Abstract
Predicting the blood–brain barrier (BBB) permeability of small-molecule compounds using a novel artificial intelligence platform is necessary for drug discovery. Machine learning and a large language model on artificial intelligence (AI) tools improve the accuracy and shorten the time for new drug development. The primary goal of this research is to develop artificial intelligence (AI) computing models and novel deep learning architectures capable of predicting whether molecules can permeate the human blood–brain barrier (BBB). The in silico (computational) and in vitro (experimental) results were validated by the Natural Products Research Laboratories (NPRL) at China Medical University Hospital (CMUH). The transformer-based MegaMolBART was used as the simplified molecular input line entry system (SMILES) encoder with an XGBoost classifier as an in silico method to check if a molecule could cross through the BBB. We used Morgan or Circular fingerprints to apply the Morgan algorithm to a set of atomic invariants as a baseline encoder also with an XGBoost classifier to compare the results. BBB permeability was assessed in vitro using three-dimensional (3D) human BBB spheroids (human brain microvascular endothelial cells, brain vascular pericytes, and astrocytes). Using multiple BBB databases, the results of the final in silico transformer and XGBoost model achieved an area under the receiver operating characteristic curve of 0.88 on the held-out test dataset. Temozolomide (TMZ) and 21 randomly selected BBB permeable compounds (Pred scores = 1, indicating BBB-permeable) from the NPRL penetrated human BBB spheroid cells. No evidence suggests that ferulic acid or five BBB-impermeable compounds (Pred scores < 1.29423E−05, which designate compounds that pass through the human BBB) can pass through the spheroid cells of the BBB. Our validation of in vitro experiments indicated that the in silico prediction of small-molecule permeation in the BBB model is accurate. Transformer-based models like MegaMolBART, leveraging the SMILES representations of molecules, show great promise for applications in new drug discovery. These models have the potential to accelerate the development of novel targeted treatments for disorders of the central nervous system.
Keywords: Blood–brain barrier (BBB) permeability, Machine learning, Artificial intelligence (AI), Natural Products Research Laboratories (NPRL)
Subject terms: Computational biology and bioinformatics, Mathematics and computing
Introduction
The blood–brain barrier (BBB) is a customized capillary bed that separates the brain from the circulatory system. It can protect the brain from pathogens, such as bacteria and viruses1–4. BBB-penetrating drugs are commonly used to treat central nervous system (CNS) disorders, such as Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, brain tumors (glioblastoma), and CNS infections (e.g., Neisseria meningitides infection) using antibiotic agents, such as meningitis agents1,5–7. The BBB, with tight junction and efflux transporter proteins, prevents the entry of therapeutic agents into the brain, resulting in unsuccessful therapy for brain and CNS diseases8–10. Alternatively, compounds with targets in peripheral tissues should be investigated for their BBB permeability to prevent CNS adverse drug reactions, such as drowsiness, respiratory depression, nausea, vomiting, dizziness, trance, and anxiety11. Through the development of this model and rapid screening of the compound database, new compounds for treating CNS diseases can be developed, and unknown compounds can be predicted for absorption, distribution, metabolism, excretion, and toxicity12–17.
Developing a practical and accurate model for predicting the BBB permeability of compounds is important for brain and neuron therapeutic new drug discovery in silico13,18. These compounds have known BBB permeable compounds. A widely used database is LightBBB, which contains 7162 compounds with 5453 BBB permeable compounds (BBB +) and 1709 BBB impermeable compounds (BBB-)19. These 1155 compounds had LogBB (logarithm of drug concentration in the brain by the concentration in the blood) values (accession date: 2/20/2023). Another database is B3DB, which includes 7807 compounds with 4956 BBB permeable compounds (BBB+) and 2851 BBB impermeable compounds (BBB−), and the 1058 compounds are with LogBB values20. LightBBB has been included in the new B3DB database. DeePred-BBB collects 3605 compounds, including 2607 BBB permeable compounds (BBB+) and 998 BBB impermeable compounds (BBB−)21–24.
Inspired by natural language processing, transformer-based architectures for solving chemo-informatics tasks have become increasingly popular in recent years25–27. Because chemical structures are in a simplified molecular input line entry system (SMILES) format, they are similar to their own language28. Thus, SMILES strings can be trained using transformers for transformer models to learn different characteristics of chemical data, such as chemical properties and its structures28–31. Chemical data are often complex and high-dimensional, making it difficult to train a model from scratch using limited data28. Pre-training on abundant data using techniques that do not require labeling, such as pre-training through the use of auto-encoders, can help the model learn general representations that can be transferred to downstream tasks, leading to improved performance and faster convergence32–34. MegaMolBART35 is a small-molecule language model pre-trained using a bidirectional and autoregressive transformer (BART) architecture on the ZINC-15 dataset36. The encoder of the model can be used to extract molecular features for down-stream predictive models. MegaMolBART was implemented using NVIDIA’s NeMo Toolkit, which is a Python framework agnostic toolkit for creating artificial intelligence (AI) applications through reusability, abstraction, and composition35. The MegaMolBART framework is open source and extends the NeMo Toolkit’s functionalities to add chemistry-specific functions, such as SMILES masking and RDkit functionalities for training augmentation37,38. Previous research on predicting blood–brain barrier (BBB) permeability for small molecules has employed various features and machine learning techniques11,20,39. Physicochemical properties were calculated using software toolkits like Dragon and PaDEL40,41. Additionally, molecular fingerprints, substructure fingerprints, and 2D compound images generated by the RDKit package were utilized as input features42,43. These features were then used to train both traditional machine learning algorithms such as support vector machines (SVMs)44,45, k-nearest neighbors (kNNs)46,47, random forests48,49, and naive Bayes classifiers50–52, as well as deep learning methods including dense neural networks (DNNs)53,54, 1D convolutional neural networks (CNNs), and 2D CNNs21,38,55.
In this study, we hypothesized that a deep learning model can provide a quick method to determine if a novel compound design can cross the BBB. To achieve this, we used MegaMolBART as the SMILES encoder to identify if a molecule passes through the BBB. We compared the results with those of traditional molecular similarity methods called fingerprinting. Here, we use Morgan or Circular Fingerprints which apply the Morgan algorithm to a set of atom invariants56,57. We will also verify these results using newly created natural product compound libraries that are not currently included in any database, such as the Compound Library of the Natural Products Research Laboratories (NPRL) of China Medical University Hospital (CMUH) in Taiwan58. Furthermore, an in vitro liquid chromatography and mass spectrometry (LC–MS/MS) study was conducted to assess the integrity of BBB spheroids and the permeability of compounds from NPRL.
