Abstract
The human herpesvirus 8 (HHV-8) latency-associated nuclear antigen (LANA) is expressed in all latently HHV-8 infected cells and in HHV-8-associated tumors, including primary effusion lymphoma (PEL). To better understand the contribution of LANA to tumorigenesis and to the PEL phenotype, we performed a yeast two-hybrid screen which identified the corepressor protein SAP30 as a LANA binding protein. SAP30 is a constituent of a large multicomponent complex that brings histone deacetylases to the promoter. Glutathione S-transferase affinity assays confirmed interaction between LANA and SAP30 and also demonstrated interactions between LANA and two other members of the corepressor complex, mSin3A and CIR. The corepressors bound to the amino-terminal 340-amino-acid domain of LANA. In transient expression assays, this same domain of LANA mediated repression when targeted to a 5×Gal4tk-CAT reporter as a GAL4-LANA fusion. PEL cells have the unusual feature that they are frequently dually infected with both HHV-8 and Epstein-Barr virus (EBV). We found that EBV EBNA-1 expression is downregulated in PEL cells at both the RNA and protein levels. In transient expression assays, LANA repressed activated expression from the EBV Qp and Cp latency promoters. Reduction of endogenous Qp activity could also be demonstrated in EBV-infected Rael cells transfected with a LANA expression plasmid. In contrast to the effect of LANA on EBV latency promoters, LANA activated expression from its own promoter. The data indicate that LANA can mediate transcriptional repression through recruitment of an mSin3 corepressor complex and further that LANA-mediated repression is likely to contribute to the low level of EBV latency gene expression seen in dually infected PEL cells.
Human herpesvirus 8 (HHV-8) or Kaposi's sarcoma-associated herpesvirus (KSHV), is a recently identified human herpesvirus that is associated with Kaposi's sarcoma, a proportion of multicenteric Castleman's disease, and primary effusion lymphoma (PEL) (7, 9, 11, 16, 17, 48, 64). PEL occurs predominantly in human immunodeficiency virus-infected patients and has distinctive clinical, immunophenotypic, and genetic characteristics (6, 25, 50). PEL cells are genotypically B cells with plasmacytic features, and analyses of the immunoglobulin locus have identified somatic mutations indicating that most PEL cells are of germinal-center or post-germinal-center origin (42, 43, 50). PEL cells lack c-myc rearrangements and have the unusual feature that the tumor cells are frequently dually infected with both HHV-8 and Epstein-Barr virus (EBV) (10, 21, 50). In PEL cell lines, both HHV-8 and EBV genomes exist as episomes, and the viruses maintain a predominantly latent infection in the absence of chemical manipulation (10, 44, 49, 58, 61).
The HHV-8 latency protein LANA (latency-associated nuclear antigen) is a marker for HHV-8 infection (20, 30). LANA is a 222- to 234-kDa protein that is encoded within open reading frame (ORF) 73 (19, 29, 57). In a screen of a cDNA expression library derived from a PEL cell line with Kaposi's sarcoma patient sera, the immunodominant epitope was mapped to the C-terminal domain of ORF 73 (31). The generation of polyclonal and monoclonal antibodies against ORF 73 has allowed definitive demonstration that LANA is consistently expressed in Kaposi's sarcoma, multicentric Castleman's disease, and PEL tumor cells (17, 28, 32, 54). Two transcripts encoding LANA have been described. These transcripts, which initiate from the same promoter and are coterminal, differ only in that one is unspliced and the other is spliced to remove 5′ noncoding information (15, 62, 67). A third mRNA, which encodes ORF K13 (vFLIP) and ORF 72 (vCYC) but splices out LANA, is expressed from the same promoter and is also 3′ coterminal. In HHV-8-infected cells, LANA has a punctate nuclear distribution and associates with mitotic chromosomes (29, 57, 65). LANA and HHV-8 DNAs have been shown to colocalize in the punctate spots, and LANA mediates the persistence of episomes containing a segment of the HHV-8 DNA from the left-hand terminus of the HHV-8 genome (3, 14). The punctate structures containing LANA and HHV-8 DNA do not colocalize with other recognized nuclear bodies (66). LANA is likely to be multifunctional. LANA interacts with a protein, RING3, which may mediate phosphorylation of the C-terminal domain of LANA (56) and has also been found to interact with p53 (18). The interaction with p53 has been shown to inhibit p53-mediated apoptosis, and thus LANA may have a role in promoting the survival of HHV-8-infected cells.
In dually infected PEL cell lines, EBV gene expression is limited. The latency promoter Qp is used to express EBNA-1 (22, 65). Expression of LMP-1 and LMP-2A transcripts have also been detected, but the Wp and Cp promoters are silent and the EBNA-2 and EBNA-3 family genes are not expressed (8, 22, 45, 65). Expression of EBNA-1 in the absence of EBNA-2 and EBNA-3 is a mark of Qp promoter usage. The dual infection of PEL cells with EBV and HHV-8 raises the possibility of intervirus interactions that might modulate the course of infection and potentially contribute to the unique PEL phenotype. To gain further insight into the contribution of LANA to HHV-8 pathogenesis, we performed a yeast two-hybrid screen to identify LANA binding proteins. This screen identified the mSin3-associated corepressor SAP30 as a LANA-interacting protein. We demonstrate here that the N-terminal domain of LANA mediates transcriptional repression through tethering of an mSin3-containing corepressor complex. In transfected cells, LANA reduced Qp-mediated EBNA-1 expression and abolished activated Cp expression, suggesting that LANA may be responsible for the reduced levels of EBV latency gene expression seen in dually infected PEL cells.
MATERIALS AND METHODS
Plasmids.
The yeast expression plasmids Gal4ACT-SAP30, CIR-Gal4ACT, and Gal4ACT have been described previously (24). The LANA insert in Gal4DBD-LANA (pDY20) was present in a NarI-AccI fragment derived from a cloning intermediate, pDY15, which in turn was derived from the BCBL lambda clone 3-2b (52). For expression in mammalian cells, Gal4DBD fusion proteins were generated in the pGH250 vector, which has a simian virus 40 promoter. LANA fusions (with plasmid name in parentheses) were derived from the pDY15 derivative pDY22: Gal4DBD-LANA (pGL11), Gal4DBD-LANA(d341-939) (pDH338), Gal4DBD-LANA(1-340) (pDH339), and Gal4DBD-LANA (940-1177) (pDH341). Glutathione S-transferase (GST)-LANA fusions were constructed in the pGEX (Promega)-derived plasmid pGH413 with LANA sequences derived from the corresponding GAL4-LANA fusions: GST-LANA(d341-939) (pGL9), GST-LANA(1-340) (pGL13), GST-LANA(940-1177) (pGL12), and GST-LANA(341-1177) (pMF8). GST-Zta (pDH237) was derived from pYNC76 (12) as a BamHI-EcoRI fragment. GST-EBNA2(1-58) (pPDL116) was derived as an EcoRI fragment from pPDL32A (40).
