Abstract
Lactococcus lactis is a lactic acid bacterium of major importance for food fermentation and biotechnological applications. The ability to manipulate its genome quickly and easily through competence for DNA transformation would accelerate its general use as a platform for a variety of applications. Natural transformation in this species requires the activation of the master regulator ComX. However, the growth conditions that lead to spontaneous transformation, as well as the regulators that control ComX production, are unknown. Here, we identified the carbon source, nitrogen supply, and pH as key factors controlling competence development in this species. Notably, we showed that these conditions are sensed by three global regulators (i.e., CcpA, CodY, and CovR), which repress comX transcription directly. Furthermore, our systematic inactivation of known signaling systems suggests that classical pheromone-sensing regulators are not involved. Finally, we revealed that the ComX-degrading MecA-ClpCP machinery plays a predominant role based on the identification of a single amino-acid substitution in the adaptor protein MecA of a highly transformable strain. Contrasting with closely-related streptococci, the master competence regulator in L. lactis is regulated both proximally by general sensors and distantly by the Clp degradation machinery. This study not only highlights the diversity of regulatory networks for competence control in Gram-positive bacteria, but it also paves the way for the use of natural transformation as a tool to manipulate this biotechnologically important bacterium.
Author summary
Lactic acid bacteria play important roles in our daily lives as microbiota members or fermentation starters. Understanding the natural horizontal gene transfer mechanisms that shape their genomes will allow us to better control and understand their evolution over time. This study identified the physiological conditions that activate competence for DNA transformation in Lactococcus lactis, one of the most important lactic acid bacteria. We also unveiled the guardians of the master competence regulator, ComX. In this species, global carbon and nitrogen regulators, as well as a general stress system, directly repressed its production instead of its activation by classical cell-to-cell communication systems as reported in streptococci. Furthermore, it was discovered that ComX degradation, orchestrated by the Clp machinery, plays a dominant role in the strict control of competence activation. This alternative regulation model could inspire future work aiming to uncover competence regulation networks in other Gram-positive bacteria with important medical or biotechnological implications.
Introduction
Lactococcus lactis is one of the most studied species among lactic acid bacteria for both basic and applied research. Initially, the food industry has used its ability to produce lactic acid for centuries to preserve a wide variety of fermented foods (e.g., cheese, buttermilk, and sour cream) [1]. In addition, L. lactis is now largely exploited as a safe production platform for recombinant proteins due to their easy production and downstream purification [2]. Finally, this species is of high interest as a vector for therapeutics or vaccine antigens [3,4]. Because of all of these possible applications, L. lactis has become a powerful living tool of tremendous interest across the scientific community in just a few decades [1].
One of the commonalities shared by this large subset of applications is the ability to easily modify the lactococcal genome. Aside from commonly used techniques such as electroporation, conjugation, or transduction, the use of competence for DNA transformation is an appealing alternative. Natural transformation is a horizontal gene transfer process in which the bacterium is able to capture, internalize, and recombine a naked DNA fragment present in the extracellular environment [5]. This process is thus of major interest for a variety of reasons. First, natural transformation does not necessitate the involvement of a third party as required for conjugation or transduction (i.e., another cell or a bacteriophage, respectively). Second, the donor DNA could simply be a linear fragment obtained by polymerase chain reaction and added to the culture medium. Third, natural transformation provides a quick and accurate genome editing tool [6].
Competence activation is typically divided into two distinct phases (i.e., early and late). During the early phase, a stimulus provokes the expression of the central regulator of competence. The latter, in association with the RNA polymerase, will then trigger the expression of the competence (com) regulon, which corresponds to the late phase [7–9]. This regulon codes for all proteins responsible for DNA transformation, from DNA capture outside the cell to recombination into the genome (for recent reviews, see [6,10]). While the late-phase development and associated mechanisms are well conserved across prokaryotes, stimuli and regulators responsible for the expression of the central regulator of competence are highly species-specific (for a review, see [9]).
In the closely-related streptococci, competence was shown to be controlled by a pheromone quorum-sensing system that directly activates the central competence regulator ComX (σX) [8]. In four out of the six groups of streptococci (i.e, salivarius, mutans, pyogenes, and suis), the intracellular sensor ComR of the Rgg family was identified, in combination with its secreted and re-internalized communication peptide XIP (ComX-Inducing Peptide, ComRS system), as the main trigger of competence activation [8,11,12]. In the two other groups (i.e., mitis and anginosus), a different communication system is in charge of competence activation [9]. This system is based on a phospho-relay that is activated by the secreted communication peptide CSP (Competence-Stimulating Peptide, ComCDE system). The peptide is not re-internalized in this case but instead interacts with the extracellular part of the histidine kinase from the two-component system (TCS) ComDE [13]. After a phosphorylation cascade, the response regulator ComE~P triggers comX expression [14]. In Bacillus subtilis, a similar mechanism was identified with the ComXAP system, in which the communication peptide ComX is secreted in the extracellular environment to activate the TCS ComAP [15]. This leads to the production of ComS, a small intracellular peptide that disrupts the interaction between the central regulator of competence ComK and the adaptor protein MecA [16–18]. This loss of interaction prevents ComK from being degraded by the ClpCP machinery, allowing its accumulation and expression of the late com regulon (for a review, see [19]).
So far, no such mechanisms of early-phase activation have been identified in Lactococcus species. However, a systematic analysis of all sequenced and complete genomes of L. lactis revealed that ~25% of the strains have a complete and intact set of late com genes along with the comX gene, and that this relatively small proportion of strains is primarily isolated from plants [6]. Interestingly, two strains containing the entire set of late com genes, Lactococcus cremoris KW2 and L. lactis KF147, were challenged for their ability to be transformed by exogenous DNA upon artificial ComX overproduction [20,21]. In both cases, the controlled induction of ComX allowed transformation events, demonstrating the capacity of L. cremoris and L. lactis to develop and carry out natural transformation [20,21]. Besides, spontaneous natural transformation has never been observed in those two species [20,21]. While an increase in comX expression along with some late com genes was reported under carbon starvation conditions in L. lactis KF147, transformation attempts with linear plasmid DNA failed [22]. In L. cremoris KW2 containing the comX-overexpression plasmid in non-induced conditions, low spontaneous transformation events were observed in mutant strains lacking MecA, ClpC, or ClpP [20]. However, this was attributed to a weak leakage of the inducible promoter and the cell’s inability to degrade ComX via the MecA-ClpCP machinery, resulting in ComX accumulation above the competence activation threshold [20].
In this work, we report the first observations of spontaneous competence in multiple L. lactis strains after optimization of DNA transformation conditions. Notably, we identified three global nutritional/stress regulators that repress comX transcription directly. Furthermore, we discovered a plant-derived L. lactis strain capable to transform at high level due to a single amino-acid substitution in the adaptor protein MecA of the Clp machinery. Together, these findings show that competence development in L. lactis is negatively controlled by (i) global regulators sensing nutritional and environmental conditions to modulate ComX abundance and (ii) the Clp machinery degrading ComX to limit its availability, both levels of regulation ensuring tight control of competence activation in this species.
Results
Spontaneous transformation of L. lactis is strain-dependent
An in silico screening of an in-house database of ~300 genomes of L. lactis allowed the identification of 18 strains (16 of plant origin) harboring the complete and supposedly intact set of com genes required for DNA transformation (comX and 16 late genes) (S1 Table) [20,23]. These 18 strains were initially tested for their potential ability to perform natural transformation. For this purpose, we used a plasmid-based system that allows comX overexpression by xylose induction (pGhPxylTcomX, carrying comX from strain IO-1) [20]. This plasmid was successfully electroporated in 16 out of the 18 selected strains. As negative control, we electroporated the plasmid in strain 1AA59 of L. lactis that contains a mutation in the comFA-FC operon. This mutation generates a fusion product between ComFA and ComFC that should alter the functionality of the transformasome (S1 Table). Transformation assays were performed in M17GX (0.1% glucose-1% xylose) with a mutated version of the rpsL gene (rpsL*, RpsL-K56R) as donor DNA, which confers streptomycin resistance [20]. While strain 1AA59 could not be transformed, 15 out of the 16 strains harboring an intact set of com genes displayed a transformation rate between ~10−7 and ~10−2 (S1A Fig). Those results highlight a high correlation between the presence of a complete set of competence genes and the functionality of the natural transformation machinery.
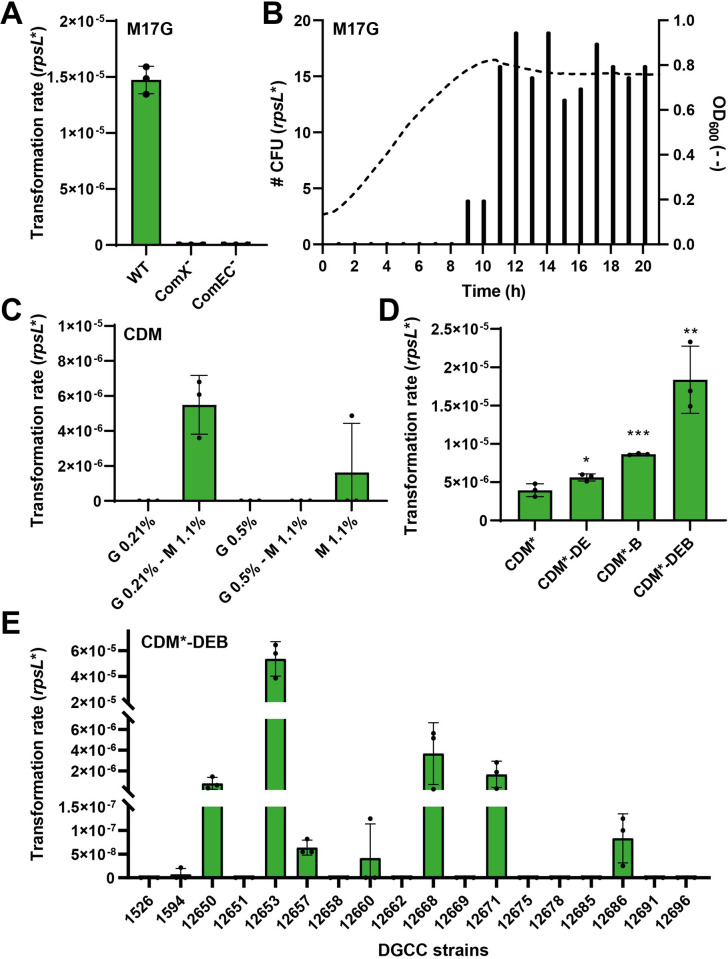
After this validation step, we assessed these strains for spontaneous transformation in rich medium M17G (0.5% glucose) with rpsL* DNA. Notably, strain DGCC12653 of plant origin was the only one to show spontaneous transformation events in those conditions (S1B Fig). We validated that natural transformation took place through the transformation machinery since a deficiency in ComX or in the DNA channel ComEC abolished transformability (Fig 1A). Moreover, a time-lapse experiment showed that transformation occurred at the entry of the stationary growth phase (Fig 1B), contrasting with competence development in streptococci that takes place during the early logarithmic phase [7,8], yet similar to this phenomenon in B. subtilis [19].
Fig 1. Effects of medium composition on spontaneous natural transformation.
(A) Effects of the inactivation of ComX or ComEC on DGCC12653 transformability in M17G medium. (B) Timing of transformation events (bars, number of streptomycin-resistant CFUs) during cell growth (dotted line) in M17G medium. (C) Effects of glucose [G]-maltose [M] concentrations (%, w/v) on transformation rate in CDM. (D) Effects of the omissions of aspartate and glutamate [-DE], nitrogen bases[-B], and their combination [-DEB] on transformation rate in CDM* (CDM Glu 0.21%—Mal 1.1%). The optimized competence-inducing medium was named CDM*-DEB. Statistical t test was performed for each omission in comparison to the CDM* medium (n = 3; *, P < 0.05, **, P < 0.01, and ***, P < 0.001). (E) Transformation rates of 18 L. lactis strains harboring the complete set of competence genes in CDM*-DEB. All transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture (except in panel B). Dots show the values for biological triplicates (panel A) or technical triplicates (panels C, D, and E); mean values ± standard deviations.
Together, these results showed that spontaneous transformation of L. lactis is highly strain-dependent in commonly-used rich medium and develops at the entry of the stationary growth phase.
