Abstract
A subgroup of patients with atopic dermatitis (AD) suffers from recurrent, disseminated herpes simplex virus skin infection, termed eczema herpeticum. To determine the transcriptional mechanisms of the skin and immune system pathobiology that underlie development of AD with eczema herpeticum (ADEH), we performed RNA-sequencing analysis of nonlesional skin (epidermis, dermis) from AD patients with and without a history of ADEH (ADEH+, n = 15; ADEH−, n = 13) along with healthy controls (n = 15). We also performed RNA sequencing on participants’ plasmacytoid dendritic cells infected in vitro with herpes simplex virus 1. ADEH+ patients exhibited dysregulated gene expression, limited in the dermis (14 differentially expressed genes) and more widespread in the epidermis (129 differentially expressed genes). ADEH+-upregulated epidermal differentially expressed genes were enriched in type 2 cytokine (IL4R, CCL22, CRLF2, IL7R), interferon (CXCL10, ICAM1, IFI44, IRF7), and IL-36γ (IL36G) inflammatory gene pathways. All ADEH+ participants exhibited type 2 cytokine and inteferon endotypes, and 87% were IL36G-high. In contrast, these endotypes were more variably expressed among ADEH− participants. ADEH+ skin also had dysregulated epidermal differentiation complex gene expression of the late-cornified envelope, S100A, and small proline-rich gene families, which are involved in skin barrier function and antimicrobial activities. Plasmacytoid dendritic cell transcriptional responses to herpes simplex virus 1 infection were unaltered by ADEH status. The study concluded that the pathobiology underlying ADEH+ risk is associated with a unique, multifaceted epidermal inflammation that accompanies dysregulation of epidermal differentiation complex genes. These findings will help direct future studies that define how these inflammatory patterns may drive risk of eczema herpeticum in AD.
Keywords: AD, ADEH, HSV, RNA sequencing, Skin disease
Graphical abstract
Introduction
Atopic dermatitis (AD) is a common chronic inflammatory disease of the skin, with a complex etiology driven by interacting genetic and environmental factors, culminating in both immune and skin barrier dysfunction. Despite these broad uniting factors, the clinical phenotype of AD is heterogeneous, with patients exhibiting different levels of severity, age of onset, symptoms, and duration of illness as well as differing in their comorbidities and disease complications (Czarnowicki et al, 2019).
One important AD complication is increased susceptibility to bacterial and viral skin infections (Ong and Leung, 2016; Wang et al, 2021). Aside from Staphylococcus aureus, the most common pathogen for AD skin is herpes simplex virus (HSV) 1 (Ong and Leung, 2016; Wang et al, 2021), which can lead to disseminated HSV1 infection, termed eczema herpeticum (EH) (Beck et al, 2009). EH is a devastating exacerbation of AD that requires immediate antiviral therapy. The etiology of EH is an epidemiological quandary: whereas 62% of adults are HSV1 seropositive, only 3% of patients with AD develop EH-like symptoms (Bin et al, 2021; Leung et al, 2011). Moreover, there is a remarkable bimodality in the recurrence of HSV infection; most individuals have only a single episode, but a small subgroup of patients with AD is susceptible to multiple recurrent episodes (AD with EH [ADEH]–positive patients). The discrepancy between probable HSV1 exposure and EH emergence suggests that intrinsic host factors may determine the risk for EH development and that those with ADEH+ disease may exhibit unique AD pathobiology.
Multiple studies support intrinsic differences in the pathobiology of ADEH+ patients compared with that of patients without a history of EH (ADEH−). For example, studies have found that ADEH+ patients have more severe disease, with early age disease onset, more frequent history of other atopic disorders, and high rate of skin S aureus infections (Beck et al, 2009). In addition, ADEH+ patients have been shown to exhibit higher frequencies of both FLG, TSLP, and IL7R genetic risk variants than ADEH− patients (Bin et al, 2021; Gao et al, 2010, 2009). A study of PBMCs and CD8+ T cells from ADEH+ individuals also found diminished capacity to produce interferon-gamma (IFN-γ) in response to HSV1 compared with that in ADEH− individuals, suggesting that systemic immune responses may differ in ADEH+ patients (Bin et al, 2014).
Few studies have evaluated skin dysregulation in ADEH+ patients, but the occurrence of EH regardless of humoral immunity to HSV suggests that skin innate immunity may play a critical role in ADEH+ susceptibility. For example, type 2 cytokine (T2) inflammation is a strong regulator of innate immunity, and biomarkers of T2 inflammation (blood eosinophils and serum IgE) have been found to be upregulated in ADEH+ patients (Beck et al, 2009). Moreover, multiple skin transcriptomic studies of AD have documented different inflammatory AD endotypes, including those driven by type 2, type 1, type 17, and IL-36γ cytokines (Brunner et al, 2017; Chan et al, 2018; Czarnowicki et al, 2019; Dyjack et al, 2018; Noda et al, 2015; Sanyal et al, 2019). Of particular interest are the dysregulated processes, such as those related to innate immunity, that are present in the skin of ADEH+ patients, which predispose their skin to this pathology.
Thus, to investigate the intrinsic pathobiological risk factors and susceptibility mechanisms that predispose patients with AD to developing ADEH+ disease, we characterized the molecular differences between the skin of ADEH+ individuals with a prior history of multiple EH episodes and the skin of ADEH− individuals on the basis of transcriptomic profiling of nonlesional skin biopsies from these patient groups, matched by disease severity. Nonlesional skin was chosen for characterization because it has been recently shown that AD nonlesional skin exhibits inflammation and early pathobiology, allowing a window into the mechanisms that underlie predisposition to the development of disease (Dyjack et al, 2018; Leung et al, 2019; Pavel et al, 2021; Suárez-Fariñas et al, 2011). In doing so, we assayed both epidermal tissue, the primary origin of barrier function in the skin, and dermal tissue, wherein many tissue-resident immune cells reside. On the basis of these data, we define the distinct inflammatory pathways that underlie ADEH+ disease risk. We also examined HSV1-stimulated blood-circulating plasmacytoid dendritic cells (pDCs) from these individuals, which secrete type I and type III interferons (IFNs) and play an important role in antiviral host responses in the skin and thus may contribute to the increased susceptibility to infection exhibited by ADEH+ patients.
Results
ADEH+ patients exhibit more severe dysregulation of common AD skin disease pathobiology
To identify ADEH disease mechanisms, we generated whole-transcriptome sequencing on skin samples and blood-derived pDCs from ADEH+ (n = 15), ADEH− (n = 13) and healthy control (HC) (n = 13) participants. First, exploring the mechanisms related to dysregulated HSV1 viral responses, we examined whole-transcriptome sequencing from patient pDCs stimulated with live HSV1 virus or untreated cells (mock control). We then modeled pDC gene expression as a function of HSV1 treatment, testing the interaction between infection and ADEH+ status. Although we found 9079 differentially expressed genes (DEGs) in pDCs by HSV1 infection (Supplementary Table S1), there were no significant interactions with ADEH status. We also performed weighted gene coexpression network analysis on the pDC samples, finding that 19 of 23 pDC coexpression networks were associated with HSV1 infection (Supplementary Figure S1 and Supplementary Table S2). However, similar to the single-gene differential expression analysis, HSV1-associated changes in the expression of these 19 networks were not significantly different on the basis of ADEH status (Supplementary Figure S1). In summary, we found no evidence that pDC response to HSV1 infection is unique among ADEH+ patients.
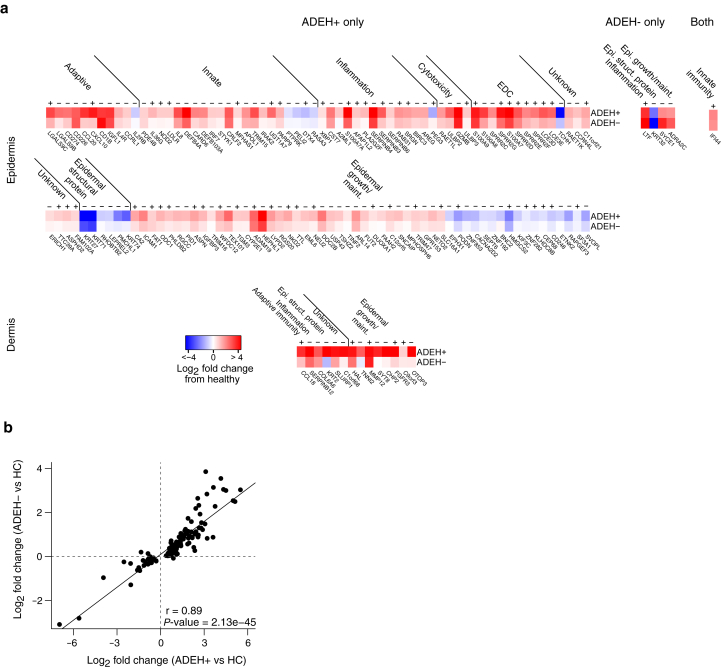
We next explored whether ADEH+ skin exhibits gene expression changes, performing whole-transcriptome sequencing on nonlesional epidermal and dermal skin (separated from skin biopsies) of the same ADEH+, ADEH−, and HC participants (Supplementary Figure S2). To examine this, we performed single-gene differential expression analysis between ADEH+ and HC participants, finding 14 ADEH+ DEGs in the dermis (Figure 1a and Supplementary Table S3). Upregulated dermal DEGs included known AD genes: CCL18 chemokine, the inflammatory tissue protease, matrix metalloproteinase 12 MMP12, and fibroblast growth factor receptor FGFR3. Lesser known dermal DEGs included collagen (COL6A6) and cytokeratin (KRT2) genes and a serine protease inhibitor gene (SERPINB12). Interestingly, we found 2 genomically adjacent (11p11) genes, TNNI2 and SYT8, both upregulated in ADEH+ participants. Comparing epidermal expression in ADEH+ with that in HC participants, we observed 129 DEGs (Figure 1a and Supplementary Table S3), many of which have known roles in innate and adaptive immunity, inflammation, and cytotoxic immune functions as well as contain multiple epidermal differentiation complex (EDC) genes and genes involved in epidermal growth and maintenance (Figure 1a).
Figure 1.
