Abstract
CD8+ T cells are end effectors of cancer immunity. Most forms of effective cancer immunotherapy involve CD8+ T cell cytotoxic function. Here we review the current understanding of T cell function in cancer, focusing on key CD8+ T cell subtypes and states. We discuss factors that influence CD8+ T cell differentiation and function in cancer through a framework that incorporates the classic three signal model and a signal 4 – metabolism - and consider also the impact of the tumor microenvironment from a T cell perspective. We argue for the notion of immunotherapies as “pro-drugs” that act to augment or modulate T cells, which ultimately serve as the drug in vivo, and for the importance of overall immune health in cancer treatment and prevention. The progress in understanding T cell function in cancer will enable improved harnessing of the immune system across broader tumor types so as to benefit more patients.
CD8+ T cells are “end effectors” of cancer immunity. Wherry, Kaech and colleagues review the current understanding of CD8+ T cell differentiation and function in cancer, discussing recent advance within a four-signal framework that incorporates T cell metabolism and the impact of the tumor microenvironment.
Introduction
Our understanding of the Cancer Immunity Cycle1 has advanced substantially in the past 10 years. These advances are nowhere more evident than in our knowledge of and ability to therapeutically target T cells for enhancing anti-tumor immunity. Major drivers of this field are the unprecedented successes of immune checkpoint blockade (ICB) therapies together with breakthroughs in engineered cellular therapies. The widespread use of such T cell-based therapeutics provides an extraordinary opportunity to apply science in the clinical setting and directly link efforts from preclinical models to observed realities in human disease. Furthermore, advances in high dimensional cytometry and single cell genomics have greatly expanded our understanding of T cell differentiation states. Such studies, including in cancer patients, have highlighted the central role of exhausted CD8+ T (Texh) cells and other CD8+ T cell subtypes in cancer immunity and in response to ICB therapies.
One useful framework for discussing advances in T cell immunity to cancer is a three-signal model for effective T cell priming and differentiation, incorporating TCR signaling by antigens presented by MHC molecules (signal 1); co-stimulation (signal 2), and cytokines (signal 3). It is only with all three optimal signals that naïve T cells are effectively primed, differentiate into functional effector CD8+ T (Teff) cells, and form high quality memory CD8+ T (Tmem) cells. There have been major advances in our understanding of all three signals in cancer immunity. Considerable efforts have been directed towards understanding the nature of antigens that can activate T cell responses in cancer and harnessing such antigens for treatments like cancer vaccines and cellular therapies. The nature of the checkpoints CTLA-4 and PD-1 also reframes the concept of signal 2 and highlights the “negative” co-stimulation exploited by tumors for immune evasion. Signal 3 was initially conceptualized as the direct sensing of inflammation through cytokines necessary for full T cell activation and differentiation. The absence of such inflammatory signals may partly underly the poor priming environments of many tumors resulting in “tolerance” to tumor antigens. However, recent advances also highlight the role of chronic inflammation as a potential negative regulator of T cell immunity in cancer. Moreover, the ability to exploit cytokines to augment T cell immunity to cancer, and the expansion of this concept to include growth factor-type cytokines (e.g. IL-2), remains of considerable interest. In addition to this classic three signal model, recent work in cancer immunology has highlighted the role of a signal 4, the nutrients and metabolites that fuel T cell metabolism. We now appreciate the importance of how T cells are metabolically reprogrammed and how changes in nutrients, oxygen, and other environmental signals constrain T cell responses to tumors. Lastly, the influence of the physical structure and the other cell types within the tumor microenvironment (TME) on the T cell response is beginning to be better characterized, and will be discussed in this review. Together, these advances provide a framework for understanding the major signals that influence T cell differentiation and how these signals may be therapeutically manipulated to improve T cell functions for anti-tumor immunity.
Here, we first discuss our current understanding of CD8+ T cell differentiation as it relates to cancer, then explore variations in signals 1, 2, 3 and 4 in the Cancer Immunity Cycle, and lastly consider how additional factors in the TME alter T cell state and function. Although this review is mainly focused on CD8+ T cells, work over the past decade has also highlighted a critical role for CD4+ T cells in cancer biology and response to immunotherapy. CD4+ T cells can have pro- or anti-tumor effects, including providing “help” for CD8+ T cells, influencing myeloid and other cells within the TME, and mediating either direct cytotoxicity or regulatory effects. For further detail, we refer the reader to other excellent reviews discussing the CD4+ T cell compartment 2, 3, 4.
CD8+ T Cell Differentiation States
Over the past decade, an increasingly comprehensive and detailed understanding of CD8+ T cell differentiation in tumors has begun to emerge. This knowledge has come from an array of human studies and mouse models of cancer and chronic infection. While technological advances have enabled deep immune profiling directly in human patients, precisely controlled mouse models have generated the foundational framework with which to interpret these new data. Here, we synthesize information across all these sources to discuss our current understanding of CD8+ T cell differentiation as it relates to the Cancer Immunity Cycle.
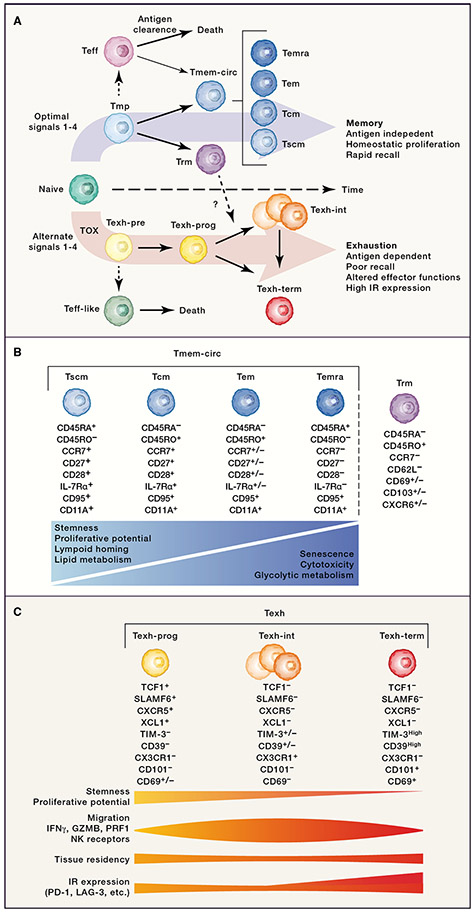
Upon activation, CD8+ T cell differentiation follows two main developmental pathways that ultimately results in the formation of either Tmem cells or Texh cells, with branches of shorter-lived effector-type cells (Figure 1). In settings with optimal signals 1-4, such as acutely-resolving infections or vaccinations, CD8+ T cells initially differentiate into Teff cells or memory-precursors (Tmp) cells, and as antigen is cleared, long-lived Tmem cells emerge 5, 6, 7. The Tmem population is heterogeneous with multiple subsets defined by surface phenotype, function, differentiation potential, and location. The most commonly used classification scheme for the circulating Tmem (Tmem-circ) population include stem cell memory (Tscm), central memory (Tcm), effector memory (Tem), and effector memory CD45RA+ (Temra) 8; however, alternative schemes have also been proposed 9. A distinct subset of Tmem cells reside within tissues, termed “resident memory” (Trm). These Trm cells likely arise from Tmp cells that enter tissues and acquire additional programs of long-term tissue residency that include the ability to provide local protective immunity 10, 11, 12. Among Trm cells there is additional heterogeneity, in part reflecting tissue-specific signals 13, 14. Within the TME, it is often possible to identify cells that phenotypically resemble Tem, Temra, and Trm cells, with Tscm and Tcm cells being less common.
Figure 1: CD8+ T cell differentiation: Two pathways.
A. Upon activation, naïve CD8+ T cells differentiate down two main developmental pathways which are regulated, in part, by four signals. B. Conventional classification scheme for human memory CD8+ T cells. Below each subset are expression profiles commonly used for identifying these subsets within Tmem population. Key functional characteristics are depicted by relative expression across the memory CD8+ T cell differentiation spectrum. These comparisons focus on later stage memory fate-committed subsets. C. The three major Texh subtypes with expression profiles commonly used for identification within Texh population. The relative expression of various traits, functions, and expression profiles across subsets within Texh population are represented below. These comparisons focus on later stage exhaustion fate-committed subsets.
In contrast to the development of Tmem cells, persistent stimulation, particularly through the TCR (signal 1) 15, 16, 17 leads to the development of T cell exhaustion, a branch of CD8+ T cell differentiation epigenetically coordinated by the transcription factor (TF) TOX 18, 19, 20, 21, 22. When exactly the Texh branch diverges from the Tmem branch is an active area of investigation (Box 1). Regardless, the majority of CD8+ T cells within many human tumors are likely Texh cells based on surface protein or gene expression, though the frequency can vary among cancer types and between patients 23. Although Texh cells were originally identified in a mouse model of chronic infection 24, 25, it is now clear that this cell fate arises in many settings of persistent stimulation including infections such as HIV, HCV, and HBV, and human cancers 23, 26, 27. Texh cells have been traditionally characterized by high co-expression of multiple inhibitory receptors (IRs), such as PD-1, CD39 and LAG-3, as well as reduced production of effector cytokines, including IFN-γ and TNF, and also IL-2 27, 28, 29. We now know that Texh cells are defined by a distinct transcriptional and epigenetic program compared to naïve, Teff, and Tmem cells 30, 31, 32, 33, 34, 35, 36. A frequent misconception is that Texh cells are non-functional. Rather, Texh cells have altered effector functions and are adapted to withstand chronic stimulation associated with persisting infections and tumors 37, 38, 39, 40. During chronic infection, Texh cells can maintain moderate viral control while avoiding excessive immunopathology 16, 41, 42. Indeed, loss of Texh cells in chronic SIV infection results in disease progression 43, 44, and Texh cells may drive epitope escape in persisting HIV and HCV infection 45, 46. Furthermore, the association of ICB efficacy and Texh cells within the TME indicates functional activity of these cells in cancer 47, 48. These data highlight the attenuated, yet biologically important functional capacity of Texh cells. Despite Texh cell activity in cancer, an immune-tumor stalemate, or equilibrium, is often unstable and eventually results in failure of immune control, tumor escape and disease progression 49. Major regulation of this balance is through IRs. Indeed, blocking IRs through ICB therapies, such as αPD-1, enhances Texh cell effector functions, boosts anti-tumor immunity, and has produced long-lasting cures in many patients.
Box 1: When does exhaustion start?
When exactly development of Texh cells diverges from that of Teff/Tmem cells, when the Texh differentiation state becomes irreversibly established, and whether there are early changes before the Texh epigenetic commitment to exhaustion occurs are all central questions with implications for immunotherapy. CD8+ T cell exhaustion is progressive, becoming more severe with higher concentrations and/or a longer duration of antigen stimulation 15, 27, 58, 273, 274, 275. Many studies have shown that once the mature Texh population is formed, these cells cannot be de/re-differentiated into the Teff/Tmem states 15, 31, 274, 276, 277, 278. Mouse models of cancer and chronic infection have revealed a potential early divergence (~5-8 days) of exhausted precursors (Texh-pre) from cell states such as Tmp and conventional Teff that arise during acute-resolving infections 15, 18, 34, 74, 279. Recently, it has been suggested that this bifurcation may occur as early as the first cell division 273 280. However, the “inflection” point for this transition to irreversible exhaustion appears to be ~2-3 weeks of chronic stimulation 15, 276, 277, 278. This switch is associated with major epigenetic and transcriptional changes 32, 34, 41, 74.
One key reason that a progressive model of T cell exhaustion has emerged is that the early Texh-pre cells in mouse models of cancer and chronic infection retain fate-flexibility whereas later Texh do not. If Texh-pre are removed from chronic antigen stimulation and adoptively transferred into an antigen-free host, these cells can recover and differentiate into Tmem cells 15, 277. In the setting of chronic stimulation, these early fate-flexible Texh-pre cells seed the fully committed Texh population by differentiating into exhausted progenitors (Texh-prog) 21, 41, 74, 279. Within the mature Texh population, these Texh-prog give rise to the other Texh subsets. Committed Texh-prog cells are transcriptionally and epigenetically distinct from the fate-flexible Texh-pre cells 74; however, both Texh-pre and Texh-prog express TCF-1 and CXCR5 74. Numerous human studies have used TCF-1 and/or CXCR5 to identify Texh-prog 51, 52, 53, 54, 55, 56, 59, but the distinction between Texh-pre and Texh-prog is unclear based on these two markers alone. Indeed, not all TCF-1+ CD8+ T cells are the same. This TF is also expressed by naïve T cells, as well as Tmem and Tmp cells during acute-resolving infection. Furthermore, additional complexity within the Texh-prog population has been proposed, with high expression of CD62L marking cells with enhanced stem-like capacity 281 similar to a CD62L+ Tmp population282. In contrast, CCR6 is expressed by TCF-1+ Texh-prog cells that respond poorly to ICB 90. Identifying developmentally “young” Texh-pre within the TME through the discovery of additional robust biomarkers could have considerable utility because these cells may be more amenable to therapeutic manipulation compared to fate-committed mature Texh cells. However, disentangling these relatively similar cell types in heterogenous and evolving human tumors is challenging without knowing when each T cell clone was initially activated.
During chronic infections, in addition to the Texh-pre subset, a second population of short-lived effector-like (Teff-like) cells is generated that is distinguished from bona fide Teff cells in acute infection 21, 41, 55, 74, 279. This population is terminal, dies off quickly after the first week in the presence of persisting stimulation, and does not contribute substantially to the mature Texh population 41. It is currently unclear whether an analogous subset arises in developing cancers.