For eukaryotic expression, LANA was cloned into the SG5 (Stratagene)-based vectors pJH253 [Flag-LANA (pDY52) and Flag-LANA(1-275) (pMF23)] and pRTS2 (pDY17). pEBO-LANA (pDY43) was generated in the vector pEBO-pLPP, which carries EBV oriP and the EBNA-1 coding region and is maintained as an episome in transfected cells (41). Myc-SAP30 (pAK1) was generated by moving Myc epitope-tagged SAP30 from pDY33 into a modified SG5 vector, pRTS2. Expression vectors have been described previously for Myc-mSin3A (36), CIR-Flag (pJH518) (24), EBNA-2 (pPDL151) (39), and JAK-1 (71) and for the reporter plasmids 5×GAL4DBDtk-CAT, tk-luciferase, Cp-CAT, and Qp-CAT (13, 23, 40). The LANAp-luciferase reporter was generated in pGL2basic (Promega) and contained HHV-8 sequences 127,981 to 127,126 (47, 60). The HHV-8 insert was generated by PCR using the primers described by Sarid et al. (62) and BC-2 genomic DNA.
Yeast assays.
The yeast two-hybrid screen was performed using Saccharomyces cerevisiae HF7c transformed with Gal4DBD-LANA (pDY20) and a commercial B-cell Gal4ACT library (Clontech). A total of 3.7 × 106 independent transformants were screened for growth on His− medium in the presence of 50 mM 3-aminotriazole followed by selection for induction of beta-galactosidase activity. Subsequent interaction assays were performed in S. cerevisiae Y190. Beta-galactosidase activity was measured in two independent cotransformants using 2-nitrophenol-β-d-galactopyranoside as a substrate. The amount of 2-nitrophenol liberated after 4 h of incubation was measured by absorbance at 420 nm.
GST affinity assay.
GST and GST fusion proteins were induced by growth for 4 h at 37°C in medium containing 0.1 mM isopropyl-β-d-thiogalactopyranoside. Pelleted bacteria were resuspended in 50 mM Tris-HCl (pH 7.5)–0.5 mM EDTA–100 mM NaCl–5 mM MgCl–5% glycerol–0.1 mM phenylmethylsulfonyl fluoride (PMSF)–1 μg of aprotinin/ml–1 μg of pepstatin/ml–0.5% Nonidet P-40 and then sonicated. Cell debris was removed by centrifugation at 10,000 × g for 10 min. The supernatant was incubated at 4°C overnight with glutathione Sepharose 4B beads (Sigma, St. Louis, Mo.) and then washed three times in lysis buffer (50 mM Tris-HCl [pH 7.4], 1 mM EDTA, 200 mM NaCl, 0.5 mM MgCl2, 5% glycerol, 0.1 mM PMSF, 1 μg of aprotinin/ml, 1 μg of pepstatin/ml, 0.2% Nonidet P-40) without the protease inhibitors and Nonidet P-40. The amount of protein bound to the beads was determined by Coomassie blue staining of proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Equal amounts of each GST protein were used in the affinity assays.
Three 100-mm-diameter dishes of HeLa cells (1.5 × 106 cells per dish) were transfected with 12 μg of DNA per dish, and the cells were harvested 48 h after transfection. The cell pellet was resuspended in 6 ml of lysis buffer and sonicated. Cell debris was removed by centrifugation at 10,000 × g for 10 min. The supernatant was incubated overnight at 4°C with the bead-bound GST fusion proteins, after which the complex was washed six times with lysis buffer. The complex was dissociated from the beads by boiling for 5 min in 6× SDS-PAGE loading buffer (2% SDS, 10% glycerol, 100 mM dithiothreitol, 60 mM Tris [pH 6.8], 0.2% bromophenol blue), and the proteins were subjected to electrophoresis through an SDS–9% PAGE gel. The separated proteins were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.), and the interacting proteins were detected by incubation with mouse anti-Myc (Santa Cruz) or rabbit anti-Flag (Sigma) antibody (1:200) and visualized using the enhanced chemiluminescence reaction (Amersham Life Sciences, Little Chalfont, Buckinghamshire, England).
Immunofluorescence assay.
Vero cells were seeded at 8 × 104 per well in two-well slide chambers (LabTek). Cells were transfected using the calcium phosphate procedure with Myc-SAP30 (1.0 μg), Flag-LANA (2.0 μg), or Myc-SAP30 plus Flag-LANA. The total amount of transfected DNA was equalized using SG5 vector DNA. The transfected cells were incubated in Dulbecco modified Eagle medium plus 10% fetal bovine serum for 24 h at 35°C in 3% CO2, followed by a medium change and incubation for a further 24 h at 37°C in 5% CO2. Cells were washed in 1× phosphate-buffered saline (PBS; 0.144 g of KH2PO4, 9.0 g of NaCl, and 0.795 g of Na2HPO4 · 7H2O per liter of H2O), fixed with 1% paraformaldehyde in PBS for 5 min at room temperature, washed in 1× PBS, permeabilized for 20 min on ice in 2% Triton X-100 in PBS, and washed for 5 min on ice in 1× PBS. Cells were incubated with the primary antibody for 45 min at 37°C, followed by three washes for 15 min each on ice in 1× PBS. Incubation with the secondary antibody was carried out for 30 min at 37°C, followed by two washes for 10 min each on ice in 1× PBS. In single transfections, the primary antibodies were rabbit anti-Myc (Santa Cruz) and rabbit anti-Flag (Sigma) and the secondary antibody was donkey anti-rabbit IgG conjugated with rhodamine (Chemicon). In cotransfections, the primary antibodies were mouse anti-Myc (Santa Cruz) and rabbit anti-Flag and the secondary antibodies were donkey anti-mouse IgG conjugated with fluorescein isothiocyanate (FITC) (Chemicon) and donkey anti-rabbit IgG conjugated with rhodamine.