Multitissue transcriptome analysis of nonlesional skin from subjects with AD with and without EH reveals dysregulated epidermal expression among subjects. (a) Heat maps showing LFC in expression between ADEH+ (top row) or ADEH− (bottom row) participants compared with HCs for genes differentially expressed in 1 or the other disease group or both (from left to right) in epidermis (top) or dermis (bottom). The genes are organized into broad functional groups. Symbols above heat maps indicate whether (+) or not (−) each gene has been previously associated with AD in nonlesional skin. (b) Scatter plot comparing LFCs between ADEH− and HC epidermal skin with those between ADEH+ and HC epidermal skin on the basis of genes with significantly modified expression in ADEH+ epidermis compared with that in HCs. Pearson correlation between the 2 sets of LFCs is indicated and demonstrates that genes dysregulated in ADEH+ skin are similarly dysregulated in ADEH− individuals. AD, atopic dermatitis; ADEH, atopic dermatitis with eczema herpeticum; EDC, epidermal differentiation complex; EH, eczema herpeticum; HC, healthy control; LFC, log2 fold change.
In contrast, when comparing skin gene expression in ADEH− with that in HC participants, we observed no significant DEGs in dermal tissue and only 5 significant DEGs in the epidermis, where only 1 epidermal DEG (IFI44) was shared between ADEH+ and ADEH− groups (Figure 1a and Supplementary Table S3). However, when comparing fold changes in expression of ADEH+ DEGs between ADEH+ and HC and ADEH− and HC analyses, the direction of effect (upregulation or downregulation) for the ADEH+ and ADEH− analyses was concordant for 98 and 86% of the epidermal and dermal DEGs, respectively. Furthermore, there was a strong positive correlation between ADEH+ DEG fold changes and fold changes observed for these genes in the ADEH− group (Pearson r = 0.89, P = 2.13e-45) (Figure 1b), with consistently less pronounced dysregulation observed among ADEH− participants. To further explore whether ADEH+ pathobiology is shared with ADEH− patients, we examined the overlap between our ADEH+ DEGs and AD genes reported in 5 published RNA-sequencing studies of AD (Cole et al, 2014; Dyjack et al, 2018; He et al, 2020; Nattkemper et al, 2018; Sanyal et al, 2019). In this study, we found that the majority of ADEH+ epidermal DEGs (55 of 129 or 43% for nonlesional AD skin and 111 of 129 or 86% for lesional AD skin) and dermal DEGs (5 of 14 or 36% for nonlesional skin and 12 of 14 or 86% for lesional AD skin) have been previously associated with AD in at least 1 of the 5 studies (Figure 1a). Together, these results suggest that skin pathobiology underlying AD complicated by EH is similar in kind but more severe in form than that exhibited by patients with AD without EH history.
Dysregulation of EDC gene expression in the skin of ADEH+ patients corresponds with T2 inflammation
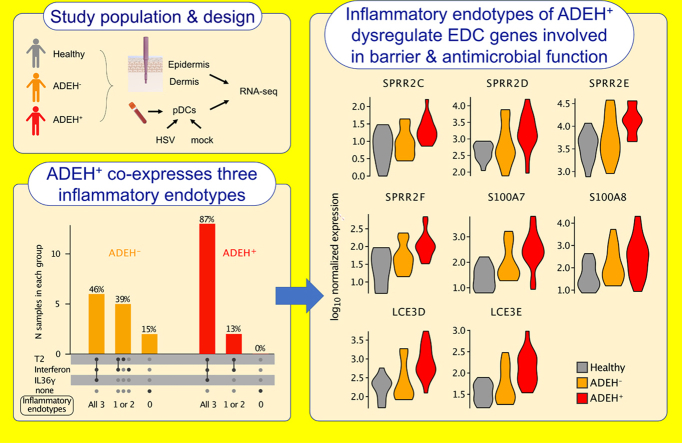
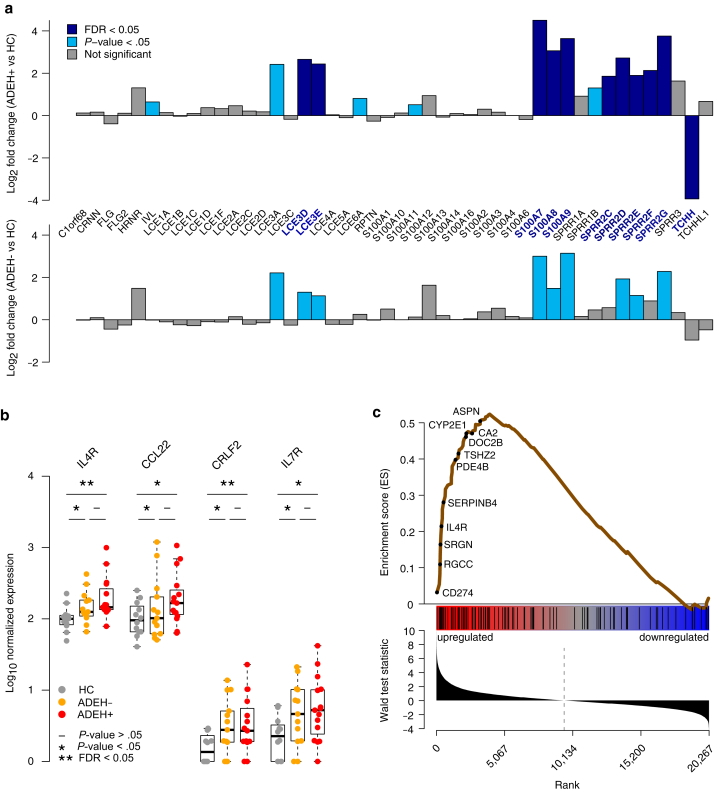
Examining the functional basis of ADEH+ differential expression observed in the epidermis, we found strongly increased expression of terminal skin differentiation pathways. Specifically, we found 10 EDC genes with significantly higher expression in the ADEH+ group, including 5 among the top 10 ADEH+ DEGs (on the basis of false discovery rate [FDR]). These upregulated EDC genes included 5 small proline-rich protein genes (SPRR2C, SPRR2D, SPRR2E, SPRR2F, and SPRR2G), 2 late-cornified envelope genes (LCE3D and LCE3E), and 3 S100A-type calcium binding protein genes (S100A7, S100A8, and S100A9) (Figure 2a). We also found strong downregulation of the epidermal structural gene, trichohyalin, TCHH, among ADEH+ subjects. Although none of these 10 EDC DEGs exhibited a discordant direction of effect between ADEH+ and ADEH− differential expression comparisons, the magnitude of dysregulation was consistently lower, although not significantly, in the ADEH− group (Figure 2a).
Figure 2.
Dysregulation of EDC gene expression in ADEH+ corresponds with T2 inflammation. (a) Bar plots of LFCs for EDC genes, comparing epidermal ADEH+ with HC skin (top) and ADEH− with HC skin (bottom). Teal = nominally significant differences (P < .05); dark blue = significant differences (FDR < 0.05). Many dysregulated EDC genes in ADEH+ skin are similarly dysregulated in ADEH− skin. (b) Box plots of epidermal expression for canonical T2 genes in HC (gray), ADEH− (orange), and ADEH+ (red) samples. Asterisk (∗) denotes nominally significant differences (P < .05); asterisks (∗∗) denote significant differences (FDR < 0.05). (c) ES plot from GSEA visualizing enrichment of ADEH+ epidermal DEGs within a gene set preranked on the basis of DE analysis between T2-high and T2-low AD. Top: running-sum ES profile (brown line) across T2-ranked genes, where the left-side peak of positive ES values indicates particular enrichment of ADEH+ DEGs among the most T2-upregulated genes. A subset of genes belonging to the leading edge of enrichment (appearing at/before the ES maximum and most strongly contributing to enrichment) are indicated. Middle: vertical black bars denote positions of ADEH+ DEGs along the T2-ranked list of genes. Bottom: bar plot showing the ordered distribution of the gene ranking statistic (Wald test statistic from DESeq2). AD, atopic dermatitis; ADEH, atopic dermatitis with eczema herpeticum; DE, differential expression; DEG, differentially expressed gene; EDC, epidermal differentiation complex; ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; HC, healthy control; LFC, log2 fold change; T2, type 2 cytokine.
Potentially relevant to this upregulation of EDC genes is previous work showing that T2 inflammation, particularly IL-4/IL-13 stimulation of keratinocytes, can elicit characteristic gene signatures in the skin, including dysregulated expression of EDC genes (Howell et al, 2009). Supporting this, we found that 2 of the strongest skin biomarkers of T2 inflammation, IL4R and CCL22, were both upregulated in the epidermis of ADEH+ participants (Figure 2b and Supplementary Table S3). In addition, we found that both gene subunits of the TSLP receptor (CRLF2 and IL7R) were significantly or nearly significantly upregulated in the dermis and epidermis of ADEH+ participants (Figure 2b and Supplementary Table S3). To further explore ADEH+ upregulation in T2 inflammation, we used gene set enrichment analysis (GSEA) to test whether the 99 upregulated ADEH+ DEGs were enriched among genes previously found to characterize subjects with T2-high AD (Dyjack et al, 2018) (Supplementary Table S4), finding strong enrichment of ADEH+ DEGs among genes most upregulated in T2-high subjects (P < .001) (Figure 2c). Notably, 8 of the 10 EDC genes upregulated with ADEH+ disease were among the leading-edge genes responsible for the T2-high enrichment. Together, these findings show that T2 inflammation is elevated among ADEH+ participants as a group and that this increased inflammation may contribute to observed dysregulation in EDC expression in the skin of ADEH+ patients.
