Abstract
Continued advances in metabolic engineering are increasing the number of small-molecules being targeted for microbial production. Pathway yields and productivities, however, are often suboptimal and strain improvement remains a persistent challenge given that the majority of small-molecules are both difficult to screen for and their biosynthesis does not improve host fitness. In this work, we have developed a generalized approach to screen or select for improved small-molecule biosynthesis using transcription factor-based biosensors. Using a tetracycline resistance gene 3′ of a small-molecule inducible promoter, host antibiotic resistance, and hence growth rate, was coupled to either small-molecule concentration in the growth medium or a small-molecule production phenotype. Biosensors were constructed for two important chemical classes, dicarboxylic acids and alcohols, using transcription factor-promoter pairs derived from Pseudomonas putida, Thauera butanivorans, or E. coli. Transcription factors were selected for specific activation by either succinate, adipate, or 1-butanol, and we demonstrate product-dependent growth in E. coli using all three compounds. The 1-butanol biosensor was applied in a proof-of-principle liquid culture screen to optimize 1-butanol biosynthesis in engineered E. coli, identifying a pathway variant yielding a 35% increase in 1-butanol specific productivity through optimization of enzyme expression levels. Lastly, to demonstrate the capacity to select for enzymatic activity, the 1-butanol biosensor was applied as synthetic selection, coupling in vivo 1-butanol biosynthesis to E. coli fitness, and an 120-fold enrichment for a 1-butanol production phenotype was observed following a single round of positive selection.
The potential for microbes to convert inexpensive sugar feedstocks into structurally diverse, industrially important products is a well-recognized opportunity in metabolic engineering1. Technical hurdles slowing proof-of-principle biosynthetic process demonstration are rapidly being removed by advances in metabolic pathway design and construction. For example, the accelerating pace of genome sequencing, gene annotation, and enzyme characterization has enabled assembly of biological databases2–5 that functionally serve as catalogs of enzyme-catalyzed reactions; along the same lines, pathway design algorithms6–8 facilitate assembly of enzyme activities into de novo biosynthetic routes to novel compounds. Attendant to advances in pathway design are a decreased cost of DNA synthesis and improved cloning techniques9–11, both of which enable rapid translation of in silico pathway designs to proof of principle small-molecule biosynthesis.
As is near universally the case, the initial pathway and host strain require extensive optimization with regard to product yields, productivities, and titers before being industrially relevant. In contrast to synthetic chemistry, in which catalysis is optimized in a well-defined system, optimization of microbial transformations is severely hindered by the complexity of the living biological system. Directed evolution, in which a non-natural selective pressure is applied to a diverse pool of target genetic sequences to identify a desired trait, is a hallmark of metabolic engineering efforts to improve biological transformations. The success of any directed evolution strategy is contingent upon the effectiveness of two key steps: first, generating large, diverse genotypic libraries; and second, effectively identifying the desired phenotype from a heterogeneous population. To date, the ability to generate genotypic diversity far outstrips the ability to efficiently and effectively interrogate the resulting library. In vitro methods for incorporation of either random or targeted mutations into user-specified plasmid DNA sequences are numerous and well-developed12; library size is limited only by the host transformation efficiency and can exceed 1010 variants. Complementing traditional, plasmid-based libraries are advances in genome engineering techniques, including multiplex genome engineering (MAGE)13, global transcription machinery engineering (gTEM)14, and multiscale analysis of library enrichment (SCALEs)15. These next-generation technologies have greatly expanded the capacity to generate genomic diversity and investigate the role of both individual genes as well as combinatorial effects when numerous loci are varied simultaneously.
The potential for genetic diversity to yield process improvements, however, is only realized when the target phenotype can be screened or selected for. For this reason advances in directed evolution approaches within the context of metabolic engineering have been near exclusively applied to improving host resistance to a toxic product16,17 or overproduction of the natural chromophore lycopene13,18,19. In contrast, the majority of small-molecules being targeted for overproduction today – including fatty acids20, diols and diamines21,22, and short-chain alcohols23 among others – are inconspicuous and cannot be directly screened or selected for. More universally applicable gas and liquid chromatography techniques are well utilized in the field, but the throughputs (102-103 variants per machine per day) fall far short of levels necessary for effective interrogation of large genetic libraries.
In nature, the need for sensitive, specific, small-molecule detection and response has been addressed, in part, through evolution and selection for ligand-responsive transcription factors and their cognate promoters. Transcription factor-promoter pairs are archetypal genetic devices within the synthetic biology paradigm24,25; abundant in nature, highly modular, and capable of being evolved or re-engineered, transcription factor-based devices are well suited for a broad range of applications. While engineered transcription factor-based biosensors have been employed for detection of exogenous environmental pollutants – thoroughly reviewed elsewhere26 – this work has only recently been explored in the context of metabolic engineering27,28.
We seek herein to apply these concepts to construct and characterize transcription factor-based small-molecule screens and synthetic selections (defined herein as non-natural selections mediated through activity of an engineered genetic device). We demonstrate the robustness of the approach by characterizing biosensor mediated coupling of small-molecule product concentration to host growth rate using three biosensors from different genetic backgrounds to detect either medium-chain (C4-C7) alcohols or dicarboxylic acids. We then focus on a heterologously expressed 1-butanol biosynthetic pathway for proof-of-principle demonstration of biosensor-based screening and synthetic selection in E. coli. Biosynthetic pathways to medium-chain alcohols proceeding through either reduction of acetoacetyl-CoA29–32 or decarboxylation and reduction of 2-keto acids33–35 have been the subjects of intensive research because of the end products’ roles as next-generation biofuel alternatives to ethanol. Furthermore, high-throughput detection of alcohols remains a persistent challenge in the field. Screens for detection of aldehydes have been reported36,37 and could serve as indirect alcohol assays, but these methods require enzymatic conversion of the alcohol to the more reactive aldehyde, are highly non-specific, and difficult to scale. Biosensor-mediated product detection provides an alternative, flexible method to assay for 1-butanol, or other small-molecule, production phenotypes.
Results and Discussion
Engineering 1-butanol-dependent growth in E. coli
To couple intracellular 1-butanol concentration to E. coli growth rate we first constructed a biosensor based on a 1-butanol responsive transcription factor-promoter pair controlling expression of a tetracycline resistance reporter protein. While no native alcohol-responsive transcription factor has been characterized in E. coli to date, a putative σ54-transcriptional activator (BmoR) and a σ54-dependent, alcohol-regulated promoter (PBMO) were previously identified in Pseudomonas butanovora (later reclassified as Thauera butanivorans sp. nov.38) 5′ of an n-alkane catabolic operon39. BmoR contains the three archetypal domains common to members of the σ54 enhancer-binding protein family, namely the sensory, ATPase, and helix-turn-helix DNA-binding domains40. The exact mechanism of BmoR ligand detection and signal transduction, however, was unknown. Numerous σ54-dependent promoters are regulated by two-component systems, requiring co-expression of a sensory histidine kinase and cytoplasmic regulatory protein for promoter activation. Two-component systems can be identified by the presence of a phosphorylation site, comprised of three acidic amino acids, in the sensory domain of the cytoplasmic regulatory protein40. Sequence alignment of BmoR against both E. coli and Pseudomonas two-component system σ54-transcriptional activators indicated an absence of this phosphorylation motif (Supplementary Fig. 1), leading us to conclude BmoR functions independent of a sensory histidine kinase. Supporting lines of analysis include absence of an annotated sensory histidine kinase proximal to the n-alkane catabolic operon, and observation that medium-chain alcohols are membrane permeable, thereby negating a physical requirement for signal transduction.