The early bifurcation between the Texh and Teff/Tmem branches suggests alterations during priming such as increased signal 1 (TCR stimulation), suboptimal signal 2 (low co-stimulation/high co-inhibition), altered signal 3 (low or high inflammatory cytokines), or different signal 4 (nutrients and fuel) may drive these distinct CD8+ T cell fates. A potentially relevant aspect of cancer is TME evolution during tumorigenesis which may provide different contextual settings for T cell priming over the course of cancer development. However, direct comparison of mature Texh populations in cancer and chronic infection has revealed central transcriptional and epigenetic principles, pathways, and specific drivers of Texh differentiation, while also revealing potential disease-specific features related to microenvironment, such as inflammatory signals and diverse regulatory pathways 56. Indeed, differences between cancer types, such as cold/non-inflamed versus hot/inflamed tumors, are also likely to influence CD8+ T cell priming and differentiation.
Within the fate-committed mature Texh population, there is additional heterogeneity. Initially, this complexity was conceptualized as a progenitor-like subset capable of responding to PD-1 blockade and a terminal subset that could not 50. These progenitor and progeny subsets were subsequently identified by many groups in mouse models of chronic infection 51, 52, 53, 54, 55, mouse models of cancer 53, 55, 56, 57, 58, and human tumors 55, 59, 60. Additional studies point to at least three major subtypes within the mature Texh population: exhausted progenitor cells (Texh-prog), intermediate exhausted T cells that possess some effector function (Texh-int), and terminal exhausted T cells (Texh-term) 61, 62, 63, 64, 65. Texh-prog cells have both exhausted and stem-like qualities. This subset expresses TCF-1, has enhanced proliferative capacity, and gives rise to the other mature Texh subsets. Texh-prog are often localized to secondary lymphoid organs 51, 52, 56, 57, 58, 61, 64, 66, 67 but are also found within the TME 48, 57, 68, 69, 70 where they reside within specialized niches near antigen-presenting cells (APCs) or tertiary lymphoid structures (TLSs) 60, 71, 72, 73. Texh-prog cells can express chemokines and/or chemokine receptors such as XCL1, CCR7, and CXCR5 that might facilitate these interactions 51, 52, 53, 54, 55, 56, 59, 74. The Texh-int subset expresses both exhaustion-related molecules, such as TOX and IRs, as well as effector-associated molecules. This subtype exhibits functional heterogeneity that likely depends on specific environmental signals. For example, a subset expressing NK receptors (or killer-cell immunoglobulin-like receptors, KIRs) is observed in chronic infection and some human cancers 23, 74, 75, 76. Texh subpopulations expressing interferon stimulated genes (ISGs), granzyme molecules, heat shock proteins, IL-17, the oxidative phosphorylation pathway, or other specific programs may also fall into this Texh-int subset 23, 67, 69, 74, 76. Although this subset has been found in models of chronic infection 61, 62, 63, 74, 76 and human cancer 23, 26, 64, 65, 67, 69, 77, diverse definitions and nomenclature, combined with considerable biologic heterogeneity have made it difficult to consistently identify and compare these cells across data sets. Indeed, despite an underlying epigenetic program of exhaustion, Texh-int cells may appear as a type of effector-like population in some settings based on high mRNA (or protein) expression of individual granzymes or other genes. Lastly, Texh-term cells have the lowest proliferative capacity and highest IR expression. Some Texh-term cells express GZMA 74 and thus may contribute to direct killing. It is also tempting to speculate whether these cells play a role in local immune regulation. They produce large amounts of beta chemokines, specifically CCL3, CCL4, and CCL5, which can contribute to both inflammation and tissue repair through recruitment of other leukocytes 74, 78. Moreover, this subset has high expression of CD39 and may exert regulatory effects through the production of adenosine 79. These three major Texh subsets all share a core exhaustion epigenetic program and depend on TOX 18, 19, 20, 21, 22. Together, they form a proliferative hierarchy that resembles those observed in regenerating or self-sustaining tissues enabling the Texh population to persist even in stressful conditions, such as chronic antigen stimulation, while retaining some functionality. Defining the lineage relationships between Texh subsets remains an active area of investigation. Current data suggest that both linear (Texh-prog → Texh-int → Texh-term) and branched (Texh-prog → Texh-int and Texh-prog → Texh-term) paths exist, and these trajectories may be regulated, at least in part, by strength of TCR signaling (signal 1) 76, 80 and IFN signaling (signal 3) 80. Furthermore, how these exhaustion trajectories intersect with those of Trm populations is also of considerable interest (see Box 2).
Box 2: Texh or Trm?
There is increasing interest in defining the relationship between Texh and Trm cells because both populations can be present in non-lymphoid tissues, and these cell types can display partially overlapping phenotypes 14, 275, 283. Texh-term cells display features of residency, such as CD69 expression 61, and Trm cells share some characteristics with Texh cells, including IR expression even in healthy antigen-free tissues 284, 285, 286. However, Trm cells can also form in tissues where antigen persists 287, 288, 289, but they may have distinct phenotypes in these settings compared to in healthy tissues 290. Indeed, it is currently unclear whether restimulated Trm cells can migrate from adjacent tissues into the TME and become exhausted, or whether Texh cells (or Texh-pre or Texh-prog cells) migrate from circulation and upregulate residency programs upon entry to the TME. Trajectory analyses from human cancer suggest that both differentiation pathways can occur with some tumor types predominated by T cells employing one pathway over the other 23, 291. It is also likely that the stage of tumorigenesis effects which pathway is most dominant. Early stage tumors that have less vascularization and exhibit less inflammation may only recruit Trm from nearby tissues, whereas later stage highly vascular tumors can draw in more CD8+ T cells from circulation with chemokines and other inflammatory cues.
Increased Trm cells and/or Trm signatures are associated with better prognosis in several cancer types 292, 293, 294, 295, 296. These data may indicate increased infiltration of Trm cells from adjacent tissues or increased expression of residency programs from circulating cells. However, mouse models have demonstrated better protection against tumor challenge when both tumor-specific tissue-resident and circulating T cells are present 297, 298, 299, 300. Regardless, enhancing migration into the TME and/or improving retention within the TME may be beneficial to enhance tumor immunity. As a result, therapeutic targeting of Trm is currently an active area of drug development. Although, it is also possible that retention within the TME could drive antigen-specific CD8+ TIL towards terminal exhaustion differentiation due to chronic stimulation and suppressive elements within the TME – if the tumor is not eradicated. The intersection between T cell exhaustion and tissue residency and how these biologic programs can be manipulated to improve tumor control will be key research areas in the future.
Immune profiling techniques, including high dimensional cytometry and next generation sequencing (NGS) assays, coupled with clinical and translational efforts to collect patient samples have revealed considerable heterogeneity of Texh and Tmem subpopulations within tumors defined by protein or gene expression. However, interpreting human profiling data poses several challenges: 1) we lack a unified nomenclature for T cell subpopulations, 2) connecting T cell phenotypes or states to antigen specificity remains difficult, and 3) it is often challenging to define temporal relationships between differentiation states. The first challenge arises, at least in part, from different naming conventions including those based on historical subpopulation names, highly expressed genes, and/or functional qualities 26. Comprehensive comparison to existing profiles, such as “reference maps” for scRNA-seq, can help provide context 81; however, these references may not capture the full spectrum of cell states that are possible within disease contexts. Furthermore, an important question is what constitutes a distinct “subset” of T cells? Data from our group and others74, 82, 83 suggest that classification strategies that use epigenetic information may resolve more distinct and stable subtypes compared to transcriptional or protein information, in part because the latter may represent transient responses to environmental signals. As more studies capture multimodal data, the ability to compare and cross-validate cellular definitions using different biologic measurements should improve, enabling a potential unified and robust classification system.
A second major challenge in interpreting T cell phenotypic data from human cancer patients is the limited information regarding antigen specificity. Within the TME, TCR clonality and local expansion of the Texh population is often used to infer tumor specificity 23, 68, 70, 77, 84, 85. However, confirming antigen specificity of individual T cell clones is technically challenging 86, and some studies indicate only a small fraction of CD8+ TIL are tumor-specific 86, 87. Recently, new strategies have profiled scRNA-seq/scTCR-seq in parallel with in vitro tumor-reactivity assays then use TCRs as barcodes to match differentiation state defined by gene expression with antigen specificity determined in vitro 70, 88. One study that tested the reactivity of highly expanded TCRs found that 83% of the tested TCRs within the Texh population were tumor-specific compared to only 10% from non- Texh cells 70. Even though the sample size was small and from a classically “hot” tumor type (melanoma), this result indicates a greater proportion of CD8+ TIL may represent on-target responses than originally suggested. Overall, T cell clones with confirmed tumor reactivity are often found within Texh subsets whereas virus-specific T cells (CMV, EBV, influenza virus, etc.) that can also be found in tumors reside in Tmem populations 28, 70, 87, 88, 89. Collectively, these data raise several questions. First, these data are consistent with tumor antigen driving T cell exhaustion. However, strength of stimulation is likely to vary substantially across different microenvironments, therefore focusing on the most expanded clones or Texh-like cells may not capture all tumor-reactive T cells. For example, T cells that experience weaker stimulation may be less clonally expanded, but also less exhausted and thus potentially useful therapeutic targets. Indeed, T cells specific for sub-dominant tumor antigens may be enriched for TCF-1+ Texh-prog cells 90. Second, these observations suggest that bystander T cells of unrelated specificities can be recruited into the TME, potentially through inflammatory signals, such as CXCL9 and CXCL10 via CXCR3, which is expressed by most Tmem subsets 91. Alternatively, reactivation of latent viruses such as EBV or CMV, or de-repression of endogenous retroviruses (ERVs), within tumor cells or other cells within the TME, may recruit antigen-specific Tmem subsets from circulation or adjacent tissues. In addition, tumor antigens may be molecular mimics of viral or other pathogenic peptides and provide antigenic stimulation for cross-reactive T cells 92. Regardless, bystander T cells that become activated have the potential to exert anti-tumor effects 93. Another T cell population that may be present in the TME are those specific for tumor antigens that have been lost by immune editing and/or loss of antigen presentation. Such tumor-specific T cells would become ignorant and, depending on their differentiation state when antigen was lost, could differentiate into Tmem or memory-like cells (e.g. if these cells where Texh-pre or possibly Texh-prog when antigen mutated). Alternatively, these clones could simply disappear if they had become fully exhausted before antigen loss. Thus, defining what is meant by tumor-specific is not necessarily straightforward.
Defining temporal relationships for tumor-reactive T cells in humans remains a third challenge. As tumors evolve, endogenous CD8+ T cells can be activated at different times and in different locations. However, most patient data only captures snapshots of these trajectories. In silico techniques used to infer temporal relationships have provided some insights, but these approaches remain challenged in their ability to accurately capture T cell trajectories. T cells can undergo cycles of activation and quiescence, and quiescent cells, such as naïve and Tmem, are often predicted to be temporally related based on transcriptome despite substantial epigenetics differences 94. Using TCR sequences as cellular barcodes and tracking individual clones through scRNA-seq data can provide additional insight regarding potential relatedness of different T cell clusters. For example, the same T cell clones have been identified in Texh-prog and Texh-term cells in human patients suggesting potential lineage relationships 67, 70. Such data aligns with the Texh developmental relationships defined in mouse models 39, 52, 54, 57, 61, 62, 74, 76, 80. However, this strategy depends on the number of cells sampled, biasing such analyses to more abundant clones and more frequent cell states. In addition, existence of a TCR sequence in multiple scRNA-seq clusters is often interpreted to mean recent clonal relatedness; however, this TCR sharing may result from distal events, including the first division after priming. Indeed, without temporal data, the exact relationship between different T cell clusters or subsets may remain challenging to determine unambiguously.
Given the clinical success of ICB in the past decade, a central question has been to understand the underlying mechanisms by which these drugs work and specifically how they alter T cell differentiation states and functions. Texh cells are thought to be a primary target of ICB due to their high expression of IRs and preclinical data on PD-1 pathway blockade 27. Indeed, pre- or early on-treatment tumor infiltration by Texh cells in humans is positively correlated with clinical benefit across diverse tumor types 95, 96, 97, 98, 99, 100. Early studies in mice showed that PD-1 blockade “reinvigorated” Texh cells, numerically increasing antigen-specific T cells and improving function 101, 102. However, PD-1 blockade does not substantially change the epigenetic landscape of fate committed Texh population 31. Rather PD-1 blockade (at least temporarily) alters the balance of Texh subsets, acting primarily on Texh-prog cells and driving differentiation towards downstream subsets including Texh-int and Texh-term cells 50, 52, 54, 56, 57. An increase in the Texh-int subset is likely responsible for much of the anti-tumor activity given the higher expression of effector molecules by this subset 61, 62, 63, 64, 65, 67, 74. Recent work also suggests that these Texh-prog cells can “self-renew” under these conditions of PD-1 pathway blockade 103. An increased frequency of Texh-prog cells is linked to improved clinical outcomes after ICB treatment 23, 48, 104. Although, as noted above, phenotypic diversity or similarity, varying nomenclature, lack of antigen specificity and/or temporal information create significant hurdles for synthesizing information across studies. Regardless, these data highlight the importance of progenitor or “stem-like” cells capable of further differentiation into progeny with anti-tumor activity in ICB treatment. Although PD-1 blockade does not re-program the CD8+ T cell differentiation state from Texh to Teff or Tmem cells, a large expansion of the Texh-int subset can be sufficient for substantial clinical benefit, and even cure if tumor burden is low 105. Treatments that boost the expansion and/or function of Texh-int cells may enhance anti-tumor efficacy. Indeed, combining PD-1 blockade with other ICBs, such as αCTLA-4 or αLAG-3, improves clinical efficacy 106, and preclinical data supports combination treatments with cytokines signals such as IL-2 to further boost T cell function 107, 108, 109.
