Abstract
Purified fusion proteins made up of a retroviral integrase and a sequence-specific DNA-binding protein have been tested in in vitro assays for their ability to direct integration into specific target sites. To determine whether these fusion proteins can be incorporated into human immunodeficiency virus type 1 (HIV-1) and are functional to mediate integration, we used an in trans approach to deliver various integrase-LexA proteins to an integrase-defective virus containing an integrase mutation at aspartate residue 64. Integrase-LexA, integrase-LexA DNA-binding domain, or N- or C-terminally truncated integrase-LexA proteins were fused to the HIV-1 accessory protein, Vpr. Coexpression of the Vpr fusion proteins and an integrase-defective HIV-1 molecular clone by a producer cell line resulted in efficient incorporation of the fusion protein into the integrase-mutated virus. In addition, each of these viruses was infectious and capable of performing integration, as determined by two independent cellular assays that measure reporter gene expression. With the exception of the N-terminally truncated integrase fused to LexA, which was at about 1%, all of the fusion proteins restored integration to a similar level, at 17 to 24% of that of the wild-type virus. The low level observed with the N-terminally truncated integrase fused to LexA is consistent with previous results implying that the N terminus of integrase is involved in multiple steps of the retroviral life cycle. These data indicate that the integrase-fusion proteins retain catalytic function in the integrase-mutated viruses and demonstrate the feasibility of incorporating integrase fusion proteins into HIV-1 for the development of site-directed retroviral vectors.
Retroviruses are highly promising vectors for gene therapy and at present are the most widely used in clinical trials (64). A critical advantage they offer is the ability to permanently and precisely insert a gene of interest into the chromosomes of a target cell. The stage of the viral life cycle responsible for this joining of a cDNA copy of the viral genome to the chromosomal DNA is integration, mediated by the viral enzyme integrase (2, 37). Integration is performed in the context of the preintegration complex (PIC), following reverse transcription and nuclear entry of an infected cell (21). The human immunodeficiency virus type 1 (HIV-1) PIC consists of a double-stranded DNA copy of the retroviral genome, the viral proteins integrase, reverse transcriptase, matrix, and Vpr, and at least one host cellular protein, HMG-I(Y) (6, 19, 20, 50).
Integration occurs via a three-step process. In the first step, 3′-end processing, integrase cleaves the terminal 2 nucleotides from each 3′ end of the retroviral DNA, exposing a highly conserved CA dinucleotide (3, 10, 24, 38, 41). Next, in 3′-end joining, integrase uses the -OH group of the newly processed 3′ ends of the viral genome to attack the phosphodiester backbone of the chromosomal DNA in a transesterification reaction (18, 29). In HIV-1, the two viral ends are joined with a spacing of 5 bp in the cellular DNA (13, 52). The final step of integration, 5′-end joining, is most probably carried out by cellular enzymes (11). It involves repair of the gapped structure created by integrase during the 3′-end processing and joining steps and results in a short duplication of the cellular DNA sequence flanking the provirus (3, 12, 33, 48, 61).
Although integration is part of the appeal of retroviruses in gene therapy, it also has a potential pitfall. The sites in the chromosomal DNA into which integration occurs are nonspecific (9, 22, 34, 56, 69). Therefore, insertional mutagenesis may result in the loss of an essential gene or in the inappropriate activation of cellular gene expression due to regulatory elements present in the viral long terminal repeats (LTRs). To develop a retroviral vector with added safety against nonspecific integration, it is desirable to produce a virus that is capable of integrating into the chromosomal DNA at specific sites and to remove sequences in the viral LTRs that may incidentally disregulate neighboring genes. Self-inactivating vectors have already been developed that eliminate regulatory elements present in the U3 region of the viral LTR (51, 76). To further reduce the risk of nonspecific integration during transduction, we are interested in developing a strategy for conferring site specificity to retroviral integrases.
In in vitro assays, using purified proteins and short annealed oligonucleotides that mimic the U5 LTR, integration can be directed toward specific sites in target DNA. Fusion of integrase to a sequence-specific DNA-binding protein, such as the DNA-binding domain (DBD) of phage lambda repressor (7) or the full-length or DBD of Escherichia coli LexA repressor (30, 36), results in a bias of integration into DNA flanking the recognition site of the sequence-specific DNA-binding protein. Although in vitro assays indicate the potential success of such an integrase fusion protein for directing integration, it has not yet been possible to incorporate these fusion proteins into an infectious retroviral vector (8, 36). One difficulty in achieving this is manipulating the 3′ end of the integrase gene so that the fusion protein is encoded in the viral genome. For HIV-1, the 3′ coding region of integrase overlaps the reading frame of vif and contains an important splice acceptor site (59). To avoid interfering with crucial elements in the integrase gene, we decided to incorporate the integrase-LexA fusion protein into HIV-1 in trans. The in trans incorporation method has been previously described and exploits the packaging properties of the viral accessory protein, Vpr. Vpr is incorporated into HIV-1 through an interaction with the LXX triplet repeat in the C terminus of the p6 protein in Gag (40, 46). By fusing a protein to the C terminus of Vpr, it can also be incorporated into HIV-1 particles via this Vpr-p6 interaction (23, 44, 45, 72, 74). Furthermore, fusing wild-type integrase to Vpr results in the delivery of integrase to viruses encoding a defective integrase gene and in provirus formation in infected cells (23, 44, 74). It is unknown, however, whether fusion of a protein to the C terminus of integrase will result in a loss of integration activity during retroviral infection. Vpr must be removed from the N terminus of integrase for it to restore integration to an integrase-mutant HIV-1 clone (23). It is possible that C-terminal fusions to integrase will likewise affect its proper function.
In this study, we showed that a variety of integrase-LexA fusion proteins could be included in HIV-1 particles by the Vpr fusion method. In addition to the full-length wild-type integrase-LexA protein, a number of truncated fusion proteins were incorporated. These include N- and C-terminal deletions of integrase, as well as a C-terminal truncation of LexA. The various integrase-LexA fusion proteins were incorporated into a replication-defective virus, which contains a mutation in the catalytic core of integrase, to reveal whether the fusion protein provided in trans was functional. In the presence of an HIV- 1 protease cleavage site for the removal of Vpr from the integrase-LexA protein after packaging, all resulting viruses containing the various fusion proteins were infectious and able to stably express a selectable marker gene. Our results demonstrate an efficient means for delivering integrase fusion proteins into virions and further reveal that the integrase fusion proteins catalyze integration during retroviral infection.