Butanol-dependent growth using the BmoR-PBMO device was first demonstrated in engineered E. coli using a previously described tetA-gfp gene fusion41 (plasmid pSelect#1, Table 1). A TetA-GFP fusion protein was chosen as the reporter in order to simultaneously acquire both growth rate- and fluorescence-based measurements of biosensor activation. The tetA gene encodes for a tetracycline/H+ antiporter and was chosen as the selectable marker for two reasons. First, as a transport protein, TetA does not catalyze antibiotic degradation and high selective pressure can be maintained over time. Second, by supplementing the growth medium with nickel-chloride, which selects against TetA activity42, in place of tetracycline the same biosensor construct can be used for negative selection against spurious promoter activation. A negative control plasmid, pBMO#1, was constructed identically to pSelect#1 with the exception that gfp, which encodes green fluorescent protein, was cloned in place of the tetA-gfp reporter 3′ of PBMO. Because pBMO#1 lacks a tetracycline resistance cassette, E. coli harboring this plasmid should not grow after addition of tetracycline to the growth medium. The performance of all biosensor constructs was tested in engineered E. coli DH1 containing an adhE deletion (ΔadhE) to mitigate the potential for unwanted biosensor activation due to host ethanol production.
Table 1.
| Strain | Genotype | Source |
|---|---|---|
| DH1 | endA1 recA1 thi-1 gyrA96 glnV44 relA1 hsdR17 (rk− mk+) λ− | ATCC |
| K12 | F’ proA+B+
lacIq ΔlacZM15/fhuA2 Δ(lac-proAB) glnV gal R(zgb-210::Tn10)TetS endA1 thi-1 Δ(hsdS-mcrB)5 |
NEB |
| DH1 ΔadhE | DH1 ΔadhE | This study |
| JAD-1 | DH1 ΔadhE (ilvD-ilvC)::neo | This study |
| JAD-2 | DH1 ΔadhE Δ(ilvD-ilvC) | This study |
| Plasmid | Description | Resistance | Origin | Source |
|---|---|---|---|---|
| pBMO#1 | PBmoR-bmoR, PBMO-gfp | CbR | ColE1 | This study |
| pSelect#1 | PBmoR-bmoR, PBMO-tetA-gfp | CbR | ColE1 | This study |
| pSelect#2 | PBmoR-bmoR, PBMO-tetA | CbR | ColE1 | This study |
| pC6Select#1 | PPcaR-pcaR | CmR | pSC101 | This study |
| pC6Select#2 | PpcaIJ-tetA | CbR | ColE1 | This study |
| pC4Select#1 | PdctA-tetA | CbR | ColE1 | This study |
| pKivD#1* | Ptrc-kivD.ADH6 | CmR | p15a | This study |
| pKivD#1-S71* | Ptrc-kivD.ADH6; optimized RBS | CmR | p15a | This study |
| pKivD#2 | PLacO1-kivD.ADH6 | CmR | pSC101 | This study |
| pPDC | PLacO1-PDC.ADH6 | CmR | pSC101 | This study |
| pPDC(I472A) | PLacO1-PDC(I472A).ADH6 | CmR | pSC101 | This study |
| pRFPA2 | Ptrc-rfp | CmR | p15a | 61 |
| pRFPS2 | PLacO1-rfp | CmR | pSC101 | 61 |
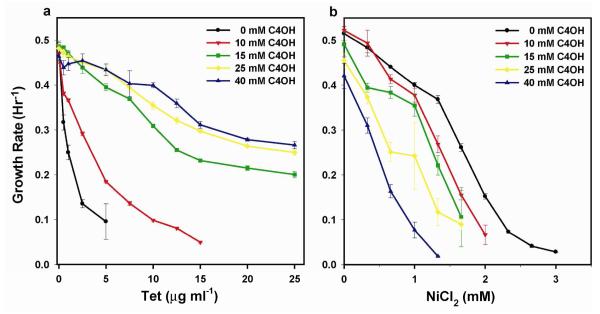
Growth rate measurements of engineered E. coli transformed with pSelect#1 or pBMO#1 were acquired in both positive (tetracycline) and negative (nickel-chloride) selection modes (Fig. 1). E. coli transformed with the GFP control plasmid pBMO#1 grew only in the absence of tetracycline and cell densities were below the detectable limit upon addition of tetracycline selective pressure; by comparison, E. coli transformed with pSelect#1 exhibited 1-butanol-dependent growth up to 40 mM concentration, at which point alcohol toxicity overwhelmed the positive selective pressure. In negative selection mode, the growth medium was supplemented with nickel-chloride, inverting the dependence between E. coli growth rate and 1-butanol concentration. In both positive and negative selection modes differences in host fitness, as measured by the change in growth rate, were amplified by increasing the selective pressure through further tetracycline or nickel-chloride supplementation, respectively. For example, at greater than 5 μg ml−1 tetracycline no measurable growth was observed in cultures absent 1-butanol supplementation; cultures with full biosensor activation, in contrast, maintained growth rates greater than 0.30 hr−1 for all tetracycline concentrations tested. Nickel-chloride proved a much more toxic selective agent to wild-type E. coli relative to tetracycline; thus fitness differences between E. coli with low and high 1-butanol supplementation were substantially decreased during negative selections. The results from both the positive and negative synthetic selection characterizations demonstrate an ability to consistently control E. coli fitness through modification of both inducer and selective agent concentrations.
Figure 1. Coupling 1-butanol-induced TetA expression to E. coli growth rate.
E. coli DH1 ΔadhE harboring tetA-gfp under control of PBMO displayed butanol-dependent changes in growth phenotype. (a) In positive selection mode, tetracycline (Tet) supplementation resulted in a positive correlation between E. coli growth rate and exogenous 1-butanol concentration. 1- butanol concentrations greater than 40 mM overwhelmed the synthetic selective pressure. (b) In negative selection mode, nickel-chloride (NiCl2) supplementation inverted the correlation between growth rate and exogenous 1-butanol concentration. Data are mean (s.d.) (n=4).
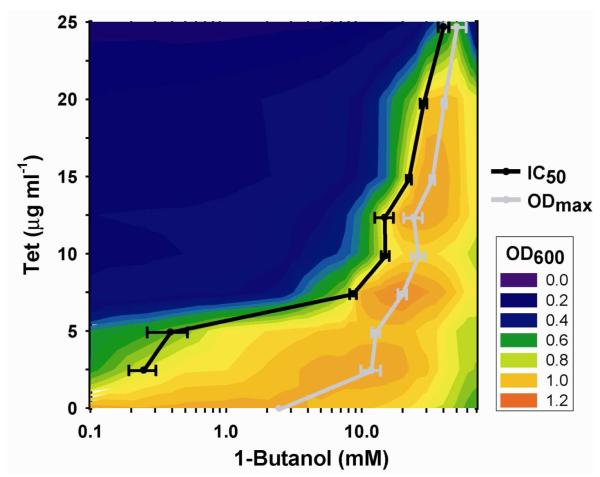
Endpoint measurements of E. coli culture density were also collected to derive biosensor transfer functions and study the effects of tetracycline concentration on high-throughput screening parameters (Fig. 2). A combined log-logistic mathematical model43 was used to fit the biphasic dose-response curves and derive values for the concentration of 1-butanol resulting in maximal (ODmax) and half-maximal () E. coli culture density. The was controlled over a three order-of-magnitude range, between 0.240 ± 0.05 mM and 38.0 ± 3.5 mM 1-butanol (n=4; mean ± s.d.), by increasing the tetracycline concentration in the culture broth. Similarly, the ODmax values increased from 11.9 ± 2.0 mM 1-butanol in the control culture absent tetracycline to 50.5 ± 8.7 mM 1-butanol under the highest tetracycline selective pressure (n=4; mean ± s.d.). The simultaneous increase in ODmax and values with increasing tetracycline concentration demonstrated an ability to control the linear range of detection. Furthermore, tetracycline supplementation proved a highly effective means of eliminating background signal, a feature of particular importance in high-throughput screening applications. Additional control over the screening parameters was realized by altering the time between addition of 1-butanol and tetracycline to the culture medium as well as the total incubation time (Supplementary Fig. 2). By modifying either the incubation time or tetracycline concentration we were able to finetune the assay to better control the linear range of detection, fold-induction, and background signal. In general, the biosensor was able to discriminate differences in 1-butanol concentration up to approximately 25 mM, at which point 1-butanol toxicity resulted in a decrease in cell density. Thus, as with more traditional physicochemical screens samples must be diluted to fall within the linear range of detection.