Chloramphenicol acetyltransferase (CAT) and luciferase assays.
HeLa cells were plated in six-well dishes at 3 × 105 cells per well 24 h before transfection, with a medium change 3 h prior to transfection. Cells were transfected by the calcium phosphate procedure with 5×Gal4BStk-CAT or tk-CAT reporters (1.5 μg), a tk-luciferase control (1.0 μg), and the Gal4DBD-LANA fusion plasmid pDH338, pDH339, pDH341, or pGL11 (1.0 μg). Vector SG5 DNA was used to equalize the amount of DNA in each transfection. For EBV promoter studies, Cp-CAT (1.0 μg) was transfected into HeLa cells alone or in the presence of EBNA-2 (0.3 μg). LANA (2 μg), or EBNA-2 plus LANA. Qp-CAT (1.0 μg) was transfected into HeLa cells alone or in the presence of JAK-1 (2.0 μg). LANA (1.0 μg), or JAK-1 plus LANA. In each case the total amount of transfected DNA was equalized with vector DNA.
Transfected cells were incubated in Dulbecco modified Eagle medium plus 10% fetal bovine serum for 24 h at 35°C in 3% CO2 and, after a medium change, for a further 24 h at 37°C in 5% CO2. CAT and luciferase activities were assayed as previously described (23, 40). CAT activity was quantified using an InstantImager (Packard).
Reverse transcription-PCR (RT-PCR) for EBNA-1.
mRNA was prepared from 107 cells each of the EBV+ B95-8 and Raji cell lines and the BC-2 and HBL-6 EBV+ HHV-8+ PEL cell lines using the Pharmacia Quickprep kit plus DNase treatment. A 1-μg portion of mRNA was reverse transcribed in a total volume of 28 μl using avian myeloblastosis virus (AMV) reverse transcriptase (Promega). Serial dilutions of 1:1, 1:10, 1:100, and 1:1,000 were made of each cDNA. The PCR was performed using the EBNA-1 primers LGH2643 and LGH2644, with denaturation at 95°C for 2 min and 29 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s. The PCR products were separated on a 2% agarose gel and photographed.
EBNA-1 protein expression.
EBV+ B95-8 cells, EBV+ HHV-8+ HBL-6 and BC-2 cells, EBV− HHV-8+ BCBL1 cells, and EBV− HHV-8− DG75 cells (4 × 106) were pelleted at 800 rpm in a Beckman GS-6R centrifuge for 5 min and washed once with ice-cold 1× PBS. Cells were resuspended in 100 μl of 6× loading buffer, sonicated, and boiled at 95°C for 5 min. Cell debris was removed by centrifugation for 2 min at 10,000 rpm in a Sorvall microcentrifuge. The supernatant (5 μl) was subjected to electrophoresis at 180 V for 3 h through an SDS–10% polyacrylamide gel, and the separated proteins were transferred to a nitrocellulose membrane. After being blocked for 1 h in 5% milk–1% Tween 20–PBS, the membrane was incubated with either a mouse anti-EBNA-1 monoclonal antibody (EBNA.OT1x) at a 1:10,000 dilution or serum from a patient with nasopharyngeal carcinoma (NPC) (1:50) in 3% bovine serum albumin–0.1% Tween 20–PBS. After two washes with 0.1% Tween 20 in PBS, the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies (1:200) and the reactive bands were visualized using the enhanced chemiluminescence system (Amersham).
Stably transfected LANA cell lines.
EBV− DG75 cells or EBV+ Rael cells (3 × 107) were electroporated with 10 μg of pEBOpLPP vector or pEBO-LANA (pDY43), and transfected cells were selected by growth in medium containing 800 μg of hygromycin B/ml.
RESULTS
Identification of SAP30 as a LANA-interacting protein in yeast.
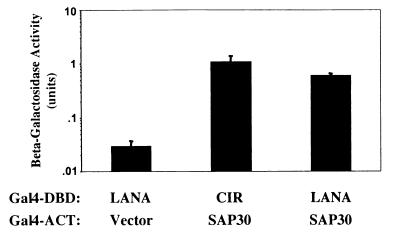
Gal4DBD-LANA was cotransformed into yeast with a B-cell cDNA library to seek cellular binding partners for LANA. This screen identified four proteins involved in transcriptional regulation, each of which interacted with LANA with similar affinity. One of these proteins was SAP30, which is a member of the mSin3-histone deacetylase (HDAC) corepressor complex (36, 73). Corepressor complexes consist of a multicomponent assembly of proteins that link HDACs to specific DNA-binding proteins. Interaction between Gal4DBD-LANA and Gal4ACT-SAP30 in cotransformed yeast (Fig. 1) was measured by induction of beta-galactosidase activity. A known SAP30-associated cellular corepressor is CIR (24). The SAP30-CIR interaction was used as a positive control in this experiment.
FIG. 1.
LANA binds to SAP30. Shown are results of a yeast two-hybrid assay in which interaction is measured by induction of beta-galactosidase activity. Yeast cells were cotransformed with Gal4DBD-LANA and Gal4ACT-SAP30, Gal4DBD-LANA plus Gal4ACT vector (negative control), or Gal4DBD-CIR plus Gal4ACT-SAP30 (positive control). Data are averages from two experiments, with the range of values indicated.
LANA binds to members of the mSin3 corepressor complex.
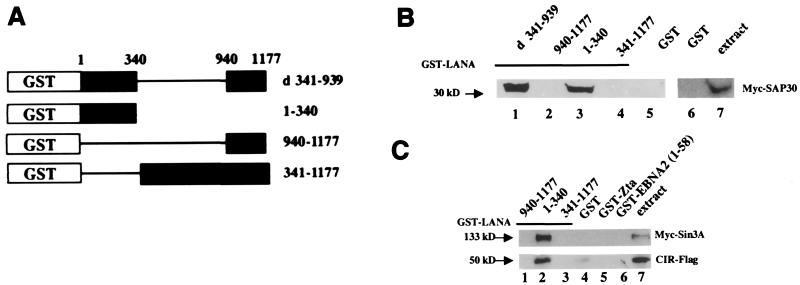
The interaction between LANA and SAP30 was confirmed using GST affinity assays. Extracts of HeLa cells transfected with a vector expressing a Myc epitope-tagged SAP30 were incubated with control GST protein and with four GST fusion proteins containing the segments of LANA shown in Fig. 2A. Expression of GST fusion proteins was examined by SDS-PAGE and Coomassie staining, and equal amounts of protein were used in each assay. As illustrated in Fig. 2B, the 30-kDa Myc-SAP30 protein interacted with the GST fusion expressing a LANA variant with the central region deleted (LANA d341-939) (lane 1) and with that expressing the N-terminal domain, LANA(1-340) (lane 3). No interaction was observed with GST fusions expressing the carboxy-terminal regions of LANA from amino acid (aa) 940 to 1177 or from aa 341 to 1177 (lanes 2 and 4). Similarly, there was no interaction with the control GST protein (lanes 5 and 6).