Results and discussions
Supplementary Figure S1 shows the training and validation loss curves of training with PyTorch using the MegaMolBART embedding connected to the MegaMolBART encoder and then connected to a classifier layer. The training showed that the loss converged quickly, with over-fitting occurring at approximately 400 epochs. Supplementary Figure S2 shows the validation best area under curve (AUC) with and without the exponential moving average (EMA); the occurred immediately before the model started to over-fit (from the loss curve). We also tested different sizes of MegaMolBART, with training on the CMUH-NPRL test set with B3DB dataset (Supplementary Table S1), and B3DB test set with CMUH-NPRL dataset (Supplementary Table S2). These models exhibited validation AUC curves similar to those shown in Supplementary Fig. S2.
Since we believe that the small dataset caused MegaMolBART to over-fit the BBB datasets, we believe that the model did not take full advantage of the pre-training done on the ZINC-15 dataset. Thus, we attempted to use regression with XGBoost. When using regression, we compared it with the Morgan Fingerprints generated using RDKit with 2048 as the number of features37–39. The regression results are shown below. We also examined the accuracy of the results by converting the predicted LogBB value into accuracy using the formula shown in the previous section. The results of the regression with XGBoost in Supplementary Fig. S3 show that MegaMolBART embeddings work significantly better compared to Morgan Fingerprints, with the larger model showing the best performance. However, when the computed accuracy was compared using the predicted LogBB, the Morgan fingerprints performed slightly worse compared to the MegaMolBART embeddings. As the classification performed worse, the data distribution was checked using t-distributed stochastic neighbor embedding (t-SNE) on the NVIDIA GPU Cloud (NGC) MegaMolBART embeddings. The t-distributed stochastic neighbor embedding (t-SNE) distribution results shown in Supplementary Fig. S4 are that the data with LogBB are closely grouped together, whereas the data without LogBB are more spread out. This indicated that more data without LogBB were required to train a better model. Finally, we train the model with the XGBoost classifier using only the B3DB dataset. The results shown in Supplementary Fig. S5 indicate a significant improvement in the test dataset. However, this model was applied to the CMUH-NPRL dataset, the accuracy decreased by approximately 50%.
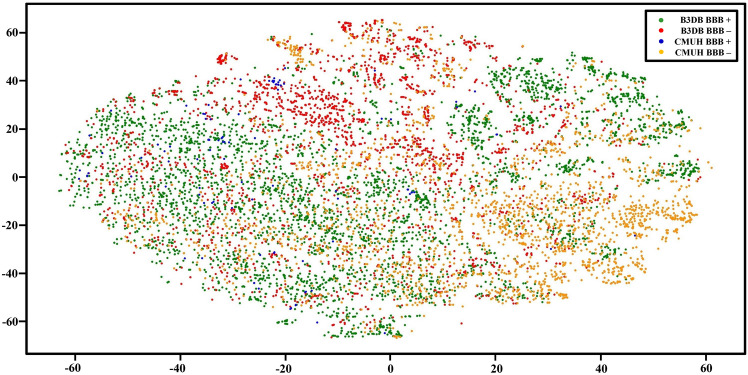
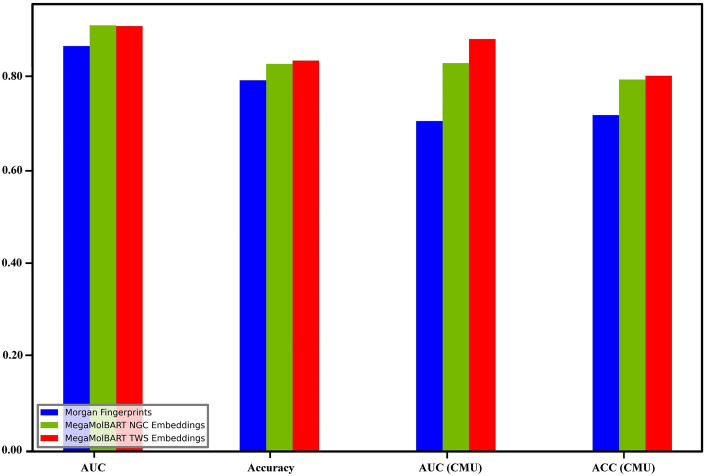
Next, we checked the distribution of the CMUH and B3DB data. Figure 1 show the t-distributed stochastic neighbor embedding (t-SNE) applied to the CMUH-NPRL and B3DB data using the NVIDIA GPU Cloud (NGC) embeddings. Our results clearly shows that the CMUH-NPRL and B3DB data are distributed far apart; therefore, the next model would involve mixing both types of datasets together for training. Finally, using 80% of both datasets for training, 10% of both datasets for validation, and 10% of both datasets for testing, we achieved an AUC of 0.88 using MegaMolBART. We also compared the same classifier with the Morgan Fingerprints and found a significant difference between the Fingerprints and Embeddings, with the larger MegaMolBART model performing slightly better (Fig. 2). Furthermore, we performed a comparative analysis of previous machine learning models that use physicochemical properties of molecules for BBB permeability classification and our MegaMolBART transformer-based. The traditional machine learning models used were the LightGBM mentioned in the LightBBB paper19 and DNN in the DeePred paper21–24. Both were trained using various physicochemical properties of the molecules, including molecular weight, lipophilicity, and hydrogen bonding potential and polar surface area, calculated using Dragon software59 and PaDEL60 respectively. Our MegaMolBART transformer-based model is a variant of the BART transformer architecture, adapted for BBB permeability classification using the SMILES representation of molecules. The model was pre-trained using the ZINC-15 database, and the BERT encoder was used to transform molecules into embeddings, which were then used to train a large dataset of molecules with known BBB permeabilities and optimized using a combination of gradient descent and back propagation. For the comparative analysis, we used the datasets provided by the respective papers, analyzed the datasets using their described tenfold cross-validation method, and reported the AUC for comparison.
Figure 1.
