office of Mongolia; https://www.en.nso.mn/, accessed on April 08, 2022)28,41.
To avoid multicollinearity and overfitting of the variables in the models, a Pearson’s correlation analysis was conducted using the SDM Toolbox v.2.070 to remove highly correlated bioclimatic variables (Pearson’s correlation coefficient >|0.8|; Table S8), and as a result, subsequent modeling was performed using seven variables, namely annual mean temperature (BIO1), mean diurnal range (BIO2), isothermality (BIO3), temperature seasonality (BIO4), annual precipitation (BIO12), precipitation during the driest month (BIO14), and precipitation seasonality (BIO15).
The current and future distributions of the three dung beetles were inferred using maximum entropy modeling71. The maximum entropy model (MaxEnt) is one of the most popular SDMs and uses presence-only data to estimate species distribution and habitat suitability72. The ′′ENMeval′′ R package73 was used to select the best models for each species based on the optimization of two parameters: regularization multiplier (RM) and feature combination (FC). SDMs were generated using tenfold cross-validation with 10,000 background points. The models were optimized using RM values ranging from 0.5 to 4.0 (in increments of 0.5) and six feature combinations (L, LQ, H, LQH, LQHP, and LQHPT), which resulted in a total of 48 parameter combinations of SDMs for each species. The k-fold cross-validation approach was used instead of the delete-one-jackknife approach, which performs better for small sample sizes74,75. The optimized SDMs were evaluated using the difference between the training and testing AUCs (AUCdiff) and 10% training omission rate (OR10), which indicate the degree of overfitting, and the Akaike information criterion corrected for small sample sizes (AICc), which reflects model complexity14,74,76. The optimal parameter combination for each species was selected based on the lowest delta.AICc value. To maintain a balance between model complexity and predictive accuracy, the values of OR10, AUCdiff, and AUCtest were considered collectively14,77.
All SDMs were generated using the ′′ENMTools′′ R package78, and MaxEnt v.3.4.471.
Predicting distribution and genetic diversity under climate change scenarios
To simulate the future distributions and genetic diversities of the three dung beetle species under three climatic scenarios (SSP245, SSP370, and SSP585) at two time points (2050 and 2070), the predicted suitability across Mongolia was transformed into binary maps (presence/absence) based on the 10% training presence logistic threshold on the established final SDMs for each species. Changes in distribution were calculated as the difference in range size between current distributions and those predicted at each time point under each climatic scenario. We assumed that only populations occurring in areas with suitability greater than the present threshold would persist and, thus, contribute to the gene pool of subsequent generations39. The number of haplotypes, haplotype diversity, and nucleotide diversity were only calculated for the populations remaining at each time point under each climatic scenario. The loss of haplotypes under different scenarios was visualized in the phylogenetic tree for each species.
Supplementary Information
Acknowledgements
We are grateful to members of the National University of Mongolia (NUM) and Korea University (KU) for their assistance with field investigations. We thank Kijong Cho (KU) and Jinsol Hong (KU) for their valuable input regarding distribution modeling. This study was supported by a National Research Foundation (NRF) grant funded by the Ministry of Education of Korea (Grant Number 2022R1A2C1009024).
Author contributions
Conceptualization: C.L., J.H.K., B.B., Y.J.B. Sampling: C.L., B.B., Y.J.B. Experiments and Data analysis: C.L., J.H.K., Writing: C.L., J.H.K. Y.J.B. Review and editing: C.L., J.H.K., B.B., Y.J.B.
Data availability
The sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession number OR363454–OR363626, OR363219–OR363370, and OR364414–OR364436. The geographical coordinates of three model dung beetles occurrences are provided in Table S2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-66260-1.
References
- 1.Walther GR, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- 2.Keogan K, et al. Global phenological insensitivity to shifting ocean temperatures among seabirds. Nat. Clim. Change. 2018;8:313–318. doi: 10.1038/s41558-018-0115-z. [DOI] [Google Scholar]
- 3.Dawson TP, Jackson ST, House JI, Prentice IC, Mace GM. Beyond predictions: Biodiversity conservation in a changing climate. Science. 2011;332:53–58. doi: 10.1126/science.1200303. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- 5.Razgour O, et al. An integrated framework to identify wildlife populations under threat from climate change. Mol. Ecol. Resour. 2018;18:18–31. doi: 10.1111/1755-0998.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prince SD, Wessels KJ, Tucker CJ, Nicholson SE. Desertification in the Sahel: A reinterpretation of a reinterpretation. Glob. Change Biol. 2007;13:1308–1313. doi: 10.1111/j.1365-2486.2007.01356.x. [DOI] [Google Scholar]
- 7.IPCC. Climate change and land, in An IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems. https://www.ipcc.ch/srccl (2019).