Designing rational treatment combinations that act on a cellular target of choice and elicit a specific change in differentiation state or functional capacity is becoming a reality and will be an important goal for the next decade. T cells are a prime target in the immune system for such therapies. As discussed further below, it is now possible to therapeutically intervene in many key signals that shape T cell activation and differentiation, including signals 1-4. These pathways can be targeted extrinsically with drugs that can act on the endogenous immune response and/or targeted intrinsically through cellular engineering and adoptive cell therapy.
Signal 1: TCR Signaling and Antigens
The first step of T cell activation is recognition of antigen presented by MHC on a professional APC, typically a dendritic cell (DC), through the TCR on the T cell. One of the historically challenging areas of tumor immunology has been identifying tumor antigens capable of providing signal 1 for T cell activation (Figure 2). Although somatic mutation is hallmark of tumorigenesis, not all tumors produce sufficient immunogenic antigens to induce a robust endogenous immune response. These tumors may represent a failure in the first step of the Cancer Immunity cycle due to insufficient antigen-specific priming.
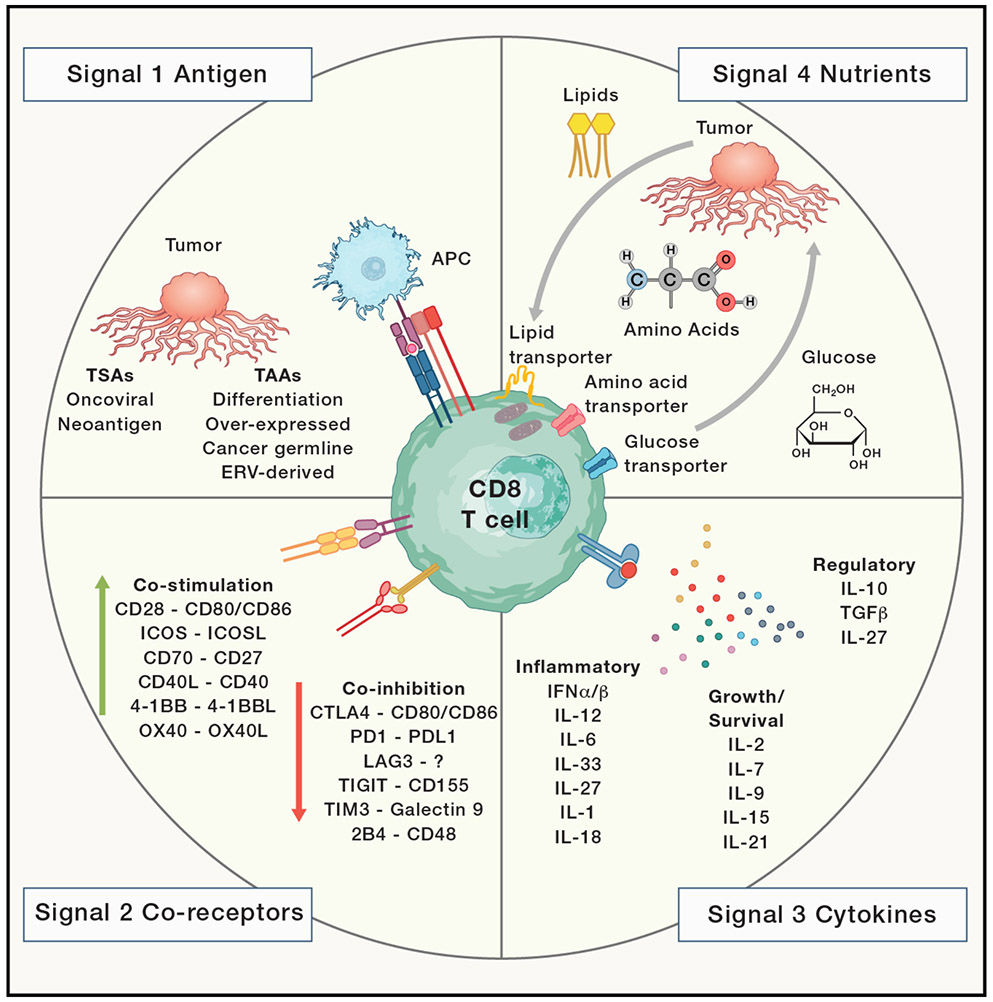
Figure 2: Four signal model of T cell activation and differentiation.
Signal 1 is TCR recognition of peptides presented on MHC molecules. In cancer, these antigens are classified as tumor-associated antigen (TAAs) which can be expressed by both tumor and healthy tissue or tumor-specific antigens (TSAs) which are exclusively expressed by cancer cells. Signal 2 encompasses co-stimulatory and co-inhibitory signals, the balance of which influences T cell activation status and effector functions. Signal 3 includes signaling received through cytokines. The 3 main classes of cytokines include inflammatory, growth/survival, and regulatory. Signal 4 includes nutrient sensing and usage as well as internal changes in metabolic lifestyle.
Tumor antigens can be broadly classified as tumor-associated antigens (TAAs), expressed by both tumor and healthy tissues, or tumor-specific antigens (TSAs), exclusively expressed by tumor cells. The four main classes of TAAs include differentiation, over-expressed, cancer germline or testis antigens, and ERV-derived antigens 110. As a result of thymic selection, T cells targeting TAAs may express TCRs with lower affinity 111. On the other hand, TSAs are essentially foreign antigens and thus TSA-specific T cells are not subject to central tolerance. TSAs can be either viral proteins (e.g. from oncogenic viruses) or neoantigens. Infection with specific viruses can cause cellular transformation resulting in expression of viral proteins detectable by the immune system 112. In contrast, neoantigens have novel amino acid sequences derived from the host. Neoantigens can be produced at any point during peptide production, including DNA mutations, transcription, such as alternative splicing or fusion events, distinct post-translational protein modifications, or changes due to proteasomal processing 110.
Not all tumor mutations produce a neoantigen. To generate signal 1 for T cell activation, a mutation must be expressed, presented by MHC, and stimulate a T cell response. Identifying such immunogenic antigens has been a major effort in the field for the past decade. DNA sequencing followed by in silico prediction is the most widely used approach due to its high throughput nature and is often combined with mass spectrometry and/or T cell reactivity assays 86. Many different neoantigen prediction pipelines have been developed that incorporate different kinds of data, use different assumptions, and employ distinct algorithms. Recently, a consortium-based benchmarking study compared 25 neoantigen prediction pipelines and identified key parameters for predicting immunogenic peptides, including strong and stable MHC binding, high tumor abundance, and immunogenicity quantified as distance to self (“agretopicity”) or similarity to pathogens (“foreignness”) 113. Such advances in neoantigen prediction are key for the successful design of many immunotherapy strategies as discussed below.
The goal of antigen-targeted therapies is to increase tumor-specific T cells by providing antigens for signal 1 or bypassing this step in vivo by directly transferring activated tumor-specific T cells. These therapies have two primary considerations: the target antigen(s) and the type of therapy. Antigen-directed therapies include cellular therapy (CAR T cells, TCR-engineered, and TIL) and vaccination. Engineered cells can introduce new specificities or higher affinities compared to the endogenous response. Cellular therapies have shown curative potential, especially in hematological malignancies 114, but can also have considerable side effects including cytokine release syndrome (CRS) arising in part from artificially increasing the magnitude of the T cell response to a disseminated target such as CD19 for B cell malignancies. Despite substantial success in treating liquid tumors, identifying appropriate tumor antigen targets (and overcoming exhaustion) for solid cancer-targeting CAR T cells remains challenging 115. Numerous synthetic biology engineering strategies using, for example, logic gates that require recognition of more than one antigen or other strategies have the potential to unlock other tumor targeting possibilities and enhance safety 115. In addition to cellular therapy, TAAs are targets in vaccine trials aiming to enhance the endogenous T cell response. Results from early efforts were mixed 116, but new technologies, including mRNA vaccine platforms, have renewed interest in TAAs as therapeutic targets 117, 118.
In contrast to TAAs, neoantigens are tumor-specific and thus excellent potential targets for immunotherapy. The first evidence that T cells could recognize neoantigens was reported over 20 years ago 119, 120. However, most neoantigens are patient-specific 86, requiring individually designed treatments. Advancements in NGS and neoantigen prediction pipelines over the past decade are making such personalized vaccine treatments a reality. Although many different types of cancer vaccines have been tested (reviewed in 117, 118), mRNA-based platforms are attractive options in part due to advances made during the accelerated production of the SARS-CoV2 vaccines. A recently completed phase I trial for pancreatic ductal adenocarcinoma (PDAC) treated patients with personal neoantigen vaccines using uridine mRNA-lipoplex nanoparticles 121. Half of the patients (8/16) were classified as responders. None of these responders experienced recurrence at the time of publication, whereas the non-responders had a median recurrence free survival of 13.4 months. Furthermore, a recent trial (mRNA-4157-P201/KEYNOTE-942) demonstrated that mRNA personalized neoantigen vaccines could be combined with pembrolizumab (αPD-1) to increase recurrence-free survival in patients with resected high-risk melanoma 122. These studies highlight the feasibility of manufacturing personalized cancer vaccines in real time and the ability of such vaccines to elicit novel T cell responses, including in a cancer type that is traditionally considered immunologically cold.
One major potential consequence of strong T cell pressure on tumors, predicted by the Cancer Immunity Cycle, is immune editing. Originally defined in mouse models where T cells, largely through IFN-γ, shape the antigenicity of tumors 49, immune editing is now appreciated to have a considerable impact on the human cancer landscape. On-target T cell responses can lead to the elimination of tumor subpopulations expressing immunogenic antigens, resulting in eventual tumor escape 1. Designing tumor antigen-directed therapies can proactively consider predictions about tumor-immune evolution. One example is targeting multiple antigens, exemplified by CD19 and CD22 CAR T cells, allowing for tumor recognition even if one antigen is lost 123. Indeed, endogenous single TCRs have been found that recognize multiple TAAs which could also protect the host against antigen escape 124. Another strategy is to target clonal driver mutations that control tumor cell growth or viability. Some driver mutations occur frequently at specific genomic locations called "hotspots" 125, such as amino acid 12 in the KRAS oncogenic protein 126. These mutations in oncogenic drivers can sometimes produce shared neoantigens, providing the opportunity to generate therapies that can be used across different patients; such efforts are underway targeting KRAS mutations 127. A third method is engineering tumor cells to express specific antigen(s), such as CD19 encoded by an oncolytic virus 128 or membrane-inserting tags 129, that are paired with specificity matched CAR T cells. Lastly, it is also possible to target non-tumor host cells that are critical for a tumor-supportive local environment through targets such as Fibroblast Associated Protein (FAP) which is expressed by cancer-associated fibroblasts 130. Ultimately, longitudinal tracking of both tumor evolution and the immune response can inform effective design and application of antigen-directed immunotherapy. Vaccines and cellular therapies built upon optimal antigens have the potential to introduce new T cell responses that can help overcome hurdles at the beginning or later states of the Cancer Immunity Cycle, such as T cell exhaustion.
Signal 2: Co-stimulation and Co-inhibition
Signal 2 was originally defined as co-stimulation from CD80/CD86 ligands on the APC, received through the CD28 receptor on the T cell. During infections, these co-stimulatory ligands are upregulated on APCs in response to pattern recognition receptor activation and/or inflammation. Signaling through CD28 facilitates survival, metabolism and cytokine production by T cells, ensuring full activation 131. Since these original concepts were defined for CD28 and CD80/86, a variety of co-stimulatory molecules have been identified (e.g. CD40L, OX40, 4-1BB, ICOS, CD27, etc.) that can have a role in priming or at later stages of T cell differentiation and be delivered by many cell types including help from CD4+ T cells 132. In cancer, a non-inflammatory or suppressive TME may prevent robust upregulation of co-stimulatory molecules on APCs resulting in sub-optimal T cell activation. Several strategies have been developed to stimulate APCs and thus enhance T cell activation, such as αCD40 133 and soluble FLT3 ligand 134. In addition, agonists that target co-stimulatory molecules directly on CD8+ T cells are currently being tested 135.
Following proper activation, T cells increase expression of CTLA-4 which eventually outcompetes CD28 for binding to CD80/CD86 and transmits inhibitory signals that regulate the extent and duration of T cell activation 131 . Beyond CTLA-4, many other "negative signal 2" molecules have been identified including PD-1, LAG-3, 2B4, TIGIT, TIM-3, and others 132. The balance of positive (co-stimulation) and negative (co-inhibition) signals, akin to “gas” and “breaks”, regulates T cell activation and differentiation. The inhibitory aspect of the signal 2 concept has received considerable attention in the past decade, primarily due to the clinical success of ICB, first from αCTLA-4 (ipilimumab and tremelimumab), then αPD-1 and αPD-L1 (nivolumab, pembrolizumab, atezolizumab, etc.). In cancer, Texh cells express high levels of multiple co-inhibitory molecules 27. PD-1 pathway blockade provides a temporary release from this inhibitory signal, shifting the balance toward activation and driving proliferation 136. Notably, this αPD-1 effect requires signaling through CD28 137, highlighting the interplay between positive and negative signal 2s for T cells in cancer biology.
Although the importance of these co-stimulatory and co-inhibitory molecules in regulating T cell function is now well-recognized, the specific downstream signaling pathways involved remain incompletely understood. It is clear that signals through different receptors can have different functional consequences. For example, CAR T cells with 4-1BB domains exhibit enhanced durability compared to those with CD28 domains that generate more effector and exhaustion features 115. Furthermore, blocking different IRs, such as PD-1 and LAG-3, have distinct functional outcomes 138. However, defining proximal downstream targets, impact on cellular function, and whether these effects are conserved or context-specific remains an area of active investigation. This mechanistic knowledge should enable better rational design of next generation combination treatments. Moreover, the patterns of co-stimulatory and co-inhibitory molecules expressed by T cells change at different stages of effector, memory, or exhaustion development. In fact, CD28 is expressed on Texh-prog, but not Texh-term cells 52, 74 which may contribute to the ability of Texh-prog cells to selectively respond to ICB. Thus, it may be possible to selectively tune endogenous T cell responses by appropriate targeting of these co-stimulatory and co-inhibitory pathways.
