Abstract
Previous studies have indicated that the E4orf4 protein of human adenovirus type 2 (Ad2) induces p53-independent apoptosis. We believe that this process may play a role in cell death and viral spread at the final stages of productive infection. E4orf4 may also be of therapeutic value in treating some diseases, including cancer, through its ability to induce apoptosis when expressed individually. The only previously identified biochemical function of E4orf4 is its ability to associate with the Bα subunit of protein phosphatase 2A (PP2A). We have used a genetic approach to determine the role of such interactions in E4orf4-induced cell death. E4orf4 deletion mutants were of only limited value, as all were highly defective. We found that E4orf4 proteins from most if not all adenovirus serotypes induced cell death, and thus point mutations were introduced that converted the majority of highly conserved residues to alanines. Such mutants were used to correlate Bα-subunit binding, association with PP2A activity, and cell killing following the transfection of appropriate cDNAs into p53-null H1299 or C33A cells. The results indicated that binding of the Bα subunit is essential for induction of cell death, as every mutant that failed to bind efficiently was totally defective for cell killing. This class of mutations (class I) largely involved residues between amino acids 51 and 89. Almost all E4orf4 mutant proteins that associated with PP2A killed cancer cells at high levels; however, several mutants that associated with significant levels of PP2A were defective for killing (class II). Thus, binding of E4orf4 to PP2A is essential for induction of p53-independent apoptosis, but E4orf4 may possess one or more additional functions required for cell killing.
Successful, productive infection of human cells by adenoviruses involves a complex interplay between the induction and suppression of apoptosis (43a). Proteins encoded by early region 1A (E1A) transactivate early viral gene expression and stimulate cells to enter S phase to enhance viral DNA synthesis. One consequence of E1A expression by human adenovirus type 5 (Ad5) is the stabilization of p53 (11, 29) resulting from complex formation with members of either the retinoblastoma tumor suppressor or the p300/CBP families of cell cycle regulators (8, 42). Adenoviruses prevent apoptosis or growth arrest by p53 through the action of two E1B products, the 55-kDa protein that binds to and inhibits p53 (51, 58) and the 19-kDa polypeptide that suppresses apoptosis via a mechanism analogous to the cellular Bcl-2 protein (6, 18). The turnover of p53 is also enhanced through the action of complexes between the Ad5 E1B 55-kDa protein and a product of E4, E4orf6 (13, 41). In addition, E1A enhances cell susceptibility to killing by tumor necrosis factor (14, 46, 57), an effect that is also inhibited by the E1B 19-kDa protein (38, 40) as well as E3 products (reviewed in reference 53).
In previous studies it was noted that E1B-defective Ad5 also induces apoptosis in p53-null cells through the transactivation of another early viral product (52) that was subsequently mapped to E4, which encodes seven polypeptides (30). Using a series of Ad5 E4 mutants, we identified E4orf4 as the E4 product that induces p53-independent apoptosis (31). These studies indicated that the expression of Ad2 E4orf4 alone in the absence of other viral products results in p53-independent cell death. Similar results have been obtained by another group (46a). Furthermore, we established cell lines expressing Ad2 E4orf4 under an inducible promoter and showed that such cells die rapidly upon induction, exhibiting classic apoptotic features, including DNA degradation, chromatin condensation, and the presence of phosphatidylserine on the outer cell membrane as determined by annexin V staining (28). We believe that one of the roles of E4orf4 may be to cooperate in the killing of infected cells at the end of the infectious cycle. An E4orf4-null mutant was originally described as being more cytotoxic than the wild-type virus, as mutant-infected cells were observed to detach from the plastic dishes at early times (35); however, we found that such cells were still viable and survived longer than those infected by the wild-type virus (31). Mutants defective in the E3-11.6K protein also exhibit a similar phenotype (54), and thus, this protein may cooperate with E4orf4 in cell killing. Apoptosis is a mechanism used by a number of viruses as the host inflammatory response is diminished, and progeny are protected from host antibodies and proteases because they are released in apoptotic membrane-bound vesicles (reviewed in references 43a and 50).
The biological activity of E4orf4 was first revealed through studies showing a synergistic effect on transcription factor AP-1 by E1A products and cyclic AMP (15, 16, 36). Under these conditions E4orf4 caused a decrease in phosphorylation of both the E1A protein and c-Fos, the latter causing decreased AP-1 transcriptional activity. The kinetics of this effect implied that E4orf4 was acting to inhibit a cellular kinase involved in phosphorylation of these proteins (35). E4orf4 was additionally shown to suppress JunB and c-Fos protein production at both transcriptional and translational levels (35). A major insight came with the discovery that E4orf4 associates with the Bα subunit of cellular protein phosphatase 2A (PP2A) (26). The effects of E4orf4 on AP-1 are believed to be caused by PP2A-dependent dephosphorylation and inactivation of a mitogen-activated protein kinase, which can act as a PP2A substrate and is believed to play a role in the regulation of AP-1 (1, 4, 17, 22). This effect also has direct implications in the regulation of E4 gene expression, which is dependent on transcription factor E4F, which in turn is activated by E1A-induced phosphorylation (2, 3, 43). E4orf4 inhibits E4 expression, but treatment with okadaic acid, an inhibitor of PP2A, results in upregulation, suggesting that E4orf4-induced PP2A activity may cause the dephosphorylation of E4F (2). We have also identified two mitogen-activated protein kinase-dependent sites in the E1A protein that must be phosphorylated for E1A-mediated activation of expression of E4 but not of E3 (56). This form of autoregulation may limit the levels of E4orf4 and thus prevent early cell death. In addition, the E4orf4-PP2A complexes appear to induce the hypophosphorylation of SR splicing factors involved in spliceosome assembly and splice site recognition, to promote usage of a secondary splice site required for late viral gene expression (25).
PP2A is an abundant cellular serine/threonine phosphatase which exists as a trimer of the C catalytic subunit and A and B regulatory subunits. The A subunit is a 65-kDa rodlike protein comprised of 15 nonidentical repeats (7, 20, 27, 55). The catalytic C subunit is a 37-kDa protein that binds to repeats 11 to 15, whereas B subunits bind to repeats 1 to 10 of the A subunit (7, 20, 23, 24, 27, 33, 44, 45, 55). Thus far, about 20 B-subunit variants have been cloned and exist in three classes. The B class comprises at least four members of about 55 kDa, termed Bα, Bβ, and Bγ (19, 39a, 47a, 60, 61). The B′ (B56) class contains at least 13 isoforms (10, 33, 34, 48, 49, 59). The B" class contains two forms produced by alternative splicing of about 72 and 130 kDa (21). The various B subunits, which share limited or no homology, are believed to function not only in defining the substrate specificity of the PP2A holoenzyme (5) but also in intracellular targeting, in tissue specificity, and as binding partners for interacting proteins and second messengers. PP2A targets in mammalian and yeast cells include proteins involved in basal metabolism, DNA replication, cell proliferation, and cell cycle regulation (9). Substrate specificity is altered considerably depending on the form of B subunit in the holoenzyme (12, 32, 47). The AC (core) dimer is also present in cells at reasonable levels but seems to lack substrate specificity (27). Complex formation with the B subunit increases core phosphatase activity (60). As described above, E4orf4 may bind selectively to the Bα subunit (26), although interactions with other B subunits have not been rigorously tested.