ADEH+ skin exhibits enhanced IFN and IL-36γ inflammation
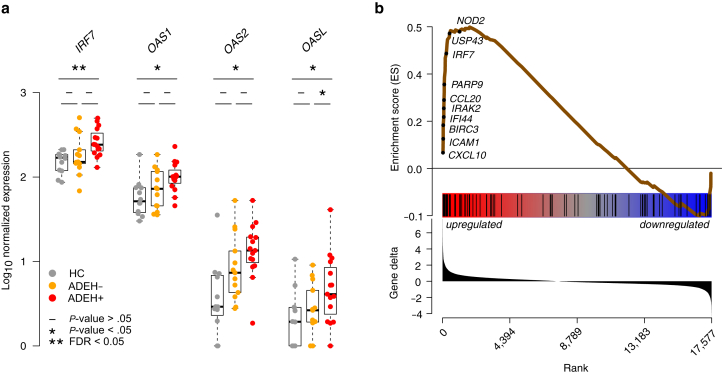
Further examination of ADEH+-upregulated DEGs revealed multiple genes known to be upregulated by the epidermis in response to viral infection or IFN stimulation (Figure 3a). Included among these was IRF7, which drives the expression of type I IFNs and other viral response genes, and 3 members of the 2′-5′-oligoadenylate synthetase gene family (OAS1, OAS2, and OASL), which are IFN-inducible antiviral and innate immunity genes (Figure 3a). To specifically address whether ADEH+ epidermal DEGs were more enriched in viral infection response genes, we leveraged published data identifying the transcriptomic response of keratinocytes to the viral mimetic, polyinosinic:polycytidylic acid (Poly[I:C]) (Zhu et al, 2017) (Supplementary Table S4). GSEA revealed strong enrichment of ADEH+-upregulated epidermal DEGs among the genes most induced by Poly(I:C) treatment of keratinocytes (P < .001) (Figure 3b), including CXCL10, ICAM1, IFI44, and IRF7. Overall, these results support significant antiviral/IFN inflammation in the skin of ADEH+ patients.
Figure 3.
ADEH skin exhibits enhanced viral/IFN inflammation. (a) Box plots of epidermal expression for canonical viral response genes in HC (gray), ADEH− (orange), and ADEH+ (red) participants. The asterisk (∗) denotes nominally significant differences (P < .05); the asterisks (∗∗) denote significant differences (FDR < 0.05). (b) ES plot from GSEA visualizing enrichment of ADEH+ epidermal DEGs within a gene set preranked on the basis of expression within poly(I:C)-stimulated keratinocytes compared with the controls. Top: running-sum ES profile (brown line) across the poly(I:C)-ranked genes. A subset of leading-edge genes is shown. Middle: positions of ADEH+ DEGs along the poly(I:C)-ranked gene list. Bottom: ordered distribution of the gene-ranking statistic (gene deltas). ADEH, atopic dermatitis with eczema herpeticum; DEG, differentially expressed gene; ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; HC, healthy control; Poly(I:C), polyinosinic:polycytidylic acid.
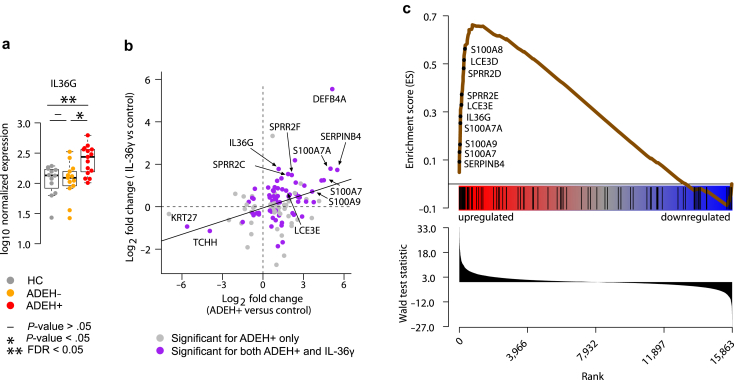
In addition, ADEH+ patients exhibited upregulation in expression of the inflammatory cytokine gene, IL36G (FDR = 0.01) (Figure 4a). IL-36γ is a member of the IL-1 family and has been reported to be prominently upregulated in psoriatic skin but also in some AD cohorts, where it can drive the expression of other chemokines and disrupt skin cornification. To capture the transcriptional consequences of IL-36γ stimulation on the epidermis, we used an in vitro human epidermal keratinocyte, neonatal (HEKn) culture system to differentiate keratinocytes in the presence and absence of IL-36γ protein. Whole-transcriptome RNA-sequencing analyses revealed that IL-36γ treatment altered the expression of 5596 genes (FDR < 0.05) (Supplementary Table S4) in differentiated keratinocytes. Of 129 genes modified in ADEH+ epidermal skin, over half (53%) were among these IL-36γ–stimulated DEGs, and log fold changes associated with ADEH+ disease and IL-36γ stimulation were significantly correlated (P = 2.02e-5, r = 0.38) (Figure 4b). Remarkably, among the genes strongly upregulated in both datasets were all 10 of the ADEH+-upregulated EDC genes (LCE3D and LCE3E; SPRR2C, SPRR2D, SPRR2E, SPRR2F, and SPRR2G; and S100A7, S100A8, and S100A9). Moreover, on the basis of GSEA, we found strong enrichment of the set of ADEH+-upregulated DEGs among the genes most induced by IL-36γ treatment of keratinocytes (P < .001) (Figure 4c). All 10 ADEH+-upregulated EDC genes were among the 33 leading-edge genes identified in the IL-36γ enrichment analysis (Figure 4c). The only EDC gene that was downregulated among ADEH+ subjects, TCHH, was also strongly downregulated by IL-36γ treatment (FDR = 1.36e-34) (Figure 4b). Taken together, these results strongly suggest that both IFN and IL-36γ–driven inflammation are elevated in the epidermis of ADEH+ subjects.
Figure 4.
ADEH skin exhibits enhanced IL-36γ–driven inflammation. (a) Box plots of epidermal expression for IL36G in HC (gray), ADEH− (orange), and ADEH+ (red). Asterisk (∗) denotes nominally significant differences (P < .05); asterisks (∗∗) denote significant differences (FDR < 0.05). (b) Scatter plot comparing LFCs for epidermal ADEH+ versus HC DEGs (x-axis) with LFCs of these same genes on the basis of comparing IL-36γ–stimulated keratinocytes with controls (y-axis). Gray = DEG only in the ADEH+ versus HC comparison; purple = DEG in both comparisons. (c) ES plot from GSEA visualizing enrichment of ADEH+ epidermal DEGs within a gene set preranked on the basis of IL-36γ–stimulated keratinocytes versus controls. Top: running-sum ES profile (brown line) across the IL-36γ–ranked genes. A subset of leading-edge genes is shown. Middle: positions of ADEH+ DEGs along the IL-36γ–ranked gene list. Bottom: ordered distribution of the gene-ranking statistic (Wald test statistic from DESeq2). ADEH, atopic dermatitis with eczema herpeticum; DEG, differentially expressed gene; ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; HC, healthy control; LFC, log2 fold change.
ADEH+ disease is characterized by coexpression of multiple inflammatory skin endotypes
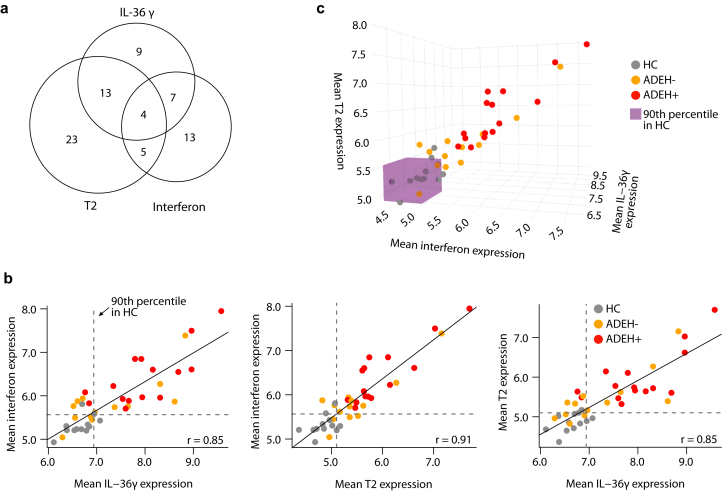
We next investigated the shared genic aspects of the epidermal T2, IFN, and IL-36γ inflammatory expression signatures, finding that 39% of signature genes were within 2 or more of the 3 inflammatory programs (Figure 5a). Notably, the T2/IL-36γ overlapping genes included 8 of the 10 ADEH+-upregulated EDC genes. Despite this shared inflammatory character, we also identified 9, 13, and 23 genes specific to the IL-36γ, IFN, and T2 inflammatory programs, respectively (Figure 5a).
Figure 5.
Subjects with ADEH coexpress multiple inflammatory skin endotypes. (a) Venn diagram of leading-edge genes detected in distinct GSEAs that test for enrichment of epidermal ADEH+ DEGs within preranked genes on the basis of T2-high, viral-induced interferon (poly(I:C)-stimulation), and IL-36γ–stimulated inflammation. (b) Pairwise scatter plots of mean expression for distinct leading-edge genes from the 3 inflammatory endotypes (T2, interferon, and IL-36γ). Gray = HC, orange = ADEH−, red = ADEH+. Dashed lines = the 90th percentiles of mean expression for HC samples, solid line = slope of the linear relationship, r = Pearson correlation coefficient. (c) The 3D scatter plot of mean expression for distinct leading-edge genes from the 3 inflammatory endotypes. Point colors are as in b. The purple box subsumes all values that are below the 90th percentile threshold of mean expression for HCs. 3D, 3-dimensional; ADEH, atopic dermatitis with eczema herpeticum; DEG, differentially expressed gene; GSEA, gene set enrichment analysis; HC, healthy control; Poly(I:C), polyinosinic:polycytidylic acid; T2, type 2 cytokine.
Although T2, IFN, and IL-36γ inflammatory signatures were upregulated on a group level among ADEH+ patients, it is unclear whether this upregulation is uniform across all ADEH+ patients or driven by a subset of participants with high expression levels (ie, an inflammatory endotype group). Supporting this possibility, prior studies have found significant inflammatory heterogeneity across the AD population (Brunner et al, 2017; Chan et al, 2018; Czarnowicki et al, 2019; Dyjack et al, 2018; Noda et al, 2015; Sanyal et al, 2019). Therefore, we leveraged mean expression of the T2, IFN, and IL-36γ–specific gene sets to dichotomously classify or endotype all participants for these inflammation patterns, defining participants as inflammatory endotype positive or negative on the basis of whether their mean expression levels were greater or less than the 90th percentile of the metric among HCs, respectively. Strikingly, we found that all 15 ADEH+ subjects were positive for the T2 and IFN endotypes, whereas 87% were also positive for the IL-36γ endotype (Figure 5b). In contrast, the presence of these endotypes was much more variable among ADEH− patients, with 69, 69, and 46% of this group positive for the T2, IFN, and IL-36γ endotypes, respectively (Figure 5b). These patterns show that multiple inflammatory endotypes co-occur simultaneously in nonlesional AD skin, particularly among ADEH+ patients, where all 3 endotypes are nearly universally present, potentially underlying the more severe AD disease among ADEH+ patients. Moreover, we found that the expression levels for the pairwise combinations of inflammatory metrics were all strongly positively correlated (Figure 5b and c) (r ≥ 0.85), further suggesting a strong relationship among these different inflammation types.
