Abstract
A major determinant for transcriptional incompetence of murine leukemia virus (MLV) and MLV-derived vectors in embryonal cells is located at the proline primer binding site (PBS). The mechanism of silencing is unknown, yet the effect is capable of spreading to adjacent promoters. Based on a retroviral vector containing an internal promoter and the escape mutant B2 PBS with expressional capacity in embryonal cells, we have developed an assay to test the ability of putative insulators to shield the silencer at the PBS. Since the B2 PBS reverts to the wild-type PBS at high frequency, a shielding ability of a putative insulator can be assessed from the ratio of expressing B2 PBS to proline PBS proviruses in the target embryonal carcinoma cell population as measured by primer extension. Our results show that none of the possible insulators, scs, BEAD-1, or HS4, is able to shield an internal promoter from the repressive effect of the silencer at the PBS region when inserted between the silencer and the promoter.
Infection by murine leukemia viruses (MLV) and MLV-based vectors is impeded in embryonal carcinoma (EC) (15, 63, 66) and embryonal stem (ES) cells (24, 32) as well as in cells of the early embryo (27, 73) which are nonpermissive for proviral transcription. In Moloney MLV (MoMLV), the major determinant for this repression has been mapped to the primer binding site (PBS) region of the 5′ untranslated region (UTR), which binds a proline (Pro) tRNA primer for initiation of reverse transcription. By employing a retroviral vector to select for escape mutants in EC cells, a single G-to-A mutation at position 15 in the PBS (the B2 PBS) was identified and found to be responsible for alleviating the silencing (3). The inhibitory element has been delineated to overlap the Pro PBS, and virtually any nucleotide changes within the sequence appear to relieve repression (32, 39, 49). The PBS sequence from the revertant MoMLV dl587rev (14) matching a glutamine (Gln) tRNA has an A at the position characterizing the B2 mutation and is also proficient for expression in both ES (24) and EC (49) cells. The mechanisms of action of the silencer are unknown but presumably are mediated at the DNA level, distinct from the roles of the PBS in reverse transcription, as it functions when placed in an intron (49), when 5′ of the long terminal repeat (LTR), outside of the transcriptional unit (39), and in transfection assays in the absence of retroviral proteins (3, 17, 37, 38, 39). Binding of a putative repressor to a probe spanning the Pro PBS but not the B2 PBS was detected in exonuclease III protection (39) and bandshift experiments (49), suggesting that a protein, factor A, enriched in nuclear extract from EC as compared to differentiated cells (77) mediates the repression via the silencer in the PBS region. Notably, the silencer is sufficient to repress transcription from heterologous promoters placed internally in a retroviral vector (49), although the effect has been reported to attenuate as the distance to the target promoter increases (7, 32).
Furthermore, nonfunction of the enhancer caused by lack of transcriptional activators (17, 35, 36, 37, 62) or by binding of negative acting trans factors has been implicated in embryonic silencing of MLV LTR-directed transcription (1, 11, 22, 56, 67). In accordance with this, as found for expanded-host-range mutants of Moloney murine sarcoma virus (18, 26), expressional competence in embryonal cells correlates with a point mutation in the recognition site for an EC-specific negative factor (1) and with the acquisition of a functional binding site for the transcription factor Sp1 (51). However, activating mutations in the LTR are not sufficient to overcome repressor activity (11) by the silencing element of the PBS region.
Although no causal relationship has been inferred, an inverse correlation between transcriptional activity and methylation of the 5′ part of the provirus is well established (7, 28, 34, 55, 56, 75), with provirus integration also capable of inducing de novo methylation of 1 to 2 kb of flanking chromosomal DNA (7, 28). This suggests that methylation plays a role in silencing by means of binding MeCP2, which may recruit histone deacetylases proficient for repressing transcription (29, 45). However, the primary immediate block to provirus expression in undifferentiated cells appears to be independent of methylation, which is detected only at later time points (20, 47, 63). In accordance with this, differentiation of EC cells permits de novo infection, but for proviruses integrated prior to differentiation to be activated, treatment with a demethylating agent is also required (47, 63). In a study pursuing the issue of secondary mechanisms maintaining provirus repression in a permissive background, two cis-acting mechanisms were proposed to regulate provirus expression: (i) a partial repression in undifferentiated cells accompanied by methylation and (ii) extinction during early stages of differentiation, independent of changes in methylation, but with the surrounding chromatin structure being a key determinant in repression (34).
How the silencing element in the PBS region functions to inhibit transcription has not been recognized, nor has it been discerned if the element can be insulated in cis to impede the spreading of the effect to a promoter placed downstream. Insulators or boundary elements are DNA sequences thought to play a role in the independent regulation of genes by preventing inappropriate interactions between transcriptional elements in separate chromatin domains (65, 68). Insulators from a variety of organisms have been described. Among the best characterized are the specialized chromatin structures scs and scs′, which flank the hsp70 genes of the 87A7 heat shock locus in Drosophila melanogaster (69), and, from vertebrates, the 5′ hypersensitive site HS4 from the chicken β-globin locus (30). These elements are able to protect a stably integrated reporter gene from position effects, i.e., to protect against stimulatory endogenous enhancers or neighboring repressing chromatin (13, 30, 50) and to disrupt enhancer-promoter interactions when positioned in the intervening sequence (10, 12, 13, 31). The finding that some insulators function in highly diverse species, i.e., chicken HS4 in Drosophila (13) and Drosophila scs in Xenopus laevis (16) and humans (71, 79), indicates evolutionary conservation of insulator properties. The insulating activity operates with the element in any orientation but is strictly dependent on the position in the specific constructs, and since it is not associated with any enhancer or silencer activity in itself, it prompted the idea that insulators are neutral boundaries of a chromosomal domain. Such a role is supported by their presumed function in their natural contexts—scs and scs′ possibly delimiting the domain of decondensation appearing after heat shock induction of the hsp70 genes (69) and HS4 (at the periphery of a condensed region of generalized DNase I inaccessibility and low levels of histone acetylation) providing a boundary assuring independent regulation of two flanking loci during erythroid development (25, 52). Analysis of enhancer blocking by the Drosophila gypsy insulator (59) has emphasized a functional rather than a structural role for insulators, suggesting that some elements operate more as transcriptional decoys intercepting the enhancer signal before it reaches the promoter (59). Recently, for both scs (19) and HS4 (5), a minimal core element and corresponding binding protein proficient for enhancer blocking activity have been defined, the latter case extending the notion of boundaries as conserved components of gene regulation, since identical sites for the highly conserved zinc finger protein CTCF were identified in HS4 and in the less-characterized vertebrate insulators BEAD-1 from the human T-cell receptor α/δ locus and Xenopus RO (5).
The frequently employed assays for insulator function utilize transgenic flies or a colony assay in stably transfected cell lines to address blocking of enhancer or position effects. To provide simpler means of studying the elements, eliminating an ill-defined contribution from flanking chromatin or the effects of tandemly integrated multiple copies, insulators have been tested in transient assays (16, 54, 78). In these, both scs (16) and HS4 (54) were found to exert blocking activity independent of integration into the genome, while scs′ had no activity under transient conditions (78). Homologous recombination to study insulation of enhancer action at a few genomic locations (74), increased probability of expression from a retroviral vector carrying HS4 in the LTRs (55), and blocking of repressive effects mediated by a number of chromatin-associated repressors, such as PcG proteins, mHP1, and MeCP2 (60, 71), have provided further evidence for insulators' ability to shield against negative effects, yet have also underscored the existence of different classes of both insulators and repressive mechanisms.
We were interested in the ability of these putative insulators to shield spreading of the negative effect of the silencer residing at the Pro PBS to a promoter placed downstream in a retroviral vector. For this purpose, we have established an assay of EC cells which measures differences in the amount of expressing B2 PBS and Pro PBS vectors in EC cells as an indication of the degree of repression exerted toward the wild-type (wt) PBS provirus. We show here that neither scs, HS4, nor the BEAD-1 element shields the silencer at the Pro PBS.
MATERIALS AND METHODS
Plasmid construction and oligonucleotides.
