Abstract
The A9L open reading frame of vaccinia virus was predicted to encode a membrane-associated protein. A transcriptional analysis of the A9L gene indicated that it was expressed at late times in vaccinia virus-infected cells. Late expression, as well as virion membrane association, was demonstrated by the construction and use of a recombinant vaccinia virus encoding an A9L protein with a C-terminal epitope tag. Immunoelectron microscopy revealed that the A9L protein was associated with both immature and mature virus particles and was oriented in the membrane with its C terminus exposed on the virion surface. To determine whether the A9L protein functions in viral assembly or infectivity, we made a conditional-lethal inducible recombinant vaccinia virus. In the absence of inducer, A9L expression and virus replication were undetectable. Under nonpermissive conditions, viral late protein synthesis occurred, but maturational proteolytic processing was inhibited, and there was an accumulation of membrane-coated electron-dense bodies, crescents, and immature virus particles, many of which appeared abnormal. We concluded that the product of the A9L gene is a viral membrane-associated protein and functions at an early stage in virion morphogenesis.
Vaccinia virus is a complex enveloped DNA virus that replicates within the cytoplasm of a wide variety of cell types. Viral DNA replication, intermediate and late transcription, structural protein accumulation, and virion formation all occur in the viral factory areas that are characteristically located near the nucleus. Early studies using transmission electron microscopy provided the basic pathway of virion assembly (12, 21, 32). The first discrete structures are the crescent-shaped membranes that develop into spherical particles and acquire a DNA nucleoid before sealing. These particles, known as immature virions (IV), are not thought to be infectious. Infectivity is acquired through an incompletely understood maturation process that is evident by the cleavage of a variety of structural proteins (29, 33) and a concomitant change in particle morphology. Processing occurs at a consensus AGX motif and is presumably mediated by a virally encoded protease (62, 63, 66). The majority of the resultant brick-shaped intracellular mature virions (IMV) remain inside the cell until lysis occurs. Egress from the intact cell is accomplished by a population of the IMV that become enwrapped by a double membrane derived from a late Golgi or early endosomal compartment (22, 52, 58). In the wrapping process, viral particles acquire an additional complement of viral proteins to become intracellular enveloped virions (IEV). The IEV are transported through the cytoplasm to the cell periphery, where they fuse with the plasma membrane and may either remain as cell-associated virions (CEV) or become detached as extracellular enveloped virions (EEV). It is thought that CEV and EEV are responsible for cell-to-cell and long-range spread, respectively (1, 6, 9, 38).
Six proteins unique to the IEV or extracellular particles have been identified. Five are integral membrane proteins and are encoded by the open reading frames (ORFs) A56R, B5R, A33R, A34R, and A36R (17, 19, 26, 35, 36, 39, 46). The sixth, encoded by the F13L ORF (23), is a cytoplasmically oriented protein that appears to be associated with the membrane via a palmitate group that is essential for its function (22). Deletion of genes encoding any of these proteins, except A56R, yields recombinant vaccinia viruses with a small-plaque phenotype. Analysis of deletion or other mutants has revealed roles for the B5R (19, 69) and F13L proteins (5, 53) in EEV formation. With both the B5R and F13L deletion mutants, the defect occurs at the level of wrapping and IEV formation. Although the A33R and A34R mutants have abnormalities in the wrapping process, increased amounts of EEV are released compared with that for wild-type virus (17, 47). The A36R mutant also makes both IEV and EEV despite its small-plaque phenotype (49, 72). The principal defect of the A33R, A34R, and A36R mutants appears to be the failure of actin tails to form on virus particles, a process that greatly enhances cell-to-cell spread of wild-type virus (47, 49, 70, 72). The A36R protein appears to be directly involved in interaction with cellular proteins that are involved in the nucleation of the actin tails (20).
The mechanism of formation of IMV membranes has not been elucidated, although models have been proposed (11, 54, 55). Thus far, 12 proteins have been shown to be associated with the IMV membrane (4, 27, 56). Molecular genetic analysis has revealed that at least three of these, encoded by the D13L, A14L, and A17L ORFs, and a serine/threonine kinase encoded by the F10L ORF have an early role in the process of viral assembly (41, 59, 64, 71). The A14L and A17L ORFs encode membrane proteins that form a complex with the product of the A27L ORF (42) and are substrates for the F10L kinase (3, 16). Although phosphorylation and proteolytic processing are important for morphogenesis, their roles in the mechanism of crescent membrane formation are still not understood.
An understanding of viral morphogenesis depends on the identification of the critical proteins involved in the process. Preliminary observations in our laboratory (J. Granek, E. W. Wolffe, and B. Moss, unpublished data) had suggested that a protein encoded by the A9L ORF might be involved in an early stage of vaccinia virus morphogenesis. Here we describe the initial characterization of the A9L protein and show that it is a component of the IMV membrane and is required for virion morphogenesis.
MATERIALS AND METHODS
Cells and viruses.
Cells were maintained, and vaccinia virus strain WR (ATCC Vr119) and recombinant vaccinia viruses vT7lacOI (65) and vA9L-HA were propagated in HeLa cells, as previously described (18). For some experiments, virus particles were purified by sedimentation through a 36% sucrose cushion and banding once on a 25 to 40% sucrose gradient as previously described (18).
Antibodies.
Rabbit antiserum to the A9L protein was generated using the maltose binding protein (MBP) fusion protein system (New England Biolabs, Beverly, Mass.). DNA encoding the predicted C-terminal 40-amino-acid hydrophilic domain of the A9L protein was cloned into the fusion vector, pMAL-c, so as to be in frame with the MBP sequence. Oligonucleotide primers CF-76 (GGGGATATCTCTGAACAAGATGACAAG [EcoRV site underlined]) and CF-78 (GGGAAGCTTTTACTTAGGACTGGAGT [with the HindIII site underlined) were used to generate, by PCR, a fragment containing the A9L ORF. This PCR product was digested with EcoRV and HindIII and inserted into pMAL-c, generating pMAL-c/A9L. The plasmid was sequenced to ensure that no mistakes were introduced during the PCR. Escherichia coli TB1 competent cells (New England Biolabs) were transformed with pMAL-c/A9L, and synthesis of the recombinant MBP fusion protein was induced with isopropyl-β-d-thiogalactopyranoside (IPTG). The fusion protein was affinity purified using amylose resin according to the manufacturer's instructions and injected into New Zealand White rabbits.
The monoclonal antibody (MAb) MHA.11 and the polyclonal antibody PHA.11 (BabCo/Covance, Berkeley, Calif.), both of which recognize the 9-amino-acid influenza hemagglutinin (HA) peptide (YPYDVPDYA), were used in accordance with the manufacturer's instructions. MAb C3 against the vaccinia A27L protein was the kind gift of Mariano Esteban (43).
Generation of recombinant viruses.
