Abstract
In the replication cycle of hepadnavirus DNA, the double-stranded linear form of viral DNA is generated as a minor replicative intermediate, which is efficiently converted to covalently closed circular DNA (cccDNA) by intramolecular recombination (W. Yang and J. Summers, J. Virol. 69:4029–4036, 1995). We previously found a binding site of transcription factor Yin and Yang 1 (YY1) in one terminal region of the double-stranded linear replicative hepatitis B virus (HBV) DNA (M. Nakanishi-Matsui, Y. Hayashi, Y. Kitamura, and K. Koike, J. Virol. 74:5562–5568, 2000). However, it is not known whether the YY1-binding site is required for the intramolecular recombination of HBV DNA. In this study, we established an HBV-producing system in which the cccDNA appeared to be generated from the transfected linear DNA or the linear replicative DNA by nonhomologous end joining (NHEJ) or by both NHEJ and homologous recombination between terminally repeated sequences, respectively. When the YY1-binding site in the terminal region of transfected linear viral DNA was mutated, the cccDNA was generated merely by NHEJ. Results suggest that the YY1-binding site in the terminal region of linear replicative HBV DNA is required for intramolecular recombination between terminally repeated sequences.
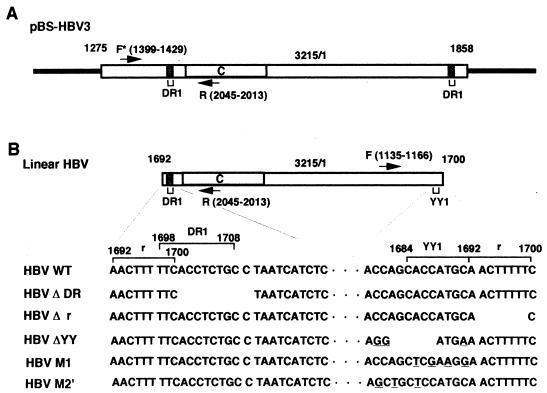
Hepadnavirus DNA is a partially single-stranded circular duplex molecule with the minus strand covering the entire genome and the plus strand containing a partial genomic region. Initiation of hepadnavirus DNA replication begins with the conversion of partially single-stranded circular DNA to covalently closed circular duplex DNA (cccDNA) in the nuclei of infected cells (2). Recently, it was demonstrated by Yang and Summers (28) that, in the minor pathway of DNA replication, the double-stranded linear form of duck hepatitis virus DNA (the almost genome-sized DNA molecule with some overlapping nucleotide sequences at both ends) was produced as a result of a failure to prime plus-strand DNA synthesis at the correct location; however, this linear replicative DNA was efficiently converted to the cccDNA by illegitimate recombination (termed NHEJ for nonhomologous end joining in this paper) between both ends of the linear replicative DNA. Yang et al. (27) suggested the same mechanism to be operative in the replication cycle of the mammalian hepadnavirus woodchuck hepatitis virus. It seems likely that some fraction of human hepatitis B virus (HBV) DNA replicates via a similar recombination mechanism. It has been described that the 5′ and 3′ ends of the HBV minus strand are located within or near the 11-bp direct repeat 1 (DR1) region, with some overlapping nucleotides (2, 12, 16), and the linear HBV DNA appears to have part of DR1 in one end and a complete DR1 with a 9-bp terminally repeated nucleotide sequence (termed r) in the other end (Fig. 1B).
FIG. 1.
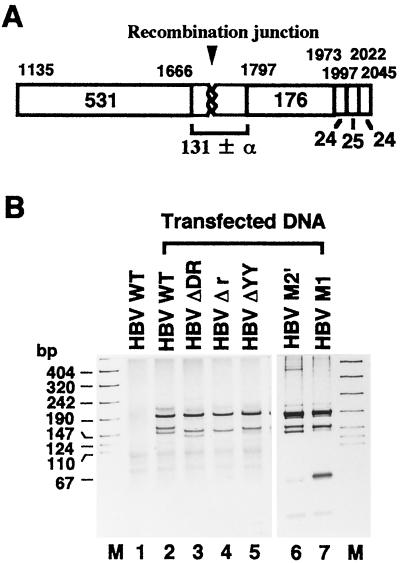
Schematic representation of pBS-HBV3 and linear HBV DNA. (A) Schematic representation of pBS-HBV3 DNA. The open rectangles show the inserted whole HBV genome (3,215 bp) with the 584-bp overlapping DNA region (nt 1275 to 1858). Numbers above the diagram show the nucleotide numbers of the HBV subtype adr sequence (6). The shaded regions indicate DR1 (nt 1698 to 1708). C indicates the open reading frame of HBcAg. The arrows indicate the locations of the PCR primers F* (nt 1399 to 1429) and R (nt 2045 to 2013). The solid bars indicate the vector sequence. (B) Schematic representation of linear HBV DNA. Numbers and a region referred as C are shown as in panel A. Arrows indicate the locations of the PCR primers F (nt 1135 to 1166) and R. The DNA sequences of both terminal regions of WT HBV, HBV ΔDR, HBV Δr, HBV ΔYY, HBV M1, and HBV M2′ are enlarged; the deleted sequences are missing, the mutated sequences are underlined, and the dots indicate the common internal HBV DNA sequence. The terminally repeated r sequence (nt 1692 to 1700), the YY1-binding site (nt 1684 to 1692), and DR1 are also mapped. Note that the promoter region for the 3.6-kb mRNA is located at the terminal region distant from the open reading frame of HBcAg (2).
