Abstract
Two activities of retroviral integrase, 3′ processing and DNA strand transfer, are required to integrate viral cDNA into a host cell chromosome. Integrase activity has been analyzed in vitro using purified protein and recombinant DNA substrates that model the U3 and U5 ends of viral cDNA or by using viral preintegration complexes (PICs) that form during virus infection. Numerous studies have investigated changes in integrase or viral DNA for effects on both 3′ processing and DNA strand transfer activities using purified protein, but similar analyses have not been carried out using PICs. Here, we analyzed PICs from human immunodeficiency virus type 1 (HIV-1) strain 604del, an integration-defective mutant lacking 26 bp of U5, and revE1, a revertant of 604del containing an additional 19-bp deletion, for levels of 3′ processing activity that occurred in infected cells and for levels of in vitro DNA strand transfer activity. Whereas revE1 supported one-third to one-half of the level of wild-type DNA strand transfer activity, the level of 604del DNA strand transfer activity was undetectable. Surprisingly, integrase similarly processed the 3′ ends of 604del and revE1 in vivo. We therefore conclude that 604del is blocked in its ability to replicate in cells after the 3′ processing step of retroviral integration. Whereas Western blotting showed that wild-type, revE1, and 604del PICs contained similar levels of integrase protein, Mu-mediated PCR footprinting revealed only minimal protein-DNA complex formation at the ends of 604del cDNA. We propose that 604del is replication defective because proteins important for DNA strand transfer activity do not stably associate with this cDNA after in vivo 3′ processing by integrase.
Retroviral DNA integration requires a series of DNA cutting and joining reactions catalyzed by the viral integrase protein. Integrase recognizes and acts on the viral DNA attachment (att) site, which is comprised of U3 and U5 sequences at the ends of viral cDNA. In an initial 3′ processing reaction, integrase cleaves each DNA end adjacent to the conserved sequence CA. In the case of human immunodeficiency virus type 1 (HIV-1), 2 nucleotides are removed from each end, yielding 3′-hydroxyl groups. In the subsequent DNA strand transfer step, integrase uses these 3′-OHs to make a double-stranded staggered cut in chromosomal DNA, which at the same time joins the viral 3′ ends to 5′-phosphates at the site of integration. The mechanism of the final step of retroviral integration, gap repair, remains to be elucidated and may require either viral proteins, cellular machinery, or both (see reference 2 for a detailed review of retroviral integration).
Retroviral integrase activity can be analyzed in vitro using purified protein and recombinant DNA substrates that model the U3 and U5 ends of viral cDNA (6, 9, 18, 20, 22, 33, 37, 38). Alternatively, strand transfer of endogenous cDNA can be analyzed using integrase-containing preintegration complexes (PICs) following their isolation from infected cells (3–5, 7, 8, 12, 16, 17, 25, 27, 39). Levels of 3′ processing activity promoted by integrase as an in vivo PIC component can be quantified by digesting isolated deproteinized cDNA with restriction enzymes and probing each 3′ end for loss of 2 nucleotides (4, 17, 25, 27–29, 31). Whereas numerous integrase, U3, and U5 mutations have been analyzed for their effects on the 3′ processing and DNA strand transfer activities of purified integrase proteins (6, 9–11, 15, 19–24, 32, 34–37), such mutations have not been analyzed for effects on both 3′ processing and DNA strand transfer as catalyzed by the PICs that mediate viral integration in vivo. By doing so, we identify in this report an HIV-1 att site mutant that is blocked in its ability to replicate in cells after the 3′ processing step of retroviral integration.
