Abstract
The cortex has a characteristic layout with specialized functional areas forming distributed large-scale networks. However, substantial work shows striking variation in this organization across people, which relates to differences in behavior. While most prior work treats all individual differences as primarily linked to boundary shifts between the borders of regions, here we show that cortical ‘variants’ also occur at a distance from their typical position, forming ectopic intrusions. Both ‘border’ and ‘ectopic’ variants are common across individuals, but differ in their location, network associations, properties of subgroups of individuals, activations during tasks, and prediction of behavioral phenotypes. Border variants also track significantly more with shared genetics than ectopic variants, suggesting a closer link between ectopic variants and environmental influences. This work argues that these two dissociable forms of variation – border shifts and ectopic intrusions – must be separately accounted for in the analysis of individual differences in cortical systems across people.
1. Introduction
The cortex shows a characteristic organization, with distinct functional areas linking together to form distributed large-scale systems (i.e., networks1). While following a stereotyped general pattern, this organization is not uniform, varying both across and within species. Comparative neuroanatomy studies demonstrate that variations in cortical organization appear in a constrained set of possible forms, including changes in the relative size, connectivity, or functional characteristics of cortical fields2, 3. Notably, variation in cortical organization within a species can be influenced by both genetic and developmental factors4, 5 and relates to differences in phenotypic characteristics and behavior2. Thus, variation in cortical architecture in humans may provide insights into the sources of varying behavioral traits relevant to cognition and disease.
The regional and systems-level organization of the human brain can be mapped non-invasively using functional connectivity MRI (fcMRI; correlations in the fMRI activity patterns between different regions). FcMRI can be used to identify functionally homogenous regions6-8 and distinct systems9, 10 that correspond to patterns detected with task activation methods. For large groups of individuals, a “canonical” average pattern of distributed functional systems emerges that is reproducible across studies and maps onto differences in motor, sensory, and higher-level processing9, 10.
However, recent work has also highlighted that any given person differs from this group pattern, at least in some locations11-20. We characterized locations, that we call network variants, where an individual’s functional connectivity pattern differs markedly from the typical group average15,. Network variants are stable over time and across task states21 and relate to individual differences in task-related brain activations and behavioral measures15, with consistent differences across sub-groups of individuals. We hypothesize that network variants reflect trait-like variations in the organization of functional brain areas across individuals15.
While many studies have identified individual differences in brain network organization11, 12, 15-17, 19, 20, less work has investigated if variants take different forms and how these forms link to sources and mechanisms observed in past comparative studies of cortical neuroanatomy in other species2. Many studies, including our past work, treat all forms of cortical variation uniformly, often assuming that variants are driven by boundary shifts in the borders between systems: a functional region may expand, contract, or be slightly offset relative to the typical pattern (e.g., as has been documented for V1, which can differ more than 2-fold in size across individuals22, 23). These border shifts likely represent important individual differences linked to variation in behavior19, 24, 25, and could potentially be addressed through functional alignment methods that allow for individual-specific regions to be locally displaced (e.g., refs.24, 26).
However, brain organization may also differ more markedly from the stereotypical pattern, with islands of idiosyncratic fcMRI occurring in locations remote from their canonical system organization27. We call these shifts in brain organization ectopic intrusions to reflect the abnormal locations of these variants (see Fig. 1). Dramatic shifts in the function or connectivity of primary sensory/motor areas can be seen with systematic deprivations in development5, 28, 29, but it is less well understood how common these shifts are in higher-level areas in neurotypical humans – despite some prior observations that they occur11, 15. Ectopic intrusions are not easily explained by expansion mechanisms and will be poorly addressed by functional alignment techniques that assume only local displacements in brain architecture, confounding group studies.
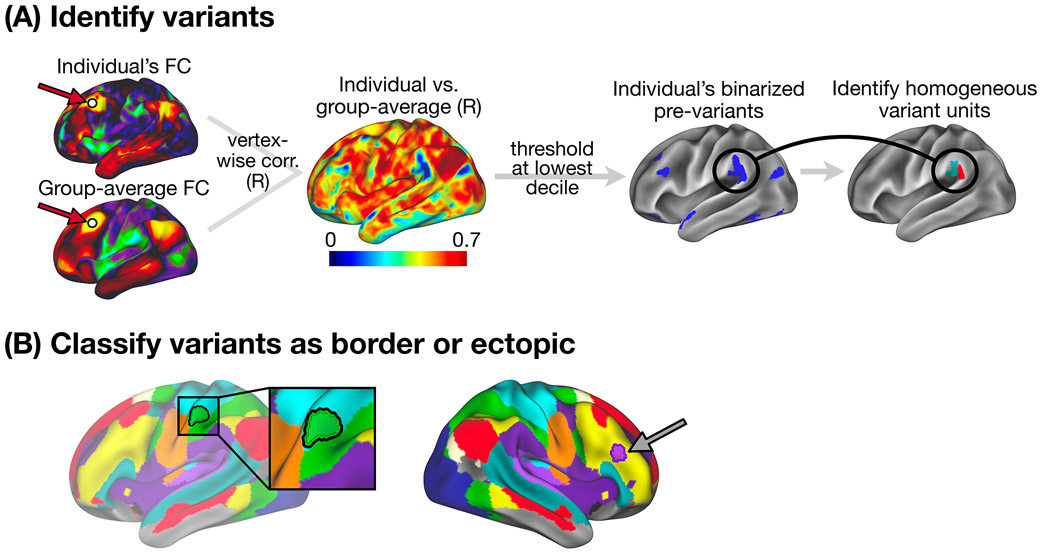
Figure 1: Variant definition, splitting, and classification as border or ectopic.
(A) Following Seitzman et al.15, we used spatial correlation to compare the seedmap at a given location between an individual and an independent group average (left) to generate an individual-to-group “similarity map” (middle). This similarity map was thresholded and binarized to identify locations with low similarity to the group (right) that we call “pre-variants” in this work (Note: these were also thresholded to remove small areas and areas of low signal - see Methods). We then further refined these pre-variants to create homogeneous units for border vs. ectopic variant classification (see Methods and Supp. Fig. 1 for a description of the variant definition process). (B) Each variant was classified as either a border shift or an ectopic intrusion based on its edge-to-edge distance from the nearest same-network boundary in the group-average network map. Here, we display an example of a border shift variant (left, green region with black outline) and an ectopic variant (right, purple region with dark outline), overlayed on the group average network map. Distances > 3.5 mm were classified as ectopic; distances < 3.5 mm were classified as border (see Fig. 2B and Supp. Fig. 8 for exploration of additional distance criteria). We also employed a secondary method for defining border and ectopic variants that did not rely on a group-level network parcellation (see description in Methods and Supp. Fig. 2).
The goal of this study was to examine the relative prevalence of ectopic intrusions and border shift variants in humans, and to contrast the properties of these two forms of variation. Separating these forms will deepen our understanding of individual differences in human brain organization and their sources and consequences.
2. Results
Using a combination of the large HCP dataset (N = 374 unrelated individuals used for primary analyses, and N = 793 individuals used for analyses of behavior and similarity among twin samples) and the precision MSC dataset (N = 9, scanned 10 times each), we investigated “border” shift variants and “ectopic” intrusions – more remote islands of functional networks not adjacent to their typical layout variants (Fig. 1). The two datasets were used in combination to leverage their relative strengths in terms of sample size and data quantity per individual, respectively, and to demonstrate replicability of this work.
We first examined how common ectopic intrusions were relative to border shifts variants across participants. We next asked how the two variant forms compare in their spatial occurrence, the networks they are affiliated with, and how they respond during tasks. We contrasted variants in familial samples to estimate how they are affected by genetic similarity and we examined whether subgroups of individuals showed similar forms of border and ectopic variants. Finally, we examine how border and ectopic variants predict behavioral phenotypes.
2.1. Border and ectopic variants are common in humans
Our first goal was to establish how frequently border and ectopic variants occur. We used two methods for defining border shifts and ectopic intrusions. Our primary method (Fig. 1) defines border shifts and ectopic intrusions based on their distance to pre-defined canonical (i.e., published group-average) network boundaries11. Our secondary method (Supp. Fig. 2) used a parcellation-free approach to define these two forms of variants, based on a continuous measure of similarity to nearby locations in the group-average. This approach has the advantage that it does not depend on a specific group parcellation for networks (e.g., group networks from ref. 11 vs. ref. 10) or network resolution (e.g., 7 vs. 17 networks from Yeo et al.10). Importantly, results were largely convergent across the two methods; see Supp. Results.
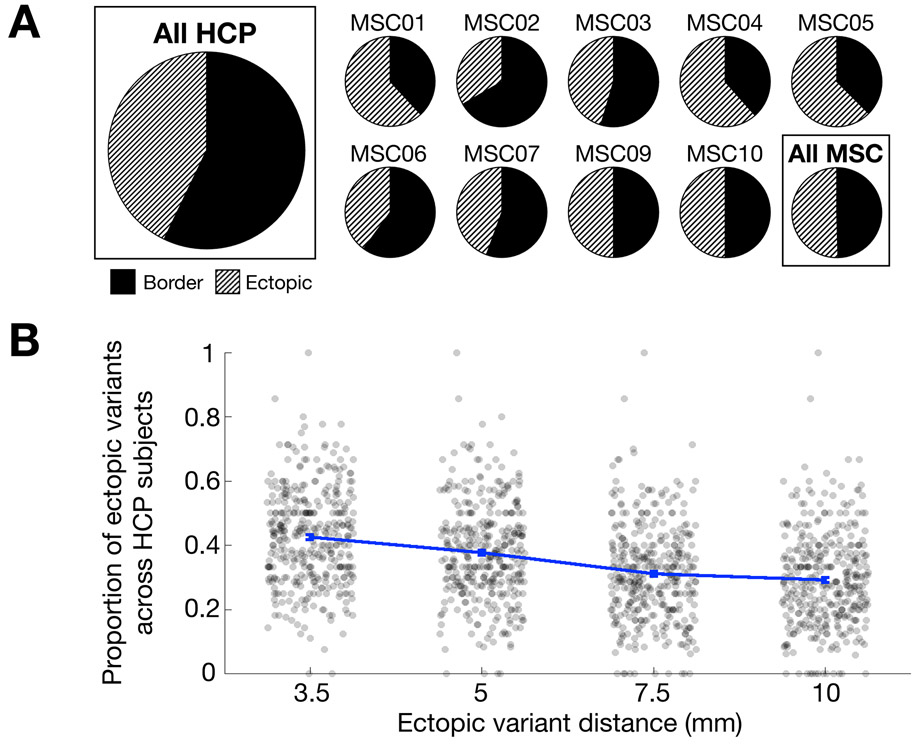
Using either method, both ectopic and border variants were consistently identified in almost all individuals: at least one ectopic variant and at least one border variant were observed in 9/9 MSC and 371/374 HCP subjects (> 99%). In the MSC, an average of 49.9% [±10.4%] of an individual’s network variants were ectopic; in the HCP, an average of 42.6% [±14.9%] of an individual’s network variants were ectopic (Fig. 2A; see Supp. Fig. 3 for similar results with our secondary method and Supp. Fig. 4 for a breakdown of ectopic proportions in the HCP). Increasing the distance criteria (from 3.5 to 5, 7.5, or 10 mm) still left a large proportion of ectopic variants, with roughly 30% of variants classified as ectopic at 10 mm (Fig. 2B). Indeed, the median distance between ectopic variants and their own network was > 15 mm (Supp. Fig. 5). Results were consistent when using our secondary method for defining border and ectopic variants (Supp. Fig. 6). Similar results were also obtained when ectopic variants were defined relative to a participant’s own network boundaries (Supp. Fig. 7, Supp. Table. 1). Thus, distant ectopic variants are not rare phenomena and, along with border variants, appear to be relatively ubiquitous across people.
Figure 2: Prevalence of border and ectopic variants across individuals.
(A) The panel displays the proportion of border and ectopic variants across all subjects in the HCP dataset (far left), and within subjects in the MSC dataset (two rows on right). Both ectopic variants and border variants were consistently identified in almost all individuals in both the HCP and the MSC datasets. (B) Proportions of ectopic variants at other distances (error bars represent SEM across participants; each dot represents a single subject). As the required minimum distance for a variant to be classified as ectopic increases to 5, 7.5, and 10 mm, ectopic variants continue to comprise a sizable percentage of all variants in the HCP dataset, nearly 30%, even at a distance of 10 mm. Indeed, the median distance for ectopic variants from a network border was > 15 mm (Supp. Fig. 5). Similar results are seen with our secondary parcellation-free method of defining network variants (Supp. Fig. 3, 6).