Figure 2. Modifying screening parameters through control of 1-butanol- and tetracycline-dependent E. coli growth.
Biosensor transfer functions in a 96-well plate, liquid culture screen were characterized, showing an increasing in half-maximal, (black line), and maximal, ODmax (grey line), cell density as tetracycline was supplemented to the growth medium. Increasing the tetracycline supplementation in the assay eliminated background E. coli growth and shifted the linear range of detection. and ODmax curves are mean (s.d.) (n=4). Heat plot of OD600 values are an average of 4 replicate cultures (%CV < 10% for all data points).
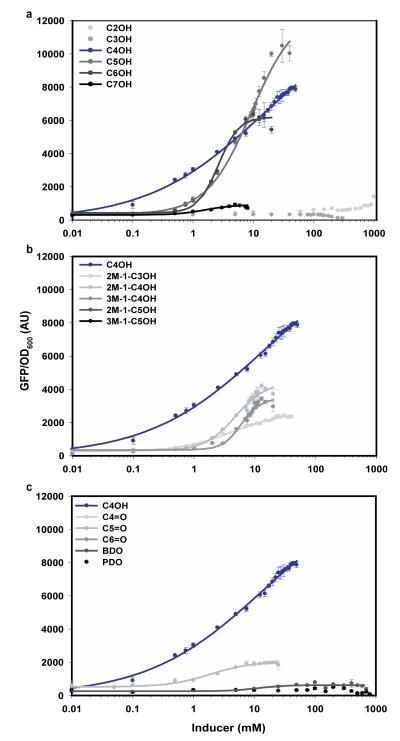
GFP fluorescence data resulting from TetA-GFP reporter expression was acquired concomitant to measurement of E. coli cell density with the goal of providing a secondary measure of promoter activation. In all conditions tested a positive correlation between GFP expression and 1-butanol concentration was observed under conditions of low tetracycline supplementation (less than or equal to 2.5 μg ml−1); however, further increases in tetracycline supplementation resulted in a loss of GFP fluorescence (Supplementary Fig. 3). Normalized fluorescence readings were also an order-of-magnitude lower than when using a GFP reporter alone. Fluorescence data were instead collected using the previously described plasmid pBMO#1, harboring a GFP reporter in place of the TetA-GFP selection device, and were used to derive biosensor performance features for a range of exogenously added small-molecule inducers, including linear- and branched-chain alcohols, aldehydes, and diols (Fig. 3). Small-molecule toxicity was observed for all compounds tested, resulting in a decrease in culture densities at high solvent concentrations (Supplementary Fig. 4). For this reason, only data up to the toxicity limit for each inducer were collected; data were fit with the Hill Equation to mathematically describe biosensor transfer functions, or dose-response curves, and derive biosensor performance features (Table 2). Key biosensor performance features include specificity (the slope of the transfer function in the linear region), dynamic range (the difference in output signal between high and low states), and linear range of detection. With regard to application as a high-throughput screen, a biosensor transfer function with a large dynamic range and high specificity enables more accurate discrimination of incremental increases in target analyte concentration. Biosensor transfer functions for the C4 to C6 linear alcohols yielded the largest dynamic range and specificity of all compounds tested; and in the case of 1-butanol, the linear range of detection exceeded three orders-of-magnitude. The Z’-factor (refer to Supplementary Materials and Methods) – a commonly used gauge of high-throughput screen quality based on both the variability within, and difference in signal between, positive and negative control samples – was 0.93 for 1-butanol; a Z’-factor greater than 0.50 indicates an excellent high-throughput screen44, and is reflective of the greater than forty standard deviations separating the two control populations. In addition to a broad linear range of detection and large dynamic range, the biosensor assay was also highly selective for C4 to C6 linear alcohols. Neither 1-propanol nor ethanol elicited a detectable increase in normalized GFP fluorescence, and biosensor response to the branched-chain alcohols and butyraldehyde was substantially lower than for their cognate linear alcohols.
Figure 3. Biosensor transfer functions show responsiveness to (a) linear alcohols, (b) branched-chain alcohols, and (c) aldehydes or diols.
The fluorescent response of E. coli DH1 ΔadhE harboring plasmid pBMO#1 was shown to be highly selective for C4-C6 linear alcohols and C3-C5 branched-chain alcohols. No response was observed for short-chain (C2-C3) linear alcohols or butyraldehyde, and only a slight increase in normalized fluorescence was observed with 1-heptanol. The linear range of detection for 1-butanol was 100 μM to 40 mM, the broadest of all inducers tested. Abbreviations: C2OH, ethanol; C3OH, 1-propanol; C4OH, 1-butanol; C5OH, 1-pentanol; C6OH, 1-hexanol; C7OH, 1-heptanol; 2M-1-C3OH, 2-methyl-1-propanol; 2M-1-C4OH, 2-methyl-1-butanol; 3M-1-C4OH, 3-methyl-1-butanol; 3M-1-C5OH, 3-methyl-1-pentanol; 4M-1-C5OH, 4-methyl-1-pentanol; C4=0, butyraldehyde; BDO, 1,4-butanediol; PDO, 1,5-pentanediol. Data are mean (s.d.) (n=3).
Table 2. Biosensor performance features.
Transfer functions were fit to the Hill Equation and used to derive biosensor performance features. High biosensor dynamic range and sensitivity facilitate resolution of small differences in inducer concentration. Km, the inducer concentration resulting in half-maximal induction, provides an indication of the linear range of detection. Selectivity, defined as the ratio of the 1-butanol Km relative to the target ligand Km, is a measure of whole-cell biosensor cross-reactivity. The Z’-factor (see Supplementary Materials for a complete description) provides a metric to gauge the effectiveness of a high-throughput screen; a Z-factor greater than 0.5 is generally regarded as an excellent screen.
| Compound | Dynamic range (GFPmax) |
Km (mM) |
Sensitivity | Linear range of detection (mM) |
Selectivity | Z’- Factor |
|---|---|---|---|---|---|---|
| C4OH | 8000 | 3.83 | 0.78 | 0.01 – 40 | 1.00 | 0.93 |
| C5OH | 10200 | 9.13 | 1.18 | 0.1 – 20 | 0.42 | 0.85 |
| C6OH | 5700 | 2.63 | 2.54 | 0.1 – 7.5 | 1.45 | 0.86 |
| C7OH | 600 | 1.62 | 1.86 | 0.5 – 3.0 | 2.36 | 0.76 |
| 2M-1-C3OH | 2400 | 4.34 | 1.01 | 0.1 – 20 | 0.88 | 0.91 |
| 2M-1-C4OH | 4100 | 4.82 | 1.74 | 0.01 – 10 | 0.79 | 0.80 |
| 3M-1-C4OH | 3100 | 6.00 | 2.84 | 0.1 – 11 | 0.64 | 0.88 |
| 3M-1-C5OH | 2800 | 3.93 | 1.26 | 1.0 – 4.0 | 0.97 | 0.85 |
| 4M-1-C5OH | 2300 | 3.97 | 1.67 | 1.0 – 5.0 | 0.96 | 0.84 |
| C4=0 | 1500 | 1.90 | 1.90 | 1.0 – 7.5 | 2.01 | 0.60 |
| BDO | 400 | 11.49 | 1.87 | 10 – 100 | 0.33 | −0.28 |
In addition to ligand selectivity, small-molecule screen performance is dictated by the biosensor transfer function and its associated features, including linear range of detection, dynamic range, sensitivity, and signal background. In the case of 1-butanol, the BmoR-PBMO transfer function was ideally suited for use as a high-throughput screen. A linear increase in reporter output was observed over a three order-of-magnitude 1-butanol concentration range, and that large dynamic range enabled accurate discrimination of differences in 1-butanol concentration as low as 100 μM.