MATERIALS AND METHODS
Cells and antibodies.
HeLa-CD4 (47) and 293T cells, obtained from the National Institutes of Health (NIH) Reference and Reagent Program and Richard Sutton at the Baylor College of Medicine, respectively, were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 10% fetal bovine serum (Gemini), 100 U of penicillin per ml, and 0.1 mg of streptomycin (Sigma) per ml. HeLa-CD4-LTR–β-gal cells (39), received from the NIH Reference and Reagent Program, were grown in DMEM supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 0.1 mg of streptomycin per ml, 0.1 mg of G418 (Sigma) per ml, and 0.05 mg of hygromycin B (Sigma) per ml. The polyclonal antibody directed against HIV-1 integrase residues 23–34 (31) was provided by the NIH Reference and Reagent Program. Vpr antiserum was donated by Nathaniel Landau of the Salk Research Institute, HIV-1 immune serum was purchased from Scripps, and LexA antiserum was provided by John Little at the University of Arizona.
HIV-1 proviral clones and recombinant expression plasmids.
The packaging construct used for producing virions used in the multinuclear activation of a galactosidase indicator (MAGI) cell assay was an HIV-1 NL4-3-derived clone made by cutting wild-type pNL4-3 with SalI and SpeI to remove the integrase coding region and ligating with the integrase sequence of a similarly cut HIVNLluc-env containing a D64E mutation in integrase, obtained from Irvin S. Y. Chen at the University of California, Los Angeles, Calif. (49).
The plasmids used for generating viruses for the hygromycin resistance assay, which contain the HXB2 strain of HIV-1 with the hygromycin resistance gene in place of envelope and either the wild-type (pHXB-Hygro) or D64V-mutated (pHXB-IN64-Hygro) form of integrase, were generously provided by Andrew Leavitt of the University of California, San Francisco, Calif. (54). The construct for expression of amphotropic murine leukemia virus (MLV) envelope (SV-A-MLVenv) was generously supplied by Mark Muesing at the Aaron Diamond AIDS Research Center (68).
The recombinant expression plasmids for the various integrase-LexA fusions were each cloned into a pLR2P construct containing a Vpr integrase (R-PC-IN) gene, kindly provided by Beatrice Hahn of the University of Alabama, Birmingham, Ala. (23). The pLR2P expression construct contains the HIV-2 LTR and Rev response element at the 5′ and 3′ ends, respectively, of the Vpr-integrase fusion protein gene to drive its expression and ensure that the full-length transcript is exported out of the nucleus for translation (72). An HIV-1 protease cleavage site is also present following Vpr, because Vpr-integrase fusion proteins are inactive at mediating integration and the removal of Vpr from integrase is necessary for integration (23).
To construct the R-PC-IN/LA clone, PCR on the pT7-7 E. coli protein expression construct containing a full-length integrase-LexA gene was performed with primers against integrase at residue 50, to exploit a unique NsiI site, and the 3′ end of lexA (30). All primers were obtained from Operon Technologies Inc. The primer used to amplify the integrase gene at residue 50 was 5′-CCAGTGATGCATGGACAAGTAGACTGTAGT-3′ (HIV5NsiCAS), and the 3′ primer against lexA was 5′CAGTCACTCGAGTTACAGCCAGTCGCCGTT-3′ (LexA3-Xho). Underlined nucleotides indicate the NsiI and XhoI sites, respectively. After amplification, the product was digested with NsiI and XhoI, purified with the Qiagen gel extraction kit, and then ligated into the pLR2P R-PC-IN construct that had previously been cut with NsiI and XhoI. The pLR2P R-PC-IN/LADBD construct was prepared in an identical fashion to the previous construct, except that the LexADBDXho primer, 5′-ATTCTCGAGTTATGGTTCACCGGCAGC-3′, was used as a 3′ primer to amplify the lexA gene from residue 88 (the XhoI site is underlined).
pLR2P R-PC-IN1–234/LA was prepared by amplifying the integrase 1–234-LexA gene from a pT7-7 expression construct (30) with the HIV5NsiCAS and LexA3-Xho primers previously discussed. All further cloning steps were performed as described above. The R-PC-IN50–288/LA gene was cloned into pLR2P by amplifying the full-length integrase-LexA gene from the pT7-7 expression construct with the LexA3-Xho primer and DIN50-ScaI (5′TCAAGTACTAATGCATGGACAAGTA-3′) (the ScaI site is underlined), to begin the integrase coding region at residue 50. The resulting PCR product was digested with ScaI and XhoI and purified with the Qiagen gel extraction kit as above. The digested fragment was then ligated into the pLR2P R-PC-IN construct that was previously digested with XhoI and partially digested with ScaI to remove the entire integrase coding region. The sequences of all clones were verified by Sanger DNA sequencing (60).
pLR2P R-PC-IN1–234/LADBD was prepared by cutting pLR2P R-PC-IN1–234/LA with XhoI and then partially digesting with KpnI to remove the full-length lexA gene. The pLR2P R-PC-IN/LADBD expression construct was then fully digested with XhoI and KpnI, and the fragment containing the lexA DBD was gel purified by Qiagen gel extraction and ligated into the purified pLR2P DNA encoding IN1–234.
Virus preparation.
All viral stocks were prepared by standard calcium phosphate transfection of monolayers of 293T cells with 20 μg of DNA in 75-cm2 flasks (1). Control viruses used in the MAGI cell assay were produced by transfection with 20 μg of pNL4-3 or 20 μg of pNL4-3 harboring the D64E mutation pNL-IND64E. The virus containing the integrase-LexA fusion protein was obtained by transfection at a 5:1 ratio with pLR2P R-PC-IN/LA and the integrase-mutated pNL4-3.
