Summary
Although up to 80% small cell lung cancer (SCLC) patients’ response is good for first-line chemotherapy regimen, most patients develop recurrence of the disease within weeks to months. Here, we report cytostatic effect of leflunomide (Leflu) and teriflunomide (Teri) on SCLC cell proliferation through inhibition of DRP1 phosphorylation at Ser616 and decreased mitochondrial fragmentation. When administered together, Teri and carboplatin (Carbo) act synergistically to significantly inhibit cell proliferation and DRP1 phosphorylation, reduce abundance of intermediates in pyrimidine de novo pathway, and increase apoptosis and DNA damage. Combination of Leflu&Carbo has anti-tumorigenic effect in vivo. Additionally, lurbinectedin (Lur) and Teri potently and synergistically inhibited spheroid growth and depleted uridine and DRP1 phosphorylation in mouse tumors. Our results suggest combinations of Carbo and Lur with Teri or Leflu are efficacious and underscore how the relationship between DRP1/DHODH and mitochondrial plasticity serves as a potential therapeutic target to validate these treatment strategies in SCLC clinical trials.
Subject areas: physiology, molecular biology, cell biology, cancer
Graphical abstract

Highlights
-
•
Leflu and Teri are preferably bound at the DRP1 GDP-binding site
-
•
Teri inhibited DRP1 phosphorylation at Ser616 and mitochondrial fragmentation
-
•
Combination of Teri and Leflu with Carbo or Lur accelerated SCLC cell death
Physiology; Molecular biology; Cell biology; Cancer.
Introduction
Small cell lung cancer (SCLC) is an aggressive cancer that is often fatal.1 First-line treatments have response rates up to 80%, but many patients relapse within 6 months, and the 5-year survival rate remains 6%.1,2,3 Standard chemotherapy treatment options for recurrent tumors are limited and ineffective, resulting in poor clinical outcomes.4 Mitochondrial morphology plays a crucial role in cellular functions, including metabolism, cell proliferation, and apoptosis.5,6 DRP1 regulates mitochondrial fission by increasing phosphorylation at Ser616 and drives mitochondrial dynamics toward fission.7 Overexpression of fission proteins, including DRP1, has been correlated with resistance to drug treatment and uncontrolled proliferation.8,9 Mitochondrial fragmentation presents a potential target for the development of effective drugs for SCLC. Several studies have associated mitochondrial fragmentation with tumor progression in multiple cancers, such as mesothelioma and ovarian, breast, lung (non-small cell lung cancer [NSCLC]), and pancreatic cancer,8,10,11 but not specifically in SCLC. However, the rapid proliferation of cancer cells resulting from their constant growth and metabolic shifts triggers mitochondrial fragmentation.12 DNM1L encodes DRP1 protein, which plays a central role in regulating mitochondrial fission and fusion. Notably, DRP1 phosphorylation at Ser616 leads to its recruitment to mitochondrial fission receptors and activation of mitochondrial fission, whereas phosphorylation at Ser637 (fusion) leads to its inactivation and results in mitochondrial fusion.13 Growing evidence indicates that mitochondrial fission and fusion are therapeutic targets for cancer.13 To this end, studies have reported the effect of the DHODH inhibitors Leflu and Teri on mitochondrial fusion/fission dynamics, highlighting the role of phenotypic plasticity in cancer.14,15,16 As well, Leflu and Teri are approved DHODH inhibitors used as anti-inflammatory drugs to treat autoimmune and infectious diseases.17 DHODH is a mitochondrial inner membrane enzyme and regulates de novo pyrimidine biosynthesis, which is necessary for cell proliferation.14 Leflu inhibits DHODH catalytic activity for conversion of dihydroorotate into orotate.18 It has also been shown that Leflu and Teri have potent anticancer effects on human bladder cancer and NSCLC cells.19,20
Inhibition of transcription is another strategy for targeting SCLC. For example, several SCLC cell lines were observed to be sensitive to treatment focused on transcription inhibition and promoting the degradation of elongating RNA.21 Lur is an inhibitor of RNA pol II, which is commonly hyperactivated in SCLC. Lur treatment results in massive transcription inhibition in tumor cells.22 Indeed, Lur has shown promise in treating SCLC patients after failure to first-line systemic therapy.23
We hypothesized that combining Leflu and Teri with Carbo or Lur chemotherapeutic agents may be a potent therapeutic strategy for SCLC. In the present study, we demonstrated the cytostatic effect of Leflu and Teri on SCLC cell proliferation through inhibition of DRP1 phosphorylation at Ser616 and decreased mitochondrial fragmentation. The combination of Teri&Carbo significantly inhibited cell proliferation and DRP1 phosphorylation, reduced the abundance of several substances in the pyrimidine de novo pathway, and increased apoptosis and DNA damage in vitro. Furthermore, combination of Leflu&Carbo also significantly decreased tumor growth in mice through inhibition of DRP1 fission activity. Additionally, Teri&Lur synergistically inhibited SCLC cell spheroid growth. Teri&Lur combination depleted uridine and DRP1 phosphorylation at Ser616 and increased cell-cycle arrest and DNA damage in SCLC spheroids. To our knowledge, the present study revealed synergistic interaction of Teri and Leflu with chemotherapeutics on mitochondrial plasticity in SCLC the first time, paving the way for additional medical modalities for this deadly disease.
Results
DRP1 in lung cancer: Leflu and Teri are putative inhibitors of DRP1 and their cytotoxicity in SCLC cells
To investigate the prevalence of DRP1 among the cancer types, we analyzed DNML1 TCGA dataset. The expression DRP1 was high in lung tissue (Figure 1A). Analysis of the correlation between the fission protein DRP1 and patient survival employing the TCGA database showed a poorer overall survival rate (p = 0.0033) for patients with high expression of DNM1L gene (Figure 1B). Next, we used a publicly available dataset to compare DRP1 mRNA expression in normal lung tissue (GSE19945) and SCLC (149507) tissue. The DRP1 mRNA level was significantly higher in malignant lung tissue than in adjacent normal lung tissue (Figure 1C). Next, eight SCLC cell lines and nonmalignant BEAS 2B cells were examined for DRP1 expression and phosphorylation. Elevated levels of DRP1 and pDRP1 Ser616 were detected in all SCLC cell lines tested (Figure 1D). In addition, high DHODH expression was observed in SCLC cells compared with BEAS 2B cells. Previous studies have shown robust anticancer activity of DHODH inhibitors in SCLC preclinical models.24 Based on these findings, we proposed that DHODH inhibitors can inhibit DRP1 activity.
Figure 1.
DRP1 expression and targeting by Leflu and Teri directly in SCLC
(A) DNM1L RNA expression in cancer (TCGA dataset).
(B) TCGA data for the survival probability of DRP1 in LUAD. Graph shows the survival curves for two groups, low and high level of DNM1L gene expression in tumor tissue. Dataset is available on website: https://doi.org/10.1016/j.neo.2017.05.002 (UALCAN).
(C) DRP1 mRNA expression in normal and SCLC lung tissue (NCBI/GEO/GSE149507). Bar graphs represent mean ± SEM, and p value was calculated by Student’s t test, ∗∗∗p < 0.001.
(D) Representative immunoblots showing expression of DHODH and DRP1 in SCLC cell lines and BEAS 2B cells.
(E) Binding AA docking poses of Leflu, Teri, and MDV1 with DRP1. Binding mode of Leflu and Teri at the GDP-binding site of DRP1 protein. MDV1 binds at another site with low score.
(F) 2D representation of the binding pose. Red arrows represent hydrogen bonds.
(G) Effect of Teri and Leflu on SBC3 and SBC5 cells proliferation was recorded by the Sartorius IncuCyte S3 live-cell analysis system (n = 6, biologically independent samples). Data are represented as mean ± SD. (D) and (G) were repeated independently three and two times with similar results, accordingly.
Leflu and Teri were used to identify the inhibitor GTP-binding pocket in DRP1 by AA docking.25 We found that Leflu and Teri preferably bind at the DRP1 GDP-binding site, which produced higher docking scores. Teri showed the highest docking score (−8.02 kcal/mol) in the GDP-binding pocket. Leflu had a moderate docking score of −5.6 kcal/mol. MDV1 showed the lowest docking score (−3.66 kcal/mol), indicating a weaker interaction with DRP1 (Figure 1E). The amino acids involved in the interactions between Leflu, Teri, MDV1, and DRP1 are shown in Figure 1F.
The in vitro cytotoxic effects of Teri and Leflu were tested in a panel of SCLC cell lines; the half-maximal inhibitory combination (IC50) values are reported in Table S1. SCLC cell lines were treated with increasing concentrations of each drug for 72 h. Teri active metabolite of Leflu had potent inhibitory effect on five SCLC cell lines with IC50 values less than 11 μM. Teri sensitive SBC3 and least sensitive SBC5 cells, in which Teri exhibited IC50 values of 2 μM and 33 μM, respectively, were used for additional experiments. Treatment of SBC3 and SBC5 cells with Leflu or Teri inhibited cell proliferation in a dose-dependent manner, with the inhibitory effect of Teri being stronger in both cell lines (Figure 1G). These data suggest that treatment with Teri and Leflu remarkably inhibited SCLC cells growth in vitro.
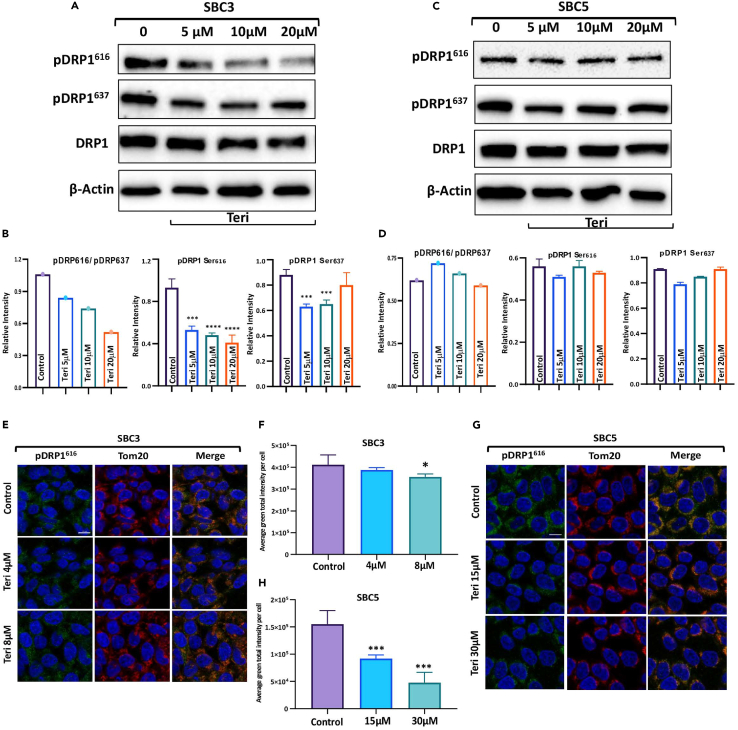
Teri&Leflu treatment inhibits DRP1 fission activity
Since activation of DRP1 is regulated by phosphorylation, we investigated the effect of Teri on the phosphorylation of DRP1. In SBC3 cells, Teri treatment significantly inhibited DRP1 phosphorylation at Ser616 and Ser637, and the ratio of pDRP1 (Ser616) to pDRP1 (Ser637) significantly decreased; however, the total DRP1 level was unaffected (Figures 2A and 2B). Western Blot bar graphs showed that Teri treatment reduced the levels of Fis1 and OPA1 in SBC3 cells but did not change the MFN2 level (Figure S1A). Teri showed dose-dependent inhibition of mitochondrial dynamic proteins in SBC3 cells. The expression of DRP1 and the ratio of pDRP1 (Ser616) to pDRP1 (Ser637) in SBC5 cells were not affected by Teri treatment (Figures 2C and 2D). Levels of fission Fis1 and two fusion MFN2 and OPA1 proteins were nonsignificant compared with control in SBC5 cells (Figure S1B).
Figure 2.
Teri affects the phosphorylation of DRP1 at Ser616 and mitochondrial/nuclear morphology
(A) Effect of Teri on DRP1 phosphorylation; SBC3 cell lysates were subjected to immunoblotting with DRP1, pDRP1 Ser616 (activated form), and Ser637 (inactivated form) antibodies.
(B) The ratio of pDRP1 Ser616 to pDRP1 Ser637 in SBC3 cells. Expression level of pDRP1 Ser616 to pDRP1 Ser637 was calculated via densitometric analysis of each blot using ImageJ software and normalized to actin level.
(C and D) DRP1 phosphorylation in SBC5 cells after Teri dose and calculation of the expression ratio of pDRP1 Ser616 to pDRP1 Ser637.
(E and G) Immunofluorescence staining of SBC3 and SBC5 cells with pDRP1 Ser616 and Tom20 antibodies. Scale bar: 10 μm (n = 3, biologically independent samples).
(F and H) Calculating pDRP1 Ser616 fluorescence signal by QuPath software (see STAR Methods).