Engineering dicarboxylic acid-dependent growth in E. coli
To demonstrate the broad applicability of our transcription factor mediated screening and selection strategy we expanded the repertoire of biosensors to include two dicarboxylic acid responsive transcription factor-promoter pairs. We selected the linear dicarboxylic acids succinate and adipate as our target ligands; as with 1-butanol, both molecules are industrially important chemicals that are difficult to screen for using existing technologies. Both succinate and adipate are subjects of metabolic engineering efforts to optimize their biosynthesis from renewable feedstocks45–47, and a method to selectively detect these diacids would facilitate development of these biological processes. Adipate is industrially manufactured from benzene, a toxic petroleum derived feedstock, and the manufacturing process releases nitric oxide, a greenhouse gas several orders-of-magnitude more potent than carbon dioxide48.
The β-ketoadipate responsive PcaR-PpcaIJ transcription factor-promoter pair from Pseudomonas putida, which has been previously characterized49,50, was chosen for the construction of the adipate biosensor. In the case of succinate, wild type E. coli possesses a C4-dicarboxylate responsive signal transduction pathway mediated by the DcuR/DcuS two-component system; we chose the previously characterized51, DcuR/DcuS-regulated dctA promoter (PdctA) for our biosensor design.
Both the adipate and succinate biosensors were built using the requisite transcription factor under control of its native promoter and a tetA reporter gene under control of the dicarboxylic acid regulated promoter. In particular, the adipate biosensor, comprising plasmids pC6select#1 and pC6select#2 (Table 1), was constructed using the native P. putida PcaR transcription factor, its native promoter, and the PcaR-regulated pcaIJ promoter (PpcaIJ) 5′ of tetA. The succinate biosensor comprises only one plasmid, pC4select#1, harboring tetA under control of PdctA. Thus, as with the 1-butanol biosensors, both adipate and succinate biosensors use tetracycline resistance to couple dicarboxylic acid concentration to E. coli growth.
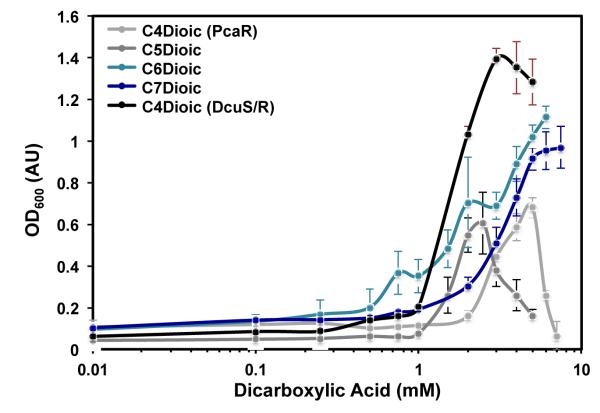
E. coli transformed with either the adipate or succinate biosensor exhibited dicarboxylic acid-dependent growth in the presence of positive tetracycline selective pressure (Fig. 4). Both biosensors were tested against C4 through C7 linear dicarboxylic acids (i.e., succinate, glutarate, adipate, and pimelate) under 25 μg/ml tetracycline selective pressure. The biosensors were highly responsive to their cognate ligand, and in the case of the PcaR-based biosensor, a strong response was also observed for pimelate. No E. coli growth was measured above background levels under any conditions tested when a negative control plasmid absent the tetA reporter cassette was assayed. Furthermore, the DcuR/DcuS-based biosensor was only responsive to succinate, and higher-chain length dicarboxylic acids did not enable E. coli growth. Both biosensors were responsive to their cognate dicarboxylic acid inducers over approximately an order-of-magnitude range (0.75-6 mM), and fold-induction varied from 6- to 15-fold for DcuR/DcuS- and PcaR-mediated dicarboxylic acid detection, respectively. While further optimization of the biosensor design may decrease basal tetA transcription and result in improved fold-induction measurements, the already low background signal should enable effective discrimination between non-producing and high-producing strains in a liquid culture assay format. As with the 1-butanol biosensor, we also found the dicarboxylic acid dose-response curves to be malleable (Supplementary Fig. 5), and by adjusting the tetracycline selective pressure the linear range of detection, fold-induction, and background signal could be defined.
Figure 4. Coupling dicarboxylic acid concentration to E. coli growth.
C4-C7 dicarboxylic acid biosensor transfer functions were obtained in liquid culture medium under 25 μg/ml tetracycline selective pressure. The PcaR biosensor produced the highest cell densities using adipate or pimelate; the weakest response was observed using succinate (sample C4Dioic (PcaR)). In the case of adipate, the linear range of detection was between 0.5 – 6 mM, and a 12-fold increase in cell density was observed between the maximally induced and uninduced samples. The E. coli-derived DcuS/DcuR two-component system only enabled growth upon addition of succinate to the growth medium (sample C4Dioic (DcuS/R)). Differences in response decreases at high succinate concentrations were observed comparing the DcuS/R- and PcaR-based biosensors and may be due to the increase in cell stress using the two plasmid PcaR system. Abbreviations: C4Dioic, succinate; C5Dioic, glutarate; C6Dioic, adipate; C7Dioic, pimelate. For succinate, parentheticals are used to indicate use of either the PcaR or DcuS/R based biosensors. Samples are mean (s.d.) (n=3).
In general, the dicarboxylic acid biosensors produced dose-response curves similar to those observed when assaying for 1-butanol but with a smaller linear range of detection and thus steeper slope within the linear range. These distinctions may be due to differences in small-molecule physico-chemical properties (e.g., membrane permeability) or inherent differences in the activation kinetics of each transcription factor-promoter pair. All biosensor designs, however, enabled E. coli growth to be coupled to exogenous small-molecule concentration.
Increasing E. coli 1-butanol biosynthesis using a biosensor-mediated screen
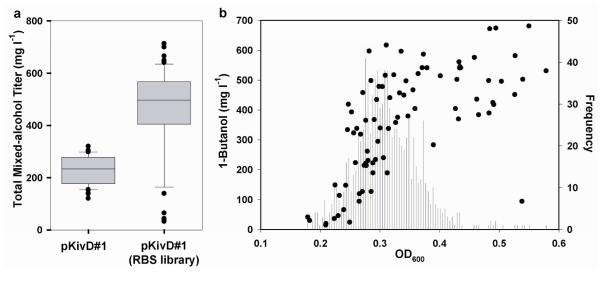
To demonstrate the applicability of the biosensors under high-throughput screening conditions we applied the 1-butanol biosensor to directed evolution of a heterologous biosynthetic pathway in E. coli. We chose the 2-keto acid-derived mixed alcohol production platform33–35,52 for alcohol biosynthesis in engineered E. coli. Through activity of the substrate promiscuous Lactococcus lactis 2-keto acid decarboxylase KivD and S. cerevisiae alcohol dehydrogenase ADH6, a variety of mixed alcohols can be diverted from native amino acid biosynthesis (Scheme 1). We assembled the synthetic, codon-optimized genes encoding for KivD and ADH6 on a vector containing an IPTG-inducible promoter and medium copy origin of replication (plasmid pKivD#1; Table 1). Upon transformation of E. coli DH1 ΔadhE with plasmid pKivD#1 accumulation of 2-keto acid-derived mixed alcohols, including 1-propanol, 1-butanol, and 3-methyl-1-butanol, was observed after 24 hours growth. In order to validate our high-throughput screen we desired a heterogeneous population exhibiting broad diversity in alcohol productivity and titer, therefore we mutated the KivD and ADH6 ribosome binding site (RBS) sequences on construct pKivD#1. The resulting total mixed-alcohol titers ranged from 32 to 713 mg l−1 (median of 497 mg l−1, n=50) in the RBS library population; comparatively, total mixed alcohol titers using the original pKivD#1 design ranged between 120 and 320 mg l−1 (median of 233 mg l−1, n=50; Fig. 5). Interestingly, the median mixed alcohol titer observed for the library population was substantially higher than for the original construct, indicating sub-optimal synthetic RBS sequences were used in the original vector design.