For the wild-type and D64V integrase-mutated virus stocks used in the hygromycin resistance assay, the hygromycin resistance construct with or without the integrase mutation (pHXB-Hygro or pHXB-IN64-Hygro) was cotransfected with the SV-A-MLVenv construct at a 1:1 ratio. The viruses provided with the integrase-LexA fusion proteins in trans were generated by cotransfection of pHIV-IN64-Hygro, SV-A-MLVenv, and the appropriate pLR2P expression construct at a ratio of 1:1:5. The medium containing the virus after transfection was harvested after 48 h and filtered through 0.45-μm-pore-size filters (Corning) via gravity drip. The virus titer was then determined by an enzyme-linked immunosorbent assay against the HIV-1 p24 antigen, and the stocks were aliquoted and stored at −80°C until use.
Western blot analysis.
Viruses were prepared for immunoblot analysis by ultracentrifugation of the filtered transfectant medium at 120,000 × g for 2 h at 4°C. The resulting pellets were resuspended in lysis buffer (62.5 mM Tris-HCl [pH 6.8], 0.2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 10% glycerol) (44, 72), and normalized for p24 content. A 12-ng p24 equivalent of each virus was loaded onto SDS–12% polyacrylamide gels for separation and then transferred to nitrocellulose membranes (pore size, 0.45 μm; Micron Separations Inc.) for probing. Western blot analysis was carried out with an alkaline phosphatase detection kit as specified by the manufacturer (Bio-Rad).
MAGI cell infectivity assay.
HeLa-CD4-LTR–β-gal cells were seeded at 4 × 104 cells per well of 24-well plates. The following day, each of 50-, 5-, and 0.5-ng p24 equivalents of either control virus (wild type or integrase-mutated NL-IND64E) or integrase-LexA-containing virus (NL-IND64E with R-PC-IN/LA) was used to infect each well for 2 h, in duplicate. Approximately 48 h after infection, the cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as previously described (39). Cells containing a blue nucleus were then counted.
Hygromycin resistance infectivity assay.
Viruses collected from double (pHXB-Hygro or pHXB-IN64-Hygro and SV-A-MLVenv) or triple (pHXB-IN64-Hygro, SV-A-MLVenv and a pLR2P expression construct) transfections were used to infect 106 HeLa-CD4 cells in 100-mm plates at concentrations of 50-, 5-, and 0.5-ng equivalents of p24. At 4 h postinfection, the medium containing the virus was removed and nonselective medium, DMEM, was added to the plates. The cells were maintained in nonselective medium for an additional 40 h. The DMEM was then exchanged for a medium containing 200 μg of hygromycin B per ml. Selection was continued for 14 days, and the colonies were then stained with 0.2% crystal violet in 10% phosphate-buffered formalin (pH 7.0) and counted.
RESULTS
Experimental design.
The HIV-1 integrase-LexA fusion protein was examined in this study because it retains integration activity to the same level as wild-type HIV-1 integrase in all functional assays and is capable of biasing integration into the DNA regions flanking the LexA operator in vitro (30).
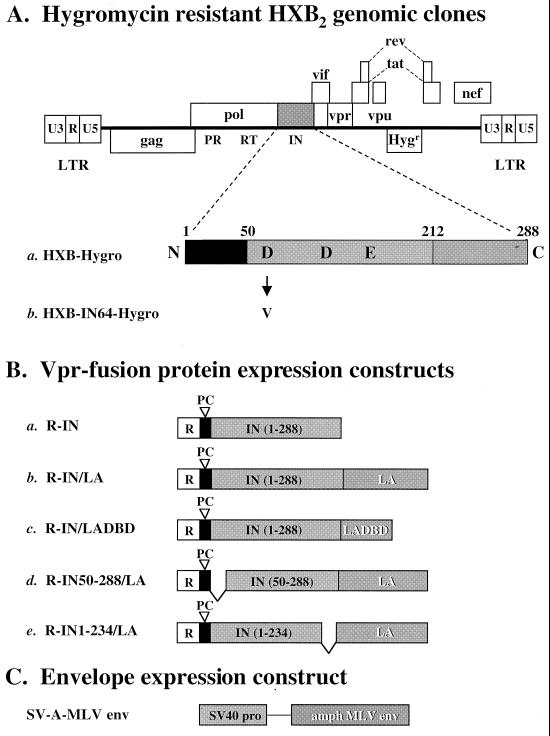
The viral clones used to generate the various virus stocks contain an aspartate-to-valine or aspartate-to-glutamic acid mutation at residue 64 of integrase (D64V [Fig. 1A, clone b]; D64E [Table 1]). These mutated clones were chosen for two reasons. First, the catalytic function of the integrase in the viral genome was removed to determine if the integrase-LexA fusion proteins provided in trans were active and able to restore integration. Second, the D64 integrase mutations result solely in a defect in integration and do not affect other critical stages of the virus life cycle (16, 42, 68).
FIG. 1.
DNA constructs used for in trans incorporation of various integrase-LexA fusion proteins. (A) Representation of the HXB2 clones containing a hygromycin resistance gene (Hygr) in place of envelope (pHXB-Hygro and pHXB-IN64-Hygro). The construct is not drawn to scale. HXB2 contains a defective vpr gene due to an insertion of a T nucleotide at position 5771 (71a). The integrase coding region is magnified. The two viral clones are identical, except that pHXB-Hygro encodes a wild-type integrase gene (a) while pHXB-IN64-Hygro is mutated in one of the catalytic core triad residues of integrase, aspartate 64 to valine, D64V (b). (B) Fusion protein constructs provided in trans to the IND64V viral clone. Each construct encodes the Vpr protein at the N terminus (R, open boxes) for packaging of the integrase and integrase-LexA fusion proteins into the viruses. The solid boxes denote HIV-1 protease cleavage sites (PC), which are required for removal of Vpr from the integrase-LexA fusion protein after packaging (23). The integrase (IN) coding segment is denoted by the lightly shaded boxes. The numbers in parentheses indicate the residues included in each fusion protein (wild-type integrase is 288 amino acids). The full-length LexA (202 amino acids) or DBD of LexA (LADBD, residues 1 to 87) is fused to the C terminus of integrase (constructs b to e) and is denoted by the darkly shaded boxes with white lettering. All constructs are cloned into the pLR2P expression plasmid (72). (C) Amphotropic MLV envelope expression construct. Each virus was pseudotyped with an amphotropic MLV envelope protein driven by the simian virus 40 promoter (68). Viruses were generated by double (pHXB-Hygro or pHXB-IND64-Hygro plus SV-A-MLVenv) or triple (pHXB-IN64-Hygro with SV-A-MLVenv and a pLR2P expression plasmid) transfection and then normalized for p24 content.