2.2. Variant forms show differences in their spatial distribution
Next, we asked whether ectopic and border variants occur in different locations. We examined the spatial overlap of ectopic variants across people and compared it to the overlap of border variants (Fig. 3A). Both forms of variants generally follow the distribution previously described of higher prevalence in association regions of cortex16, 17, 19, especially in lateral frontal cortex, superior frontal cortex and near the temporoparietal junction15. However, direct contrasts suggest significant differences between border and ectopic variants. Similar distributions were seen in the HCP and MSC datasets (Supp. Fig. 8).
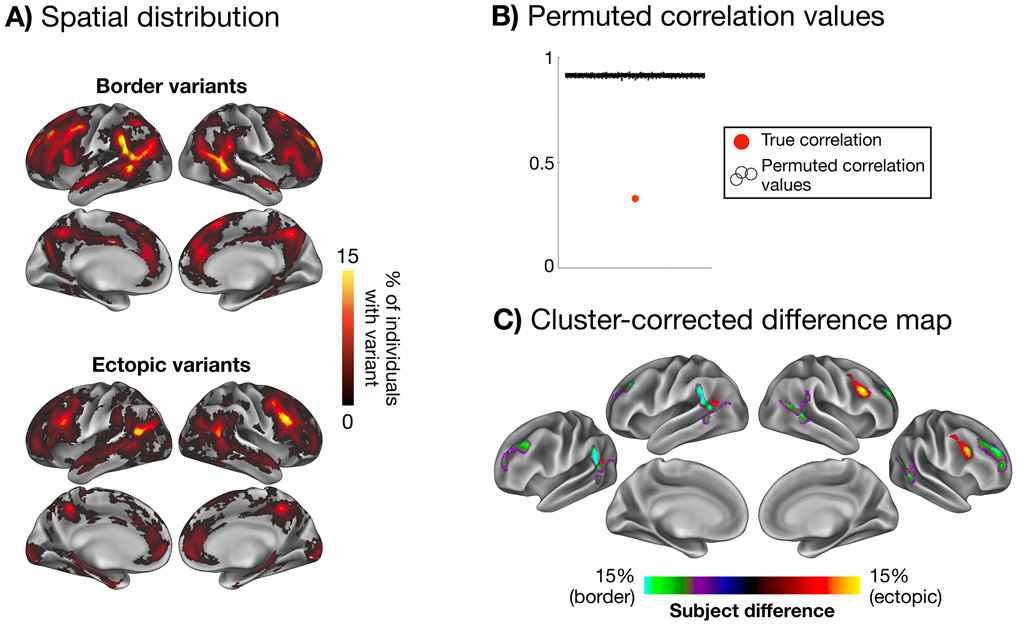
Figure 3: Spatial distributions of ectopic and border variants.
The maps in (A) show the spatial distribution of border variants and ectopic variants, overlapped across participants in the HCP. (B) These two spatial distributions differ significantly more than expected by chance from random permutations. Permuted correlation values are jittered across the x-axis for visualization. (C) A cluster-corrected difference map is shown highlighting regions with a significantly higher occurrence of border variants (green/purple) and ectopic variants (yellow/red; p < 0.05 cluster-corrected for multiple comparisons based on permutation testing). Ectopic variants were more prevalent in the right posterior inferior frontal sulcus and left posterior TPJ regions, while border variants were more prevalent in dorsal and ventral portions of the anterior TPJ and superior rostral frontal regions. See Supp. Fig. 9 for evidence of similar results for ectopic variants at greater distances, Supp. Fig. 10 for similar results using our secondary parcellation-free variant definition method, and Supp. Fig. 11 for border and ectopic overlap maps for each network individually.
The true ectopic and border variant maps were significantly more dissimilar than expected by chance (p < 0.001 based on permutation testing, see Methods; Fig. 3B). Ectopic variants appear more frequently in dorsolateral frontal regions of the right hemisphere, whereas border variants are more frequent around rostral portions of temporoparietal junction and superior rostral frontal regions in both hemispheres (p<0.05 based on cluster-corrected permutation testing). Similar results were seen with ectopic variants defined at longer distances (Supp. Fig. 9) and our secondary parcellation-free definition method (Supp. Fig. 10).
2.3. Network assignments differ across variant forms
We next examined the network associations of each form of variant. Variants were “assigned” to a network by identifying the template (Supp. Fig. 12) that best fit that variant’s seedmap (a note on terminology: variants of a given network assignment are henceforth referred to as, e.g., “DMN variants” referring to locations that are not in typical DMN regions but have a seedmap that matches canonical DMN distributions; regions found in canonical DMN locations are referred to in that way without any shortening). Variants were frequently assigned to the default mode, fronto-parietal, and cingulo-opercular networks for both forms and across both datasets, consistent with previous reports15 (Fig. 4A, Supp. Fig. 13).
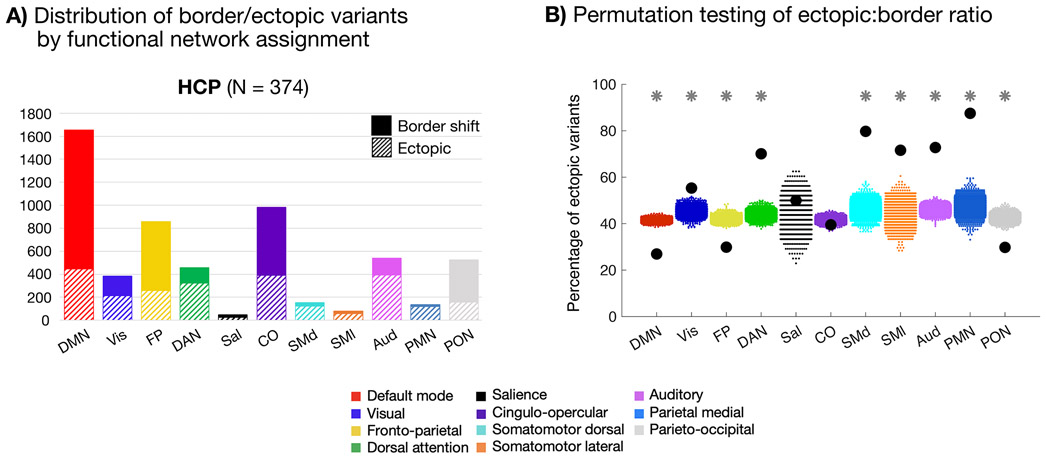
Fig. 4: Network linkages of border and ectopic variants.
(A) Network distributions of border and ectopic variants in the HCP dataset. Variants of both forms are commonly associated with the DMN, FP, and CO networks, as reported in past work15. Similar results were seen in the MSC dataset (Supp. Fig. 13) and using the parcellation-free approach to defining border and ectopic variants (Supp. Fig. 14) (B) Plot depicting permutation testing of the ectopic:border ratio in the HCP dataset. For all networks with the exception of salience and cingulo-opercular, the true proportion of ectopic variants (black dots) was significantly different from permuted proportions (colored dots, 1000 random permutations of shuffled labels) at p<0.001 (*; FDR corrected for multiple comparisons). DMN, FP, and PON variants were more likely to be border shifts, while sensorimotor, DAN, and PMN variants were more likely to be ectopic. Notably, ectopic variants were commonly found in all systems. See Supp. Fig. 12 for cortical depiction of each listed network.
When contrasted with one another, we find that the relative prevalence of border and ectopic variants differs by network. Indeed, a mixed-effects generalized linear model analysis revealed a significant interaction between variant form and network (p<0.001), suggesting that the influence of variant form on subject-level variant frequencies varies significantly across networks.
Using our primary variant definition method in the HCP dataset, border variants were more commonly assigned to the default mode (DMN), fronto-parietal (FP), and parieto-occipital (PON) networks. Ectopic variants were relatively more linked to the dorsal attention (DAN), parietal memory (PMN), and sensory/motor networks (see also similar results using our secondary method, Supp. Fig. 14). Permutation testing of variant labels in the HCP dataset confirmed these observations (Fig. 4B). As may be expected, ectopic variants were relatively more abundant in smaller or more spatially local networks, as it is easier to be distant from these network boundaries. However, ectopic variants were still often associated with large networks such as the DMN, FP, cingulo-opercular (CO), DAN, and Visual systems, emphasizing their common nature. Distributions of variant network assignments in the MSC are shown in Supp. Fig. 13. Although there were fewer variants in the MSC overall (leading to greater variability in per-network proportions of variant forms), the findings were consistent with the HCP dataset such that ectopic variants were associated with many large (and small) networks. The parcellation-free classification method resulted in comparable network distributions for most networks, although with some differences in ectopic:border ratios (Supp. Fig. 14).
Interestingly, combining information from the location and network-assignment analysis, one can find that border and ectopic variants exhibited different patterns of “swaps” in their territory relative to the canonical structure (Supp. Fig. 15). For example, border variants found in canonical (i.e., group average) regions of the FP network most often are re-assigned to DMN, CO, or DAN. Thus, while idiosyncratic, both forms of variants show constraints in how they vary.
While it is beyond the scope of this manuscript to fully explore the interactions between variant form (border, ectopic) and network assignment, it will be useful in future work to examine this question in more detail. To aid in this endeavor, Supp. Fig. 11 provides border and ectopic variant maps for each network individually.
2.4. Border and ectopic variants exhibit shifted task responses
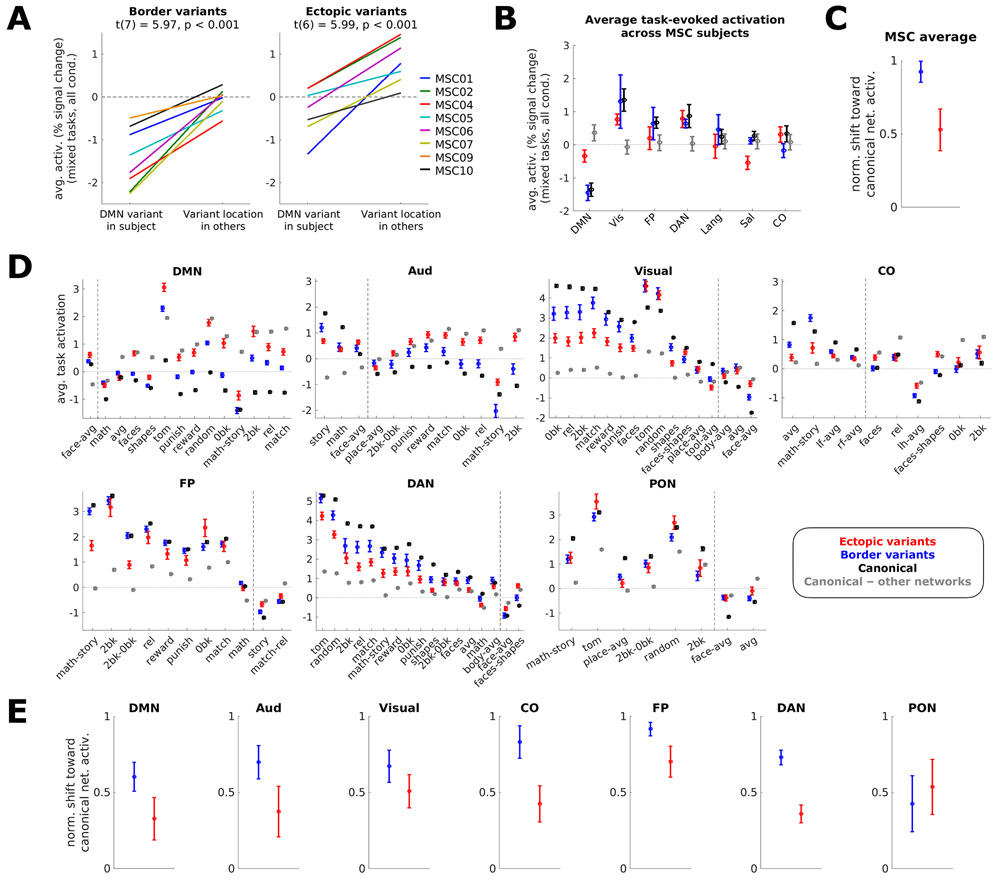
Next, using both datasets, we asked if both forms of variants show altered task responses, consistent with their idiosyncratic network affiliation. Following Seitzman et al.15, we first focused on the mixed design task from the MSC (see details in Methods). Average activations were contrasted with baseline, a contrast that elicits negative activations in DMN regions typically, and positive activations in the FP, DAN, and visual systems15, 30.
In the MSC, ectopic and border variants exhibit shifted task activation responses in the direction expected based on their network assignment (Fig. 5B). Border variants are shifted further toward the expected activation based on their new assignment than ectopic variants. While we observe a strong task deactivation for border DMN variants (t(7) = −5.38, p = 0.001 for 8 MSC participants with border DMN variants), aligning closely with canonical DMN deactivation, ectopic DMN variants exhibit a relatively weak (but still significant) deactivation (t(6) = −2.76, p = 0.033 for 7 MSC participants with ectopic DMN variants). Similar patterns are seen with “task positive” networks like the visual, FP, and DAN networks; the CO, language, and salience networks were not strongly modulated by this contrast.