Signal 3: Cytokines
Signal 3 was initially conceptualized as the inflammatory cytokine signal perceived by T cells during priming that instructed proper effector differentiation 139. This paradigm linked the innate immune system directly to T cell activation (i.e. in addition to the role in activating APC) and allowed T cells to calibrate the magnitude and quality of the effector response to the severity of infection. Signal 3 was initially thought to comprise IL-12 and type I interferons. However, numerous cytokines, including IL-1, IL-18, IL-33, IL-6, and IL-27, as well as growth factors and regulatory cytokines like IL-10 and TGF-β, are now recognized to contribute to T cell responses 140. These cytokines orchestrate aspects of T cell differentiation that are non-redundant with signals 1 and 2 and can also influence bystander memory T cells, an effect that may be relevant to anti-tumor responses in some settings.
Cytokines that can function as growth and survival factors also have a critical role as a form of signal 3, primarily members of the common γ-chain family (IL-2, IL-7, IL-9, IL-15, and IL-21). Although the effect of each cytokine varies based on receptor expression and differentiation state, these cytokines collectively regulate cell cycle, survival, and/or homeostasis. In cancer immunology, these cytokines, especially IL-2, have long been appreciated and exploited 141. Combining PD-1 blockade with IL-2 has a potent effect on modulating T cell exhaustion and enhancing anti-tumor immunity 107, 108, 109, 142, 143 and numerous ongoing trials are aimed at exploiting the biology of IL-2 in cancer immunity 144. In contrast, Texh cells typically express lower IL-7R than Tmem cells resulting in suboptimal responses to IL-7 compared to other competing T cells in vivo 27. Nevertheless, IL-7 treatment during chronic viral infection in mice can expand the Texh population and moderately improve function 145, 146, including in combination with PD-1 blockade 31. IL-15 may be another important common γ-chain cytokine for Texh cells, and targeting this cytokine may achieve some similar effects to IL-2 based on the shared use of IL-2Rβ. IL-21R is necessary for Texh cells to maintain functionality and control during chronic viral infection 147, 148, 149, and IL-21 can augment ex vivo expansion of tumor-specific CD8 T cells 150. This effect is likely due to a requirement for IL-21 produced by CD4 T cells to promote generation of the Texh-int subset which contains more functional cells within the exhausted hierarchy 63. Indeed, treatment with a tumor-targeting IL-21 fusion protein expanded cytokine-producing PD1IntTim-3neg CD8+ T cells resulting in tumor control 151 and IL-21 combined with αCTLA-4 may have beneficial effects in patients 152. Cellular engineering approaches have also exploited common γ-chain cytokines to influence T cell state. The use of IL-7, IL-15, and IL-21 during CAR T cell production in vitro can promote memory differentiation and maintain proliferative capacity in infusion products 153. In addition, some CAR T cells are being designed to secrete specific cytokines to augment their function in an autocrine manner but also enhance anti-tumor immunity through effecting in the local TME 154. Despite the potential of common γ-chains to modulate (and improve) T cell function and differentiation, consistent clinical benefit has been challenging to achieve on a broad scale, likely due to complexities in drugging these cytokines and their receptors selectively on the T cell subsets of interest. Nevertheless, common γ-chain cytokines remain prime targets for therapeutic manipulation.
Particularly important for T cell biology in cancer are cytokines with regulatory properties, such as TGF-β and IL-10. These cytokines can be produced by a variety of cells, including Treg cells and suppressive myeloid cells, and are often found at high levels within the TME. Although TGF-β and IL-10 are generally considered to suppress CD8+ T cell proliferation and function140, they can also have potentially pro-immune/anti-tumor effects. For example, in chronic infection Texh cells express high levels of TGF-β and downstream phosphorylation of SMADs. Attenuation of TGF-β results in decreased expression of the proapoptotic protein BIM resulting in increased cell numbers155. In addition, TGF-β can promote Texh-term differentiation156 and enhance PD-1 expression on CD8+ TIL via SMAD3 157. These data demonstrate a generally suppressive effect of TGF-β. However, TGF-β is also required for maintaining the stem-like capacity of Texh-prog cells through suppression of mTOR signaling and preservation of cellular metabolism 158. TGF-β also plays a major role in the formation and maintenance of CD103+ Trm cells 10, 12. Thus, tumor-derived TGF-β could enhance tissue-residency programs which has the potential to have pro-immune and anti-tumor effects. For example, ex vivo treatment of CAR T cells with TGF-β increased stem-like and residency properties resulting in increased tumor infiltration 159. However, TGF-β also promotes the retention of Texh-prog cells in secondary lymphoid organs by regulating migration-related proteins 160 which can prevent migration from tumor-draining lymph nodes (TDLNs) to tumor after vaccination 161. Paradoxical roles for IL-10 have also been observed. IL-10 is typically considered to be immunosuppressive, but treatment with PEGylated IL-10 can activate Texh cells and induce tumor regression in cancer patients 162. This effect may be due, at least in part, to promoting OXPHOS in Texh-term subset 163. These data highlight the complex effects of IL-10 and TGF-β beyond traditional roles in regulation and draw attention their influence altering metabolism and ultimately T cell function. Nevertheless, there are also strategies to block these regulatory cytokines 164 or convert them to positive activation signals 165.
Overall, it is now clear that signal 3 is required for optimal T cell differentiation. However, the net effect of cytokine signaling on CD8+ T cell differentiation is complex, temporally regulated, and likely interactive with other signals. This complexity is highlighted by the example of IFN signaling. Although IFN-α/β serves as a critical positive signal for CD8+ T cells during priming, in Texh cells IFN-α/β may drive the differentiation to terminal exhaustion and prevent tumor clearance 55, 80. In addition, IFN-α/β can promote cell death through the Fas/FasL pathway 166. Furthermore, persistent IFN-α/β and IFN-γ signaling induces negative regulators of T cell function, such as PD-L1, on various cell types including tumor cells, Treg cells, and suppressive myeloid cells 167. As a result, type I and II IFN represents a double-edged sword with both positive and negative effects on tumor immunity 168. These complexities highlight the need for a nuanced understanding of the role of signal 3 and cytokines on CD8+ T cells to optimize immunotherapeutic strategies and overcome resistance mechanisms.
Signal 4: Nutrients
Over the past decade, the existence of a crucial fourth set of signals that regulate T cell function and differentiation has become increasingly apparent – metabolic fuels and nutrients, like glucose, amino acids (AAs) and lipids. The importance of this set of signals becomes particularly evident in the TME. Here, the metabolic activities of many cells, especially the tumor cells, alter the nutrient landscape, and these changes can metabolically suppress T cell function and enhance exhaustion. This signal 4 also presents a new opportunity for therapeutic intervention, and treatments that metabolically rewire T cells, tumor cells, or other cells in the TME have the potential to improve anti-tumor immunity.
In a healthy setting, the nutrients that a T cell consumes and the metabolic pathways it employs change throughout differentiation to meet its energetic needs. These changes not only affect T cell bioenergetics, but also alter gene expression via epigenetic and post-translational protein modifications that, in turn shape T cell effector function and differentiation state. Naive T cells primarily rely on mitochondrial respiration driven by IL-7 dependent glucose use 169. As T cells become activated, they need to generate biomass to support activation, proliferation, and logarithmic increase in cell numbers, as well as effector functions. This metabolic switch is driven largely by the combined effects of signals 2 and 3 that stimulate signaling pathways, such as PI3K → AKT→ MTORC1, and is regulated by TFs, including HIF1, MYC and STAT5 170, 171, 172. This anabolic phase is characterized by robust glycolytic activity, one-carbon metabolism, fatty acid synthesis and mitochondrial respiration and biogenesis, and is fueled by the large consumption of nutrients like glucose, glutamine, leucine, arginine and imported fatty acids 173. As T cells return to quiescence and develop into Tmem cells, they switch metabolic activities to synthesizing and storing triglycerides and burning fatty acids and ketones 174, 175, 176. Mitochondrial fitness therefore is a key feature of long-lived Tmem cells.
In contrast, Texh cells have an altered metabolic profile. These alterations include decreased mitochondrial mass, membrane potential, and oxygen consumption, whereas ROS production is increased 177, 178, 179, 180, 181. Texh-prog cells, however, have improved mitochondrial function compared to Texh-term cells, consistent with the lack of proliferative capacity of the latter subset 158. Mitochondrial cristae in Texh cells also appear damaged, diffuse and swollen, but increasing mitochondrial biogenesis via PGC-1α can overcome some of the poor metabolic features associated with T cell exhaustion 178, 179, 182. The decreased mitochondrial function of Texh cells is coupled to PD-1 driven impairment of glycolysis, an effect that is associated with a greater reliance of Texh cells on lipids as they become more catabolic and autophagic 183, 184. In tumors, TILs can exhibit increased lipid import, particularly of oxidized lipids, through the expression of the scavenger receptor CD36 185, 186, 187. This increased lipid uptake leads to increased lipid peroxidation, p38 signaling and ferroptosis, effects that can be reversed by overexpression of glutathione peroxidase 4 leading to improved TIL survival and function 186, 187. Additionally, pharmacologically improving fatty acid use by TILs with peroxisome proliferator-activated receptor agonists enhances anti-tumor immunity and responses to ICB 188, 189. There is also evidence that T cells can use the microbiota-derived metabolites inosine or acetate, which can improve anti-tumor efficacy and responsiveness to ICB 190, 191, 192.
The Tumor Metabolic Landscape
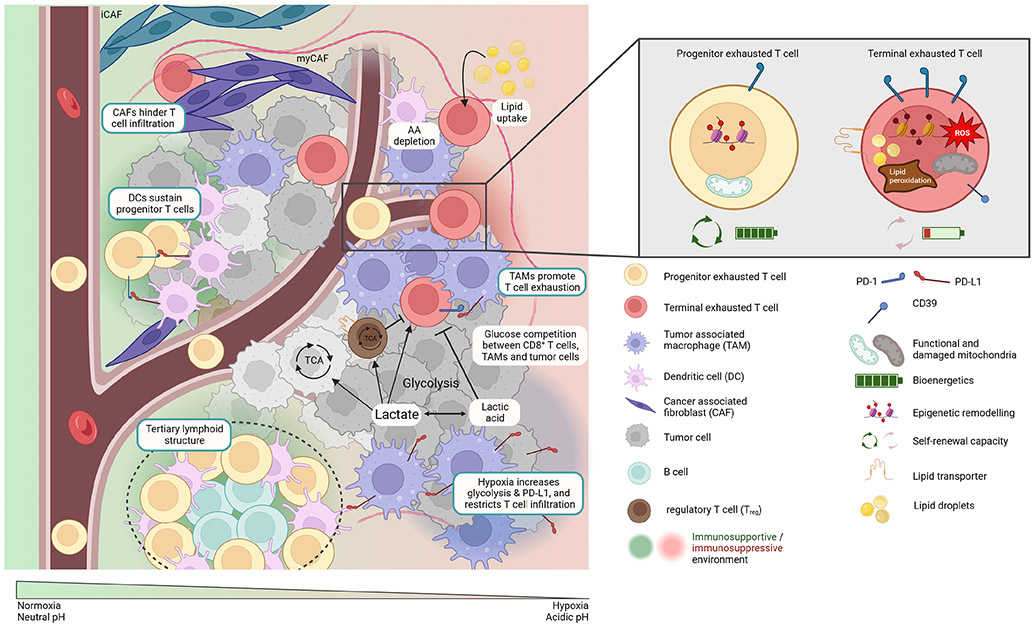
The metabolic states of different cell types and their fuel preferences can create different metabolic ecosystems within the TME (Figure 3). Tumor cells have high nutrient demands to support their rapid proliferation, and their aberrant consumption and production of metabolites alters the metabolic composition of the TME. Although recent work suggests that primary tumors might not be as hypermetabolic as once thought 193, the metabolic cross-talk between tumor cells, TILs, and other cell types within the TME can further influence the metabolic state of CD8+ TILs, that in turn, affects their differentiation and function. In healthy organs, resident immune cells including Trm cells likely develop cooperative or complementary metabolic relationships with neighboring tissue cells. In contrast, in tumors, a metabolic tug-of-war occurs between the tumor cells, stromal cells, and immune cells with competition for key nutrients, such as glucose, that forces TILs to adopt metabolic states that limit tumor control. As tumors evolve, specific metabolic states may be selected for that not only promote tumor growth and invasion, but also suppress or evade T cell attack. Indeed, nutrient availability, oxygen levels, pH, and immunosuppressive metabolites in tumors can all affect CD8+ T cell infiltration, survival, and antitumor functions, presenting challenges for rejuvenating immune responses through immunotherapy.
Figure 3: Metabolic ecosystems in the TME.
The metabolic states of different cell types and their fuel preferences, as well as histological landmarks in the tumor microenvironment (TME) such as vasculature can create metabolic niches. These niches in combination with the cellular interactions of T cells with other immune and non-immune cell types generate immunosupportive (green) or immunosuppressive (red) conditions, that can influence T cell differentiation and function. Progenitor exhausted and more terminal exhausted T cells display unique metabolic characteristics.