- 8.D’Odorico P, Bhattachan A, Davis KF, Ravi S, Runyan CW. Global desertification: Drivers and feedbacks. Adv. Water Resour. 2013;51:326–344. doi: 10.1016/j.advwatres.2012.01.013. [DOI] [Google Scholar]
- 9.Liang X, et al. Research progress of desertification and its prevention in Mongolia. Sustainability. 2021;13:6861. doi: 10.3390/su13126861. [DOI] [Google Scholar]
- 10.Han J, Dai H, Gu Z. Sandstorms and desertification in Mongolia, an example of future climate events: A review. Environ. Chem. Lett. 2021;19:4063–4073. doi: 10.1007/s10311-021-01285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, et al. Modelling the potential distribution and shifts of three varieties of Stipa tianschanica in the eastern Eurasian Steppe under multiple climate change scenarios. Glob. Ecol. Conserv. 2018;16:e00501. doi: 10.1016/j.gecco.2018.e00501. [DOI] [Google Scholar]
- 12.Qin A, et al. Predicting the current and future suitable habitats of the main dietary plants of the Gobi Bear using MaxEnt modeling. Glob. Ecol. Conserv. 2020;22:e01032. doi: 10.1016/j.gecco.2020.e01032. [DOI] [Google Scholar]
- 13.Ye X, et al. Impacts of future climate and land cover changes on threatened mammals in the semi-arid Chinese Altai Mountains. Sci. Tot. Environ. 2018;612:775–787. doi: 10.1016/j.scitotenv.2017.08.191. [DOI] [PubMed] [Google Scholar]
- 14.Warren DL, Seifert SN. Ecological niche modeling in Maxent: The importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011;21:335–342. doi: 10.1890/10-1171.1. [DOI] [PubMed] [Google Scholar]
- 15.Elith J, Kearney M, Phillips S. The art of modelling range-shifting species. Methods Ecol. Evol. 2010;1:330–342. doi: 10.1111/j.2041-210X.2010.00036.x. [DOI] [Google Scholar]
- 16.Pacifici M, et al. Assessing species vulnerability to climate change. Nat. Clim. Change. 2015;5:215–224. doi: 10.1038/nclimate2448. [DOI] [Google Scholar]
- 17.Carvalho SB, Brito JC, Crespo EG, Watts ME, Possingham HP. Conservation planning under climate change: Toward accounting for uncertainty in predicted species distributions to increase confidence in conservation investments in space and time. Biol. Conserv. 2011;144:2020–2030. doi: 10.1016/j.biocon.2011.04.024. [DOI] [Google Scholar]
- 18.Thuiller W, et al. A road map for integrating eco-evolutionary processes into biodiversity models. Ecol. Lett. 2013;16:94–105. doi: 10.1111/ele.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvanovic M, Kennedy JD, Nogués-Bravo D, Marske KA. Persistence of genetic diversity and phylogeographic structure of three New Zealand forest beetles under climate change. Divers. Distrib. 2019;25:142–153. doi: 10.1111/ddi.12834. [DOI] [Google Scholar]
- 20.Willi Y, van Buskirk J, Hoffmann AA. Limits to the adaptive potential of small populations. Ann. Rev. Ecol. Evol. Syst. 2006;37:433–458. doi: 10.1146/annurev.ecolsys.37.091305.110145. [DOI] [Google Scholar]
- 21.Moritz C. Conservation units and translocations: Strategies for conserving evolutionary processes. Hereditas. 1999;130:217–228. doi: 10.1111/j.1601-5223.1999.00217.x. [DOI] [Google Scholar]
- 22.Nichols E, et al. Ecological functions and ecosystem services provided by Scarabaeinae dung beetles. Biol. Conserv. 2008;141:1461–1474. doi: 10.1016/j.biocon.2008.04.011. [DOI] [Google Scholar]
- 23.Davis AJ, et al. Dung beetles as indicators of change in the forests of northern Borneo. J. Appl. Ecol. 2001;38:593–616. doi: 10.1046/j.1365-2664.2001.00619.x. [DOI] [Google Scholar]
- 24.Bicknell JE, et al. Dung beetles as indicators for rapid impact assessments: Evaluating best practice forestry in the neotropics. Ecol. Indic. 2014;43:154–161. doi: 10.1016/j.ecolind.2014.02.030. [DOI] [Google Scholar]
- 25.Numa C, et al. The Conservation Status and Distribution of MEDITERRANEAN DUNG BEETLES. IUCN; 2020. [Google Scholar]
- 26.Davis ALV. Dung beetle diversity in South Africa: Influential factors, conservation status, data inadequacies and survey design. Afr. Entomol. 2002;10:53–65. [Google Scholar]
- 27.Menéndez R, González-Megías A, Jay-Robert P, Marquéz-Ferrando R. Climate change and elevational range shifts: Evidence from dung beetles in two European mountain ranges. Glob. Ecol. Biogeogr. 2014;23:646–657. doi: 10.1111/geb.12142. [DOI] [Google Scholar]
