Extract
We echo the latest calls that have been made to increase the capacity for antimicrobial susceptibility testing (AST) for bedaquiline for the Mycobacterium tuberculosis complex [1, 2]. However, we would like to highlight the limitations of using insufficiently standardised or validated phenotypic AST methods and breakpoints as the reference standard for bedaquiline AST. Moreover, we advocate for adoption of a composite reference standard that considers genotypic AST results to minimise false-susceptible results for borderline/low-level resistance mechanisms and avoid confusion during clinical decision-making.
Shareable abstract
A composite reference standard minimises false-susceptible AST results for bedaquiline https://bit.ly/3wAVvFm
To the Editor:
We echo the latest calls that have been made to increase the capacity for antimicrobial susceptibility testing (AST) for bedaquiline for the Mycobacterium tuberculosis complex [1, 2]. However, we would like to highlight the limitations of using insufficiently standardised or validated phenotypic AST methods and breakpoints as the reference standard for bedaquiline AST. Moreover, we advocate for adoption of a composite reference standard that considers genotypic AST results to minimise false-susceptible results for borderline/low-level resistance mechanisms and avoid confusion during clinical decision-making.
For pragmatic reasons, Perumal et al. [2] and the World Health Organization [3] have used a critical concentration (CC) of 0.25 mg·L−1 for a lyophilised broth microdilution (BMD) plate for bedaquiline AST, even though this breakpoint has not been endorsed by any breakpoint committee. This CC has been called into question as potentially too high, thereby increasing the rate of misclassification of borderline bedaquiline resistance mutations such as mmpR5 (Rv0678) M146T (see below), and WHO requested methodological improvements to lyophilised BMD plates [4, 5]. In fact, even the WHO-endorsed bedaquiline CCs for Middlebrook 7H11 and the MGIT system were set based on limited evidence [6]. This underlines the importance of following the guidelines by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) to consider minimum inhibitory concentration (MIC), clinical and pharmacokinetic/pharmacodynamic data to define a quality control (QC) range/target, epidemiological cut-off (ECOFF), clinical breakpoints and, if warranted, an area of technical uncertainty for the EUCAST reference method (figure 1) [5, 7, 8]. Moreover, other methods should be calibrated against the reference method, as is ongoing for a lyophilised bedaquiline product for MGIT, so that outcome data from multiple studies using those methods can be pooled to reach sufficient statistical power to assess whether mmpR5 mutants that correlate with elevated MICs increase the likelihood of failure, for which the evidence is mounting [7, 9].
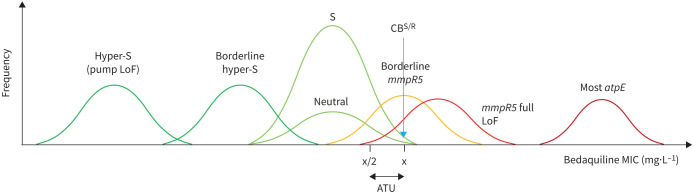
FIGURE 1.
Illustrative plot of different bedaquiline minimum inhibitory concentration (MIC) distributions, where the clinical breakpoint (CB)S/R corresponds to the epidemiological cut-off (ECOFF) [8]. Because the dosing of bedaquiline is fixed and no regulator has endorsed that higher exposures at particular sites overcome modest MIC increases, there is no “intermediate” or “susceptible, increased exposure” range for bedaquiline, which means that “intermediate” should not be used to refer to mutations either [2, 8]. The relative frequency of the different MIC distributions is not representative (e.g. atpE resistance mutations are rarer than those in mmpR5) and the relative MIC increases were chosen for illustrative purposes given that the different mechanisms have never been tested in the same study using the same method under controlled conditions (e.g. with an on-scale quality control (QC) strain in every batch and with sufficiently low antibiotic concentrations to obtain untruncated MICs for all distributions [3, 13, 15, 17]). Most atpE resistance mutations confer large MIC increases (>16-fold), meaning that these mutations test reliably as resistant [3, 13]. Full loss-of-function (LoF) mmpR5 mutants that cause maximal overexpression of the mmpL5-mmpS5 efflux pump confer smaller relative MIC increases (4- to 8-fold) that would be expected to be even more modest for mmpR5 mutants that retain some repressor activity [13]. Given the inherent technical variability of MIC testing, the reproducibility of the latter borderline mutants would be particularly poor at the CBS/R. In fact, the overlap between the susceptible (S) MIC distribution of mmpR5 borderline/full LoF mutants is likely exacerbated by