Discussion
In this study, we considered whether molecular dysfunction in the skin (epidermis and dermis) and/or in pDCs (an immune cell population critical to antiviral responses) is a predisposing factor for ADEH+ disease. We uncovered significant transcriptional dysregulation in the epidermis of ADEH+ patients. The ADEH+ disease profile, which included upregulation of EDC genes, was similar in form to that observed for ADEH− patients (in this study and in previously published studies [Cole et al, 2014; Dyjack et al, 2018; He et al, 2020; Nattkemper et al, 2018; Sanyal et al, 2019]) but was more intense in degree. These alterations in EDC expression were accompanied by epidermal inflammation consistent with stimulation by T2, IFN, and IL-36γ cytokines. Each of these inflammatory endotypes was observed among subgroups of ADEH− patients; however, ADEH+ patients were unique in their propensity to exhibit either 2 or all 3 of these inflammatory endotypes simultaneously. These findings suggest that ADEH+ disease is characterized by complex, multifaceted inflammation of the epidermis, accompanied by a dysregulated epidermal skin layer.
AD has traditionally been known as a T2-associated disease (Bieber, 2008; Weidinger and Novak, 2016), supported by many studies (Dyjack et al, 2018; Möbus et al, 2021; Tsoi et al, 2019). However, recent genomic profiling of skin samples from patients with AD has revealed significant heterogeneity in disease mechanisms, where T2-dominated inflammation is only observed in a subset of patients with AD (Brunner et al, 2017; Chan et al, 2018; Dyjack et al, 2018; Noda et al, 2015; Sanyal et al, 2019). In this study, we found that only a subgroup of ADEH− patients was T2-high, whereas all 15 ADEH+ patients were T2-high, exhibiting increased expression of the skin T2 biomarker genes IL4R and CCL22 and coreceptor genes for TSLP (an epithelial T2 alarmin previously associated with AD [Soumelis et al, 2002]) (CRLF2 and IL7R). These findings suggest that excessive T2 inflammation may play a significant role in the development of ADEH+ disease, possibly through dysregulation of the normal innate immune mechanisms that protect skin from viral infection. Supporting this, patients with food allergy who have AD with T2 skin inflammation exhibit enhanced transepidermal water loss (Leung et al, 2019). Moreover, IL-4 and IL-13 stimulation of the skin downregulates loricrin and involucrin expressions, critical skin barrier genes, and increases S aureus infection risk (Kim et al, 2008; Leyva-Castillo et al, 2013). In addition, T2 cytokines decrease skin production of multiple antimicrobial peptides, including human β-defensin 2 and 3 and the cathelicidin LL37 (Howell et al, 2006; Ong et al, 2002).
In this study, we found that 8 of the ADEH+-upregulated DEGs were EDC genes, all previously shown to be upregulated among T2-high patients with AD (Dyjack et al, 2018). These EDC genes (LCE3D, LCE3E, SPRR2C, SPRR2D, SPRR2E, SPRR2F, S100A8, and S100A9) encode proteins that contribute to structural stability of the cornified envelope and function as skin antimicrobial peptides. LCE genes have long been recognized to contribute to the formation of a healthy skin barrier because the proteins they encode are cross-linked into the cornified epithelium (Niehues et al, 2022). The 18 human LCE genes are arranged into 6 groups on the basis of sequence and expression similarity. LCE genes in groups 1, 2, 5, and 6 are expressed in normal skin (Bergboer et al, 2011), whereas group 3 LCE genes, which we found to be upregulated among ADEH+ patients (LCE3D/3E), are induced by injury and are strongly upregulated in lesional psoriatic skin (Bergboer et al, 2011). Furthermore, multiple genetic studies have confirmed that LCE3B/3C deletion is associated with psoriasis risk (Liu et al, 2008; Zhang et al, 2009). Importantly, LCE proteins have recently been shown to exert potent antimicrobial activity, including against S aureus and Pseudomonas aeruginosa; the species selectivity and strength of this antimicrobial activity vary by LCE group (Niehues et al, 2022). Further supporting this differential antimicrobial activity of LCE group proteins, patients with the LCE3B/3C genetic deletion exhibit a unique microbiome profile (Niehues et al, 2022). In addition, among the ADEH+-upregulated EDC genes were the inflammatory alarmins S100A7, S100A8 and S100A9. S100A8 and S100A9 proteins form a heterodimer called calprotectin that exerts significant antimicrobial skin activity through the sequestration of calcium (Abtin et al, 2010; Christmann et al, 2020). Although these genes have been found to be upregulated in AD, they are better known as psoriasis biomarkers and are induced by IL-17 stimulation (Christmann et al, 2020; Matsunaga et al, 2021). The third set of ADEH+-upregulated EDC genes, SPRR2 genes, are known to cross-link the cornified envelope of the skin (Tarcsa et al, 1998) but more recently were found to act as antimicrobial peptides by disrupting bacterial membranes (Zhang et al, 2022). Moreover, mice lacking the Sprr2a gene were found to be more susceptible to S aureus and P aeruginosa infections (Zhang et al, 2022). We hypothesize that the selective upregulation of these EDC genes may alter both the barrier function and skin microbiome of ADEH+ patients, rendering their epidermis more susceptible to viral infection. We note that in 1-year follow-up cumulative transepidermal water loss area under the curve measures collected on a subset of these patients (n = 23), ADEH+ patients trended toward higher values (worse barrier integrity) than ADEH− and healthy patients, but this trend was not statistically significant (Supplementary Figure S3).
We also observed strong upregulation of IL36G expression in the epidermis of ADEH+ patients, along with significant enrichment of ADEH+ DEGs for genes upregulated by IL-36γ stimulation of keratinocytes, including all 8 of the ADEH+-upregulated EDC genes. IL-36γ belongs to the IL-1 cytokine superfamily and is strongly associated with development of psoriasis (Ding et al, 2018), being elevated in skin lesions of patients with psoriasis (D'Erme et al, 2015) and underlying T helper 17–driven mouse models of psoriasis (Liu et al, 2017; Nakagawa et al, 2017). Moreover, loss-of-function variants in IL36RN (an IL-36 receptor antagonist) are strong risk factors for development of pustular psoriasis (Farooq et al, 2013; Onoufriadis et al, 2011; Sugiura et al, 2013). In comparison with psoriasis, only a few studies have implicated IL-36γ inflammation in AD, showing increased IL-36γ in AD skin lesions (Brunner et al, 2018; Otobe et al, 2018). One possible reason for elevated IL-36γ in AD relates to S aureus infection, a well-known feature of AD (Callewaert et al, 2020; Kim et al, 2019; Leonard et al, 2020) that is also elevated in ADEH+ disease (Beck et al, 2009). Recent studies using topical application of S aureus on the skin surface of Il36r-knockout mice demonstrated that IL36R-mediated signaling was essential to skin inflammatory responses (Liu et al, 2017; Nakagawa et al, 2017), suggesting that IL-36 cytokines may be critical in S aureus–induced skin inflammation in ADEH+ patients. We note that in our dataset, although S aureus positivity trended slightly higher among ADEH+ patients than among ADEH− patients (73.3 vs 61.5%), these differences were not statistically significant. Supporting the profound influence of IL-36γ signaling on skin biology, we found that IL-36γ stimulation of keratinocytes led to dysregulated expression of thousands of genes, including those involved in antimicrobial defense and epidermal barrier function. Together, our findings support an important role for IL-36γ–driven inflammation in ADEH+ pathobiology.
On the basis of the previous observation that PBMCs from ADEH+ subjects produced fewer type I and type II IFNs upon HSV1 stimulation ex vivo (Bin et al, 2014; Leung et al, 2011), we hypothesized that we might also observe diminished IFN programming in (i) pDCs, one of the major type I and type III IFN-producing peripheral immune cells (Swiecki and Colonna, 2015), and/or (ii) the epidermis/dermis of ADEH+ individuals. However, we observed no reduced IFN signaling among ADEH+ patients in either cell/tissue type, with (pDCs only) or without HSV1 stimulation. Surprisingly, we instead observed significantly increased IFN activation in ADEH+ versus ADEH− epidermis. As a first line of defense against virus infection (Samuel, 2001), an enhanced IFN program would be expected to diminish viral infection risk. However, in the context of coactivation with T2 and/or IL-36γ programs, IFN responses to future viral infection may nonetheless be slowed or diminished (Durrani et al, 2012). In addition to the unknown effects of this IFN inflammation on skin function, it remains unclear by what mechanism this inflammation is provoked. We note that IFN inflammation has been observed in the epithelium of another atopic disease: the airway epithelium of a subset of patients with asthma (Bhakta et al, 2018). In fact, T2 inflammation may itself promote IFN inflammation, as suggested by reports that IFN inflammation and cellular stress are induced in epithelial cells stimulated with IL-13 (Jackson et al, 2020). Similarly, whereas acute AD is most associated with elevated T2 inflammation, chronic AD may invoke an elevated IFN inflammatory profile (Bieber, 2010). Another intriguing possibility is that recurrent HSV1 skin infections in ADEH+ patients may epigenetically orient the epithelium toward an IFN program, often referred to as inflammatory memory (Larsen et al, 2021). For example, in the gut, inflammatory epithelial remodeling originally prompted by a single infection persists, even after the infection resolves (Fonseca et al, 2015). We speculate that such inflammatory memory could be encoded within skin stem cells, which then gives rise to keratinocytes primed to express the IFN endotype. This phenomenon was demonstrated in mice, where inflammation-challenged epithelial skin stem cells showed epigenetic memory of exposures by exhibiting enhanced responses to subsequent injury (Naik et al, 2017). Moreover, dysregulated inflammatory memory resulting from prior exposure in ADEH+ patients specifically was supported by persistent long-term abnormalities in the sphingosine-1-phosphate signaling system in the skin and plasma of these patients (Berdyshev et al, 2022). Such epigenetic imprinting may underlie the heightened inflammatory milieu of ADEH+ disease and calls for further study.