All vectors are shown in Fig. 1. pProPLTneo-TATA− is contained in a pUC19 plasmid backbone and consists of the LTRs of Akv MLV (70) flanking 262 bp of the 5′ UTR from Akv MLV with an insertion of a 23-bp polylinker (5′-ATCGATTTAAATCTAGACAATTG-3′) 51 bp downstream of the LTR, 321-bp simian virus 40 (SV40) early promoter-enhancer sequences (from StuI to PvuII at nucleotide [nt] 3434 of pSV2neo [61]), a 1,493-bp Tn5 fragment containing the neomycin phosphotransferase-encoding gene (neo) (4), and 565 nt including part of Akv MLV env and 3′ UTR. The 5′-TATAAA-3′ sequence of the 3′ LTR TATA box in the plasmid was mutated to a SacI 5′-GAGCTC-3′ site. To generate pB2PLTneo-TATA−, Pro PBS was replaced by B2 PBS (5′-TGGGGGCTCGTCCGAGAT-3′, the single nucleotide G-to-A mutation shown in bold [3]). LJB2-AdMLPEnh−, contained in a pBR322 plasmid backbone, is derived from the MoMLV-based LJ-PAdMLPEnh− (49) (provided by E. Barklis) by exchange of the Pro PBS with B2 PBS obtained by PCR amplification from B2BAG (7) (provided by E. Barklis). All mutations, including the polylinker insertion, were introduced by standard PCR (57)-mediated mutagenesis procedures essentially as described previously (40). Briefly, for mutagenesis procedures on plasmid templates, PCR amplification was generally performed in 100 μl of PCR buffer containing 100 to 150 ng of the respective template, 2.5 U of Taq polymerase, 1 U of Pfu polymerase, 0.2 mM deoxynucleoside triphosphates, and 10 pmol of biotinylated and 25 pmol of unbiotinylated primers (with restriction enzyme sites for subsequent cloning of amplified fragments) in 12 cycles at 94°C for 1 min, at 55 to 60°C for 1 min, and at 73°C for 1 min per ∼1,000 bp. All PCR-amplified sequences were verified by sequencing. PCR-amplified control and insulator sequences described below were cloned into the polylinker immediately upstream of the adenoviral major late promoter (AdMLP) in pLJB2-AdMLPEnh−. Numbers and sequences of oligonucleotides employed for amplification are given in parentheses, with X denoting biotinylation. pLJB2-680-control contains a 680-bp subfragment of the green fluorescence protein (GFP) gene (in reverse orientation) amplified from pIRES-EGFP (Clontech Laboratories, Inc., Palo Alto, Calif.) (no. 1, 5′-XAGACCTGGATCCCGCTTTACTTGTACAGCTCGTCCATGC-3′; no. 2, 5′-AAGGAAAAAGCGGCCGCCCTGGTCGAGCTGGACGGCGACGTAA-3′). pLJB2-1.9-control contains an additional 1,272-bp reverse-oriented cDNA sequence of the murine transcription factor ALF1 gene amplified from pUHD-ALF1b (46) (no. 3, 5′-TCTACCGGGCCCAGGTTTCCAATTGATGCATTATGGG-3′; no. 4, 5′-CGCGGATCCGCGGGCAGGTATGGATGAGCG-3′). pLJB2-680scs contains the 850- to 1,530-bp fragment (16) from D. melanogaster scs (provided by H. Cai) (no. 5, 5′-XAGACCTGGATCCTGAAAACATAAACAGAATCACTTGTTG-3′; no. 6, 5′-AAGGAAAAAAGCGGCCGCAGTTCGAATATGCTCTTTAAATCCCA-3′). LJB2-BEAD-1 contains the 1,970-bp BEAD-1 fragment from the human T-cell receptor α/δ locus, obtained from pRN (79) (provided by M. S. Krangel) (no. 7, 5′-CCTGGATCCCAGAAATCTTTGATTTCAGATGCTTGAG-3′; no. 8, 5′-ATAGTTTAGCGGCCGCCACTCTTAGCCATTATACTGCATTGCTGT-3′). In pLJB2-BEAD-1rev, the BEAD-1 insert was cloned in the opposite orientation, and pLJB2-BEAD-A contains a 120-bp subfragment of BEAD-1 identified by Bell et al. (5) (no. 9, 5′-GCCTGGATCCTGGAAGAGG GATGTTGAGGGCCCAGGGGCTGCCTTGCCGGTGCATTGGCTGCCCA GGCCTGCACTGCCGCCTGCCGGCAGGGGTCCAGTCCACGAGAC CCAGCTCCCTGCTGGCGGAAGGCGGCCGCGGGCCCTAAAC-3′, made double stranded with no. 10, 5′-GTTTAGGGCCCGCGGCCGC-3′). pLJB2-FII contains the 42-bp binding site sequence for CTCF (5) (no. 11, 5′-GCCTGGA TCCCCCAGGGATGTAATTACGTCCCTCCCCCGCTAGGGGGCAGCA GCGGCCGCGGGCCCTAAAC-3′ made double stranded with primer no. 10 shown above). pLJB2-FIIrev harbors the FII sequence cloned in the opposite orientation, and pLJB2-1.2HS4 contains the 1.2-kb chicken β-globin insulator from pJC13-1 (13) (provided by G. Felsenfeld) (no. 12, 5′-CCTGGATCCGAGCTCACGGGGACAGCCCCC-3′; no. 13, 5′-GTTTAGGGCCCGCGGCCGCAATATTCTCACTGACTCCGTC-3′). All oligonucleotides were obtained from DNA Technology A/S, Aarhus, Denmark.
FIG. 1.
Retroviral vectors. (A) Vectors based on Akv MLV (grey LTR) or MoMLV (white LTR) containing either SV40-neo, AdMLP-neo expression cassettes, or neo and a Pro PBS (PBSpro, black box), B2 PBS (PBSB2, grey box), or Gln PBS (PBSgln, white box). LJ-P, LJ-Q, LJ-PAdMLPEnh−, and LJ-QAdMLPEnh− were obtained from Petersen et al. (49). PBS-Pro was from Lund et al. (40). PL, polylinker; small cross, mutation of the TATA box; large cross, deletion of the enhancer. (B) Vectors based on LJB2-AdMLPEnh− with insertion of control sequences or putative insulators. revGFP, 680-bp reverse-oriented cDNA sequence from the GFP gene; revALFGFP, a total of 1.9 kb of cDNA sequence from the GFP gene and the ALF1 gene in reverse orientation; scs, 680-bp specialized chromatin structures from Drosophila; BEAD-1, 1,970-bp BEAD-1 from the human T-cell receptor α/δ gene in forward (→) or reverse (←) orientation; BEAD-A, 120-bp subfragment of BEAD-1; FII, 42-bp binding site sequence for CTCF in forward (→) or reverse (←) orientation; HS4, 1.2-kb hypersensitive site 4 of the chicken β-globin locus. Not drawn to scale.
Cell cultures, transfection, and transduction.
The human kidney-derived BOSC 23 packaging cell line (48) was passaged three or four times in HAT-Dulbecco's modified Eagle's medium (DMEM) with Glutamax-1 selective medium (48) to ensure expression of the env gene. Subsequent maintenance was in DMEM supplemented with 10% fetal calf serum (FCS) (Gibco BRL, Life Technologies), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The mouse EC cell line F9 (6, 64) was grown on 0.1% gelatin-coated culture flasks in DMEM supplemented with 10% FCS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. NIH 3T3 mouse fibroblasts were grown in DMEM with 10% FCS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. For transduction of target cells, BOSC 23 cells, seeded at 7 × 104 cells per cm2 the day prior to transfection, were transfected with 10 μg of vector by the calcium phosphate method (23) without a glycerol shock, medium was renewed after 12 to 16 h, and virus supernatant was harvested 48 to 72 h posttransfection, filtered through a sterile 0.45-μm-pore-size filter, diluted 3- to 10-fold (for subsequent primer extension analysis) or serially diluted (for titer determination), and transferred to F9 and NIH 3T3 target cells (seeded at [F9] 3 × 103 and [NIH 3T3] 5 × 103 or 1 × 104 cells per cm2 the day prior to transduction) in the presence of 5 and 6 μg of Polybrene (Aldrich Chemical Co., Inc.)/ml, respectively. G418 (Sigma, St. Louis, Mo.)-containing selective media were added 24 h posttransduction at 600 μg (active compound)/ml for fibroblasts, at 400 μg/ml for F9 cells, or at graduated levels as indicated for the respective experiments, and resistant colonies were counted after 10 to 14 days of selection.
Northern blot analysis.