HIV-1 strain 604del is a replication-defective deletion mutant lacking 26 bp of U5 upstream of the conserved CA dinucleotide. During tissue culture passage, a revertant of 604del, designated revE1, that lacks an additional 19 bp extending upstream from the original change was isolated (35). Cells infected with wild type, 604del, and revE1 contained similar levels of unintegrated HIV-1 DNA (35). Since wild-type, 604del, and revE1 virions were released similarly from cells following transfection and contained similar levels of viral RNA, oligonucleotide substrates that model the different U5 ends of these viruses were used to assess the in vitro 3′ processing and DNA strand transfer activities of purified HIV-1 integrase protein (35). Whereas 604del substrates supported about 10 and 4% of wild-type 3′ processing and DNA strand transfer, respectively, revE1 was about 25 and 22% active, respectively. Thus, these results were consistent with the notion that 604del was integration defective in infected cells and that the novel U5 end present in revE1 repaired this replication defect (35). There are instances, however, where results of in vitro integration assays do not reflect what occurs in vivo. For example, in vitro 3′ processing and DNA strand transfer activities of purified HIV-1 integrase were undetectable using a U3 oligonucleotide substrate with changes at the conserved CA dinucleotide (23), and yet these changes reduced the integration of a U3 mutant virus only 2.5-fold in infected cells (26). Also, simian immunodeficiency virus (SIV) integrase containing the substitution of Lys for Glu-136 (E136K) restored an in vivo replication defect to SIV att site mutant strain 7, but purified SIV E136K integrase did not show any preference over wild-type integrase for synthetic mutant 7 substrates in vitro (11). With these results in mind, we further characterized the in vivo replication block of HIV-1 mutant 604del by analyzing PICs derived from infected cells.
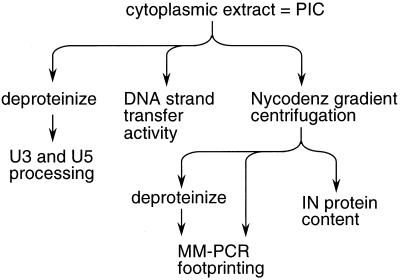
Wild-type, 604del, and revE1 virus stocks were produced by transfecting 293T cells, and C8166 T cells were infected with these stocks, as previously described (8). Eight hours postinfection, cells were lysed and cytoplasmic extract was prepared as previously described (8). This cell extract, which contains HIV-1 PICs in their native form, was analyzed using a variety of techniques to determine levels of integrase catalytic activities and associated protein content (Fig. 1). Specifically, the crude cytoplasmic extract was examined by indirect end labeling to detect the structures of U3 and U5 ends and quantitate in vivo 3′ processing activity and in in vitro integration assays to detect levels of unintegrated HIV-1 cDNA and quantify DNA strand transfer activity. To examine the extent of protein-DNA complex formation at the ends of HIV-1 cDNA, PICs first purified by Nycodenz gradient centrifugation were analyzed by Mu-mediated PCR (MM-PCR) footprinting as previously described (8). Purified PICs were also analyzed by Western blotting to determine levels of integrase protein (Fig. 1).
FIG. 1.
Experimental strategy to assess integrase catalytic activities and PIC protein content. Cytoplasmic extract of HIV-1-infected cells was processed in the indicated ways to determine levels of integrase 3′ processing activity that occurred in infected cells, levels of in vitro DNA strand transfer activity, and levels of PIC-associated proteins as detected by MM-PCR footprinting and Western blotting. IN, integrase.
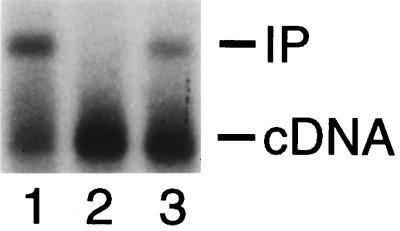
To measure the level of PIC-associated DNA strand transfer activity, cytoplasmic extract (0.25 ml) was reacted with φX174 target DNA as described previously (8). Integration reactions were terminated and analyzed by Southern blotting also as previously described (8). DNA levels were quantified either with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) or by densitometry (IS-1000 Digital Imaging System; Alpha Innotech Corp., San Leandro, Calif.), and integration activity was calculated as the percentage of the 9.7-kb HIV-1 cDNA substrate converted into the 15.1-kb integration product. Cells infected with wild type, 604del, and revE1 contained similar levels of HIV-1 cDNA (Fig. 2), in agreement with the previous report that 604del and revE1 each support the wild-type level of reverse transcription following infection (35). In repeated experiments, revE1 PICs supported 30 to 50% of the level of wild-type DNA strand transfer activity (Fig. 2, compare lane 3 to lane 1; Table 1). 604del PICs, in contrast, did not support detectable levels of DNA strand transfer activity (Fig. 2, lane 2; Table 1). Thus, these results are consistent with the interpretation that 604del is blocked at the integration step in vivo and that the novel U5 end in the revE1 revertant virus rescues this integration defect (35).