TABLE 1.
Production of HXB2 viruses complemented with various Vpr-integrase and Vpr-integrase-LexA derivative proteins
| Virus construct | pLR2P construct | p24 antigen (ng/ml)a |
|---|---|---|
| pHXB-Hygro | None | 670 |
| pHXB-IN64-Hygro | None | 873 |
| pHXB-IN64-Hygro | R-IN | 27 |
| pHXB-IN64-Hygro | R-IN/LA | 81 |
| pHXB-IN64-Hygro | R-IN/LADBD | 207 |
| pHXB-IN64-Hygro | R-IN50-288/LA | 35 |
| pHXB-IN64-Hygro | R-IN1-234/LA | 30 |
HIV-1 p24 antigen concentration of a single representative experiment after harvesting media from calcium phosphate-cotransfected 293T cells. IN, integrase; LA, LexA; R, Vpr.
The constructs used to provide the various integrase and integrase-LexA fusion proteins to the integrase-defective viral clone are shown in Fig. 1B. As a positive control for the in trans incorporation method, a Vpr-integrase (R-IN) expression construct was used (Fig. 1B, construct a). Providing this protein in trans complements viruses containing an integrase gene mutation, restoring their infectivity to about 20% that of wild-type virus (23, 44, 74). The integrase-LexA fusion protein constructs used are also shown in Fig. 1B, (constructs b to e). The fusion protein consisting of full-length HIV-1 integrase fused to LexA serves as the prototype (Fig. 1B, construct b). An integrase-LexA DBD and an N- or C-terminal truncated integrase-LexA fusion protein were also examined (constructs c to e). These were chosen to aid in determining the critical criteria for including an integrase fusion protein in a retroviral vector and to allow us to understand the domain requirements of integrase that are necessary to restore integration to an integrase mutant retroviral clone.
In each of the integrase-LexA fusion protein constructs, LexA was fused to the C-terminus of integrase. This was done because previous data have demonstrated that these fusion proteins are catalytically active in vitro (30) and because fusing proteins to the N terminus of HIV-1 integrase adversely affects their ability to restore infectivity to an integrase core-mutated viral clone (23). These experiments will also delineate whether the same detrimental effect of fusing a protein to the C terminus of integrase is observed.
In trans incorporation of various integrase-LexA fusion proteins into HIV-1.
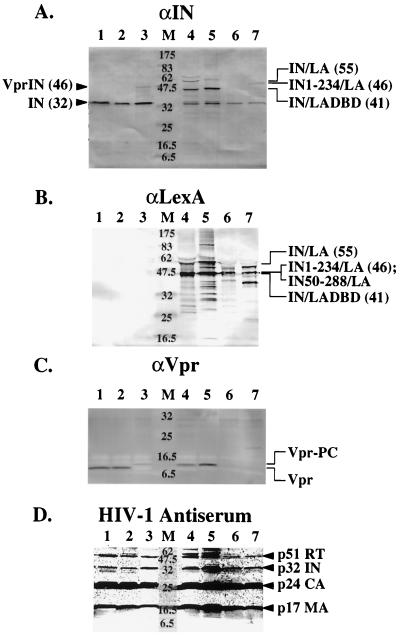
Various integrase-LexA fusion proteins were incorporated into HIV-1 by using the in trans incorporation protocol because it is highly efficient in delivering proteins into HIV-1 and because supplying wild-type integrase in trans to viruses containing a mutant integrase gene restores provirus formation (23, 44, 74). Immunoblot analysis of viruses produced by cotransfection of packaging cells with an HXB2 viral clone, an amphotropic MLV envelope, and the various pLR2P expression constructs was performed to assess the inclusion of each integrase-LexA fusion protein into HIV-1. For each of the viruses generated by the in trans method, the protein encoded by the pLR2P construct was efficiently incorporated (Fig. 2A to C). Antibodies directed against integrase and LexA cross-reacted with proteins of the expected size for each of the Vpr-integrase-LexA fusion proteins (Fig. 2A and B). In addition, the majority of each of the fusion proteins was processed appropriately by HIV-1 protease. The integrase and LexA immunoblots detected proteins about 14 kDa smaller than the unprocessed Vpr-integrase-LexA fusion proteins (Fig. 2A and B). Also, the Vpr antibody cross-reacted with bands running slightly above the size of Vpr in the positive wild-type HXB2 and negative HXB2-IND64V control lanes, corresponding to the size of Vpr-PC (Fig. 2C). None of the full-length Vpr-integrase-LexA fusion proteins were detected, since the sensitivity of the Vpr antibody is poor relative to those of integrase and LexA. Only slight cross-reactivity was found for the N- and C-terminally truncated integrases fused to LexA at the size of the Vpr-PC band. This is consistent with the lower detection levels for these fusion proteins in the integrase and LexA immunoblots (Fig. 2A and B, lanes 6 and 7), possibly indicating lower incorporation levels into these viruses. Similar to observations in other studies, cleavage by either HIV-1 or cellular proteases did occur at additional sites in some of the incorporated fusion proteins (23, 44, 45, 72–74) (Fig. 2A and B).
FIG. 2.
Vpr-integrase-LexA and derivative fusion proteins are incorporated into HIV-1 by the in trans method. Immunoblot analysis was performed with antibodies against integrase residues 23 to 34 (A), LexA (B), Vpr (C), or HIV-1 immune antiserum (D). After concentration for 2 h at 120,000 × g and resuspension in lysis buffer (see Materials and Methods), 12-ng p24 equivalent of each virus was separated by SDS-polyacrylamide gel electrophoresis (12% polyacrylamide). Proteins were then transferred to nitrocellulose membranes (Micron Separations, Inc.), and Western blot analysis was carried out as described in Materials and Methods. Lanes are the same for each immunoblot: 1, wild-type HXB2; 2, HXB-IND64V; 3, HXB-IND64V plus R-IN; 4, HXB-IND64V plus R-IN/LA; 5, HXB-IND64V plus R-IN/LADBD; 6, HXB-IND64V plus R-IN50-288/LA; 7, HXB-IND64V plus R-IN1-234/LA; M, molecular weight standards (in thousands) (New England Biolabs). The Vpr encoded by HXB2 contains an insertion of a T nucleotide at position 5771. The HXB2 Vpr is truncated and unstable, which may account for its low detectability on the immunoblot (71a). Arrowheads denote bands corresponding to the expected size for the indicated proteins, and numbers in parentheses refer to their predicted molecular weights (in thousands).