To construct vA9L-HA, standard overlap PCR methodology was used to make a DNA fragment containing (i) the coding sequence of the A9L gene with a C-terminal HA tag, (ii) the gus gene under the control of the p7.5 promoter, and (iii) flanking sequences of the A10L and A8R ORFs to allow for subsequent homologous recombination into the viral genome. To generate the recombinant virus, BS-C-1 cells were infected with vaccinia virus WR and transfected with 5 μg of the PCR product. After 48 h, cells were harvested and diluted lysates were used to infect fresh BS-C-1 cell monolayers. The infected cells were overlaid with agar, incubated for 2 days, and then overlaid with a second layer of agar containing 200 μg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc; Clontech Laboratories, Palo Alto, Calif.)/ml. After 2 more days, blue plaques containing recombinant viruses expressing gus were picked and used to infect fresh monolayers of BS-C-1 cells. In this way, the recombinant virus vA9L-HA was purified by three successive rounds of plaque isolation.
The recombinant virus vT7lacOI-A9L (referred to below as vA9i) was made in a similar fashion by generating a final PCR fragment of 3,585 bp. This DNA contained (i) the A9L ORF under the control of the bacteriophage T7 promoter with E. coli lacO inserted between the promoter and the coding sequence, (ii) a gus gene under the control of the vaccinia virus P11 promoter between the A10L and A9L ORFs, and (iii) flanking sequences from the A10L and A8R genes. The transfection and plaque purification procedures were similar to those described for vA9L-HA except that 100 μM IPTG was included in the first agar overlay.
DNAs from candidate recombinant viruses were screened by PCR and sequenced by fluorescence dideoxy-termination methods using an Applied Biosystems 310A genetic analyzer to confirm that no errors were introduced into A9L and neighboring genes.
Plaque-purified vA9i and vA9L-HA were amplified in HeLa cells in the presence or absence of 100 μM IPTG, respectively.
RNA purification.
BS-C-1 cells were mock infected or infected with vaccinia virus WR at a multiplicity of 15 PFU/cell and were harvested at various times. Total RNA was purified using an RNAqueous kit (Ambion, Austin, Tex.).
Northern blot analysis.
RNA samples were treated with formaldehyde, and 3.5 μg of total RNA per lane was separated according to size by electrophoresis on a 1% agarose gel in denaturing buffer (Ambion). The RNA was transferred to a BrightStar-Plus membrane (Ambion) using a Transblot electroblotter (Bio-Rad, Hercules, Calif.). The blots were then prehybridized in Zip-Hybridization solution (Ambion), hybridized with a radiolabeled probe, washed, and subjected to autoradiography.
A 350-bp PCR fragment was used as the template to prepare a uniformly labeled complementary A9L RNA probe by in vitro transcription using T7 RNA polymerase (Ambion). A 350-bp PCR fragment containing the entire region of C11R, encoding the early viral growth factor, was used to generate a positive-control DNA probe. A random prime kit (Promega, Madison, Wis.) was used to generate radiolabeled DNA probes using the standard manufacturer's protocol.
RNase protection assay.
PCR was used to synthesize a DNA containing a T7 promoter attached to a segment encompassing 72 bp before and 162 bp after the putative A9L RNA start site. A radiolabeled riboprobe complementary to the coding sequence was made and gel purified using standard protocols. The radiolabeled probe was hybridized in 80% deionized formamide at 45°C overnight with 15 to 25 μg of total RNA collected at various times after infection. Unhybridized probe was digested in a 1:150 dilution of the nuclease mixture containing S1 nuclease, RNase A, and RNase T1. Nuclease-protected products were resolved by electrophoresis on a denaturing 8% polyacrylamide sequencing gel and subjected to autoradiography.
Detergent extraction of purified vA9L-HA virus.
Purified vA9L-HA virus was extracted with 50 mM Tris-HCl buffer (pH 7.4) containing 1% Nonidet P-40 (NP-40) detergent in the presence or absence of 50 mM dithiothreitol (DTT) for 1 h at 4°C. The extract was separated into soluble (S) and insoluble (P) pellet fractions by centrifugation at 12,000 × g for 30 min. These fractions were then diluted in Tricine sample buffer, run on a 10% polyacrylamide Tricine gel (51), and transferred onto a nitrocellulose membrane as previously described.
Phase separation of A9L from vA9L-HA-infected cells.
For phase separation studies, BS-C-1 cells were infected with vaccinia virus WR or vA9L-HA at a multiplicity of 10. After overnight incubation, cells were harvested and extracted for 30 min at 4°C in buffer containing 1% precondensed Triton X-114 (Calbiochem, San Diego, Calif.). Insoluble material was collected by centrifugation at 12,000 × g, and the supernatant fraction was subjected to phase separation as previously described (8). Proteins in the resulting aqueous and detergent phases were analyzed by electrophoresis on a 10% polyacrylamide Tricine gel, transferred to a membrane, and detected with MAb HA.11.
Immunoprecipitations and Western blot analysis.
Cells were labeled overnight with 100 μCi of [35S]methionine/ml added to methionine-free medium and were then incubated for 20 min in phosphate-buffered saline containing 1% NP-40 and protease inhibitors. Lysates were centrifuged at 30,000 rpm for 60 min in a Beckman 42.2 Ti rotor, incubated with preimmune serum followed by protein A-Sepharose, and then washed.
Radioimmunoprecipitations were carried out as previously described (37) using either MAb HA.11 or anti-MBP-A9L sera. The antigen-antibody complexes bound to protein A-Sepharose beads were resuspended in Tricine sample buffer in the presence of sodium dodecyl sulfate (SDS) and DTT and were boiled for 3 min before application to a 10 to 20% gradient polyacrylamide gel. After electrophoresis, the gels were dried and subjected to autoradiography.
For Western blotting, cell lysates or purified virions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the resolved protein bands were electrophoretically transferred onto nitrocellulose. Membranes were blocked by incubation in 5% nonfat dry milk in phosphate-buffered saline–0.2% Tween and were then incubated with a 1:1,000 dilution of MAb HA.11. After being washed with phosphate-buffered saline–Tween, the nitrocellulose membrane was incubated with horseradish peroxide-conjugated anti-mouse immunoglobulin G (IgG). Bound IgG was visualized using the Supersignal chemiluminescent substrate (Pierce, Rockford, Ill.).
Analysis of [35S]methionine-labeled polypeptides by SDS-PAGE.
For pulse-labeling, infected BS-C-1 cells were incubated in methionine-free medium for 15 min and then labeled for 30 min in methionine-free medium containing 5% dialyzed fetal bovine serum and 100 μCi of [35S]methionine (Dupont, NEN, Boston, Mass.)/ml. For pulse-chase experiments, the labeling medium was removed at 12 h and then replaced with medium containing an excess of unlabeled methionine for a further 12 h.
Electron microscopy.
BS-C-1 cells were grown in 60-mm-diameter dishes and infected at a multiplicity of 10. After 24 h, the cells were prepared for transmission electron microscopy as previously described (10), except that samples were postfixed in reduced osmium. For immunoelectron microscopy, cells were fixed and prepared for freezing as previously described (71). Ultrathin sections were cut using a Leica/Reichert Ultracut FSC and stained on Formvar-coated microscope grids using standard protocols. The primary antibody used was MAb HA.11. Grids were subsequently incubated with a secondary antibody, rabbit IgG fraction against whole mouse IgG (Cappel-ICN Pharmaceuticals, Aurora, Ohio), followed by protein A conjugated to 10-nm-diameter colloidal gold (Department of Cell Biology, Utrecht University School of Medicine, Utrecht, The Netherlands). Purified virus was adsorbed onto Formvar-coated copper mesh grids and incubated with the polyclonal anti-HA serum PHA.11 or MAb C3, followed by protein A conjugated to 10-nm-diameter colloidal gold. The virions were stained briefly with uranyl acetate and examined. Stained ultrathin plastic sections and cryosections were viewed using a Philips CM100 transmission electron microscope.