the modest collateral hyper-susceptibility conferred by katG mutations (i.e. such mutants have approximately 2-fold lower bedaquiline MICs) [20]. The misclassification of those mutants as susceptible could be minimised by setting an area of technical uncertainty (ATU) that corresponds to the CBS/R. By contrast, some mutations in resistance genes do not affect the phenotype and the C-11A mmpR5 promoter mutation correlates with a borderline hyper-susceptible phenotype [17]. Such neutral and modest hyper-susceptible mutations are not distinguished in the World Health Organization (WHO) mutation catalogue and would be classified as group 4/5 “not associated with resistance (interim)” [3]. Lastly, LoF mutations in either subunit of the mmpL5-mmpS5 efflux pump should result in a more marked hyper-susceptible phenotype that must be considered for genotypic AST (i.e. group 1/2 mmpR5 mutations cannot confer bedaquiline resistance if genetically linked to a LoF mutation in either subunit, but WHO did not endorse epistasis for mmpS5 as the available dataset lacked clinical mmpS5 mutants [3, 17]). pepQ mutations likely confer similar MIC increases to mmpR5 but appear to be much rarer and are not affected by LoF mutations in mmpL5-mmpS5 [3].
All approved bedaquiline CCs correspond to ECOFFs that are used as surrogates of clinical breakpoints to report phenotypically wildtype (pWT) strains as susceptible [6, 8]. In this context, the choice of the percentile of the pWT distribution (i.e. 97.5th, 99th or 99.9th) can have important consequences [10]. Using the 97.5th percentile as the ECOFF may reduce the misclassification of borderline resistance mechanisms but result in a lower positive predictive value (PPV) in settings with low bedaquiline resistance rates due to rare random false-resistant results [11, 12]. Choosing the 99.9th percentile may increase the PPV at the expense of more false-susceptible results, which could be reduced with an area of technical uncertainty (figure 1) [8, 10]. Given that the pWT distribution has not been studied adequately to date, it is not known precisely to which percentile each of the current bedaquiline ECOFFs correspond and what the expected PPVs are, particularly if only the ECOFF is tested instead of a broader concentration range to monitor the technical variability using a QC strain [5]. However, considering that the pooled baseline resistance prevalence in the study reported by Perumal et al. [2] is only 2.4% (95% CI 1.7–3.5%), the PPV of the phenotypic AST results in this study is unlikely to be very high. Using such datasets to assess the performance of genotypic AST would likely result in the sensitivity of resistance mutations being underestimated [2, 3]. Repeat phenotypic AST of strains that appear to be phenotypically resistant but lack plausible resistance mutations is rarely done for routine clinical practice, but capacity for high-quality MIC testing with stringent QC at reference laboratories should be established for such discrepancies to periodically monitor the quality of initial AST results and identify novel resistance mechanisms, including changes in known resistance genes that might be missed by some analysis pipelines, such as IS6110 insertions and large genomic rearrangements [3, 12, 13].
Its limited sensitivity notwithstanding, genotypic AST using targeted next-generation sequencing is the only viable option to obtain rapid results for bedaquiline directly from clinical samples. In this context, mutations must be interpreted carefully given that some are genuinely neutral or can even correlate with a hyper-susceptible phenotype (figure 1) [3]. Therefore, it is not appropriate to refer to all mutations in bedaquiline resistance genes or their regulator regions as “resistance-associated variants”, as this might deprive patients from receiving bedaquiline or lead to the inappropriate conclusion that baseline mmpR5 mutations do not predict treatment outcomes [2, 14]. Instead, “resistance-associated variants” should only be used for changes that were associated with resistance in at least some genetic backgrounds according to clear criteria, such as the group 1/2 “associated with resistance (interim)” mutations in the WHO mutation catalogue (figure 1) [3].
Crucially, genotypic AST can reliably detect known resistance mechanisms conferring modest MIC increases (figure 1). A good example is mmpR5 M146T, a group 2 resistance mutation that is frequently missed by MGIT because the mode of its MIC distribution is close to the CC [3]. Notably, this mutation has been found in approximately one-third of rifampicin-resistant strains in Eswatini [15, 16].