The carbon source and diauxic shift are triggers of spontaneous transformation
In order to explore the influence of various compounds in the growth medium on competence activation, we investigated transformation of strain DGCC12653 in chemically-defined medium (CDM) [24]. As the carbon source was previously shown to have a strong impact on spontaneous transformation in many bacteria [25], we tested a large set of sugars using phenotype microarrays (Biolog plates). While no transformation was observed in CDM with glucose (0.5%) as the sole carbon source, we obtained transformants at low levels (~10−8, < 10 transformants) with alternative sugars mainly originating from plants (i.e. maltose, xylose, cellobiose, galactose, and arabinose). As glucose starvation was previously reported to trigger the expression of competence genes in L. lactis [22,26], we attempted to combine these two effects by testing the influence of a diauxic shift on transformability. Transformation assays were performed in CDM supplemented with glucose and maltose in different ratios (from 0 to 1%). A design of experiment (DoE; central composite plane) was set up and the surface response highlighted the most efficient mix between these two sugars (0.21% glucose-1.1% maltose) (Figs 1C and S1C). This competence-inducing medium (named CDM*) allowed a reproducible transformation rate of ~5 × 10−6 with rpsL*. We also investigated the effect of amino acid and nitrogen base (purines and pyrimidines) omissions on spontaneous transformation. By removing aspartate, glutamate and nitrogen bases from the CDM medium (named CDM*-DEB), the transformation rate was ~5-fold increased, reaching ~2.0 × 10−5 (Fig 1D). Finally, transformation assays (rpsL* DNA) in this optimized medium were performed with the 18 L. lactis strains harboring the complete set of late com genes. In those conditions, spontaneous transformation was observed in 8 out of the 18 wild-type strains, with a transformation rate ranging from ~10−8 to ~10−5. Among these strains, DGCC12653 remained the most efficient spontaneous transformer (Fig 1E).
Together, these results highlight that a sugar diauxic shift improves the transformation rate of a range of L. lactis strains, showing that growth on alternative carbon sources to glucose is a key physiological trait to activate spontaneous transformation in this species.
The carbon catabolite regulator CcpA is a direct repressor of comX
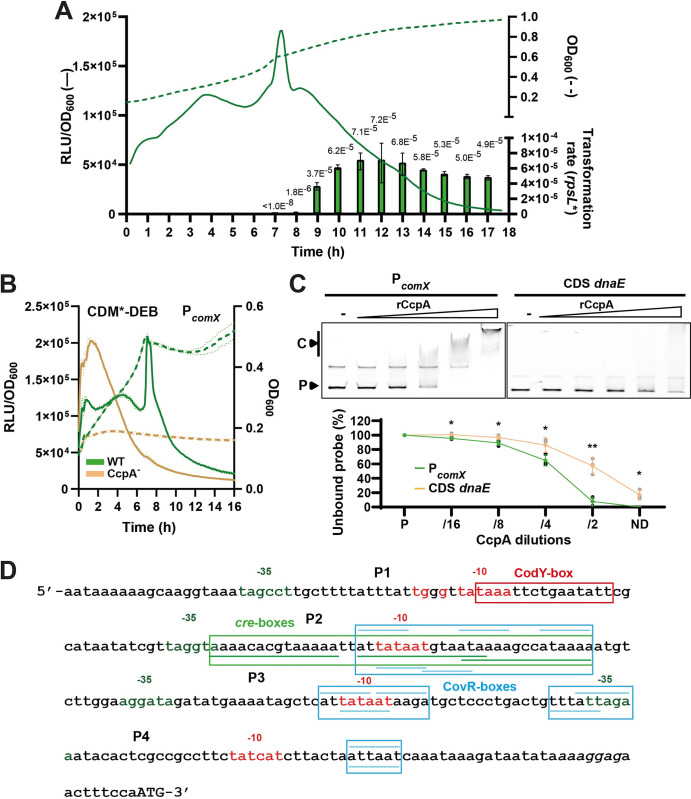
To further investigate the role of the glucose-maltose diauxic shift on spontaneous competence of strain DGCC12653, we monitored the expression of the comX gene. For this purpose, a luminescence reporter system was designed by cloning the luciferase genes luxAB under the control of the comX promoter (PcomX) in a low-copy number plasmid (pGhPcomX-luxAB). Luminescence assays showed that comX expression was boosted after seven hours of culture, which corresponds to the diauxic shift between glucose starvation and a second growth consuming maltose (Figs 2A and S1D). Interestingly, fluorescence microscopy analyses (plasmid pGhPcomX-gfpsf) revealed a heterogeneity in competence activation at the diauxic shift since only a subpopulation of cells activate the PcomX-gfpsf fusion at detectable levels (S2 Fig). Parallelly, we performed transformation assays by adding rpsL* DNA at the beginning of the culture and monitored the transformation rate every hour after DNA addition. While no transformation was observed before seven hours of culture, transformation events were detected one hour after the diauxic shift and increased until ten hours post DNA addition to reach a maximum transformation rate of ~6 × 10−5 (Fig 2A). This revealed that the diauxic shift stimulates ComX production at a sufficient level to allow competence activation.
Fig 2. Involvement of CcpA in competence regulation.
(A) Timing of transformation rate (green bars) during cell growth (green dotted line; OD600) and kinetics of PcomX specific luciferase activity (green solid line; RLU/OD600) for DGCC12653 WT in CDM*-DEB. Transformation assays were performed as reported in Fig 1. Bars show mean values of technical triplicates ± standard deviations. The mean value is indicated on the top each bar. (B) Growth (dotted lines; OD600) and kinetics of PcomX specific luciferase activity (solid lines; RLU/OD600) monitored over time for DGCC12653 WT (green) and CcpA- (light brown) strains. Solid and dotted dark lines are representative of the mean of technical and biological triplicates (WT and CcpA-, respectively), and light lines are representative of standard deviation. (C) EMSAs (top panels) performed with a gradient of purified CcpA (rCcpA) on PcomX (left) and a 150-bp fragment of the CDS of dnaE as negative control (right). Lanes without rCcpA are indicated by a minus sign. C and P indicate the rCcpA-DNA complex and the unbound probe, respectively. Gels displayed are representative of technical triplicates. The percentage of unbound probe (bottom panel) for PcomX (green line) and CDS dnaE (orange line) was calculated using the Amersham Typhoon analysis software. ND and P stand for non-diluted rCcpA and probe-alone condition, respectively. Statistical t test was performed for each dilution in comparison to the negative control (n = 3; *, P < 0.05; **, P < 0.01). (D) Mapping of regulation elements identified in PcomX (intergenic region). CodY-, cre- and CovR-boxes are surrounded in red, green, and blue, respectively. Top and bottom blue lines correspond to CovR boxes identified on the top and bottom strands, respectively. Putative -35 and -10 boxes of mapped vegetative promoters (P1 to P4) are colored in red and dark green, respectively. The Shine-Dalgarno (italics) and start codon (capitals) of comX are mapped at the 3’end.
As we observed a stimulating effect of the diauxic shift on competence development, we questioned the role of the global transcriptional regulator CcpA (carbon catabolite control protein A) in this phenomenon. CcpA is involved in carbon catabolite repression (CCR) in Gram-positive bacteria and plays a role in the diauxic shift between the preferred glucose catabolism and alternative sugar utilization pathways in the closely-related species L. cremoris [27]. A ccpA-deleted mutant of strain DGCC12653 was constructed and transformed with the PcomX-luxAB reporter system. We observed an increased expression of comX in the early growth phase (~2-fold) along with a major growth defect for the ccpA mutant compared to the wild-type in optimized conditions (Fig 2B). We also performed transformation assays in the same conditions that showed a decreased transformation rate when ccpA was deleted (S3A Fig). Although comX expression was slightly unleashed in the ccpA mutant, the constitutive CcpA deficiency has a global impact on growth and carbon metabolism that could be incompatible with the transformation process (e.g., alterations of cell-wall, ATP availability, or cell cycle).
To test whether CcpA is directly involved in competence regulation by binding PcomX, we performed electrophoretic mobility shift assays (EMSAs) with purified CcpA of strain DGCC12653 (S3B Fig). The purified protein (CcpA with N-terminal 6-His tag; named rCcpA) was incubated with fluorescent Cy3-labelled PcomX (Cy3PcomX) as a DNA probe (Fig 2C), which corresponds to the intergenic region located upstream of the comX gene. The results showed that rCcpA significantly interacts with PcomX compared to the control probe (CDS of dnaE) (Fig 2C). Thanks to in silico analyses and different DNA probes (S3C-S3E Fig), we were able to identify three potential CcpA-binding sites (cre-box consensus, WGWAARCGYTWWMA) (Fig 2D) [28]. We also searched in silico for vegetative promoters in the PcomX region and identified four candidates (named P1 to P4) (Fig 2D). Those putative promoters display similar features with a highly conserved -10 box (5 or 6 conserved positions with TATAAT) and a less conserved -35 box (3 conserved positions with TTGACA), separated by 16–18 nucleotides. The three cre-boxes overlap the putative vegetative promoter P2 in the upstream intergenic region of comX (Fig 2D).
Together, these results show that a sugar diauxic shift activates comX expression at the transcriptional level. They also suggest that CcpA directly represses PcomX in glucose conditions, a repression effect that is potentially alleviated on alternative carbon sources such as maltose.
The nitrogen global regulator CodY is a direct repressor of comX
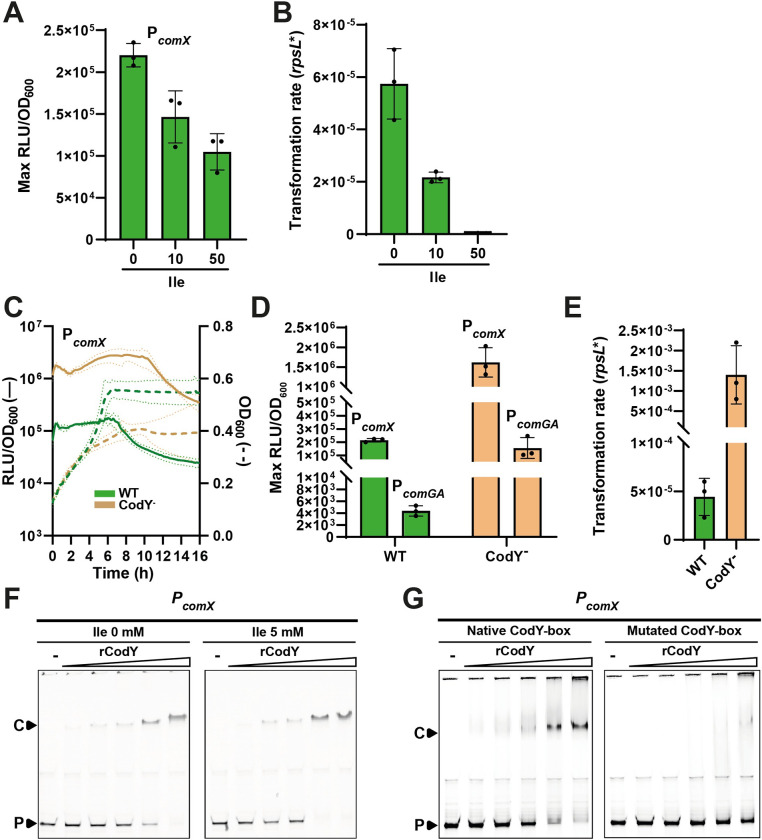
During the optimization process of CDM composition, we observed that omission of branched-chain amino acids (BCAAs; Leu, Val, and Ile) inhibited growth of strain DGCC12653 (increase in OD600 < 0.05 during 6 hours post-inoculation) but that an excess of isoleucine decreased comX expression and spontaneous transformation in optimized conditions (Fig 3A and 3B). As BCCAs are well known to control the activity of the nitrogen transcriptional regulator CodY in L. lactis and L. cremoris [28–31], we investigated its potential role in the control of competence. For this purpose, we generated a codY-deleted mutant of strain DGCC12653 that was transformed with the PcomX-luxAB reporter system. CodY inactivation resulted in a nearly 10-fold increase in comX expression at the diauxic shift (Fig 3C and 3D) and a drastic improvement of the transformation rate (maximum of ~ 10−3) compared to the wild type in optimized conditions (Fig 3E). Moreover, fluorescence microscopy analyses performed at the diauxic shift revealed a higher PcomX activation, which also takes place in a larger part of the cell population than observed in the wild type (> 97% vs. 70% of cells, respectively) (S2 Fig). We also used PcomGA as a proxy of the activation of the late com phase, as comGA is a well-established gene under ComX control in lactococci [20,32]. The activation of the PcomGA-luxAB fusion in the codY mutant was ~50-fold increased compared to the wild type (Fig 3D). In addition, a RNAseq analysis was performed to compare the wild type and the codY mutant using total RNAs extracted during the diauxic shift (S2 Table). This analysis confirmed the higher expression of comX along with the complete late com regulon in the CodY-deficient strain, corroborating the ComX regulon previously reported in L. cremoris [20]. These results suggest that CodY is somehow repressing competence development by downregulating comX expression and in cascade late com genes.