In the present study we have used a genetic approach to determine if binding of E4orf4 to the Bα subunit is required for E4orf4-induced p53-independent apoptosis. Our results from using a large series of point mutants indicated that such is clearly the case. However, there is a second class of mutants that exhibit reduced cell killing, even though they are able to bind the Bα subunit at high levels. Thus, although binding to PP2A is essential for inducing p53-independent apoptosis, E4orf4 may possess one or more additional functions required for killing.
MATERIALS AND METHODS
Cell lines.
Human C33A (ATCC HTB-31) and H1299 cells (ATCC CRL-5803), which are both deficient in p53 expression, were cultured in Falcon tissue culture dishes in α-minimal essential medium (Bio-Whittaker) supplemented with 10% fetal bovine serum (Bio-Whittaker), 100 U of penicillin and streptomycin/ml, and 0.292 mg of l-glutamine/ml.
Plasmids.
A cDNA expressing Ad2 E4orf4 (39) was subcloned into the mammalian expression vector pcDNA3 (Invitrogen) by using BamHI-XhoI digestion to produce pcDNA3-E4orf4. The E4orf4 sequence was excised from pcDNA3-E4orf4 by using PCR with the additional introduction of flanking BamHI (5′) and EcoRI (3′) sites. The forward oligonucleotide was GC GGA TCC ATG GTT CTT CCA GCT C, and the reverse oligonucleotide was CGA ATT CTA CTG TAC GGA GTG (both 5′ to 3′). The digested PCR product was cloned into a pcDNA3 variant in which a hemagglutinin (HA) epitope tag had been previously inserted upstream of the multicloning cassette. This process fused the HA tag to the 5′ end of the E4orf4 sequence with glycine and serine residues between them, producing pcDNA3-HAE4orf4. The final product was confirmed by DNA sequencing. cDNAs expressing E4orf4 from other adenovirus serotypes were cloned as follows. For those from Ad9, Ad12, and Ad40, the E4orf4 coding region was cloned from virally infected KB cells by PCR using published sequences of the E4 regions, adding 5′ BamHI and 3′ EcoRI sites for cloning into pcDNA3. For Ad3 and Ad4, the region between the fiber coding sequence and the inverted terminal repeat was cloned by PCR and the entire region was sequenced. Sequences encoding E4orf4 were then cloned by PCR and inserted into pcDNA3 as above.
Mutant construction.
As we found that the products of most E4orf4 deletion mutants were highly unstable when expressed in mammalian cells (data not shown), E4orf4 deletion mutants were made as fusion products with HA-tagged maltose binding protein (HA-MBP) present at the amino terminus. Various in-frame deletions and carboxy-terminal truncations were created by using PCR in the E4orf4 coding sequence. The E4orf4 point mutant alanine substitutions were generated by using standard three-oligonucleotide PCR-based mutagenesis with pcDNA3-E4orf4 as the template. As described above, BamHI and EcoRI sites were inserted at either end of the coding sequence and the PCR products were cloned into the pcDNA3 construct with the HA epitope. All mutants were confirmed by DNA sequencing. The point mutants were named by using the original residue code, the residue location, and then the substituted residue code.
Protein expression.
To verify protein expression from wild-type and mutant E4orf4 plasmids, Lipofectamine PLUS (Gibco-BRL) was used together with 1.25 μg of plasmid DNA and lipofected into 60-mm plates of H1299 or C33A cells. Cell extracts were prepared in a volume of 100 μl at 24 h postlipofection (0.1% Nonidet P-40 [IGEPAL; Sigma], 0.4 M KCl, 50 mM HEPES [pH 7.9], 10% glycerol, 0.2 M EDTA, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride). Protein levels were quantified using the Bio-Rad protein assay reagent, and 10 μg of total protein per lane was resolved by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis (SDS–15% PAGE). Separated proteins were transferred to nitrocellulose and probed with mouse anti-HA antibody (HA.11; BAbCO) at a 1/2,500 dilution. Visualization was completed using a goat anti-mouse immunoglobulin G Fc fragment-specific antibody linked to horseradish peroxidase (Jackson ImmunoResearch) at a 1/100,000 dilution followed by enhanced chemiluminescence (ECL) detection (NEN Life Science Products). Protein expression was in some cases also quantified using an 125I-conjugated secondary antibody with HA.11, followed by analysis using a Fuji phosphorimager.
Preparation of construct expressing a FLAG-tagged Bα subunit.
The DNA sequence encoding the Bα subunit of PP2A was excised from pPP2A55(7-1) (39a) by using PCR with the additional introduction of flanking KpnI (5′) and EcoRI (3′) sites. The forward oligonucleotide was CGG GGT ACC GTC GAC TAT GGC AGG AGC TGG AGG AGG G, and the reverse oligonucleotide was CGA ATT CTA ATT CAC TTT GTC TTG AAA TAT ATA C (both 5′ and 3′). The digested PCR product was cloned into a pcDNA3 variant in which the FLAG epitope tag had been previously inserted upstream of the multicloning cassette. This placed the FLAG tag at the 5′ end of the Bα sequence, followed by six residues from the cloning cassette (Ser-Leu-Val-Pro-Ser-Thr) and then the Bα coding region, to produce pcDNA3-FLAGPP2ABα. The final product was confirmed by DNA sequencing.
PP2A assay.
Phosphatase activity was assessed following the introduction of 1.25 μg of wild-type or mutant pcDNA3-HAE4orf4 together with 1.25 μg of pcDNA3-FLAGPP2ABα into 60-mm plates of H1299 or C33A cells using Lipofectamine PLUS (Gibco-BRL). One milliliter of cell extracts prepared in 50 mM Tris-HCl (pH 7.5) containing 0.5% IGEPAL, 0.1% Triton X-100, 250 mM NaCl, and 5 mM EDTA was clarified and immunoprecipitated using 2 μl of HA.11 together with protein A Sepharose CL-4B beads (Pharmacia Biotech). The immunoprecipitated complexes were washed three times with extraction buffer, washed twice with 20 mM MOPS (morpholinepropanesulfonic acid) (pH 7.5) containing 60 mM β-mercaptoethanol, 0.1 M NaCl, and 0.1 mg of bovine serum albumin/ml, and were then assayed for phosphatase activity with the malachite green-based Ser/Thr phosphatase assay kit (UBI). A synthetic phosphopeptide containing a generic PP2A consensus site (KRpTIRR) was used as the substrate.
E4orf4-PP2A binding assay.