Historically, considerable attention was directed towards glucose use by tumor cells, largely due to the seminal work of Otto Warburg. However, glucose partitioning in tumors based on FDG-PET-tracing reveals that tumor associated macrophages (TAMs) also consume substantial amounts of glucose 194, suggesting a rethinking of glucose use in tumors. Nonetheless, TMEs with high glycolytic activity are often associated with reduced T cell infiltration, reduced IFN-γ production, and poor response to TIL therapy 195, 196, 197, 198, 199. One possibility is direct competition between cancer cells, TAMs, and T cells for the limited supply of glucose in the TME 196, 197. Indeed, deprivation of glucose and/or poor metabolism downstream of glucose uptake are hallmarks of Texh cells and poor TIL functionality. In contrast to Teff or Texh cells, Treg cells remain functional (ie., suppressive) under the low glucose conditions in the TME 200, 201. This imbalance in Treg:Teff functions in highly glycolytic tumors likely contributes to their noteworthy resistance to ICB and TIL therapy 199, 202, 203. Notably, hypoxia is another tumor feature linked to ICB resistance 204. In addition to increasing glycolysis, hypoxia also increases expression of PD-L1 on both immune and tumor cells, and restricts T cell infiltration into hypoxic zones 205, 206, 207, 208. Finding ways to overcome immunosuppressive hypoxic ecosystems in tumors to increase ICB responsiveness will be key. Mitochondrial toxins, like metformin, or anti-angiogenics, like VEGF-inhibitors, aim to achieve this goal and have demonstrated beneficial outcomes with increased TILs in certain cancer types like kidney, liver, and lung cancer, but not in others 209.
A byproduct of increased glycolysis is the production of lactate and lactic acid. Lactic acid secretion acidifies the TME, an effect that may exacerbate T cell exhaustion 199, 210, 211, 212, 213. However, tumors and T cells can also use lactate as a carbon source for oxidative metabolism, and LDH-A is necessary for Teff function 214, 215, 216. Quantifying the contribution of different TME cell populations to tumor glycolysis together with other approaches such as spatial metabolomics may provide new opportunities to better understand the glycolysis-lactate-hypoxia axis in solid tumors.
Tumors further consume large amounts of AAs like glutamine, and the regulation of other AA availability and/or metabolism in the TME can impact T cell biology. For example, excessive uptake of glutamine by cancer cells can interfere with optimal T cell responses since T cells require glutamine to provide carbon and nitrogen for anabolic metabolism upon activation 217. However, whereas cancer cells tend to be detrimentally affected by glutamine blockade, T cells display metabolic plasticity and can shift their metabolism to retain anti-tumor function 218, 219. Glutamine and glucose metabolism are linked, and increased glutamine uptake can suppress glucose metabolism whereas under glutamine restriction, T cells increase glucose uptake 194. The link between these metabolic pathways highlights a potentially important immunoregulatory metabolic axis in the TME. Additionally, immunoregulatory immune cells also shape the intratumoral metabolic environment. For example, TAMs import AAs and express enzymes like arginase or indoleamine 2,3-dioxygenase (IDO) that hydrolyze AAs into T cell suppressive derivatives 220. Thus, both tumor cells and other immune cells can contribute to a metabolically hostile immune environment through AA-dependent processes including AA consumption and conversion into immunosuppressive molecules.
While some nutrients are limited in the TME, others like lipids, especially oxidized lipids, appear more abundant 186. Increased lipid uptake and peroxidation can limit T cell function and survival, yet foster Treg activity in tumors 186, 187, 221, 222, 223. On the other hand, tumors that excessively burn lipids through fatty acid oxidation may be sensitized to ICB 224, and perhaps such activities limit the availability of oxidized lipids to T cells. The lipid Prostaglandin E2 signals via the receptors EP2 and EP4 to suppress survival and function of T and NK cells. Indeed, reducing PGE2 production via Celecoxib synergizes with ICB225, 226, 221, 228. Notably, PGE2 production can be affected by dietary lipids, suggesting a potential mechanism contributing to the increased risk of cancer caused by obesity and diets rich in sugars and fats229.
Tumor-Immune Ecosystems Within the TME
In addition to the diverse metabolic ecosystems, microenvironments of cellular interactions exist within the TME (also discussed in this issue 230). Besides cancer cells and T cells, the TME is comprised of many other immune and non-immune cells, including stromal cells (like cancer associated fibroblasts (CAFs)), blood vessels, neurons, and extracellular matrix (ECM). The interactions between these diverse cell types give rise to unique ecosystems, niches within tumors, which will likely emerge as regulators of tumor growth, invasion, and T cell suppression. It is becoming increasingly clear, that pathological landmarks such as fibrosis, hypoxia, necrosis, vascularization, tumor cell heterogeneity, and TLSs are associated with differences in local TIL composition and functions. High resolution spatial mapping of cell types and non-cellular structures within the TME is revealing conserved cellular networks and transcriptional programs and these insights will be crucial for advancing our comprehension of tumor immunology and tailoring effective therapeutic strategies.
Distinct tumor “archetypes”, “ecosystems”, or “hubs” which are organized cellular neighborhoods composed of variations in T cells, APCs, and stromal cells have been proposed that collectively impact antitumor T cell responses and consequently patient survival and response to therapy 231, 232, 233, 234, 235, 236, 237. Immune-rich archetypes are dominated by CD8+ T cells and further divided into macrophage-enriched (associated with Texh cells) or NK and conventional type 1 dendritic cell (cDC1)-enriched subtypes (associated with Teff cells) or CD4+ T cell and conventional type 2 dendritic cell (cDC2)-enriched archetypes 85, 231, 237, 238. Tumors can exhibit multiple ecosystems simultaneously, that can communicate with each other 231, and can change over time and in response to therapeutic interventions. While the dominant ecosystem in tumors is typically immunosuppressive, ecosystems that are immuno-stimulatory, such as TLSs, can exist within the same tumor. ICB and other immune-based interventions aim to shift the TME from a pro-tumorigenic environment to a more immunosupportive environment. Collectively, these data indicate that aside from the frequency of certain immune cell types, it is their spatial localization within the TME that is relevant for the generation of productive anti-tumor immune responses. Understanding the spatial dimensions of the TME will allow us to move beyond simplistic classifications of “hot” and “cold” tumors based solely on the quantity of intratumoral TILs.
APC-T cell interactions within the TME determine the efficiency of T cell responses by influencing signals 1 and 2 (and perhaps also 3 and 4), and the type of APC can determine the resulting T cell response 239. APCs exist along a spectrum, ranging from T cell suppressive TAMs to specific subsets of conventional dendritic cells (cDCs) (also discussed in this issue 240) that support anti-tumor T cells in some cases and tolerize in others. APC-T cell interactions can be geographically categorized into three primary locations: (1) local interactions within the tumor bed, (2) interactions within peritumoral areas including TLSs, and (3) distal interactions in the TDLNs.
Within solid tumors, the most prevalent APCs are typically TAMs derived from either tissue-resident macrophages or circulating monocytes that mature intratumorally241. TAMs are heterogeneous and can adapt to their surrounding environment. Antigen-specific T cells generally reside in TAM-rich regions of the TME and T cell-TAM interactions contribute to T cell exhaustion programs or prevent TIL infiltration into the tumor nests 242, 243, 244 In contrast, cDCs, although more rare, are a prognostic marker and correlate with the number of stem-like Texh-prog cells 60, 245, 246. cDCs can be divided into cDC1s that predominantly promote CD8+ T cell responses and cDC2s that support CD4+ T cell responses, though cDCs can likely support either T cell type 247, 248. Although both TAMs and cDCs express PD-L1, it appears that cDC-derived PD-L1 suppresses TILs more dominantly than TAM-derived PD-L1, making cDCs an important target of PD-L1 blockade249. cDC1-T cell interactions also help maintain Texh-prog, and NK cells can help recruit cDC1s via Flt3, with both cell-cell interactions linked to ICB responsiveness 250, 251. In addition to cDC1 interactions, intratumoral Texh-prog differentiation into Texh-int subsets can be enhanced by CD4+ T cells and mreg DCs (dendritic cells enriched in maturation and regulatory molecules) in distinct intra-tumoral niches 63, 65, 104. Structural cells like MHCII+ CAFs can also serve as APCs. Other CAF subtypes including myofibroblastic CAFs and inflammatory CAFs can develop different spatio-functional relationships around tumor nests 252. CAFs can impair T cell responses via multiple mechanisms, such as altering ECM density and composition, preventing TIL trafficking into tumor beds (often called TIL-‘excluded’ tumors), and promoting T cell exhaustion and apoptosis 253, 254, 255, 256, 257.
Peritumoral regions can often be more immune rich than deeper intratumoral areas. TLSs in these regions appear to be critical for facilitating intratumoral immune responses. Indeed, TLSs support Texh-prog biology and may enable Texh-prog cell activation and differentiation upon ICB. In fact, these structures often correlate with better prognosis and/or response to immunotherapy. Specifically, the presence of PD-1+CXCL13+ CD8 T cells is associated with responses to PD-1 blockade 70, 95, 236, 258, 259. In addition to TLSs, proliferating Texh-prog-like cells can co-cluster with APCs in semi-organized structures, and this clustering can be associated with better disease outcome 60, 260. It is likely that these organized or semi-organized lymphoid aggregates require additional support from stroma. Indeed, some CAFs may be involved in orchestrating formation of TLSs 261.
Immunotherapy can also activate immune responses distally in TDLNs 262. Texh-prog-like cells that are clonally related to Texh cells in the tumor migrate from the TDLNs to the tumor, sustain the anti-tumor T cell response, and eventually undergo differentiation to terminal exhaustion 58, 64, 66, 263, 264. Although some data from animal models using FTY720, a drug that blocks T cell migration, suggest that responses to ICB can occur without contribution from TDLNs 265, in most settings, it appears that some contribution from regional LN is important 64, 262, 264, 266. TDLNs are often surgically removed due to the risk of metastasis. Given this emerging geographic understanding of Texh-prog biology and the role of TDLNs, re-evaluating how to maintain optimal anti-tumor T cell responses from TDLNs during the initiation of immunotherapy may be warranted in some cases 64, 267. Tumor metastases in the TDLNs, however, impair ICB responses by altering the TDLN architecture and promoting the generation of Texh-term cells 64, 268. Targeting TDLNs suggests new therapeutic opportunities including strategies to promote migration of tumor-specific T cells from TDLNs, or cancer vaccines that target TDLNs to improve activation of the lymphoid anti-tumor immune response. Thus, the appreciation that the systemic immune response might contribute to anti-tumor immunity not only broadens the opportunities for therapeutic interventions, but is also consistent with the ability to detect anti-tumor T cell responses in blood in many settings 239. During the last decade, we have significantly improved the geographic and temporal resolution of our understanding of the TME and note the importance of incorporating space and time into the concept of signals 1-4.
New concepts for CD8 T cells in the Cancer Immunity Cycle
Over the past decade, in part due to the unparalleled progress in immunotherapy, significant strides have been made in understanding the role of the immune system role in cancer. The successes of T cell-based immunotherapies provoke three new concepts that could frame the next advances in harnessing T cells for treatment, cure, and prevention of cancer.
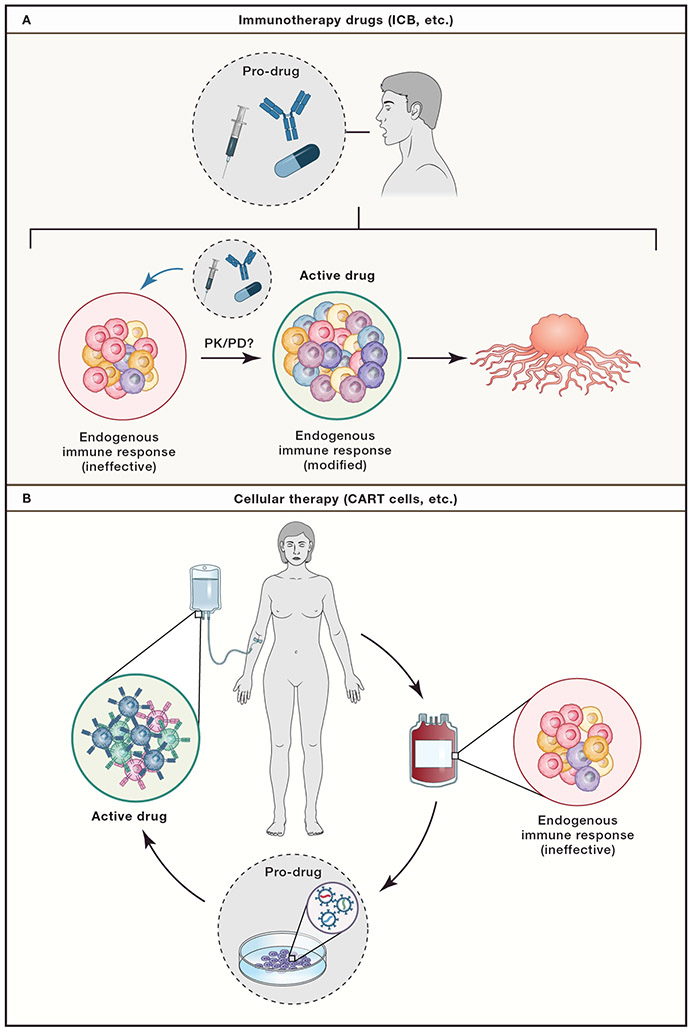
First, the increased mechanistic knowledge regarding the actions of many immunotherapies suggests that the therapies delivered to patients are “pro-drugs” (Figure 4). The T cells (and perhaps other immune cells) that are reinvigorated, induced, or otherwise manipulated are the actual in vivo drug. For engineered cellular therapies, the “pro-drug” phase has been skipped - or at least performed ex vivo. Such a concept of pro-drug and drug has implications for immune-pharmacodynamics. This concept highlights the need to examine and define pharmacological principles (pharmacokinetic/pharmacodynamic, PK/PD) for the T cell targets themselves in addition to the pro-drug molecules (i.e. ICB antibodies). Defining the kinetics, magnitude, and duration of the target T cell population (such as Texh cells) as well as specific anti-tumor effector functions (cytokine production, etc.), can then be used to further optimize properties of the pro-drugs. For example, the burst of CD8+ T cell reinvigoration that occurs following PD-1 pathway blockade typically peaks at ~2-4 weeks. Such a pharmacodynamics suggest that combination therapies intended to capitalize on changes to the in vivo drug (i.e. the reinvigorated CD8+ T cells) should be delivered during this window of response. Dose regimens and combination therapies could be re-imagined if immune-pharmacodynamics were known in detail.
