Abstract
The gut microbiome has come to prominence across research disciplines, due to its influence on major biological systems within humans. Recently, a relationship between the gut microbiome and hematopoietic system has been identified and coined the gut-bone marrow axis. It is well established that the hematopoietic system and gut microbiome separately alter with age; however, the relationship between these changes and how these systems influence each other demands investigation. Since the hematopoietic system produces immune cells that help govern commensal bacteria, it is important to identify how the microbiome interacts with hematopoietic stem cells (HSCs). The gut microbiota has been shown to influence the development and outcomes of hematologic disorders, suggesting dysbiosis may influence the maintenance of HSCs with age. Short chain fatty acids (SCFAs), lactate, iron availability, tryptophan metabolites, bacterial extracellular vesicles, microbe associated molecular patterns (MAMPs), and toll-like receptor (TLR) signalling have been proposed as key mediators of communication across the gut-bone marrow axis and will be reviewed in this article within the context of aging.
Keywords: Aging, Microbiome, Hematopoietic stem cells, Stem cell regulation, Hematologic disorders, Gut dysbiosis, Inflammation
Highlights
-
•
Changes to the microbiome during aging can impact age-associated functional decline.
-
•
Microbially derived molecules can influence and regulate hematopoietic stem cells.
-
•
Key microbial mediators play a role in communication via the gut-bone marrow axis.
-
•
Extracellular vesicles pose a potential mechanism to transport microbial molecules.
1. Introduction
This review aims to investigate the relationship between the microbiota and hematopoietic system and address how age impacts this relationship. The microbiota consists of a collection of bacteria, fungi, archaea, and viruses that naturally inhabit the human body in a commensal relationship [1,2]. Each individual's microbiota is dependent on age, diet, medication, lifestyle, and physical environment [3]. Microbes release various metabolites and molecules that can directly enter the circulation to induce cellular changes. Although the hematopoietic system sustains blood and immune cell populations, the microbiota plays a significant role in the development of innate and adaptive immunity during the lifetime of the individual [4]. Because the microbiome releases molecules directly into circulation, the microbiota has the potential to contribute to the development of age-associated changes within hematopoietic stem cells (HSCs) and mature hematopoietic populations. Murine studies investigating the microbiome and hematopoietic system are limited with respect to extrapolating findings to humans, due to differences between mouse and human hematopoietic and microbiome compositions. Despite the variation between mouse and human, the effects of microbial influence on hematopoiesis in murine models provide a preliminary basis for modelling the gut-bone marrow axis. The age-associated changes observed in the gut microbiome and their possible role in the functional decline of the hematopoietic system will be reviewed in this article, with the goal of outlining the current understanding of the gut-bone marrow axis.
2. Changes in the gut microbiome throughout life and markers of healthy aging
The human microbiota is composed of diverse communities of bacteria, fungi, archaea, and viruses that are proven to systemically influence physiological processes, with the gut biota being the most densely populated and most studied [5]. The diversity and composition of microbial populations is dynamic and changes throughout life (Fig. 1). It has been shown that the chronological age of healthy mice can be accurately estimated from bacterial RNA-seq data of their feces [6]. Theoretically age-based models could be developed using human datasets considering the composition of the gut microbiome is also known to be indicative of age. However, a more thorough characterization of human bacterial gut populations during aging is required [7].
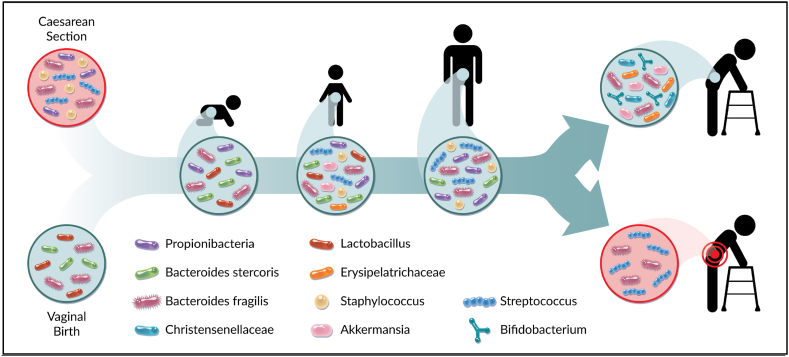
Fig. 1.
Microbiota composition throughout life. The human microbiota is influenced by various factors unique to an individual, including differences in diet, environment, and exposure to antibiotics resulting in increases and decreases in microbial diversity throughout life. However, certain bacterial species confer greater protection against age-related decline. At birth, the infant's microbiota is initially influenced by the mother's microbiota and by the mode of birth. Through adolescence and adult life, the microbiota is altered by environmental changes during an individual's life. The microbiota is stable throughout adulthood unless there is a pressure that causes dysbiosis. The later stages of aging correspond to a decrease in diversity within an individual, but an increased diversity between elderly individuals. Considering the gut-bone marrow axis, the composition of an aged microbiota may influence the onset of age-related decline within HSCs and the hematopoietic system. Figure made by Adobe Suite.
At birth, the human microbiome is significantly influenced by maternal microbiota, through childbirth and breastfeeding [8]. Infants born vaginally vs Caesarian-section have significantly different microbiomes [9,10]. Infants born vaginally have microbiomes enriched for Lactobacillus species introduced from the vaginal microbiota of the mother, while infants born by Caesarian-section have microbiomes enriched for organisms known to colonize the skin including Staphylococcus, Streptococcus, and Cutibacterium [8]. Development of the human gut microbiome has also been shown to be influenced by feeding modality, with breast-fed infants possessing increased Bifidobacterium and Lactobacillus populations [11]. It is clear from these studies that the early intimate physical environment provides the opportunity for initial microbial colonization. Therefore, events in life where there is close physical contact can drive development of the human microbiome. Although delivery method does not measurably influence the composition of the microbiota beyond 6 months of age, dysbiosis endured during early life may impact the development of disease at later stages [12].
In the first year of life an infant's microbiome is less complex, but often similar to the maternal microbiome, and begins to rapidly shift when solid food is introduced [13]. Despite infants having less individual complexity (alpha-diversity) compared to adults, there is greater diversity between infants (beta-diversity) than between adults [14]. At around age three, children start to develop similar microbial communities and variability of species as found in adults. Alpha-diversity of microbial species inhabiting the gut tends to increase throughout adulthood and beta-diversity tends to decrease from childhood to adulthood [15]. There are known sex differences in community profiles of the human gut microbiome, with certain bacterial populations being differentially enriched between sexes [16]. In a study on 2301 healthy individuals from Ukraine, it was determined that the female microbiome had proportionally more Firmicutes and Actinobacteria, and proportionally less Bacteroides compared to the male microbiome [16].
The adult gut can be influenced by various factors including their diet, lifestyle, environment, disease state, and treatment with antibiotics [17]. Disruption of the gut microbiota with infection and subsequent antibiotic treatment can occur many times throughout every human's life, which challenges the gut microbiome resulting in dysbiosis. Eventually the human gut recolonizes following the end of antibiotic treatment, but the time it takes to return to normal depends on alpha-diversity before treatment, the health of the individual, and their chronological stage of life [18]. Because some individuals vary at recovery from antibiotics, the use of narrow spectrum antibiotics are considered over broad-spectrum treatments to minimize dysbiosis [19,20].
The influence of diet on the composition of the human gut microbiome has been well characterized using studies documenting eating habits from different cultures and geographic locations. The western diet ranks as one of the poorest diets for promoting gut health, with a high fat and low fiber intake characteristic of American cuisine [21]. Individuals that immigrate from Asia to the United States have been shown to rapidly lose microbial diversity in the gut after immigrating, as well as an increased risk for obesity [22]. Characterizing specific commensal bacterial species comprising a healthy human microbiome, is a major focus in the field [15,18]. Bacteria that have been found to be consistent and important inhabitants across many different host species are known as keystone species. Examples of human keystone species include Bacteroides fragilis and Bacteroides stercosis, known to influence the macro structure of the gut microbiome and are found across most age groups and geographical areas [23]. Keystone species are essential in the reconstitution of the microbiome after events like antibiotic treatment, that cause depletion of species susceptible to certain drugs [23]. Most keystone species are highly efficient at degrading complex polysaccharides in the gut, which is thought to play a protective role by providing alternative energy sources for other bacterial species after an event of dysbiosis [24].