- 28.Bayartogtokh B, Kim JI, Bae YJ. Lamellicorn beetles (Coleoptera: Scarabaeoidea) in Korea and Mongolia. Entomol. Res. 2012;42:211–218. doi: 10.1111/j.1748-5967.2012.00468.x. [DOI] [Google Scholar]
- 29.Bayartogtokh B, Otgonjargal E. Assemblages of coprophilous beetles (Insecta: Coleoptera) in the pastureland of Central Mongolia. Mong. J. Biol. Sci. 2009;7:19–27. [Google Scholar]
- 30.Jargalsaikhan P, et al. Communities of dung beetles (Coleoptera: Scarabaeoidea) in the steppe of Mongolia. Mong. J. Biol. Sci. 2021;19:17–27. [Google Scholar]
- 31.Kang JH, et al. Historical domestication-driven population expansion of the dung beetle Gymnopleurus mopsus (Coleoptera: Scarabaeidae) from its last refuge in Mongolia. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-22182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauls SU, Nowak C, Bálint M, Pfenninger M. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 2013;22:925–946. doi: 10.1111/mec.12152. [DOI] [PubMed] [Google Scholar]
- 33.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Ziani S, Gharakhloo MM. Further records of Scarabaeoidea found inside burrows of rodents in Iran (Coleoptera) Fragm. Entomol. 2011;43:57–74. doi: 10.4081/fe.2011.53. [DOI] [Google Scholar]
- 35.Lv X, et al. Continental refugium in the Mongolian Plateau during Quaternary glacial oscillations: Phylogeography and niche modelling of the endemic desert hamster, Phodopus roborovskii. PLoS One. 2016;11:e0148182. doi: 10.1371/journal.pone.0148182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekiné K, Bayartogtokh B, Bae YJ. Post-glacial distribution of a Mongolian mayfly inferred from population genetic analysis. Freshw. Biol. 2017;62:102–110. doi: 10.1111/fwb.12853. [DOI] [Google Scholar]
- 37.Duan RY, Kong XQ, Huang MY, Varela S, Ji X. The potential effects of climate change on amphibian distribution, range fragmentation and turnover in China. PeerJ. 2016;4:e2185. doi: 10.7717/peerj.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler CJ, Larson M. Climate change winners and losers: The effects of climate change on five palm species in the Southeastern United States. Ecol. Evol. 2020;10:10408–10425. doi: 10.1002/ece3.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abreu-Jardim TP, Jardim L, Ballesteros-Mejia L, Maciel NM, Collevatti RG. Predicting impacts of global climatic change on genetic and phylogeographical diversity of a Neotropical treefrog. Divers. Distrib. 2021;27:1519–1535. doi: 10.1111/ddi.13299. [DOI] [Google Scholar]
- 40.Kawecki TJ. Adaptation to marginal habitats. Ann. Rev. Ecol. Evol. Syst. 2008;39:321–342. doi: 10.1146/annurev.ecolsys.38.091206.095622. [DOI] [Google Scholar]
- 41.Lim C, et al. Morphometric analysis of dung beetle (Gymnopleurus mopsus: Scarabaeidae: Coleoptera) populations from two different biomes in Mongolia. Biol. J. Linnean Soc. 2020;131:369–383. doi: 10.1093/biolinnean/blaa110. [DOI] [Google Scholar]
- 42.Cuesta E, Mingarro M, Lobo JM. Between locality variations in the seasonal patterns of dung beetles: The role of phenology in mitigating global warming effects. Ecol. Entomol. 2021;46:592–600. doi: 10.1111/een.13005. [DOI] [Google Scholar]
- 43.Macagno AL, Zattara EE, Ezeakudo O, Moczek AP, Ledón-Rettig CC. Adaptive maternal behavioral plasticity and developmental programming mitigate the transgenerational effects of temperature in dung beetles. Oikos. 2018;127:1319–1329. doi: 10.1111/oik.05215. [DOI] [Google Scholar]
- 44.Riddell EA, Odom JP, Damm JD, Sears MW. Plasticity reveals hidden resistance to extinction under climate change in the global hotspot of salamander diversity. Sci. Adv. 2018;4:eaar5471. doi: 10.1126/sciadv.aar5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liepe KJ, van der Maaten E, van der Maaten-Theunissen M, Liesebach M. High phenotypic plasticity, but low signals of local adaptation to climate in a large-scale transplant experiment of Picea abies (L.) Karst. in Europe. Front. For. Glob. Change. 2022;5:804857. doi: 10.3389/ffgc.2022.804857. [DOI] [Google Scholar]