Figure 5: Task-evoked activation of variant locations in the MSC and HCP.
(A) Average activation in the MSC of DMN-assigned variants in an individual vs. average activation in that location in all other individuals for border (left) and ectopic (right) variants; 8 of 9 subjects with a border DMN variant and 7 of 9 subjects with an ectopic DMN variant were included. Colors represent different MSC participants. Two-tailed t-tests showed that border (p=0.00056) and ectopic (p=0.00098) variants exhibited functional activation shifted from the typical responses of their location toward the response expected based on their network associations. Similar results were seen with our secondary method of variant classification (Supp. Fig. 16). (B) Average activation (z) across all task conditions in a set of mixed-design tasks from the MSC (N=9) for variants (red = ectopic, blue = border), canonical locations of the listed network (black), or canonical locations of other networks (gray). Error bars represent the standard error of the mean across subjects. (C) Normalized shift of border (blue) and ectopic (red) variants in the MSC (N=9) toward listed networks. Values represent the average normalized value across listed networks (calculated as (variant–other networks)÷(canonical network–other networks)); error bars represent standard error of the mean across networks. (D) Average task activation across HCP subjects (N=358). For each network, contrasts were included if the activation in canonical regions of the network exceeded that of other networks by at least 0.5% signal change. Dashed lines delineate contrasts where a given canonical network’s activation is greater than other networks’ activation from contrasts where other networks’ activation is greater. Error bars represent standard error of the mean across subjects for a given contrast; see Supp. Table 2 for more information on contrasts. (E) Normalized shift of border (blue) and ectopic (red) variants toward each listed network in the HCP (N=358). Again, border and ectopic variants exhibit shifted responses during tasks, with border variants consistently shifting more strongly toward the canonical activation of their assigned network (with the exception of PON variants). Error bars represent standard error of the mean normalized value across contrasts within a network.
This finding is further examined in Fig. 5A for each MSC participant by comparing the task activation of DMN variants in a given subject to that same location in other subjects. This analysis showed that – in every individual participant – similar prominent decreases in activations were observed relative to what is expected for that location in both border variants (t(7) = 5.97, p<0.001) and ectopic variants (t(6) = 5.99, p<0.001). However, ectopic DMN variants appear in locations that have a more positive typical response pattern in comparison to border variants, and thus do not reach as strong a level of deactivation. We found similar results with the secondary parcellation-free classification method (Supp. Fig. 16).
We extended these results to additional tasks and participants in the HCP. Analysis-level task fMRI maps from the HCP dataset were used to query all contrasts across 7 tasks (see Methods and Supp. Table 2 for contrast names). Fig. 5D shows these results for each contrast by network. Again, border variants are shifted further toward the expected direction for canonical regions of the same network than are ectopic variants. The consistency of this result was confirmed by calculating the average activation shift of border or ectopic variants toward the response of regions of its canonical network, normalized by the response of regions of all other networks (Fig. 5E). The same analysis was also applied to the MSC task results for consistency (Fig. 5C). In all cases, border and ectopic variants exhibited shifts in their task activations toward canonical network responses. For all but one network in the HCP, border variants consistently shift more strongly toward their network’s canonical response than ectopic variants.
Jointly, these findings demonstrate that both ectopic and border variants are associated with altered task activations, shifting toward the expected activations for the variant’s assigned network. Border variants associated with the default mode network retain a task activation similar to canonical locations of their assigned network, while ectopic variants show an intermediate task activation response.
2.5. Variant forms exhibit differing genetic influences
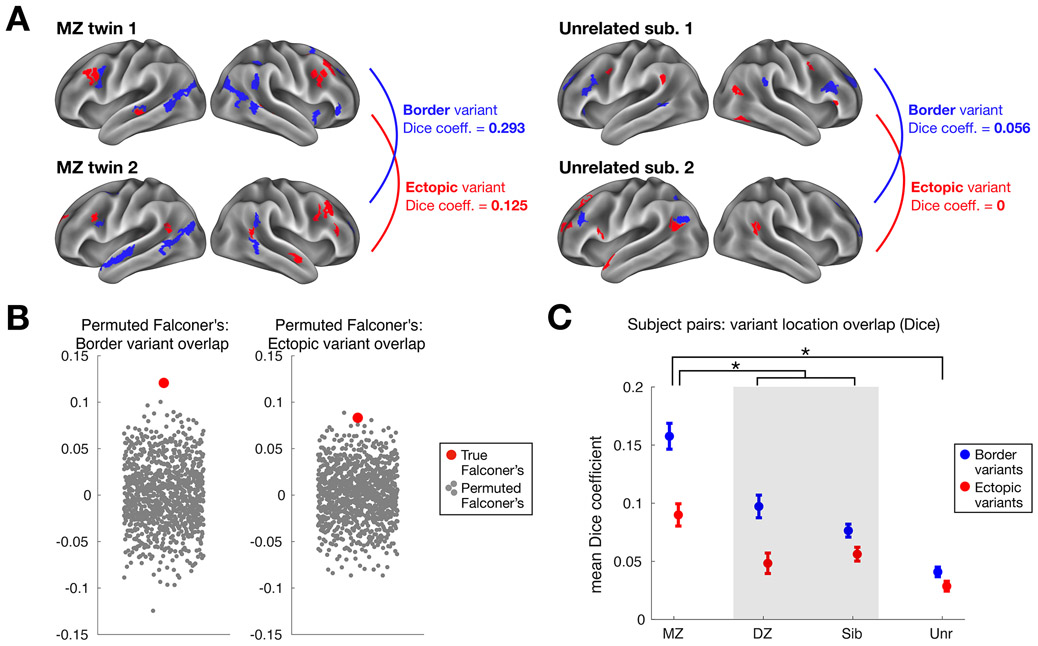
To better understand factors contributing to the formation of network variants, we examined their similarity in twin samples using an expanded subset of HCP subjects with familial relationships (Fig. 6A): monozygotic twins (N = 88 pairs), dizygotic twins (N = 45 pairs), siblings (N = 137 pairs), and unrelated individuals (N = 122 randomly selected pairs). Network variants were most similar in location among monozygotic twins, with intermediate similarity among dizygotic twins and siblings, and with the highest dissimilarity among unrelated individuals (Fig. 6C). This pattern is indicative of a genetic influence in network variant locations. Indeed, estimates of similarity among twin samples based on Falconer’s formula31, 32 were significantly higher for both border and ectopic variants relative to permuted null distributions (p<0.001 and p<0.002 respectively; Fig. 6B). Similar twin sample similarity results were also found when using the parcellation-free variant method (Supp. Fig. 17). These findings are consistent with past reports that functional network organization is heritable4, 33, but here are extended to demonstrate genetic influence specifically for the locations of idiosyncratic variation of the brain.
Fig. 6: Estimating the genetic influence of border and ectopic variants across individuals in the HCP dataset.
(A) The similarity of border (blue) and ectopic (red) variant locations was measured for monozygotic (MZ) and dizygotic (DZ) twin pairs, siblings, and unrelated individuals. This schematic shows an example pair of MZ twins (left) and unrelated individuals (right). The MZ twins exhibit a greater Dice coefficient of overlap between their border variants (in blue) and ectopic variants (in red) than a pair of unrelated subjects. (B) This observation was confirmed by estimating genetic influence with Falconer’s formula, which compares similarity in MZ and DZ twins. Both border and ectopic variants (red dots) exhibited significant similarity (p<0.001 and p<0.002 respectively) relative to a permuted null (gray dots) where MZ and DZ labels were randomly shuffled (*note, our use of Dice instead of R-values in Falconer’s formula does not produce heritability estimates, but does provide a valid way to assess non-zero genetic influence via permutation testing, randomly shuffling MZ and DZ labels). (C) Average similarity among border and ectopic variants is shown for pairs of HCP participants. For both forms of variants, MZ twins (N= 88 pairs) showed the highest similarity, DZ twins (N=45 pairs) and siblings (N=137 pairs) showed intermediate similarity, and unrelated individuals (N=122 pairs) showed the lowest similarity, a pattern consistent with an influence on genetics on variant locations. A two-way mixed-effects ANOVA revealed a significant interaction between variant group and form (p<0.001), with unpaired t-tests (assuming unequal variance) revealing a larger difference between variant forms in MZ pairs compared to DZ/non-twin sibling pairs (p=0.015) and to unrelated individuals (p<0.001). Bars represent standard error across subject pairs, with mean Dice coefficient across pairs at the center.
Intriguingly, border variants appear to be more similar across identical twins than ectopic variants (Fig. 6C; a significant interaction was found between variant form and group, p<0.001). Monozygotic twins show a greater difference between variant forms compared to dizygotic and non-twin sibling pairs (p=0.015), and to unrelated individuals (p<0.001, Bonferroni-corrected). Paired t-tests comparing border and ectopic variant similarity within each group revealed a significant difference for all but unrelated individuals (p<0.001 each for MZ pairs and DZ/sibling pairs, Bonferroni-corrected). Thus, border variants track more closely with shared genetics than ectopic variants, suggesting that they may have different underlying sources.
2.6. Subgroups diverge between border and ectopic variants
Given this evidence, was asked if different people share variant properties. Previously15, we found that network variants differed across people in a categorical manner, and could be used to identify subgroups of individuals with similar forms of idiosyncratic brain organization. In that work, we found two large reproducible clusters of individuals: those with variants more associated with the DMN and those with variants more associated with control and sensorimotor processing systems. This information may help to constrain theories on how individual differences in brain organization arise across the population. Here, we examined the extent to which similar subgroups would be evident for ectopic and border variants when examined separately.
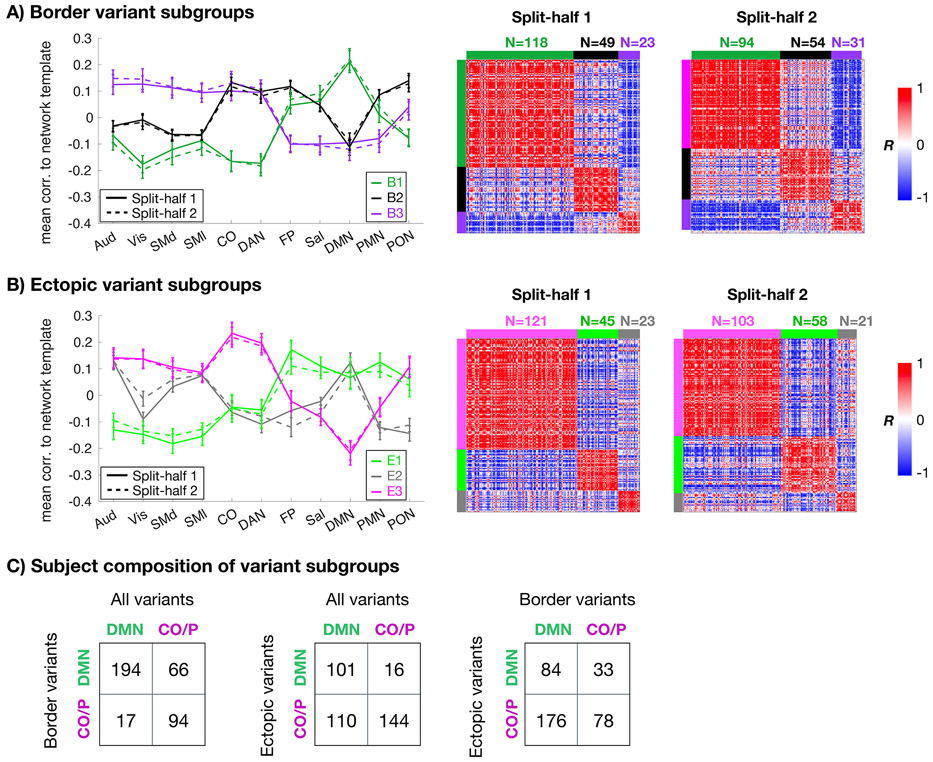
The 374 unrelated individuals in the HCP dataset were grouped based on variants’ similarity to each of 11 network templates and then clustered into subgroups (see Methods). Analyses were conducted in two split-halves (as in ref. 15) to test for replicability. Three consistent subgroups were present for both border and ectopic variants (Fig. 7A/B). These subgroups consisted of differences in how variants linked to control networks, default (and related) networks, and sensory/motor networks (described further in Supp. Results). Assignents were robust, with high consistency of subgroup assignments across sessions within an individual (>80%; Supp. Fig. 18) and further validated against null models (Supp. Results).
Fig. 7: Similarity of border and ectopic variants across subgroups of individuals.