Data distribution of the molecule embeddings visualized using the t-distributed stochastic neighbor embedding (t-SNE) color coded by dataset and BBB+/BBB−.
Figure 2.
Classification AUC and accuracy of the test set from B3DB and CMUH and classification AUC and accuracy of only the CMUH test set.
The results of our comparative analysis showed that on the LightBBB dataset, the AUC of our model was 0.93 compared to the LightBBB reported AUC of 0.94 (Supplementary Table S3). For the DeePred dataset, the AUC of our model was 0.96 compared to the DeePred dataset, which reported an AUC of 0.99 (Supplementary Table S4). However, the transformer-based model does not require pre-computation of SMILE features using other software tools. Calculating physicochemical properties of molecules requires significant computational resources and can be time-consuming61–63. Moreover, many properties may not be easily interpreted or available for all molecules64. This means that these models may be unsuitable for large-scale drug discovery applications in which the number of molecules considered can be in the millions. In contrast, our MegaMolBART transformer-based model, can handle large and diverse sets of molecules without requiring extensive feature engineering or computationally intensive calculations (Supplementary Fig. S6). SMILES is a widely used standard for representing molecular structures as strings of characters that can be easily input into a transformer-based model65–69. Furthermore, using SMILES allows for greater flexibility and generalization of the input data because it can capture various molecular structures and properties66,67,70. This makes the transformer-based models more robust and adaptable to new and diverse sets of molecules, which are critical for new drug discovery71–73. Another advantage of transformer-based models is their ability to learn complex patterns and relationships in the input data, which may not be easily captured through calculations of physicochemical properties or fingerprints64,74. Transformers use a self-attention mechanism that allows them to selectively attend to different parts of the input sequence and capture long-range dependencies and complex relationships among different parts of the SMILES sequence75–77.
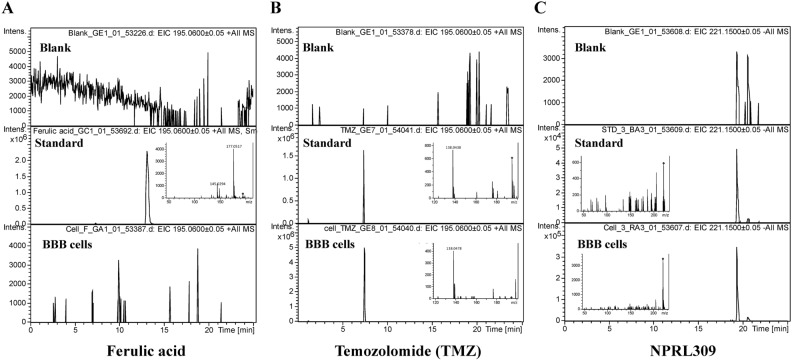
Using LC–MS/MS to assess BBB integrity has become an advanced technology in recent years78–80. We used LC–MS/MS on human BBB spheroid cells (consisting of human brain microvascular endothelial cells, brain vascular pericytes, and astrocytes) to analyze BBB permeability in vitro. We selected, at random, 21 (Pred scores = 1, indicating BBB-permeable compounds) and five (Pred scores < 1.29423E−05, indicating BBB-impermeable compounds) compounds of NPRL to be verified in vitro. Figure 3 and Supplementary Fig. S9 demonstrate TMZ and 21 BBB permeable compounds (BBB +) (predicted by NVIDIA’s NeMo Toolbox to be BBB-permeable) of NPRL penetrated human BBB spheroid cells. Ferulic acid and five BBB-impermeable compounds (Pred scores < 1.29423E−05) predicted by the NPRL were inaccessible to human BBB spheroid cells. To the best of our knowledge, this was the first study on the BBB permeability of compound libraries using abundant databases. Our method offers a novel cellular model for BBB permeability measurements. The results summarized in Table 1 provide evidence that the BBB permeable compounds (BBB+) of NPRL, predicted by NVIDIA’s NeMo Toolkit, can penetrate human brain microvascular endothelial cells and reach human BBB spheroid cells. The permeability coefficients validated these findings. The Natural Products Research Laboratories (NPRL) compound library was established by Professor Kuo-Hsiung Lees (The University of North Carolina at Chapel Hill) from China Medical University Hospital (CMUH) to determine the bioactivity of these treasured natural products and their synthesized derivatives58,81. Our research provides a fast and highly specific in silico and in vivo methods and a new bioactivity assay for NPRL compounds. This study provides a novel research method for building platforms for compound laboratories with large databases. In the future, we aim to use a human brain endothelial cell model (hCMEC/D3 human BBB cells) to further explore molecular and pharmacologic transport mechanisms of novel compounds entering the BBB82.
Figure 3.
Human BBB spheroid cells were analyzed by LC–MS/MS, which shows that TMZ, ferulic acid, and NPRL-309 have standard peaks.
Table 1.
In vitro permeability assay and in silico prediction outcomes for BBB spheroid cells.