Figure 4: The immune system is the active drug component of immunotherapy.
A. Immunotherapies, such as ICB, represent pro-drugs that are taken by patients. These pro-drugs (i.e. αPD-1) act on cells in the endogenous immune system, such as ineffective Texh cells. These modified endogenous cells (such as “reinvigorated” Texh cells) then represent the active drug which mediates the anti-tumor activity. In this scenario, traditional pharmacology concepts such pharmacokinetic/pharmacodynamic (PK/PD) measurements and modeling can be applied to the active drug, the modified immune cells, instead of the pro-drug. In this case, the kinetics, magnitude, and duration of the T cell response would be treated as the measurements of interest. B. Cellular therapy, such as CAR T cells, or other engineered cells including TIL, are manufactured from a pro-drug (ineffective and unmanipulated endogenous CD8+ T cells) to an active drug in vitro (i.e. CART19) which are then transferred in vivo to mediate tumor killing.
Second, we have witnessed a major shift in human immunology research from primarily observational studies to attempting true mechanistic science in patients and healthy donors. This transformation can be traced, in part, to major investments and advancements made during the height of the HIV crisis in the last century but has also been accelerated by the ever-increasing capacity for high dimensional analysis, and more recently advancements spurred on by the SARS-CoV-2 pandemic. Studies in human cancer patients receiving immunotherapies have significantly contributed to these advances in human immunology by providing opportunities to capture temporal immune changes associated with therapeutic interventions, such as ICB. The ability to capture the window of immune dynamics upon initiation of immune-based treatments has been a key to some of these insights, for example, in neoadjuvant ICB studies where changes in the intratumoral T cell response can be examined shortly after treatment initiation. These data have led to transformational insights into the mechanisms of CTLA-4 and PD-1, the action of CAR T cells and TILs. Thus, a tremendous opportunity for the future is to continue to capture these interventions and deeply interrogate the mechanisms not only of clinical response to the intervention, but the broader impact of these immunotherapies on the immune system.
Third, a major emerging concept is that overall “immune health”, including immune history and lifestyle, plays a substantial role in shaping potential immune responsiveness to future interventions. It is becoming increasingly clear that biologic sex, hormones, and age can have a substantial impact on baseline immune health status and ability to respond to perturbations such as cancer or infections. In addition, we now appreciate that many other non-genetic factors influence future immune responses. There is strong evidence for major immunological events including previous infections 269, chemotherapy treatment, or immunosuppression impacting responses to vaccines or other immune-based therapies. There is also emerging data that concomitant medication may shape immune responses to cancer. Similarly, the immunological role of the microbiome in cancer immunotherapy has received considerable attention, provoking obvious questions about the use of antibiotics with cancer immunotherapy 270, 271. Such data also highlight the role of diet, and emerging evidence supports dietary influences, including fiber intake, on ICB outcomes through effects on the microbiome 272. The human immune system is shaped by a lifetime of interactions with the microbial and external antigen world, many of which form durable imprints (i.e. immunological memory) that shape future responses. More broadly, however, lifestyle factors including obesity, fasting behavior, circadian biology, chronological versus biological age and many other variables are likely to influence immune fitness. Together, these durable and/or malleable features of individual immune systems form the basis of how an individual patient will respond to a future intervention. A major opportunity for the future is to assess how immune health characteristics of patients relate to T cell responses to immunotherapy and use this information to tailor our interventions to the needs of individuals.
Concluding Remarks
Our ability to deeply profile both the immune and the non-immune cells in tumors can help identify which part(s) of the Cancer Immunity Cycle require intervention to improve tumor control. Examining such information from individual patients in the context of the signals 1-4 that control T cell differentiation and function should provide guidance for personalized immune-based therapies. For example, a dearth of activated T cells in a tumor might suggest that providing additional signal 1 through antigen-targeted therapies such as vaccination could be useful. Absence of inflammatory DCs could indicate αCD40L, perhaps in combination with vaccination, could help activate APCs and provide more optimal signals 1,2 and even 3 to induce an endogenous tumor-specific T cell response. In contrast, if a tumor is highly infiltrated with Texh cells, ICB such as αPD-1 alone or in combination with other therapies, such as αLAG-3 or IL-2 treatment (further decreasing inhibitory signal 2 or augmenting signal 3) could stimulate these tumor-specific T cells to enhance tumor cell killing. Moreover, therapeutically addressing signal 4 from metabolism and nutrients provides additional opportunities for individual or combination immunotherapy. A major goal for the future will be to integrate broader data about overall immune health and immune fitness with detailed tumor immunity and tumor genetics data, at baseline and on-treatment to guide treatment choices.
Thus, the last decade has seen a true transformation of our understanding of CD8+ T cells not only as key targets for our immunotherapies, but as critical in vivo drugs. Our knowledge of CD8+ T cell differentiation and developmental biology, while still incomplete, has improved our understanding of how immunotherapies like ICB function, and has also enabled considerable progress in engineering CAR T cells and TCR engineered cellular therapies. A conceptual framework of a 4-signal model of T cell activation together with an ever improving map of the differentiation trajectories of CD8+ T cells should continue to provide a foundation for understanding and improving cancer immunity.
Acknowledgments
This work was supported by grants from the NIH AI105343, AI082630, AI108545, AI155577, and AI149680 (to EJW); HL143613, T32 CA009140, Cancer Research Institute-Mark Foundation Fellowship, and the Parker Institute for Cancer Immunotherapy (to JRG, EJW); R01 CA230275, P01 AG073084, and the Mark Foundation (to SMK), P30 CA014195 (R. Shaw); NOMIS Center Postdoctoral Fellowship and German Research Foundation (Deutsche Forschungsgemeinschaft, GL 991/1-1, to AMG). Work in the Wherry laboratory is supported by the Parker Institute for Cancer Immunotherapy.
Footnotes
Declaration of Interests
EJW is a member of the Parker Institute for Cancer Immunotherapy. EJW is an advisor for Danger Bio, Janssen, New Limit, Marengo, Pluto Immunotherapeutics Related Sciences, Santa Ana Bio, Synthekine, and Surface Oncology. EJW is a founder of and holds stock in Surface Oncology, Danger Bio, and Arsenal Biosciences. JRG is a consultant for Arsenal Biosciences. SMK is on the scientific advisory boards and has equity in EvolveImmune Therapeutics, Affini-T Therapeutics, Arvinas, and Pfizer. AMG has no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen D & Mellman I Oncology meets immunology: The cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Plitas G & Rudensky AY Regulatory T Cells in Cancer. Annual Review of Cancer Biology 4, 459–477 (2020). [Google Scholar]
- 3.Togashi Y, Shitara K & Nishikawa H Regulatory T cells in cancer immunosuppression — implications for anticancer therapy. Nature Reviews Clinical Oncology 16, 356–371 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Speiser DE, Chijioke O, Schaeuble K & Münz C CD4(+) T cells in cancer. Nat Cancer 4, 317–329 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Kaech SM et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Huster KM et al. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc Natl Acad Sci U S A 101, 5610–5615 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi NS et al. Inflammation Directs Memory Precursor and Short-Lived Effector CD8(+) T Cell Fates via the Graded Expression of T-bet Transcription Factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jameson SC & Masopust D Understanding Subset Diversity in T Cell Memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buggert M, Price DA, Mackay LK & Betts MR Human circulating and tissue-resident memory CD8(+) T cells. Nat Immunol 24, 1076–1086 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Mackay LK et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat Immunol 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Gebhardt T, Palendira U, Tscharke DC & Bedoui S Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol Rev 283, 54–76 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Sheridan BS et al. Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity 40, 747–757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konjar Š, Ficht X, Iannacone M & Veldhoen M Heterogeneity of tissue resident memory T cells. Immunology Letters 245, 1–7 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Masopust D. et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med 207, 553–564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schietinger A. et al. Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 45, 389–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blattman JN, Wherry EJ, Ha SJ, van der Most RG & Ahmed R Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J Virol 83, 4386–4394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Utzschneider DT et al. High antigen levels induce an exhausted phenotype in a chronic infection without impairing T cell expansion and survival. J Exp Med 213, 1819–1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan O. et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott AC et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfei F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Yao C. et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat Immunol 20, 890–901 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proceedings of the National Academy of Sciences 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Zajac AJ et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188, 2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallimore A. et al. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med 187, 1383–1393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Leun AM, Thommen DS & Schumacher TN CD8+ T cell states in human cancer: insights from single-cell analysis. Nature Reviews Cancer 20, 218–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLane LM, Abdel-Hakeem MS & Wherry EJ CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Baitsch L. et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. The Journal of Clinical Investigation 121, 2350–2360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmadzadeh M. et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114, 1537–1544 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wherry EJ et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Pauken KE et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sen DR et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott-Browne JP et al. Dynamic Changes in Chromatin Accessibility Occur in CD8+ T Cells Responding to Viral Infection. Immunity 45, 1327–1340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philip M. et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doering TA et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mognol GP et al. Exhaustion-associated regulatory regions in CD8(+) tumor-infiltrating T cells. Proc Natl Acad Sci U S A 114, E2776–e2785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virgin HW, Wherry EJ & Ahmed R Redefining chronic viral infection. Cell 138, 30–50 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Wherry EJ T cell exhaustion. Nat Immunol 12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Paley MA et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speiser DE et al. T cell differentiation in chronic infection and cancer: functional adaptation or exhaustion? Nat Rev Immunol 14, 768–774 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Chen Z. et al. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51, 840–855.e845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frebel H. et al. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J Exp Med 209, 2485–2499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz JE et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283, 857–860 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Jin X. et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189, 991–998 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasprowicz V. et al. Hepatitis C virus (HCV) sequence variation induces an HCV-specific T-cell phenotype analogous to spontaneous resolution. J Virol 84, 1656–1663 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamieson BD et al. Epitope escape mutation and decay of human immunodeficiency virus type 1-specific CTL responses. J Immunol 171, 5372–5379 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Huang AC et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med 25, 454–461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sade-Feldman M. et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 176, 404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunn GP, Old LJ & Schreiber RD The Three Es of Cancer Immunoediting. Annual Review of Immunology 22, 329–360 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Blackburn SD, Shin H, Freeman GJ & Wherry EJ Selective expansion of a subset of exhausted CD8 T cells by aPD-L1 blockade. Proc Natl Acad Sci U S A 105, 15016–15021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He R. et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Im SJ et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leong YA et al. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nature Immunology 17, 1187–1196 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Utzschneider Daniel T. et al. T Cell Factor 1-Expressing Memory-like CD8+ T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Wu T. et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller BC et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siddiqui I. et al. Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e110 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Schenkel JM et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1(+) CD8(+) T cells in tumor-draining lymph nodes. Immunity 54, 2338–2353.e2336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brummelman J. et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J Exp Med 215, 2520–2535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jansen CS et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beltra JC et al. Developmental Relationships of Four Exhausted CD8(+) T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841.e828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hudson WH et al. Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity 51, 1043–1058.e1044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zander R. et al. CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042.e1024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahim MK et al. Dynamic CD8(+) T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes. Cell 186, 1127–1143.e1118 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magen A. et al. Intratumoral dendritic cell-CD4(+) T helper cell niches enable CD8(+) T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat Med 29, 1389–1399 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Connolly KA et al. A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Science Immunology 6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pai JA et al. Lineage tracing reveals clonal progenitors and long-term persistence of tumor-specific T cells during immune checkpoint blockade. Cancer Cell 41, 776–790.e777 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Li H. et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789.e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveira G. et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duraiswamy J. et al. Myeloid antigen-presenting cell niches sustain antitumor T cells and license PD-1 blockade via CD28 costimulation. Cancer Cell 39, 1623–1642.e1620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaglia G. et al. Lymphocyte networks are dynamic cellular communities in the immunoregulatory landscape of lung adenocarcinoma. Cancer Cell 41, 871–886.e810 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen JH et al. Spatial analysis of human lung cancer reveals organized immune hubs enriched for stem-like CD8 T cells and associated with immunotherapy response. bioRxiv (2023). [Google Scholar]