For border variants (A) and ectopic variants (B), we separated individuals into subgroups based on the average network similarity vector of their variants (left). Matrices on the right show across-subject similarity (correlation) of variant profiles for each split-half in the HCP. Color blocks at the edges of the matrices denote the subgroup identities. The two variant forms produced three subgroups each with high similarity across matched split-halves of the HCP data. However, the subgroups differed between the two forms. Error bars in (A) and (B) represent standard error of correlation values across subjects within a given subgroup/split-half. (C) Contingency tables show the composition of subgroups in which each individual’s variant profile (all variants, border variants only, and ectopic variants only) was forced to sort into either a DMN-like subgroup or a control/processing subgroup. Note that ectopic and border variant subgroup labels had poor association with one another.
Notably, it appeared that subjects were not consistently sorted into the same subgroup across variant forms (i.e., a subject whose border variants assign them to the DMN-like B1 subgroup was not necessarily assigned to the DMN-like E1 subgroup based on ectopic variants). To confirm this discrepancy, an additional analysis was performed in which each subject was forced into either DMN or control/processing clusters, as these were the most consistently identified across analyses, using a template approach (see Methods). All HCP subjects with at least one border and one ectopic variant were included in this analysis (N=371/374). Cluster grouping was then compared (Fig. 7C). While an individual’s subgroup based on border variants was somewhat related to its overall (all variants) subgroup (adjusted Rand index = 0.30), subgroups based on ectopic variants were independent of the subgroups based on border (adjusted Rand index = −0.008) or all variants (adjusted Rand index = 0.099). These differences in clustering across forms were substantially larger than those seen across sessions (Supp. Fig. 18). This suggests that border and ectopic variant forms are relatively independent, appearing in distinct patterns across individuals.
2.7. Variant forms differ in their prediction of behavior
In a final exploratory analysis, we examined whether border and ectopic variants can predict broad behavioral phenotypes measured outside of the scanner, using the same network-affiliation measures as were used in our clustering analysis. While these relationships are likely to be small19, 24, 34, 35, they may begin to identify subtle differences in the links between these measures of brain organization and measures of cognition and psychopathology.
We used 10-fold cross-validation with support vector regression to predict out-of-scanner HCP behavioral measures (modeled after ref. 19) from variant features (see Methods). Consistent with past results19, 24 we found that it is possible to predict behavioral phenotype from fcMRI– in this case even in relatively sparse measures restricted to the most idiosyncratic network locations in each individual (Supp. Fig. 19, Supp. Table 3). The network associations (i.e., the same measures used to sub-group individuals) of both border and ectopic variants predicted behavioral variables to a small degree (average border prediction r = 0.012, p < 0.04; ectopic prediction r = 0.030, p < 0.001). The strongest predictions were associated with a range of affective, cognitive, and quality of life measures (Supp. Fig. 19A), with ectopic variants showing stronger predictions than border shifts (Supp. Fig. 19B).
Importantly, border and ectopic variants predicted distinct behavioral variables and no behavioral variables were predicted by both variant forms. Predictions between border and ectopic variants correlated at r = −0.126 (Supp. Fig. 19C). Similar differences between border and ectopic variants were seen when the locations of variants were used as features for prediction instead of their network associations (Supp. Fig. 20). Again, no behavioral phenotypes were predicted by both border and ectopic variants (predictions between border and ectopic variants correlated at r = −0.09). Note that, in this case, only border variants were significantly predictive of behavioral phenotypes on average (average r = 0.015, p < 0.007) with the strongest predictions seen for cognitive variables. Similar results were obtained when border vs. ectopic features were defined using the secondary parcellation-free approach (Supp. Fig. 21, 22).
Jointly, these findings suggest that using brain network features to predict behavioral measures is possible, but these relationships are complex and relatively weak on average. The evidence that border and ectopic variants predict different outcomes suggests that simple summary measures (e.g., grouping across all forms of individual differences in brain networks) will mix together distinct brain endophenotypes and muddy interpretations. To test this idea , we examined whether joining border and ectopic features together would improve behavioral prediction. Interestingly, despite doubling the number of features in analysis, we found that prediction results were relatively comparable between the joint model and the best-performing individual models (e.g., for network affiliation, ectopic prediction r = 0.030 and joint prediction was r = 0.030). Prediction values were correlated between ectopic and the joint model at r = 0.73. Thus, at least in this case, joining border and ectopic features together does not lead to gains in prediction, but adds ambiguity to the sources of prediction performance.
3. Discussion
Here, we document that individual differences in functional brain networks come in two forms: shifts in the borders between adjacent systems and ectopic intrusions, islands of an atypical system at a distance from its usual location. Separation of these two forms is likely to provide new insights into the sources of individual differences and their implications for normative and pathological behavior.
3.1. Ectopic islands are common forms of variation
A wealth of recent studies has provided evidence of individual differences in brain organization linked to cognitive variation19, 25, 36. In these studies individual differences are typically treated equivalently, frequently with the assumption that they reflect differences in the boundaries between systems – e.g., a region taking over territory in nearby locations. However, our work suggests that variant brain locations also appear as ectopic intrusions. Ectopic variants are common, comprising ~40-50% of variants across both datasets. Even at greater distances (>10mm from a canonical network), ~30% of variants were ectopic. Thus, not only are ectopic variants common, but many are fairly remote from their expected network boundaries. Different theories and approaches will be needed to address ectopic variation.
Prior to this paper, the existence of ectopic variants had been hinted at in previous work. Ref. 15 and ref. 11 observed regions where an individual’s functional network organization differs from the group-average, including distant from typically observed boundaries (e.g., DMN variants in Fig. 4B of ref. 15 and island of CO appearing in Fig. 7 of ref. 11). Glasser et al.37 noted several instances of sizable spatial displacements in individual-level topography relative to a group-average representation. We build on these observations to systematically characterize the prevalence of ectopic variants and determine how they differ from border shifts.
Notably, ectopic variants were defined relative to a group average, under the assumption that the group average represents the prototypical network structure across individuals. This approach is also relevant in terms of understanding the consequences of individual variation relative to many analysis strategies that are based on group averages. However, an alternate question is how these variants are positioned relative to a given person’s network structure. When investigated in detail, we find that the majority of ectopic variants defined relative to the group also consisted of islands, separate from the same network within individuals, and these island regions have similar basic properties to ectopic variants (Supp. Fig. 7). However, future research will be needed to provide a more detailed examination of island variants relative to a particular person’s networks.
3.2. Border and ectopic variants are distinct forms of brain variation
Comparative neuroanatomy studies have shown that cortical functional architecture can differ in a variety of ways across mammals, including differences in the cortical area size/position, number, organization, and connectivity5. Although not as often discussed, many of these differences are also seen across individuals within a species2. Past work suggests that even in the typical population, there is substantial variation in the size and position of human cortical areas (e.g., in V1 size22 with potential links to functional differences in vision38-40 and in the position of Broca’s area41, 42). Linking to this work, we hypothesized15, 43 that variants represent a combination of border shifts (expansions, contractions, or displacements relative to the canonical cortical area layout, which will result in differences adjacent to their typical locations) and ectopic intrusions (islands of altered connectivity and function of a region at a distance from the canonical organizational structure).
Here we demonstrate that both forms of variation occur commonly across people and many brain regions, but that they differ along a number of dimensions: spatial location, network assignment, task activations, genetic influence, prediction of behavioral variables, and subgrouping. Together, these observations suggest that border and ectopic variants may link to differing underlying sources.
A number of developmental factors influence how cortical functional systems are organized. Gradients in the expression of transcription factors in patterning centers of the cortex control the size and position of many cortical areas44. It has been proposed that intrinsic genetic factors create “proto-areas” whose boundaries are refined and sharpened through experience-dependent mechanisms45. Similar principles have been theorized to underlie the development of distributed cortical systems, starting from a proto-organization that is refined, fractionated, and sharpened with experience46, and groups have reported evidence for heritability in functional brain networks4, 33, 47-52. Here we present evidence that the locations of variants are influenced by genetics. However, variants were still far from identical between monozygotic twins, indicating a large contribution of environmental factors. Interestingly, border variants were significantly more similar than ectopic variants in monozygotic twins, suggesting that border shifts may be relatively more linked to genetic factors.
Indeed, while some properties are genetically determined, profound differences in experience (e.g., sensory deprivation during critical periods) can substantially alter cortical area size, layout, and connectivity in rodents5. Similarly, experience with faces is critical to the formation of face selective areas in macaques, although basic retinotopic organization remains28. In humans, congenitally blind individuals show intact internal topography of visual areas, but differences in interareal connectivity29, while those born with only one hand show cortical expansions of regions representing motor functions of other body parts53. The lower similarity of ectopic variants among identical twins suggests that these variants may be more influenced by experience-dependent mechanisms.
These studies in humans and non-human animal models help to explain how cortical organization can both demonstrate substantial commonalities across individuals, but also punctate locations of differences within a species – some associated with local changes (e.g., due to changes in area sizes, which would likely result in border variants), and others that could be linked to more distant alterations (e.g., strong changes in connectivity/function of a (sub)-region that may underlie ectopic variants). Network variants provide a robust and high-throughput approach to identify variations in brain organization across the human brain, helping to constrain theories of the sources and consequences of cortical area variation. Future studies can systematically test the hypotheses raised by this work, using a combination of network modeling methods (for example, testing cascading interaction models for the evolution of network activity54), studies of variants in different age populations (to determine how border and ectopic properties may change over the course of the lifespan), and longitudinal study designs to determine how border and ectopic variants change over time within an individual.
3.3. Impact for basic research studies
At present, many resting-state and task-based fMRI studies aggregate or compare data across participants based on spatial normalization, assuming that the spatial layout of brain systems is conserved across individuals55. However, widespread individual differences in the localization of brain regions and systems may lead to detrimental effects when performing group-level analyses, including loss of sensitivity and functional resolution56, prediction accuracy of task functional connectivity57 and task-evoked signals58, 59, and prediction of behavior from resting networks19, 24, 60, 61. These limitations lead group studies to fall short of providing comprehensive explanations of brain function and cognition as a whole.
Alignment across individuals may be improved by more accurate anatomical registration, such as via surface-based mapping methods62, 63. However, the resulting functional overlap may vary depending on brain area23, 64, with a bias toward enhancing the concordance of regions supporting sensory/motor functions65. As an alternative, approaches based on the alignment of functional signals themselves may increase cross-subject correspondence. This includes collecting functional localizer task data from each individual subject55, 66-68, adopting fcMRI methods to define brain systems and areas from subjects with large quantities of data12, 69, or hyperalignment-based techniques to increase functional correspondence26, 58, 59. Other methods developed to address functional alignment include template-matching techniques13, 14, 70, multi-modal functional/anatomical registration37, and hierarchical functional parcellation approaches19, 24 to identify brain systems and regions in individuals even with more modest amounts of data.
Each of these methods has demonstrated great utility in cross-subject comparisons, including improved definition of default systems69, 71, 72, language systems68, 73, 74, and sensory-biased frontal regions75, 76. However, many approaches rely on adjusting regions within a relatively restricted spatial extent. While this is likely appropriate in some cases, imposing a strict distance criterion will not be optimized for detecting ectopic variants that are more distant from their typical network boundaries (Supp. Fig. 5; nearly one-third of ectopic variants occur >30 mm from their same-network boundaries). Our results argue for using procedures that (a) conduct individual-level region identification (e.g., individual localizers) or (b) that allow for longer-distance displacements of regions (e.g., with enlarged spotlight procedures). It is also possible that individual functional features will link with variable fine-scale anatomical features (e.g., tertiary sulci77, 78) that can be used for alignment.
Adjusting for ectopic variants will be more relevant in some brain locations than others. Ectopic variants are relatively more prevalent in lateral frontal cortex, a region believed to play a role in task control30, 79, 80, language68, 73, and sensory-biased attention and working memory75, among other high-level processes. The prevalence of ectopic variants in lateral frontal cortex suggest that researchers should be particularly cautious in interpreting group-level results in these regions, unless a cross-subject functional alignment method is performed that accounts for non-local deviations. Advances in our understanding of lateral frontal cortex will likely be spurred by studies using improved functional alignment methods, which enable separation of multifunctional regions from specialized regions in the face of cross-subject heterogeneity73, 81 (see Smith et al.82). In contrast, border variants were relatively more common in the temporoparietal junction and rostral superior frontal regions, which have been linked to shifting attention83 and theory of mind84-86 among other functions, suggesting that studies focusing on these domains and areas may benefit from alignment approaches which impose distance-constrained changes.
In considering the impact of border and ectopic variants on cognitive neuroscience studies, one intriguing question is the extent to which variant functional connectivity is likely to be changed across task contexts. In our prior work87 as well as those of others88, 89, we have typically seen that task and rest fcMRI share substantial commonalities, with only relatively subtle changes associated with task states. Similarly, we have demonstrated that variants are largely stable in across various tasks21. However, subtle differences in functional connectivity do occur in tasks87, 90, and can be used to predict task state57, 91, 92, even from individuals57. Thus, a fruitful avenue of investigation will be to establish how tasks affect border and ectopic variants separately.