- 46.Benito-Garzón M, Robson TM, Hampe A. ΔTrait SDMs: Species distribution models that account for local adaptation and phenotypic plasticity. New Phytol. 2019;222:1757–1765. doi: 10.1111/nph.15716. [DOI] [PubMed] [Google Scholar]
- 47.Simon C, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 48.Folmer O, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 49.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozas J, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 52.Petit RJ, Mousadik AE, Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 1998;12:844–855. doi: 10.1111/j.1523-1739.1998.96489.x. [DOI] [Google Scholar]
- 53.Teacher AGF, Griffiths DJ. HapStar: Automated haplotype network layout and visualization. Mol. Ecol. Resour. 2011;11:151–153. doi: 10.1111/j.1755-0998.2010.02890.x. [DOI] [PubMed] [Google Scholar]
- 54.Leigh JW, Bryant D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 55.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 56.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li WLS, Drummond AJ. Model averaging and Bayes factor calculation of relaxed molecular clocks in Bayesian phylogenetics. Mol. Biol. Evol. 2012;29:751–761. doi: 10.1093/molbev/msr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mlambo S, Sole CL, Scholtz CH. A molecular phylogeny of the African Scarabaeinae (Coleoptera: Scarabaeidae) Arthropod Syst. Phylogeny. 2015;73:303–321. doi: 10.3897/asp.73.e31806. [DOI] [Google Scholar]
- 62.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trifinopoulos J, et al. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoang DT, et al. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambaut, A. FigTree-v1.4.2. 2014. http://tree.bio.ed.ac.uk/software/figtree (2014).
- 66.Fick SE, Hijmans RJ. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017;37:4302–4315. doi: 10.1002/joc.5086. [DOI] [Google Scholar]
- 67.ESRI. ArcGIS Desktop Release 10.8. Redlands: Environmental Systems Research Institute (2020).
- 68.Wu T, et al. The Beijing climate center climate system model (BCC-CSM): The main progress from CMIP5 to CMIP6. Geosci. Model Dev. 2019;12:1573–1600. doi: 10.5194/gmd-12-1573-2019. [DOI] [Google Scholar]
- 69.Barve N, et al. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol. Model. 2011;222:1810–1819. doi: 10.1016/j.ecolmodel.2011.02.011. [DOI] [Google Scholar]
- 70.Brown JL, Bennett JR, French CM. SDMtoolbox 2.0: The next generation Python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. PeerJ. 2017;5:e4095. doi: 10.7717/peerj.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phillips SJ, Anderson RP, Dudík M, Schapire RE, Blair ME. Opening the black box: An open-source release of Maxent. Ecography. 2017;40:887–893. doi: 10.1111/ecog.03049. [DOI] [Google Scholar]
- 72.Elith J, et al. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011;17:43–57. doi: 10.1111/j.1472-4642.2010.00725.x. [DOI] [Google Scholar]
- 73.Kass JM, et al. ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol. Evol. 2021;12:1602–1608. doi: 10.1111/2041-210X.13628. [DOI] [Google Scholar]
- 74.Pearson RG, Raxworthy CJ, Nakamura M, Townsend Peterson A. Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. J. Biogeogr. 2007;34:102–117. doi: 10.1111/j.1365-2699.2006.01594.x. [DOI] [Google Scholar]
- 75.Shcheglovitova M, Anderson RP. Estimating optimal complexity for ecological niche models: A jackknife approach for species with small sample sizes. Ecol. Model. 2013;269:9–17. doi: 10.1016/j.ecolmodel.2013.08.011. [DOI] [Google Scholar]
- 76.Radosavljevic A, Anderson RP. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2014;41:629–643. doi: 10.1111/jbi.12227. [DOI] [Google Scholar]
- 77.Velasco JA, González-Salazar C. Akaike information criterion should not be a “test” of geographical prediction accuracy in ecological niche modelling. Ecol. Inform. 2019;51:25–32. doi: 10.1016/j.ecoinf.2019.02.005. [DOI] [Google Scholar]
- 78.Warren DL, et al. ENMTools 1.0: An R package for comparative ecological biogeography. Ecography. 2021;44:504–511. doi: 10.1111/ecog.05485. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession number OR363454–OR363626, OR363219–OR363370, and OR364414–OR364436. The geographical coordinates of three model dung beetles occurrences are provided in Table S2.