(I and J) The mitochondrial and nuclear surface areas of SBC3 and SBC5 cells were obtained with Imaris Cells module. Individual cell data are plotted and organized by treatment group. The minimal number of SBC3 cells analyzed for each group was 103 (M1 10 μM) and the maximal was 383 (Control) (I). The minimal number of SBC5 cells analyzed for each group was 357 (MDIVI1 10 μM) and the maximal was 570 (Control) (J). (I) and (G) (right) are representative immunofluorescence (IF) images ([red] Tom 20 antibody with Hoechst 33342 [blue]) of SBC3 and SBC5 mitochondria after treatments. Scale bar: 20 μm. Bar graphs represent means ±SEM in (B), (D), (F), (H), (I), and (J). p values were calculated by ANOVA with Tukey post-test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 in (B), (D), (F), (H), (I), and (J). (A), (C), (E), and (G) were repeated independently three times and (I) and (J) two times.
Additionally, SBC3 and SBC5 cells were stained with an pDRP1 Ser616 antibody, and the phosphorylation level was analyzed with QuPath 0.2.0M9 software. Representative images of SBC3 cells showed a decrease in pDRP1 Ser616 levels after Teri treatment (Figure 2E). Quantification of the green IF signal showed a significant decrease in signal intensity in SBC3 cells (Figure 2F). SBC5 cells, least sensitive to Teri, were treated with higher concentrations of Teri (1/2 IC50 = 15 μM and IC50 = 30 μM) than SBC3 cells; quantification of the pDRP1 Ser616 IF signal by QuPath revealed the strong inhibitory effect of Teri on this phosphorylation event in both cell lines (Figures 2G and 2H).
To understand how mitochondrial fission dynamics are altered by Teri and Leflu, we performed immunofluorescent staining of the mitochondrial network and obtained whole-cell mitochondrial and nuclear area measurements from Imaris Cells. MitoTracker Red and Hoechst 33342 staining was used to visualize mitochondria and nuclei in SBC3 and SBC5 cells. The compounds M1 and MDIVI1 were used as positive controls because they promote mitochondrial fusion. Mitochondria in SBC3 and SBC5 control cells were closely packed around the nuclei, whereas Teri treatment rearranged the mitochondrial network and extended the area from the nucleus (Figures S2 and S3). The mitochondrial and nuclear area was larger with Teri treatment compared to the control in SBC3 cells (Figure 2I). However, Leflu, M1, and MDIVI1 did not affect the mitochondrial or nuclear area in SBC3 cells. In SBC5 cells, Teri had no significant effect on mitochondrial area, whereas MDIVI1 treatment significantly reduced size of mitochondrial area. A significant increase in nuclear area was observed in SBC5 cells treated with Teri and Leflu (Figure 2J). Nuclear area is associated with cancer cell drug response,26 and both SBC3 and SBC5 cell nuclear areas became large with Teri treatment.
Altogether, these data suggest that targeting DRP1 with Teri decreases DRP1 phosphorylation at Ser616 and affects mitochondrial morphology and nuclear area in SBC3 and SBC5 cells. Therefore, we chose to investigate the effects of Teri and Leflu as valuable additions to SCLC therapy and tested the effect of chemotherapeutic drugs on mitochondrial morphology and cell survival.
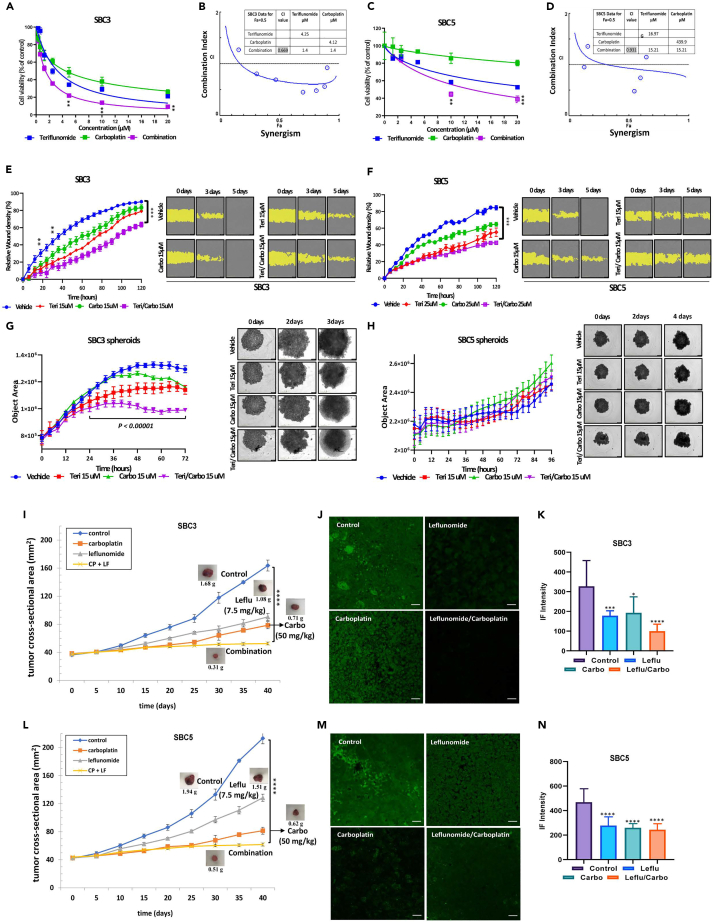
Teri&Carbo combination inhibits cell viability, migration, and spheroid growth: effect of Teri&Carbo on tumor growth
To define the functional effects of Teri administration in combination with Carbo on SCLC cells, we tested two-dimensional (2D) and three-dimensional (3D) models. SBC3 cell viability decreased upon combination treatment with Teri and Carbo at several doses (dose range ∼4–10 μM) (Figure 3A). The Chou-Talalay method was used for detecting and quantifying drug combination index (CI) of Teri and Carbo. An isobologram and CI values were calculated to examine the potential for synergism and indicated concentrations of Teri (4.24 μM) or Carbo (4.12 μM) were approximately 3-fold higher (4.12 μM Teri and Carbo) than for combination treatment (Teri&Carbo 1.4 μM) and a CI value of 0.669, indicating the synergistic effect in SBC3 cells (Figure 3B). Teri&Carbo had an additive effect on Teri-less sensitive SBC5 cells and significantly enhanced the cytotoxicity (15.2 μM Teri&Carbo, CI = 0.931) compared with treatment alone (Figures 3C and 3D).
Figure 3.
Effect of Teri and Leflu on in vitro and in vivo models in combination with Carbo
SBC3 (A) and SBC5 (C) cells were treated with different concentrations of Teri and Carbo for 72 h to detect cell viability (n = 3, biologically independent samples).
(B and D) Combination index (CI) was calculated using the Chou-Talalay method to find synergism. IC50 isobologram after SBC3 and SBC5 cells treatment with Teri/Carbo. CI values at the 50% inhibition of cell proliferation were below 1, indicating a synergistic effect of Teri/Carbo on SBC3 (0.562) and an additive effect on SBC5 (1.02) cells.
(E and F) SBC3/SBC5 relative wound density at different time points (left panels, % mean values ± SD, monolayer wound measurements). Representative IncuCyte brightfield images of wounds on different days masked in yellow (right panels) (n = 3, biologically independent samples); experiment was repeated independently two times.
(G and H) Live spheroid images analysis and IncuCyte brightfield images of SBC3 and SBC5 spheroids. Left panels show data as mean ± SD (n = 2, four technical replicates).
(I and K) Athymic nude mice were implanted with SBC3 and SBC5 cells and treated with vehicle (control), Carbo (50 mg/kg), Leflu (7.5 mg/kg), and Carbo&Leflu, five mice per group.
(J and M) Control and treated FFPE mouse tumor sections were stained with phospho-DRP1 (Ser616). Displayed are representative single channel Airyscan images for each group. Scale bar 20 μm. Multichannel images of pDRP1, EpCAM, and nuclei IF are available (Figure S6).
(K and N) Mean signal intensities were measured from confocal images with ZEN 2.3 Lite (see STAR methods). Data are represented as means ± SEM in (A), (C), (G), (H), (I), (K), (L), and (M). p values were calculated by Student’s t test, ∗∗p < 0.01; ∗∗∗∗p < 0.0001 vs. control in (A), (C), (E), (F), (I), and (L); and by ANOVA with Tukey post-test, ∗p < 0.05; ∗∗p < 0.01;∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 vs. control in (K) and (N). (A), (C), (E), (F), (G), and (H) were repeated independently two times with similar results.
The IncuCyte Live Cell Imaging system was used to evaluate live SBC3 cell migration in the presence of Teri and Carbo. After 24 h, the wound closure rate of the treated SBC3 cells was significantly decreased compared to that of the control cells (Figure 3E, left panel). Remarkable differences in the wound closure rate were observed between the cells treated with a single drug and the control cells at later time points over a period of 60 h. A similar response was observed in SBC5 cells (Figure 3F, left panel). Data shown in Figures 3E and 3F clearly demonstrate Teri&Carbo’s significant inhibition of wound closure in SBC3 and SBC5 cells.
3D spheroid cultures consisting of SBC3 or SBC5 cells were then used to assess the effect of the drugs on cell proliferation and death. SBC3 and SBC5 spheroid size was evaluated with time-lapse images. Images of SBC3 spheroids after 3-day treatment with Teri&Carbo indicated the detachment of dead and fragmented cells from the surface, and quantitative analysis demonstrated significant inhibition of spheroid size after Teri, Carbo, and Teri&Carbo treatment (Figure 3G).
Longer drug treatment of SB5 spheroids (4 days) did not affect growth or morphology of spheroids, though cell viability was reduced in 2D culture (Figure 3H). These data confirm that Teri&Carbo reduced SBC3 and SBC5 spheroid cell viability.
To define the functional effects of Leflu administration in combination with Carbo on SCLC cells, we used mouse xenograft model of SCLC. We injected SBC3 and SBC5 cells in the flanks of athymic mice and randomized the animals to four groups once tumors became palpable. Mice were treated with drug for 40 days. The effectiveness of Carbo treatment in reducing tumor growth was comparable to that of Leflu treatment in the SBC3 xenografts. The average tumor weight at the end of the study (day 40) was significantly lower in the treated animals than in the control animals (control 1.67 ± 0.12 g; vs. treated: Carbo 0.75 ± 0.10 g, Leflu 1.01 ± 0.14 g, and Leflu&Carbo 0.32 ± 0.04 g) (Figure 3I). Single treatment with Carbo and Leflu caused a ∼58% or ∼36% reduction in tumor size, respectively. A much higher reduction of ∼82% in tumor size and weight was observed with combined treatment (Figure 3I). Leflu&Carbo significantly inhibited primary tumor growth compared with control (Figure S5). Next, based on our in vitro results that Teri inhibited cell proliferation and pDRP1616 phosphorylation, we investigated Leflu effect on DRP1 activity in mouse tumors. Tumor sections were stained with an anti-pDRP1 antibody. Quantitative analysis of immunofluorescence staining data showed high level of DRP1 phosphorylation at Ser616 in the untreated tumors. A significant decrease in pDRP1(Ser616) was observed after Leflu and Carbo treatment. Importantly, combination treatment more effectively inhibited DRP1 phosphorylation (Figures 3J and 3K). Additionally, analysis of SBC5 mouse xenografts demonstrated the inhibition of tumor growth after treatment with Leflu, Carbo, or both (control 2.01 ± 0.19 g; vs. treated: Carbo 0.64 ± 0.10 g, Leflu 1.49 ± 0.20 g, and Leflu&Carbo 0.54 ± 0.06 g) (Figure 3L). Immunofluorescence staining of DRP1 phosphorylated at Ser616 showed levels comparable to those in SBC3 xenografts. Combined Leflu&Carbo treatment had a greater inhibitory effect than the control or single-drug treatment (Figures 3M and 3N). EpCAM Abs were used for detection of tumor cells. Representative images for pDRP1 (Ser616) and EpCAM staining were shown on Figure S6. Together, these results confirm the significant inhibitory effect of Teri&Carbo on SCLC cells viability and migration. Moreover, Leflu&Carbo exerted remarkable antitumor inhibition of mouse tumors.
Combination treatment with Teri and Carbo promotes apoptosis and DNA damage and disrupts the purine-pyrimidine pool. KO of DRP1 inhibited SBC3 and SBC5 cell proliferation
To understand if inhibition of proliferation by Teri&Carbo is associated with DRP1 phosphorylation, and how Teri&Carbo affects apoptotic and DNA damage response, we evaluated their protein levels in SBC3 and SBC5 cells. Combination treatment of SBC3 cells inhibited the phosphorylation of DRP1 at Ser616 and non-significantly reduced DRP1 phosphorylation in SBC5 cells; levels of total DRP1 or pDRP1-Ser637 were unchanged after Teri&Carbo treatment in SBC3 and SBC5 cells (Figures 4A and 4B). Then, Teri&Carbo promoted apoptosis by increasing cleaved PARP in SBC3 and SBC5 cells (Figures 4C and 4D). Alteration of p53 signaling is the most common genetic modification in SCLC.27 Several studies have shown that p53 phosphorylation is integral to apoptosis and p53 translocation in the mitochondria during cell death.28,29 p53 phosphorylation at Ser15 was increased in SBC3 and SBC5 cells treated with Teri&Carbo (Figures 4C and 4D). We detected high levels of DNA damage proteins (p-Chk1 and γ-H2AX) in Teri&Carbo-treated SBC3 and SBC5 cells (Figures 4E and 4F). Thus, treatment of SBC3 and SBC5 cells with Teri&Carbo upregulated p53 phosphorylation, increased DNA damage, and inhibited DRP1 activity.