3.4. Impact for studies of individual differences
One notable difference between the two variant forms is in how they co-vary across participants. In both cases, subgroups of individuals showed similar patterns of variants: for example, with both forms of variants, one subgroup had variants with strong links to the DMN, while another subgroup had variants with stronger links to top-down control networks. However, the subgroups differed in their specific variant profiles (e.g., whether the fronto-parietal network was grouped with the DMN or CO subgroup). Perhaps most notably, the two variant forms appeared independent: that is, a person in the “DMN” subgroup based on border variants could easily be in the “CO” subgroup based on ectopic variants. Other individual differences were also seen in variant properties, including in the relative proportion of each variant form. These findings beg the question of how variants related to individual differences in brain function and behavior.
We demonstrated that border and ectopic variant properties are related to robust differences in task activations, both in the MSC and a range of task activations in the HCP. Border shifts strongly matched the task responses of their associated network, despite their differing location. Ectopic variants, in contrast, showed a more intermediate profile, suggesting that they may be sites of intermediate functional processing.
Consistent with the idea that border and ectopic variants are associated with altered brain function, we also find that these variants can be used to predict behavioral measures. While prediction levels were low, they were significant in cross-validated samples. In our previous work, we demonstrated that network variants have high stability across sessions, even up to a year15, and across task states21. These trait-like characteristics of network variants, as well as its link to differences in functional responses during tasks15, suggest that they may serve as markers of individual differences in behavior in both the neurotypical population15 and in cases of psychiatric and neurological disorders43. Other studies have also shown that individual differences in functional networks relate to behavior, including links between individual-level network topography and measures of cognition and emotion19, associations between changes in functional network topography and a variety of behavioral factors36, and links between cognitive ability and maturation of networks supporting executive function25, 93. For example, Kong et al.19, 24 have shown that individual network topography and individualized parcellations predict behavior at a level higher than that shown by traditional group parcellation approaches.
Intriguingly, we find that border and ectopic variants predict different behavioral phenotypes (with low correlations in their prediction performance). Ectopic variants showed the most robust prediction based on the network associations of variants, and linked to a range of affective and cognitive variables. Border shifts, in contrast, exhibited better prediction based on their locations and were most tightly linked to cognitive measures. Importantly, prediction was not materially improved by joining border and ectopic variant features together. These differences suggest that deeper insights may be provided if these two forms of variation are separated, or integrated in a more sophisticated manner that acknowledges that they can contribute differing sources of information. This understanding will allow for improved theories about the mechanistic links between individual differences in network variation and behavior, likely critical to using this information to guide clinical practice and interventions.
Despite these observations, we note that the brain-behavior predictions reported in this study were relatively small. These results are not surprising given recent findings on the small size of brain-behavior correlations24, 34, 94 , and our prediction levels are in line with past work that uses similar number/types of features95. It may be interesting to speculate on why we find a limited relationship to behavioral measures, and a closer correspondence to differences in task activations. One possibility is limitations in measures of brain networks and behavior34, 35, 94, 96. Another is that individual differences in brain organization can be degenerate, producing similar behavioral outcomes (an phenomenon previously termed “behavioral phenocopy”97). An important future avenue of research will be to establish principles for how and when individual differences in brain organization relate to individual differences in behavioral performance.
4. Conclusion
Here, we demonstrate that two distinct forms of individual variation in brain networks are both common, but differ in their spatial positions, network assignments, task response patterns, and subgrouping characteristics. Both show evidence of genetic influence, but border shifts are significantly more similar in identical twins, suggesting they may be more tightly linked to genetic variables. Finally, the two forms of variants predict distinct behavioral variables. These different properties must be accounted for in the study of cortical system organization and its links to behavior.
METHODS
Datasets and Overview
Network variants were investigated using data from two separate publicly available datasets: the Midnight Scan Club (MSC) dataset12, and a subset of individuals from the Human Connectome Project (HCP)98. The MSC dataset is a “precision” fMRI dataset consisting of 10 highly sampled subjects (5 female; average age 29.3 years; 1 participant excluded due to high motion and sleep12) with over 154 minutes of low-motion rest fMRI data and task fMRI data across 3 conditions (mixed, memory, motor – in this manuscript we focus on the results of the mixed design tasks). From the larger HCP dataset, we primarily analyzed 384 unrelated subjects (210 female; average age 28.4 years as in ref. 15, selected to be unrelated and have a minimum of 45 min. of low motion resting-state fMRI; see SI Table 1 in ref. 15 for details on exclusion criteria for this dataset). Of these 384 subjects, 374 were retained for analysis after removing subjects with exceptionally low spatial correspondence to the group-average indicating low quality data (see Defining network variants; note however that results remain the same prior to their removal). For analysis of behavioral links and genetic influence on variant locations, a larger subset of the full HCP dataset was used. This group consisted of 823 individual subjects (793 after quality control), including those with familial relationships, each with a minimum of 40 min. of low-motion resting-state fMRI data. No statistical methods were used to pre-determine sample sizes; the analyses in this work are based on publicly accessible datasets. Data collection and analysis were not performed blind to the conditions of the experiments.
In compliance with ethical regulations, informed consent was obtained from all participants. Study protocol for the MSC dataset was approved by the Washington University School of Medicine Human Studies Committee and Institutional Review Board, and protocol for the HCP dataset was approved by the Washington University Institutional Review Board. Compensation was provided to all participants.
In both datasets, previously defined idiosyncratic locations of functional connectivity (“network variants”)15 were divided into homogeneous segments (see criteria below) and segregated into ectopic intrusions (“ectopic variants”) and border shifts (“border variants”) based on the criteria described below. Several features of each variant form were then examined. First, we quantified the prevalence of both variant forms across the two datasets. Second, we characterized the spatial location and idiosyncratic (individual-specific) network assignment of these regions. Third, we examined the task responses of both variant forms using task data from the MSC dataset. Fourth, we examined if there were common profiles of border shift and ectopic variants (as we have seen in past data for variants as a whole15), by clustering individuals into subgroups based on their variant characteristics.
Preprocessing
Imaging data from the MSC and HCP subjects used in the present analyses were preprocessed identically to ref. 15. Full details on acquisition parameters, preprocessing, FC processing, and volume-to-surface mapping can be found in that manuscript, but are outlined briefly below.
Functional data from both datasets were preprocessed to remove noise and artifacts, following ref. 99. For the HCP dataset, we began with the dataset as processed following the minimal preprocessing pipelines100. Procedures included field map distortion correction of the functional images, slice-timing correction (for the MSC dataset only), mode-1000 normalization, motion correction via a rigid body transformation, affine registration of functional data to a T1-weighted image, and affine alignment into stereotactic atlas space (MNI for HCP (Montreal Neurological Institute, Montreal, QC, Canada); Talairach for MSC101).
Following this, resting-state fMRI data was further denoised for functional connectivity analysis, including regression of white matter, cerebrospinal fluid, and whole brain signals, six rigid-body parameters and their derivatives, and their expansion terms102. High-motion frames (calculated as framewise displacement; FD103) were censored; frames with FD > 0.2 were censored for the MSC data12 and frames with filtered FD > 0.1 were censored from the HCP data following ref. 104 to address respiration contamination of motion parameters (filtered FD = low-pass filtering at <0.1 Hz of the original motion parameters prior to FD calculation; note that two participants in the MSC dataset – MSC03 and MSC10 – with strong respiratory contamination of their motion parameters also used the filtered FD measure12, 87). As in ref. 105, 5 frames at the start of each run along with any segments < 5 frames long were also removed. These censored frames were interpolated over using a power-spectral matched interpolation. Subsequent to this, a temporal bandpass filter was applied to the data from 0.009 to 0.08 Hz.
Following this processing, BOLD data were mapped to each subject’s native cortical surface as generated by FreeSurfer from the atlas-registered T1106. Data were registered into fs_LR space64 and were aligned to the surface following Gordon et al.7, producing a CIFTI file with a BOLD timeseries for each functional run. From this point on, all analyses were conducted on the cortical surface (at the vertex-level).
Data on the cortical surface was spatially smoothed with the application of a geodesic smoothing kernel (σ = 2.55; FWHM = 6 mm). Finally, high motion frames (that were previously interpolated) were removed from analysis. Note that participants were required to have at least 40 min. of data total to be retained in analysis (see refs. 15, 21 for evidence that ~40 minutes of data is necessary to achieve reliable network variant measures). Some additional improved reliability is seen beyond 40 min., however. This factor motivated our use and replication of results in both the HCP and MSC datasets (when possible), in order to balance the advantages of large numbers of participants with the added reliability of extended amounts of data.
Defining network variants
Network variants are defined as locations in an individual that show strong differences in their functional network patterns relative to the group average. In the current manuscript we began with the same set of variants as originally presented in ref. 15. Briefly, for each subject, each vertex’s seedmap (i.e., its cortex-wide connectivity map) was correlated with the same vertex’s seedmap from a group-average reference dataset, composed of an independent 120 young adults (the WashU 120107). After repeating this procedure for all cortical vertices, this produced one individual:group similarity map per person (with a spatial correlation value for each vertex). HCP subjects whose average individual:group spatial correlation value was greater than 2 standard deviations below the mean (r = 0.436) were excluded from further analyses (10 out of the primary subset of 384 subjects; 30 out of the larger pool of 823 subjects used for analyses of similarity within twin samples and behavior). Note that for these analyses we focused on cortex-wide seedmaps, not separating edges from within and between network connections. In our experience, focusing on whole brain maps and top within-network connections produces similar results70. It will be interesting in future work to more fully explore the differences between defining variants on each form of edge.
Similarity maps were thresholded to include only the lowest decile of correlation values (i.e., to identify the 10% of locations where the individual was least similar from the group-average) and were then binarized. The decile criterion was originally selected because it represents the approximate inflection point on a histogram of individual-to-group similarity, suggesting that it may serve as a natural criterion for identifying points with strong differences from the group average. In past work, we have tested other methods for selecting network variants (based on absolute rather than relative criteria or different relative thresholds15, 21), with similar overall results. As in ref. 15, a vertex-wise map of low-signal regions was used to mask out potential variant locations. Clusters of at least 50 neighboring vertices in size were flagged as pre-variants for further analysis (see Fig. 1A).
Following this original variant definition, a series of steps was taken to further refine the set of pre-variants to generate the final variants that were used in the border/ectopic analyses, given observations that some pre-variants were large and irregularly shaped, suggesting they might consist of separate units. To divide pre-variants, previously defined contiguous units were divided into segments with the goal of minimizing heterogeneity in the connectivity of individual pre-variants. This procedure consisted of a two-fold check of (1) the variance explained by the first principal component of the pre-variant, resulting from a principal component analysis on variants’ vertex-wise seed maps (i.e., ‘homogeneity’ 7) and (2) the proportion of the variant’s territory that is dominated by a single network in the individual’s subject-specific vertex-wise network map. Note that while we have made efforts to identify homogeneous network variants, each network variant is not necessarily equivalent to a full cortical area (for example, border shift variants could arise because an area within an individual has been expanded or displaced across a border to subsume territory of another region). Using our methods, only the non-overlapping segment will be labeled a variant. Therefore, even if functional connectivity were a perfect proxy for brain area divisions, these variants might represent only a subunit within the area. In subjects' vertex-wise maps, each cortical vertex is individually assigned to a network using a template-matching procedure (see refs. 13, 70, 107 which matches each vertex’s thresholded seedmap to each network’s thresholded seedmap (each thresholded at the top 5% of values) and assigns the vertex to the network with the best fit (measured via the Dice coefficient; similar to ref. 13).
Using the MSC as a pilot dataset, two independent raters visually assessed all subjects’ variants by examining connectivity seedmaps of vertices within a pre-variant. Based on the apparent homogeneity or inhomogeneity of seedmaps at various locations within the pre-variant, the raters evaluated whether each pre-variant should be flagged to be divided. Based on these results, an algorithm was developed that best matched the hand ratings from reviewers, based on a combination of template-match network representation and homogeneity thresholds. The thresholds were then applied to the broader set of MSC and HCP participants. These final thresholds were set at 66.7% homogeneity and 75% network dominance in the individual network map. The thresholds were then applied to the broader set of MSC and HCP participants. Flagged pre-variants were split along the network boundaries of the vertex-wise network, resulting in final “split” – but each contiguous – variants into homogeneous regions. Clusters smaller than 30 contiguous vertices were removed. See Fig. 1A for a schematic representation of the splitting procedure and examples of split variants. Following the combination of variant definition, size and SNR exclusion, and pre-variant homogeneity refinement, approximately 2% of vertices were defined as variants in each individual.