Fig 3. Involvement of CodY in competence regulation.
(A) Effect of isoleucine excess on PcomX activity in DGCC12653 WT. Data show maximum specific luciferase activity (Max RLU/OD600) observed at the diauxic shift for different concentrations of isoleucine (Ile; 0, 10, and 50 mM) added to the initial concentration (0.76 mM) in CDM*-DEB. (B) Effect of isoleucine excess (conditions as in panel A) on transformability of DGCC12653 WT after overnight culture with donor DNA (C) Growth (dotted lines; OD600) and kinetics of PcomX specific luciferase activity (solid lines, RLU/OD600) monitored over time for WT (green) and CodY- (light brown) strains in CDM*-DEB. Solid and dotted dark lines are representative of the mean of technical and biological triplicates (WT and CodY-, respectively), and light lines are representative of standard deviation. (D) Effect of codY deletion on activation of early (PcomX) and late (PcomGA) competence phases. Data show maximum specific luciferase activity (Max RLU/OD600) of PcomX-luxAB and PcomGA-luxAB reporter fusions, observed at the diauxic shift for WT (green) and CodY- (light brown) strains in CDM*-DEB. (E) Effect of codY deletion on transformability. Data show transformation rates observed for WT (green) and CodY- (light brown) strains in CDM*-DEB after overnight culture with donor DNA. Dots show the values for biological triplicates (CodY-) or technical triplicates (WT), mean values ± standard deviations. (F) Effect of isoleucine on CodY binding to PcomX. EMSAs performed with a gradient of purified CodY (rCodY) on PcomX without (left) or with 5 mM isoleucine (right). (G) Identification of the CodY-box in PcomX. EMSAs comparing rCodY binding to native (left) and mutated (right) CodY-boxes. In the mutated CodY-box, the two central nucleotides CG were swapped with AA. Lanes without rCodY are indicated by a minus sign. C and P indicate the rCodY-DNA complex and the unbound probe, respectively.
To question the possible direct interaction between CodY and PcomX, we purified CodY of strain DGCC12653 (CodY with N-terminal 6-His tag; named rCodY) for binding assays (S4A Fig). EMSAs performed with Cy3PcomX confirmed the direct binding of rCodY to PcomX. Subsequent analyses revealed that rCodY binding was favored by the presence of BCAAs with the strongest effect observed with isoleucine (5 mM), but not by GTP as previously reported in L. lactis [30] (Figs 3F and S4B). We also identified a CodY-binding motif (CodY-box TAAATTC T GAATATT, conserved positions underlined; [29]) that overlaps the -10 box of the putative vegetative promoter P1 in the upstream intergenic region of comX (Fig 2D). To confirm the CodY-box, we mutated the two central nucleotides CG into two AA (TAAATTA T AAATATT), and showed that rCodY binding was drastically reduced (Fig 3G). In addition, the mapping of RNAseq reads on PcomX displays a strong increase of transcripts around the CodY box for the CodY-deficient strain compared to the wild type. However, most of those reads result from a transcriptional readthrough from the upstream ezrA gene (S4C Fig).
These results show that CodY represses competence development by downregulating comX expression through a direct binding on PcomX in L. lactis. This highlights that the central competence gene comX of this species is regulated by the cell nutritional status through the sensing of both carbon and nitrogen growth substrates.
The environmental stress sensor CovRS is a direct repressor of comX
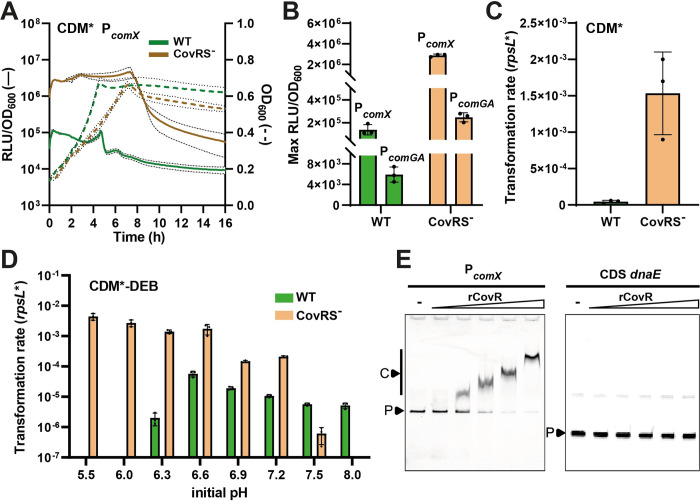
In order to determine if competence in L. lactis is controlled by a cell-to-cell communication system or stress sensor(s), as reported in other Gram-positive bacteria [7–9], we systematically inactivated all members of the Rgg family and TCSs. Ten Rgg and eight complete TCS (response regulator [RR] and histidine kinase [HK] in tandem) encoding genes were identified in the genome of strain DGCC12653 (S3 Table). All these genes were successfully inactivated by deletion, except the VicRK (WalRK) system that was previously reported to be essential in Gram-positive bacteria [33]. The collection of 17 mutants was tested for comX expression (PcomX-luxAB) and spontaneous transformation in the optimized medium. All ten rgg mutants behaved as the wild-type strain, with less than a two-fold impact on transformation and luminescence (S5A and S5B Fig). Regarding the seven TCS mutants (tandem inactivation of RR/HK), similar analyses revealed that CovRS (DGCC12653_01910–01915) inactivation allowed both an increase in comX expression (up to 15-fold) and transformation rate (up to ~ 10−3), while all other deletion mutants did not significantly differ from the wild-type strain (Figs 4A-4C, S5C and S5D). Moreover, fluorescence microscopy analyses confirmed a high PcomX activation in a large part of the cell population (> 97%) for the CovRS-deficient strain (S2 Fig). A RNAseq analysis was also performed to compare the transcriptomes of the covRS mutant with the wild type. As the CovRS-deficient strain displayed a strong growth defect in CDM*-DEB, both the wild type and covRS mutant were propagated in CDM*. Total RNAs were extracted from cells collected during the diauxic shift, when spontaneous transformation was previously shown to take place. This experiment confirmed the increase of comX expression, as well as the complete set of late com genes, in the covRS mutant compared to the wild type (S2 Table). In addition, the PcomGA-luxAB reporter system of the late competence phase was strongly induced (Fig 4B), confirming RNAseq data and high transformation rate.
Fig 4. Involvement of CovRS in competence regulation.
(A) Growth (dotted lines, OD600) and kinetics of PcomX specific luciferase activity (solid lines, RLU/OD600) monitored over time for DGCC12653 WT (green) and CovRS- (brown) strains in CDM*. Solid and dotted dark lines are representative of the mean of technical and biological triplicates (WT and CovRS-, respectively), and light lines are representative of standard deviation. (B) Effect of covRS deletion on activation of early (PcomX) and late (PcomGA) competence phases. Data show maximum specific luciferase activity (Max RLU/OD600) of PcomX-luxAB and PcomGA-luxAB reporter fusions, observed at the diauxic shift for WT (green) and CovRS- (light brown) strains in CDM*. (C) Effect of covRS deletion on transformability. Data show transformation rates observed for WT (green) and CovRS- (light brown) strains in CDM* after overnight culture with donor DNA. (D) Effect of the initial pH on transformability. Data show transformation rates observed for WT (green) and CovRS- (light brown) strains after overnight culture with donor DNA in CDM*-DEB buffered at different initial pHs. Dots show the values for biological triplicates (CovRS-) or technical triplicates (WT), mean values ± standard deviations. (E) EMSAs performed with a gradient of purified CovR (rCovR) on PcomX (left) and the CDS of dnaE as negative control (right). Lanes without rCovR are indicated by a minus sign. C and P indicate the rCovR-DNA complex(es) and the unbound probe, respectively. Gels displayed are representative of technical triplicates.
As CovRS and CodY are dominant repressors of comX expression, we also generated a double covRS codY mutant to evaluate additive or synergic effects. Although transformability and maximum comX expression were not increased, comX expression became constitutive during growth (S6 Fig). This suggests that these two regulatory systems are controlling comX with a different temporality, synergistically during early logarithmic growth and then dominantly by CovRS until the entry into the stationary growth phase (S6 Fig). As CovRS was previously shown to respond to pH in streptococci [34–36], we monitored the transformation rate in a range of initial pH values (pH 5.5 to 8.0). While spontaneous transformation in the wild-type strain was abolished below pH 6.6, the covRS mutant remained transformable in a much larger range of low pH values (Fig 4D). In addition to the nutritional sensors CcpA and CodY, these results show that the stress sensor CovRS represses comX expression and thus transformation efficiency in L. lactis DGCC12653.
To question the possibility that CovR directly regulates competence by binding PcomX, we purified CovR from strain DGCC12653 (CovR with N-terminal 6-His tag; named rCovR) (S7A Fig) and performed EMSAs with PcomX as a probe. EMSAs showed that CovR was able to bind PcomX, a binding that was improved when CovR was in vitro-phosphorylated (Figs 4E and S7B). EMSAs assays with different DNA probes allowed us to identify multiple CovR-binding regions covering putative promoters P2, P3, and P4 (Figs S7C-S7E and 2D). In silico analysis of these regions revealed multiple CovR-boxes (ATTARA) that were previously shown to be CovR-binding sites in streptococci (Fig 2D) [37,38]. We also showed that the mapping of RNAseq reads on PcomX displays a strong increase of transcripts downstream of the promoter P2 for the CovRS-deficient strain compared to the wild type, corroborating EMSA results (S7F Fig).
Altogether, these results indicate that classical cell-to-cell communication systems (Rgg and TCS) reported for competence control in Gram-positive bacteria [8] are not involved in comX activation in L. lactis. Conversely, these results confirm that comX expression is under the direct control of multiple global regulators, including the general stress sensor system CovRS, which responds to a range of environmental stimuli in closely-related species [34–36,39,40].
The Clp degradation machinery plays a central role in competence repression
In order to extend the role of global regulators in the control of natural transformation in other L. lactis strains, we generated codY and covRS mutants in three additional L. lactis strains (DGCC12651, DGCC12671 and DGCC12678) whose natural transformation was validated by the artificial comX overexpression system (S1A Fig). The comX expression was similarly increased in those mutants compared to strain DGCC12653, corroborating the high conservation of the PcomX sequence in a range of L. lactis strains (S8 and S9 Figs), Surprisingly, none of these mutant strains (neither codY nor covRS deleted mutant) showed an increase in spontaneous transformation with rpsL* DNA in optimized conditions. These results suggest an additional layer of regulation at the post-translational level between ComX production and transcriptional activation of the late com genes.
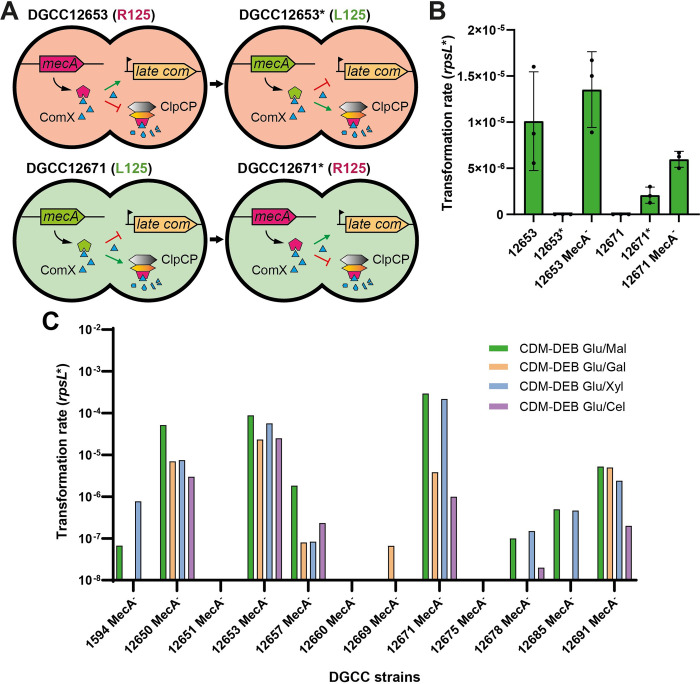
As we previously reported that the ComX-degrading MecA-ClpCP machinery negatively affects artificially-induced natural transformation of L. cremoris [20], we compared the protein sequences of MecA, ClpC, and ClpP of these three strains with those from strain DGCC12653. While alignments of ClpC and ClpP proteins showed no specific amino acid variation, the adaptor protein MecA displayed a single amino acid change (L125R) that is only present in strain DGCC12653 (S10 Fig). To test whether the L125R substitution could be responsible for the spontaneous transformability of DGCC12653, we reciprocally swapped mecA genes between strains DGCC12653 (MecA-R125; rare) and DGCC12671 (MecA-L125; common) (Fig 5A). We also generated mecA deletion mutants (MecA-) of both genetic backgrounds. Notably, we observed that DGCC12653 that produces the common version of MecA (L125) lost its capacity to transform exogenous DNA. Conversely, the production of the rare version of MecA (R125) in a non-spontaneously transformable strain (DGCC12671) activated natural transformation at a rate comparable to a MecA-deficient strain (Fig 5B). In addition, we were able to delete mecA in 12 out of the 18 L. lactis strains initially selected. Natural transformation assays performed in four different competence-inducing media showed that mecA deletion allowed transformation events to different extents in eight out of the 12 MecA- strains (Fig 5C).