Despite these exciting findings, we note several limitations to this study. First, as a cross-sectional analysis performed in the context of established disease, we are unable to determine the sequence by which these inflammatory and epidermal profiles occurred over the course of ADEH+ disease development. Both longitudinal skin profiling studies of ADEH+ patients and mouse models of ADEH+ disease development are needed to delineate the stepwise ADEH pathogenesis process. Second, although our results in this study are significant, the cohort studied is limited in numbers because ADEH is a rare AD endotype; therefore, additional studies are needed to replicate our findings. Furthermore, although our study implicates potential changes in skin barrier function and microbial dysbiosis underlying ADEH+ disease, mechanistic studies are needed to directly investigate whether these hypotheses are correct. In addition, molecular risk factors for ADEH development that occur distinctly in lesional skin might not have been captured by our study. We focused on nonlesional skin given that it has been shown that mechanisms underlying AD lesions can be studied in nonlesional tissue, which is unobstructed from gene expression patterns present in lesional skin that result from tissue damage and do not underlie the initial development of the pathobiology (Leung et al, 2019). Furthermore, although EH eruptions often initiate at the site of a current skin lesion (Wollenberg et al, 2003), EH can also infect or spread to nonlesional skin (DermNet. Eczema herpeticum, 2023; Santmyire-Rosenberger and Nigra, 2005), which is the ultimate source of all skin lesions, and indeed can occur in individuals without any history of skin disease (Traidl et al, 2021). Thus, the ADEH+ expression profiles reported are systemic inflammatory risk factors in skin that predispose patients to developing ADEH and which are likely present in both nonlesional and lesional skin. Finally, because our study was designed to determine the pathobiological mechanisms predisposing patients with AD to develop EH skin eruptions, we explicitly matched ADEH− and ADEH+ patients by eczema severity (as measured by Eczema Area and Severity Index [EASI] and SCORing Atopic Dermatitis [SCORAD]). Thus, our dataset is not particularly amenable to investigating how AD severity may contribute to pathobiological activation in ADEH+ patients. However, when we investigated genes that may be activated in patients with relatively high EASI or SCORAD disease severity scores, no genes were significantly upregulated among patients with high SCORAD scores (6 below vs 9 above SCORAD = 50), but there were 18 genes upregulated among 4 patients with EASI > 20 compared with those among 5 patients with EASI < 11; among them were proinflammatory genes RAB31, GZMB, and IGFBP3, all of which were among DEGs upregulated in ADEH+ versus HC participants. Finally, when we divided patients by those relatively low (<3, n = 8) and relatively high (>3, n = 7) in variance stabilized transformation expression of TARC (CCL17), a biomarker of AD severity (Stutte et al., 2012), we found 127 genes upregulated among the TARC-high group. These genes were enriched in antiviral pathways such as IFN response (eg, IFNAR2, B2T2, IFI44, IFIT3, CD47, APOL6, GBP4, HLA-DPA1) and neutrophil chemotaxis (eg, CXCL11, CCL4, CCL19, ITGB2, S100A9, S100A8) and in T2 activation genes (eg, TSLP, IL13RA1, IL7R, ALOX15, CST7) as well as the group 3 LCE gene LCE3A. These genes were also enriched in leading-edge genes from each of the 3 GSEA analyses (T2, P = 2.33e-14; viral, P = 8.40e-05; and IL36G, P = 1.06e-04). In contrast, 68 genes downregulated in the TARC-high groups were enriched for skin development and keratinization pathways (eg, KAP11-1, K31, K9, LCE2A, LCE2C, LCE6A, SCEL, KLK7). These findings suggest that among ADEH+ patients, those at risk for more severe AD disease may more highly express ADEH+ pathobiological signatures and have more pronounced inhibition of epidermal differentiation signatures, although more samples are needed to confirm this.
Despite these limitations, our study advances the understanding of ADEH+ disease pathobiology, uncovering a multifaceted epidermal inflammation underlying this severe AD phenotype group, which is associated with dysregulated expression of cornified envelope genes critical to epidermal innate defense. Given that a similar, albeit diminished, pathobiology also characterizes ADEH− skin, understanding of additional molecular and environmental factors that may govern how and when these pathways may drive ADEH+ disease awaits further study. Moreover, additional research is needed to determine how these pathways may be further altered in lesional ADEH+ skin. This work nevertheless helps to reorient ADEH+ research toward these pathways and genes and also suggests that ADEH+ patients may benefit from therapeutics directed against these pathways.
Materials and Methods
Human subject recruitment
Nonlesional skin samples were obtained from 15 adult patients with AD with physician-confirmed history of recurrent EH (≥3 episodes) (ADEH+ patients) (6 male and 9 female patients; age [mean ± SD] = 33.1 ± 10.5 years), 13 adult patients with AD with no history of EH (ADEH− patients) (4 male and 9 female patients; age [mean ± SD] = 37.7 ± 11.4 years), and 13 nonatopic HC subjects (3 male and 10 female subjects; age [mean ± SD] = 41.0 ± 8.4 years) with neither a personal nor family history of atopy and no skin disease. To identify characteristic ADEH+ traits, ADEH+ and ADEH− participants were matched by disease severity and were anti-HSV1 and anti-HSV2 IgG seropositive. Furthermore, none of the patients had active HSV infection at the time of evaluation. At the time of skin sample collection, EASI, SCORAD, and Rajka–Langeland total scores were collected, and serum IgE levels were measured. All study subjects were HSV seropositive (per past documentation or current HSV test performed for this study). Skin cultures were examined for the presence of S aureus. Study subjects’ characteristics are summarized in Table 1 (Supplementary Table S5 presents the full subject metadata). Study subjects did not receive topical corticosteroids, topical calcineurin inhibitors, or topical/oral antibiotics for 1 week before enrollment. Patients were not treated with systemic immunosuppressive medications for >1 month before enrollment in this study. None of the patients in the study reported being on biologics. The study was approved by the Western Institutional Review Board (protocol number 20161695) and was conducted at National Jewish Health (Denver, CO). All subjects provided written informed consent before participation in the study.
Table 1.
Clinical Characteristics of the Study Subjects
| Characteristic | ADEH+ n = 15 | ADEH− n = 13 | HC n = 13 | P-Value, Overall |
|---|---|---|---|---|
| Age, y | ||||
| Mean (SD) | 33.1 (10.5) | 37.7 (11.4) | 41.0 (8.4) | .134 |
| Gender, n (%) | ||||
| Female | 9 (60.0) | 9 (69.2) | 10 (76.9) | .929 |
| Male | 6 (40.0) | 4 (30.8) | 3 (23.1) | |
| Ethnicity, n (%) | ||||
| Hispanic or Latino | 1 (6.7) | 1 (7.7) | 1 (7.7) | .993 |
| Non-Hispanic or Latino | 14 (93.3) | 12 (92.3) | 12 (92.3) | |
| Race, n (%) | ||||
| White | 13 (86.7) | 11 (84.6) | 10 (76.9) | .777 |
| Black or African American | 2 (13.3) | 2 (15.4) | 3 (23.1) | |
| EASI score | ||||
| Mean (SD) | 17.5 (11.0) | 15.9 (14.3) | NA | .749 |
| SCORAD | ||||
| Mean (SD) | 50.8 (13.8) | 43.1 (18.7) | NA | .224 |
| Rajka–Langeland total score | ||||
| Mean (SD) | 7.3 (1.2) | 6.8 (1.8) | NA | .459 |
| Serum IgE | ||||
| Median (1st, 3rd quartile] | 1100 (118, 2400)1,2 | 300 (91.7, 841)3 | 29 (7.5, 44.5) | .0002 |
| Skin Staphylococcus aureus culture positive, n (%) | 11 (73.3) | 8 (61.5) | 0 (0) | .0002 |
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; EASI, Eczema Area and Severity Index; NA, not applicable; SCORAD, SCORing Atopic Dermatitis.
P = .0003 compared with HC.
P = .08 compared with ADEH− participants.
P = .003 compared with HC.
Skin biopsy collection and RNA extraction
Skin punch biopsy specimens of 2 mm were collected from the nonlesional skin area of the upper extremities. Nonlesional skin enables characterization of the overall skin subinflammatory state in ADEH+ patients, not just terminal inflammation reflected in lesional skin. The biopsy specimens were dissected into the epidermis and dermis after 1 hour of digestion at 37 °C in 5 U/ml dispase diluted in PBS (catalog number 354235). Isolated epidermal and dermal samples were immediately submerged into RLT buffer with β-mercaptoethanol (Qiagen) and frozen at −80 °C for future RNA isolation. RNeasy Micro Kits (Qiagen) were used according to the manufacturer’s instructions to isolate RNA from the epidermis and dermis.
Peripheral blood pDCs isolation and HSV stimulation
Heparinized venous peripheral blood was collected from each study participant. PBMCs were isolated using density gradient centrifugation in Ficoll. Immediately after purification, 5 × 107 PBMCs were subjected to pDC purification using EasySep Human pDC Isolation kit (STEMCELL Technologies) according to the manufacturer’s instructions. pDCs were plated at 5 × 104 cells/100 μl in complete RPMI 1640 medium (RPMI 1640, 10% fetal calf serum, supplemented with L-glutamine, 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid, and antibiotics) in 96-well plates, and mock or HSV1 treated (multiplicity of infection of 1) for 20 hours. After treatment, cells were harvested for RNA extraction. HSV1 (VR-733) strain was purchased from ATCC.