Network variant locations are present even after surface-based normalization62,63,108 that align data across people by large-scale sulcal features. Individual differences in fcMRI are not well related to variations in anatomical metrics (refs. 7, 15; although it is possible that they relate to finer-scale anatomical features, e.g., see ref. 77). This dissociation from gross anatomical features, together with correspondence to task responses, suggests that variants may relate more closely to differences in the positions of functional brain areas or systems, which can vary relative to anatomical landmarks23, 64.
Functional network assignment of variants
Variants were then assigned to a best-fitting canonical functional network by a procedure which matched each variant to its best-fitting functional network template as in ref. 15. To assign variants to networks in the MSC dataset we used group-average network templates that were generated in previous work from 14 networks using data from the WashU 120 (refs. 13, 15; see Supp. Fig. 12A). Networks used for MSC analyses included default mode (DMN), visual, fronto-parietal (FP), dorsal attention (DAN), language (Lang.; note this has been referred to as the ventral attention network in our past work but we have now reclassified as language based on its correspondence with language localizers74), salience, cingulo-opercular (CO), somatomotor dorsal (SMd), somatomotor lateral (SMl), auditory, temporal pole (Tpole), medial temporal lobe (MTL), parietal medial (PMN), and parieto-occipital (PON). For terminology, variants whose best-fitting functional network is DMN based on comparisons to all network templates, for example, are termed “DMN variants.”
For the HCP dataset (given differences in dataset resolution and acquisition parameters98; see also ref. 70), a dataset-specific network template was generated (Supp. Fig. 12B; see ref. 15 for template generation procedure). The networks included in HCP-specific analyses were similar to those for the MSC, but did not include the language, MTL, or T-pole networks as these networks did not emerge consistently across edge density thresholds from the data-driven group-average network identification procedure (Infomap109). In both cases, the average seedmap for each variant in an individual was compared with each of the network templates (after binarizing both to the top 5% of connectivity values (as in ref. 13) and assigned to the template with the highest Dice coefficient overlap. Network variants were removed from further analysis if they did not match to any functional network (Dice coefficient of zero) or if over 50% of their vertices overlapped with the group-average network for that location.
Classification of variants as ectopic intrusions or border shifts
Two methods were used to classify variants as either ectopic intrusions or border shifts. The primary method was implemented to identify variants which lay adjacent or at a distance from the canonical group-average regions of the same network. First, using the MSC as a pilot dataset, all variants were manually classified as ectopic variants by examining whether they visually appeared to be spatial extensions of existing network features or whether they appeared to arise unconnected from other same-network locations. Next, we tested different geodesic distance criteria to optimize agreement of computed border/ectopic classifications with manual classifications of variants as border/ectopic based on our visual inspection of the MSC participant variants. The final procedure specified that all variants further than 3.5 mm (edge-to-edge distance) away from a same-network cluster were classified as ectopic variants; variants closer than 3.5 mm were classified as border variants. This procedure was then applied to the independent HCP dataset (see Fig. 1 for schematic representation). As a check to determine whether the 3.5 mm distance threshold we specified would significantly impact the proportion of ectopic variants in our sample, we also classified variants as ectopic or border based on a distance criterion of 5, 7.5, and 10 mm and computed the proportion of ectopic variants at each. Finally, we also quantified the distance between each final ectopic variant and canonical regions of their assigned network.
A secondary, parcellation-free method to classify variants was also implemented for use in a subset of analyses (reported in the supplement). Rather than relying on a group parcellation to make a border-ectopic distinction, this technique classified a variant based on whether its average connectivity seedmap reached high similarity to the group average seedmap (i.e., high variant-to-group R) at brain locations near the variant (a border shift), or whether the variant’s similarity to the group did not approach a peak in correlation at any brain location near the variant (an ectopic intrusion). This procedure specified that if the seedmap of a variant reached at least 90% of its peak correlation to the group average at a distance within 10 mm (edge-to-edge distance), then the variant was classified as a border shift. Conversely, a variant whose seedmap similarity to the group did not approach at least 90% of its peak correlation within a distance of 10 mm was classified as ectopic. In both cases, the “peak” variant-to-group R was defined as the most similar seedmap comparison within 150 mm of the variant. For a schematic illustration of this method, see Supp. Fig. 2.
Of note, each of these variant classification methods relies on a comparison between an individual and a group-average description (a group-average network map in the primary method, or a group-average connectivity map in the secondary method). The motivation for this approach was twofold: first, we sought to understand how group-level representations may err, both in terms of proximal adjustments (border shifts, which can be more straightforwardly addressed by available techniques) and for more distant, individually idiosyncratic locations (ectopic intrusions, which arise further away than expected based on group priors). Second, if we are to approach the question from a theoretical standpoint, we may assume that an individual may deviate from a “standard” template of brain organization in various ways. Some mechanisms may result in relatively proximal border shifts (e.g., differences in the transcription factor gradients which mediate a standard set of developmental pathways), whereas others may operate over longer distances (e.g., experience-dependent competitive mechanisms). Thus, it is important to characterize each of these possible routes for individual variation. However, to contextualize an individual’s variants within their own individually defined network map, we provide an exploration of variants in the MSC dataset with ectopic variants classified into 3 sub-types based on how isolated or connected the variants were to their individual-specific networks; see Supp. Fig. 7 and legend for an illustration, and Supp. Table 1 for classification results.
Examining differences in spatial distribution between ectopic and border variants
To visualize the spatial distribution of border and ectopic network variants, a spatial overlap map was generated by summing network variant maps (separated by form) across individuals. This produced an overlap map highlighting regions of high and low occurrence of variants in both the primary and parcellation-free classification method.
These spatial overlap maps were quantitatively contrasted at two levels. First, an omnibus map-wise permutation analysis was run. For this permutation test and all others in this work, assumptions of normality are not required; rather, exchangeability is assumed. For ANOVAs/t-tests, data distributions were assumed to be normal but were not formally tested. Our null hypothesis was that the distribution of border and ectopic variants was no more different than would be expected by chance. We used a permutation approach to address this hypothesis. This was achieved by (1) shuffling variant classification labels (ectopic vs. border) randomly at the subject level (i.e., flipping variant labels within a subject’s variants map 50% of the time) to create pseudo-ectopic and pseudo-border variants, (2) summing variant locations across subjects to generate a cross-subject overlap map for pseudo-ectopic variants and a cross-subject overlap map for pseudo-border variants, (3) calculating the similarity (spatial correlation) between the pseudo-ectopic and pseudo-border overlap maps, and (4) repeating steps 1-3 1000 times for different permuted labels. We then compared the distribution of permuted similarity values with the true similarity between ectopic and border spatial distribution maps. We calculated significance as a p-value based on the proportion of permutations in which the permuted correlation value exceeded the true correlation value. This procedure was used to determine whether spatial distributions, as a whole, differed between ectopic intrusions and border shifts. Permuting border and ectopic labels at the subject level allowed us to preserve spatial relationships and local spatial auto-correlation110 within each subject’s variant map, in turn allowing us to directly test the null hypothesis that the distributions of border versus ectopic variants across subjects are no more different than would be expected by chance.
Second, to locate specific regions where the distribution of ectopic variants was significantly distinct from border variants, a cluster-size-based permutation analysis was run. The first two steps were the same as before: (1) shuffling ectopic and border labels within each subject’s variant map to create pseudo-ectopic and pseudo-border variants in the same proportion and (2) summing variant locations across subjects to create an overlap map for pseudo-ectopic and pseudo-border variants for each of 1000 permutations. Using these permuted overlap maps, we (3) generated 1000 difference maps of the pseudo-ectopic variants distribution minus the pseudo-border variants distribution, (4) thresholded these pseudo-difference maps to only keep locations with differences of at least 5% of participants (19 subjects), and (5) calculated the size (number of vertices) of retained clusters. This procedure produced 1000 permuted pseudo cluster size calculations. This distribution of cluster sizes was used to define a cluster threshold that corresponded to the top 5% of permuted (random-chance) clusters (p < 0.05 cluster-corrected). Finally, the true ectopic–border difference map was also thresholded to only keep locations with a difference of at least 5% of participants, and all clusters composed of fewer vertices than the cluster-correction threshold (229 vertices) were removed, and the resulting cluster-corrected difference map is displayed.
Comparing network assignments between variant forms
In addition to its spatial location, each variant has an idiosyncratic functional network assignment (see Functional network assignment of variants section above; e.g., a network variant located in a typical DMN region may have functional connectivity more closely associated with FP, causing it to be assigned to that system). In the next analysis, we used a permutation approach to quantify differences in border and ectopic variants’ functional network assignments.
For this analysis: (1) we permuted the label of each variant as border or ectopic within participants in the HCP dataset (permutations were done per participant), (2) we then calculated the proportion of pseudo-ectopic to pseudo-border variants for each network, (3) repeated steps 1-2 1000 times. We then compared the true ectopic to border proportion for each network to the permuted distribution of proportions. Significance was assessed via p-values calculated as the proportion of permutations in which a network’s true percentage of ectopic variants was less or greater than (two-tailed) all permuted percentages after FDR correcting for multiple comparisons across networks. This analysis was also performed using the parcellation-free variant labels. To further test whether frequencies of each variant form varied by network, a mixed-effects generalized linear model analysis was performed. Factor effects were defined for variant form (border or ectopic) and variant network, with one level per subject recording counts of each observed combination. The interaction model was performed to measure the extent to which the influence of variant form on variant frequencies varied across networks, while accounting for within-subject dependence.
Finally, we determined the “swaps” of network territory occupied by border and ectopic variants (e.g., a variant located in canonical cingulo-opercular territory that “swaps” its network assignment to the fronto-parietal network). To do so, we defined each variant’s consensus network assignment as the modal network across variant vertices in the pre-defined group-average system map, compared this with the variant’s assigned network, and tabulated the frequency of all cross-network swaps.
Examining task activation of variants
In addition to defining FC features of network variants, we also examined how these regions responded during tasks. Following Seitzman et al.15, we first focused on fMRI task activations during the mixed-design tasks in the MSC dataset (semantic and coherence), given strong a-priori hypotheses about the responses of different networks during these tasks. The semantic task involved participants indicating whether presented words were nouns or verbs, and the coherence task required participants to indicate whether an array of white dots on a black background111 were displayed in a concentric (as opposed to random) arrangement. Within each task block, a short cue signaled the onset of the block, with a series of individual trials presented with jittered timing. Another short cue signaled the end of the block, and task blocks were separated by fixation periods of 44 seconds (see ref. 12 for more details on task design).
Task fMRI data from the MSC dataset underwent the same basic preprocessing as listed in the Preprocessing section (i.e., field map correction, slice timing correction, motion correction, alignment, and normalization, registration to the cortical surface and smoothing). These tasks were then analyzed using a general linear model (GLM). For each event (cues, correct and error trials of each type), eight separate timepoints were modeled in a finite impulse response modeling approach99); for each task block, a block regressor was modeled to estimate sustained activations. The tasks and analysis streams are described in further detail by refs. 12 and 87.
In this study, we examined the activation image across all conditions (start/end cues, trials, and sustained activations across semantic and coherence tasks) versus baseline to interrogate network variant locations to examine whether forms of variants exhibited differences in task activations. A series of comparisons were conducted following ref. 15: (1) comparisons (two-tailed t-tests) of the task activation of DMN variant locations in a given subject relative to the same location in other subjects, (2) a comparison of task activations of variant locations in each network relative to canonical regions of their assigned network, and (3) a comparison of task activations of variant locations in each network relative to canonical regions of other networks. Task activations were examined separately for ectopic variants and border variants; this analysis was repeated using the parcellation-free variant labels.
Task fMRI data from the HCP dataset were also used to query the activation properties of border and ectopic variant regions. We used the MSM-Sulc registered, 4mm-smoothing versions of task contrast images from the publicly available analysis-level data from all 7 tasks (emotional processing, gambling, language, motor, relational processing, social cognition, and working memory), originally processed following the HCP preprocessing pipeline100. All tasks used a blocked design to model each contrast; see ref. 112 for details. Of the primary set of 374 subjects, 358 who had both forms of variants and data available for all tasks were retained for the task activation analyses. Again, we compared task activations of variant locations in each network relative to both canonical regions of their assigned network and canonical regions of other networks (this time separately per contrast and per network, given the expanded number of contrasts). Networks with fewer than 200 variants overall were excluded to increase the stability of the results.
To determine the extent to which each form of variant exhibited shifted task activations relative to the typical response expected for canonical regions of their assigned network, we calculated the proportion of this shift (normalized by the average activation seen across all other networks) as follows:
In order to assess the variants’ relative shift, contrasts were only included in this analysis if they showed a difference in the given network’s activation compared to all other networks’ activation; only contrasts with a difference of at least 0.5% signal change were considered. This analysis was performed in both datasets: in the MSC summarized across networks, and in the HCP summarized across contrasts within a network.