FIG. 2.
LANA interacts with the corepressors SAP30, mSin3A, and CIR in GST affinity assays. (A) Diagram of the GST-LANA constructs used in the affinity assay. (B) Extracts from HeLa cells transfected with Myc-SAP30 were incubated with the indicated GST-LANA fusion proteins (lanes 1 to 4) or with control GST protein (lanes 5 and 6). Transfected cell extract (15 μl) was loaded in lane 7. A longer exposure (lanes 6 and 7) was needed to detect Myc-SAP30 in the cell extract. (C) Extracts from HeLa cells transfected with Myc-mSin3A or CIR-Flag were incubated with the indicated GST-LANA fusion proteins (lanes 1 to 3) or with control GST protein, control GST-Zta, or GST-EBNA2 protein (lanes 4 to 6). Transfected cell extract (15 μl) was loaded in lane 7. Bound protein was separated by SDS-PAGE and subjected to Western blot analysis with either an anti-Myc or an anti-Flag antibody. Reactive proteins were visualized by chemiluminescence.
SAP30 was originally identified as a partner of the corepressor mSin3A (36), and an interaction between SAP30 and the CBF1-associated corepressor CIR has also been reported (24). Since SAP30 is closely associated with mSin3A and CIR, evidence for a functional interaction between LANA and a corepressor complex would be strengthened by a demonstration that LANA also bound, either directly or indirectly through SAP30, to these other members of the complex. Binding of LANA to mSin3A and to CIR was examined using GST-LANA fusion proteins and extracts of HeLa cells transfected with vectors expressing either Myc-mSin3A or CIR-Flag (Fig. 2C). The amino-terminal domain (aa 1 to 340) of LANA bound both the 133-kDa Myc-mSin3A and the 50-kDa CIR-Flag protein (lane 2). There was no interaction between the corepressors and the GST fusions expressing LANA carboxy-terminal aa 940 to 1177 (lane 1) or the extended carboxy terminus, aa 341 to 1177 (lane 3). Minimal or no interaction was seen with the control protein GST (lane 4), GST-Zta (lane 5), or GST-EBNA2(1-58) (lane 6). Taken together, these results indicate that LANA, and specifically the N-terminal 340-aa domain, interacts with recognized components of the mSin3-HDAC corepressor complex.
Colocalization of LANA and SAP30 in cotransfected cells.
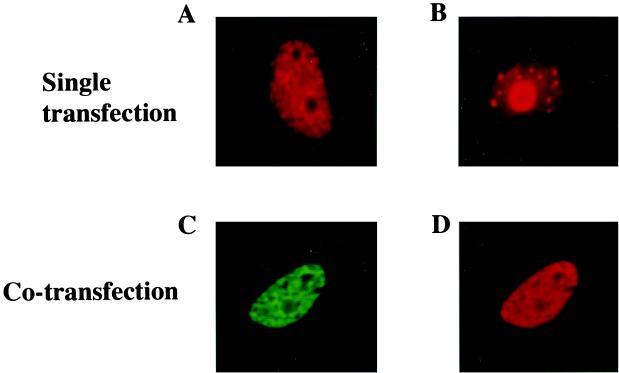
To investigate whether the localization of LANA and SAP30 in mammalian cells was compatible with interaction between them, immunofluorescence assays were performed on Vero cells transfected with Flag-LANA and Myc-SAP30 (Fig. 3). In individually transfected cells, Myc-SAP30 was detected using a rabbit anti-Myc antibody and rhodamine-conjugated anti-rabbit secondary antibody, and Flag-LANA was detected using a rabbit anti-Flag antibody and a rhodamine-conjugated secondary antibody. Flag-LANA gave patchy nuclear staining with nucleolar sparing (Fig. 3A). (This staining pattern is typical of LANA in the absence of HHV-8 genomes.) Myc-SAP30 localized as discrete spots within the nuclei of transfected cells and was also present in the nucleoli (Fig. 3B). In cotransfected cells, LANA retained the staining pattern seen in the individually transfected cells (Fig. 3D). Myc-SAP30, which was detected using a mouse anti-Myc antibody and a fluorescein-conjugated anti-mouse secondary antibody, was redistributed into the LANA staining pattern in the cotransfected cells (Fig. 3C). The colocalization of SAP30 with LANA in the dually transfected cells is consistent with their participating in the same functional complex.
FIG. 3.
SAP30 colocalizes with LANA in transfected cells. An indirect immunofluorescence assay was performed on Vero cells transfected with Flag-LANA (A), Myc-SAP30 (B), or Myc-SAP30 plus Flag-LANA (C and D). In singly transfected cells, Flag-LANA (A) exhibited micropunctate staining with nucleolar sparing and Myc-SAP30 (B) exhibited nuclear punctate plus diffuse nucleolar staining. The doubly transfected cells were stained for Myc-SAP30 (C) (green) or Flag-LANA (D) (red). In cotransfected cells, Myc-SAP30 (C) exhibited the same micropunctate distribution as Flag-LANA (D). Myc-SAP30 (green) was detected using a rabbit anti-Myc primary antibody and FITC-conjugated donkey anti-rabbit immunoglobulin secondary antibody. Flag-LANA (red) was detected using a mouse monoclonal anti-Flag primary antibody and a rhodamine-conjugated donkey anti-mouse secondary antibody.
LANA functions as a transcriptional repressor.