Total RNA was extracted from cells with RNA Isolator (Genosys) following the instructions provided by the manufacturer. Northern blot analysis was done by standard procedures (58) on a 1% agarose gel. The neo gene-specific probe was the 1,325-bp HindIII-EcoRI fragment from pLJ-PAdMLPEnh− (49).
DNA preparation and primer extension analysis.
For primer extension analysis of transduced proviruses in F9 and NIH 3T3 cells, a preferable minimum number of 100 G418-resistant colonies was expanded, and genomic DNA was isolated from confluent T80 flasks by lysis with DNAzol, following the protocol provided by the manufacturer (Molecular Research Center, Inc., Cincinnati, Ohio). Specific PCR amplification of the transduced vector provirus was obtained by employing a primer (no. 14, 5′-GTCGACCGGTCGACCCTAGAGAAC-3′) spanning the deletion of the enhancer in the U3 region of the LJB2AdMLPEnh−-derived vectors together with a primer specific for the adenovirus major late promoter (AdMLP) (no. 15, 5′-XGACGCGAGCCTTTGTCTCAGAGTGG-3′) or a primer annealing to the 5′ UTR upstream of the insulator insertion site (no. 16, 5′-XCCGAACTCGTCAGTTCCACCAC-3′). PCR amplification, performed on 1 μg of DNA template in a standard reaction buffer with 2.5 U of TaqGold polymerase, was at 95°C for 10 min to activate the enzyme followed by 94°C for 1 min, 55 to 60°C for 1 min, and 73°C for 3 min in 40 cycles. Volumes of 50 to 70 μl of PCR-amplified product were purified with Dynabeads (Dynal M-280), and the NaOH-denatured biotinylated strand was employed in primer extension analysis with modified T7 DNA polymerase (Sequenase version 2.0; Amersham Pharmacia Biotech) essentially as previously described (42). Briefly, the end-labeled 18-mer extension primer (no. 17, 5′-TTTCATTTGGGGGCTCGT-3′) was annealed to the template in 10 μl of the extension buffer supplied with the enzyme by heating to 65°C for 2 min and cooling slowly to room temperature. Extension was carried out for 10 min at 37°C in the presence of 5 μl of extension mix (1 mM ddATP, 0.1 mM concentrations [each] of dCTP, dGTP, and dTTP, and 3.25 U of modified T7 DNA polymerase) and was terminated by the addition of 10 μl of formamide loading buffer and heated to 95°C for 2 min. The samples were analyzed on 20% polyacrylamide gels and exposed in a Personal Molecular Imager Fx, and the radioactivity was measured by using Quantity One.
Nuclear extract and electrophoretic mobility shift assay.
For preparation of nuclear protein extracts, pelleted cells were lysed by resuspension in 3 volumes of buffer A (20 mM HEPES [pH 7.9], 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 10% glycerol, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 5 μg of aprotinin/ml) and then centrifuged, and the nuclear pellet was resuspended in 3 volumes of buffer B (20 mM HEPES [pH 7.9], 420 mM NaCl, 20% glycerol, 10 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 5 μg of aprotinin/ml) and extracted by rotation for 30 min at 4°C. The nuclear extract supernatant was collected after centrifugation and diluted in 3 volumes of buffer Bminus (buffer B without NaCl). The 67-bp F1 probe (5′-GAATTCCATGAAGAAATTGAGACCTCTACAGGATAGCTATGGTATTTACATGTCTTTTTGCCTTAAG-3′) (provided by L. Burke), which binds CTCF (9), is a sequence upstream of the chicken lysozyme gene (2) and was synthesized as complementary oligonucleotides 61 nt in length, which were annealed for approximately 16 h and labeled with [α-32P]dATP during extension with Klenow enzyme. The unspecific probe (U) is 31 bp long (5′-CTAGGGCCCGGGACTAGGGCCAAGAACAGAT-3′). Binding reactions were performed with 10 μg of nuclear protein extract in 40 μl of binding buffer (20 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 0.5% Triton X-100) supplemented with 0.5 μg of poly(dI-dC) and 0.5 μg of sheared salmon sperm DNA. After 15 min of preincubation on ice, 20,000 cpm (Cerenkov counting) of radiolabeled F1 probe was added, together with a 0-, 25-, 100-, 500-, or 1,000-fold molar excess of cold competitor (F1 or unspecific probe U) as indicated for each experiment, and the reaction mixture was incubated for 15 min at room temperature. Glycerol at a concentration of up to 10% was added, and DNA-protein complexes were analyzed on 5% nondenaturing polyacrylamide gels.
RESULTS
An assay for insulator activity in a retroviral vector.
MLVs are transcriptionally silenced in undifferentiated embryonal cells, such as the EC cell line F9. A mutated PBS known as B2, with a single base pair change from G to A at the 15th position of the PBS, alleviates the repression of the vector in embryonic cells, while the expression in differentiated cells like NIH 3T3 fibroblasts is unaffected (3).
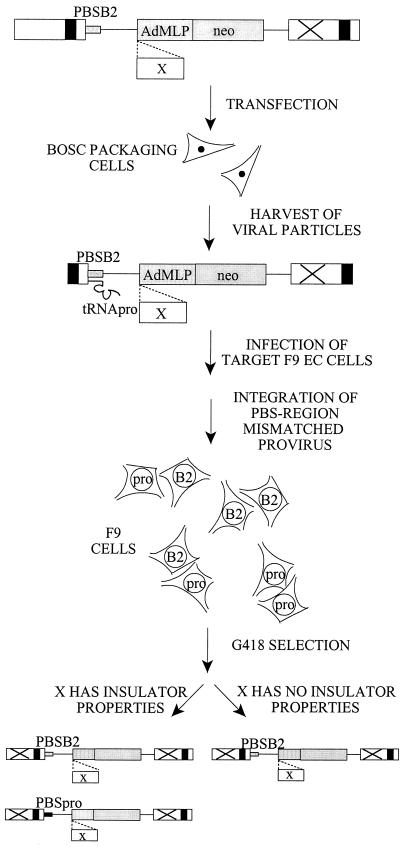
We were interested in the ability of the silencer in the wt Pro PBS region of MLVs to be shielded by putative insulators, such as scs, BEAD-1, and HS4. Since the inhibitory effect on the viral LTR can be expanded to repress transcription from heterologous promoters placed internally in a retroviral vector (49), we developed an assay (delineated in Fig. 2) for the shielding of an internal promoter from the repression of the PBS silencer by insertion of putative insulators between the promoter and the PBS in a retroviral vector. The vector contains the B2 PBS (3), a linker for insertion of putative insulators or control sequences in front of an internal promoter, the G418-selectable neo reporter gene, and an impaired 3′ LTR, which in the provirus of the transduced target cells is copied to the 5′ LTR, assuring transcription of the neo gene from the internal promoter in order to avoid interference with transcripts from the LTR (Fig. 1). Since the viral B2 PBS and the annealed Pro tRNA primer are both copied during the process of reverse transcription, a PBS region-mismatched virus will occur in the target cells (7). The mismatch may be corrected before or after integration, according to either of the template nucleotides, or DNA replication and cell division may occur before repair of the mismatch. In either case, the B2 PBS reverts to the wt Pro sequence with a frequency of approximately 50% (7). Hence, an unselected F9 or NIH 3T3 target cell population contains a mixture of both B2 PBS and Pro PBS proviruses (data not shown). After G418 selection for vector expression, only proviruses with a PBS B2 will be present in F9 target cells (Fig. 2, right), while a functional insulator able to shield the internal promoter from the repressive effect of the Pro PBS region will result in a population of target cells harboring both Pro PBS and B2 PBS proviruses (Fig. 2, left). To avoid any impediments on transcription of the vector in the packaging cells that might result from the presence of an insulator and its putative interactions with the nuclear scaffold when stably integrated in the genome, we employed viral supernatant harvested after transient production in the BOSC 23 packaging cell line for transduction of target cells. Transductions were made at a low multiplicity of infection (MOI) to assure single-copy integration of the vector while at the same time achieving a minimum number of 100 colonies to avoid stochastic fluctuations in the reversion of the B2 PBS. Since F9 and NIH 3T3 cells are not equally transducible even by the B2 PBS vectors (Table 1), based on the F9 titer, F9 cells were transduced with an MOI of approximately 0.001, but with an MOI estimate of 0.2 based on the NIH 3T3 cell titer. NIH 3T3 fibroblasts transduced in parallel served as controls in all experiments, since they have no restrictions toward viral expression and contain both Pro PBS and B2 PBS proviruses regardless of selection or the presence of a functional insulator.