The immunoprecipitates prepared as described above were separated by SDS–8% PAGE. Proteins were transferred to nitrocellulose and were then probed with the M2 mouse anti-FLAG antibody (Kodak/Sigma-Aldrich) at a dilution of 1/1,000. Proteins were visualized by using a goat anti-mouse immunoglobulin G F(ab′)2 fragment-specific antibody linked to horseradish peroxidase (Jackson ImmunoResearch) at a 1/100,000 dilution, followed by ECL detection (NEN Life Science Products).
E4orf4 cell death assays.
Wild-type or mutant pcDNA3-HAE4orf4 (0.45 μg) was introduced into H1299 or C33A cells at 50% confluence in 12-well plates using Lipofectamine PLUS (Gibco-BRL). Two days after transfection the cells were trypsinized and 1/100 of the cells in each well was replated in each of three 60-mm plates. The cells were cultured in the presence of 750 μg of G418 (Gibco-BRL)/ml for 2 weeks to select for neomycin-resistant cell growth, as pcDNA3 contains the neo resistance marker. At the end of this period, the cells were trypsinized and viable cells were counted with a hemocytometer. In some experiments, cell colonies were fixed in methanol-acetic acid (3:1), stained with 0.15 mg of Giemsa stain/ml of phosphate-buffered saline, and scored visually.
RESULTS
Analysis of cell killing by E4orf4 products of different adenovirus serotypes.
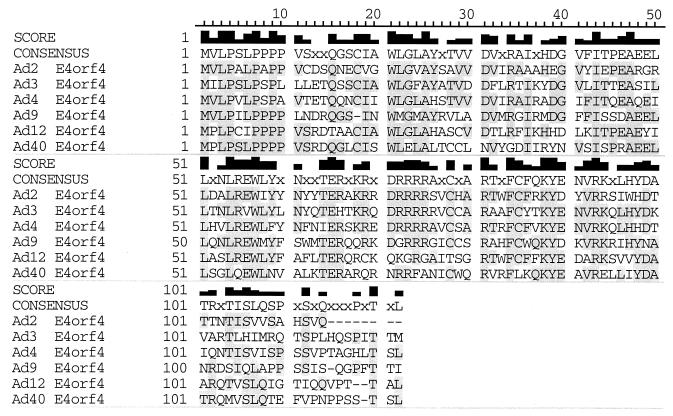
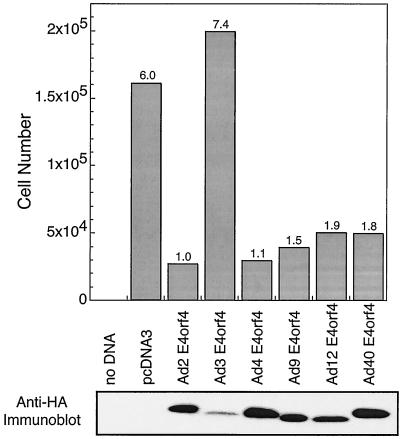
It was shown previously that E4orf4 interacts with the Bα subunit of PP2A (26) and that this interaction leads to decreased phosphorylation of adenovirus E1A products (26, 56), c-Fos (35), transcription factor E4F (2), and SR proteins (25). These results suggested that the association of E4orf4 with PP2A may either activate or redirect the activity of this phosphatase. To determine if such interactions are also required for E4orf4-induced p53-independent apoptosis (28, 30, 31, 52), a genetic approach was used to define the regions of E4orf4 involved in these functions. Furthermore, we wanted to determine if E4orf4 regions apart from those required for PP2A binding were involved in cell killing. As a first approach, the E4orf4 products encoded by several classes of human adenoviruses were tested, as they represent a series of natural E4orf4 mutants. Figure 1 shows that considerable homology exists among E4orf4 products of different groups of human adenoviruses, with differences evident primarily at the carboxy terminus. cDNAs containing E4orf4 coding sequences from representative members of each adenovirus group were cloned by PCR from viral genomic DNA and fused with a sequence encoding an HA epitope tag. Such cDNAs were used in colony inhibition assays similar to those of a previous study (31), in which the killing of transfected human H1299 lung carcinoma cells was measured by the reduction in colony formation following selection with G418 (as described in Materials and Methods). Figure 2 shows that although Ad2 E4orf4, (group C) was the most efficient at reducing colony formation, Ad4 E4orf4, (group E), Ad9 E4orf4, (group D), Ad12 E4orf4, (group A), and Ad40 E4orf4 (group F) all yielded substantially fewer colonies than control cells transfected with pcDNA3 DNA alone. Thus, all of these E4orf4 molecules appeared to induce cell death with similar efficiencies, suggesting that highly conserved residues within the E4orf4 coding sequences may be of particular importance in the killing potential of E4orf4. The clear exception was Ad3 E4orf4 (group B), which exhibited no colony inhibitory activity. Western blotting analysis using anti-HA antibody indicated that the level of expression of Ad3 E4orf4 was considerably lower than that of the other adenovirus E4orf4 products (Fig. 2, bottom). As discussed below and elsewhere in more quantitative studies (R. C. Marcellus, H. Chang, D. Paquette, D. Boivin, G. C. Shore, and P. E. Branton, unpublished data), it appears that the expression of a certain critical level of E4orf4 is required to induce significant cell death. Thus, either Ad3 E4orf4 was defective for cell killing, or its level of expression was insufficient. As the promoter constructs used to express all of the E4orf4 products were identical, it is likely that the Ad3 E4orf4 protein may be more unstable than the products of other serotypes.
FIG. 1.
E4orf4 coding sequence for different human adenovirus serotypes. The protein sequences for members of various classes of human adenoviruses are presented. The alignment was performed by using the CLUSTAL V multiple-sequence-alignment algorithm (21a) and Gene Inspector (Textco Inc.). The score is based on amino acid identity.
FIG. 2.
Cell killing by various adenovirus E4orf4 proteins. H1299 cells were lipofected with plasmid DNA expressing neo and wild-type or mutant E4orf4 and then trypsinized, replated, and allowed to grow in the presence of G418. The results represent the averages of two separate experiments, each involving three plates per sample. The standard error in such experiments was less than 30% (top). At the time of replating, some cells were examined for E4orf4 expression by Western blotting using anti-HA antibodies (bottom).
As discussed in detail below, one of the objectives of the present study was to determine the importance in cell killing of interactions between E4orf4 and the Bα subunit of PP2A. Binding studies were therefore carried out exactly as described (see Fig. 5) using HA-tagged versions of E4orf4 from the different human adenovirus serotypes in cells expressing FLAG-Bα. The results indicated that E4orf4 from all serotypes, including Ad3, interacted with comparable levels of Bα when corrected for expression levels (data not shown). These data confirmed the universality of E4orf4 function among human adenoviruses.