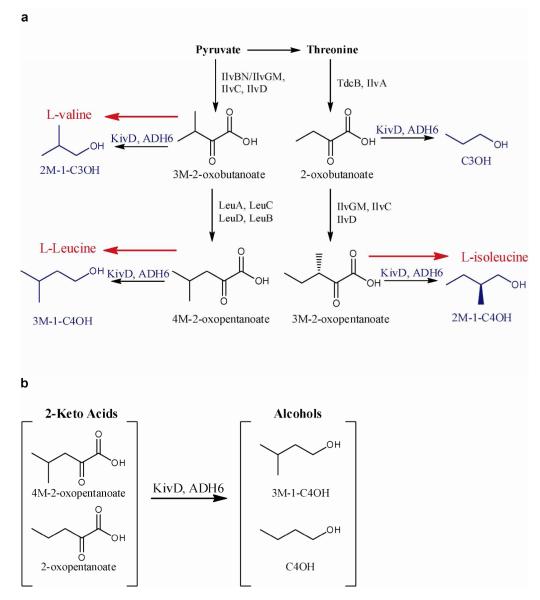
Scheme 1. 2-keto acid-derived alcohol production in E. coli.
(a) Biosensor-relevant 2-keto acid-derived alcohols (blue) produced in engineered E. coli are rooted in high-flux amino acid biosynthetic pathways (red). Deletion of the ilvDAYC operon yielded a valine, isoleucine, and leucine auxotroph incapable of producing biosensor-inducing alcohols without 2-keto acid supplementation. 2-oxopentanoate, the 2-keto acid precursor to 1-butanol, is not naturally produced in E. coli. (b) Heterologously expressed L. lactis KivD and S. cerevisiae ADH6 were used to produce user-defined alcohols by medium supplementation with the cognate 2-keto acid substrate.
Figure 5. BmoR-PBMO biosensor screen for improved 1-butanol biosynthesis.
(a) The mean total mixed alcohol titer in E. coli DH1 ΔadhE harboring pKivD#1 was significantly lower (t-test; unpaired, p=1×10−11) as compared to a heterogeneous population containing mutated kivD and ADH6 ribosome binding site (RBS) sequences, a result suggesting the initial RBS was non-optimal. The RBS library population produced a broad range of alcohol titers (n=50; box and whisker plot depicts 10th, 25th, median, 75th and 90th percentiles) suitable for characterization of the high-throughput screen. (b) The biosensor response (OD600) to spent production medium from a 960-member library of mutated kivD and ADH6 RBS sequences was distributed around OD600=0.31. Gas chromatography-mass spectrometry was used to confirm 1-butanol titers for 10% of the sample population, demonstrating a positive correlation between biosensor response and 1-butanol titer.
To simplify analysis – given differences in biosensor responsiveness to the various alcohols – we also wanted to limit biosynthesis to a single, user-defined alcohol. This was accomplished by incorporating a single genetic deletion, Δ(ilvD–ilvC), into an E. coli DH1 ΔadhE background. The resulting strain, JAD-2, is auxotrophic for valine, leucine and isoleucine; furthermore, pKivD#1-mediated production of all C4 to C6, 2-keto acid-derived alcohols was eliminated in this host (Supplementary Fig. 6). Supplementing the growth medium with 1 g l−1 of 2-oxopentanoate resulted in production of a single alcohol, 1-butanol, at over 95% yield following 12 hours incubation. Notably, deletion of threonine dehydratase (TdcB), whose activity can complement an ilvA gene deletion, was found to be unnecessary in our engineered production strain. Supporting this observation are published studies reporting TdcB expression only during anaerobic growth in the absence of glucose53,54.
The pKivD#1 RBS library was transformed into strain JAD-2, and 960 colonies (ten 96-well plates) were randomly selected for growth in autoinduction medium supplemented with 1 g l−1 2-oxopentanoic acid. 1-Butanol titers were assayed using the TetA-based whole-cell biosensor following addition of 7.5 μgml−1 tetracycline selective pressure, and the final cell densities measured after 24 hours growth (Fig. 5). Biosensor output was distributed around OD600=0.31, and thirteen samples (1.35% of the population) exhibited outputs three standard deviations above the mean, which we identified as positive hits. 1-Butanol titers were confirmed by gas chromatography-mass spectrometry (GC-MS) for both the positive hits, and an additional 10% of the population selected at equal increments from across the range of biosensor outputs. The average 1-butanol concentration for the positive hits, 493 mg l−1 (n=13), was significantly higher (p=0.005; one-tailed, unpaired t-test) than the average for those samples falling within one standard deviation of the mean, 345 mg l−1 (n=42). These data indicated the high-throughput screen was statistically sensitive and able to accurately identify strains exhibiting improved 1-butanol titers from a heterogeneous population exhibiting diverse 1-butanol productivities. The small number of false positives observed was attributed to carryover of production strain cell material into the biosensor assay well, resulting in an artificial increase in biosensor output signal.
To determine if the screen provided a functional increase in 1-butanol biosynthesis, the top positive hit identified from our screen of the ribosome binding site library was sequenced and analyzed in greater detail. This mutant variant contained four point mutations in the KivD and AHD6 ribosome binding sites (plasmid pKivD#1-S71; Table 1, see Supplementary Materials for sequences). To confirm improvement in alcohol productivity, we repeated the 2-keto acid feeding experiment using either 2-oxopentanoate or 4-methyl-2-oxopentanoate, yielding 1-butanol and 3-methyl-1-butanol, respectively. Product titers were monitored over time by GC-MS and used to calculate specific productivities; results were also compared to a negative control strain harboring a red fluorescent protein (RFP) plasmid (plasmid pRFPA2; Table 1, Supplementary Fig. 7). The negative control strain failed to convert either of the 2-keto acid substrates to the corresponding alcohols. Plasmids pKivD#1 and pKivD#1-S71 exhibited 3-methyl-1-butanol–specific productivities of 33.8 ± 1.7 mg l−1 hr−1 OD−1 and 40.3 ± 3.5 mg l−1 hr−1 OD−1, respectively (n=3; mean ± s.d.). Production of 1-butanol occurred more slowly, at rates of 13.5 ± 1.3 mg l−1 hr−1 OD−1 for pKivD#1 and 18.6 ± 1.0 mg l−1 hr−1 OD−1 for pKivD#1-S71 (mean ± s.d.; n=3); thus, the mutant variant isolated using the genetic screen displayed a nearly 35% increase in 1-butanol specific productivity. While in our proof-of-principle demonstration 103 colonies were screened by a single user, with automation 105 samples per user per day can readily be achieved in 96-well plate format27, a substantial improvement over existing chromatography-based techniques.
From a screening standpoint, transcription factor-based biosensors possess numerous advantages over their synthetic chemistry-based counterparts. Transcription factor-based screens employ an environmentally-benign, self-renewing catalyst; a versatile, modular design framework adaptable to different input ligands and desired output signals; and, perhaps most importantly, exhibit high ligand selectivity. Complex three-dimensional protein folds and the breadth of amino acid chemistries contained within – typically heralded as an advantage of enzymatic over synthetic catalysts – impart transcription factors with superior ligand selectivity over their synthetic counterparts. In the case of BmoR, removal of a single aliphatic carbon, assaying for 1-propanol as compared to 1-butanol, eliminated alcohol-induced reporter expression; of particular importance with regard to 1-butanol biosynthesis, the biosensor was also unresponsive to ethanol and exhibited only a weak response to butyraldehyde. Butyraldehyde is the penultimate intermediate in all 1-butanol biosynthetic pathways explored to date, and ethanol is a major byproduct in engineered microbes – resulting from alcohol dehydrogenase substrate promiscuity – observed at between 3- to 25-fold higher titers relative to 1-butanol29–31. Published, synthetic chemistry-based, alcohol screens have been reported36,37, but the assays are indirect, requiring oxidation of the terminal alcohol to the aldehyde, and can neither discriminate between the different chain-length alcohols nor between an alcohol and its cognate aldehyde. Thus, this work highlights one advantage of a transcription factor-mediated assay over alternative screening approaches.