Figure 4.
Teri&Carbo affected DRP1 phosphorylation, apoptotic signaling, and DNA damage response and disturbed purine/pyrimidine pools
DRP1 phosphorylation response to drug treatment in SBC3 (A) and SBC5 cells (B) and the average relative density of pDRP1 Ser616 and Ser637.
(C and D) Western blot images and band densitometry analysis of levels of apoptotic markers in SBC3 and SBC5 cells.
(E and F) Western blot images and band densitometry analysis of levels of DNA damage regulatory proteins in SBC3 and SBC5 cells. GAPDH was used as an internal control in western blot panels. Experiments were repeated independently three times with similar results.
(G) Schematic diagram displays the effect of Teri&Carbo treatment on purine and pyrimidine pools in SBC3 cells.
(H) Statistical comparisons of drug treatments were performed to evaluate the synergistic effect of Teri&Carbo on the concentrations of pyrimidine/purine bases. The table includes synergistically affected substances and p-values for three treatments of SBC3 cells.
(I–K) Pyrimidine, purine, and purine/pyrimidine substance concentrations measured by LC-MS/MS in SBC3 cells after 24 h (n = 3, biologically independent samples). Bar graphs show concentration of synergistically significant substances from table (H). Concentrations of measured substances are available in the Table S2 and https://doi.org/10.17632/zfmb6dtw78.1. p values were calculated and were assessed with a linear model (ANOVA). See STAR methods, ∗p < 0.05; ∗∗p < 0.01.
(L) Total clone number for SBC3 (DOX−/DOX+) and SBC5(DOX−/DOX+) cells. Each CRISPR-edited cell sample was sorted with single cell with DAPI into 3 × 96 well plates. The total visible clones were counted after 14 days incubation without DOX.
(M) The NanoString differential expression analysis of gene patterns for SBC3 and SBC5 (DOX+ vs. DOX−). The dot plots were generated using the R ggplot2 package.
(N) SBC3 and SBC5 expression pattern of public dataset GSE1037. The normalized data from NIH Gene Expression Omnibus (GEO). SBC3, SBC5, and 19 normal samples from GSE1037 are used for the analysis to identify the different expression pattern of SBC3 and SBC5 (vs. normal) using the R LIMMA package (v3.42.2). The differentially expressed genes are identified with FDR<0.05. (M and N) The dot size represents the p value, and the color represents the expression levels (log2-fold change). Bar graphs represent means ± SEM in (A)–(L). p values were calculated by ANOVA with Tukey post-test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 vs. control in (A), (B), (C), (D), (E), (F), and (L).
Levels of pyrimidines and purines in SBC3 cells after Teri&Carbo were measured using LC-MS assay. Summary of Teri&Carbo effect on purine, pyrimidine, and purine&pyrimidine pools is presented on a schematic diagram and shows significantly altered metabolites (Figure 4G).
The abundance of 10 substances was found to be significantly and synergistically altered upon combination treatment (Figure 4H). The synthesis of three pyrimidine substances was significantly inhibited with Teri&Carbo treatment. While the UTP concentration significantly decreased (Figure 4I; Table S2), Teri&Carbo showed remarkable inhibitory effect on five purine substances (Figure 4J). Furthermore, inhibition of DHODH with Teri&Carbo significantly depleted several components of the ATP pool (Figure 4K). More specifically, Teri&Carbo treatment inhibited the de novo synthesis of the pyrimidine UTP in SBC3 cells, accounting for the depletion of ATP-dependent substance pools. Additionally, the combination of Teri&Carbo reduced levels of the purine nucleotides in SBC3 cells.
To further understand the role of DRP1 in regulation of SBC3 and SBC5 function, we performed gene KO (knockout) by CRISPR in these cells (see the STAR methods for the CRISPR‒Cas9 protocol). First, we elected to target the DRP1 EXON7 and EXON8 of the total 19 exons, as these two exons translate the protein located within the major functional domain of DRP1 (https://www.ncbi.nlm.nih.gov/gene) (Figures S7A and S7B). While both SBC3 and SBC5 have a related long cell doubling time and CRISPR KO is induced in M phase in cell cycle, the proper sampling time of the experiment needs to be evaluated before the gene functional assay. Next, we tested the CRISPR KO efficiency at different time points by NGS analyzing in cell pools. We found that doxycycline (DOX) treatment for 72 and 96 h induced less than 5% gene editing outcomes in all cells. However, DOX treatment for 7 or 14 days induced gene editing at rates of 53.2% (Exon7)/31.6% (Exon8) and 47.36% (Exon7)/31.56% (Exon8), respectively, in SBC3 cells. Similar gene editing rates were observed in SBC5 cells (37.4% (Exon7)/24.2% (Exon8) and 35.2% (Exon7)/24.3% (Exon8), respectively). While the 7-day DOX(+)-treated samples retained the highest mutation rate and exhibited the slowest SBC3 and SBC5 cell growth, we elected to perform single-cell clone isolation in the 7-day DOX(+)-treated samples. CRISPR/sgRNA-integrated SBC3 and SBC5 cells were sorted into single-cell clones and cultured for an additional two weeks without DOX. After two weeks of culture, we noticed that DOX treatment reduced colony formation efficiency by 80% (SBC3 cells) and 40% (SBC5 cells) (Figure 4L). Among the visible colonies, two types of growth patterns were apparent in the population: a normal growth pattern (as a negative control) and a significantly slower-growing population. We selected clones of both types for PCR and subsequent sequencing to evaluate the relationship between CRISPR editing in the targeted region of DRP1 and growth rate. Slow-growing clones carried CRISPR-mediated mutations in both sites or strong editing in one site (SBC3-C9 (99.54%/40.21%), G5 (99%/0%), A9 (99.65%/0%), A10 (0%/99.63%), SBC5-D3 (99.78%/99.6%), SBC5-E3 (99.26%, 99.74%), and SBC5-F4 (NA/0%) (Table S3).
To further investigate the function of DRP1, we performed a NanoString assay to analyze the panel of genes associated with fission/fusion (Table S4). Due to the scarcity of KO clones, RNA samples used for the assay contained pooled RNA isolated from 10 clones. NanoString gene expression analysis and the untreated cells served as a negative control. NanoString analysis identified 16 differentially expressed genes in both SBC3 and SBC5 cells (DOX+ vs. DOX–), shown in Figure 4M. Among those 16 different expressed genes, several mitochondrial-related genes were downregulated after treatment. DNM1L expression was significantly decreased in both SBC3 and SBC5 cells after CRISPR KO. More genes, such as MYC, MFN1, MAPK1, COX411, CAD, and ATP5F, were downregulated in SBC3 cells than in SBC5 cells. We also checked the expression patterns of those 16 differentially expressed genes using in silico analysis with the public dataset GSE1037, which covers 11 of those 16 genes we identified in the NanoString experiment. The expression of most of these genes was increased in SBC3 and SBC5 cells compared with normal human lung tissue in GSE1037. Figure 4N shows the differential expression profiles of these 11 genes of SBC3 and SBC5 cells in GSE1037. SBC5 cells showed highly upregulated expression of those 11 genes compared with health normal lung samples. These results, together with the NanoString experiment, showed that DRP1 KO treatment significantly downregulates the expression of mitochondria-related genes. The pattern of high level of upregulation of mitochondrial-related genes observed in SBC5 cells was reduced after DOX treatment. In the case of the sample with moderate upregulation pattern for those mitochondrial-related genes (SBC3 cells), the change in expression pattern was reversed after treatment.
Together, our data demonstrate that inhibition of DRP1 by Teri in combination with Carbo induced apoptotic signaling and DNA damage response. Moreover, Teri&Carbo synergistically inhibits purines and pyrimidines through two potential targets, DRP1 and DHODH. Finally, DRP1 KO significantly inhibited SCLC proliferation through downregulation of mitochondrial-related genes.
Teri/Leflu treatment increases the efficacy of Lur in vitro and in vivo
In this study, we tested the combination of Lur with Teri to determine whether Teri&Lur would reduce SCLC cell viability. RNA pol II expression and phosphorylation were increased in SCLC cell lines compared with nonmalignant BEAS 2B cells (Figure S8A). We evaluated the synergistic effects of Teri and Lur in a panel of SCLC cell lines. A cell viability assay showed that the addition of Teri to Lur did not affect Lur-induced cytotoxicity in H69, H82, H446, SBC3, and SBC5 cells (Figure S8B).
We used multicellular tumor spheroids as a second model to validate the synergistic effect of Teri&Lur treatment on SCLC cell viability and apoptosis. In addition, we used an LDH cytotoxicity assay to monitor therapeutic efficacy in 3D culture systems to assess potential synergy between Teri and Lur. Treatment of H446 spheroids with Lur or Teri&Lur significantly increased the percentage of dead cells in a dose-dependent manner, and less cytotoxicity effect was observed after Teri treatment (Figure 5A, left panel). The CI was calculated using the Chou-Talalay method and showed synergistic effect on H446 spheroids. The CI value for combination treatment with 4315 nM Teri and 4315 p.m. Lur was 3-fold lower than that for single treatment. The CI for combination treatment was 0.3, indicating the synergistic effects of Teri&Lur (Figure 5A, right panel). Treatment of SBC3 spheroids with Teri&Lur identified low-dose, strongly synergistic effect on spheroid viability (CI = 0.0072) (Figure 5B).
Figure 5.
Spheroid response for drug treatment
(A and B) LDH cytotoxicity/death assay. The synergy of Teri&Lur was calculated by Chou-Talalay method. CI isobologram analysis indicated synergy for Teri&Lur doses below midline. Insert table shows CI of treatment.
(C and D) H446 and SBC3 spheroid cell lysates were subjected to immunoblotting with RNA poly II and DNA damage marker antibodies (left panel) and DRP1, MFN2, and ERK antibodies (middle panel). (Right panel) Cell-cycle regulatory antibody MDM2, p21, E2F1, CDK4, and cyclin D1 were used for western blotting detection after 72 h of drug treatment. Experiments were repeated independently three times with similar results.
(E) H446 spheroid MitoSOX production was detected and quantified with IncuCyte system (n = 3, biologically independent samples). Bar graphs show the maximal level of MitoSOX in H446 spheroids after 44 h of treatment. Bottom MitoSOX/brightfield images of spheroids. Scale bar: 100 μm.
(F) Dose-response relative ATP measurement in H446 spheroids after 72 h treatment with Teri, Lur, and Teri-Lur (n = 3, biologically independent samples). Significance between Lur and T-Lur treatments was assessed by Student’s t test; ∗∗p < 0.01, ∗∗∗∗p < 0.0001 (right panel).
(G) Mitochondrial representative images: EL (elongated), R (round), ITM (intermediate), and CL (cluster) with Imaris Surface detections of individual mitochondria. Scale bar: 0.3 μm.
(H) Panels show the percentage of mitochondria from the total number based on length ratios of round, elongated, and ITM groups. The average number of cells was 344–575 cells per group.
(I) In vivo efficacy of Leflu&Lur in mouse xenograft. Each value in graph line is the mean ± SD from five mice per group (control vs. combination, ∗∗∗∗p < 0.0001, Student’s t test).
(J) Control and treated FFPE mouse tumor sections were stained with phospho-DRP1 (Ser616). Displayed are representative single-channel Airyscan images for each group. Scale bar 20 μm. Multichannel images of pDRP1, EpCAM, and nuclei IF are available (Figure S11).
(K) Mean signal intensities were measured from confocal images with ZEN 2.3 Lite (see STAR methods).
(L) Schematic diagram displays the effect of Leflu&Lur combination on purine/pyrimidine pools in mouse xenograft.
(M and N) Purine and pyrimidine substances measured by LC-MS/MS in mouse tumor tissue at the end of the study. All substance concentrations are available in Table S5 and https://doi.org/10.17632/ghzcbd74tz.1. p values were calculated and were assessed with a linear model (ANOVA) see STAR methods, ∗∗p < 0.01, ∗∗∗p < 0.001. Bar graphs represent means ± SEM in (E), (F), (H), (K), (M), and (N). p values were calculated by ANOVA with Tukey post-test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 vs. control in (E), (H), (L), (M), and (N).
To further study spheroid sensitivity to Teri&Lur, we evaluated apoptosis, dynamic mitochondria protein levels, and the cell cycle in 3D spheroids. Treatment of H446 spheroids with Teri (800; 1,600; and 3,200 nM) or Lur (800; 1,600; and 3,200 pM) had a very small effect on the level of total or phosphorylated RNA pol II. In contrast, the combination of Teri&Lur inhibited RNA pol II activity and increased the levels of proteins related to DNA damage and replication stress, such as γH2AX and pChk1(Ser345) (Figure 5C). Next, we tested the effect of Teri and Lur on mitochondrial plasticity. H446 spheroid lysates were probed with DRP1 and MFN2 antibodies. The dose-dependent inhibitory effect of Teri (1,600 and 3,200 nM) on DRP1 phosphorylation at Ser616 in the 3D spheroids was comparable to our observations in 2D culture. Interestingly, the Lur-mediated reduction in DRP1 phosphorylation correlated with decreased ERK phosphorylation, indicating inhibition upstream of ERK, but there was no change in MFN2 levels. The inhibition of DRP1 phosphorylation increased upon combination Teri&Lur treatment (Figure 5C, middle panel).