FIG. 2.
Assay for shielding of the PBS silencer by insulators. A retroviral vector with a B2 PBS and an internal AdMLP driving neo expression is transfected into transient-packaging cells. Virus particles contain a B2-Pro mismatch at the PBS region in the RNA genome, resulting in both Pro PBS- and B2 PBS-containing proviruses in the target cell population. Due to restriction of the Pro PBS vector in EC cells, only the B2 PBS vector will be present after selection for neo expression (right), while a DNA element (X) capable of shielding the internal promoter from the silencer gives rise to a cell population of both Pro and B2 proviruses (left).
TABLE 1.
Vector transduction efficienciesa
| Vector | Expt no. | Titer (CFU/ml)
|
NIH 3T3/F9 ratio | Avg ratiob | Old NIH 3T3/ F9 ratioc | |
|---|---|---|---|---|---|---|
| F9 | NIH 3T3 | |||||
| PBS-Pro | 1 | 0 | 3.7 × 105 | NVd | 66,346 | |
| 2 | 0 | 5.4 × 104 | NV | |||
| 5 | 4 | 4.6 × 105 | 115,000 | |||
| 7 | 26 | 4.6 × 105 | 17,692 | |||
| ProPLTneo-TATA− | 1 | 2.3 × 103 | 7.8 × 104 | 34 | 62 | |
| 2 | 4.9 × 102 | 4.8 × 104 | 98 | |||
| 3 | 1.2 × 103 | 1.0 × 105 | 83 | |||
| 5 | 7.4 × 103 | 2.5 × 105 | 34 | |||
| B2PLTneo-TATA− | 1 | 3.6 × 103 | 8.6 × 104 | 24 | 33 | |
| 3 | 1.5 × 103 | 9.0 × 104 | 60 | |||
| 5 | 1.9 × 104 | 2.9 × 105 | 15 | |||
| LJ-P | 4 | 1.3 × 103 | 2.2 × 105 | 169 | 274 | 6,651 |
| 5 | 6.6 × 102 | 2.5 × 105 | 379 | |||
| LJ-Q | 4 | 1.0 × 104 | 2.7 × 105 | 27 | 25 | 80 |
| 5 | 1.4 × 104 | 3.3 × 105 | 24 | |||
| LJ-PAdMLPEnh− | 4 | 6 | 6.0 × 103 | 1,000 | 1,891 | 390 |
| 5 | 9 | 2.9 × 104 | 3,222 | |||
| 6 | 5 | 6.7 × 103 | 1,340 | |||
| 7 | 2 | 3.0 × 103 | 1,500 | |||
| LJ-QAdMLPEnh− | 4 | 1.0 × 102 | 8.0 × 103 | 80 | 52 | 9 |
| 5 | 2.9 × 102 | 1.8 × 104 | 62 | |||
| 6 | 4.8 × 102 | 7.0 × 103 | 15 | |||
| LJ-B2AdMLPEnh− | 6 | 1.1 × 102 | 2.3 × 103 | 21 | 21 | |
| 6 | 1.0 × 102 | 2.6 × 103 | 26 | |||
| 7 | 0.9 × 102 | 1.5 × 103 | 17 | |||
NIH 3T3 and F9 cells were transduced in parallel with dilutions of virus particles from transiently transfected BOSC 23 cells.
Average of NIH 3T3/F9 ratio.
Data are from Petersen et al. (49).
NV, not valid.
Quantification of B2 PBS and Pro PBS proviruses in target cells.
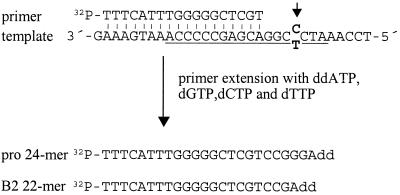
The composition of proviruses in the resulting cell population was measured in a primer extension assay (42) (Fig. 3) employing an 18-mer extension oligonucleotide whose 3′ end anneals four residues away from the variable site in the PBS. Extension in the presence of ddATP, dGTP, dCTP, and dTTP results in either of two radiolabeled products, a 24-mer from a wt template or a 22-mer from a B2 template, because the reaction terminates at the site of incorporation of the dideoxy analog.
FIG. 3.
Primer extension assay for analysis of transduced proviruses. The PBS (underlined) and surrounding sequences are shown with the 18-mer end-labeled extension primer annealed. The arrow points to the position of the divergent nucleotide (bold), T in B2 PBS and C in Pro PBS. Extension was terminated by incorporation of a ddATP analog, resulting in a 22-mer extension product from a B2 PBS template and a 24-mer product from a Pro PBS template. The products were resolved by polyacrylamide gel electrophoresis.
Akv MLV-based vectors with internal SV40 promoter.
For establishment of the assay, we employed the vectors ProPLTneo-TATA− and B2PLTneo-TATA− (Fig. 1) consisting of an Akv MLV retroviral vector backbone, a Pro PBS or B2 PBS, respectively, a polylinker for insertion of putative insulators 51 bp downstream of the PBS, an internal SV40 promoter-neo gene cassette, and a mutation of the TATA box in the LTR, disabling any transcription from the LTR in the target cells (44). For determination of transduction efficiencies, F9 and NIH 3T3 cells were transduced with serially diluted supernatant and selected in 0.4 and 0.6 mg of G418/ml, respectively. The titers on NIH 3T3 cells were similar for the two constructs (Table 1), but unexpectedly, on F9 cells the difference was only two- to threefold in favor of B2PLTneo-TATA− in repeated experiments. As the absolute titers of different virus stocks may vary, the ratio of the NIH 3T3 titer versus the F9 titer, the restriction index (32), is used to compare EC restriction of different constructs within the same experiment. Data given as the average of three independent transductions (Table 1) show that the mean difference in restriction between the B2 PBS and the Pro PBS vector was only twofold, indicating that the internal SV40 promoter of the Akv MLV-based vector was not significantly repressed by the silencer in the Pro PBS region. Since most studies concerning silencing in embryonic cells have been performed employing MoMLV or MoMLV-derived vectors, it is possible that vectors based on the related Akv MLV virus might not be subject to the same degree of silencing. Similarly, differentiation of the F9 cells could account for the lack of silencing of ProPLTneo-TATA−. However, an Akv MLV-based vector, PBS-Pro (40), in which the neo gene is transcribed from the LTR was efficiently silenced in parallel transductions (Table 1), confirming the undifferentiated state of the F9 cells and indicating that the silencing mechanism in embryonic cells also applies to vectors based on Akv MLV.
MoMLV-based vectors with internal SV40 or AdMLP.