RESULTS
Analysis of the A9L ORF and its potential regulatory sequences.
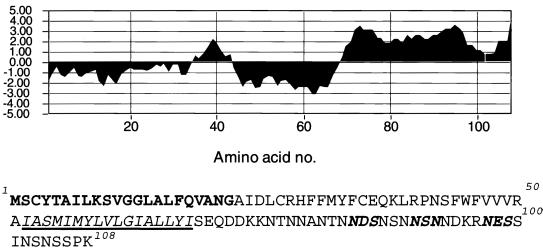
The A9L ORF encodes a protein of 108 amino acids with a predicted molecular mass of 12,108 Da. Analysis of the amino acid sequence of the A9L protein revealed a predicted N-terminal cleavable signal peptide (amino acids 1 to 22) and a central transmembrane domain (amino acids 52 to 68) defined by using Klein's method for transmembrane region allocation (30). In addition, putative N-linked glycosylation sites were encoded at amino acids 84, 89, and 97 (Fig. 1). Two well-characterized essential genes abut the A9L ORF: A10L and A8R encode a late viral core protein (61, 68) and an early protein that functions as a subunit of the intermediate gene transcription factor vITF-3 (50), respectively. The proximity of the neighboring ORFs indicated that regulatory elements of the A9L gene lie within the preceding A10L ORF. Inspection of this region revealed possible early and late A9L promoter consensus sequences (13, 14, 48) within the A10L ORF. The closest early termination signal (TTTTTNT) (45, 73) was 433 nucleotides past the A9L stop codon.
FIG. 1.
Sequence analysis of the A9L protein. A hydrophilicity plot and the corresponding sequence of the A9L ORF are shown. The predicted signal peptide is boldfaced, the transmembrane domain is italicized and underlined, and the potential sites of N-linked glycosylation are indicated by bold italics.
Transcriptional analysis of the A9L gene.
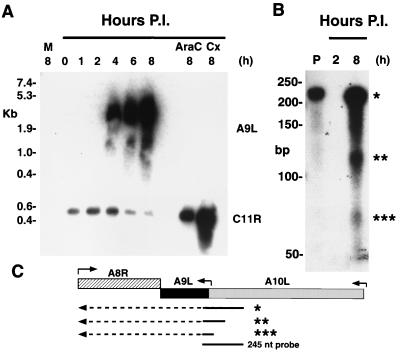
A transcriptional analysis was carried out to determine the time of A9L gene expression and to facilitate genetic engineering at a later stage of this study. BS-C-1 cells were infected with vaccinia virus, and total cellular RNA was isolated between 0 and 8 h after adsorption. The RNA species were separated by electrophoresis according to size, transferred to a nylon membrane, and hybridized to a 32P-labeled RNA probe that was complementary to the A9L coding sequence. The probe hybridized to RNAs of approximately 1,000 to 6,000 nucleotides that were collected at 4, 6, and 8 h after infection. During this period, the signal increased in intensity (Fig. 2A). Both the timing and the broadness of the bands were characteristics of late RNAs. Furthermore, no signal was detected in samples from the 0-, 1-, and 2-h time points, mock-infected cells, or cells in which cycloheximide or cytosine arabinoside (araC) had been used to accumulate early mRNAs and prevent late transcription (Fig. 2A). We tested the integrity of the RNA collected at early time points and in the presence of drugs by stripping the blot and reprobing it with a 32P-labeled single-stranded DNA specific for the C11R gene. C11R encodes the protein VGF, which is synthesized at early times during infection (7). In contrast to the results obtained with A9L, the C11R probe hybridized to a discrete message of 550 nucleotides that was most abundant between 2 and 4 h after infection (Fig. 2A). Furthermore, an intensified signal of the same size was seen in both the araC- and cycloheximide-treated samples (Fig. 2A). Thus, we concluded that the A9L gene was transcribed exclusively at late times.
FIG. 2.
Transcriptional analysis of the A9L gene. (A) Northern blot. BS-C-1 cells were infected with vaccinia virus and harvested after 1 to 8 h. Additional cells were harvested after 8 h of infection in the presence of araC or cycloheximide (Cx). RNAs were resolved by electrophoresis on a denaturing agarose gel, transferred to a nylon membrane, and hybridized with RNA (upper blot) or DNA (lower blot) probes specific for the A9L and C11R ORFs, respectively. The sizes of RNA markers are shown on the left. Abbreviations: M, mock-infected cells; P.I., postinfection. (B) RNase protection assay of RNAs from infected cells harvested at 2 and 8 h after infection. RNAs were incubated with a uniformly 32P-labeled RNA probe complementary to a sequence overlapping the 5′ end of the A9L ORF and the 3′ end of the A10L ORF. After digestion with a mixture of RNases, the material was resolved on a polyacrylamide gel and autoradiographed. Sample P is the full-length undigested probe. Asterisks indicate protected products. (C) Schematic showing the probe and RNA species that could generate the protected products. Arrows above the ORFs indicate the approximate locations of promoters and the direction of transcription. Arrows below the ORFs represent RNAs, with the solid parts protecting the probe and the dashed parts extending beyond. Asterisks correspond to the bands in panel B.
To determine the RNA start site more precisely and to ensure that the transcripts did not consist solely of read-through RNAs from neighboring genes, we carried out RNase protection assays. A 245-nucleotide uniformly labeled complementary RNA probe that overlapped the start sites of the putative promoter sequences of A9L was hybridized to total cellular RNA from cells infected with vaccinia virus for either 2 or 8 h. After RNase treatment of the hybridized 8-h sample, three resistant RNA species were resolved by electrophoresis and visualized by autoradiography (Fig. 2B). The largest RNA represents the protected full-length probe, and it could be derived from transcripts starting from the upstream A10L gene. The band of 72 nucleotides corresponded to the size expected for a transcript from the predicted TAAAT RNA start site immediately preceding the A9L gene. We also detected an additional species of 120 nucleotides that could arise by hybridization with mRNA initiating within a second TAAAT 40 bp upstream of the first. Translation of this message, however, would yield only a 12-amino-acid peptide. The relatively low intensity of the 72-nucleotide band might reflect competition from the larger RNAs for hybridization to the probe. No protection of the probe occurred when the RNA was collected at 2 h after infection, confirming our previous finding that there is no early A9L message.
Addition of an HA tag to the A9L ORF.
Antisera to peptides derived from the predicted amino acid sequence of the A9L ORF and to an MBP-A9L fusion protein reacted with multiple bands on Western blots, making it difficult to identify the A9L species. Therefore, we constructed a recombinant vaccinia virus, vA9L-HA, in which 9 codons from the influenza virus HA protein were inserted just before the stop codon of the A9L ORF. The chimeric gene remained under the control of the native A9L late promoter. The recombinant vA9L-HA virus was readily isolated, grew with normal kinetics, and produced standard-size plaques (data not shown), suggesting that the addition of the tag onto the C terminus of the A9L protein had no deleterious effect.