| Sample number | Sample | In silico prediction | In vitro study | |
|---|---|---|---|---|
| PRED SCORES | PRED LABEL | LC–MS/MS | ||
| Control | – | – | – | – |
| Negative control | Ferulic acid | BBB impermeable | ||
| Positive control | TMZ | BBB permeable | ||
| 1 | NPRL309 | 1.00 | BBB + | BBB permeable |
| 2 | NPRL358 | 1.00 | BBB + | BBB permeable |
| 3 | NPRL588 | 1.00 | BBB + | BBB permeable |
| 4 | NPRL818 | 1.00 | BBB + | BBB permeable |
| 5 | NPRL833 | 1.00 | BBB + | BBB permeable |
| 6 | NPRL835 | 1.00 | BBB + | BBB permeable |
| 7 | NPRL836 | 1.00 | BBB + | BBB permeable |
| 8 | NPRL842 | 1.00 | BBB + | BBB permeable |
| 9 | NPRL1089 | 1.00 | BBB + | BBB permeable |
| 10 | NPRL1185 | 1.00 | BBB + | BBB permeable |
| 11 | NPRL1188 | 1.00 | BBB + | BBB permeable |
| 12 | NPRL1192 | 1.00 | BBB + | BBB permeable |
| 13 | NPRL1195 | 1.00 | BBB + | BBB permeable |
| 14 | NPRL1241 | 1.00 | BBB + | BBB permeable |
| 15 | NPRL1958 | 1.00 | BBB + | BBB permeable |
| 16 | NPRL2026 | 1.00 | BBB + | BBB permeable |
| 17 | NPRL2029 | 1.00 | BBB + | BBB permeable |
| 18 | NPRL2051 | 1.00 | BBB + | BBB permeable |
| 19 | NPRL2059 | 1.00 | BBB + | BBB permeable |
| 20 | NPRL2148 | 1.00 | BBB + | BBB permeable |
| 21 | NPRL3767 | 1.00 | BBB + | BBB permeable |
| 22 | NPRL2359 | 1.38848E−05 | BBB− | BBB impermeable |
| 23 | NPRL2576 | 1.49735E−05 | BBB− | BBB impermeable |
| 24 | NPRL2646 | 1.40275E−05 | BBB− | BBB impermeable |
| 25 | NPRL3098 | 1.29423E−05 | BBB− | BBB impermeable |
| 26 | NPRL3183 | 1.74123E−05 | BBB− | BBB impermeable |
Our study shows that pre-training can significantly accelerate the convergence of down-stream task models. The Large MegaMolBART pretrained on the ZINC-15 dataset shows the most promise and best accuracy on B3DB (Fig. 2), although more pre-training may be required to obtain a better accuracy score, and more LogBB data are required for a better regression accuracy score. The current distribution of the B3DB data is uneven. In addition, the classification of B3DB can reach up to 0.90 of AUC with our Taiwan Web Service (TWS) embedding and XGBoost regression (Supplementary Fig. S3). Classification can reach up to 90% AUC with TWS embeddings and XGBoost classification (Supplementary Fig. S5). The results of the classification can also been seen through the confusion matrices and evaluation metrics of the test set found in Supplementary Fig. S7. Additionally, in vitro experiments confirmed the accuracy of the in silico prediction of the small-molecule BBB permeation model (Supplementary Fig. S8). Our results in this studies demonstrated that the Transformer-based models that use SMILES representations of molecules offer several advantages over traditional machine learning models that rely on physicochemical properties. These advantages include greater computational efficiency, flexibility in handling diverse sets of molecules, and the ability to learn complex patterns and relationships from the input data. Supplementary Table S5 showed the raw data of MegaMolBART analysis on blood brain barrier (BBB) permeability of NPRL compounds. Therefore, these models are promising for drug discovery and can accelerate the development of new treatments for CNS disorders.
In conclusion, our study underscores the benefits of large language models like MegaMolBART over traditional machine learning approaches. A key advantage is the ability to predict blood–brain barrier (BBB) permeability directly from SMILES molecular representations, circumventing the need for additional physicochemical property calculations. Such calculations can be computationally expensive and time-consuming processes.
Material and methods
In silico study
For our dataset, we used a collection of molecules curated by Natural Products Research Laboratories (NPRL) from China Medical University Hospital (CMUH), which consisted of drugs approved by the Food and Drug Administration (FDA) that either cross or do not cross the BBB, with more than 512 characters removed and converted to their canonical forms. We also included an open source BBB database (B3DB) and similarly converted SMILES to their canonical forms (URL: https://github.com/theochem/B3DB). After preprocessing, the CMUH dataset consisted of 105 molecules that crossed the BBB (BBB+) and 2394 that did not (BBB−), whereas the B3DB dataset consisted of 4956 BBB+ molecules and 2851 BBB− molecules. First, we attached the MegaMolBART embedding and encoder layers to different classifiers in PyTorch, such as a linear and other 1D CNN-based classifiers. We pulled the pre-trained MegaMolBART model available on NVIDIA GPU Cloud (NGC)35 which was trained with data parallelism on 64 V100 GPUs (4 nodes × 16 GPUs) for eight epochs (approximately 160 k iterations or ~ 80 wall-clock hours), using a batch size of 32 molecules per GPU (micro batch) (URL: https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/models/megamolbart). The Noam scheduler was used with peak learning rate values of 0.0005 and 8000 warm-up steps. FusedAdam optimization was used with the following parameters: beta 1 = 0.9; beta 2 = 0.999. Categorical cross-entropy loss is used to train the models. The model is trained using the ZINC-15 dataset. We experimented with different hyper-parameters, such as freezing the MegaMolBART parts and allowing them to undergo fine-tuning. For datasets, we split the B3DB into 80% training, 10% validation, and 10% testing and used the CMUH dataset as the test set, as well as combining both datasets with 80% + 80% train, 10% + 10% validation, and 10% + 10% testing. The results were all fairly similar, with the area under the receiver operating characteristic curve (AUC) ranging from 0.57 to 0.63. To improve the performance of the MegaMolBART model, we collaborated with the Taiwan Web Service (TWS) operated by ASUS, which operates the TAIWANIA-2 cluster. We obtained eight nodes × eight V100 GPUs for a total of 64 GPUs and ran the large MegaMolBART configuration, allowing every other configuration and dataset to be consistent with the one that had been pre-trained on NGC. We ran the model for approximately 1 week, which lasted for three epochs (compared to the eight epochs above). Finally, once we had the large MegaMolBART pre-trained model that was trained on TWS, we again attempted to combine the embedding and encoder layers into a classifier in PyTorch (URL: https://pytorch.org/), but we could not obtain results better than an AUC score of 0.63. From there, we took a step back and examined the different MegaMolBART downstream task resources and used an XGBoost regressor through the embeddings from MegaMolBART and compared with Morgan Fingerprints. For this portion of the study, we found that only 1058 samples in the B3DB dataset had LogBB values that could be used for the regression analysis. A LogBB value that is ≥ − 1 means that the molecule was able to cross the BBB. Supplementary Figure S6 shows the calculated LogBB values in our model.
Cbrain: Concentration of the molecule in the brain, Cblood: Concentration of the molecule in blood.
We connected an XGBoost Regressor to all three feature types: Morgan Fingerprints, NGC MegaMolBART Embeddings, and TWS MegaMolBART Embeddings. The B3DB dataset with log BB was divided into 80% training, 10% validation for early stopping, and 10% testing groups. The mean square error (MSE) and R-square (R2) values were calculated with the 10% test set, whereas the accuracy was calculated with the inferred LogBB of the 6749 samples without LogBB and the 2499 CMUH dataset and converted to BBB+ or BBB−, depending on the inferred LogBB value. Next, because we required more training samples, we used the existing pipeline of MegaMolBART embeddings and replaced the XGBoost Regressor with an XGBoost classifier. For the next experiment, we used all B3DB and CMUH datasets split into 80% training, 10% validation, and 10% testing.