FIG. 2.
In vitro DNA strand transfer activity of wild-type, 604del, and revE1 PICs. The integration reactions contained wild-type PICs (lane 1), 604del PICs (lane 2), and revE1 PICs (lane 3). cDNA, 9.7-kb HIV-1 integration substrate; IP, 15.1-kb integration product.
TABLE 1.
Quantitation of wild-type, 604del, and revE1 DNA strand transfer and 3′ processing activities
| Virus | DNA strand transfera
|
3′ processinga
|
||||
|---|---|---|---|---|---|---|
| Expt 1 | Expt 2b | U5
|
U3
|
|||
| Expt 1 | Expt 3b | Expt 1 | Expt 3b | |||
| Wild type | 32 | 40 | 77 | 88 | 60 | 90 |
| 604del | NDc | ND | 72 | 72 | 34 | 34 |
| revE1 | 14 | 20 | 40 | NOd | 54 | 57 |
DNA strand transfer and 3′ processing activities were quantified and expressed as percentages as described in the text.
Experiments 2 and 3 were conducted with two different sets of infected cells.
ND, not detected (<2% activity).
NO, not observed. The revE1 U5 plus strand was electrophoresed off the gel during this experiment.
To analyze the extent of 3′ processing that occurred during virus infection, cytoplasmic extract (1 ml) was deproteinized by treating it overnight at 56°C with 50 μl of a 10-mg/ml solution of proteinase K, 60 μl of a 10% (wt/vol) solution of sodium dodecyl sulfate (SDS), and 12 μl of 0.5 M EDTA. Following extraction with phenol, phenol-chloroform (1:1), and chloroform, DNA was recovered by precipitation with ethanol. The DNA was reacted with 20 U each of restriction enzymes HaeIII and HindIII in buffer (50 μl) as recommended by the manufacturer (New England Biolabs, Inc., Beverly, Mass.). Following digestion with RNase A (10 μg per ml for 30 min at 37°C), DNA was recovered by precipitation with ethanol and resuspended in 6 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The sample was heated at 68°C for 30 min, 6 μl of sequencing stop buffer (95% formamide, 20 mM EDTA, 0.05% [wt/vol] bromophenol blue, 0.05% [wt/vol] xylene cyanol) was added, and the sample was reheated at 68°C for 30 min, followed by boiling for 3 min. DNA (6 μl) was electrophoresed at 60 W through a 6% denaturing polyacrylamide gel until the bromophenol blue dye reached the bottom of the gel. The DNA was transferred to Duralon-UV Membrane (Stratagene, La Jolla, Calif.) in 44.5 mM Tris base–44.5 mM borate–1 mM EDTA (pH 8.3) for 1 h at 12 V, 390 mA, using a Genie Electrophoretic Blotter (Idea Scientific, Minneapolis, Minn.).
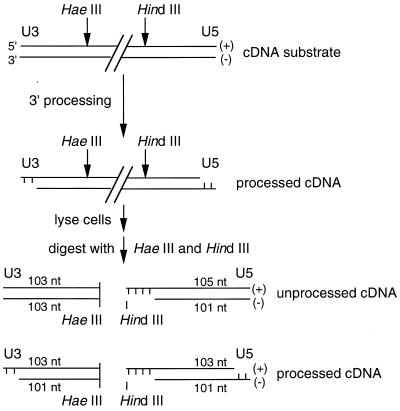
The U3 minus strand and U5 plus strand of retroviral cDNA are processed by integrase (Fig. 3). The other two strands, the U3 plus and U5 minus, which are not processed by integrase, are referred to here as nonprocessed strands. The structures of the processed and nonprocessed U3 and U5 strands were detected using strand-specific riboprobes (Fig. 3). For the nonprocessed strands, U3- and U5-specific riboprobes were synthesized from EcoRV-digested pMM104 and BstYI-cut pMM106 plasmid DNA (27), respectively, using T3 RNA polymerase as recommended by the manufacturer (Promega Corp., Madison, Wis.). Similarly, U3- and U5-specific riboprobes for detecting processed HIV-1 strands were synthesized from BamHI-cut pMM104 and HindIII-digested pMM105 (27), respectively, using T7 RNA polymerase. In this way, wild-type nonprocessed U3 plus and U5 minus strands were detected as 103- and 101-base fragments, respectively (Fig. 3). Whereas the U3 minus strands were detected as 103- and 101-base fragments depending on the extent of integrase 3′ processing, the wild-type U5 plus strand was detected as 105- and 103-base fragments (Fig. 3). Membranes hybridized at 68°C for 1 h in QuikHyb hybridization solution (Stratagene) containing 0.3 mg of tRNA per ml and 2 × 106 to 5 × 106 cpm of riboprobe per ml were washed twice for 15 min at room temperature in 300 mM NaCl–30 mM sodium citrate–0.1% SDS, followed by a wash for 30 min at 60°C in 15 mM NaCl–1.5 mM sodium citrate–0.1% SDS. Levels of 3′ processing activity were detected by autoradiography, quantified by densitometry, and expressed as the percentage of the U3 minus strand or U5 plus strand that was converted into the −2 cleavage product.