The antiproliferative effect of Teri is related to cell-cycle arrest at G1/S.19 Thus, we also investigated the effect of Teri/Lur on the spheroid cell cycle after drug treatment. Figure 5C (right panel) shows the expression of MDM2, p21, and E2F1, which are regulators of p53 and cell-cycle cyclins, in H446 spheroids. MDM2 and E2F1 were downregulated with combination Teri&Lur treatment. Expression of the cell-cycle arrest protein p21 was strongly elevated with Teri and Teri&Lur treatment. Additionally, the cyclin-dependent kinase CDK4 expression decreased after Lur or Teri&Lur treatment, but the level of cyclin D1 was not affected.
SBC3 spheroids were treated with the same doses of Teri and Lur as H446 spheroids. Teri&Lur treatment had a more potent effect on SBC3 spheroids and was effective at lower concentrations (800 and 1,600 Teri/nM, Lur/pM). Combination Teri&Lur treatment slightly reduced the phosphorylation of RNA pol II and increased the level of γH2AX and pChk1(Ser345) (Figure 5D, left panel). Like the effects of Teri&Lur treatment on H446 spheroids, pDRP1 (Ser616) pERK levels were reduced in SBC3 spheroids after Teri&Lur (Figure 5D, middle panel). Additionally, SBC3 spheroids were analyzed for cell-cycle protein levels. Teri&Lur treatment decreased the expression of MDM2 and E2F1. Both Lur and Teri&Lur treatment upregulated p21. The activation of p21 with Lur and Teri&Lur inhibited cyclin D1 (Figure 5D, right panel).
Next, mitochondrial dysfunction was evaluated in H446 spheroids by measuring mitochondrial superoxide levels. Superoxide levels were unaffected by Teri or Lur treatment. However, a significant increase was observed after 40 h of Teri&Lur treatment (Figure 5E, left panel). Teri&Lur at two doses (1,600 and 3,200 nM/pM) led to an increase in mitochondrial ROS, as measured by an increase in MitoSOX intensity, in H446 spheroids (Figure 5E, right panel). ATP was then measured to test the effect of Teri and Lur on spheroid cytotoxicity and cell death; the ATP level of treated spheroids was normalized to that of the control. Treatment with Teri&Lur (1,600 nM/pM) significantly decreased the ATP level in the spheroids to 50% compared with that of the control. Single-drug treatment with the same concentration of Teri or Lur reduced ATP levels to approximately 80% and 60%, respectively. There was also a significant difference in ATP level upon treatment with Lur (1600 p.m.) and Teri&Lur (1600 nM/pM) compared to control treatment (Figure 5F). Teri and Lur demonstrated significant effectiveness of drug combination on mitochondria of H446 spheroids that induced an increase of mitochondrial superoxide decreased ATP production.
3D Imaris Surface objects were detected from mitochondria signal and displayed in Figure 5G as representations of the morphology categories. 2D subsets of mitochondrial fluorescent images were analyzed in Imaris to evaluate the effect of Teri&Lur on mitochondrial morphology. Bounding box OO lengths were generated from surface detection of individual mitochondria (Figure S9), and mitochondria were categorized into three groups: round, elongated, and intermediate. The percentage of round/fragmented mitochondria significantly decreased with Teri (1,600 nM) or Lur (1,600 p.m.) treatment or treatment with Teri&Lur at two doses (800; 1,600 nM/pM). The percentage of elongated mitochondria significantly increased only with 1,600 nM/pM Teri&Lur treatment. The percentage of ITM mitochondria was unaffected (Figure 5H). Thus, a significant decrease of round mitochondria after Teri&Lur is associated with inhibition of mitochondrial fission.
Finally, the efficacy of Teri&Lur was tested in SBC3 mouse xenograft model. Both Leflu (∼22%) and Lur (∼39%) alone, as well as the combination of Leflu&Lur (∼84%), significantly reduced tumor growth over time compared to the control (Figure 5I, Figure S10). Leflu&Lur also inhibited DRP1 phosphorylation at Ser616 (Figures 5J and 5K). Leflu, Lur, and Leflu&Lur treatment suppressed tumor growth by inhibiting DRP1activation. Results of immunofluorescence double staining of EpCAM with pDRP1 (Ser 616) are shown on Figure S11.
The inhibition of tumor growth can be regulated by interrupting active purine or pyrimidine pools.14,17 To investigate the effect of Leflu and Lur on purine and pyrimidine pathways, we evaluated the levels of purines and pyrimidines in mouse tumor tissue (Table S5). A schematic diagram shows the effect of Leflu&Lur on purine and pyrimidine pools (Figure 5L). The mathematical model (see STAR methods) was used to evaluate the potential synergistic effects of Leflu and Lur on the concentrations of purines and pyrimidines. Leflu&Lur synergistically affected the concentration of only one pyrimidine base, uridine, which is generated by the salvage pathway. The levels of dUTP and m5UTM, two substances generated from the de novo pyrimidine pathway, were significantly lower in Leflu&Lur than in the control (Figure 5M). Leflu&Lur treatment strongly reduced the concentrations of the purines TDP/dTDP and dGTP (Figure 5N). The mean levels of the five affected substances are shown in Table S6. Pharmacological inhibition of DRP1 and RNA pol II with Leflu&Lur significantly reduced tumor growth of SCLC cells in mouse xenograft.
These data indicate that Teri&Lur inhibited the cell cycle, as decreased numbers of H446 and SBC3 spheroid cells passed through G1 into S phase, thus blocking DNA replication and leading to diminished spheroid growth. Teri&Lur induced changes in mitochondrial morphology by notably reducing fragmented mitochondria in H446 spheroids. Leflu&Lur suppressed tumor growth by inhibiting DRP1 phosphorylation and mitochondrial fission and interrupting purine and pyrimidine pathways.
RNA-seq analysis of SBC3 spheroids
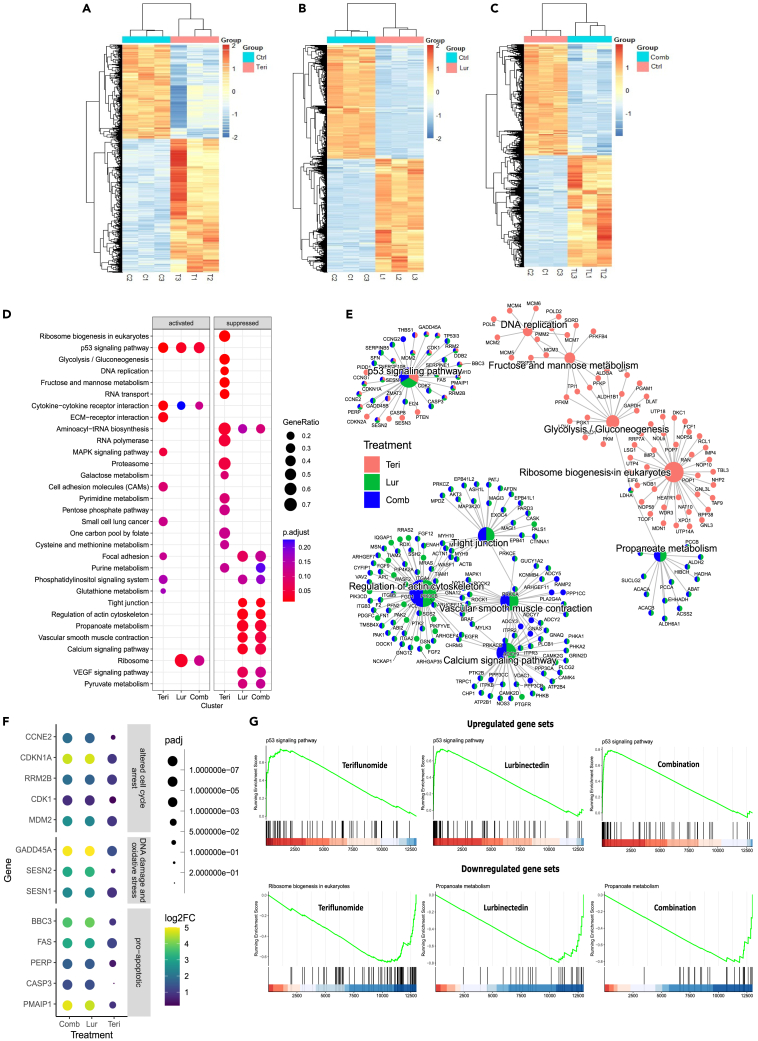
Next, we employed RNA sequencing to identify genes and pathways whose expression and activation, respectively, were significantly affected by 24 h of treatment with Teri or Lur alone or in combination in SBC3 spheroids. The results showed unique different gene expression profiles for each treatment compared with control treatment. Lur and Teri&Lur treatment caused almost 10 times more differentially expressed genes (FDR<0.05) than with Teri treatment (Lur 8724 genes, Teri&Lur 8098 genes, and Teri 832 genes) (Figures 6A, 6B, and 6C). The top upregulated and downregulated genes with log2-fold change and adjusted p values for each treatment are listed in Table S7. The functions and related pathways of these drug-treatment-specific different expressed genes were analyzed by gene set enrichment analysis (GSEA) with KEGG pathway database. The results demonstrated that the treatments regulated multiple signaling pathways. The p53 signaling pathway was activated in each drug treatment group. Excess activation of p53 induced cellular stress mechanisms, including DNA damage, cell-cycle arrest, and apoptosis (Figure 6D). The percentage of genes in the p53 signaling pathway with altered expression was 34% for Teri treatment, 38% for Lur, and 43% for Teri&Lur (Table S8, columns L and M). Teri treatment upregulated additional pathways, such as metabolic processes (the MAPK signaling pathway, phosphatidylinositol signaling system, and glutathione metabolism) and cellular communication pathways (ECM-receptor interactions, small cell lung cancer, and cell adhesion molecules) (Figure 6D). All drug-treated groups exhibited activated cytokine-cytokine receptor interactions; additionally, Lur and combination Teri&Lur treatment promoted ribosome signaling.
Figure 6.
RNA-seq-based gene expression profiles in response to different treatment
(A–C) The RNA-seq differentially expressed gene profile for Teri, Lur, and combination treatment (vs. control) (GSE267928).
(D) The comparison of top enriched KEGG pathways for the different treatment expression profiles.
(E) The network plot of top enriched KEGG pathways comparing these three different treatments and the significant alternated genes. The proportion of treatment in the pie chart is determined by the number of different expressed genes in each treatment expression profile.
(F) The dot plot for these different expressed genes in altered cell-cycle arrest, DNA damage and oxidative stress, and pro-apoptotic gene sets. The color of the dot represents the log2-fold change compared between the treatment and control. The dot size represents the adjusted p value.
(G) The GSEA plots for the top activated and top suppression KEGG pathways for each treatment.
We investigated the inhibitory effects of Teri, Lur, and combination treatment on signaling pathways. Aminoacyl-tRNA biosynthesis, which is responsible for protein synthesis and is involved in tumorigenesis,30 was suppressed by Teri, Lur, and Teri&Lur treatment (Figure 6D). Teri alone displayed robust metabolic effects and downregulated ribosome biogenesis in eukaryotes, glycolysis/gluconeogenesis, fructose/mannose and galactose metabolism, pyrimidine and purine metabolism, the pentose phosphate pathway, and cysteine/methionine metabolism (Figure 6D). In addition, Lur alone and in combination with Teri suppressed two other metabolic pathways, propanoate and pyruvate metabolism, which could contribute to the inhibition of disseminating metastatic cells.31,32 Lur alone or in combination with Teri negatively impacted cellular interactions in SBC3 spheroids through the inhibition of focal adhesion, tight junctions, the actin cytoskeleton, and calcium signaling pathways (Figure 6D). Two other drivers of tumor angiogenesis, the VEGF pathway and vascular smooth muscle contraction, were also suppressed by Lur and combination Teri&Lur treatment.