Similarity of network variants in twin samples
We next asked about potential sources for these different forms of idiosyncratic variation in functional networks. We used the familial design of the HCP data to interrogate this question. Thus, unlike in previous analyses, we included all N = 793 participants who passed our quality control criteria (see sections Preprocessing and Defining network variants), not excluding those with familial relationships.
Of these, 784 subjects with at least one border variant and at least one ectopic variant were carried forward in the twin analyses (note: only MZ and DZ twin pairs whose twin status was confirmed via genotyping were included as twins).
Next, we estimated the variant similarity among twin samples by contrasting the Dice overlap in monozygotic (MZ) and dizygotic (DZ) twins using Falconer’s formula for broad sense heritability32: h2 = 2(DMZ – DDz). The formula is derived from the model that MZ twins share 100% of their genes and DZ twins share 50%, while both share common environmental variables. Note that our input values to Falconer’s formula were Dice overlap coefficients and not r values, and as such they cannot be directly contrasted with Falconer’s estimates based on correlation, but still provide a valid means to test non-zero genetic influence. To assess the significance of our estimate of genetic influence, we used a non-parametric permutation approach, in which MZ and DZ labels were randomly shuffled before Falconer’s formula was re-computed. This was done 1000 times to create a null distribution. P-values were then calculated relative to this null distribution. These analyses were repeated using variant labels from the parcellation-free variant classification method.
Finally, we then compared the locations of border and ectopic variants across individuals with different familial relationships (monozygotic twins, dizygotic twins, siblings, and unrelated individuals) using the Dice overlap coefficient. A two-way mixed effects ANOVA was performed using SPSS Statistics (version 29) to test for an interaction between variant form and group, with groups associated with 100% genetic overlap (MZ twins), 50% genetic overlap (DZ and non-twin siblings; note that for this analysis, Dice values from DZ and non-twin sibling pairs were combined due to approximately equivalent genetic relatedness and the smaller N of DZ pairs alone), and 0% genetic overlap (unrelated individuals). Following the ANOVA, two-sample t-tests were performed, assuming unequal variance, to assess between-group differences of border minus ectopic Dice coefficients (e.g., MZborder–ectopic vs. combined DZ/sibling border–ectopic vs. unrelated border–ectopic). Bonferroni correction was applied to these three tests. Finally, a paired t-test was performed to test whether, in each group, there was a difference between border and ectopic variant similarity among pairs. Again, Bonferroni correction was applied to these three tests.
Identifying subgroups of ectopic and border variants across individuals
We next sought to investigate whether there were commonalities across subgroups of individuals in their network variant characteristics. To this end, we conducted a similar analysis as in ref. 15 to examine the subgrouping potential of ectopic and border variants. In this case, data was restricted to the sample of N = 374 unrelated individuals to avoid confounding subgrouping analyses with potential familial relationships examined in the previous section.
As in ref. 15, we split the HCP dataset into 2 matched samples consisting of 192 subjects each (further pared down to 183 and 191 subjects, as described in the Defining network variants section), allowing us to search for data-driven findings that replicate across split-halves. In every individual in the HCP dataset, the mean similarity between all variant vertices and each network was produced by calculating the correlation between the variants’ seedmap and each of 11 network template seedmaps. This created an 11 x 1 vector for each subject containing information on the network similarity of variant locations. These vectors were correlated between all subjects in each split half, producing a subject adjacency matrix. A data-driven approach, the Infomap clustering algorithm109, was then conducted on the cross-subject correlation matrix in one split-half; the algorithm was applied after thresholding the correlation matrix across a wide range of density thresholds (5% to 50%, in increments of 1%, requiring a minimum subgroup size of 20 subjects). Subgroups were defined based on consistent results across a range of Infomap thresholds. Sub-group assignments were validated in the second split-half of subjects by correlating each subject’s network vector with one of the resulting subgroup average network vectors; a minimum correlation of 0.3 was required for assignment. In this analysis, we opted to use Infomap as our method of community detection as it has been shown to out-perform other methods – including modularity-maximization approaches – on benchmark testing for community detection113. Infomap better addresses issues that arise from resolution limits, with an improved ability to identify modules of different sizes.
After running Infomap, three primary subgroups were consistently produced across a broad range of thresholds; also note that additional subgroups can be identified at sparser thresholds). Subgroups of individuals with similar network similarity profiles were identified in the first (discovery) split-half, and the subgroups were replicated in the second (validation) split-half as described above. Variant network connectivity patterns of resulting subgroups of individuals were subsequently examined. The subgrouping analysis was performed twofold: first operating solely on ectopic variants, then operating solely on border variants.
Next, we examined to what extent an individual’s subgroup assignment was consistent when grouped by all of their variants, their border variants only, and their ectopic variants only. All HCP subjects with at least one of each form of network variant were included in this analysis. The network similarity vectors of two previously identified stable subgroups (a DMN subgroup and a control/processing subgroup) were identified by clustering individuals’ network vectors across all variant forms within each split-half, and averaging across split-halves. The DMN and control/processing network similarity vectors were then used as templates with which each individual’s network variant profiles were correlated (separately across all variants, border variants only, and ectopic variants only). Similar network similarity results were found in original analyses based on all variants15; see Supp. Fig. 22 for DMN and control/processing profiles. Following these “forced” subgroupings, the adjusted Rand index was calculated three-fold to investigate any similarity between an individual’s subgroup assignment between (1) all variants and border variants only, (2) all variants and ectopic variants only, and (3) border variants and ectopic variants.
We next investigated the robustness of an individual’s subgroup assignment, asking whether this assignment was consistent within a subject when their resting-state data was divided in two. For this analysis, a participant’s data was split into two parts, one for each session, and an 11 x 1 network-similarity vector containing information on border and ectopic variant locations (as described above) was produced for each session’s data. The data-driven subgroup profiles (network-similarity vectors, each sub-group profile averaged across split-halves as in Fig. 7A/B) were correlated with each subject’s network-similarity vector (separately for border and ectopic variants). For each subject, the subgroup profile with the highest correlation was designated the match for a given session’s data, and a contingency table was produced to illustrate the proportion of subjects whose split-session data yielded an equivalent subgroup assignment (see Supp. Fig. 18).
Finally, we sought to evaluate the quality of our Infomap clustering in two ways. First, we randomized the 1x11 vector of network associations for each subject, and used the randomized subject-to-subject correlation matrix to cluster individuals. This process was repeated 1000 times to produce a null distribution of 1000 permuted modularity values of the clustering solution, which were compared to the modularity of the true clustering solution. Second, we used the null_model_und_sign function from the Brain Connectivity Toolbox (www.brain-connectivity-toolbox.net; version 2019-03-03) to create a subject-to-subject adjacency matrix matched in degree, weight, and strength distributions to the original input matrix (using the default of 5 swaps of each edge); this was again repeated 1000 times and compared to the true modularity.
Predicting behavioral phenotypes from network variants
An additional set of analyses modeled after ref. 19 were conducted to determine how border and ectopic variants predicted behavioral measures collected outside of the scanner. For this set of analyses we used the full set of HCP participants that passed our low-motion threshold and quality control criteria (N = 784, the same subset used for twin analyses described above), including all twin and non-twin siblings, in order to maximize our sample size given recent evidence that cross-sectional brain-behavior associations require large samples to be robust34.
Our primary analyses focused on using the network affiliations of variants (as in the section above) to predict 58 behavioral variables in the HCP as in ref. 19; see Supp. Table 3 for a full list of variables. Prediction features from this analysis were based on the average affiliation of variants to 11 template networks. That is, for each person, we estimated the extent to which their variants (on average) were correlated with the 11 template networks, producing 11 continuous feature values that ranged from −1 to 1. This same affiliation measure was used to sub-group individuals in the analyses described in the section above, connecting with that work. Separate prediction analyses were conducted for each form of variants (border and ectopic), and for both forms together. Supplemental analyses also examined how the location of border and ectopic variants (vectorized binary map) predicted behavioral performance.
We used 10-fold cross-validation for prediction, accounting for familial status in the creation of folds (i.e., related individuals – based on either father or mother ID -- were kept in the same fold to ensure independence across folds). Within each fold, we used support vector regression in MATLAB (fitrsvm) with default parameters to identify a relationship between brain features and behavioral variables; this relationship was then tested on the left-out test fold. As in ref. 19, we regressed out age, sex, BMI, mean FD, and DVARS from the behavioral measures prior to prediction as these measures correlate with scanner motion114 (regression was carried out independently in the training data and then applied to the test data to prevent leakage across folds19). Prediction accuracy is reported as the correlation between the predicted and true behavioral measures in independent test data. Correlations were measured per fold and averaged over folds.
We used permutation testing to assess the significance of predictions. For each permutation, behavioral variables were randomly reordered across subjects, thereby breaking the link between variant measures and behavioral outcomes in each participant. Then, the same support vector regression with 10-fold cross validation approach was carried out, resulting in the average correlation between true and predicted values in permuted data. Permutations were repeated 1000 times (500 times for supplemental data based on variant locations) to generate a null distribution that was used as a benchmark for the predictions in the true data. These analyses were repeated using variant labels from the parcellation-free variant classification method.
Supplementary Material
Acknowledgements:
Funding was provided by NIH grant R01MH118370 (CG), the James S. McDonnell Foundation (SEP), NIH grant R01MH111640 (MN), NIH grant T32MH100019 (ANN), the Therapeutic Cognitive Neuroscience Fund (DMS), and NIH grant K01AA030083 (ASH).
Footnotes
Competing interests statement:
The authors declare no competing interests.
Data availability
Data from the Midnight Scan Club is publicly available at https://openneuro.org/datasets/ds000224. Imaging data from the Human Connectome Project (1200 Subjects Release) can be accessed at https://db.humanconnectome.org/; some data elements utilized in this work (e.g., family structure, behavioral measures) require second-tier permissions from the HCP for access. Data associated with the WashU-120 is available at https://openneuro.org/datasets/ds000243/versions/00001.
Code availability
Code for original network variant definition and primary analyses is available at https://github.com/MidnightScanClub/SeitzmanGratton-2019-PNAS. Code used to analyze border and ectopic variants is available at https://github.com/GrattonLab/Dworetsky_BorderEctopicVariants.