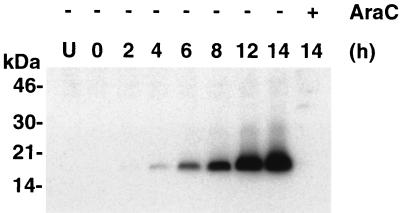
The HA tag was used to determine the time of synthesis of the A9L protein. Cells were infected with vA9L-HA in the presence or absence of araC and harvested at the times indicated. Lysates were subjected to SDS-PAGE and analyzed by Western blotting using MAb HA.11 against the HA tag (Fig. 3). The tagged A9L protein had an estimated molecular mass of approximately 18 kDa, slightly larger than predicted. The A9L protein was detected at 4 h postinfection and increased in intensity throughout the 14-h time course. These kinetics, as well as failure to detect the HA-tagged protein in the presence of araC, were in agreement with our transcriptional analysis.
FIG. 3.
Late expression of the HA-tagged A9L protein. Uninfected cells or cells infected with vA9L-HA in the presence (+) or absence (−) of araC were harvested between 0 and 14 h. Proteins in total-cell extracts were resolved by SDS-PAGE and detected by Western blotting with a MAb to the HA tag (MAb HA.11). The positions of migration and molecular masses (in kilodaltons) of marker proteins are indicated on the left.
Membrane association of the A9L protein.
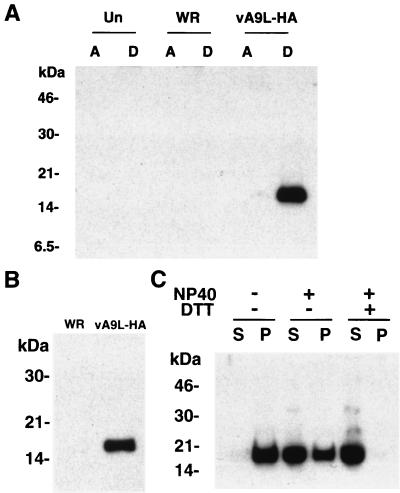
To test whether the hydrophobicity of the A9L protein was sufficient for membrane association, we extracted vA9L-HA-infected cells with Triton X-114 and subjected the clarified lysates to phase separation as described by Bordier (8). After phase separation, the proteins of the aqueous and detergent phases were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with MAb HA.11. The A9L-HA protein from vA9L-HA-infected cells partitioned entirely into the detergent phase, as shown in Fig. 4A, consistent with membrane association.
FIG. 4.
Hydrophobicity and virion association of the A9L protein. (A) Uninfected BS-C-1 cells or cells infected with either vaccinia virus WR or vA9L-HA were harvested 24 h after infection. Triton X-114 extracts were prepared and subjected to phase separation. The A9L protein in the aqueous (A) and detergent (D) phases was analyzed by SDS-PAGE and Western blotting using MAb HA.11. (B) Sucrose gradient-purified vaccinia virus WR and vA9L-HA virions were solubilized directly in Laemmli gel-loading buffer and separated by SDS-PAGE on a 10% polyacrylamide gel. Proteins were transferred to nitrocellulose and subjected to Western blot analysis using MAb HA.11. (C) Purified vA9L-HA virions were incubated at 4°C for 1 h in 50 mM Tris-HCl buffer (pH 7.4) or in the same buffer containing either 1% NP-40 or 1% NP-40 with 50 mM DTT. Soluble material (S) and insoluble material (P) were collected by centrifugation and mixed with Laemmli sample buffer containing DTT, and the proteins were separated by electrophoresis on an SDS–16% polyacrylamide gel. Separated proteins were transferred to nitrocellulose and detected using MAb HA.11. The positions of migration and molecular masses (in kilodaltons) of marker proteins are indicated on the left.
Next we wanted to determine whether the A9L protein was associated with sucrose gradient-purified virions. SDS-PAGE and Western blot analysis using MAb HA.11 revealed a band of the expected molecular mass from vA9L-HA virions but not from control vaccinia virus WR (Fig. 4B). To ensure that similar amounts of viral protein were loaded and that the virus preparations were not contaminated with membranes, the same amounts were probed with an antiserum to the mature N terminus of the A17L protein. A single major band of the expected molecular mass for the processed virion-associated form of A17L was of equal intensity in WR and vA9L-HA virions (data not shown). In addition, samples of purified virus were examined by electron microscopy and were found to be largely free of contaminating membranes.
Typically, membrane-associated proteins can be released from virions with a non-ionic detergent, although in some cases DTT is required. When purified vA9L-HA virions were treated with NP-40 in the absence of DTT and centrifuged, the A9L HA protein was largely in the soluble fraction, but some remained insoluble. Inclusion of 50 mM DTT resulted in nearly complete recovery of the protein in the supernatant fraction (Fig. 4C). In the absence of detergent, the protein remained associated with the particles.
Localization of the A9L protein within the virion.
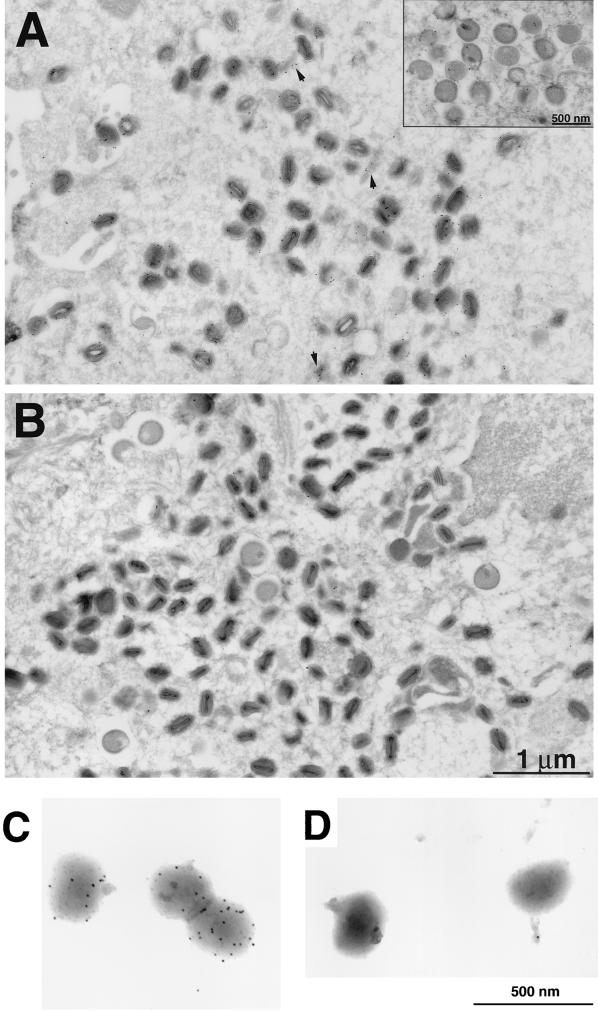
Indirect immunofluorescence examination of cells infected with vA9L-HA revealed that the HA antibodies localized predominantly within the cytoplasmic factory areas delineated by Hoechst staining of viral DNA, which are the sites of assembly of immature viral particles (data not shown). Specific antibody binding was not observed in uninfected cells. To determine which viral forms were stained with the antibody, we carried out immunoelectron microscopy on ultrathin sections of cells infected with either vA9L-HA or the control virus vT7lacOI. Relatively few gold grains were found in cells infected with the control virus (Fig. 5B). Both immature (Fig. 5A inset) and mature vA9L-HA particles (Fig. 5A) were decorated with gold, although grains appeared to be more numerous on mature forms. There was relatively little labeling of the cytoplasmic matrix, but some gold grains were associated with unidentified viral or cellular structures (Fig. 5A). Many of the gold grains overlay the IMV membrane, though we could not determine whether the HA tag was on the inner or the outer surface. To answer the latter question, freshly purified and unfrozen preparations of vA9L-HA or control vaccinia virus virions were analyzed. Purified vA9L-HA virions that appeared structurally intact were decorated with gold on their surfaces (Fig. 5C), whereas the non-HA-tagged virions were not (Fig. 5D), demonstrating that the C-terminally oriented tag was accessible to antibody. A control antibody, MAb C3, which reacts with the 14-kDa fusion protein encoded by the A27L ORF, was also used. With this antibody, heavy staining was observed on the majority of particles in each preparation, suggesting that the virions were intact and the preparations were largely composed of IMV (data not shown). These data indicated that the A9L protein is oriented with the C terminus on the surface of the virion.