We then performed functional enrichment analyses to detect the top genes that regulate key signaling pathways after Teri, Lur, or Teri&Lur treatment (Figure 6E). In the p53 signaling pathway, Teri, Lur, and the combination of Teri&Lur altered cell-cycle arrest genes (MDM2, CDK1, RRM2B, CDKN1A, and CCNE2), DNA damage and oxidative stress genes (SESN1, SESN2, and GADD45A), and proapoptotic genes (PMAIP1, CASP3, PERP, FAS, and BBC3) (Figure 6F). In the ribosome signaling pathway, only the LDHA gene was targeted by all three treatments; this gene is associated with propanoate metabolism, which was affected by only treatment with Lur or Teri&Lur. Lur and Teri&Lur treatment had comparable effects on the expression of genes related to tight junctions, regulation of the actin cytoskeleton, vascular smooth muscle contraction, and calcium signaling pathways. These four pathways were not affected by Teri treatment (Figure 6E). GSEA detected upregulated genes related or critical to the p53 signaling pathway after Teri, Lur, or Teri&Lur treatment, leading to cycle arrest. Furthermore, the GSEA plot obtained after combination treatment indicated that Teri sensitized SCLC spheroids to Lur and mediated apoptosis by activating the p53 signaling pathway (Figure 6G). Additionally, combination treatment downregulated propanoate metabolism. These metabolic changes impaired mitochondrial oxidation and morphology. The RNA-seq results indicated the robust effect of combination Teri&Lur treatment on critical cancer pathways in an SBC3 spheroid model. Applying Teri and Lur at a low concentration range for 24 h increased cellular cytotoxicity, cell-cycle arrest, apoptosis, and DNA damage and suppressed cell-cell interactions and cellular metabolism. Functional enrichment analysis of the RNA-seq data yielded potential key targets critical to understanding the molecular mechanisms of Teri&Lur in SCLC. These data suggest that pharmacological inhibition of DRP1 activity and mitochondrial plasticity upregulate p53 signaling pathway and induce apoptosis in SCLC.
Discussion
SCLC patients often develop recurrence and disease progression after receiving regiments of chemotherapy, and the dismal survival rate has not changed in the last decades.33 Lack of response to available therapies warrants new therapeutic strategies to improve the prognosis of SCLC. A recent study showed that mRNA DRP1 levels were increased 2-fold in SCLC tissue compared with control tissue and reported a correlation between high expression of DRP1 and poor survival probability for males and females.34 Increasing evidence suggests that targeting DRP1 to suppress mitochondrial fission may be a potential strategy to treat SCLC. MDIVI1, a putative inhibitor of mitochondrial fission and the GTPase activity of DRP1, induces mitochondrial elongation in cells.35 Moreover, Parida et al. reported that targeting mitochondrial plasticity by knocking down DRP1 expression and inhibiting MDIVI1 attenuates oncosphere formation and reduces the brain metastatic potential of Lat and M-BM cells in preclinical models.36 In addition, several studies have reported that FDA-approved drugs Leflu and Teri promoted mitochondrial fusion15,20,37 and can influence DRP1. Our molecular modeling of the interaction of DRP1 with Leflu, Teri, and MDIVI1 revealed that Teri had the highest binding score, Leflu showed a less robust interaction, and MDIVI1 showed the lowest binding score. We found higher sensitivity of SCLC cells to Teri that can be explained by Leflu conversion to Teri; upon oral administration of Leflu over 80% of the drug is converting to Teri.38
The mitochondrial plasticity manifested by its fusion/fission cycle is a highly coordinated event regulated by a shared environment, communication, group formation, synchronization of behavior, and contact with other organelles.39,40 Our data indicate that Teri treatment was associated with an increase in mitochondrial area and correlated with mitochondrial elongation per cell. Teri treatment shifted the balance from mitochondrial fission to mitochondrial fusion in SBC3 and SBC5 cells and decreased mitochondrial fragmentation and cell proliferation. Mitochondrial fragmentation is activated by Ca2+/CaMKII/ERK/FAK pathway and is required for lamellipodia formation, leading to more efficient cell migration and metastasis.41 We found the migration and motility of SBC3 and SBC5 cells was significantly inhibited with Teri. Similarly, Teri treatment induced a significant decrease in DRP1 phosphorylation at Ser616 and reduced the pDRP1Ser616/637 ratio in a dose-dependent manner in SBC3 cells. Our results are consistent with the finding that pharmacological or genetic manipulation of DRP1 phosphorylation at sites Ser616/637 reduced tumor growth.7,10 However, the regulation of fusion activity by Teri is unclear, and future investigation is required to identify molecular mechanisms.
We observed that Teri&Carbo enhanced chemotherapy-induced cell cytotoxicity and apoptosis in SBC3 and SBC5 cells. Recent studies have reported that mitochondria contribute to cell chemotherapeutic resistance by creating mitochondrial dynamic imbalance.6,16,42 Consistent with these reports, we found that Teri directly targets DRP1; the Teri&Carbo treatment inhibited DRP1 phosphorylation at Ser616. Furthermore, combination treatment increased cleaved PARP and p-p53 levels and modulated DNA damage responses including ChK1 and histone phosphorylation. And accumulation of p53 in mitochondria can induce proapoptotic Bax and Bak activation during the process of cell death.28,29 Thus, Teri&Carbo combination treatment presents an exciting treatment option for SCLC patients with DRP1 overexpression.
Leflu and Teri inhibit the mitochondrial enzyme DHODH, which regulates pyrimidine nucleotide biosynthesis. SBC3 and SBC5 cells have elevated levels of DHODH compared with those of nonmalignant BEAS2B cells. The measurement of pyrimidines in SBC3 cells after Teri&Carbo treatment showed a significant synergistic reduction in the levels of deoxyribonucleotides. There was no significant effect on SBC5 cells; however, we observed a decrease in the levels of several molecules upon Teri&Carbo treatment. Our findings showing the inhibition of DRP1 phosphorylation at Ser616 and a reduction in purine and pyrimidine concentrations in SBC3 cells suggest crosstalk between pyrimidines with mitochondrial fission proteins in SCLC. Interestingly, there appears a correlation between pyrimidine metabolism and mitochondrial dynamics in an MFN1-/MFN2-dependent manner.14 Thus, our data underscore the potential involvement of mitochondrial plasticity protein DRP1 in regulation of purine and pyrimidine pathways.
Knockdown DRP1 in SBC3 and SBC5 cells significantly suppressed cell proliferation. The expression patterns of 16 metabolic and mitochondrial genes correlated with the knockdown of DNM1L, which significantly decreased the expression of nine genes in SBC3 cells: the mitochondrial genes MFN1, COX4IL, COX14, and ATP51D and the metabolic genes MYC, MAPK1, CAD, and TIGAR. Downregulation of CAD expression by DNM1L-KO confirmed the possibility of crosstalk between the de novo pyrimidine generation pathway and mitochondrial fission in SBC3 cells. Interestingly, mitochondrial fusion is required for innate immune signaling and is regulated through DHODH inhibition,43 and viral infection is known to limit the interferon (IFN) response by promoting DRP1 phosphorylation and mitochondrial fission.44 Furthermore, ATP5B KO cells contain mitochondria without cristae and modulate the mitochondrial morphology apoptosis-related pathways.45,46 Nonetheless, it remains unknown how DRP1 accelerates mitochondrial morphological alterations and functions of metabolic and mitochondria genes during mitochondrial dynamics in SCLC, and further investigations are required for clarification.
In parallel with Teri&Carbo treatment, we also tested combination treatment with Teri&Lur in 2D and 3D SCLC cell cultures. Interestingly, we observed a synergistic effect in only the SCLC spheroids. Only a few studies have compared the effects of combination drug treatment in these two culture formats. The concentrations of Lur (∼0.8–3.2 μM) used in combination treatment are 100 times less than the peak Lur plasma concentrations.47 But even at the lowest concentration of Teri&Lur combination the level of DRP1 phosphorylation at Ser616 was significantly inhibited in H446 and SBC3 spheroids. Furthermore, Teri&Lur reduced RNA pol II activity and increased the levels of DNA damage and replication stress proteins such as γH2AX and pChk1(Ser345) consistent with other studies using combination treatment of ceralasertib/berzosertib with lurbinectedin48 or ONC201&Lur.49
Mitochondrial dynamics respond to various stressors and regulate reactive oxygen species (ROS), pro-apoptotic molecules, metabolic intermediates, and adenosine triphosphate (ATP). Mitochondrial ROS are known to have dual actions in cancer, as elevated ROS concentrations affect cell survival and increase cell death, and at lower concentrations, ROS act as signaling molecules that mediate redox signaling events beneficial to tumor progression. ROS are DNA damage mediators whose accumulation induces mitochondrial DNA degradation. We found the amplification of mitochondrial superoxide levels in H446 spheroids due to mitochondrial depolarization and cell death induced by Teri&Lur treatment. We also observed that Lur and Teri synergistically decreased the ATP concentration more effectively than treatment with only Lur, the effects of which were recently published.50 Further investigations are needed to understand how the Teri&Lur combination treatment influences mitochondrial ROS.
Finally, we detected decreased mitochondrial fragmentation in H446 spheroids treated with Teri and Teri&Lur using Imaris Surface for SCLC cells. As well, Leflu&Lur treatment inhibited mouse tumor growth by inhibiting DRP1 activation and the pyrimidine salvage pathway. Phenotypic synergy between Teri and Lur was confirmed by GSEA. Furthermore, RNA-seq results using a cancer hallmark collection showed the significant upregulation of apoptosis-related genes through activation of the p53 pathway and downregulation of genes involved in cellular communication and motility, angiogenesis, and metabolic function.
Despite the therapeutic potential of our findings, the present study has some limitations. For example, the effect of Carbo or Lur with Leflu combination was tested only in mouse xenograft model but efficacy needs to be determined in metastatic lung cancer model to inspire additional confidence. Similarly, in vivo pyrimidine metabolism and mitochondrial dynamics in SCLC tumors are likely to be different from those observed in SCLC cell lines. Perhaps, live imaging of mitochondria in mouse and patient tissue samples would help to evaluate the direct effects of combination treatment on mitochondrial morphology.
Nonetheless, taken together, the results of the present study indicate that (1) targeting the fragmented mitochondrial network in SCLC tumors, which have high DRP1 expression, can re-sensitize resistant tumors and (2) targeting of mitochondrial plasticity pharmacologically is a potential therapeutic approach to suppress SCLC progression. These results underscore the role of phenotypic plasticity of cancer cells and pave the way for clinical trials in the future to validate these treatment strategies in patients with SCLC.