During the latest stages of life, the alpha-diversity of the gut microbiome tends to decrease, while the trajectory of beta-diversity also reverses and increases for the remainder of life [25]. The genus’ Akkermansia and Erysipelotrichaceae have been shown to be more abundant in healthy aging individuals, while the Streptococcus genus has been shown to be more abundant in individuals with an unhealthy aging phenotype [26]. Diseases associated with aging, such as cancer, cardiovascular disease, pulmonary disease, and diabetes are associated with the colonization of more pathogenic microbiota [26]. Individuals that live past the age of 105 i.e. supercentenarians, are often enriched in Akkermansia, Bifidobacterium, and Christensenellaceae [27]. In addition, Akkermansia muciniphia has been shown in mouse models to reduce age-related decline through anti-inflammatory effects on various tissues [28].
The gut microbiome and metabolites such as short chain fatty acids (SCFA), can serve as markers of healthy aging. Acetate, butyrate, and propionate are produced by the gut microbiota and can act as markers of physical frailty [29]. Other SCFA metabolites are known to play a role in inflammation and immunity [30]. For example, butyrate produced by the commensal class of bacteria Clostridia, positively correlate to the number of regulatory T-cells in the colon of mouse models [31]. A proposed mechanism by which certain SCFAs (acetate propionate, and butyrate) promote healthy aging, includes the ability to influence signalling pathways involved in redox homeostasis pathways mediated by nuclear erythroid 2-related factor 2 (Nrf2), which may be protective against oxidative stress [32].
Lactate has a dual role as a metabolite produced by specific colonic bacteria, as well as an important factor in the production of SCFAs, thereby making lactate vital in the homeostasis of the microbiota and colonization of key microbial species. It is important to note that a healthy microbiome is reliant on diverse microbiota to balance lactate production and consumption [33]. When an over abundance of lactate occurs, this is associated with increased inflammation and reduced production of butyrate and propionate.
Indole, another metabolite of the microbiome, is important in maintaining gut health through the activation of the aromatic hydrocarbon receptor (AhR). Indole is involved in the production of AhR ligands and consequently plays a role in the regeneration of intestinal epithelial cells after injury. AhR maintains homeostasis by protecting the stem cell niche, through restriction of proliferation via transcriptional regulators [34]. Microbial production of indole also plays a role in the AhR/IL-22 axis. Activation of this axis increases IL-22 transcription and balances mucosal responses allowing the development of a diverse microbiome [35].
Lastly, despite not being a metabolite, iron is another factor key to induction of cellular signaling events triggered by the microbiota. Iron plays a vital role influencing the composition of the microbiota, while inversely aiding in iron storage within intestinal cells [36]. Several studies have identified that the microbiota and iron regulate each other; this crosstalk has been observed in infants and adults and its regulation is vital to prevent anemia across multiple age groups [[37], [38], [39]]. These studies highlight that many metabolites of the microbiome play an important role, both directly and indirectly, in regulating gut health and are perturbed with age-associated changes to the microbiota.
3. Changes in the hematopoietic system through life
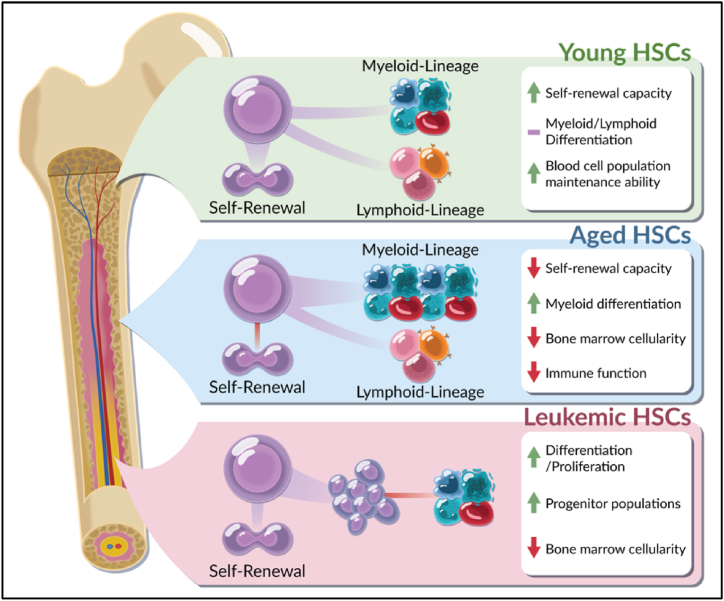
Biological aging can be defined as time dependent changes resulting in a functional decline of molecular mechanisms, at all levels of cellular organization. Although aging is associated with specific biological alterations, not all cellular systems experience functional decay at the same rate. Biological age refers to the phenotypic presentation of age-related decline, while chronological age refers to the time that has passed from birth to a given date. Individuals with decreased biological aging can be defined as super-agers and are typified by a delay in age-related decline such as myeloid differentiation, loss of self-renewal, and decreased HSC fitness [40] (Fig. 2). In contrast, accelerated agers are associated with a more rapid decline in cellular mechanisms and are more prone to earlier disease onset [40].
Fig. 2.
Changes within young, old and leukemic hematopoietic stem cells. During aging, hematopoietic stem cells (HSCs) undergo changes that affect function. Young HSCs display balanced myeloid and lymphoid differentiation and an ability to maintain homeostatic mature blood cell populations. In contrast, old HSCs are associated with decreased self-renewal and the onset of myeloid-biased differentiation. Leukemic HSCs exhibit specific changes characteristic of malignant transformation, however, these are more consistent with aged HSCs than young HSCs. Myeloid malignancies are characterized by a reduction in lymphoid progenitors and mature cells with concurrent over-expansion of myeloid progenitor populations. Figure made by Adobe Suite.
Hematopoietic stem cells exist as long-term HSC (LT-HSC) and short-term HSC (ST-HSC) populations. Through a process of asymmetric cell division, LT-HSCs can self-renew to sustain the stem cell pool or differentiate into short-term HSCs (ST-HSCs) that undergo proliferation and differentiation to produce terminally differentiated, functional hematopoietic cells [41]. ST-HSCs or multipotent progenitors (MPPs) sustain hematopoiesis in the short term, while the LT-HSCs are required to maintain the stem cell pool over the lifespan of the organism [42]. Since the probability of acquiring mutations increases as individuals age, LT-HSCs of elderly individuals are more likely to harbor mutations.
Aging impacts hematopoietic stem and progenitor cells (HSPCs) and as a result, the absolute number of HSCs increase [43]. HSCs also display reduced functionality and are less capable of producing mature cells. Changes to quantity and function of these populations subsequently lead to the dysfunction of homeostatic local and systemic immune response mechanisms. Aging is associated with the acquisition of a subset of mutations in HSCs that results in the expansion of a single hematopoietic stem cell clone. This phenomenon, known as clonal hematopoiesis (CH), increases as individuals age and is detected in 10–20 % of individuals over 70 years [44].
Aged HSCs can be characterized by the mutations they carry, resulting in loss of function or the transcriptional upregulation of genes involved in cell-cycle, cell proliferation, and hematopoietic development. The most common CH mutations are deletions in epigenetic regulators (DNMT3A and TET2) and are often the earliest acquired mutations in acute myeloid leukemia (AML) patients [45]. Mutations in CH genes ultimately result in increased self-renewal capacity, and a differentiation bias toward myeloid lineages [46]. Aged HSCs exhibit altered cellular fitness through the acquisition of other mutations resulting in deficiencies in DNA damage repair pathways (PPM1D, TP53, ATM, CHEK2), 3’ splicing, (SF3B1, SRSF2, U2AF1) and cell cycle activity (DNMT3A, TET2, ASXL1) [47]. These mutations are acquired exponentially in an age-associated manner [48]. The rate of somatic mutation acquisition in humans increases with chronological age, putting the individual at a greater risk of decline. For example, deficiencies in DNA damage repair ultimately allow for an increase in mutational burden, which results in the loss of stem cell function. Mutations in these genes correspond to an increased frequency of immunophenotypic HSCs observed in aged mice and humans, that produce less functional mature blood cells [49].