We previously found the binding site of transcription factor Yin and Yang 1 (YY1) in one terminal region containing part of DR1 (11). However, it has not been determined whether the YY1-binding site in the terminal region is required for HBV DNA recombination. Since we (4, 26) and others (1, 15, 19, 24) had established an efficient HBV-producing system using a human hepatoblastoma cell line and cloned HBV DNA, we tried to analyze HBV replication by transfection of linear HBV DNA into cultured hepatoblastoma cells. In this system, the cccDNA was generated from transfected linear HBV DNA or linear replicative DNA by NHEJ or by both NHEJ and homologous recombination between terminally repeated r sequences, respectively. The necessity of the YY1-binding site in the terminal region of transfected viral DNA for the formation of cccDNA will be described.
MATERIALS AND METHODS
Plasmids.
The HBV expression plasmid, designated pBS-HBV3 (Fig. 1A), was constructed by cloning the HindIII-SalI fragment of pHBV3 (25) into pBluescript II (Stratagene). pCMVβ (Clontech) was cotransfected to monitor transfection efficiency.
Linear HBV DNA for DNA transfection.
The linear HBV DNA used in this study, as shown in Fig. 1B, was obtained from linearized pBS-HBV3 DNA by PCR amplification (20 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 8 min), using Pfu DNA polymerase (Stratagene). The linear HBV DNA products from 10 to 15 independent reactions were pooled and treated with DpnI at 37°C for 1 h to remove template DNA. The resulting DNA was methylated at 37°C for 1 h with Dam methylase (New England Biolabs), and then unmethylated DNA was eliminated by MboI treatment at 37°C for 1 h. The methylated linear HBV DNA was gel purified with a QIAquick gel extraction kit (Qiagen) and then used for transfection. DNA primers used for PCR amplification were as follows. For wild-type (WT) HBV, the forward primer was 5′-AACTTTTTCACCTCTGCCTAATCATCTCATGTTCATG-3′ and the reverse primer was 5′-GAAAAAGTTGCATGGTGCTGGTGAACAGACCATTATG-3′; for HBV with DR1 deleted (HBV ΔDR), the forward primer was 5′-AACTTTTTCTAATCATCTCATGTTCATG-3′; for HBV with r deleted (HBV Δr), the reverse primer was 5′-GTGCATGGTGCTGGTGAACAGACCAATTTATG-3′; for HBV with YY1 deleted (HBVΔYY), the reverse primer was 5′-GAAAAAGTTTCATCCTGCTGGTGAACAGACCAATTTATG-3′; for HBV M1, the reverse primer was 5′-GAAAAAGTTCCTTCGAGCTGGTGAACAGACCATTATG-3′; and for HBV M2′, the reverse primer was 5′-GAAAAAGTTGCATGGAGCAGCTGAACAGACCATTATG-3′.
Cell culture and DNA transfection.
HepG2 cells, derived from a human hepatoblastoma cell line (5), were cultured in DM-160AU medium (Kyokuto) supplemented with 10% fetal calf serum and 60 μg of kanamycin per ml at 37°C in 5% CO2. For DNA transfection, cells were trypsinized and suspended in suspension medium (RPMI 1640 without NaHCO3–50 mM HEPES–10 mM glucose–0.1 mM dithiothreitol). Five micrograms of pBS-HBV3 or linear HBV DNA (WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY) and 1 μg of pCMVβ were mixed with 0.5 × 107 to 1 × 107 cells in 300 μl of suspension medium. DNA transfection was carried out by electroporation (Gene Pulser; Bio-Rad) at 250 V and 960 μF, and the transfected cells thus obtained were plated into 60- and 100-mm-diameter dishes. The cells in 60-mm-diameter dishes were cultured for 2 days after transfection and subjected to a β-galactosidase assay (Promega) to monitor transfection efficiency. The β-galactosidase assay was carried out according to the manufacturer's protocol.
Preparation and blot analysis of RNA.
Two days after transfection, total RNA was extracted from the cells in 100-mm-diameter dishes using an RNeasy kit (Qiagen), subjected to electrophoresis in a formaldehyde-agarose gel, and then transferred to nitrocellulose filter paper (Schleicher & Schuell) as described previously (23). The RNA-blotted filter papers were hybridized with a DNA probe at 65°C overnight in 6× SSC (0.9 M NaCl–90 mM sodium citrate [pH 7.0]) containing 0.02% Ficoll, 0.02% polyvinylpyrrolidone, 0.02% bovine serum albumin, 0.5% sodium dodecyl sulfate (SDS), and 100 μg of sonicated salmon sperm DNA per ml. The 3.2-kb whole HBV genomic probe and the 0.5-kb human β-actin DNA probe were prepared by nick translation (13), using [α-32P]dCTP as a substrate. After hybridization, the filter papers were washed with 2× SSC–0.1% SDS at room temperature for 20 min and then with 0.1× SSC–0.5% SDS at 65°C for 1 h. Hybridized signals were visualized by autoradiography.
Preparation of HBV or core particles and assay of HBV antigens.
Preparation of HBV or core particles was carried out as described previously (4, 26). Briefly, cell extracts were prepared from the cells in 100-mm-diameter dishes by homogenizing cells with hypotonic buffer (20 mM Tris-HCl [pH 7.5]–50 mM NaCl–5 mM MgCl2–0.1% 2-mercaptoethanol–0.5 mM phenylmethylsulfonyl fluoride). After being assayed for HBV core antigen (HBcAg) and HBV e antigen (HBeAg) with a COBAS CORE HBeAg enzyme immunoassay II kit (Roche), the extract was subjected to 30% sucrose zone centrifugation at 35,000 rpm for 16 h in a Beckman SW 50.1 rotor. The pellet thus obtained was used as the core particles. HBV particles secreted into the culture medium were collected by 20% sucrose zone centrifugation.