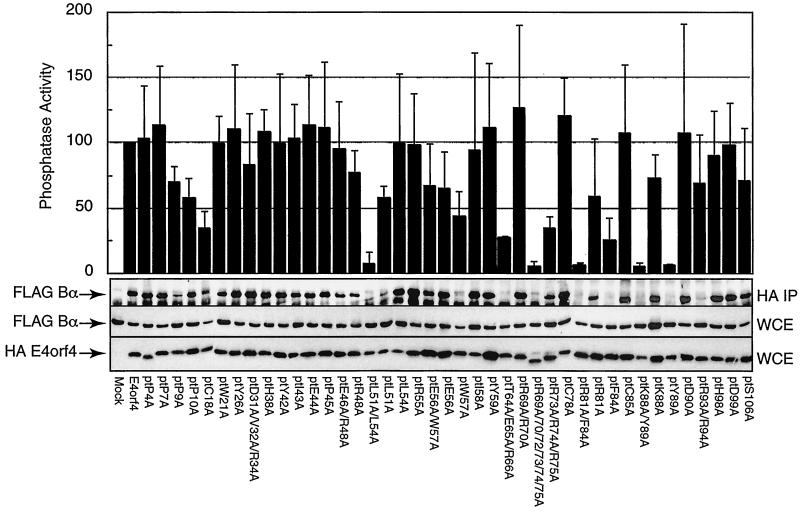
FIG. 5.
Analysis of Bα-subunit binding and associated phosphatase activity of E4orf4 mutants. H1299 cells were lipofected with the HAE4orf4 mutants together with FLAG-tagged Bα. Cell extracts were prepared 24 h postlipofection, and anti-HA immunoprecipitations were performed. Immunoprecipitates were assayed for PP2A activity using a synthetic phosphopeptide as the substrate, as described in Materials and Methods, and results of the average of four separate experiments were plotted relative to those for wild-type E4orf4, which were set at 100% (top). The standard error in these assays was less than 30%. Immunoprecipitates (IP) or whole-cell extracts (WCE) were separated by SDS-PAGE, transferred to nitrocellulose, and then immunoblotted against the FLAG epitope to determine Bα-subunit levels. In addition, blots containing whole-cell extracts were immunoblotted to detect the level of HAE4orf4 by using a murine anti-HA monoclonal antibody. Results from one of four such studies are presented.
Analysis of PP2A Bα-subunit binding using E4orf4 deletion mutants.
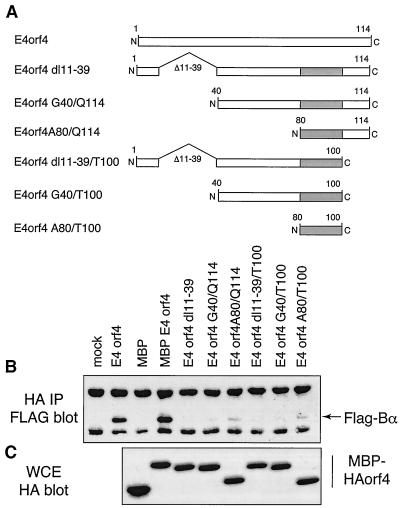
Deletion mutagenesis was attempted as an approach to study E4orf4 interactions with the Bα subunit. Initial mutants containing amino- or carboxy-terminal truncations and 10 to 15 amino acid internal deletions in the Ad2 E4orf4 protein largely yielded highly unstable products when expressed in mammalian cells (data not shown). To avoid such problems, deletion-containing and truncated E4orf4 coding sequences were constructed as fusion products with HA-MBP (i.e., HA-MBP-Ad2 E4orf4), as illustrated in Fig. 3A. cDNAs encoding these HA-tagged mutant E4orf4 products were cotransfected into cells along with a construct expressing a FLAG-tagged Bα subunit. Figure 3C shows that in all cases, high levels of expression were obtained, as determined by Western blotting analysis of whole-cell extracts using anti-HA antibody. Cell extracts were immunoprecipitated with anti-HA antibody; following separation by SDS-PAGE, proteins were transferred and analyzed by Western blotting using anti-FLAG antibody to determine the level of binding of the Bα subunit. Figure 3B shows that the levels of binding with all mutants were extremely low or undetectable in comparison to those observed with full-length E4orf4 or MBP-E4orf4. The highest levels of Bα-subunit binding among the mutants were with constructs expressing only residues 80 to 100 (A80/T100) and 80 to 114 (A80/Q114), suggesting that residues 80 to 100 may be involved in the interaction with the Bα subunit; however, the deletion of residues 11 to 39 (dl11–39) eliminated binding, suggesting that this region may be required for the interaction in some fashion. These data suggested that maintenance of the integrity of the E4orf4 protein is critical for efficient complex formation with the Bα subunit and that the E4orf4 protein structure is not tolerant of large-scale alterations. In addition, none of these deletion constructs was capable of inducing significant cell death in colony inhibition assays similar to those shown in Fig. 2 (data not shown), suggesting that deletion analysis is of limited value in analyzing the structure and function of E4orf4.
FIG. 3.
Analysis of Bα-subunit binding by E4orf4 deletion mutants. A series of E4orf4 deletions were produced and expressed as fusion products with HA-MBP, as described in Materials and Methods. Such DNA constructs were lipofected into H1299 cells along with a cDNA expressing a FLAG-tagged human Bα subunit. Aliquots of the cell extracts were immunoprecipitated with anti-HA antibody. After SDS-PAGE and transfer, they were immunoblotted with anti-FLAG antibody to detect associated Bα binding. Other aliquots were separated directly by SDS-PAGE and after transfer were immunoblotted with anti-HA antibody to detect levels of HA-MBP-E4orf4 expression. (A) Map of E4orf4 deletion mutants; (B) Bα-subunit binding; (C) HA-MBP-E4orf4 expression in whole-cell extracts (WCE).
Construction of E4orf4 point mutants.
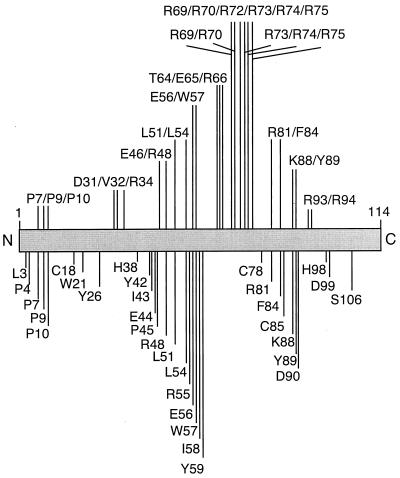
The alignments shown in Fig. 1 for E4orf4 from different adenovirus serotypes were used to identify highly conserved residues that were then targeted for mutagenesis. Thus, a series of HA-tagged single-point mutants as well as some groupings of point mutants for Ad2 E4orf4 was produced, all with alanine substitutions (Fig. 4). To verify that these mutants were stably expressed at high levels, corresponding cDNAs were introduced by lipofection into H1299 cells. Figure 5 shows that most of these mutants yielded high levels of stable products comparable to those obtained with wild-type E4orf4. The only exceptions were mutants L3A and P7A/P9A/P10A, which were shown by quantitative immunoblotting using 125I-labeled antibody to be expressed at less than one-third of wild-type levels. These mutants therefore were excluded from the present study. Protein expression levels were also determined in the human cervical carcinoma line C33A. Some differences in stability were evident between H1299 and C33A cells, which are discussed further, below.