FIG. 5.
Localization of the A9L-HA protein by immunoelectron microscopy. BS-C-1 cells were infected with either vA9L-HA (A) or vT7lacOI (B) for 22 h, fixed in paraformaldehyde, cryosectioned, and incubated with MAb HA.11 followed by rabbit anti-mouse IgG and then protein A conjugated to 10-nm-diameter colloidal gold. Electron micrographs of these samples are shown with a 1-μm and a 500-nm marker (inset). Arrowheads point to unidentified structures with associated gold grains. Grids containing purified intact vA9L-HA (C) or WR (D) virions were stained as in panels B and C.
Generation of a conditional-lethal recombinant vaccinia virus expressing an inducible copy of the A9L ORF.
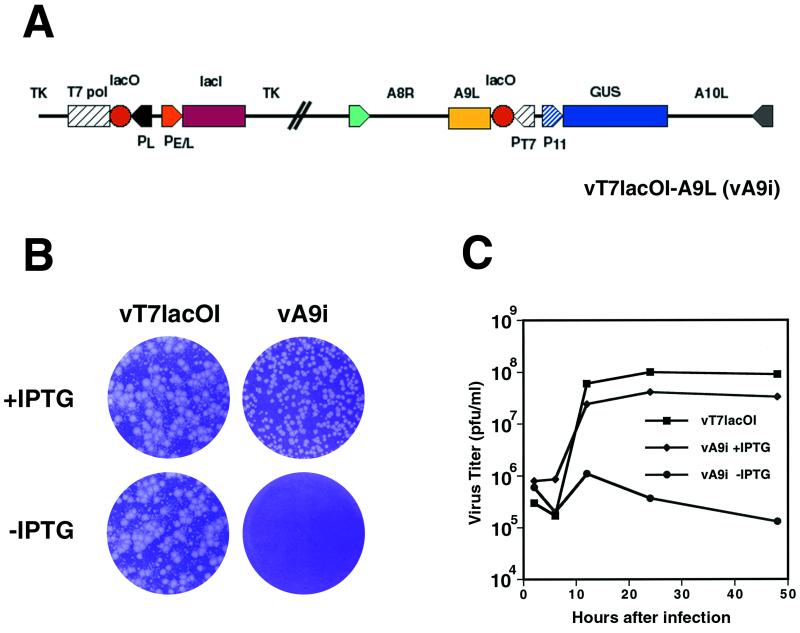
To study the role of the A9L protein in the virus life cycle, we made a recombinant vaccinia virus in which the native A9L gene was regulated by an inducible T7 promoter using a simplified scheme devised by T. Senkevich (T. Senkevich and B. Moss, in unpublished data). The recombinant virus, vA9i, depicted in Fig. 6A, was isolated in the presence of inducer (100 μM IPTG) and identified by Gus expression as described in Materials and Methods. After three rounds of plaque purification, the viral DNA was sequenced to ensure that no PCR errors had been introduced into the A9L, A8R, or A10L gene. The parental virus formed plaques in the presence or absence of IPTG, whereas vA9i required IPTG for plaque formation (Fig. 6B). The plaques formed by vA9i appeared slightly smaller than those formed by the parental virus. This phenomenon was not due to overexpression of A9L, as plaque sizes increased from 20 to 100 μM and then remained constant from 200 to 500 μM (data not shown). Virus yields were determined under one-step growth conditions. In the presence of inducer, the yields and kinetics of vA9Li were similar to those of the parental virus, vT7lacOI (Fig. 6C). Importantly, vA9i did not replicate in the absence of IPTG.
FIG. 6.
(A) Diagram of the vA9i genome. The expression of the A9L ORF is under the control of the T7 promoter and is IPTG inducible. Abbreviations: PT7, T7 promoter; P11, vaccinia virus late promoter; PE/L, vaccinia early/late promoter; TK, thymidine kinase locus; T7 pol, T7 polymerase ORF; GUS, β-glucuronidase ORF; lacI, E. coli lac repressor ORF; lacO, E. coli lac operator element. (B) Effect of IPTG on formation of vA9i plaques. Monolayers of BS-C-1 cells were infected with either vT7lacOI or vA9i in the presence or absence of 100 μM IPTG as indicated. At 48 h after infection, plaques were visualized by staining with crystal violet. (C) Effect of IPTG on virus yields over time. BS-C-1 cells were infected with vaccinia virus T7LacOI or vA9i at a multiplicity of 5 in the presence or absence of 100 μM IPTG and were harvested at 6, 12, 24, and 48 h after infection. Virus titers for each sample were determined by plaque assay on BS-C-1 cells in the presence of IPTG.
Repression of A9L expression.
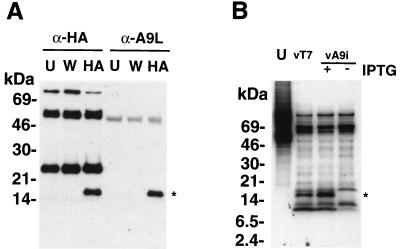
Because the inducible A9L protein did not contain a HA tag, a polyclonal antiserum to an MBP-A9L fusion protein (anti-MBP-A9L) was used to determine the efficiency of repression. However, this antibody did not work in Western blots, and multiple bands were detected by radioimmunoprecipitation. To prove that this antibody could recognize the A9L protein, cells were infected with vA9L-HA, extracts were incubated either with the anti-HA MAb HA.11 or with rabbit anti-MBP-A9L, and the bound proteins were analyzed by Western blotting with MAb HA.11. As seen in Fig. 7A, the MBP-A9L antiserum and MAb HA.11 were equally efficient at immunoprecipitating the 18-kDa HA-tagged vA9L-HA protein. Additional bands of approximately 50 and 25 kDa represent the heavy and light chains of mouse IgG, respectively.