The mSin3 corepressor complex is part of a larger complex that recruits HDAC to DNA-bound factors to mediate transcriptional repression (34). To investigate whether LANA could mediate transcriptional repression when tethered to DNA through the Gal4 DNA binding domain (DBD), intact LANA (aa 1 to 1177) and three truncated LANA variants were generated as Gal4DBD-LANA fusions in a mammalian expression vector (Fig. 4A). These constructions were then cotransfected into HeLa cells with a 5×Gal4BS-tkCAT reporter and a tk-luciferase control plasmid. Cotransfection of Gal4DBD-LANA(1-1177) markedly repressed expression of the CAT reporter (Fig. 4B). The carboxy-terminal domain in Gal4DBD-LANA(940-1177) had minimal repressive activity. In contrast, Gal4DBD-LANA(1-340) was as effective a repressor as intact LANA. Deletion of the central region of LANA resulted in a fusion protein, Gal4DBD-LANA(d341-939), that was less effective at repressing CAT expression than either Gal4DBD-LANA(1-1177) or Gal4DBD-LANA(1-340). It is possible that the large internal deletion affected protein folding. Cotransfection of the Gal4DBD-LANA fusions with a tk-CAT reporter did not result in repression of CAT expression, indicating that repression required targeting of LANA to the promoter (data not shown). The localization of a transcriptional repression domain within aa 1 to 340 of LANA is consistent with the results of the protein-protein interaction experiments, which demonstrated that the amino terminus of LANA interacted with corepressor proteins.
FIG. 4.
LANA acts as a repressor of gene expression. (A) Diagram of the Gal4-LANA fusion constructs used in the reporter assay. (B) Transient expression assay in which HeLa cells were transfected with the tk-luciferase control plasmid (1.0 μg) and a 5×Gal4BS-tkCAT reporter (1.5 μg) alone or in the presence of the indicated Gal4-LANA fusion plasmids (1 μg). Fusion proteins containing LANA aa 1 to 340 repressed reporter CAT expression as efficiently as full-length LANA(1-1177). The assay was repeated three times with similar results.
EBV EBNA-1 expression in dually infected PEL cell lines.
In PEL cell lines dually infected with HHV-8 and EBV, the EBV Qp is used to express EBNA-1 and Cp is not active (22, 65). The expression of EBV EBNA-1 also appears to be reduced compared to the levels seen in latently EBV infected B-cell lines (8). For example, when extracts from the EBV B95-8 B-cell line and from three dually infected PEL cell lines were probed in an immunoblot with an anti-EBNA-1 monoclonal antibody, EBNA-1 was detected only in the B95-8 extract (Fig. 5A). The sensitivity of detection in this case may be biased by the derivation of the monoclonal antibody, which was raised against EBNA-1 from B95-8. Western blot analysis using NPC serum as the probe detected weak EBNA-1 expression in the dually infected BC-2 PEL cells but not in HBL-6 cells (Fig. 5B). The EBNA-1 expression in BC-2 cells was much reduced compared to that in the latently EBV infected Raji B-cell line or in the Qp-using Akata cell line. Note that polymorphisms in the Gly-Gly-Ala repeat region of EBNA-1 lead to characteristic differences in the migration of EBNA-1 from the different cell lines.
FIG. 5.
EBV EBNA-1 protein expression is reduced in dually infected PEL cells. Shown are Western blot analyses comparing the expression of EBNA-1 in EBV-positive cell lines and HHV-8 and EBV dually infected cell lines. B95-8, Raji, and Akata are EBV+ B-cell lines. HBL-6 and BC-2 are HHV-8+ EBV+ PEL cell lines. BCBL-1 is an HHV-8+ EBV− PEL cell line, and DG75 is negative for both viruses. EBNA-1 was detected by using the anti-EBNA-1 monoclonal antibody EBNA.OT1x (A) or NPC serum (B). Reactive proteins were visualized by using chemiluminescence.
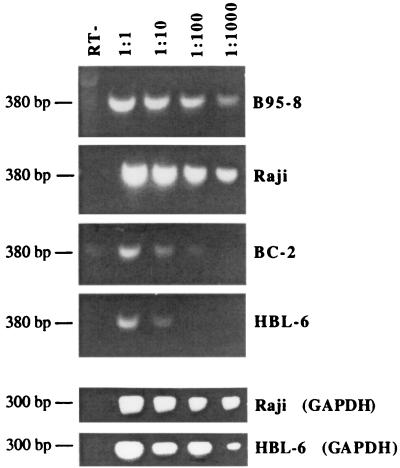
The low levels of EBNA-1 protein in BC-2 and HBL-6 PEL cells correlated with a reduction in the amount of EBNA-1 mRNA detectable in these cells compared to that in EBV+ B-cell lines. A semiquantitative RT-PCR analysis of EBNA-1 transcripts revealed significantly reduced levels of EBNA-1 mRNA in BC-2 PEL cells and severely reduced levels of EBNA-1 mRNA in HBL-6 cells (Fig. 6). RT-PCR for cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was performed on all samples to ensure equal RNA quality and quantity. Results of the control reactions for Raji and HBL-6 cells are presented in Fig. 6. The relative amounts of EBNA-1 mRNA detected in the BC-2 and HBL-6 cells also correlated with the EBNA-1 protein levels in these cells, with EBNA-1 protein being detectable in BC-2 cells but below the detection level in HBL-6 cells (Fig. 5B). The RNA analysis suggests that the reduced levels of EBNA-1 in PEL cells derive from repression at the transcriptional level.
FIG. 6.
EBNA-1 mRNA expression is also reduced in PEL cells. A semiquantitative RT-PCR assay for EBNA-1 transcripts was performed using the indicated serial 10-fold dilutions of the template cDNA. RT-PCR products were separated by agarose gel electrophoresis, stained with ethidium bromide, and photographed. B95-8 and Raji are EBV+ B-cell lines; HBL-6 and BC-2 are EBV+ HHV-8+ PEL cell lines. Control RT-PCR results for cellular GAPDH mRNA are shown for the Raji and HBL-6 cell lines.
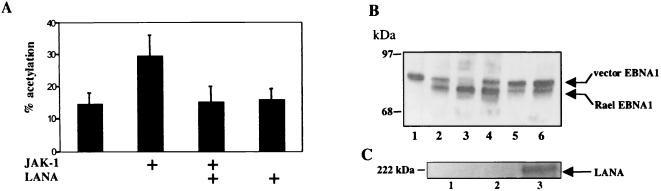
To determine whether LANA had a role in the low-level expression of EBNA-1 from Qp in PEL cells, two approaches were taken. First, LANA was cotransfected with a Qp-CAT reporter into HeLa cells (Fig. 7A). Qp is positively regulated by the JAK/STAT pathway (13), and CAT expression from Qp-CAT was upregulated in the presence of transfected JAK-1. The addition of LANA had no effect on basal Qp-CAT expression but blocked JAK-1 upregulation of Qp.
FIG. 7.