Fig 5. Involvement of MecA in competence regulation.
(A) Schematic view of the swap of mecA genes between strains DGCC12653 (salmon) and DGCC12671 (light green). Genes encoding MecA-R125 (rare) and MecA-L125 (common) are colored in pink and green, respectively. Green and red lines illustrate activated or repressed pathways, respectively. Black lines display gene expression. (B) Effects of MecA-L125R and -R125L substitutions on transformability. Data show transformation rates observed for DGCC12653 WT (MecA-R125), DGCC12653* (MecA-L125), DGCC12671 WT (MecA-L125), DGCC12671* (MecA-R125), and MecA- (mecA deletion) strains after overnight culture with donor DNA in CDM*-DEB. Dots show the values for biological triplicates (mutant strains) or technical triplicates (WT), mean values ± standard deviations. (C) Effects of mecA deletion (MecA-) on spontaneous transformability in 12 L. lactis strains. Data show transformation rates in four different media composed of CDM-DEB with glucose (Glu) 0.21% (w/v) and a second plant sugar at 1.1% (w/v) (maltose [Mal], green; galactose [Gal], light brown; xylose [Xyl], blue; and cellobiose [Cel], purple).
Together, these results showed that MecA plays a crucial role in competence repression and that the single substitution MecA-L125R explains the unique transformation behavior of strain DGCC12653. Contrasting with the low impact of the Clp degradation machinery on competence control in streptococci [8,41,42], this machinery plays a dominant role at the global level of competence regulation in L. lactis.
Discussion
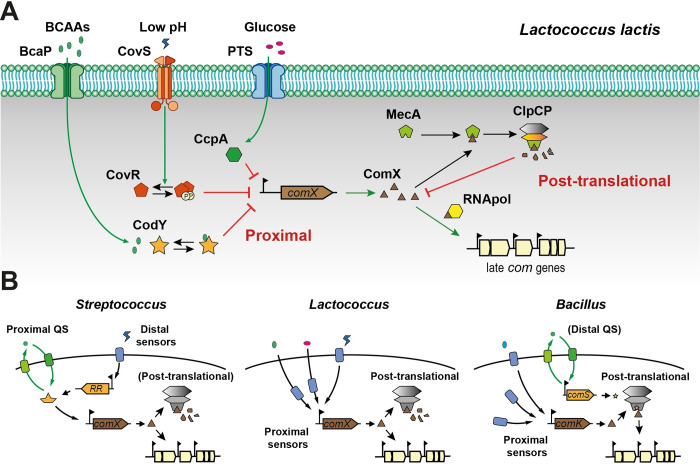
In this work, we highlighted a different regulatory network controlling competence in L. lactis when compared to closely-related streptococci (Fig 6). Although ComX is the common central regulator of competence in both, the proximal regulation of its activation does not appear to be controlled by a cell-to-cell communication module like in streptococci, but rather by global regulators sensing cellular nutritional status (CcpA and CodY) or stress conditions (CovRS) (Fig 6A). By comparison, global regulators are distantly controlling ComX by modulating the communication module in streptococci (for a review, see [8]) (Fig 6B). Moreover, the post-transcriptional layer of regulation through ComX degradation by the MecA-ClpCP machinery plays a predominant role in L. lactis, while it is more seen as a secondary locking device in streptococci [8,41–45]. In addition, the kinetics of competence activation during growth on a single carbon source are totally different: stationary phase in L. lactis versus transitory during early logarithmic growth for streptococci, controlled by the on/off switch of the communication module [7,8]. Notably, the timing of competence activation and the network hierarchy share some similarities with competence regulation in bacilli, where the central transcriptional regulator ComK is proximally modulated by global regulators and primarily controlled through degradation, which may, or may not, be regulated by a cell-to-cell signaling system. (Fig 6B) [19,46].
Fig 6. Competence control integrating proximal-distal transcriptional and post-translational regulation of the central regulator in Lactococcus lactis and closely-related species.
(A) Model of competence regulation in L. lactis. The transcription of comX (proximal regulation) is repressed by three regulators, sensing either the cellular nutritional status (CcpA and CodY, green and orange, respectively) or stress conditions (CovRS, red). When environmental/stress conditions are appropriate, ComX (brown) production is unrepressed. Then, ComX can interact with the adaptor protein MecA (light green) for its degradation by the ClpCP machinery (light orange and gray, respectively), keeping ComX concentration at a low level (post-translational regulation). When ComX abundance overcomes the degradation machinery (probably in a subpopulation), it can interact with the RNA polymerase (yellow) to trigger the expression of late com genes and, in turn, natural transformation. (B) Comparison of competence regulation at the global level between Lactococcus, Streptococcus, and Bacillus. In Streptococcus (left panel), the proximal regulation of comX is performed by a quorum sensing (QS) system (ComRS or ComCDE, green). Additionally, distal regulation systems (e.g., TCSs, blue) responding to environmental cues are modulating the expression of the QS response regulator/sensor instead of comX as found in L. lactis. Finally, the post-translational regulation system (grey) plays a minor role in ComX degradation. In Bacillus (right panel), the proximal regulation of ComK is achieved by a range of transcriptional regulators (e.g., CodY, DegU, Rok, and AbrB). However, a distal regulation involving a QS system (ComXAP) triggers the production of ComS to inhibit ComK degradation in B. subtilis. In Lactococcus (middle panel), no QS system has been identified so far and proximal regulation of comX seems to be exclusively influenced by environmental conditions. Moreover, post-translational regulation was shown to be dominant in competence repression.
Among the competence triggers in L. lactis, we identified carbon nutritional status as a key signal. While spontaneous transformation in rich medium with glucose is activated at the entry of the stationary phase, it is abolished in CDM-glucose conditions (Fig 1B and 1C). However, growth on alternative sugars mostly found in plants reactivates spontaneous competence at a low level in those conditions (Fig 1C). This makes sense as strain DGCC12653 is a maize isolate with a high potential to catabolize alternative non-PTS (phosphoenolpyruvate:carbohydrate phosphotransferase system) sugars such as maltose, xylose, and arabinose [47]. This is also reminiscent of the activation of competence by non-PTS sugars in Gram-negative bacteria [48]. For instance, competence in Vibrio cholerae is induced by chitin oligomers but repressed by PTS-sugars such as glucose through CCR via the cAMP-activated CRP protein [25]. In Gram-positive bacteria, the impact of sugars and the implication of CcpA-mediated CCR on competence development have not been deeply studied [49–51]. Notably, our results showed that a diauxic shift between glucose and the non-PTS sugar maltose activates comX expression and transformability at the growth shift between the two sugars (Figs 2A and S1D). They also showed that glucose starvation and the presence of maltose are both required to maximize spontaneous transformation (Fig 1C). This corroborates previous observations showing that glucose starvation alone was unable to activate natural transformation in L. lactis, although a range of competence genes (including comX) were upregulated [22,26]. In addition, growth on maltose could be strongly delayed after glucose starvation from one experiment to the other without major effect on transformability. We hypothesize that the second sugar catabolism is required to maintain the energy demand needed for natural transformation [52]. Interestingly, fluorescence microscopy analyses revealed a bimodality in competence activation at the diauxic shift (S2 Fig). This suggests a bet-hedging strategy for cells that are developing alternative physiological adaptations, as previously reported for diauxic growth with L. cremoris [27] or during competence activation in B. subtilis [53]. In L. cremoris, CcpA-mediated CCR was previously shown to be involved in the diauxic shift due to glucose exhaustion [27]. The inactivation of CcpA in strain DGCC12653 drastically alters growth and the dynamics of comX expression, with a two-fold upregulation at an early growth stage compared to the wild-type (Fig 2B), potentially due to an absence of CcpA binding to the comX promoter. Similarly, a two-fold increase in comX expression was observed in a CcpA-negative strain of Streptococcus oligofermentans [50]. In this case, CcpA was shown to indirectly control comX due to a post-transcriptional effect on the expression of the communication system ComCDE [50]. However, CcpA inactivation in strain DGCC12653 leads to reduced transformability, as previously reported in different streptococci [51,54,55]. CcpA is a pleiotropic regulator that controls, among others, the central carbon metabolism and a range of cell envelope components in lactococci and streptococci [28,55,56], whose constitutive inactivation could negatively alter the transformation process independently of an upshift in comX expression.
Besides the impact of the carbon source, the nitrogen status sensed by the global regulator CodY has a major impact on competence development in L. lactis. In this species, CodY is a global regulator involved in nitrogen metabolism (e.g., peptide and amino acid uptake, peptide degradation, de novo biosynthesis of amino acids, purine biosynthesis) that senses the intracellular pool of BCAA (mostly isoleucine) [29–31]. We showed that an excess of isoleucine in the growth medium repressed comX expression and spontaneous transformation (Fig 3A and 3B). We also identify that a lack of aspartate and glutamate in the growth medium improves transformability (Fig 1D). As it was previously reported that both amino acids could be catabolized for their conversion into BCAA [57], we hypothesized that their removal could reduce the intracellular pool of BCAA and consequently CodY-mediated repression. In addition, CodY inactivation has strong effects, with a ~10-fold higher activation of comX, a higher proportion of fluorescent cells post-diauxic shift, and a ~100-fold increase in transformability (Figs 3C-3E and S2). This resembles the impact of CodY on the direct transcriptional repression of comK [58] and the importance of the amino-acid balance on transformability in B. subtilis [59]. Finally, we showed that CodY directly binds the comX promoter, a region that was also identified by CHIPseq experiments as bound by CodY in L. lactis but without being further investigated [60]. Although previously suggested in L. lactis [22], these results demonstrate that nitrogen sensing through CodY is central for the metabolic control of competence in this species.
In our search for competence-related quorum-sensing systems, we first scanned the genome for orthologous systems known in other Gram-positive species (S3 Table). As no such system was found, we systematically knocked out every Rgg (ten genes) and TCS (eight pairs of genes) and identified CovRS (TcsA in L. cremoris) as a major player in competence regulation in L. lactis. As found for CodY inactivation, its deletion dramatically increases comX expression, the proportion of fluorescent cells post-diauxic shift, and natural transformation (Figs 4A-4C and S2). We also showed that CovRS directly controls comX transcription in L. lactis, as confirmed by EMSA and RNAseq experiments (Figs 4E and S4). Interestingly, we recently showed that this system indirectly controls comX expression in salivarius streptococci by modulating the ComRS signaling system via comR repression [34]. CovRS is a well-known regulatory system of virulence genes in pathogenic streptococci, responding to various stimuli (e.g., [Mg++], defensins, osmotic stress, pH) [35,36,39,40,61]. In L. cremoris, CovR (LlrA) was shown to be involved in acid stress resistance through arginine catabolism [62]. Interestingly, we showed that competence sensitivity to low pH was abolished in the CovRS-deficient strain (Fig 4D), corroborating its involvement in acid stress resistance. Besides CovRS, no other TCS or Rgg is involved in the repression or activation of the comX gene in strain DGCC12653 (S5A-S5D Fig). As all identified quorum-sensing systems regulating competence in Gram-positive bacteria belong to those two families [7–9], this questioned the presence of a cell-to-cell communication system for the control of comX expression in L. lactis. A similar situation was observed in Bacillus licheniformis, where competence regulation does not involve the ComXAP-like quorum-sensing system found in B. subtilis but seems to rely solely on global regulators for the control of comK expression [46,63,64].