In vitro study
Supplementary Figure S8 shows the in vitro experimental design. ScienCellTM (cat. no. Cat. #SP3D-8768; ScienCell Research Laboratories, Inc., CA, USA) supplied normal human BBB spheroids consisting of human brain microvascular endothelial cells, brain vascular pericytes, and astrocytes in a 1:1:1 ratio to simulate intracellular interactions at the BBB. These spheroids consisted of human microvascular endothelial cells, brain vascular pericytes, and astrocytes. The spheroids were cultured in the 3D-BBB spheroid medium (3D-BBBSpM; Cat. #3D-8701) supplemented with 3D-BBB spheroids (3D-BBBSpS; Cat. #3D-8752), and fetal bovine serum (FBS; cat. #0010; ScienCell Research Laboratories, Inc., CA, USA), 100 U/mL penicillin, and 100 g/mL streptomycin in 96 well round bottom ultralow attachment plates (Corning; Cat. #CLS7007) under a humidified atmosphere with 5% CO2 at 37 °C83. Spheroids from normal human BBB were cultured in 96-well round-bottom ultralow attachment plates. Spheroid cells were treated with 10 g/mL of Temozolomide (TMZ; positive control), ferulic acid (negative control), and NPRL compounds. They were collected and washed twice with phosphate-buffered saline; subsequently, acetone precipitation was used to remove the detritus and centrifuged for 10 min at 12,000 rpm. The supernatant was collected and vacuum-dried. For the MS analysis, the sample was re-dissolved in 20 μL of a solvent containing MeOH/H2O/FA (1:1:0.001 v/v/v), and the supernatant was directly used for the LC–MS/MS analysis. With an orthogonal electrospray ionization (ESI) source, a UHPLC system (Ultimate 3000; Dionex, Germany) equipped with a C18 reversed‐phase column (2.1 × 150 mm, 3 μm, T3; Waters, Milford, MA, USA) was coupled to a hybrid QTOF mass spectrometer (maXis impact; Bruker Daltonics, Bremen, Germany). The initial flow rates were 0.25 mL/min of 99% for solvent A (0.1% formic acid) and 1% for solvent B (acetonitrile with 0.1% formic acid). A sample volume of 5 µL was injected. Within 1 min of the injection, the solvent B concentration was maintained at 1%, increased to 40% over 15 min, increased to 99% over 3 min, and maintained for 3 min before returning to its initial concentration for 4.5 min. The MS was operated in positive and negative ion modes with an m/z range of 50 ~ 1000 at 1 Hz. The capillary voltage of the ion source was set at + 3600 V and − 3000 V, and the endplate offset was 500 V. The nebulizer gas flow was one bar, and the drying gas flow was 8 L/min. A temperature of 200 °C was set for drying. The radiofrequency (RF) power in Funnels 1 and 2 was 200 Vpp. The RF for the hexapole was 200 Vpp and the low mass cutoff for the quadrupole was 100 m/z. A data-dependent analysis mode was used to obtain the data. The four most intense precursor ions were selected for the MS/MS analysis, excluded after two spectra, and released after 0.5 min. The total cycle time was 1.8–2.3 s84,85.
Supplementary Information
Acknowledgements
We sincerely thank the Taiwan Web Service (TWS) to provide AIHPC for Large language model (LLM) training on Taiwania-2 for providing assistance and equipment for the present study. We thank Dr. Chao-Jung Chen and Miss Yu-Ning Lin and Yu‑Ning Juan (Proteomics Core Laboratory, Department of Medical Research, China Medical University Hospital) for their support. The experiments and data analysis were performed in part at the Medical Research Core Facilities Center, Office of Research & Development, China Medical University, Taichung, Taiwan.
Author contributions
E.T.C.H. and F.J.T. contributed to designing the study. F.J.T., K.Y.K.L., W.C.W.T., and J.S.Y. performed the experiments. K.Y.K.L., W.C.W.T., C.K.L., and J.S.Y. analyzed the data. E.T.C.H., F.J.T., K.Y.K.L., W.C.W.T., and J.S.Y. wrote the manuscript. M.G., C.C., S.S., modified the article. All authors contributed to revising the manuscript. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-66897-y.