FIG. 4.
Summary of Ad2 point mutants. A series of E4orf4 point mutants were produced as described in Materials and Methods. The residues below the bar are the sites for a series of single-point mutations. Above the bar are the locations of mutations for multiple-point mutants. Single-letter amino acid codes were used to show the original residue, and all mutants were constructed with alanine substitutions.
Association of the Bα subunit and PP2A activity with E4orf4 mutant proteins.
E4orf4 binds to the Bα subunit of the PP2A holoenzyme, which also contains the catalytic C and A subunits (reviewed in reference 37). Such binding has been shown to link E4orf4 to the PP2A holoenzyme (26). The abilities of wild-type and mutant E4orf4 proteins to associate with functional PP2A activity were determined in H1299 cells which had been colipofected with cDNAs expressing exogenous FLAG-tagged Bα and HA-tagged E4orf4. In the present and previous studies (31), no evidence that these molecular tags have any effect on the biological activities of E4orf4 or the Bα subunit has been detected. Cell extracts were immunoprecipitated with anti-HA antibodies, and the precipitates were then resuspended in reaction buffer and assayed in vitro for phosphatase activity by using a synthetic peptide containing a universal phosphoserine phosphatase substrate site. Phosphate release was determined spectrophotometrically using a malachite green-based assay. Figure 5 shows that high levels of phosphatase activity were present in precipitates containing wild-type E4orf4, indicating that the HA tag did not interfere with the association with PP2A. Figure 5 also shows that the expression of the Bα subunit was comparable in all cell extracts. For most of the mutants, the level of phosphatase activity was similar to that present with wild-type E4orf4; however, a number of mutants were defective in coprecipitating active phosphatase, including L51/54A, R69A/R70A/R72/73/74/75A, R81A/F84A, K88A/Y89A, and Y89A, and to some extent, C18A, T64A/E65A/R66A, R73/74/75A, and F84A. As expected, these levels of phosphatase activity corresponded well to the quantities of FLAG-Bα bound to E4orf4 as detected by immunoblotting (Fig. 5), suggesting that these nine mutants were either partially or almost completely defective in binding to the Bα subunit. The analyses of binding of Bα in these and previous (46b) studies were only semiquantitative because of the use of the ECL method of detection. Thus relative phosphatase levels were used in further correlations as quantitative measurements of the interaction of E4orf4 with the Bα of PP2A. This approach provides a more quantitative estimate of Bα binding, and our extensive analyses have indicated that the binding of E4orf4 has no effect on PP2A activity against this universal substrate (data not shown). It was noted that the apparent correlation between phosphatase activity and Bα binding (Fig. 5) of two mutants, F84A and R93/94A, appeared to be somewhat at variance. It should be remembered that the phosphatase data were derived from four separate experiments, whereas the binding data represented a single Western blotting analysis which may not be representative of all experiments. For both of these mutants, other analyses indicated somewhat higher levels of binding of Bα, in keeping with the phosphatase activities presented in Fig. 5.
Correlation between Bα binding and induction of cell death using E4orf4 mutants.
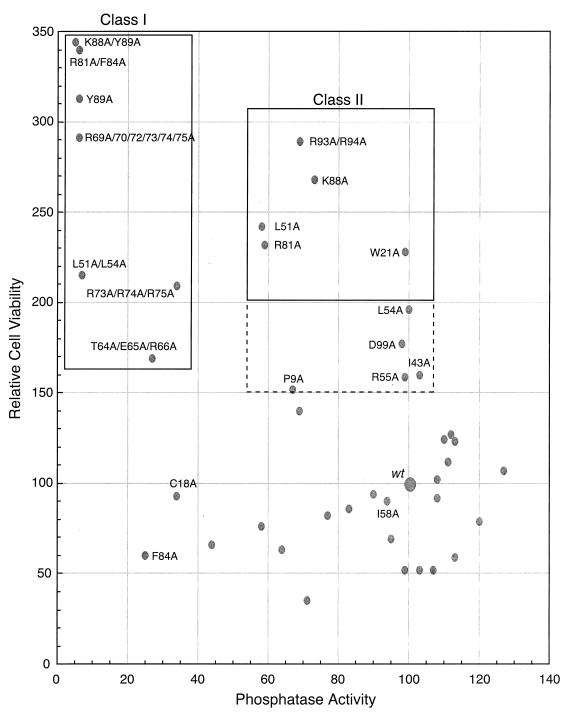
To determine the importance of Bα binding in the induction of cell death, colony formation was assessed following the lipofection of H1299 cells with plasmid DNA coexpressing wild-type or mutant E4orf4 and the neo marker and selection by G418. In such experiments, the number of colonies obtained with wild-type E4orf4 was arbitrarily set at 100. The mean colony formation index derived from four separate experiments, each containing three plates per mutant, was determined. The overall variation for each analysis in such studies was about 30%. Most of the mutants inhibited colony formation to about the same extent as wild-type E4orf4; however, with several mutants a considerable increase in the number of colonies was noted, thus indicating reduced cell killing. To correlate cell killing determined in these experiments with the binding of the Bα subunit, relative cell viability (i.e., colony-forming ability) was plotted against the phosphatase activity found to be associated with each mutant E4orf4 protein. Figure 6 indicates that for most mutants, the inhibition of colony formation was similar to that of wild-type E4orf4. This effect was seen not only with mutants that associated efficiently with PP2A but also with three or four mutants that bound less than 60% of wild-type levels. Remarkably, mutants F84A and C18A associated with only 25 and 34% of wild-type levels yet still killed as well as or better than the wild type. These results suggested that cell killing was induced if binding exceeded a critical lower threshold. Nonetheless, most mutants that associated with lower amounts, including L51/54A, R69A/R70A/R72/73/74/75A, R81A/F84A, K88A/Y89A, and Y89, were greatly defective for killing. These have been termed class I mutants (Fig. 6 and Table 1). Two additional mutants, T64A/E65A/R66A and R73/74/75A, which associated with PP2A at less than 34% of wild-type levels and were considerably defective in killing, are probably also class I mutants that interact at levels lower than the threshold. These results indicated that a reduction in association with PP2A blocks cell killing and thus links PP2A binding to the induction of apoptosis. A second class of mutants was also apparent, including W21A, L51A, R81A, K88A, and R93/94A, in which substantive levels of PP2A binding were evident but cell killing was greatly impaired. These mutants have been termed class II mutants. Five additional mutants displayed more modest reductions in cell killing yet interacted with PP2A normally, including P9A, I43A, L54A, R55A, and D99A. These results suggested that although PP2A binding was necessary for the induction of cell killing, it was not sufficient.
FIG. 6.
Analysis of cell killing versus associated phosphatase activity for E4orf4 mutants. The ability of E4orf4 mutants to associate with PP2A and to kill H1299 cells was assessed. The number of colonies and phosphatase activity detected with wild-type (wt) E4orf4 were arbitrarily set at 100. Class I mutants that failed to bind and to induce cell death are indicated in a box at the top left, whereas class II mutants that bound Bα but failed to kill efficiently are indicated in a box at the top right. Mutants that may be of the class II type are presented in the box bordered by dotted lines. The experimental error in each of these assays was less than 30%.