Hematopoietic stem cells and progenitors are also susceptible to time dependent changes. As HSPCs differentiate, they commit to either lymphoid or myeloid clonal-level lineage commitment pathways resulting in lymphopoiesis or myelopoiesis. Within the lymphoid compartment, aging induces disruption of T-cell function and a reduction of B-cell production. T-cells are central to adaptive immunity and migrate from the bone marrow to develop and mature within the thymus before contributing to immunological processes. T-cells aid in the immune response by activating B-cells and by directly killing infected cells. During aging, T-cells display reduced T-cell receptor (TCR) diversity [50]. Decreased TCR-antigen presenting cell (APC) signaling leads to defects in the recruitment of the supramolecular activation cluster (SMAC), also known as the immunological synapse. The immunological synapse is the site where the TCR is triggered by antigen ligands [51]. Dysfunction in SMAC recruitment results in a reduction in activated T-cells. The decreased functionality of T cells is a major contributor to the age-related changes observed in mature blood cell populations.
The other lymphoid lineage of mature blood cells known to change as humans age are B-cells, which initially develop within bone marrow, migrate, and eventually produce antibodies in response to pathogens. B-cell activation and maturation can be T-cell dependent or independent and occurs within germinal centers of secondary lymphoid tissues; lymph nodes, spleen, and tonsils [52]. Aging leads to reduced B-cell maturation, as well as a decreased induction of germinal center formation [53]. Both of these processes rely on CD4+ helper T cells and subsequent impairments in T-cell function correspond to direct alterations in B-cell function. Dysfunctional B-cells observed with aging are associated with altered immunoglobulin levels [54]. Similar to T cells, there is general decline in the functional capacity of B-cells as humans age, contributing to the overall decline in immunological function.
In contrast to lymphoid populations, myeloid cells increase in number throughout aging, a process referred to as myeloid skewing, whereby a greater proportion of HSCs commit to the myeloid lineage. However, despite the increase in quantity, the overall functionality of myeloid cells decrease. Neutrophils display reduced phagocytosis and cell killing in older individuals [55]. Macrophages have been shown to functionally decline similar to neutrophils, where older mice display a reduction in phagocytic activity [56]. Monocytes isolated from older humans have been shown to have both attenuated immune responses when stimulated by toll like receptor (TLR) signaling and a decreased cytotoxicity against tumor cells due to decreased activation status [57]. Similar to other myeloid cells, erythrocytes and erythrocyte parameters, such as hemoglobin levels, display an age-related decline in both males and females [58,59]. Further supporting this observation, it has been observed that 10–15 % of individuals older than 65 years have anemia due to age-related dysfunction, rather than nutritional deficiencies. This figure increases to 24.2 % and 39.5 % in females and males respectively when older than 85 years of age [58]. The prevalence of anemia is higher in populations of lower-middle income countries [60].
The bone marrow microenvironment or milieu is also profoundly affected by time. Cell types that comprise the bone marrow milieu include endothelial cells, mesenchymal stem cells (MSC), fibroblasts, adipocytes, chondrocytes, osteoblasts, megakaryocytes, CXCL12-abundant reticular (CAR) cells, pericytes, smooth muscle cells, sympathetic neurons, Schwann cells and macrophages, all of which contribute to HSC maintenance during the lifetime of the individual [61,62]. The composition of the bone marrow milieu also influences the functionality of HSCs [43]. This is demonstrated by experiments transplanting HSCs to young or old bone marrow environments. Transplantation of old HSCs into young mice (8–10 weeks) resulted in the rejuvenation of old HSCs, which were functionally equivalent to young HSCs. Transplantation of young HSCs within old mice (19–24 weeks), resulted in a decrease in HSC function. The restoration of function to old HSCs were attributed to factors provided by the young bone marrow milieu [63]. Bone marrow milieu extracellular factors such as stem cell factor (SCF) and CXCL12 are highly expressed by osteoprogenitors and CAR cells which additionally influence HSC activity [64]. The accessibility and regulation of these bone marrow factors are aided by MSCs, skeletal cells, and stromal cells. Within the bone marrow it has been shown that LT-HSCs bind to cadherin positive cells which are frequently located near homogenous perivascular regions of the bone marrow [65]. Therefore, both perivascular regions and cadherin positive cells in the bone marrow are important in the age-related changes observed in the regulation and maintenance of HSCs.
Bone marrow aging is associated with the activation of pro-inflammatory responses that contributes to a systemic low-grade inflammation, known as inflamm-aging [66]. The induction of inflamm-aging is activated by the release of pro-inflammatory cytokines (IL-1, IL-6, IL-11, TNF-a, IFN-a, IFN-y) as well as an increase in inflammatory markers: C-reactive protein, MIP-2, NF-κB, and RANTES [66]. Additional alterations to inflammatory signaling can contribute to more significant inflammation, a common characteristic associated with the development of hematologic disorders, and disease in general. Long-term inflammation contributes to HSC depletion and potential changes to differentiation of downstream progenitors [67]. Lastly, aging within the bone marrow milieu is characterized by the overall loss of cellularity over time. Decreased cellularity results in the replacement of hematopoietic tissues with adipocytes, leading to reduced hematopoietic regeneration [68]. The depletion of bone marrow cellularity begins at birth and continues to decline with age. At 50 years of age the average cellularity is 50 %; however, wide ranges of 30–70 % have been observed [69]. This highlights the importance of the bone marrow milieu in the aging hematopoietic system.
4. Messengers of the gut-bone marrow axis
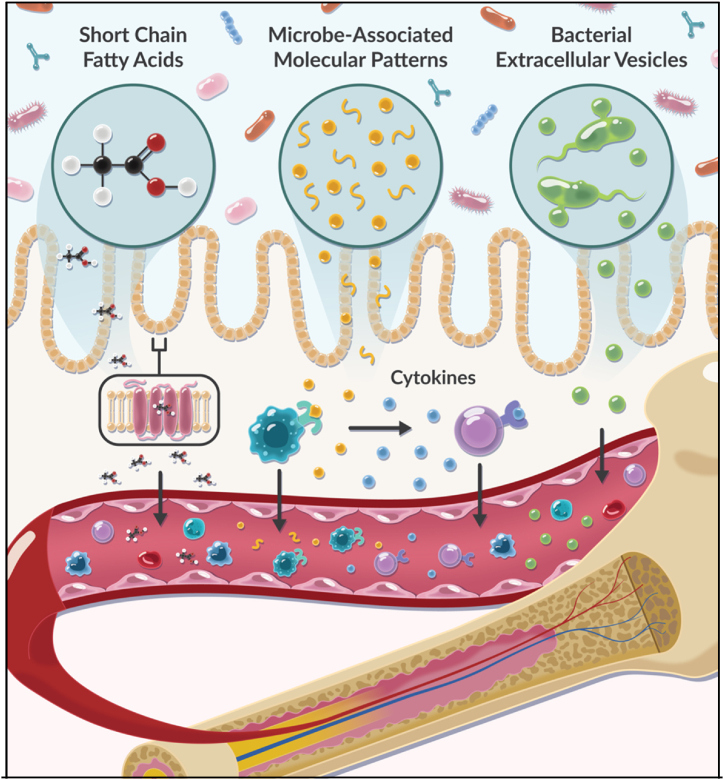
The commensal relationship between the microbiota and the intestine results in the release of SCFAs, lactate, iron, tryptophan metabolites, bacterial extracellular vesicles and microbial molecules (microbe-associated molecular patterns (MAMPs)). These molecules can enter the bloodstream via interaction with specialized receptors or extravasate between gaps in the intestinal epithelium and act as regulating effectors on hematopoiesis via the gut-bone marrow axis [70] (Fig. 3). Alternatively, they can also act on intestinal cells to initiate signaling pathways that additionally affect hematopoiesis. It has been suggested that microbial sourced molecules contribute to inflamm-aging, through the perpetual release of pro-inflammatory mediators. This is also implicated in age-related inflammation within the hematopoietic system; evidence suggests that pro-inflammatory cytokine release increases throughout an individual's life [71]. Due to its importance in aging, inflammation may play a significant role in mediating the gut-bone marrow axis [72]. Although links between the hematopoietic system and microbially derived molecules have been identified, exact mechanisms and resulting effects on hematopoiesis have yet to be fully appreciated [[73], [74], [75], [76]].