References
- 1.Khor SLQ, Ng KY, Koh RY, Chye SM. Blood–brain barrier and neurovascular unit dysfunction in Parkinson's disease: From clinical insights to pathogenic mechanisms and novel therapeutic approaches. CNS Neurol. Disord. Drug Targets. 2023 doi: 10.2174/1871527322666230330093829. [DOI] [PubMed] [Google Scholar]
- 2.Harris WJ, et al. In vivo methods for imaging blood–brain barrier function and dysfunction. Eur. J. Nucl. Med. Mol. Imaging. 2023;50:1051–1083. doi: 10.1007/s00259-022-05997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence JM, Schardien K, Wigdahl B, Nonnemacher MR. Roles of neuropathology-associated reactive astrocytes: A systematic review. Acta Neuropathol. Commun. 2023;11:42. doi: 10.1186/s40478-023-01526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suprewicz L, et al. Blood-brain barrier function in response to SARS-CoV-2 and its spike protein. Neurol. Neurochir. Pol. 2023;57:14–25. doi: 10.5603/PJNNS.a2023.0014. [DOI] [PubMed] [Google Scholar]
- 5.Ailioaie LM, Ailioaie C, Litscher G. Photobiomodulation in Alzheimer's disease-a complementary method to state-of-the-art pharmaceutical formulations and nanomedicine? Pharmaceutics. 2023 doi: 10.3390/pharmaceutics15030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Critchley BJ, Gaspar HB, Benedetti S. Targeting the central nervous system in lysosomal storage diseases: Strategies to deliver therapeutics across the blood-brain barrier. Mol. Ther. 2023;31:657–675. doi: 10.1016/j.ymthe.2022.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang R, et al. Blood–brain barrier integrity damage in bacterial meningitis: The underlying link, mechanisms, and therapeutic targets. Int. J. Mol. Sci. 2023 doi: 10.3390/ijms24032852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okura T, Higuchi K, Deguchi Y. The blood–brain barrier transport mechanism controlling analgesic effects of opioid drugs in CNS. Yakugaku Zasshi. 2015;135:697–702. doi: 10.1248/yakushi.14-00234-2. [DOI] [PubMed] [Google Scholar]
- 9.Ueno M. Mechanisms of the penetration of blood-borne substances into the brain. Curr. Neuropharmacol. 2009;7:142–149. doi: 10.2174/157015909788848901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss N, Miller F, Cazaubon S, Couraud PO. Blood–brain barrier part III: Therapeutic approaches to cross the blood–brain barrier and target the brain. Rev. Neurol. (Paris) 2010;166:284–288. doi: 10.1016/j.neurol.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PA, Andersson O, Hansen SH, Simonsen KB, Andersson G. Models for predicting blood–brain barrier permeation. Drug Discov. Today. 2011;16:472–475. doi: 10.1016/j.drudis.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Racz A, Bajusz D, Miranda-Quintana RA, Heberger K. Machine learning models for classification tasks related to drug safety. Mol. Divers. 2021;25:1409–1424. doi: 10.1007/s11030-021-10239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remtulla R, Das SK, Levin LA. Predicting absorption-distribution properties of neuroprotective phosphine-borane compounds using in silico modeling and machine learning. Molecules. 2021 doi: 10.3390/molecules26092505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. In silico prediction of blood–brain barrier permeability of compounds by machine learning and resampling methods. ChemMedChem. 2018;13:2189–2201. doi: 10.1002/cmdc.201800533. [DOI] [PubMed] [Google Scholar]
- 15.Montanari F, Ecker GF. Prediction of drug-ABC-transporter interaction—Recent advances and future challenges. Adv. Drug Deliv. Rev. 2015;86:17–26. doi: 10.1016/j.addr.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varadharajan S, et al. Exploring in silico prediction of the unbound brain-to-plasma drug concentration ratio: Model validation, renewal, and interpretation. J. Pharm. Sci. 2015;104:1197–1206. doi: 10.1002/jps.24301. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Winiwarter S, Friden M, Antonsson M, Engkvist O. In silico prediction of unbound brain-to-plasma concentration ratio using machine learning algorithms. J. Mol. Graph. Model. 2011;29:985–995. doi: 10.1016/j.jmgm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Guntner AS, Bogl T, Mlynek F, Buchberger W. Large-scale evaluation of collision cross sections to investigate blood–brain barrier permeation of drugs. Pharmaceutics. 2021 doi: 10.3390/pharmaceutics13122141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaker B, et al. LightBBB: Computational prediction model of blood–brain-barrier penetration based on LightGBM. Bioinformatics. 2021;37:1135–1139. doi: 10.1093/bioinformatics/btaa918. [DOI] [PubMed] [Google Scholar]
- 20.Meng F, Xi Y, Huang J, Ayers PW. A curated diverse molecular database of blood–brain barrier permeability with chemical descriptors. Sci. Data. 2021;8:289. doi: 10.1038/s41597-021-01069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R, et al. DeePred-BBB: A blood brain barrier permeability prediction model with improved accuracy. Front. Neurosci. 2022;16:858126. doi: 10.3389/fnins.2022.858126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao YH, et al. Predicting penetration across the blood–brain barrier from simple descriptors and fragmentation schemes. J. Chem. Inf. Model. 2007;47:170–175. doi: 10.1021/ci600312d. [DOI] [PubMed] [Google Scholar]
- 23.Shen J, Cheng F, Xu Y, Li W, Tang Y. Estimation of ADME properties with substructure pattern recognition. J. Chem. Inf. Model. 2010;50:1034–1041. doi: 10.1021/ci100104j. [DOI] [PubMed] [Google Scholar]
- 24.Roy D, Hinge VK, Kovalenko A. To pass or not to pass: Predicting the blood–brain barrier permeability with the 3D-RISM-KH molecular solvation theory. ACS Omega. 2019;4:16774–16780. doi: 10.1021/acsomega.9b01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osipenko S, Botashev K, Nikolaev E, Kostyukevich Y. Transfer learning for small molecule retention predictions. J. Chromatogr. A. 2021;1644:462119. doi: 10.1016/j.chroma.2021.462119. [DOI] [PubMed] [Google Scholar]
- 26.Woo S, Shenvi RA. Natural product synthesis through the lens of informatics. Acc. Chem. Res. 2021;54:1157–1167. doi: 10.1021/acs.accounts.0c00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lampa S, Dahlo M, Alvarsson J, Spjuth O. SciPipe: A workflow library for agile development of complex and dynamic bioinformatics pipelines. Gigascience. 2019 doi: 10.1093/gigascience/giz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Przybylak KR, et al. Characterisation of data resources for in silico modelling: Benchmark datasets for ADME properties. Expert Opin. Drug Metab. Toxicol. 2018;14:169–181. doi: 10.1080/17425255.2017.1316449. [DOI] [PubMed] [Google Scholar]
- 29.Afantitis A, et al. NanoSolveIT Project: Driving nanoinformatics research to develop innovative and integrated tools for in silico nanosafety assessment. Comput. Struct. Biotechnol. J. 2020;18:583–602. doi: 10.1016/j.csbj.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minkiewicz P, Iwaniak A, Darewicz M. Annotation of peptide structures using SMILES and other chemical codes-practical solutions. Molecules. 2017 doi: 10.3390/molecules22122075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munteanu CR, Gonzalez-Diaz H, Garcia R, Loza M, Pazos A. Bio-AIMS collection of chemoinformatics web tools based on molecular graph information and artificial intelligence models. Comb. Chem. High Throughput Screen. 2015;18:735–750. doi: 10.2174/1386207318666150803140950. [DOI] [PubMed] [Google Scholar]
- 32.Irwin R, Dimitriadis S, He J, Bjerrum EJ. Chemformer: A pre-trained transformer for computational chemistry. Mach. Learn Sci. Technol. 2022;3:015022. doi: 10.1088/2632-2153/ac3ffb. [DOI] [Google Scholar]
- 33.Ullah Z, Usman M, Gwak J. MTSS-AAE: Multi-task semi-supervised adversarial autoencoding for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2023;216:119475. doi: 10.1016/j.eswa.2022.119475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulamali FF, et al. Autoencoders for sample size estimation for fully connected neural network classifiers. NPJ Digit. Med. 2022;5:180. doi: 10.1038/s41746-022-00728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NVIDIA. Nvidia/MegaMolBART: A deep learning model for small molecule drug discovery and cheminformatics based on smiles. GitHub. Retrieved February 20, 2023, from https://github.com/NVIDIA/MegaMolBART. (2022).