Synthetic selection for 1-butanol biosynthesis in engineered E. coli
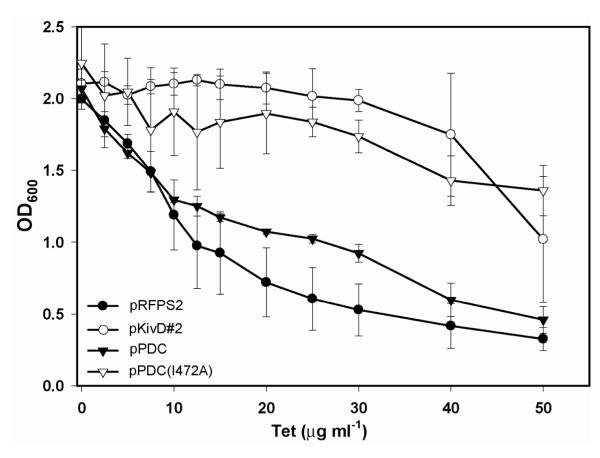
While the ability to select for improved small-molecule production phenotypes has been a long-standing goal of metabolic engineering, overproduction of only a small minority of metabolites is naturally selected for. Transcription factor-promoter pairs provide a means through which the scope of selectable small-molecule production phenotypes can be substantially broadened. To demonstrate this application we tested the 1-butanol biosensor under conditions of concomitant in vivo alcohol production and synthetic selection. For these experiments, plasmid pSelect#2 was constructed containing tetA under control of PBMO (Table 1). Additionally, a set of alcohol production plasmids and controls were constructed using low-copy number, pSC101-based vectors with PLacO1 promoters. A low copy vector was chosen to control for heterogeneity in a population’s production phenotype due to variations in biosynthetic pathway copy number between different host cells. Plasmid pRFPS2 harboring the gene encoding red fluorescent protein under control of PlacO1 was used to control for heterologous protein expression; similarly, plasmid pKivD#2 contains the KivD.ADH6 1-butanol biosynthetic pathway on an identical plasmid backbone. In an attempt to further understand the effect of concomitant pathway expression and biosensor activation on host fitness we also included in our analysis a second 2-keto acid decarboxylase. The wild type Zymomonas mobilis pyruvate decarboxylase, PDC, has negligible activity toward 2-oxopentanoate, but addition of a single point mutation (I472A) into the PDC coding sequence increases specific activity 160-fold55. Thus, to a first approximation, differences in E. coli fitness between hosts harboring wild-type and mutant Z. mobilis PDC are due to pathway activity and not pathway enzyme overexpression. The resulting plasmids, pPDC and pPDC(I472A) (Table 1), were each co-transformed with biosensor plasmid pSelect#2 into E. coli JAD#2 and tested in parallel with pKivD#2 and pRFPS2. Using the redesigned vectors, a correlation between 1-butanol production and final cell culture density was observed following growth in selective medium (Fig. 6). The two negative control strains harboring either pRFPS2 or pPDC exhibited decreased fitness relative to the 1-butanol production strains harboring pKivD#2 or pPDC(I472A). As was also observed in the exogenous 1-butanol characterization assays, fitness differences between production and non-production phenotypes were amplified through increased tetracycline supplementation.
Figure 6. Demonstration of a synthetic selection for in vivo 1-butanol biosynthesis.
E. coli JAD#2 co-transformed with the pSelect#2, TetA-based synthetic selection device and either an RFP control plasmid (pRFPS2) or a non-functional alcohol biosynthetic pathway with a Z. mobilis pyruvate decarboxylase (pPDC) displayed poor growth upon addition of tetracycline selective pressure. Use of the L. lactis promiscuous 2-keto acid decarboxylase pathway (pKivD#2) or introduction of a single point mutation into the Z. mobilis pyruvate decarboxylase (pPDCI472A), imparting 2-oxobutanoate decarboxylase activity, resulted in improved fitness relative to the negative control strains. Data are mean (s.d.) (n=3).
Lastly, we demonstrated the ability to utilize differences in host fitness to select for E. coli possessing a 1-butanol production phenotype. Fold-enrichment was determined by performing assays using two background strains. E. coli JAD-1 contains an (ilvD-ilvC)::neo insertion and is kanamycin resistant, while E. coli JAD-2 contains a Δ(ilvD-ilvC) deletion and is kanamycin sensitive. The strains have identical amino acid auxotrophies, and when transformed with pKivD#2 both are capable of converting 2-oxopentanoate to 1-butanol. E. coli JAD-1 was transformed with plasmid pKivD#2 and diluted to a frequency of 1×105 in a population of JAD-2 harboring control plasmid pRFPS2. Following a single round of positive selection for 1-butanol production, the ratio of kanamycin resistant to sensitive cells was measured, demonstrating an average 120-fold enrichment for pKivD#2 (n=4). These results indicate that BmoR-PBMO can be used to construct a synthetic selection for a 1-butanol production phenotype; moreover, because fold-enrichment is intrinsically linked to the fitness differences between two strains, these results suggest further improvements can be realized through additional control and modification of the biosensor transfer function.
In conclusion, using 1-butanol biosynthesis as a case study, we have shown that a transcription factor-based biosensor can effectively address two persistent challenges in metabolic engineering: construction of robust, high-throughput, small-molecule screens and development of synthetic selections coupling in vivo small-molecule production to host growth rate. The ability to select for user-defined, small-molecule production phenotypes has been a persistent, but unrealized, goal in the field; transcription factor-based biosensors provide one method of coupling a small-molecule production phenotype to cell growth. In this study we demonstrated the ability to couple 1-butanol production to increased tetracycline resistance and synthetically select for host cells engineered for 1-butanol production. Of note, we observed an increase in negative control strain background signal during in vivo production assays relative to exogenous alcohol supplementation experiments. We attributed this finding to the extended culture time necessary to achieve biosynthetic pathway expression, resulting in an accumulation of TetA, the tetracycline transporter. Decreasing the background noise resulting from leaky reporter transcription would result in further amplification of differences in the 1-butanol-dependent growth rate and substantially improve fold-enrichments. Indirect approaches to decrease the background signal, apart from engineering the transcription factor-promoter pair itself, may be found by decreasing translation initiation rate or increasing reporter degradation rate. Likewise, future optimization of a transcription factor-promoter selection device, including the ability to discriminate between strains exhibiting different small-molecule productivities, or an ability to alter the linear range of detection can be approached through additional modification of, and control over, the biosensor transfer function. For example, using the biosensor as a selection the linear range of detection caps the scale of improvement in small-molecule production that can be effectively selected for. This limitation could theoretically be circumvented through introduction of targeted genetic modifications resulting in an increase in transcription factor-ligand binding Kd, shifting the dose-response curve to the right58.
In general, transcription factor-based screens and selections can be used to accelerate metabolic engineering and test hypotheses requiring construction of large genetic libraries. These efforts are greatly facilitated by techniques for the design and reengineering of transcription factor-promoter devices with user-defined transfer functions and altered ligand selectivities. For example, robust mathematical models of promoter activation56,57 can be used to guide the reengineering of existing biosensors and tune device transfer functions for specific screening and selection applications. Similarly, transcription factor ligand selectivity can be reengineered to alter an existing device’s input signal28,58; however, this approach is generally limited to only chemically and structurally similar small molecules. One approach, which we demonstrate here, is identification of native transcriptional activators and heterologous expression in an E. coli host. In cases were the only responsive protein is a repressor protein, a genetic inverter provides a useful, but more complex, method of wiring a biosensor to couple small-molecule concentration to microbe growth rate. We hypothesize that transcription factors for detection of most small-molecule ligands – either naturally occurring or anthropogenic – found in substantial concentrations in the environment have been naturally selected for, and we demonstrate these transcription factors-promoter pairs can be readily rewired to enable small-molecule dependent growth phenotypes.