-
(i)
Microbial Translocation:
Fig. 3.
Gut translocation of microbially-derived molecules influences signaling pathways within the bone marrow. Molecules derived from the gut microbiota are implicated in the induction of changes and regulation of HSCs and the hematopoietic system. SCFAs bind various receptors that influence hematopoiesis/erythropoiesis in healthy individuals. MAMPs translocate to the bone marrow and activate TLR signaling on mature blood cells. This interaction can generate a microbiota-induced release of cytokines that activate signaling pathways involved in inflammation. Bacterially-derived extracellular vesicles (EVs) can also be secreted by bacteria and mobilize into the circulation (including the bone marrow). Once in the circulation, bacterial EVs locate to distant tissues and activate cellular signaling through the shuttling of bioactive cargo. Figure made by Adobe Suite.
Gut permeability is associated with age and can be characterized by a disruption of intestinal barrier integrity paired with increased translocation of microbial products and the dysregulation of cytokines, intestinal stem cell regeneration, and tight junctions [77]. The loss of integrity alters the membrane's ability to mediate the movement of microbial compounds [77]. When membrane integrity is perturbed, there is a marked increase in the translocation of microbial effector molecules into the bloodstream, which can then circulate to the bone marrow, thus participating in the gut-bone marrow axis [78]. If humans have greater intestinal permeability as they age, then it can be assumed that the dynamics of the gut-bone marrow axis are more prominent in older individuals.
-
(ii)
Short Chain Fatty Acids:
An essential part of commensal bacteria inhabiting the human gut is to ferment undigested carbohydrates into smaller molecules, including SCFAs. These molecules can bind SCFA receptors allowing transport across the intestinal epithelium and have been implicated in regulating HSC fate and promoting hematopoiesis and erythropoiesis [73,74]. An important SCFA in the gut-bone marrow axis is butyrate, which has a role in regulating hematopoiesis by increasing local iron levels, through macrophage activity. Increased macrophage activity enhances phagocytosis of red blood cells (RBCs) and elevates iron availability in the bone marrow [73]. The presence of iron in the bone marrow aids in the maintenance of homeostatic lineage regeneration by HSCs. In addition to iron-related regulation, supplementation with butyrate helps maintain bone marrow cellularity and lineage differentiation by providing rapid access of iron for regeneration of HSCs [73]. In contrast, the depletion of butyrate results in dramatic expansion of HSPCs but with a significant reduction in long-term repopulating activity, likely due to the lack of iron available to regenerating HSCs leading to dysfunctional differentiation capacity [73]. Therefore, the presence of SCFAs within circulation plays a role in the maintenance of hematopoiesis.
Age-related changes in SCFA levels remain controversial in the literature. Some studies demonstrate that age alone is sufficient to result in decreased SCFA concentrations [79], while other studies have shown elevation of peripheral blood SCFA levels with age [80]. Potential changes in SCFA levels in aging individuals may be due to reduced SCFA-producing bacteria colonizing the gut microbiome or alternatively an increased prevalence of microbial translocation with age [81]. Age-related changes in SCFA concentrations are also impacted by the microbiome makeup and expression of the SCFA receptor Gpr41 [80]. Therefore, changes in the specific microbiota with age have a complex relationship with physiological SCFA concentrations that in turn impact the age-associated changes within the hematopoietic system.
-
(iii)
Lactate:
Lactate, a primary messenger in the gut-bone marrow axis, is a biproduct of both human endogenous glycolysis and microbial metabolism. Similar to SCFAs in structure and production, lactate is formed through the fermentation of glucose. Lactate has been implicated in the regulation of hematopoiesis, from studies analyzing the effects of lactate depletion on HSC differentiation. It has been observed that lactate dehydrogenase and corresponding lactate levels affect differentiation of HSCs and HSPCs [82]. In relation to the microbiome, alteration of microbially-derived lactate levels from Lactobacillus plantarum, promotes both hematopoiesis and erythropoiesis [74]. Lactate aids in SCF expression by leptin receptor-expressing (LepR+) MSCs and accelerates SCF secretion. Increased SCF levels are associated with bone marrow cellularity through the interaction with Gpr81 signals [74]. Lactate requires TLR signaling to initiate Gpr81-dependent intracellular signaling pathways that influence HSC differentiation. The connection between lactate and cell differentiation highlights the importance of lactate-producing bacteria in a healthy gut microbiome.
In the context of aging, lactate levels in blood remain variable between individuals as they age [83]. Inconsistent with blood lactate levels, lactate concentration increases in the brain with age [84]. Interestingly, the presence of hyperlactatemia is suggested to increase with age due to mitochondrial dysfunction and is associated with poorer prognosis in multiple diseases. Although there remain limited investigations into the connection between lactate and age, these findings highlight the importance of a homeostatic gut microbiome to maintain a metabolic balance between oxidative phosphorylation and glycolysis and subsequent lactate levels.
-
(iv)
Iron:
Iron has been suggested as an alternative signaling molecule due to its influence on hematopoiesis, while also playing a role in controlling microbial composition and gut activity [36,85]. Luminal concentrations of iron regulate the microbiota, as demonstrated by the supplementation of iron resulting in altered microbial composition (increases in Bifidobacterium, Succinivibrio, Turicibacter, and Clostridium) compared to iron depletion [86]. Iron homeostasis is important in regulating and maintaining self-renewal of HSCs via FBXL5 [87]. FBXL5 acts as a mediator of iron regulatory protein 2 (IRP2) by regulating IRP2 stability, which plays a role iron homeostasis and metabolism. Downregulation of FBXL5 is associated with dysfunction of hematopoiesis, common in myelodysplastic syndrome (MDS). Deficiencies in local iron availability within the bone marrow leads to abnormal activation and cell cycle induction of HSCs. Macrophages, a major contributor to this axis, coordinate iron availability in the regulation of HSC dynamics. Bone marrow macrophages control local iron availability to HSPCs ready for differentiation [73,88]. Varying iron levels initiate tissue-specific alterations with age, for example, in older individuals, the brain has been documented to possess increased iron levels [89,90]. However, within peripheral blood, iron levels typically decrease with increasing age. This is likely due to elevated circulating hepcidin, a protein that inhibits iron transport through gut enterocytes and traps iron within the gut. This decrease in iron levels corresponds to the increased prevalence of anemia, a disease coupled to iron depletion, within the elderly population [91].
-
(v)
Indole and other Tryptophan Metabolites:
Tryptophan metabolites, particularly indole, play a crucial role in regulating hematopoietic functions by targeting the self-renewal of hematopoietic stem cells. Zeng et al. identified that fecal microbiota transplantation (FMT) from young donor mice can rejuvenate aging HSCs through the production of tryptophan metabolites. Specifically, treatment with tryptophan, indole, or indole-3-butyric enhances the population of LT-HSCs. When transplanted, these LT-HSCs show improved engraftment efficiency and clonogenic capacity within the bone marrow. Moreover, they significantly reduce serum levels of specific inflammatory mediators, such as IL-1β, IL-5, and CXCL1, in aged mice [92]. This suggests the potential role of the microbiota in regulating HSC function through the metabolism of endogenous and exogenous tryptophan.
Alternative indole derivatives have demonstrated regulatory effects on different HSC populations. Incubating umbilical cord blood cells with pyrimidoindole derivatives that target the aryl hydrocarbon receptor pathways results in the expansion of HSC-enriched populations. This expansion coincides with the maintenance of primitive LT-HSC populations while enhancing the expansion of progenitor populations [93]. Pyrimidoindole derivatives do not affect the rate of division of primitive populations but enhances LT-HSC function. Engraftment studies with pyrimidoindole treated umbilical cord blood cells resulted in a three-fold increase in engraftment potential [93]. Additionally, incubation suppresses mature cell differentiation. This study suggests that bacteria capable of metabolizing tryptophan into indole derivatives could offer a promising avenue for regulating and maintaining HSCs.