Preparation and blot analysis of viral DNA from HBV or core particles.
The HBV or core particle fraction, prepared as described above, was treated with 1 mg of proteinase K (Boehringer Mannheim Biochemicals) per ml and 1% SDS at 37°C for 2 h and then directly subjected to 1% agarose gel electrophoresis in Tris-acetate-EDTA buffer. The resultant DNA was transferred to nitrocellulose filter paper as described previously (17), followed by hybridization with an α-32P-labeled HBV DNA probe as described above. After the filter paper was washed, hybridized signals were visualized by autoradiography.
Preparation of viral cccDNA.
The viral cccDNA was prepared as described previously (7, 18). Briefly, the transfected cells in 100-mm-diameter dishes were lysed at 37°C for 5 min in 1 ml of lysis buffer (50 mM Tris-Cl [pH 8.0]–10 mM EDTA–150 mM NaCl–1% SDS). Cell lysate was collected with a cell scraper, and protein-detergent complex was precipitated by treatment with 0.25 volumes of 2.5 M KCl (final concentration, 0.5 M KCl) at 4°C for 5 min. The insoluble materials containing viral replicative intermediates and most of the cellular DNA were removed by centrifugation, and the resultant supernatant containing viral cccDNA was extracted with phenol. To remove the transfected linear viral DNA, the nucleic acid obtained was treated with Exonuclease III (New England Biolabs) at 37°C for 1 h, followed by treatment with mung bean nuclease (New England Biolabs) at 37°C for 30 min. The resultant cccDNA fraction was subjected to recombination junction analysis as described in the following section.
Analysis of the recombination junction in cccDNA.
To separately analyze the recombination junction in the cccDNA derived from the transfected linear or the linear replicative HBV DNA, the cccDNA fraction was treated with MboI or DpnI, respectively. The MboI-resistant fraction was used as the cccDNA generated from transfected linear HBV DNA, whereas the DpnI-resistant fraction was used as the cccDNA generated from linear replicative DNA. After treatment with Exonuclease III and mung bean nuclease, the cccDNA thus obtained was linearized at the ApaI site (nucleotide [nt] 2473). The DNA region containing the recombination junction was amplified by PCR (35 cycles of 98°C for 20 s and 68°C for 3 min). The primers used were the F primer (nt 1135 to 1166; 5′-CCGATCCATACTGCGGAACTCCTAGCAGCTTG-3′) and the R primer (nt 2045 to 2013; 5′-CATTGACATAGCTGACTACTAATTCCCTGGATG-3′) (Fig. 1B). To amplify the DNA from pBS-HBV3, the F* primer (nt 1399 to 1429; 5′-GCACCTCTCTTTACGCGGTCTCCCCGTCTG-3′) (Fig. 1A) was used instead of the F primer. The PCR products were digested with Tsp509I at 65°C for 1 h and then subjected to 15 to 25% gradient polyacrylamide gel electrophoresis in Tris-glycine buffer to analyze the recombination junction. The resultant DNA was visualized by staining with silver (Dai-ichi Chemicals). PCR products were cloned into pGEM (Invitrogen) and subjected to DNA sequencing using a T7 sequenase version 2.0 DNA sequencing kit (Amersham Pharmacia Biotech).
RESULTS
Detection of cccDNA derived from transfected HBV DNA.
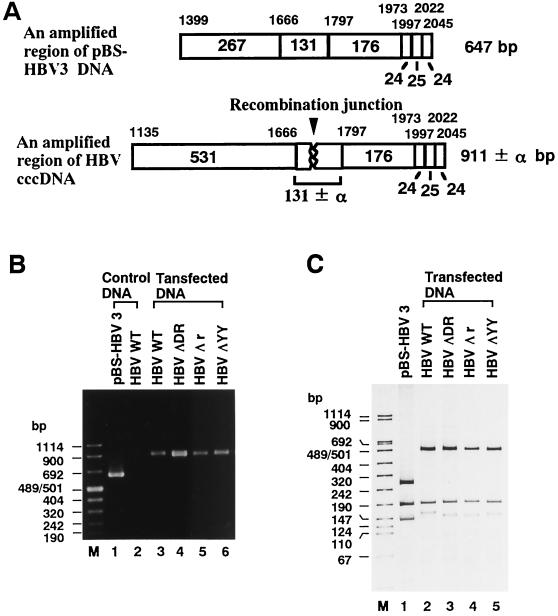
Based on the indication of Yang and Summers (28) that the double-stranded linear form of HBV DNA was a precursor of the cccDNA in the viral DNA replication cycle, we attempted to establish an HBV-producing system by transient transfection of linear HBV DNA (Fig. 1B). As will be described in the following sections, the transfected linear HBV DNA can be converted to cccDNA, which is subsequently utilized as a template for pregenomic RNA transcription. RNA transcription is followed by viral DNA synthesis in HBV or in core particles. To address the issue of whether the nucleotide deletion in the terminal region of transfected linear HBV DNA affects the formation of cccDNA, we prepared the cccDNA fraction by subjecting it to MboI treatment to digest the replicated DNA and then subjected it to further digestion with Exonuclease III and mung bean nuclease to eliminate linear DNA, as described in Materials and Methods. The cccDNA fraction thus obtained was linearized and then subjected to PCR to amplify the recombination junction (Fig. 2A). As shown in Fig. 2B, a single 647-bp band was amplified from pBS-HBV3 DNA with the F* (nt 1399 to 1429) and R (nt 2045 to 2013) primers (lane 1) whereas no band was amplified with the F (nt 1135 to 1166) and R primers (lane 2) from the linear HBV DNA. As expected, the 911-bp band was detected with the F and R primers in the cccDNA fractions prepared from the cells transfected with linear DNAs of WT HBV, HBV ΔDR, HBV Δr, and HBV ΔYY (lanes 3 to 6, respectively). To further analyze the recombination junction of the cccDNA, the amplified band was digested with Tsp509I and subjected to electrophoresis using a 15 to 25% gradient polyacrylamide gel (Fig. 2C). When the 647-bp band amplified from control pBS-HBV3 DNA was digested with Tsp509I, three bands (267, 176, and 131 bp) were detected in the polyacrylamide gel (lane 1). As for the 911-bp band from the cccDNA fraction of the cells transfected with WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY, three bands that migrated around 531, 176, and 131 bp were detected (lanes 2 to 5, respectively) by the same enzyme digestion. Data indicate that the nucleotide deletion in the terminal region of transfected linear HBV DNA does not affect cccDNA formation.