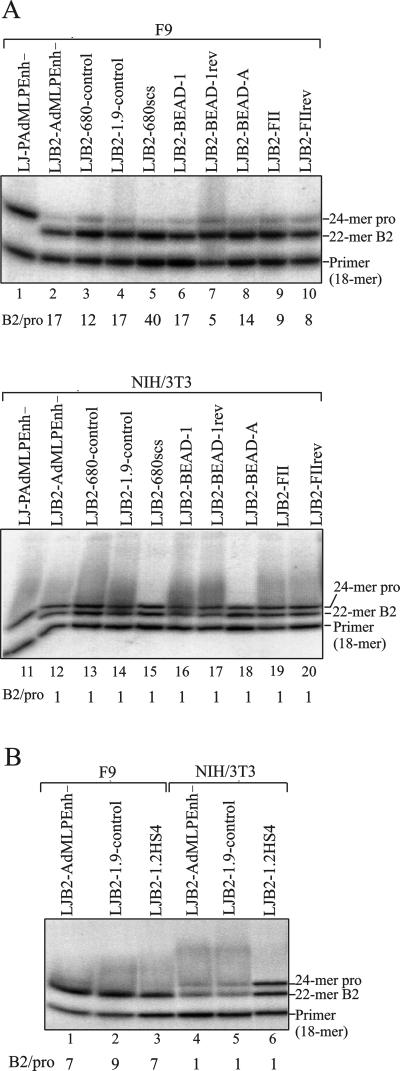
In the MoMLV-based vectors LJ-P and LJ-PAdMLPEnh− (49) (Fig. 1A), an internal SV40 or AdMLP, respectively, were found to be efficiently silenced in F9 cells, while similar vectors with a Gln PBS were unrestricted (49). When testing these vectors, we obtained titers analogous to those found by Petersen et al. (49) in both F9 and NIH 3T3 cells (compare restriction indices in Table 1), except that we did not see as profound an effect on LJ-P in F9 cells. Hence, we achieved a restriction index of 274 for LJ-P as opposed to 6,651 found by Petersen et al. (49). LJ-P is thus only 10 times more restricted than LJ-Q, while the difference between LJ-PAdMLPEnh− and LJ-QAdMLPEnh− is approximately 40-fold (Table 1). LJ-PAdMLPEnh− and LJ-QAdMLPEnh− contain an 178-bp deletion encompassing most of the enhancer repeats in U3 of the LTR and therefore, in general, have lower titers on both target cells compared to the other vectors. However, since LJ-PAdMLPEnh− is efficiently silenced in F9 cells with a titer of 6, we sought to employ this vector for our purpose by replacing the wt PBS with the B2 PBS, obtaining LJB2-AdMLPEnh−, and testing the transduction efficiency as compared to that of LJ-PAdMLPEnh−. LJB2-AdMLPEnh− transduces both NIH 3T3 and F9 cells with efficiencies similar to those of LJ-QAdMLPEnh− (Table 1); i.e., LJ-PAdMLPEnh− is approximately 50 times more restricted than LJB2-AdMLPEnh− in F9 cells. When the transduced LJB2-AdMLPEnh− colonies were analyzed with a primer extension assay, this difference in restriction was reflected as a B2 band that was 17-fold more intense than the Pro band in F9 (Fig. 4A, lane 2), while in fibroblasts both Pro PBS and B2 PBS versions of the provirus were present in equal amounts (Fig. 4A, lane 12). As a control, cells transduced with LJP-AdMLPEnh− only contained provirus with a Pro PBS (Fig. 4A, lanes 1 and 11). Based on the difference in restriction indices between LJ-PAdMLPEnh− and LJB2-AdMLPEnh− when measured in titer experiments (Table 1), a more pronounced ratio was expected in the primer extension assay. Readthrough of the first A in primer extension on a B2 template and termination at the subsequent A (Fig. 3) could account for an overestimation of the Pro band in the population. We do not consider this very likely, however, since only on very rare occasions did we see imperfect termination with the modified DNA polymerase, revealed by an additional, weak 30-nt band from a Pro PBS template (data not shown). No imperfect termination is seen for LJ-PAdMLPEnh− in the assay shown here (Fig. 4A, lanes 1 and 11).
FIG. 4.
Primer extension analysis of transduced vector proviruses with putative insulators. Genomic DNA from G418-selected populations of F9 and NIH 3T3 cells transduced in parallel with vectors as indicated above each lane were employed for PCRs with primers specifically amplifying the transduced provirus. Amplified fragments were subjected to primer extension dideoxy termination analysis with an end-labeled primer as shown in Fig. 3 to analyze the presence of Pro PBS and B2 PBS proviruses. Extension products were resolved by denaturing polyacrylamide gel electrophoresis, visualized after exposure in a Personal Molecular Imager Fx, and quantified by using Quantity One software. The ratio of B2 PBS to Pro PBS band intensities is given below each lane. Panels A and B are from two independent experiments.
During optimization of the assay, we tested a range of selection levels on the target cells. If the resistant colonies primarily reflect the transcriptional strength of the vector itself, it could be argued that the applied selection level of 0.4 mg/ml may be too low, thereby allowing expression of LJ-PAdMLPEnh−; conversely, the differences between the vectors may be blurred if the selection level is too high, resulting in colonies arising primarily due to a positive influence on transcription by the surrounding chromatin. To test these possibilities, F9 cells were split into four dishes the day after transduction, selected in 0.2, 0.4, 1, or 2 mg of G418/ml, and resistant colonies were subjected to primer extension analysis. No major change in the relative intensities of the Pro and B2 bands were seen under the different selection schemes (B2/Pro ratios of 8 to 26 were obtained with the vectors LJB2-AdMLPEnh−, LJB2-680-control, and LJB2-680scs [Fig. 1] at G418 levels from 0.2 to 1 mg/ml [data not shown]). Only at 2.0 mg of G418/ml was the Pro PBS band absent from most analyses, but it also severely reduced the titer to a level providing only 30 to 40 colonies for the analysis. A final level of 0.4 mg of G418/ml was chosen.
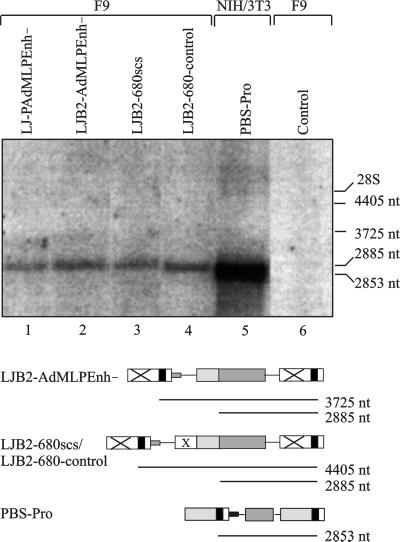
Since the TATA box is retained in the LTR in the enhancer-deleted vectors, we wanted to be sure that the difference in restriction could not be attributed to LTR transcription from LJB2-AdMLPEnh−. For this purpose, we made a Northern blot with a neo-specific probe of the two parental vectors and of the vectors LJB2-scs and LJB2-680-control (Fig. 1B) with 680-bp inserts of scs and control sequences, respectively (Fig. 5). The 2,885-nt band arising from the internal promoter was present in all vectors (Fig. 5, lanes 1 to 4), while no bands of 4,405 or 3,725 nt, indicative of LTR-promoted transcription of vectors with or without a 680-bp insert, respectively, are seen, confirming that neo expression is driven by the internal promoter. This is in accordance with results for a similar vector harboring an internal SV40 instead of AdMLP (49).
FIG. 5.
Proviral neo transcripts in F9 cells. RNA was extracted from G418-selected populations transduced by viral supernatant from transfected BOSC 23 cells. Twenty micrograms of total RNA was resolved by denaturing agarose gel electrophoresis, blotted onto Zeta-Probe membranes, and hybridized with a neo-specific probe. The blot was exposed in a PhosphorImager Fx and visualized by using ImageQuant software. The expected sizes of transcripts initiated from the LTR or the internal promoter are delineated below each vector, using the graphic representation from Fig. 1. Position of bands in the gel were inferred from the location of 28S (4.7 kb) and 18S (1.9 kb) ribosomal bands seen on ethidium bromide staining of the agarose gel. NIH 3T3 cells transduced with PBS-Pro (lane 5) were included as a size marker. Control, untransduced F9 cells.
Test of putative insulators in promoter shielding assay.
To test if repression of the internal promoter by the silencer at the Pro PBS could be shielded, we cloned some of the putative insulators in LJB2-AdMLPEnh− between the PBS and the internal promoter (Fig. 1B). LJB2-680scs contains a 680-bp subfragment of the scs insulator element from D. melanogaster (30, 69). This subfragment containing the DNase I-hypersensitive and -resistant regions of scs has been found to be as effective as the entire 1.8-kb element in blocking enhancer-activated transcription of the Xenopus rRNA promoter (16). LJB2-BEAD-1 contains the 1,970-bp BEAD-1 fragment from the human T-cell receptor α/δ locus (79), which in LJB2-BEAD-1rev is cloned in the opposite orientation. A 120-bp subfragment, BEAD-A (5), encompassing the CTCF binding site was cloned in LJB2-BEAD-A. LJB2-1.2HS4 harbors the 1.2-kb DNA element corresponding to 5′ DNase I-hypersensitive site 4 of the chicken β-globin locus (12, 13). In LJB2-FII, and in the reverse orientation in LJB2-FIIrev, is the 42-bp CTCF binding site sequence derived from the chicken insulator that is sufficient to account for most of its enhancer blocking ability (5). A 680- or 1,900-bp length of presumed noninsulating DNA was cloned in the vectors LJB2-680-control and LJB2-1.9-control, respectively, to test for a potential effect of increased distance between the silencer and the internal promoter on repression. For all transduced vectors except the controls, DNA from 125 to 300 resistant F9 or NIH 3T3 colonies was used for primer extension analysis (Fig. 4). Both of the vectors LJB2-680-control and LJB2-1.9-control had lower titers than the others and gave rise to between 9 and 45 colonies (data not shown).