FIG. 7.
Production of the A9L protein in vA9i-infected cells is dependent on inducer. (A) To test the specificity of the anti-A9L antiserum, uninfected BS-C-1 cells (U) or cells infected for 24 h with vaccinia virus WR (W) or vA9L-HA (HA) were lysed in radioimmunoprecipitation assay buffer. Lysates were incubated with either MAb HA.11 (α-HA) or anti-MBP-A9L (α-A9L) followed by protein A-Sepharose. The immune complexes were collected by centrifugation and washed, and the proteins were resolved by SDS-PAGE. The proteins were transferred to nitrocellulose membranes and probed with MAb HA.11. (B) BS-C-1 cells were infected with vaccinia virus vT7LacOI (vT7) or vA9i in the presence or absence of 100 μM IPTG at a multiplicity of 10. After 6 h, infected and uninfected (U) cells were labeled for 18 h with [35S]methionine. Lysates of labeled cells were incubated with anti-MBP-A9L followed by protein A-Sepharose, and immunoprecipitated proteins were separated by SDS-PAGE and visualized by autoradiography. The positions of migration and molecular masses (in kilodaltons) of marker proteins are indicated on the left. The position of migration of the A9L protein is indicated by an asterisk.
Having proved that the MBP-A9L antiserum recognized the A9L protein, we used it to analyze extracts of [35S]methionine-labeled mock-infected cells, cells infected with vT7lacOI, and cells infected with vA9i in the presence or absence of IPTG. SDS-PAGE and autoradiography revealed the presence of a band of approximately 17 kDa, the size expected for the A9L protein without the HA tag, in cells infected with vA9i in the presence but not in the absence of inducer (Fig. 7B). A band of 8 kDa was present in reduced amounts in the absence of inducer, but its relationship to A9L is unclear at this time.
Effect of A9L repression on the synthesis and processing of late viral proteins.
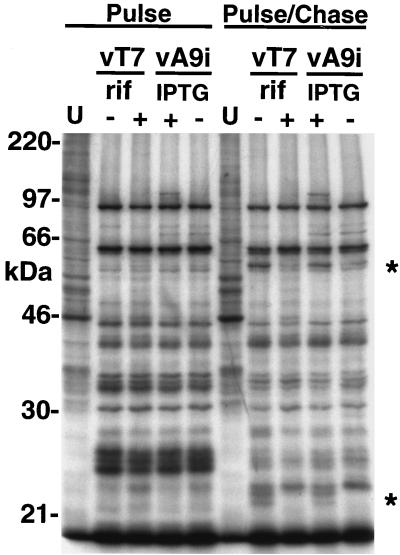
Next, we determined whether repression of A9L affected the synthesis or processing of viral late proteins. Cells were left uninfected or were infected either with vT7lacOI in the presence or absence of rifampin or with vA9i in the presence or absence of IPTG. After 12 h, cells were labeled with [35S]methionine for 1 h. These cells were lysed directly into sample buffer and analyzed by SDS-PAGE (Fig. 8). A duplicate set of cells was subsequently incubated with an excess of cold methionine for a further 12 h and treated in the same way (Fig. 8). An analysis of the autoradiogram of the pulse-labeled cells revealed a similar pattern of late proteins under all conditions. After the chase, however, rifampin and repression of A9L synthesis produced similar defects in the proteolytic processing of certain late structural proteins (Fig. 8), suggesting a block at an early stage of viral assembly.
FIG. 8.
Late protein synthesis was normal in cells infected with vA9i, but maturational processing was inhibited. Uninfected BS-C-1 cells (U) or cells infected either with vT7lacOI (vT7) in the presence (+) or absence (−) of rifampin (rif; 100 μg/ml) or with vA9Li in the presence (+) or absence (−) of 100 μM IPTG were labeled for 1 h with [35S]methionine at 12 h after infection. Cells were either harvested immediately into sample buffer and analyzed by SDS-PAGE (Pulse) or incubated in medium with excess cold methionine for 12 h, harvested, and then analyzed (Pulse/Chase). Asterisks mark positions of processed forms of proteins. The masses of marker proteins (in kilodaltons) are shown on the left.
Repression of vA9i blocks maturation of IV.
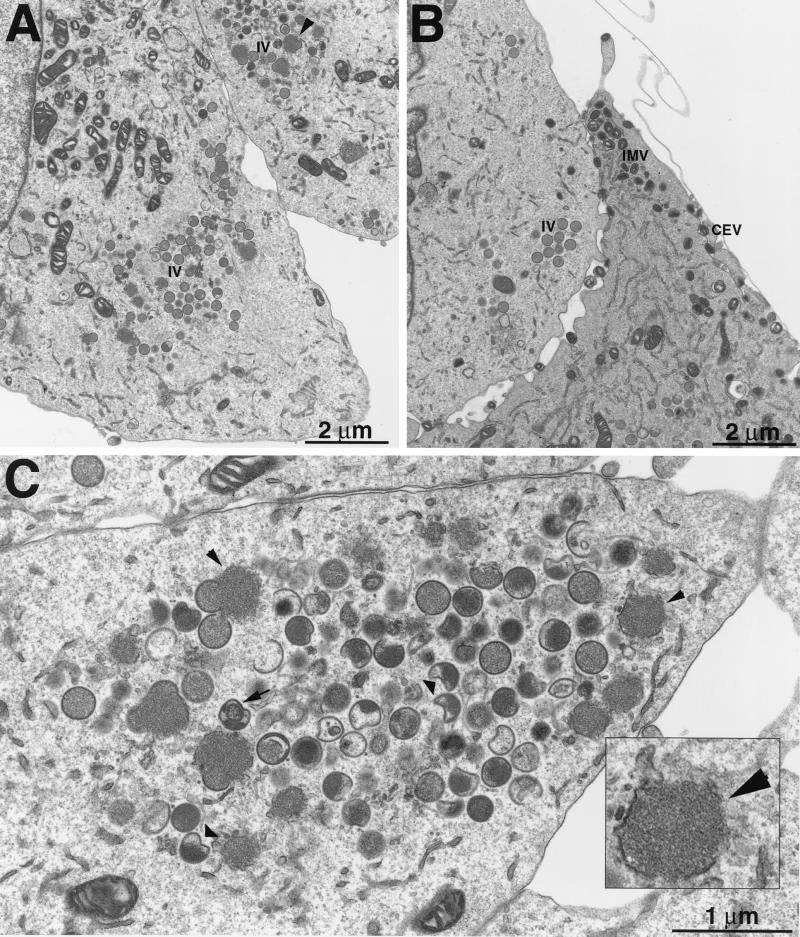
Transmission electron microscopy of sections of infected cells was used to determine the stage at which virus replication was blocked in the absence of A9L expression. Cells infected with vA9i in the presence of IPTG showed the expected range of virus structures (Fig. 9B), which were indistinguishable from those of the parental virus (data not shown). Such cells contained crescent membranes, IV with and without nucleoids, IMV, IEV, and extracellular forms. In contrast, we did not observe any mature virions in cells infected with vA9i in the absence of inducer. Instead, viroplasm with loosely associated amorphous membrane structures reminiscent of small rifampin bodies (Fig. 9C inset), crescents, IV occasionally with nucleoid bodies, and many aberrant IV structures were observed (Fig. 9A and C).