Besides the proximal transcriptional regulation of comX by global sensors, competence in L. lactis is tightly controlled at the post-translational level through ComX degradation. Indeed, codY or covRS deletion in three L. lactis strains other than DGCC12653 activates comX transcription at a high level (S8 Fig) but without unleashing natural transformation. We identified a natural substitution (L125R) in the adaptor protein MecA of strain DGCC12653 that is responsible for its unique behavior regarding spontaneous transformability in laboratory conditions. Moreover, a systematic analysis of more than 600 L. lactis genomes confirmed that this MecA variant is unique to this specific strain. This corroborates previous results on strains developing high levels of spontaneous transformation in laboratory conditions that are frequently harboring mutated competence regulators such as CovR in S. thermophilus [34] or global repressors in B. licheniformis [63]. The L125R substitution is located in the linker region between the N- and C-terminal macrodomains of MecA. We previously reported that this region was required for the interaction between MecA and ComX in S. thermophilus [42]. Thus, we hypothesize that the presence of a charged residue at that position in MecA impairs the interaction with ComX and its subsequent degradation by the MecA-ClpCP machinery. This layer of control through ComX degradation is of major importance for competence development in L. lactis, as MecA inactivation in a range of strains boosted spontaneous transformation (Fig 5C). In B. subtilis, the ComS peptide encoded by the srf operon activates competence by blocking MecA-ComK interaction, thus avoiding ComK degradation by ClpCP [16–18,65,66]. If a similar mechanism is at work in L. lactis, it remains to be discovered.
To conclude, our work highlights a different model of competence regulation in L. lactis (and possibly L. cremoris) than in closely-related streptococci. In this model, the accumulation of the master competence regulator ComX required for natural transformation is under the direct negative control of global nutritional/stress sensors and the Clp degradation machinery. This work paves the way towards a better understanding of the diversity of regulatory networks controlling competence development in Gram-positive bacteria and offers opportunities to use natural transformation as a tool for genome engineering in this lactic acid bacterium of major biotechnological importance.
Materials and methods
Bacterial strains, plasmids, and oligonucleotides
Bacterial strains, plasmids, and oligonucleotides used in this study are listed and described in supplemental material (S4–S6 Tables).
Growth conditions
L. lactis strains were cultivated in M17 (Difco Laboratories, Detroit, MI) or CDM (adapted from [24]) supplemented with 0.5% (w/v) glucose (M17G and CDMG, respectively), or supplemented with 0.21% (w/v) glucose and 1.1% (w/v) secondary sugar (CDM*) when required, at 30°C without agitation. Glutamate, aspartate, xanthine, adenine, uracil, and guanine were omitted in the competence-optimized CDM (CDM*-DEB). Escherichia coli EC1000 was cultivated at 37°C in LB (Lysogeny Broth) medium with agitation. Solid agar plates were prepared by adding 2% (w/v) agar to the medium. When required, 5 μg ml-1 of erythromycin, 1 mg ml-1 of streptomycin, 0.5 mg ml-1 of spectinomycin, and/or 10 μg ml-1 of chloramphenicol were added to the medium for L. lactis; and 250 μg ml-1 of erythromycin, 250 μg ml-1 of ampicillin, and 10 μg ml-1 of chloramphenicol for E. coli.
DNA techniques and electrotransformation
Electrotransformations of L. lactis [67] and E. coli [68] were performed as previously described. Chromosomal DNA of L. lactis used as template for PCR was extracted as previously described [69]. PCRs were performed using the Q5 DNA polymerase (NEB) in a GeneAmp PCR system 2400 (Applied Biosystems).
Isolation of a rpsL* mutant conferring resistance to streptomycin
Spontaneous streptomycin-resistant IL1403 mutants were isolated on 1 mg ml-1 streptomycin-containing plates. After the sequencing of the rpsL gene with primers BlD-RpsLUnivUp/BlD-RpsLUnivDown, one spontaneous mutant containing the substitution K56R into the ribosomal protein S12, which was previously shown to confer resistance to streptomycin, was selected [20]. A 3.7-kb fragment containing the rpsL mutated gene (strA2 allele, named rpsL*) was amplified by PCR with primers BlD-LLcfusARpsL/BlD-LLldacARpsL and cloned into the pGEM-T Easy vector (Promega), yielding plasmid pGEM-rpsL*. This plasmid was used as template to generate the 3.7-kb PCR product with primers BlD-LLcfusARpsL/BlD-LLldacARpsL that was used as donor DNA in natural transformation assays.
Xylose-induced comX expression and natural transformation
The L. lactis strains containing plasmid pGhPxylT-comX (PxylT and comX from L. lactis IO-1) were grown overnight at 30°C. Cells were washed twice in distilled water, and OD600 was adjusted to 0.1 in M17 supplemented with 1% (w/v) xylose and erythromycin. Typically, 2 μg of DNA were added to 100 μl of inoculated transformation medium, and the culture was further incubated during 24 hours at 30°C. Cells were then spread on M17G agar plates supplemented with appropriate antibiotics, and CFUs were counted after 48 hours of incubation at 30°C. The transformation frequency was calculated as the number of antibiotic-resistant CFU ml-1 divided by the total number of viable CFU ml-1.
Spontaneous transformation assays
After an overnight preculture in M17G at 30°C of L. lactis DGCC12653 (and other strains or mutants), 100 μl of the preculture was diluted in 900 μl of fresh M17G to restart the culture. After 2 hours of growth, cells were washed twice in distilled water and inoculated to a final OD600 of 0.1 in competence-activating medium (CDM* or CDM*-DEB). Typically, 2.0 μg of donor DNA were added to 100 μl of transformation medium, and the culture was further incubated for 20 hours at 30°C. Cells were then spread on M17G agar plates supplemented or not with appropriate antibiotics and CFUs were counted after 48 hours of incubation. The transformation frequency was calculated as reported above. The transfer of the mutation conferring streptomycin resistance was confirmed by DNA sequencing of the rpsL gene after its amplification by PCR using primers BlD-RpsLUnivUp/BlD-RpsLUnivDown.
Detection of absorbance and luminescence
Overnight precultures in M17G were washed with sterile water and inoculated in the adequate medium to a final OD600 of 0.1. After inoculation in 300 μl-culture volumes and before luminescence analysis, nonyl-aldehyde was diluted 100 times in mineral oil, and 50 μl of this preparation was disposed between the wells of a white 96-well plate with a transparent bottom (Greiner, Alphen a/d Rijn, The Netherlands). Growth (OD600) and luciferase activity (Lux) were monitored at 5-minute intervals in a Hidex Sense microplate reader (Hidex, Lemminkäisenkatu, Finland) for a maximum of 24 hours. The luciferase activity is expressed in relative light units (RLU) and the specific luciferase activity in RLU OD600-1.
Construction of competence reporter plasmids
The PcomX-luxAB reporter plasmid for luminescence assays was constructed as follows. The comX promoter was amplified by PCR from chromosomal DNA of L. lactis DGCC12653 with primers FT886_PcomX_fw/FT887_PcomX_rec_rv. The plasmid containing the luxAB genes pGhPcomGA[MG]-luxAB [20] was amplified by PCR using primers FT884_pGhPcomXlux_fw/FT925_PcomXlux_rec_rv. Both PCR fragments were joined by the Gibson assembly method [70] and the resulting plasmid was named pGhPcomX-luxAB. The PcomGA-luxAB reporter plasmid for luminescence assays was constructed as follows. The comGA promoter was amplified by PCR from chromosomal DNA of L. lactis IO-1 using primers LuxcIOF1_XhoI/LuxIOR1. The plasmid pGhPcomGA[MG]-luxAB was amplified by PCR using primers LuxcIOR2_XhoI/LuxIOF2. Both PCR fragments were joined by overlap PCR, digested by XhoI, and self-ligated using T4 DNA ligase. The resulting plasmid was named pGhPcomGA[IO]-luxAB. The PcomX-gfpsf reporter plasmid for fluorescence microscopy was constructed as follows. The comX promoter was amplified by PCR from chromosomal DNA of L. lactis DGCC12653 using primers FT886_PcomX_fw/FT968_sfGFP_rec. The gfpsf gene was amplified by PCR from plasmid DNA pDR111_sfGFP(Bs) [71] using primers FT967_sfGFP_fw/FT969_sfGFP_rv_SacII. The plasmid pG+host9 was amplified using primers FT970_pGh_SacII/FT925_PcomX_rec_rv. The three PCR fragments were joined by overlapping PCR, digested using SacII, and self-ligated using T4 DNA ligase. The resulting plasmid was named pGhPcomX-gfpsf.
Construction of deletion mutants in L. lactis strains
The comX, comEC, ccpA, codY, covRS, and mecA genes as well as Rgg and TCS encoding genes were similarly inactivated by the exchange of their coding sequences with either P32-cat or spc resistance cassette (conferring chloramphenicol or spectinomycin resistance, respectively) using double crossing-over events. For this purpose, overlapping PCR products containing the resistance cassette flanked by two recombination arms of ~1.2 kb (upstream and downstream homologous regions) were generated as previously reported [20]. Briefly, upstream, downstream, and resistance-cassette fragments were separately amplified by PCR, purified, mixed in equimolecular amount, and assembled by overlapping PCR by using the most external primers (see S6 Table). Typically, 5 μg of the obtained overlapping PCR product were used as donor DNA for xylose-induced natural transformation of the L. lactis strains harboring plasmid pGhPxylT-comX. The correct insertion of the resistance cassette in each targeted locus of the transformants was validated by PCR (see S6 Table). To obtain the final mutant strains, the curing of the thermosensitive vector pGhPxylT-comX was performed by growing the cells 16 hours at 37°C without erythromycin. The cultures were subsequently diluted and plated on M17G agar without erythromycin at 30°C. The resulting colonies were streaked in parallel on M17G plates with and without erythromycin. The absence of plasmid pGhPxylT-comX in erythromycin-sensitive clones was validated by PCR. To generate the double codY covRS mutant, codY was inactivated by exchange with the spc cassette. Then, the high transformability of the ΔcovRS::cat mutant was used to transfer the ΔcodY::spc mutation in order to obtain the double mutant.
Swapping of mecA genes
The mecA mutants (ΔmecA::cat) of strains DGCC12653 and DGCC12671 harboring plasmid pGhPxylT-comX were used for this exchange as they display high transformation rates. The mecA genes from strains DGCC12653 and DGCC12671 were amplified by PCR with primers FT1319/FT1320, generating ~5-kb fragments used as donor DNA. Replacement of the ΔmecA::cat deletion by the counterpart mecA gene was performed by xylose-induced natural transformation as described above. Five hundred isolated colonies were streaked in parallel on M17G-agar plates containing or not chloramphenicol, searching for antibiotic-sensitive transformants. Then, the swapping of mecA genes from sensitive clones was validated by PCR and sequencing. Finally, the curing of the pGhPxylT-comX plasmid was performed as described above.
Construction of strain DGCC12653 nisRK for nisin-induced overexpression
The nisRK genes from strain IO-1 were inserted between genes DGCC12653_06785 and DGCC12653_06790 using double crossing-over events. For this purpose, an overlapping PCR product containing nisRK genes associated with the spc cassette in opposite orientation and flanked by two recombination arms of ~1.2 kb was generated as reported above (see S6 Table). The obtained overlapping PCR product (5 μg) was used as donor DNA for natural transformation of strain DGCC12653 harboring plasmid pGhPxylT-comX. The correct insertion of nisRK genes in the targeted locus of the transformants was validated by PCR. To obtain the final mutant strain, the curing of the thermosensitive vector pGhPxylT-comX was performed as described above.
Purification of transcriptional regulators
The overexpression plasmids were constructed as follows. The ccpA, codY, and covR genes flanked by an N-terminal 6-His tag encoding sequence were amplified by PCR from strain DGCC12653. For regulator overexpression, pBAD (for ccpA) or pNZ8048 (for codY and covR) was amplified by PCR and joined to the regulator-encoding genes by the Gibson assembly method (see S6 Table) [70]. The final constructs, pBAD-6his-ccpA, pNZ8048-6his-codY, and pNZ8048-6his-covR were validated by sequencing. The 6His-CcpA (rCcpA) protein was overexpressed in E. coli Top10. The 6His-CodY (rCodY) and 6His-CovR (rCovR) were overexpressed in L. lactis DGCC12653 nisRK (nisin induction with 1 ng ml-1). The recombinant proteins were purified using Ni-NTA resin according to the manufacturer’s instructions (ProBond Resin, Novex).