- 36.Sterling T, Irwin JJ. ZINC 15—Ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadukova M, Chupin V, Grudinin S. Docking rigid macrocycles using Convex-PL, AutoDock Vina, and RDKit in the D3R Grand Challenge 4. J. Comput. Aided Mol. Des. 2020;34:191–200. doi: 10.1007/s10822-019-00263-3. [DOI] [PubMed] [Google Scholar]
- 38.Landrum, G. RDKit Documentation. Release 2011.12.1. (2012).
- 39.Plisson F, Piggott AM. Predicting blood–brain barrier permeability of marine-derived kinase inhibitors using ensemble classifiers reveals potential hits for neurodegenerative disorders. Mar. Drugs. 2019 doi: 10.3390/md17020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jillella GK, Ojha PK, Roy K. Application of QSAR for the identification of key molecular fragments and reliable predictions of effects of textile dyes on growth rate and biomass values of Raphidocelis subcapitata. Aquat. Toxicol. 2021;238:105925. doi: 10.1016/j.aquatox.2021.105925. [DOI] [PubMed] [Google Scholar]
- 41.Jillella GK, Khan K, Roy K. Application of QSARs in identification of mutagenicity mechanisms of nitro and amino aromatic compounds against Salmonella typhimurium species. Toxicol. In Vitro. 2020;65:104768. doi: 10.1016/j.tiv.2020.104768. [DOI] [PubMed] [Google Scholar]
- 42.Zulfiqar M, Gadelha L, Steinbeck C, Sorokina M, Peters K. MAW: The reproducible Metabolome Annotation Workflow for untargeted tandem mass spectrometry. J. Cheminform. 2023;15:32. doi: 10.1186/s13321-023-00695-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimadiev T, et al. CGRdb2.0: A python database management system for molecules, reactions, and chemical data. J. Chem. Inf. Model. 2022;62:2015–2020. doi: 10.1021/acs.jcim.1c01105. [DOI] [PubMed] [Google Scholar]
- 44.Donmazov S, Saruhan EN, Pekkan K, Piskin S. Review of machine learning techniques in soft tissue biomechanics and biomaterials. Cardiovasc. Eng. Technol. 2024 doi: 10.1007/s13239-024-00737-y. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y, Zhang YQ, Chawla NV, Krasser S. SVMs modeling for highly imbalanced classification. IEEE Trans. Syst. Man Cybern. B Cybern. 2009;39:281–288. doi: 10.1109/TSMCB.2008.2002909. [DOI] [PubMed] [Google Scholar]
- 46.Orel E, et al. An automated literature review tool (LiteRev) for streamlining and accelerating research using natural language processing and machine learning: Descriptive performance evaluation study. J. Med. Internet. Res. 2023;25:e39736. doi: 10.2196/39736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hassaballah M, Wazery YM, Ibrahim IE, Farag A. ECG heartbeat classification using machine learning and metaheuristic optimization for smart healthcare systems. Bioengineering (Basel) 2023 doi: 10.3390/bioengineering10040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohlmann A, Mostafa J, Kumar M. Machine learning and medication adherence: Scoping review. JMIRx Med. 2021;2:e26993. doi: 10.2196/26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo W, et al. Review of machine learning and deep learning models for toxicity prediction. Exp. Biol. Med. (Maywood) 2023;248:1952–1973. doi: 10.1177/15353702231209421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aldhoayan MD, Aljubran Y. Prediction of ICU patients' deterioration using machine learning techniques. Cureus. 2023;15:e38659. doi: 10.7759/cureus.38659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan MA, et al. A deep learning-based intrusion detection system for MQTT enabled IoT. Sensors (Basel) 2021 doi: 10.3390/s21217016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afzal M, Alam F, Malik KM, Malik GM. Clinical context-aware biomedical text summarization using deep neural network: Model development and validation. J. Med. Internet. Res. 2020;22:e19810. doi: 10.2196/19810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ju W, et al. A comprehensive survey on deep graph representation learning. Neural Netw. 2024;173:106207. doi: 10.1016/j.neunet.2024.106207. [DOI] [PubMed] [Google Scholar]
- 54.Hajek P, Barushka A, Munk M. Neural networks with emotion associations, topic modeling and supervised term weighting for sentiment analysis. Int. J. Neural Syst. 2021;31:2150013. doi: 10.1142/S0129065721500131. [DOI] [PubMed] [Google Scholar]
- 55.Hudson IL. Data integration using advances in machine learning in drug discovery and molecular biology. Methods Mol. Biol. 2021;2190:167–184. doi: 10.1007/978-1-0716-0826-5_7. [DOI] [PubMed] [Google Scholar]
- 56.Johnson GW, et al. Localizing seizure onset zones in surgical epilepsy with neurostimulation deep learning. J. Neurosurg. 2023;138:1002–1007. doi: 10.3171/2022.8.JNS221321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng S, et al. Application of machine learning and deep learning methods for hydrated electron rate constant prediction. Environ. Res. 2023;231:115996. doi: 10.1016/j.envres.2023.115996. [DOI] [PubMed] [Google Scholar]
- 58.Yang J-S, et al. In silico de novo curcuminoid derivatives from the compound library of natural products research laboratories inhibit COVID-19 3CLpro activity. Nat. Prod. Commun. 2020;15:1934578x20953262. doi: 10.1177/1934578x20953262. [DOI] [Google Scholar]
- 59.Mauri A, Consonni V, Pavan M, Todeschini R. Dragon software: An easy approach to molecular descriptor calculations. Math. Comput. Chem. 2006;56:237–248. doi: 10.1111/j.1467-9280.1995.tb00298.x. [DOI] [Google Scholar]
- 60.Yap CW. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 61.Janke JJ, et al. Computational screening for mAb colloidal stability with coarse-grained, molecular-scale simulations. J. Phys. Chem. B. 2024;128:1515–1526. doi: 10.1021/acs.jpcb.3c05303. [DOI] [PubMed] [Google Scholar]
- 62.Shimamura K, Takeshita Y, Fukushima S, Koura A, Shimojo F. Computational and training requirements for interatomic potential based on artificial neural network for estimating low thermal conductivity of silver chalcogenides. J. Chem. Phys. 2020;153:234301. doi: 10.1063/5.0027058. [DOI] [PubMed] [Google Scholar]