Inclusion of the various integrase-LexA fusion proteins into HXB2 and the HXB-IND64V mutation did not interfere with viral protein production or with virus maturation following transfection of packaging cells. The titers after the viruses were harvested from transfected 293T cells were diminished for several viruses (Table 1), presumably because the trans-complemented virions result from the transfection of three, rather than two, expression plasmids. However, once normalized by p24 antigen, immunoblotting with HIV-1 immune serum showed the presence of all viral proteins to similar amounts in each virus with respect to the wild-type control (Fig. 2D). Therefore, the Western blot results show that the Vpr-integrase-LexA fusion proteins can be incorporated into HIV-1 by the in trans incorporation method. The proteins are mostly processed correctly between Vpr and integrase, and normal virus protein composition and maturation are maintained.
Integrase-LexA and integrase-LexA DBD fusion proteins are capable of restoring integration to integrase-mutated viruses.
Many recombinant viruses, although efficiently produced after transfection, are not infectious. A MAGI cell assay was initially performed to determine whether the prototype virus, containing the integrase-LexA fusion protein, was capable of provirus formation and expression of a reporter gene (Table 2). Each MAGI cell assay performed with the integrase-LexA-complemented NL-IND64E virus yielded approximately 31% the number of positively stained blue nuclei as did the assay with the wild-type NL4-3 control.
TABLE 2.
MAGI cell infectivity of viruses complemented with Vpr-IN or Vpr-IN/LA
| Virus | pLR2P construct | % Infectivitya |
|---|---|---|
| pNL4-3 | None | 100 |
| pNL-IND64E | None | 2 |
| pNL-IND64E | R-IN | 52 |
| pNL-IND64E | R-IN/LA | 31 |
Data are the average of three independent experiments using 50, 5, and 0.5 ng of p24 equivalents of the indicated virus, performed in duplicate. All values are expressed as a percentage of the wild-type (NL4-3) virus, which produced an average of 65 blue nuclei per ng of p24.
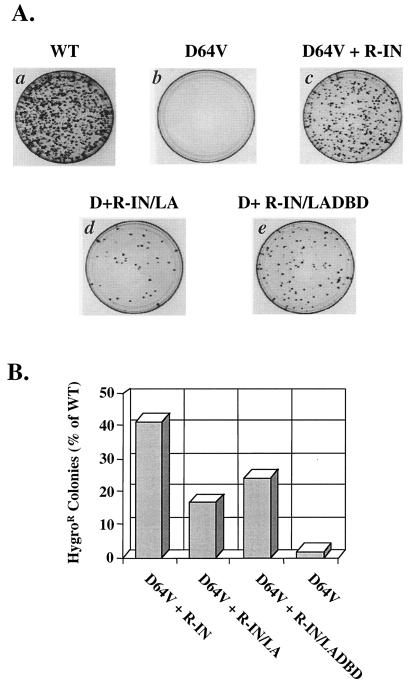
To further examine the ability of each virus containing a fusion protein to mediate integration, we used a hygromycin resistance assay, which utilizes a viral clone that contains the hygromycin resistance gene in place of envelope (Fig. 1). The hygromycin resistance assay offers several advantages over other assays that test for viral infectivity. First, because the cells infected are maintained in the presence of hygromycin B for 2 weeks following infection, cell survival requires stable gene expression from proviruses, producing extremely low background. Viruses that contain mutations in the catalytic core of integrase, such as HXB-IND64V, typically yield infectious viral titers that are 103- to 104-fold lower than those of wild-type viruses (28, 42). In contrast, other infectivity assays, such as the MAGI assay, can detect infectious viral titers from integrase-mutated virions as high as 10 to 20% that of wild-type virus, believed to be driven by Tat expression from two-LTR circles, an unintegrated form of the viral cDNA (16, 68). Second, cells that do not have an integrated provirus are unable to sustain growth under antibiotic selection for this period. Therefore, HeLa-CD4 cells were infected with pseudotyped viruses with or without integrase-LexA fusion proteins and selected with hygromycin B. Under these conditions, both the integrase-LexA- and integrase-LexA DBD-containing viruses were capable of integration. Hygromycin-resistant colonies were consistently found on plates of cells infected with these viruses (Fig. 3A, plates d and e). In contrast, virtually no colonies were found on plates of cells infected with the negative control virus, HXB-IND64V (Fig. 3A, plate b), containing only the catalytically inactive integrase. When the HXB-IND64V-mutated virus that also contained the integrase-LexA protein was used for infection, colonies were produced at 17% of the level of those in cells infected with the wild-type HXB2 clone or 43% of the level of the HXB-IND64V virus provided with the wild-type integrase protein in trans (Fig. 3B). The integrase-LexA DBD fusion protein was better at restoring integration to the HXB-IND64V, producing 24% of the number of colonies produced by wild-type virus or 61% of the number produced by HXB-IND64V provided with the wild-type integrase in trans (Fig. 3B). The presence of the LexA or LexA DBD at the C terminus of integrase did not significantly affect the ability of the in trans proteins to restore infectivity to the catalytically inactive integrase viruses, signifying that sequence-specific DNA-binding proteins can be fused to the C-terminus of integrase for targeting integration.
FIG. 3.
The integrase-LexA and integrase-LexA DBD fusion proteins are able to restore integration activity to an HXB-IND64V viral clone. (A) Representative plates of HeLa-CD4 cells infected with 50-ng p24 equivalent of the indicated virus in the hygromycin resistance assay. One million cells were infected for 4 h with viruses carrying the IND64V mutation and containing either R-IN (c), R-IN/LA (d), or R-IN/LADBD (e) fusion proteins. Viruses carrying a wild-type integrase gene served as a positive control (a), and viruses mutated at the D64 residue of integrase were used as a negative control (b). Dark spots on each plate are colonies that grew after selection with medium plus 200 μg of hygromycin B per ml for 12 days, beginning 2 days postinfection. The colonies are a result of provirus formation and stable expression of the hygromycin resistance gene. (B) Relative integration levels of the HXB-IND64V virus complemented with the R-IN/LA and R-IN/LADBD proteins. The data are the average for three independent experiments, each performed with 50-, 5-, and 0.5-ng p24 equivalents of virus in the hygromycin resistance assay. The average number of colonies counted on plates of cells infected with 50-ng p24 equivalent of the wild-type control virus was approximately 1,650.