Electromobility shift assays (EMSA)
The whole promoter region of comX (PcomX, intergenic region) from DGCC12653 was amplified by PCR using fluorescent-labeled primers (FT851_Cy3_PcomX_up/FT853_Cy5_PcomX_dw). Shorter versions of PcomX were amplified by PCR using either FT851_Cy3_PcomX_up or FT853_Cy5_PcomX_dw in combination with a non-fluorescent primer located inside PcomX (see S6 Table). The PcomX fragments harboring the mutated or native version of the CodY-box were amplified by PCR using primer pairs FT1369_PcomX_codY_mute/FT853_Cy5_PcomX_dw and FT1368_PcomX_codY_control/FT853_Cy5_PcomX_dw, respectively. In each assay, the purified proteins were incubated in 2-fold serial dilutions with a constant quantity of labeled probe (2.5 ng μl-1). As a negative control, a 150-bp DNA sequence located in the CDS region of the dnaE gene was amplified by PCR using primers AK350/AK303 [34]. CodY-binding assays were performed in CodY buffer (Tris-HCl 20 mM pH 7.5, NaCl 150 mM, BSA 1 mg ml-1, EDTA 1 mM, glycerol 10% [w/v]) at 30°C for 20 minutes. CcpA-binding assays were performed in CodY buffer supplemented with glucose-6-phosphate (25 mg ml-1) and poly-dIdC (2 μg ml-1) as reported above. CovR-binding assays were performed in Buffer-CovR (NaPO4 50 mM pH 6.5, NaCl 50 mM, MgCl2 1 mM, CaCl2 1 mM, DTT 1 mM, poly-dIdC (2 μg ml-1), glycerol 10% [w/v]) at 30°C for 15 minutes. Prior to incubation with the DNA probe, rCovR was incubated in CovR buffer supplemented with 50 mM acetyl phosphate for 30 minutes at 20°C. Samples were migrated in native poly-acrylamide gels in non-denaturing MOPS buffer, and revealed by the Amersham Typhoon device.
Microscopy
M17G overnight cultures (containing adequate antibiotics) were diluted (1:10) in fresh M17G at 30°C for 2 hours. Cells were then washed in water and inoculated at an OD600 of 0.1 in CDM*-DEB until reaching the comX-expression peak, determined in parallel with similar clones harboring the pGhPcomX-luxAB reporter plasmid. Cells were then centrifuged, resuspended in 50 μl of PBS and observed on agarose pads composed of 1% agarose and PBS buffer [34]. Images were obtained using an Axio I inverted microscope (Zeiss) equipped with a Plan-Apochromat objective (100 ×/1.46 oil differential interference contrast [DIC] M27) (Zeiss), a HXP 120 C lighting unit (Zeiss), and a C10600 ORCA-R2 camera (Hamamatsu). GFP fluorescence was detected with filter set 38 HE, displaying bandpass excitation at 470/40 nm and bandpass emission at 525/50 nm (Zeiss). Images were analyzed using ZenPro software (Zeiss) and MicrobeJ [72].
RNA sequencing of L. lactis mutants
RNA extraction and sequencing were performed on the wild type, ΔcodY mutant, and ΔcovRS mutant of strain DGCC12653. Each mutant and the wild-type control were grown in their respective efficient competence-activating medium (CDM*-DEB and CDM* for ΔcodY and ΔcovRS, respectively) and harvested at their respective diauxic shift. RNA was extracted using the RNeasy plus bacteria kit (Qiagen) according to manufacturer indications. Total RNA was checked for integrity with an RNA Nano chip (Agilent Technologies) and sent for Illumina sequencing (GeneWiz). Raw data were processed on the Galaxy server (use.galaxy.org) using Bowtie2 algorithm to yield BAM files containing the read coordinates and Seqmonk to count the number of reads per coding sequence (CDS). The dataset was exported into an Excel file for further analyses. First, the dataset was standardized to CDS-mapped reads per ~10 million overall reads. Then, we estimated the ratio of CDS-mapped reads in mutants vs. wild type.
Supporting information
(A) Transformability upon ComX overexpression. Data show transformation rates of 16 DGCC strains and strain 1AA59 (negative control) harboring the pGhPxylT-comX plasmid in M17GX (0.1% glucose-1% xylose). All transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture. Dots show the values for technical replicates (n = 3) ± standard deviations. (B) Transformability of 18 DGCC strains in M17G medium. Conditions for transformation assays as in panel A. Dots show the values for technical replicates (n = 3) ± standard deviations. (C) Effects of glucose-maltose combinations on spontaneous transformation of strain DGCC12653 WT in CDM. A design of experiment (composite central plan) was setup using the JMP Pro software. Sugar concentrations were varied from 0 to 2% (w/v) in 12 different conditions (glucose %/Maltose %; 0.01/0, 0.05/0.1, 0.3/0.1, 0/0.25, 0.2/0.25, 0.1/0.5, 0.5/0.5, 0.05/1.0, 0.3/1.0, 0/2.0, 0.2/2.0, and 0.4/2.0) containing donor DNA. Compilation of transformation rates was treated with JMP Pro and a response surface was generated. (D) Growth curves (OD600) of strain DGCC12653 WT in CDM-DEB supplemented with glucose 0.5% [G 0.5%], glucose 0.21% [G 0.21%], maltose 1.1% [M 1.1%], and a mixture of glucose 0.21% and maltose 1.1% [G0.21%-M 1.1%].
(TIF)
(A) Density plot of single-cell fluorescence intensity (arbitrary unit [AU]) for WT, CodY-, CovRS- strains harboring the reporter fusion PcomX-gfpsf (green, orange and blue, respectively) and the negative control (NC) harboring the empty plasmid (gray). Cells were cultured in CDM*-DEB, harvested at the diauxic shift, and analyzed by epifluorescence microscopy. The fluorescence of more than 1,200 individual cells was examined in each experiment. A minimum threshold of significant fluorescence (2.81 AU) was determined as the maximum signal reached by the negative control. Percentages in the top of the plot indicate the percentage of the cell population displaying a fluorescence signal higher than 2.81 for each strain. (B) Representative images of experiments depicted in panel A for two channels (left, phase contrast [PC] and right, sfGFP signal).
(JPG)
(A) Effect of ccpA deletion on transformability. Data show transformation rates observed for WT (green) and CcpA- (light brown) strains in CDM*-DEB after overnight culture with donor DNA. Transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture. Dots show the values for biological triplicates (CcpA-) or technical triplicates (WT), mean values ± standard deviations. The mean value is indicated on the top each bar. (B) CcpA purification. SDS-PAGE of the elution step [E] of 6His-CcpA (rCcpA) purified from E. coli. MW, molecular weight (kDa). The star indicates the enriched rCcpA. (C) Mapping of fluorescent (Cy3_PcomX_up, Cy5_PcomX_dw) and non-fluorescent (X5 Rv, X1 Fw, X3 Fw, X2 Fw, X3 Fw, and X4 Fw) primers designed in PcomX (complete intergenic region). The region containing three cre-boxes (underlined) is surrounded in green. The alignment of the three cre-boxes with the consensus is shown with conserved positions in capitals. (D) Mapping of the different probes used for EMSAs. Green and red lines indicate the presence or absence of a band shift. (E) EMSAs performed with a gradient of purified CcpA (rCcpA) on the different probes shown in panel D. Lanes without rCcpA are indicated by a minus sign. C and P indicate the CcpA-DNA complex(es) and the unbound probe, respectively.
(TIF)
(A) CodY purification. SDS-PAGE of the elution step [E] of the 6His-CodY (rCodY) purified from L. lactis. MW, molecular weight (kDa). The star indicates the enriched rCodY. (B) EMSAs performed with a gradient of purified CodY (rCodY) on PcomX in absence (left side of each panel) or presence (right side of each panel) of 5 mM of GTP (top left panel), Ile (top right panel), Leu (lower left panel) or Val (lower right panel). Lanes without rCodY are indicated by a minus sign. C and P indicate rCodY-PcomX complex and unbound probe, respectively. (C) Mapping of RNAseq reads on PcomX, from WT (top panel) and CodY- (lower panel) strains. Cells were harvested at the diauxic shift in CDM*-DEB. The PcomX (complete intergenic region) is illustrated as a black line, while CodY-, cre- and CovR-boxes are localized and displayed in red, green and blue, respectively.
(TIF)
(A and B) Effects on transformability (A) and PcomX activity (B) of the independent inactivation of 10 Rgg from DGCC12653. (C and D) Effects on transformability (C) and PcomX activity (D) of the independent inactivation of 7 TCS from DGCC12653 (labels indicate the first gene of the TCS). Data on transformability (panels A and C) show transformation rates observed for WT and mutant strains in CDM*-DEB after overnight culture with donor DNA. Transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture. Data on PcomX activity (panels B and D) show maximum specific luciferase activity (Max RLU/OD600) observed at the diauxic shift in CDM*-DEB. Dots (panels A to D) show the values for biological triplicates (Rgg- and TCS- strains) or technical triplicates (WT), mean values ± standard deviations. Statistical t test was performed for each mutant strain in comparison to the WT (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
(TIF)
Growth (dotted lines–OD600) and kinetics of PcomX specific luciferase activity (continuous lines–RLU/OD600) monitored over time for CodY- (blue), CovRS- (red) and CodY- CovRS- (brown) mutant strains. Continuous and dotted dark lines are the mean values of biological triplicates, and light lines are standard deviations.
(TIF)
(A) CovR purification. SDS-PAGE of the elution step [E] of the 6His-CovR (rCovR) purified from L. lactis. MW, molecular weight (kDa). The star indicates the enriched rCovR. (B) EMSAs performed with a gradient of purified CovR (rCovR) without (left) or with (right) in vitro phosphorylation with acetyl-P on PcomX. Lanes without rCovR are indicated by a minus sign. C and P indicate the rCovR-DNA complex(es) and the unbound probe, respectively. (C) Mapping of fluorescent (Cy3_PcomX_up, Cy5_PcomX_dw) and non-fluorescent (X5 Rv, X6 Rv, X2 Fw, X3 Fw, and X4 Fw) primers designed in PcomX (complete intergenic region). The regions containing CovR-boxes (underlined) are surrounded in blue. (D) Mapping of the different probes used for EMSAs. Green and red lines indicate the presence or absence of a band shift. (E) EMSAs performed with a gradient of purified CovR (rCovR) on the different probes shown in panel C. Lanes without rCovR are indicated by a minus sign. C and P indicate the rCovR-DNA complex(es) and the unbound probe, respectively. (F) Mapping of RNAseq reads on PcomX, from WT (top panel) and CovRS- (lower panel) strains. Cells were harvested at the diauxic shift in CDM*. The PcomX (complete intergenic region) is illustrated as a black line, while CodY-, cre- and CovR-boxes are localized and displayed in red, green, and blue, respectively.
(TIF)
The effects of these deletions (CodY- in CDM*-DEB, light red bars and CovRS- in CDM*, blue bars) were compared to their respective WT in the same culture conditions (WT in CDM*, dark green bars and CDM*-DEB, light green bars). Data show maximum specific luciferase activity (Max RLU/OD600) observed at the diauxic shift either in CDM*-DEB or CDM*. Dots show the values for biological triplicates (CodY- and CovR- strains) or technical triplicates (WT), mean values ± standard deviations.
(TIF)
The nucleotide sequences correspond to the intergenic regions located upstream of comX (PcomX) in the genomes of the 18 DGCC strains used in this work, the laboratory strain IL1403, and strain KF147 shown to be transformable by comX overexpression. Strains DGCC12653, DGCC12651, DGCC12671 and DGCC12678 in red were selected for codY and covRS inactivation. Red bars indicate putative -10 boxes of vegetative promoters (P1 to P4). CodY-, cre- and CovR-boxes are surrounded in red, green, and blue, respectively. The alignment was performed using Clustal Omega.
(PDF)
The L125R substitution is in red and underlined in MecA from strain DGCC12653. Strains DGCC12651, DGCC12671 and DGCC12678 contain the common MecA-L125 residue. The alignment was performed using Clustal Omega.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We warmly thank the technical assistance of Marie-Christine Eloy and Sylvie Derclaye for microscopy experiments. We thank Jan-Willem Veening for the Addgene depository of plasmid pDR111_sfGFP(Bs).
Data Availability
The draft genome sequence of L. lactis subsp. lactis DGCC12653 has been deposited at DDBJ/ENA/GenBank under accession number JAZICS000000000. All RNAseq data were deposited in the GEO database under accession number GSE255719. Large scale datasets have been released and all numerical data (including statistical analyses) are available in S1 data.
Funding Statement
This study was funded by IFF-Danisco France SAS (https://bioscience.iff.com/; funds allocated to Pa.H). This work was also supported by the PDR grant T.0110.18 (awarded to Pa.H) from the Belgian National Fund for Scientific Research (FNRS, https://www.frs-fnrs.be/en/) and the Concerted Research Actions (ARC) grant 17/22-084 (awarded to Pa.H) from Federation Wallonia-Brussels (FWB, http://www.recherchescientifique.be/). The funders had no role in study design, data collection and analysis, or preparation of the manuscript. The authors declare that IFF-Danisco France SAS as main funder authorized the publication of this work.