Limitations of the study
Although we have exercised caution in interpreting our results, it is possible that the absence of uridine in the medium may lead to an overestimation of in vitro results since pyrimidine synthesis depends solely on DHODH inhibited by Teri. It will be interesting to test the addition of uridine in future in vitro studies to explore the precise beneficial effects of teriflunomide on SCLC cells. It would also be worthwhile to test the effect of Teri/Carbo and Teri/Lur on DHODH activity. Secondly, a study exploring Teri and Leflu in combination with Carbo and Lur using SCLC PDX models, though imminent, was beyond the scope of the present study. Finally, demonstrating interaction between DRP1 and DHODH and mitochondrial plasticity would lend further credence to the present study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Actin, Pan Antibody, Unconjugated | Cell Signaling Technology | Cat# 4968; RRID:AB_2313904 |
| Rabbit Anti-Human DRP1, phospho (Ser616) Polyclonal Antibody, Unconjugated | Cell Signaling Technology | Cat# 3455; RRID:AB_2085352 |
| Phospho-DRP1 (Ser637) Antibody | Cell Signaling Technology | Cat# 4867; RRID:AB_10622027 |
| Histone H2B (V119) Antibody | Cell Signaling Technology | Cat# 8135; RRID:AB_10891053 |
| Phospho-Histone H2A.X (Ser139) Antibody | Cell Signaling Technology | (1) Cat# 2577; RRID:AB_2118010 |
| Phospho-p53 (Ser15) Antibody | Cell Signaling Technology | Cat# 9284; RRID:AB_331464 |
| p53 (1C12) Mouse mAb | Cell Signaling Technology | Cat# 2524; RRID:AB_331743 |
| Cleaved PARP (Asp214) (D64E10) XP Rabbit mAb | Cell Signaling Technology | Cat#5625; RRID:AB_10699459 |
| Phospho-Chk1 (Ser345) (133D3) Rabbit mAb | Cell Signaling Technology | Cat# 2348; RRID:AB_331212 |
| p21 Waf1/Cip1 (12D1) Rabbit monoclonal antibody | Cell Signaling Technology | Cat# 2947; RRID:AB_823586 |
| Anti-β-Actin Antibody | Sigma | Cat# A5441; RRID:AB_476744 |
| Phospho RNA Polymerase II (S2) Antibody | Thermo Fisher Scientific | Cat# A300-654A; RRID:AB_519341 |
| Purified anti-RNA Polymerase II | BioLegend | Cat# 904001; RRID:AB_2565036 |
| DHODH (E−8) | Santa Cruz Biotechnology | Cat# sc-166348; RRID:AB_2091729 |
| DRP1 (C-5) | Santa Cruz Biotechnology | Cat# sc-271583; RRID:AB_10659110 |
| GAPDH Antibody (0411) | Santa Cruz Biotechnology | Cat# sc-47724; RRID:AB_627678 |
| c-Myc Antibody (9E10) | Santa Cruz Biotechnology | (2) Cat# sc-40; RRID:AB_627268 |
| Tom20 (F-10) | Santa Cruz Biotechnology | Cat# sc-17764; RRID:AB_628381 |
| Fis1 (B-5) | Santa Cruz Biotechnology | Cat# sc-376447; RRID:AB_11149382 |
| OPA1 (D-9) | Santa Cruz Biotechnology | Cat# sc-393296; RRID: AB_3101815 |
| Mitofusin 2 antibody | Abcam | (3) Cat# ab56889; RRID:AB_2142629 |
| Chk1 | Proteintech | Cat# 60277-1-Ig; RRID:AB_2881396 |
| MDM2 antibody | Proteintech | Cat# 27883-1-AP; RRID:AB_2881003 |
| CDK4 antibody | Proteintech | Cat# 11026-1-AP; RRID:AB_2078702 |
| Cyclin D1 antibody | Proteintech | Cat# 60186-1-Ig; RRID:AB_10793718 |
| E2F1 antibody | Proteintech | Cat# 66515-1-Ig; RRID:AB_2881878 |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640, 1X with Glutamine | Corning® | Cat# 10-040-CV |
| Bovine Serum Albumin, lyophilized powder, ≥96% (agarose gel electrophoresis) | Sigma | Cat# A2153-100G |
| Dry Milk Powder; Non-Fat | Research Products International | Cat# M17200–500.0 |
| Fetal Bovine Serum Premium Select, Heat Inactivated | R&D Systems | Cat# S11550H |
| MitoSOX™ Mitochondrial Superoxide Indicators, for live-cell imaging | Thermo Fisher Scientific | Cat# M36008 |
| Teriflunomide | TOCRIS | Cat# 5069 |
| Leflunomide | TOCRIS | Cat# 2228 |
| Lurbinectedin (Synonyms: PM01183; LY-01017) | MedChemExpress | Cat# HY-16293 |
| Critical commercial assays | ||
| Cell Counting Kit-8 (CCK-8) | DOJINDO | Cat# CK04-05 |
| MycoAlert® Mycoplasma Detection Kit (100 Tests) | Lonza | Cat# LT07-318 |
| CyQUANT™ LDH Cytotoxicity Assay | Thermo Fisher Scientific | Cat #C20300 |
| CellTiter-Glo® 3D Cell Viability Assay | Promega | Cat# G9683 |
| LC-MS analysis | Creative Proteomics (Shirley, NY) | https://metabolomics.creative-proteomics.com/pyrimidine-metabolism-service.htm |
| Experimental models: Cell lines | ||
| NCI-H524 [H524] | ATCC | Cat# CRL-5831 ™ |
| NCI-H526 [H526] | ATCC | Cat # CRL-5811 ™ |
| NCI-H69 [H69] | ATCC | Cat# HTB-119 ™; RRID:CVCL_1579 |
| DMS 114 | ATCC | Cat# CRL-2066 ™ |
| BEAS-2B | ATCC | Cat# CRL-3588; RRID:CVCL_0168 |
| DMS 273 | Park et al. (gift from Nakamura Lab) | Sigma; Cat# 95062830 |
| SBC-3 | Park et al. (gift from Nakamura Lab) | Glow Biologics; Cat# GBTC-1122B; RRID:CVCL_1678 |
| SBC-5 | Park et al. (gift from Nakamura Lab) | Glow Biologics; Cat# GBTC-1121B; RRID:CVCL_1679 |
| Experimental models: Organisms/strains | ||
| Female NCI Ath/nu MOUSE | Charles River | Cat # 553NCIATH/NU |
| Recombinant DNA | ||
| pCW-Cas9 (Plasmid #50661) Standard format: Plasmid sent in bacteria as agar stab | ADDGENE | Cat# 50661; RRID:Addgene_50661 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software https://graphpad.com/ |
v8; RRID:SCR_002798 |
| Incucyte Software | Sartorious https://www.sartorius.com/en/products/live-cell-imaging-analysis/live-cell-analysis-software | RRID:SCR_017316 |
| nSolver Analysis Software | NanoString https://nanostring.com/ |
v4.0; RRID:SCR_003420 |
| Imaris | Bitplane, AG https://imaris.oxinst.com/ |
v9.9.0; RRID:SCR_007370 |
| QuPath | QuPath: Open Software for Bioimage Analysis https://qupath.github.io/ |
v0.2.0M9; RRID:SCR_018257 |
| R Project for Statistical Computing | The R project for Statistical Computing https://www.r-project.org/ |
RRID:SCR_001905 |
| ggplot2 | ggplot2 package in R | v3.4.0; RRID:SCR_014601 |
| LIMMA | LIMMA package in R | RRID:SCR_010943 |
| featureCounts | program in Subread package in R | RRID:SCR_012919 |
| DESeq2 | DESeq2 package in R | v1.38.2; RRID:SCR_015687 |
| Heatmap | heatmap package in R | v1.0.12 |
| clusterProfiler | clusterProfiler package in R | v4.6; RRID:SCR_016884 |
| ZEISS ZEN Microscopy Software | Carl Zeiss https://www.zeiss.com/microscopy/en/products/software/zeiss-zen-lite.html |
v2.3 Lite; RRID:SCR_013672 |
| Deposited data | ||
| LC-MS result for the synergistic effect of Teri&Carbo on the concentrations of pyrimidine/purine bases in SBC3 cells | Mendeley database | https://doi.org/10.17632/zfmb6dtw78.1 |
| LC-MS result for the effect of Leflu&Lur combination on purine/pyrimidine pools in SBC3 mouse xenograft | Mendeley database | https://doi.org/10.17632/ghzcbd74tz.1 |
| SBC3 RNA-Seq Sequences data | Gene Expression Omnibus | GSE267928 |
Resource availability
Lead contact
Further information and requests for resources and reagents is available from the lead contact, Dr. Ravi Salgia (rsalgia@coh.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon reasonable request.
This paper does not report the original code.
Further details or additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Mouse model
Athymic nude nu/nu mice (8-weeks-old female) were obtained from Charles River, Wilmington, MA, and were acclimated for one week before beginning the experiment. All animal experiments were carried out using a protocol approved by the Institutional Animal Care and Use Committee COH (IACUC # 16004). For treatments, mice were divided into 4 groups of five mice each and subcutaneously injected in one flank with suspensions of 2 × 106 SBC3 or SBC5 cells in 100 μL of PBS. Treatment started on the 14th day after cell injection when tumors became palpable. Two in vivo studies were performed: 1) SBC3 and SBC5 xenografted mice were treated with i) vehicle control, ii) Carbo 50 mg/kg, iii) Leflu 7.5 mg/kg, or iv) Carbo/Leflu; and 2) SBC3 xenografted mice were treated with i) vehicle control, ii) Lur 0.5 mg/kg, iii) Leflu 3.8 mg/kg, or iv) Lur/Leflu. Treatments were administered to the mice via intraperitoneal injection twice a week for 6 weeks. Animals were examined daily for signs of tumor growth. Tumors were measured in two dimensions using calipers, and body weight was recorded. At the end of the study, the mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Tumors were removed on day 40 and photographed. A portion of the tumor was fixed in 10% buffered formaldehyde solution and paraffin-embedded for Formalin-fixed, paraffin-embedded (FFPE). Tumor sections were deparaffinized and rehydrated, followed by heat-mediated antigen retrieval (pH 6, Dako). After permeabilization and blocking, tumor sections were stained with pDRP1 Ser616 antibody overnight, washed, incubated with Alexa 488 secondary antibody for 1 h, mounted and observed on an LSM 880 with a 10×/0.45NA objective (Carl Zeiss). Immunofluorescent confocal images were acquired for each treatment (magnification: 100×). Mean immunofluorescence intensities were calculated with ZEN 2.3 Lite software.
Cell culture
H524, H526, H69, DMS114 SCLC cells and BEAS 2B, normal epithelial cells, were purchased from American Type Culture Collection (Manassas, VA). DMS273, SBC3 and SBC5 SCLC cells were received from Dr. Yusuke Nakamura (University of Chicago). Cells were maintained in RPMI 1640 medium (Thermo Fisher Scientific Waltham, MA) supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) penicillin/streptomycin (Corning Life Science) and l-glutamine at 37°C with 5% CO2. Cells were routinely screened for mycoplasma (MycoAlert kit, Lonza, Basel, Switzerland).
Method details
Immunoblotting
Total SCLC cell lysates were prepared using radioimmunoprecipitation assay (RIPA) lysis buffer. For immunoblotting, 25–50 μg protein was separated on 4–15% SDS-PAGE gels and transferred onto Immobilon membranes (Millipore, MO) then blocked in 5% BSA or TBST-milk for 1–2 h. Membranes were washed incubated with primary antibody for overnight at 4 °C on a shaker. After three times of wash (10 min, TBS-T) membranes were incubated with secondary antibodies for 1 h, washed three times and were developed using a Bio-Rad imager and Medical film processor. Representative WB panels for Figures 1, 2, 4, and 5 with weight/size markers are shown in Supplemental data (Data S1).
CCK 8 assay
Cells were plated in 96 well plates at a density of 10, 000 cells per well and treated with Tei, Carbo and combination or with Teri, Lur and combination for 3 days. Cell cytotoxicity was evaluated using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD). The absorbance of 450 nm was determined by a microplate reader (Tecan Spark 10, Salzburg, Austria).
pDRP1 immunofluorescence and analysis
SBC3 and SBC5 cells were grown on glass coverslips and treated with various optimized concentrations of Teri overnight for immunofluorescent staining. Next, the cells were fixed in 4% paraformaldehyde, permeabilized, and blocked before incubation with the pDRP1 (Ser616) antibody overnight, followed by incubation with the Alexa 488 secondary antibody and staining with the Tom20 (F-10) antibody. A far-red dye was used for cytoplasmic staining. After washing with TBS-T the coverslips were mounted on slides using Vectashield with DAPI. Images were acquired on an Axio Observer 7 inverted microscope (Carl Zeiss, Jena, Germany). All imaging was performed using a 40x/0.95NA Plan-Apochromat objective with a final per pixel scaling of 0.147 by 0.147 microns. Within each 3 × 3 field of view (>200 cells per image). QuPath 0.2.0M9 was used to detect cells from the presence of both nuclei and cytoplasmic signal. The analysis area was further defined by a far-red (cytoplasmic) signal intensity threshold through the subcellular detection command. The mean pDRP1 signal intensity was then measured within that area. The green intensity sum of these areas was calculated per cell and then averaged for each sample image.
Cell line generation using CRISPR‒Cas9
DRP1 KO sgRNA was designed using the GPP sgRNA Designer, after confirming the CRISPR editing efficiency by the surveyor nuclease assay, two sgRNAs, hDRP1-EX7-CR1 (5′-CAAGTTTAGTGATTACAGCT-3′) and hDRP1-EX8-CR1 (5′-TCTTGTTGTTAATATCTAGC-3′), were selected for use in the project (Figure S4B). In the KO experiments, the SBC3 and SBC5 cell lines were transfected with pCW-Cas9-Puro (Addgene #50661), a Tet-mediated Cas9 lentiviral vector with a puromycin-selectable marker. Transfected cells were selected with 1 μg/mL puromycin and then transfected with LV-Blast-DRP1-dCREX7-EX8-CRs expressing both sgRNAs hDRP1-EX7-CR1and hDRP1-EX8-CR1, followed by blasticidin screening. After selection, the SBC3 KO and SBC5 KO cell pools which transduced with vCW-Cas9-Puro and vLV-Blast-DRP1-dCREX7-EX8-CRs virus were analyzed by NGS) to evaluate KO efficiency. CRISPR experiments were performed by The Gene Editing and Viral Vector Core at City of Hope (Duarte, CA).
NanoString differential expression analysis
The expression of a custom panel of 20 genes was assessed using a NanoString nCounter platform (NanoString Technologies, Inc., Seattle, WA). All experimental procedures were performed following the manufacturer’s protocol with minor modifications. Briefly, 100 ng of RNA extracted from SBC3 and SBC5 cells in the presence or absence of doxorubicin (DOX) was hybridized with the Reporter CodeSet and the Capture ProbeSet at 65°C for 18 h. Next, all reagents were transferred, magnetic beads separated, and molecular complexes immobilized on the sample cartridge using the Prep Station (nCounter MAX/FLEX). Subsequently, the cartridge was scanned using an nCounter Digital Analyzer, and finally, the data were analyzed using nSolver Analysis Software 4.0. Each experiment was conducted in triplicate. Genes with a normalized gene expression level below the background threshold were excluded from the differential expression analysis. Expression patterns are presented in dot plots prepared using the R ggplot2 package. The assay and data analysis were performed by the Research Molecular Pathology facility (City of Hope, Duarte, CA).
In silico analysis
A cDNA microarrays (CHUGAI 41K platform) gene expression dataset (GSE1037) containing 19 unrelated normal human lung tissue samples (GSM17287∼GSM17305), and data from SBC3 (GSM17313) and SBC5 (GSM17314) cells were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO). The array annotation files were downloaded from GEMMA. Differential gene expression analysis was performed using GEO2R with the R LIMMA package. The array data was log2 transformed and quantile normalized. The sample data were grouped based on the sample type and were fit to the linear model using lmFit function from R LIMMA package. The different expression genes were analyzed using Empirical Bayes Statistics for Differential Expression function from R LIMMA package at an FDR<0.05. The data are presented in dot plots prepared using the R ggplot2 package. Most genes have multiple clones on this array platform. Each row represents a clone labeled with a gene symbol and probe ID.