FIG. 3.
Strategy for detecting 3′ processing activity in infected cell extracts. HaeIII and HindIII cleave HIV-1 cDNA approximately 100 bp from the U3 and U5 ends, respectively. 3′ processing by integrase shortens the U3 minus strand and U5 plus strand by 2 nucleotides (nt). Following cell lysis and cleavage with HaeIII and HindIII, both processed and nonprocessed strands were detected by indirect end labeling using strand-specific riboprobes.
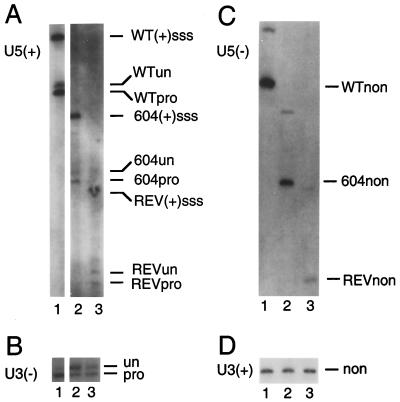
Similar to the results of Southern blotting presented in Fig. 2, levels of unintegrated linear HIV-1 cDNA as detected by indirect end labeling were identical in cells infected with wild type, 604del, and revE1 (Fig. 4). The U5 ends of wild type, 604del, and revE1 migrated during electrophoresis to positions that were consistent with the 26- and 47-bp deletions in 604del and revE1, respectively (Fig. 4A and C). Apart from integrase-specific 3′ processing of the U3 minus and U5 plus strands, all four cDNA strands of wild type, 604del, and revE1 were intact, indicating that the cDNAs in these PICs were not subject to cellular exonuclease activities in vivo (Fig. 4). Integrase processed about 77 and 60% of the wild-type U5 and U3 ends, respectively (Fig. 4A and B, lanes 1; Table 1). We were surprised to find that integrase cleaved the U5 end of 604del with an efficiency similar to that of wild type: about 72% of the U5 end and about 34% of U3 604del were cleaved (Fig. 4A and B, lanes 2; Table 1). In the case of the revE1 revertant, about 40% of U5 and 54% of U3 were processed by integrase (Fig. 4A and B, lanes 3; Table 1).
FIG. 4.
Structure of wild-type, 604del, and revE1 cDNA ends and detection of in vivo 3′ processing activity. (A) The processed U5 plus strand. Lane 1, DNA from wild-type-infected cells; lane 2, DNA from 604del-infected cells; lane 3, DNA from revE1-infected cells. WT(+)sss, cleavage product of wild-type plus-strand strong-stop DNA; WTun, the unprocessed wild-type strand (105 nucleotides); WTpro, the processed wild-type strand (103 nucleotides); 604(+)sss, cleavage product of 604del plus-strand strong-stop DNA; 604un, the unprocessed 604del strand (79 nucleotides); 604pro, the processed 604del strand (77 nucleotides); REV(+)sss, cleavage product of revE1 plus-strand strong-stop DNA; REVun, the unprocessed revE1 strand (60 nucleotides); REVpro, the processed revE1 strand (58 nucleotides). (B) The processed U3 minus strand. The samples in lanes 1 to 3 were the same as those in panel A. un, the 103-nucleotide unprocessed minus strand; pro, the processed 101-nucleotide product. (C) The nonprocessed U5 minus strand. The samples in lanes 1 to 3 were the same as those in panel A. The wild-type, 604del, and revE1 nonprocessed strands are indicated as WTnon, 604non, and REVnon, respectively. The higher-molecular-weight bands in lanes 1 to 3 display mobilities consistent with the minus-strand strong-stop products of each of the viruses. (D) The nonprocessed U3 plus strand. The samples in lanes 1 to 3 were the same as those in panel A. non, the 103-nucleotide nonprocessed strand. For panels A and C, the DNA levels in lanes 2 and 3 appear lower than the levels in lanes 1 because the 604del and revE1 deletions removed U5 sequences that are present in these wild-type-derived riboprobes. The sizes of the unprocessed, processed, and nonprocessed U5 and U3 strands were confirmed by comparing their migration distances to those of an M13 sequencing ladder (31).