FIG. 2.
Detection of the recombination junction in MboI-resistant cccDNA. (A) Schematic representation of PCR products (911 ± α bp: nt 1135 to 2045), containing the recombination junction (arrowhead), amplified from MboI-resistant cccDNA with the F and R primers. The DNA region (647 bp; nt 1399 to 2045) amplified from pBS-HBV3 DNA, using the F* and R primers, is also shown on the top. The locations of the three primers are shown in Fig. 1. Numbers above the open rectangles show the nucleotide numbers of Tsp509I recognition sites in the HBV DNA sequence (6), and the numbers in or under the open rectangles indicate the sizes (in base pairs) of Tsp509I-digested DNA fragments. (B) Detection of PCR products. Shown are PCR products generated by 2% agarose gel electrophoresis (lanes 3 to 6) from MboI-resistant cccDNA in cells transfected with WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY (Fig. 1B). Products generated with pBS-HBV3 (lane 1) and WT HBV DNA (lane 2) by the same treatment are also shown as controls. Lane M and numbers at the left indicate the size markers in base pairs. (C) Detection of the DNA fragment containing the recombination junction. The PCR products obtained in panel B were digested with Tsp509I and subjected to 15 to 25% gradient polyacrylamide gel electrophoresis. Lanes 2 to 5 contain the samples from MboI-resistant cccDNA in WT-HBV-, HBV ΔDR-, HBV Δr-, and HBV ΔYY-transfected cells, respectively. The PCR product generated from pBS-HBV3 DNA by the same treatment was also loaded as the size marker control (lane 1).
Nucleotide deletion in the terminal region of transfected linear HBV DNA does not affect HBV mRNA expression or production of HBV antigen.
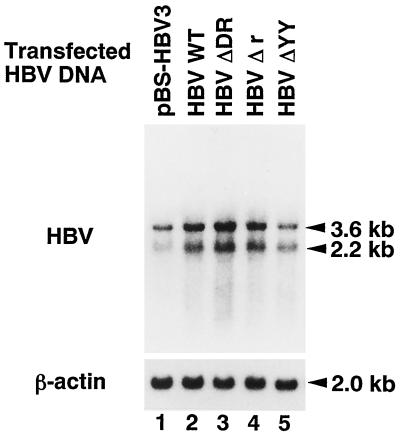
To examine whether the nucleotide deletion in the terminal region of transfected linear HBV DNA affects HBV mRNA expression, total cellular RNA was prepared from the cells transfected with pBS-HBV3 or linear HBV DNA (WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY) and subjected to Northern blot hybridization (Fig. 3). When cellular RNA from the pBS-HBV3-transfected cells was hybridized with the HBV DNA probe, two major mRNA transcripts of 3.6 and 2.2 kb were detected (lane 1), as described previously (4, 26). In the WT-HBV-transfected cells, two major transcripts were detected in similar amounts (lane 2), though the promoter region for the 3.6-kb mRNA was located at the terminal region distant from the transcription start site (2). The results are consistent with the previous observation (28) that transfected linear DNA is efficiently converted to cccDNA. Two major transcripts were also detected in the cells transfected with HBV ΔDR, HBV Δr, or HBV ΔYY (lanes 3 to 5), the relative amounts of which were estimated to be 1.5-, 1.5-, or 0.9-fold the amount of the transcripts detected in WT-HBV-transfected cells, respectively. Data indicate that the nucleotide deletion in the terminal region of transfected linear HBV DNA slightly affects HBV mRNA expression.
FIG. 3.
Northern blot analysis of HBV mRNA. Total RNA obtained from the cells transfected with pBS-HBV3, WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY (lanes 1 to 5, respectively) was subjected to 1% agarose gel electrophoresis, transferred to nitrocellulose filter paper, and then hybridized with an HBV DNA probe. Two major transcripts (3.6 and 2.2 kb) were detected. The filter paper was rehybridized with a β-actin DNA probe to show the 2.0-kb transcript.
The effect of nucleotide deletion in the terminal region of transfected linear HBV DNA on production of HBV antigen was also analyzed by measuring the levels of HBcAg and HBeAg in the cytoplasm by using the extract from the cells transfected with pBS-HBV3 or linear HBV DNA (WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY). As summarized in Table 1, the nucleotide deletion in the terminal region of transfected linear HBV DNA brought about no inhibitory effect on HBV antigen production.
TABLE 1.
Production of HBcAg and HBeAg in cells transfected with linear HBV DNA
| Transfected DNA | % of HBcAg and HBeAg absorbed
|
|
|---|---|---|
| Meana | SD | |
| WT HBV | 100 | |
| HBV ΔDR | 145 | 16 |
| HBV Δr | 134 | 4.5 |
| HBV ΔYY | 107 | 4.5 |
| pBS-HBV3 | 110 | 9 |
The numbers in this column indicate the relative percentage of absorbance of each sample against the absorbance of WT-HBV-transfected cells, which was considered 100%.