LANA attenuates Qp-driven expression of EBNA-1. (A) Transient expression assay in which HeLa cells were transfected with a Qp-CAT reporter (1 μg) alone or with JAK-1 (2 μg) and in the presence or absence of cotransfected LANA (1 μg). The results shown are averages from three experiments, with the standard deviations indicated. (B) LANA reduces Qp-driven EBNA-1 protein expression in Rael cells. Shown is a Western blot examining EBNA-1 expression in EBV-negative DG75 cells carrying the pEBO vector, which expresses EBNA-1 (lane 1), EBV-positive Rael cells carrying the pEBO vector and grown for 11 weeks in selection medium containing hygromycin B (lane 2), Rael cells (lane 3), Rael pEBO-LANA cells grown for 3 weeks in selection medium (lane 4), Rael pEBO-LANA cells after 7 weeks of selection (lane 5), and Rael pEBO-LANA cells after 11 weeks of selection (lane 6). The pEBO-encoded EBNA-1 differs in mobility from the endogenous Rael EBNA-1. EBNA-1 was detected using NPC serum and a chemiluminescence visualization protocol. (C) LANA expression in transfected Rael cells. Shown is a Western blot in which LANA was detected using a rat anti-LANA monoclonal antibody (ABI). Lane 1, Rael pEBO cells after 11 weeks of selection; lane 2, Rael cells; lane 3, Rael pEBO-LANA cells after 11 weeks of selection. Rael pEBO-LANA cells express LANA, unlike the parental Rael or control Rael pEBO cells.
To examine whether the negative modulatory effect of LANA seen in the reporter assays was likely to be relevant to EBNA-1 expression levels in established cell lines, a pair of stably transfected cell lines was created. Rael is an EBV-positive B-cell line that utilizes Qp for EBNA-1 expression. Rael cells were transfected with the pEBO vector or with pEBO-LANA, and stably transfected cell lines were selected by growth in hygromycin B. The pEBO vector expressed an EBNA-1 of a different size from the endogenous Rael EBNA-1, and the two proteins can be distinguished by their migration in denaturing gels. The pEBO EBNA-1 migrates more slowly through the gel than the endogenous Rael EBNA-1. After 3, 7, and 11 weeks of selection, the amount of EBNA-1 in the transfected cells was examined by Western analysis using NPC serum (Fig. 7B). pEBO uses a heterologous promoter to express EBNA-1, and we could therefore compare the ratio of vector-expressed EBNA-1 to that of Qp-driven Rael EBNA-1 as a measure of the effect of LANA on Qp activity. After 11 weeks of selection, the Rael cells transfected with the pEBO vector were expressing approximately twofold more Rael EBNA-1 than pEBO EBNA (Fig. 7B, lane 2). However, expression of Rael EBNA-1 decreased relative to that of pEBO EBNA-1 with increased passaging in the presence of coexpressed LANA. After 11 weeks of selection in the presence of LANA, Rael cells exhibited a reversed ratio, with Qp-driven EBNA-1 being approximately fourfold less abundantly than pEBO EBNA-1 (Fig. 7B, lane 6). It is noteworthy that the total amounts of EBNA-1 expressed in the parental Rael cells and in the Rael pEBO-LANA cells are comparable. Expression of LANA in the Rael cells after 11 weeks of selection is shown in Fig. 7C. These data suggest that LANA may have a role in the reduced EBNA-1 expression seen in dually infected PEL cell lines.
LANA represses EBV Cp activity.
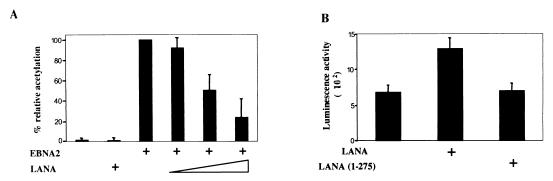
The EBV Cp, which drives expression of the EBNA-1, EBNA-2, and EBNA-3 family proteins during primary infection and in the majority of EBV-positive lymphoblastoid cell lines, is not used in dually infected PEL cells. To address whether LANA might contribute to the downregulation of Cp, transient expression assays were performed in which the effect of cotransfected LANA on basal and EBNA-2-activated expression from a Cp-CAT reporter was examined (Fig. 8A). As expected, cotransfection of EBNA-2 strongly activated expression of Cp-CAT. LANA had no effect on basal expression of the Cp-CAT reporter but ablated EBNA-2 activation in a dose-responsive manner.
FIG. 8.
In transient assays LANA represses EBV Cp-driven expression but activates the HHV-8 LANAp. CAT and luciferase reporter assays were performed in transfected HeLa cells. Results shown are averages from three experiments, with the standard deviations indicated. (A) Cells were transfected with the Cp-CAT reporter (1.0 μg), EBNA-2 (0.3 μg), LANA (2.0 μg), or EBNA-2 (0.3 μg) plus increasing amounts of LANA (0.5, 1.0, or 2.0 μg) as indicated. LANA did not affect basal Cp activity but abolished EBNA-2-induced expression. (B) Cells were transfected with the LANAp-luciferase reporter (1.0 μg), alone or together with LANA (2.0 μg) or LANA(1-275) (2.0 μg) as indicated. LANA positively regulated its own promoter.
To provide some context for the negative regulation of the EBV Qp and Cp latency promoters, the effect of cotransfected LANA on an HHV-8 latency promoter was also examined. A LANAp-luciferase reporter containing 924 bp of the LANA promoter (LANAp) from −773 to +30 was generated (62). Cotransfection of full-length LANA resulted in approximately twofold activation of the LANAp (Fig. 8B). A truncated amino-terminal derivative of LANA did not mediate this effect. Thus, LANA appears to differentially regulate EBV and HHV-8 latency genes, with the EBV latency Qp and Cp promoters being repressed while LANA potentially stimulates its own expression.
DISCUSSION
The consistent detection of LANA in all HHV-8-infected cell lines and tumor specimens examined strongly suggests that LANA contributes either to HHV-8 latency or to growth proliferation. The demonstration that LANA colocalizes with HHV-8 genomes on metaphase chromosomes and is sufficient to maintain HHV-8 episomal DNA in dividing cells (3) provides experimental evidence for a role for LANA in the segregation of HHV-8 genomes and in the maintenance of HHV-8 latent infection. LANA may also contribute to tumor development, and the ability of LANA to interfere with p53-mediated apoptosis by negatively regulating p53 transcriptional activity may represent one such function (18). Our demonstration that LANA binds to members of the mSin3 corepressor complex suggests that LANA may have a more global effect on cellular gene expression.