Two activities of retroviral integrase, 3′ processing and DNA strand transfer, are required to covalently join the 3′ ends of viral cDNA to 5′-phosphates at the site of integration in chromosomal DNA (2). Numerous groups previously analyzed changes in integrase and DNA substrates for their effects on the in vitro 3′ processing and DNA strand transfer activities of purified integrase protein (6, 9–11, 15, 19–24, 32, 34–37). The majority of these changes were found to similarly affect 3′ processing and DNA strand transfer (6, 9, 10, 15, 19, 21–24, 32, 34–37), which is taken as evidence that a single active site in the integrase protein catalyzes both enzyme activities (2). The effects of mutations in either integrase or the DNA substrate on both 3′ processing and DNA strand transfer activities as catalyzed by the PICs that form in infected cells, however, have not been previously reported. In the case of HIV-1, this was in part due to the lack of a system with which to initiate efficient tissue culture infections from molecular DNA clones. Our recent description of such a system (8) has allowed us for the first time to assess the effects of att site changes on both 3′ processing and DNA strand transfer as promoted by the integrase-containing PIC that mediates viral integration in vivo.
Results of in vitro integration assays using purified recombinant HIV-1 integrase and synthetic oligonucleotide substrates previously suggested that novel U5 sequences in revE1 restored an integration defect to replication-defective 604del (35). Indeed, our results using PICs support this contention. Consistent with the results of that report, we found that cells infected with wild type, 604del, and revE1 contained similar levels of unintegrated HIV-1 cDNA (Fig. 2 and 4). Whereas 604del PICs did not display detectable levels of in vitro DNA strand transfer activity, revE1 PICs supported 30 to 50% of the level of wild-type activity (Fig. 2 and Table 1). Since revE1 originated in cells infected with 604del, 604del must integrate in vivo at some frequency. We speculate that this level of DNA strand transfer activity is below the limit of detection of our Southern blotting system. In contrast to the results of the DNA strand transfer assays, we were surprised to find that there were only slight differences in the efficiencies with which the 3′ ends of wild type, 604del, and revE1 were processed by integrase in vivo. The approximate 87% efficiency with which the U5 end of 604del was processed in infected cells compared to that in wild type (Table 1) differs significantly from the previously reported 10% efficiency using purified integrase protein and oligonucleotide substrate DNA (35). This result emphasizes the importance of examining mutant viral integration phenotypes in vivo as well as in vitro. Although the U3 end of 604del was processed about 1.7-fold less efficiently than was the corresponding revE1 end, this difference would not seem to account for the rather large difference (≥10-fold) in in vitro DNA strand transfer activities supported by these PICs (Table 1). We therefore conclude that 604del is blocked in its ability to replicate in cells at a step that is after 3′ processing but either before or at integrase-catalyzed strand transfer of endogenous cDNA into a host cell chromosome.
It is unclear what aspect(s) of PIC biology might be defective in 604del-infected cells. One function that PICs most likely perform after 3′ processing but before DNA strand transfer is transporting retroviral cDNA from the cell cytoplasm into the nucleus (4, 17, 25, 27) (Fig. 4). Whereas simpler oncoretroviruses such as Moloney murine leukemia virus require mitosis for their PICs to gain access to chromosomal DNA for integration (30), lentiviruses such as HIV-1 can be actively transported into the nuclei of nondividing cells by an energy-dependent process (5). The C8166 cells used in this study, however, were rapidly dividing, so 604del PICs could readily gain access to chromosomal DNA during mitosis when the nuclear membrane breaks down. Because the cytoplasmic extract of 604del-infected cells did not support a detectable level of in vitro DNA strand transfer activity (Fig. 2), the integration defect appears to manifest itself prior to nuclear localization.