Deletion of DR1 or the YY1-binding site in the terminal region of transfected linear HBV DNA reduces viral DNA synthesis in HBV or core particles.
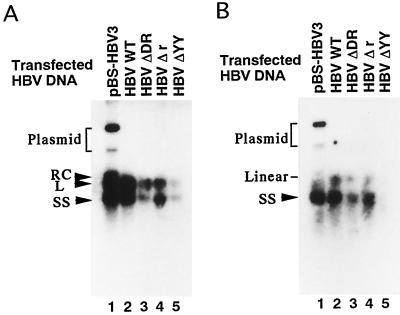
We next examined whether the nucleotide deletion in the terminal region of transfected linear HBV DNA affects HBV DNA synthesis. We analyzed the amounts of HBV DNA in viral and core particles prepared from equal amounts of culture medium and the cytoplasm, respectively, of cells transfected with pBS-HBV3 or linear HBV DNA (WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY). When HBV DNA was prepared from viral particles in pBS-HBV3-transfected cells and subjected to Southern blot analysis, two bands (the upper band corresponds to a partially double-stranded relaxed circular molecule, RC, and the lower band corresponds to a single-stranded molecule, SS) were detected (lane 1 in Fig. 4A), as previously described (4, 26). The DNA in viral particles from the cells transfected with linear HBV DNA (WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY) was similarly analyzed, in which the partially double-stranded linear molecule, L, was observed as a band that migrated slightly faster than that of the RC molecule, along with that of the SS molecule (lanes 2 to 5 in Fig. 4A). The difference in mobilities between the RC and the L molecules has been confirmed by 1.25% agarose gel electrophoresis in Tris-borate-EDTA buffer (3, 8, 10). In the case of the DNA prepared from the core particles of transfected cells, the SS molecule was detected by Southern blot analysis (Fig. 4B). Although internal DNA controls were not available, the amount of HBV DNA in viral or core particles reproducibly decreased in several independent experiments using cells transfected with HBV ΔDR or HBV ΔYY DNA compared to that in the cells transfected with pBS-HBV3, WT HBV, or HBV Δr DNA (lanes 1 to 5). Data suggest that the deletion of DR1 or the YY1-binding site inhibits HBV DNA synthesis, consistent with the previous observation that some mutation around the initiation site (nt 1700) for minus-strand synthesis inhibits DNA replication in core particles (9, 14, 22).
FIG. 4.
Southern blot analysis of HBV DNA in viral or core particles. (A) HBV DNA in viral particles. HBV particles secreted into the culture medium of the cells transfected with pBS-HBV3, WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY (lanes 1 to 5) were treated with 1 mg of proteinase K per ml and 1% SDS and then directly subjected to 1% agarose gel electrophoresis. The resultant DNA was blotted to the filter paper and hybridized with an HBV DNA probe. Arrowheads indicate the positions corresponding to three different forms of HBV DNA (RC, L, and SS) and the bracket shows the position of transfected plasmid DNA. (B) HBV DNA in core particles was treated as described for panel A. Lanes 1 to 5 contain the samples from pBS-HBV3-, WT-HBV-, HBV ΔDR-, HBV Δr-, and HBV ΔYY-transfected cells, respectively. The arrowhead indicates the position of the SS form of HBV DNA. The positions of the transfected plasmid and linear DNA are also indicated.
cccDNA formation from linear replicative HBV DNA.
It is known that the cytoplasmic core particles bearing the replicative viral DNA can be shunted to the nucleus by an intracellular pathway (2). To address the issue of whether the nucleotide deletion in the terminal region of transfected linear DNA affects the formation of cccDNA derived from the linear replicative HBV DNA, we analyzed the cccDNA prepared by DpnI treatment to digest transfected DNA and then subjected the cccDNA to further digestion with Exonuclease III and mung bean nuclease to eliminate the linear DNA. The resulting DpnI-resistant cccDNA was used as a template for PCR to amplify the recombination junction of cccDNA (Fig. 5A). The single 911-bp band obtained was digested with Tsp509I and then subjected to electrophoresis using a 15 to 25% gradient polyacrylamide gel (Fig. 5B). Under these conditions, linear HBV DNA was unable to generate any significant bands in the polyacrylamide gel (lane 1). As expected, two bands that migrated around 176 and 131 bp were detected in the cccDNA fraction prepared from the cells transfected with linear HBV DNA (WT HBV, HBV ΔDR, HBV Δr, or HBV ΔYY) (lanes 2 to 5, respectively). An additional band that migrated slightly faster than the band around 131 bp was detected in the cells transfected with WT HBV (lane 2) or HBV ΔDR (lane 3) but not in HBV Δr-transfected cells (lane 4) or HBV ΔYY-transfected cells (lane 5). To further demonstrate the association of the YY1-binding site with cccDNA formation, two mutant linear HBV DNAs (HBV M1 and HBV M2′ in Fig. 1B) with nucleotide substitutions were transfected. HBV M1 contains four nucleotide changes which reduce the binding activity of YY1 (11). On the other hand, HBV M2′ contains three nucleotide substitutions at the YY1-binding site as in M2 (11) which do not affect YY1-binding activity (11). When the recombination junction of cccDNA derived from the linear replicative DNA in the polyacrylamide gel was analyzed, the additional band was seen to have migrated slightly faster than the band around 131 bp that was detected in the HBV M2′-transfected cells (lane 6). In HBV M1-transfected cells, however, the intensity of the additional band decreased (lane 7). Data suggest that the r sequence and the YY1-binding site are required for the reaction to yield the additional band.