TABLE 1.
Comparison of mutant E4orf4-induced killing of H1299 and C33A cellsa
| Mutant | Mutant phenotype for:
|
|
|---|---|---|
| H1299 cells | C33A cells | |
| L51/54A | Class I | Low expression |
| T64A/E65A/R66A | Probable class I | Class I |
| R69A/R70A/R72/73/74/75A | Class I | Low expression |
| R73/74/75A | Probable class I | Class I |
| R81A/F84A | Class I | Class I |
| F84A | Wild-type | Class I |
| K88A/Y89A | Class I | Low expression |
| Y89A | Class I | Low expression |
| W21A | Class II | Class II |
| L51A | Class II | Class I |
| I58A | Wild-type | Class II |
| R81A | Class II | Probable class II |
| K88A | Class II | Class II |
| R93/94A | Class II | Class II |
To strengthen the interpretation of these results, a similar set of studies was carried out on C33A cells, which, like H1299 cells, are killed by E4orf4. This analysis yielded data that were largely similar to those presented for H1299 cells (data not shown). There were, however, a few differences that may be of importance. Table 1 compares results in the two cell lines for class I and class II mutants, which were defined by the criteria described for Fig. 6. Although we have not detected any differences in the stability of wild-type E4orf4 in H1299 and C33A cells, several of the mutants were expressed at lower levels in the latter. These results suggest that C33A cells are less tolerant of E4orf4 proteins that are not folded properly. T64A/E65A/R66A, R73/74A, and R81A/F84A appeared to be class I mutants in both cell types. F84A, which killed at wild-type levels in H1299 cells, was clearly a class I mutant in C33A cells, suggesting that the low level of PP2A binding that it exhibited in both cell types was just above the threshold level for H1299 but below the threshold for C33A. Almost all of the class II mutants identified in H1299 cells were also class II in C33A cells. The only exception was L51A, which associated with less PP2A activity in C33A cells and thus was more similar to a class I mutant.
DISCUSSION
E4orf4 products encoded by most if not all human adenovirus serotypes are highly efficient in inducing cell death. It is our belief that one part of this function is to play a role in the death of infected cells at the late stage of lytic infection.
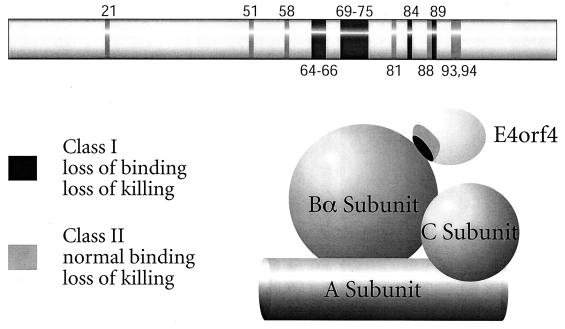
Figure 7 summarizes the results of our genetic analysis of E4orf4 function. The present results demonstrate that PP2A binding is clearly necessary for induction of cell death by E4orf4. This possibility was also suggested in a recent study, but that work employed a smaller number of E4orf4 mutants and semiquantitative analysis (46b). In the present study, all E4orf4 class I mutants that failed to bind Bα and associate with PP2A activity were highly defective for induction of cell death. This genetic approach suggested that a region in E4orf4 including residues 51 to 89 may be of some importance in binding of the Bα subunit. This region could be involved directly in binding or could represent a portion of the E4orf4 molecule that regulates binding at another site; however, class I mutants were not found in any other regions. Analysis of E4orf4 deletion mutants suggested that residues 80 to 100 may be directly involved in binding, and two class I mutations, K88A/Y89A and Y89A, as well as a class II mutation, R93/94A, were identified in this region. These possibilities were also strengthened by the results of a previous study, which showed that mutants containing S95P and G19A/N103I mutations had phenotypes similar to those of our class I mutants, as did mutant R55A/E56A/W57A/C78R (46b). It should be noted that one of the class II mutations, W21A, is located within residues 11 to 39, which were shown by using deletion mutants to play a role in binding when the amino terminus of E4orf4 is present. Further detailed, structural information will be required to assess this situation. Nonetheless, binding to PP2A is clearly essential for cell killing by E4orf4.
FIG. 7.
Summary of E4orf4 residues involved in PP2A binding and cytotoxicity. The mutations that gave rise to E4orf4 proteins deficient in cell killing, PP2A binding, or both are indicated. E4orf4 is shown binding to the PP2A Bα subunit.
The other important outcome of the present study was the identification of the class II mutants that bind the Bα subunit at normal levels but are defective for killing. These results suggested that association with PP2A is necessary but not sufficient for killing. It is unclear whether this additional requirement implies a function distinct from Bα binding or if it reflects the need for a highly precise interaction between Bα and E4orf4. For example, a class II mutant E4orf4 protein may be competent to interact with and remain bound to Bα during the isolation and immunoprecipitation procedures but fail to exert a biological effect on PP2A. Apart from W21A, class II mutants were found close to and sometimes immediately adjacent to residues shown in class I mutants to be required for binding. In addition, although mutants L51A and L54A were individually of the class II type, when the mutations were present in combination (i.e., L51/54A) the mutant was class I. These observations suggest but do not prove that the class II mutants may fail to kill because the binding which they exhibit is not sufficiently exact to exert a biological effect. Again, structural analysis could resolve this issue.
What might be the consequences of the interaction between Bα and E4orf4? A reasonable interpretation of earlier work on E4orf4 is that binding activates PP2A activity (16, 26, 35, 36). Although we have not noted any overall change in PP2A activity against the universal peptide substrate as a consequence of interaction with E4orf4, it is highly possible that E4orf4 alters activity against specific substrates that are normally regulated by the B subunit. The other possibility is that E4orf4 provides an additional function required for cell killing, such as altering the intracellular localization of PP2A, altering the binding of other Bα-interacting proteins, or linking PP2A with critical substrates. Such effects on PP2A would not be detected in the present in vitro assays. Studies to address these possibilities are under way.
ACKNOWLEDGMENTS
We are indebted to Goran Akusjärvi for the E4orf4 cDNA.
This work was supported in part by grants to P.E.B. from the National Cancer Institute of Canada.