To further investigate this integration defect, wild-type, 604del and revE1 PICs purified by Nycodenz gradient centrifugation were analyzed by MM-PCR footprinting as previously described (8). The purified PICs were divided into two fractions, and one of these fractions was deproteinized to serve as naked DNA control during DNA footprinting (Fig. 1). MM-PCR uses Mu transpososomes as the DNA footprinting reagent (8, 39). Proteins important for integration protect the ends of retroviral cDNA from attack by Mu, revealing footprints in the native protein-DNA samples that are absent from deproteinized controls (8, 39). In addition to these approximately 200-bp footprinted regions at the U3 and U5 ends of retroviral cDNA, hot spots of Mu insertion occur near the very ends of the viruses (1, 8, 39). This native protein-cDNA complex in retroviral PICs, typified by these end-specific transpositional footprints and enhanced regions, is referred to as the retroviral intasome (8, 39). Numerous analyses of retroviruses by MM-PCR footprinting, including integrase and att site mutant PICs (8, 39, 40), functional reconstitution of wild-type PICs using host cell extracts or purified proteins after treatment with high concentrations of salt (7, 39, 40), and functional interference footprinting (39), have established a central role for the intasome in the integration of endogenous retroviral cDNA.
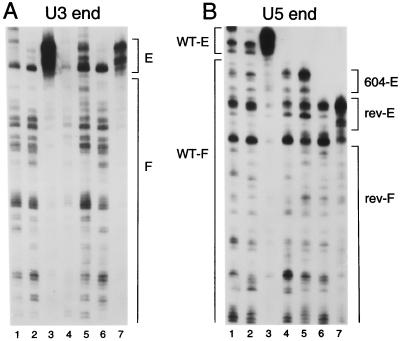
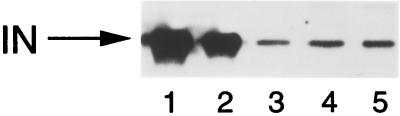
Oligonucleotides AE347 (1) and AE459 (8) were used in second PCR rounds to analyze the U3 and U5 ends, respectively, by MM-PCR. As previously shown, the frequency and distribution of Mu transposition into deproteinized HIV-1 cDNA were nearly identical to those for a naked plasmid DNA control (Fig. 5A and B, compare lanes 2 to lanes 1). Also as expected, the U3 and U5 ends of native wild-type PICs displayed the end-specific transpositional footprinted and enhanced regions that define the retroviral intasome (Fig. 5A and B, compare lanes 3 to lanes 2). Native revE1 PICs also displayed the intasome structure, although in this case the transpositional enhancements were less pronounced than those for wild-type U3 and U5 (Fig. 5A and B, compare lanes 7 to lanes 3). This lower level of end-specific Mu transposition may reflect the lower level of DNA strand transfer activity supported by revE1 PICs than by wild type (Fig. 2 and Table 1) (7). In contrast to the results obtained with revE1, native 604del PICs displayed only a minimal protein-DNA structure. In repeated experiments, regions of protein footprints were not detected, and only slight transpositional enhancements were observed (Fig. 5A and B, compare lanes 5 to lanes 4). It therefore seems that the majority of the protein factors responsible for the wild-type HIV-1 intasome as detected by MM-PCR footprinting either were absent from 604del PICs following Nycodenz gradient centrifugation or were loosely bound such that in vitro transposition readily displaced them from the cDNA. To investigate whether this protein-DNA structure defect might be due to the loss of the majority of PIC-associated proteins that occurred during sample preparation and/or purification, gradient-purified PICs were analyzed for integrase content by Western blotting essentially as previously described (8). The results of this experiment revealed that wild-type, 604del, and revE1 PICs contained similar levels of integrase protein (Fig. 6). Thus, the vast majority of HIV-1 integrase remained associated with 604del cDNA during gradient purification.
FIG. 5.