Methods
Characterization of butanol- and dicarboxylic acid-dependent E. coli growth
Selective pressure, as measured by cell growth rates, was determined in 96-well plates. E. coli DH1 ΔadhE or MG1655 harboring biosensor plasmid(s) pSelect#1, pSelect#2, pC6Select#1/pC6Select#2, or pC4Select#1 were cultured as follows. E. coli harboring the appropriate biosensor plasmids were cultured overnight in Luria-Bertani (LB) medium (200 rpm, 30°C) supplemented with 50 μg ml−1 carbenicillin (Cb50) and 50 μg ml−1 chloramphenicol (Cm50), as appropriate. Cultures were inoculated 1% v/v into either fresh LB medium or EZ Rich medium (Teknova; 0.5% w/v glucose) containing appropriate antibiotic and grown to a final cell density of OD600=0.20-0.80 (200 rpm, 30°C). Cultures were then diluted 1:4 in fresh LB (dicarboxylic acid assays) or EZ Rich medium (1-butanol assays; 0.5% w/v glucose, Cb50) supplemented with inducing small-molecule in 96 deep well plates (2 mL well volume; Corning). For the 1-butanol assays, following dilution into induction medium the cultures were grown for 1 hr before addition of tetracycline or nickel-chloride. For dicarboxylic acid assays, tetracycline was added to the induction medium immediately following inoculation with E. coli cultures.
For experiments requiring measurement of strain growth rate the cultures were incubated in a 96-well plate reader (30°C), and OD600 measurements taken every 15 minutes for a total culture time of 20 hours. Growth rates were determined by first normalizing each curve to the starting cell density and fitting to a modified Gompertz Equation (Equation (1)) for microbial growth59:
| (1) |
where A is the maximum cell density ln(N/No), λ is the lag period (hr), t is the time (hr), and μm is the maximum specific growth rate (hr−1).
For endpoint-based characterization of liquid culture screening parameters using the TetA-based biosensor, cultures were prepared as described above with the exception that assays were performed in deep-well plates (2-ml well volume, Corning, Corning, NY). Briefly, 150 μl of biosensor strain at OD600=0.20 was added to 450 μl fresh medium containing 1-butanol or dicarboxylic acid at the indicated concentration. For 1-butanol assays, cultures were incubated in inducing medium for a period of 1 hour, unless indicated otherwise (Supplementary Fig. 2 describes results using alternative incubation times), before supplementation with 0, 2.5, 5, 7.5, 10, 12.5, 15, 20, or 25 μg/ml tetracycline. For dicarboxylic acid assays, the cultures were supplemented with 0, 10, 25, 30, or 40 μg/ml tetracycline immediately following inoculation of the growth medium. Following application of selective pressure, cultures were incubated for 16-18 hrs (30°C, 300 rpm) at which point cell density and fluorescence (for strains harboring pSelect#2) were measured. A mathematical model based on a combined log-logistic function (Equation (2)) was used to describe the biphasic butanol response curves observed43:
| (2) |
Here OD600 at a given concentration of butanol ([C4OH]) is described by the parameters α and ω, the horizontal asymptotes as the butanol concentration approaches 0 and positive infinity, respectively. Additional parameters include the slopes of the rising (β Up) and falling (β Dn) sides of the biphasic relationship as well as the half-maximal response due to tetracycline-induced () and butanol-induced () toxicities.
Characterization of GFP-based biosensor
E. coli DH1 ΔadhE harboring the appropriate biosensor plasmid(s) were cultured overnight in Luria-Bertani (LB) medium (200 rpm, 30°C) supplemented with 50 μg ml−1 carbenicillin (Cb50). Cultures were inoculated 1% v/v into fresh EZ Rich medium (Teknova; 0.5% w/v glucose, Cb50) and grown to a final cell density of OD600=0.20 (200 rpm, 30°C). Cultures were subsequently diluted 1:4 in fresh EZ Rich medium (0.5% w/v glucose, Cb50) supplemented with inducing small-molecule in 96 deep well plates (2 mL well volume; Corning). Cultures were incubated for 16 hrs (200 rpm, 30°C), and the fluorescence and absorbance signals measured. GFP (GFPuv) fluorescence was measured using an excitation wavelength of 400 nm and an emission wavelength of 510 nm. Optical density measurements were performed at 600 nm (OD600). GFP fluorescence values were first normalized to OD600 (GFP/OD600) and E. coli auto-fluorescence subtracted using a standard curve of GFP auto-fluorescence versus E. coli optical density. Fold-induction, where indicated, was calculated as the difference between the averages of the induced and un-induced GFP fluorescence measurements normalized to the un-induced GFP measurement. Derivation of biosensor performance features and Z’-score calculations are described in Supplementary Materials and Methods online.
Liquid culture screening assays
For butanol production strains, colonies of engineered E. coli harboring the indicated production plasmid were inoculated into 600 μL LB medium supplemented with glucose (2% v/v) and chloramphenicol (50 μg ml−1, Cm50) and grown overnight (200 rpm, 37°C). Strains were then sub-cultured 1% v/v into fresh M9 minimal medium (Cm50) containing 2X autoinduction sugars60 (1.0% w/v glycerol, 0.1% w/v glucose, 0.4% w/v lactose). Cultures containing E. coli JAD-2 were additionally supplemented with 1 g l−1 L-valine, L-isoleucine, L-leucine and 1 g l−1 of either 2-oxopentanoate for 1-butanol production or 4-methyl-2-oxopentanoate for 3-methyl-1-butanol production. Cultures were incubated for 24 hrs (30°C, 300 rpm), centrifuged (3000 × g, 4 min), and 150 μl of the supernatant was used for biosensor-based assay of 1-butanol titers.
Biosensor strains were prepared as described previously. When assaying 1-butanol concentrations in spent production medium, 150 μL of biosensor culture (OD600 = 0.20) was added to 150 μL spent production medium and 300 μL 2X EZ Rich medium (0.5% w/v glucose, Cb50; Teknova) in 96 deep-well plates (Corning). Assay samples were incubated for 1 hr and then supplemented with 7.5 μg ml−1 tetracycline. Cultures were then grown for an additional 16 hrs (200 rpm, 30°C), and the cell density measured as described previously. Alcohol titers were confirmed for a subset of samples by gas chromatography-mass spectrometry as described in the Supplementary Material and Methods online.
1-butanol biosynthesis selection assays
E. coli JAD-2 was co-transformed with biosensor plasmid pSelect#1 and the appropriate 1-butanol biosynthesis plasmid. Individual colonies were inoculated into LB medium (0.5% w/v glucose, Cm50, Cb50), cultured overnight (30°C, 200 rpm), and subsequently inoculated 1% v/v into 50 mL fresh M9 minimal medium (Cm50, Cb50) supplemented with 2X autoinduction sugars (1.0% w/v glycerol, 0.1% w/v glucose, 0.4% w/v lactose) and 1 g l−1 amino acids (L-valine, L-leucine, L-isoleucine). Cultures were then incubated for 24 hours (30°C, 200 rpm) to induce heterologous 1-butanol biosynthetic pathway expression, and then supplemented with 1 g l−1 of 2-oxopentanoate. Following a 1-hr incubation, tetracycline resistance was measured by inoculating strains 1% v/v into 600 μL fresh EZ Rich medium (0.5% w/v glucose, Cb50; Teknova) supplemented with the indicated tetracycline concentration. Cultures were incubated overnight (30°C, 300 rpm), and the final cell densities (OD600) measured. Enrichment experiments are described in the Supplementary Materials and Methods online.
Supplementary Material
Acknowledgements
This work was part of the DOE Joint BioEnergy Institute supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research through the contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. J.A.D acknowledges a fellowship from the Siebel Scholars Foundation, funding from the NIH Applied Biology and Bioprocess Engineering Training Grant, and a QB3 Biocatalyst Grant. The dicarboxylic acid material is based upon work supported by the Department of Energy under Award Number DE-SC0006469. We thank W. Holtz, J. Carothers, and J. Gilmore for critical reading of the manuscript.
Footnotes
Supporting Information Available This information is available free of charge via the Internet at http://pubs.acs.org/
Author Contributions J.A.D. conceived the project, cloned 1-butanol biosensor and pathway genes, performed experiments, and wrote the manuscript. D.S. cloned 1-butanol biosensor and pathway genes, and performed experiments. A.A. performed dicarboxylic acid experiments. J.D.K. directed the project and wrote the manuscript.