Keratinocyte culture and stimulation
HEKns from a single donor were purchased from Thermo Fisher Scientific and maintained in EpiLife medium containing 0.06 mM calcium chloride and S7 supplement in 5% carbon dioxide at 37 °C. For HEKn differentiation, in triplicate, cells were cultured in EpiLife medium containing 1.3 mM calcium chloride for 3 days and then treated with 200 ng/ml of IL-36γ (R&D Systems) for 2 days. After treatment, the cells were harvested for RNA extraction using a Qiagen RNeasy Kit according to the manufacturer’s instructions.
RNA transcriptome gene expression and quality control
For skin biopsies and pDCs, RNA-sequencing libraries were constructed and barcoded using the Ion AmpliSeq Transcriptome Human Gene Expression Kit. Barcoded RNA-sequencing libraries were pooled and sequenced on the Ion Torrent Proton sequencer using P1 chips. Sequencing reads were mapped to AmpliSeq transcriptome target regions with the Torrent Mapping Alignment Program and quantified with the Ion Torrent ampliSeqRNA plugin using the uniquely mapping option. Duplicated sequences were removed from the FASTA file, and incorrect amplicon locations were corrected as previously reported (Poole et al, 2014). Two dermal baseline samples that had <6 million assigned reads and 1 epidermal baseline sample that exhibited a clear dermal expression signature on the basis of multidimensional scaling analysis were removed from the dataset.
For HEKn cultures, whole-transcriptome libraries were constructed using the KAPA mRNA HyperPrep library kit (Roche Sequencing and Life Science, Kapa Biosystems) from 250 ng of total input RNA according to the manufacturer’s instructions. Barcoded libraries were pooled and sequenced using 150 bp paired-end reads on the Illumina HiSeq 2500 system (Illumina). Raw-sequencing reads were trimmed using skewer (version 0.2.2) with the following parameters: end quality = 15, mean quality = 25, min = 30. Read alignment to the human reference genome GRCh38 was performed using HISAT2 (version 2.1.0) with default parameters. Gene quantification was performed using htseq-count (version 0.9.1) and GRCh38 Ensemble (version 84) gene transcript model with the following parameters: stranded = reverse, a = 20, and mode = intersection-nonempty.
Analysis of bulk RNA-sequencing data
To compare gene expression among disease groups in vivo, we performed gene-level differential expression analysis using the DESeq2 R package (version 1.3.0) (Love et al, 2014). We adjusted P-values to control for FDR using the Benjamini–Hochberg method. DEGs were those with FDR < 0.05. Long intergenic noncoding genes and genes of uncertain function (genes with an "LOC" prefix) as well as lowly expressed genes (not reaching at least 10 counts in 3% of samples) were excluded from analysis. To compare gene expression between paired pDC samples, which were mock or HSV1 stimulated, we performed variance stabilized transformation normalization of counts using DESeq2 and then used the lmerSeq R package (version 0.1.6) (Vestal et al, 2022) to run a linear mixed model that predicts variance stabilized transformation expression as a function of treatment, including a random intercept for each subject and an interaction term between treatment and disease status to test whether response to treatment differs by disease status. To compare keratinocyte samples stimulated with or without IL-36γ, we used DESeq2. Pathway enrichment analysis was performed throughout using the EnrichR API (Chen et al, 2013) (libraries: Kyoto Encyclopedia of Genes and Genomes, 2021; Reactome, 2016; and Gene Ontology, 2021). Plots of gene expression were based on log10 counts normalized by size factor using DESeq2.
We carried out weighted gene coexpression network analysis on ex vivo stimulated pDC samples to detect functional networks of coexpressed genes (Zhang and Horvath, 2005) that may respond differently to HSV1 on the basis of disease status. For analysis, we used variance stabilized transformation–normalized counts, excluding genes expressing <10 reads in 10% of samples. We ran weighted gene coexpression network analysis using the WGCNA R package (version 1.70) (Langfelder and Horvath, 2008) on the basis of signed Pearson correlations, a soft-thresholding power of 37, a minimum network size of 30, maxCoreScatter = 0.70, minGap = 0.30, and cutHeight = 1.
For GSEA, we ranked genes on the basis of Wald statistic as calculated using DESeq2 for the T2 and IL-36γ–based rankings. We obtained these values for T2 (Dyjack et al, 2018) and generated them ourselves for IL-36γ on the basis of the transcriptomic responses of the IL-36γ–stimulated HEKn cultures. For rankings based on poly(I:C), we downloaded expression values for HEKns treated with either poly(I:C) or vehicle from Gene Expression Omnibus (Zhu et al, 2017) (accession GSE92646) and then ranked genes on the basis of shift in expression between vehicle and poly(I:C) (gene deltas). For each GSEA, we ran 1000 permutations using GSEA (version 4.1.0) (Mootha et al, 2003).
Ethical Statement
This study was approved by the Western Institutional Review Board (protocol number 20161695) under which all subjects provided written informed consent.
Data Availability Statement
Participant metadata used in this study as well as full differential expression results have been included as supplementary tables. All raw and QCed AmpliSeq counts for dermal and epidermal skin samples and plasmacytoid dendritic cells are available at Open Science Framework at the http://osf.io/ under DOI 10.17605/OSF.IO/2N4K5. Raw and processed RNA-sequencing data from the IL-36γ–stimulation experiments are available at the National Center for Biotechnology Information/Gene Expression Omnibus under repository accession number Gene Expression Omnibus (GSE262071; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE262071).
ORCIDs
Nathan D. Jackson: http://orcid.org/0009-0008-2675-021X
Nathan Dyjack: http://orcid.org/0000-0001-6944-6252
Elena Goleva: http://orcid.org/0000-0002-4685-9318
Lianghua Bin: http://orcid.org/0000-0002-3786-0106
Michael T. Montgomery: http://orcid.org/0000-0002-6748-2329
Cydney Rios: http://orcid.org/0000-0003-3965-9059
Jamie L. Everman: http://orcid.org/0000-0002-1935-4672
Patricia Taylor: http://orcid.org/0000-0002-6719-0449
Caroline Bronchick: http://orcid.org/0000-0002-3522-3058
Brittany N. Richers: http://orcid.org/0009-0001-8504-7734
Donald Y. M. Leung: http://orcid.org/0000-0002-0177-3844
Max A. Seibold: http://orcid.org/0000-0002-8685-4263
Conflict of Interest
MAS consults on eczema and asthma drug development for Escient Pharmaceuticals. DYML consults for Sanofi-Genzyme Pharma and Leo Pharma. EG has obtained research funding from Sanofi. The remaining authors have no conflicts of interest to declare.
Acknowledgments
This study was funded by National Institutes of Health grants U19-AI117673, 1UM1AI151958, and 1U01AI152037. The authors thank all the volunteers who participated in these studies.
Author Contributions
Conceptualization: MAS, DYML, EG; Formal Analysis: NDJ, ND, EG; Investigation: NDJ, ND, EG, LB, MTM, CR, JLE; Methodology: MAS, NDJ, ND, EG, LB, DYML; Resources: EG, LB, MTM, CR, JLE, PT, CB, BNR; Writing - Original Draft Preparation: NDJ, MAS, ND, EG, DYML; Writing - Review and Editing: NDJ, MAS, ND, EG, DYML, LB, MTM, CR, JLE, PT, CB, BNR
Declaration of Generative Artificial Intelligence (AI) or Large Language Models (LLMs)
No artificial intelligence or large language model tools were used to prepare this manuscript.
accepted manuscript published online XXX; corrected published online XXX
Footnotes
Cite this article as: JID Innovations 2024.100279
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.xjidi.2024.100279.
Supplementary Materials
Differentially Expressed Genes and Pathways in pDCs Stimulated with HSV1
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; FDR, false discovery rate; HC, healthy control; HSV1, herpes simplex virus 1; pDC, plasmacytoid dendritic cell.
Shown are output tables from lmerSeq, giving gene-specific differences in HSV1-stimulated compared with mock-stimulated pDCs in HC (tab 1), ADEH− patients (tab 2), and ADEH+ patients (tab 3) and averaged across all 3 groups (tab 4). Tabs 5–7 give estimates for the interaction between HSV1 stimulation and disease status. The last 2 tabs give results from EnrichR, with enriched terms and pathways of upregulated (tab 8) and downregulated (tab 9) genes based on the overall effect of virus across disease groups (tab 4). For each of the differential expression summary tables (tabs 1–7), contrast estimates, standard errors, degrees of freedom (denoted as df), t-test statistics, 95% confidence intervals, P-values, and adjusted P-values (FDR based on the Benjamini–Hochberg method) are given. For each of the enrichments in the enrichment tables (tabs 8–9), the originating enrichment library, enriched term, gene overlap (number of overlapping genes is before the underscore, and number of genes in the pathway is after the underscore), P-value, adjusted P-value (Benjamini–Hochberg method), combined score (ln[P-value] × z-score), and itemized overlapping genes are given.
WGCNA Network Genes and their Enriched Pathways Based on Blood pDCs Stimulated and Mock Stimulated with HSV1
Abbreviation: HSV1, herpes simplex virus 1; pDC, plasmacytoid dendritic cell; WGCNA, weighted gene co-expression network analysis.
The first tab gives genes for each network, and the subsequent tabs give EnrichR results for each network. Enrichment tables are as described above for Supplementary Table S1.
Result Tables for Differential Expression Analysis in DESeq2, Comparing Pairwise Disease Groups (HC, ADEH−, and ADEH+) in Epidermis (Tabs 1–3) and Dermis (Tabs 4–6)
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; HC, healthy control.
Each table gives genes, mean expression in the baseline group (listed second in the tab title), log2 fold change, Wald statistic, P-value, and adjusted P-value (Benjamini–Hochberg method).
Tables with Gene Rankings Used for GSEA
Abbreviations: AD, atopic dermatitis; GSEA, gene set enrichment analysis; Poly(I:C), polyinosinic:polycytidylic acid; T2, type 2 cytokine.
Shown are ranking of genes for GSEA on the basis of differential expression analysis between T2-high and T2-low subjects with AD (tab 1) (Dyjack et al, 2018), shifts in expression between keratinocytes stimulated with poly(I:C) and vehicle (tab 2) (Zhu and Garza, 2017), and differential expression analysis between keratinocytes stimulated and mock stimulated with IL-36γ (tab 3) (this study).
Subject Metadata Underlying Table 1
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; EASI, Eczema Area and Severity Index; HC, healthy control; SCORAD, SCORing Atopic Dermatitis.