FIG. 5.
Detection of the recombination junction in DpnI-resistant cccDNA. (A) Schematic representation of the PCR product (nt 1135 to 2045) amplified from DpnI-resistant cccDNA with the F and R primers. Numbers above the open rectangles show the Tsp509I recognition sites in the HBV DNA sequence (6), and the numbers in or under the open rectangles indicate the sizes (in base pairs) of Tsp509I-digested fragments. (B) Detection of the DNA fragment containing the recombination junction. PCR products amplified from DpnI-resistant cccDNA in the cells transfected with WT HBV, HBV ΔDR, HBV Δr, HBV ΔYY, HBV M2′, or HBV M1 (lanes 2 to 7, respectively) were digested with Tsp509I and subjected to 15 to 25% gradient polyacrylamide gel electrophoresis. The bands that migrated around 176 and 131 bp are shown, but the 531-bp band is omitted. The PCR product generated from WT HBV DNA (lane 1) by the same treatment is shown as a control. Lane M and the numbers at the left indicate the size markers in base pairs.
The cccDNA derived from transfected linear DNA or linear replicative DNA appears to be generated by NHEJ or by both NHEJ and recombination between terminally repeated r sequences, respectively.
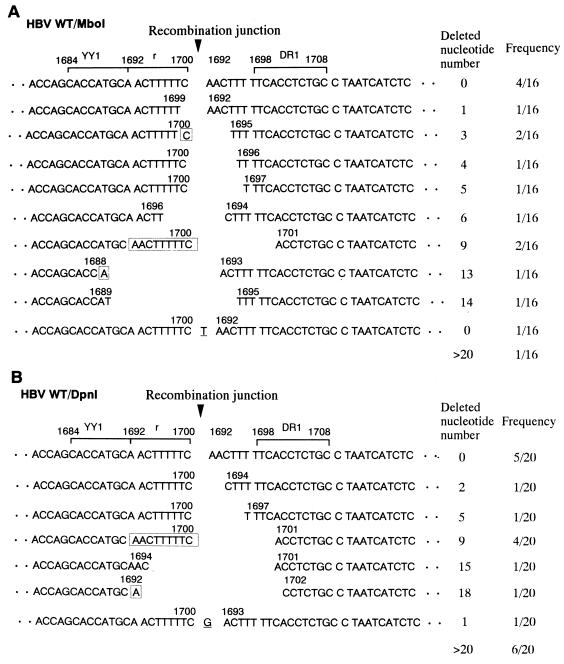
To understand the mechanism involved in the generation of the recombination junction of cccDNA derived from transfected linear DNA or linear replicative DNA in the cells transfected with WT HBV, we cloned and sequenced the junction of MboI- or DpnI-resistant cccDNA, respectively. When the 16 cloned recombination junctions of MboI-resistant cccDNA were sequenced and aligned according to their numbers of nucleotide deletions (Fig. 6A), four clones were found to have the same nucleotide sequence at the recombination junction without any nucleotide deletion. Two clones had the same nucleotide sequence at the recombination junction with a 3-nt deletion, and the other two clones had a 9-nt deletion. The remaining eight clones had different nucleotide sequences at the recombination junction. Data indicate that the cccDNA derived from transfected linear DNA appears to be generated by NHEJ and explain why, in Fig. 2C, the junction-containing band in WT-HBV-transfected cells is 9 bp longer than the 131-bp product of pBS-HBV3 and why it therefore migrates more slowly (compare lane 2 with lane 1). On the other hand, when 20 recombination junctions of DpnI-resistant cccDNA were sequenced and aligned (Fig. 6B), five clones had the same nucleotide sequence at the recombination junction with no nucleotide deletion. Four clones had the 9-nt deletion in the r sequence. The remaining 11 clones had varied nucleotide deletions at the recombination junction in each case. Data indicate that the cccDNA derived from linear replicative DNA appears to be generated not only by NHEJ but also by recombination between 9-bp terminally repeated r sequences.
FIG. 6.
Sequence alignment of the recombination junction of cccDNA formed in WT-HBV-transfected cells. (A) Sequence alignment of the recombination junction of MboI-resistant cccDNA. The 16 junction clones obtained were sequenced and aligned in order of the number of nucleotides deleted. When exactly the same sequence was obtained, the numbers of clones are written in the frequency column. The sequence with more than 20 nt deleted is omitted. The numbers above the DNA sequences correspond to the nucleotide numbers of the HBV DNA sequence (6). The possible nucleotide sequence involved in the joining of two terminal regions of viral DNA is boxed. Underlined nucleotides are not derived from the HBV DNA sequence. The YY1-binding site (nt 1684 to 1692), the r sequence (nt 1692 to 1700), and DR1 (nt 1698 to 1708) are indicated with brackets. The arrowhead indicates the recombination junction. (B) Sequence alignment of the recombination junction of DpnI-resistant cccDNA. The 20 junction clones obtained were sequenced and aligned as described for panel A.
The YY1-binding site in the terminal region of linear replicative DNA is required for intramolecular recombination between terminally repeated r sequences.