- 74.Giles JR et al. Shared and distinct biological circuits in effector, memory and exhausted CD8(+) T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat Immunol 23, 1600–1613 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casado JG et al. CD8 T cells expressing NK associated receptors are increased in melanoma patients and display an effector phenotype. Cancer immunology, immunotherapy : CII 54, 1162–1171 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daniel B. et al. Divergent clonal differentiation trajectories of T cell exhaustion. Nature Immunology 23, 1614–1627 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Crawford A, Angelosanto JM, Nadwodny KL, Blackburn SD & Wherry EJ A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog 7, e1002098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vignali PDA et al. Hypoxia drives CD39-dependent suppressor function in exhausted T cells to limit antitumor immunity. Nature Immunology 24, 267–279 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kasmani MY et al. Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection. Journal of Experimental Medicine 220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giles JR et al. Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 55, 557–574.e557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshida H. et al. The cis-Regulatory Atlas of the Mouse Immune System. Cell 176, 897–912.e820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng C. et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356.e1316 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Braun DA et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 39, 632–648.e638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schumacher TN, Scheper W & Kvistborg P Cancer Neoantigens. Annu Rev Immunol 37, 173–200 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Simoni Y. et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Caushi JX et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126–132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lowery FJ et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science 375, 877–884 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burger ML et al. Antigen dominance hierarchies shape TCF1(+) progenitor CD8 T cell phenotypes in tumors. Cell 184, 4996–5014.e4926 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lanitis E, Dangaj D, Irving M & Coukos G Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann Oncol 28, xii18–xii32 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Chiou S-H et al. Global analysis of shared T cell specificities in human non-small cell lung cancer enables HLA inference and antigen discovery. Immunity 54, 586–602.e588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meier SL, Satpathy AT & Wells DK Bystander T cells in cancer immunology and therapy. Nat Cancer 3, 143–155 (2022). [DOI] [PubMed] [Google Scholar]
- 94.Hudson WH & Wieland A Technology meets TILs: Deciphering T cell function in the - omics era. Cancer Cell 41, 41–57 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thommen DS et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 24, 994–1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumagai S. et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat Immunol 21, 1346–1358 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Daud AI et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 126, 3447–3452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Terranova-Barberio M. et al. Exhausted T cell signature predicts immunotherapy response in ER-positive breast cancer. Nat Commun 11, 3584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chalabi M. et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med 26, 566–576 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Attrill GH et al. Detailed spatial immunophenotyping of primary melanomas reveals immune cell subpopulations associated with patient outcome. Front Immunol 13, 979993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barber DL et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006). [DOI] [PubMed] [Google Scholar]
- 102.Wong RM et al. Programmed death-1 blockade enhances expansion and functional capacity of human melanoma antigen-specific CTLs. Int Immunol 19, 1223–1234 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Gill AL et al. PD-1 blockade increases the self-renewal of stem-like CD8 T cells to compensate for their accelerated differentiation into effectors. Science Immunology 8, eadg0539 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu B. et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nature Cancer 3, 108–121 (2022). [DOI] [PubMed] [Google Scholar]
- 105.Huang AC et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carlino MS, Larkin J & Long GV Immune checkpoint inhibitors in melanoma. Lancet 398, 1002–1014 (2021). [DOI] [PubMed] [Google Scholar]
- 107.Codarri Deak L. et al. PD-1-cis IL-2R agonism yields better effectors from stem-like CD8(+) T cells. Nature 610, 161–172 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hashimoto M. et al. PD-1 combination therapy with IL-2 modifies CD8(+) T cell exhaustion program. Nature 610, 173–181 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beltra J-C et al. Enhanced STAT5a activation rewires exhausted CD8 T cells during chronic stimulation to acquire a hybrid durable effector like state. bioRxiv, 2022.2010.2003.509766 (2022). [Google Scholar]
- 110.Haen SP, Löffler MW, Rammensee H-G & Brossart P Towards new horizons: characterization, classification and implications of the tumour antigenic repertoire. Nature Reviews Clinical Oncology 17, 595–610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aleksic M. et al. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur J Immunol 42, 3174–3179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mui UN, Haley CT & Tyring SK Viral Oncology: Molecular Biology and Pathogenesis. J Clin Med 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wells DK et al. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell 183, 818–834.e813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Melenhorst JJ et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 602, 503–509 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weber EW, Maus MV & Mackall CL The Emerging Landscape of Immune Cell Therapies. Cell 181, 46–62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Melero I. et al. Therapeutic vaccines for cancer: an overview of clinical trials. Nature reviews. Clinical oncology 11, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 117.Lin MJ et al. Cancer vaccines: the next immunotherapy frontier. Nature Cancer 3, 911–926 (2022). [DOI] [PubMed] [Google Scholar]
- 118.Liu Z. et al. Engineering neoantigen vaccines to improve cancer personalized immunotherapy. Int J Biol Sci 18, 5607–5623 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wölfel T. et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269, 1281–1284 (1995). [DOI] [PubMed] [Google Scholar]
- 120.Coulie PG et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci U S A 92, 7976–7980 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rojas LA et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Khattak A. et al. Distant metastasis-free survival results from the randomized, phase 2 mRNA-4157-P201/KEYNOTE-942 trial. Journal of Clinical Oncology 41, LBA9503–LBA9503 (2023). [Google Scholar]
- 123.Furqan F & Shah NN Multispecific CAR T Cells Deprive Lymphomas of Escape via Antigen Loss. Annual review of medicine 74, 279–291 (2023). [DOI] [PubMed] [Google Scholar]
- 124.Dolton G. et al. Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy. Cell 186, 3333–3349.e3327 (2023). [DOI] [PubMed] [Google Scholar]
- 125.Pearlman AH et al. Targeting public neoantigens for cancer immunotherapy. Nature Cancer 2, 487–497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Asimgil H. et al. Targeting the undruggable oncogenic KRAS: the dawn of hope. JCI Insight 7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bear AS et al. Biochemical and functional characterization of mutant KRAS epitopes validates this oncoprotein for immunological targeting. Nat Commun 12, 4365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Park AK et al. Effective combination immunotherapy using oncolytic viruses to deliver CAR targets to solid tumors. Science Translational Medicine 12, eaaz1863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang AQ et al. Universal redirection of CAR T cells against solid tumours via membrane-inserted ligands for the CAR. Nature Biomedical Engineering (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bughda R, Dimou P, D’Souza RR & Klampatsa A Fibroblast Activation Protein (FAP)-Targeted CAR-T Cells: Launching an Attack on Tumor Stroma. ImmunoTargets and Therapy 10, 313–323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alegre ML, Frauwirth KA & Thompson CB T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol 1, 220–228 (2001). [DOI] [PubMed] [Google Scholar]
- 132.Chen L & Flies DB Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vonderheide RH CD40 Agonist Antibodies in Cancer Immunotherapy. Annual review of medicine 71, 47–58 (2020). [DOI] [PubMed] [Google Scholar]
- 134.Cueto FJ & Sancho D The Flt3L/Flt3 Axis in Dendritic Cell Biology and Cancer Immunotherapy. Cancers (Basel) 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choi Y. et al. T-cell agonists in cancer immunotherapy. J Immunother Cancer 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Freeman GJ et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 192, 1027–1034 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hui E. et al. T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Blackburn SD et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Curtsinger JM & Mescher MF Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol 22, 333–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cox MA, Kahan SM & Zajac AJ Anti-viral CD8 T cells and the cytokines that they love. Virology 435, 157–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rosenberg SA IL-2: the first effective immunotherapy for human cancer. J Immunol 192, 5451–5458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blattman JN et al. Therapeutic use of IL-2 to enhance antiviral T-cell responses in vivo. Nat Med 9, 540–547 (2003). [DOI] [PubMed] [Google Scholar]
- 143.West EE et al. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J Clin Invest 123, 2604–2615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Raeber ME, Sahin D, Karakus U & Boyman O A systematic review of interleukin-2-based immunotherapies in clinical trials for cancer and autoimmune diseases. eBioMedicine 90, 104539 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nanjappa SG, Kim EH & Suresh M Immunotherapeutic effects of IL-7 during a chronic viral infection in mice. Blood 117, 5123–5132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pellegrini M. et al. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell 144, 601–613 (2011). [DOI] [PubMed] [Google Scholar]
- 147.Elsaesser H, Sauer K & Brooks DG IL-21 is required to control chronic viral infection. Science 324, 1569–1572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yi JS, Du M & Zajac AJ A Vital Role for Interleukin-21 in the Control of a Chronic Viral Infection. Science 324, 1572–1576 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fröhlich A. et al. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science 324, 1576–1580 (2009). [DOI] [PubMed] [Google Scholar]
- 150.Li Y, Bleakley M & Yee C IL-21 Influences the Frequency, Phenotype, and Affinity of the Antigen-Specific CD8 T Cell Response1. The Journal of Immunology 175, 2261–2269 (2005). [DOI] [PubMed] [Google Scholar]
- 151.Deng S. et al. Targeting tumors with IL-21 reshapes the tumor microenvironment by proliferating PD-1intTim-3-CD8+ T cells. JCI Insight 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chapuis AG et al. Combined IL-21-primed polyclonal CTL plus CTLA4 blockade controls refractory metastatic melanoma in a patient. J Exp Med 213, 1133–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dwyer CJ et al. Fueling Cancer Immunotherapy With Common Gamma Chain Cytokines. Front Immunol 10, 263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chmielewski M & Abken H TRUCKS, the fourth-generation CAR T cells: Current developments and clinical translation. ADVANCES IN CELL AND GENE THERAPY 3, e84 (2020). [Google Scholar]
- 155.Tinoco R, Alcalde V, Yang Y, Sauer K & Zuniga EI Cell-intrinsic transforming growth factor-beta signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity 31, 145–157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hu Y. et al. TGF-β regulates the stem-like state of PD-1+ TCF-1+ virus-specific CD8 T cells during chronic infection. J Exp Med 219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Park BV et al. TGFβ1-Mediated SMAD3 Enhances PD-1 Expression on Antigen-Specific T Cells in Cancer. Cancer Discov 6, 1366–1381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gabriel SS et al. Transforming growth factor-β-regulated mTOR activity preserves cellular metabolism to maintain long-term T cell responses in chronic infection. Immunity 54, 1698–1714.e1695 (2021). [DOI] [PubMed] [Google Scholar]
- 159.Jung IY et al. Tissue-resident memory CAR T cells with stem-like characteristics display enhanced efficacy against solid and liquid tumors. Cell Rep Med 4, 101053 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ma C. et al. TGF-β promotes stem-like T cells via enforcing their lymphoid tissue retention. J Exp Med 219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li G. et al. TGF-β-dependent lymphoid tissue residency of stem-like T cells limits response to tumor vaccine. Nature Communications 13, 6043 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Naing A. et al. PEGylated IL-10 (Pegilodecakin) Induces Systemic Immune Activation, CD8(+) T Cell Invigoration and Polyclonal T Cell Expansion in Cancer Patients. Cancer Cell 34, 775–791.e773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo Y. et al. Metabolic reprogramming of terminally exhausted CD8+ T cells by IL-10 enhances anti-tumor immunity. Nature Immunology 22, 746–756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kloss CC et al. Dominant-Negative TGF-β Receptor Enhances PSMA-Targeted Human CAR T Cell Proliferation And Augments Prostate Cancer Eradication. Mol Ther 26, 1855–1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Burga RA et al. Engineering the TGFβ Receptor to Enhance the Therapeutic Potential of Natural Killer Cells as an Immunotherapy for Neuroblastoma. Clin Cancer Res 25, 4400–4412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kaser A, Nagata S & Tilg H Interferon alpha augments activation-induced T cell death by upregulation of Fas (CD95/APO-1) and Fas ligand expression. Cytokine 11, 736–743 (1999). [DOI] [PubMed] [Google Scholar]
- 167.Odorizzi PM & Wherry EJ Immunology. An interferon paradox. Science 340, 155–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Benci JL et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 167, 1540–1554.e1512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rathmell JC, Farkash EA, Gao W & Thompson CB IL-7 enhances the survival and maintains the size of naive T cells. J Immunol 167, 6869–6876 (2001). [DOI] [PubMed] [Google Scholar]
- 170.Klein Geltink RI et al. Mitochondrial Priming by CD28. Cell 171, 385–397.e311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sinclair LV et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol 14, 500–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ma EH et al. Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8(+) T Cells. Immunity 51, 856–870.e855 (2019). [DOI] [PubMed] [Google Scholar]
- 173.Buck MD, Sowell RT, Kaech SM & Pearce EL Metabolic Instruction of Immunity. Cell 169, 570–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cui G. et al. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell 161, 750–761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pearce EL et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van der Windt GJW et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences 110, 14336–14341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Staron MM et al. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity 41, 802–814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bengsch B. et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8+ T Cell Exhaustion. Immunity 45, 358–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Scharping, Nicole E. et al. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 45, 374–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vardhana SA et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat Immunol 21, 1022–1033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yu YR et al. Disturbed mitochondrial dynamics in CD8(+) TILs reinforce T cell exhaustion. Nat Immunol 21, 1540–1551 (2020). [DOI] [PubMed] [Google Scholar]
- 182.Lontos K. et al. Metabolic reprogramming via an engineered PGC-1α improves human chimeric antigen receptor T-cell therapy against solid tumors. J Immunother Cancer 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Patsoukis N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat Commun 6, 6692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xu X. et al. Autophagy is essential for effector CD8 T cell survival and memory formation. Nature immunology 15, 1152–1161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ao YQ et al. Tumor-infiltrating CD36(+)CD8(+)T cells determine exhausted tumor microenvironment and correlate with inferior response to chemotherapy in non-small cell lung cancer. BMC Cancer 23, 367 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xu S. et al. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8(+) T cells in tumors. Immunity 54, 1561–1577.e1567 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ma X. et al. CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell Metab 33, 1001–1012.e1005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Y. et al. Enhancing CD8(+) T Cell Fatty Acid Catabolism within a Metabolically Challenging Tumor Microenvironment Increases the Efficacy of Melanoma Immunotherapy. Cancer Cell 32, 377–391.e379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Saibil SD et al. Activation of Peroxisome Proliferator-Activated Receptors α and δ Synergizes with Inflammatory Signals to Enhance Adoptive Cell Therapy. Cancer Res 79, 445–451 (2019). [DOI] [PubMed] [Google Scholar]
- 190.Mager LF et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489 (2020). [DOI] [PubMed] [Google Scholar]