REFERENCES
- 1.Anderson N G, Maller J L, Tonks N K, Sturgill T W. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 2.Bondesson M, Öhman K, Mannervik M, Fan S, Akusjärvi G. Adenovirus E4 open reading frame 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bondesson M, Svensson C, Linder S, Akusjärvi G. The carboxy-terminal exon of the adenovirus E1A protein is required for the E4F-dependent transcription activation. EMBO J. 1992;11:3347–3354. doi: 10.1002/j.1460-2075.1992.tb05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braconi-Quintaje S B, Church D J, Rebsamen M, Valloton M B, Hemmings B A, Lang U. Role of protein phosphatase 2A in the regulation of mitogen-activated protein kinase activity in ventricular cardiomyocytes. Biochem Biophys Res Commun. 1996;221:539–547. doi: 10.1006/bbrc.1996.0632. [DOI] [PubMed] [Google Scholar]
- 5.Cegielska A, Shaffer S, Derua R, Goris J, Virshup D M. Different oligomeric forms of protein phosphatase 2A activate and inhibit simian virus 40 DNA replication. Mol Cell Biol. 1994;14:4616–4623. doi: 10.1128/mcb.14.7.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Branton P E, Yang E, Korsmeyer S J, Shore G C. Adenovirus E1B 19-kDa death suppressor protein interacts with Bax but not with Bad. J Biol Chem. 1996;271:24221–24225. doi: 10.1074/jbc.271.39.24221. [DOI] [PubMed] [Google Scholar]
- 7.Chen P-L, Scully P, Shew J-Y, Wang J Y J, Lee W-H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cell differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 8.Chiou S-K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 10.Csortos C, Zolnierowicz S, Bako E, Durbin S D, DePaoli-Roach A A. High complexity in the expression of the B′ subunit of protein phosphatase 2A0. Evidence for the existence of at least seven novel isoforms. J Biol Chem. 1996;271:2578–2588. doi: 10.1074/jbc.271.5.2578. [DOI] [PubMed] [Google Scholar]
- 11.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 12.DePaoli-Roach A A, Park I K, Cerovsky V, Csortos C, Durbin S D, Kuntz M J, Sitikov A, Tang P M, Verin A, Zolnierowicz S. Serine/threonine protein phosphatases in the control of cell function. Adv Enzyme Reg. 1994;34:199–224. doi: 10.1016/0065-2571(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 13.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 14.Duerksen-Hughes P J, Hermiston T W, Wold W S M, Gooding L R. The amino-terminal portion of CD1 of the adenovirus E1A proteins is required to induce susceptibility to tumor necrosis factor cytolysis in adenovirus-infected mouse cells. J Virol. 1991;65:1236–1244. doi: 10.1128/jvi.65.3.1236-1244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel D A, Hardy S, Shenk T. cAMP acts in synergy with E1A protein to activate transcription of the adenovirus genes E4 and E1A. Genes Dev. 1988;2:1517–1528. doi: 10.1101/gad.2.12a.1517. [DOI] [PubMed] [Google Scholar]
- 16.Engel D A, Müller U, Gedrich R W, Eubanks J S, Shenk T. Induction of c-fos mRNA and AP-1 DNA binding activity by cAMP in cooperation with either the adenovirus 243- or the adenovirus 289-amino acid E1A protein. Proc Natl Acad Sci USA. 1991;88:3957–3961. doi: 10.1073/pnas.88.9.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez N, Cohen P. Dissection of the protein kinase cascade by which nerve growth factor activates MAP kinases. Nature. 1991;353:170–173. doi: 10.1038/353170a0. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Sabbatini P, Perez D, Rao L, Modha D, White E. The E1B 19K protein blocks apoptosis by interacting with and inhibiting the p53-inducible and death-promoting Bax protein. Genes Dev. 1996;10:461–477. doi: 10.1101/gad.10.4.461. [DOI] [PubMed] [Google Scholar]
- 19.Healy A M, Zolnierowicz S, Stapleton A E, Goebl M, DePaoli-Roach A A, Pringle J R. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol. 1991;11:5767–5780. doi: 10.1128/mcb.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemmings B A, Adams-Pearson C, Maurer F, Muller P, Goris J, Merlevede W, Hofsteenge J, Stone S R. α- and β-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry. 1990;29:3166–3173. doi: 10.1021/bi00465a002. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix R E, Mayer-Jaekel P, Cron J, Goris J, Hofsteenge W, Merlevede W, Hemmings B A. Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A. J Biol Chem. 1993;268:15267–15276. [PubMed] [Google Scholar]
- 21a.Higgins D D, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 22.Jaiswal R K, Murphy M B, Landreth G E. Identification and characterization of a nerve growth factor-stimulated mitogen-activated protein kinase activator in PC12 cells. J Biol Chem. 1993;10:7055–7063. [PubMed] [Google Scholar]
- 23.Kamibayashi C, Estes R, Lickteig R L, Yang S I, Craft C, Mumby M C. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J Biol Chem. 1994;269:20139–20148. [PubMed] [Google Scholar]
- 24.Kamibayashi C, Lickteig R L, Estes R, Walter G, Mumby M C. Expression of the A subunit of protein phosphatase 2A and characterization of its interactions with the catalytic and regulatory subunits. J Biol Chem. 1992;267:21864–21872. [PubMed] [Google Scholar]
- 25.Kanopka A, Mühlemann O, Petersen-Mahrt S, Estmer C, Öhrmalm C, Akusjärvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 26.Kleinberger T, Shenk T. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol. 1993;67:7556–7560. doi: 10.1128/jvi.67.12.7556-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremmer E, Ohst K, Kiefer J, Brewis N, Walter G. Separation of PP2A core enzyme and holoenzyme with monoclonal antibodies against the regulatory A subunit: abundant expression of both forms in cells. Mol Cell Biol. 1997;17:1692–1701. doi: 10.1128/mcb.17.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavoie J N, Nguyen M, Marcellus R C, Branton P E, Shore G C. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway which is not inhibited by zVAD-fmk. J Cell Biol. 1998;140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 30.Marcellus R C, Teodoro J G, Wu T, Brough D E, Ketner G, Shore G C, Branton P E. Adenovirus type 5 early region 4 is responsible for E1A-induced p53-independent apoptosis. J Virol. 1996;70:6207–6215. doi: 10.1128/jvi.70.9.6207-6215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcellus R C, Lavoie J N, Boivin D, Shore G C, Ketner G, Branton P E. The early region 4orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J Virol. 1998;72:7144–7153. doi: 10.1128/jvi.72.9.7144-7153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer-Jaekel R E, Ohkura H, Ferrigno P, Andjelkovic N, Shiomi K, Uemura T, Glover D M, Hemmings B A. Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J Cell Sci. 1994;107:2609–2616. doi: 10.1242/jcs.107.9.2609. [DOI] [PubMed] [Google Scholar]
- 33.McCright B, Virshup D M. Identification of a new family of protein phosphatase 2A regulatory subunits. J Biol Chem. 1995;270:26123–26128. doi: 10.1074/jbc.270.44.26123. [DOI] [PubMed] [Google Scholar]