In the transduced fibroblasts, all vectors showed an equal intensity of the 22-mer B2 PBS band and the 24-mer Pro PBS band (Fig. 4A, lanes 12 to 20, and Fig. 4B, lanes 4 to 6), confirming a reversion frequency of 50% for the B2 PBS (7) in a cell population with no restriction to viral expression. In F9 cells, the difference in intensity of 12 to 17 times between the bands seen with LJB2-AdMLPEnh− was retained after insertion of up to 1.9 kb of spacer DNA between the silencer and the promoter (Fig. 4A, lanes 2 to 4; Fig. 4B, lanes 1 and 2), showing repression of the internal promoter to function over large distances. Insertion of any of the putative insulators did not change this relation between the B2 band and the Pro band. The differences in intensity as measured in a Personal Molecular Imager ranged from 5 to 40 (Fig. 4A, lanes 5–10; Fig. 4B, lane 3); however, a high background in the lane showing the fivefold difference (Fig. 4, lane 7) has probably obscured the measurement in that lane. The Pro band is in no case seen at a level similar to that of the B2 band. This applies to all the tested insulators as well as to the subfragments, which in enhancer-blocking assays (5, 16) retain the activity of the full-length elements. These data are consistent with the early results obtained with the vectors LJB2-AdMLPEnh−, LJB2-680scs, and LJB2-680-control, which were tested repeatedly under various selection schemes during establishment of the assay.
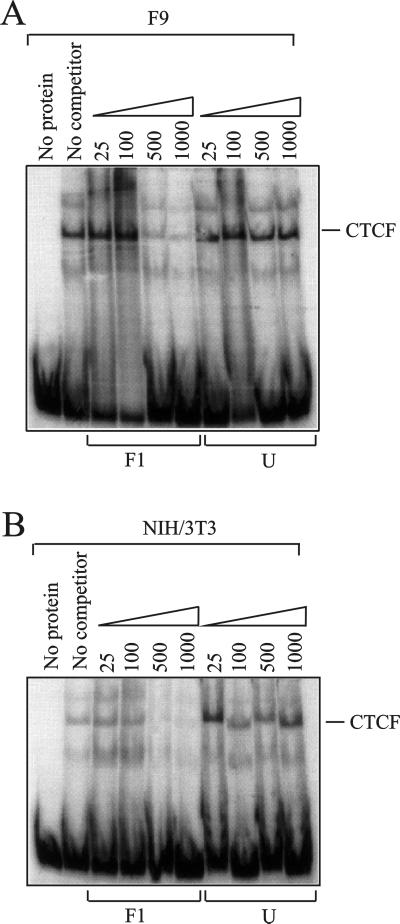
We conclude that the silencing of the internal promoter mediated by the PBS region is not shielded by any of the possible insulators, scs, BEAD-1, or HS4, although the sensitivity of the assay did not permit detection of a weak insulating capability. Since the enhancer blocking function of HS4, BEAD-1, and other chromatin insulators is mediated by the ubiquitously expressed zinc finger protein CTCF (5), we wanted to test for the presence of this protein in the F9 cells. We performed a gel shift experiment employing radioactively labeled CTCF-binding probe F1 (see Materials and Methods) which in both F9 and control NIH 3T3 cells gave rise to a major, CTCF-specific band (Fig. 6). This band could be competed by 500- and 1,000-fold molar excesses of unlabeled F1 but not by an unspecific competitor. The minor shifts may have derived from degradation of CTCF (lower band) and from Oct-1 (upper band), which also binds the F1 probe (33). Lack of insulation in our assay is thus not due to the absence of CTCF in the F9 cells.
FIG. 6.
Electrophoretic mobility shift assay with a CTCF-binding probe. A 10-μg sample of nuclear extract from F9 cells (A) or NIH 3T3 cells (B) was incubated with radiolabeled CTCF-binding probe F1 and competed with increasing amounts (25- to 1,000-fold molar excess) of either cold F1 probe (left) or cold unspecific probe (right). The gel was exposed in a Personal Molecular Imager Fx and visualized in Quantity One.
DISCUSSION
Transcriptional repression of MLV in embryonic cells has been ascribed partly to binding of a protein to a silencing element overlapping the Pro PBS of the viral genome (3, 32, 39, 49, 76, 77) and partly to malfunction of the viral enhancer caused by a lack of transcription factors (17, 35, 36, 37, 62) and/or the presence of a negative factor recognizing sequences of the enhancer (1, 22, 67). Repression has been shown also to apply to heterologous promoters placed downstream of the PBS (49), though the effect diminishes with increasing distance (32), and to correlate with methylation of the 5′ part of the provirus and flanking DNA (7, 28, 55, 56, 75).
Here we investigated the ability of the embryonic silencer residing at the Pro PBS to be shielded by putative insulators. The assay took advantage of the mutant B2 PBS conferring escape from suppression in embryonal cells. Since this point mutation of the PBS reverts at high frequency (7), a shielding ability of a putative insulator could be measured directly, revealed by the ratio of B2 PBS expressing proviruses to Pro PBS proviruses in the target EC cell population. Our results show that none of the putative insulators, scs, BEAD-1, or HS4, is able to shield an internal promoter from the repressive effect of the silencer region at the PBS when inserted between the silencer and the promoter.
Repression of an internal promoter by the PBS-associated silencer.
We were not able to confirm the pronounced repression of the internal SV40 promoter in LJ-P as reported by Petersen et al. (49), nor did we achieve strong repression of an internal SV40 promoter in an Akv MLV vector very similar to the MoMLV-based LJ-P (Table 1). Of our SV40 promoter vectors with a mutation of the TATA box in the LTR, ProPLTneoTATA− was on average only two times more repressed than the corresponding B2PLTneoTATA− when comparing transduction efficiencies on NIH 3T3 and F9 cells. This ratio was not affected when we assayed isogenic vectors without mutation of the TATA box at a range of selection levels, from 250 to 2,000 μg of G418/ml (C. Modin, F. S. Pedersen, and M. Duch, unpublished data). For LJ-P and LJ-Q, the difference in restriction index was approximately 10-fold; hence, the internal SV40 promoter escapes most of the silencing from the PBS in both an MoMLV and an Akv MLV context. In accordance with the results of Petersen et al. (49), we found the vector LJ-PAdMLPEnh− with an internal AdMLP and a wt PBS to be sensitive to silencing, since on average it is repressed 36 and 90 times more than the Gln PBS or B2 PBS counterparts, respectively. We note that LJ-PAdMLPEnh− differs from the Pro PBS internal SV40 vectors by deletion of the LTR enhancer repeats. We cannot exclude the possibility that this LTR enhancer deletion contributes to efficient repression of the internal promoter in LJ-PAdMLPEnh− by decreasing the overall amount of enhancer elements in the construct or by abolishing LTR transcription (Fig. 5), hence reducing competition between the LTR and the internal promoter for a silencer. However, since LTR-initiated transcription is also eliminated in the TATA box-mutated vector (44), lack of transcription from the LTR per se is not sufficient to direct silencing to the internal promoter. Rather, susceptibility to repression might be influenced by the strength (74) and the nature of the transcriptional elements; i.e., the SV40 promoter-enhancer might be less prone to silencing due to an increased strength compared to that of the AdMLP. In addition, the SV40 early promoter-enhancer harbors six CpG-containing sites recognized by the ubiquitous transcription factor Sp1 possibly contributing to the inability to achieve efficient silencing in F9 cells, as Sp1 sites have been implicated in conferring resistance to de novo methylation in both embryonal and nonembryonal cell lines (8, 41, 43, 53) and in the expressional capability of Moloney murine sarcoma virus escape mutants myeloproliferative sarcoma virus and PCC4 cell-passaged myeloproliferative sarcoma virus in EC cells (51).
Properties of the assay for promoter shielding.