Viruses with a catalytically inactive integrase gene are complemented with an N- or C-terminally truncated integrase fused to LexA.
To determine if a full-length integrase is required for the integrase-LexA fusion protein to restore integration to the integrase-mutated viral clone, N- and C-terminally truncated integrase-LexA proteins were incorporated in trans. Although the in vitro activity of these proteins was weak compared to that of the full-length integrase-LexA protein, integration activity directed toward the LexA operator sequence could be detected using a highly sensitive PCR-based assay (30). In addition, these proteins may be able to restore integration to the D64V integrase-mutated viral clone because of functional complementation. In vitro, integrases that contain a mutation in one of the catalytic triad residues are complemented by integrases that lack either the N- or C-terminal domain (15, 63). Also, viruses that contain mutations in the catalytic core of integrase are complemented with N- or C-terminally mutated integrases in trans, restoring infectivity to 1 and 44%, for the N and C terminus, respectively, of the level obtained with wild-type integrase provided in trans (23).
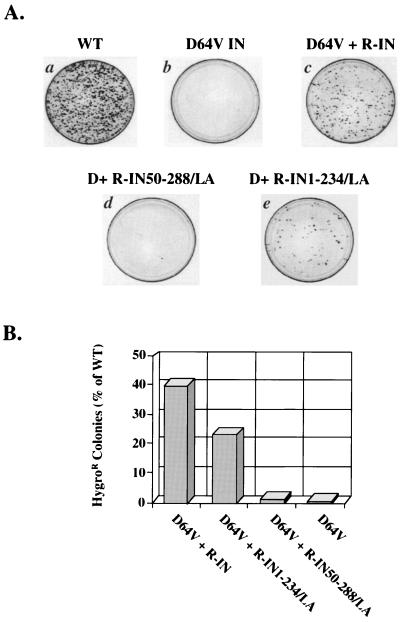
Although no colonies grew on the plates infected with the HXB-IND64V mutant virus (Fig. 4A, plate b), a high level of complementation was observed with the C-terminally truncated integrase-LexA protein provided in trans (Fig. 4A, plate e). The number of colonies that grew on plates of cells infected with this virus was approximately 24% of that on plates of cells infected with wild-type HXB2, or 57% of that for HXB-IND64V that contained wild-type integrase in trans (Fig. 4B). In contrast, the N-terminally truncated integrase fused to LexA did not mediate integration to a high degree. It consistently restored infectivity to the integrase-mutated viruses at a level of only about 1% of that of wild-type virus or 2% of the Vpr-integrase complementation control (Fig. 4A, plate d, and B). The N terminus of integrase, though, is involved in multiple stages of retroviral infection (16, 42, 49, 68, 73).
FIG. 4.
Integration by HXB-IND64V viruses that contain N- and C-terminally truncated integrase proteins fused to LexA. (A) A 50-ng p24 equivalent was used to infect each of the representative plates shown for the hygromycin resistance assay. One million HeLa-CD4 cells were infected for 4 h with viruses containing the Vpr fusion proteins R-IN (c), R-IN50-288/LA (d), or R-IN1-234/LA (e). A positive control virus (WT) (a) and a negative control virus (D64V) (b) were also assessed. At 2 days after infection, the cells were grown in 200 μg of hygromycin B per ml for 12 days. (B) Graphical representation of the average of three independent experiments for infectivity and integration by the HXB-IND64V virus containing IN50-288/LA or IN1-234/LA. Each experiment was conducted with each of 50-, 5-, and 0.5-ng p24 equivalents of the indicated virus in the hygromycin resistance assay. An average of 1,200 colonies were counted on plates infected with 50-ng p24 equivalent of the positive-control, wild-type virus.
Because the IN1-234/LA and IN/LADBD proteins were better at restoring infectivity to the HXB-IND64V viral clone in the hygromycin resistance assay, we also incorporated IN1-234/LADBD into HXB-IND64V in trans. Combining truncated versions of both integrase and LexA resulted in a slight to no increase in provirus formation (data not shown).
DISCUSSION
Integration by retroviral integrases is nonspecific and occurs throughout the genomic DNA (9, 22, 34, 57, 69). To reduce the risk of disrupting normal cell function by insertional mutagenesis during transduction, a method for directing the sites of integration is desirable. One approach to conferring site specificity is to fuse integrase to a sequence-specific DNA-binding protein. In vitro analysis of purified proteins made up of a sequence-specific DNA-binding protein and a retroviral integrase has revealed that integration can be directed into certain DNA sites (7, 30, 36). To evaluate the applicability of this strategy in vivo, it is important to develop an efficient method for incorporating functional integrase fusion proteins into infectious virions. Using the in trans method of incorporation, we were able to package a variety of integrase-LexA proteins into HIV-1. Fusing an integrase-LexA, integrase-LexA DBD, or an N- or C-terminally truncated integrase-LexA protein to Vpr results in efficient incorporation into viruses. Western blot analysis confirmed the presence of each integrase-LexA fusion protein in HIV-1. The viruses containing the integrase-LexA proteins were also infectious and able to perform integration. Although the integrase gene carried by the viruses contained a mutation in one of the catalytic triad residues, it was able to be complemented with the integrase-LexA fusion proteins and to produce a positive result in infectivity assays that require infection, integration, and reporter gene expression.