LC‒MS analysis
Before LC-MS SBC3 and SBC5 cells were treated with Teri, Carbo and combination at the concentrations presented in original results. Mouse tumor tissue samples for the assay were collected from a mouse xenograft experiment with Leflu and Lur. Cell and tissue samples were processed by Creative Proteomics (Shirley, NY). Cell samples were placed into 250 μL of 80% methanol. Tissue samples were weighed in 2-mL Eppendorf tubes, and 10 μL of 80% methanol was added per mg of raw tissue. The tissue and cell samples were then homogenized on an MM 400 mill mixer with the aid of two 4-mm metal beads at a shaking frequency of 30 Hz for 2 min, and the homogenization step was repeated twice. The samples were placed at −20°C for 1 h and then centrifuged at 21,000 × g for 10 min at room temperature. The clear supernatants were collected for LC‒MS quantification. The protein content of clear supernatants was measured using the Bradford protein assay, and these values were used to normalize metabolite concentrations in cell pellets. A stock solution containing standards of all the targeted nucleotides, nucleosides, and nucleobases was prepared at 200 nmol/mL each in an internal standard solution of 13C- and/or 15N-labeled AMP, ATP, GMP, GTP, UMP, UTP, adenine and adenosine. This solution was serially diluted with the same solution to prepare 10-point calibration solutions at a concentration range of 0.0001–50 μM. Ten microliters of the supernatant of each sample was mixed with 90 μL of the internal standard solution. Ten microliter aliquots of the sample solutions and the standard solutions were injected into a C18 LC column (2.1∗150 mm, 1.9 μm), and UPLC-MRM/MS was run on a Waters Acquity UPLC system coupled to a Sciex QTRAP 6500 Plus mass spectrometer (AB Sciex LLC, MA) operated in negative-ion mode to detect nucleotides. The mobile phase was a triethylamine buffer (A), and acetonitrile/methanol (1:1) (B) was used for binary gradient elution (5%–60% B in 25 min) at 0.25 mL/min and 55°C. For quantitation of nucleosides and nucleobases, 10-μL aliquots of the sample solutions and the standard solutions were injected onto a polar reversed-phase C18 column (2.1∗100 mm, 1.6 μm), and UPLC-MRM/MS was run on a Waters Acquity UPLC system coupled to a Sciex QTRAP 6500 Plus mass spectrometer operated in positive-ion mode. The mobile phase was 0.1% formic acid–ammonium formate buffer (A), and methanol (B) was used for binary gradient elution (0%–40% B in 15 min) at 0.25 mL/min and 30°C. Concentrations of the nucleotides, nucleosides or nucleobases detected in the UPLC-MRM/MS runs were calculated with internal standard calibration by interpolating the constructed linear regression curves of individual compounds, with the analyte-to-internal standard peak area ratios measured from the sample solutions.
Spheroid growth and staining
SBC3 and H446 cells were plated at a density of 10,000 cells per well in 96-well ULA round-bottomed plates (S-Bio, Hudson, NH) and incubated for 72 h at 37°C, 5% CO2, and 95% humidity. Spheroids were transferred to low-cell-attachment dishes, grown for two days, and treated with the appropriate drug(s). After treatment, spheroid samples were investigated by Western blot analysis. For mitochondria visualization, the spheroids were stained with 250 nM MitoTracker Red CMXRos and Hoechst (Thermo Fisher Scientific, Waltham, MA) for 2 h after drug treatment. Spheroids were washed, fixated with 4% PFA, transferred into 96-well flat glass-bottom black microplates (Cellvis, Mountain View, CA), and mounted in wells. Mitochondria were visualized with LSM 880 inverted microscope (Carl Zeiss).
H446 spheroid image acquisition
Images of mitochondria were acquired on an LSM 880 inverted microscope (Carl Zeiss) controlled with Zen software (2.3 Black) equipped with a 63×/1.4NA Plan-Apochromat Oil objective and captured with GaAsP-PMT detectors. Z-stacks were acquired in Airyscan Fast mode with a 2.0 μm interval and optical zoom of 2.0. Images had a width and height of 1900 × 1900 pixels and a spatial resolution of dx = 0.04 μm, dy = 0.04 μm and dz = 2 μm. In Airyscan Fast mode, excitation was provided by 405 nm (1.0%) and 561 nm (1.0%) lasers with the following gain and detection wavelengths: DAPI 670 V (450 nm) and Red 650 V (579 nm). 2D images were acquired with a 20×/0.8NA Plan-Apochromat objective and captured with PMT detectors. Images had a width and height of 2580 × 2580 pixels and a spatial resolution of dx = 0.08 μm and dy = 0.08 μm. Excitation was provided by 405 nm (0.3%) and 561 nm (0.2%) lasers with the following gain and detection wavelengths: DAPI 669.1 V (410–490 nm) and Red 639.1 V (569–620 nm).
Imaris method to analyze images of 2D cell monolayer and spheroid
Imaris 9.9.0 (Bitplane, AG), with the Surfaces and Cells modules, was used to obtain mitochondrial morphology data from 2D images and 2D z stack subsets. Surfaces were used to evaluate individual mitochondria from cell monolayers and spheroids. Detection parameters were optimized for visible mitochondria and nuclei in each group with varying signal intensities. Roundness ratios were calculated from surface-generated bounding box OO length measurements. Object lengths less than 0.1 μm were excluded, and objects with a length greater than 1.26 μm were separately analyzed as clusters. Thresholds were uniformly applied to roundness ratios to morphologically categorize mitochondria as elongated (≤0.599), intermediate (≥0.600–0.649), or round/fragmentated (≥0.650). A perfectly round object has a ratio of 1.0. The percentage of each mitochondria type was calculated from the total amount of mitochondria in each treatment group. The Cells module was used to detect and quantify mitochondrial and the nuclear area from cell monolayers. Identical parameters were applied to all groups with cell boundary detection based on mitochondrial signals. Imaris includes nuclei in cytoplasm measurements, so the mitochondrial area is calculated by subtracting the nucleus area from the cell cytoplasm area. Abnormal nuclei (<100 μm2, >750 μm2) were excluded.
Spheroid LDH assay
LDH activity was measured using an LDH Cytotoxicity Assay Kit following the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). Briefly, after 72 h of drug treatment, H446 and SBC3 spheroids were mixed with 10 μL of 10× lysis buffer and incubated for 45 min in a cell culture incubator. The absorbance (at 490 nm and 680 nm) by 50 μL of lysed cells from each sample was measured by a plate reader (Tecan Spark 10, Salzburg, Austria). The percentage of cell cytotoxicity/death was calculated by using the following formula:
Spheroid MitoSOX assay
The IncuCyte system recorded two days of MitoSOX Red dye (Thermo Fisher Scientific, Waltham, MA) penetration in live H446 spheroids. Spheroids were labeled with 5 μM dye, and red fluorescence was assessed by IncuCyte software. Fluorescence and brightfield images were acquired on an Axio Observer 7 (Carl Zeiss, Jena, Germany) inverted microscope equipped with a 10×/0.45NA Plan-Apochromat objective, TL halogen lamp, Colibri 7, and filter set 90 HE and captured on a Zeiss Axiocam 702 CMOS camera. The exposure times and wavelengths were as follows: 15 ms (red fluorescence), 9.4 ms (brightfield), 555 BP/30 excitation filter, and QBP 592/25 emission filter.
Spheroid ATP assay
A Cell Titer Glo/ATP assay (Promega, Madison, WI) was used to measure ATP levels in spheroids. After 72 h of drug treatment, the intracellular spheroid ATP concentration was measured by luminescence following the manufacturer’s protocol using a Tecan Spark 10 plate reader.
RNA isolation and data processing
Total RNA was extracted from SBC3 control spheroids and those treated with Teri (3200 nM), Lur (1600 p.m.) or Teri/Lur for 24 h and purified using the RNeasy Plus Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The sample libraries were sequenced on an Illumina HiSeq4000 (Illumina, San Diego, CA) to generate 101 bp paired-end reads. First, the adaptor sequences and polyA sequences were trimmed and filtered out using fastp (ver 0.23.2). Then, the RNA-seq data were aligned to a reference sequence (hg38) using STAR (ver2.7.9) with the Gencode annotation database (v38). The R Subread package with the featureCount function generated RNA-seq raw counts for quantification (ver2.0.3). Genes expressed at very low levels, with maximum expression <0.1 RPKM from all samples, were excluded from downstream differential expression analysis. Differential expression analysis on the basis of RNA-seq results was performed using the DESeq2 package (ver 1.38.2). A heatmap of differentially expressed genes was generated using R heatmap (ver 1.0.12). The genes were sorted by signed p value for GSEA, using R clusterProfiler (ver 4.6) with the KEGG pathway database (https://www.genome.jp/kegg/pathway.html). A dot plot reflecting the differentially expressed genes was generated using R ggplot2 (ver 3.4).
Quantification and statistical analysis
Statistical analysis
All data are presented as the mean ± SE or mean ± SD. Data from two sample groups were compared by unpaired, two-sided Student’s t tests. Data from more than two groups were analyzed by one-way ANOVA followed by Tukey’s multiple comparison tests. Values of p < 0.05 were considered to indicate statistical significance and are indicated as follows: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Data analysis was conducted using GraphPad Prism 8 software (La Jolla, CA). The tumor weights were compared between groups using an unpaired Student’s t test.
Statistical analysis of purine and pyrimidine concentrations
The effect of treatment on the concentration of purines and pyrimidines for both cell lines and xenografts were modeled as follows: concentration = Treatment1 + Treatment2 + Treatment1 ∗Treatment2, where Treatment1 and Treatment2 were set as factors 1/0, treatment applied or not respectively. For the cell lines, Treatments 1 and 2 were Carbo and Teri, while for the xenografts, they were Lur and Leflu. The significance of the effects was determined by calculating p values associated with the coefficients of the model. Specifically, the data were imported into the R environment and the generalized linear model function “lm” was applied to calculate the model coefficients as well as the p values. The p values were FDR corrected to account for repeated testing. The supplementary archive with R code and data are provided at
Additional resources
This study has not generated or contributed to a new website/forum and is not part of a clinical trial.
Acknowledgments
Research reported in this publication included work performed in the Shared Resource Microscopy Digital Imaging Core, Integrative Genomics Core, and Genome Editing Core and supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. Work in Dr. Salgia’s laboratory was also supported in part by the United States Department of Defense grant HT9425-23-1-0581. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Michael S Nelson, a former member of the Light Microscopy Digital Imaging Core City of Hope for method development and result analysis of mitochondrial network. We would also like to thank Eric F Medina for the support throughout RNA-seq data analysis. The authors wish to express their apologies to all those whose important work they were unable to cite due to limitations on space and the number of references permissible. Graphical abstract was created with BioRender.com.
Funding Statement: research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number 5R01CA218545-05 and CCSG—P30 CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization: T.M., S.T.R., S.S.S., P.K., and R.S. Experimental performance, data analysis, and interpretation: T.M., L.T., B.M., H.L., C-H.L., A.P., S.K.R., S.N., I.M., S.T.R., D.L.W., S.S.S., and R.S. Data curation and formal analyssis: T.M., L.T., B.M., H.L., I.M., C-H.L., A.P., S.K.R., S.N., B.A. Writing: T.M., L.T., I.M., S.S.S., P.K., R.S. Manuscript revision: T.M., L.T., B.M., H.L., C-H.L., A.P., S.K.R., S.N., I.M., B.A., J.M., L.A., M.W.N., S.K.B., S.T.R., D.L.W., S.S.S., P.K., and R.S. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
R.S., S.T.R, and T.M. report a patent pending for combination treatment of teriflunomide&leflunomide with carboplatin&lurbinectedin.
Published: May 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110132.