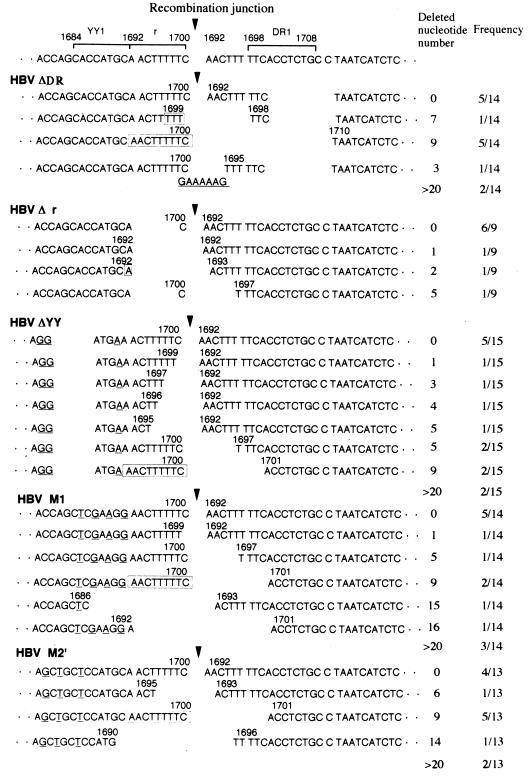
To further demonstrate whether the r sequence and the YY1-binding site in the terminal region of replicative linear DNA is required for recombination between terminally repeated r sequences, we examined the nucleotide sequence at the recombination junction of DpnI-resistant cccDNA in cells transfected with HBV ΔDR, HBV Δr, or HBV ΔYY, as well as HBV M1 or HBV M2′. Figure 7 shows a sequence alignment of the recombination junctions together with the lengths of nucleotide deletions and their frequencies. When the 14 cloned junctions of cccDNA formed in the HBV ΔDR-transfected cells were sequenced and aligned, five clones had the same sequence at the recombination junction with no nucleotide deletion. Five other clones had the same sequence with a 9-nt deletion of the r sequence. In the four remaining clones, the deleted sequences at the recombination junction were varied in size. Data indicate that the cccDNA formed in HBV ΔDR-transfected cells appears to be generated not only by NHEJ but also by recombination between 9-bp terminally repeated r sequences. On the other hand, when the nine recombination junctions of cccDNA in HBV Δr-transfected cells were analyzed, six clones had the same sequence at the recombination junction without any nucleotide deletion while the three remaining clones had different nucleotide deletions at the junction sequences. When 15 recombination junctions of cccDNA in HBV ΔYY-transfected cells were sequenced and aligned, the nucleotide sequence at the recombination junction was found to be intact (without deletion) in five clones. Two clones had the same sequence with a 5-nt deletion, and the other two clones had a 9-nt deletion at the recombination junction. In the six remaining clones, the nucleotide deletion at the junction differed in each case. Data indicate that the cccDNA formed in the HBV Δr- or HBV ΔYY-transfected cells appears to be generated merely by NHEJ. Among 14 clones of HBV M1-transfected cells, five clones were found without a nucleotide deletion. Two clones had the 9-nt deletion, and the six remaining clones had varied nucleotide deletions at the recombination junction. When the recombination junctions of 13 cccDNAs in HBV M2′-transfected cells were sequenced and aligned, no nucleotide deletion was observed in four clones while the 9-nt deletion was detected in five clones. The remaining four clones had different nucleotide deletions at the recombination junction. Taken together, these results suggest that not only the r sequence but also the YY1-binding site in the terminal region of linear replicative DNA is required for intramolecular recombination between terminally repeated sequences.
FIG. 7.
Sequence alignment of the recombination junctions of DpnI-resistant cccDNAs formed in cells transfected with HBV ΔDR, HBV Δr, HBV ΔYY, HBV M1, and HBV M2′. The 14, 9, 15, 13, or 14 junction clones of DpnI-resistant cccDNA obtained in HBV ΔDR-, HBV Δr-, HBV ΔYY, HBV M1-, or HBV M2′-transfected cells, respectively, were sequenced and aligned as described for Fig. 6.
DISCUSSION
In this study, we established an HBV-producing system by transient transfection of the linear HBV DNA to separately analyze the cccDNA molecules derived from transfected linear and linear replicative HBV DNAs by means of MboI and DpnI treatment, respectively. When the MboI-resistant cccDNA formed in the cells transfected with linear WT HBV DNA was analyzed, the single band that migrated around 131 bp was detected as the recombination junction-containing band (Fig. 2). Sequence analysis of the recombination junction (Fig. 6) revealed that the band was generated without nucleotide deletion, indicating that the cccDNA derived from transfected linear HBV DNA appears to be generated by NHEJ, as observed previously (27, 28).
On the other hand, when the DpnI-resistant cccDNA formed in the linear WT-HBV-transfected cells was analyzed, the band(s) that contains the recombination junction was detected as the doublet(s) around 131 bp (Fig. 5). Sequence analysis of the recombination junction (Fig. 6) revealed that the band with slower mobility was generated without any nucleotide deletion during the process of NHEJ but that the band with faster mobility appeared to be generated by recombination between terminally repeated r sequences. Even when DR1 was deleted from the terminal region of transfected linear DNA (HBV ΔDR), essentially the same results were obtained (Fig. 5 and 7). Since DR1 is known to be essential for the formation of the RC molecule in core particles (2), the band with faster mobility is not likely to be generated from the RC molecule recycled into the nucleus, which was circularized by a template switch between terminally repeated r sequences in core particles (8). The alternative possibility is that the band with faster mobility may be generated by a template switch of the double-stranded linear L molecule in core particles. However, the L molecule exhibits no detectable template switch activity in core particles (10). Furthermore, when the r sequence was deleted from the terminal region of transfected linear HBV DNA (HBV Δr), the cccDNA was generated merely by NHEJ (Fig. 5 and 7). These results together suggest that the cccDNA derived from linear replicative HBV DNA is formed not only by NHEJ but also by intramolecular recombination between terminally repeated sequences in the nucleus. Major difference in the products may come from the fact that the linear replicative HBV DNA, not the transfected linear DNA, binds the primer protein at one end and the primer RNA at the other end (2). It should be emphasized that the cccDNA molecule generated by intramolecular recombination between terminally repeated sequences will not be discriminated from the WT.