Competing Financial Interests Statement J.D.K. has financial interests in Amyris, LS9, and Lygos. J.A.D. and A.A. have financial interests in Lygos. D.L.S. is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
References
- 1.Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 2.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanehisa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CH. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspi R. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigo G, Carrera J, Prather KJ, Jaramillo A. DESHARKY: automatic design of metabolic pathways for optimal cell growth. Bioinformatics. 2008;24:2554–2556. doi: 10.1093/bioinformatics/btn471. [DOI] [PubMed] [Google Scholar]
- 7.Ellis LBM. The University of Minnesota Biocatalysis/Biodegradation Database: the first decade. Nucleic Acids Res. 2006;34:D517–D521. doi: 10.1093/nar/gkj076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Henry CS, Jankowski MD, Ionita JA, Hatzimanikatis V, Broadbelt LJ. Computational discovery of biochemical routes to specialty chemicals. Chem. Eng. Sci. 2004;59:5051–5060. [Google Scholar]
- 9.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 10.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Meth. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 12.Shivange AV, Marienhagen J, Mundhada H, Schenk A, Schwaneberg U. Advances in generating functional diversity for directed protein evolution. Curr. Opin. Chem. Biol. 2009;13:19–25. doi: 10.1016/j.cbpa.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alper H, Stephanopoulos G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab. Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Lynch MD, Warnecke T, Gill RT. SCALEs: multiscale analysis of library enrichment. Nat. Methods. 2007;4:87–93. doi: 10.1038/nmeth946. [DOI] [PubMed] [Google Scholar]
- 16.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 17.Warnecke TE, Lynch MD, Karimpour-Fard A, Sandoval N, Gill RTA. genomics approach to improve the analysis and design of strain selections. Metab. Eng. 2008;10:154–165. doi: 10.1016/j.ymben.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Smolke CD, Martin VJJ, Keasling JD. Controlling the metabolic flux through the carotenoid pathway using directed mRNA processing and stabilization. Metab. Eng. 2001;3:313–321. doi: 10.1006/mben.2001.0194. [DOI] [PubMed] [Google Scholar]
- 19.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 20.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 21.Qian Z, Xia X, Lee SY. Metabolic engineering of Escherichia coli for the production of cadaverine: A five carbon diamine. Biotechnol. Bioeng. 2011;108:93–103. doi: 10.1002/bit.22918. [DOI] [PubMed] [Google Scholar]
- 22.Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 23.Atsumi S, Liao JC. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr. Opin. Biotech. 2008;19:414–419. doi: 10.1016/j.copbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voigt CA. Genetic parts to program bacteria. Curr. Opin. Biotech. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 26.Su L, Jia W, Hou C, Lei Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011;26:1788–1799. doi: 10.1016/j.bios.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Dietrich JA, McKee AE, Keasling JD. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 28.Tang S, Cirino PC. Design and application of a mevalonate-responsive regulatory protein. Angew. Chem. Int. Ed. 2011;50:1084–1086. doi: 10.1002/anie.201006083. [DOI] [PubMed] [Google Scholar]
- 29.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen DR, Leonard E, Yoon S, Tseng H, Yuan C, Prather KLJ. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 2009;11:262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Berezina OV, Zakharova NV, Brandt A, Yarotsky SV, Schwarz WH, Zverlov VV. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl. Microbiol. Biot. 2010;87:635–646. doi: 10.1007/s00253-010-2480-z. [DOI] [PubMed] [Google Scholar]
- 32.Bond-Watts BB, Bellerose RJ, Chang MCY. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 33.Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. P. Natl. Acad. Sci. USA. 2008;105:20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 35.Shena CR, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 2008;10:312–320. doi: 10.1016/j.ymben.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Sawicki E, Hauser TR, Fox FT. Spectrophotometric determination of aliphatic aldehyde 2,4-dinitrophenylhydrazones with 3-methyl-2-benzothiazolinone hydrazone. Anal. Chim. Acta. 1962;26:229–234. [Google Scholar]
- 37.Dickinson RG, Jacobsen NW. A new sensitive and specific test for the detection of aldehydes: formation of 6-mercapto-3-substituted-s-triazolo[4,3-b]-s-tetrazines. J. Chem. Soc. D. 1970:1719–1720. [Google Scholar]
- 38.Dubbels BL, Sayavedra-Soto LA, Bottomley PJ, Arp DJ. Thauera butanivorans sp. nov., a C2–C9 alkane-oxidizing bacterium previously referred to as ‘Pseudomonas butanovora’. Int. J. Syst. Evol. Micr. 2009;59:1576–1578. doi: 10.1099/ijs.0.000638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurth EG, Doughty DM, Bottomley PJ, Arp DJ, Sayavedra-Soto LA. Involvement of BmoR and BmoG in n-alkane metabolism in ‘Pseudomonas butanovora’. Microbiology. 2008;154:139–147. doi: 10.1099/mic.0.2007/012724-0. [DOI] [PubMed] [Google Scholar]
- 40.Buck M, Gallegos M-T, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma 54 (sigma N) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muranaka N, Sharma V, Nomura Y, Yokobayashi Y. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res. 2009;7:e39. doi: 10.1093/nar/gkp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Podolsky T, Fong S-T, Lee BTO. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid. 1996;36:112–115. doi: 10.1006/plas.1996.0038. [DOI] [PubMed] [Google Scholar]
- 43.Parkins C, Beckon AV, Beckon WN, Maximovich A. A general approach to modeling biphasic relationships. Environ. Sci. Technol. 2008;42:1308–1314. doi: 10.1021/es071148m. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J-H, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assay. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 45.Lin H, Bennett GN, San K-Y. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab. Eng. 2005;7:116–127. doi: 10.1016/j.ymben.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Sánchez AM, Bennett GN, San K-Y. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab. Eng. 2005;7:229–239. doi: 10.1016/j.ymben.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Niu W, Draths KM, Frost JW. Benzene-free synthesis of adipic acid. Biotechnol. Prog. 2002;18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- 48.Daniel Lashof, Ahuja D. Relative contributions of greenhouse gas emissions to global warming. Nature. 1990;344:529–531. [Google Scholar]
- 49.Parales R, Harwood C. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 1993;175:5829–5838. doi: 10.1128/jb.175.18.5829-5838.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romero-Steiner S, Parales RE, Harwood CS, Houghton JE. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 1994;176:5771–5779. doi: 10.1128/jb.176.18.5771-5779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies SJ, Golby P, Omrani D, Broad SA, Harrington VL, Guest JR, Kelly DJ, Andrews SC. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 1999;181:5624–5635. doi: 10.1128/jb.181.18.5624-5635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connor MR, Liao JC. Engineering of an Escherichia coli strain for the production of 3-Methyl-1-Butanol. Appl. Environ. Microbiol. 2008;74:5769–5775. doi: 10.1128/AEM.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egan RM, Phillips AT. Requirements for induction of the biodegradative threonine dehydratase in Escherichia coli. J. Bacteriol. 1977;132:370–376. doi: 10.1128/jb.132.2.370-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hobert EH, Datta P. Synthesis of biodegradative threonine dehydratase in Escherichia coli: role of amino acids, electron acceptors, and certain intermediary metabolites. J. Bacteriol. 1983;155:586–592. doi: 10.1128/jb.155.2.586-592.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegert P, McLeish MJ, Baumann M, Iding H, Kneen MM, Kenyon GL, Pohl M. Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein Eng. Des. Sel. 2005;18:345–357. doi: 10.1093/protein/gzi035. [DOI] [PubMed] [Google Scholar]
- 56.Bintu L, Buchler N, Garcia H, Gerland U, Hwa T, Kondev J, Phillips R. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R. Transcriptional regulation by the numbers: applications. Curr. Opin. Genet. Dev. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawkins AC, Arnold FH, Hauer RSB, Leadbetter JR. Directed evolution of Vibrio fischeri LuxR for improved response to butanoyl-homoserine lactone. Appl. Environ. Microbiol. 2007;73:5775–5781. doi: 10.1128/AEM.00060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zwietering MH, Jongenburger I, Rombouts FM, Riet K. v. t. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Studier WF. Protein production by auto-induction in high-density shaking cultures. Protein Expres. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Lee TS, Krupa R, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets; a synthetic biology platform for gene expression. J. Biol. Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.