MM-PCR footprints of wild-type, 604del, and revE1 PICs. (A) The U3 end of HIV-1. In lane 1, naked plasmid DNA was used as the target for Mu transposition. Lane 2, deproteinized wild-type cDNA; lane 3, native wild-type PICs; lane 4, deproteinized 604del cDNA; lane 5, native 604del PICs; lane 6, deproteinized revE1 cDNA; lane 7, revE1 PICs. E, region of transpositional enhancements; F, footprinted regions. Although the deproteinized 604del cDNA in lane 4 did not amplify well, results of other experiments showed that this pattern of Mu transposition was indistinguishable from the wild-type and revE1 deproteinized cDNA patterns in lanes 2 and 6, respectively. (B) The U5 end. The samples in lanes 1 to 7 were the same as those in panel A. WT-E, the wild-type region of transpositional enhancements; WT-F, the wild-type footprinted region; 604-E, 604del region of weak transpositional enhancements; rev-E, the revE1 region of transpositional enhancements; rev-F, the revE1 footprinted region. The U5 ends of 604del and revE1 were shorter in panel B due to the 26- and 47-bp deletions, respectively. Due to the polarity of Mu transposition, only the very ends of the nonprocessed U3 and U5 strands can be analyzed by MM-PCR (8, 39).
FIG. 6.
Integrase protein content of gradient-purified wild-type, 604del, and revE1 PICs. Lane 1, 50 ng of recombinant HIV-1 integrase; lane 2, 25 ng of recombinant integrase; lane 3, gradient-purified wild-type PICs; lane 4, 604del PICs; lane 5, revE1 PICs. IN, integrase.
Although the U3 end of 604del has the wild-type sequence, it, like the mutant U5 end, showed little evidence of protein binding (Fig. 5). Similarly, it was previously shown that a replication-defective Moloney murine leukemia virus U3 att site mutant lacked detectable protein binding at both U3 and U5, even though in this case U5 was wild type in sequence (40). In contrast to the results reported here, that single-end att site mutation blocked 3′ processing of both viral ends in vivo (28). We therefore conclude that a mutation in just one end of the retroviral att site can influence functional protein-DNA interactions at both ends regardless of successful 3′ processing by integrase. The finding that the novel U5 end in revE1 partially restored both PIC activity (Fig. 2) and intasome structure (Fig. 5) further highlights the functional relevance of the retroviral intasome.
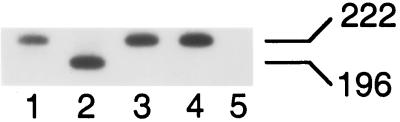
Whereas results of Southern blotting showed that cytoplasmic extracts of wild-type-, 604del-, and revE1-infected cells contained similar levels of unintegrated linear cDNA (Fig. 2), results of indirect end labeling showed that the ends of these cDNAs were intact (Fig. 4). Yet, results of MM-PCR footprinting suggested that protein factors that bind to the ends of HIV-1 cDNA and are important for in vitro integration activity were for the most part absent from gradient-purified 604del PICs (Fig. 5). Because of this, we next investigated whether the 604del replication defect might be due in part to degradation of this cDNA after it enters the cell nucleus. In addition to the linear cDNA integration substrate, two different circular forms of retroviral DNA, containing either one or two copies of the viral long terminal repeat (LTR), are found in the nuclei of infected cells (reviewed in reference 2). The two-LTR circle is readily detected by PCR because the unique LTR-LTR junction is absent from potentially contaminating plasmid DNA. Cells infected with integration-specific retroviral mutants tend to contain higher levels of the two-LTR circle than do wild-type-infected controls (reviewed in reference 14). To investigate the nuclear viability of unintegrated 604del DNA, CEM-12D7 cells infected with wild type, 604del, and integration-defective HIV-1 integrase mutants containing either the substitution of Lys for Gln-62 (Q62K) or Asn for Asp-116 (D116N) (8) were lysed and analyzed for two-LTR circle content using nested PCR essentially as previously described (1). One exception here is that U5-specific primers AE322 and AE609 (1) were used in the first and second PCR rounds, respectively, because the previously described second-round U5 primer coincided with the 604del deletion. Because of this deletion, 604del two-LTR circles displayed an electrophoretic mobility that was faster than that of wild type and integrase mutants Q62K and D116N (Fig. 7, compare lane 2 to lanes 1, 3, and 4). Importantly, cells infected with 604del contained two-LTR circles at a level that was similar to the levels detected for Q62K and D116N (Fig. 7). Thus, nucleus-associated 604del cDNA is apparently as stable as wild type and these integration-defective HIV-1 integrase mutants.