Based on the titer differences between the vectors LJP-AdMLPEnh− and LJB2-AdMLPEnh− in F9 cells, we employed the latter to establish an assay testing the ability of putative insulators to shield the silencer in the PBS region. The assay measures differences in the levels of B2 PBS and Pro PBS revertants in the target cell population by primer extension analysis and relies on (i) equal reversion of the B2 PBS to the wt sequence under conditions of no selective pressure and (ii) repression of an internal promoter in G418-selected EC cells exerted by the silencer at the Pro PBS but not by the B2 PBS. We have confirmed these conditions by analysis of populations transduced with LJB2-AdMLPEnh−. An equal amount of Pro PBS and B2 PBS proviruses seen for NIH 3T3 populations (Fig. 4A, lane 12, and Fig. 4B, lane 4) corroborates reversion of the B2 PBS to the wt sequence (7), since the fibroblasts have no restrictions toward viral expression. Conversely, repression of the Pro PBS revertant in F9 cells is reflected in the primer extension analysis, which show a B2 PBS band 7 to 17 times more intense than a Pro PBS band (Fig. 4A, lane 2; Fig. 4B, lane 1). The analysis was performed on a population of transduced cells representing a large number of different integration sites, thereby reducing the contribution from positional effects at individual sites. However, based on the difference in restriction indices between LJ-PAdMLPEnh− and LJB2-AdMLPEnh− as measured in titer experiments (Table 1), a more pronounced ratio was expected in the primer extension assay. Stochastic fluctuation in the mismatch repair accounting for this discrepancy is unlikely, since in general more than 100 colonies were analyzed for each transduction. Rather, the disproportionate increase in the amount of Pro PBS in the LJB2-AdMLPEnh−-transduced population compared to the low titer of the LJ-PAdMLPEnh−vector may derive from LJB2-AdMLPEnh− cells aiding the survival of the otherwise poorly growing, low-expressing LJ-PAdMLPEnh− revertants. The high number of colonies analyzed should also have reduced the effects of differences in colony size, which are frequently seen with the F9 cells.
Distance effects of the PBS silencer.
The repressive effect of the Pro PBS silencer has been shown to decrease with increasing distance to the internal promoter. Assaying two-gene vectors in F9 and NIH 3T3 cells, an LTR-driven luc reporter was repressed 20-fold more in a Pro vector than in a Gln vector while expression of the SV40-promoted neo gene placed 2 kb downstream of the PBS was barely distinguishable between the two vector versions (32). However, when controlling for the effect of altering the spacing between the PBS and the internal promoter, we observed that repression of the AdMLP in the Pro PBS provirus compared to the B2 PBS version was maintained after insertion of 680 bases or 1.9 kb of spacer sequences in LJB2-680-control and LJB2-1.9-control, respectively (Fig. 4A, lanes 2 to 4; Fig. 4B, lanes 1 and 2). A single-base-pair difference in the silencer at the PBS thus affects transcription from a promoter located almost 2 kb away. However, the reduced titers of these vectors in both F9 and NIH 3T3 cells may reflect a negative effect of the spacer during transduction, questioning the anticipation of functionally inert DNA sequences. The LTR enhancerless vectors may depend on stimulatory effects from the surrounding chromatin for transcription from the AdMLP; hence, the reduced titer could derive from impaired interactions with endogenous enhancers caused by the increased distance and/or properties of the specific sequence inserted. Since no reduction of the titers was observed on insertion of the enhancer blocking elements BEAD-1 and HS4, however, this result of the control vectors is most likely explained by a negative effect of the spacer sequences on vector transfer.
Lack of shielding by scs, HS4, and BEAD-1 insulator elements.
Despite several lines of evidence showing the insulators' abilities to block a variety of repressive effects (13, 30, 50, 55, 60, 71, 74), we saw in our assay no indication that the tested elements blocked the silencing mechanism residing at the PBS region in a retroviral vector. Hence, neither the full-length HS4 operative in protection against position effects (13, 50) nor BEAD-1 or the delineated elements of HS4, BEAD-1 (5, 79), and scs (16, 72) proficient for enhancer blocking had any effect in our assay (Fig. 4). The sensitivity of the assay, however, did not allow us to distinguish minor contributions from the putative insulators in shielding of the internal promoter. We cannot exclude the possibility that a blocking effect is obtained by multimerizing an element or subfragments thereof in accordance with previous results for chicken HS4 (5, 12, 13, 71), the gypsy insulator (59), and scs (19, 72). However, we refrained from testing this, since in the context of a retrovirus such repeated elements would most likely recombine and be deleted from the vector. Additionally, although they may operate in conjunction with other as-yet-unidentified elements to accomplish their role in their natural settings, the putative insulators themselves are present only as single copies, making it relevant to test them like this in the retroviral vector.
One reason for the lack of insulating properties in our assay may be that DNA-protein interactions required for insulator function are not sufficiently conserved in the murine embryonal cell line employed. In Drosophila, protein SBP has been shown to be a component of the scs insulating complex (19), and the ubiquitously expressed DNA-binding protein CTCF is required and sufficient for enhancer blocking activity of the vertebrate insulators HS4, BEAD-1, and RO (5). We show here that CTCF is also present in F9 cells (Fig. 6). However, for HS4 the ability to protect against position effects is governed by sequences outside the CTCF binding part of the element (5), making it likely that the complete activity involves multiple components, as is also the case for the gypsy insulator (21). Nevertheless, the function of some insulators seem evolutionarily conserved. Chicken HS4 blocks chromosomal position effects in Drosophila (13), and although scs is from Drosophila melanogaster, in which no methylation is operant, the element functions in enhancer blocking in Xenopus (16), as well as in human Jurkat cells (79), and in blocking repression mediated by chromatin-associated repressors in human U-2 OS cells (71). No insulator activity for scs was seen in a colony assay with human K562 cells (13). Thus, as is also evident from the description of HS4 above and from the results presented here, each assay contributes to reveal insulator function, but the understanding is hampered by lack of knowledge about enhancer action and the actual repression mechanisms studied. In the context of the retroviral vector employed here, the putative insulators did not function as mere boundaries between domains.
The nature of the silencer element at the PBS region is not understood, nor are the interactions mediating the spreading to an internal promoter. Yet from the results shown here, they appear to differ in constitution from mechanisms of enhancer-promoter interactions, positional effects, and silencing exerted by some of the described chromatin-associated repressors, such as the PcG proteins HPC2, RING1, and Su(z)2, mHP1, and the methyl-CpG-binding MeCP2.
ACKNOWLEDGMENTS
We kindly acknowledge E. Barklis for providing the vectors pLJP, pLJQ, pLJQ-AdMLPEnh−, pLJPro-AdMLPEnh−, and B2BAG, H. Cai for the scs element, P. Jørgensen for the ALF plasmid, M. S. Krangel for BEAD-1, G. Felsenfeld for HS4, L. Burke for the F1 probe and guidance on bandshift analysis, and L. Svinth for technical assistance.
This work was supported by contracts CT 95-0100 (Biotechnology) and CT 95-0675 (Biomed 2) of the European Commission, the Karen Elise Jensen Foundation, the Danish Cancer Society, the Danish Biotechnology Programme, the Danish Natural Sciences, and Medical Research Councils.