FIG. 9.
Only immature and abnormal viral particles were made when A9L was repressed. BS-C-1 cells were infected with vA9i in the presence (B) or absence (A and C) of IPTG and were prepared for electron microscopy. All viral structures were seen in the presence of IPTG (B), whereas in the absence of inducer only electron-dense viroplasm with associated membranes and immature forms developed (A). With some enlargement (C), it can be seen that many of the IV are abnormal. (Inset) Higher-magnification view of electron-dense viroplasm with associated membranes.
DISCUSSION
Preliminary studies using a screen designed to identify proteins involved in early morphogenesis (Granek, Wolffe, and Moss, unpublished) suggested that the product of the previously uncharacterized A9L gene was likely to have such a function. In this report, we show that the A9L gene is expressed late in infection and that the protein is a component of the virion membrane, is essential for virus replication, and has a role in morphogenesis.
Our initial inspection of the A9L gene sequence suggested that it might have early and late promoters. However, both Northern blotting and RNase protection assays revealed only late transcripts, even under conditions that enhanced early RNA synthesis. A late mRNA start site was mapped at the predicted TAAAT motif preceding the A9L ORF. Another RNA start was mapped at a second TAAAT site approximately 40 bp upstream, but such an mRNA would be predicted to encode only a short peptide. The inactive putative early promoter consensus lies in a highly conserved region of the A10L coding sequence, suggesting that the similarity to an early promoter may be related to A10L codon preferences.
Analysis of A9L protein synthesis was facilitated by the addition of a C-terminal epitope tag. Since the promoter sequences were untouched and the A9L gene remained in its original location, we are confident that it was expressed normally. From a practical point of view, the existing MAb and monospecific polyclonal antibodies to the epitope tag were far superior to the antisera that were generated to A9L peptides and a fusion protein. Examination of the predicted protein sequence suggested that the protein was sufficiently hydrophobic to be membrane associated. Using the HA-tagged version of A9L, we were able to show by electron microscopy of infected cells that the protein was associated with membranes of immature and mature virions. In addition, the HA antibody labeled the surface of apparently intact purified virions, indicating that the A9L protein was anchored with its hydrophilic C-terminal portion toward the cytoplasm. However, incubation of the purified virions with 1% NP-40 and DTT enhanced the antibody reactivity, suggesting that this treatment increased the availability of the epitope (data not shown). This was surprising, since most of the protein can be separated from the virions under similar conditions.
To investigate the role of the A9L protein, we constructed a conditional-lethal inducible mutant. The stringency of the original T7 inducible system (62) was enhanced by omitting the encephalomyocarditis virus leader sequence to prevent internal translation initiation while maintaining lacO to repress the vaccinia virus promoter regulating the T7 RNA polymerase and the T7 promoter regulating A9L. Replication of the mutant virus was dependent on IPTG and was blocked at the immature-virus stage of assembly in the absence of inducer. While apparently normal crescents and IV are formed, condensation of the internal matrix to form the viral core does not proceed as usual in the absence of the A9L protein. The matrix appears to be incompletely associated with the membrane, leading to the production of defective particles. Possibly the A9L protein is responsible for the association of matrix or core components with the IV membrane.
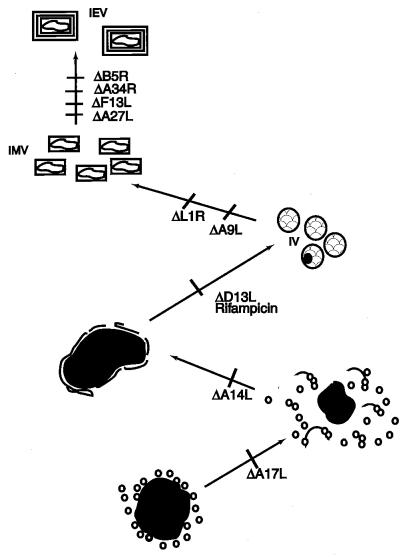
The A9L-sensitive stage relative to mutations of other membrane proteins or to drugs that have an effect on morphogenesis is illustrated in Fig. 10. The earliest defect in morphogenesis occurs with a temperature-sensitive F10L kinase mutant (59, 64); under nonpermissive conditions, late viral protein synthesis occurred but no recognizable viral membranes were detected. A similar phenotype was also described for a temperature-sensitive mutant of the H5R protein generated by clustered charge-to-alanine mutagenesis of the H5R ORF (15). When two F10L kinase substrates encoded by the A17L and A14L ORFs were repressed, electron-dense viroplasm and small vesicles accumulated (41, 44, 60, 71), as depicted in Fig. 10. However, A14L and A17L mutant viruses have slightly different phenotypes. When the A14L protein was not expressed, the vesicles accumulated at a distance from the viroplasm, and some empty crescents were formed. In cells infected with the A17L deletion virus, no crescent structures were observed, but the vesicles appeared to be more closely associated with the viroplasm. Studies with the drug rifampin led to the identification of a protein involved in crescent formation. Rifampin inhibits vaccinia virus morphogenesis at a precrescent stage without significantly affecting either protein or DNA synthesis (21, 34). A protein, p65, encoded by the D13L ORF was mapped by characterizing rifampin-resistant mutants (2, 57). When expression of this protein was inhibited, the infected cells had the same phenotype as rifampin-treated cells (74). A possible scenario is that the A14L protein is required for the interaction of the nascent viral membranes with a component of the viroplasm, perhaps a core protein such as that encoded by the F18L ORF (60). Once the membranes are associated with the viroplasm, the A17L and A14L proteins are both required for the efficient formation of contiguous membrane sheets. The D13L protein is then needed for the membranes to assume rigid crescent structures. The major block resulting from repression of A9L occurs later than that produced by rifampin or repression of D13L, but before that caused by inhibiting the expression of I1L (31), 17L (28), F18L (75), or L1R (40). Infection of cells with recombinant viruses that do not express the A5L, A8L, A32L, or D6L protein resulted in reduced amounts of mature forms and an accumulation of IV (24, 25), many of which were visibly abnormal (67) or lacked DNA (10).
FIG. 10.
Diagram indicating the effects of deleting or repressing the expression of specific membrane proteins on virion morphogenesis. In each case, the mutant with a deleted or repressed gene is indicated by a Δ next to the ORF; morphogenesis is blocked at the stage before the slash. The stage at which morphogenesis is blocked by rifampin is also indicated.
Analysis of vaccinia virus mutants has led to the identification of proteins required for early stages of morphogenesis. However, the mechanism of membrane recruitment and the protein-protein interactions involved in assembly of the crescent and IV structure are not understood. Determination of the topology, intracellular trafficking, and protein interactions of the A9L protein may help to elucidate early steps in virion morphogenesis.
ACKNOWLEDGMENTS
We thank Norman Cooper for preparing cells, Tania Senkevich for sharing her methods for making conditional-lethal inducible virus mutants, Jerry Sisler for oligonucleotides, and Andrea Weisberg for assistance with electron microscopy.
W.W.Y. was an HHMI-NIH Research Scholar and medical student at the University of Washington School of Medicine.