FIG. 7.
Two-LTR circle content of cells infected with wild type, 604del, and integrase mutants Q62K and D116N. Lane 1, lysate from cells infected with wild type; lane 2, lysate from 604del-infected cells; lane 3, lysate from Q62K-infected cells; lane 4, lysate from D116N-infected cells; lane 5, lysate from mock-infected cells. 222 and 196 indicate the sizes of PCR products for wild type and 604del, respectively, in base pairs.
Based on our observations that integrase processed the 3′ ends of 604del in vivo with an efficiency similar to that for revE1 and wild type (Fig. 4 and Table 1) and that 604del PICs were selectively defective for in vitro DNA strand activity (Fig. 2 and Table 1) and intasome structure (Fig. 5) despite containing the majority of their integrase protein (Fig. 6), we propose that protein factors important for DNA strand transfer activity either do not properly associate with 604del cDNA or dissociate from this cDNA more readily than they do from either wild type or revE1 following in vivo 3′ processing. We conclude that this is an integration-specific defect because it did not render 604del cDNA susceptible to nuclease attack by cellular enzymes (Fig. 2, 4, and 7). Although the mechanistic basis for this defect is unclear, it depends on the novel U5 sequence in 604del.
Results of in vitro integration assays using purified HIV-1 integrase protein and oligonucleotide substrate DNA revealed that the nonprocessed 5′ AC overhang produced by 3′ processing contributes to the stability of the functional integrase-viral DNA complex (13). Since this terminus is identical in wild type, 604del, and revE1, we speculate that subterminal U5 sequences might also affect the stability of functional integrase-viral DNA interactions after 3′ processing in vivo. To the best of our knowledge, 604del is the first example of a replication-defective mutant that is blocked in its ability to grow in cells at the integration step despite processing the 3′ ends of retroviral cDNA at nearly normal levels. Based on this novel finding, we speculate that it may be possible to isolate inhibitors of HIV-1 integration that block virus replication after the 3′ processing step of retroviral integration.
Acknowledgments
We thank M. Miller for plasmid DNAs and valued advice and M. Mizuuchi for purified Mu A protein.
This work was supported by NIH grant AI39394, by the G. Harold and Lelia Y. Mathers Foundation, and by the Friends of Dana-Farber Cancer Institute.
REFERENCES
- 1.Brown H E V, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Wei S-Q, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 9.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 10.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 11.Du Z, Ilyinskii P O, Lally K, Desrosiers R C, Engelman A. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J Virol. 1997;71:8124–8132. doi: 10.1128/jvi.71.11.8124-8132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison V, Abrams H, Roe T, Lifson J, Brown P O. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison V, Brown P O. A stable complex between integrase and DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman A. In vivo analysis of retroviral integrase structure and function. Adv Virus Res. 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 15.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 18.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 19.Katz R A, Mack J P G, Merkel G, Kulkosky J, Ge Z, Leis J, Skalka A M. Requirement for a conserved serine in both processing and joining activities of retroviral integrase. Proc Natl Acad Sci USA. 1992;89:6741–6745. doi: 10.1073/pnas.89.15.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafemina R L, Callahan P L, Cordingley M G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by the human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase function in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 25.Lee Y M H, Coffin J M. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency type 1 preintegration complexes: studies of organization and function. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage at both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauza C D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 30.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 32.Sherman P A, Dickson M L, Fyfe J A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleavage and joining reaction. J Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicenzi E, Dimitrov D S, Engelman A, Migone T-H, Purcell D A F, Leonard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vink C, van Gent D C, Elgersma Y, Plasterk R H A. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vora A C, Fitzgerald M L, Grandgenett D P. Removal of 3′-OH-terminal nucleotides from blunt-ended long terminal repeat termini by the avian retrovirus integration protein. J Virol. 1990;64:5656–5659. doi: 10.1128/jvi.64.11.5656-5659.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei S-Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S-Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]