HDACs are believed to mediate transcriptional repression in part by modifying chromatin structure. HDACs generally associate with multiprotein corepressor assemblies that are brought to DNA through interactions with DNA binding proteins such as the nuclear hormone receptors or CBF1 (24, 27, 51). Two distinct vertebrate corepressor complexes have been described, an mSin3-containing complex and a NuRD complex that does not contain mSin3 (34). The ability to detect mSin3A binding to LANA in GST affinity assays indicates that the mSin3 corepressor complex is the one that is likely to be involved in LANA-mediated transcriptional repression. While we have demonstrated that LANA can function as a transcriptional repressor when artificially targeted to DNA through the Gal4 DBD, the natural targeting mechanism remains unclear. In transient assays, LANA repressed activated expression from the EBV Cp and Qp but had relatively little effect on basal reporter expression. These two EBV latency promoters are responsive to different transactivators: Cp is activated by viral EBNA-2, and Qp is activated by cellular STATs. A commonality to EBNA-2 and STAT transactivation is their dependence on the p300, CBP, and P/CAF coactivators (4, 26, 35, 70). Adenovirus E1A binds to p300 and interferes with p300 function (2, 38), and it is possible that LANA may similarly target one of the coactivators. Such a mechanism would be consistent with the reported inhibition of p53 activity and with the differential effect of LANA on basal versus activated transcription that we observed.
The majority of PEL cells are dually infected with HHV-8 and EBV. Although PEL cell lines singly infected with HHV-8 have been established from PEL tumors, HHV-8 alone has not been shown to transform B cells. Incubation of primary B cells with supernatant from dually infected PEL cells led to the outgrowth of B-cell lines that were dually infected or EBV infected, but no singly HHV-8 infected lines were obtained (1, 33). The specific outgrowth of dually infected B cells in one set of experimental conditions further implies that dual infection may provide a growth advantage over EBV infection alone (33). In our analyses, EBV EBNA-1 expression was reduced at both the protein and mRNA levels in HBL-6 and BC-2 PEL cells compared to expression in EBV+ lymphoblastoid cell lines. In both transient transfections and established cell lines, LANA downregulated Qp-driven EBNA-1 expression, suggesting that LANA may be responsible for the EBNA-1 phenotype. An example of an HHV-8+ EBV+ PEL tumor giving rise to an HHV-8+ EBV− cell line has been described elsewhere (28). EBNA-1 is required for maintenance of the EBV genome (37), and it is possible that in culture EBNA-1 may become sufficiently downregulated in PEL cells to lead to loss of EBV genomes and outgrowth of an HHV-8+ EBV− cell population.
Cp is the promoter used to express the EBV EBNA proteins, including EBNA-1, during primary infection and is also frequently active in lesions developing in immunosuppressed patients with lymphoproliferative disease (53, 69, 72). Promoter switching from Cp to Qp has the effect of eliminating expression of the immunogenic EBNA-2 and EBNA-3 latency proteins and hence rendering the infected cell less susceptible to immune surveillance. Cp repression is maintained through methylation (46, 55, 59, 63, 68), but the regulation of Cp methylation repression is not fully understood. Whether LANA-mediated Cp downregulation might predispose Cp to methylation inactivation in dually infected PELs is an interesting question. One of the methyl CpG binding proteins, MeCP2, interacts with mSin3A (5), as does LANA, and it is possible that LANA-mediated tethering of mSin3 might stimulate recruitment of MeCP2.
While LANA repressed expression from the EBV Cp and Qp promoters, the HHV-8 latency promoter that regulates LANA expression was activated by LANA. The implication is that LANA autoregulates its own expression and apparently has the capacity to modulate transcription either positively or negatively. Our experiments specifically link LANA-mediated repression to the binding of LANA to the mSin3 corepressor complex and implicate LANA in the modification of EBV latency gene expression in dually infected PELs.
ACKNOWLEDGMENTS
We thank C. Laherty for the mSin3A plasmid, D. Alcindor for HHV-8 lambda 3-2 DNA, Mabel Chiu for technical assistance, and Feng Chang for help with manuscript preparation.
This work was supported by National Institutes of Health (NIH) grant RO1 CA85151 to S.D.H. A.K. and D.G. received support from NIH Anti-Cancer Drug Development training grant 5 T32 CA09243.
REFERENCES
- 1.Aguirre A J, Robertson E S. Characterization of intertypic recombinants of the Epstein-Barr virus from the body-cavity-based lymphoma cell lines BC-1 and BC-2. Virology. 1999;264:359–369. doi: 10.1006/viro.1999.0015. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 5.Bird A P, Wolffe A P. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C, Gao S-J, Healy L E, Matthews S, Thomas A J, Coignet L, Warnke R A, Strauchen J A, Matutes E, Kamel O W, Moore P S, Weiss R A, Chang Y. Establishing a KSHV negative cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 7.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infected endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 8.Callahan J, Pai S, Cotter M, Robertson E S. Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: potential switches in latent gene expression due to coinfection. Virology. 1999;262:18–30. doi: 10.1006/viro.1999.9876. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two AIDS-related lymphoma cell lines containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 11.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowlex D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y-N, Dong D L-Y, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Lee J M, Wang Y, Huang D P, Ambinder R F, Hayward S D. The Epstein-Barr virus latency Qp promoter is positively regulated by STATs and Zta interference with JAK-STAT activation leads to loss of Qp activity. Proc Natl Acad Sci USA. 1999;96:9339–9344. doi: 10.1073/pnas.96.16.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter M A, II, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 15.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupin N, Diss T L, Kellam P, Tulliez M, Du M Q, Sicard D, Weiss R A, Isaacson P G, Boshoff C. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood. 2000;95:1406–1412. [PubMed] [Google Scholar]
- 17.Dupin N, Fisher C, Kellam P, Arid S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 19.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 20.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 21.Gessain A, Briere J, Angelin-Duclos C, Valensi F, Beral H M, Davi F, Nicola M A, Sudaka A, Fouchard N, Gabarre J, Troussard X, Dulmet E, Audouin J, Diebold J, de The G. Human herpesvirus 8 (Kaposi's sarcoma herpesvirus) and malignant lymphoproliferations in France: a molecular study of 250 cases including two AIDS-associated body cavity based lymphomas. Leukemia. 1997;11:266–272. doi: 10.1038/sj.leu.2400549. [DOI] [PubMed] [Google Scholar]