We acknowledge that genotypic AST can yield systematic false-resistant results. For example, WHO endorsed an additional grading rule whereby any frameshift in mmpR5 should be interpreted as a group 2 mutation, but one frameshift at codon 141 may not confer resistance [3, 17]. To demonstrate such exceptions definitively requires multiple replicates of high-quality MICs and careful analysis of other confounders, which is beyond the capacity of most laboratories (once proven, such exceptions should be incorporated in the WHO catalogue).
In the absence of clear guidance on how to interpret genotypically resistant but phenotypically susceptible AST results, clinicians may attribute this discordance to poor quality testing, undermining their trust in AST and encouraging empiric use of bedaquiline [12]. In our view, the detection of a group 1/2 mutation should overrule a susceptible phenotypic result on a routine basis, provided that obvious human, instrument and reagent errors and, if possible, epistasis have been excluded, as current phenotypic AST methods cannot reliably confirm many mmpR5 resistance mutations (figure 1). The thresholds used for interpretation, such as the mutation frequency for genotypic AST and the critical proportion for phenotypic AST using the proportion method, need to be studied further and, ideally, correlated with treatment outcomes, although this is challenging in practice for multidrug regimens [5, 8, 18, 19]. In other words, we call for the adoption of a composite reference standard, as recommended by WHO for rifampicin, whereby bedaquiline resistance is defined as phenotypic and/or genotypic resistance using the WHO mutation catalogue [12].
Shareable PDF
Footnotes
Conflicts of interest: C.U. Köser is a consultant for Becton Dickinson, the Foundation for Innovative New Diagnostics, the TB Alliance, and the WHO Regional Office for Europe; the consultancy for Becton Dickinson involves a collaboration with Janssen and Thermo Fisher Scientific. C.U. Köser is an unpaid advisor to Cepheid and GenoScreen (GenoScreen covered related travel and accommodation expenses only), has worked as a consultant for the Stop TB Partnership and the WHO Global TB Programme, has given a paid educational talk for Oxford Immunotec, has collaborated with PZA Innovation and has been an unpaid advisor to BioVersys. D.M. Cirillo is the co-chair of the Working Group of the Stop TB Partnership New Diagnostics and is an unpaid member of EUCAST subcommittee for antimicrobial susceptibility testing of mycobacteria, the CLSI M24 mycobacterial working group, and the WHO Strategic and Technical Advisory Group for diagnostics. The remaining authors have no potential conflicts of interest to disclose.
Support statement: C.U. Köser is a visiting scientist at the Department of Genetics, University of Cambridge, and a research associate at Wolfson College, University of Cambridge. S. Niemann is supported by the German Ministry of Health through the SubSaharanSeqNet project (ZMVI1-2519GHP708), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2167 Precision Medicine in Inflammation and the Research Training Group 2501 TransEvo, and the Leibniz Science Campus Evolutionary Medicine of the Lung (EvoLUNG). L. Rigouts is partially supported by the Belgian Science Policy Office and received additional support from the Flemish government through the IntegrOmicsDR.MTB project (G0B0222N). M.R. Farhat is supported by the US National Institutes of Health (NIH) (awards NIAID R01-AI176498 and R01-AI155765). T. Schön is supported by the Heart and Lung Foundation (Oscar II Jubilee Foundation 20220148) and the Swedish Research Council (2022-00865 and 2022-05263). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of NIH. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Maurer FP, Shubladze N, Kalmambetova G, et al. Diagnostic capacities for multidrug-resistant tuberculosis in the World Health Organization European Region: action is needed by all member states. J Mol Diagn 2022; 24: 1189–1194. doi: 10.1016/j.jmoldx.2022.07.005 [DOI] [PubMed] [Google Scholar]
- 2.Perumal R, Bionghi N, Nimmo C, et al. Baseline and treatment-emergent bedaquiline resistance in drug-resistant tuberculosis: a systematic review and meta-analysis. Eur Respir J 2024; 62: 2300639. doi: 10.1183/13993003.00639-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization . Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance, 2nd ed. Geneva, World Health Organization, 2023. https://iris.who.int/handle/10665/374061