DONOR gives a unique identifier for each participant, and DISEASE_STATUS gives whether the donor had ADEH (ADEH+) or did not (ADEH−) or was a HC. Demographic traits are also included (age, sex, ethnicity, and race). EASI denotes EASI score at screening, SCORAD denotes SCORAD score at screening, RLTOT denotes total Rajka–Langeland score at screening, IGER denotes IgE result, and STAPH denotes Staphylococcus aureus test result (on the basis of detection in either lesional or nonlesional skin).
References
- Abtin A., Eckhart L., Gläser R., Gmeiner R., Mildner M., Tschachler E. The antimicrobial heterodimer S100A8/S100A9 (calprotectin) is upregulated by bacterial flagellin in human epidermal keratinocytes. J Invest Dermatol. 2010;130:2423–2430. doi: 10.1038/jid.2010.158. [DOI] [PubMed] [Google Scholar]
- Beck L.A., Boguniewicz M., Hata T., Schneider L.C., Hanifin J., Gallo R., et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–269.e2697. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdyshev E., Goleva E., Bronova I., Bronoff A.S., Streib J.E., Vang K.A., et al. Signaling sphingolipids are biomarkers for atopic dermatitis prone to disseminated viral infections. J Allergy Clin Immunol. 2022;150:640–648. doi: 10.1016/j.jaci.2022.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergboer J.G., Tjabringa G.S., Kamsteeg M., van Vlijmen-Willems I.M., Rodijk-Olthuis D., Jansen P.A., et al. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol. 2011;178:1470–1477. doi: 10.1016/j.ajpath.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakta N.R., Christenson S.A., Nerella S., Solberg O.D., Nguyen C.P., Choy D.F., et al. IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am J Respir Crit Care Med. 2018;197:313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- Bieber T. Atopic dermatitis. Ann Dermatol. 2010;22:125–137. doi: 10.5021/ad.2010.22.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin L., Edwards M.G., Heiser R., Streib J.E., Richers B., Hall C.F., et al. Identification of novel gene signatures in patients with atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2014;134:848–855. doi: 10.1016/j.jaci.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin L., Malley C., Taylor P., Preethi Boorgula M., Chavan S., Daya M., et al. Whole genome sequencing identifies novel genetic mutations in patients with eczema herpeticum. Allergy. 2021;76:2510–2523. doi: 10.1111/all.14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner P.M., Guttman-Yassky E., Leung D.Y. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol. 2017;139:S65–S76. doi: 10.1016/j.jaci.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner P.M., Israel A., Zhang N., Leonard A., Wen H.C., Huynh T., et al. Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. J Allergy Clin Immunol. 2018;141:2094–2106. doi: 10.1016/j.jaci.2018.02.040. [DOI] [PubMed] [Google Scholar]
- Callewaert C., Nakatsuji T., Knight R., Kosciolek T., Vrbanac A., Kotol P., et al. IL-4Rα blockade by dupilumab decreases Staphylococcus aureus colonization and increases microbial diversity in atopic dermatitis. J Invest Dermatol. 2020;140:191–202.e7. doi: 10.1016/j.jid.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.C., Sanyal R.D., Pavel A.B., Glickman J., Zheng X., Xu H., et al. Atopic dermatitis in Chinese patients shows TH2/TH17 skewing with psoriasiform features. J Allergy Clin Immunol. 2018;142:1013–1017. doi: 10.1016/j.jaci.2018.06.016. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann C., Zenker S., Martens L., Hübner J., Loser K., Vogl T., et al. Interleukin 17 promotes expression of alarmins S100A8 and S100A9 during the inflammatory response of keratinocytes. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.599947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Kroboth K., Schurch N.J., Sandilands A., Sherstnev A., O’Regan G.M., et al. Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J Allergy Clin Immunol. 2014;134:82–91. doi: 10.1016/j.jaci.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnowicki T., He H., Krueger J.G., Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol. 2019;143:1–11. doi: 10.1016/j.jaci.2018.10.032. [DOI] [PubMed] [Google Scholar]
- D’Erme A.M., Wilsmann-Theis D., Wagenpfeil J., Hölzel M., Ferring-Schmitt S., Sternberg S., et al. IL-36γ (IL-1F9) is a biomarker for psoriasis skin lesions. J Invest Dermatol. 2015;135:1025–1032. doi: 10.1038/jid.2014.532. [DOI] [PubMed] [Google Scholar]
- 2023. DermNet. Eczema herpeticum.https://dermnetnz.org/topics/eczema-herpeticum (accessed August 16, 2023) [Google Scholar]
- Ding L., Wang X., Hong X., Lu L., Liu D. IL-36 cytokines in autoimmunity and inflammatory disease. Oncotarget. 2018;9:2895–2901. doi: 10.18632/oncotarget.22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrani S.R., Montville D.J., Pratt A.S., Sahu S., DeVries M.K., Rajamanickam V., et al. Innate immune responses to rhinovirus are reduced by the high-affinity IgE receptor in allergic asthmatic children. J Allergy Clin Immunol. 2012;130:489–495. doi: 10.1016/j.jaci.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyjack N., Goleva E., Rios C., Kim B.E., Bin L., Taylor P., et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol. 2018;141:1298–1309. doi: 10.1016/j.jaci.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M., Nakai H., Fujimoto A., Fujikawa H., Matsuyama A., Kariya N., et al. Mutation analysis of the IL36RN gene in 14 Japanese patients with generalized pustular psoriasis. Hum Mutat. 2013;34:176–183. doi: 10.1002/humu.22203. [DOI] [PubMed] [Google Scholar]
- Fonseca D.M., Hand T.W., Han S.J., Gerner M.Y., Glatman Zaretsky A., Byrd A.L., et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell. 2015;163:354–366. doi: 10.1016/j.cell.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P.S., Rafaels N.M., Hand T., Murray T., Boguniewicz M., Hata T., et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124(507–13) doi: 10.1016/j.jaci.2009.07.034. 13 e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P.S., Rafaels N.M., Mu D., Hand T., Murray T., Boguniewicz M., et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. J Allergy Clin Immunol. 2010;125:1403–1407.e4. doi: 10.1016/j.jaci.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Suryawanshi H., Morozov P., Gay-Mimbrera J., Del Duca E., Kim H.J., et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol. 2020;145:1615–1628. doi: 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Kim B.E., Gao P., Grant A.V., Boguniewicz M., DeBenedetto A., et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2009;124(3):R7–R12. doi: 10.1016/j.jaci.2009.07.012. Suppl. 2. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Wollenberg A., Gallo R.L., Flaig M., Streib J.E., Wong C., et al. Cathelicidin deficiency predisposes to eczema herpeticum. J Allergy Clin Immunol. 2006;117:836–841. doi: 10.1016/j.jaci.2005.12.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson N.D., Everman J.L., Chioccioli M., Feriani L., Goldfarbmuren K.C., Sajuthi S.P., et al. Single-Cell and population transcriptomics reveal pan-epithelial remodeling in type 2-high asthma. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.E., Leung D.Y., Boguniewicz M., Howell M.D. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clin Immunol. 2008;126:332–337. doi: 10.1016/j.clim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim B.E., Ahn K., Leung D.Y.M. Interactions between atopic dermatitis and Staphylococcus aureus infection: clinical implications. Allergy Asthma Immunol Res. 2019;11:593–603. doi: 10.4168/aair.2019.11.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S.B., Cowley C.J., Sajjath S.M., Barrows D., Yang Y., Carroll T.S., et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell. 2021;28:1758–1774.e8. doi: 10.1016/j.stem.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A., Wang J., Yu L., Liu H., Estrada Y., Greenlees L., et al. Atopic dermatitis endotypes based on allergen sensitization, reactivity to Staphylococcus aureus antigens, and underlying systemic inflammation. J Allergy Clin Immunol Pract. 2020;8:236–247.e3. doi: 10.1016/j.jaip.2019.08.013. [DOI] [PubMed] [Google Scholar]
- Leung D.Y., Gao P.S., Grigoryev D.N., Rafaels N.M., Streib J.E., Howell M.D., et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-γ response [published correction appears in J Allergy Clin Immunol 2011;128:833] J Allergy Clin Immunol. 2011;127:965–973.e735. doi: 10.1016/j.jaci.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.Y.M., Calatroni A., Zaramela L.S., LeBeau P.K., Dyjack N., Brar K., et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aav2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Castillo J.M., Hener P., Jiang H., Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J Invest Dermatol. 2013;133:154–163. doi: 10.1038/jid.2012.239. [DOI] [PubMed] [Google Scholar]
- Liu H., Archer N.K., Dillen C.A., Wang Y., Ashbaugh A.G., Ortines R.V., et al. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-Mediated T cell responses. Cell Host Microbe. 2017;22:653–666.e5. doi: 10.1016/j.chom.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Helms C., Liao W., Zaba L.C., Duan S., Gardner J., et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga Y., Hashimoto Y., Ishiko A. Stratum corneum levels of calprotectin proteins S100A8/A9 correlate with disease activity in psoriasis patients. J Dermatol. 2021;48:1518–1525. doi: 10.1111/1346-8138.16032. [DOI] [PubMed] [Google Scholar]
- Möbus L., Rodriguez E., Harder I., Stölzl D., Boraczynski N., Gerdes S., et al. Atopic dermatitis displays stable and dynamic skin transcriptome signatures. J Allergy Clin Immunol. 2021;147:213–223. doi: 10.1016/j.jaci.2020.06.012. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Naik S., Larsen S.B., Gomez N.C., Alaverdyan K., Sendoel A., Yuan S.P., et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage [published correction appears in Nature 2018;560:E2] Nature. 2017;550:475–480. doi: 10.1038/nature24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Matsumoto M., Katayama Y., Oguma R., Wakabayashi S., Nygaard T., et al. Staphylococcus aureus virulent PSMα peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe. 2017;22:667–677.e5. doi: 10.1016/j.chom.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattkemper L.A., Tey H.L., Valdes-Rodriguez R., Lee H., Mollanazar N.K., Albornoz C., et al. The genetics of chronic itch: gene expression in the skin of patients with atopic dermatitis and psoriasis with severe itch. J Invest Dermatol. 2018;138:1311–1317. doi: 10.1016/j.jid.2017.12.029. [DOI] [PubMed] [Google Scholar]