- 22.Horenstein M G, Nador R G, Chadburn A, Hyjek E M, Inghirami G, Knowles D M, Cesarman E. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. Blood. 1997;90:1186–1191. [PubMed] [Google Scholar]
- 23.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh J J-D, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaffe E S. Primary body cavity-based AIDS-related lymphomas. Evolution of a new disease entity. Am J Clin Pathol. 1996;105:141–143. doi: 10.1093/ajcp/105.2.141. [DOI] [PubMed] [Google Scholar]
- 26.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Kao H-Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katano H, Hoshino Y, Morishita Y, Nakamura T, Satoh H, Iwamoto A, Herndier B, Mori S. Establishing and characterizing a CD30-positive cell line harboring HHV-8 from a primary effusion lymphoma. J Med Virol. 1999;58:394–401. [PubMed] [Google Scholar]
- 29.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedes D H, Oberskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 31.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 32.Kellam P, Bourboulia D, Dupin N, Shotton C, Fisher C, Talbot S, Boshoff C, Weiss R A. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J Virol. 1999;73:5149–5155. doi: 10.1128/jvi.73.6.5149-5155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kliche S, Kremmer E, Hammerschmidt W, Koszinowski U, Haas J. Persistent infection of Epstein-Barr virus-positive B lymphocytes by human herpesvirus 8. J Virol. 1998;72:8143–8149. doi: 10.1128/jvi.72.10.8143-8149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 35.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 36.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 37.Lee M A, Diamond M E, Yates J L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, Imhof A, Collingwood T N, Urnov F D, Wolffe A P. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 1999;18:5634–5652. doi: 10.1093/emboj/18.20.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling P D, Hayward S D. Contribution of conserved amino acids in mediating interaction between EBNA2 and CBF1/RBPJk. J Virol. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margolskee R F, Kavathas P, Berg P. Epstein-Barr virus shuttle vector for stable episomal replication of cDNA expression libraries in human cells. Mol Cell Biol. 1988;8:2837–2847. doi: 10.1128/mcb.8.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matolcsy A, Nador R G, Cesarman E, Knowles D M. Immunoglobulin VH gene mutational analysis suggests that primary effusion lymphomas derive from different stages of B cell maturation. Am J Pathol. 1998;153:1609–1614. doi: 10.1016/S0002-9440(10)65749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsushima A Y, Strauchen J A, Lee G, Scigliano E, Hale E E, Weisse M T, Burstein D, Kamel O, Moore P S, Chang Y. Posttransplantation plasmacytic proliferations related to Kaposi's sarcoma-associated herpesvirus. Am J Surg Pathol. 1999;23:1393–1400. doi: 10.1097/00000478-199911000-00010. [DOI] [PubMed] [Google Scholar]
- 44.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller G, Rigsby M O, Heston L, Grocan E, Sun R, Metroka C, Levy J A, Gao S-J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 46.Minarovits J, Hu L F, Imai S, Harabuchi Y, Kataura A, Minarovits-Kormuta S, Osato T, Klein G. Clonality, expression and methylation patterns of the Epstein-Barr virus genomes in lethal midline granulomas classified as peripheral angiocentric T cell lymphomas. J Gen Virol. 1994;75:77–84. doi: 10.1099/0022-1317-75-1-77. [DOI] [PubMed] [Google Scholar]
- 47.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 48.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 49.Moore P S, Gao S-J, Dominguez G, Cesarman E, Lungu O, Knowles D, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nador R G, Cesarman E, Chadburn A, Dawson D B, Ansari M Q, Sald J, Knowles D M. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpesvirus. Blood. 1996;88:645–656. [PubMed] [Google Scholar]
- 51.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 52.Nicholas J, Zong J-C, Alcendor D J, Ciufo D M, Poole L J, Sarisky R T, Chiou C J, Zhang X, Wan X, Guo H-G, Reitz M S, Hayward G S. Novel organization features, captured cellular genes and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- 53.Oudejans J J, Jiwa M, Vandenbrule A J C, Grasser F A, Horstman A, Vos W, Kluin P M, Vandervalk P, Walboomers J M M, Meijer C J L M. Detection of heterogenous Epstein-Barr virus gene expression patterns with individual post-transplantation lymphoproliferative disorders Am. J Pathol. 1995;147:923–933. [PMC free article] [PubMed] [Google Scholar]
- 54.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paulson E J, Speck S H. Differential methylation of Epstein-Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J Virol. 1999;73:9959–9968. doi: 10.1128/jvi.73.12.9959-9968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Platt G M, Simpson G R, Mittnacht S, Schulz T F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertson K D, Ambinder R F. Methylation of the Epstein-Barr virus genome in normal lymphocytes. Blood. 1997;90:4480–4484. [PubMed] [Google Scholar]
- 60.Russo J J, Bohenzky R A, Chein M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaefer B C, Strominger J L, Speck S H. Host cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol Cell Biol. 1997;17:364–377. doi: 10.1128/mcb.17.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 65.Szekely L, Chen F, Teramoto N, Ehlin-Henriksson B, Pokrovskaja K, Szeles A, Manneborg-Sandlund A, Lowbeer M, Lennette E T, Klein G. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J Gen Virol. 1998;79:1445–1452. doi: 10.1099/0022-1317-79-6-1445. [DOI] [PubMed] [Google Scholar]
- 66.Szekely L, Kiss C, Mattsson K, Kashuba E, Pokrovskaja K, Juhasz A, Holmvall P, Klein G. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J Gen Virol. 1999;80:2889–2900. doi: 10.1099/0022-1317-80-11-2889. [DOI] [PubMed] [Google Scholar]
- 67.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 68.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. Epstein-Barr virus (EBV) in endemic Burkitt's lymphoma: molecular analysis of primary tumor tissue. Blood. 1998;91:1373–1381. [PubMed] [Google Scholar]
- 69.Tao Q, Swinnen L J, Yang J, Srivastava G, Robertson K D, Ambinder R F. Methylation status of the Epstein-Barr virus major latent promoter C in iatrogenic B cell lymphoproliferative disease. Am J Pathol. 1999;155:619–625. doi: 10.1016/S0002-9440(10)65157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu X, Sun Y-L, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 72.Young L, Alfieri C, Hennessy K, Evans H, O'Hare C, Anderson K C, Ritz J, Shapiro R S, Rickinson A, Kieff E. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]