The in trans method of incorporation is a valuable tool for the development of retroviral vectors containing integrase fusion proteins. Previous attempts to produce retroviruses that contained integrase fusion proteins were unsuccessful due to loss of virus infectivity after transfection (8) or loss of fusion protein expression during viral replication owing to reversion (36). Each of these attempts encoded the integrase fusion protein as part of the viral genome, inserting the DNA sequence of the sequence-specific DNA-binding protein at the 3′ end of the integrase gene. The difficulty in encoding the fusion protein in the viral genome may lie in the fact that the 3′ end of the integrase gene overlaps the coding sequence of vif and contains a splice acceptor site. To avoid interfering with critical elements of the viral genome, we used the in trans incorporation method, which is highly efficient at including proteins in HIV-1. This method employs Vpr as a vehicle to package proteins into HIV-1 (23, 44, 45, 72–74). For each complemented virus, Western blot analysis with antibodies directed against all three portions of the fusion proteins, Vpr, integrase, and LexA, detected proteins of the appropriate size. In addition, correct protease cleavage between Vpr and integrase was observed.
We tested the ability of a variety of integrase-LexA fusion proteins to restore integration to an integrase-mutated viral clone to determine critical parameters for achieving high transduction efficiency in infected cells. The full-length LexA or the DBD of LexA was fused to integrase to examine the effect of the size and the dimerization domain of the sequence-specific DNA-binding protein on the integration activity of the fusion protein. In each hygromycin resistance assay performed, the fusion proteins were able to restore integration to the integrase-mutated viral clone, with the integrase-LexA DBD protein yielding almost 50% more colonies than the integrase-LexA delivered in trans. Both proteins were incorporated into the viruses at equally large amounts as determined by Western blots analyses, indicating that the difference in complementation efficiency is not due to a different level of fusion protein incorporation (Fig. 2). Since integrase is active as a multimer (14, 15, 63), the inclusion of the LexA dimerization domain may adversely affect the ability of integrase to achieve its correct multimeric state. It is also possible that such an alteration may have a negative effect on multiple steps of the viral infection process, ranging from binding of integrase to the viral cDNA ends to nuclear import of the PIC (5, 25–27, 32, 55, 67) and interaction with the chromosomal DNA.
We also examined whether introduced fusion proteins that contained a truncated integrase in trans were capable of complementing the integrase-defective virus. Truncation of the C- terminus of integrase in the fusion protein was able to restore integration by the HXB-IND64V mutant viral clone at a level of 24% of that for wild-type virus. Meanwhile, the N-terminally truncated integrase fused to LexA produced colonies at a level of 1%. This is the first time that a restoration of integration has been described in vivo with integrases that lack an N- or C-terminal domain. It is possible that fusion to LexA aided the truncated integrases in their ability to restore integration. However, in vivo complementation may also be taking place between different domains of integrase during virus infection. In vitro complementation data have indicated that mixing integrase monomers deleted in the N- or C-terminal domain, or both, complements integration activity with integrase monomers mutated in the catalytic core (15, 63). Furthermore, viruses harboring a catalytic core mutation in integrase are complemented with integrase proteins provided in trans that contain single-amino-acid substitutions in either the N- or C-terminal domain. The C-terminal mutant integrase restores integration to the core-mutated virus at a level of about 9%, of that of the wild-type virus, and the N-terminal mutant integrase complements integration to about 0.2% of that of the wild-type virus (23). Our result from the HXB-IND64V virus containing the truncated integrase-LexA fusion proteins is consistent with the previous data. In addition, in in vitro assays using purified proteins, complementation occurs between an N- or C-terminally truncated integrase fused to LexA and an integrase containing a D116G catalytic core mutation (data not shown). Therefore, the N- or C-terminally truncated integrase, although fused to LexA, probably restores integration by complementation with the core-mutated integrase during infection.
Based on the biochemical properties and catalytic activity of integrase, it is probable that the integrase fusion proteins provided in trans formed mixed multimers with the virally encoded integrase (15, 23, 63). To test this hypothesis, an integrase-minus virus can be provided with the integrase-LexA fusion protein in trans. The ideal viral clone for this purpose is one that lacks expression of its own integrase gene, for instance, one that contains a stop codon near the N terminus of integrase. Unfortunately, such a virus suffers from severe processing defects (4, 16), which have been more difficult to overcome, even when provided with wild-type integrase in trans (23; M. L. Holmes-Son and S. A. Chow, unpublished results). Because C-terminally truncated integrases cannot complement each other (15, 63), an alternative approach is to introduce stop codons after the integrase 234 residue of the HXB2 viral clone and test the ability of IN1-234/LA to restore integration activity in trans. We are currently preparing DNA constructs to test this hypothesis.
In contrast to the C-terminally truncated integrase fused to LexA, the fusion protein containing the N-terminally truncated integrase is much worse at restoring integration to the integrase-mutated viral clone. This may be because the N terminus of integrase plays multiple roles during retroviral infection and complementation of these functions by the core-mutated integrase encoded by the virus is difficult. Viruses containing mutations in the conserved HHCC motif of this domain have abnormal morphology, suggesting that the N terminus of integrase may be involved in maturation of particles (16). In addition, these viruses produce reduced levels of viral cDNA, implicating the N terminus of integrase as playing a role during reverse transcription (16, 42, 49, 68, 73). Integration may also be negatively affected because integrase may be prevented from multimerizing (43, 75) or recognizing the viral cDNA ends (62, 65). The C terminus of integrase, on the other hand, may play a role only during the integration step of the retroviral life cycle, which would be more easily complemented by the core-mutated integrase. In vitro assays indicate that the C-terminal domain of integrase is involved in nonspecific- DNA binding (17, 35, 53, 58, 66, 70, 71), and replacing the C-terminal domain of HIV-1 integrase with LexA, a DNA-binding protein, may enable this fusion protein to better restore integration activity than the N-terminally truncated integrase-LexA can. Therefore, although both of these fusion proteins would be able to provide a catalytic motif to the core-mutated integrase encoded by the HXB2 clone, the ability of the core-mutated integrase to restore functions provided by an N-terminally deleted integrase fused to LexA may be more difficult.
Although the DBD of LexA better restores integration to the HXB-IND64V virus than does the full-length LexA protein fused to the C terminus of integrase and although the C-terminally truncated integrase also displays high integration activity, we have not yet determined whether any of the fusion protein-containing viruses are able to confer heightened integration specificity to the LexA operator. Therefore, we have not yet evaluated whether the factors that provide increased complementation efficiency are the same as those required for integration specificity. It is also unknown if any of these viruses perform bona fide integration reactions when joining the viral cDNA to the chromosomes of the infected cells. Experiments are being conducted to determine the ability of the integrase-LexA fusion protein to direct integration into specific DNA sites and to examine whether those integration events are concerted, bona fide reactions.