We previously found the YY1-binding site in the terminal region of double-stranded linear HBV DNA (11). To answer the question of whether the YY1-binding site is necessary for HBV DNA recombination, we analyzed cccDNA formation in cultured cells by introducing linear HBV ΔYY DNA or HBV M1 DNA, in which the YY1-binding site was deleted or mutated in the terminal region. Unlike in the cells transfected with WT HBV DNA, the DpnI-resistant cccDNA was formed merely by NHEJ in the HBV ΔYY- and HBV M1-transfected cells (Fig. 5 and 7). Data indicate that the YY1-binding site is required for intramolecular recombination between terminally repeated sequences of linear replicative HBV DNA but not for NHEJ.
It has been suggested that the linear replicative hepadnavirus DNA integrates into cellular DNA by a recombination mechanism similar to that operative in cccDNA formation (27, 28). Takada et al. (20, 21) demonstrated that the virus-cell junction designated N2-7 was formed by recombination between the r sequence of HBV DNA and the 5′-AACTATTTC-3′ sequence (mismatched nucleotide is underlined) of cellular DNA. These data imply that recombination between terminally homologous r sequences may be involved not only in the formation of cccDNA (by intramolecular recombination) but also in the integration of viral DNA into cellular DNA (by intermolecular recombination).
REFERENCES
- 1.Chang C, Jeng K-S, Hu C-P, Lo S J, Su T-S, Ting L-P, Chou C-K, Han S-H, Pfaff E, Salfeld J, Schaller H. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe P M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 3.Havert M B, Loeb D D. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J Virol. 1997;71:5336–5344. doi: 10.1128/jvi.71.7.5336-5344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi Y, Koike K. Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J Virol. 1989;63:2936–2940. doi: 10.1128/jvi.63.7.2936-2940.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles B B, Howe C C, Aden D P. Human hepatocellular carcinoma cell lines secrete the major protein and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi M, Koike K. Complete nucleotide sequence of hepatitis B virus DNA of subtype adr and its conserved gene organization. Gene. 1984;30:227–232. doi: 10.1016/0378-1119(84)90124-0. [DOI] [PubMed] [Google Scholar]
- 7.Lenhoff R, Summers J. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J Virol. 1994;68:5706–5713. doi: 10.1128/jvi.68.9.5706-5713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb D D, Gulya K J, Tian R. Sequence identity of the terminal redundancies on the minus-strand DNA template is necessary but not sufficient for the template switch during hepadnavirus plus-strand DNA synthesis. J Virol. 1997;71:152–160. doi: 10.1128/jvi.71.1.152-160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb D D, Tian R. Transfer of the minus strand of DNA during hepadnavirus replication is not invariable but prefers a specific location. J Virol. 1995;69:6886–6891. doi: 10.1128/jvi.69.11.6886-6891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb D D, Tian R, Gulya K J, Qualey A E. Changing the site of initiation of plus-strand DNA synthesis inhibits the subsequent template switch during replication of a hepadnavirus. J Virol. 1998;72:6565–6573. doi: 10.1128/jvi.72.8.6565-6573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakanishi-Matsui M, Hayashi Y, Kitamura Y, Koike K. Integrated hepatitis B virus DNA preserves the binding sequence of transcription factor Yin and Yang 1 at the virus-cell junction. J Virol. 2000;74:5562–5568. doi: 10.1128/jvi.74.12.5562-5568.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieger A, Nassal M. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J Virol. 1996;70:585–589. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rigby P W J, Dieckman M A, Rhodes C, Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- 14.Seeger C, Maragos J. Identification of a signal necessary for initiation of reverse transcription of the hepadnavirus genome. J Virol. 1991;65:5190–5195. doi: 10.1128/jvi.65.10.5190-5195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sells M A, Chen M-L, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih C, Burke K, Chou M-J, Zeldis J B, Yang C-S, Lee C-S, Isselbacher K J, Wands J R, Goodman H M. Tight clustering of human hepatitis B virus integration sites in hepatomas near a triple-stranded region. J Virol. 1987;61:3491–3498. doi: 10.1128/jvi.61.11.3491-3498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Summers J, Smith P M, Horwich A L. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sureau C, Romet-Lemonne J-L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 20.Takada S, Gotoh Y, Hayashi S, Yoshida M, Koike K. Structural rearrangement of integrated hepatitis B virus DNA as well as cellular flanking DNA is present in chronically infected hepatic tissues. J Virol. 1990;64:822–828. doi: 10.1128/jvi.64.2.822-828.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada S, Koike K. Trans-activation function of a 3′ truncated X gene-cell fusion product from integrated hepatitis B virus DNA in chronic hepatitis tissues. Proc Natl Acad Sci USA. 1990;87:5628–5632. doi: 10.1073/pnas.87.15.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavis J E, Ganem D. RNA sequences controlling the initiation and transfer of duck hepatitis B virus minus-strand DNA. J Virol. 1995;69:4283–4291. doi: 10.1128/jvi.69.7.4283-4291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas P S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsurimoto T, Fujiyama A, Matsubara K. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaginuma K, Koike K. Identification of a promoter region for 3.6-kilobase mRNA of hepatitis B virus and specific cellular binding protein. J Virol. 1989;63:2914–2920. doi: 10.1128/jvi.63.7.2914-2920.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Summers J. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]