- Niehues H., van der Krieken D.A., Ederveen T.H.A., Jansen P.A.M., van Niftrik L., Mesman R., et al. Antimicrobial late cornified envelope proteins: the psoriasis risk factor deletion of LCE3B/C genes affects microbiota composition. J Invest Dermatol. 2022;142:1947–1955.e6. doi: 10.1016/j.jid.2021.11.036. [DOI] [PubMed] [Google Scholar]
- Noda S., Suárez-Fariñas M., Ungar B., Kim S.J., de Guzman Strong C., Xu H., et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol. 2015;136:1254–1264. doi: 10.1016/j.jaci.2015.08.015. [DOI] [PubMed] [Google Scholar]
- Ong P.Y., Leung D.Y. Bacterial and viral infections in atopic dermatitis: a comprehensive review. Clin Rev Allergy Immunol. 2016;51:329–337. doi: 10.1007/s12016-016-8548-5. [DOI] [PubMed] [Google Scholar]
- Ong P.Y., Ohtake T., Brandt C., Strickland I., Boguniewicz M., Ganz T., et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- Onoufriadis A., Simpson M.A., Pink A.E., Di Meglio P., Smith C.H., Pullabhatla V., et al. Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am J Hum Genet. 2011;89:432–437. doi: 10.1016/j.ajhg.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otobe S., Sugaya M., Nakajima R., Oka T., Takahashi N., Kabasawa M., et al. Increased interleukin-36γ expression in skin and sera of patients with atopic dermatitis and mycosis fungoides/Sezary syndrome. J Dermatol. 2018;45:468–471. doi: 10.1111/1346-8138.14198. [DOI] [PubMed] [Google Scholar]
- Pavel A.B., Renert-Yuval Y., Wu J., Del Duca E., Diaz A., Lefferdink R., et al. Tape strips from early-onset pediatric atopic dermatitis highlight disease abnormalities in nonlesional skin. Allergy. 2021;76:314–325. doi: 10.1111/all.14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A., Urbanek C., Eng C., Schageman J., Jacobson S., O’Connor B.P., et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133:670–678.e12. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santmyire-Rosenberger B.R., Nigra T.P. Psoriasis herpeticum: three cases of Kaposi's varicelliform eruption in psoriasis. J Am Acad Dermatol. 2005;53:52–56. doi: 10.1016/j.jaad.2005.01.140. [DOI] [PubMed] [Google Scholar]
- Sanyal R.D., Pavel A.B., Glickman J., Chan T.C., Zheng X., Zhang N., et al. Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/TH17 attenuation. Ann Allergy Asthma Immunol. 2019;122:99–110.e6. doi: 10.1016/j.anai.2018.08.024. [DOI] [PubMed] [Google Scholar]
- Soumelis V., Reche P.A., Kanzler H., Yuan W., Edward G., Homey B., et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–680. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- Stutte S., Gerbitzki N., Novak N., Förster I. In: Atopi dermatitis - disease etiology and clinical management. Esparza-Gordillo J., Dekio I., editors. IntechOpen; Maastricht, Netherlands: 2012. Expression and function of CCL17 in atopic dermatitis; pp. 81–104. [Google Scholar]
- Suárez-Fariñas M., Tintle S.J., Shemer A., Chiricozzi A., Nograles K., Cardinale I., et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127:954–964.e1. e4. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K., Takemoto A., Yamaguchi M., Takahashi H., Shoda Y., Mitsuma T., et al. The majority of generalized pustular psoriasis without psoriasis vulgaris is caused by deficiency of interleukin-36 receptor antagonist. J Invest Dermatol. 2013;133:2514–2521. doi: 10.1038/jid.2013.230. [DOI] [PubMed] [Google Scholar]
- Swiecki M., Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcsa E., Candi E., Kartasova T., Idler W.W., Marekov L.N., Steinert P.M. Structural and transglutaminase substrate properties of the small proline-rich 2 family of cornified cell envelope proteins. J Biol Chem. 1998;273:23297–23303. doi: 10.1074/jbc.273.36.23297. [DOI] [PubMed] [Google Scholar]
- Traidl S., Roesner L., Zeitvogel J., Werfel T. Eczema herpeticum in atopic dermatitis. Allergy. 2021;76:3017–3027. doi: 10.1111/all.14853. [DOI] [PubMed] [Google Scholar]
- Tsoi L.C., Rodriguez E., Degenhardt F., Baurecht H., Wehkamp U., Volks N., et al. Atopic dermatitis is an IL-13-dominant disease with greater molecular heterogeneity compared to psoriasis. J Invest Dermatol. 2019;139:1480–1489. doi: 10.1016/j.jid.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestal B.E., Wynn E., Moore C.M. lmerSeq: an R package for analyzing transformed RNA-Seq data with linear mixed effects models. BMC Bioinformatics. 2022;23:489. doi: 10.1186/s12859-022-05019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V., Boguniewicz J., Boguniewicz M., Ong P.Y. The infectious complications of atopic dermatitis. Ann Allergy Asthma Immunol. 2021;126:3–12. doi: 10.1016/j.anai.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger S., Novak N. Atopic dermatitis. Lancet. 2016;387:1109–1122. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Zoch C., Wetzel S., Plewig G., Przybilla B. Predisposing factors and clinical features of eczema herpeticum: a retrospective analysis of 100 cases. J Am Acad Dermatol. 2003;49:198–205. doi: 10.1067/s0190-9622(03)00896-x. [DOI] [PubMed] [Google Scholar]
- Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhang C., Hu Z., Lone A.G., Artami M., Edwards M., Zouboulis C.C., et al. Small proline-rich proteins (SPRRs) are epidermally produced antimicrobial proteins that defend the cutaneous barrier by direct bacterial membrane disruption. eLife. 2022;11 doi: 10.7554/eLife.76729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.J., Huang W., Yang S., Sun L.D., Zhang F.Y., Zhu Q.X., et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- Zhu A.S., Li A., Ratliff T.S., Melsom M., Garza L.A. After skin wounding, noncoding dsRNA coordinates prostaglandins and Wnts to promote regeneration. J Invest Dermatol. 2017;137:1562–1568. doi: 10.1016/j.jid.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially Expressed Genes and Pathways in pDCs Stimulated with HSV1
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; FDR, false discovery rate; HC, healthy control; HSV1, herpes simplex virus 1; pDC, plasmacytoid dendritic cell.
Shown are output tables from lmerSeq, giving gene-specific differences in HSV1-stimulated compared with mock-stimulated pDCs in HC (tab 1), ADEH− patients (tab 2), and ADEH+ patients (tab 3) and averaged across all 3 groups (tab 4). Tabs 5–7 give estimates for the interaction between HSV1 stimulation and disease status. The last 2 tabs give results from EnrichR, with enriched terms and pathways of upregulated (tab 8) and downregulated (tab 9) genes based on the overall effect of virus across disease groups (tab 4). For each of the differential expression summary tables (tabs 1–7), contrast estimates, standard errors, degrees of freedom (denoted as df), t-test statistics, 95% confidence intervals, P-values, and adjusted P-values (FDR based on the Benjamini–Hochberg method) are given. For each of the enrichments in the enrichment tables (tabs 8–9), the originating enrichment library, enriched term, gene overlap (number of overlapping genes is before the underscore, and number of genes in the pathway is after the underscore), P-value, adjusted P-value (Benjamini–Hochberg method), combined score (ln[P-value] × z-score), and itemized overlapping genes are given.
WGCNA Network Genes and their Enriched Pathways Based on Blood pDCs Stimulated and Mock Stimulated with HSV1
Abbreviation: HSV1, herpes simplex virus 1; pDC, plasmacytoid dendritic cell; WGCNA, weighted gene co-expression network analysis.
The first tab gives genes for each network, and the subsequent tabs give EnrichR results for each network. Enrichment tables are as described above for Supplementary Table S1.
Result Tables for Differential Expression Analysis in DESeq2, Comparing Pairwise Disease Groups (HC, ADEH−, and ADEH+) in Epidermis (Tabs 1–3) and Dermis (Tabs 4–6)
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; HC, healthy control.
Each table gives genes, mean expression in the baseline group (listed second in the tab title), log2 fold change, Wald statistic, P-value, and adjusted P-value (Benjamini–Hochberg method).
Tables with Gene Rankings Used for GSEA
Abbreviations: AD, atopic dermatitis; GSEA, gene set enrichment analysis; Poly(I:C), polyinosinic:polycytidylic acid; T2, type 2 cytokine.
Shown are ranking of genes for GSEA on the basis of differential expression analysis between T2-high and T2-low subjects with AD (tab 1) (Dyjack et al, 2018), shifts in expression between keratinocytes stimulated with poly(I:C) and vehicle (tab 2) (Zhu and Garza, 2017), and differential expression analysis between keratinocytes stimulated and mock stimulated with IL-36γ (tab 3) (this study).
Subject Metadata Underlying Table 1
Abbreviations: ADEH, atopic dermatitis with eczema herpeticum; EASI, Eczema Area and Severity Index; HC, healthy control; SCORAD, SCORing Atopic Dermatitis.
DONOR gives a unique identifier for each participant, and DISEASE_STATUS gives whether the donor had ADEH (ADEH+) or did not (ADEH−) or was a HC. Demographic traits are also included (age, sex, ethnicity, and race). EASI denotes EASI score at screening, SCORAD denotes SCORAD score at screening, RLTOT denotes total Rajka–Langeland score at screening, IGER denotes IgE result, and STAPH denotes Staphylococcus aureus test result (on the basis of detection in either lesional or nonlesional skin).
Data Availability Statement
Participant metadata used in this study as well as full differential expression results have been included as supplementary tables. All raw and QCed AmpliSeq counts for dermal and epidermal skin samples and plasmacytoid dendritic cells are available at Open Science Framework at the http://osf.io/ under DOI 10.17605/OSF.IO/2N4K5. Raw and processed RNA-sequencing data from the IL-36γ–stimulation experiments are available at the National Center for Biotechnology Information/Gene Expression Omnibus under repository accession number Gene Expression Omnibus (GSE262071; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE262071).