- 4.CRyPTIC Consortium . Reply: Epidemiological cut-off values for a 96-well broth microdilution plate for high-throughput research antibiotic susceptibility testing of M. tuberculosis. Eur Respir J 2023; 61: 2300426. doi: 10.1183/13993003.00426-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization . Optimized broth microdilution plate methodology for drug susceptibility testing of Mycobacterium tuberculosis complex. Geneva, World Health Organization, 2022. https://iris.who.int/handle/10665/353066
- 6.World Health Organization . Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. Geneva, World Health Organization, 2018. https://iris.who.int/handle/10665/260470
- 7.Schön T, Köser CU, Werngren J, et al. What is the role of the EUCAST reference method for MIC testing of the Mycobacterium tuberculosis complex? Clin Microbiol Infect 2020; 26: 1453–1455. doi: 10.1016/j.cmi.2020.07.037 [DOI] [PubMed] [Google Scholar]
- 8.Antimycobacterial Susceptibility Testing Group . Updating the approaches to define susceptibility and resistance to anti-tuberculosis agents: implications for diagnosis and treatment. Eur Respir J 2022; 59: 2200166. doi: 10.1183/13993003.00166-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timm J, Bateson A, Solanki P, et al. Baseline and acquired resistance to bedaquiline, linezolid and pretomanid, and impact on treatment outcomes in four tuberculosis clinical trials containing pretomanid. PLoS Glob Public Health 2023; 3: e0002283. doi: 10.1371/journal.pgph.0002283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahlmeter G, Turnidge J. Wild-type distributions of minimum inhibitory concentrations and epidemiological cut-off values-laboratory and clinical utility. Clin Microbiol Rev 2023; 36: e0010022. doi: 10.1128/cmr.00100-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaniga K, Hasan R, Jou R, et al. Bedaquiline drug resistance emergence assessment in multidrug-resistant tuberculosis (MDR-TB): a 5-year prospective in vitro surveillance study of bedaquiline and other second-line drug susceptibility testing in MDR-TB isolates. J Clin Microbiol 2022; 60: e0291920. doi: 10.1128/JCM.02919-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Köser CU, Robledo J, Shubladze N, et al. Guidance is needed to mitigate the consequences of analytic errors during antimicrobial susceptibility testing for TB. Int J Tuberc Lung Dis 2021; 25: 791–794. doi: 10.5588/ijtld.21.0428 [DOI] [PubMed] [Google Scholar]
- 13.Sonnenkalb L, Carter JJ, Spitaleri A, et al. Bedaquiline and clofazimine resistance in Mycobacterium tuberculosis: an in-vitro and in-silico data analysis. Lancet Microbe 2023; 4: e358-e368. doi: 10.1016/S2666-5247(23)00002-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelan JE, Utpatel C, Ismail N, et al. Careful classification of potential bedaquiline resistance mutations is critical when analysing their clinical impact. Int J Tuberc Lung Dis 2024; in press [ 10.5588/ijtld.24.0083]. 10.5588/ijtld.24.0083 [DOI] [PubMed] [Google Scholar]
- 15.Beckert P, Sanchez-Padilla E, Merker M, et al. MDR M. tuberculosis outbreak clone in Eswatini missed by Xpert has elevated bedaquiline resistance dated to the pre-treatment era. Genome Med 2020; 12: 104. doi: 10.1186/s13073-020-00793-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vambe D, Sibandze B, Ziyane M, et al. The benefit of targeted next-generation sequencing for the treatment of patients diagnosed with drug-resistant TB in Eswatini [abstract OA18-345-16]. Int J Tuberc Lung Dis 2023; 27: Suppl. 1, S226–S227. [Google Scholar]
- 17.Miotto P, Cirillo DM, Schön T, et al. The exceptions that prove the rule—a historical view of bedaquiline susceptibility. Genome Med 2024; 16: 39. doi: 10.1186/s13073-024-01311-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nimmo C, Brien K, Millard J, et al. Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine 2020; 55: 102747. doi: 10.1016/j.ebiom.2020.102747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park M, Satta G, Haldar P. Heteroresistance in tuberculosis: are we missing drug-resistant bacteria hiding in plain sight? Thorax 2024; 79: 599–600. [DOI] [PubMed] [Google Scholar]
- 20.Ofori-Anyinam N, Hamblin M, Coldren ML, et al. KatG catalase deficiency confers bedaquiline hyper-susceptibility to isoniazid resistant Mycobacterium tuberculosis. bioRxiv 2023; preprint [ 10.1101/2023.10.17.562707]. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00391-2024.Shareable (784.1KB, pdf)