- 191.Wang T. et al. Inosine is an alternative carbon source for CD8+-T-cell function under glucose restriction. Nature metabolism 2, 635–647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Qiu J. et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep 27, 2063–2074.e2065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bartman CR et al. Slow TCA flux and ATP production in primary solid tumours but not metastases. Nature 614, 349–357 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Reinfeld BI et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chang CH et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chang CH et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ho PC et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162, 1217–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ottensmeier CH et al. Upregulated Glucose Metabolism Correlates Inversely with CD8+ T-cell Infiltration and Survival in Squamous Cell Carcinoma. Cancer Res 76, 4136–4148 (2016). [DOI] [PubMed] [Google Scholar]
- 199.Cascone T. et al. Increased Tumor Glycolysis Characterizes Immune Resistance to Adoptive T Cell Therapy. Cell Metab 27, 977–987.e974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Angelin A. et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab 25, 1282–1293.e1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Watson MJ et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591, 645–651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cohen IJ et al. Increased tumor glycolysis is associated with decreased immune infiltration across human solid tumors. Frontiers in immunology 13, 880959 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jaiswal AR et al. Melanoma Evolves Complete Immunotherapy Resistance through the Acquisition of a Hypermetabolic Phenotype. Cancer Immunol Res 8, 1365–1380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jayaprakash P, Vignali PDA, Delgoffe GM & Curran MA Hypoxia Reduction Sensitizes Refractory Cancers to Immunotherapy. Annu Rev Med 73, 251–265 (2022). [DOI] [PubMed] [Google Scholar]
- 205.Doedens AL et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nature immunology 14, 1173–1182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liikanen I. et al. Hypoxia-inducible factor activity promotes antitumor effector function and tissue residency by CD8+ T cells. The Journal of clinical investigation 131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jayaprakash P. et al. Targeted hypoxia reduction restores T cell infiltration and sensitizes prostate cancer to immunotherapy. The Journal of clinical investigation 128, 5137–5149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Scharping NE et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nature immunology 22, 205–215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hack SP, Zhu AX & Wang Y Augmenting Anticancer Immunity Through Combined Targeting of Angiogenic and PD-1/PD-L1 Pathways: Challenges and Opportunities. Frontiers in immunology 11, 598877 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brand A. et al. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab 24, 657–671 (2016). [DOI] [PubMed] [Google Scholar]
- 211.Fischer K. et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819 (2007). [DOI] [PubMed] [Google Scholar]
- 212.Quinn WJ 3rd et al. Lactate Limits T Cell Proliferation via the NAD(H) Redox State. Cell reports 33, 108500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Haas R. et al. Lactate Regulates Metabolic and Pro-inflammatory Circuits in Control of T Cell Migration and Effector Functions. PLoS Biol 13, e1002202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kaymak I. et al. Carbon source availability drives nutrient utilization in CD8(+) T cells. Cell Metab 34, 1298–1311.e1296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Peng M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xu K. et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Carr EL et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. Journal of immunology (Baltimore, Md. : 1950) 185, 1037–1044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Leone RD et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Edwards DN et al. Selective glutamine metabolism inhibition in tumor cells improves antitumor T lymphocyte activity in triple-negative breast cancer. The Journal of clinical investigation 131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Han C, Ge M, Ho PC & Zhang L Fueling T-cell Antitumor Immunity: Amino Acid Metabolism Revisited. Cancer Immunol Res 9, 1373–1382 (2021). [DOI] [PubMed] [Google Scholar]
- 221.Yang W. et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature 531, 651–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang H. et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nature immunology 21, 298–308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lim SA et al. Lipid signalling enforces functional specialization of T(reg) cells in tumours. Nature 591, 306–311 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Harel M. et al. Proteomics of Melanoma Response to Immunotherapy Reveals Mitochondrial Dependence. Cell 179, 236–250.e218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chen JH et al. Prostaglandin E2 and programmed cell death 1 signaling coordinately impair CTL function and survival during chronic viral infection. Nat Med 21, 327–334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Miao J. et al. Prostaglandin E(2) and PD-1 mediated inhibition of antitumor CTL responses in the human tumor microenvironment. Oncotarget 8, 89802–89810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pelly VS et al. Anti-Inflammatory Drugs Remodel the Tumor Immune Environment to Enhance Immune Checkpoint Blockade Efficacy. Cancer Discov 11, 2602–2619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gorchs L. et al. Human Pancreatic Carcinoma-Associated Fibroblasts Promote Expression of Co-inhibitory Markers on CD4(+) and CD8(+) T-Cells. Frontiers in immunology 10, 847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhao S, Peralta RM, Avina-Ochoa N, Delgoffe GM & Kaech SM Metabolic regulation of T cells in the tumor microenvironment by nutrient availability and diet. Semin Immunol 52, 101485 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ghorani E, Swanton C & Quezada SA Cancer cell-intrinsic mechanisms driving aquired immune tolerance. Immunity (2023). [DOI] [PubMed] [Google Scholar]
- 231.Schürch CM et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 182, 1341–1359.e1319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jackson HW et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020). [DOI] [PubMed] [Google Scholar]
- 233.Phillips D. et al. Immune cell topography predicts response to PD-1 blockade in cutaneous T cell lymphoma. Nature communications 12, 6726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Binnewies M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chen DS & Mellman I Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 236.Pelka K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184, 4734–4752.e4720 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Combes AJ et al. Discovering dominant tumor immune archetypes in a pan-cancer census. Cell 185, 184–203.e119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wagner J. et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 177, 1330–1345.e1318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schenkel JM & Pauken KE Localization, tissue biology and T cell state - implications for cancer immunotherapy. Nat Rev Immunol (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pittet MJ, Pilato M, Garris C & Mempel TR Dendritic cells as sheperds of T cell immunity in cancer. Immunity (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Cassetta L & Pollard JW A timeline of tumour-associated macrophage biology. Nat Rev Cancer 23, 238–257 (2023). [DOI] [PubMed] [Google Scholar]
- 242.Kersten K. et al. Spatiotemporal co-dependency between macrophages and exhausted CD8(+) T cells in cancer. Cancer Cell 40, 624–638.e629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Peranzoni E. et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Natl Acad Sci U S A 115, E4041–e4050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nixon BG et al. Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity 55, 2044–2058.e2045 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Garris CS et al. Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-γ and IL-12. Immunity 49, 1148–1161.e1147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Roberts EW et al. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 30, 324–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Binnewies M. et al. Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4(+) T Cell Immunity. Cell 177, 556–571.e516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ferris ST et al. cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Oh SA et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat Cancer 1, 681–691 (2020). [DOI] [PubMed] [Google Scholar]
- 250.Barry KC et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med 24, 1178–1191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Böttcher JP et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037.e1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kennel KB, Bozlar M, De Valk AF & Greten FR Cancer-Associated Fibroblasts in Inflammation and Antitumor Immunity. Clin Cancer Res 29, 1009–1016 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Chen Y, McAndrews KM & Kalluri R Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 18, 792–804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Dominguez CX et al. Single-Cell RNA Sequencing Reveals Stromal Evolution into LRRC15(+) Myofibroblasts as a Determinant of Patient Response to Cancer Immunotherapy. Cancer Discov 10, 232–253 (2020). [DOI] [PubMed] [Google Scholar]
- 255.Krishnamurty AT et al. LRRC15(+) myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature 611, 148–154 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Elyada E. et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 9, 1102–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Huang H. et al. Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer. Cancer Cell 40, 656–673.e657 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Groeneveld CS et al. Tertiary lymphoid structures marker CXCL13 is associated with better survival for patients with advanced-stage bladder cancer treated with immunotherapy. Eur J Cancer 148, 181–189 (2021). [DOI] [PubMed] [Google Scholar]
- 259.Workel HH et al. A Transcriptionally Distinct CXCL13(+)CD103(+)CD8(+) T-cell Population Is Associated with B-cell Recruitment and Neoantigen Load in Human Cancer. Cancer Immunol Res 7, 784–796 (2019). [DOI] [PubMed] [Google Scholar]
- 260.Di Pilato M. et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530.e4522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sautès-Fridman C, Petitprez F, Calderaro J & Fridman WH Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer 19, 307–325 (2019). [DOI] [PubMed] [Google Scholar]
- 262.Spitzer MH et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502.e415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Prokhnevska N. et al. CD8(+) T cell activation in cancer comprises an initial activation phase in lymph nodes followed by effector differentiation within the tumor. Immunity 56, 107–124.e105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Dammeijer F. et al. The PD-1/PD-L1-Checkpoint Restrains T cell Immunity in Tumor-Draining Lymph Nodes. Cancer Cell 38, 685–700.e688 (2020). [DOI] [PubMed] [Google Scholar]
- 265.Spranger S. et al. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. J Immunother Cancer 2, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Huang Q. et al. The primordial differentiation of tumor-specific memory CD8(+) T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell 185, 4049–4066.e4025 (2022). [DOI] [PubMed] [Google Scholar]
- 267.Saddawi-Konefka R. et al. Lymphatic-preserving treatment sequencing with immune checkpoint inhibition unleashes cDC1-dependent antitumor immunity in HNSCC. Nature communications 13, 4298 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Reticker-Flynn NE et al. Lymph node colonization induces tumor-immune tolerance to promote distant metastasis. Cell 185, 1924–1942.e1923 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Picarda G & Benedict CA Cytomegalovirus: Shape-Shifting the Immune System. J Immunol 200, 3881–3889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Routy B. et al. The gut microbiota influences anticancer immunosurveillance and general health. Nature reviews. Clinical oncology 15, 382–396 (2018). [DOI] [PubMed] [Google Scholar]
- 271.Elkrief A, Derosa L, Kroemer G, Zitvogel L & Routy B The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: a new independent prognostic factor? Ann Oncol 30, 1572–1579 (2019). [DOI] [PubMed] [Google Scholar]
- 272.Koh A, De Vadder F, Kovatcheva-Datchary P & Bäckhed F From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 273.Rudloff MW et al. Hallmarks of CD8+ T cell dysfunction are established within hours of tumor antigen encounter before cell division. Nature Immunology (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tonnerre P. et al. Differentiation of exhausted CD8(+) T cells after termination of chronic antigen stimulation stops short of achieving functional T cell memory. Nat Immunol 22, 1030–1041 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R & Ahmed R Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77, 4911–4927 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Abdel-Hakeem MS et al. Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nature Immunology 22, 1008–1019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Angelosanto JM, Blackburn SD, Crawford A & Wherry EJ Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol 86, 8161–8170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Yates KB et al. Epigenetic scars of CD8+ T cell exhaustion persist after cure of chronic infection in humans. Nature Immunology 22, 1020–1029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Utzschneider DT et al. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat Immunol 21, 1256–1266 (2020). [DOI] [PubMed] [Google Scholar]
- 280.Quezada LK et al. Early transcriptional and epigenetic divergence of CD8+ T cells responding to acute versus chronic infection. PLoS Biol 21, e3001983 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Tsui C. et al. MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature 609, 354–360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Johnnidis JB et al. Inhibitory signaling sustains a distinct early memory CD8+T cell precursor that is resistant to DNA damage. Science Immunology 6, eabe3702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Blackburn SD et al. Tissue-specific differences in PD-1 and PD-L1 expression during chronic viral infection: implications for CD8 T-cell exhaustion. J Virol 84, 2078–2089 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hombrink P. et al. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nature Immunology 17, 1467–1478 (2016). [DOI] [PubMed] [Google Scholar]
- 285.Park SL et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 19, 183–191 (2018). [DOI] [PubMed] [Google Scholar]
- 286.Kumar BV et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Khanna KM, Bonneau RH, Kinchington PR & Hendricks RL Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity 18, 593–603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Verjans GM et al. Selective retention of herpes simplex virus-specific T cells in latently infected human trigeminal ganglia. Proc Natl Acad Sci U S A 104, 3496–3501 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Steinbach K, Vincenti I & Merkler D Resident-Memory T Cells in Tissue-Restricted Immune Responses: For Better or Worse? Front Immunol 9, 2827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Gavil NV et al. Chronic antigen in solid tumors drives a distinct program of T cell residence. Sci Immunol 8, eadd5976 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Gueguen P. et al. Contribution of resident and circulating precursors to tumor-infiltrating CD8(+) T cell populations in lung cancer. Sci Immunol 6 (2021). [DOI] [PubMed] [Google Scholar]
- 292.Djenidi F. et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol 194, 3475–3486 (2015). [DOI] [PubMed] [Google Scholar]
- 293.Wang ZQ et al. CD103 and Intratumoral Immune Response in Breast Cancer. Clin Cancer Res 22, 6290–6297 (2016). [DOI] [PubMed] [Google Scholar]
- 294.Koh J. et al. Prognostic implications of intratumoral CD103+ tumor-infiltrating lymphocytes in pulmonary squamous cell carcinoma. Oncotarget 8, 13762–13769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Savas P. et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nature Medicine 24, 986–993 (2018). [DOI] [PubMed] [Google Scholar]
- 296.Edwards J. et al. CD103(+) Tumor-Resident CD8(+) T Cells Are Associated with Improved Survival in Immunotherapy-Naïve Melanoma Patients and Expand Significantly During Anti-PD-1 Treatment. Clin Cancer Res 24, 3036–3045 (2018). [DOI] [PubMed] [Google Scholar]
- 297.Park SL et al. Tissue-resident memory CD8+ T cells promote melanoma–immune equilibrium in skin. Nature 565, 366–371 (2019). [DOI] [PubMed] [Google Scholar]
- 298.Enamorado M. et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8(+) T cells. Nat Commun 8, 16073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Gálvez-Cancino F. et al. Vaccination-induced skin-resident memory CD8(+) T cells mediate strong protection against cutaneous melanoma. Oncoimmunology 7, e1442163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Nizard M. et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nature Communications 8, 15221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]