REFERENCES
- 1.Churchland PS & Sejnowski TJ Perspectives on cognitive neuroscience. Science 242, 741–745 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Krubitzer LA & Seelke AM Cortical evolution in mammals: the bane and beauty of phenotypic variability. Proc Natl Acad Sci U S A 109 Suppl 1, 10647–10654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaas JH The evolution of neocortex in primates. Prog Brain Res 195, 91–102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson KM, et al. Heritability of individualized cortical network topography. Proc Natl Acad Sci U S A 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krubitzer LA & Prescott TJ The Combinatorial Creature: Cortical Phenotypes within and across Lifetimes. Trends Neurosci 41, 744–762 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Wig GS, Laumann TO & Petersen SE An approach for parcellating human cortical areas using resting-state correlations. Neuroimage 93 Pt 2, 276–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon EM, et al. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex 26, 288–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eickhoff SB, Yeo BTT & Genon S Imaging-based parcellations of the human brain. Nat Rev Neurosci 19, 672–686 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Power JD, et al. Functional network organization of the human brain. Neuron 72, 665–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo BT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laumann TO, et al. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron 87, 657–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon EM, et al. Precision Functional Mapping of Individual Human Brains. Neuron 95, 791–807 e797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon EM, Laumann TO, Adeyemo B & Petersen SE Individual Variability of the System-Level Organization of the Human Brain. Cereb Cortex 27, 386–399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon EM, et al. Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage 146, 918–939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seitzman BA, et al. Trait-like variants in human functional brain networks. Proc Natl Acad Sci U S A 116, 22851–22861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller S, et al. Individual variability in functional connectivity architecture of the human brain. Neuron 77, 586–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn ES, et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci 18, 1664–1671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miranda-Dominguez O, et al. Connectotyping: model based fingerprinting of the functional connectome. PLoS One 9, e111048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong R, et al. Spatial Topography of Individual-Specific Cortical Networks Predicts Human Cognition, Personality, and Emotion. Cereb Cortex 29, 2533–2551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SM, et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat Neurosci 18, 1565–1567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus BT, et al. Network variants are similar between task and rest states. Neuroimage 229, 117743 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougherty RF, et al. Visual field representations and locations of visual areas V1/2/3 in human visual cortex. J Vis 3, 586–598 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Frost MA & Goebel R Measuring structural-functional correspondence: spatial variability of specialised brain regions after macro-anatomical alignment. Neuroimage 59, 1369–1381 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Kong R, et al. Individual-Specific Areal-Level Parcellations Improve Functional Connectivity Prediction of Behavior. Cereb Cortex (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Z, et al. Individual Variation in Functional Topography of Association Networks in Youth. Neuron 106, 340–353 e348 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haxby JV, et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron 72, 404–416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon EM & Nelson SM Three types of individual variation in brain networks revealed by single-subject functional connectivity analyses. Current Opinion in Behavioral Sciences 40, 79–86 (2021). [Google Scholar]
- 28.Arcaro MJ, Schade PF, Vincent JL, Ponce CR & Livingstone MS Seeing faces is necessary for face-domain formation. Nat Neurosci 20, 1404–1412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Striem-Amit E, et al. Functional connectivity of visual cortex in the blind follows retinotopic organization principles. Brain 138, 1679–1695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dosenbach NU, et al. A core system for the implementation of task sets. Neuron 50, 799–812 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber WW Chapter 5: Genetics in Pharmacology: Twin Studies. in Pharmacogenetics 107–108 (Oxford University Press, 2008). [Google Scholar]
- 32.Falconer DS Introduction to quantitative genetics (Ronald Press Co., New York,, 1960). [Google Scholar]
- 33.Ge T, Holmes AJ, Buckner RL, Smoller JW & Sabuncu MR Heritability analysis with repeat measurements and its application to resting-state functional connectivity. Proc Natl Acad Sci U S A 114, 5521–5526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marek S, et al. Reproducible brain-wide association studies require thousands of individuals. Nature 603, 654–660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gratton C, Nelson SM & Gordon EM Brain-behavior correlations: Two paths toward reliability. Neuron 110, 1446–1449 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Bijsterbosch JD, et al. The relationship between spatial configuration and functional connectivity of brain regions. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glasser MF, et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verghese A, Kolbe SC, Anderson AJ, Egan GF & Vidyasagar TR Functional size of human visual area V1: a neural correlate of top-down attention. Neuroimage 93 Pt 1, 47–52 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Schwarzkopf DS, Song C & Rees G The surface area of human V1 predicts the subjective experience of object size. Nat Neurosci 14, 28–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson NC, Kupers ER, Barbot A, Carrasco M & Winawer J Cortical magnification in human visual cortex parallels task performance around the visual field. Elife 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Juch H, Zimine I, Seghier ML, Lazeyras F & Fasel JH Anatomical variability of the lateral frontal lobe surface: implication for intersubject variability in language neuroimaging. Neuroimage 24, 504–514 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Fedorenko E & Blank IA Broca's Area Is Not a Natural Kind. Trends Cogn Sci 24, 270–284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gratton C, et al. Defining Individual-Specific Functional Neuroanatomy for Precision Psychiatry. Biol Psychiatry 88, 28–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Leary DD, Chou SJ & Sahara S Area patterning of the mammalian cortex. Neuron 56, 252–269 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Cadwell CR, Bhaduri A, Mostajo-Radji MA, Keefe MG & Nowakowski TJ Development and Arealization of the Cerebral Cortex. Neuron 103, 980–1004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiNicola LM & Buckner RL Precision Estimates of Parallel Distributed Association Networks: Evidence for Domain Specialization and Implications for Evolution and Development. Curr Opin Behav Sci 40, 120–129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adhikari BM, et al. Comparison of heritability estimates on resting state fMRI connectivity phenotypes using the ENIGMA analysis pipeline. Hum Brain Mapp 39, 4893–4902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colclough GL, et al. The heritability of multi-modal connectivity in human brain activity. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elliott ML, et al. General functional connectivity: Shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks. Neuroimage 189, 516–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fornito A, et al. Genetic influences on cost-efficient organization of human cortical functional networks. J Neurosci 31, 3261–3270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y, et al. Genetic influences on resting-state functional networks: A twin study. Hum Brain Mapp 36, 3959–3972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glahn DC, et al. Genetic control over the resting brain. Proc Natl Acad Sci U S A 107, 1223–1228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hahamy A, et al. Representation of Multiple Body Parts in the Missing-Hand Territory of Congenital One-Handers. Curr Biol 27, 1350–1355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misic B, et al. Cooperative and Competitive Spreading Dynamics on the Human Connectome. Neuron 86, 1518–1529 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Fedorenko E The early origins and the growing popularity of the individual-subject analytic approach in human neuroscience. Current Opinion in Behavioral Sciences 40, 105–112 (2021). [Google Scholar]
- 56.Nieto-Castanon A & Fedorenko E Subject-specific functional localizers increase sensitivity and functional resolution of multi-subject analyses. Neuroimage 63, 1646–1669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porter A, Nielsen A & Gratton C Masked features of task states found in individual brain networks. bioRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haxby JV, Guntupalli JS, Nastase SA & Feilong M Hyperalignment: Modeling shared information encoded in idiosyncratic cortical topographies. Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guntupalli JS, Feilong M & Haxby JV A computational model of shared fine-scale structure in the human connectome. PLoS Comput Biol 14, e1006120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brennan BP, et al. Use of an Individual-Level Approach to Identify Cortical Connectivity Biomarkers in Obsessive-Compulsive Disorder. Biol Psychiatry Cogn Neurosci Neuroimaging 4, 27–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan YS, et al. Individual-specific functional connectome biomarkers predict schizophrenia positive symptoms during adolescent brain maturation. Hum Brain Mapp (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein A, et al. Evaluation of volume-based and surface-based brain image registration methods. Neuroimage 51, 214–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischl B FreeSurfer. Neuroimage 62, 774–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Essen DC, Glasser MF, Dierker DL, Harwell J & Coalson T Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb Cortex 22, 2241–2262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahmasebi AM, et al. Is the link between anatomical structure and function equally strong at all cognitive levels of processing? Cereb Cortex 22, 1593–1603 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Kanwisher N, McDermott J & Chun MM The fusiform face area: a module in human extrastriate cortex specialized for face perception. J Neurosci 17, 4302–4311 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saxe R, Brett M & Kanwisher N Divide and conquer: a defense of functional localizers. Neuroimage 30, 1088–1096; discussion 1097–1089 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Fedorenko E, Duncan J & Kanwisher N Broad domain generality in focal regions of frontal and parietal cortex. Proc Natl Acad Sci U S A 110, 16616–16621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braga RM & Buckner RL Parallel Interdigitated Distributed Networks within the Individual Estimated by Intrinsic Functional Connectivity. Neuron 95, 457–471 e455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dworetsky A, et al. Probabilistic mapping of human functional brain networks identifies regions of high group consensus. Neuroimage 237, 118164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gordon EM, et al. Default-mode network streams for coupling to language and control systems. Proc Natl Acad Sci U S A 117, 17308–17319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilmore AW, Nelson SM & McDermott KB Precision functional mapping of human memory systems. Current Opinion in Behavioral Sciences 40, 52–57 (2021). [Google Scholar]
- 73.Fedorenko E, Duncan J & Kanwisher N Language-selective and domain-general regions lie side by side within Broca's area. Curr Biol 22, 2059–2062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Braga RM, DiNicola LM, Becker HC & Buckner RL Situating the left-lateralized language network in the broader organization of multiple specialized large-scale distributed networks. J Neurophysiol 124, 1415–1448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Noyce AL, Cestero N, Michalka SW, Shinn-Cunningham BG & Somers DC Sensory-Biased and Multiple-Demand Processing in Human Lateral Frontal Cortex. J Neurosci 37, 8755–8766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Somers DC, Michalka SW, Tobyne SM & Noyce AL Individual subject approaches to mapping sensory-biased and multiple-demand regions in human frontal cortex. Current Opinion in Behavioral Sciences 40, 169–177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller JA, D'Esposito M & Weiner KS Using Tertiary Sulci to Map the "Cognitive Globe" of Prefrontal Cortex. J Cogn Neurosci 33, 1698–1715 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Weiner KS The Mid-Fusiform Sulcus (sulcus sagittalis gyri fusiformis). Anat Rec (Hoboken) 302, 1491–1503 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Gratton C, Sun H & Petersen SE Control networks and hubs. Psychophysiology 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duncan J & Owen AM Common regions of the human frontal lobe recruited by diverse cognitive demands. Trends Neurosci 23, 475–483 (2000). [DOI] [PubMed] [Google Scholar]
- 81.Assem M, Glasser MF, Van Essen DC & Duncan J A Domain-General Cognitive Core Defined in Multimodally Parcellated Human Cortex. Cereb Cortex 30, 4361–4380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith DM, Perez DC, Porter A, Dworetsky A & Gratton C Light through the fog: using precision fMRI data to disentangle the neural substrates of cognitive control. Current Opinion in Behavioral Sciences 40, 19–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corbetta M & Shulman GL Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci 3, 201–215 (2002). [DOI] [PubMed] [Google Scholar]
- 84.DiNicola LM, Braga RM & Buckner RL Parallel distributed networks dissociate episodic and social functions within the individual. J Neurophysiol 123, 1144–1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saxe R & Kanwisher N People thinking about thinking people. The role of the temporo-parietal junction in "theory of mind". Neuroimage 19, 1835–1842 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Saxe R Uniquely human social cognition. Curr Opin Neurobiol 16, 235–239 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Gratton C, et al. Functional Brain Networks Are Dominated by Stable Group and Individual Factors, Not Cognitive or Daily Variation. Neuron 98, 439–452 e435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krienen FM, Yeo BT & Buckner RL Reconfigurable task-dependent functional coupling modes cluster around a core functional architecture. Philos Trans R Soc Lond B Biol Sci 369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cole MW, Bassett DS, Power JD, Braver TS & Petersen SE Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yin D & Kaiser M Understanding neural flexibility from a multifaceted definition. Neuroimage 235, 118027 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Gonzalez-Castillo J, et al. Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proc Natl Acad Sci U S A 112, 8762–8767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shirer WR, Ryali S, Rykhlevskaia E, Menon V & Greicius MD Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex 22, 158–165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Satterthwaite TD, et al. Functional maturation of the executive system during adolescence. J Neurosci 33, 16249–16261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marek S, et al. Identifying reproducible individual differences in childhood functional brain networks: An ABCD study. Dev Cogn Neurosci 40, 100706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kong R, et al. Comparison between gradients and parcellations for functional connectivity prediction of behavior. Neuroimage 273, 120044 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tervo-Clemmens B, et al. Reply to: Multivariate BWAS can be replicable with moderate sample sizes. Nature 615, E8–E12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schlaggar BL, et al. Functional neuroanatomical differences between adults and school-age children in the processing of single words. Science 296, 1476–1479 (2002). [DOI] [PubMed] [Google Scholar]
- 98.Van Essen DC, et al. The Human Connectome Project: a data acquisition perspective. Neuroimage 62, 2222–2231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miezin FM, Maccotta L, Ollinger JM, Petersen SE & Buckner RL Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. Neuroimage 11, 735–759 (2000). [DOI] [PubMed] [Google Scholar]
- 100.Glasser MF, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Talairach J & Tournoux P Co-planar stereotaxic atlas of the human brain : 3-dimensional proportional system : an approach to cerebral imaging (Georg Thieme, Stuttgart ; New York, 1988). [Google Scholar]
- 102.Friston KJ, Williams S, Howard R, Frackowiak RS & Turner R Movement-related effects in fMRI time-series. Magn Reson Med 35, 346–355 (1996). [DOI] [PubMed] [Google Scholar]
- 103.Power JD, Barnes KA, Snyder AZ, Schlaggar BL & Petersen SE Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fair DA, et al. Correction of respiratory artifacts in MRI head motion estimates. Neuroimage 208, 116400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Power JD, et al. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 84, 320–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dale AM, Fischl B & Sereno MI Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 179–194 (1999). [DOI] [PubMed] [Google Scholar]
- 107.Gordon EM, et al. Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage 146, 918–939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van Essen DC A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage 28, 635–662 (2005). [DOI] [PubMed] [Google Scholar]
- 109.Rosvall M & Bergstrom CT Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A 105, 1118–1123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Markello RD & Misic B Comparing spatial null models for brain maps. Neuroimage 236, 118052 (2021). [DOI] [PubMed] [Google Scholar]
- 111.Glass L Moire effect from random dots. Nature 223, 578–580 (1969). [DOI] [PubMed] [Google Scholar]
- 112.Barch DM, et al. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fortunato S Community detection in graphs. Physics Reports 486, 75–174 (2010). [Google Scholar]
- 114.Siegel JS, et al. Data Quality Influences Observed Links Between Functional Connectivity and Behavior. Cereb Cortex 27, 4492–4502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the Midnight Scan Club is publicly available at https://openneuro.org/datasets/ds000224. Imaging data from the Human Connectome Project (1200 Subjects Release) can be accessed at https://db.humanconnectome.org/; some data elements utilized in this work (e.g., family structure, behavioral measures) require second-tier permissions from the HCP for access. Data associated with the WashU-120 is available at https://openneuro.org/datasets/ds000243/versions/00001.
Code for original network variant definition and primary analyses is available at https://github.com/MidnightScanClub/SeitzmanGratton-2019-PNAS. Code used to analyze border and ectopic variants is available at https://github.com/GrattonLab/Dworetsky_BorderEctopicVariants.