- 63.Wardecki D, Dolowy M, Bober-Majnusz K. Evaluation of the usefulness of topological indices for predicting selected physicochemical properties of bioactive substances with anti-androgenic and hypouricemic activity. Molecules. 2023 doi: 10.3390/molecules28155822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baira K, et al. Multitask quantum study of the curcumin-based complex physicochemical and biological properties. Int. J. Mol. Sci. 2022 doi: 10.3390/ijms23052832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pham T, et al. DeepARV: Ensemble deep learning to predict drug–drug interaction of clinical relevance with antiretroviral therapy. NPJ Syst. Biol. Appl. 2024;10:48. doi: 10.1038/s41540-024-00374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Correa I, Giunta PD, Marino FJ, Francesconi JA. Transformer-based representation of organic molecules for potential modeling of physicochemical properties. J. Chem. Inf. Model. 2023;63:7676–7688. doi: 10.1021/acs.jcim.3c01548. [DOI] [PubMed] [Google Scholar]
- 67.Tran T, Ekenna C. Molecular descriptors property prediction using transformer-based approach. Int. J. Mol. Sci. 2023 doi: 10.3390/ijms241511948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Ye K, van Vlijmen HWT, van Westen GJP. DrugEx v3: Scaffold-constrained drug design with graph transformer-based reinforcement learning. J. Cheminform. 2023;15:24. doi: 10.1186/s13321-023-00694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang L, et al. Transformer-based generative model accelerating the development of novel BRAF inhibitors. ACS Omega. 2021;6:33864–33873. doi: 10.1021/acsomega.1c05145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim S, Tariq S, Heo S, Yoo C. Interpretable attention-based multi-encoder transformer based QSPR model for assessing toxicity and environmental impact of chemicals. Chemosphere. 2024;350:141086. doi: 10.1016/j.chemosphere.2023.141086. [DOI] [PubMed] [Google Scholar]
- 71.Merk D, Friedrich L, Grisoni F, Schneider GD. Novo design of bioactive small molecules by artificial intelligence. Mol. Inform. 2018 doi: 10.1002/minf.201700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arus-Pous J, et al. Randomized SMILES strings improve the quality of molecular generative models. J. Cheminform. 2019;11:71. doi: 10.1186/s13321-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carracedo-Reboredo P, et al. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotechnol. J. 2021;19:4538–4558. doi: 10.1016/j.csbj.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsukiyo Y, Yamanaka C, Yamanishi YD. Novo generation of chemical structures of inhibitor and activator candidates for therapeutic target proteins by a transformer-based variational autoencoder and bayesian optimization. J. Chem. Inf. Model. 2024;64:2345–2355. doi: 10.1021/acs.jcim.3c00824. [DOI] [PubMed] [Google Scholar]
- 75.Pereira TO, Abbasi M, Arrais JP. Enhancing reinforcement learning for de novo molecular design applying self-attention mechanisms. Brief Bioinform. 2023 doi: 10.1093/bib/bbad368. [DOI] [PubMed] [Google Scholar]
- 76.Wu T, Tang Y, Sun Q, Xiong L. Molecular joint representation learning via multi-modal information of SMILES and graphs. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023;20:3044–3055. doi: 10.1109/TCBB.2023.3253862. [DOI] [PubMed] [Google Scholar]
- 77.Yan X, Liu Y. Graph-sequence attention and transformer for predicting drug-target affinity. RSC Adv. 2022;12:29525–29534. doi: 10.1039/d2ra05566j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moradi-Afrapoli F, et al. Validation of UHPLC-MS/MS methods for the determination of kaempferol and its metabolite 4-hydroxyphenyl acetic acid, and application to in vitro blood–brain barrier and intestinal drug permeability studies. J. Pharm. Biomed. Anal. 2016;128:264–274. doi: 10.1016/j.jpba.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 79.Noorani B, et al. LC-MS/MS-based in vitro and in vivo investigation of blood-brain barrier integrity by simultaneous quantitation of mannitol and sucrose. Fluids Barriers CNS. 2020;17:61. doi: 10.1186/s12987-020-00224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun L, et al. Development and validation of a highly sensitive LC-MS/MS method for determination of brain active agent dianhydrogalactitol in mouse plasma and tissues: Application to a pharmacokinetic study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018;1087–1088:90–97. doi: 10.1016/j.jchromb.2018.04.026. [DOI] [PubMed] [Google Scholar]
- 81.Wu SY, et al. NPRL-Z-1, as a new topoisomerase II poison, induces cell apoptosis and ROS generation in human renal carcinoma cells. PLoS One. 2014;9:e112220. doi: 10.1371/journal.pone.0112220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toth AE, Nielsen SSE, Tomaka W, Abbott NJ, Nielsen MS. The endo-lysosomal system of bEnd.3 and hCMEC/D3 brain endothelial cells. Fluids Barriers CNS. 2019;16:14. doi: 10.1186/s12987-019-0134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiu YJ, et al. Next-generation sequencing analysis reveals that MTH-3, a novel curcuminoid derivative, suppresses the invasion of MDA-MB-231 triple-negative breast adenocarcinoma cells. Oncol. Rep. 2021 doi: 10.3892/or.2021.8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang C-W, et al. In silico target analysis of treatment for COVID-19 using Huang-Lian-Shang-Qing-Wan, a traditional Chinese medicine formula. Nat. Prod. Commun. 2021;16:1934578X211030818. doi: 10.1177/1934578X211030818. [DOI] [Google Scholar]
- 85.Wang CH, et al. Protective effects of Jing-Si-herbal-tea in inflammatory cytokines-induced cell injury on normal human lung fibroblast via multiomic platform analysis. Tzu Chi Med. J. 2024;36:152–165. doi: 10.4103/tcmj.tcmj_267_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.