-
(vi)
Bacterial Extracellular Vesicles
Extracellular vesicles (EVs) are lipid encapsulated nanoparticles thought to be produced by all human cells and the microbiota. In humans, EVs are abundant in all bodily fluids, including blood, and have been shown to play a role in the maintenance of HSCs [[94], [95], [96]]. Bacterial EVs, known as outer-membrane vesicles (OMVs), have been documented to be present in human blood [97]. As much as one million OMVs per mL can be found in human blood; in contrast human blood contains ∼100 to a 1000 fold more eukaryotic EVs per mL of blood [97].
Although less abundant than EVs derived from human cells, OMVs have the potential to affect systems distant from the microbiota. In mouse models, OMVs display a similar bioavailability to eukaryotic EVs, and have been shown to be capable of crossing the blood brain barrier and promoting TNF-α secretion [98]. Extracellular vesicles are of major interest as mediators of intercellular communication, therefore understanding how OMVs may interact in the bone marrow milieu, and possibly influence hematopoiesis will be crucial in future studies involving the gut-bone marrow axis.
Despite limited research investigating OMVs within the context of hematopoiesis, one study has identified significant increases in circulating OMVs with age. The OMVs were identified to be increased due to dysregulation of the intestinal barrier, which was associated with aging in mice [99]. Bacterial EVs have also been shown to dysregulate inflammatory pathways and myeloid differentiation in monocytes, macrophages, and dendritic cells in the context of aging [100]. OMVs have been shown to reflect the composition of the microbiota, potentially highlighting the impact of an aging microbiome on intercellular signaling [101]. This is further supported by a study identifying patients with gastric cancer having a significantly altered alpha diversity and microbial composition compared to healthy individuals [102]. Collectively, these studies emphasize the potential impact bacterial EVs may have on HSC regulation, and how this may change during aging.
-
(vii)
Intracellular Signaling Pathways influenced by the Microbiome
Microbe-associated molecular patterns (MAMPs), foreign antigens produced by microbes, have a significant influence on hematopoiesis. These molecules directly interact with TLRs and can induce changes in HSCs and contribute to age-associated inflamm-aging [103]. Microbe-associated molecular patterns contribute to steady-state myelopoiesis and granulopoiesis, via MyD88 and TLR signaling [104]. MyD88 plays a role in innate immunity and is a canonical adaptor for inflammatory signaling pathways through its interaction with TLR and IL-1 receptor families [105]. The relationship of IL-1 receptors and MAMPs is associated with increased inflammation in older mice as shown by analysis of IL-1R1 knock-out models by Kovtonyuk et al. Knockout of IL-1 receptors in 24-month old mice restored lineage bias and enhanced engraftment potential of HSCs to levels similar to young HSCs [103]. The expression of MyD88 in myeloblast and myeloid cells is required for systemic recruitment of neutrophils and can be influenced by MAMPs. Reduced MyD88 stimulation by MAMPs can result in impaired neutrophil migration into the blood [106]. Additionally, MyD88 activation in myeloid cells through the TLR4 complex triggers the production and secretion of inflammatory cytokines [107]. Aged mice demonstrate increased expression of TLR4 on macrophages, further implicating MAMPs and TLR signaling in age-associated inflammation [108]. Therefore, MAMPs are implicated in the development of the chronic inflammatory state of aged individuals. The makeup of the microbiome and subsequent secretion of MAMPs specific to inflammatory cytokine production contributes to age-associated changes in the hematopoietic system.
The activation of other intracellular sensors by MAMPs include nucleotide-binding oligomerization domain-containing protein 1 (NOD1). NOD-1 ligands such as D-glutamyl-meso-diaminopimelic acid, maintains and/or restores HSC and precursor populations in the bone marrow [109]. Signaling of NOD1/NOD2 has also been shown to mobilize HSCs during infection through the stimulation of granulocyte colony-stimulating factor (G-CSF) [110]. These protein receptors also stimulate expression of hematopoietic associated cytokines and a pro-inflammatory response [109].
The regulation of hematopoiesis by MAMP signaling events are not limited to HSCs or HSPCs in the bone marrow. Both MAMPs and intracellular sensors like NOD-1 utilize MSCs in the bone marrow as intermediates to induce changes within the hematopoietic system. MSCs maintain both oxidative phosphorylation and glycolytic functions and additionally signal through the production of other metabolic factors [111]. This has been highlighted by assessing the impact of the microbiota on adipogenic and osteogenic factor expression in MSCs, exhibiting changes in metabolic pathways, HIF-1/inflammatory signalling, and neurodegenerative pathways [112]. Alterations in MSCs by the microbiota are purported to have downstream effects on age-associated changes observed in HSCs. It is known that the mesenchymal compartment supports HSCs and progenitor populations through physical contacts and by the secretion of soluble factors [113]. The microbiota thereby regulates MSCs by regulating proliferation and differentiation capacities of HSPCs which in turn promotes cytokine secretion and maintains the immunomodulatory properties of these cell populations.
5. The role of the microbiome in hematologic malignancies
The gut microbiota has been implicated in increasing the risk of developing cancer and impacting disease outcome. The most studied interaction of the microbiome in cancer is the influence of gut health on immunotherapy, where certain microbial species are associated with better responses to therapy. For example, the presence of commensal Bifidobacterium in the gut was shown to be closely associated with more favourable outcomes in mice treated with anti-PDL-1 immunotherapy [114]. Specifically, oral administration of Bifidobacterium alone was shown to have anti-tumour effects on melanoma [114]. These findings illustrate how gut health, and the diversity of gut flora may be key in the treatment of particular cancers.
Recent studies into the gut-bone marrow axis have shown how the gut microbiome also plays a role in hematologic disorders. When receiving treatment for leukemia, patients are often treated with prophylactic antibiotics to reduce the risk of infection as they become immunocompromised. Antibiotic treatment causes dysbiosis of the gut microbiota, which has been shown to negatively impact disease progression of leukemia in murine models [115]. This effect was shown to be reversable with fecal transplant from healthy controls, highlighting a possible therapeutic target [115]. In humans, it has been shown that the treatment of acute myeloid leukemia (AML) with chemotherapeutic agents is more likely to be successful in patients with a greater diversity of gut microbial species, where greater baseline alpha-diversity and less microbial loss during treatment are associated with positive outcomes [116]. Another study using autologous fecal microbiota transfer (AFMT) demonstrated that repopulation of pre-treatment microbiota significantly improved the outcomes of treatment for AML [117]. When comparing AML and chronic myelogenous leukemia (CML) to healthy controls, both myeloid leukemias demonstrated very similar microbiota compositions at the phylum level, however, patients exhibit slight differences between CML and AML at the genus level [118].
Similar findings have been observed in chronic lymphocytic leukemia (CLL), where patients with lower microbial diversity, presented with more advanced and aggressive forms of disease [119]. This was linked to a reduction in bacteria associated with better health, specifically Prevotella copri, Dorea longicatena, or Bifidobacterium adolescentis. Interestingly, patients with acute leukemias have been shown to have increased beta-diversity, coinciding with enrichment of Prevotella and Alistipes linking the impact of Prevotella on chronic and acute lymphoid leukemias [120]. When analyzing pediatric populations, the microbiome has been implicated in the onset of leukemia. Variations in the oral and gut microbiomes of children diagnosed with acute lymphoblastic leukemia (ALL) are detectable at birth and further dysregulated throughout childhood [121,122]. Consistent with other blood cancers, patients with multiple myeloma (MM), exhibit significant differences in microbial diversity between different disease states, with the greatest microbial diversity in patients with complete remission. The enrichment of Agathobacter, a SCFA producing bacteria, was associated with better overall survival [123]. In addition to predicting survival outcome in (MM), the composition of the microbiota can also be used to potentially assess MM-related risk; Acidaminococcaceae, Bacteroidales, and Porphyromonadaceae have been positively correlated with MM [124].