References
- 1.Song AA, In LLA, Lim SHE, Rahim RA. A review on Lactococcus lactis: from food to factory. Microb Cell Fact. 2017. Apr 4; 16(1):55. 10.1186/s12934-017-0669-x [pii];669 [pii]; doi: 10.1186/s12934-017-0669-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morello E, Bermudez-Humaran LG, Llull D, Sole V, Miraglio N, Langella P et al. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol. 2008; 14(1–3):48–58. 000106082 [pii]; doi: 10.1159/000106082 [DOI] [PubMed] [Google Scholar]
- 3.Bahey-El-Din M, Gahan CG. Lactococcus lactis: from the dairy industry to antigen and therapeutic protein delivery. Discov Med. 2010. May; 9(48):455–61. [PubMed] [Google Scholar]
- 4.Tavares LM, de Jesus LCL, da Silva TF, Barroso FAL, Batista VL, Coelho-Rocha ND et al. Novel Strategies for Efficient Production and Delivery of Live Biotherapeutics and Biotechnological Uses of Lactococcus lactis: The Lactic Acid Bacterium Model. Front Bioeng Biotechnol. 2020; 8517166. doi: 10.3389/fbioe.2020.517166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004. Mar; 2(3):241–9. nrmicro844 [pii]; doi: 10.1038/nrmicro844 [DOI] [PubMed] [Google Scholar]
- 6.Di Giacomo S., Toussaint F, Ledesma-Garcia L, Knoops A, Vande CF, Fremaux C et al. Expanding natural transformation to improve beneficial lactic acid bacteria. FEMS Microbiol Rev. 2022. Jul 20; 46(4). 6543703 [pii];fuac014 [pii]; doi: 10.1093/femsre/fuac014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006; 60451–75. doi: 10.1146/annurev.micro.60.080805.142139 [DOI] [PubMed] [Google Scholar]
- 8.Fontaine L, Wahl A, Flechard M, Mignolet J, Hols P. Regulation of competence for natural transformation in streptococci. Infect Genet Evol. 2015. Jul; 33343–60. S1567-1348(14)00328–1 [pii]; doi: 10.1016/j.meegid.2014.09.010 [DOI] [PubMed] [Google Scholar]
- 9.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014. Mar; 12(3):181–96. nrmicro3199 [pii]; doi: 10.1038/nrmicro3199 [DOI] [PubMed] [Google Scholar]
- 10.O’Connell LM, Kelleher P, van Rijswijck IMH, de WP, van Peij NNME, Mahony J et al. Natural Transformation in Gram-Positive Bacteria and Its Biotechnological Relevance to Lactic Acid Bacteria. Annu Rev Food Sci Technol. 2022. Mar 25; 13409–31. doi: 10.1146/annurev-food-052720-011445 [DOI] [PubMed] [Google Scholar]
- 11.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P et al. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol. 2010. Mar; 192(5):1444–54. JB.01251-09 [pii]; doi: 10.1128/JB.01251-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010. Nov; 78(3):589–606. doi: 10.1111/j.1365-2958.2010.07361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996. Aug; 21(4):853–62. doi: 10.1046/j.1365-2958.1996.501417.x [DOI] [PubMed] [Google Scholar]
- 14.Martin B, Soulet AL, Mirouze N, Prudhomme M, Mortier-Barriere I, Granadel C et al. ComE/ComE~P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol Microbiol. 2013. Jan; 87(2):394–411. doi: 10.1111/mmi.12104 [DOI] [PubMed] [Google Scholar]
- 15.Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J Bacteriol. 1993. May; 175(10):3182–7. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Souza C, Nakano MM, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci U S A. 1994. Sep 27; 91(20):9397–401. doi: 10.1073/pnas.91.20.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura M, Liu L, Lacelle M, Nakano MM, Zuber P. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol Microbiol. 1999. May; 32(4):799–812. doi: 10.1046/j.1365-2958.1999.01399.x [DOI] [PubMed] [Google Scholar]
- 18.Prepiak P, Dubnau D. A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP. Mol Cell. 2007. Jun 8; 26(5):639–47. S1097-2765(07)00311-5 [pii]; doi: 10.1016/j.molcel.2007.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003. Jan; 149(Pt 1):9–17. [DOI] [PubMed] [Google Scholar]
- 20.David B, Radziejwoski A, Toussaint F, Fontaine L, de Frahan MH, Patout C et al. Natural DNA Transformation Is Functional in Lactococcus lactis subsp. cremoris KW2. Appl Environ Microbiol. 2017. Aug 15; 83(16). AEM.01074-17 [pii];01074–17 [pii]; doi: 10.1128/AEM.01074-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder J, Wels M, Kuipers OP, Kleerebezem M, Bron PA. Unleashing Natural Competence in Lactococcus lactis by Induction of the Competence Regulator ComX. Appl Environ Microbiol. 2017. Oct 15; 83(20). AEM.01320-17 [pii];01320–17 [pii]; doi: 10.1128/AEM.01320-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ercan O, Wels M, Smid EJ, Kleerebezem M. Genome-wide transcriptional responses to carbon starvation in nongrowing Lactococcus lactis. Appl Environ Microbiol. 2015. Apr; 81(7):2554–61. AEM.03748-14 [pii]; doi: 10.1128/AEM.03748-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol. 2000. Nov; 182(21):6192–202. 0552 [pii]; doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci U S A. 1999. Aug 3; 96(16):8985–90. 1367 [pii]; doi: 10.1073/pnas.96.16.8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012. Aug; 14(8):1898–912. doi: 10.1111/j.1462-2920.2011.02689.x [DOI] [PubMed] [Google Scholar]
- 26.Redon E, Loubiere P, Cocaign-Bousquet M. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J Bacteriol. 2005. May; 187(10):3589–92. 187/10/3589 [pii];1209–04 [pii]; doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solopova A, van GJ, Weissing FJ, Bachmann H, Teusink B, Kok J et al. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci U S A. 2014. May 20; 111(20):7427–32. 1320063111 [pii];201320063 [pii]; doi: 10.1073/pnas.1320063111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol. 2007. Feb; 189(4):1366–81. JB.01013-06 [pii];1013–06 [pii]; doi: 10.1128/JB.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J, Kuipers OP. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem. 2005. Oct 7; 280(40):34332–42. S0021-9258(20)79023-X [pii]; doi: 10.1074/jbc.M502349200 [DOI] [PubMed] [Google Scholar]
- 30.Guedon E, Serror P, Ehrlich SD, Renault P, Delorme C. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol Microbiol. 2001. Jun; 40(5):1227–39. mmi2470 [pii]; doi: 10.1046/j.1365-2958.2001.02470.x [DOI] [PubMed] [Google Scholar]
- 31.Guedon E, Sperandio B, Pons N, Ehrlich SD, Renault P. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology (Reading). 2005. Dec; 151(Pt 12):3895–909. doi: [DOI] [PubMed] [Google Scholar]
- 32.Wydau S, Dervyn R, Anba J, Dusko ES, Maguin E. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol Lett. 2006. Apr; 257(1):32–42. FML141 [pii]; doi: 10.1111/j.1574-6968.2006.00141.x [DOI] [PubMed] [Google Scholar]
- 33.Takada H, Yoshikawa H. Essentiality and function of WalK/WalR two-component system: the past, present, and future of research. Biosci Biotechnol Biochem. 2018. May; 82(5):741–51. doi: 10.1080/09168451.2018.1444466 [DOI] [PubMed] [Google Scholar]
- 34.Knoops A, Vande CF, Fontaine L, Verhaegen M, Mignolet J, Goffin P et al. The CovRS Environmental Sensor Directly Controls the ComRS Signaling System To Orchestrate Competence Bimodality in Salivarius Streptococci. mBio. 2022. Jan 4; 13e0312521. doi: 10.1128/mbio.03125-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SE, Jiang S, Wessels MR. CsrRS and environmental pH regulate group B streptococcus adherence to human epithelial cells and extracellular matrix. Infect Immun. 2012. Nov; 80(11):3975–84. IAI.00699-12 [pii];00699–12 [pii]; doi: 10.1128/IAI.00699-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santi I, Grifantini R, Jiang SM, Brettoni C, Grandi G, Wessels MR et al. CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J Bacteriol. 2009. Sep; 191(17):5387–97. JB.00370-09 [pii];0370–09 [pii]; doi: 10.1128/JB.00370-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Churchward G. The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci. Mol Microbiol. 2007. Apr; 64(1):34–41. MMI5649 [pii]; doi: 10.1111/j.1365-2958.2007.05649.x [DOI] [PubMed] [Google Scholar]
- 38.Federle MJ, Scott JR. Identification of binding sites for the group A streptococcal global regulator CovR. Mol Microbiol. 2002. Mar; 43(5):1161–72. 2810 [pii]; doi: 10.1046/j.1365-2958.2002.02810.x [DOI] [PubMed] [Google Scholar]
- 39.Froehlich BJ, Bates C, Scott JR. Streptococcus pyogenes CovRS mediates growth in iron starvation and in the presence of the human cationic antimicrobial peptide LL-37. J Bacteriol. 2009. Jan; 191(2):673–7. JB.01256-08 [pii];1256–08 [pii]; doi: 10.1128/JB.01256-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gryllos I, Grifantini R, Colaprico A, Jiang S, Deforce E, Hakansson A et al. Mg(2+) signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol Microbiol. 2007. Aug; 65(3):671–83. MMI5818 [pii]; doi: 10.1111/j.1365-2958.2007.05818.x [DOI] [PubMed] [Google Scholar]
- 41.Tian XL, Dong G, Liu T, Gomez ZA, Wahl A, Hols P et al. MecA protein acts as a negative regulator of genetic competence in Streptococcus mutans. J Bacteriol. 2013. Nov; 195(22):5196–206. JB.00821-13 [pii]; doi: 10.1128/JB.00821-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahl A, Servais F, Drucbert AS, Foulon C, Fontaine L, Hols P. Control of natural transformation in salivarius Streptococci through specific degradation of sigmaX by the MecA-ClpCP protease complex. J Bacteriol. 2014. Aug; 196(15):2807–16. JB.01758-14 [pii]; doi: 10.1128/JB.01758-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boutry C, Wahl A, Delplace B, Clippe A, Fontaine L, Hols P. Adaptor protein MecA is a negative regulator of the expression of late competence genes in Streptococcus thermophilus. J Bacteriol. 2012. Apr; 194(7):1777–88. JB.06800-11 [pii]; doi: 10.1128/JB.06800-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong G, Tian XL, Gomez ZA, Li YH. Regulated proteolysis of the alternative sigma factor SigX in Streptococcus mutans: implication in the escape from competence. BMC Microbiol. 2014. Jul 9; 14183. 1471-2180-14-183 [pii]; doi: 10.1186/1471-2180-14-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Son M, Kaspar J, Ahn SJ, Burne RA, Hagen SJ. Threshold regulation and stochasticity from the MecA/ClpCP proteolytic system in Streptococcus mutans competence. Mol Microbiol. 2018. Dec; 110(6):914–30. MMI13992 [pii]; doi: 10.1111/mmi.13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakobs M, Meinhardt F. What renders Bacilli genetically competent? A gaze beyond the model organism. Appl Microbiol Biotechnol. 2015. Feb; 99(4):1557–70. doi: 10.1007/s00253-014-6316-0 [DOI] [PubMed] [Google Scholar]
- 47.Cavanagh D, Fitzgerald GF, McAuliffe O. From field to fermentation: the origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiol. 2015. May; 4745–61. S0740-0020(14)00261–5 [pii]; doi: 10.1016/j.fm.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 48.Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev. 2013. May; 37(3):336–63. doi: 10.1111/j.1574-6976.2012.00353.x [DOI] [PubMed] [Google Scholar]
- 49.Underhill SAM, Shields RC, Burne RA, Hagen SJ. Carbohydrate and PepO control bimodality in competence development by Streptococcus mutans. Mol Microbiol. 2019. Nov; 112(5):1388–402. doi: 10.1111/mmi.14367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Cai J, Shang N, Zhu L, Shao N, Dong X et al. The carbon catabolite repressor CcpA mediates optimal competence development in Streptococcus oligofermentans through post-transcriptional regulation. Mol Microbiol. 2019. Aug; 112(2):552–68. doi: 10.1111/mmi.14274 [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Zhang J, Xiao J, Wang H, Yang R, Guo X et al. comCDE (Competence) Operon Is Regulated by CcpA in Streptococcus pneumoniae D39. Microbiol Spectr. 2023. Jun 15; 11(3):e0001223. 00012–23 [pii];spectrum.00012-23 [pii]; doi: 10.1128/spectrum.00012-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaccaria E, Wells JM, van BP. Metabolic Context of the Competence-Induced Checkpoint for Cell Replication in Streptococcus suis. PLoS One. 2016; 11(5):e0153571. PONE-D-15-49063 [pii]; doi: 10.1371/journal.pone.0153571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leisner M, Stingl K, Radler JO, Maier B. Basal expression rate of comK sets a ’switching-window’ into the K-state of Bacillus subtilis. Mol Microbiol. 2007. Mar; 63(6):1806–16. MMI5628 [pii]; doi: 10.1111/j.1365-2958.2007.05628.x [DOI] [PubMed] [Google Scholar]
- 54.Tong H, Zhu B, Chen W, Qi F, Shi W, Dong X. Establishing a genetic system for ecological studies of Streptococcus oligofermentans. FEMS Microbiol Lett. 2006. Nov; 264(2):213–9. FML453 [pii]; doi: 10.1111/j.1574-6968.2006.00453.x [DOI] [PubMed] [Google Scholar]