REFERENCES
- 1.Akgün E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J Virol. 1991;65:382–388. doi: 10.1128/jvi.65.1.382-388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baniahmad A, Steiner C, Köhne A C, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–514. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- 3.Barklis E, Mulligan R C, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 4.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 5.Bell A C, West A D, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 6.Bernstine E G, Hooper M L, Grandchamp S, Ephrussi B. Alkaline phosphatase activity in mouse teratoma. Proc Natl Acad Sci USA. 1973;70:3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berwin B, Barklis E. Retrovirus-mediated insertion of expressed and non-expressed genes at identical chromosomal locations. Nucleic Acids Res. 1993;21:2399–2407. doi: 10.1093/nar/21.10.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandeis M, Frank D, Keshet I, Siegfred Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 9.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova G N, Lobanenkov V V, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 11.Challita P-M, Skelton D, El-Khoueiry A, Yu X-J, Weinberg K, Kohn D B. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung J H, Bell A D, Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung J H, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 14.Colicelli J, Goff S P. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1987;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Auriol L, Yang W K, Tobaly J, Cavaleiri F, Peries J, Emanoil-Ravicovitch R. Studies on the restriction of ecotropic murine retrovirus replication in mouse teratocarcinoma cells. J Gen Virol. 1981;55:117–122. doi: 10.1099/0022-1317-55-1-117. [DOI] [PubMed] [Google Scholar]
- 16.Dunaway M, Hwang J Y, Xiong M, Yuen H-L. The activity of the scs and scs′ insulator element is not dependent on chromosomal context. Mol Cell Biol. 1997;17:182–189. doi: 10.1128/mcb.17.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feuer G, Taketo M, Hanecak R C, Fan H. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J Virol. 1989;63:2317–2324. doi: 10.1128/jvi.63.5.2317-2324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franz T, Hilberg F, Seliger B, Stocking C, Ostertag W. Retroviral mutants efficiently expressed in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1986;83:3292–3296. doi: 10.1073/pnas.83.10.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaszner M, Vasquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautsch J W, Wilson M C. Delayed de novo methylation in teratocarcinoma suggests additional mechanisms for controlling gene expression. Nature. 1983;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- 21.Gdula D A, Gerasimova T I, Corces V G. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman C M, Rigby P W J, Lane D P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985;42:519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- 23.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 24.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilberg F, Stocking C, Ostertag W, Grez M. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1987;84:5232–5236. doi: 10.1073/pnas.84.15.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jähner D, Stuhlmann H, Stewart C L, Harbers K, Löhler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 28.Jähner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 29.Jones P L, Venstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 30.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 31.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempler G, Freitag B, Berwin B, Nanassy O, Barklis E. Characterization of the Moloney leukemia virus stem cell-specific repressor binding site. Virology. 1993;193:690–699. doi: 10.1006/viro.1993.1177. [DOI] [PubMed] [Google Scholar]
- 33.Köhne A C, Baniahmad A, Renkawitz R. A ubiquitous transcription factor synergizes with v-ERBA in transcriptional silencing. J Mol Biol. 1993;232:747–755. doi: 10.1006/jmbi.1993.1428. [DOI] [PubMed] [Google Scholar]
- 34.Laker C, Meyer J, Schopen A, Friel J, Heberlein C, Ostertag W, Stocking C. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol. 1998;72:339–348. doi: 10.1128/jvi.72.1.339-348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linney E, Davis B, Overhauser J, Chao E, Fan H. Non-function of a Moloney murine leukemia virus regulatory sequence in F9 embryonal carcinoma cells. Nature. 1984;308:470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- 36.Linney E, Neill S D, Prestridge D S. Retroviral vector gene expression in F9 embryonal carcinoma cells. J Virol. 1987;61:3248–3253. doi: 10.1128/jvi.61.10.3248-3253.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh T P, Sievert L L, Scott R W. Proviral sequences that restrict retroviral expression in mouse embryonal carcinoma cells. Mol Cell Biol. 1987;7:3775–3784. doi: 10.1128/mcb.7.10.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh T P, Sievert L L, Scott R W. Negative regulation of retrovirus expression in embryonal carcinoma cells mediated by an intragenic domain. J Virol. 1988;62:4086–4095. doi: 10.1128/jvi.62.11.4086-4095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loh T P, Sievert L L, Scott R W. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol Cell Biol. 1990;10:4045–4057. doi: 10.1128/mcb.10.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lund A H, Duch M, Lovmand J, Jørgensen P, Pedersen F S. Mutated primer binding sites interacting with different tRNAs allow efficient murine leukemia virus replication. J Virol. 1993;67:7125–7130. doi: 10.1128/jvi.67.12.7125-7130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 42.Modin C, Pedersen F S, Duch M R. Comparison of DNA polymerases for quantification of single nucleotide differences by primer extension analysis. BioTechniques. 2000;28:48–50. doi: 10.2144/00281bm08. [DOI] [PubMed] [Google Scholar]
- 43.Mummaneni P, Yates P, Simpson J, Rose J, Turker M S. The primary function of a redundant Sp1 binding site in the mouse aprt gene promoter is to block epigenetic gene inactivation. Nucleic Acids Res. 1998;26:5163–5169. doi: 10.1093/nar/26.22.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajima K, Ikenaka K, Nakahira K, Morita N, Mikoshiba K. An improved retroviral vector for assaying promoter activity. Analysis of promoter interference in pTP211 vector. FEBS Lett. 1993;315:129–133. doi: 10.1016/0014-5793(93)81148-s. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng H-H, Jones C A, Laherty C D, Turner B M, Eisenman R N, Bird A D. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen A L, Pallisgaard N, Pedersen F S, Jørgensen P. Murine helix-loop-helix transcriptional activator proteins binding to the E-box motif of the Akv murine leukemia virus enhancer identified by cDNA cloning. Mol Cell Biol. 1992;12:3449–3459. doi: 10.1128/mcb.12.8.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 48.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol. 1991;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prince V E, Rigby P W J. Derivatives of Moloney murine sarcoma virus capable of being transcribed in embryonal carcinoma stem cells have gained a functional SpI binding site. J Virol. 1991;65:1803–1811. doi: 10.1128/jvi.65.4.1803-1811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prioleau M-N, Nony P, Simpson M, Felsenfeld G. An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qu G, Ehrlich M. Demethylation and expression of methylated plasmid DNA stably transfected into HeLa cells. Nucleic Acids Res. 1999;27:2332–2338. doi: 10.1093/nar/27.11.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Recillas-Targa F, Bell A C, Felsenfeld G. Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc Natl Acad Sci USA. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivella S, Callegari J A, May C, Tan C W, Sadelain M. The chicken HS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J Virol. 2000;74:4679–4687. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robbins P B, Yu X-J, Skelton D S, Pepper K A, Wasserman R M, Zhu L, Kohn D B. Increased probability of expression from modified retroviral vectors in embryonal stem cells and embryonal carcinoma cells. J Virol. 1997;71:9466–9474. doi: 10.1128/jvi.71.12.9466-9474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H-A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Scott K S, Taubman A D, Geyer P K. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 1999;153:787–798. doi: 10.1093/genetics/153.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigrist C J A, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 62.Speck N A, Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987;7:1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart C L, Stuhlmann H, Jähner D, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strickland S. Mouse teratocarcinoma cells: prospects for the study of embryogenesis and neoplasia. Cell. 1981;24:277–278. doi: 10.1016/0092-8674(81)90313-5. [DOI] [PubMed] [Google Scholar]
- 65.Sun F-L, Elgin S R. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 66.Teich N M, Weiss R A, Martin G R, Lowy D R. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 67.Tsukiyama T, Niwa O, Yokoro K. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol Cell Biol. 1989;9:4670–4676. doi: 10.1128/mcb.9.11.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Udvardy A. Dividing the empire: boundary chromatin elements delimit the territory of enhancers. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 70.Van Beveren C, Coffin J, Hughes S. Nucleotide sequences complemented with functional and structural analysis. In: Weiss R, Teich H, Varmus H, Coffin J, editors. RNA tumour viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1985. pp. 790–805. [Google Scholar]
- 71.van der Vlag J, den Blaauwen J L, Sewalts R G A B, van Driel R, Otte A P. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J Biol Chem. 2000;275:697–704. doi: 10.1074/jbc.275.1.697. [DOI] [PubMed] [Google Scholar]
- 72.Vasquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vernet M, Cebrian J. cis-acting elements that mediate the negative regulation of Moloney murine leukemia virus in mouse early embryos. J Virol. 1996;70:5630–5633. doi: 10.1128/jvi.70.8.5630-5633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walters M C, Fiering S, Bouhassira E E, Scalzo D, Goeke S, Magis W, Garrick D, Hitelaw E, Martin D I K. The chicken β-globin 5′HS4 boundary element blocks enhancer-mediated suppression of silencing. Mol Cell Biol. 1999;19:3714–3726. doi: 10.1128/mcb.19.5.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Robbins P B, Carbonaro D A, Kohn D B. High-resolution analysis of cytosine methylation in the 5′ long terminal repeat of retroviral vectors. Hum Gene Ther. 1998;9:2321–2330. doi: 10.1089/hum.1998.9.16-2321. [DOI] [PubMed] [Google Scholar]
- 76.Weiher H, Barklis E, Ostertag W, Jaenisch R. Two distinct sequence elements mediate retroviral gene expression in embryonal carcinoma cells. J Virol. 1987;61:2742–2746. doi: 10.1128/jvi.61.9.2742-2746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamauchi M, Freitag B, Kan C, Berwin B, Barklis E. Stem cell factor binding to retrovirus primer binding site silencers. J Virol. 1995;69:1142–1149. doi: 10.1128/jvi.69.2.1142-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao K, Hart C M, Laemmli U K. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 79.Zhong X-P, Krangel M S. An enhancer-blocking element between α and δ gene segments within the human T cell receptor α/δ locus. Proc Natl Acad Sci USA. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]