The microbiome has also been shown to play a potential role in a pediatric and adult lymphoma. In a study comparing adolescent Hodgkin lymphoma patients and their unaffected twins, lymphoma patients demonstrated lower levels of species-level diversity, with even greater differences between randomly paired participants [125]. Although differences were not significant in adolescents, adults with Hodgkin's lymphoma, demonstrate significantly different microbiota compositions compared to healthy controls, highlighting the impact of age on the microbiome and its induction of different signaling cascades [126]. In contrast, adults with diffuse large B-cell lymphoma (DLBCL) possessed a significantly lower alpha diversity and different microbial composition. Disease was correlated with higher abundance of Enterobacteriaceae members [127]. A diverse microbiome was shown to provide protection against pathogenic bacteria prone to promoting lymphoma. For example, H. pylori infection is highly associated with lymphoma of mucosa-associated lymphoid tissue (MALT) and curing the infection can regress high-grade disease [128]. It is clear from these studies that dysbiosis of the gut does occur in patients with hematologic malignancies, and can be potentially addressed to improve disease outcomes, strongly supporting the relevance of the gut-bone marrow axis in cancer treatment.
The gut microbiome has also been identified to contribute to the initiation of leukemia. Treatment with antibiotics alone is sufficient to augment leukemia development in PAX5+/− mice genetically predisposed to precursor B-cell acute lymphoblastic leukemia (pB-ALL), with the absence of commensal bacterial confirmed to drive disease development [129]. Pre-leukemic myeloproliferation is a hematologic disorder that can occur due to mutations in genes such as TET2 and has been shown to be inducible and dependant on microbial IL-6 signalling from the gut microbiome in mice [130]. The influence of the gut microbiome on the development of leukemia in mouse models suggest that maintaining a healthy gut microbiome may aid in the prevention of hematologic malignancies.
6. Conclusion
This review highlights the relationship between the microbiota and hematopoietic system in the context of aging. It has been identified that an individual's microbial composition varies over time, and the altered production of microbial derived molecules, have the potential to impact the gut-bone marrow axis and subsequently contribute to the development of hematological malignancies and other age-related diseases. The findings outlined in this review highlight the important role microbial-derived molecules play in regulating both healthy and age-associated decline observed in HSPCs. Interactions between microbial-derived molecules and the hematopoietic system are vital in the maintenance and regulation of HSPCs. Understanding the relationship between an aging microbiome and aging HSCs may provide valuable insight into targetable mechanisms in hematologic malignancies. This review highlights that alterations in the microbiome, specifically decreases in biodiversity, are intricately linked to a variety of hematologic malignancies. A diverse microbiota of specific composition, offers protection against the onset of disease. A future goal will be to identify connections between the influence of aging on the composition of the microbiome and the specific microbes and microbial messengers that have the most impact on HSC regulation. Understanding this link will also lead to highlighting mechanisms associated with disease onset and whether the supplementation of particular microbiota and or factors may offer an alternative route of prevention against hematologic diseases [131].
Funding
Canadian Institutes of Health Research grant 394568 (SAA). The Terry Fox New Frontiers Program Project Grant 6039065 (SAA). Terry Fox New Investigator Grant 1133 (SAA)
Data availability statement
Has data associated with your study been deposited into a publicly available repository?
No, no data was used for the research described in the article.
CRediT authorship contribution statement
Christopher Wells: Writing – review & editing, Writing – original draft, Conceptualization. Tristan Robertson: Writing – review & editing, Writing – original draft, Conceptualization. Prameet Sheth: Writing – review & editing. Sheela Abraham: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
There are no competing interests by the authors associated with the writing of this review. This review was written with no influence or interests by external parties or institutions involved.
Contributor Information
Christopher Wells, Email: 16cjw2@queensu.ca.
Sheela Abraham, Email: sa183@queensu.ca.
References
- 1.Cheng J., et al. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016;10(4):1002–1014. doi: 10.1038/ismej.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suau A., et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. : AEM. 1999;65(11):4799. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasan N., Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gensollen T., et al. How colonization by microbiota in early life shapes the immune system. Science. 2016;352(6285):539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grice E.A., Segre J.A. The human microbiome: our second genome. Annu. Rev. Genom. Hum. Genet. 2012;13(1):151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Low A., et al. Host age prediction from fecal microbiota composition in male C57BL/6J mice. Microbiol. Spectr. 2022;10(3) doi: 10.1128/spectrum.00735-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Skin Oral, and gut microbiomes predict chronological age. mSystems. 2020;5(1) doi: 10.1128/mSystems.00630-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamburini S., et al. The microbiome in early life: implications for health outcomes. Nat. Med. 2016;22(7):713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 9.Shao Y., et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574(7776):117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez-Bello M.G., et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J., et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imoto N., et al. Administration of β-lactam antibiotics and delivery method correlate with intestinal abundances of Bifidobacteria and Bacteroides in early infancy, in Japan. Sci. Rep. 2021;11(1):6231. doi: 10.1038/s41598-021-85670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ihekweazu F.D., Versalovic J. Development of the pediatric gut microbiome: impact on health and disease. Am. J. Med. Sci. 2018;356(5):413–423. doi: 10.1016/j.amjms.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti P., et al. Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24(1):133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatsunenko T., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koliada A., et al. Sex differences in the phylum‐level human gut microbiota composition. BMC Microbiol. 2021;21(1):131. doi: 10.1186/s12866-021-02198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David L.A., et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Recovery of gut microbiota of healthy adults following antibiotic exposure. Nature Microbiology. 2018;3(11):1255. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 19.Lee K., et al. Population-level impacts of antibiotic usage on the human gut microbiome. Nat. Commun. 2023;14(1) doi: 10.1038/s41467-023-36633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jernberg C., et al. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1(1):56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 21.Malesza I.J., et al. High-fat, western-style diet, systemic inflammation, and gut microbiota: a narrative review. Cells. 2021;10(11):3164. doi: 10.3390/cells10113164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vangay P., et al. US immigration westernizes the human gut microbiome. Cell. 2018;175(4):962–972.e10. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher C.K., Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keystone taxa indispensable for microbiome recovery. Nature Microbiology. 2020;5(9):1067. doi: 10.1038/s41564-020-0783-0. [DOI] [PubMed] [Google Scholar]
- 25.Martino C., et al. Microbiota succession throughout life from the cradle to the grave. Nat. Rev. Microbiol. 2022;20:707–720. doi: 10.1038/s41579-022-00768-z. [DOI] [PubMed] [Google Scholar]
- 26.Singh H., et al. Gastro-intestinal and oral microbiome signatures associated with healthy aging. GeroScience. 2019;41(6):907–921. doi: 10.1007/s11357-019-00098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biagi E., et al. Gut microbiota and extreme longevity. Curr. Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Cerro E.D.-D., et al. Daily ingestion of Akkermansia mucciniphila for one month promotes healthy aging and increases lifespan in old female mice. Biogerontology. 2022;23(1):35–52. doi: 10.1007/s10522-021-09943-w. [DOI] [PubMed] [Google Scholar]
- 29.Castro‐Mejía J.L., et al. Physical fitness in community‐dwelling older adults is linked to dietary intake, gut microbiota, and metabolomic signatures. Aging Cell. 2020;19(3) doi: 10.1111/acel.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez de Aguero M., et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 31.Furusawa Y., et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 32.González-Bosch C., et al. Short-chain fatty acids as modulators of redox signaling in health and disease. Redox Biol. 2021;47 doi: 10.1016/j.redox.2021.102165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S.P., et al. Pivotal roles for pH, lactate, and lactate-utilizing bacteria in the stability of a human colonic microbial ecosystem. mSystems. 2020;5(5) doi: 10.1128/mSystems.00645-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metidji A., et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018;49(2):353–362.e5. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zelante T., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Deschemin J.C., et al. The microbiota shifts the iron sensing of intestinal cells. Faseb. J. 2016;30(1):252–261. doi: 10.1096/fj.15-276840. [DOI] [PubMed] [Google Scholar]