- 55.Zheng L, Chen Z, Itzek A, Herzberg MC, Kreth J. CcpA regulates biofilm formation and competence in Streptococcus gordonii. Mol Oral Microbiol. 2012. Apr; 27(2):83–94. doi: 10.1111/j.2041-1014.2011.00633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One. 2011; 6(10):e26707. PONE-D-11-13661 [pii]; doi: 10.1371/journal.pone.0026707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dressaire C, Redon E, Gitton C, Loubiere P, Monnet V, Cocaign-Bousquet M. Investigation of the adaptation of Lactococcus lactis to isoleucine starvation integrating dynamic transcriptome and proteome information. Microb Cell Fact. 2011. Aug 30; 10 Suppl 1(Suppl 1):S18. 1475-2859-10-S1-S18 [pii]; doi: 10.1186/1475-2859-10-S1-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serror P, Sonenshein AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996. Oct; 178(20):5910–5. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968. Apr; 95(4):1439–49. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu H, Tian K, Feng J, Qi H, Qiao J. Systems-Level Analysis of the Global Regulatory Mechanism of CodY in Lactococcus lactis Metabolism and Nisin Immunity Modulation. Appl Environ Microbiol. 2022. Mar 8; 88(5):e0184721. 01847–21 [pii];aem.01847-21 [pii]; doi: 10.1128/AEM.01847-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004. Jun; 186(12):3928–37. 186/12/3928 [pii];1611–03 [pii]; doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Connell-Motherway M, van SD, Morel-Deville F, Fitzgerald GF, Ehrlich SD, Morel P. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology (Reading). 2000. Apr; 146 (Pt 4)935–47. doi: 10.1099/00221287-146-4-935 [DOI] [PubMed] [Google Scholar]
- 63.Jakobs M, Hoffmann K, Grabke A, Neuber S, Liesegang H, Volland S et al. Unravelling the genetic basis for competence development of auxotrophic Bacillus licheniformis 9945A strains. Microbiology (Reading). 2014. Oct; 160(Pt 10):2136–47. doi: 10.1099/mic.0.079236-0 [DOI] [PubMed] [Google Scholar]
- 64.Jakobs M, Hoffmann K, Liesegang H, Volland S, Meinhardt F. The two putative comS homologs of the biotechnologically important Bacillus licheniformis do not contribute to competence development. Appl Microbiol Biotechnol. 2015. Mar; 99(5):2255–66. doi: 10.1007/s00253-014-6291-5 [DOI] [PubMed] [Google Scholar]
- 65.Hamoen LW, Eshuis H, Jongbloed J, Venema G, van SD. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol Microbiol. 1995. Jan; 15(1):55–63. [DOI] [PubMed] [Google Scholar]
- 66.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998. Nov 16; 17(22):6730–8. doi: 10.1093/emboj/17.22.6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holo H, Nes IF. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989. Dec; 55(12):3119–23. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988. Jul 11; 16(13):6127–45. doi: 10.1093/nar/16.13.6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferain T, Hobbs JN Jr., Richardson J, Bernard N, Garmyn D, Hols P et al. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J Bacteriol. 1996. Sep; 178(18):5431–7. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009. May; 6(5):343–5. nmeth.1318 [pii]; doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 71.Overkamp W, Beilharz K, Detert Oude WR, Solopova A, Karsens H, Kovacs A et al. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl Environ Microbiol. 2013. Oct; 79(20):6481–90. AEM.02033-13 [pii];02033–13 [pii]; doi: 10.1128/AEM.02033-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016. Jun 20; 1(7):16077. nmicrobiol201677 [pii]; doi: 10.1038/nmicrobiol.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Transformability upon ComX overexpression. Data show transformation rates of 16 DGCC strains and strain 1AA59 (negative control) harboring the pGhPxylT-comX plasmid in M17GX (0.1% glucose-1% xylose). All transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture. Dots show the values for technical replicates (n = 3) ± standard deviations. (B) Transformability of 18 DGCC strains in M17G medium. Conditions for transformation assays as in panel A. Dots show the values for technical replicates (n = 3) ± standard deviations. (C) Effects of glucose-maltose combinations on spontaneous transformation of strain DGCC12653 WT in CDM. A design of experiment (composite central plan) was setup using the JMP Pro software. Sugar concentrations were varied from 0 to 2% (w/v) in 12 different conditions (glucose %/Maltose %; 0.01/0, 0.05/0.1, 0.3/0.1, 0/0.25, 0.2/0.25, 0.1/0.5, 0.5/0.5, 0.05/1.0, 0.3/1.0, 0/2.0, 0.2/2.0, and 0.4/2.0) containing donor DNA. Compilation of transformation rates was treated with JMP Pro and a response surface was generated. (D) Growth curves (OD600) of strain DGCC12653 WT in CDM-DEB supplemented with glucose 0.5% [G 0.5%], glucose 0.21% [G 0.21%], maltose 1.1% [M 1.1%], and a mixture of glucose 0.21% and maltose 1.1% [G0.21%-M 1.1%].
(TIF)
(A) Density plot of single-cell fluorescence intensity (arbitrary unit [AU]) for WT, CodY-, CovRS- strains harboring the reporter fusion PcomX-gfpsf (green, orange and blue, respectively) and the negative control (NC) harboring the empty plasmid (gray). Cells were cultured in CDM*-DEB, harvested at the diauxic shift, and analyzed by epifluorescence microscopy. The fluorescence of more than 1,200 individual cells was examined in each experiment. A minimum threshold of significant fluorescence (2.81 AU) was determined as the maximum signal reached by the negative control. Percentages in the top of the plot indicate the percentage of the cell population displaying a fluorescence signal higher than 2.81 for each strain. (B) Representative images of experiments depicted in panel A for two channels (left, phase contrast [PC] and right, sfGFP signal).
(JPG)
(A) Effect of ccpA deletion on transformability. Data show transformation rates observed for WT (green) and CcpA- (light brown) strains in CDM*-DEB after overnight culture with donor DNA. Transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture. Dots show the values for biological triplicates (CcpA-) or technical triplicates (WT), mean values ± standard deviations. The mean value is indicated on the top each bar. (B) CcpA purification. SDS-PAGE of the elution step [E] of 6His-CcpA (rCcpA) purified from E. coli. MW, molecular weight (kDa). The star indicates the enriched rCcpA. (C) Mapping of fluorescent (Cy3_PcomX_up, Cy5_PcomX_dw) and non-fluorescent (X5 Rv, X1 Fw, X3 Fw, X2 Fw, X3 Fw, and X4 Fw) primers designed in PcomX (complete intergenic region). The region containing three cre-boxes (underlined) is surrounded in green. The alignment of the three cre-boxes with the consensus is shown with conserved positions in capitals. (D) Mapping of the different probes used for EMSAs. Green and red lines indicate the presence or absence of a band shift. (E) EMSAs performed with a gradient of purified CcpA (rCcpA) on the different probes shown in panel D. Lanes without rCcpA are indicated by a minus sign. C and P indicate the CcpA-DNA complex(es) and the unbound probe, respectively.
(TIF)
(A) CodY purification. SDS-PAGE of the elution step [E] of the 6His-CodY (rCodY) purified from L. lactis. MW, molecular weight (kDa). The star indicates the enriched rCodY. (B) EMSAs performed with a gradient of purified CodY (rCodY) on PcomX in absence (left side of each panel) or presence (right side of each panel) of 5 mM of GTP (top left panel), Ile (top right panel), Leu (lower left panel) or Val (lower right panel). Lanes without rCodY are indicated by a minus sign. C and P indicate rCodY-PcomX complex and unbound probe, respectively. (C) Mapping of RNAseq reads on PcomX, from WT (top panel) and CodY- (lower panel) strains. Cells were harvested at the diauxic shift in CDM*-DEB. The PcomX (complete intergenic region) is illustrated as a black line, while CodY-, cre- and CovR-boxes are localized and displayed in red, green and blue, respectively.
(TIF)
(A and B) Effects on transformability (A) and PcomX activity (B) of the independent inactivation of 10 Rgg from DGCC12653. (C and D) Effects on transformability (C) and PcomX activity (D) of the independent inactivation of 7 TCS from DGCC12653 (labels indicate the first gene of the TCS). Data on transformability (panels A and C) show transformation rates observed for WT and mutant strains in CDM*-DEB after overnight culture with donor DNA. Transformation assays were performed with rpsL* as donor DNA (20 μg ml-1), added at time zero. Cells were spread after ~24 hours of culture. Data on PcomX activity (panels B and D) show maximum specific luciferase activity (Max RLU/OD600) observed at the diauxic shift in CDM*-DEB. Dots (panels A to D) show the values for biological triplicates (Rgg- and TCS- strains) or technical triplicates (WT), mean values ± standard deviations. Statistical t test was performed for each mutant strain in comparison to the WT (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
(TIF)
Growth (dotted lines–OD600) and kinetics of PcomX specific luciferase activity (continuous lines–RLU/OD600) monitored over time for CodY- (blue), CovRS- (red) and CodY- CovRS- (brown) mutant strains. Continuous and dotted dark lines are the mean values of biological triplicates, and light lines are standard deviations.
(TIF)
(A) CovR purification. SDS-PAGE of the elution step [E] of the 6His-CovR (rCovR) purified from L. lactis. MW, molecular weight (kDa). The star indicates the enriched rCovR. (B) EMSAs performed with a gradient of purified CovR (rCovR) without (left) or with (right) in vitro phosphorylation with acetyl-P on PcomX. Lanes without rCovR are indicated by a minus sign. C and P indicate the rCovR-DNA complex(es) and the unbound probe, respectively. (C) Mapping of fluorescent (Cy3_PcomX_up, Cy5_PcomX_dw) and non-fluorescent (X5 Rv, X6 Rv, X2 Fw, X3 Fw, and X4 Fw) primers designed in PcomX (complete intergenic region). The regions containing CovR-boxes (underlined) are surrounded in blue. (D) Mapping of the different probes used for EMSAs. Green and red lines indicate the presence or absence of a band shift. (E) EMSAs performed with a gradient of purified CovR (rCovR) on the different probes shown in panel C. Lanes without rCovR are indicated by a minus sign. C and P indicate the rCovR-DNA complex(es) and the unbound probe, respectively. (F) Mapping of RNAseq reads on PcomX, from WT (top panel) and CovRS- (lower panel) strains. Cells were harvested at the diauxic shift in CDM*. The PcomX (complete intergenic region) is illustrated as a black line, while CodY-, cre- and CovR-boxes are localized and displayed in red, green, and blue, respectively.
(TIF)
The effects of these deletions (CodY- in CDM*-DEB, light red bars and CovRS- in CDM*, blue bars) were compared to their respective WT in the same culture conditions (WT in CDM*, dark green bars and CDM*-DEB, light green bars). Data show maximum specific luciferase activity (Max RLU/OD600) observed at the diauxic shift either in CDM*-DEB or CDM*. Dots show the values for biological triplicates (CodY- and CovR- strains) or technical triplicates (WT), mean values ± standard deviations.
(TIF)
The nucleotide sequences correspond to the intergenic regions located upstream of comX (PcomX) in the genomes of the 18 DGCC strains used in this work, the laboratory strain IL1403, and strain KF147 shown to be transformable by comX overexpression. Strains DGCC12653, DGCC12651, DGCC12671 and DGCC12678 in red were selected for codY and covRS inactivation. Red bars indicate putative -10 boxes of vegetative promoters (P1 to P4). CodY-, cre- and CovR-boxes are surrounded in red, green, and blue, respectively. The alignment was performed using Clustal Omega.
(PDF)
The L125R substitution is in red and underlined in MecA from strain DGCC12653. Strains DGCC12651, DGCC12671 and DGCC12678 contain the common MecA-L125 residue. The alignment was performed using Clustal Omega.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The draft genome sequence of L. lactis subsp. lactis DGCC12653 has been deposited at DDBJ/ENA/GenBank under accession number JAZICS000000000. All RNAseq data were deposited in the GEO database under accession number GSE255719. Large scale datasets have been released and all numerical data (including statistical analyses) are available in S1 data.