The feasibility of directing integration has been tested in vitro, demonstrating that fusing integrase to a sequence-specific DNA-binding protein biases integration toward certain sites on target DNA (7, 30, 36). Evidence that these proteins are functional in retroviral vectors, however, has been lacking (8, 36). Using the in trans method of incorporation, we have shown that integrase-LexA proteins are functional to mediate integration in infected cells, resulting in stable expression of a reporter gene. To increase the applicability of retroviral vectors in gene transfer practices and their safety in gene therapy, it is desirable to control integration sites in the chromosomal DNA. These data present an efficient method for incorporating integrase fusion proteins into infectious viral particles and will allow such a strategy to be tested in vivo.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant CA68859 to S.A.C. M.L.H.-S. was supported by an Esther Hays Student Research Award from the UCLA AIDS Institute (NIH grant AI28697) and a predoctoral fellowship from the Universitywide AIDS Research Program, University of California.
We thank the UCLA AIDS core facility and Yao Wang for technical assistance, David Twomey and Diane Martin for graphic support, and Michael Emerman, Beatrice Hahn, and Andrew Leavitt for helpful discussions.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. [Google Scholar]
- 2.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–204. [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukovsky A, Gottlinger H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol. 1996;70:6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman F D, Miller M D. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau S, Hoffmann C, Bushman F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 11.Daniel R, Katz R A, Skalka A M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 12.Dhar R, McClements W L, Enquist L W, Vande Woude G F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci USA. 1980;77:3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison V, Abrams H, Roe T, Lifson J, Brown P O. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 15.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutants in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman A, Hickman A B, Craigie R. The core and carboxy-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 19.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 20.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald M L, Grandgenett D P. Retroviral integration: in vitro host site selection by avian integrase. J Virol. 1994;68:4314–4321. doi: 10.1128/jvi.68.7.4314-4321.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher T M, Soares M A, McPhearson S, Huxiong H, Wiskerchen M, Muesing M, Shaw G M, Leavitt A D, Boeke J D, Hahn B H. Complementation of integrase function in HIV-1 virions. EMBO J. 1997;16:5123–5138. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 25.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;17:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 28.Gaur M, Leavitt A D. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J Virol. 1998;72:4678–4685. doi: 10.1128/jvi.72.6.4678-4685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerton J L, Herschlag D, Brown P O. Stereospecificity of reactions catalyzed by HIV-1 integrase. J Biol Chem. 1999;274:33480–33487. doi: 10.1074/jbc.274.47.33480. [DOI] [PubMed] [Google Scholar]
- 30.Goulaouic H, Chow S A. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grandgenett D P, Goodarzi G. Folding of the multidomain human immunodeficiency virus type-1 integrase. Protein Sci. 1994;3:888–897. doi: 10.1002/pro.5560030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes S H, Mutschler A, Bishop J M, Varmus H E. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. Proc Natl Acad Sci USA. 1981;78:4299–4303. doi: 10.1073/pnas.78.7.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes S H, Shank P R, Spector D H, Kung H-J, Bishop J M, Varmus H E. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated into many sites. Cell. 1978;15:1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- 35.Kahn E, Mack J P G, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz R A, Merkel G, Skalka A M. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 37.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 38.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo E, Gottlinger H G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S P, Xiao J, Knutson J R, Lewis M S, Han M K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Wu X, Xiao H, Conway J A, Kappes J C. Incorporation of functional human immunodeficiency virus type 1 integrase into virions independent of the gag-pol precursor protein. J Virol. 1997;71:7704–7710. doi: 10.1128/jvi.71.10.7704-7710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Wu X, Xiao H, Kappes J C. Targeting human immunodeficiency virus (HIV) type 2 integrase protein into HIV type 1. J Virol. 1999;73:8831–8836. doi: 10.1128/jvi.73.10.8831-8836.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Y-L, Bennett R P, Wills J W, Goulick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 48.Majors J E, Varmus H E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981;289:253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- 49.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller M D, Wang B, Bushman F. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous strand are competent to integrate in vitro. J Virol. 1995;69:3938–3944. doi: 10.1128/jvi.69.6.3938-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyoshi H, Blomer U, Takahashi M, Gage F H, Verma I M. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muesing M A, Smith D H, Cabradilla C D, Benson C V, Lasky L A, Capon D J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 53.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popov S, Rexach M, Zybarth G, Reiling N, Lee M-A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pryciak P M, Muller H P, Varmus H E. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc Natl Acad Sci USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pryciak P M, Sil A, Varmus H E. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puras Lutzke R A, Vink C, Plasterk R H A. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 61.Shimotohno K, Mizutani S, Temin H M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980;285:550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- 62.van den Ent F, Vos A, Plasterk R H A. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J Virol. 1999;73:3176–3183. doi: 10.1128/jvi.73.4.3176-3183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Gent D, Vink C, Oude Groeneger A A M, Plasterk R H A. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 65.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vink C, Oude Groeneger A A M, Plasterk R H A. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes, Proc. Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiskerchen M, Muesing M A. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Withers-Ward E S, Kitamura Y, Barnes J P, Coffin J M. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 1994;8:1473–1487. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- 70.Woerner A M, Klutch M, Levin J G, Markus-Sekura C J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:2433–2437. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 71.Woerner A M, Marcus-Sekura C J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71a.Wong-Staal F, Chanda P K, Ghrayeb J. Human immunodeficiency virus: the eighth gene. AIDS Res Hum Retroviruses. 1987;3:33–39. doi: 10.1089/aid.1987.3.33. [DOI] [PubMed] [Google Scholar]
- 72.Wu X, Hongmei L, Hongling X, Justin K, Partha S, Georges N, Boeke J D, Hahn B H, Kappes J C. Targeting foreign proteins to human immunodeficiency virus particles via fusion with vpr and vpx. J Virol. 1995;69:3389–3398. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu X, Liu H, Xiao H, Conway J A, Hehl E, Kalpana G V, Prasad V, Kappes J C. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol. 1999;73:2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu X, Liu H, Xiao H, Conway J A, Hunter E, Kappes J C. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zufferey R, Dull T, Mandel R J, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]