- 34.McCright B, Rivers A M, Audlin S, Virshup D M. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J Biol Chem. 1996;271:22081–22089. doi: 10.1074/jbc.271.36.22081. [DOI] [PubMed] [Google Scholar]
- 35.Müller U, Kleinberger T, Shenk T. Adenovirus E4orf4 protein reduces phosphorylation of c-Fos and E1A proteins while simultaneously reducing the level of AP-1. J Virol. 1992;66:5867–5878. doi: 10.1128/jvi.66.10.5867-5878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller U, Roberts M P, Engel D A, Doerfler W, Shenk T. Induction of transcription factor AP-1 by adenovirus E1A protein and cAMP. Genes Dev. 1989;3:1991–2002. doi: 10.1101/gad.3.12a.1991. [DOI] [PubMed] [Google Scholar]
- 37.Mumby M C, Walter G. Protein serine/threonine phosphatases: structure, regulation, and functions in cell growth. Physiol Rev. 1993;73:673–700. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen M, Branton P E, Roy S, Nicholson D W, Alnemri E S, Yeh W-C, Mak T W, Shore G C. E1A-induced processing of procaspase-8 can occur independently of FADD and is inhibited by Bcl-2. J Biol Chem. 1998;273:33099–33102. doi: 10.1074/jbc.273.50.33099. [DOI] [PubMed] [Google Scholar]
- 39.Ohman K, Nordqvist K, Akusjarvi G. Two adenovirus proteins with redundant activities in virus growth facilitates tripartite leader mRNA accumulation. Virology. 1993;194:50–58. doi: 10.1006/viro.1993.1234. [DOI] [PubMed] [Google Scholar]
- 39a.Pallas D C, Weller W, Jaspers S, Miller T B, Lane W S, Roberts T M. The third subunit of protein phosphatase 2A (PP2A), a 55-kilodalton protein which is apparently substituted for by T antigens in complexes with the 36- and 63-kilodalton PP2A subunits, bears little resemblance to T antigens. J Virol. 1992;66:886–893. doi: 10.1128/jvi.66.2.886-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez D, White E. E1B 19K inhibits Fas-mediated apoptosis through FADD-dependent sequestration of FLICE. J Cell Biol. 1998;141:1255–1266. doi: 10.1083/jcb.141.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Querido E, Teodoro J G, Branton P E. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychaudhuri P, Bagchi S, Nevins J R. DNA-binding activity of the adenovirus-induced E4F transcription factor is regulated by phosphorylation. Genes Dev. 1989;3:620–627. doi: 10.1101/gad.3.5.620. [DOI] [PubMed] [Google Scholar]
- 43a.Roulston A, Marcellus R C, Branton P E. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 44.Ruediger R, Hentz M, Fait J, Mumby M C, Walter G. Molecular model for the A subunit of protein phosphatase 2A: interaction with other subunits and tumor antigens. J Virol. 1994;68:123–129. doi: 10.1128/jvi.68.1.123-129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruediger R, Roeckel D, Fait J, Bergqvist A, Magnusson G, Walter G. Identification of binding sites on the regulatory A subunit of protein phosphatase 2A for the catalytic C subunit and for tumor antigens of simian virus 40 and polyomavirus. Mol Cell Biol. 1992;12:4872–4882. doi: 10.1128/mcb.12.11.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shisler J, Duerksen-Hughes P, Hermiston T M, Wold W S M, Gooding L R. Induction of susceptibility to tumor necrosis factor by E1A is dependent on binding to either p300 or p105-Rb and induction of DNA synthesis. J Virol. 1996;70:68–77. doi: 10.1128/jvi.70.1.68-77.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Shtrichman R, Kleinberger T. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol. 1998;72:2975–2982. doi: 10.1128/jvi.72.4.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46b.Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc Natl Acad Sci USA. 1999;96:10080–10085. doi: 10.1073/pnas.96.18.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sontag E, Nunbhakdi-Craig V, Bloom G S, Mumby M C. A novel pool of protein phosphatase 2A is associated with microtubules and is regulated during the cell cycle. J Cell Biol. 1995;128:1131–1144. doi: 10.1083/jcb.128.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Strack S, Chang D, Zaucha J A, Colbran R J, Wadzinski B E. Cloning and characterization of B delta, a novel regulatory subunit of protein phosphatase 2A. FEBS Lett. 1999;460:462–466. doi: 10.1016/s0014-5793(99)01377-0. [DOI] [PubMed] [Google Scholar]
- 48.Tanabe O, Nagase T, Murakami T, Nozaki H, Usui H, Nishito Y, Hayashi H, Kagamiyama H, Takeda M. Molecular cloning of a 74-kDa regulatory subunit (B" or delta) of human protein phosphatase 2A. FEBS Lett. 1996;379:107–111. doi: 10.1016/0014-5793(95)01500-0. [DOI] [PubMed] [Google Scholar]
- 49.Tehrani M A, Mumby M C, Kamibayashi C. Identification of a novel protein phosphatase 2A regulatory subunit highly expressed in muscle. J Biol Chem. 1996;271:5164–5170. doi: 10.1074/jbc.271.9.5164. [DOI] [PubMed] [Google Scholar]
- 50.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teodoro J G, Halliday T, Whalen S G, Takayesu D, Graham F L, Branton P E. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J Virol. 1994;68:776–786. doi: 10.1128/jvi.68.2.776-786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teodoro J G, Shore G C, Branton P E. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- 53.Tollefson A E, Hermiston T W, Lichtenstein D L, Colle C F, Tripp R A, Dimitrov T, Toth K, Wells C E, Doherty P C, Wold W S M. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature. 1998;392:726–730. doi: 10.1038/33712. [DOI] [PubMed] [Google Scholar]
- 54.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter G, Ferre F, Espiritu O, Carbone-Wiley A. Molecular cloning and sequence of cDNA encoding polyoma medium tumor antigen-associated 61-kDa protein. Proc Natl Acad Sci USA. 1989;86:8669–8672. doi: 10.1073/pnas.86.22.8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whalen S G, Marcellus R C, Whalen A, Ahn N G, Ricciardi R P, Branton P E. Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J Virol. 1997;71:3545–3553. doi: 10.1128/jvi.71.5.3545-3553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White E, Sabbatini P, Debbas M, Wold W S M, Kusher D I, Gooding L R. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor α. Mol Cell Biol. 1992;12:2570–2580. doi: 10.1128/mcb.12.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y, Boguslawski G, Zitomer R S, DePaoli-Roach A A. Saccharomyces cerevisiae homologues of mammalian B and B subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J Biol Chem. 1997;272:8256–8262. doi: 10.1074/jbc.272.13.8256. [DOI] [PubMed] [Google Scholar]
- 60.Zolnierowicz S, Csortos C, Bondor J, Verin A, Mumby M C, DePaoli-Roach A A. Diversity in the regulatory B-subunits of protein phosphatase 2A: identification of a novel isoform highly expressed in brain. Biochemistry. 1994;33:11858–11867. doi: 10.1021/bi00205a023. [DOI] [PubMed] [Google Scholar]
- 61.Zolnierowicz S, van Hoof C, Andjelkovic N, Cron P, Stevens I, Merlevede W, Goris J, Hemmings B A. The variable subunit associated with protein phosphatase 2A defines a novel multimember family of regulatory subunits. Biochem J. 1996;317:187–194. doi: 10.1042/bj3170187. [DOI] [PMC free article] [PubMed] [Google Scholar]