- 37.Jaeggi T., et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut. 2015;64(5):731–742. doi: 10.1136/gutjnl-2014-307720. [DOI] [PubMed] [Google Scholar]
- 38.Paganini D., et al. Prebiotic galacto-oligosaccharides mitigate the adverse effects of iron fortification on the gut microbiome: a randomised controlled study in Kenyan infants. Gut. 2017;66(11):1956–1967. doi: 10.1136/gutjnl-2017-314418. [DOI] [PubMed] [Google Scholar]
- 39.Balamurugan R., et al. Low levels of faecal lactobacilli in women with iron-deficiency anaemia in south India. Br. J. Nutr. 2010;104(7):931–934. doi: 10.1017/S0007114510001637. [DOI] [PubMed] [Google Scholar]
- 40.Khan S.S., Singer B.D., Vaughan D.E. Molecular and physiological manifestations and measurement of aging in humans. Aging Cell. 2017;16(4):624–633. doi: 10.1111/acel.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossi L., et al. Methods in Molecular Biology. Humana Press; 2011. Hematopoietic stem cell characterization and isolation; pp. 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ergen A.V., et al. Basic Cell Culture Protocols. Humana Press; 2013. Isolation and characterization of mouse side population cells; pp. 151–162. [DOI] [PubMed] [Google Scholar]
- 43.Farahzadi R., et al. Targeting the stem cell niche micro-environment as therapeutic strategies in aging. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1162136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young A.L., et al. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papaemmanuil E., et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper J.N., Young N.S. Clonality in context: hematopoietic clones in their marrow environment. Blood. 2017;130(22):2363–2372. doi: 10.1182/blood-2017-07-794362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arends C.M., et al. Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia. 2018;32(9):1908–1919. doi: 10.1038/s41375-018-0047-7. [DOI] [PubMed] [Google Scholar]
- 48.Welch S.John, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pang W.W., et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 2011;108(50):20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han S., et al. Age-associated remodeling of T cell immunity and metabolism. Cell Metabol. 2023;35(1):36–55. doi: 10.1016/j.cmet.2022.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez-Martín N., et al. T cell receptor internalization from the immunological synapse is mediated by TC21 and RhoG GTPase-dependent phagocytosis. Immunity. 2011;35(2):208–222. doi: 10.1016/j.immuni.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao W., et al. Characterization of T-dependent and T-independent B cell responses to a virus-like particle. J. Immunol. 2017;198(10):3846–3856. doi: 10.4049/jimmunol.1601852. [DOI] [PubMed] [Google Scholar]
- 53.Xie X., et al. B‐cell capacity for differentiation changes with age. Aging Cell. 2021;20(4) doi: 10.1111/acel.13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Listì F., et al. A study of serum immunoglobulin levels in elderly persons that provides New insights into B cell immunosenescence. Ann. N. Y. Acad. Sci. 2006;1089(1):487–495. doi: 10.1196/annals.1386.013. [DOI] [PubMed] [Google Scholar]
- 55.Wenisch C., et al. Effect of age on human neutrophil function. J. Leukoc. Biol. 2000;67(1):40–45. doi: 10.1002/jlb.67.1.40. [DOI] [PubMed] [Google Scholar]
- 56.Kim O.-H., et al. Impaired phagocytosis of apoptotic cells causes accumulation of bone marrow-derived macrophages in aged mice. BMB Reports. 2017;50(1):43–48. doi: 10.5483/BMBRep.2017.50.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pirabe A., et al. Age related differences in monocyte subsets and cytokine pattern during acute COVID-19—a prospective observational longitudinal study. Cells. 2021;10(12):3373. doi: 10.3390/cells10123373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seitz A.E., Eberhardt M.S., Lukacs S.L. Anemia prevalence and trends in adults aged 65 and older: U.S. National health and nutrition examination survey: 2001–2004 to 2013–2016. J. Am. Geriatr. Soc. 2018;66(12):2431–2432. doi: 10.1111/jgs.15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahlknecht U., Kaiser S. Age-related changes in peripheral blood counts in humans. Exp. Ther. Med. 2010;1(6):1019–1025. doi: 10.3892/etm.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kinyoki D., et al. Anemia prevalence in women of reproductive age in low- and middle-income countries between 2000 and 2018. Nat. Med. 2021;27(10):1761–1782. doi: 10.1038/s41591-021-01498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Méndez-Ferrer S., et al. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(7186):442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 62.Chow A., et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 2011;208(2):261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guidi N., et al. An aged bone marrow niche restrains rejuvenated hematopoietic stem cells. Stem Cell. 2021;39(8):1101–1106. doi: 10.1002/stem.3372. [DOI] [PubMed] [Google Scholar]
- 64.Sugiyama T., et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 65.Chen J.Y., et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530(7589):223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kovtonyuk L.V., et al. Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front. Immunol. 2016;7:502. doi: 10.3389/fimmu.2016.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solimando A.G., et al. The bone marrow niche landscape: a journey through aging, extrinsic and intrinsic stressors in the haemopoietic milieu. Journal of Cancer Metastasis and Treatment. 2022;8 [Google Scholar]
- 68.Naveiras O., et al. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460(7252):259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Eekelen L., et al. Using deep learning for quantification of cellularity and cell lineages in bone marrow biopsies and comparison to normal age-related variation. Pathology. 2022;54(3):318. doi: 10.1016/j.pathol.2021.07.011. [DOI] [PubMed] [Google Scholar]
- 70.Clarke T.B., et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alvarez-Rodriguez L., et al. Aging is associated with circulating cytokine dysregulation. Cell. Immunol. 2012;273(2):124. doi: 10.1016/j.cellimm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Sayed N., et al. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nature Aging. 2021;1(7):598–615. doi: 10.1038/s43587-021-00082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang D., et al. The microbiota regulates hematopoietic stem cell fate decisions by controlling iron availability in bone marrow. Cell Stem Cell. 2022;29(2):232–247.e7. doi: 10.1016/j.stem.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee Y.-S., et al. Microbiota-derived lactate promotes hematopoiesis and erythropoiesis by inducing stem cell factor production from leptin receptor+ niche cells. Exp. Mol. Med. 2021;53(9):1319–1331. doi: 10.1038/s12276-021-00667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balmer M.L., et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 2014;193(10):5273–5283. doi: 10.4049/jimmunol.1400762. [DOI] [PubMed] [Google Scholar]
- 76.Khosravi A., et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15(3):374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thevaranjan N., et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21(4):455–466.e4. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matteini F., Florian M.C. The gut-bone marrow axis: a novel player in HSC aging. Blood. 2022;139(1):3–4. doi: 10.1182/blood.2021014134. [DOI] [PubMed] [Google Scholar]
- 79.Lee J., et al. Young versus aged microbiota transplants to germ-free mice: increased short-chain fatty acids and improved cognitive performance. Gut Microb. 2020;12(1) doi: 10.1080/19490976.2020.1814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi S.I., et al. Changes in cecal microbiota and short-chain fatty acid during lifespan of the rat. Journal of Neurogastroenterology and Motility. 2021;27(1):134–146. doi: 10.5056/jnm20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salazar N., et al. Age-associated changes in gut microbiota and dietary components related with the immune system in adulthood and old age: a cross-sectional study. Nutrients. 2019;11(8):1765. doi: 10.3390/nu11081765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y.-H., et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158(6):1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cannon C., et al. Age-adjusted and expanded lactate thresholds as predictors of all-cause mortality in the emergency department. West. J. Emerg. Med. 2020;21(5) doi: 10.5811/westjem.2020.5.46811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Datta S., Chakrabarti N. Age related rise in lactate and its correlation with lactate dehydrogenase (LDH) status in post-mitochondrial fractions isolated from different regions of brain in mice. Neurochem. Int. 2018;118:23–33. doi: 10.1016/j.neuint.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 85.Rusu I.G., et al. Iron supplementation influence on the gut microbiota and probiotic intake effect in iron deficiency—a literature-based review. Nutrients. 2020;12(7):1993. doi: 10.3390/nu12071993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Werner T., et al. Depletion of luminal iron alters the gut microbiota and prevents Crohn's disease-like ileitis. Gut. 2011;60(3):325–333. doi: 10.1136/gut.2010.216929. [DOI] [PubMed] [Google Scholar]