REFERENCES
- 1.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Baldick C J, Moss B. Resistance of vaccinia virus to rifampicin conferred by a single nucleotide substitution near the predicted NH2 terminus of a gene encoding an Mr 62,000 polypeptide. Virology. 1987;156:138–145. doi: 10.1016/0042-6822(87)90444-2. [DOI] [PubMed] [Google Scholar]
- 3.Betakova T, Wolffe E J, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J Virol. 1999;73:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betakova T, Wolffe E J, Moss B. The vaccinia virus A14.5L gene encodes a hydrophobic 53-amino-acid virion membrane protein that enhances virulence in mice and is conserved among vertebrate poxviruses. J Virol. 2000;74:4085–4092. doi: 10.1128/jvi.74.9.4085-4092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomquist M C L, Hunt L T, Barker W C. Vaccinia virus 19-kilodalton protein: relationship to several mammalian proteins, including two growth factors. Proc Natl Acad Sci USA. 1984;81:7363–7367. doi: 10.1073/pnas.81.23.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 9.Boulter E A, Appleyard G. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog Med Virol. 1973;16:86–108. [PubMed] [Google Scholar]
- 10.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dales S, Mosbach E H. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- 12.Dales S, Siminovitch L. The development of vaccinia virus in Earles L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davison A J, Moss B. The structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 14.Davison A J, Moss B. The structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 15.DeMasi J, Traktman P. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J Virol. 2000;74:2393–2405. doi: 10.1128/jvi.74.5.2393-2405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrien M, Punjabi A, Khanna M, Grubisha O, Traktman P. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 19.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 20.Frischknecht F, Moreau V, Rottger S, Gonfloni S, Reckmann I, Superti-Furga G, Way M. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signaling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 21.Grimley P M, Rosenblum E N, Mims S J, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Carroll L J, Wolffe E J, Moss B. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu X, Wolffe E J, Weisberg A S, Carroll L J, Moss B. Repression of the A8L gene, encoding the early transcription factor 82-kilodalton subunit, inhibits morphogenesis of vaccinia virions. J Virol. 1998;72:104–112. doi: 10.1128/jvi.72.1.104-112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Ashford T, Mann M, Griffiths G, Krijnse Locker J. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kane E M, Shuman S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J Virol. 1993;67:2689–2698. doi: 10.1128/jvi.67.5.2689-2698.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz E, Moss B. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc Natl Acad Sci USA. 1970;6:677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 31.Klemperer N, Ward J, Evans E, Traktman P. The vaccinia virus I1 protein is essential for the assembly of mature virions. J Virol. 1997;71:9285–9294. doi: 10.1128/jvi.71.12.9285-9294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan C, Ellison S, Rose H, Moore D. Structure and development of viruses observed in the electron microscope. II. Vaccinia and fowl pox viruses. J Exp Med. 1954;100:301–310. doi: 10.1084/jem.100.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss B, Rosenblum E N. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J Mol Biol. 1973;81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 34.Moss B, Rosenblum E N, Katz E, Grimley P M. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature. 1969;224:1280–1284. doi: 10.1038/2241280a0. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr 43-50 K protein in the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 36.Payne L. Polypeptide composition of extracellular enveloped vaccinia virus. J Virol. 1978;27:28–37. doi: 10.1128/jvi.27.1.28-37.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne L G. Characterization of vaccinia virus glycoproteins by monoclonal antibody preparations. Virology. 1992;187:251–260. doi: 10.1016/0042-6822(92)90313-e. [DOI] [PubMed] [Google Scholar]
- 38.Payne L G. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J Gen Virol. 1980;31:147–155. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 39.Payne L G, Norrby E. Presence of haemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 40.Ravanello M P, Hruby D E. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virus assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez D, Esteban M, Rodriguez J-R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez D, Rodriguez J-R, Esteban M. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the protein encoded by the vaccinia virus A17L gene. J Virol. 1993;67:3435–3440. doi: 10.1128/jvi.67.6.3435-3440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez J F, Janeczko R, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodriguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohrmann G, Moss B. Transcription of vaccinia virus early genes by a template-dependent soluble extract of purified virions. J Virol. 1985;56:349–355. doi: 10.1128/jvi.56.2.349-355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roper R L, Wolffe E J, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. 1998;72:4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–439. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanderson C M, Frischknecht F, Way M, Hollinshead M, Smith G L. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- 50.Sanz P, Moss M. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc Natl Acad Sci USA. 1999;96:2692–2697. doi: 10.1073/pnas.96.6.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 52.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J Virol. 1994;68:103–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmutz C, Payne L G, Gubser J, Wittek R. A mutation in the gene encoding the vaccinia virus 37,000-Mr protein confers resistance to an inhibitor of virus envelopment and release. J Virol. 1991;65:3435–3442. doi: 10.1128/jvi.65.7.3435-3442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sodeik B, Doms R W, Ericcson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stern W, Dales S. Biogenesis of vaccinia: concerning the origin of the envelope phospholipids. Virology. 1974;62:293–306. doi: 10.1016/0042-6822(74)90393-6. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 57.Tartaglia J, Piccini A, Paoletti E. Vaccinia virus rifampicin-resistance locus specifies a late 63,000 Da gene product. Virology. 1986;150:45–54. doi: 10.1016/0042-6822(86)90264-3. [DOI] [PubMed] [Google Scholar]
- 58.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 59.Traktman P, Caliguri A, Jesty S A, Liu K, Sankar U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Traktman P, Liu K, DeMasi J, Rollins R, Jesty S, Unger B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J Virol. 2000;74:3682–3695. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Meir E, Wittek R. Fine structure of the vaccinia virus gene encoding the precursor of the major core protein 4a. Arch Virol. 1988;102:19–27. doi: 10.1007/BF01315559. [DOI] [PubMed] [Google Scholar]
- 62.VanSlyke J K, Franke C A, Hruby D E. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J Gen Virol. 1991;72:411–416. doi: 10.1099/0022-1317-72-2-411. [DOI] [PubMed] [Google Scholar]
- 63.VanSlyke J K, Whitehead S S, Wilson E M, Hruby D E. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology. 1991;183:467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- 64.Wang S, Shuman S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J Virol. 1995;69:6376–6388. doi: 10.1128/jvi.69.10.6376-6388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitehead S S, Hruby D E. Differential utilization of a conserved motif for the proteolytic maturation of vaccinia virus proteins. Virology. 1994;200:154–161. doi: 10.1006/viro.1994.1174. [DOI] [PubMed] [Google Scholar]
- 67.Williams O, Wolffe E J, Weisberg A S, Merchlinsky M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J Virol. 1999;73:4590–4599. doi: 10.1128/jvi.73.6.4590-4599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wittek R, Richner B, Hiller G. Mapping of the genes coding for the two major vaccinia virus core polypeptides. Nucleic Acids Res. 1984;12:4835–4848. doi: 10.1093/nar/12.12.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolffe E J, Katz E H, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolffe E J, Weisberg A S, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;244:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]
- 73.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Moss B. Inducer-dependent conditional-lethal mutant animal viruses. Proc Natl Acad Sci USA. 1991;88:1511–1515. doi: 10.1073/pnas.88.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]