Supplemental information
References
- 1.Koinis F., Kotsakis A., Georgoulias V. Small cell lung cancer (SCLC): no treatment advances in recent years. Transl. Lung Cancer Res. 2016;5:39–50. doi: 10.3978/j.issn.2218-6751.2016.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin C.M., Ismaila N., Hann C.L., Malhotra N., Movsas B., Norris K., Pietanza M.C., Ramalingam S.S., Turrisi A.T., Giaccone G. Treatment of small-cell lung cancer: American Society of Clinical Oncology endorsement of the American College of Chest Physicians guideline. J. Clin. Onco. 2015;33:4106–4111. doi: 10.1200/JCO.2015.63.7918. [DOI] [PubMed] [Google Scholar]
- 3.Society A.C. American Cancer Society; 2015. Cancer Facts & Figures 2015. [Google Scholar]
- 4.Subbiah S., Nam A., Garg N., Behal A., Kulkarni P., Salgia R. Small cell lung cancer from traditional to innovative therapeutics: building a comprehensive network to optimize clinical and translational research. J. Clin. Med. 2020;9 doi: 10.3390/jcm9082433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y., Wang L., Jia R. The role of mitochondrial dynamics in human cancers. Am. J. Cancer Res. 2020;10:1278–1293. [PMC free article] [PubMed] [Google Scholar]
- 6.Lennon F.E., Salgia R. Mitochondrial dynamics: biology and therapy in lung cancer. Expert Opin. Investig. Drugs. 2014;23:675–692. doi: 10.1517/13543784.2014.899350. [DOI] [PubMed] [Google Scholar]
- 7.Hu J., Zhang H., Li J., Jiang X., Zhang Y., Wu Q., Shen L., Shi J., Gao N. ROCK1 activation-mediated mitochondrial translocation of Drp1 and cofilin are required for arnidiol-induced mitochondrial fission and apoptosis. J. Exp. Clin. Cancer Res. 2020;39:37. doi: 10.1186/s13046-020-01545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grieco J.P., Allen M.E., Perry J.B., Wang Y., Song Y., Rohani A., Compton S.L.E., Smyth J.W., Swami N.S., Brown D.A., Schmelz E.M. Progression-mediated changes in mitochondrial morphology promotes adaptation to hypoxic peritoneal conditions in serous ovarian cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson G.R., Wardell S.E., Cakir M., Yip C., Ahn Y.R., Ali M., Yllanes A.P., Chao C.A., McDonnell D.P., Wood K.C. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat. Commun. 2018;9:1677. doi: 10.1038/s41467-018-04033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennon F.E., Cianci G.C., Kanteti R., Riehm J.J., Arif Q., Poroyko V.A., Lupovitch E., Vigneswaran W., Husain A., Chen P., et al. Unique fractal evaluation and therapeutic implications of mitochondrial morphology in malignant mesothelioma. Sci. Rep. 2016;6 doi: 10.1038/srep24578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Mirzapoiazova T., Carol Tan Y.H., Pang K.M., Pozhitkov A., Wang Y., Wang Y., Mambetsariev B., Wang E., Nasser M.W., et al. Inhibiting crosstalk between MET signaling and mitochondrial dynamics and morphology: a novel therapeutic approach for lung cancer and mesothelioma. Cancer Biol. Ther. 2018;19:1023–1032. doi: 10.1080/15384047.2018.1472193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashatus D.F. The regulation of tumor cell physiology by mitochondrial dynamics. Biochem. Biophys. Res. Commun. 2018;500:9–16. doi: 10.1016/j.bbrc.2017.06.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuyoshi H., Orisaka M., Fujita Y., Asare-Werehene M., Tsang B.K., Yoshida Y. Prognostic impact of Dynamin related protein 1 (Drp1) in epithelial ovarian cancer. BMC Cancer. 2020;20:467. doi: 10.1186/s12885-020-06965-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miret-Casals L., Sebastian D., Brea J., Rico-Leo E.M., Palacin M., Fernandez-Salguero P.M., Loza M.I., Albericio F., Zorzano A. Identification of new activators of mitochondrial fusion reveals a link between mitochondrial morphology and pyrimidine metabolism. Cell Chem. Biol. 2018;25:268–278.e4. doi: 10.1016/j.chembiol.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Yu M., Nguyen N.D., Huang Y., Lin D., Fujimoto T.N., Molkentine J.M., Deorukhkar A., Kang Y., San Lucas F.A., Fernandes C.J., et al. Mitochondrial fusion exploits a therapeutic vulnerability of pancreatic cancer. JCI Insight. 2019;5 doi: 10.1172/jci.insight.126915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L., Zhou T., Xie Y., Bode A.M., Cao Y. Mitochondria-shaping proteins and chemotherapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.769036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fragoso Y.D., Brooks J.B.B. Leflunomide and teriflunomide: altering the metabolism of pyrimidines for the treatment of autoimmune diseases. Expert Rev. Clin. Pharmacol. 2015;8:315–320. doi: 10.1586/17512433.2015.1019343. [DOI] [PubMed] [Google Scholar]
- 18.Alamri R.D., Elmeligy M.A., Albalawi G.A., Alquayr S.M., Alsubhi S.S., El-Ghaiesh S.H. Leflunomide an immunomodulator with antineoplastic and antiviral potentials but drug-induced liver injury: A comprehensive review. Int. Immunopharmacol. 2021;93 doi: 10.1016/j.intimp.2021.107398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng L., Wang H., Wang Z., Huang H., Zhuo D., Lin J. Leflunomide inhibits proliferation and induces apoptosis via suppressing autophagy and PI3K/Akt signaling pathway in human bladder cancer cells. Drug Des. Devel. Ther. 2020;14:1897–1908. doi: 10.2147/DDDT.S252626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L., Zhang W., Li W., Ling C., Jiang M. Anti-inflammatory drug, leflunomide and its metabolite teriflunomide inhibit NSCLC proliferation in vivo and in vitro. Toxicol. Lett. 2018;282:154–165. doi: 10.1016/j.toxlet.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Christensen C.L., Kwiatkowski N., Abraham B.J., Carretero J., Al-Shahrour F., Zhang T., Chipumuro E., Herter-Sprie G.S., Akbay E.A., Altabef A., et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santamaria Nunez G., Robles C.M., Giraudon C., Martinez-Leal J.F., Compe E., Coin F., Aviles P., Galmarini C.M., Egly J.M. Lurbinectedin specifically triggers the degradation of phosphorylated RNA polymerase II and the formation of DNA breaks in cancer cells. Mol Cancer Ther. 2016;15:2399–2412. doi: 10.1158/1535-7163.MCT-16-0172. [DOI] [PubMed] [Google Scholar]
- 23.Dumoulin D.W., Cantini L., Cornelissen R., Vink M., Klaase L., Slooff K., Tebayna N., Mankor J.M., Baart S.J., Hendriks R., et al. Lurbinectedin shows clinical activity and immune-modulatory functions in patients with pre-treated small cell lung cancer and malignant pleural mesothelioma. Eur. J. Cancer. 2022;172:357–366. doi: 10.1016/j.ejca.2022.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Ng S.R., Colón C.I., Drapkin B.J., Hsu P.P., Li Z., Nabel C.S., Lewis C.A., Romero R., Mercer K.L., et al. Identification of DHODH as a therapeutic target in small cell lung cancer. Sci. Transl. Med. 2019;11:eaaw7852. doi: 10.1126/scitranslmed.aaw7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu D.D., Andrali S.S., Li H., Lin M., Huang W., Forman B.M. Novel FXR (farnesoid X receptor) modulators: potential therapies for cholesterol gallstone disease. Bioorg. Med. Chem. 2016;24:3986–3993. doi: 10.1016/j.bmc.2016.06.039. [DOI] [PubMed] [Google Scholar]
- 26.Tollis S., Rizzotto A., Pham N.T., Koivukoski S., Sivakumar A., Shave S., Wildenhain J., Zuleger N., Keys J.T., Culley J., et al. Chemical Interrogation of Nuclear Size Identifies Compounds with Cancer Cell Line-Specific Effects on Migration and Invasion. ACS Chem. Biol. 2022;17:680–700. doi: 10.1021/acschembio.2c00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbons D.L., Byers L.A., Kurie J.M. Smoking, p53 mutation, and lung cancer. Mol. Cancer Res. 2014;12:3–13. doi: 10.1158/1541-7786.mcr-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W., Wang F., Tang J., Liu X., Yuan Z., Nie C., Wei Y. Proapoptotic protein Smac mediates apoptosis in cisplatin-resistant ovarian cancer cells when treated with the anti-tumor agent AT101. J. Biol. Chem. 2012;287:68–80. doi: 10.1074/jbc.M111.271205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Guo W., Zhou H., Luo N., Nie C., Zhao X., Yuan Z., Liu X., Wei Y. Mitochondrial p53 phosphorylation induces Bak-mediated and caspase-independent cell death. Oncotarget. 2015;6:17192–17205. doi: 10.18632/oncotarget.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z., Sun B., Nie A., Yu D., Bian M. Roles of aminoacyl-tRNA synthetases in cancer. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.599765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q., Xiong D., Pan J., Wang Y., Hardy M., Kalyanaraman B., You M. Chemoprevention of lung cancer with a combination of mitochondria-targeted compounds. Cancers. 2022;14 doi: 10.3390/cancers14102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes A.P., Ilter D., Low V., Drapela S., Schild T., Mullarky E., Han J., Elia I., Broekaert D., Rosenzweig A., et al. Altered propionate metabolism contributes to tumour progression and aggressiveness. Nat. Metab. 2022;4:435–443. doi: 10.1038/s42255-022-00553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong J., Salgia R. Managing Patients With Relapsed Small-Cell Lung Cancer. J. Oncol. Pract. 2018;14:359–366. doi: 10.1200/JOP.18.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu L., Xiao Z., Tu H., Tong B., Chen S. The expression and prognostic significance of Drp1 in lung cancer: a bioinformatics analysis and immunohistochemistry. Medicine. 2019;98 doi: 10.1097/MD.0000000000018228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehman J., Zhang H.J., Toth P.T., Zhang Y., Marsboom G., Hong Z., Salgia R., Husain A.N., Wietholt C., Archer S.L. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parida P.K., Marquez-Palencia M., Ghosh S., Khandelwal N., Kim K., Nair V., Liu X.Z., Vu H.S., Zacharias L.G., Gonzalez-Ericsson P.I., et al. Limiting mitochondrial plasticity by targeting DRP1 induces metabolic reprogramming and reduces breast cancer brain metastases. Nat. Cancer. 2023;4:893–907. doi: 10.1038/s43018-023-00563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu S., Yan X., Xiang Z., Ding H.F., Cui H. Leflunomide reduces proliferation and induces apoptosis in neuroblastoma cells in vitro and in vivo. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bar-Or A., Pachner A., Menguy-Vacheron F., Kaplan J., Wiendl H. Teriflunomide and its mechanism of action in multiple sclerosis. Drugs. 2014;74:659–674. doi: 10.1007/s40265-014-0212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirzapoiazova T., Li H., Nathan A., Srivstava S., Nasser M.W., Lennon F., Armstrong B., Mambetsariev I., Chu P.G., Achuthan S., et al. Monitoring and determining mitochondrial network parameters in live lung cancer cells. J. Clin. Med. 2019;8 doi: 10.3390/jcm8101723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picard M., Sandi C. The social nature of mitochondria: implications for human health. Neurosci. Biobehav. Rev. 2021;120:595–610. doi: 10.1016/j.neubiorev.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boulton D.P., Caino M.C. Mitochondrial Fission and Fusion in Tumor Progression to Metastasis. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.849962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yapa N.M.B., Lisnyak V., Reljic B., Ryan M.T. Mitochondrial dynamics in health and disease. FEBS Lett. 2021;595:1184–1204. doi: 10.1002/1873-3468.14077. [DOI] [PubMed] [Google Scholar]
- 43.Bahat A., MacVicar T., Langer T. Metabolism and innate immunity meet at the mitochondria. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.720490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasukawa K., Kinoshita D., Yaku K., Nakagawa T., Koshiba T. The microRNAs miR-302b and miR-372 regulate mitochondrial metabolism via the SLC25A12 transporter, which controls MAVS-mediated antiviral innate immunity. J. Biol. Chem. 2020;295:444–457. doi: 10.1074/jbc.RA119.010511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bajzikova M., Kovarova J., Coelho A.R., Boukalova S., Oh S., Rohlenova K., Svec D., Hubackova S., Endaya B., Judasova K., et al. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 2019;29:399–416.e10. doi: 10.1016/j.cmet.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo H., Lee I., Chung H.S., Bae G.U., Chang M., Song E., Kim M.J. ATP5B regulates mitochondrial fission and fusion in mammalian cells. Anim. Cells Syst. 2016;20:157–164. doi: 10.1080/19768354.2016.1188855. [DOI] [Google Scholar]
- 47.Trigo J., Subbiah V., Besse B., Moreno V., López R., Sala M.A., Peters S., Ponce S., Fernández C., Alfaro V., et al. Lurbinectedin as second-line treatment for patients with small-cell lung cancer: a single-arm, open-label, phase 2 basket trial. Lancet Oncol. 2020;21:645–654. doi: 10.1016/S1470-2045(20)30068-1. [DOI] [PubMed] [Google Scholar]
- 48.Kundu K., Cardnell R.J., Zhang B., Shen L., Stewart C.A., Ramkumar K., Cargill K.R., Wang J., Gay C.M., Byers L.A. SLFN11 biomarker status predicts response to lurbinectedin as a single agent and in combination with ATR inhibition in small cell lung cancer. Transl. Lung Cancer Res. 2021;10:4095–4105. doi: 10.21037/tlcr-21-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liguori N.R., Sanchez Sevilla Uruchurtu A., Zhang L., Abbas A.E., Lee Y.S., Zhou L., Azzoli C.G., El-Deiry W.S. Preclinical studies with ONC201/TIC10 and lurbinectedin as a novel combination therapy in small cell lung cancer (SCLC) Am. J. Cancer Res. 2022;12:729–743. [PMC free article] [PubMed] [Google Scholar]
- 50.Xie W., Forveille S., Iribarren K., Sauvat A., Senovilla L., Wang Y., Humeau J., Perez-Lanzon M., Zhou H., Martínez-Leal J.F., et al. Lurbinectedin synergizes with immune checkpoint blockade to generate anticancer immunity. OncoImmunology. 2019;8 doi: 10.1080/2162402X.2019.1656502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon reasonable request.
This paper does not report the original code.
Further details or additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.