- 87.Muto Y., et al. Essential role of FBXL5-mediated cellular iron homeostasis in maintenance of hematopoietic stem cells. Nat. Commun. 2017;8(1) doi: 10.1038/ncomms16114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caiado F., Manz M.G. A microbiome-macrophage-iron axis guides stressed hematopoietic stem cell fate. Cell Stem Cell. 2022;29(2):177. doi: 10.1016/j.stem.2022.01.009. [DOI] [PubMed] [Google Scholar]
- 89.Hahn P., et al. Age-dependent and gender-specific changes in mouse tissue iron by strain. Exp. Gerontol. 2009;44(9):594–600. doi: 10.1016/j.exger.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu J., et al. Iron accumulation with age, oxidative stress and functional decline. PLoS One. 2008;3(8):e2865. doi: 10.1371/journal.pone.0002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stauder R., et al. Prevalence and possible causes of anemia in the elderly: a cross-sectional analysis of a large European university hospital cohort. Clin. Interv. Aging. 2014:1187. doi: 10.2147/CIA.S61125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeng X., et al. Fecal microbiota transplantation from young mice rejuvenates aged hematopoietic stem cells by suppressing inflammation. Blood J, Am. Soci. Hematol. 2023;141(14):1691–1707. doi: 10.1182/blood.2022017514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fares I., et al. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345(6203):1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grenier‐Pleau I., et al. Blood extracellular vesicles from healthy individuals regulate hematopoietic stem cells as humans age. Aging Cell. 2020;19(11) doi: 10.1111/acel.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.tiRNA signaling via stress-regulated vesicle transfer in the hematopoietic niche. Cell Stem Cell. 2021;28(12):2090. doi: 10.1016/j.stem.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goloviznina N.A., et al. Mesenchymal stromal cell-derived extracellular vesicles promote myeloid-biased multipotent hematopoietic progenitor expansion via toll-like receptor engagement. J. Biol. Chem. 2016;291(47):24607–24617. doi: 10.1074/jbc.M116.745653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tulkens J., De Wever O., Hendrix A. Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat. Protoc. 2020;15(1):40–67. doi: 10.1038/s41596-019-0236-5. [DOI] [PubMed] [Google Scholar]
- 98.Han E.C., et al. Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF‐α production in human macrophages and cross the blood‐brain barrier in mice. Faseb. J. 2019;33(12):13412–13422. doi: 10.1096/fj.201901575R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ou Z., et al. Single‐particle analysis of circulating bacterial extracellular vesicles reveals their biogenesis, changes in blood and links to intestinal barrier. J. Extracell. Vesicles. 2023;12(12) doi: 10.1002/jev2.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gul L., et al. Extracellular vesicles produced by the human commensal gut bacterium Bacteroides thetaiotaomicron affect host immune pathways in a cell-type specific manner that are altered in inflammatory bowel disease. J. Extracell. Vesicles. 2022;11(1) doi: 10.1002/jev2.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoo J.Y., et al. 16S rRNA gene-based metagenomic analysis reveals differences in bacteria-derived extracellular vesicles in the urine of pregnant and non-pregnant women. Exp. Mol. Med. 2016;48(2) doi: 10.1038/emm.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park J.-Y., et al. Bacteria-derived extracellular vesicles in urine as a novel biomarker for gastric cancer: integration of liquid biopsy and metagenome analysis. Cancers. 2021;13(18):4687. doi: 10.3390/cancers13184687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kovtonyuk L.V., et al. IL-1 mediates microbiome-induced inflammaging of hematopoietic stem cells in mice. Blood. 2022;139(1):44–58. doi: 10.1182/blood.2021011570. [DOI] [PubMed] [Google Scholar]
- 104.Weaver L.K., et al. Microbiota-dependent signals are required to sustain TLR-mediated immune responses. JCI Insight. 2019;4(1) doi: 10.1172/jci.insight.124370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deguine J., Barton G.M. MyD88: a central player in innate immune signaling. F1000Prime Reports. 2014;6:97. doi: 10.12703/P6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karmarkar D., Rock K.L. Microbiota signalling through MyD88 is necessary for a systemic neutrophilic inflammatory response. Immunology. 2013;140(4):483–492. doi: 10.1111/imm.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diomede F., et al. MyD88/ERK/NFkB pathways and pro-inflammatory cytokines release in periodontal ligament stem cells stimulated by Porphyromonas gingivalis. Eur. J. Histochem. 2017;61(2):2791. doi: 10.4081/ejh.2017.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barrett J.P., et al. Bone marrow-derived macrophages from aged rats are more responsive to inflammatory stimuli. J. Neuroinflammation. 2015;12(1) doi: 10.1186/s12974-015-0287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iwamura C., et al. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 2017;129(2):171–176. doi: 10.1182/blood-2016-06-723742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burberry A., et al. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and toll-like receptor signaling. Cell Host Microbe. 2014;15(6):779–791. doi: 10.1016/j.chom.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pattappa G., et al. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J. Cell. Physiol. 2011;226(10):2562–2570. doi: 10.1002/jcp.22605. [DOI] [PubMed] [Google Scholar]
- 112.Xiao E., et al. Microbiota regulates bone marrow mesenchymal stem cell lineage differentiation and immunomodulation. Stem Cell Res. Ther. 2017;8(1):213. doi: 10.1186/s13287-017-0670-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crippa S., Bernardo M.E. Mesenchymal stromal cells: role in the BM niche and in the support of hematopoietic stem cell transplantation. HemaSphere. 2018;2(6):e151. doi: 10.1097/HS9.0000000000000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sivan A., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang R., et al. Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat. Commun. 2022;13(1) doi: 10.1038/s41467-022-30240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Galloway-Peña J.R., et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer. 2016;122(14):2186–2196. doi: 10.1002/cncr.30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malard F., et al. Gut microbiota diversity after autologous fecal microbiota transfer in acute myeloid leukemia patients. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-23376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu D., et al. Profiling of gut microbial dysbiosis in adults with myeloid leukemia. FEBS Open Bio. 2021;11(7):2050–2059. doi: 10.1002/2211-5463.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Faitova T., et al. The diversity of the microbiome impacts chronic lymphocytic leukemia development in mice and humans. Haematologica. 2024 doi: 10.3324/haematol.2023.284693. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vázquez X., et al. Study of the intestinal microbiota composition and the effect of treatment with intensive chemotherapy in patients recovered from acute leukemia. Sci. Rep. 2024;14(1) doi: 10.1038/s41598-024-56054-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang R., et al. Cesarean section and risk of childhood acute lymphoblastic leukemia in a population-based, record-linkage study in California. Am. J. Epidemiol. 2017;185(2):96–105. doi: 10.1093/aje/kww153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Amitay E.L., Keinan-Boker L. Breastfeeding and childhood leukemia incidence. JAMA Pediatr. 2015;169(6) doi: 10.1001/jamapediatrics.2015.2643. [DOI] [PubMed] [Google Scholar]
- 123.Rodríguez-García A., et al. Short-chain fatty acid production by gut microbiota predicts treatment response in multiple myeloma. Clin. Cancer Res. 2024;30(4):904–917. doi: 10.1158/1078-0432.CCR-23-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feng Z., et al. Exploring the causal relationship between gut microbiota and multiple myeloma risk based on Mendelian randomization and biological annotation. Front. Microbiol. 2024;15 doi: 10.3389/fmicb.2024.1310444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cozen W., et al. Fecal microbiota diversity in survivors of adolescent/young adult Hodgkin lymphoma: a study of twins. Br. J. Cancer. 2013;108(5):1163–1167. doi: 10.1038/bjc.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li B., et al. The causal relationship between gut microbiota and lymphoma: a two-sample Mendelian randomization study. Front. Immunol. 2024;15 doi: 10.3389/fimmu.2024.1397485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yoon S.E., et al. The influence of microbial dysbiosis on immunochemotherapy-related efficacy and safety in diffuse large B-cell lymphoma. Blood. 2023;141(18):2224–2238. doi: 10.1182/blood.2022018831. [DOI] [PubMed] [Google Scholar]
- 128.Gong E.J., et al. Helicobacter pylori eradication therapy is effective as the initial treatment for patients with H. Pylori-Negative and disseminated gastric mucosa-associated lymphoid tissue lymphoma. Gut Liver. 2016;10(5):706–713. doi: 10.5009/gnl15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vicente-Dueñas C., et al. An intact gut microbiome protects genetically predisposed mice against leukemia. Blood. 2020;136(18):2003–2017. doi: 10.1182/blood.2019004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meisel M., et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 2018;557(7706):580–584. doi: 10.1038/s41586-018-0125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heshiki Y., et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Has data associated with your study been deposited into a publicly available repository?
No, no data was used for the research described in the article.